Highlights
-
•
In the 2021-2022 influenza season, a delayed A(H3N2) epidemic was observed in Italy.
-
•
A marked genetic diversity of the hemagglutinin of A(H3N2) viruses in the 2021-2022 influenza season was observed.
-
•
The predicted vaccine efficacy estimated by analysing HA sequences by a sequence-based model was nearly −28.9%.
Keywords: Influenza, A(H3N2), Phylogenetic analysis, Predicted vaccine analyses, Selective pressure analysis
Abstract
Aims
To assess influenza viruses (IVs) circulation and to evaluate A(H3N2) molecular evolution during the 2021-2022 season in Italy.
Materials and methods
12,393 respiratory specimens (nasopharyngeal swabs or broncho-alveolar lavages) collected from in/outpatients with influenza illness in the period spanning from January 1, 2022 (week 2022-01) to May 31, 2022 (week 2022-22) were analysed to identify IV genome and were molecularly characterized by 12 laboratories throughout Italy. A(H3N2) evolution was studied by conducting an in-depth phylogenetic analysis of the hemagglutinin (HA) gene sequences. The predicted vaccine efficacy (pVE) of vaccine strain against circulating A(H3N2) viruses was estimated using the sequence-based Pepitope model.
Results
The overall IV-positive rate was 7.2% (894/12,393), all were type A IVs. Almost all influenza A viruses (846/894; 94.6%) were H3N2 that circulated in Italy with a clear epidemic trend, with 10% positivity rate threshold crossed for six consecutive weeks from week 2022-11 to week 2022-16. According to the phylogenetic analysis of a subset of A(H3N2) strains (n=161), the study HA sequences were distributed into five different genetic clusters, all of them belonging to the clade 3C.2a, sub-clade 3C.2a1 and the genetic subgroup 3C.2a1b.2a.2. The selective pressure analysis of A(H3N2) sequences showed evidence of diversifying selection particularly in the amino acid position 156. The comparison between the predicted amino acid sequence of the 2021-2022 vaccine strain (A/Cambodia/e0826360/2020) and the study strains revealed 65 mutations in 59 HA amino acid positions, including the substitution H156S and Y159N in antigenic site B, within major antigenic sites adjacent to the receptor-binding site, suggesting the presence of drifted strains. According to the sequence-based Pepitope model, antigenic site B was the dominant antigenic site and the p(VE) against circulating A(H3N2) viruses was estimated to be -28.9%.
Discussion and conclusion
After a long period of very low IV activity since public health control measures have been introduced to face COVID-19 pandemic, along came A(H3N2) with a new phylogenetic makeup. Although the delayed 2021-2022 influenza season in Italy was characterized by a significant reduction of the width of the epidemic curve and in the intensity of the influenza activity compared to historical data, a marked genetic diversity of the HA of circulating A(H3N2) strains was observed. The identification of the H156S and Y159N substitutions within the main antigenic sites of most HA sequences also suggested the circulation of drifted variants with respect to the 2021-2022 vaccine strain. Molecular surveillance plays a critical role in the influenza surveillance architecture and it has to be strengthened also at local level to timely assess vaccine effectiveness and detect novel strains with potential impact on public health.
1. Introduction
Seasonal influenza viruses evolve to evade pre-existing immunity and gain competitive advantage through surface protein mutations which yield new antigenic variants (Petrova and Russell, 2018 Jan) that cause annual epidemics on average accounting for infections in 5–15% of the global population and up to 650,000 deaths a year (Word Health Organization (OMS) 2022). Three main features contribute to the rapid evolution of influenza viruses: large populations, short generation times, and high mutation rates (Shao et al., 2017 Aug). Influenza viruses undergo antigenic drift by mutation in the hemagglutinin (HA) gene, which encodes the main protein target for immune responses. Accumulation of these mutations can result in the emergence of antigenically distinct groups if certain amino acid substitutions are introduced into the HA glycoprotein (Petrova and Russell, 2018 Jan, Weis et al., 1988 Jun). The globular head of HA includes the receptor binding site (RBS) (Weis et al., 1988 Jun) that – although usually conserved - may be exposed to mutations that evade antibody recognition (Thyagarajan and Bloom, 2014 Jul, Lee and Wilson, 2015). The pace of antigenic selection varies over time for influenza A virus (IAV) subtypes and influenza B virus (IBV) lineages mainly due to population-level fluctuations in immune pressure, thus confounding vaccine strain selection, which relies on the anticipation of antigenic evolution (Barrat-Charlaix et al., 2021 06 25). Among human IAVs, H3N2 subtypes are those with the highest mutation rate; after their introduction into the human population in 1968, they started circulating displaying a rapid turn-over of the viral population, with the appearance of new antigenic variants every 2-5 years, usually generating epidemics characterized by high morbidity and mortality, and reducing influenza vaccine efficacy (Allen and Ross, 2018). The constant evolution to evade host immune pressure is achieved through the addition of N-glycosylation sites, antigenic drift, and charged amino acid substitutions near the RBS (Petrova and Russell, 2018 Jan, Allen and Ross, 2018). Specifically, the emergence of new H3N2 variants has been associated with the accumulation of amino acid substitutions at five antigenic sites (designated as A-E and incorporating more than 100 amino acid positions) on the globular head of H3. The substitution of a single amino acid in only one of seven specific amino acid positions adjacent to the RBS may cause major antigenic changes during the evolution of IAVs (Koel et al., 2013 Nov).
Population density and regional interconnectedness play an important role in maintaining viral populations (Ebranati et al., 2015, Russell et al., 2008 Apr 18, Bahl et al., 2011 Nov 29). However, the genetic and antigenic diversity of seasonal influenza has been severely impacted since the onset of the COVID-19 pandemic in March 2020. Since then, most countries have seen historically low seasonal influenza virus circulation (Sullivan et al., 2020, Olsen et al., 2020 Sep 18, Istituto Superiore di Sanità (ISS) 2022) attributable to non-pharmaceutical interventions (NPIs), such as travel restrictions, social distancing, school and workplace closures, mask wearing, and enhanced hygiene. NPIs have similarly disrupted the circulation of other common respiratory viruses such as respiratory syncytial virus and human metapneumovirus (Baker et al., 2020, Adenaiye et al., 2022 Aug 24, Leung et al., 2020, Tang et al., 2021 Jul, Gomez et al., 2021) by limiting opportunities for reintroduction and local transmission. As the use of NPIs to limit COVID-19 pandemic has been gradually declining and international travels have been returning to pre-pandemic levels, a resurgence of influenza virus circulation with an increased severity (due to reduced population immunity over the last couple of years) are expected.
The objectives of this study were i) to describe IVs detection and distribution during the 2021-2022 season in Italy, and ii) to conduct an in-depth phylogenetic analysis of the HA gene of influenza A(H3N2) viruses identified in Italy during the 2021-2022 influenza season in order to evaluate the evolution of these viruses after a long period of very low activity.
2. Materials and methods
2.2. Clinical samples and IAVs/IBVs detection and subtyping methods
Respiratory specimens (nasopharyngeal swabs or broncho-alveolar lavages) collected from in/outpatients with influenza illness in the period spanning from January 1, 2022 (week 2022-01) to May 31, 2022 (week 2022-22) were analysed to detect IV genome by 12 laboratories located in 8 Italian regions belonging to 4 macro-areas (according to NUTS classification (NUTS 2022)): North-West (Lombardy), North-East (Emilia Romagna, Trentino Alto-Adige, Veneto), Centre (Lazio, Marche), and South (Campania, Calabria). Laboratory names and their location by region and macro-area are detailed in Table 1.
Table 1.
