Highlights
-
•
Updated global analysis of movement and nuclear shuttle proteins from RefSeq.
-
•
RaXML trees show deep split between Old World and New World begomoviruses.
-
•
Shannon entropy reveals low sequence diversity in new world movement protein.
-
•
No statistical evidence for recombination within these proteins.
Abstract
Approximately half of the characterized begomoviruses have bipartite genomes, but the second genomic segment, the DNA-B, is understudied relative to the DNA-A, which is homologous to the entire genome of monopartite begomoviruses. We examined the evolutionary history of the two proteins encoded by the DNA-B, the genes of which make up ∼60% of the DNA-B segment, from all bipartite begomovirus species. Our dataset of 131 movement protein (MP) and nuclear shuttle protein (NSP) sequences confirmed the deep split between Old World (OW) and New World (NW) species, and showed strong support for deep, congruent branches among the OW sequences of the MP and NSP. NW sequences were much less diverse and had poor phylogenetic resolution; over half of nodes in both the NSP and MP NW clades were supported by <50% bootstrap support. This poor resolution hampered our ability to detect incongruent phylogenies between the MP and NSP datasets, and we found no statistical evidence for recombination within our MP and NSP datasets. Finally, we quantified the sequence diversity between the NW and OW proteins, showing that the NW MP has particularly low diversity, suggesting it has been subject to different evolutionary pressures than the NW NSP.
1. Introduction
Geminiviruses are among the most abundant emerging plant viruses on the planet (Seal et al., 2006). As of 2021, there are 14 genera and 520 species in this family (Fiallo-Olivé et al., 2021). The genus Begomovirus, in the family Geminiviridae, is the largest and most diverse viral genus in the world with 445 different species (Fiallo-Olivé et al., 2021). Begomoviruses are concentrated and are an emergent problem in tropical and sub-tropical regions (Kenyon et al., 2014; Varma and Malathi, 2003). Infection by begomoviruses can cause leaf curling, mosaic patterns on leaves, vein yellowing, stunted plant growth and/or stunted fruit growth (Leke et al., 2015). The resulting crop loss affects the livelihood and nutrition of billions of people across all inhabited continents (Rojas et al., 2018). For one instance, mosaic disease of cassava, caused by begomoviruses, is responsible for over 2 billion dollars in crop losses annually and threatens the food security of more than 800 million people in sub-Saharan Africa each year (Patil and Fauquet, 2009).
The two genomic segments in bipartite begomoviruses are called DNA-A and DNA-B. The two ∼2.7 kb segments are separately encapsidated (Hesketh et al., 2018; Fiallo-Olivé et al., 2021). DNA-A is an ambisense segment that contains at least five open reading frames (ORFs) that largely overlap, and which encode the genes necessary for assisting in replication, capsid formation and countering antiviral defense (Fondong, 2013; Hanley-Bowdoin et al., 2013; Fig. 1). DNA-B is also ambisense, but typically has only two non-overlapping functional ORFs. BV1, on the virion sense strand, encodes the nuclear shuttle protein NSP, which binds both ssDNA and dsDNA and mediates intracellular transport from the nucleus to the cytoplasm (Hehnle et al., 2004; Martins et al., 2020). BC1, in the complementary sense, encodes the movement protein MP, which interacts with NSP intracellularly and can enlarge the size exclusion limit of plasmodesmata to translocate the begomovirus genome between cells (Fondong, 2013; Hanley-Bowdoin et al., 2013). Both proteins have been associated with begomovirus pathogenicity (Ingham et al., 1995; Hussain et al., 2005). The two genetic components share a ‘common region’ of around 200 nucleotides that allows both segments to be recognized by the Replication-associated protein encoded on the DNA-A segment. The common region is centered around the origin of replication for rolling circle replication, at which individual complete segments are cleaved and ligated to yield covalently closed circular ssDNA molecules that are encapsidated. Outside of this common region, the DNA-A and DNA-B do not have any nucleotide sequence similarity (Briddon et al., 2010).
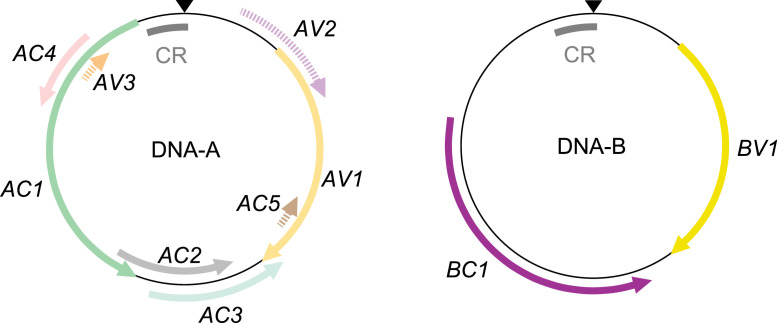
Fig. 1.
Begomovirus DNA-A and DNA-B segments and open reading frames (ORFs). Both genomic segments are ambisense: some genes are encoded on the virus sense strand (V, the strand that is encapsidated), and others are encoded on the opposite complementary sense strand (C). Bipartite begomovirus DNA-As contain at least five functional ORFs. Only one of the required five genes is encoded in the virion sense—AV1 encodes the coat protein. In the complementary sense, AC1 encodes the replication-associated protein, AC2 encodes the transcriptional activator protein, AC3 encodes the replication enhancing protein and AC4 encodes an RNA-silencing suppressor. Some DNA-A molecules have additional ORFs, shown with dashed lines to indicate that they are not always present or functional. The AV2 ORF present in OW begomoviruses encodes a pre-coat protein that functions in movement. AC5 and AV3 are inconsistently annotated ORFs that encode silencing suppressors (Gong et al., 2021). The extent to which AC5 and AV3 function is conserved across the begomovirus phylogeny is unknown (Zhao et al., 2022; Li et al., 2015). DNA-B is also ambisense. BV1, on the virion sense strand, encodes the nuclear shuttle protein NSP and BC1, on the complementary strand, encodes the movement protein MP (Fondong, 2013; Hanley-Bowdoin et al., 2000). An additional BV1-overlapping ORF (BV2), thus far only described in a single species (Chiu et al., 2022) is omitted. The homologous common region containing the origin of replication nick site (black triangle) is denoted CR.
