Highlights
-
•
An aptamer with high affinity and specificity for 3ABC protein of FMDV was obtained.
-
•
The FMDV1 aptamer was selected by Capillary Electrophoresis SELEX (CE-SELEX).
-
•
The FMDV1 aptamer presents potential for FMD diagnosis by 3ABC detection.
Keywords: Aptamer, 3ABC protein, Foot-and-mouth disease, CE-SELEX
Abstract
Foot-and-mouth disease (FMD) is a devastating livestock disease caused by foot-and-mouth disease virus (FMDV), a member of the Picornaviridae family. The 3ABC is a non-structural protein of FMDV, produced during viral replication and absent from inactivated FMD vaccines. Nucleic acid aptamers are DNA or RNA oligonucleotides capable of binding with high specificity and affinity to a molecular target. The aim of this study was to obtain DNA aptamers specific for 3ABC protein with a view of their application in the FMD diagnosis.
Aptamers are usually obtained through SELEX (Systematic Evolution of Ligands by EXponential enrichment) procedure. In this study, an aptamer (termed FMDV1) was selected by a variation of this technique called Capillary Electrophoresis SELEX (CE-SELEX). The FMDV1 aptamer showed high binding affinity to the 3ABC protein with Kd value in the nano molar range: 22.69 ± 1.79 nM. The FMDV1 aptamer binding to 3ABC was significantly higher when compared with the BSA protein, used as control, demonstrating its specificity.
1. Introduction
Foot-and-mouth disease (FMD) is a devastating livestock disease caused by foot-and-mouth disease virus (FMDV), a member of the Aphthovirus genus of the Picornaviridae family. The virus infects a wide range of wild and domesticated cloven-footed mammals. FMD outbreaks in a country always result in economic losses to livestock industry due to the immediate imposition of trade embargo.
Rapid and accurate diagnoses are imperative to virus control. Diagnoses based on clinical disease signs are highly unreliable, because several other diseases share similar FMD signs (Fernández et al., 2008; Smith et al., 2012). Hence, confirmatory laboratory diagnosis is vital. Differentiating Infected from Vaccinated Animals (DIVA) assays are also essential to improve the efficiency of the control strategy. Vaccination prevents clinical manifestation of the disease, but does not necessarily prevent the sub-clinical infection or virus persistence (Hutber et al., 1998). Therefore, the vaccinated animals that were infected by FMDV may become carriers to spread the virus to other susceptible animals (Arzt et al., 2018).
Virus isolation test and conventional immunological techniques such as complement fixation test, virus neutralization test, and enzyme-linked immunosorbent assay (ELISA) are routinely used to diagnose FMD in clinical samples. Molecular techniques, through detection of the viral RNA, have also been used to rapid and accurate FMD diagnoses (Jamal and Belsham, 2013). However, these methods are time-consuming; need sophisticated instruments and skilled personnel, being not suitable for in field analysis. Development of chromatographic strip tests, portable reverse transcription polymerase chain reaction (RT-PCR) and reverse transcription loop-mediated isothermal amplification (RT-LAMP) devices are efforts performed in this direction. Nevertheless, applications of these assays in the field remain limited (Wong et al., 2020). Consequently, there remains strong interest for developing new swift point-of-care biosensing systems for early FMD diagnosis with high sensitivity and specificity (Vidic et al., 2017).
The FMDV genome is translated into a single large polyprotein and cleaved by virally encoded proteases to produce structural (VP1, VP2, VP3, and VP4) and non-structural (2A, 2B, 2C, 3A, 3B, 3Cpro, and 3Dpol) proteins. Non-structural proteins (NSPs) are usually used for developing DIVA assays, since these proteins are produced only during viral replication and inactivated vaccines are devoid of NSPs, containing only structural proteins (SP). Among all NSPs, 3ABC protein is considered the best target to this purpose due to its high immunogenicity and low concentration during vaccine production (Clavijo et al., 2004). The 3ABC is a polyprotein that is auto-processed to 3A, three copies of 3B and 3Cpro by 3Cpro protease (Sariya et al., 2011). Different modes of 3ABC-ELISA, based on the detection of anti-3ABC antibodies, were developed to discriminate among infected and vaccinated animals and to detect subclinical cases (Wong et al., 2020).
