Highlights
-
•
New emerging swine and avian influenza viruses could infect tree shrews and cause mild symptoms without adaptation.
-
•
Animal influenza viruses induced effective innate and adaptive immune responses in tree shrews.
-
•
Tree shrews may be a suitable mammalian model to study the cross-species transmission of animal influenza A viruses.
Keywords: Tree shrew, Low pathogenic, Cross-species infection, Swine influenza virus, Avian influenza virus, Anti-infection immunity, Transmissibility
Abstract
Animal influenza viruses can spread across species and pose a fatal threat to human health due to the high pathogenicity and mortality. Animal models are crucial for studying cross-species infection and the pathogenesis of influenza viruses. Tupaia belangeri (tree shrew) has been emerging as an animal model for multiple human virus infections recently because of the close genetic relationship and phylogeny with humans. So far, tree shrew has been reported to be susceptible to human influenza virus subtype H1N1, avian influenza viruses subtype H9N2, subtype H5N1, and subtype H7N9. However, the pathogenicity, infection, and immunity of swine and land avian influenza viruses with low pathogenicity and the potential to jump to humans remain largely unexplored in the tree shrew model. Previously, our team has successfully isolated the newly emerging swine influenza virus subtype H3N2 (A/Swine/GX/NS2783/2010, SW2783) and avian influenza virus subtype H6N6 (A/CK/ZZ/346/2014, ZZ346). In this study, we observed the pathogenicity, immune characteristics, and cross-species infection potential ability of SW2783 and ZZ346 strains in tree shrew model with 50% tissue culture infective dose (TCID50), hematoxylin and eosin (HE) staining, immunohistochemistry (IHC), real-time quantitative PCR (qRT-PCR) and other experimental methods. Both animal-borne influenza viruses had a strong ability on tissue infection in the turbinate and the trachea of tree shrews in vitro, in which SW2783 showed stronger replication ability than in ZZ346. SW2783 and ZZ346 both showed pathogenic ability with infected tree shrews model in vivo without prior adaptive culture, which mainly happened in the upper respiratory tract. However, the infection ability was weak, the clinical symptoms were mild, and the histopathological changes in the respiratory tract were relatively light. Furthermore, innate immune responses and adaptive immunity were observed in the tree shrew model after the infection of SW2783 and ZZ346 strains. We observed that the unadapted SW2783 and ZZ346 virus could transmit among tree shrews by direct contact. We also observed that SW2783 virus could transmit from tree shrews to guinea pigs. These results indicated that both animal-borne influenza viruses could induce similar pathogenicity and immune response to those caused by human-common influenza viruses. Tree shrews may be an excellent animal model for studying the interaction between the influenza virus and the host and the cross-species infection mechanism of the animal influenza virus.
1. Introduction
Influenza A viruses (IAVs) have a wide range of hosts, frequent variation, multiple subtypes, and rapid spread. On one hand, the recombination between human and animal stains may lead to the emergence of seasonal outbreaks and even a pandemic since the general lack of immunity to the recombination strain in the population. On the other hand, IAVs are circularly transmitted among animal hosts, and some of the viruses can cross the species barrier and jump to humans, and result in zoonosis, which might trigger high mortality and morbidity pandemic risks. Therefore, IAVs pose a persistent threat to human health (Short et al., 2015; Richman et al., 2017; Zheng et al., 2019). Although the pathogenic mechanism and characteristics of IAVs, and the underlying mechanism of cross-species transmission, have been well studied, it is not fully understood. The gap between pathogenic and transmission has a profound impact on the precise implementation of influenza prevention and control strategies. With the characteristics of gene subtypes diversity and strong adaptability, IAVs can cross the species barrier and cause cross-species infections. In addition, avian influenza viruses (AIVs) with low pathogenic (LPAIV) can turn into highly pathogenic avian influenza virus (HPAIV) strains through gene mutation or recombination events, thus gaining the functions of effective transmission among humans. Based on the above reasons, it is tough to predict the next infectious phenotype of the influenza virus that will spread among human populations. Furthermore, the influenza pandemic depends on both the viral ecosystem and the virological factors (Monne et al., 2014; Long et al., 2019), so it is necessary to stay high alert for those strains with a high pandemic potential (Petrova and Russell, 2018; Zhao et al., 2019; Zhu et al., 2019). The research results from the global IVA community emphasized that coordinated global surveillance of zoonotic influenza viruses in animals and humans is needed. The further research should focus on epidemiological investigations, virology, pathogenesis, mechanism of cross-species transmission, and new prevention and treatment strategies to provide a scientific basis for risk assessment and early warning of a potential influenza pandemic (Wang et al., 2021), research efforts can also provide an important tool to respond to the challenges of the continuous evolution of IAVs and the changes of ecological environment.
Animal models have unique advances for studying the infection and transmission processes, the pathogenesis of diseases, and host immune response. They are also crucial for evaluating the efficacy and safety of new drugs and vaccines. Several animal models of the influenza virus have been established, such as mice, guinea pigs, pigs, ferrets, non-human primates, and each has its advantages and disadvantages (Margine and Krammer, 2014). For example, mice, the most widely used animal model in IAVs studies currently, can usually be infected by IAVs with adaptive cultivation, and no human-influenza-like symptoms, such as fever, cough, and runny nose were observed after infection (Matsuoka et al., 2009). Ferrets and non-human primates are ideal animal models for IVAs research (Bouvier, 2015). However, it is hard to investigate the molecular mechanism in ferrets during IAVs infection with lack of well-annotated genomic sequence information to characterise immune responses; and the high maintenance cost has limited its application (Wong, 2019). As for non-human primates, clinical symptoms caused by IAVs infection and receptors distribution in the respiratory tract are different from those of humans. The high maintenance cost, strict feeding requirements, and ethical problems make non-human primates the most ideal but difficult model for influenza research (Margine and Krammer, 2014).
The tree shrew (Tupaia belangeri) belongs to the Scandentia. It has genetically closer to humans than rodents (Kumar and Hedges, 1998; Roberts et al., 2011; Xiao et al., 2017). Being a cousin of primates, tree shrews are emerging as potential animal models of various human diseases with several unique characteristics, including highly identical neurodevelopment, immune, biochemistry, and metabolism; in addition, the small body size (about 100∼150 g), fast growth and reproduction circle (about six weeks), and low feeding cost make it an ideal animal mode (Fan et al., 2013; Xiao et al., 2017; Yao, 2017; Kayesh et al., 2021). Study results showed that the distribution of IAV sialic acid receptors of respiratory in tree shrews is analogous to that of humans and ferrets (Yang et al., 2013). Tree shrews would present various pathological changes and clinical symptoms after being infected with pathogens such as human H1N1, avian influenza viruses H9N2, H5N1, and H7N9 (Yang et al., 2013; Li et al., 2018; Sanada et al., 2019; Xu et al., 2019); the release of pro-inflammatory cytokines and specific antibody was observed (Li et al., 2018; Sanada et al., 2019; Xu et al., 2019). These studies suggest that tree shrews may be one of the helpful mammalian models for influenza virus research.
To date, limited studies have shown up on the pathogenicity, infection, and immune characteristics of low-pathogenic swine and land bird influenza viruses that potentially to infect humans with the tree shrew model. To evaluate the availability of tree shrews as animal models for influenza viruses, in the current study, we selected two animal-born IAV virus strains that were proven to be potentially infective to humans by our team previously: the newly emerged H3N2 subtype and chicken H6N6 subtype avian influenza viruses. The genome of the H3N2 subtype of swine influenza virus integrates the gene fragment of the 2009 new H1N1 pandemic influenza (pdm/09), and the chicken H6N6 subtype avian influenza virus are mainly prevalent in domestic poultry. Tree shrews were used as experimental animal models to observe the ability of infection, pathogenicity, immunological characteristics, and potential cross-species transmission of the two zoonotic influenza viruses.
2. Materials and methods
2.1. Ethics and biosafety statements
Animal experiments were approved and conducted following the protection and testing rules of the experimental animal's ethical committee of Guangxi Medical University. The operation of virus infection and infectious materials involved in this project was carried out in the biosafety level 2 (BSL2) laboratory of Guangxi Medical University (registration number: Nanwei Laboratory [2019]00003). The experimental operations of virology in this study referred to the standard operating procedures of the National Influenza Center and were optimized according to the actual conditions.
2.2. Animals
Healthy adult tree shrews of the Western Yunnan subspecies (female/male, 100∼140g of body weight) were purchased from the Experimental Animal Center of Kunming Medical University. Six-week-old Hartley strain guinea pigs (female, 250∼300 g) were purchased from Vital River Co. Ltd., Guangdong, China. All animals were housed in ventilated cages and were allowed free access to food and water. They were confirmed both sero-negative to A/Swine/GX/NS2783/2010 (H3N2) and A/CK/ZZ/346/2014 (H6N6) influenza viruses by hemagglutination inhibition (HI) assay and culture negative of isolated influenza virus before used in the experiment.
2.3. Viruses and cells
A/Swine/GX/NS2783/2010 (H3N2) (abbreviation: SW2783) is a new H3N2 subtype swine influenza virus isolated by our research group in 2010, whose genome integrates the gene fragment of the 2009 new influenza H1N1 pandemic virus (pdm/09) and has the ability of cross-species infection. A/CK/ZZ/346/2014 (H6N6) (abbreviation: ZZ346) is a subtype AIV of chicken-origin H6N6 isolated from Zhangzhou, Fujian province. A/DK/ST/3208/2012 (H9N2) (abbreviation: ST3208) is a subtype AIV of duck-origin H9N2 isolated from Shantou, Guangdong province. A/California/07/2009 (H1N1) (abbreviation: CA09) is the vaccine strain of the 2009 new H1N1 pandemic virus. The above virus strains were expanded using 9∼11 days old embryonated chicken eggs with specified pathogens free (SPF) and stored in the -70℃ for subsequent experiments. Madin-Darby canine kidney (MDCK) cells donated by Prof. Guan Yi from the University of Hong Kong were cultured with MEM medium containing 10% FBS and 1% mixture of penicillin, streptomycin in 37℃, 5% CO2 condition.
2.4. In vitro studies
Healthy adult Western Yunnan tree shrews were anesthetized and dissected. The respiratory tract, nasal turbinate, trachea, and lung tissues were taken for culture in vitro (Zhang et al., 2010). In short, the tissues were infected with 106 TCID50/ mL influenza viruses SW2783, ZZ346, ST3208, and 105 TCID50/ mL CA09, respectively. Meanwhile, PBS treatment as the control group and tissues were incubated in 37℃, 5% CO2 incubator for one hour, then discarded the supernatant, and the tissue was repeatedly rinsed with sterile PBS. Afterward, the virus growth solution (serum-free F-12K medium containing 0.2% TPCK trypsin and 1% penicillin-streptomycin) was added and cultured in 37℃, 5% CO2 incubator. At 12 h, 24 h, 36 h, 48 h, and 72 h after infection, the culture medium was collected for TCID50 assay. The tissue was ground in cold virus preservation solution, and after centrifuged, the supernatant was collected for TCID50 assay. Another piece of tissue was fixed in 4% paraformaldehyde, and observed for histopathological changes by HE staining and IHC.