Not all begomoviruses are bipartite: 56% of recognized begomovirus species are monopartite (Virus Metadata Resource #18, 2021-10-19, https://ictv.global/vmr). Monopartite begomoviruses are homologous to the DNA-A of bipartite begomoviruses, and obligately include the V2 gene for the pre-coat protein which helps in cell-to-cell movement within a plant (Padidam et al., 1996). This gene is called AV2 when it is found on the DNA-A of bipartite begomoviruses from the Old World (Hanley-Bowdoin et al., 2013; Fig. 1). The pre-coat protein is thought to serve the same function as the MP from the DNA-B, though there is no homology between the genes (Priyadarshini et al., 2011), and the MP is considered a 30K protein that may descend from an RNA virus (Koonin et al., 1992). Monopartite geminiviruses in the recently established genus Citlodavirus also encode a 30K-type MP (Loconsole et al., 2012; Roumagnac et al., 2022). The NSP is thought to have evolved from the begomovirus coat protein due to protein sequence similarity (Kikuno et al., 1984; Martins et al., 2020), and the coat protein can complement the function of NSP when NSP is disabled by mutations (Qin et al., 1998).
The rising prevalence of monopartite begomoviruses, the species definition of begomoviruses only requiring characterization of a DNA-A-like segment (Brown et al., 2015) and the ability of DNA-A segments to function in plants with multiple genetically distinct DNA-B segments (Sicard et al., 2016; Lucía-Sanz and Manrubia, 2017) has led research on the DNA-B to be deprioritized. However, DNA-B is an essential component of the vast majority of New World begomoviruses, which are bipartite and lack the AV2 ORF. DNA-B segments also affect viral host range (Idris et al., 2011) and determine symptoms and symptom severity (Von Arnim and Stanley, 1992; Jyothsna et al., 2013), even for an otherwise monopartite begomovirus (Ouattara et al., 2022). The DNA-B segment also appears to dominate reproduction in infected plants; quantitative PCR studies have shown that plants infected with bipartite begomoviruses have higher DNA-B accumulation than DNA-A accumulation (Péréfarres et al., 2012; Naseem and Winter, 2016).
When researchers have examined the evolution of the DNA-B, typically they compare its evolutionary history to that of its accompanying DNA-A segments (Briddon et al., 2010; Xavier et al., 2021). An important phylogeographic study from 2010 found that both segments separated into Old World (OW, from Africa and Asia) and New World (NW, from the Americas and the Caribbean) clades (Briddon et al., 2010). NW begomoviruses are thought to have emerged from migration from Asia to the Americas, and that these viruses evolved separately since their geographic divide until the past few decades (Rybicki, 1994; Lefeuvre et al., 2011). The overwhelming majority of NW begomoviruses are bipartite and appear to have diversified from a common ancestor that came from the OW (Xu et al., 2008; Torres-Herrera et al., 2019; Ho et al., 2014).
In this study, we focus on the evolutionary history of the two proteins encoded on the DNA-B segment (MP and NSP) across all bipartite begomoviruses. As it can be difficult to align nucleotide sequences from DNA-B segments even in related groups of bipartite begomoviruses (Crespo-Bellido et al., 2021) we focused on amino acid sequences to conduct this global analysis. We slightly revise the phylogeographic clades previously found (Briddon et al., 2010), find that the evolutionary history of the MP and NSP are largely congruent in the Old World and report evidence for greater variation maintained in the NSP compared to the MP, especially in the New World. We found little or no evidence for recombination within or between MP or NSP.
2. Methods
Begomovirus MP and NSP RefSeq sequences were downloaded via the NCBI Taxonomy Browser (https://www-ncbi-nlm-nih-gov/Taxonomy/Browser/wwwtax.cgi), using RefSeq accession numbers for DNA-B genomic segments from the ICTV Virus Metadata Resource (Virus Metadata Resource #18, 2021-10-19, https://ictv.global/vmr). From these DNA-B nucleotide segment accessions, the amino acid sequences for MP and NSP were obtained. Species that had an annotated MP sequence but no NSP sequence, or vice versa, were excluded from the datasets. Several RefSeq sequences were misannotated in NCBI, most commonly with the MP and NSP mislabeled as the other. We confirmed the identity of the ORF products by BLAST (Altschul et al., 1990) and swapped the protein sequences for the following nine species (virus name abbreviation followed by corrected protein and its accession number): MerMPRV (MP: YP_004429247; NSP: YP_004429246), RhMMV (YP_004429241; YP_004429240), PHYVV (NP_597687; NP_040354), PYMV/To (NP_047242; NP_047241), BGYMV (YP_009507974; YP_009507973), ToGMoV (YP_619879; YP_619878), EACMZV (NP_808890; NP_808889), ICMV/Ker (NP_047236; NP_047235), BGMV/Ala (NP_660086; NP_660085). Because several species names changed after their submission to GenBank, the dataset includes two ICTV exemplars of Merremia mosaic virus (MerMV/PR; MerMV/VE). In total, our MP and NSP datasets contained amino acid sequences from 131 begomovirus species (Supplementary File 1).
The amino acid sequences were then renamed in the following format: “virus abbreviation_[MP/NSP]_[OW/NW]_accession number.” The virus name abbreviations were obtained from the ICTV Virus Metadata Resource. OW/NW indicates whether the sequence is from the Old World or New World geographic region. The viral sequence names from NCBI often include the region where the sequence was isolated and sufficient location information is often supplied in the GenBank file. Our OW and NW classifications were concordant with previously published delineations (Briddon et al., 2010).
