Abstract
Cyclopropane fatty acids (CFAs) are generally synthesized as bacterial cultures enter stationary phase. In Escherichia coli, the onset of CFA synthesis results from increased transcription of cfa, the gene encoding CFA synthase. However, the increased level of CFA synthase activity is transient; the activity quickly declines to the basal level. We report that the loss of CFA activity is due to proteolytic degradation dependent on expression of the heat shock regulon. CFA synthase degradation is unaffected by mutations in the lon, clpP, and groEL genes or by depletion of the intracellular ATP pools. It seems likely that CFA synthase is the target of an unidentified energy-independent heat shock regulon protease. This seems to be the first example of heat shock-dependent degradation of a normal biosynthetic enzyme.
Cyclopropane fatty acids (CFAs) are very widely distributed among the bacteria and are formed by addition of a methylene group, derived from the methyl group of S-adenosyl-l-methionine (AdoMet), across the carbon-carbon double bond of unsaturated fatty acids (UFAs) (11). Methylene addition does not involve free fatty acids or intermediates of phospholipid biosynthesis, but rather, uses mature phospholipid molecules previously incorporated into and functioning within membrane bilayers. CFAs are typically produced at the onset of stationary phase in bacterial cultures, and CFA formation can thus be considered a conditional, postsynthetic modification of bacterial membrane lipid bilayers. This modification is catalyzed by a soluble enzyme, despite the fact that one of the substrates, the UFA double bond, is normally sequestered deep within the hydrophobic interior of the phospholipid bilayer. This and other properties imply topologically novel protein-lipid interactions in the biosynthesis of CFAs. No physiological role for CFA formation had been ascribed until we recently reported that Escherichia coli strains lacking CFA survive poorly when exposed to pH 3 (2).
Only a small fraction of UFAs are converted to CFAs in exponentially growing cultures of E. coli and other bacteria (11). However, as cultures reach stationary phase, the UFAs of the membrane phospholipid bilayers are rapidly and almost quantitatively converted to CFA. Wang and Cronan (28) showed that the growth-phase dependence of CFA synthesis is due to activation of an RpoS-dependent cfa promoter. The cfa gene encodes CFA synthase, the 43-kDa enzyme catalyzing the modification (29), and is transcribed from two promoters, P1 and P2 (28). Promoter P1 is a standard RpoD (ς70) promoter and is active throughout the growth curve, whereas function of promoter P2 requires the stationary-phase sigma factor RpoS (ς38). The onset of stationary phase results in efficient conversion of phospholipid UFA moieties to CFAs by two mechanisms, the accumulation of RpoS gives increased cfa expression, and the decreased phospholipid synthetic rate allows CFA synthesis to catch up to phospholipid synthesis (CFA synthase must no longer cope with a continually increasing substrate pool) (28). However, these studies also gave the unexpected result that CFA synthase activity did not plateau at the higher level but instead rapidly decreased to the level seen in exponentially growing cultures. This abrupt decrease in CFA synthase activity suggested that CFA synthase is extremely sensitive to proteolysis, and consistent with this suggestion, a CFA fusion protein carrying a C-terminal tag was found to be very unstable in vivo (28). However, the presence of the abnormal fusion junction could have destabilized the CFA synthase portion of the fusion protein, and thus these results might not be a valid indication of CFA synthase degradation. We have now examined the stability of E. coli CFA synthase by immunoprecipitation of the radioactively labeled protein. We report that native CFA synthase is a short-lived protein in vivo and its degradation is dependent on expression of the heat shock regulon.
MATERIALS AND METHODS
Bacterial strains and media.
All bacterial strains are derivatives of E. coli K-12 and are listed in Table 1. Genetic markers were transferred among the strains by phage P1vir transduction. Plasmid pAYW19, which carries the cfa gene, was constructed previously (29). The liquid medium used was minimal medium E (27) plus either glucose (0.4%) or glycerol (0.4%) as carbon source. l-Methionine (45 μg/ml) was added to the media of methionine auxotrophs. The concentrations of antibiotics and amino acids used in media (in milligrams per liter) were tetracycline hydrochloride, 10; sodium ampicillin, 100; chloramphenicol, 30; l-tryptophan, 20.5; l-leucine, 50; l-cystine, 60; and l-threonine, 50. l-Methionine supplementation was as given below.
TABLE 1.
E. coli strains used
| Strain | Relevant genotype | Source or derivation |
|---|---|---|
| BL21(DE3) | Phage T7 RNA polymerase | F. Studier |
| CAG178 | thr leu | C. Gross |
| CAG9310 | thr leu groEL | C. Gross |
| CAG18475 | metC::Tn10 | C. Gross |
| SC122 | supCts lac(Am) trp(Am) pho(Am) rpsL mal(Am) relA1 spoT1 | F. Neidhardt |
| K165 | rpoH(Am) of SC122 | F. Neidhardt |
| MG1655 | Wild type | Coli Genetic Stock Center |
| SG1095 | lon::mini-Tn10 | S. Gottesman |
| SG22093 | clpP::Cm | S. Gottesman |
| ZK126 | Wild type | R. Kolter |
| YYC1168 | cfa::Kan recA::Tn10 of MG1655 | This study |
| YYC1272 | cfa::Kan of ZK126 | This study |
| YYC1309 | metC::Tn10 of ZK126 | ZK126 × P1(CAG18475) |
| YYC1314 | metC::Tn10 lac::Kan of ZK126 | This study |
| YYC1317 | metC::Tn10 of YYC1272 | This study |
| YYC1318 | YYC1314/pAYW19 | This study |
| YYC1321 | BL21(DE3)/pAYW58 | This study |
| YYC1322 | metC::Tn10 of SC122 | SC122 × P1(CAG18475) |
| YYC1323 | metC::Tn10 of K165 | K165 × P1(CAG18475) |
| YYC1324 | YYC1322/pAYW19 | This study |
| YYC1325 | YYC1323/pAYW19 | This study |
| YYC1326 | Tets of YYC1309 | This study |
| YYC1327 | lon::mini-Tn10 of YYC1326 | YYC1326 × P1(SG1095) |
| YYC1328 | YYC1326/pAYW19 | This study |
| YYC1329 | YYC1327/pAYW19 | This study |
| YYC1331 | clpP::Cm of YYC1327 | YYC1327 × P1(SG22093) |
| YYC1332 | YYC1331/pAYW19 | This study |
| YYC1336 | YYC1340/pAYW19 | This study |
| YYC1340 | metC::Tn10 of CAG178 | CAG178 × P1(CAG18475) |
| YYC1338 | YYC1342/pAYW19 | This study |
| YYC1342 | metC::Tn10 of CAG9310 | CAG9310 × P1(CAG18745) |
Pulse-chase experiments.
