Abstract
A mobilizable suicide vector, pSUP5011, was used to introduce Tn5-mob in a new facultative sulfur lithotrophic bacterium, KCT001, to generate mutants defective in sulfur oxidation (Sox−). The Sox− mutants were unable to oxidize thiosulfate while grown mixotrophically in the presence of thiosulfate and succinate. The mutants were also impaired in oxidizing other reduced sulfur compounds and elemental sulfur as evident from the study of substrate oxidation by the whole cells. Sulfite oxidase activity was significantly diminished in the cell extracts of all the mutants. A soxA gene was identified from the transposon-adjacent genomic DNA of a Sox− mutant strain. The sequence analysis revealed that the soxA open reading frame (ORF) is preceded by a potential ribosome binding site and promoter region with −10- and −35-like sequences. The deduced nucleotide sequence of the soxA gene was predicted to code for a protein of 286 amino acids. It had a signal peptide of 26 N-terminal amino acids. The amino acid sequence showed similarity with a putative gene product of Aquifex aeolicus, soluble cytochrome c551 of Chlorobium limicola, and the available partial SoxA sequence of Paracoccus denitrificans. The soxA-encoded product seems to be a diheme cytochrome c for KCT001 and A. aeolicus, but the amino acid sequence of C. limicola cytochrome c551 revealed a single heme-binding region. Another transposon insertion mutation was mapped within the soxA ORF. Four other independent transposon insertion mutations were mapped in the 4.4-kb soxA contiguous genomic DNA region. The results thus suggest that a sox locus of KCT001, essential for sulfur oxidation, was affected by all these six independent insertion mutations.
Chemolithotrophy, discovered by Sergei N. Winogradsky in 1887, originated from the observation of sulfur droplets in the filaments of Beggiatoa growing in the presence of hydrogen sulfide (18, 48). Among the few inorganic substrates used by bacteria in the chemolithotrophic process, a comparatively larger variety of reduced inorganic sulfur species support lithotrophic growth of a large number of phylogenetically diverse groups of bacteria and archaea (14). However, chemolithotrophic growth on sulfur was thought to be a conserved genetic trait and was used as the key taxonomic characteristic for the genus Thiobacillus (16, 17). As a result, a variety of physiologically and genetically unrelated eubacteria were classified as Thiobacillus species (14). Phylogenetic analyses based on 5S or 16S ribosomal DNA sequences had shown that the sulfur lithotrophs including the Thiobacillus species belong to the α, β, and γ subclasses of Proteobacteria (21, 22, 32).
The knowledge of the biochemistry and molecular biology of sulfur lithotrophy in microbes must be considered important in understanding the genetic relatedness within the members of Thiobacillus and the relationship of this genus with other sulfur lithotrophs. Extensive biochemical investigations of the oxidative dissimilatory metabolism of sulfur compounds were reported previously (6, 24, 25, 29, 34, 35, 42). Even so, the mechanism involving the specific enzymes, proteins, or accessory factors is rather poorly understood. The element sulfur enjoys a wide range of oxidation states, −2 to +6, and sulfur lithotrophs are not necessarily similar in using specific sulfur species in their lithotrophic processes. Consequently, distinct biochemical pathways have been proposed for different sulfur lithotrophs (14, 16, 36).
Thiosulfate is the common oxidizable substrate that is most suitable for the investigations of sulfur lithotrophic processes. For Paracoccus versutus (formerly Thiobacillus versutus), a thiosulfate-oxidizing periplasmic multienzyme system comprising enzyme A, enzyme B, and multiheme cytochromes was characterized (24, 25). The proposed mechanism is designated the Paracoccus sulfur oxidation (PSO) pathway (18). The function of enzyme A or enzyme B was not demonstrated. In Paracoccus denitrificans, a DNA region essential for sulfur oxidation (Sox) was identified (31). The sequence analysis revealed a partial open reading frame (ORF), soxA, and five additional ORFs (soxBCDEF) downstream of soxA (45, 46). soxB seems essential in sulfur lithotrophy, and the product SoxB appears similar to enzyme B of P. versutus (45). soxC encodes a sulfite dehydrogenase, the requirement for which in thiosulfate-dependent lithotrophic growth in P. denitrificans was experimentally verified (46). The products of soxD and soxE were suggested to be c-type cytochromes. The partial sequence available for soxF exhibits significant similarity with the flavoprotein of Chromatium vinosum, a thiosulfate-oxidizing phototrophic chemolithoautotroph (7, 14). The thiobacilli and other sulfur lithotrophs, phylogenetically close to P. denitrificans or P. versutus (α-3 subgroup [22]), may have acquired this PSO pathway (14, 18, 31) in the sulfur lithotrophic process.
Neither P. versutus nor P. denitrificans uses tetrathionate, an oxidizable substrate commonly used to support lithotrophic growth of many species of Thiobacillus (14, 16). An alternative mechanism of thiosulfate oxidation via the formation of tetrathionate, coupled with the electron transport at the level of cytochrome b instead of cytochrome c, was proposed for obligately lithoautotrophic, moderately thermophilic Thiobacillus tepidarius (26, 47). The tetrathionate-utilizing sulfur lithotrophs such as Thiobacillus thiooxidans or Thiobacillus ferrooxidans, belonging to the β subclass of Proteobacteria and closely related to T. tepidarius (21, 22), may follow this tetrathionate intermediate pathway (16, 18). Further, cleavage of thiosulfate to sulfite and sulfur by rhodanese was demonstrated to be the primary reaction in the process of lithotrophy of thiosulfate by Thiobacillus novellus (6, 14, 16, 34). However, this process of sulfur lithotrophy, apparently distinct from the PSO or tetrathionate intermediate pathway, is yet to be investigated for other sulfur lithotrophs.
Several facultative sulfur lithotrophs, KCT001, KCT002, AS001, and AS002, have been recently isolated and characterized by this laboratory. KCT002, AS001, and AS002 are classified as strains of Paracoccus, and the strain KCT001 is phylogenetically distinct from known strains of Thiobacillus, other sulfur lithotrophs, and other bacterial species of the α subclass of Proteobacteria (unpublished observation; C. Deb, E. Stackebrandt, A. Saha, and P. Roy, unpublished data). In the present study, transposon Tn5-mob insertional mutagenesis in KCT001 was performed to generate mutants impaired in the oxidation of sulfur compounds. A soxA gene was identified from transposon-adjacent genomic DNA of a thiosulfate oxidation-negative (Sox−) mutant. Two primers designed from this soxA gene and one from Tn5 were used in a PCR-based method to walk down the genome of transposon insertion Sox− mutants. We have shown that six independent insertion mutations were mapped within a DNA region of 4.4 kb in the genome of KCT001.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The bacterial strains and plasmids used in this study are listed in Table 1.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s) | Reference and/or source |
|---|---|---|
| Escherichia coli | ||
| SM10 | Neorthi thr leu SuIII RP4-2-Tc::Mu | 39 |
| C600 | Nxrthr leu hsd | 3 |
| XL1-Blue | recA1 lac endA1 gyrA96 thi hsdR17 supE44 relA1 [F′ proAB laclqZΔM15 Tn10(Tetr)] | 5; Stratagene |
| Sulfur chemolithotrophs | ||
| KCT001 | Wild type, Sox+a | Unpublishedb |
| KCT001S | Sox+ Smr (spontaneous) | This study |
| KCT001SR | Sox+ Smr Rifr (spontaneous) | This study |
| KO2-1 | KCT001SR soxA::Tn5-mob Sox− Smr Rifr Neor | This study |
| KO2-2 | KCT001SR soxA::Tn5-mob Sox− Smr Rifr Neor | This study |
| KO2-3 through KO2-12 | KCT001SR sox::Tn5-mob Sox− Smr Rifr Neor | This study |
| P. versutus (= T. versutus) | Sox+; type strain | ATCC 25364 |
| T. novellus | Sox+; type strain | ATCC 8093 |
| T. ferrooxidans | Sox+; type strain | ATCC 23270 (NCIB8455) |
| AS001 | Wild type; Sox+ | Unpublishedb |
| AS002 | Wild type; Sox+ | Unpublishedb |
| KCT002 | Wild type; Sox+ | Unpublishedb |
| Plasmids | ||
| RP4 | Neor Tcr Apr IncP Tra+ | 8 |
| pBluescript KS(+) | AprlacZ′; T7φ10 promoter f1 ori | Stratagene |
| pSUP5011 | pBR325(Bam−)::Tn5-mob Apr Neor Cmr | 38, 39 |
| pKTn-1 | 1.1-kb soxA::Tn5-mob insert of strain KO2-1 in pBluescript KS(+); Apr Neor | This study |
| pPM-1 | 3.7-kb BamHI fragment of pKTn-1 inserted in pBluescript KS(+); Apr | This study |
| pPM-2 | 6.2-kb BamHI fragment of pKTn-1 religated; Apr Neor | This study |
Ability to oxidize reduced sulfur compounds and chemolithotrophic growth is denoted by Sox+, and inability to do the same is denoted by Sox−.