After renaming the sequences, we separately aligned MP and NSP datasets with MUSCLE (Edgar, 2004). We then manually optimized each alignment in AliView 1.28 (Larsson, 2014). The final alignments are publicly available (DOI: 10.5281/zenodo.6985414). We then used ProtTEST 3 to determine the best model for describing amino acid substitution in the datasets, in preparation for phylogenetic analysis (Darriba et al., 2011). We then created bootstrapped (1000 replicates) maximum likelihood trees using RaxML 8.2.12 (Stamatakis, 2014) on the CIPRES portal (http://phylo.org). Resulting trees were visualized in FigTree 1.44 (http://tree.bio.ed.ac.uk/software/figtree/).
To visualize evolutionary relationships without the imposition of strict bifurcation, we built unrooted phylogenetic networks for MP and NSP using the default settings in SplitsTree 4.0 (Huson and Bryant, 2006). The SplitsTree phi test, which is a conservative test for presence/absence of recombination, was applied to both datasets (Bruen et al., 2006). A tanglegram of the maximum likelihood trees was created in R (phanghorn 2.8.1, Schliep et al., 2017). The tanglegram was disentangled using the method side2side in the dendextend package (Galili, 2015) in R 4.2.1 (R Core Team, 2020). Shannon entropies of the aligned protein datasets were calculated with Python scripts (https://github.com/dd886/Begomovirus-DNA-B-protein-evolution). Two corchorus viruses from Vietnam, CoYVV(MP: YP_115514; NSP: YP_115513) and CoGMV/IN (MP: YP_001333690; NSP: YP_001333689), which were always excluded from the OW versus NW clades, were also excluded from Shannon entropy calculations because they do not neatly fit into either the OW clade or the NW clade (Ha et al., 2006).
3. Results
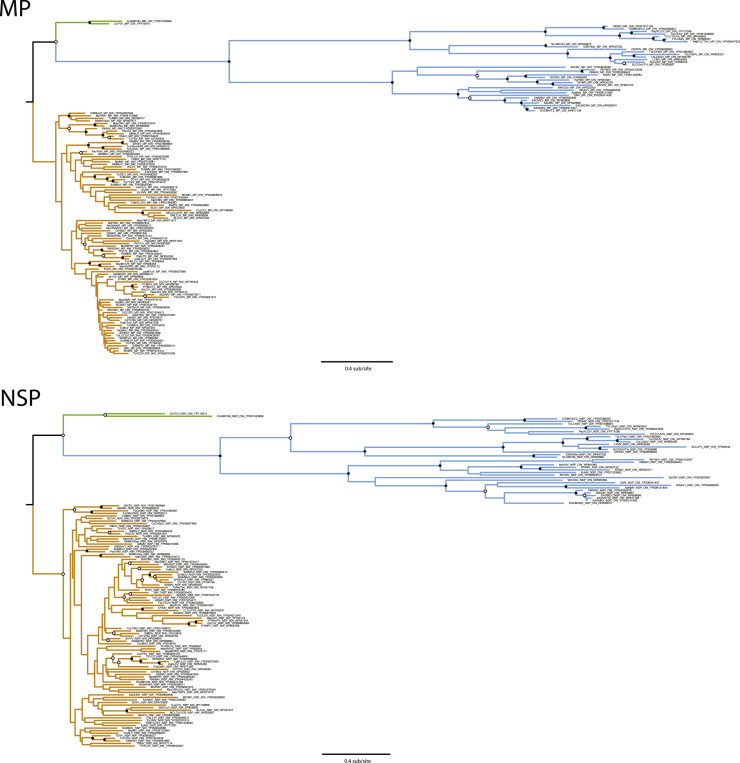
The maximum likelihood (ML) phylogeny of MP sequences, constructed using the JTT substitution model, is shown in Fig. 2. The data splits up into three clades: Old World (OW), New World (NW), and the two corchorus sequences from Vietnam (CoYVV and CoGMV/IN) being intermediate between the larger clades. The corchorus viruses have been previously described as viruses present in the OW that have features of NW begomoviruses (Ha et al., 2006): they do not have an AV2 gene and are more phylogenetically similar to NW viruses. These sequences were not considered part of either OW or NW clades for all subsequent analyses. There is strong bootstrap support (≥90% support) for most of the branching patterns seen in the OW clade, and poor support (<70% support) within the NW clade.
Fig. 2.
Begomovirus MP (top) and NSP (bottom) maximum likelihood trees. The OW clade is shown in blue, the NW clade is shown in orange, and the corchorus virus clade is shown in green. Bootstrap support is denoted by circles at supported nodes: a filled circle indicates at least 90% bootstrap support and an open circle indicates 70 to 90% bootstrap support. The scale bar represents amino acid substitutions per site.
Consistent with the MP tree, the NSP protein sequences separate into OW and NW clades with an intermediate corchorus virus group, and there is much higher bootstrap support for branching events in the OW clade (Fig. 2). The well-supported branching patterns in the OW clades appear similar in both trees, suggesting that these two proteins encoded on the DNA-B share a similar evolutionary history. One notable exception is the placement of soybean chlorotic blotch virus (SbCBV) from Nigeria, which groups with other African sequences in the NSP tree (with 55% bootstrap support) but groups with sequences of other viruses of legumes from Thailand, India and Pakistan in the MP tree (54% bootstrap support). Regardless of the placement of SbCBV, the lack of strong bootstrap support means that in neither tree did all sequences from Africa, nor all OW viruses of legumes, form a well-supported clade.
In both trees, the OW clade has deeper branches (longer horizontal lengths between nodes, indicating sequence divergence) than the NW viruses. Both trees support the notion that there is significantly more protein diversity in the MP and NSP in the OW compared to the NW. We confirmed the greater diversity in the OW proteins compared to NW proteins by measuring Shannon Entropy (Table 1). In aggregate and in both the OW and NW, NSP is more diverse compared to MP. However, the most notable result was the very low Shannon Entropy calculated for the NW MP dataset (0.07), which was much lower than the NW NSP value (0.52). This indicates that the forces shaping sequence diversity in the NW are not acting equally on both the MP and NSP proteins – the MP sequences are much more similar to each other.