These experiments were carried out using two different protocols. In protocol A, an overnight culture was diluted 1:20 into methionine-supplemented minimal medium E plus either glucose or glycerol as carbon source (the overnight cultures were grown on the same medium). Ampicillin was added when required to select plasmid maintenance. The cultures were grown to mid-exponential phase (A600 of 0.5 to 0.6) at the temperature given, centrifuged, and washed once with medium E, and the cells were resuspended in the same medium except that the methionine concentration was decreased to 2.5 μg/ml to permit efficient radioactive labeling. A mixture of l-[35S]methionine and l-[35S]cysteine (70:25 molar ratio; American Radiolabeled Chemicals, Inc.) was added to 50 μCi/ml (specific activity of 1,175 Ci/mmol) when the labeling was performed on strains carrying plasmids or 100 μCi/ml when strains carrying a single cfa gene copy were labeled. The labeling period was usually 30 to 40 min. Labeling was terminated by addition of nonradioactive l-methionine (45 μg/ml), and samples were taken following growth for various lengths of time. In protocol B, radioactive labeling was done as in protocol A except that l-cystine (60 μg/ml) was included in all media. Labeling was halted by centrifuging the cells, washing once with minimal medium E, and resuspension in minimal medium plus glucose, l-methionine (45 μg/ml), and l-cystine (60 μg/ml).
The samples were processed by harvesting the cells by centrifugation followed by washing once with 0.1 M potassium phosphate buffer (pH 7.5). The cell pellets were then suspended in 1/10 of the original sample volume of phosphate buffer, lysed by sonication, and centrifuged at 30,000 × g for 1 h. The resulting supernatants were treated with preimmune serum to decrease the nonspecific binding before immunoprecipitation with a polyclonal rabbit antiserum prepared against purified E. coli CFA synthase. Usually, 0.1 ml of the cell extracts was treated with 2 μl of the CFA synthase antiserum and incubated for 1 h at 0°C, and 16 μl of protein A-Sepharose CL-4B beads (Pharmacia Inc.) was added. The protein A-Sepharose CL-4B beads were prepared as a 1:1 (vol/vol) suspension in bead buffer containing 10 mM Tris-HCl (pH 8.0), 10 mM EDTA, 150 mM NaCl, 1% bovine serum albumin, and 0.4% (wt/vol) Triton X-100. After incubation for 30 min on ice, the Sepharose beads were washed twice with the bead buffer, once with 0.1 M Tris-HCl (pH 8.0), and finally once with 0.01 M Tris-HCl (pH 8.0). To the washed beads, 20 μl of sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer was added, and the sample was boiled for 2 min. The samples were centrifuged, and the supernatants were loaded on SDS–10% polyacrylamide gels. Following SDS-PAGE, the gels were dried and scanned with a PhosphorImager (Molecular Dynamics, Inc.). The software for data analyzing and storage was ImageQuaNT (Molecular Dynamics). The gels were also exposed to X-ray film for permanent records. Although we used equal amounts of sample radioactivity for each immunoprecipitation, the total radioactivity in each gel lane varied somewhat, probably due to the multiple steps of the procedure. Therefore, we normalized the radioactivity present in the CFA synthase band to the total radioactivity present in the lane. A strain carrying a cfa null mutation (strain YYC1317) was used to determine the background radioactivity in the gel section where CFA synthase migrated.
The anti-CFA synthase antibody nonspecifically precipitated numerous other proteins, and thus the CFA synthase band was identified by use of the cfa null mutant plus an 35S-labeled CFA synthase standard prepared from strain YYC1321, which expresses CFA synthase from a phage T7 promoter. This strain was grown to mid-log phase, and CFA synthase production was induced (0.4 mM isopropyl-β-d-thiogalactopyranoside) for 30 min at 37°C followed by addition of rifampin (200 μg/ml) to block expression of chromosomal genes. After 5 min, the culture was labeled with radioactive methionine-cysteine, and the proteins were extracted as described above. Samples of the extracts were loaded on the gels without immunoprecipitation to serve as a CFA synthase marker. As expected from the work of Studier and Moffatt (25), CFA synthase was the major labeled protein.
Antibody preparation.
CFA synthase was purified as described previously (29). The purified CFA synthase was further purified by on a preparative SDS-polyacrylamide gel (BioRad model 491 Prep Cell; 14-cm length and 2.8-cm diameter; 10% running and 4% stacking gel). Fractions containing only CFA synthase were pooled and dialyzed against 0.1 M potassium phosphate buffer (pH 7.5). The antiserum was prepared by injection of rabbits with 150 μg of the protein dispersed in complete Freund's adjuvant and boosted after 2 and 4 weeks with 50 μg of CFA synthase dispersed in incomplete Freund's adjuvant.