Methods used for molecular detection of IVs by each GLIViRe center.
| Laboratory | Region (Macro-area) | IAVs/IBVs RNA detection method | IAVs subtyping method | |
|---|---|---|---|---|
| 1 | Department of Biomedical Sciences for Health, University of Milan, Milan, Italy |
Lombardy (North-West) |
Home-made (Word Health Organization (WHO) 2011) | Home-made [(Word Health Organization (WHO) 2011, Centers for Disease Control and Prevention (CDC) 2022)] |
| 2 | Microbiology and Virology Department, Fondazione IRCCS Policlinico San Matteo, Pavia, Italy | Lombardy (North-West) |
Home-made (Centers for Disease Control and Prevention (CDC) 2022) |
Home-made (Word Health Organization 2022) |
| 3 | Virology Unit, Clinical Laboratory, Fondazione IRCCS Ca' Granda Ospedale Maggiore Policlinico, Milan, Italy | Lombardy (North-West) |
AllplexTM Respiratory Panel Assays on All-in-One Platform (Seegene); Alinity mResp-4-Plex assay (Abbott) |
AllplexTM Respiratory Panel Assays on All-in-One Platform (Seegene) |
| 4 | Ospedale di Circolo e Fondazione Macchi, ASST Sette Laghi, Varese, Italy | Lombardy (North-West) |
Alinity M Resp-4-Plex AMP Kit (Abbott) | Home-made [(Word Health Organization (WHO) 2011, Centers for Disease Control and Prevention (CDC) 2022)] |
| 5 | Laboratorio Aziendale di Microbiologia e Virologia, Hospital of Bolzano (SABES-ASDAA), Bolzano-Bozen, Italy | Trentino Alto-Adige (North-East) |
AllplexTM Respiratory Panel Assays on All-in-One Platform (Seegene) | AllplexTM Respiratory Panel Assays on All-in-One Platform (Seegene) |
| 6 | Microbiology Unit, Azienda ULSS2 Marca Trevigiana, Treviso, Italy | Veneto (North-East) |
AllplexTM Respiratory Panel Assays on All-in-One Platform (Seegene) | AllplexTM Respiratory Panel Assays on All-in-One Platform (Seegene) |
| 7 | Microbiology Unit, IRCCS Azienda Ospedaliero-Universitaria di Bologna, Bologna, Italy | Emilia Romagna (North-East) |
Simplexa Flu A/B & RSV Direct Kit (Diasorin) | Home-made (The European Centre for Disease Prevention and Control (ECDC) 2022) |
| 8 | Virology Laboratory, Azienda Ospedaliera Ospedali Riuniti di Ancona, Ancona, Italy | Marche (Centre) |
Alinity M-Resp-4-Plex AMP Kit (Abbott) | Home-made [(Word Health Organization (WHO) 2011, Centers for Disease Control and Prevention (CDC) 2022)] |
| 9 | Department of Diagnostic and Laboratory Medicine, Unit of Microbiology and Diagnostic Immunology, Bambino Gesù Children Hospital IRCCS, Rome, Italy | Lazio (Centre) |
AllplexTM Respiratory Panel Assays on All-in-One Platform (Seegene) | AllplexTM Respiratory Panel Assays on All-in-One Platform (Seegene) |
| 10 | Istituto Nazionale per le Malattie Infettive Lazzaro Spallanzani, Rome, Italy | Lazio (Centre) |
Home-made (Centers for Disease Control and Prevention (CDC) 2022) |
Home-made (Word Health Organization 2022) |
| 11 | Microbiology and Virology, Cotugno Hospital AORN dei Colli, Naples, Italy | Campania (South) |
AllplexTM Respiratory Panel Assays on All-in-One Platform (Seegene) | AllplexTM Respiratory Panel Assays on All-in-One Platform (Seegene) |
| 12 | Microbiology & Virology Unit, Annunziata Hub Hospital, Azienda Ospedaliera di Cosenza, Cosenza, Italy. | Calabria (South) |
AllplexTM Respiratory Panel Assays on All-in-One Platform (Seegene) | AllplexTM Respiratory Panel Assays on All-in-One Platform (Seegene) |
Respiratory samples were collected from outpatients with the symptoms of influenza-like illness (ILI) or from hospitalised patients with symptoms ranging from mild to severe respiratory syndromes such as, acute respiratory infection (ARI), severe acute respiratory infection (SARI) and acute respiratory distress syndrome (ARDS). Clinical samples were analysed by means of specific real-time PCR assays according to protocols of each participating laboratory. The methods used by each laboratory are detailed in Table 1.
2.3. A(H3N2) influenza viruses sequencing and phylogenetic analysis
A representative subgroup of influenza A(H3N2) positive samples were molecularly characterised by means of the sequence analysis of the complete HA gene (nt. 1-1663). A one-step RT-PCR was performed from 15 µl of extracted RNA in a final reaction volume of 60 µl by using the kit SuperScriptTM III One-Step RT-PCR System with PlatinumTM Taq DNA Polymerase (Thermofisher) in order to obtain the entire influenza A virus genome. The amplification of the complete HA gene was carried out by an in-house nested-PCR split into two reaction mixtures in order to amplify two overlapped fragments of 970 nt. (nt. 1-969) and 813 nt. (nt. 851-1663) (Galli et al., 2020). The HA amplicons were purified and sequenced with both forward and reverse primers by means of the Sanger method.
All HA nucleotide sequences were obtained directly from clinical specimens and submitted to GISAID database (GISAID 2022) under the accession numbers provided in Supplementary Table 1. Sequences were aligned with reference sequences, retrieved from the online repository GISAID (GISAID 2022), by using ClustalW program implemented in BioEdit software (Hall, 1999). The alignment was used to construct the phylogenetic tree by means of the Neighbor-Joining method and the Kimura 2-parameter model with the bioinformatic programme MEGA6 (Tamura et al., 2013 Dec). A bootstrap analysis with 1000 replicates was conducted and bootstrap values ≥70% were considered significant.
Mean nucleotide identities and mean amino acid similarities were calculated by using the Sequence Identity Matrix tool of BioEdit software (Hall, 1999) for intra-group sequence analysis and between study and reference sequences, including the vaccine reference strain of Northern hemisphere for 2021-2022 influenza season (A/Cambodia/e0826360/2020_egg-derived; EPI_ISL_806547).The mean values are expressed as crude rate with the respective range. The genetic distance among sequences of the same genetic group was calculated by means of the p-distance model using MEGA6 program (Tamura et al., 2013 Dec) and it was expressed as mean value and the respective standard deviation (DS).
The predicted amino acid HA sequences were obtained by the Toggle translation tool implemented in BioEdit (Hall, 1999) and amino acid residues were numbered according to the H3 numbering (Lindstrom et al., 1996). Predicted amino acid sequences of the study strains were compared with that of the vaccine strain of Northern hemisphere for 2021-2022 influenza season (A/Cambodia/e0826360/2020; EPI_ISL_806547) to identify amino acid changes, focusing on mutations within the 5 HA antigenic sites of A(H3N2) strains (Wiley and Skehel, 1987), particularly at major antigenic sites within the RBS (Yang et al., 2015 Mar).
