Highlights
-
•
HERV-K(HML8) group is composed by 78 proviruses, 23 newly discovered.
-
•
HML8 loci have a biased genomic distribution and are often peri- and centromeric.
-
•
HML8 elements can be subdivided into 3 clusters: MER11A, MER11B, and MER11C.
-
•
HML8 types are characterized by recurrent deletions as compared to the reference.
-
•
MER11C-associated proviruses are significantly younger respect to MER11A and MER11B.
Keywords: HERV, HERV-K, HML8, Endogenous retroviruses, Retrotransposons
Abstract
Around 8% of the human genome is composed by Human Endogenous Retroviruses (HERVs), ancient viral sequences inherited from the primate germ line after their infection by now extinct retroviruses. Given the still underexplored physiological and pathological roles of HERVs, it is fundamental to increase our information about the genomic composition of the different groups, to lay reliable foundation for functional studies. Among HERVs, the most characterized elements belong to the beta-like superfamily HERV-K, comprising 10 groups (HML1-10) with HML2 being the most recent and studied one. Among HMLs, the HML8 group is the only one still lacking a comprehensive genomic description. In the present work, we investigated HML8 sequences’ distribution in the human genome (GRCh38/hg38), identifying 23 novel proviruses and characterizing the overall 78 HML8 proviruses in terms of genome structure, phylogeny, and integration pattern. HML8 elements were significantly enriched in human chromosomes 8 and X (p<0.005) while chromosomes 17 and 20 showed fewer integrations than expected (p<0.025 and p<0.005, respectively). Phylogenetic analyses classified HML8 members into 3 clusters, corresponding to the three LTR types MER11A, MER11B and MER11C. Besides different LTR types, common signatures in the internal structure suggested the potential existence of three different ancestral HML8 variants. Accordingly, time of integration estimation coupled with comparative genomics revealed that these three clusters have a different time of integration in the primates’ genome, with MER11C elements being significantly younger than MER11A- and MER11B associated proviruses (p<0.005 and p<0.05, respectively). Approximately 30% of the HML8 elements were found co-localized within human genes, sometimes in exonic portions and with the same orientation, deserving further studies for their possible effects on gene expression. Overall, we provide the first detailed picture of the HML8 group distribution and variety among the genome, creating the backbone for the specific analysis of their transcriptional activity in healthy and diseased conditions.
Abbreviations
- HERV
human endogenous retrovirus
- HML
human MMTV-like
- LTR
long terminal repeats
- MER
MEdium Reiteration frequency interspersed repeat
- IFN
interferon
- lncRNA
long non-coding RNA
- O.C.A.
oldest common ancestor
- Mya
million years ago
1. Introduction
HERVs, acronym for “Human Endogenous Retroviruses”, are proviral fossils deriving from exogenous retroviruses that infected the germ cells of primates millions of years ago. This led to the stable acquisition of HERVs into the genomes, which have been piling up during the course of evolution until currently covering approximately 8% of the human genome. HERVs have been categorized as human transposable elements, belonging to retrotransposons provided with Long Terminal Repeat (LTR). It has been noticed that the molecular evolution of the genomic sequences led to the domestication of certain HERV loci, which currently play remarkable functions in the human genome. One of the most significant roles is the one carried out by two HERV envelope proteins during placental development and homeostasis (Blond et al., 2000; Lavialle et al., 2013; Mangeney et al., 2007; Sha et al., 2000). Moreover, ever growing findings demonstrate the important role of HERVs as drivers of genomic innovation and major evolutionary shapers of transcription pathways, including the major innate immunity branch of interferon (IFN)(Ferrari et al., 2021; Grandi and Tramontano, 2018a,2018b). In line with this, HERV expression is known to be influenced by innate immunity, and vice versa, and such an interplay is currently a main field of HERV investigation (Pisano et al., 2021). Beyond that, HERV expression is also highly investigated for its possible role in several complex human disorders, such as autoimmunity and cancer, even if no strict correlation has been proved yet (Grandi et al., 2019). This is due, in part, to the still lacking knowledge of the individual HERV integrations inside the human genome, which excludes the investigation of those loci (Grandi and Tramontano, 2017; Liu et al., 2020). HERVs have been classified according to the similarity to their exogenous counterpart, thus divided in Class I, Class II and Class III, since similar respectively to Gamma- and Epsilon-, Beta-, and Spumaretroviruses (Bannert and Kurth, 2006; Subramanian et al., 2011). Nevertheless, this approach has proved to be fallacious given that this nomenclature, designed for exogenous retroviruses, reflects some phenotypical characteristics not applicable to HERV(Bannert and Kurth, 2006). More recently, further classification methods have been applied, although relying to conflicting criteria and resulting hence in more confusion (Grandi et al., 2016). In addition, the great majority of HERV groups are still lacking key information about the total number of members, their genomic distribution and nucleotide structure (Mayer et al., 2011). Recently, a new global classification has been made through the software RetroTector, performing deep research of conserved retroviral motifs in vertebrate genomes to reconstruct each proviral insertion (Sperber et al., 2007). Then, the sequences were classified using a multi-step approach including Simage (Similarity Image Analysis) that considers the proviral integrity and composition. This allowed the cataloguing of 31 canonical and 39 non-canonical HERV groups that showed a high amount of mosaicism derived by recombination events (Vargiu et al., 2016). This automated characterization was then implemented with a group-by-group BLAT research, leading to the inclusion of some proviruses previously missed by the software due to their defective structure or the presence of mutations in the main recognition sites. Accordingly, several HERV groups have been widely described, among which HERV-W (Grandi et al., 2020a, 2018, 2016), HERV-H (Jern et al., 2005), and most of the members of the HERV class II. The latter is divided in 10 groups, composed by betaretrovirus-like elements named HML (Human MMTV-like) from 1 to 10 accordingly to their similarity to the exogenous Mouse Mammary Tumor Virus (MMTV)(Subramanian et al., 2011; Vargiu et al., 2016). Among the HMLs, the HML2 group is the one that arouses the greatest interest (Subramanian et al., 2011), being in fact the only one including human-specific integrations even polymorphic in the modern population (Marchi et al., 2014). The wide knowledge of this group made it to be the most investigated in the pathological scenario, with several studies focusing on its possible contribution to different cancers, neurological and autoimmune diseases (Garcia-Montojo et al., 2018). Similarly, a dedicated classification has been done for other 9 HMLs (Broecker et al., 2016; Flockerzi et al., 2005; Grandi et al., 2017a; Lavie et al., 2004; Mayer and Meese, 2002; Pisano et al., 2019; Seifarth et al., 1998), being still lacking for the last HML group, namely HML8. The latter has been taken into account in the above general classification of around 3200 HERVs by the software RetroTector, which identified a total of 58 HML8 sequences further classified into 34 canonical members (59%) and 24 non-canonical sequences (41%) that showed higher degree of mosaicism (Vargiu et al., 2016). Beside the partial identification in Vargiu et al., literature search reported only one study considering HML8 elements, as conducted by Chang et al. (2019). Also in this case, the study was not specifically dedicated to the group but aimed to search for an association between HERVs single nucleotide variations (SNV) and human cancers. They identified the HML8 group as the one with the highest number of somatic mutations among Alpha- and Beta- related HERVs in the chromosomal non-coding regions, suggesting that they might have a role in carcinogenesis, especially if affecting regulatory regions (Chang et al., 2019). Despite such potential relevance, the group remains without a comprehensive description at the genomic level.
The present study provides the first complete characterization of all the 78 HML8 elements present in the latest refined version of human genome assembly (hg38), with a particular attention on their genomic distribution, context of insertion, phylogeny, proviral structure, and age of integration estimation. Thus, the present analysis brings to completion the long process of classification and characterization of the HML groups, laying the bases for forthcoming studies on their possible role in the human organism, either physiological or pathological.
2. Materials and method
2.1. Identification of HERV-K(HML8) sequences in the human genome
To achieve a complete database of all the HML8 proviral sequences dispersed in the human genome, we started from the 58 sequences already identified in a previous classification work of the most intact HERVs in genome assembly GRCh37/hg19 (Vargiu et al., 2016) through the bioinformatic tool RetroTector (Sperber et al., 2007). To confirm and refine the actual position of these sequences and detect eventual other HML8 loci missed by RetroTector, we did a search in the hg38 genome assembly with the Human BLAT Search tool provided by UCSC Genome Browser (Haeussler et al., 2019). In particular, we have performed this analysis using the reference sequence for HML8 internal region (HERV-K11) as a query, associating it with each of the three different variants of LTRs (MER11A, MER11B, and MER11C) as annotated in Dfam database of repetitive elements (Hubley et al., 2016). For each genomic positions identified by BLAT search, we took advantage of the RepeatMasker annotations to check the element identity, and we subsequently downloaded all HML8 sequences adding 500 nucleotides flanking 5’ and 3’ sides. To ascertain the presence of the complete HERV locus, we aligned the retrieved sequences with the reference, refining their coordinates when needed. The same BLAT search has been used to identify the HML8 solitary LTRs and to assign them to the respective MER11A, MER11B, and MER11C types. Also in this case, each sequence has been downloaded with 500 additional flanking nucleotides at both sides and then aligned to the reference. Alignments have been generated using Geneious Prime software, version 2020.1.1 (Biomatters Ltd., Auckland, New Zealand) with MAFFT algorithms FFT-NS-I x1000 and G-INS-I (Katoh and Standley, 2013).
2.2. Chromosomal distribution and integration context
To understand if the genomic distribution of the HML8 elements is random among human chromosomes, we calculated the expected distribution with the formula “e=Cl∗n/Tl”, in which e stands for the number of expected integrations in the chromosome, Cl for the length of the chromosome, n for the total number of the HML8 loci, and Tl for the sum of the length of all chromosomes. Given that this kind of test works better with a larger number of sequences, in addition to the HML8 proviruses (78) we decided to include in the analysis also all the identified solitary LTRs (504). The analysis has been performed for each chromosome and results have been verified applying the chi-square test and assessing statistical significance with p-value calculation. Additionally, the position within each chromosome has been evaluated using UCSC Genome Browser Data Integrator tool, to intersect HML8 coordinates with the annotation of chromosomal loci. In this way, we also evaluated the proportion of HML8 elements located in centromeric or peri-centromeric regions, considering as peri-centromeres the two loci flanking the centromere with the only exception of chromosome 6, for which we have consider also the q12 section as peri-centromeric given the small dimensions of the q11.2 locus. The above Data Integrator tool has been similarly used to intersect HML8 coordinates with Gencode set of annotations (Harrow et al., 2012), to evaluate each HML8 sequence genomic context of integration and identify which of them was co-localized with cellular genes.
2.3. Phylogenetic analyses
To infer HML8 phylogeny we performed different phylogenetic analyses taking into consideration the complete proviral sequence of all HML8 loci as well as their individual genic portions and LTRs. Alignments for tree building were performed with Geneious Prime software, version 2020.1.1 (Biomatters Ltd., Auckland, New Zealand) using MAFFT algorithms FFT-NS-I x1000 and G-INS-I (Katoh and Standley, 2013). The different HML8 genes were identified according to the Dfam annotations for HERV-K11 reference: gag (∼156-2297), pro (∼2063-3082), pol (∼3037-5785) and env (∼5624-7951). Moreover, every alignment also included a consensus sequence for each HERV-K group (HML1 to 10) as a benchmark. All phylogenetic trees were built with MEGA-X software (version 10.1) using neighbour joining (NJ) method and applying p-distance model. Resulting phylogenies were tested by bootstrap method with 1000 replicates (Kumar et al., 2018). Phylogenetic analyses also allowed to classify HML8 elements into the three types: MER11A, MER11B, and MER11C. MEGA X has been also used to infer MER11A, MER11B, and MER11C ancestral proviruses sequences. Briefly, the best model for ML analysis of LTRs and proviral genes has been determined and used to infer the ancestral sequences in each tree. The ancestral node for HML8 elements has been selected and the correspondent consensus sequence extracted. Ancestral consensus sequences for each portion have then been combined to obtain the complete proviral consensus sequence for each HML8 subtype, assessing the conservation of all the ORFs.