RESULTS
E. coli CFA synthase is unstable in vivo.
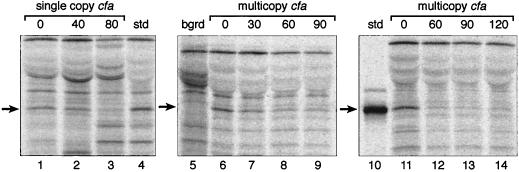
In preliminary studies of the degradation of E. coli CFA synthase, we tested several different growth and labeling protocols. We first attempted to label cultures in early stationary phase, since that is the time CFA synthase levels are maximal. However, labeling under these conditions was very inefficient such that CFA synthase, a minor protein, could not be detected. We therefore labeled mid-log-phase cultures and allowed the culture to enter stationary phase during the chase period. Glycerol-grown cultures of E. coli strain YYC1314, which contains only the chromosomal copy of cfa, were first examined. The intensity of the labeled CFA synthase band decreased with time during the chase with unlabeled methionine (Fig. 1). After chase for 40 or 80 min with excess unlabeled methionine, only 63 or 35%, respectively, of the original radioactivity of CFA synthase remained. Similar results obtained when this strain was grown with glucose as carbon source. The half-life of the protein calculated from these experiments was in the range of 40 to 60 min (the doubling time for this strain under these conditions was about 60 min). Strain YYC1318, which contains a high-copy-number plasmid encoding cfa (Fig. 1), was also examined. About half of the radioactivity in CFA synthase disappeared after 30 min of chase at 37°C (Fig. 1), and most of the remaining label was subsequently lost. In several repeats of this experiment in glucose medium, the CFA synthase half-life varied within 30 to 60 min. Therefore, the half-life of the CFA synthase encoded by multiple copies of the cfa gene was similar to that obtained for a strain having one copy of the gene. All subsequent experiments used strains carrying this plasmid to take advantage of the increased sensitivity resulting from increased cfa expression.
FIG. 1.
Proteolytic degradation of CFA synthase. Autoradiograms show SDS-polyacrylamide gel separations of immunoprecipitates of pulse-chase experiments with various strains. Three independent gels are shown (lanes 1 to 4, 5 to 9, and 10 to 14). Lanes 1 to 3 are extracts of strain YYC1314 (which contains only the chromosomal cfa gene) growing with glycerol as carbon source. Lane 1, extracts from the strain labeled for 40 min without chase; lanes 2 and 3, the same as lane 1, but chases were for 40 and 80 min, respectively. Lane 4 is strain YYC1168 carrying plasmid pAYW19 and was used as a CFA synthase standard (Materials and Methods). Lanes 5 to 9 are extracts of strains grown with glucose as carbon source. Lane 5, cfa null mutant, YYC1317; lanes 6 to 9, extracts of the cfa plasmid-containing strain YYC1318 labeled for 30 min and chased for 0, 30, 60, and 90 min, respectively. Lanes 11 to 14 are extracts from experiments done on strain YYC1318 grown with glucose and labeled for 30 min followed by chase periods of 0, 60, 90, and 120 min, respectively. Lane 10 is a CFA synthase marker in which CFA synthase was labeled in the presence of rifampin in strain YYC1321 (Materials and Methods). The arrows indicate the protein band corresponding to CFA synthase (note that the standard in lane 4 differs from that used in lane 10 and elsewhere in this study). The experiments were done with protocol A (Materials and Methods). Note that residual labeling with cysteine occurred during the chase (see text). Abbreviations: bgrd, background (cfa null mutant); std, standard.
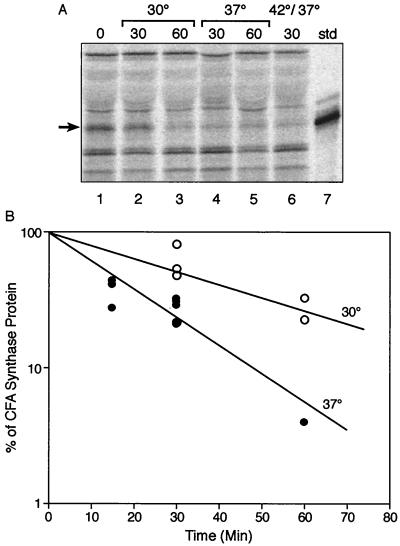
In the early experiments, comparison with the null mutant background control showed that some radioactivity remained in the CFA synthase band even after extended chase periods (e.g., 90 or 120 min). It should be noted that the 35S-labeled methionine source used was a mixture containing 70% methionine and 25% cysteine plus 5% miscellaneous amino acids. We had not added cysteine (or cystine) to our chase medium because we expected that endogenous cysteine synthesis would provide an effective chase. To test this assumption, the method was modified by including exogenous cystine (cysteine is toxic to E. coli K-12 at high concentrations) in all of the media. With this modification, the half-life (15 min) of CFA synthase obtained was shorter than that measured in the absence of a source of exogenous cysteine (30 to 60 min), and the CFA synthase band completely disappeared upon prolonged chase (Fig. 2). We therefore added cystine to all the media used in subsequent experiments. These results were obtained from cultures grown at 37°C beginning from mid-exponential phase (A600 of 0.5 to 0.6) (Fig. 2). Similar results were obtained for late-log-phase cultures (A600 of 1.0), whereas stationary-phase cultures could not be sufficiently labeled to detect CFA synthase.
FIG. 2.