These strains are the new isolates described in the text; these will be reported elsewhere.
Media and growth conditions.
Luria-Bertani broth (LB) and Luria-Bertani agar media (30) were used to cultivate Escherichia coli (at 37°C) and facultative neutrophilic sulfur lithotrophs (at 30°C). For physiological and biochemical studies related to sulfur lithotrophic functions, the cells were grown in different medium formulations based on a modified basal mineral salts (MS) solution (9). The MS solution contained the following (per liter of distilled water): NH4Cl, 1.0 g; K2HPO4, 4.0 g; KH2PO4, 1.5 g; MgSO4 · 7H2O, 0.5 g; trace metal solution (44), 5.0 ml. Mixotrophic medium (MSTSY) contained the following: Na2S2O3 · 5H2O, 10.0 g; sodium succinate, 5.0 g; and yeast extract, 5.0 g per liter of MS. Heterotrophic medium (MSSY) contained the same constituents as MSTSY except thiosulfate. Autotrophic medium (MST) and minimal succinate medium (MSS) contained the following: Na2S2O3 · 5H2O, 5.0 g; or sodium succinate, 5.0 g per liter of MS. Yeast extract (50.0 mg) or a vitamin mixture (10.0 mg each of nicotinic acid, pantothenic acid, pyridoxine, thiamine, para-aminobenzoic acid, riboflavin, and biotin) was added in each liter of MST or MSS. The pH of each medium was adjusted to 7.5 to 8.0 with 2 N NaOH. Phenol red was added (20 mg/liter) as a pH indicator to thiosulfate-containing medium, whenever required. To prepare solid medium, agar was added to a final concentration of 20 g per liter.
Isolation of transposon insertion mutants.
The plasmid vector pSUP5011 (38), which is nonsegregative in nonenteric gram-negative hosts by virtue of a narrow-host-range (ColE1) replicon, carrying the transposon Tn5 with the broad-host-range mobilization sequence (mob), was used to deliver the transposon in the strain KCT001SR. Transconjugants were generated by the plate mating method as described previously (10). Fresh LB-grown cultures of the donor SM10(pSUP5011) and the recipient KCT001SR were mixed at a 1:5 ratio to a final volume of 6 ml and centrifuged to pellet the cells. The cell pellet, resuspended in 200 μl of LB, was spread on a Luria-Bertani agar plate and incubated overnight at 30°C. The conjugate growth on the mating plate was suspended in LB and plated after appropriate dilutions onto the selective medium (streptomycin [200 μg/ml] plus neomycin [20 μg/ml]). The plates thus obtained were incubated for development of single colonies of transconjugants. Transconjugants were picked to prepare master plates, incubated for 2 to 3 days at 30°C, and replica plated on autotrophic agar (MSTA) and mixotrophic agar (MSTSYA) containing phenol red indicator and on a minimal succinate agar (MSSA) plate. The colonies which failed to change the color of phenol red in MSTA and MSTSYA were selected as Sox−, and those which could not grow on MSSA were identified as the auxotrophic mutants.
Substrate-dependent oxygen consumption.
KCT001 (wild type) or KCT001SR was grown in MSTSY (induced condition) and MSSY (noninduced condition) media. The Tn5-mob insertion mutants were grown only in MSTSY medium. Exponentially growing cells at late log phase were harvested by centrifugation, washed, and resuspended in sodium phosphate buffer (100 mM, pH 8.0). The sulfur-oxidizing (Sox) activity of whole cells was determined polarographically with a biological oxygen monitor having a Clark-type oxygen electrode (Yellow Springs Instrument Co., Inc., Yellow Springs, Ohio). The assay mixture contained 300 μmol of sodium phosphate buffer (pH 8.0), cell suspension (300 μg of protein), an aliquot of a substrate solution, and water to a final volume of 3.0 ml. Concentrations of different substrates were as follows: sodium thiosulfate, 100 μmol; potassium tetrathionate, 100 μmol; sodium sulfite (dissolved in 5 mM EDTA), 8 μmol; sodium sulfide, 13 μmol; elemental sulfur (S0), as water-insoluble amorphous powder, 100 mg. Oxygen consumption rates were corrected for chemical oxidation and endogenous respiration.
Enzyme assays.
Freshly grown cells were harvested by centrifugation, resuspended in potassium phosphate (100 mM, pH 7.5) or Tris-HCl (100 mM, pH 7.5) buffer, and broken by sonication. Cell debris was removed by centrifugation (12,000 × g, 15 min), and the supernatant (cell extract [CFE]) was used for the enzyme assay. The activity of thiosulfate oxidase was determined by following the method described by Trudinger (42, 43). The reaction mixture (2.5 ml) contained 250 μmol of potassium phosphate buffer (pH 7.5), 3 μmol of potassium ferricyanide, CFE (300 or 600 μg of protein), water, and 20 μmol of sodium thiosulfate to start the reaction. Sulfite oxidase was assayed by a method slightly modified from that of Charles and Suzuki (6). The assay mixture contained 300 μmol of Tris-HCl (pH 7.9), 3 μmol of potassium ferricyanide, CFE (300 or 600 μg of protein), water, and 10 μmol of sodium sulfite in 5 mM EDTA to start the reaction. Enzyme activities were determined spectrophotometrically by measuring the rate of ferricyanide reduction at 420 nm. The activity of the thiosulfate-cleaving enzyme rhodanese was measured by the rate of thiocyanate formation from thiosulfate and cyanide by following the method described by Oh and Suzuki (34) with minor modifications. The reaction mixture contained 150 μmol of Tris-HCl (pH 7.5), 20 μmol of potassium phosphate buffer (pH 7.5), 150 μmol of thiosulfate, 100 μmol of potassium cyanide, CFE (300 or 600 μg of protein), and water to a final volume of 2.5 ml and was incubated for 10 min. Adding 0.5 ml of 38% (vol/vol) formaldehyde solution stopped the reaction, and formation of thiocyanate was measured at 460 nm after addition of 2 ml of 20% ferric nitrate solution.
Analytical methods.
The level of thiosulfate in the medium was estimated by iodometric titration. Protein concentrations of soluble extract and whole cells were estimated with the Folin phenol reagent of Lowry et al. (23) and the modified Lowry method (15), respectively.
Recombinant DNA techniques.