By comparing the predicted amino acid sequences of the study and vaccine strains, the predicted vaccine efficacy (pVE) against circulating A(H3N2) viruses was estimated by using a sequence-based model, named Pepitope model, as previously described by others (Bonomo and Deem, 2018 Sep, Gupta et al., 2006 May, Bonomo et al., 2019 May 27). Pepitope is a mathematical model that allows to measure the antigenic distance between the predominant circulating strains and the vaccine virus by considering the amino acid substitutions observed within the residues of the five A(H3N2) antigenic sites. The antigenic site with the highest Pepitope value is considered as the dominant antigenic site and is used to estimate the pVE applying the following formula: (-3.32 × Pepitope(dominant antigenic site) + 0.66) × 100%. When there is a perfect match between circulating strain and vaccine strain, the Pepitope is null and the pVE is 66% (maximum pVE that could be estimated by this sequence-based model) (Bonomo and Deem, 2018 Sep). A negative value of pVE suggests a suboptimal vaccine efficacy against the circulating strains.
2.4. Selective pressure analysis
In order to evaluate the HA evolution due to immunological pressure, a series of probabilistic models of codon substitution were used. In detail, tests for positive selection were conducted using single-likelihood ancestor counting (SLAC), fixed-effects likelihood (FEL), the mixed-effects model of evolution (MEME), fast unconstrained Bayesian approximation (FUBAR), adaptive Branch-Site Random Effects Likelihood (aBSREL), and Branch-site Unrestricted Statistical Test for Episodic Diversification (BUSTED) methods on the Datamonkey 2.0 server (Weaver et al., 2018 Mar 01). To avoid an excessive false-positive rate, sites with SLAC, FEL, MEME and aBSREL p-values <0.05 and a FUBAR posterior probability >0.95 were accepted as candidates for selection.
2.5. Statistical analysis
Statistical analysis was performed using the Open Source Epidemiologic Statistics for Public Health OpenEpi, version 3.03 (Dean et al., 2022). The frequency of positive samples was expressed as a crude proportion, with the corresponding 95% confidence interval (95% CI) calculated by the Mid-P exact test, assuming a normal distribution. The inter-quartile range (IQR) was computed as the difference between the first and third quartiles of the age distribution. The positivity rate was calculated as the number of laboratory-confirmed infections out of the total number of samples. Proportions between groups were compared using the Mid-P exact test based on binomial distribution. For continuous variables, the paired t-test was used.
We estimated influenza seasonal characteristics, including season onset (or start), duration, peak and offset (or end) applying the RS10 method (Midgley et al., 2017), which defines the start of epidemic season as the first 2 consecutive weeks when virus detection exceeds 10% of virus‐positivity rate.
A p-value <0.05 was considered significant (two-tailed test).
3. Results
IVs detection and distribution during the 2021-2022 season in Italy
Overall, 12,393 respiratory specimens were tested for IVs detection. Of these, 894 tested positive to IVs, resulting in an overall positivity rate of 7.2%. IV-positivity rate by center ranged from 1.1% to 14.6% in samples collected in hospital setting and it was 17.3% in an outpatient care setting. IV positivity rates by macro-area are summarized in Table 2. The IVs positivity rate was 7.8% in the centers of North-West Italy, 7.7% in those of North-East Italy, and 1.6% in participating centers of Central and Southern Italy.
Table 2.
Influenza virus positivity rate by type/subtype and by macro-area.
| Macro-area | IVs positivity rate % |
IAVs positivity rate % |
IBVs positivity rate % |
A(H3N2) positivity rate % |
A(H1N1) positivity rate % |
|---|---|---|---|---|---|
| North-West Italy | 7.8% | 7.8% | 0% | 99.6% | 0.4% |
| North-East Italy | 7.7% | 7.7% | 0% | 99.5% | 0.5% |
| Central Italy | 1.6% | 1.6% | 0% | 97.9% | 2.1% |
| Southern Italy | 1.6% | 1.6% | 0% | 100% | 0% |
| Total | 7.2% | 7.2% | 0% | 94.6% | 5.4% |
All the 894 IV-positive samples were IAVs: 94.6% (846/894) of those were H3N2 and 5.4% (48/894) belonged to the H1N1 subtype (Table 2).
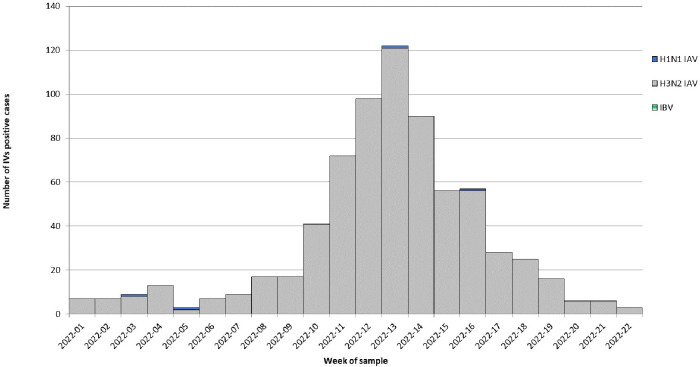
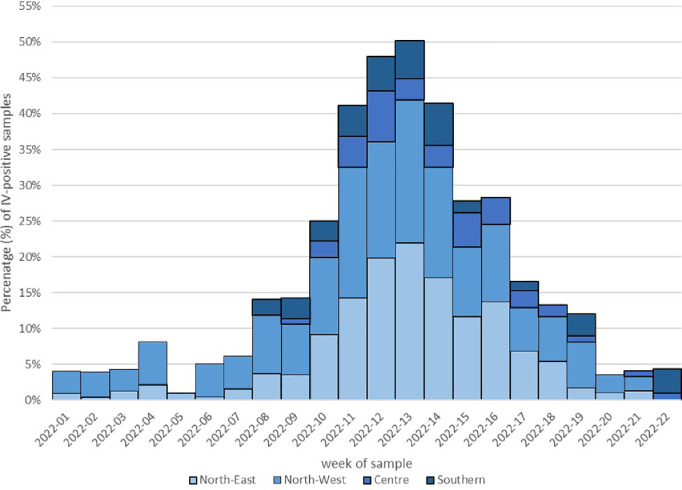
During the study period (from week 2022-01 to week 2022-22), A(H3N2) detection in respiratory samples had a clear epidemic trend, crossing the 10% positivity rate threshold for six consecutive weeks (from week 2022-11 to week 2022-16). In fact, A(H3N2) epidemic wave started in week 2022-11, peaked in week 2022-13 and ended in week 2022-16 (Fig. 1). During the peak, the overall positivity rate reached 20% (117/578). Temporal distribution showed a geographical pattern from North-West to North-East Italy. Considering results by center in each Italian macro-area, A(H3N2) epidemic was evident in centers of both North-West and North-East Italy, whereas A(H3N2) started to be detected from week 2022-08 and 2022-09 in centers from Southern and Central Italy, respectively, never crossing the 10% positivity rate threshold (Fig. 2).
Fig. 1.
Influenza viruses positive samples by type/subtype and by week of sample collection.
Fig. 2.
Temporal distribution of IV-positivity rate by week (from week 2022-01 to week 2022-22) and by macro-area.
3.1. Phylogenetic analysis of A(H3N2) IVs
Overall, 161 A(H3N2) circulating strains were molecularly characterised by sequencing and their HA sequences were phylogenetically analysed. The H3N2 viruses considered in this study were identified in 161 individuals (median age: 9 years; IQR: 29 years; range: 0-93 years; 52% males); 110/161 (68%) were inpatients from hospital settings and 51/161 (32%) were outpatients from ambulatory care settings.