Table 1.
Shannon Entropy estimates for the full and for geographic subsets of the aligned MP and NSP datasets.
|
MP |
NSP |
||
| 0.21 |
0.62 |
||
| OW | NW | OW | NW |
| 0.60 | 0.07 | 0.82 | 0.52 |
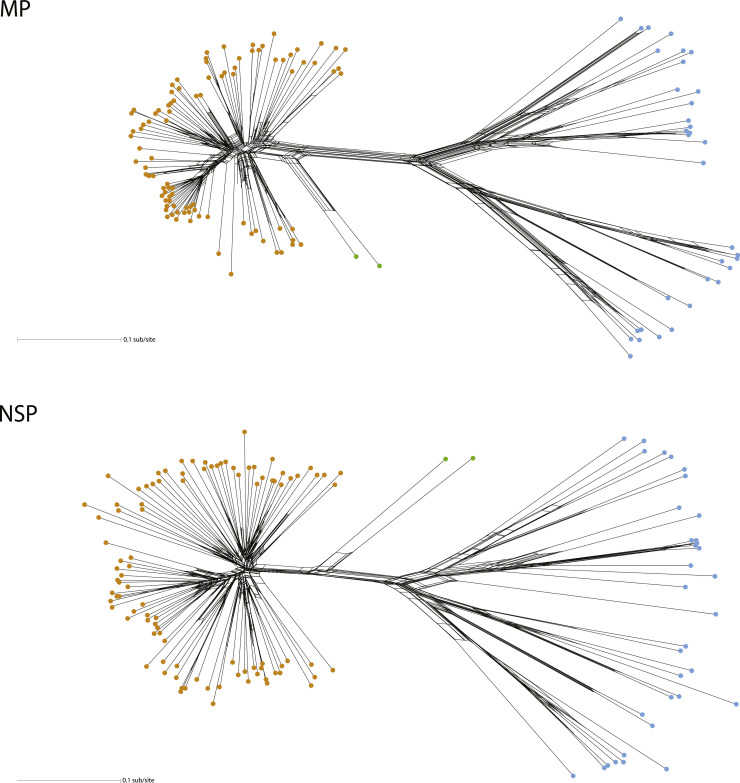
Next, we determined if recombination contributed to the diversity of these proteins. We used SplitsTree to create unrooted phylogenetic networks from the same alignments used for construction of ML trees (Fig. 3). All three clades (NW, OW, corchorus) have some reticulation but the phi test did not support a significant effect of recombination in the evolution of MP (p = 0.107) or NSP (p = 0.255). Again, the branch lengths of the network for the OW virus group are longer than the branch lengths for NW, reflecting the larger genetic diversity illustrated in ML phylogenies and quantified by Shannon Entropy.
Fig. 3.
Reticulate networks of relatedness for MP (top) and NSP (bottom) datasets. Viruses with blue dots are OW viruses and viruses with orange dots are NW viruses. The corchorus viruses CoYVV and CoGMV are green dots. The scale bar represents amino acid substitutions per site.
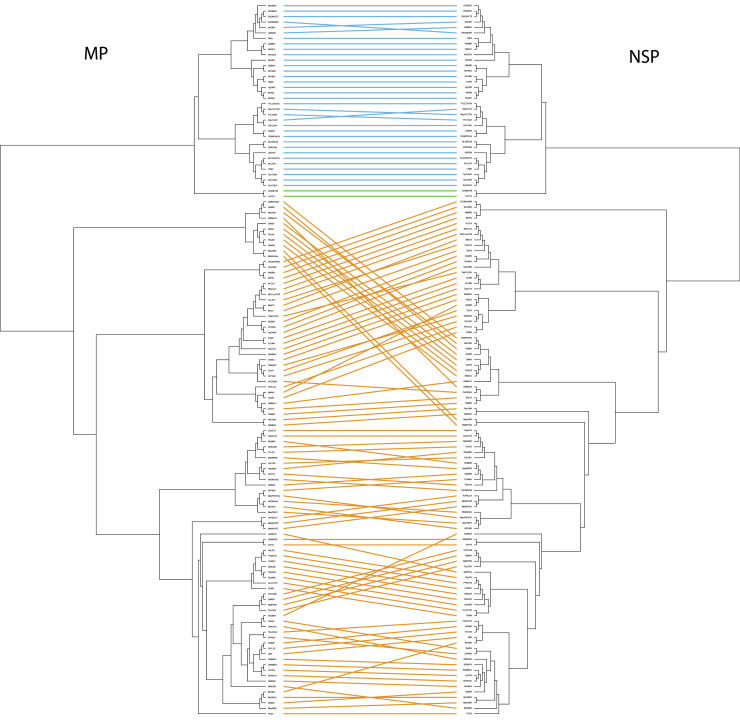
We then examined the extent of recombination that may have occurred between the two protein-coding regions on the DNA-B using a tanglegram (Venkatachalam et al., 2010). A tanglegram showed that there was no gene exchange across clades that were well-supported in the phylogenies (e.g., between OW and NW, or between subclades in the OW, Fig. 4). There were two crossed lines among OW viruses indicating that one of the ORFs may have recombined between two cassava mosaic begomoviruses, and between two pepper viruses. There are substantially more crossed lines in the NW than the OW, even after adjusting for the increased representation of sequences from the NW. While this could indicate increased recombination in the NW, we think it is more likely that this reflects the poor bootstrap support of the NW clade in the two ML trees (MP: 63% of nodes have <50% support; NSP: 53% of nodes have <50% support). When we looked for phylogenetic support (≥70% in both the MP and NSP) for viruses that have different sister taxa on the two trees we failed to find a single example from the NW viruses that supports recombination causing the crossed lines – the poor phylogenetic resolution explained them all.
Fig. 4.
Tanglegram connecting the same species’ MP (left) and NSP (right) sequences on the two phylogenies. Sequences connected by the blue lines are OW viruses. Sequences connected by the orange lines are NW viruses. The corchorus viruses CoYVV and CoGMV are shown as the green lines between the OW and NW clades.