2.5. In vivo study
Healthy adult Western Yunnan tree shrews were anesthetized with 3% sodium pentobarbital through intraperitoneal injection. 105 TCID50/ mL SW2783, ZZ346, and PBS were infected through a nasal drip (200 μL), conjunctiva drip (20 μL), and pharyngeal tonsil drip (30 μL). The status of tree shrews was recorded regularly after infection (∼14th day), including mental status, activity, food intake, infection-related symptoms, body temperature, and weight. On the 1st, 3rd, 5th, and 7th day after infection (1, 3, 5, and 7-day post-infection (dpi)), swab samples from nasal cavities, pharyngeal cavities, and conjunctival eyes were collected and inoculated with MDCK cells, consequent upon measuring virus titer by TCID50 assay. On the 3rd, 5th, and 7th day after infection (3, 5, 7 dpi), four tree shrews were randomly selected from each infection group. After euthanasia, sera were collected for HI assay. At the same time, the appearance of tissues such as turbinate, trachea, lung, thymus, spleen, and kidney were recorded and weighed. Organ coefficient (%) was calculated (organ weight (g))/body weight (g)×100). The trachea and lung tissues of tree shrews were divided into three parts. One tissue block was ground in cold virus preservation solution, and the virus titer of the supernatant was assessed by TCID50 assay. Another tissue block was fixed with 4% paraformaldehyde for histopathological and immunohistochemical examination. The last tissue block was used to determine immune molecules by real-time quantitative PCR.
2.6. Histopathological and immunohistochemical staining
The tissues were fixed with 4% paraformaldehyde, embedded in paraffin, and selected with a thickness of 4 μm. The pathological changes and nucleoprotein (NP) expression in infected tissues were observed by HE and IHC staining, respectively (Zhang et al., 2010; Zhu et al., 2018). In brief, after dewaxing and hydration, the tissue sections were repaired antigen, obstructed by endogenous peroxidase inhibitor for 10 min, then blocked by non-specific serum of normal goat for 10 min, followed by mouse anti-influenza virus nucleoprotein (NP) monoclonal antibody (primary antibody), and incubated overnight at 4℃. On the next day, we added biotinylated goat anti-mouse IgG (secondary antibody) and streptavidin-peroxidase to the section, coloration with DAB, and restain with hematoxylin. Results were observed and recorded under the microscope.
2.7. Total RNA extraction and real-time quantitative PCR
Total RNA was extracted by the TRIzol method, and cDNA was synthesized by reverse transcription (RT-PCR) with PrimeScriptTM RT Reagent Kit with gDNA Eraser (Takara, Japan). TB Green Premix Ex TaqTM II (Tli RNasH Plus) (Takara, Japan) was used for fluorescence real-time quantitative PCR (qRT-PCR) detection according to instructions, and mRNA relative expression of innate immune molecules were calculated by 2−△△C (Livak and Schmittgen, 2001). NCBI primer Blast was used to design qRT-PCR primers online. Primer information is shown in Supplementary table 1.
2.8. Influenza virus specific antibody measurement
We measured specific antibodies against the influenza virus in serum by modified HI assay . In short, serum samples were inactivated at 56℃ for 30 min and diluted with PBS in a U-shaped bottom microtitration plate, followed by the addition of 4 agglutinating units of viral antigen, incubation at room temperature for 1 h. Subsequently, added an equal volume of 0.6% Turkey red blood cell suspension, and determined antibody titer after 30 min of incubation at room temperature. The maximum dilution of the serum that completely inhibited erythrocyte agglutination was defined as HI titer (the detection limit was 1).
2.9. Intra- or inter-species transmission study
For the intra-species transmission study, three tree shrews or guinea pigs (a well-difined transmission model for influenza virus as a control) were inoculated through a nasal drip (200 μL) and conjunctiva drip (20 μL) with 105 TCID50 of the test virus. After 24 h post-infection, three guinea pigs or tree shrews were cohoused in the same cage with the inoculated animals. For the inter-species transmission study, three tree shrews or guinea pigs were inoculated through a nasal drip (200 μL) and a conjunctiva drip (20 μL) with 105 TCID50 of test virus, and three animals of the other species (guinea pigs or tree shrews) were cohoused in the same cage at 24 h post-infection. Body weights of the experimental animals were recorded regularly after infection (∼21th day). Swab samples from nasal cavities and conjunctival eyes were collected from all animals at 2-day intervals, starting on day two post-infection or one day post-exposure, and first kept at −80°C, consequent upon measuring virus titer by TCID50 assay. After euthanasia, sera were collected from each animal on day two before infection and day 21th post-infection for HI assay.
2.10. Statistical analysis
The data were processed and analyzed by GraphPad Prism 8.0 software. The Student's unpaired t test or Mann-Whitney U test was used to compare the difference of virus titer and mRNA expression between groups. P values <0.05 was considered significant.
3. Results
3.1. Effectively replication in respiratory tissues in vitro of tree shrew model
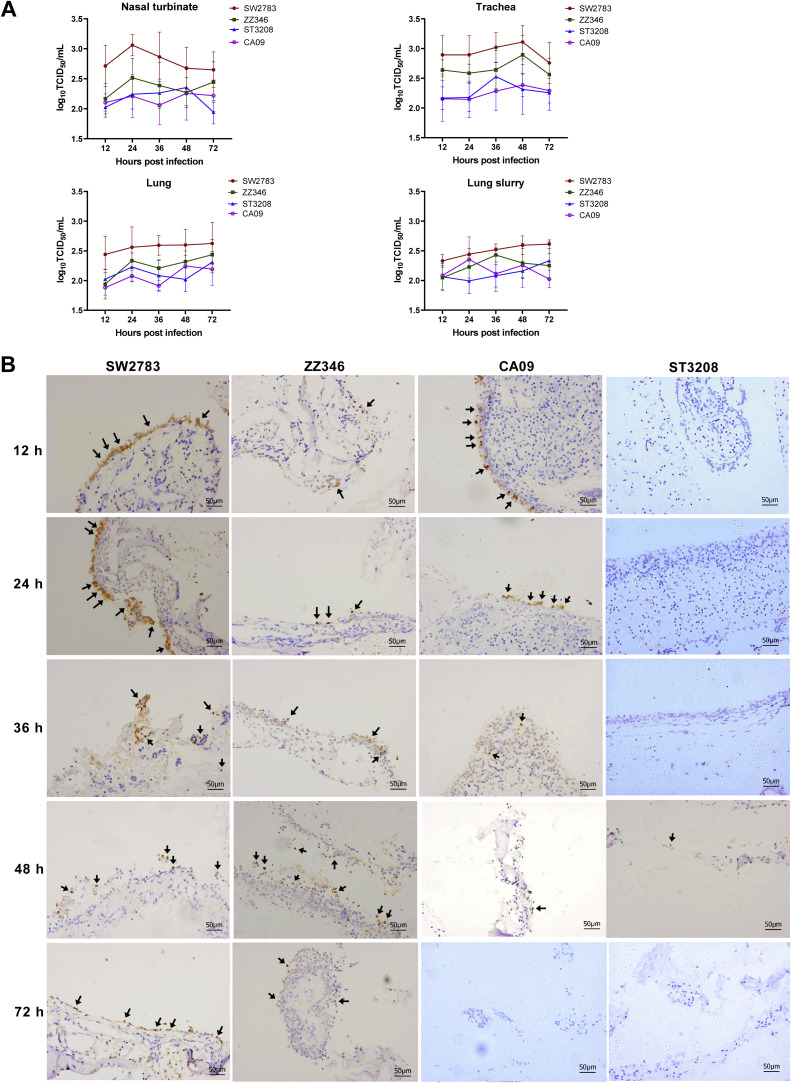
Newly emerged H3N2 subtype swine influenza virus SW2783, H6N6 subtype chicken influenza virus ZZ346, H9N2 subtype duck influenza virus ST3208, and H1N1 subtype human influenza virus CA09) were selected to infect nasal turbinate, trachea, and lung of tree shrews in vivo. Results showed that all four influenza viruses could infect and replicate in the respiratory tract of the tree shrew model without adaptive cultivation. Although the difference in infection ability in various tissues and at timepoints was not significant among different experimental groups, the trends of virus titers were not the same. Among the tissue culture supernatant, SW2783 had the highest titer, followed by ZZ346, while ST3208 and CA09 were relatively lower, indicating SW2783 had strong replication ability in the respiratory tract of tree shrews. In the lung tissue abrasive solution, the replication trend of SW2783 and ST3208 strains was similar to the trend in the tissue culture supernatant, showing an increasing trend in the late stage of culture. However, ZZ346 and CA09 showed a decreasing trend in the late stage of culture (Fig. 1A).
Fig. 1.
Susceptibility of four influenza virus strains in ex vivo cultures of tree shrew respiratory tract tissues. A. Growth kinetics in ex vivo culture of nasal turbinate, trachea, and lung (n=3 per time point) were infected with 106 TCID50 /mL of influenza virus (SW2783, ZZ346, or ST3208), or 105 TCID50 /mL of CA09. Viral titers were determined by TCID50 assay. Data are show as means ± SD. B. Immunohistochemical staining in ex vivo cultures of tree shrew nasal turbinate tissues infected with four influenza virus strains. Nasal turbinate tissues were harvested at 12 h, 24 h, 36 h, 48 h, and 72 h post-infection, prepared for IHC staining to detect the expression of NP protein in influenza viruses. Black arrows indicate positive staining. Bar, 50 μm.
In addition, respiratory tract tissue samples inoculated with the virus were prepared for IHC to detect the expression of NP protein in influenza viruses. IHC staining showed that distinctive NP proteins were expressed in four influenza viruses in the respiratory turbinate, trachea, and lung tissues (Fig. 1B, Supplementary Fig. 1, and Supplementary Fig. 2). The NP protein tested positive in the turbinate, trachea, and lung tissues of tree shrews, and was still detectable at 24h, 48h, 36h, and 72h post-infection. In the respiratory tract tissues of infected tree shrew models, NP protein was highest expressed in turbinate tissues, followed by trachea tissues, and relatively less expressed in lung tissues.
The total number of NP protein-expressed cells from tree shrew respiratory tract tissue infected with SW2783 was significantly higher than with ZZ346. Moreover, at the early stage of SW2783 infection, more NP protein-positive cells detected in the nasal turbinate tissue and trachea tissue. Our results indicated that both SW2783 and ZZ346 influenza viruses could effectively replicate in the respiratory tract of tree shrews but replicated mainly in the upper respiratory tract. SW2783 has a stronger ability for cross-species transmission compared with ZZ346. Upon CA09 infection, NP protein-expressed various in the trachea and lung tissues of tree shrews. Interestingly, the NP protein in turbinate tissues mainly presented at 12h and 24h after infection, and in trachea tissues, it was still detectable at 12 h and 48h after infection, while in lung tissues, few NP protein positive cells were observed at each time points after 24 h of infection. It demonstrated that the replication ability of the CA09 virus strain in the tissues of the respiratory tract was relatively weak. Furthermore, only a limited amount of NP protein was detected after the ST3208 infection. Some NP protein-expressed cells were detected merely at 48 h post-infection in turbinate, at 36 h in trachea tissue, and 48 h and 72 h in lung tissue, illustrating that ST3208 was barely to infect the respiratory tract tissue of tree shrews.