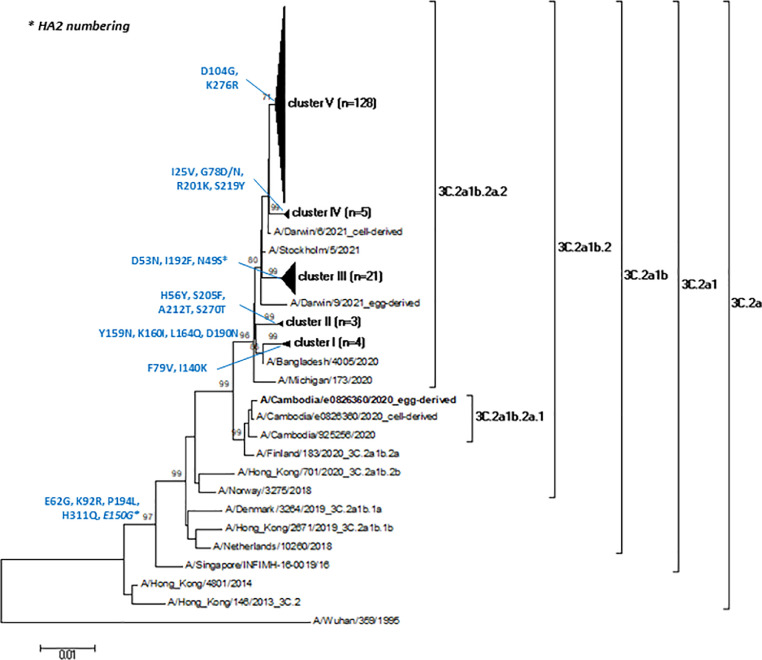
The study strains showed a mean nucleotide identity of 99.3% (range: 98.2%-100%) and a mean amino acid similarity of 99.2% (range: 97.5%-100%). As shown in Fig. 3, all A(H3N2) strains of this study belonged to the clade 3C.2a, sub-clade 3C.2a1 sharing a mean nucleotide identity of 97% (range: 96.6%-97.3%) and a mean amino acid similarity of 95.7% (range: 95.3%-96.1%) to the reference strain A/Singapore/INFIMH-16-0019/2016.
Fig. 3.
Phylogenetic tree of the 161 HA nucleotide sequences (1479 nt.) of A(H3N2) strains identified in this study. The vaccine A(H3N2) strain of the Northern hemisphere for 2021-2022 influenza season (A/Cambodia/e0826360/2020) is in bold. For reasons of clarity, interior branches representing the study sequence clusters are compressed into elongated triangles, whose height is proportional to the number of taxa condensed and whose width is proportional to the maximum distance between taxa. Amino acid substitutions characterising the main branches are detailed close to each node. Only bootstrap values >70% are displayed.
Study sequences were furtherly characterised by the amino acid substitutions E62G (in antigenic site E), K92R (in antigenic site E), P194L (in antigenic site B) and H311Q (in antigenic site C) in HA1 and E150G in HA2, defining the genetic group 3C.2a1b (bootstrap 99%) and sharing a mean nucleotide identity of 97.5% (range: 97.2%-97.9%) and a mean amino acid similarity of 95.9% (range: 95.5%-96.3%) to the reference strain A/Netherlands/10260/2018. The 3C.2a1b genetic group also includes the 2021-2022 vaccine reference strain A/Cambodia/e0826360/2020. However, while the vaccine strain belonged to the genetic subgroup 2a.1, the A(H3N2) strains of this study were characterised by the amino acid mutations Y159N, K160I, L164Q and D190N (all in the antigenic site B of HA1), and segregated into the genetic subgroup 2a.2, displaying a mean nucleotide identity of 99.1% (range: 98.7%-99.5%) and a mean amino acid similarity of 98.9% (range: 98.5%-99.3%) to the reference strain A/Bangladesh/4005/2020. In more detail, study sequences showed a mean intra-group p-distance of 0.006 (standard deviation, SD=0) and they were furtherly distributed in 5 different clusters. By comparing the study sequences with the A/Bangladesh/4005/2020 reference strain, our sequences segregated into different clusters (described below), each of them characterized by specific amino acid mutations. Cluster I (4/161 sequences; 2.5%), characterized by the amino acid substitutions F79V and I140K (in antigenic site A) in HA1, showed a mean intra-group nucleotide identity of 99.7% (range: 99.5% -99.9%) and a mean intra-group amino acid similarity of 99.6% (range: 99.3%-100%). Cluster II (3/161 sequences; 1.9%), characterized by the amino acid mutations H56Y, S205F, A212T (in antigenic site D) and S270T in HA1, showed a mean intra-group nucleotide identity of 99.8% (range: 99.7% -99.9%) and a mean intra-group amino acid similarity of 99.8% (range: 99.7%-100%). Cluster III (21/161 sequences; 13%), characterized by the amino acid substitutions D53N (in antigenic site C) and I192F (in antigenic site B) in HA1 and N49S in HA2, showed a mean intra-group nucleotide identity of 99.6% (range: 99.2%-100%) and a mean intra-group amino acid similarity of 99.6% (range: 98.9%-100%). Cluster IV (5/161 sequences; 3.1%), characterized by the amino acid mutations I25V, G78D/N, R201K (in antigenic site D) and S219Y (in antigenic site D) in HA1, showed a mean intra-group nucleotide identity of 99.8% (range: 99.7%-100%) and a mean intra-group amino acid similarity of 99.8% (range: 99.5%-100%). Cluster V (128/161 sequences; 79.5%), characterized by the amino acid substitutions D104G and K276R (in antigenic site C) in HA1, showed a mean intra-group nucleotide identity and amino acid similarity of 99.7% (range: 98.9%-100%). Finally, no significant difference was observed in the HA sequences distribution by type of setting (ambulatory or hospital) and by geographical area.
3.2. Comparison between study and vaccine strains and predicted vaccine efficacy
The 161 A(H3N2) HA sequences of this study showed a mean nucleotide identity of 98.4% (range: 97.9%-98.7%) and a mean amino acid similarity of 97.5% (range: 97.1%-97.9%) to the A(H3N2) vaccine strain of the Northern hemisphere for the 2021-2022 influenza season, A/Cambodia/e0826360/2020(H3N2).
The comparison between the predicted amino acid sequences of A(H3N2) study viruses and the 2021-2022 vaccine reference strain A/Cambodia/e0826360/2020 revealed 65 mutations in 59 HA amino acid positions; particularly, 9/65 (14%) amino acid substitutions were observed in >80% of study sequences, and all of them were within an antigenic site (antigenic site B: 7/9, 78%; C: 1/9, 11%; D: 1/9, 11%). The list of mutations by amino acid position is presented in Supplementary Table 2. Overall, 54% (35/65) of amino acid mutations was observed within an antigenic site (designated as A-E). All of the five antigenic sites had at least one mutated amino acid position. Particularly, 55% of the substitutions are located in the antigenic sites D (29%, 10/35) and B (26%, 9/35). 17% (6/35) of the mutations are in the antigenic site A, 17% (6/35) in the antigenic site C, and 11% in the antigenic site E.
Among the 7 major antigenic sites (amino acid positions: 145, 155, 156, 158, 159, 189 and 193) adjacent to the RBS, two amino acid positions were characterised by a substitution: H156S shared by 154/161 (96%) study sequences belonging to cluster III-V, and Y159N in all the study strains (161/161; 100%).
The comparison between the predicted amino acid sequence of the 2021-2022 vaccine reference strain A/Cambodia/e0826360/2020 (genetic subgroup 3C.2a1b.2a.1) and the circulating A(H3N2) strains of this study (genetic subgroup 3C.2a1b.2a.2) showed the following changes in 100% HA study sequences: N171K in antigenic site D and Y159N, K160I, L164Q, R186D, D190N and P198S in antigenic site B (Supplementary Table 2). According to the Pepitope model, the antigenic site B was the dominant antigenic site, revealing a Pepitope value of 0.286. The pVE of vaccine strain against the A(H3N2) viruses circulating during the 2021-2022 influenza season was estimated to be -28.9%.