2.4. HML8 proviruses structural characterization
To gain a detailed knowledge of each HML8 provirus’ nucleotide structure and to detect the presence of mutations, deletions, and insertions, we aligned every sequence with the respective consensus, as assembled with the internal region HERV-K11 flanked by MER11A, MER11B, or MER11C LTRs depending on the subset of sequences considered. Alignments were generated through Geneious software, and all the mutations, integrations and deletions were annotated in each sequence. Furthermore, we used the online resource NCBI Conserved Domain (Marchler-Bauer et al., 2015) to evaluate the presence of relevant retroviral motifs.
2.5. Time of integration
The integration time of every HML8 locus has been estimated using the formula T=D/0.2%, in which the value 0.2% represents the spontaneous substitution rate of the human genome, as expressed in mutations per nucleotide per million years. D stands instead for the percentage of divergent nucleotides of each element with respect to a reference, as calculated with the software MEGA-X (Kumar et al., 2018) through the pairwise deletion option and without considering CpG dinucleotides. Integration time analysis has been carried out for each HML8 element considering individual genes and LTRs in comparison to a consensus sequence generated by aligning all the HML8 group members. Moreover, the analysis has been applied to the two LTRs of the same provirus, known to be identical at the time of the integration, and accumulating then mutations independently over time. The results of both methods have been considered to obtain the final estimated age by calculating their average and excluding the values with a standard deviation >20%.
To further confirm age estimations, we checked each HML8 sequence presence among primate's species (both Catarrhini and Platyrrhini parvorders) through the comparative genomic annotations provided by UCSC Genome Browser, paying particular attention to the flanking regions to ensure the actual presence of the same HML8 locus. In this way, we were able to assign the oldest common ancestor (O.C.A) to every HML8 member. With this multiple approach, in the absence of a reliable age estimation, the range of the integration time was based on the O.C.A. in which the HML8 sequence has been firstly found.
3. Results
3.1. Identification of all HERV-K(HML8) across human genome assembly hg38
Aiming to achieve a comprehensive characterization of all HML8 sequences dispersed within the latest human genome assembly hg38, we performed a BLAT search on UCSC Genome Browser. For this survey, we used as a query the HML8 reference for the internal proviral portion (HERV-K11) that was associated to the three different LTRs reported for the group in Dfam (MER11A, MER11B, and MER11C), to ensure the detection of all HML8 types. Thereafter, we downloaded all the matching sequences incorporating extra 5’ and 3’ regions of 500 nucleotides each, to avoid artificial truncation. The retrieved HML8 candidates have then been aligned to the Dfam reference to verify their integrity and discard those with an identity <90%. All the proviral sequences having sufficient identity to the reference have been refined manually to reach accurate coordinates and nucleotide sequence. In total, we identified and validated 78 HERV-K(HML8) sequences including 55 (out of 58) elements already classified as HML8 by RetroTector analysis (Vargiu et al., 2016) plus 23 newly discovered members (Table 1). In addition, 3 other sequences initially classified as HML8 by RetroTector were removed from the group members in the present work: 4432 was a portion of the already present 4431, while 5465 and 5466 were non-specific matches with no HERV sequence in the corresponding positions (IDs refer to the rvnr number assigned by RetroTector). As shown in Table 1, each HML8 member has been named referring to its genomic locus of integration, distinguishing two or more sequences located in the same locus adding an alphabet letter at the end of the name, following the coordinate's increasing number.
Table 1.
Characteristics of the 78 HML8 elements identified in the human genome (hg38).
| Locus | Strand | Coordinates | Age (My) | O.C.A. | Subtype | Reference |
|---|---|---|---|---|---|---|
| 1p13.3 | + | chr1:109702615-109709644 | 21.1 (20-17) | Orangutan | MER11A | Vargiu et. al |
| 1p21.1 | + | chr1:106159133-106162062 | 30-20 | Gibbon | MER11C | Vargiu et. al |
| 1p33 | + | chr1:46900689-46908067 | 21.7 (20-17) | Orangutan | MER11A | This study |
| 1p35.1 | + | chr1:33065126-33068378 | 48.0 (20-17) | Orangutan | - | Vargiu et. al |
| 1q23.3 | - | chr1:160686556-160703551 | 28.9 (30-20) | Gibbon | MER11C | Vargiu et. al |
| 1q25.3 | + | chr1:181245995-181255232 | 28.5 (30-20) | Gibbon | MER11A | Vargiu et. al |
| 2p14 | + | chr2:63988445-63997145 | 35.8 (43-30) | Rhesus | MER11C | This study |
| 2q11.2P | - | chr2:100361988-100365940 | 42.7 (30-20) | Gibbon | MER11B | Vargiu et. al |
| 2q14.2 | - | chr2:119373755-119376781 | 38 (43-30) | Rhesus | MER11A | Vargiu et. al |
| 2q34a | - | chr2:210424462-210428103 | 30.3 (30-20) | Gibbon | MER11C | This study |
| 2q34b | + | chr2:213446552-213449649 | 53.7 (43-30) | Rhesus | MER11A | This study |
| 3p12.3 | + | chr3:79051663-79060597 | 40 (43-30) | Rhesus | MER11C | Vargiu et. al |
| 3q13.13 | + | chr3:111524145-111530464 | 35.5 (43-30) | Rhesus | MER11A | Vargiu et. al |
| 3q22.1 | - | chr3:130443999-130453404 | 26.5 (30-20) | Gibbon | MER11C | Vargiu et. al |
| 4p16.3 | - | chr4:4042123-4049160 | 17.2 (20-17) | Orangutan | MER11C | This study |
| 4q13.1 | - | chr4:64142025-64148090 | 42.3 (43-30) | Rhesus | MER11A | This study |
| 4q13.2 | + | chr4:69191808-69199233 | 46.9 (30-20) | Gibbon | MER11A | Vargiu et. al |
| 4q13.3 | - | chr4:73949403-73954209 | 48.7 (30-20) | Gibbon | MER11B | This study |
| 4q21.21 | - | chr4:79824404-79827356 | 24.3 (30-20) | Gibbon | MER11C | Vargiu et. al |
| 4q31.1 | + | chr4:139632366-139639045 | 30-20 | Gibbon | MER11A | Vargiu et. al |
| 4q32.3 | + | chr4:164748875-164753249 | 42.2 (43-30) | Rhesus | MER11A | Vargiu et. al |
| 5p13.1 | + | chr5:40102909-40112787 | 24.7 (30-20) | Gibbon | MER11C | Vargiu et. al |
| 5q14.1 | + | chr5:81899808-81901603 | 30-20 | Gibbon | MER11C | This study |
| 5q35.1 | + | chr5:172396993-172402677 | 26.4 (30-20) | Gibbon | MER11B | Vargiu et. al |
| 6p11.2P | + | chr6:58399942-58403965 | 9-7 | Chimp | - | Vargiu et. al |
| 6q11.1C | + | chr6:61274495-61278513 | 20-17 | Orangutan | - | Vargiu et. al |
| 6q14.1 | - | chr6:76487320-76496698 | 42 (43-30) | Rhesus | MER11B | Vargiu et. al |
| 6q25.3 | - | chr6:158296613-158303611 | 39.3 (30-20) | Gibbon | MER11A | Vargiu et. al |
| 7p12.1 | + | chr7:50569934-50583960 | 30-20 | Gibbon | MER11C | Vargiu et. al |
| 8p11.1C | + | chr8:43892563-43899366 | 42.6 (17-9) | Gorilla | MER11C | Vargiu et. al |
| 8p23.1a | - | chr8:7152780-7159818 | 20-17 | Orangutan | MER11C | Vargiu et. al |
| 8p23.1b | + | chr8:8127655-8134678 | 20-17 | Orangutan | MER11C | Vargiu et. al |
| 8p23.1c | - | chr8:12531005-12538032 | 20-17 | Orangutan | MER11C | This study |
| 8q11.1C | + | chr8:46580301-46590040 | 20-17 | Orangutan | MER11C | Vargiu et. al |
| 8q21.3 | - | chr8:88670474-88676583 | 30-20 | Gibbon | MER11C | This study |
| 9p21.1 | + | chr9:31770537-31778849 | 34.7 (43-30) | Rhesus | MER11A | Vargiu et. al |
| 9q32 | - | chr9:112391953-112400106 | 37.6 (30-20) | Gibbon | MER11A | Vargiu et. al |
| 10p11.1C | + | chr10:39042475-39051569 | 17-9 | Gorilla | MER11A | Vargiu et. al |
| 10q21.3 | - | chr10:65105943-65112846 | 30-20 | Gibbon | MER11B | This study |
| 10q23.1 | - | chr10:81429499-81431222 | 30-20 | Gibbon | MER11C | This study |
| 10q24.32 | - | chr10:102838109-102845149 | 34.2 (43-30) | Rhesus | MER11A | Vargiu et. al |
| 11p11.12aP | + | chr11:50438109-50447947 | 9-7 | Chimp | MER11B | Vargiu et. al |
| 11p11.12bP | - | chr11:50629499-50637560 | 39.1 (17-9) | Gorilla | MER11C | Vargiu et. al |
| 11p11.12cP | - | chr11:50487842-50495622 | 17-9 | Gorilla | MER11B | Vargiu et. al |
| 11p15.2 | - | chr11:15062966-15070542 | 31.1 (43-30) | Rhesus | MER11B | Vargiu et. al |
| 11q11C | + | chr11:54758703-54766192 | 42.8 (9-7) | Chimp | MER11B | Vargiu et. al |
| 11q13.2 | + | chr11:67698550-67705411 | 40 (43-30) | Rhesus | MER11A | Vargiu et. al |
| 11q22.1 | - | chr11:101211205-101221382 | 31.3 (43-30) | Rhesus | MER11A | Vargiu et. al |
| 12p11.1C | + | chr12:34522067-34530751 | 37.7 (9-7) | Chimp | MER11C | Vargiu et. al |
| 12q15 | - | chr12:69380691-69382268 | 30-20 | Gibbon | MER11C | Vargiu et. al |
| 12q21.31 | - | chr12:84102187-84109415 | 28.5 (30-20) | Gibbon | MER11C | This study |
| 12q23.3 | + | chr12:105299228-105314444 | 44.5 (43-30) | Rhesus | MER11B | Vargiu et. al |
| 13q22.3 | - | chr13:78056549-78058995 | 47.9 (20-17) | Orangutan | MER11A | This study |
| 14q32.11 | + | chr14:90951493-90954217 | 48.7 (43-30) | Rhesus | MER11A | This study |
| 15q15.1 | - | chr15:40686062-40690093 | 30-20 | Gibbon | MER11C | This study |
| 16q21 | - | chr16:59621926-59625900 | 39.6 (30-20) | Gibbon | MER11C | This study |
| 17q11.2P | - | chr17:27686099-27696448 | 28.2 (20-17) | Orangutan | MER11C | Vargiu et. al |
| 18q21.1 | - | chr18:50220146-50223833 | 54.5 (43-30) | Rhesus | MER11A | This study |
| 19p11aC | + | chr19:24242724-24251824 | 20-17 | Orangutan ° | MER11B | Vargiu et. al |
| 19p11bC | + | chr19:24270246-24273528 | 20-17 | Orangutan * | MER11B | Vargiu et. al |
| 19p11cC | + | chr19:24320890-24325976 | 20-17 | Orangutan ° | - | Vargiu et. al |
| 19p12aP | - | chr19:20500234-20509675 | 30.3 (30-20) | Gibbon | MER11A | Vargiu et. al |
| 19p12bP | - | chr19:23847319-23857439 | 31.7 (43-30) | Rhesus | MER11A | Vargiu et. al |
| 19q11aC | + | chr19:27578693-27582287 | 17-9 | Gorilla | MER11C | Vargiu et. al |
| 19q11bC | - | chr19:27703454-27713310 | 35.1 (20-17) | Orangutan ° | MER11C | This study |
| 22q11.21P | + | chr22:19934438-19940686 | 40.4 (43-30) | Rhesus | MER11A | Vargiu et. al |
| Xp11.21P | + | chrX:56942413-56948730 | 20-17 | Orangutan | - | Vargiu et. al |
| Xp11.3 | - | chrX:46486519-46490277 | 9-7 | Chimp | MER11C | This study |
| Xp11.4a | + | chrX:41644222-41648694 | 30.2 (20-17) | Orangutan | MER11C | Vargiu et. al |
| Xp11.4b | - | chrX:42161343-42164806 | 38.7 (30-20) | Gibbon | MER11B | This study |
| Xp21.1 | + | chrX:34796024-34803445 | 21.6 (30-20) | Gibbon | MER11B | Vargiu et. al |
| Yp11.2P | - | chrY:7991874-7998823 | 48.2 (9-7) | Chimp | MER11C | Vargiu et. al |
| Yq11.221 | - | chrY:14374864-14381444 | 9-7 | Chimp | MER11A | Vargiu et. al |
| Yq11.222a | + | chrY:17684505-17688179 | 9-7 | Chimp | MER11B | Vargiu et. al |
| Yq11.222b | - | chrY:18217835-18221509 | 9-7 | Chimp | MER11B | Vargiu et. al |
| Yq11.223 | - | chrY:22829524-22834840 | 30-20 | Gibbon | MER11C | Vargiu et. al |
| Yq11.23a | - | chrY:24463282-24468583 | 30-20 | Gibbon | MER11C | This study |
| Yq11.23b | + | chrY:25201520-25206820 | 43-30 | Rhesus | MER11C | This study |
My = million years, O.C.A. = Oldest Common Ancestor
= centromeric
= peri-centromeric, °= lacking the orthologous in Gorilla, *=converted to solitary LTRs in Chimp. In the “Locus” column, clusters of sequences being the result of segmental duplication (e.g. 8p23.1) are included in a unique cell. In the “Age” column, calculated time of integration (in million years, my) is provided as the average of the estimates resulted from the different approaches (see materials and methods), excluding the values with a standard deviation >20%. In the absence of a reliable estimation or for the above clusters of duplicated sequences, only the age range based on comparative genomics in non-human primates is indicated. The latter has been added within brackets also for those HML8 elements with an estimated age. In the “Subtype” column, sequences lacking both LTRs could not be classified and were hence reported as “- “.