Effect of temperature on degradation of CFA synthase. (A) Autoradiograms of SDS-polyacrylamide gel separations of immunoprecipitates of pulse-chase experiments with various strains are shown. The cfa plasmid carrying strain YYC1318 was grown on glucose minimal medium supplemented with methionine and cysteine and labeled as described in Materials and Methods. In each case, the cultures were labeled with radioactive methionine for 30 min at 37°C before initiation of the chase period. In lanes 2 and 3, the chase experiments were done at 30°C. Lane 1, no chase; lane 2, 30-min chase; lane 3, 60-min chase. Lanes 4 and 5 were chased at 37°C for 30 and 60 min, respectively. Lane 6 was chased for 5 min at 42°C and then for 30 min at 37°C. Lane 7 is the CFA synthase marker. Arrows indicate the CFA synthase band. std, standard. (B) Turnover of CFA synthase of strain YYC1318 at 30°C (open circles) and 37°C (filled circles). Each point represented the result from one lane of an experiment such as that of panel A. The experiments were done with protocol B (Materials and Methods).
The effect of temperature on CFA synthase degradation was also tested. When strain YYC1318 was grown at 30°C, the half-life of CFA synthase was 30 min (Fig. 2), about twice that obtained at 37°C, whereas at 42°C the half-life was too rapid to measure accurately but was considerably less than 15 min. Therefore, E. coli CFA synthase was metabolically unstable and the rate of degradation increased with temperature.
Degradation of E. coli CFA synthase depends on induction of the heat shock regulon.
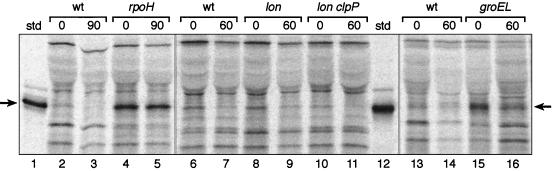
The temperature-dependent degradation of E. coli CFA synthase suggested that heat shock proteases may be involved in the instability of this enzyme. For example, Lon and the Clp family of proteases are well-known members of the heat shock regulon (9, 12). The genes of this regulon are specifically transcribed by RNA polymerase directed by the RpoH sigma factor (ς32). Therefore, the effect of a mutation in rpoH on degradation of E. coli CFA synthase was tested by use of strain K165 (21), which carries an rpoH nonsense mutation plus a temperature-sensitive suppressor tRNA. The temperature-sensitive suppressor tRNA is inactive at 42°C and suppresses poorly at lower temperatures (1, 5). Strain K165 grows fairly well at 30°C, whereas higher temperatures block cell division and are lethal. To avoid these complications, we chose to examine cells deficient in (rather than devoid of) this transcription factor (1, 5). We first transduced strains K165 and its parent, strain SC122, to methionine auxotrophy followed by introduction of the cfa plasmid pAYW19 to give strains YYC1325 and YYC1324, respectively. These strains were then grown at 30°C, and the half-lives of CFA were determined for both strains (Fig. 3 and Table 2). In the parental strain the CFA synthase half-life was about 45 min, whereas no detectable loss of radioactive CFA synthase occurred in the RpoH-deficient strain during a 90-min chase. Therefore, it seems that degradation of the CFA synthase of E. coli requires high level expression of RpoH.
FIG. 3.
Effect of mutations in rpoH, lon, clpP, or groEL on degradation of CFA synthase. Autoradiograms of SDS-polyacrylamide gel separations of immunoprecipitates of pulse-chase experiments with various strains are shown. Three gels are shown (lanes 1 to 5, 6 to 12, and 13 to 16). Lanes 2 and 3 are extracts of strain YYC1324 (wild type [wt]) labeled with [35S]methionine for 40 min at 30°C and then chased for 0 (lane 2) or 90 (lane 3) min at 30°C. Lanes 4 and 5 are identical to lanes 2 and 3, respectively, except that the strain labeled was YYC1325 (rpoH). Lanes 6 and 7 are strain YYC1328 (wild type), lanes 8 and 9 are strain YYC1329 (lon), and lanes 10 and 11 are strain YYC1332 (lon clpP). In each case, the strain was labeled for 30 min at 37°C followed by no chase (lanes 6, 8, and 10) or by chase for 60 min at 37°C (lanes 7, 9, and 11). In lanes 13 to 16, the experiments were done at 30°C. Lanes 13 and 14 were strain YYC1336 (wild type), whereas lanes 15 and 16 were from strain YYC1338 (groEL). In each case, the odd-numbered lane received no chase period, whereas the even-numbered lane was chased for 60 min (labeling was done for 30 min). Lanes 1 and 12 contain the CFA synthase marker, and the arrows denote the CFA synthase protein band. The experiments were done using protocol A (Materials and Methods). std, standard.
TABLE 2.
Fraction of labeled CFA synthase remaining in various strains after chase
| Strain | Defective gene | Time of chase (min) | % of labeled CFA synthase remaininga |
|---|---|---|---|
| YYC1324 | None | 90 | 38.6 (3) |
| YYC1325 | rpoH | 90 | 108.6 (6) |
| YYC1328 | None | 60 | 57.3 (4) |
| YYC1329 | lon | 60 | 57.7 (6) |
| YYC1332 | lon clpP | 60 | 55 (2) |
| YYC1336 | None | 60 | 55 (2) |
| YYC1338 | groEL | 60 | 60 (2) |
Average of several experiments. The number of experiments done is given in parentheses. In each case, the radioactivity in the sample that was not chased was taken as 100%. The strains were labeled with radioactive methionine-cysteine for either 30 or 40 min prior to chase. The experiments were done using protocol A (Materials and Methods).