4. Discussion
We confirmed the overall pattern of strong separation of OW and NW clades in the DNA-B observed by Briddon et al. (2010) using an updated dataset of 131 sequences (twice as many species represented). We further saw the two OW subclades of that previous work to be well-supported, into what we would call “Asian non-legumoviruses” and “African viruses and legumoviruses.” Briddon et al. (2010) did not report support values for their DNA-B phylogeny, so we cannot compare relative strength of support for various patterns, but we saw the incongruous placement of SbCBV between our two trees (Fig. 2) as a signal that neither the African viruses nor the legumoviruses formed a supported monophyly in both trees. As migration is regularly documented for begomoviruses it is not surprising that increased sampling has begun to break down clading patterns that correlated well to geography (Duffy and Holmes, 2007; Idris et al., 2006; Paximadis et al., 1999).
While recombination on the DNA-B segment is understudied compared to the DNA-A, there is ample evidence that recombination occurs between DNA-B segments (Crespo-Bellido et al., 2021; Xavier et al., 2021), and the common region is a hotspot of recombination on the DNA-B (Lefeuvre et al., 2009). Recombinants between the common region of DNA-A and DNA-B is also readily observed, which can lead to high fitness reassortants (Aimone et al., 2021; Briddon et al., 2010; Pita et al., 2001). In our protein-focused analysis, which necessarily excluded the non-protein coding common region, we found little evidence of recombination within genes and limited evidence of it between genes. The potential recombination events seen in the OW in the tanglegram (Fig. 4) occurred between closely related viruses and did not substantially disrupt the congruent relationships between the MP and NSP (i.e., Pepper yellow leaf curl Indonesia (PepYLCIV) moved locations between the two trees in Fig. 2, South African cassava mosaic virus (SACMV) and East African cassava mosaic Zanzibar virus (EACMZV) were still part of the same supported group of five cassava mosaic begomoviruses in both trees). There were many incongruities between the MP and NSP trees for the NW viruses, which could reflect significant recombination, but our ability to detect this is hampered by the poor resolution of the two protein trees (most nodes are not supported with >50% bootstrap support). While the protein level was appropriate for studying the evolution of the global population of the DNA-B, the shorter timescale of evolution of the NW begomoviruses means that recombination analyses should be conducted at the nucleotide level (e.g., Xavier et al., 2021). However, the lack of detected recombination in our dataset is consistent with a general pattern of a low number of recombination events found within genes on the DNA-B when examined on the nucleotide level (Lefeuvre et al., 2009).
Previous research based on the full nucleotide sequence of the DNA-B segment demonstrated that DNA-B is more variable than the DNA-A segment (Briddon et al., 2010). This was rigorously confirmed by a nucleotide diversity analysis of DNA-A and DNA-B segments from 5 different NW begomovirus datasets: DNA-B sequences were more diverse than DNA-A sequences in 4 of 5 species (Xavier et al., 2021). This enhanced diversity is especially notable because it is thought that DNA-B segments do not reassort as readily in the NW as in the OW (Briddon et al., 2010), and bioinformatic analyses suggest that NW DNA-B ORFs are under more purifying selection than their OW counterparts, and even most ORFs on the DNA-A (Ho et al., 2014). While our study cannot speak to the relative diversity of MP and NSP compared to the proteins encoded on the DNA-A, we confirmed that the OW proteins are more diverse than those in the NW, consistent with the NW viruses having a more recent common ancestor than the OW viruses (Rybicki, 1994; Lefeuvre et al., 2011). Our Shannon Entropy results are consistent with greater diversity implied by the low pairwise sequence comparison values for OW DNA-B segment sequences compared to those in the NW (Briddon et al., 2010). Building upon previous work comparing the selection pressures on the OW and NW ORFs (Ho et al., 2014), we show there is a large difference in protein diversity between the MP and NSP proteins in the NW (Table 1), indicating that the bottleneck of introduction into the NW and subsequent evolutionary pressures are affecting the two proteins differently. The very low diversity of NW MP proteins is consistent with the previous suggestion of high purifying selection pressure on MP accompanying the loss of the pre-coat protein and the possible emergence of novel phosphorylation sites (Ho et al., 2014), while the NSP can be complemented by the coat protein produced by the DNA-A (Qin et al., 1998). Importantly, an overlapping ORF, BV2, was recently suggested to be functional in a narrow OW clade that includes tomato yellow leaf curl Thailand virus (TYLCTHV) (Chiu et al., 2022). BV2-BV1 overlap is expected to constrain the evolution of NSP, whereas the evolution of MP should be unaffected. It is possible that additional overlapping functional ORFs remain to be discovered in specific begomovirus lineages (Chiu et al., 2022; Gong et al., 2021). There is no obvious genomic explanation for the lower diversity of the MP protein, especially in the New World, and it is likely due to its unique evolutionary ecology.
The increased sequencing of DNA-B segments offers a complementary window into the evolutionary history of bipartite begomoviruses, one that can help clarify patterns in DNA-A evolution (e.g., Crespo-Bellido et al., 2021). Our updated analysis confirmed most of the conclusions of Briddon et al. (2010) and suggests that recombination is not responsible for the evolutionary patterns or sequence diversity in the movement and nuclear shuttle proteins.
CRediT authorship contribution statement
Divya Dubey: Conceptualization; Data Curation; Formal Analysis; Visualization; Writing J. Steen Hoyer: Formal Analysis; Visualization; Writing Siobain Duffy: Funding acquisition; Project Administration; Writing/
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was funded by a grant from the National Science Foundation (OIA 1545553, to SD). We would like to thank the members of the Duffy lab, especially Alvin Crespo-Bellido for his help with the tanglegram and Erik Lavington for assistance with his python script for calculating Shannon Entropy.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.virusres.2022.198959.
Appendix. Supplementary materials
Data availability
Data was obtained from GenBank (public), our alignments are public (zenodo record 6985414) , code is on github..