These results in vitro showed that all four influenza viruses could infect the respiratory tract tissues of tree shrews, but the infection ability was different. The new emerging swine-derived SW2783 and chicken-origin ZZ346 virus strain have a good infection capacity. In contrast, human pdm/09 vaccine strain CA09 and duck-origin ST3208 virus strain showed limited and poor replication ability. Thus, we selected SW2783 and ZZ346 virus strains to observe pathogenicity, immune characteristics, and cross-species infection potential with tree threw in vivo study.
3.2. New emerging swine SW2783 and avian ZZ346 could infect tree shrews in vivo and cause mild clinical symptoms without adaptive cultivation
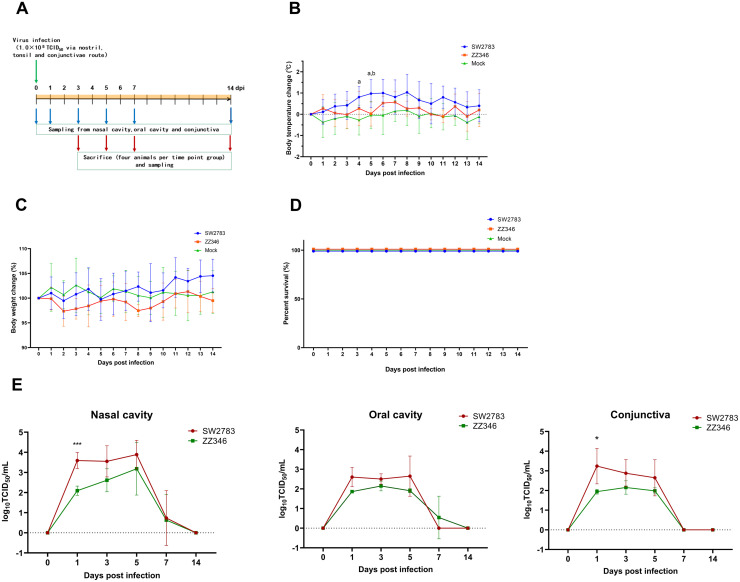
To further investigate the infection ability of the two influenza strains in vivo, we inoculated swine SW2783 and avian ZZ346 (105 TCID50/ mL) with tree shrews through the nasal cavity, pharynx cavity, and conjunctiva, respectively.
We first observed the general situation and symptoms in virus-infected tree shrews: on the 1st day of the infection, decreased appetite, reduced activity, and increased nasopharyngeal secretions were observed and lasted eight days. The symptoms described above were severe in models infected with the SW2783 group than in the ZZ346 group, but no severe symptoms or animal death were observed. The body temperature and the body weight of tree shrews were affected by the infection (Fig. 2B, C). Tree shrews’ body temperature first increased gradually after the SW2783 infection and then decreased slowly after the 8th day of infection. Especially, body temperature on the 4th and 5th days was significantly higher than those in the control group (P<0.05), and it was also significantly higher than those in the ZZ346 group on the 5th day. The body temperature of tree shrews after ZZ346 infection was slightly higher than that of the control group, but there was no significant difference (As the body temperature curve showed).
Fig. 2.
Clinical symptoms and growth kinetics of viruses in tree shrews after infection with swine and avian influenza viruses. A. Study design and experimental schedule. Tree shrews (n = 4 per group) were inoculated with swine SW2783, avian ZZ346 (105 TCID50/ mL) and PBS via nasal cavity, pharynxcavity, and conjunctiva, respectively. Changes of body temperature (B), body weight (C) and survival curve (D) in tree shrews infected with SW2783 and ZZ346 influenza viruses. E. Changes of viral titers in swab samples. Viral titers in nasal cavity, pharynxcavity, and conjunctival swap samples harvested from tree shrews infected with influenza viruses at 0, 1, 3, 5, 7, and 14 dpi (days post-infection). Viral titers were determined by TCID50 assay. Data are shown as means ± SD. For panels (B) and (C), “a” indicates a significant difference between SW2783 and Mock groups, P<0.05. and “b” indicates a significant difference between SW2783 and ZZ346 groups, P<0.05. For panels (E), asterisks indicate significant difference between SW2783 and ZZ346 groups. *P<0.05, ***P<0.001.
Compared with the control group, despite the weight changes, tree shrews showed no significant difference in SW2783 and ZZ346 infected groups, the weight of tree shrews decreased slightly on the 2nd day after infection, and the ZZ346 infected group showed a light and continuous decrease until 10th day after infection. Interestingly, the tree shrews gained weight in the later stage of SW2783 infection. After SW2783 and ZZ346 virus infection, no animal died before the scheduled sacrifice (Fig. 2D). Our results emphasize that both influenza viruses SW2783 and ZZ346 can cause disease to tree shrews. Nevertheless, no obvious clinical symptoms were observed in infected tree shrew models.
Next, we access the replication and release characteristics of SW2783 and ZZ346 influenza virus strains in tree shrew respiratory tissues (Fig. 2E). High titers of the virus in the nasal cavity, pharyngeal cavity, and conjunctival eye of tree shrews were detected on 1st-day post-infection, and the high titers maintained until 5th day of infection. In detail, virus titers in nasal and pharyngeal swabs peaked on the 5th day of infection, while those titers in conjunctiva eye swabs peaked on the 1st day and dropped after the first day of infection; the virus titer dropped sharply on day 7 of infection, it was undetectable in some tree shrews by then; and the titers cannot detect in all tree shrews since 14th day. The results indicated that both viruses could replicate and spread well in tree shrews; the replication capacity of SW2783 was stronger than that of ZZ346, especially on the first day of infection (P<0.05).
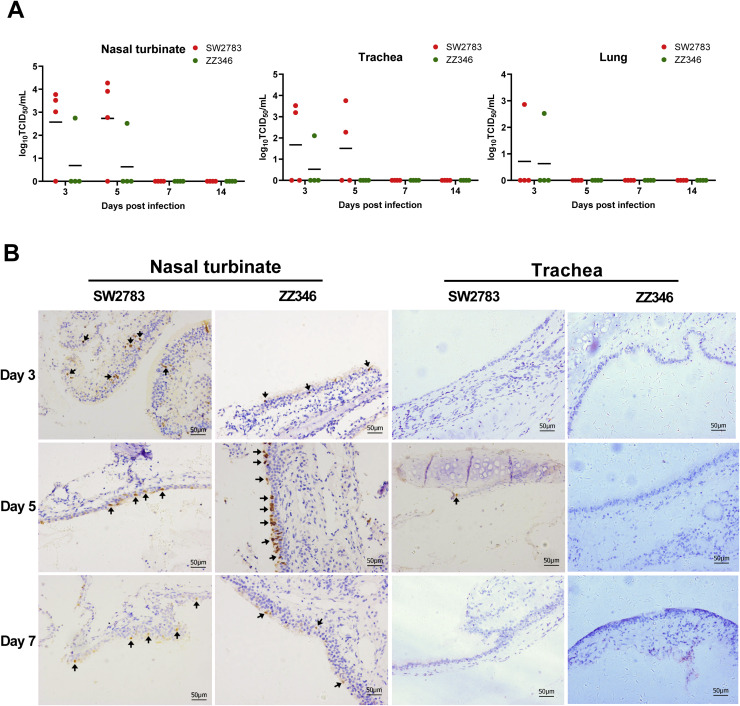
Then, the replicative infection ability of the two influenza viruses in respiratory tissues, such as nasal turbinate, tracheal, and lung tissues of the tree shrews were further evaluated (Fig. 3). The virus titer of nasal turbinate tissue from SW2783-infected tree shrews were relatively high on the 3rd day post-infection, and then peak on the 5th day. The trends of the titers were similar to those replicated in nasal swabs. For the infection in tracheal tissue, virus titers were detected in two tree shrews on the 3rd and 5th day of post-infection. As for lung tissue infection, the virus was detected only in one tree shrew on 3rd day. The replication ability of ZZ346 in the tree shrew respiratory tract was weaker than that of SW2783, presenting the virus replication was only detected in one of the models on day three and day five of infection. On the 7th and 14th day, virus titers were undetectable in nasal turbinate, tracheal, and lung tissues (Fig. 3A). The results emphasized again that both the influenza virus trains could infect and replicate in tree shrews without adaptation. Although there was no statistical difference in virus titer at each time-point, the replication ability of the two strains was distinct in different tissues, and the newly emerged swine SW2783 has a much stronger replication ability than that of avian ZZ346.
Fig. 3.
Replication and release characteristics of SW2783 and ZZ346 influenza viruses in tree shrew respiratory tissues. A. Viral titers from different tissues of the respiratory tract in tree shrews infected with SW2783 and ZZ346 influenza viruses. Tree shrews (n = 4 per group) were infected with 105 TCID50 /mL of swine SW2783 and avian ZZ346 influenza virus, respectively. The kinetics and time courses of viral load in nasal turbinate, trachea, and lung tissues harvested from infected tree shrews. Viral titers were determined by TCID50 assay. The dots represent the viral titer of individual tree threw; the thick horizontal bars indicate the mean values in each inoculated group. B. Immunohistochemical staining in tree shrew respiratory tract tissues infected with SW2783 and ZZ346 influenza viruses. Nasal turbinate and trachea tissues were harvested at 3, 5, 7, and 14 dpi, prepared for IHC staining to detect the expression of NP protein in influenza viruses. Black arrows indicate positive staining. Bar, 50 μm.
The infectious ability and characteristics of the two influenza viruses were further evaluated by investigating the expression and distribution of NP protein in tree shrew respiratory tract tissues using the IHC staining method (Fig. 3B and Table 1). NP protein was detected positive in the turbinate tissues on the 3rd, 5th, and 7th days after virus infection, which was relatively high on the 3rd and the 5th days. The NP protein expression in the SW2783 infected turbinate tissues was decreased on the 7th day after the infection, while it declined on 5th day in the ZZ346 infected turbinate tissues. The NP protein expression was detected only in one turbinate tissue on the 7th day of post-infection infected with ZZ346, no NP protein was detected in the two influenza viruses infected turbinate tissue of tree shrew on the 14th day. During the observation period, NP protein was undetectable in the trachea and lung tissues except one trachea tissue; NP protein was expressed weakly in trachea tissue on the 5th day after the SW2783 infection. These results again emphasized that both virus strains could infect tree shrews in vivo, of which SW2783 was much contagious. However, the infection was restricted to the upper respiratory tract; expression peaked at 3-5 days and gradually recovered at 7-14 days. The infectious process consisted with the infection of human influenza.
Table 1.