An analogue BLAT search has been applied using the sole MER11A/B/C to identify 504 HML8 solitary LTRs, which emerged from recombination processes between the LTRs of the same provirus, leading to the displacement of the internal region (Supplementary Table 1).
3.2. Chromosomal distribution and co-localization with cellular genes
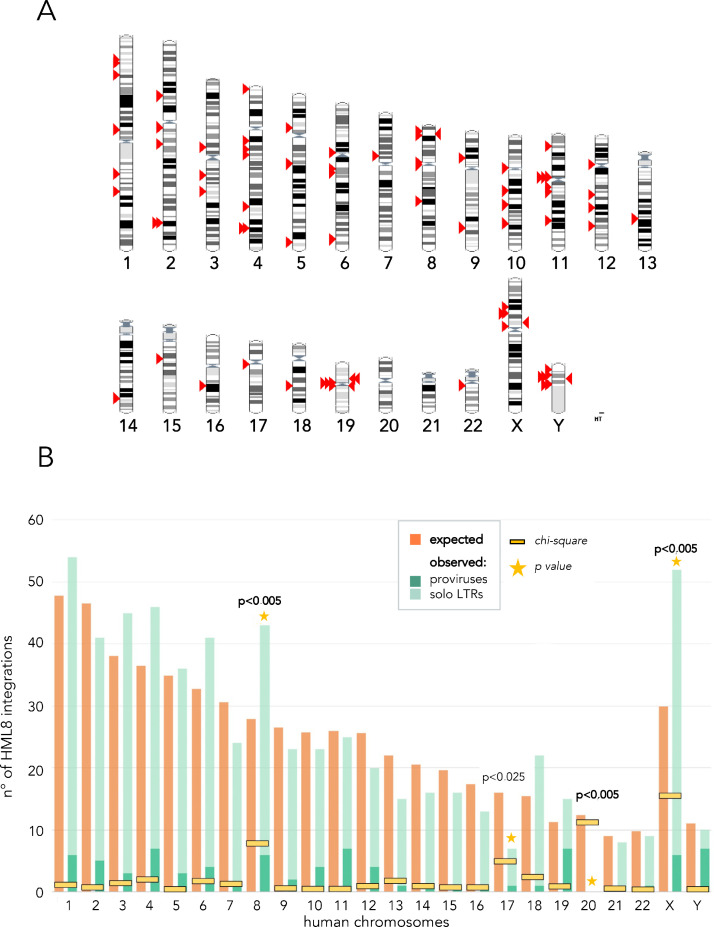
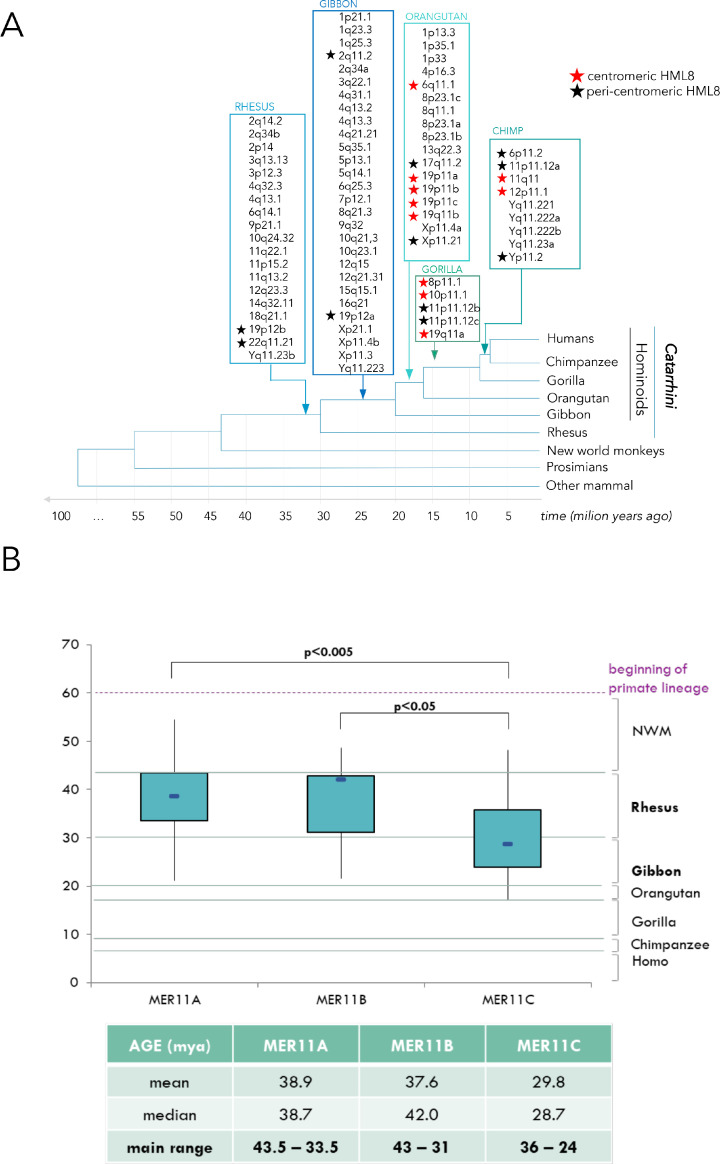
Having a detailed map of all HML8 proviruses and solitary LTRs in the human genome, we asked whether their spread among chromosomes was random or had some biases. We hence compared the observed HML8 distribution with the theoretically estimated one applying the chi-square test and calculating the p-value. In this comparison, the expected number of HML8 integrations (e) was obtained with the formula “e=Cl∗n/Tl”, which considers the length of each chromosome multiplied by the total number of HML8 loci then divided by the sum of the length of every chromosome. We assessed that the genomic distribution of the HML8 members is not random, showing an enrichment within chromosomes 8 and X (p<0.005) whereas chromosomes 17 (p<0.025) and 20 (p<0.005) present fewer integrations than expected (Fig. 1). Particularly, chromosome 20 did not hold any HML8 integration, neither proviruses nor solitary LTRs, while chromosome 21 did not present any proviral integrations but only solitary LTRs (Fig. 1).
Fig. 1.
Chromosomal distribution of HML8 elements.
A. Position of each HML8 provirus (red arrows) in human chromosomes. B. Statistical comparison between expected and observed amount of HML8 integrations per chromosome (also considering solitary LTRs)
Beside the above distribution bias among chromosomes, HML8 proviruses showed an enrichment within centromeric and peri-centromeric regions. In fact, 22 out of 78 sequences were found inside the centromere (11) or within the peri-centromeric regions (11) of the respective chromosomes, representing the 28.2% of the total HML8 insertions (Table 1). Such percentage is higher than other HERV-K groups, such as HML2 (15.4%), HML6 (16.7%), and HML7 (21.7%), and with respect to the well-characterized Gamma-like HERV-W group (9.9%) (data not shown). An opposite situation has been found regarding the HML8 solitary LTRs, with just 7.5% of them found in a centromeric or pericentromeric region. Such proportion was similar to the one of the HERV-W group (7.3%), while the above other Beta-like groups have around 13% (data not shown).
Moreover, the genomic context of integration has been examined to evaluate the colocalization of HML8 proviruses with cellular genes. Across the 78 HML8 elements, 26 (33%) were found to be intragenic, being inserted into 31 human genes (Table 2) that were either protein-coding (18) or producing long non-coding RNAs (lncRNAs, 13), showing the same orientation in 9 cases. Of note, 7 of the intragenic HML8 showed an exonic localization, being integrated in exons of protein coding genes (2, of which one with the same orientation) or lncRNAs (5, of which 4 with the same orientation) (Table 2). The most relevant colocalized genes are commented in the discussion section.
Table 2.
HML8 loci co-localized with human cellular genes.