3.3. Selective pressure analysis
Overall, one site in position 156 was identified as being under diversifying selection by site-specific analyses in the HA of study A(H3N2) IVs alignment by at least three of the methods used (SLAC, FEL, REL, FUBAR and MEME) (Table 3). The aBSREL analyses showed evidence of episodic diversifying selection for a branch of tree including 6 strains, with Y159N, T160I, L164Q, N171K, S186D, D190N, P198S changes (A/Bolzano/24/2022, A/Varese/04/2022, A/PoliclinicoMilano/22/2022, A/Milano/04/2022, A/Milano/21/2022, and A/Milano/63/2022) as compared to the vaccine strain. In three of them (A/Bolzano/24/2022, A/Varese/04/2022, A/Policlinico_Milano/22/2022), additional changes were observed (E50K, F79V and I140), while A/Milano/04/2022, A/Milano/21/2022, and A/Milano/63/2022 were characterized by the presence of other additional changes (H56Y, S205F, A212T and S270T). On these branches, BUSTED analyses evidenced that at least one site on at least one test branch experienced diversifying selection (LRT, p-value=0.033 p-value ≤0.05).
Table 3.
Selected sites of HA for A(H3N2) IV strains identified in this study.
| Methods | H3 codons or tree branch | |
|---|---|---|
| Diversifying selection |
Negative selection |
|
| SLAC | None | 10, 24, 239 |
| FEL | 53, 156, 192, 201, 219, 378 | None |
| FUBAR | none | 10, 24, 35, 56, 66, 65. 90, 108, 142, 214, 239, 321, 403, 436, 470, 496 |
| MEME | 156 | None |
| aBSREL | Branch with 6 strainsa (H156, D186, D225). | None |
Positions selected by at least 3 methods are reported in bold.
A/Bolzano/24/2022, A/Varese/04/2022, A/Policlinico_Milano/22/2022, A/Milano/04/2022, A/Milano/21/2022, and A/Milano/63/2022
4. Discussion
Following the worldwide abrupt halt of influenza circulation caused by the emergence and widespread of SARS-CoV-2, scientists have been worrying about increased IVs activity and new viral phylogenetic makeup. In fact, COVID-19 pandemic restrictions such as lockdowns, school closure, facemask use and social distancing helped keep respiratory viruses at bay so much that influenza largely disappeared until early 2022 [(Istituto Superiore di Sanità (ISS) 2022, The European Centre for Disease Prevention and Control (ECDC) 2022, Word Health Organization (WHO) 2022)]. Data from the Southern Hemisphere and in particular from the Australia's Department of Health and Aged Care have showed an unusual influenza activity in 2022, spiking and dropping earlier than usual with the laboratory-confirmed influenza rate higher than the five-year average (Australian Influenza 2022).
Our study aimed at describing IVs distribution during the 2021-2022 season in Italy conducting an in-depth phylogenetic analysis of the HA gene of A(H3N2) influenza viruses. According to our findings, even if there was a clear evidence of influenza epidemic in 2022 in Italy, with the epidemic threshold of 10% positivity crossed for six consecutive weeks, the overall influenza positivity rate was 7.2%, significantly lower than that observed during the pre-COVID-19 seasons [(Istituto Superiore di Sanità (ISS) 2022, The European Centre for Disease Prevention and Control (ECDC) 2022, The European Centre for Disease Prevention and Control (ECDC) 2022, Pellegrinelli et al., 2022)]. In fact, according to the ECDC annual reports, a percentage of influenza virus positive specimens ranging between 40% (2016-2017) and 49% (2017-2018) was observed in the framework of ILI and ARI sentinel consultations from 2016–2017 to 2018-2019 influenza season [(The European Centre for Disease Prevention and Control (ECDC) 2022, The European Centre for Disease Prevention and Control (ECDC) 2022, The European Centre for Disease Prevention and Control (ECDC) 2022)]. The percentage of respiratory samples positive for influenza went across the 10% threshold, indicating the beginning of the epidemic, in week 2022-11, later than observed in previous seasons when the epidemic usually started between week 48 and week 52 [(Istituto Superiore di Sanità (ISS) 2022, The European Centre for Disease Prevention and Control (ECDC) 2022, The European Centre for Disease Prevention and Control (ECDC) 2022, Pellegrinelli et al., 2022, The European Centre for Disease Prevention and Control (ECDC) 2022)]. The percentage of influenza virus positive specimens at its peak (week 2022-13) was 20%, much lower than what observed during the 2018-2019 season when the percentage of influenza viruses positive samples reached up to 62% (The European Centre for Disease Prevention and Control (ECDC) 2022). In Italy, the temporal distribution did not show a clear geographical pattern, probably in consideration to the low number of influenza viruses detected in Central and Southern Italian macro-area (influenza positive rate of 1.6%).
Overall, our data underlined that the 2021-2022 influenza season in Italy was characterized by a significant reduction of the width of the epidemic curve (6 weeks versus 12-19 weeks observed between 2014-2015 to 2018-2019 season; [(Istituto Superiore di Sanità (ISS) 2022, Pellegrinelli et al., 2022)]) and in the intensity of the influenza activity compared to previous seasons that can be undoubtedly related to the emergence of SARS-CoV-2, the actions introduced to control the COVID-19 pandemic and the increased number of testing.
As observed in Europe and in Southern hemisphere countries (Australian Influenza 2022, The European Centre for Disease Prevention and Control (ECDC) March 2022), no influenza B viruses were identified in Italy and among type A influenza viruses, H3N2 subtype largely dominated over H1N1.
To investigate the molecular and evolutionary characteristics of the influenza A(H3N2) viruses circulating during the 2021-2022 season, a phylogenetic analysis of the HA gene sequences of nearly 20% of A(H3N2) viruses detected during the study was conducted. All A(H3N2) study strains belonged to the clade 3C.2a, sub-clade 3C.2a1 and the genetic group 3C.2a1b, sharing a high mean nucleotide identity (99.3%) and a high mean amino acid similarity (99.2%). Our results mirror the European data of influenza virus molecular characterisation during the 2021-2022 season [(The European Centre for Disease Prevention and Control (ECDC) 2022, Istituto Superiore di Sanità 2022)]. Despite belonging to the same phylogenetic branch, the HA sequence analysis of study A(H3N2) strains pointed out the constant tendency of influenza viruses to evolve. In fact, A(H3N2) viruses in the clade 3C.2a were dominant since the 2014-2015 influenza season with the 3C.2a1b viruses predominating over the course of the 2019-2020 season (The Francis Crick Institute - Worldwide Influenza Centre lab 2022). However, the A(H3N2) strains of this study, as observed for the majority of the 2021-2022 strains in other Northern hemisphere countries [(The European Centre for Disease Prevention and Control (ECDC) 2022, Istituto Superiore di Sanità 2022)], segregated into the genetic subgroup 3C.2a1b.2a.2, circulating since October 2020 and named during the 2021-2022 season (The Francis Crick Institute - Worldwide Influenza Centre lab 2022). The 3C.2a1b.2a.2 viruses result in the loss of the glycosylation site at residues 158-160 in HA1, and the acquisition of which had been a defining feature of clade 3C.2a viruses (e.g. A/Hong Kong/4801/2014). Moreover, the 161 HA study sequences were further distributed into five different genetic clusters, each of them marked by specific amino acid substitutions; this genetic diversity confirms the influenza viruses’ continuous ability to mutate. This observation was also emphasized by the fact that the selective pressure analysis showed evidence that some amino acid positions (particularly amino acid 156) are under diversifying selection, therefore resulting more prone to evolve.