References
- Aimone C.D., Lavington E., Hoyer J.S., Deppong D.O., Mickelson-Young L., Jacobson A., Kennedy G.G., Carbone I., Hanley-Bowdoin L., Duffy S. Population diversity of cassava mosaic begomoviruses increases over the course of serial vegetative propagation. J. Gen. Virol. 2021;102(7) doi: 10.1099/jgv.0.001622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Briddon R.W., Patil B.L., Bagewadi B., Nawaz-ul-Rehman M.S., Fauquet C.M. Distinct evolutionary histories of the DNA-A and DNA-B components of bipartite begomoviruses. BMC Evol. Biol. 2010;10(1):1–17. doi: 10.1186/1471-2148-10-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown J.K., Zerbini F.M., Navas-Castillo J., Moriones E., Ramos-Sobrinho R., Silva J.C.F., Fiallo-Olivé E., Briddon R.W., Hernández-Zepeda C., Idris A., Malathi V.G., Martin D.P., Rivera-Bustamante R., Ueda S., Varsani A. Revision of Begomovirus taxonomy based on pairwise sequence comparisons. Arch. Virol. 2015;160(6):1593–1619. doi: 10.1007/s00705-015-2398-y. [DOI] [PubMed] [Google Scholar]
- Bruen T., Philippe H., Bryant D. A simple and robust statistical test for detecting the presence of recombination. Genetics. 2006;172:2665–2681. doi: 10.1534/genetics.105.048975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu C.W., Li Y.R., Lin C.Y., Yeh H.H., Liu M.J. Translation initiation landscape profiling reveals hidden open-reading frames required for the pathogenesis of tomato yellow leaf curl Thailand virus. Plant Cell. 2022;34(5):1804–1821. doi: 10.1093/plcell/koac019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crespo-Bellido A., Hoyer J.S., Dubey D., Jeannot R.B., Duffy S. Interspecies recombination has driven the macroevolution of cassava mosaic begomoviruses. J. Virol. 2021;95(17) doi: 10.1128/JVI.00541-21. e00541-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darriba D., Taboada G.L., Doallo R., Posada D. ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics. 2011;27(8):1164–1165. doi: 10.1093/bioinformatics/btr088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffy S., Holmes E.C. Multiple Introductions of the Old World Begomovirus Tomato yellow leaf curl virus into the New World. Appl. Environ. Microbiol. 2007;73(21):7114–7117. doi: 10.1128/AEM.01150-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R.C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32(5):1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiallo-Olivé E., Lett J.-M., Martin D.P., Roumagnac P., Varsani A., Zerbini F.M., Navas-Castillo J. ICTV virus taxonomy profile: geminiviridae 2021. J. Gen. Virol. 2021;102(12) doi: 10.1099/jgv.0.001696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fondong V.N. Geminivirus protein structure and function. Mol. Plant Pathol. 2013;14(6):635–649. doi: 10.1111/mpp.12032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galili T. dendextend: an R package for visualizing, adjusting and comparing trees of hierarchical clustering. Bioinformatics. 2015;31(22):3718–3720. doi: 10.1093/bioinformatics/btv428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong P., Tan H., Zhao S., Li H., Liu H., Ma Y., Zhang X., Rong J., Fu X., Lozano-Durán R., Li F., Zhou X. Geminiviruses encode additional small proteins with specific subcellular localizations and virulence function. Nat. Commun. 2021;12(1):4278. doi: 10.1038/s41467-021-24617-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha C., Coombs S., Revill P., Harding R., Vu M., Dale J. Corchorus yellow vein virus, a new world geminivirus from the old world. J. Gen. Virol. 2006;87(4):997–1003. doi: 10.1099/vir.0.81631-0. [DOI] [PubMed] [Google Scholar]
- Hanley-Bowdoin L., Bejarano E.R., Robertson D., Mansoor S. Geminiviruses: masters at redirecting and reprogramming plant processes. Nat. Rev. Microbiol. 2013;11(11):777–788. doi: 10.1038/nrmicro3117. [DOI] [PubMed] [Google Scholar]
- Hanley-Bowdoin L., Settlage S.B., Orozco B.M., Nagar S., Robertson D. Geminiviruses: models for plant DNA replication, transcription, and cell cycle regulation. Crit. Rev. Biochem. Mol. Biol. 2000;35(2):105–140. doi: 10.1080/07352689991309162. [DOI] [PubMed] [Google Scholar]
- Hehnle S., Wege C., Jeske H. Interaction of DNA with the movement proteins of geminiviruses revisited. J. Virol. 2004;78(14):7698–7706. doi: 10.1128/JVI.78.14.7698-7706.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hesketh E.L., Saunders K., Fisher C., Potze J., Stanley J., Lomonossoff G.P., Ranson N.A. The 3.3 Å structure of a plant geminivirus using cryo-EM. Nat. Commun. 2018;9(1):2369. doi: 10.1038/s41467-018-04793-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho E.S., Kuchie J., Duffy S. Bioinformatic analysis reveals genome size reduction and the emergence of tyrosine phosphorylation site in the movement protein of new world bipartite begomoviruses. PLoS One. 2014;9(11) doi: 10.1371/journal.pone.0111957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huson D.H., Bryant D. Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 2006;23(2):254–267. doi: 10.1093/molbev/msj030. [DOI] [PubMed] [Google Scholar]