Positive results of immunohistochemical staining in tree shrew respiratory tract tissues infected with SW2783 and ZZ346 influenza viruses.
| Viruses | Tissues | Days post-infection | |||
|---|---|---|---|---|---|
| 3 dpi | 5 dpi | 7 dpi | 14 dpi | ||
| SW2783 | Nasal turbinate | 4/4 | 4/4 | 4/4 | 0/4 |
| Trachea | 0/4 | 1/4 | 0/4 | 0/4 | |
| Lung | 0/4 | 0/4 | 0/4 | 0/4 | |
| ZZ346 | Nasal turbinate | 4/4 | 3/4 | 1/4 | 0/4 |
| Trachea | 0/4 | 0/4 | 0/4 | 0/4 | |
| Lung | 0/4 | 0/4 | 0/4 | 0/4 | |
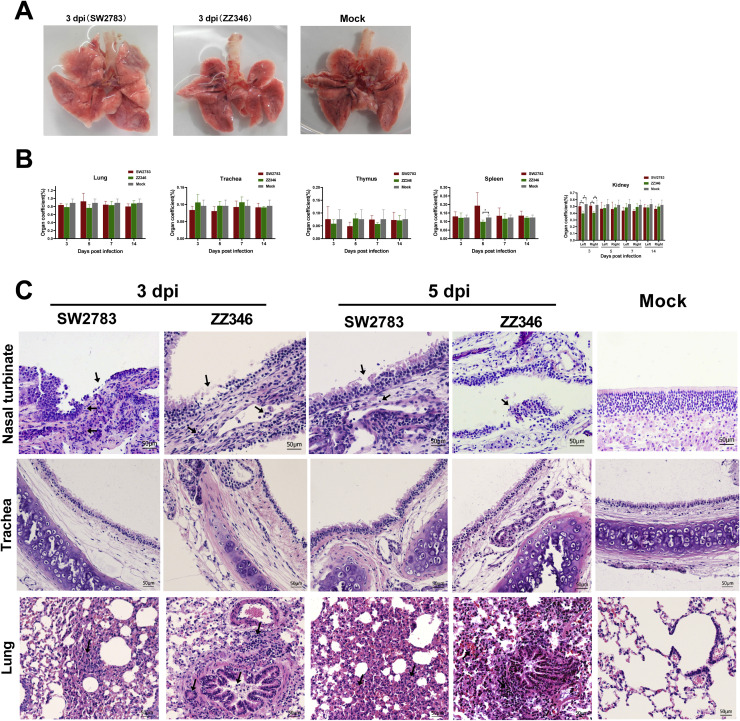
Additionally, four tree shrews were weighed and executed on the 3rd, 5th, 7th, and 14th days after infection of SW2783 and ZZ346, respectively. Organ indexes were calculated on the trachea lung, thymus, spleen, and kidney. The turbinate, trachea, and lung tissues were stained with HE staining to study the histopathological changes after the viruses’ infection. The results showed no significant differences in the lung, trachea, and thymus gland (Fig. 4A, B). The spleen index on the 5th day after ZZ346 infection was lower than that of the control group (P<0.05), while no difference was observed at other time points. The left and right renal indices on the 3rd day after the ZZ346 infection were lower than those in the control group and in the SW2783 group (P<0.05). However, there were no significant differences between the two virus infection groups and the control group at other time points (Fig. 4B). Results of HE staining showed that the nasal turbinate and lung tissues from the two viral strains infected models had various degrees of histopathological changes, with a small number of cell necrosis and inflammatory cell infiltration in the mucosal epithelium of the nasal turbinate (Fig. 4C). The infiltration of inflammatory cells was around parabronchus, and alveolar septal congestion was observed in the lung tissues (Fig. 4C). There were no pathological changes in the tracheal tissues after infection (Fig. 4C). The Mock group (PBS treated) showed complete structures of turbinate, trachea, and lung tissues with no structural damage.
Fig. 4.
The pathological changes in tree shrews after infection with swine and avian influenza viruses. A. Representative gross appearance of lungs from tree shrews inoculated with SW2783, ZZ346 influenza viruses and PBS. B. The organ coefficient of different organs from tree shrews (n = 4 per group) infected with influenza viruses. Data are show as means ± SD. Asterisks indicate significant differences, *P<0.05. C. Histopathology of tree shrew respiratory tract tissues infected with SW2783 and ZZ346 influenza viruses. Tree shrews (n = 4 per group) were infected with 105 TCID50 /mL of swine SW2783 and avian ZZ346 influenza virus, respectively. Representative pathological images (HE staining) of nasal turbinate, trachea, and lung tissues were harvested on 3 and 5 dpi. Black arrows indicate infiltration of inflammatory cells, cell necrosis, sloughed epithelial cells and alveolar septal congestion. Bar, 50 μm.
3.3. SW2783 and ZZ346 induced effective innate and adaptive immune responses in tree shrews
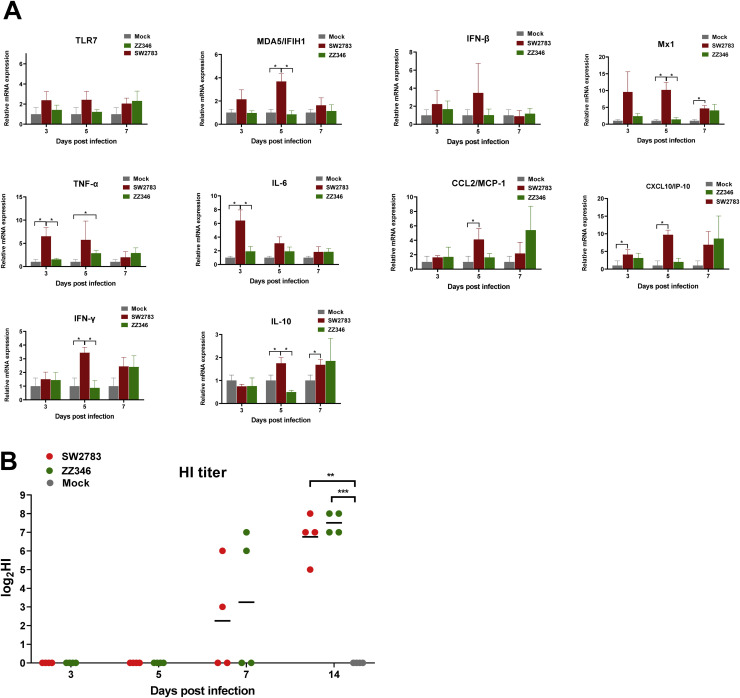
To understand the immune response, and the antibody dynamics after SW2783 and ZZ346 infection, qRT-PCR was used to measure the mRNAs of ten innate immunity molecules, such as innate immune pattern recognition molecules (TLR7, MDA5/IFIH1), the antiviral protein IFN-β, the interferon-induced antiviral protein Mx1, the pro-inflammatory cytokine (TNF-α, IL-6, IFN-γ, CCL2/MCP-1, CXCL10/IP-10), and immunomodulatory molecular IL-10, et al. in the turbinate mucosa in tree shrews. Viral hemagglutination inhibition (HI) assay was used to test the antibody in serum.
Results showed that except for innate immune pattern recognition molecules TLR7 and antiviral protein IFN-β, other molecules have been up-regulated after the two stains infection. For SW2783, TNF-α and IL-6 peak on the 3rd day after infection; the degree was higher than those in the ZZ346 group (P<0.05). On the 5th day after infection, MDA5, Mx1, CCL2/ McP-1, CXCL10/IP-10, IFN-γ, and IL-10 expression achieved the peak, and MDA5, Mx1, IFN-γ, IL-10 were significantly higher than the ZZ346 group (P<0.05), besides IL-10, was still at a high level on 7th day after infection (Fig. 5A). These results demonstrated that tree shrews infected with the two influenza stains could stimulate early antiviral immunity effectively and regulate the response degrees.
Fig. 5.
Host immune responses of tree shrew against infection of SW2783 and ZZ346 influenza viruses. A. Gene expression levels of innate immune molecular in tree shrew nasal turbinate tissues (n = 4 per group) inoculated with SW2783, ZZ346 influenza viruses and PBS at 3, 5, and 7 dpi (days post-infection). The mRNA expression levels of pattern recognition receptor (TLR7, MDA5/IFIH1), antiviral molecules (IFN-β, Mx1), inflammatory molecules (TNF-α, IL-6, CCL2/MCP-1, CXCL10/IP-10), and immunomodulatory molecules (IFN-γ, IL-10) were measured by qRT-PCR (real-time quantitative PCR). The relative expression values were normalized to those of tree shrew GAPDH and the expression level of mock group was set to one. Data are shown as means ± SD. B. Antibody response in tree shrew against influenza viruses’ infection. Serum antibody titer were determined by HI (hemagglutination inhibition) assay to SW2783 and ZZ346 influenza viruses with tree shrews’ serum collected at 3, 5, 7, and 14 dpi. The dots were presented HI titer of individual animal; the thick horizontal bars indicate the mean values in each inoculated group. *P<0.05, **P<0.01, and ***P<0.001.
In addition, the HI assay results proved that no specific influenza virus antibody was detected on the 3rd and 5th day after infection. However, virus antibodies were detected in two tree shrews in each group on the 7th day after infection. On the 14th day after infection, all tree shrews infected with the two virus strains were detected with antibodies against the influenza virus (Fig. 5B). It highlighted that antiviral immunity could gradually initiate in tree shrews after influenza virus infection.
3.4. New emerging swine SW2783 influenza viruses and chicken-origin ZZ346 influenza virus are transmissible in tree shrews, and SW2783 could also transmit from tree shrews to guinea pigs
To evaluate the intra- or inter-species transmissibility of two animal influenza viruses among tree shrews or guinea pigs, as well as between tree shrews and guinea pigs by using a direct-contact approach, we collected swab samples from nasal cavity and conjunctiva of tree shrews and guinea pigs infected with either swine SW2783 or avian ZZ346 influenza virus. We measured the viral titers by TCID50 assay. Sera were collected from each animal on day two before infection and on day 21 post-infection, the influenza virus-specific antibodies were measured by HI assay.
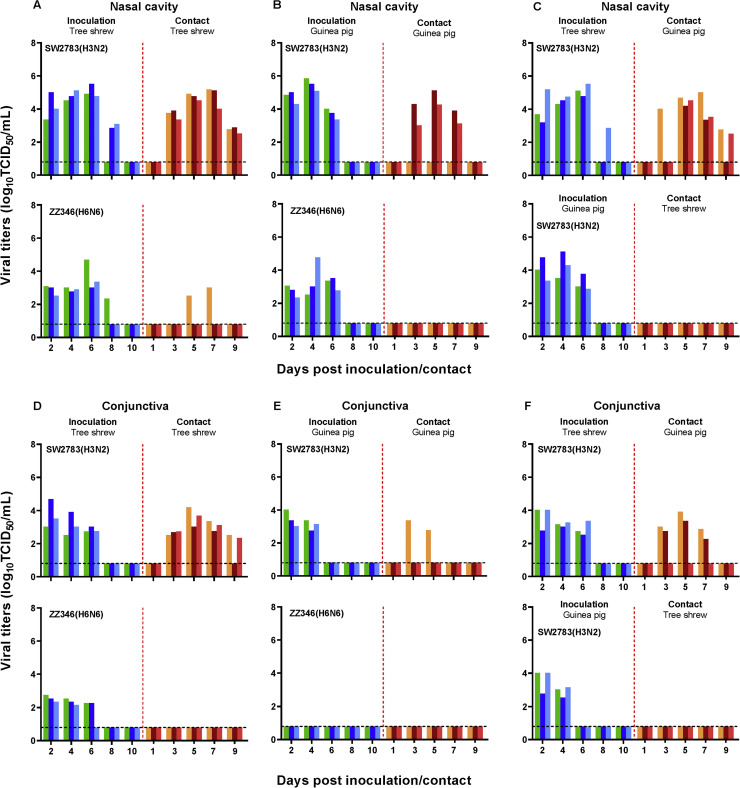
In the intra-species transmission experiment for swine SW2783 virus, surprisingly, SW2783 virus was readily transmitted from the three inoculated tree shrews to the three contacts. The viral load in the nasal and conjunctiva swap samples peaked at 6 and 7 dpi, respectively. The detectable virus persisted in nasal and conjunctiva swap samples from exposed tree shrews until 9 dpi, respectively (Fig. 6A, D). Sera from all inoculated and exposed tree shrews were seroconverted with higher antibody titers (Table 2). Viral titers was detected only in two of three exposed guinea pigs in the nasal swap samples at 3, 5 and 7 days post-exposure (Fig. 6B). Serum antibodies from two of three exposed guinea pigs were seroconverted, whereas all inoculated guinea pigs were seroconverted high antibody titers (Table 2).