Finally, from the comparison between the HA sequences of the A(H3N2) strains of this study and the vaccine strain of the Northern hemisphere for the 2021-2022 season ( A/Cambodia/e0826360/2020), a mean nucleotide identity of 98.4% and a mean amino acid similarity of 97.5% were observed. Although the vaccine strain A/Cambodia/e0826360/2020 belongs to the same genetic group (3C.2a1b) of our study sequences, it differs from them for the genetic subgroup (2a.1 for vaccine strain vs. 2a.2 for study sequences). Our analysis of the predicted amino acid sequences of the HA gene of study A(H3N2) strains revealed numerous amino acid substitutions (65 mutations in 59 sites) compared to the HA sequence of the 2021-2022 vaccine strain. In particular, most of the study sequences had the H156S (96%) and Y159N (100%) mutations, which are located within the major antigenic sites of the receptor binding site. The receptor binding site is generally conserved, but it may also be exposed to selective pressure which determines the introduction of new mutations in order to evade the antibody recognition (Allen and Ross, 2018). The amino acid substitutions in the major antigenic sites (particularly positions 145, 155, 156, 158, 159, 189 and 193) are mutations that, more than others, lead to antigenic changes (Koel et al., 2013 Nov). Thus, the presence of H156S and Y159N mutations in the study sequences suggests the circulation of HA drifted A(H3N2) strains compared to the 2021-2022 vaccine strain; this means that circulating strains could be able to evade the recognition of vaccine-induced antibodies. Overall, more than half (54%) of HA amino acid substitutions was observed within an antigenic site and, among them, 55% were located in the antigenic site D (29%) and B (26%). According to the Pepitope model (Bonomo and Deem, 2018 Sep), the predicted vaccine efficacy of the 2021-2022 vaccine strain against the circulating A(H3N2) viruses was estimated to be -28.9%. Pepitope is a mathematical model that accounts for immunological diversity, modularity, and hierarchy during human antibody recognition of influenza antigens, previously described by Bonomo et al. (Bonomo et al., 2019 May 27). This is a sequence-based model which allows to estimate the antigenic distance between the A(H3N2) predominant circulating strains and the vaccine virus and it can be used only to estimate the vaccine efficacy and not the vaccine effectiveness. Therefore, the Pepitope model cannot replace the test-negative design studies that remain the gold standard to estimate the vaccine effectiveness (Chua et al., 2020 Jan). The pVE estimated in our study by the Pepitope model suggests a suboptimal vaccine efficacy against A(H3N2) circulating in the 2021-2022 influenza season. Although the pVE calculated in this study is the result of a sequence-based analysis only, a suboptimal vaccine effectiveness against circulating A(H3N2) has also been reported by interim analyses of 2021-2022 seasonal influenza vaccine effectiveness through observational studies conducted in the US (Chung et al., 2022 Mar 11), in Denmark (Emborg et al., 2022, Kim et al., 2022), and in Canada (Emborg et al., 2022, Kim et al., 2022). Vaccine effectiveness estimates from these studies have suggested that the low vaccine effectiveness could be due to the circulation of A(H3N2) drifted variants. Surely, a limitation of our study is that no data on the antigenic characteristics of circulating viruses were available, therefore the interpretation of our results is limited to genotypic characteristics of the viruses, and does not consider any phenotypic alterations. However, considering the antigenic analyses of circulating influenza viruses provided by the WHO (The Francis Crick Institute - Worldwide Influenza Centre lab 2022), we can complement our HA gene sequence analysis and speculate that the circulating strains identified in this study are antigenically different from the A(H3N2) vaccine strain, so much that, for the following influenza season, the vaccine composition was updated and the A(H3N2) vaccine strain was changed.
This study has some limitations in terms of heterogeneity in sampled populations among the centres and, partially, in methods used to detect influenza viruses in respiratory samples.
5. Conclusion
As influenza viruses have the potential to emerge with new phylogenetic makeup and in consideration that this study has uncovered the introduction of A(H3N2) HA drifted variants after a long period of very low influenza activity in Italy, it is critical to further strengthen molecular surveillance at local level to promptly assess vaccine effectiveness and detect any novel strains with potential impact on public health.
Working group: Others GLIViRe members and collaborators are listed as follow: Alessandra Pierangeli, Guido Antonelli (Virology Laboratory, Department of Molecular Medicine, "Sapienza" University, Rome, Italy); Francesca Rovida (Department of Clinical, Surgical, Diagnostic and Pediatric Sciences, University of Pavia, Pavia, Italy and Microbiology and Virology Department, Fondazione IRCCS Policlinico San Matteo); Massimo Oggioni (Virology Unit, Clinical Laboratory, Fondazione IRCCS Ca' Granda Ospedale Maggiore Policlinico, Milan, Italy); Tiziana Lazzarotto (Microbiology Unit, IRCCS Azienda Ospedaliero-Universitaria di Bologna, Italy; Section of Microbiology, Department of Experimental, Diagnostic and Specialty Medicine, University of Bologna, Italy); Eva Caterina Borgatti (Section of Microbiology, Department of Experimental, Diagnostic and Specialty Medicine, University of Bologna, Italy); Stefano Menzo (Department of Biomedical Sciences and Public Health, Università Politecnica delle Marche, Ancona, Italy and Virology Laboratory, Azienda Ospedaliera Ospedali Riuniti di Ancona, Ancona, Italy); Luana Coltella (Department of Diagnostic and Laboratory Medicine, Unit of Microbiology and Diagnostic Immunology, Bambino Gesù Children Hospital IRCCS, Rome, Italy); Guido Scalia, Concetta Ilenia Palermo (U.O.C. Laboratory Analysis Unit, A.O.U. ‘Policlinico-Vittorio Emanuele’, University of Catania, Catania, Italy) Ferreri Monica Lucia (UOC Laboratorio Analisi G.B. Grassi - ASL Roma 3), Angelo Paolo Genoni (Ospedale di Circolo e Fondazione Macchi, ASST Sette Laghi, Varese, Italy).
Others GLIViRe members and collaborators
Alessandra Pierangeli, Guido Antonelli (Virology Laboratory, Department of Molecular Medicine, "Sapienza" University, Rome, Italy); Elizabeth Iskandar (Microbiology and Virology Department, Fondazione IRCCS Policlinico San Matteo, Pavia, Italy), Francesca Rovida (Department of Clinical, Surgical, Diagnostic and Pediatric Sciences, University of Pavia, Pavia, Italy and Microbiology and Virology Department, Fondazione IRCCS Policlinico San Matteo); Tiziana Lazzarotto (Microbiology Unit, IRCCS Azienda Ospedaliero-Universitaria di Bologna, Italy; Section of Microbiology, Department of Experimental, Diagnostic and Specialty Medicine, University of Bologna, Italy); Eva Caterina Borgatti (Section of Microbiology, Department of Experimental, Diagnostic and Specialty Medicine, University of Bologna, Italy); Stefano Menzo (Department of Biomedical Sciences and Public Health, Università Politecnica delle Marche, Ancona, Italy and Virology Laboratory, Azienda Ospedaliera Ospedali Riuniti di Ancona, Ancona, Italy); Luana Coltella (Department of Diagnostic and Laboratory Medicine, Unit of Microbiology and Diagnostic Immunology, Bambino Gesù Children Hospital IRCCS, Rome, Italy); Guido Scalia, Concetta Ilenia Palermo (U.O.C. Laboratory Analysis Unit, A.O.U. ‘Policlinico-Vittorio Emanuele’, University of Catania, Catania, Italy) Ferreri Monica Lucia (UOC Laboratorio Analisi G.B. Grassi - ASL Roma 3), Angelo Paolo Genoni (Ospedale di Circolo e Fondazione Macchi, ASST Sette Laghi, Varese, Italy).