| HML8 locus | Co-localized gene | intron/exon | Gene function | Associated diseases |
|---|---|---|---|---|
| 1p13.3 (+) | GSTM2, GSTM1 (+) | intron and exon | detoxification of electrophilic compounds | Asbestosis and Oral Leukoplakia |
| 1q23.3 (-) | CD48 (-) | intron | immunoglobulin-like receptors | Lymphoproliferative Syndrome, X-Linked, 1 and Cone-Rod Dystrophy 6 |
| 2q34a (-) | LANCL1-AS1 (+) | intron and exon | lncRNA – antisense to LANCL1 (that in turn protects cells from oxidative stress, and promotes cell proliferation) | |
| 2q34b (+) | SPAG16 (+) | intron | proteins that associate with the axoneme of sperm tail and the nucleus of postmeiotic germ cells | Pulmonary Subvalvular Stenosis and Ciliary Dyskinesia, Primary |
| 2p14 (+) | VPS54 (-) | intron | part of a trimeric vacuolar-protein-sorting complex (from prevacuoles to the late Golgi) | Amyotrophic Lateral Sclerosis 17 and Spermatogenic Failure 9 |
| 3q22.1 (-) | COL6A5 (+) | intron | collagen superfamily of proteins | Chronic Dacryoadenitis and Dermatitis |
| 3p12.3 (+) | ROBO1 (-) | intron | integral membrane protein that functions in axon guidance and neuronal precursor cell migration | Pituitary Stalk Interruption Syndrome and Dyslexia |
| 4q13.2 (+) | AC111000.4 (+) | intron and exon | lncRNA – antisense to UGT2B11 (UDP glucuronosyltransferase major role in conjugation and elimination of xenobiotics) | |
| 5q35.1 (+) | SH3PXD2B (-) | intron | podosome formation | |
| 6q25.3 (-) | TULP4 (+) | intron and exon | mediates the ubiquitination | |
| 8p23.1b (+) | AC105233.4 (+) | intron | lncRNA – uncharacterized | |
| FAM85B (-) | intron | lncRNA – family with sequence similarity 85 member B | ||
| 8p23.1c (-) | AC068587 (-) | exon | lncRNA – uncharacterized | |
| LOC100506990 (+) | intron | lncRNA – uncharacterized | ||
| 8q11.1 (+) | ASNSP1 (-) | intron | Asparagine synthetase pseudogene 1 | |
| 9q32 (-) | HSDL2 (+) | intron | Hydroxysteroids dehydrogenase like (apparently, no activity) | |
| C9orf147 (-) | exon | lncRNA - antisense to HSDL2 | ||
| 11p15.2 (-) | CALCB, CALCA (+) | intron | calcium regulation, induces vasodilation | Reflex Sympathetic Dystrophy and Spinal Stenosis. |
| 12q15 (-) | YEATS4 (+) | intron | Putative transcription factor, amplified in tumors | Macular Dystrophy, Patterned, 3 and Cellular Myxoid Liposarcoma |
| 12q23.3 (+) | KCCAT198 (-) | intron | lncRNA – renal clear cell carcinoma associated transcript 198 | Phelan-Mcdermid Syndrome |
| 13q22.3 (-) | OBI1-AS1 (+) | intron | lncRNA – antisense to OBI1 (coding for E3 ubiquitin ligase, acts as a replication origin selector during S-phase) | Waardenburg Syndrome, Type 4A and Hirschsprung Disease 2 |
| 14q32.11 (+) | RPS6KA5 (-) | intron | transferase for phosphorus-containing groups, protein tyrosine kinase activity | Septic Myocarditis and Coffin-Lowry Syndrome |
| 15q15.1 (-) | RAD51-AS1 (-) | exon and intron | lncRNA – antisense to RAD51 (recombinase involved in homologous recombination and repair of DNA) | |
| 19p12a (-) | AC008554.1 (+) | intron | lncRNA – uncharacterized | Spermatogenic Failure, X-Linked |
| 19p12b (-) | AC139769.2 (-) | intron | lncRNA – uncharacterized | Pediatric Germ Cell Cancer |
| 22q11.21 (+) | TXNRD2 (-) | intron | pyridine nucleotide-disulfide oxidoreductase family | Glucocorticoid Deficiency 5 and Familial Glucocorticoid Deficiency |
| Xp11.4a (+) | CASK (-) | intron | calcium/calmodulin-dependent serine protein kinase | |
| Xp11.3 (-) | KRBOX4 (+) | intron | zinc finger protein with an N-terminal KRAB domain found in transcriptional repressors | |
| Yq11.23b (+) | TTTY17C (-) | intron | lncRNA – testis-specific transcript, Y-linked 17C (3 copies on chr Y: this is the most telomeric one) |
The strand of HML8 proviruses and co-localized genes is reported between brackets. Gene association with human diseases are based on OMIM annotations.
3.3. Phylogeny of HML8 LTR sequences
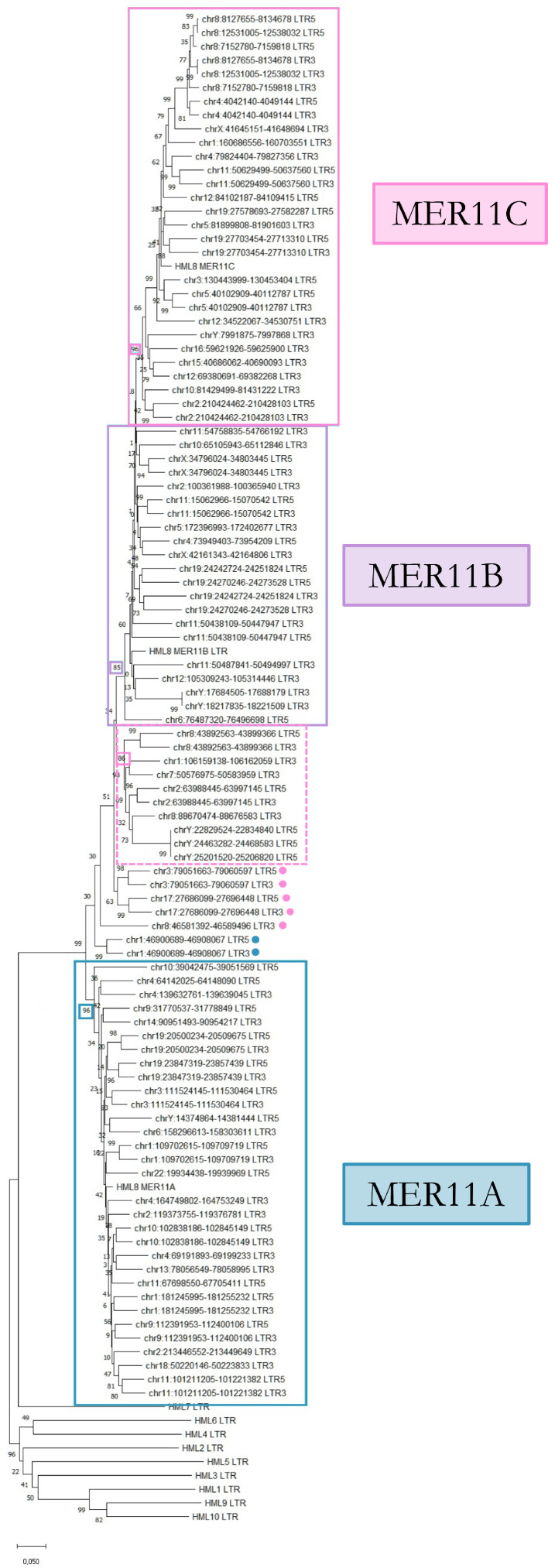
As for the other HERVs, the knowledge of HERV-K(HML8) group phylogenesis is fundamental to unravel the possible relationship between the members thereof, but also with respect to the other HML groups, in case of closer relations with a subset of them or the presence of mosaic sequences between two or more groups. Moreover, given that HML8 internal sequence can be associated to three alternative types of LTR, the analysis allowed to classify HML8 members and to assess the actual differences among them. For this purpose, we carried out a neighbour-joining (NJ) analysis with the 5’ and 3’ LTRs of each provirus, when present (Fig. 2). It is clearly visible that the sequences create 3 phylogenetic clusters, each corresponding to the three Dfam LTR types and supported by high value of bootstrap (96% for both MER11A and MER11C, and 85% for MER11B). In addition, a fourth phylogenetic group (dashed square in Fig. 2) made by additional 8 MER11C elements was not included in the main MER11C cluster due to several mutations accumulated in the LTRs of its members (Fig. 2). The LTR sequences not included in any phylogenetic cluster have been further analysed with respect to the Dfam consensus in order to assign them in one of the groups laying on sequence similarity. Finally, 5 HML8 provirus (1p35.1, 19p11c, 6p11.2, 6q11.1 and Xp11.21) could not be included in the analysis due to the loss of both LTRs, lacking a phylogenetic classification.
Fig. 2.
Phylogenetic tree of HML8 elements based on 5’ and 3’ LTRs.
Phylogeny was inferred with neighbor joining (NJ) method applying p-distance model and tested by bootstrap analysis with 1000 replicates. The three main phylogenetic clusters of HML8 LTRs -corresponding to LTR types MER11A, MER11B, and MER11C - are indicated with coloured rectangles (blue, violet, and pink, respectively). An additional MER11C supported cluster not included in the main one is indicated with a dashed rectangle. Finally, the same colour labels indicate the subgroup of belonging of a few sequences not included in the above main clusters.
Finally, the tree includes some well-supported clusters grouping together the 5’ LTRs of different HML8 elements, as do the 3’ LTRs, suggesting that these integrations are likely the result of segmental duplication events. These clusters include both HML8 elements within the same locus (8p23.1a, b, and c; 19p11a and b; Yq11.222a and b) or in different loci of the same chromosome (6p11.2 and 6q11.1; Yq11.223 and Yq11.23a and b). Overall, the elements within each cluster share several sequence features, such as insertions/deletions and single nucleotide substitutions as compared to the group reference. They also have high nucleotide identity (overall 96.3%: 95.8% for 6p11.2/6q11.1, 99% for 8p23.1 cluster, 87% for 19p11 cluster, >99.9% for Yq11.222 and Yq11.223/Yq11.23 clusters). Accordingly, we found that the 1 kb of cellular genome flanking each member of the same cluster in 5’ and 3’ share evident sequence identity as well, further suggesting that they arose by past duplication events.
3.4. Structural characterization
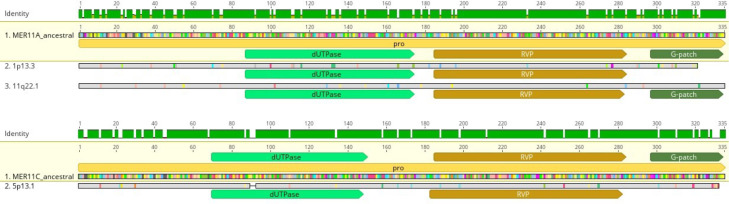
The structural characterization of HML8 proviruses would allow us to describe the uniqueness of each single member – through the analysis of mutation, deletion, and integration events - and to have a detailed picture of the possible state of activity of the group based on their genic conservation. For such purpose, we have created three consensus sequences, having in common the internal reference region of the group (HERV-K11), annotated with the retroviral genes: gag (∼156-2297), pro (∼2063-3082), pol (∼3037-5785), and env (∼5624-7951). HERV-K11 was then associated to 5’ and 3’ sides by each LTR type (MER11A, MER11B, and MER11C), to represent the three group variants. Each consensus has been aligned with the related HML8 members, as classified based on the above phylogenetic analysis.
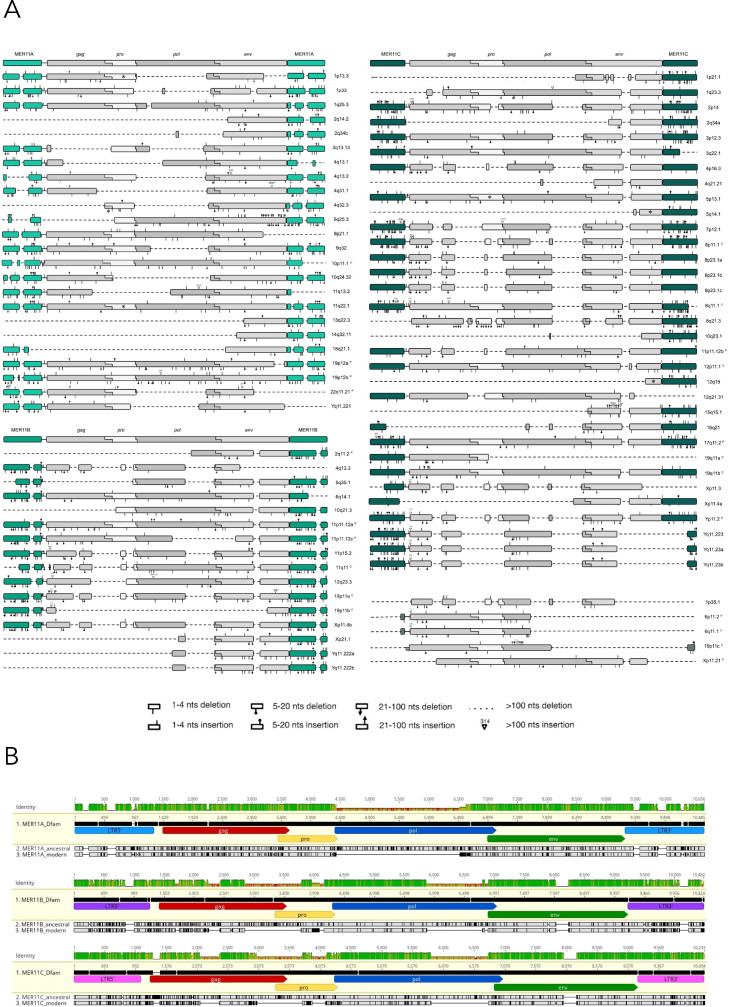
Overall, HML8 sequences show a biased nucleotide content, in line with what was previously reported (Vargiu et al., 2016). Particularly, we confirm a reduction in G content (19,1%) and an enrichment in T (30,6 %) in addition to a slight increase in A (27,7%) (data not shown). Focusing on the structural features of the LTRs, some specific nucleotide portions present in Dfam reference sequence are instead absent in the LTRs of every member of the same subgroup (Fig. 3, panel A). For example, both 5’ and 3’ LTRs of the 24 MER11A proviruses lack a region of 141 nucleotides corresponding to bases 549-689 of Dfam MER11A reference, indicating that the latter is not representative of the actual nucleotide structure. Similarly, none of the MER11B 5’ and 3’ LTRs contain a 140 bp region that is instead present in Dfam MER11B LTR reference (nucleotides 846-985). Otherwise, MER11C LTRs do not present major portions being not represented in the corresponding Dfam reference, being nevertheless still divergent in nucleotide sequence due to several mutations.