Fig. 6.
Transmission of SW2783 and ZZ346 influenza viruses in tree shrews and guinea pigs. Intra-species transmission of SW2783 and ZZ346 viruses in tree shrews (A, D) or guinea pigs (B, E), and inter-species transmission of SW2783 viruses between guinea pigs and tree shrews (C, F). Groups of tree shrews or guinea pigs seronegative for influenza viruses (n=3) were inoculated with 105 TCID50 /mL of swine SW2783 or avian ZZ346 influenza virus, respectively. The next day, for the intra-species transmission experiment, the inoculated animals were co-housed with three guinea pigs or tree shrews, and for C and F, the inoculated tree shrews were co-housed with three guinea pigs, and vice versa. After infection with the test virus, the nasal and conjunctiva swabs of tree shrews and guinea pigs were collected at the indicated dpi. Virus titers were determined by TCID50 assay. Each color bar represents a value from an individual animal. The dashed black lines indicate the lower limit of detection.
Table 2.
Seroconversion of tree shrews and guinea pigs inoculated or contact with SW2783 and ZZ346 influenza viruses.
| Viruses | Inoculated | Direct contact | ||
|---|---|---|---|---|
| animals | HI titersa | animals | HI titersa | |
| SW2783 | tree shrews | 3/3 (8, 9, 8) | tree shrews | 3/3 (7, 7, 8) |
| guinea pigs | 3/3 (6, 7, 6) | guinea pigs | 2/3 (4, 6) | |
| tree shrewsb | 3/3 (7, 8, 8) | guinea pigsb | 3/3 (7, 6, 5) | |
| guinea pigsb | 3/3 (6, 7, 7) | tree shrewsb | 0/3 | |
| ZZ346 | tree shrews | 3/3 (7, 6, 8) | tree shrews | 1/3 (5) |
| guinea pigs | 3/3 (4, 4, 5) | guinea pigs | 0/3 | |
| tree shrewsb | 3/3 (7, 7, 6) | guinea pigsb | 0/3 | |
| guinea pigsb | 3/3 (6, 5, 5) | tree shrewsb | 0/3 | |
Seroconversion of the infected or exposed animals was tested by HI assay on 21 days post inoculation or post exposed. Seroconversion: no. positive/no. total (HI titers, log2).
Animals were used for the inter-species transmission study.
However, in the intra-species transmission experiment for the avian ZZ346 influenza virus, only one of the three tree shrews replicated the virus at 5 and 7 days post contact (Fig. 6 A), and seroconverted at 21 days post contact (Table 2). ZZ346 virus did not transmit in guinea pigs by direct contact in the present study (Fig. 6 B).
Based on the intra-species transmission experiment, we observed whether the swine SW2783 and avian ZZ346 viruses could transmit from tree shrew to guinea pig or vice versa. Viral shedding of SW2783 virus was detected at 3, 5, 7, and 9 (in nasal swap samples) and at 3, 5 7 (in conjunctiva swap samples) days post contact from the three exposed guinea pigs housed with the inoculated tree shrews (Fig. 6C), but the virus was not detected in the exposed tree shrews housed with the inoculated guinea pigs (Fig. 6C). The chicken-origin influenza virus ZZ346 could not transmit from tree shrews to guinea pigs, or vice versa (Supplementary Fig. 3, Table 2). Consistent with the above, in the four groups of exposed animals, only sera from three of the exposed guinea pigs co-housed with the inoculated tree shrews were seroconverted (Table 2).
Together, these results indicated that the swine SW2783 virus can transmit among tree shrews or guinea pigs, it can also spread from tree shrews to guinea pigs via direct contact. The transmissibility among tree shrews is much stronger than among guinea pigs. On contrast, avian ZZ346 virus only transmitted among tree shrews with low transmission efficiency. The swine SW2783 virus can use eyes as entry portals in tree shrew and guinea pig models.
4. Discussion
Transmission of influenza A virus (IAV) may mainly focus on species. However, the IAV can conquer the species barrier and directly jump to humans under certain conditions (Kuiken et al., 2006; Short et al., 2015). Humans infected with avian influenza viruses, such as H5N1, H6N1, H7N9, H9N2, H5N8, and H10N3, etc., and with swine influenza viruses H1N1, H2N2, and H3N2, etc. have been reported recently (Brown, 2000; Van Reeth, 2007; Freidl et al., 2014; Richman et al., 2017; Sun et al., 2018; Zheng et al., 2019; Pyankova et al., 2021; Qi et al., 2022). The new emerging virus has posed a fatal threat to human health. Two LPAIVs used in this study, SW2783 and ZZ346 were isolated from pigs and landfowls, respectively. SW2783 is a new H3N2 subtype swine influenza virus recombined with M and NP gene fragments of pdm/09 human influenza A H1N1 pandemic strain in 2009 (Zhu et al., 2011; Liang et al., 2014), whose gene sequence is highly correlated with the H3N2 triple-reassortant virus (TRV) strain (Vietnam/302/11) isolated from a Vietnamese girl with influenza (Liang et al., 2014). Studies have proved that the M gene of pdm/09 contributes to the effective transmission of the pdm/09 virus in animal models (Chou et al., 2011; Ma et al., 2012), and swine influenza virus recombined human genes H3, N2 with pdm /09 M genes may gain the function of human infection (Takemae et al., 2017). It found that the H3N2 subtype variant (H3N2v) from swine IAV which caused outbreaks in the United States contained the M gene of pdm/09 H1N1 (Jhung et al., 2013). In addition, genes of the pdm/09 virus can be re-introduced into pigs and produce new IAV variants in pigs, which results in the persistence and prevalence of IAV in humans (Liang et al., 2014; Rajao et al., 2017; Takemae et al., 2017). Our data from the transmission experiment also showed that the new emerging swine SW2783 (H3N2) virus can cross the species barrier from tree shrews to guinea pigs. Therefore, it is concerned that new emerging H3N2v recombined with internal gene fragments of pdm/09 H1N1 would cause a new surge. The landfowl H6N6 subtype of IVA is the dominant epidemic strain of poultry currently (Wang et al., 2014; Zou et al., 2019), which has a wide range of hosts spectrum, the subtype can infect not only birds but also mice, pigs, and other mammals without adaptive culture (Zhao et al., 2013; Wu et al., 2016; Zou et al., 2019). Wang and other researchers (Wang et al., 2014) found that 68.6% of the H6N6, a strain isolated from live poultry markets in Southern China, could bind to human SAα-2,6-Gal receptors. Recent studies proved that H5N6, the recombination of H6N6 and H5 subtype of HPAIV, can spread in humans (Pan et al., 2016; Yang et al., 2017; Wang et al., 2021). Researchers (Xin et al., 2015) found the positive rate of H6 antibody was 0.4% in a serological investigation with 15,689 Chinese participants, which implies that the H6 subtype of IAV may have crossed the species barrier and have infected mammals, including humans (Zou et al., 2019). All these findings suggest that the newly emerged swine influenza virus SW2783 and landfowls influenza virus ZZ346 may potentially infect humans.
As close relatives of primates, studies have reported that the infection of human H1N1, avian H9N2, and H7N9 in tree shrews will cause mild or moderate symptoms and pathological changes in respiratory tissues. The infection of H5N1 led to severe pneumonia with fever and weight loss (Sanada et al., 2019); furthermore, infection in tree shrews have induced pro-inflammatory cytokines and specific antibodies (Li et al., 2018; Sanada et al., 2019; Xu et al., 2019), it demonstrates that tree shrews may be a excellent model for influenza virus research. Therefore, we used tree shrews as an animal model to describe the cross-species infection ability, pathogenic characteristics, and induced immune response of LPIAV stain of newly emerged swine SW2783 and landfowl H6N6 subtype ZZ346.
The capacity of hemagglutinin glycoprotein HA in the IAV envelope binding with the sialic acid (SA) receptor of the host cell is the determinant of IAV infection (Glaser et al., 2005; Imai and Kawaoka, 2012; de Graaf and Fouchier, 2014; Greber, 2016). Generally, human influenza viruses mainly bind to SAα-2,6-Gal receptors expressed in host cells, whereas avian influenza viruses always bind to SAα-2,3-Gal receptors (Rogers and Paulson, 1983; de Graaf and Fouchier, 2014). Respiratory tract tissue of swine expresses both human SAα-2,6-Gal and avian SAα-2,3-Gal receptors, which means human and avian influenza viruses can infect swine and provides an ideal host for gene recombination of these viruses. Therefore, as the influenza virus gene mixer, swine is the critical intermediate host in influenza virus evolution and transmission (Ito et al., 1998; Richman et al., 2017). Other researchers have confirmed that the sialic acid receptors, including Saα-2, 3-Gal, Saα-2, and 6-Gal, are expressed in the respiratory tract tissues of landfowls, turkeys, and quails (Wan and Perez, 2006; Kuchipudi et al., 2009; Kimble et al., 2010; Yu et al., 2011). It indicates that the avian and human influenza virus can replicate in landfowls, thus becoming adaptive hosts for the AIV receptor conversion and potential intermediate hosts for the AIV spread to humans and other mammals (Imai and Kawaoka, 2012). It's concerned that frequent recombination with multiple species-borne IAV subtypes in intermediate hosts, persistent transmission, and epidemic may increase the binding affinity for human receptors, eventually evolving into new recombinant virus strains which can cross the species barrier and pose a fatal threat to the human health.