Bacterial genomic DNA was isolated by the method of Marmur (27). Preparation of plasmid DNA, restriction analysis, ligation, and Southern hybridization were carried out as described by Sambrook et al. (37). DNA fragments from an agarose gel were vacuum transferred to a nylon membrane with a Vacuogene blotter (Pharmacia, Uppsala, Sweden). For Southern hybridization, Bio-Nick labeling and Blue-Gene nonradioactive DNA detection systems (Gibco BRL, Gaithersburg, Md.) were used as described by the manufacturer.
Nucleotide sequence determination and sequence analysis.
Sequencing the Tn5-mob-adjacent KO2-1 genomic DNA was done from the subclones pPM-1 and pPM-2 according to the manufacturer's specifications for Taq DNA polymerase-initiated cycle sequencing reactions using fluorescently labeled dideoxynucleotide terminators with an ABI PRISM 377 automated DNA sequencer (Perkin-Elmer Applied Biosystems, Inc.). The following primers were used for sequencing: TnG (5′-GTTAGGAGGTCACATGG-3′; nucleotides 63 to 79 [complementary] and 5740 to 5756 of Tn5; GenBank accession no. U00004 L19385 [4]), Sox internal primer (5′-CGGCTCCGCGCCCATGTT-3′), and T3 universal primer.
Sequence data were analyzed by using the Genetics Computer Group (version 9-UNIX) software package (12). Deduced amino acid sequence data were compared to the available databases by using the BLAST search programs (1). Molecular mass, isoelectric point (pI), and transmembrane prediction analyses were done through the ExPASy server of the Swiss Institute of Bioinformatics (http://www.expasy.ch). Identifications of signal peptides and predictions of cleavage sites were made using the Signal P program (33). Hydropathy analysis was done according to the algorithm of Kyte and Doolittle (20).
PCR.
The following primers were used: soxAF (5′-TGGGAGAAGGGCAAGGAGCT-3′), soxAR (5′-GTCACATAGACCTCAAGCGC-3′), and TnG. The PCRs were performed using ELONGase (Gibco BRL) with 50 ng of genomic DNA in 50 μl containing 200 μM (each) deoxynucleoside triphosphates and 500 nM (each) primer and buffer with 1.5 mM MgSO4. The reactions were programmed as follows: 94°C for 5 min and then 30 cycles each consisting of 30 s at 94°C, 30 s at 55°C, and 3 min at 68°C and finally 7 min at 68°C to complete the primer extension. The PCR products were electrophoresed on agarose gel in Tris-acetate-EDTA.
Nucleotide sequence accession number.
The EMBL nucleotide sequence accession number for soxA is AJ404005.
RESULTS
Transposon mutagenesis.
The soil isolate KCT001 is a gram-negative, rod-shaped, neutrophilic, and facultatively sulfur lithotrophic bacterium. Sulfur lithotrophic characteristics such as oxidative activities in the presence of sulfur compounds and thiosulfate and/or sulfite oxidase or rhodanese activities of KCT001 (Tables 2 to 4) are comparable to the lithotrophic characteristics of typical facultative sulfur lithotrophs such as T. novellus, P. versutus, or P. denitrificans (6, 13, 34, 41). However, chemotaxonomic and phylogenetic analyses and DNA-DNA hybridization with physiologically similar facultative sulfur lithotrophs revealed that KCT001 is distinct from any of the known sulfur lithotrophs (C. Deb et al., unpublished data). KCT001SR, the streptomycin- and rifampin-resistant spontaneous mutant of KCT001, is a good recipient of the broad-host-range IncP1 plasmid RP4, and the three antibiotic resistance phenotypes of RP4 (8) were expressed in the transconjugants (data not shown). Mobilization of Tn5-mob from the suicide vector pSUP5011 to the recipient cells occurred at a frequency of 10−5 per donor cell. Twelve Sox− mutants were isolated from the screening of 10,000 neomycin-resistant transconjugants from five independent mating experiments. About 0.3% of the total transconjugants failed to grow on MS succinate-glucose medium and were likely to be auxotrophic mutants (data not shown), which were not included for further study.
TABLE 2.
Thiosulfate consumption and decrease of medium pH after 48 h of growth at 30°C
| Straina | Type of growth condition | Thiosulfate consumed (μg atom of S ml−1) | Final pH of spent mediumb |
|---|---|---|---|
| KCT001SR | Autotrophic | 36.3 (90%)c | 6.0 |
| Mixotrophic | 30.0 (76%) | 6.5 | |
| KO2-1 through KO2-12 | Autotrophic | 0 | 8.0 |
| Mixotrophic | 1.6 to 4.0 (4 to 10%)d | 7.5 |
KCT001 or KCT001SR was tested for wild-type activity; the two are similar in terms of general metabolic properties. KO2-1 through KO2-12 are Sox− transposon insertion mutants. KO2-11 was not included in this study.
Initial pH of each medium was 8.0.
Values in parentheses are percents thiosulfate consumed.
Values are ranges of thiosulfate consumption for all the mutants.
TABLE 4.
Activities of enzymes related to oxidation of sulfur compoundsa
| Strainb | Type of growth condition | Sp act of enzymec:
|
||
|---|---|---|---|---|
| Thiosulfate oxidase | Sulfite oxidase | Rhodanese | ||
| KCT001SR | Mixotrophic | 23 (±5) | 320 (±60) | 625 (±125) |
| Heterotrophic | 18 (±3) | 25 (±5) | 150 (±50) | |
| KO2-1 through KO2-12 | Mixotrophic | 12 (±6) | 19 (±13) | 600 (±100) |
Values are means of three experiments; standard deviations are shown in parentheses.
KCT001 or KCT001SR was tested for wild-type activity; the two are similar in terms of general metabolic properties. KO2-1 through KO2-12 are Sox− transposon insertion mutants. KO2-11 was not included in this study.
Specific activities of thiosulfate oxidase and sulfite oxidase are expressed as nanomoles of ferricyanide reduced per minute per milligram of protein, and specific activities of rhodanese are expressed as nanomoles of thiocyanate produced per minute per milligram of protein.
Physiological characterization of sulfur oxidation-negative mutants.
In the mixotrophic medium, the wild-type strain KCT001 (or KCT001SR) consumed 76% of the thiosulfate (20 mM initial concentration) in growth medium after 48 h of incubation and the production of acid (SO42−) effected a decrease of the pH of the medium from 8.0 to 6.5 (Table 2). The Sox− mutants growing in the same medium utilized only 4 to 10% thiosulfate. Similarly, in the autotrophic medium, the wild-type strain consumed 90% thiosulfate, while the mutants were completely defective in utilizing thiosulfate in this medium (Table 2). KCT001SR grown in mixotrophic medium consumed oxygen at a significant rate in the presence of any one of the substrates thiosulfate, tetrathionate, sulfite, sulfide, and elemental sulfur, but the heterotrophically grown cells exhibited very low or negligible levels of oxidation activity of the sulfur compounds. The transposon insertion mutant strains grown in the presence of thiosulfate under mixotrophic conditions showed levels of oxygen consumption similar to those of the heterotrophic cultures of the wild-type strain (Table 3). Mixotrophically grown wild-type cells showed sulfite oxidase activity 10 times higher than that of the heterotrophically grown cells, whereas the Sox− mutants exhibited sulfite oxidase activity at a level comparable with those of noninduced heterotrophic wild-type cells. The wild-type and mutant strains grown under mixotrophic and heterotrophic conditions showed thiosulfate oxidase activity at a lower level. The noninduced wild-type cells showed significant rhodanese activity albeit at a lower level than that of the mixotrophically grown cells, while rhodanese remained unaffected in all the mutants (Table 4). The extent of heterotrophic growth of the wild-type KCT001 and the Sox− mutants in the MSS were similar. Average cell yield after 24 h of growth was 75 (±5) μg of protein/ml (optical density at 660 nm increased to 0.17 [±0.02] from ∼0.01).