Fig. 3.
Structural characterization of HML8 proviruses.
A. The 78 HML8 proviruses have been divided according to their phylogenetic classification and compared to the respective reference sequences. All insertions and deletions have been annotated, as reported in the figure legend. B. Alignment of the ancestral and modern HML8 consensus sequences, as generated for MER11A, MER11B, and MER11C subgroups, with the corresponding Dfam reference sequences (made with HERVK11 internal portion flanked with MER11A/MER11B/MER11C LTR types). The presence of intact ORFs (0 shifts and 0 stops) has been annotated with a star in the relative gene.
Beside the LTRs, also the internal proviral region showed similar differences, but in this case they seems to be due to recurrent deletions that removed a certain region from the majority of elements of the same HML8 type (Fig. 3, panel A). MER11A subgroup presents larger deletions with respect to the other two, including some defective proviruses composed solely by the env gene and the 3’ LTR (6 out of 24). Despite this, the great majority of the subgroup contains an almost intact env region, and many of them also harbour a complete gag-pro portion, while the pol gene is lost in most of the members, being present only in 8 of them (Fig. 3, panel A). Across the MER11B elements (16), two major deletions are notable: the larger one (925 bp) is shared by half of them (8) and falls inside the pol gene, leading to the loss of the integrase domain; while the smaller one (204 bp) is shared by every member and is located inside the env portion (Fig. 3, panel A). These MER11B major deletions are also common to many elements belonging to the MER11C subgroup: pol one was found in 16 out of 33 members, while the smaller env deletion in all sequences except for Xp11.3. Further, 13 MER11C members shared a 313 nucleotides deletion within gag gene (Fig. 3, panel A). Also, several MER11B and MER11C proviruses showed two neighbouring deletions of 938 and 219 nucleotides spanning gag and pro genes. Finally, the 5 HML8 elements lacking a classification due to the absence of both LTRs did not show any characteristic internal deletion except for 1p35.1 that presents the above-mentioned deletions at the integrase domain and the gag-pro portion.
Intriguingly, the above recurrent deletions were lowly represented in the HML8 proviruses integrated within centromeric and peri-centromeric regions (11 and 11, respectively), which showed a more complete structure (Fig. 3, panel A). Accordingly, 19 out of 22 of them do not present most of the identified deletions except for the one present inside the env gene.
To better summarize the major features of each HML8 subgroup - also given their high divergence with respect to the existing Dfam reference sequence - we propose here two sets of new HML8 consensus sequences (Fig. 3, panel B; Supplementary file 1). One set includes the putative ancestral consensus sequences for MER11A, MER11B, and MER11C and was generated through a phylogenetic approach; while the other includes the corresponding “modern” consensus sequences as inferred from the alignment of all members of each subgroup, thus representing their current nucleotide structure (i.e. after their prolonged persistence in the host genome).
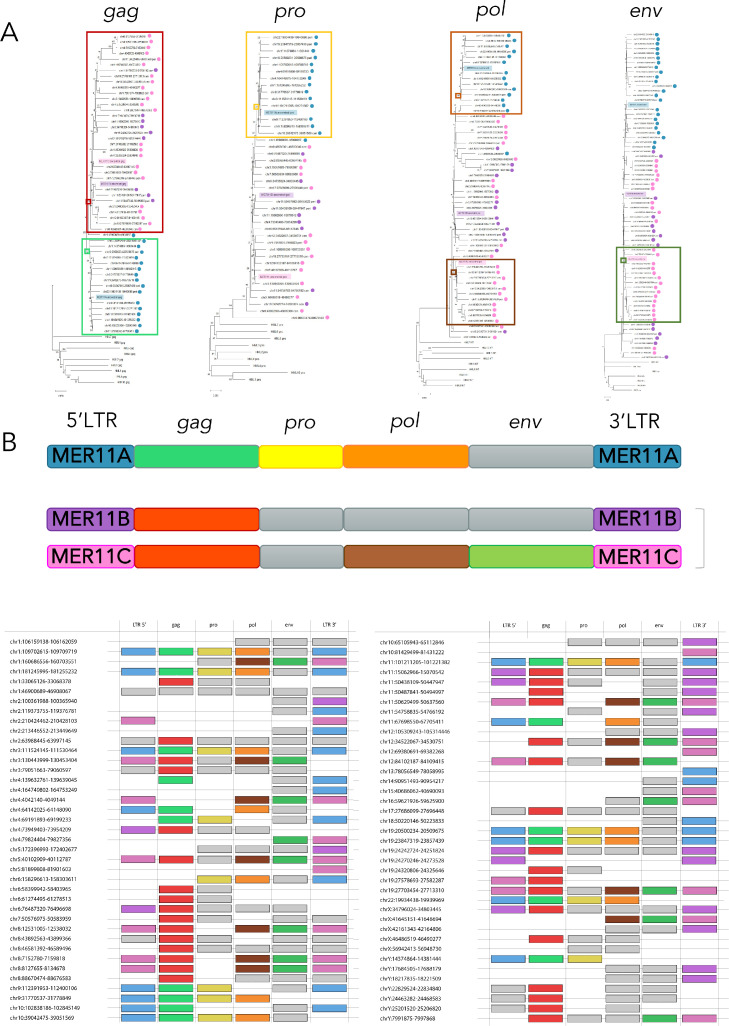
3.5. Phylogeny of HML8 genes and their combination with MER11 LTR types
In the light of the above-described nucleotide diversity among the three HML8 subgroups, which was not limited to the LTR portions, we combined the structural characterization of the single sequences to the phylogenetic analysis of the individual retroviral genes, to assess the existence of supported variants. NJ trees for gag, pro, pol, and env genes were built also including the corresponding genes from all HML groups, to evaluate their actual belonging to HML8. In all trees, the totality of HML8 sequences clustered with the three ancestral HML8 consensus sequences with 100% (gag, and pol), 99% (env), or 94% (pro) of bootstrap support, being clearly divided from other HMLs. Within each main HML8 cluster, a subset of sequences formed one (pro and env) or two (gag and pol) supported subcluster, indicating some additional similarities of these variants as compared to the rest of elements (Fig. 4, panel A). To understand how such genic variants were associated among themselves and with the three MER11 LTR types, we represented their combination in a second structure image that visually shows the composition of every HML8 member with reference to the different phylogenetic subclusters (Fig. 4, panel B). The presence of three structural patterns is recognizable, each attributable to one LTR type. MER11A pattern is composed by MER11A LTRs associated with pro variant, gag variant as identified in the lower subcluster, and pol variant as identified in the upper subcluster. MER11B pattern combines MER11B LTRs and gag variant as identified in the upper subcluster. Such gag variant is common also to MER11C pattern, in which MER11C LTRs are combined with it and with pol variant as identified in the lower subcluster followed by the only env variant present in the tree (Fig. 4, panel B).
Fig. 4.
Phylogeny of HML8 genes and structural pattern in association to MER11 LTR types.
Phylogeny of HML8 genes (A) was inferred with neighbour joining (NJ) method applying p-distance model and tested by bootstrap analysis with 1000 replicates: supported phylogenetic clusters are indicated with solid-coloured rectangles. The HML8 subgroup of each sequence is also indicated with coloured dots corresponding to LTR types MER11A, MER11B, and MER11C (blue, violet, and pink, respectively). The existing HML8 structural patterns (B) arose from the combination between MER11 LTR types and the identified gene clusters are represented, with colours corresponding to the annotations of panel A. Genes coloured in grey were part of the main cluster but did not form any supported subcluster.
3.6. Estimated time of integration and comparative genomics across the primates
Providing an estimation of the time of integration of every HML8 member is a key point to understand the evolution of the group across primates and its dynamics of distribution until humans. The most used method to perform this kind of analysis is the estimation of the nucleotide divergence between the two LTRs of each provirus, which are identical at the moment of integration and start then to accumulate mutations according to the host genome substitution rate. Given that, the formula provides an estimation of time of integration (T) dividing the percentage of divergent nucleotide (D) by the spontaneous substitution rate of the human genome (0.2%) (T=D/0.2%). The obtained T is then divided by 2, because the two LTRs accumulates mutations independently. However, this method has some limitations: beside the fact that the rate of substitution could be different across the different region of the genome, with such approach all those HML8 elements lacking one or both LTRs could obviously not be included in the analysis (Grandi et al., 2016). To overcome this problem, we decided to apply a multiple age estimation approach that - in addition to the standard LTR vs LTR comparison - calculates the divergence between each proviral portion (LTR, gag-pro, pol, and env) and a consensus sequence built aligning all the HML8 sequences. The latter allowed us to perform the analysis also on the internal region of the group, thus obtaining a more detailed age of integration estimation. In this way, T is expressed as the average of the multiple calculation results. The obtained T has further been validated by the identification of the O.C.A. among non-human primates, relying hence on their known time of evolutionary split to delimit HML8 period of invasion (Table 1 and Fig. 5, panel A). To this purpose, a comparative genomics approach has been used to identify the orthologous of each HML8 member in the various primates, also revealing that in some instances the sequence was absent in some Catarrhini species. This was the case of 19p11a, 19p11c and 19q11b HML8 loci that are present in Orangutan and Chimp but not in Gorilla; while 19p11b is present until Orangutan but was converted into two solitary LTRs in Chimp, with the removal of the whole internal region.
Fig. 5.
Overview of HML8 time of integration among primates.
(A) Dynamics of acquisition of HML8 group members by primate species based on comparative genomics of each HML8 integration. Each HML8 locus is reported based on the first primate (among the ones with available genome assembly) in which it was found. Nodes indicate estimated period of evolutionary split of each primate species. (B) Period of distribution of HML8 members according to the subgroup of belonging: age estimates (in mya) are based on a multiple approach of divergence calculation integrated with the above comparative genomics search. p-values are calculated applying the independent two-tailed t-test. The x axis reports the time of evolutionary divergence of the considered primate's species: for instance, the lineages originating Rhesus and Gibbon species had diverged around 30 mya, the lineages originating Gibbon and Orangutan species had diverged around 20 mya, and so on.
Our T calculation showed that the major wave of integration of HML8 elements took place between 43 and 17 million years ago (mya): accordingly, the highest number of integrations occurred in Rhesus (n=19, 43-30 mya), Gibbon (n=28, 30-20 mya), and Orangutan (n=17, 20-17 mya) (Fig. 5, panel A). Beside this main period of acquisition, the group maintained a residual integration activity also later, leading to a subset of latest integrations in Gorilla (n=5, 17-9 mya) and Chimpanzee (n=9, 9-7 mya) (Fig. 5, panel A).