Studies have confirmed that the nasal turbinate and trachea tissues of the upper respiratory tract of tree shrews mainly express human receptor SAα-2,6-Gal. In the lung tissues of the lower respiratory tract, non-ciliated cuboid epithelial cells of terminal bronchioles mainly express SAα-2,6-Gal, while alveolar epithelial cells and macrophages mainly express SAα-2,3-Gal, suggesting that the distribution of influenza virus receptor in the respiratory tissues of tree shrews is akin to that of human and ferrets (Stevens et al., 2006; Nicholls et al., 2007; Yang et al., 2013; Byrd-Leotis et al., 2019). In this study, we observed that two animal influenza A viruses (SW2783 and ZZ346) could both infect tree shrews without adaptation, and SW2783 had a much stronger infection and replication ability than ZZ346, suggesting that the two animal influenza A viruses could bind sialic acid (SA) receptor and enters tree shrew cells for effective replication. Among them, the newly emerged swine influenza A virus SW2783 enabled strong infect the turbinate and trachea of tree shrews, indicating that SW2783 had a much stronger ability to bind the human receptor. While the landfowl influenza virus ZZ346 infects both upper and lower respiratory tract tissues of tree shrews, it indicates that ZZ346 may have evolved into the ability to recognize both human and avian influenza virus receptors. The details need further verified. Furthermore, the results of our transmission experiment showed that the new emerging swine SW2783 (H3N2) virus can transmit among tree shrews or guinea pigs and from tree shrews to guinea pigs via direct contact, and the chicken-origin influenza virus ZZ346 (N6N6) could also transmit among tree shrews. Our data indicate that the two zoonotic influenzas A viruses, especially swine SW2783, have the potential to cross the species barrier to infect mammals and humans. And a previous report (Xu et al., 2019) has found that H7N9 viruses could transmit among tree shrews and transmit from tree shrews to naïve guinea pigs by direct contact. These results suggest that the tree shrew would be another useful animal model for the pathogenesis and transmission of animal-origin IAVs research.
Interestingly, the two viruses mainly caused upper respiratory infection in vivo in the tree shrew model; the result was consistent with that in vitro. However, unlike in vitro, no NP antigen was detected in the trachea except one and lung tissues in vivo infection, and different degrees of inflammatory response were observed in the lung tissues. The result was in line with the results reported by Li (Li et al., 2018) after the infection of tree shrews with the avian influenza virus H9N2. Our results suggest that the complex mechanism of host-virus interactions impacts the ability and outcome of viral infection.
Slight symptoms such as decreased appetite and increased nasopharyngeal secretions in tree shrews were observed after SW2783 and ZZ346 infection through nasopharyngeal and eye pathways, which gradually improved after eight days of infection. Symptoms of SW2783 infection were more apparent than ZZ346. With the SW2783 infection, the body temperature of tree shrews showed an increasing trend and peaked on the 4th day and the 5th day and then decreased gradually after the 8th day of infection. While the ZZ346 infection barely caused body temperature change. Compared with the control groups, there was no significant difference in the weight change in the tested groups. The results were similar to the symptoms of human seasonal influenza (Richman et al., 2017; Krammer et al., 2018) and similar to the findings of waterfowl influenza virus H9N2 infected with tree shrews (Li et al., 2018). Our observation proved that both strains had great infectivity and replication activity in tree shrew respiratory tract tissue. However, the strains mainly caused self-limited upper respiratory tract infection in turbinate tissue. Our findings indicated that the replication and clinical symptoms caused by the two animals LPAIV strain were closely related to each other and were similar to those of human seasonal influenza viruses (H1N1, H3N2), especially with pandemic A (H1N1) (Ip et al., 2016; Richman et al., 2017; Krammer et al., 2018), demonstrating that tree shrew is a suitable model to study human LPAIV.
In addition, replication of influenza viruses SW2783 and ZZ346 in the conjunctiva was also observed in our experiments. The observation was consistent with the previous study, which reported that H5N1 and H7N9 could replicate in the conjunctiva of tree shrews (Sanada et al., 2019). However, some avian influenza virus subtypes, such as H7 and H5, can effectively replicate in human conjunctiva and corneal epithelial cells in vitro (Belser et al., 2009; Chan et al., 2010), which was different from human influenza viruses H1N1 and H3N2. The AIV H7 and H5 subtypes, and pandemic strain pdm/09 H1N1, can also use eyes as entry portals in mouse models, which always leads to a systemic and fatal infections. Our data from the transmission experiment showed that the swine SW2783 virus could use eyes as entry portals in the tree shrew model. These results suggest that animal-borne influenza viruses, especially newly emerged influenza viruses (such as swine H3N2 SW2783 strain) that incorporate the internal genes of pandemic pdm/09 H1N1, which has gained the function of the potential to infect humans through eyes, and tree shrews may present as a good model for studying the risk of cross-species transmission of viruses through eyes.
The outcome of influenza A virus infection depends on the complex interaction between multiple factors of the host, the virus, and the environment (Mishra et al., 2017). The strength of the host immune response induced by virus infection plays a crucial role (Peiris et al., 2009; Oslund and Baumgarth, 2011; Chen et al., 2018; Gu et al., 2019). We tested ten molecules involved in innate immune recognition, antiviral, inflammatory response, and immune regulation in infected tree shrew turbinate tissue. Except for TLR7 and IFN-β, Mx1, MDA5, TNF-α, IL-6, MCP-1, IP-10, IFN-γ, and IL-10 have up-regulated after infection with the two strains of virus. Pro-inflammatory cytokines peaked on the 5th day after infection, while the TNF-α and IL-6 were expressed most on the 3rd day. These results matched the viral replication trends and clinical symptom phenotypes. It highlights that with the enhancement of viral replication, the host initiated corresponding innate immune responses, which have played an important role in antiviral activities. However, IFN-β, the critical antiviral protein, was not significantly increased in our study, which was similar to the swine influenza virus H1N1 subtype infected pig lung tissue (Li et al., 2011). It may relate to the early production of IFN-β (before 3rd day), or the LPAIV used in this study may not provide enough stimulation. Besides, TLR7, which can recognize the virus ssRNA in the cell, was not significantly increased either, suggesting that the recognition of the two virus nucleic acids by tree shrews mainly rely on RIG-like receptor MDA5/IFIH1. Moreover, the expression of IFN-γ and IL-10 up-regulated on days 5 and 7 after infection. The two cytokines have served as crucial intermedium in the differentiation and development of Th cells and thus play a key regulatory role in innate and adaptive immunity (Schoenborn and Wilson, 2007; Couper et al., 2008). The results indicate that an appropriate adaptive immune response was initiated in the middle and late stages of infection to promote the recovery of tree shrews. The results were verified mutually with the detection of coagulation antibody production on day seven after infection. Several studies reported expressions of IFN-α, IFN-γ, IL-6, TNF-α, and IL-10 increased in the nasal cavity and blood in the early stage of humans infected with the influenza virus. The IL-6 was correlated with disease progression, while the IFN-γ in the nasal cavity was correlated with the virus titer decreasing. Virus-specific serum antibodies will present in first week of illness in adults (Richman et al., 2017). The changes of cytokines and virus-specific antibodies in this study were consistent with those reported above, suggesting that the tree shrew model of influenza infection had consistency with the human influenza process in terms the cytokines expression and antibody production.
The IAV gradually adapted to the host and continuously broke through the species barrier and limited factors to spread cross-species and to cause a new host pandemic, which included binding, entering, and replicating in host cells, escaping the host innate immune response, and transmitting between hosts (Long et al., 2019). The emerging IAVs with human-adaptive potential that might lead to the next pandemic (Thompson and Paulson, 2021). Our results showed that the newly emerged swine influenza virus SW2783 and chicken-origin influenza virus ZZ346 could directly infect tree shrews and replicate effectively without adaptation. After virus infected tree shrews, the antiviral innate immunity and humoral immune response are activated at the right time to limit virus infection and promote recovery. We also observed that the newly emerged swine influenza virus SW2783 could transmit among tree shrews through direct contact, and from tree shrews to guinea pigs across the species barrier. Yet, the specific mechanisms may need further elaboration. We will further identify of the interaction between the two animal H3N2 and H6N6 influenza viruses HA and SA receptors of the host, and the genetic, and phenotypic changes required for host adaptation and transmission of these two animal influenza viruses in future studies.
In conclusion, based on the evidence of our study, we propose that tree shrews can surpassingly simulate the pathogenicity, transmissibility, and immune process of human infection with the influenza virus. It is considered a suitable mammalian model to study the pathogenicity and cross-species transmission of animal influenza A viruses.
CRediT authorship contribution statement
Conceived, designed the study and funding acquisition: ZFZ, JL and XHF. Performed the experiment: QHW, XZ, LL, XHW, ZPL, ZHL, XQH, and CXY. Analyzed the data: QHW, XZ, LXG and ST. Writing-original draft: QHW. Writing-review & editing: QHW, ZFZ, JL and XHF. All authors read and approved the final manuscript.
Conflicts of Interest
The authors declare no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This study was supported by the National Natural Science Foundation of China (grants 81560556, 31660040, 31460243, 31160190). We thank Zhigang Zheng, Yu Zheng, Xiafeng Peng, Chenyi Li for excellent technical assistance. We also thank Xiaofang Yuan, Chun Yang for excellent animal husbandry assistance.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.virusres.2022.199027.
Contributor Information
Zengfeng Zhang, Email: zfzhangphd@163.com.
Jing Leng, Email: lj986771558@163.com.
Xiaohui Fan, Email: fanxiaohui63@163.com.