Funding information
No funding.
CRediT authorship contribution statement
Cristina Galli: Data curation, Formal analysis, Writing – original draft. Laura Pellegrinelli: Data curation, Formal analysis, Writing – original draft, Writing – review & editing. Federica Giardina: Methodology, Writing – review & editing. Guglielmo Ferrari: Methodology, Writing – review & editing. Sara Colonia Uceda Renteria: Investigation, Writing – review & editing. Federica Novazzi: Investigation, Writing – review & editing. Elisa Masi: Investigation, Writing – review & editing. Elisabetta Pagani: Validation, Writing – review & editing. Giulia Piccirilli: Investigation, Writing – review & editing. Maria Vittoria Mauro: Investigation, Writing – review & editing. Sandro Binda: Validation, Writing – review & editing. Benedetta Corvaro: Investigation, Writing – review & editing. Claudia Tiberio: Investigation, Writing – review & editing. Eleonora Lalle: Investigation, Writing – review & editing. Fabrizio Maggi: Validation, Writing – review & editing. Cristina Russo: Investigation, Writing – review & editing. Stefania Ranno: Investigation, Writing – review & editing. Elisa Vian: Investigation, Writing – review & editing. Elena Pariani: Conceptualization, Validation, Writing – original draft, Writing – review & editing. Fausto Baldanti: Conceptualization, Supervision, Writing – review & editing. Antonio Piralla: Conceptualization, Writing – original draft, Writing – review & editing.
Conflict of interest
Nothing to declare.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.virusres.2022.199033.
Appendix. Supplementary materials
Data availability
Data will be made available on request.
References
- Petrova VN, Russell CA. The evolution of seasonal influenza viruses. Nat. Rev. Microbiol. 2018 Jan;16(1):47–60. doi: 10.1038/nrmicro.2017.146. [DOI] [PubMed] [Google Scholar]
- Word Health Organization (OMS). Influenza seasonal. https://www.who.int/health-topics/influenza-seasonal#tab=tab_1. Last Accessed: 29/11/2022.
- Shao W, Li X, Goraya MU, et al. Evolution of influenza A virus by mutation and Re-assortment. Int. J. Mol. Sci. 2017 Aug;18(8):E1650. doi: 10.3390/ijms18081650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weis W, Brown JH, Cusack S, et al. Structure of the influenza virus haemagglutinin complexed with its receptor, sialic acid. Nature. 1988 Jun;333(6172):426–431. doi: 10.1038/333426a0. [DOI] [PubMed] [Google Scholar]
- Thyagarajan B, Bloom JD. The inherent mutational tolerance and antigenic evolvability of influenza hemagglutinin. Elife. 2014 Jul;3:e03300. doi: 10.7554/eLife.03300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee PS, Wilson IA. Structural characterization of viral epitopes recognized by broadly cross-reactive antibodies. Curr. Top. Microbiol. Immunol. 2015;386:323–341. doi: 10.1007/82_2014_413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrat-Charlaix P, Huddleston J, Bedford T, et al. Limited predictability of amino acid substitutions in seasonal influenza viruses. Mol. Biol. Evol. 2021 06 25;38(7):2767–2777. doi: 10.1093/molbev/msab065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen JD, Ross TM. H3N2 influenza viruses in humans: viral mechanisms, evolution, and evaluation. Hum. Vaccin. Immunother. 2018;14(8):1840–1847. doi: 10.1080/21645515.2018.1462639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koel BF, Burke DF, Bestebroer TM, et al. Substitutions near the receptor binding site determine major antigenic change during influenza virus evolution. Science. 2013 Nov;342(6161):976–979. doi: 10.1126/science.1244730. [DOI] [PubMed] [Google Scholar]
- Ebranati E, Pariani E, Piralla A, et al. Reconstruction of the evolutionary dynamics of A(H3N2) influenza viruses circulating in Italy from 2004 to 2012. PLoS One. 2015;10(9) doi: 10.1371/journal.pone.0137099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell CA, Jones TC, Barr IG, et al. The global circulation of seasonal influenza A (H3N2) viruses. Science. 2008 Apr 18;320(5874):340–346. doi: 10.1126/science.1154137. [DOI] [PubMed] [Google Scholar]
- Bahl J, Nelson MI, Chan KH, et al. Temporally structured metapopulation dynamics and persistence of influenza A H3N2 virus in humans. Proc. Natl. Acad. Sci. U.S.A. 2011 Nov 29;108(48):19359–19364. doi: 10.1073/pnas.1109314108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan SG, Carlson S, Cheng AC, et al. Where has all the influenza gone? The impact of COVID-19 on the circulation of influenza and other respiratory viruses, Australia, March to September 2020. Euro Surveill. 2020;25(47) doi: 10.2807/1560-7917.ES.2020.25.47.2001847. 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen SJ, Azziz-Baumgartner E, Budd AP, et al. Decreased influenza activity during the COVID-19 pandemic - United States, Australia, Chile, and South Africa, 2020. MMWR Morb. Mortal. Wkly. Rep. 2020 Sep 18;69(37):1305–1309. doi: 10.15585/mmwr.mm6937a6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Istituto Superiore di Sanità (ISS) Epicentro. Influnet. 2022 https://www.epicentro.iss.it/influenza/influnet Available at. Last access: 29/11/ [Google Scholar]
- Baker RE, Park SW, Yang W, et al. The impact of COVID-19 nonpharmaceutical interventions on the future dynamics of endemic infections. Proc. Natl. Acad. Sci. U. S. A. 2020;117(48):30547–30553. doi: 10.1073/pnas.2013182117. 12 01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adenaiye OO, Lai J, Bueno de Mesquita PJ, et al. Infectious severe acute respiratory syndrome Coronavirus 2 (SARS-CoV-2) in exhaled aerosols and efficacy of masks during early mild infection. Clin. Infect. Dis. 2022 Aug 24;75(1):e241–e248. doi: 10.1093/cid/ciab797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung NHL, Chu DKW, Shiu EYC, et al. Respiratory virus shedding in exhaled breath and efficacy of face masks. Nat. Med. 2020;26(5):676–680. doi: 10.1038/s41591-020-0843-2. 05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang JW, Bialasiewicz S, Dwyer DE, et al. Where have all the viruses gone? Disappearance of seasonal respiratory viruses during the COVID-19 pandemic. J. Med. Virol. 2021 Jul;93(7):4099–4101. doi: 10.1002/jmv.26964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez GB, Mahé C, Chaves SS. Uncertain effects of the pandemic on respiratory viruses. Science. 2021;372(6546):1043–1044. doi: 10.1126/science.abh3986. 06 04. [DOI] [PubMed] [Google Scholar]
- NUTS - Nomenclature of territorial units for statistics. Availabe at: https://ec.europa.eu/eurostat/web/nuts/background. Last accessed: 29/11/2022.
- Galli C, Orsi A, Pariani E, et al. In-depth phylogenetic analysis of the hemagglutinin gene of influenza A(H3N2) viruses circulating during the 2016-2017 season revealed egg-adaptive mutations of vaccine strains. Expert Rev. Vaccines. 2020;19(1):115–122. doi: 10.1080/14760584.2020.1709827. 01. [DOI] [PubMed] [Google Scholar]
- GISAID. Global initiative on sharing all influenza data. Available at https://www.gisaid.org/. Last accessed: 29/11/2022.