TABLE 3.
Oxidation of sulfur compounds by washed whole cellsa
| Strainb | Type of growth condition | Oxygen consumption (nmol of O2 consumed min−1 mg of protein−1) in the presence of:
|
||||
|---|---|---|---|---|---|---|
| Thiosulfate | Tetrathionate | Sulfite | Sulfide | Elemental sulfur | ||
| KCT001SR | Mixotrophic | 225 (±25) | 210 (±10) | 245 (±40) | 220 (±20) | 15 (±5) |
| Heterotrophic | 2.5 (±2.5) | 0 | 15 (±10) | 12 (±8) | 0 | |
| KO2-1 through KO2-12 | Mixotrophic | 2 (±2) | 0 | 22 (±18) | 6 (±5) | 6 (±3) |
Values are means of three experiments; standard deviations are shown in parentheses.
KCT001 or KCT001SR was tested for wild-type activity; the two are similar in terms of general metabolic properties. KO2-1 through KO2-12 are Sox− transposon insertion mutants. KO2-11 was not included in this study.
Southern hybridization of mutant strains.
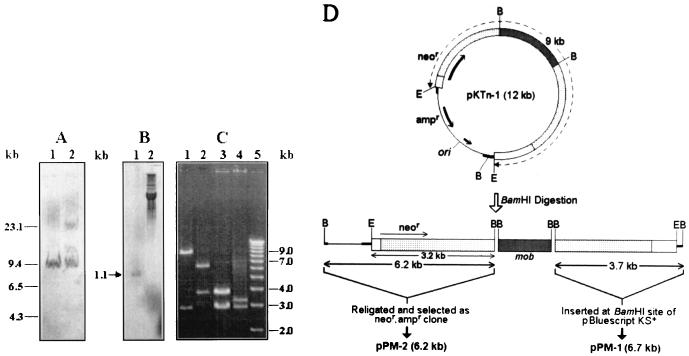
Southern hybridization of the EcoRI-digested genomic DNA of Tn5-mob insertion mutants indicated the presence of a single copy of the transposon in the genome of KO2-1 and KO2-2 (Fig. 1A) and other mutants, except in KO2-3, which had two copies of the transposon (data not shown). No hybridization was detected when the wild-type DNA was probed with pSUP5011 (data not shown). Nine mutants were grouped in three classes according to the size of the EcoRI fragment hybridized with the transposon. An 11-kb EcoRI fragment of four mutants, KO2-7, KO2-8, KO2-10, and KO2-12; a 9-kb fragment of two mutants, KO2-1 and KO2-2 (Fig. 1A); and a 15-kb fragment of three mutants, KO2-4, KO2-5, and KO2-9, contained Tn5-mob in the genome. The mutant KO2-6 produced a 22-kb hybridized band, while the two copies of Tn5-mob present in the genome of KO2-3 resulted in two hybridized bands of 8.5 and 20 kb.
FIG. 1.
(A) Southern hybridization analysis of EcoRI-restricted genomic DNA of Tn5-mob insertion mutants probed with biotin-labeled pSUP5011. A biotin-labeled HindIII digest of phage λ DNA was included as the molecular size marker (not shown). Lanes: 1, KO2-1; 2, KO2-2. (B) Southern hybridization analysis probed with pKTn-1. Lanes: 1, KCT001 genomic DNA restricted with EcoRI; 2, plasmid pKTn-1 as the positive control. (C) Agarose gel electrophoresis analysis of endonuclease-restricted plasmid constructs. Lanes: 1, EcoRI-restricted pKTn-1; 2, BamHI-restricted pKTn-1; 3, BamHI-restricted pPM-1; 4, EcoRI- and BamHI-restricted pPM-2; 5, 1-kb DNA ladder. (D) Physical and genetic map of recombinant constructs. The circular map of pKTn-1 shows the insertion of the 9-kb EcoRI fragment (dashed circular line with arrow at the ends) of Tn5-mob flanked with genomic DNA cloned from the mutant KO2-1. Linear maps show the construction of the subclones pPM-1 and pPM-2. Thick black lines adjacent to the insert ends denote the multiple cloning site (restriction sites which were not used in this study are not shown), and the thin line denotes the part of the vector pBluescript. Grey, dotted, and empty boxes depict mob, Tn5, and genomic DNA of KO2-1, respectively. B, BamHI; E, EcoRI.
Cloning and sequencing of Tn5-mob-adjacent genomic DNA of KO2-1.
The recombinant, pKTn-1, and the two subclones, pPM-1 and pPM-2, were constructed as follows. EcoRI-restricted genomic DNA of KO2-1 cloned in pBluescript KS(+) resulted in the 12-kb recombinant plasmid pKTn-1. The 9-kb insert of pKTn-1 contained Tn5-mob flanked with genomic DNA of KO2-1 (Fig. 1D). This was evident when wild-type DNA digested with EcoRI produced a 1.1-kb band in a Southern hybridization blot probed with pKTn-1 (Fig. 1B). This probe did not show any detectable hybridization signal with the genomic DNA of E. coli C600 as well as diverse species of sulfur lithotrophs: T. novellus, T. ferroxidans, AS001, AS002, KCT002, and P. versutus (data not shown). The 6.2-kb BamHI fragment of pKTn-1 was religated to construct pPM-2 (which confers resistance to neomycin in addition to ampicillin), and the 3.7-kb BamHI fragment was cloned to generate pPM-1 (Fig. 1C and D). Transposon-adjacent genomic DNA of KO2-1, present in the two subclones, was sequenced. The sequences from these two subclones, connected by a 9-bp common sequence (AACTTATGG) generated at the transposon insertion site, resulted in a total of 1,124 bp of wild-type genome (Fig. 2A).
FIG. 2.
(A) Nucleotide and deduced amino acid sequences of soxA. Arrow length denotes the respective primer sequences, and arrowheads indicate direction of extension from each primer. Two potential Shine-Dalgarno (SD) sequences, −10 and −35, of the putative promoter for soxA are shown. Downstream from soxA, another ORF with a potential SD sequence is shown. Start codons are shown in boldface. The 9-bp shared sequence of pPM-1 and pPM-2 is boxed. (B) Multiple alignment of KCT001 soxA-encoded protein (KCT) with the available partial SoxA sequence (PD) of P. denitrificans GB17 (EMBL X79242), a putative protein sequence (AE) of A. aeolicus (AE000757), and a protein sequence of cytochrome c551 of C. limicola (CL [19]). Conserved residues are shaded in grey. The consensus heme-binding domains and the cleavage sites of signal sequences are shown within boxes and with arrowheads, respectively.
Nucleotide sequence analysis.
The 1,124-bp sequence (Fig. 2A) revealed a potential ORF, designated soxA (based on homology search analysis), predicted to start at nucleotide 154 and to end at nucleotide 1014. The calculated G+C content (60%) of the coding sequence of soxA resembles the average G+C content (60%) of KCT001 estimated from the melting temperature of the genomic DNA (data not shown). The ORF is preceded by a DNA region (nucleotides 1 to 112) of low G+C content (54%) having −10- and −35-like sequences. The sequence between nucleotides 113 and 153, bearing potential ribosome binding sites, is highly GC rich (78%). Another ORF downstream of soxA initiating at nucleotide 1,104 with potential ribosome binding sites could be the second structural gene in this putative sox locus.