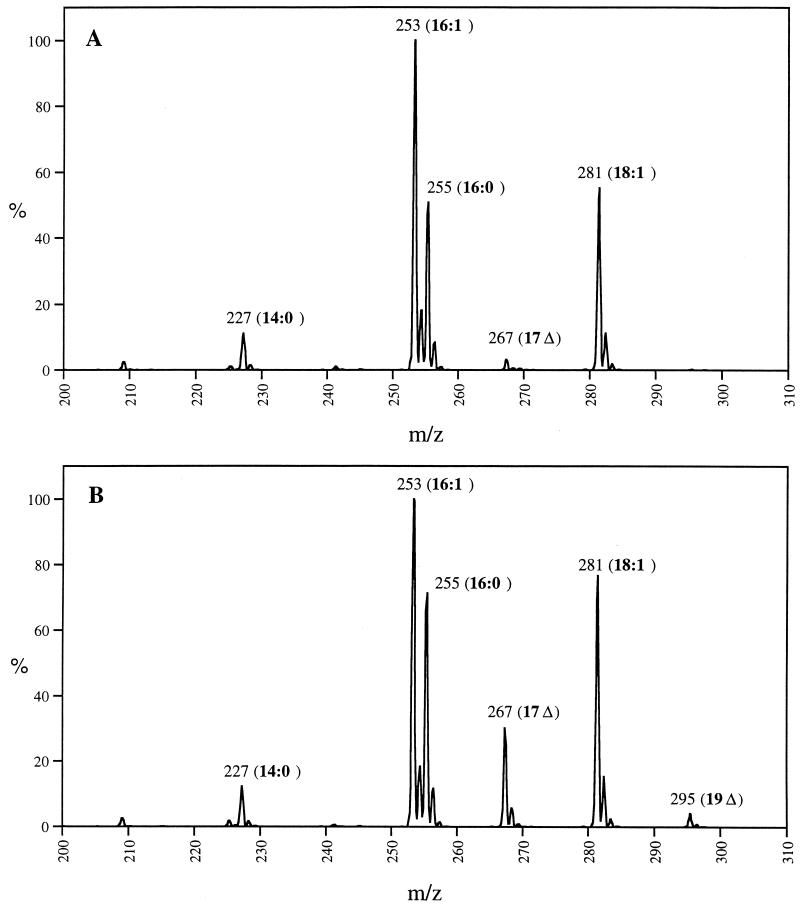
The lack of CFA synthase turnover in the RpoH-deficient strain should result in higher levels of the protein, and if this protein is fully functional, higher levels of CFA should be present in this strain than in the wild-type strain. To test this implication, we isolated phospholipids from mutant strain K165 and parental strain SC122 grown at 30°C. The lipids were analyzed by collision-induced dissociation electrospray mass spectroscopy (26) (Fig. 4). We found that the levels of synthesis of the C17 and C19 CFAs in the RpoH-deficient strain were eight- and fivefold higher, respectively, than the levels of these acids present in the parental strain. Moreover, CFA synthesis began earlier in the growth curve of the RpoH-deficient strain than in the parental strain (e.g., at an optical density of 1.3, the RpoH-deficient strain contained about threefold more CFA than the wild-type strain). These results indicated that the ς32 deficiency stabilized CFA synthase in vivo and the protein was enzymatically active. The low levels of CFA in these strains can be attributed to the relA1 and spoT1 mutations present in these strains. These mutations decrease RpoS levels and thereby inhibit CFA accumulation (4). This was of advantage in the present experiments because the supply of UFA moieties did not become limiting.
FIG. 4.
Effects of RpoH deficiency on cellular CFA content. (A) Acyl chain region of the spectrogram obtained by collision-induced dissociation electrospray mass spectroscopy of phospholipids of the wild-type strain SC122; (B) parallel analysis of the phospholipids of the RpoH-deficient strain K165. Stationary-phase cultures (optical density of 3) grown at 30°C were analyzed. Fatty acids: 14:0, myristic acid; 16:0, palmitic acid; 16:1, palmitoleic acid; 17Δ, cis-9,10-methylenehexadecanoic acid (C17 CFA); 18:1, cis-vaccenic acid; 19Δ, cis-11,12-methyleneoctadecanoic acid (C19 CFA). CFAs are formed by methyleneation of the UFAs palmitoleic and cis-vaccenic acids.
Mutations in lon, clpP, or groEL do not affect the degradation of E. coli CFA synthase.
To determine if a known heat shock protease was involved in the CFA synthase degradation, we first tested the Lon protease, which plays an important role in general protein degradation (7–9, 12, 18). We constructed an isogenic pair of strains in which strain YYC1328 carried the wild-type lon allele whereas strain YYC1329 carried a lon null mutation. The strains were methionine auxotrophs and contained the high-copy-number cfa plasmid pAYW19 (Table 1). The strains were labeled with radioactive methionine and then chased with nonradioactive methionine for 60 min at 37°C (Fig. 3). The rates of CFA synthase degradation were the same in both strains (Fig. 3 and Table 2), and thus we concluded that Lon protease was not responsible for CFA synthase degradation.
We then proceeded to test clpP and groEL mutants. Both ClpP and GroEL are heat shock proteins. ClpP (7–9, 12, 18) is an active protease when complexed with either ClpA or ClpX, whereas GroEL is a chaperone that plays an important role in protein folding and assembly but can also facilitate proteolysis of certain proteins (14, 15). We constructed strain YYC1332 carrying both lon and clpP null mutations and found that CFA synthase turnover in this strain was the same as in the isogenic wild-type strain (YYC1328) and in the single lon mutant strain YYC1329 (Fig. 3 and Table 2). Likewise, we tested the groEL strain YYC1338 and found that the rate of CFA synthase turnover was identical to that of the isogenic wild-type strain YYC1336 (Fig. 3 and Table 2). Therefore, CFA synthase degradation does not require the function of any of these heat shock proteins.