Appendix. Supplementary materials
References
- Belser J.A., Wadford D.A., Xu J., Katz J.M., Tumpey T.M. Ocular infection of mice with influenza A (H7) viruses: a site of primary replication and spread to the respiratory tract. J. Virol. 2009;83(14):7075–7084. doi: 10.1128/JVI.00535-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouvier N.M. Animal models for influenza virus transmission studies: a historical perspective. Curr. Opin. Virol. 2015:13101–13108. doi: 10.1016/j.coviro.2015.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown I.H. The epidemiology and evolution of influenza viruses in pigs. Vet. Microbiol. 2000;74(1-2):29–46. doi: 10.1016/s0378-1135(00)00164-4. [DOI] [PubMed] [Google Scholar]
- Byrd-Leotis L., Jia N., Dutta S., Trost J.F., Gao C., Cummings S.F., Braulke T., Müller-Loennies S., Heimburg-Molinaro J., Steinhauer D.A., Cummings R.D. Influenza binds phosphorylated glycans from human lung. Sci. Adv. 2019;5(2):v2554. doi: 10.1126/sciadv.aav2554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan M.C., Chan R.W., Yu W.C., Ho C.C., Yuen K.M., Fong J.H., Tang L.L., Lai W.W., Lo A.C., Chui W.H., Sihoe A.D., Kwong D.L., Wong D.S., Tsao G.S., Poon L.L., Guan Y., Nicholls J.M., Peiris J.S. Tropism and innate host responses of the 2009 pandemic H1N1 influenza virus in ex vivo and in vitro cultures of human conjunctiva and respiratory tract. Am. J. Pathol. 2010;176(4):1828–1840. doi: 10.2353/ajpath.2010.091087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Liu S., Goraya M.U., Maarouf M., Huang S., Chen J.L. Host immune response to influenza A virus infection. Front. Immunol. 2018:9:320. doi: 10.3389/fimmu.2018.00320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou Y.Y., Albrecht R.A., Pica N., Lowen A.C., Richt J.A., Garcia-Sastre A., Palese P., Hai R. The M segment of the 2009 new pandemic H1N1 influenza virus is critical for its high transmission efficiency in the guinea pig model. J. Virol. 2011;85(21):11235–11241. doi: 10.1128/JVI.05794-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couper K.N., Blount D.G., Riley E.M. IL-10: the master regulator of immunity to infection. J. Immunol. 2008;180(9):5771–5777. doi: 10.4049/jimmunol.180.9.5771. [DOI] [PubMed] [Google Scholar]
- de Graaf M., Fouchier R.A.M. Role of receptor binding specificity in influenza A virus transmission and pathogenesis. EMBO J. 2014;33(8):823–841. doi: 10.1002/embj.201387442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Y., Huang Z., Cao C., Chen C., Chen Y., Fan D., He J., Hou H., Hu L., Hu X., Jiang X., Lai R., Lang Y., Liang B., Liao S., Mu D., Ma Y., Niu Y., Sun X., Xia J., Xiao J., Xiong Z., Xu L., Yang L., Zhang Y., Zhao W., Zhao X., Zheng Y., Zhou J., Zhu Y., Zhang G., Wang J., Yao Y. Genome of the Chinese tree shrew. Nat. Commun. 2013;4(1):1426. doi: 10.1038/ncomms2416. [DOI] [PubMed] [Google Scholar]
- Freidl G.S., Meijer A., de Bruin E., de Nardi M., Munoz O., Capua I., Breed A.C., Harris K., Hill A., Kosmider R., Banks J., von Dobschuetz S., Stark K., Wieland B., Stevens K., van der Werf S., Enouf V., van der Meulen K., Van Reeth K., Dauphin G., Koopmans M. Influenza at the animal-human interface: a review of the literature for virological evidence of human infection with swine or avian influenza viruses other than A(H5N1) Euro Surveill. 2014;19(18):8–26. doi: 10.2807/1560-7917.es2014.19.18.20793. [DOI] [PubMed] [Google Scholar]
- Glaser L., Stevens J., Zamarin D., Wilson I.A., García-Sastre A., Tumpey T.M., Basler C.F., Taubenberger J.K., Palese P. A single amino acid substitution in 1918 influenza virus hemagglutinin changes receptor binding specificity. J. Virol. 2005;79(17):11533–11536. doi: 10.1128/JVI.79.17.11533-11536.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greber U.F. Virus and host mechanics support membrane penetration and cell entry. J. Virol. 2016;90(8):3802–3805. doi: 10.1128/JVI.02568-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Y., Hsu A.C., Pang Z., Pan H., Zuo X., Wang G., Zheng J., Wang F. Role of the innate cytokine storm induced by the influenza A virus. Viral Immunol. 2019;32(6):244–251. doi: 10.1089/vim.2019.0032. [DOI] [PubMed] [Google Scholar]
- Imai M., Kawaoka Y. The role of receptor binding specificity in interspecies transmission of influenza viruses. Curr. Opin. Virol. 2012;2(2):160–167. doi: 10.1016/j.coviro.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ip D., Lau L., Chan K.H., Fang V.J., Leung G.M., Peiris M., Cowling B.J. The dynamic relationship between clinical symptomatology and viral shedding in naturally acquired seasonal and pandemic influenza virus infections. Clin. Infect. Dis. 2016;62(4):431–437. doi: 10.1093/cid/civ909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T., Couceiro J.N., Kelm S., Baum L.G., Krauss S., Castrucci M.R., Donatelli I., Kida H., Paulson J.C., Webster R.G., Kawaoka Y. Molecular basis for the generation in pigs of influenza A viruses with pandemic potential. J. Virol. 1998;72(9):7367–7373. doi: 10.1128/JVI.72.9.7367-7373.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jhung M.A., Epperson S., Biggerstaff M., Allen D., Balish A., Barnes N., Beaudoin A., Berman L., Bidol S., Blanton L., Blythe D., Brammer L., D'Mello T., Danila R., Davis W., de Fijter S., Diorio M., Durand L.O., Emery S., Fowler B., Garten R., Grant Y., Greenbaum A., Gubareva L., Havers F., Haupt T., House J., Ibrahim S., Jiang V., Jain S., Jernigan D., Kazmierczak J., Klimov A., Lindstrom S., Longenberger A., Lucas P., Lynfield R., McMorrow M., Moll M., Morin C., Ostroff S., Page S.L., Park S.Y., Peters S., Quinn C., Reed C., Richards S., Scheftel J., Simwale O., Shu B., Soyemi K., Stauffer J., Steffens C., Su S., Torso L., Uyeki T.M., Vetter S., Villanueva J., Wong K.K., Shaw M., Bresee J.S., Cox N., Finelli L. Outbreak of variant influenza A(H3N2) virus in the United States. Clin. Infect. Dis. 2013;57(12):1703–1712. doi: 10.1093/cid/cit649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayesh M., Sanada T., Kohara M., Tsukiyama-Kohara K. Tree shrew as an emerging small animal model for human viral infection: a recent overview. Viruses. 2021;13(8) doi: 10.3390/v13081641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimble B., Nieto G.R., Perez D.R. Characterization of influenza virus sialic acid receptors in minor poultry species. Virol. J. 2010;7(1):365. doi: 10.1186/1743-422X-7-365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krammer F., Smith G., Fouchier R., Peiris M., Kedzierska K., Doherty P.C., Palese P., Shaw M.L., Treanor J., Webster R.G., Garcia-Sastre A. Influenza. Nat. Rev. Dis. Primers. 2018;4(1):3. doi: 10.1038/s41572-018-0002-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuchipudi S.V., Nelli R., White G.A. Differences in influenza virus receptors in chickens and ducks: implications for interspecies transmission. J. Mol. Genet. Med. 2009;03(01):143–151. doi: 10.4172/1747-0862.1000026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuiken T., Holmes E.C., McCauley J., Rimmelzwaan G.F., Williams C.S., Grenfell B.T. Host species barriers to influenza virus infections. Science. 2006;312(5772):394–397. doi: 10.1126/science.1122818. [DOI] [PubMed] [Google Scholar]
- Kumar S., Hedges S.B. A molecular timescale for vertebrate evolution. Nature. 1998;392(6679):917–920. doi: 10.1038/31927. [DOI] [PubMed] [Google Scholar]
- Li R., Yuan B., Xia X., Zhang S., Du Q., Yang C., Li N., Zhao J., Zhang Y., Zhang R., Feng Y., Jiao J., Peiris M., Zhong N., Mok C.K.P., Yang Z. Tree shrew as a new animal model to study the pathogenesis of avian influenza (H9N2) virus infection. Emerg. Microbes Infect. 2018;7(1):1–11. doi: 10.1038/s41426-018-0167-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Zhou H., Wen Z., Wu S., Huang C., Jia G., Chen H., Jin M. Transcription analysis on response of swine lung to H1N1 swine influenza virus. BMC Genom. 2011;12(1):398. doi: 10.1186/1471-2164-12-398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang H., Lam T.T., Fan X., Chen X., Zeng Y., Zhou J., Duan L., Tse M., Chan C.H., Li L., Leung T.Y., Yip C.H., Cheung C.L., Zhou B., Smith D.K., Poon L.L., Peiris M., Guan Y., Zhu H. Expansion of genotypic diversity and establishment of 2009 H1N1 pandemic-origin internal genes in pigs in China. J. Virol. 2014;88(18):10864–10874. doi: 10.1128/JVI.01327-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Long J.S., Mistry B., Haslam S.M., Barclay W.S. Host and viral determinants of influenza A virus species specificity. Nat. Rev. Microbiol. 2019;17(2):67–81. doi: 10.1038/s41579-018-0115-z. [DOI] [PubMed] [Google Scholar]
- Ma W., Liu Q., Bawa B., Qiao C., Qi W., Shen H., Chen Y., Ma J., Li X., Webby R.J., Garcia-Sastre A., Richt J.A. The neuraminidase and matrix genes of the 2009 pandemic influenza H1N1 virus cooperate functionally to facilitate efficient replication and transmissibility in pigs. J. Gen. Virol. 2012;93(Pt 6):1261–1268. doi: 10.1099/vir.0.040535-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margine I., Krammer F. Animal models for influenza viruses: implications for universal vaccine development. Pathogens. 2014;3(4):845–874. doi: 10.3390/pathogens3040845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka Y., Lamirande E.W., Subbarao K. The mouse model for influenza. Curr. Protoc. Microbiol. 2009;13(1):13G–15G. doi: 10.1002/9780471729259.mc15g03s13. [DOI] [PubMed] [Google Scholar]
- Mishra A., Vijayakumar P., Raut A.A. Emerging avian influenza infections: current understanding of innate immune response and molecular pathogenesis. Int. Rev. Immunol. 2017;36(2):89–107. doi: 10.1080/08830185.2017.1291640. [DOI] [PubMed] [Google Scholar]
- Monne I., Fusaro A., Nelson M.I., Bonfanti L., Mulatti P., Hughes J., Murcia P.R., Schivo A., Valastro V., Moreno A., Holmes E.C., Cattoli G. Emergence of a highly pathogenic avian influenza virus from a low-pathogenic progenitor. J. Virol. 2014;88(8):4375–4388. doi: 10.1128/JVI.03181-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholls J.M., Chan M.C.W., Chan W.Y., Wong H.K., Cheung C.Y., Kwong D.L.W., Wong M.P., Chui W.H., Poon L.L.M., Tsao S.W., Guan Y., Peiris J.S.M. Tropism of avian influenza A (H5N1) in the upper and lower respiratory tract. Nat. Med. 2007;13(2):147–149. doi: 10.1038/nm1529. [DOI] [PubMed] [Google Scholar]