Amino acid sequence analysis of SoxA.
soxA potentially encodes a protein of 286 amino acids. BLAST search results showed an overall similarity (29% identity and 48% similarity) of KCT-SoxA with a putative protein (AE-SoxA) of Aquifex aeolicus (gi/2984105 [AE000757] [11]), though maximum homology (52% identity and 64% similarity) was noted between the C-terminal 166 residues of KCT-SoxA and the available partial (165-residue) SoxA sequence of P. denitrificans (emb/CAA55827/X79242 [46]). Further, the amino acid sequence of a soluble cytochrome c of Chlorobium limicola (19) exhibited 28% identity to that of KCT-SoxA or SoxA of A. aeolicus (Fig. 2B) and 32% identity with the available C-terminal SoxA sequence of P. denitrificans (Fig. 2B) (19). Thus, soxA gene products of three chemolithotrophs, KCT001, P. denitrificans, and A. aeolicus, and the cytochrome c of a photolithotroph, C. limicola, showed moderate homology, and 13% of the amino acids were found to be conserved (Fig. 2B). The N-terminal 26 and 22 amino acid residues of KCT001 and A. aeolicus, respectively, appeared as signal sequences, indicating that the processed proteins of about 29 and 28 kDa, respectively, in molecular mass were located in the periplasm. The latter is a basic protein with an estimated pI of 9.97 in contrast to the pI of 4.62 of the KCT-SoxA protein.
Mapping of the transposon insertion Sox mutations relative to the position of soxA.
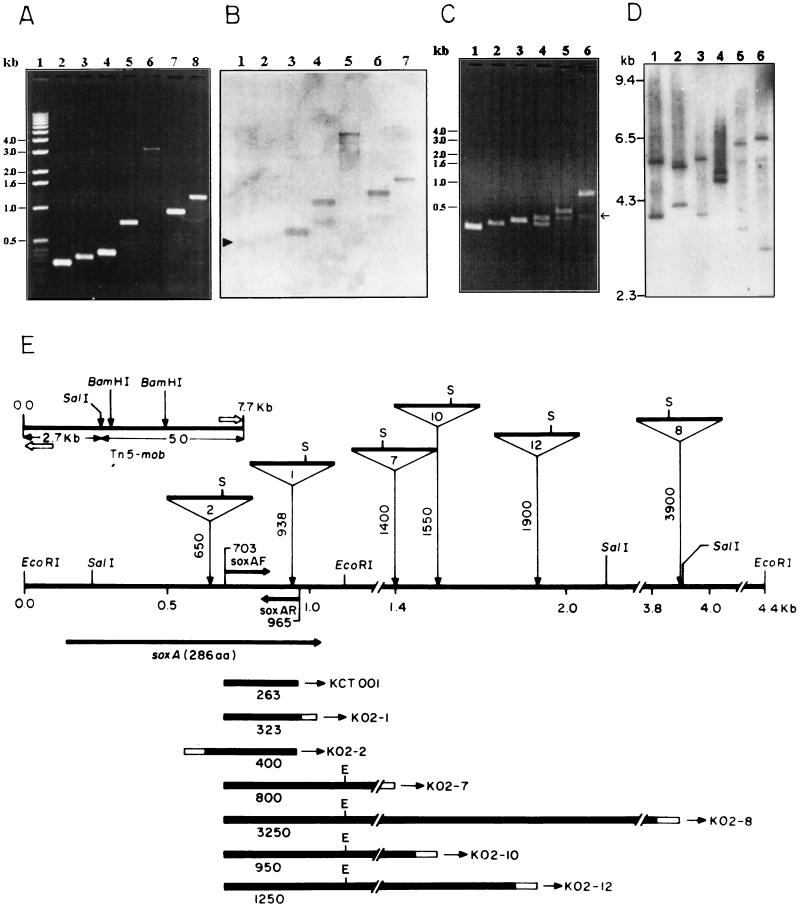
It was shown from the sequence analysis (Fig. 2A) that the mutant KO2-1 had the Tn5-mob insertion within the soxA ORF. It is unlikely that the Sox− phenotype in this mutant was due only to the mutation in soxA. A polar effect on the genes linked with soxA, distal to this insertion mutation, may explain the observed phenotype. Further, all the Sox− mutants were phenotypically similar. We developed a PCR-based method to verify that at least a few of these mutant strains contained independent transposon insertions adjacent to soxA in a common sox locus. Two primers designed from soxA at nucleotides 703 to 722 (soxAF) and 946 to 965 (complementary, soxAR) of the 1,124-bp genome sequence (Fig. 2A) were used separately with a Tn5-specific primer (TnG) in PCRs with the DNA of Sox− mutants. The wild-type DNA produced an expected PCR amplicon of 263 bp corresponding to the genomic region between nucleotides 703 and 965 with soxAF plus soxAR (Fig. 3A, lane 2). In KO2-1, the transposon insertion site at nucleotide position 938 (Fig. 2A) can predict a PCR product of 323 bp with soxAF plus TnG (Fig. 3A, lane 3). The same primer pair generated PCR products of distinct sizes from the DNA of four other mutants, KO2-7, KO2-8, KO2-10, and KO2-12 (Fig. 3A). The other primer pair, soxAR with TnG, did not produce any detectable amplicon from the DNA of these five mutants (data not shown), but a product of about 400 bp was observed from KO2-2 (Fig. 3A, lane 4). The size of a PCR amplicon provided the information to determine the specific site of an independent Tn5-mob insertion relative to the position of the genomic region corresponding to the respective soxAF or soxAR primer sequence. These results, along with the Southern hybridization data (Fig. 1A) for the EcoRI-restricted genomic DNA of the mutants probed with Tn5-mob, described earlier (data not shown), suggest that all these six independent insertion mutations could be clustered within a 4.4-kb DNA region having two contiguous EcoRI fragments of 1.1 and 3.3 kb (Fig. 3E).
FIG. 3.
Mapping of Tn5-mob insertion mutations. (A) Agarose gel electrophoresis analysis of the PCR amplicons generated with different sets of primer combinations (in parentheses). Lanes: 1, 1-kb DNA ladder; 2, KCT001 (soxAF plus soxAR); 3, KO2-1 (soxAF plus TnG); 4, KO2-2 (TnG plus soxAR); 5, KO2-7 (soxAF plus TnG); 6, KO2-8 (soxAF plus TnG); 7, KO2-10 (soxAF plus TnG); 8, KO2-12 (soxAF plus TnG). (B) Southern hybridization analysis of PCR amplicons (description as given for panel A) probed with the 263-bp PCR product from wild-type strain KCT001. Lanes: 1, KCT001; 2, KO2-1; 3, KO2-2; 4, KO2-7; 5, KO2-8; 6, KO2-10; 7, KO2-12. The arrowhead indicates the hybridized band of the 263-bp PCR product from KCT001, which has been used as the probe. Size markers are not shown; compare relative migration in panel A. (C) Agarose gel electrophoresis analysis of the EcoRI-restricted PCR amplicons (description as given for panel A). Lanes: 1, KCT001; 2, KO2-1; 3, KO2-2; 4, KO2-7; 5, KO2-10; 6, KO2-12 (KO2-8 also generated the expected EcoRI restriction product, but this is not shown). The arrow indicates the shared 422-bp EcoRI product. (D) Southern hybridization analysis of SalI-restricted genomic DNA of Tn5-mob insertion mutants probed with pSUP5011. Lanes: 1, KO2-1; 2, KO2-2; 3, KO2-7; 4, KO2-8; 5, KO2-10; 6, KO2-12. (E) Map of six independent Tn5-mob insertion mutations in a sox locus of KCT001. The inset at the upper left corner is the restriction map of Tn5-mob. The open arrows indicate the position and direction of Tn5-specific primer TnG (not to scale). The insertion position of Tn5-mob in each mutant is shown with vertical arrows coming down from a triangle. The respective nucleotide position of the Tn5-mob insertion is noted along the line of the vertical arrow in landscape layout. The thick horizontal line of each triangle represents the Tn5-mob, and S is SalI. The number inside the triangle denotes the mutant strain as follows: 2, KO2-2; 1, KO2-1; 7, KO2-7; 10, KO2-10; 12, KO2-12; 8, KO2-8. The PCR amplicons (as described for panel A) are shown with thick horizontal bars aligned below the insertion map. The number below each bar indicates the size of the amplicon (in base pairs). The hollow ends indicate the part of the amplicon coming from the Tn5 sequence when the TnG primer was used.