Degradation of CFA synthase is not energy dependent.
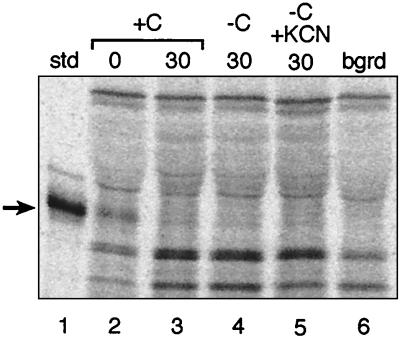
The function of the known heat shock proteins involved in proteolysis is energy dependent since ATP is required for activity of these proteases and chaperones (8, 9). We tested the energy dependence of the protease responsible for the CFA synthase turnover by depleting the intracellular ATP pool during the chase period. ATP depletion was done by carbon source starvation alone or together with cyanide treatment (17, 24). To ascertain that the decrease in intracellular ATP levels was sufficient to inhibit ATP-dependent proteolysis, we examined the degradation of the incomplete proteins formed by premature translation termination in the presence of puromycin. Degradation of such abnormal peptides is strongly (albeit not completely) dependent on ATP-dependent proteases, with Lon playing the major role (5, 9, 24). Cultures were treated with puromycin (0.3 mg/ml) for 15 min at 37°C and then labeled with [35S]methionine-cysteine for 5 min at 37°C. After labeling, the cells were washed free of [35S]methionine-cysteine and suspended either in minimal medium containing glucose or in minimal medium lacking glucose and supplemented with 1 mM potassium cyanide (KCN) for 30 min at either 37 or 30°C. We then measured the radioactivity present in the trichloroacetic acid-soluble supernatants (a measure of the proteolytic degradation of the puromycyl-modified proteins) at both temperatures. At 37°C the presence of KCN inhibited the degradation of puromycyl protein fragments by 67%, whereas an inhibition of 44% was seen at 30°C. The energy dependence of CFA synthase degradation was then examined under these conditions. We found that the rate of CFA synthase degradation was unchanged by the KCN-plus-glucose starvation treatment (Fig. 5). Therefore, ATP-dependent proteases such as those we have tested and the newly described HslU-HslV (ClpQ-ClpY) protease (16, 19, 22) are not involved in CFA synthase degradation. The lack of energy dependence also argues that an ATP-dependent chaperone such as DnaK is not required to present CFA synthase to a protease responsible for degradation.
FIG. 5.
Degradation of E. coli CFA synthase is independent of energy. Autoradiograms of SDS-polyacrylamide gel separations of immunoprecipitates of pulse-chase experiments are shown. Lane 1 is the CFA synthase marker, lanes 2 to 5 were from strain YYC1318, and lane 6 is the cfa null strain YYC1317. Strain YYC1318 was pulse-labeled for 30 min without chase (lane 2) or chased for 30 min in the presence of glucose (lane 3). Lanes 4 and 5 were the same as lane 3 except that the chase was done in the absence of glucose without or with 1 mM KCN, respectively. Arrows denote the CFA synthase protein band. The experiments were done with protocol B (Materials and Methods). Abbreviations are as in Fig. 1.
DISCUSSION
Our results show that the loss of CFA synthase activity early in stationary phase is due to proteolysis rather than to another process that inactivates the enzyme (e.g., covalent modification or tight binding of an inhibitory molecule). Our finding that CFA synthase degradation is dependent on induction of the heat shock regulon was unexpected since the known targets of heat shock-dependent protein degradation are abnormal proteins or proteins that play regulatory rather than metabolic roles (8, 9, 18).
The first question raised by these results is, Why is CFA synthase rapidly degraded while most other E. coli proteins are stable? Preferential proteolysis of a normally folded protein in E. coli has often been found to play a regulatory role, for example, the Lon-dependent degradation of the cell division inhibitor SulA during recovery from the SOS response (18). However, since we know of no regulatory role for either CFA synthase or CFA, it seems more likely that CFA synthase degradation is of some physiological advantage to E. coli. Note that the decrease in CFA synthase activity occurs after virtually all of the membrane phospholipid UFA moieties have been converted to CFA (28), and since cyclopropanation is not reversible (3), degradation of CFA synthase does not affect the level of CFA formation. The most straightforward rationale for degradation would be to prevent high levels of CFA formation when the cells reenter exponential growth and resume synthesis of UFA-containing phospholipids. This rationale predicts that cells containing CFA-modified phospholipids might grow more poorly than those containing UFA-containing lipids. However, strains carrying cfa plasmids have normal exponential-phase growth rates despite efficient conversion of the phospholipid UFA moieties to CFA (10). Moreover, UFA-auxotrophic strains supplemented with exogenous CFAs (in place of UFAs) grow well (3). Therefore, CFA-containing phospholipids do not prevent rapid growth. Another possibility arises from the fact that CFA synthesis is the major consumer of AdoMet in E. coli. Each cyclopropanation reaction consumes a molecule of AdoMet, and an E. coli cell contains 22 million phospholipid molecules, each of which contains at least one UFA to be converted to CFA. In contrast, the total consumption of AdoMet in the synthesis of other molecules (chiefly in methylation of nucleic acids and in the synthesis of spermidine) is about 10-fold less (calculated from the data of reference 19). Thus, degradation of CFA synthase may redirect AdoMet to the synthesis of the other molecules during exponential growth. Note that synthesis of these nonlipid molecules is essentially complete when cells enter stationary phase, and thus there is little or no competition for AdoMet at the time when CFA synthesis is maximal. Therefore, we propose that degradation of CFA synthase is a cellular optimization strategy designed for efficient use of a metabolically expensive intermediate (three ATPs are expended in the synthesis of each AdoMet molecule). The delay of CFA synthesis until entry into stationary phase seems appropriate since the only physiological function of CFA formation presently known is to protect bacteria from acid shock (2), and it is in stationary phase where cells are most likely to encounter an acid environment in nature (13, 23).