- Hall T. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999:95–98. [Google Scholar]
- Tamura K, Stecher G, Peterson D, et al. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013 Dec;30(12):2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindstrom S, Sugita S, Endo A, et al. Evolutionary characterization of recent human H3N2 influenza A isolates from Japan and China: novel changes in the receptor binding domain. Arch. Virol. 1996;141(7):1349–1355. doi: 10.1007/BF01718836. [DOI] [PubMed] [Google Scholar]
- Wiley DC, Skehel JJ. The structure and function of the hemagglutinin membrane glycoprotein of influenza virus. Annual Review of Biochemistry. 1987;56:p365–p394. doi: 10.1146/annurev.bi.56.070187.002053. [DOI] [PubMed] [Google Scholar]
- Yang H, Carney PJ, Chang JC, et al. Structure and receptor binding preferences of recombinant human A(H3N2) virus hemagglutinins. Virology. 2015 Mar;477:18–31. doi: 10.1016/j.virol.2014.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonomo ME, Deem MW. Predicting Influenza H3N2 vaccine efficacy from evolution of the dominant epitope. Clin. Infect. Dis. 2018 Sep;67(7):1129–1131. doi: 10.1093/cid/ciy323. [DOI] [PubMed] [Google Scholar]
- Gupta V, Earl DJ, Deem MW. Quantifying influenza vaccine efficacy and antigenic distance. Vaccine. 2006 May;24(18):3881–3888. doi: 10.1016/j.vaccine.2006.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonomo ME, Kim RY, Deem MW. Modular epitope binding predicts influenza quasispecies dominance and vaccine effectiveness: application to 2018/19 season. Vaccine. 2019 May 27;37(24):3154–3158. doi: 10.1016/j.vaccine.2019.03.068. [DOI] [PubMed] [Google Scholar]
- Weaver S, Shank SD, Spielman SJ, et al. Datamonkey 2.0: a modern web application for characterizing selective and other evolutionary processes. Mol. Biol. Evol. 2018 Mar 01;35(3):773–777. doi: 10.1093/molbev/msx335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean AG, Sullivan KM, Soe MM. OpenEpi: open source epidemiologic statistics for Public Health, Versione. Available at: www.OpenEpi.com. Last access: 29/11/2022.
- Midgley CM, Haynes AK, Baumgardner JL, et al. Determining the seasonality of respiratory syncytial virus in the United States: the impact of increased molecular testing. J. Infect. Dis. 2017;216(3):345–355. doi: 10.1093/infdis/jix275. 08 01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The European Centre for Disease Prevention and Control (ECDC) FluNews Europe. 2022 https://flunewseurope.org/ Available at: Last access: 29/11/ [Google Scholar]
- Word Health Organization (WHO) Global influenza program. Influenza updates. 2022 https://www.who.int/teams/global-influenza-programme/surveillance-and-monitoring/influenza-updates/influenza-updates-archive Available at: Last access: 29/11/ [Google Scholar]
- Australian Influenza Surveillance Report and Activity Updates –2022. Available at: https://www1.health.gov.au/internet/main/publishing.nsf/Content/cda-ozflu-2022.htm. Last Accessed: 29/11/2022.
- The European Centre for Disease Prevention and Control (ECDC). Seasonal influenza 2018–2019. Annual Epidemiological Report. Available at: https://www.ecdc.europa.eu/sites/default/files/documents/AER_for_2018_seasonal-influenza-corrected.pdf. Last Accessed: 29/11/2022.
- The European Centre for Disease Prevention and Control (ECDC). Seasonal influenza 2017–2018. Annual Epidemiological Report. Available at: https://www.ecdc.europa.eu/sites/default/files/documents/AER_for_2018_seasonal-influenza-corrected.pdf. Last Accessed: 29/11/2022.
- Pellegrinelli L, Galli C, Bubba L, et al. Respiratory syncytial virus in pediatric influenza-like illness cases in Lombardy, Northern Italy, during seven consecutive winter seasons (from 2014-2015 to 2020-2021) Influenza Other. Respir. Viruses. 2022;16(3):481–491. doi: 10.1111/irv.12940. 05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The European Centre for Disease Prevention and Control (ECDC). Seasonal influenza. Annual Epidemiological Report for 2016. Available at: https://www.ecdc.europa.eu/sites/default/files/documents/AER_for_2016-influenza-seasonal.pdf. Last Accessed: 29/11/2022.
- The European Centre for Disease Prevention and Control (ECDC) Influenza virus characterisation - Summary Europe. March 2022 https://www.ecdc.europa.eu/sites/default/files/documents/Influenza-characterisation-report-march-2022.pdf Available at: Last Accessed: 29/11/2022. [Google Scholar]
- The European Centre for Disease Prevention and Control (ECDC). Influenza Virus Characterisation Reports, summary Europe. Available at: https://www.ecdc.europa.eu/en/seasonal-influenza/surveillance-and-disease-data/influenza-virus-characterisation. Last accessed: 29/11/2022.
- Istituto Superiore di Sanità. ISS: Sorveglianza virologica dell'influenza (rete InfluNet). Available at: https://www.iss.it/sorveglianza-virologica-dell-influenza-influnet. Last access: 29/11/2022.
- The Francis Crick Institute - Worldwide Influenza Centre lab. Annual and interim reports. Available at: https://www.crick.ac.uk/research/platforms-and-facilities/worldwide-influenza-centre/annual-and-interim-reports.Last access: 23/11/2022.
- Chua H, Feng S, Lewnard JA, et al. The use of test-negative controls to monitor vaccine effectiveness: a systematic review of methodology. Epidemiology. 2020 Jan;31(1):43–64. doi: 10.1097/EDE.0000000000001116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung JR, Kim SS, Kondor RJ, et al. Interim estimates of 2021-22 Seasonal influenza vaccine effectiveness - United States, February 2022. MMWR Morb. Mortal. Wkly. Rep. 2022 Mar 11;71(10):365–370. doi: 10.15585/mmwr.mm7110a1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emborg HD, Vestergaard LS, Botnen AB, et al. A late sharp increase in influenza detections and low interim vaccine effectiveness against the circulating A(H3N2) strain, Denmark, 2021/22 influenza season up to 25 March 2022. Euro Surveill. 2022;27(15) doi: 10.2807/1560-7917.ES.2022.27.15.2200278. 04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Chuang ES, Sabaiduc S, et al. Influenza vaccine effectiveness against A(H3N2) during the delayed 2021/22 epidemic in Canada. Euro Surveill. 2022;27(38) doi: 10.2807/1560-7917.ES.2022.27.38.2200720. 09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Word Health Organization (WHO). Global influenza surveillance network. Manual for the laboratory diagnosis and virological surveillance of influenza. Available at: http://whqlibdoc.who.int/publications/2011/9789241548090_eng.pdf. 2011. Last accessed: 29/11/2022.
- Centers for Disease Control and Prevention (CDC). National Center for Immunization and Respiratory Diseases (NCIRD). Virology, surveillance, and diagnosis branch. Available at: https://www.cdc.gov/ncird/flu.html. Last accessed: 29/11/2022.
- Centers for Disease Control and Prevention (CDC). Influenza SARS - CoV - 2 (Flu SC2) M ultiplex assay primers and probes. Available at: https://www.cdc.gov/coronavirus/2019-ncov/downloads/lab/multiplex-primers-probes-printer.pdf. Last accessed: 29/11/2022.
- Word Health Organization. WHO information for the molecular detection of influenza viruses. Available at: https://cdn.who.int/media/docs/default-source/influenza/molecular-detention-of-influenza-viruses/protocols_influenza_virus_detection_feb_2021.pdf?sfvrsn=df7d268a_5. Last accessed: 29/11/2022.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.