- Oslund K.L., Baumgarth N. Influenza-induced innate immunity: regulators of viral replication, respiratory tract pathology & adaptive immunity. Future Virol. 2011;6(8):951–962. doi: 10.2217/fvl.11.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan M., Gao R., Lv Q., Huang S., Zhou Z., Yang L., Li X., Zhao X., Zou X., Tong W., Mao S., Zou S., Bo H., Zhu X., Liu L., Yuan H., Zhang M., Wang D., Li Z., Zhao W., Ma M., Li Y., Li T., Yang H., Xu J., Zhou L., Zhou X., Tang W., Song Y., Chen T., Bai T., Zhou J., Wang D., Wu G., Li D., Feng Z., Gao G.F., Wang Y., He S., Shu Y. Human infection with a novel, highly pathogenic avian influenza A (H5N6) virus: virological and clinical findings. J. Infect. 2016;72(1):52–59. doi: 10.1016/j.jinf.2015.06.009. [DOI] [PubMed] [Google Scholar]
- Peiris J.S., Cheung C.Y., Leung C.Y., Nicholls J.M. Innate immune responses to influenza A H5N1: friend or foe? Trends Immunol. 2009;30(12):574–584. doi: 10.1016/j.it.2009.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrova V.N., Russell C.A. The evolution of seasonal influenza viruses. Nat. Rev. Microbiol. 2018;16(1):47–60. doi: 10.1038/nrmicro.2017.118. [DOI] [PubMed] [Google Scholar]
- Pyankova O.G., Susloparov I.M., Moiseeva A.A., Kolosova N.P., Onkhonova G.S., Danilenko A.V., Vakalova E.V., Shendo G.L, Nekeshina N.N., Noskova L.N., Demina J.V., Frolova N.V., Gavrilova E.V., Maksyutov R.A., Ryzhikov A.B. Isolation of clade 2.3.4.4b A(H5N8), a highly pathogenic avian influenza virus, from a worker during an outbreak on a poultry farm, Russia, December 2020. Euro Surveill. 2021;26(24) doi: 10.2807/1560-7917.ES.2021.26.24.2100439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi X., Qiu H., Hao S., Zhu F., Huang Y., Xu K., Yu H., Wang D., Zhou L., Dai Q., Zhou Y., Wang S., Huang H., Yu S., Huo X., Chen K., Liu J., Hu J., Wu M., Bao C. Human Infection with an Avian-Origin Influenza A (H10N3) Virus. N. Engl. J. Med. 2022;386(11):1087–1088. doi: 10.1056/NEJMc2112416. [DOI] [PubMed] [Google Scholar]
- Rajao D.S., Walia R.R., Campbell B., Gauger P.C., Janas-Martindale A., Killian M.L., Vincent A.L. Reassortment between swine H3N2 and 2009 pandemic H1N1 in the United States resulted in influenza A viruses with diverse genetic constellations with variable virulence in pigs. J. Virol. 2017;91(4) doi: 10.1128/JVI.01763-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richman D.D., Whitley R.J., Hayden F.G. Clinical Virology. 4th ed. ASM Press; Washington, DC: 2017. pp. 1009–1058. [Google Scholar]
- Roberts T.E., Lanier H.C., Sargis E.J., Olson L.E. Molecular phylogeny of treeshrews (Mammalia: Scandentia) and the timescale of diversification in Southeast Asia. Mol. Phylogenet. Evol. 2011;60(3):358–372. doi: 10.1016/j.ympev.2011.04.021. [DOI] [PubMed] [Google Scholar]
- Rogers G.N., Paulson J.C. Receptor determinants of human and animal influenza virus isolates: differences in receptor specificity of the H3 hemagglutinin based on species of origin. Virology. 1983;127(2):361–373. doi: 10.1016/0042-6822(83)90150-2. [DOI] [PubMed] [Google Scholar]
- Sanada T., Yasui F., Honda T., Kayesh M.E.H., Takano J., Shiogama Y., Yasutomi Y., Tsukiyama-Kohara K., Kohara M. Avian H5N1 influenza virus infection causes severe pneumonia in the Northern tree shrew (Tupaia belangeri) Virology. 2019:529: 101–110. doi: 10.1016/j.virol.2019.01.015. [DOI] [PubMed] [Google Scholar]
- Schoenborn J.R., Wilson C.B. Regulation of interferon-gamma during innate and adaptive immune responses. Adv. Immunol. 2007 doi: 10.1016/S0065-2776(07)96002-2. 96: 41-101. [DOI] [PubMed] [Google Scholar]
- Short K.R., Richard M., Verhagen J.H., van Riel D., Schrauwen E.J., van den Brand J.M., Manz B., Bodewes R., Herfst S. One health, multiple challenges: the inter-species transmission of influenza A virus. One Health. 2015:1: 1–13. doi: 10.1016/j.onehlt.2015.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens J., Blixt O., Tumpey T.M., Taubenberger J.K., Paulson J.C., Wilson I.A. Structure and receptor specificity of the hemagglutinin from an H5N1 influenza virus. Science. 2006;312(5772):404–410. doi: 10.1126/science.1124513. [DOI] [PubMed] [Google Scholar]
- Sun X., Pulit-Penaloza J.A., Belser J.A., Pappas C., Pearce M.B., Brock N., Zeng H., Creager H.M., Zanders N., Jang Y., Tumpey T.M., Davis C.T., Maines T.R. Pathogenesis and transmission of genetically diverse swine-origin H3N2 variant influenza A viruses from multiple lineages isolated in the United States, 2011–2016. J. Virol. 2018;92(16):e618–e665. doi: 10.1128/JVI.00665-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takemae N., Harada M., Nguyen P.T., Nguyen T., Nguyen T.N., To T.L., Nguyen T.D., Pham V.P., Le V.T., Do H.T., Vo H.V., Le Q.V.T., Tran T.M., Nguyen T.D., Thai P.D., Nguyen D.H., Le A.Q.T., Nguyen D.T., Uchida Y., Saito T. Influenza A viruses of swine (IAV-S) in Vietnam from 2010 to 2015: multiple introductions of A(H1N1)pdm09 viruses into the pig population and diversifying genetic constellations of enzootic IAV-S. J. Virol. 2017;91(1):e1416–e1490. doi: 10.1128/JVI.01490-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson A.J., Paulson J.C. Adaptation of influenza viruses to human airway receptors. J. Biol. Chem. 2021 doi: 10.1074/jbc.REV120.013309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Reeth K. Avian and swine influenza viruses: our current understanding of the zoonotic risk. Vet. Res. 2007;38(2):243–260. doi: 10.1051/vetres:2006062. [DOI] [PubMed] [Google Scholar]
- Wan H., Perez D.R. Quail carry sialic acid receptors compatible with binding of avian and human influenza viruses. Virology. 2006;346(2):278–286. doi: 10.1016/j.virol.2005.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Zhu W., Yang L., Shu Y. The epidemiology, virology, and pathogenicity of human infections with avian influenza viruses. CSH Perspect. Med. 2021;11(4):a38620. doi: 10.1101/cshperspect.a038620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G., Deng G., Shi J., Luo W., Zhang G., Zhang Q., Liu L., Jiang Y., Li C., Sriwilaijaroen N., Hiramatsu H., Suzuki Y., Kawaoka Y., Chen H. H6 influenza viruses pose a potential threat to human health. J. Virol. 2014;88(8):3953–3964. doi: 10.1128/JVI.03292-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong J., Layton D., Wheatley A.K., Kent S.J. Improving immunological insights into the ferret model of human viral infectious disease. Influenza Other Respir. Viruses. 2019;13(6):535–546. doi: 10.1111/irv.12687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H., Lu R., Peng X., Peng X., Cheng L., Jin C., Lu X., Xie T., Yao H., Wu N. Isolation and genetic characterization of novel reassortant H6N6 subtype avian influenza viruses isolated from chickens in eastern China. Arch. Virol. 2016;161(7):1859–1872. doi: 10.1007/s00705-016-2861-4. [DOI] [PubMed] [Google Scholar]
- Xiao J., Liu R., Chen C.S. Tree shrew (Tupaia belangeri) as a novel laboratory disease animal model. Zool. Res. 2017;38(3):127–137. doi: 10.24272/j.issn.2095-8137.2017.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin L., Bai T., Zhou J.F., Chen Y.K., Li X.D., Zhu W.F., Li Y., Tang J., Chen T., Qin K., Shi J.H., Gao R.B., Wang D.Y., Chen J.M., Shu Y.L. Seropositivity for avian influenza H6 virus among humans, China. Emerg. Infect. Dis. 2015;21(7):1267–1269. doi: 10.3201/eid2107.150135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S., Li X., Yang J., Wang Z., Jia Y., Han L., Wang L., Zhu Q. Comparative pathogenicity and transmissibility of pandemic H1N1, Avian H5N1, and Human H7N9 influenza viruses in tree shrews. Front. Microbiol. 2019;10 doi: 10.3389/fmicb.2019.02955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L., Zhu W., Li X., Bo H., Zhang Y., Zou S., Gao R., Dong J., Zhao X., Chen W., Dong L., Zou X., Xing Y., Wang D., Shu Y. Genesis and dissemination of highly pathogenic H5N6 avian influenza viruses. J. Virol. 2017;91(5):e2116–e2199. doi: 10.1128/JVI.02199-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z., Zhao J., Zhu Y., Wang Y., Liu R., Zhao S., Li R., Yang C., Li J., Zhong N. The tree shrew provides a useful alternative model for the study of influenza H1N1 virus. Virol. J. 2013;10(1):111. doi: 10.1186/1743-422X-10-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Y. Creating animal models, why not use the Chinese tree shrew (Tupaia belangeri chinensis)? Zool Res. 2017;38(3):118–126. doi: 10.24272/j.issn.2095-8137.2017.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J.E., Yoon H., Lee H.J., Lee J.H., Chang B.J., Song C.S., Nahm S. Expression patterns of influenza virus receptors in the respiratory tracts of four species of poultry. J. Vet. Sci. 2011;12(1):7. doi: 10.4142/jvs.2011.12.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Zhang Z., Fan X., Liu Y., Wang J., Zheng Z., Chen R., Wang P., Song W., Chen H., Guan Y. 2009 pandemic H1N1 influenza virus replicates in human lung tissues. J. Infect. Dis. 2010;201(10):1522–1526. doi: 10.1086/650544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao G., Chen C., Huang J., Wang Y., Peng D., Liu X. Characterisation of one H6N6 influenza virus isolated from swine in China. Res. Vet. Sci. 2013;95(2):434–436. doi: 10.1016/j.rvsc.2013.06.013. [DOI] [PubMed] [Google Scholar]
- Zhao P., Sun L., Xiong J., Wang C., Chen L., Yang P., Yu H., Yan Q., Cheng Y., Jiang L., Chen Y., Zhao G., Jiang Q., Xiong C. Semiaquatic mammals might be intermediate hosts to spread avian influenza viruses from avian to human. Sci. Rep. 2019;9(1):11641. doi: 10.1038/s41598-019-48255-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Z., Lu Y., Short K.R., Lu J. One health insights to prevent the next HxNy viral outbreak: learning from the epidemiology of H7N9. BMC Infect. Dis. 2019;19(1):138. doi: 10.1186/s12879-019-3752-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H., Zhou B., Fan X., Lam T.T.Y., Wang J., Chen A., Chen X., Chen H., Webster R.G., Webby R., Peiris J.S.M., Smith D.K., Guan Y. Novel reassortment of Eurasian avian-like and pandemic/2009 influenza viruses in swine: infectious potential for humans. J. Virol. 2011;85(20):10432–10439. doi: 10.1128/JVI.05352-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W., Yang L., Shu Y. Did the highly pathogenic avian influenza A(H7N9) viruses emerged in china raise increased threat to public health? Vector-Borne Zoonotic Dis. 2019;19(1):22–25. doi: 10.1089/vbz.2018.2299. [DOI] [PubMed] [Google Scholar]
- Zhu Y.C., Zhang B., Sun Z.H., Wang X.J., Fan X.H., Gao L.X., Liang Y., Chen X.Y., Zhang Z.F. Replication and pathology of duck influenza virus subtype H9N2 in chukar. Biomed. Environ. Sci. 2018;31(4):306–310. doi: 10.3967/bes2018.039. [DOI] [PubMed] [Google Scholar]
- Zou S., Gao R., Zhang Y., Li X., Chen W., Bai T., Dong L., Wang D., Shu Y. Molecular characterization of H6 subtype influenza viruses in southern China from 2009 to 2011. Emerg. Microbes Infect. 2019;5(1):1–8. doi: 10.1038/emi.2016.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.