It is not clear why CFA synthase degradation is part of the heat shock response. However, it should be noted that RpoH null mutants grow poorly even at very low growth temperatures, and this phenotype indicates that some level of expression of the heat shock regulon is present at all growth temperatures (12). Heat shock is not a general stress response and has not been reported to be induced by entry into stationary phase or by acid shock (12, 13, 23). One possibility is that the presence of CFA in membrane phospholipids may hinder growth at high temperatures, but this does not appear to be the case (see above). However, high-level production of misfolded proteins induces heat shock in exponentially growing cells (6), and if misfolded proteins accumulate during entry to stationary phase, some induction of the heat shock regulon could occur. Nevertheless, we have no evidence that the rate of CFA synthase degradation increases as cultures enter stationary phase, although given the inherently imprecise nature of decay measurements (the decrements at successive time points are small), only large changes in degradation rates would be detected. The most straightforward interpretation of our data is that a protease transcribed from an RpoH-dependent promoter degrades CFA synthase. We favor this interpretation because degradation of the protein is not energy dependent. The lack of dependence on cellular ATP argues against other interpretations such as a heat shock chaperone being required to fold a constitutively expressed protease or to present CFA synthase to a constitutive protease.
The timing of CFA synthesis is controlled by two mechanisms involving sigma factors, the RpoS-dependent P2 promoter and RpoH-dependent proteolysis. Can these two elements adequately explain the spike in CFA synthase activity observed as cultures enter stationary phase? In many cases low levels of a protein are maintained by a rate of degradation that closely matches the rate of synthesis (7, 18). Upon an environmental stimulus, the rate of synthesis of the protein is increased and exceeds the capacity of the degradation apparatus. As pointed out by Miller (18), this regulatory strategy has several advantages. The protein can be maintained at a very low level, but when the synthetic rate is increased such that it no longer matches the rate of degradation, the levels of protein can increase dramatically. Moreover, when the protein has completed its task and the rate of synthesis has decreased, proteolysis results in a rapid return to normal levels of the protein without the need for dilution by cell growth. In the case of CFA synthase, the rate of degradation seems closely matched to the level of enzyme produced from the constitutive RpoD (ς70)-dependent P1 promoter. Upon entry into stationary phase, the large increase in RpoS levels gives activation of the strong P2 promoter (28), which increases the synthetic rate of CFA synthase above the rate of degradation, thus producing the large increase in activity observed. The enzyme then proceeds to modify the membrane lipids and can subsequently be degraded. Although transcription of cfa continues in stationary phase (28), we were unable to radioactively label the CFA synthase protein in stationary-phase cultures (see above). It therefore seems likely that degradation plus the generally low translation rate in stationary-phase cells is responsible for the observed loss of CFA synthase activity.
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health grant AI15650 and by the Deutsche Forschungsgemeinschaft (SFB197, project A7).
REFERENCES
- 1.Baker T A, Grossman A D, Gross C A. A gene regulating the heat shock response in Escherichia coli also affects proteolysis. Proc Natl Acad Sci USA. 1984;81:6779–6783. doi: 10.1073/pnas.81.21.6779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chang Y Y, Cronan J E., Jr Membrane cyclopropane fatty acid content is a major factor in acid resistance of Escherichia coli. Mol Microbiol. 1999;33:249–259. doi: 10.1046/j.1365-2958.1999.01456.x. [DOI] [PubMed] [Google Scholar]
- 3.Cronan J E, Jr, Nunn W D, Batchelor J G. Studies on the biosynthesis of cyclopropane fatty acids in Escherichia coli. Biochim Biophys Acta. 1974;348:63–75. doi: 10.1016/0005-2760(74)90093-9. [DOI] [PubMed] [Google Scholar]
- 4.Eichel J, Chang Y Y, Riesenberg D, Cronan J E., Jr Effect of ppGpp on Escherichia coli cyclopropane fatty acid synthesis is mediated through the RpoS sigma factor (ςS) J Bacteriol. 1999;181:572–576. doi: 10.1128/jb.181.2.572-576.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goff S A, Casson L P, Goldberg A L. Heat shock regulatory gene htpR influences rates of protein degradation and expression of the lon gene in Escherichia coli. Proc Natl Acad Sci USA. 1984;81:6647–6651. doi: 10.1073/pnas.81.21.6647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goff S A, Goldberg A L. Production of abnormal proteins in E. coli stimulates transcription of lon and other heat shock genes. Cell. 1985;41:587–595. doi: 10.1016/s0092-8674(85)80031-3. [DOI] [PubMed] [Google Scholar]
- 7.Gottesman S. Proteases and their targets in Escherichia coli. Annu Rev Genet. 1996;30:465–506. doi: 10.1146/annurev.genet.30.1.465. [DOI] [PubMed] [Google Scholar]
- 8.Gottesman S, Wickner S, Jubete Y, Singh S K, Kessel M, Maurizi M. Selective, energy-dependent proteolysis in Escherichia coli. Cold Spring Harbor Symp Quant Biol. 1995;60:533–548. doi: 10.1101/sqb.1995.060.01.057. [DOI] [PubMed] [Google Scholar]