Moreover, having three different HML8 variants, we asked whether they might be associated to different waves of acquisition by primates. Analysis of the range of integration time confirmed rather different period of distribution of HML8 elements according to the HML8 subtype. MER11C-related proviruses resulted significantly younger with respect to both MER11A (p<0.005) and MER11B (p<0.05) subtypes (p-value calculated applying the independent two-tailed t-test) (Fig. 5, panel B). This scenario was also confirmed by the fact that MER11A-related proviruses were integrated mainly in Rhesus (57%, main acquisition between 43.5 and 33.5 mya), confirming the elder age of the subgroup. Instead, the youngest MER11C members have been integrated mostly between Gibbon (51%) and Orangutan (24%) (main acquisition: 36-24 mya); while MER11B seems to have an intermediate distribution, with most of the insertions occurred in Rhesus and Gibbon genomes (main acquisition: 43-31 mya) (Fig. 5, panel B). The analysis also confirmed that none of the HML8 elements is present in Platyrrhini primates, limiting the group colonization to the sole Catarrhini parvorder (data not shown).
3.7. Analysis of conserved HML8 Open Reading Frames (ORFs)
To conclude our characterization of HML8 elements in the human genome assembly hg38, we assessed weather any of the MER11A, B, and C HML8 loci retained any ORF with coding potential. To this purpose, we extracted and bioinformatically translated the nucleotide sequence of HML8 genes and compared the putative proteins to the ones obtained from the respective ancestral reference. Overall, most HML8 genes lost their coding capacity due to mutations and indels, accumulating internal stop codons and frameshifts. Only 12 HML8 loci (15%) showed a single ORF with either no shifts and stops (0/0) or no shifts and a single internal stop (0/1) (Table 3). Of these, 7 belonged to MER11A, 4 were MER11C and 1 was classified as MER11B. More in details, 3 MER11A loci showed putative gag ORFs with 0/0 (4q32.3) or 0/1 (3q13.13 and 4q13.1) shifts and stop: in all three cases, the putative protein was shorter than the reference (703 aa) due to deletions and/or internal stops (Table 3). Similarly, only HML8 locus 4q31.1 (MER11A) had a 0/1 ORF for pol gene, interrupted by an internal stop at aa 46 (Table 3). For env gene, 3 ORFs were identified, having either 0/0 (5q14.1 and 12q15, both MER11C) or 0/1 (4q13.3, MER11B) shifts and stops: also in this case, none of the putative proteins was full length due to deletions affecting the 5’ (5q14.1 and 12q15) or 3’ (4q13.3) portions of the env gene. In general, the most conserved ORF was pro one, with 3 HML8 loci devoid of shifts and stops, hence potentially encoding full-length (11q22.1, MER11A: 335 aa) or near full-length (1p13.3 MER11A and 5p13.1 MER11C: 321 and 329 aa, respectively) (Table 3). We have analysed the corresponding Pro putative proteins in terms of conserved functional domains, confirming that all of them retain recognizable motifs for trimeric dUTPase and pepsin-like aspartyl protease (Fig. 6). In addition, the full-length Pro at locus 11q22.1 also shows the C-terminal G-patch domain (Fig. 6).
Table 3.
HML8 loci most conserved Open Reading Frames.
| gag | pro | pol | env | |||||
|---|---|---|---|---|---|---|---|---|
| shift/stop | aa lenght | shift/stop | aa lenght | shift/stop | aa lenght | shift/stop | aa lenght | |
| 1p13.3 | 0/0 | 321/335 | ||||||
| 3q13.13 | 0/1 | 55/703 (*25) | ||||||
| 4q13.1 | 0/1 | 179/703 (*25) | ||||||
| 4q13.3 | 0/1 | 223/706 (*222) | ||||||
| 4q31.1 | 0/1 | 136/910 (*46) | ||||||
| 4q32.3 | 0/0 | 80/703 | ||||||
| 5p13.1 | 0/0 | 329/335 | ||||||
| 5q14.1 | 0/0 | 237/708 | ||||||
| 11q22.1 | 0/0 | 335/335 | ||||||
| 12q15 | 0/0 | 164/708 | ||||||
| 19q11a | 0/1 | 160/335 (*58) | ||||||
| Yq11.221 | 0/1 | 264/335 (*196) | ||||||
HML8 ORFs showing either no shifts and internal stop codons (0/0, in bold) or one single internal stop codon (0/1) are reported with the aa length of the correspondent putative protein with respect to the ancestral reference. When present, the position of internal stops (*) is reported between brackets.
Fig. 6.
Focus on HML8 pro ORF with coding potential.
Full length and near full-length pro ORFs with no frameshifts and internal stop codons were translated and analysed for conserved functional domains as compared to the respective ancestral consensus proteins. The aa substitution with respect to the latter are highlighted in the alignment with coloured residues, while common aa are in grey.
4. Discussion
Differently from the other HMLs, the HERV-K(HML8) group is almost unaccounted in HERV literature. In fact, while no studies have been specifically dedicated to the group, two previous works took it into consideration together with other HERV elements. A single publication by Ting-Chia et al. reported it as the one containing the higher number of somatic mutations among canonical Alpha-/Beta-retrovirus-related HERVs, hypothesizing a possible role in carcinogenesis if those mutations affect regulatory regions (Chang et al., 2019). This study mentioned the HML8 group as general entity, without specifying any precise member. The second study, even if not focused on the sole HML8 group, provided a partial information about its composition and distribution. In fact, within our previous classification work of around 3200 most intact HERV integrations in hg19 as performed with the software RetroTector, we classified 58 elements as HML8, further dividing them into 34 canonical members (59%) and 24 non-canonical mosaic sequences (41%) (Vargiu et al., 2016). Starting from this preliminary identification, our work aimed to provide the first comprehensive identification and characterization of all the genomic sequences attributable to the HERV-K(HML8) group present in human genome assembly hg38. Even if we cannot exclude the presence of additional integrations that are polymorphic in the human population and thus not represented in genome assemblies, as seen for HML2 group (Subramanian et al., 2011), our work represents the most comprehensive description of HML8 group in the human genome up to date. Through BLAT searches in the hg38 genome assembly with the Dfam consensus sequences of the group – characterized by the same HERV-K11 internal region associated with three different LTR types (MER11A, MER11B, and MER11C) – we were able to identify a total of 78 HML8 proviruses (Table 1). In particular, we confirmed 55 out of the original 58 HML8 elements identified by RetroTector and added further 23 HML8 elements that were not reported in our preliminary classification work (Vargiu et al., 2016). The latter was performed in an automated way with the software RetroTector, which scans the genome searching for retroviral motifs for HERV detection (Sperber et al., 2007). Hence, the observed difference with respect to the present work is likely due to the lack of recognition of defective proviruses due to the loss of those retroviral features. Beside the 78 HML8 proviruses, the human genome harbours also ∼500 HML8 solitary LTRs, arisen from the displacement of the internal genic portion as a consequence of past recombination among the two LTRs. The ratio between the number of HML8 proviral sequences and solo LTRs, about 1:7, is in line with the one of other HMLs such as HML6 (1:6) (Pisano et al., 2019) and HML7 (1:7) (Grandi et al., 2021), while HML2 group showed a higher rate of solitary LTR formation (1:10) (Subramanian et al., 2011) likely reflecting its prolonged period of activity, which led even to human-specific and polymorphic integrations (Grandi et al., 2021; Thomas et al., 2018). Such values are rather different from Class I gamma-like HERVs, at least when considering the HERV-W group that showed a 1:2.5 ratio if comparing proviruses and solitary LTR abundance (Grandi et al., 2016). Interestingly, the genomic distribution of HML8 elements among human chromosomes is not random, since chromosomes 8 and X are significantly enriched (p<0.005), while chromosomes 17 (p<0.025) and 20 (p<0.005) present a lower number of insertions compared to estimates (Fig. 1). Even if the reason for such chromosomal bias is unclear, a possible speculation is that in some chromosomes more integrations were originally present and might have been removed in very early phases by negative selection due to their harmful effect on the host, an event that is difficult to evaluate after millions of years. Such possibility has been partially addressed by the inclusion of solitary LTRs in the analysis, accounting for previous proviruses that are now converted in this form, but other mechanisms of removal of ancestral HML8 integrations cannot be evaluated at present. Similarly, the enrichment in HML8 elements could be linked to their presence in genomic regions that are not under selective pressure. In addition to the above chromosomal bias, we identified various HML8 proviruses inserted in centromeric (11) and peri-centromeric (11) regions (Table 1, Fig. 5). Representing already the 28.2% of the group, the actual number of centromeric and peri-centromeric HML8 might be even higher, considering that these chromosomal locations are still not completely sequenced and annotated due to the condensed structure of chromatin and to their repetitive nature. This has already been shown for HML2 group that, in addition to the elements reported in the human genome assembly, is known to include hundreds of copies of two members (K111 and K222) spread across centromeres and peri-centromeres by recombination events (Contreras-Galindo et al., 2013; Zahn et al., 2015). Similarly, the peri- and centromeric HML8 sequences might have been exposed to recombination events leading to their propagation in those regions. In line with a potential residual activity, such elements show a remarkably intact structure as compared to most of the other HML8 proviruses (Fig. 3), as reflected also by the higher mean length of peri-centromeric and centromeric sequences (7433 nucleotides vs 6090 for the others). In other instances, centromeric/peri-centromeric localization could have accounted for an opposite effect, leading to the removal of these HML8 elements in some intermediate primates. Accordingly, 19p11a, 19p11c and 19q11b proviruses infected Orangutan germline but were lost in the Gorilla genome, whereas 19p11b has been converted into two solitary LTRs during Chimpanzee speciation. These occurrences could be explained by their localization inside the chromosome centromere, a region full of repeated satellite DNA in rapid evolution (Arunkumar and Melters, 2020), which could have exerted a selective negative pressure on these sequences. Furthermore, among the 78 HML8 proviruses identified in the study, 12 elements have likely been interested by segmental duplication events also involving the surrounding genome region (Table 1). These elements form accordingly 5 well supported phylogenetic clusters (6p11.2/6q11.1, 8p23.1, 19p11, Yq11.222, and Yq11.223/Yq11.23) in the LTR tree (Fig. 2) and were probably duplicated along primate evolution. Particularly, the cluster in chromosome 6 includes a pericentromeric and a centromeric element (6p11.2 and 6q11.1, respectively) that were not classified in any subgroup due to the lack of both LTRs. While 6q11.1 was found in primate genomes from Orangutan to humans, 6p11.2 is present in Chimpanzee and humans only, suggesting that it was acquired later on due to the duplication of the former. Yq11.223/Yq11.23 cluster includes 3 HML8 elements: one is found in rhesus genome (Yq11.23b), while the other two (Yq11.23a and Yq11.223) were duplicated presumably around the split of rhesus and gibbon (∼30 mya), given that they are found in gibbon but do not have a corresponding provirus in rhesus, being identical except for one single nucleotide substitution. Of note, the most ancient of the three (Yq11.23b) is integrated within the human gene TTTY17C that is also known to be present in multiple copies in Y chromosome, producing a testis-specific transcript. These additional copies of TTTY17C are in proximity to the other two elements of the cluster, at a distance of 16,7 Kb, likely suggesting that their duplication involved also the nearby HML8 integrations, in line with the fact that the distal portion of the Y-chromosome shows propensity for non-allelic homologous recombination, resulting in deletions, duplications, and inversions (Bansal et al., 2016). Interestingly, the expansion of HML2 elements in the Xq28 locus had a similar dynamic, corresponding to the duplication of the cancer testis antigen 1 (CTAG1) (Subramanian et al., 2011). Cluster 8p23.1 also includes 3 HML8 sequences - all found starting from Orangutan genome assembly - and could account at least in part for the observed enrichment of HML8 insertions in this chromosome. In this case, 8p23.1a should probably represent the original integration, since 8p23.1b and c share the same additional substitutions as compared to its nucleotide sequence. The two members of cluster 19p11 are both found in Orangutan as well but are structurally divergent due to extensive deletions affecting 19p11b HML8 element, which lacks >60% of the internal proviral sequence as compared to 19p11a. It could hence be possible that 19p11a has been duplicated by the retrotransposition of its spliced RNA, leading to the integration of 19p11b copy. In addition, the latter was converted into a solitary LTR in Chimpanzee genome after the split between Chimpanzee and Humans, losing the rest of the internal portions and one of the LTRs. Similarly, 19p11a has been deleted from Gorilla genome. The last cluster, Yq11.222, is found in Chimpanzee and Humans only and its two members differ for a single nucleotide substitution, indicating a more recent acquisition (less than ∼9 mya). Overall, HML8 elements were duplicated during a rather long period along primate evolution - forming new copies in Gibbon, Orangutan, and Chimpanzee genome – differently from the massive human-specific duplication observed for the HML2 group (Subramanian et al., 2011).