- Hussain M., Mansoor S., Iram S., Fatima A.N., Zafar Y. The nuclear shuttle protein of tomato leaf curl New Delhi virus is a pathogenicity determinant. J. Virol. 2005;79(7):4434–4439. doi: 10.1128/JVI.79.7.4434-4439.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Idris A.M., Abdel-Salam A., Brown J.K. Introduction of the new world squash leaf curl virus to Squash (Cucurbita pepo) in Egypt: a potential threat to important food crops. Plant Dis. 2006;90(9) doi: 10.1094/PD-90-1262B. 1262–1262. [DOI] [PubMed] [Google Scholar]
- Idris A.M., Shahid M.S., Briddon R.W., Khan A.J., Zhu J.-K., Brown J.K.Y. An unusual alphasatellite associated with monopartite begomoviruses attenuates symptoms and reduces betasatellite accumulation. J. Gen. Virol. 2011;92(3):706–717. doi: 10.1099/vir.0.025288-0. [DOI] [PubMed] [Google Scholar]
- Ingham D.J., Pascal E., Lazarowitz S.G. Both bipartite geminivirus movement proteins define viral host range, but only BL1 determines viral pathogenicity. Virology. 1995;207(1):191–204. doi: 10.1006/viro.1995.1066. [DOI] [PubMed] [Google Scholar]
- Jyothsna P., Rawat R., Malathi V.G. Predominance of tomato leaf curl Gujarat virus as a monopartite begomovirus: association with tomato yellow leaf curl Thailand betasatellite. Arch. Virol. 2013;158(1):217–224. doi: 10.1007/s00705-012-1468-7. [DOI] [PubMed] [Google Scholar]
- Kenyon L., Tsai W.-S., Shih S.-L., Lee L.-M. Emergence and diversity of begomoviruses infecting solanaceous crops in East and Southeast Asia. Virus Res. 2014;186:104–113. doi: 10.1016/j.virusres.2013.12.026. [DOI] [PubMed] [Google Scholar]
- Kikuno R., Toh H., Hayashida H., Miyata T. Sequence similarity between putative gene products of geminiviral DNAs. Nature. 1984;308(5959) doi: 10.1038/308562a0. 562–562. [DOI] [PubMed] [Google Scholar]
- Koonin E.V., Mushegian A., Ryabov E., Dolja V.V. Diverse groups of plant RNA and DNA viruses share related movement proteins that may possess chaperone-like activity. J. Gen. Virol. 1992;72(Pt. 12):2895–2903. doi: 10.1099/0022-1317-72-12-2895. [DOI] [PubMed] [Google Scholar]
- Larsson A. AliView: a fast and lightweight alignment viewer and editor for large datasets. Bioinformatics. 2014;30(22):3276–3278. doi: 10.1093/bioinformatics/btu531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefeuvre P., Harkins G.W., Lett J.-M., Briddon R.W., Chase M.W., Moury B., Martin D.P. Evolutionary time-scale of the begomoviruses: evidence from integrated sequences in the nicotiana genome. PLoS One. 2011;6(5):e19193. doi: 10.1371/journal.pone.0019193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefeuvre P., Lett J.-M., Varsani A., Martin D.P. Widely conserved recombination patterns among single-stranded DNA viruses. J. Virol. 2009;83(6):2697–2707. doi: 10.1128/JVI.02152-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leke W.N., Mignouna D.B., Brown J.K., Kvarnheden A. Begomovirus disease complex: emerging threat to vegetable production systems of West and Central Africa. Agric. Food Secur. 2015;4(1):1. doi: 10.1186/s40066-014-0020-2. [DOI] [Google Scholar]
- Li F., Xu X., Huang C., Gu Z., Cao L., Hu T., Ding M., Li Z., Zhou X. The AC5 protein encoded by Mungbean yellow mosaic India virus is a pathogenicity determinant that suppresses RNA silencing-based antiviral defenses. New Phytol. 2015;208(2):555–569. doi: 10.1111/nph.13473. [DOI] [PubMed] [Google Scholar]
- Loconsole G., Önelge N., Potere O., Giampetruzzi A., Bozan O., Satar S., De Stradis A., Savino V., Yokomi R.K., Saponari M. Identification and characterization of citrus yellow vein clearing virus, a putative new member of the genus mandarivirus. Phytopathology®. 2012;102(12):1168–1175. doi: 10.1094/PHYTO-06-12-0140-R. [DOI] [PubMed] [Google Scholar]
- Lucía-Sanz A., Manrubia S. Multipartite viruses: adaptive trick or evolutionary treat? NPJ Syst. Biol. Appl. 2017;3(1):1–11. doi: 10.1038/s41540-017-0035-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins L.G.C., Raimundo G.A.S., Ribeiro N.G.A., Silva J.C.F., Euclydes N.C., Loriato V.A.P., Duarte C.E.M., Fontes E.P.B. A Begomovirus nuclear shuttle protein-interacting immune hub: hijacking host transport activities and suppressing incompatible functions. Front. Plant Sci. 2020;11:398. doi: 10.3389/fpls.2020.00398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naseem S., Winter S. Quantification of African cassava mosaic virus (ACMV) and East African cassava mosaic virus (EACMV-UG) in single and mixed infected Cassava (Manihot esculenta Crantz) using quantitative PCR. J. Virol. Methods. 2016;227:23–32. doi: 10.1016/j.jviromet.2015.10.001. [DOI] [PubMed] [Google Scholar]
- Ouattara A., Tiendrébéogo F., Becker N., Urbino C., Thébaud G., Hoareau M., Allibert A., Chiroleu F., Vernerey M.-S., Traoré E.V., Barro N., Traoré O., Lefeuvre P., Lett J.-M. Synergy between an emerging monopartite begomovirus and a DNA-B component. Sci. Rep. 2022;12(1):695. doi: 10.1038/s41598-021-03957-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padidam M., Beachy R.N., Fauquet C.M. The role of av2 (“precoat”) and coat protein in viral replication and movement in tomato leaf curl geminivirus. Virology. 1996;224(2):390–404. doi: 10.1006/viro.1996.0546. [DOI] [PubMed] [Google Scholar]
- Patil B.L., Fauquet C.M. Cassava mosaic geminiviruses: actual knowledge and perspectives. Mol. Plant Pathol. 2009;10(5):685–701. doi: 10.1111/j.1364-3703.2009.00559.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paximadis M., Idris A.M., Torres-Jerez I., Villarreal A., Rey M.E.C., Brown J.K. Characterization of tobacco geminiviruses in the old and new world. Arch. Virol. 1999;144(4):703–717. doi: 10.1007/s007050050537. [DOI] [PubMed] [Google Scholar]