Confirmation of insertion mutation mapping and determination of Tn5-mob orientation.
The PCR product from KO2-1 had the genomic region between nucleotides 703 and 946 (the last base of the 9-bp duplication at the site of transposon insertion) excluding the 79 bases from the Tn5 sequence. But from the DNA of other mutants, except KO2-2, the PCRs were extended beyond the EcoRI site situated at the right end of the 1,124-bp sequence (Fig. 2A) of the genome. In KO2-2, the PCR amplicon contained the genomic region between nucleotide positions 965 and 650. Thus, a minimum genomic region between nucleotide positions 703 and 946 was assumed to be common in all the six PCR products (Fig. 3E). This proved true in a Southern hybridization experiment probed with the PCR amplicon generated from the wild-type DNA using two sox primers (Fig. 3B). Furthermore, a common EcoRI site in all the four PCR products (but not in the PCR products of KCT001, KO2-1, and KO2-2) was expected (Fig. 3E) to generate a fragment of 422 bp upon restriction with EcoRI (Fig. 3C; data for KO2-8 not shown).
The transposon insertion mutation sites as shown in Fig. 3E were further confirmed by Southern hybridization of the SalI-restricted DNA of the mutants probed with Tn5-mob. The SalI site of Tn5-mob (4, 38) bisected the transposon into two unequal portions of 2.7 and 5 kb (Fig. 3E). The 1,124-bp sequence revealed a unique SalI site at nucleotide position 240. Thus, two bands of about 5.7 and 3.9 kb obtained from KO2-1 (Fig. 3D, lane 1) may be explained exclusively for a particular orientation of Tn5-mob in this mutant (Fig. 3E). This would be possible if another SalI site at around 2,150 bp (in the 4.4-kb DNA region of the wild-type genome [Fig. 3E]) downstream of soxA could be predicted. It seems that a 1.9-kb SalI fragment of the wild-type genome suffered the independent Tn5-mob insertions, resulting in the development of five mutants, KO2-1, KO2-2, KO2-7, KO2-10, and KO2-12. Southern hybridization data with the SalI-restricted DNA of these five mutants (Fig. 3D) perfectly conformed with this prediction, and the results not only determined the orientation of the transposon in the genome of each mutant but also confirmed our mapping of the mutations. The transposon insertion mutation site in KO2-8 was outside this 1.9-kb SalI DNA region. The sizes of the two SalI bands (Fig. 3D, lane 4) from this mutant DNA indicated a possible orientation of Tn5-mob and the existence of another SalI site adjacent to the site of transposon insertion.
DISCUSSION
We have conducted transposon insertion mutagenesis in a facultative sulfur lithotrophic bacterium, KCT001, and described a simple method of mapping a group of mutations, which identified a DNA region essential for dissimilatory thiosulfate oxidation.
Twelve Tn5-mob insertion mutants impaired in thiosulfate oxidation (Sox−) were phenotypically indistinguishable in the biochemical characteristics associated with sulfur lithotrophy (Table 2). The heterotrophic growth of the mutants in MSS or complex rich medium (LB) did not show any difference from that of wild type. It indicates that the organotrophic nutrition had not been affected due to the mutation impairing sulfur lithotrophy. A soxA gene was identified from the transposon-adjacent genomic DNA of a mutant, KO2-1. The insertion mutation in KO2-2 was 300 bases upstream of the KO2-1 mutation and within the soxA ORF, and the four other transposon insertion mutations were mapped downstream of this soxA gene.
The PCR-based method used to map a group of Sox mutations had been possible because a Tn5-specific primer, TnG, directed DNA synthesis outward from both ends of the transposon. But a successful PCR product was possible from only one end of the transposon when suitably paired with the soxAF or soxAR primer. The method, in fact, has been found effective in genome walking of a bacterium using transposon insertion mutants, and it is worth pursuing with other poorly characterized genetic systems.
The amino acid sequence similarity of KCT-SoxA to a putative protein of A. aeolicus and SoxA of P. denitrificans is highly significant and needs to be explained. There are several conserved regions including the two heme-binding sites of cytochrome c, although only one such cytochrome c motif is apparent in SoxA of P. denitrificans (Fig. 2B). A. aeolicus is a hyperthermophilic, hydrogen-oxidizing obligate chemolithoautotroph (11), whereas P. denitrificans is a hydrogen- and thiosulfate-oxidizing facultative chemolithoautotroph (13, 14). Although sulfur lithotrophy of the recently described A. aeolicus is not known, the presence of a SoxA homolog in this bacterium is intriguing. More importantly, the genome sequence (11) search of A. aeolicus revealed that the respective putative gene (Aq 1807, EMBL accession no. AE000757) is clustered with a putative cytochrome c, a SoxB homolog of P. denitrificans, two other putative genes, and a rhodanese. The homology in the C-terminal residues of KCT-SoxA with the published 165-residue sequence of P. denitrificans indicates similar functions of soxA in sulfur lithotrophy of these two species. The identical nature of these two soxA gene products was evident when hydropathy profiles of these two sequences were superimposed (data not shown). On the other hand, the SoxA homolog of A. aeolicus is not very similar to that of KCT001 or P. denitrificans and has a transmembrane helix domain (between 116 and 135 residues) which is not found in the other two SoxA proteins. Further, the higher level of charged residues (32%) in this protein compared to those of the SoxA protein of KCT001 (27%) is a salient characteristic of thermophiles (11). This extremophile is one of the earliest to diverge in the eubacterial tree (11) and evolved in an environment highly dominated by archaea. At least two hyperthermophilic archaea were shown to have lithotrophic growth on sulfur compounds and hydrogen (14). Moreover, it was suggested that A. aeolicus had acquired several genes from archaebacteria by means of lateral gene transfer (2). Thus, though the putative soxA gene products appear as a diheme cytochrome c, the origin of soxA in A. aeolicus may be distinct from origins in the two other facultative sulfur lithotrophs.
The phototrophic green sulfur bacterium C. limicola can use thiosulfate as a photosynthetic electron donor (28), and cytochrome c551 was thought to be the electron acceptor in this photosynthetic process of thiosulfate oxidation (19). The homology between this cytochrome c551 and KCT-SoxA (or SoxA of P. denitrificans and A. aeolicus) is surprising because the former protein is a monoheme cytochrome c (19). Therefore, it is unlikely that these two cytochromes are related or have a similar function in the oxidative metabolism of thiosulfate by the photolithotroph C. limicola and the chemolithotroph KCT001 or P. denitrificans. P. denitrificans along with other phylogenetically close sulfur lithotrophs, such as P. versutus (14, 22), AS001, AS002, and KCT002 (unpublished observation; C. Deb et al., unpublished data), cluster in the Rhodobacter subgroup of the α subclass of Proteobacteria, whereas the strain KCT001, also a member of the α subclass of Proteobacteria, clusters in the Rhizobium-Agrobacterium subgroup (unpublished observation; C. Deb et al., unpublished data). However, the photosynthetic green sulfur bacterium C. limicola is a member of a different division, Chlorobiaceae, of the phylogenetic eubacterial tree (22). However, a moderate homology between a monoheme cytochrome c of a photosynthetic green sulfur bacterium, C. limicola, and the diheme cytochrome c of chemolithotrophic bacteria, KCT001 and P. denitrificans, indicates a common ancestral origin of Sox genetic systems.