Besides the overall chromosomal location, the specific genomic context in which every HML8 element is integrated plays a crucial role in the understanding of its possible impact on cellular processes. In fact, depending on their insertion sites and orientation, HERV elements can modulate the expression of the surrounding genes: this is particularly relevant for HERV LTRs, including regulatory sequences that can act as alternative enhancers, transcription factor binding sites and splicing acceptors or donors (Grandi and Tramontano, 2017). Such signatures have even been co-opted by the host physiology, to provide tissue specific expression to a cellular gene (Ting et al., 1992) or even to regulate and shape complex transcriptional networks: for instance, MER41 LTRs dispersed in the promoter regions of immune-related genes were found to act as inducible enhancers for INF-γ pathway (Chuong et al., 2016). Given that, we evaluated the genetic neighbourhood of each HML8 locus to understand its position with respect to cellular genes, finding that one third of the HML8 proviruses is inserted within 31 human genes (Table 2). Of these, 18 genes were protein coding and the remaining 13 produced long non-coding RNAs, holding the HERV within exons in 7 cases. Of note, 5 out of these 7 intragenic exonic HERVs are present in the same orientation of the harbouring genes: 1p13.3 with GSTM2/GSTM1 (the only protein-coding), 4q13.2 with AC111000.4, 8p23.1c with AC068587, 9q32 with C9orf147 and 15q15.1 with RAD51-AS1 (Table 2). Having a clear information about these specific loci localization and nucleotide sequence makes possible to evaluate their actual expression and eventual interplay with the co-localized genes in cellular transcriptomes (Pisano et al., 2020). For example, we know that more than 25% of HML8 loci is expressed in PBMC under physiological conditions (Pisano et al., 2020b)and that the specific HML8 locus Xp11.3 was found to be downregulated in HIV-1 infected cells (Grandi et al., 2020b).
Preliminary phylogenetic analysis confirmed the subdivision of HML8 in three subtypes based on LTR clustering: in such tree, 24 HML8 proviruses clustered with MER11A, 16 with MER11B and 33 with MER11C, all with high bootstrap values (96%, 85%, and 96%, respectively) (Fig. 2). Such classification has been further confirmed by the structural characterization of individual HML8 sequences, which revealed specific features shared among the members of the same subgroup (Fig. 3). While we expected differences within the LTR sequences, in line with the known existence of three MER11 LTR types, many nucleotide variations were also present in the associated proviral genes. This divergence in HML8 genes was indeed more unexpected, given that a unique reference sequence is reported in Dfam for the internal portion: contrarily, also this region had some characteristics variations that led us to generate two sets of new HML8 consensus sequences, both for the ancestral and modern nucleotide structure of MER11A, B, and C elements (Fig. 3, panel B; Supplementary file 1). In particular, the comparison between subgroup-specific ancestral and modern consensus is useful to evaluate which differences - with respect to Dfam reference - were acquired during the persistence in the genome and which were likely already present in the ancestral virus. For example, those LTR regions that are present in Dfam reference but absent in all the members of that subgroup were likely not present in the ancestral LTR sequence as well. Contrarily, most of the genic portions lacking as compared to Dfam reference seem to have been lost due to recurrent deletions over time, since a minority of members retained them (and were used for the ancestral reconstruction).
From the above structural comparison, we noticed that the MER11A group is the most defective in terms of structure, in line with its older acquisition. Most of its members are characterized by a recurrent deletion affecting the whole polymerase portion, often showing by contrast intact gag and env genes (Fig. 3). MER11B and MER11C sequences also present common deletions within gag, pro, pol and env genes, found in most sequences. As observed in HML7 group, some HERV genic portions might have been lost after the endogenization process because providing functions not anymore needed by the virus (Grandi et al., 2021). Alternatively, the deletion might have been acquired by an original sequence that was then copied multiple times across the germ line: in this case, removal of some genic portions may even enhance intragenomic spread by the loss of extracellular replication. This has been already reported for the env gene (Magiorkinis et al., 2012) and can possibly explain also the frequent loss of the pol gene integrase domain, which has been removed in 24 MER11B and MER11C proviruses out of 50 (Fig. 3). After the loss, such enzyme might even have been provided in trans by other retroviral elements, including non-LTR retrotransposons that in some instances contributed to HERV amplification within the genome (Grandi et al., 2016, Pavlíček et al., 2002). In addition, we evaluated if the observed recurrent mutations within env gene can indicate the presence of alternatively spliced variants, such as the ones producing the oncogenic Np9 and Rec accessory proteins. The latter were in fact reported as produced from HML2 env based on the presence or absence of a typical 292 bp deletion, respectively (Subramanian et al., 2011), and Rec one has been recently reported in HML10 sequences as well (Grandi et al., 2017b). In this case, however, NCBI Conserved Domain tool did not detected the presence of neither Rec nor Np9 domains in any HML8 sequence (data not shown). All HML8 elements were also analysed in terms of residual coding potential (i.e. 0/0 or 0/1 shifts/internal stop codons), identifying overall 12 loci with putative ORFs for gag (3), pro (5), pol (1), and env (3) (Table 3). Of these, the only potentially able to produce full-length or near full-length proteins were three pro ORFs at loci 11q22.1 (MER11A, 335 aa out of 335), 1p13.3 (MER11A, 321 aa out of 335) and 5p13.1 (MER11C, 329 aa out of 335) (Table 3). Of note, all the three retain functional motifs for trimeric dUTPase and pepsin-like aspartyl protease, with full-length Pro at locus 11q22.1 holding C-terminal G-patch domain as well (Fig. 6).
To obtain the most reliable time of integration estimation, we combined the traditional divergence calculation between the LTRs of the same provirus with the comparison of each proviral portion (LTRs and genes) with respect to a consensus sequence. This multiple approach led us to have more precise time indications and to include those sequences that did not possess both LTRs. To further validate the obtained age estimations, for each HML8 provirus we checked the presence of the orthologous HML8 sequences in the corresponding genomic position of non-human primates’ genome, until the O.C.A.. The latter should represent the first primate species – at least among the ones having a public genome assembly - in which that element has been found. Results showed that primates’ lineage was enriched by HML8 integrations in a main period of acquisition spanning from 43 to 17 mya (including hence the evolutionary split of Rhesus, Gibbon, and Orangutan from their common ancestor), with a residual integration activity until Chimpanzee (i.e. until 7 mya) (Fig. 5). Further, we showed that the three HML8 subtypes exhibit different time of integration, with MER11C being significantly younger with respect to the older MER11A (p<0.005) and MER11B (p<0.05) (Fig. 5). Accordingly, 57% of MER11A-associate proviruses were found in primates’ genome since Rhesus and most of MER11B insertions occurred in Rhesus and Gibbon genomes, while MER11C members were principally acquired later on, by Gibbon (51%) and Orangutan (24%) with latest integrations in Gorilla and Chimpanzee.
Overall, the existence of three HML8 phylogenetic subgroups characterized by recurrent structural features and different period of acquisition by primates lead us to consider the possible existence of three ancestral exogenous variants of the same ancient retrovirus, which could have infected primates’ germline during different moment of their evolution. Such HML8 variants could have eventually accounted for recombination events in the last period of their distribution, given the presence of genic patterns shared between the youngest MER11C subgroup and MER11B one, making them both rather divergent as compared to the elderly MER11A (Fig. 4).
5. Conclusion
The present study provides the first exhaustive characterization of the 78 HML8 proviruses, 23 newly identified, integrated in the human genome, adding this last HML group to the ones being described in terms of composition, structure, phylogeny, and dynamics of distribution along primates’ evolution. The dataset provided constitutes hence an essential map to finally assess the impact of individual HML8 members on human transcriptome, which is in turn crucial to investigate their contribution to human physiopathology (Pisano et al., 2020). This would also open the possibility to evaluate selected HML8 candidates as targets for innovative therapeutic strategies, as already reported in the field of human cancer (Chiappinelli et al., 2015; Díaz-Carballo et al., 2021).
CRediT authorship contribution statement
Sante Scognamiglio: Formal analysis, Investigation, Data curation, Writing – original draft, Visualization. Nicole Grandi: Conceptualization, Methodology, Investigation, Data curation, Writing – review & editing, Visualization. Eleonora Pessiu: Formal analysis, Investigation, Visualization. Enzo Tramontano: Conceptualization, Writing – review & editing, Supervision, Project administration.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.virusres.2022.198976.
Appendix. Supplementary materials
Data availability
Data will be made available on request.