- Péréfarres F., Thierry M., Becker N., Lefeuvre P., Reynaud B., Delatte H., Lett J.-M. Biological invasions of geminiviruses: case study of TYLCV and bemisia tabaci in reunion Island. Viruses. 2012;4(12):3665–3688. doi: 10.3390/v4123665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pita J.S., Fondong V.N., Sangaré A., Otim-Nape G.W., Ogwal S., Fauquet C.M. Recombination, pseudorecombination and synergism of geminiviruses are determinant keys to the epidemic of severe cassava mosaic disease in Uganda. J. Gen. Virol. 2001;82(3):655–665. doi: 10.1099/0022-1317-82-3-655. [DOI] [PubMed] [Google Scholar]
- Priyadarshini P., C G., Ambika M.V., Tippeswamy R., Savithri H.S. Functional characterization of coat protein and V2 involved in cell to cell movement of cotton leaf curl kokhran virus-dabawali. PLoS One. 2011;6(11):e26929. doi: 10.1371/journal.pone.0026929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin S., Ward B.M., Lazarowitz S.G. The bipartite geminivirus coat protein aids BR1 function in viral movement by affecting the accumulation of viral single-stranded DNA. J. Virol. 1998;72(11):9247–9256. doi: 10.1128/JVI.72.11.9247-9256.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team . R Foundation for Statistical Computing; Vienna, Austria: 2020. R: A language and environment for statistical computing.https://www.R-project.org/ URL. [Google Scholar]
- Rojas M.R., Macedo M.A., Maliano M.R., Soto-Aguilar M., Souza J.O., Briddon R.W., Kenyon L., Rivera Bustamante R.F., Zerbini F.M., Adkins S., Legg J.P., Kvarnheden A., Wintermantel W.M., Sudarshana M.R., Peterschmitt M., Lapidot M., Martin D.P., Moriones E., Inoue-Nagata A.K., Gilbertson R.L. World management of geminiviruses. Annu. Rev. Phytopathol. 2018;56(1):637–677. doi: 10.1146/annurev-phyto-080615-100327. [DOI] [PubMed] [Google Scholar]
- Roumagnac P., Lett J.-M., Fiallo-Olivé E., Navas-Castillo J., Zerbini F.M., Martin D.P., Varsani A. Establishment of five new genera in the family geminiviridae: citlodavirus, maldovirus, mulcrilevirus, opunvirus, and topilevirus. Arch. Virol. 2022;167(2):695–710. doi: 10.1007/s00705-021-05309-2. [DOI] [PubMed] [Google Scholar]
- Rybicki E.P. A phylogenetic and evolutionary justification for three genera of Geminiviridae. Arch. Virol. 1994;139(1):49–77. doi: 10.1007/BF01309454. [DOI] [PubMed] [Google Scholar]
- Schliep K., Potts A.J., Morrison D.A., Grimm G.W. Intertwining phylogenetic trees and networks. Methods Ecol. Evol. 2017;8(10):1212–1220. doi: 10.1111/2041-210X.12760. [DOI] [Google Scholar]
- Seal S.E., vandenBosch F., Jeger M.J. Factors influencing begomovirus evolution and their increasing global significance: implications for sustainable control. Crit. Rev. Plant Sci. 2006;25(1):23–46. doi: 10.1080/07352680500365257. [DOI] [Google Scholar]
- Sicard A., Michalakis Y., Gutiérrez S., Blanc S. The strange lifestyle of multipartite viruses. PLoS Pathog. 2016;12(11) doi: 10.1371/journal.ppat.1005819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30(9):1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres-Herrera S.I., Romero-Osorio A., Moreno-Valenzuela O., Pastor-Palacios G., Cardenas-Conejo Y., Ramírez-Prado J.H., Riego-Ruiz L., Minero-García Y., Ambriz-Granados S., Argüello-Astorga G.R. A lineage of begomoviruses encode rep and AC4 proteins of enigmatic ancestry: hints on the evolution of geminiviruses in the new world. Viruses. 2019;11(7):644. doi: 10.3390/v11070644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varma A., Malathi V.G. Emerging geminivirus problems: a serious threat to crop production. Ann. Appl. Biol. 2003;142(2):145–164. doi: 10.1111/j.1744-7348.2003.tb00240.x. [DOI] [Google Scholar]
- Venkatachalam B., Apple J., John K.S., Gusfield D. Untangling tanglegrams: comparing trees by their drawings. IEEE/ACM Trans. Comput. Bio. Bioinform. 2010;7(4):588–597. doi: 10.1109/TCBB.2010.57. [DOI] [PubMed] [Google Scholar]
- Von Arnim A., Stanley J. Determinants of tomato golden mosaic virus symptom development located on DNA B. Virology. 1992;186(1):286–293. doi: 10.1016/0042-6822(92)90083-2. [DOI] [PubMed] [Google Scholar]
- Xavier C.A.D., Godinho M.T., Mar T.B., Ferro C.G., Sande O.F.L., Silva J.C., Ramos-Sobrinho R., Nascimento R.N., Assunção I., Lima G.S.A., Lima A.T.M., Murilo Zerbini F. Evolutionary dynamics of bipartite begomoviruses revealed by complete genome analysis. Mol. Ecol. 2021;30(15):3747–3767. doi: 10.1111/mec.15997. [DOI] [PubMed] [Google Scholar]
- Xu X., Liu Q., Fan L., Cui X., Zhou X. Analysis of synonymous codon usage and evolution of begomoviruses. J. Zhejiang Univ. Sci. B. 2008;9(9):667–674. doi: 10.1631/jzus.B0820005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S., Gong P., Ren Y., Liu H., Li H., Li F., Zhou X. The novel C5 protein from tomato yellow leaf curl virus is a virulence factor and suppressor of gene silencing. Stress Biol. 2022;2(1):19. doi: 10.1007/s44154-022-00044-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data was obtained from GenBank (public), our alignments are public (zenodo record 6985414) , code is on github..