- 9.Gottesman S, Wickner S, Maurizi M R. Protein quality control: triage by chaperones and proteases. Genes Dev. 1997;11:815–823. doi: 10.1101/gad.11.7.815. [DOI] [PubMed] [Google Scholar]
- 10.Grogan D W, Cronan J E., Jr Cloning and manipulation of the Escherichia coli cyclopropane fatty acid synthase gene: physiological aspects of enzyme overproduction. J Bacteriol. 1984;158:286–295. doi: 10.1128/jb.158.1.286-295.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grogan D W, Cronan J E., Jr Cyclopropane ring formation in membrane lipids of bacteria. Microbiol Mol Biol Rev. 1997;61:429–441. doi: 10.1128/mmbr.61.4.429-441.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gross C A. Function and regulation of the heat shock proteins. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella typhimurium: cellular and molecular biology. 2nd ed. Vol. 1. Washington, D.C.: American Society for Microbiology; 1996. pp. 1382–1399. [Google Scholar]
- 13.Hengge-Aronis R. Regulation of gene expression during entry into stationary phase. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella typhimurium: cellular and molecular biology. 2nd ed. Vol. 1. Washington, D.C.: American Society for Microbiology; 1996. pp. 1497–1512. [Google Scholar]
- 14.Kandror O, Busconi L, Sherman M, Goldberg A L. Rapid degradation of an abnormal protein in Escherichia coli involves the chaperones GroEL and GroES. J Biol Chem. 1994;269:23575–23582. [PubMed] [Google Scholar]
- 15.Kandror O, Sherman M, Goldberg A. Rapid degradation of an abnormal protein in Escherichia coli proceeds through repeated cycles of association with GroEL. J Biol Chem. 1999;274:37743–37749. doi: 10.1074/jbc.274.53.37743. [DOI] [PubMed] [Google Scholar]
- 16.Kessel M, Wu W, Gottesman S, Kocsis E, Steven A C, Maurizi M R. Six-fold rotational symmetry of ClpQ, the E. coli homolog of the 20S proteasome, and its ATP-dependent activator, ClpY. FEBS Lett. 1996;398:274–278. doi: 10.1016/s0014-5793(96)01261-6. [DOI] [PubMed] [Google Scholar]
- 17.Maurizi M R, Trisler P, Gottesman S. Insertional mutagenesis of the lon gene in Escherichia coli: lon is dispensable. J Bacteriol. 1985;164:1124–1135. doi: 10.1128/jb.164.3.1124-1135.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miller C G. Protein degradation and proteolytic modification. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella typhimurium: cellular and molecular biology. 2nd ed. Vol. 1. Washington, D.C.: American Society for Microbiology; 1996. pp. 938–954. [Google Scholar]
- 19.Missiakas D, Schwager F, Betton J M, Georgopoulos C, Raina S. Identification and characterization of HsIV HsIU (ClpQ ClpY) proteins involved in overall proteolysis of misfolded proteins in Escherichia coli. EMBO J. 1996;15:6899–6909. [PMC free article] [PubMed] [Google Scholar]
- 20.Neidhardt F C, Umbarger H E. Chemical composition of Escherichia coli. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella typhimurium: cellular and molecular biology. 2nd ed. Vol. 1. Washington, D.C.: American Society for Microbiology; 1996. pp. 13–16. [Google Scholar]
- 21.Neidhardt F C, VanBogelen R A, Lau E T. Molecular cloning and expression of a gene that controls the high- temperature regulon of Escherichia coli. J Bacteriol. 1983;153:597–603. doi: 10.1128/jb.153.2.597-603.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rohrwild M, Coux O, Huang H C, Moerschell R P, Yoo S J, Seol J H, Chung C H, Goldberg A L. HslV-HslU: a novel ATP-dependent protease complex in Escherichia coli related to the eukaryotic proteasome. Proc Natl Acad Sci USA. 1996;93:5808–5813. doi: 10.1073/pnas.93.12.5808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Slonczewski J L, Foster J W. pH-regulated genes and survival at extreme pH. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella typhimurium: cellular and molecular biology. 2nd ed. Vol. 1. Washington, D.C.: American Society for Microbiology; 1996. pp. 1539–1549. [Google Scholar]
- 24.Straus D B, Walter W A, Gross C A. Escherichia coli heat shock gene mutants are defective in proteolysis. Genes Dev. 1988;2:1851–1858. doi: 10.1101/gad.2.12b.1851. [DOI] [PubMed] [Google Scholar]
- 25.Studier F W, Moffatt B A. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol. 1986;189:113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- 26.Sweetman G, Trinei M, Modha J, Kuselm F P, Fishov J I, Joseleau-Petit D, Redman C, Farmer P, Norris V. Electrospray ionization mass spectrometric analysis of phospholipids of Escherichia coli. Mol Microbiol. 1996;20:233–234. doi: 10.1111/j.1365-2958.1996.tb02504.x. [DOI] [PubMed] [Google Scholar]
- 27.Vogel H J, Bonner D M. Acetyl-ornithinase of Escherichia coli: partial purification and some properties. J Biol Chem. 1956;218:97–106. [PubMed] [Google Scholar]
- 28.Wang A Y, Cronan J E., Jr The growth phase-dependent synthesis of cyclopropane fatty acids in Escherichia coli is the result of an RpoS(KatF)-dependent promoter plus enzyme instability. Mol Microbiol. 1994;11:1009–1017. doi: 10.1111/j.1365-2958.1994.tb00379.x. [DOI] [PubMed] [Google Scholar]
- 29.Wang A Y, Grogan D W, Cronan J E., Jr Cyclopropane fatty acid synthase of Escherichia coli: deduced amino acid sequence, purification, and studies of the enzyme active site. Biochemistry. 1992;31:11020–11028. doi: 10.1021/bi00160a011. [DOI] [PubMed] [Google Scholar]