References
- Arunkumar G., Melters D.P. Centromeric transcription: a conserved swiss-army knife. Genes. 2020;11:1–23. doi: 10.3390/GENES11080911. (Basel) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannert N., Kurth R. The evolutionary dynamics of human endogenous retroviral families. Annu. Rev. Genom. Hum. Genet. 2006;7:149–173. doi: 10.1146/annurev.genom.7.080505.115700. [DOI] [PubMed] [Google Scholar]
- Blond J.L., Lavillette D., Cheynet V., Bouton O., Oriol G., Chapel-Fernandes S., Mandrand B., Mallet F., Cosset F.L. An envelope glycoprotein of the human endogenous retrovirus HERV-W is expressed in the human placenta and fuses cells expressing the Type D mammalian retrovirus receptor. J. Virol. 2000;74:3321–3329. doi: 10.1128/JVI.74.7.3321-3329.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broecker F., Horton R., Heinrich J., Franz A., Schweiger M.R., Lehrach H., Moelling K. The intron-enriched HERV-K(HML-10) family suppresses apoptosis, an indicator of malignant transformation. Mob. DNA. 2016;7:1–17. doi: 10.1186/S13100-016-0081-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang T.C., Goud S., Torcivia-Rodriguez J., Hu Y., Pan Q., Kahsay R., Blomberg J., Mazumder R. Investigation of somatic single nucleotide variations in human endogenous retrovirus elements and their potential association with cancer. PLoS One. 2019;14 doi: 10.1371/JOURNAL.PONE.0213770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiappinelli K.B., Strissel P.L., Desrichard A., Li H., Henke C., Akman B., Hein A., Rote N.S., Cope L.M., Snyder A., Makarov V., Buhu S., Slamon D.J., Wolchok J.D., Pardoll D.M., Beckmann M.W., Zahnow C.A., Mergoub T., Chan T.A., Baylin S.B., Strick R. Inhibiting DNA methylation causes an interferon response in cancer via dsRNA including endogenous retroviruses. Cell. 2015;162:974–986. doi: 10.1016/J.CELL.2015.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuong E.B., Elde N.C., Feschotte C. Regulatory evolution of innate immunity through co-option of endogenous retroviruses. Science. 2016;351:1083. doi: 10.1126/SCIENCE.AAD5497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contreras-Galindo R., Kaplan M.H., He S., Contreras-Galindo A.C., Gonzalez-Hernandez M.J., Kappes F., Dube D., Chan S.M., Robinson D., Meng F., Dai M., Gitlin S.D., Chinnaiyan A.M., Omenn G.S., Markovitz D.M. HIV infection reveals widespread expansion of novel centromeric human endogenous retroviruses. Genome Res. 2013;23:1505–1513. doi: 10.1101/GR.144303.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Díaz-Carballo D., Saka S., Acikelli A.H., Homp E., Erwes J., Demmig R., Klein J., Schröer K., Malak S., D'Souza F., Noa-Bolaño A., Menze S., Pano E., Andrioff S., Teipel M., Dammann P., Klein D., Nasreen A., Tannapfel A., Grandi N., Tramontano E., Ochsenfarth C., Strumberg D. Enhanced antitumoral activity of TLR7 agonists via activation of human endogenous retroviruses by HDAC inhibitors. Commun. Biol. 2021;4:276. doi: 10.1038/S42003-021-01800-3. –276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari R., Grandi N., Tramontano E., Dieci G. Retrotransposons as drivers of Mammalian brain evolution. Life. 2021;11 doi: 10.3390/LIFE11050376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flockerzi A., Burkhardt S., Schempp W., Meese E., Mayer J. Human endogenous retrovirus HERV-K14 families: status, variants, evolution, and mobilization of other cellular sequences. J. Virol. 2005;79:2941–2949. doi: 10.1128/JVI.79.5.2941-2949.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Montojo M., Doucet-O'Hare T., Henderson L., Nath A. Human endogenous retrovirus-K (HML-2): a comprehensive review. Crit. Rev. Microbiol. 2018;44:715–738. doi: 10.1080/1040841X.2018.1501345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandi N., Cadeddu M., Blomberg J., Mayer J., Tramontano E. HERV-W group evolutionary history in non-human primates: Characterization of ERV-W orthologs in Catarrhini and related ERV groups in Platyrrhini. BMC Evol. Biol. 2018;18 doi: 10.1186/S12862-018-1125-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandi N., Cadeddu M., Blomberg J., Tramontano E. Contribution of type W human endogenous retroviruses to the human genome: characterization of HERV-W proviral insertions and processed pseudogenes. Retrovirology. 2016;13 doi: 10.1186/S12977-016-0301-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandi N., Cadeddu M., Pisano M.P., Esposito F., Blomberg J., Tramontano E. Identification of a novel HERV-K(HML10): Comprehensive characterization and comparative analysis in non-human primates provide insights about HML10 proviruses structure and diffusion. Mob. DNA. 2017;8 doi: 10.1186/S13100-017-0099-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandi N., Cadeddu M., Pisano M.P., Esposito F., Blomberg J., Tramontano E. Identification of a novel HERV-K(HML10): comprehensive characterization and comparative analysis in non-human primates provide insights about HML10 proviruses structure and diffusion. Mob. DNA. 2017;8 doi: 10.1186/s13100-017-0099-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandi N., Pisano M.P., Demurtas M., Blomberg J., Magiorkinis G., Mayer J., Tramontano E. Identification and characterization of ERV-W-like sequences in Platyrrhini species provides new insights into the evolutionary history of ERV-W in primates. Mob. DNA. 2020;11 doi: 10.1186/s13100-020-0203-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandi N., Pisano M.P., Pessiu E., Scognamiglio S., Tramontano E. HERV-K(HML7) integrations in the human genome: comprehensive characterization and comparative analysis in non-human primates. Biology. 2021;10:439. doi: 10.3390/BIOLOGY10050439/S1. (Basel) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandi N., Pisano M.P., Scognamiglio S., Pessiu E., Tramontano E. Comprehensive analysis of HERV transcriptome in HIV+ cells: absence of HML2 activation and general downregulation of individual HERV Loci. Viruses. 2020;12:481 12. doi: 10.3390/V12040481. 2020Page481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandi N., Pisano M.P., Tramontano E. The emerging field of human endogenous retroviruses: understanding their physiological role and contribution to diseases. Future Virol. 2019 doi: 10.2217/fvl-2019-0061. [DOI] [Google Scholar]
- Grandi N., Tramontano E. Human endogenous retroviruses are ancient acquired elements still shaping innate immune responses. Front. Immunol. 2018;9 doi: 10.3389/FIMMU.2018.02039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandi N., Tramontano E. HERV envelope proteins: physiological role and pathogenic potential in cancer and autoimmunity. Front. Microbiol. 2018;9 doi: 10.3389/FMICB.2018.00462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandi N., Tramontano E. Type W human endogenous retrovirus (HERV-W) integrations and their mobilization by L1 machinery: Contribution to the human transcriptome and impact on the host physiopathology. Viruses. 2017;9 doi: 10.3390/V9070162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haeussler M., Zweig A.S., Tyner C., Speir M.L., Rosenbloom K.R., Raney B.J., Lee C.M., Lee B.T., Hinrichs A.S., Gonzalez J.N., Gibson D., Diekhans M., Clawson H., Casper J., Barber G.P., Haussler D., Kuhn R.M., Kent W.J. The UCSC genome browser database: 2019 update. Nucleic Acids Res. 2019;47:D853–D858. doi: 10.1093/NAR/GKY1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrow J., Frankish A., Gonzalez J.M., Tapanari E., Diekhans M., Kokocinski F., Aken B.L., Barrell D., Zadissa A., Searle S., Barnes I., Bignell A., Boychenko V., Hunt T., Kay M., Mukherjee G., Rajan J., Despacio-Reyes G., Saunders G., Steward C., Harte R., Lin M., Howald C., Tanzer A., Derrien T., Chrast J., Walters N., Balasubramanian S., Pei B., Tress M., Rodriguez J.M., Ezkurdia I., van Baren J., Brent M., Haussler D., Kellis M., Valencia A., Reymond A., Gerstein M., Guigó R., Hubbard T.J. GENCODE: the reference human genome annotation for the ENCODE project. Genome Res. 2012;22:1760–1774. doi: 10.1101/GR.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubley R., Finn R.D., Clements J., Eddy S.R., Jones T.A., Bao W., Smit A.F.A., Wheeler T.J. The Dfam database of repetitive DNA families. Nucleic Acids Res. 2016;44:D81–D89. doi: 10.1093/NAR/GKV1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jern P., Sperber G.O., Ahlsén G., Blomberg J. Sequence variability, gene structure, and expression of full-length human endogenous retrovirus H. J. Virol. 2005;79:6325–6337. doi: 10.1128/JVI.79.10.6325-6337.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K., Standley D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/MOLBEV/MST010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Stecher G., Li M., Knyaz C., Tamura K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018;35:1547–1549. doi: 10.1093/MOLBEV/MSY096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavialle C., Cornelis G., Dupressoir A., Esnault C., Heidmann O., Vernochet C., Heidmann T. Paleovirology of “syncytins”, retroviral env genes exapted for a role in placentation. Philos. Trans. R. Soc. B Biol. Sci. 2013;368 doi: 10.1098/RSTB.2012.0507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavie L., Medstrand P., Schempp W., Meese E., Mayer J. Human endogenous retrovirus family HERV-K(HML-5): status, evolution, and reconstruction of an ancient betaretrovirus in the human genome. J. Virol. 2004;78:8788–8798. doi: 10.1128/JVI.78.16.8788-8798.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C.H., Grandi N., Palanivelu L., Tramontano E., Lin L.T. Contribution of human retroviruses to disease development—a focus on the HIV– and HERV–cancer relationships and treatment strategies. Viruses. 2020;12:852. doi: 10.3390/V12080852. 2020Page12, 852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangeney M., Renard M., Schlecht-Louf G., Bouallaga I., Heidmann O., Letzelter C., Richaud A., Ducos B., Heidmann T. Placental syncytins: genetic disjunction between the fusogenic and immunosuppressive activity of retroviral envelope proteins. Proc. Natl. Acad. Sci. USA. 2007;104:20534–20539. doi: 10.1073/PNAS.0707873105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchi E., Kanapin A., Magiorkinis G., Belshaw R. Unfixed endogenous retroviral insertions in the human population. J. Virol. 2014;88:9529–9537. doi: 10.1128/JVI.00919-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A., Derbyshire M.K., Gonzales N.R., Lu S., Chitsaz F., Geer L.Y., Geer R.C., He J., Gwadz M., Hurwitz D.I., Lanczycki C.J., Lu F., Marchler G.H., Song J.S., Thanki N., Wang Z., Yamashita R.A., Zhang D., Zheng C., Bryant S.H. CDD: NCBI's conserved domain database. Nucleic Acids Res. 2015;43:D222–D226. doi: 10.1093/NAR/GKU1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer J., Blomberg J., Seal R.L. A revised nomenclature for transcribed human endogenous retroviral loci. Mob. DNA. 2011;2 doi: 10.1186/1759-8753-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer J., Meese E.U. The human endogenous retrovirus family HERV-K(HML-3) Genomics. 2002;80:331–343. doi: 10.1006/GENO.2002.6839. [DOI] [PubMed] [Google Scholar]
- Pavlíček A., Paces J., Elleder D., Hejnar J., et al. Processed pseudogenes of human endogenous retroviruses generated by LINEs: their integration, stability, and distribution. Genome Res. 2002;12:391–399. doi: 10.1101/gr.216902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisano M.P., Grandi N., Cadeddu M., Blomberg J., Tramontano E. Comprehensive characterization of the human endogenous retrovirus HERV-K(HML-6) group: overview of structure, phylogeny, and contribution to the human genome. J. Virol. 2019;93 doi: 10.1128/JVI.00110-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisano M.P., Grandi N., Tramontano E. Human endogenous retroviruses (HERVs) and mammalian apparent Ltrs retrotransposons (MALRs) are dynamically modulated in different stages of immunity. Biology. 2021;10 doi: 10.3390/BIOLOGY10050405/S1. (Basel) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisano M.P., Grandi N., Tramontano E. High-throughput sequencing is a crucial tool to investigate the contribution of human endogenous retroviruses (HERVs) to human biology and development. Viruses. 2020;12 doi: 10.3390/V12060633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisano M.P., Tabone O., Bodinier M., Grandi N., Textoris J., Mallet F., Tramontano E. RNA-Seq transcriptome analysis reveals long terminal repeat retrotransposon modulation in human peripheral blood mononuclear cells after in vivo lipopolysaccharide injection. J. Virol. 2020;94 doi: 10.1128/JVI.00587-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seifarth W., Baust C., Murr A., Skladny H., Krieg-Schneider F., Blusch J., Werner T., Hehlmann R., Leib-Mösch C. Proviral structure, chromosomal location, and expression of HERV-K-T47D, a novel human endogenous retrovirus derived from T47D particles. J. Virol. 1998;72:8384–8391. doi: 10.1128/JVI.72.10.8384-8391.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sha M., Lee X., Li X.ping, Veldman G.M., Finnerty H., Racie L., LaVallie E., Tang X.Y., Edouard P., Howes S., Keith J.C., McCoy J.M. Syncytin is a captive retroviral envelope protein involved in human placental morphogenesis. Nature. 2000;403:785–789. doi: 10.1038/35001608. [DOI] [PubMed] [Google Scholar]
- Sperber G.O., Airola T., Jern P., Blomberg J. Automated recognition of retroviral sequences in genomic data - RetroTector©. Nucleic Acids Res. 2007;35:4964–4976. doi: 10.1093/NAR/GKM515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian R.P., Wildschutte J.H., Russo C., Coffin J.M. Identification, characterization, and comparative genomic distribution of the HERV-K (HML-2) group of human endogenous retroviruses. Retrovirology. 2011;8:90. doi: 10.1186/1742-4690-8-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas J., Perron H., Feschotte C. Variation in proviral content among human genomes mediated by LTR recombination 06 Biological Sciences 0604 Genetics. Mob. DNA. 2018;9 doi: 10.1186/S13100-018-0142-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ting C.N., Rosenberg M.P., Snow C.M., Samuelson L.C., Meisler M.H. Endogenous retroviral sequences are required for tissue-specific expression of a human salivary amylase gene. Genes Dev. 1992;6:1457–1465. doi: 10.1101/gad.6.8.1457. [DOI] [PubMed] [Google Scholar]
- Vargiu L., Rodriguez-Tomé P., Sperber G.O., Cadeddu M., Grandi N., Blikstad V., Tramontano E., Blomberg J. Classification and characterization of human endogenous retroviruses mosaic forms are common. Retrovirology. 2016;13 doi: 10.1186/S12977-015-0232-Y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahn J., Kaplan M.H., Fischer S., Dai M., Meng F., Saha A.K., Cervantes P., Chan S.M., Dube D., Omenn G.S., Markovitz D.M., Contreras-Galindo R. Expansion of a novel endogenous retrovirus throughout the pericentromeres of modern humans. Genome Biol. 2015;16 doi: 10.1186/S13059-015-0641-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.