The requirement for soxA in sulfur lithotrophy of P. denitrificans was not tested (46). However, the soxA gene product of P. denitrificans has been claimed to be a 29-kDa subunit of cytochrome c (14), which is yet to be authenticated. Two Sox− mutant strains, KO2-1 and KO2-2, had Tn5-mob insertion within the soxA ORF, and hence a requirement for soxA in thiosulfate oxidation by KCT001 is evident. The phenotype of soxA mutant KO2-1 or KO2-2 cannot be determined since Tn5-mob insertion in soxA is likely to impart a polar effect in the expression of a gene(s) downstream from soxA. Four other Sox− mutations were clustered with KO2-1 and KO2-2 mutations in a 4.4-kb genomic DNA region of the wild type. The results thus indicate that a sox locus was affected by all these six independent insertion mutations. All the selected Sox− mutants of KCT001 were defective in the oxidation of sulfur compounds including tetrathionate. So, presumably the sox locus of KCT001 is distinct from that of the soxABCDEF cluster of P. denitrificans, which does not oxidize tetrathionate (14).
From the Southern blot analysis of EcoRI-restricted DNA of three mutants, KO2-4, KO2-5, and KO2-9, it is apparent that these mutations may be within an 8-kb EcoRI fragment of the genome, but the two other mutants, KO2-3 and KO2-6, showed different sites of transposon insertion in the genome. Twelve thiosulfate oxidation-negative mutant strains of KCT001, apparently mapped in distinct loci of the genome, could not be distinguished phenotypically, in terms of oxidation of sulfur compounds and thiosulfate or sulfite oxidase and rhodanese activity (Tables 2 to 4). It was claimed earlier from the study of T. novellus, P. versutus (formerly T. versutus), and P. denitrificans (formerly Thiosphaera pantotropha) that Sox mutations were mapped in distinct regions of genomic DNA as evident from Southern hybridization analysis of transposon insertional mutants (31, 40; P. Roy, X. Dai, and A. O. Summers, Abstr. Annu. Meet. Am. Soc. Microbiol. 1987, abstr. I-119, p. 192, 1987). The existing knowledge about the sulfur lithotrophic process, evolved in diverse microbial species, is unable to define the mutations with specific functions. In other words, mutations affecting distinct loci leading to a particular phenotype, viz., sulfur lithotrophy, are not distinguishable from the biochemical parameters used in this study or by previous authors. In fact, the nature of the enzymes, proteins, intermediates, and products of the individual enzymatic reactions, leading to the final oxidation product, sulfate, in any of the known sulfur lithotrophic processes remains to be understood.
Finally, we note that our identification of a sox locus in a new sulfur lithotrophic bacterium, KCT001, in conjunction with the already reported finding of a tentative sox operon of P. denitrificans and the known presence of a region in the genomic sequence of A. aeolicus having a putative gene cluster with a potential Sox-related function, will hopefully lead to further characterization that will come up with a novel operon(s) in order to uncover the molecular biology of sulfur lithotrophy, the widely distributed and earliest-discovered chemolithotrophic process.
ACKNOWLEDGMENTS
The early phase of this work was partly supported by the Department of Biotechnology, Government of India, under the project DBT/BT/TF/06/02/89. We thank P. K. Ray, Bose Institute, for providing a fellowship to C.L.
Keya Banerjee provided technical assistance in transposon mutagenesis. We thank Ranadhir Chakraborty and Anupama Saha for their help in characterizing mutants. Tusharmouli Mukherjee and Tapas C. Ghosh of the Distributed Information Center helped in analyzing the sequence. Debashis Majumder rendered excellent service in the preparation of documents. The strain SM10(pSUP5011) was kindly provided by R. Simon. The kind cooperation of S. Roychoudhury and Rajat Banerjee of IICB, Calcutta, in DNA sequencing is gratefully acknowledged. We are indebted to Sourin Bhattacharya, Mrinal Kanti Dasgupta, and Ramkrishna Bhattacharya for their kind help in going through the manuscript and appropriate suggestions for correction of the language. Finally, we thank A. O. Summers of the University of Georgia for her encouragement in the initiation of this work.
REFERENCES
- 1.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aravind L, Tatusov R L, Wolf Y I, Walker D R, Koonin E V. Evidence for massive gene exchange between archaeal and bacterial hyperthermophiles. Trends Genet. 1998;14:442–444. doi: 10.1016/s0168-9525(98)01553-4. [DOI] [PubMed] [Google Scholar]
- 3.Bachmann B J. Pedigrees of some mutant strains of Escherichia coli K-12. Bacteriol Rev. 1972;36:525–557. doi: 10.1128/br.36.4.525-557.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berg D E. Transposon Tn5. In: Berg D E, Howe M M, editors. Mobile DNA. Washington, D.C.: American Society for Microbiology; 1989. pp. 185–210. [Google Scholar]
- 5.Bullock W O, Fernandez J M, Short J M. XL1-Blue: a high efficiency plasmid transforming recA Escherichia coli strains with beta-galactosidase selection. BioTechniques. 1987;5:376–378. [Google Scholar]
- 6.Charles A M, Suzuki I. Mechanism of thiosulfate oxidation by Thiobacillus novellus. Biochim Biophys Acta. 1966;128:510–521. [Google Scholar]
- 7.Chen Z-W, Koh M, Driessche G V, Beeumen J V, Bartsch R G, Meyer T E, Cusanovich M A, Mathews F C. The structure of flavocytochrome c sulfide dehydrogenase from a purple phototrophic bacterium. Science. 1994;266:430–432. doi: 10.1126/science.7939681. [DOI] [PubMed] [Google Scholar]
- 8.Datta N, Hedges R W, Shaw E J, Sykes R B, Richmond M H. Properties of an R factor from Pseudomonas aeruginosa. J Bacteriol. 1971;108:1244–1249. doi: 10.1128/jb.108.3.1244-1249.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davidson M S, Summers A O. Wide-host-range plasmids function in the genus Thiobacillus. Appl Environ Microbiol. 1983;46:565–572. doi: 10.1128/aem.46.3.565-572.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davidson M S, Roy P, Summers A O. Transpositional mutagenesis of Thiobacillus novellus and Thiobacillus versutus. Appl Environ Microbiol. 1985;49:1436–1441. doi: 10.1128/aem.49.6.1436-1441.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deckert G, Warren P V, Gaasterland T, Young W G, Lenox A L, Graham D E, Overbreek R, Snead M A, Keller M, Aujay M, Huber R, Feldman R A, Short J M, Olsen G J, Swanson R V. The complete genome of the hyperthermophilic bacterium Aquifex aeolicus. Nature. 1998;392:353–358. doi: 10.1038/32831. [DOI] [PubMed] [Google Scholar]
- 12.Devereux J P, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Friedrich C G, Mitrenga G. Oxidation of thiosulfate by Paraococcus denitrificans and other hydrogen bacteria. FEMS Microbiol Lett. 1981;10:209–212. [Google Scholar]
- 14.Friedrich C G. Physiology and genetics of sulfur-oxidizing bacteria. Adv Microb Physiol. 1998;39:235–289. doi: 10.1016/s0065-2911(08)60018-1. [DOI] [PubMed] [Google Scholar]