Nucleic acid aptamers are single-stranded DNA or RNA oligonucleotides that bind to target molecules with high affinity and specificity. They are selected by an in vitro selection process termed SELEX (Systematic Evolution of Ligands by EXponential Enrichment) through repeated rounds of partitioning and PCR amplification from large random synthetic oligonucleotide libraries (Ellington and Szostak, 1990; Tuerk and Gold, 1990). Usually, six to as many of 15 rounds have been reported to allow the best binders selection. The PCR products of the last round are cloned into a vector and sequenced for identification of the binding sequences. Over the past three decades, numerous SELEX variations have been reported in an attempt to reduce processing time, generate aptamers with novel designs and functions, or increase the process throughput (Wang et al., 2019). CE-SELEX is a SELEX variation in which capillary electrophoresis (CE) is used to separate oligonucleotides that have bound to the target (Mendosa et al., 2004; Berezovski et al., 2005). As the molecules remain in solution during the CE separation process, there is no steric impairment for aptamers binding to the target (Gao et al., 2019). In addition, the protein targets retain their native conformation. The CE separation efficiency allows high affinity aptamers to be obtained with only 3 to 5 selection rounds.
The aim of the present study was to obtain DNA aptamers for the 3ABC protein using CE-SELEX with a view of their application in the FMD diagnosis via in vitro aptamer based diagnostic techniques (Drolet et al., 1996; Lee et al., 2017), especially by aptasensors. (Song et al., 2008).
2. Material and methods
2.1. Proteins
The recombinant 3ABC protein was produced as previously described (Clavijo et al., 2006) and obtained from National Centre for Foreign Animal Disease of Canada. Bovine serum albumin (molecular biology grade) was obtained from Sigma Aldrich (cod: B6917).
2.2. Oligonucleotide library design
The 80mer library (Berezovski et al., 2006), acquired from the IDT DNA Technologies, had the following sequence: 5’-CTTCTGCCCGCCTCCTTCC-(39N)-GGAGACGAGATAGGCGGACACT-3’, where the central 39N represents random oligonucleotides based on equal incorporation of A, G, C, and T at each position.
2.3. CE-SELEX
Five CE-SELEX selection cycles were performed. To initiate in vitro selection, 800uM ssDNA was heated at 95°C for 5 min, snap-cooled on ice for 15 min and left at room temperature for 10 min. After that, it was incubated with 80pmol of the 3ABC protein in borate buffer (60mM), for 30 min, 200 rpm, at 25°C.
An incubation solution sample was submitted to separation by CE using a neutral capillary on a P/ACE MDQ capillary electrophoresis system (Beckman Coulter Inc., Fullerton, CA). Capillary had 50 μm internal diameter, 67 cm total length with 40 cm effective length to detector. The capillary was first rinsed with 0.1 M HCl for 5 min, water (DNAse/RNAse free) for 1 min, then borate buffer for 5 min under pressure of 50 psi before injection of the incubation mixture at 0.5 psi for 5 s. The mixture was separated under 26 kV (reverse polarity) in borate buffer at 25°C, and monitored under UV absorbance detection at 214 nm.
The 3ABC−aptamer complexes were collected into a vial containing 7 μL of separation buffer at the capillary outlet. After collecting material from five runs, the bound sequences were amplified by PCR.
The PCR amplification was performed using a reaction master mix containing 200 μM dNTPs, 0.2 μM forward primer (5’-CTTCTGCCCGCCTCCTTCC-3), 0.2 μM reverse primer (5’-AGTGTCCGC CTATCTCGTCTCC-3′), 1 U Taq DNA polymerase high fidelity (Invitrogen) and 50 mM MgSO4 in 1× High Fidelity PCR buffer. The PCR thermocycling parameters were 94°C for 2 min for denaturation followed by 17 cycles of denaturation at 94°C for 15 s, annealing at 72°C for 30 s (-1ºC per cycle) and extension at 68°C for 30 s. Followed by 14 cycles of denaturation at 94°C for 15 s, annealing at 55°C for 30 s and extension at 68°C for 30 s. A final extension step at 68°C for 2 min followed the last cycle. PCR reactions were carried out in a Veriti Thermal Cycler (Applied Biosystems, Foster City, CA).