- 15.Herbert D, Phipps P J, Strange R E. Chemical analysis of microbial cells. Methods Microbiol. 1971;5-B:209–344. [Google Scholar]
- 16.Kelly D P. Physiology and biochemistry of unicellular sulfur bacteria. In: Schlegel H G, Bowien B, editors. Autotrophic bacteria. Madison, Wis: Science Tech Publishers; 1989. pp. 193–217. [Google Scholar]
- 17.Kelly D P, Harrison A P. Genus Thiobacillus Beijerinck. In: Staley J T, Bryant M P, Pfennig N, Holt J G, editors. Bergey's manual of systematic bacteriology. Vol. 3. Baltimore, Md: The Williams and Wilkins Co.; 1989. pp. 1842–1858. [Google Scholar]
- 18.Kelly D P, Shergill J K, Lu W-P, Wood A P. Oxidative metabolism of inorganic sulfur compounds by bacteria. Antonie Leewenhoek. 1997;71:95–107. doi: 10.1023/a:1000135707181. [DOI] [PubMed] [Google Scholar]
- 19.Klarskov K, Verte F, Driessche G V, Meyer T E, Cusanovich M A, Beeumen J V. The primary structure of soluble cytochrome c-551 from the phototrophic green sulfur bacterium Chlorobium limicola, strain Tassajara, reveals a novel c-type cytochrome. Biochemistry. 1998;37:10555–10562. doi: 10.1021/bi9806706. [DOI] [PubMed] [Google Scholar]
- 20.Kyte J, Doolittle R F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 21.Lane D J, Harrison A P, Jr, Stahl D, Pace B, Giovannoni S J, Olsen G J, Pace N R. Evolutionary relationships among sulfur- and iron-oxidizing eubacteria. J Bacteriol. 1992;174:269–278. doi: 10.1128/jb.174.1.269-278.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lane D J, Stahl D A, Olsen G J, Heller D J, Pace N R. Phylogenetic analysis of the genera Thiobacillus and Thiomicrospira by 5S rRNA sequences. J Bacteriol. 1985;163:75–81. doi: 10.1128/jb.163.1.75-81.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lowry O H, Rosebrough N J, Farr A L, Randall R J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 24.Lu W-P. A periplasmic location for the thiosulfate-oxidizing multienzyme system from Thiobacillus versutus. FEMS Microbiol Lett. 1986;34:313–317. [Google Scholar]
- 25.Lu W-P, Swoboda B E P, Kelly D P. Properties of the thiosulfate-oxidizing multienzyme system from Thiobacillus versutus. Biochim Biophys Acta. 1985;828:116–122. [Google Scholar]
- 26.Lu W-P, Kelly D P. Cellular location and partial purification of the ‘thiosulfate-oxidizing enzyme’ and ‘trithionate hydrolase’ from Thiobacillus tepidarius. J Gen Microbiol. 1988;134:877–886. [Google Scholar]
- 27.Marmur J. A procedure for the isolation of deoxyribonucleic acid from microorganisms. J Mol Biol. 1961;3:208–218. [Google Scholar]
- 28.Mendez-Alvarez S, Pavon V, Esteve L, Guerrero R, Gaju N. Transformation of Chlorobium limicola by a plasmid that confers the ability to utilize thiosulfate. J Bacteriol. 1994;176:7395–7397. doi: 10.1128/jb.176.23.7395-7397.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meulenberg R, Pronk J T, Hazeu W, van Dijken J P, Frank J, Bos P, Kuenen J G. Purification and partial characterization of thiosulfate dehydrogenase from Thiobacillus acidophilus. J Gen Microbiol. 1993;139:2033–2039. [Google Scholar]
- 30.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 31.Mittenhuber G, Sonomoto K, Egert M, Friedrich C G. Identification of the DNA region responsible for sulfur-oxidizing ability of Thiosphaera pantotropha. J Bacteriol. 1991;173:7340–7344. doi: 10.1128/jb.173.22.7340-7344.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moreira D, Amils R. Phylogeny of Thiobacillus cuprinus and other mixotrophic thiobacilli: proposal for Thiomonas gen. nov. Int J Syst Bacteriol. 1997;47:522–528. doi: 10.1099/00207713-47-2-522. [DOI] [PubMed] [Google Scholar]
- 33.Nielsen H, Engelbrecht J, Brunak S, von Heijne G. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 1997;10:1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- 34.Oh J K, Suzuki I. Isolation and characterization of a membrane-associated thiosulfate oxidizing system of Thiobacillus novellus. J Gen Microbiol. 1977;99:397–412. [Google Scholar]
- 35.Oh J K, Suzuki I. Resolution of a membrane-associated thiosulfate oxidizing complex of Thiobacillus novellus. J Gen Microbiol. 1977;99:413–423. [Google Scholar]
- 36.Pronk J T, Meulenberg R, Hazeu W, Bos P, Kuenen J G. Oxidation of reduced inorganic sulfur compounds by acidophilic thiobacilli. FEMS Microbiol Rev. 1990;75:293–306. [Google Scholar]
- 37.Sambrook J, Maniatis T, Fritsch E F. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 38.Simon R. High frequency mobilization of gram-negative bacterial replicons by the in vitro constructed Tn5-mob transposon. Mol Gen Genet. 1984;196:413–420. doi: 10.1007/BF00436188. [DOI] [PubMed] [Google Scholar]
- 39.Simon R, Priefer U, Pühler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram-negative bacteria. Bio/Technology. 1983;1:784–791. [Google Scholar]
- 40.Summers A O, Roy P, Davidson M S. Current techniques for the genetic manipulation of bacteria and their application to the study of sulfur-based autotrophy in Thiobacillus. Biotechnol Bioeng Symp. 1986;16:267–279. [Google Scholar]
- 41.Taylor B, Hoare D S. New facultative Thiobacillus and a reevaluation of the heterotrophic potential of Thiobacillus novellus. J Bacteriol. 1969;100:487–497. doi: 10.1128/jb.100.1.487-497.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Trudinger P A. Thiosulfate oxidation and cytochromes in Thiobacillus X. 1. Fractionation of bacterial extracts and properties of cytochromes. Biochem J. 1961;78:673–680. doi: 10.1042/bj0780673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Trudinger P A. Thiosulfate oxidation and cytochromes in Thiobacillus X. 2. Thiosulfate oxidizing enzymes. Biochem J. 1961;78:680–686. doi: 10.1042/bj0780680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vishniac W, Santer M. The thiobacilli. Bacteriol Rev. 1957;21:195–213. doi: 10.1128/br.21.3.195-213.1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wodara C, Kostka S, Egert M, Kelly D P, Friedrich C G. Identification and sequence analysis of the soxB gene essential for sulfur oxidation of Paracoccus denitrificans GB17. J Bacteriol. 1994;176:6188–6191. doi: 10.1128/jb.176.20.6188-6191.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wodara C, Bardischewsky F, Friedrich C G. Cloning and characterization of sulfite dehydrogenase, two c-type cytochromes, and a flavoprotein of Paracoccus denitrificans GB17: essential role of sulfite dehydrogenase in lithotrophic sulfur oxidation. J Bacteriol. 1997;179:5014–5023. doi: 10.1128/jb.179.16.5014-5023.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wood A P, Kelly D P. Chemolithotrophic metabolism of the newly-isolated moderately thermophilic, obligately autotrophic Thiobacillus tepidarius. Arch Microbiol. 1987;144:71–77. [Google Scholar]
- 48.Zavarzin G A. Sergei N. Winogradsky and the discovery of chemosynthesis. In: Schlegel H G, Bowien B, editors. Autotrophic bacteria. Madison, Wis: Science Tech Publishers; 1989. pp. 17–32. [Google Scholar]