Asymmetric PCR was used to obtain the single strand DNA of interest from the conventional PCR product. It was achieved using a reaction master mix containing 200 μM dNTPs, 0.2 μM forward primer, 0.02 μM reverse primer, 1 U Taq DNA polymerase high fidelity (Invitrogen) and 50 mM MgSO4 in 1× High Fidelity PCR buffer. The PCR thermocycling was performed as described above. PCR amplification was verified by electrophoresis on 2% agarose gels (in tris-borate-EDTA buffer) stained with ethidium bromide and imaged using a UV transluminator (UVP BioDoc-It Imaging System).
2.4. DNA cloning and sequencing
The ssDNA sequences obtained after the fifth CE-SELEX cycle were cloned. PCR was performed and the products were cloned into pGEM®-T Easy Vector Systems (Promega). The plasmid was transformed into Escherichia coli DH5α. After electroporation, the Luria-Bertani (LB) medium was added and the material was grown for 1 h, at 37°C, 200 rpm. The culture was then plated onto LB/agar/ampicillin/IPTG/Xgal medium and incubated overnight at 37°C. The viable white clones were then individually seeded in 10 mL of LB/ampicillin and incubated overnight at 37°C, 200 rpm.
The plasmids were extracted following the manufacturer protocol of the Wizard® Plus SV Minipreps DNA Purification System Kit (Promega) and quantified by 260 nm absorbance in a spectrophotometer. The vectors were sequenced by the automated Sanger methodology using the M13 promoter primer. The sequencing was carried out at René Rachou Institute (FIOCRUZ/Minas).
2.5. Secondary structure prediction
The aptamer secondary structure including the conserved regions was predicted using mfold algorithm (Zuker, 2003) at http://unafold.rna.albany.edu/, with input parameters of 1.0 mM Mg2+ and 60 mM Na+, at 25°C. The most likely structure was chosen based on lowest predicted free energy of formation (∆G; kcal/mol).
2.6. Dissociation Constant (Kd) determination
Different aptamer concentrations were individually incubated with the 3ABC protein (40 pmol) in the selection buffer as mentioned before. The sequences bound and unbound to the target were separated by filtration using Amicon 0.5 mL Filters for DNA Purification and Concentration - 50K (Merck) (Ramos et al., 2007) with three washes.
The amount of bound sequences in the samples were quantified by Real-time quantitative Polymerase Chain Reaction (qPCR) in a Step One Real-Time PCR System (Applied Biosystems, Foster City, CA) using SYBR green (PowerUp PCR SYBR Green Master Mix, Applied Biosystems). The assays were performed in 20 µl of reaction containing 10 µl of the Master Mix, 4.2 µL of DNAse/RNAse-free water, 5.0 µL of DNA sample and 0.4 µL (250 nM) of each of the primers, forward (5’-CTTCTGCCCGCCTCCTTCC-3) and reverse (5’-AGTGTCCGC CTATCTCGTCTCC-3′). After an initial enzyme UDG activation at 50°C for 2 min and an enzyme dual-lock DNA polymerase activation at 95°C for 2 min, 40 amplification cycles were performed using 95°C for 15 s for denaturation and 60°C for 1 min for annealing, extension, and fluorescence acquisition. A standard curve for each aptamer was performed ranging from 0.05 ng to 0.00005 ng with a dilution factor of 1:10. All samples, negative and positive controls were processed in triplicate.
A saturation curve was constructed by plotting the aptamer concentration used at each incubation point versus the mass (ng) of protein bound aptamer calculated by qPCR. The Graph Pad Prism 5 software (Graph-Pad Software, San Diego, CA, USA) was used and the dissociation constant (Kd) was calculated using the One-Site Specific Binding with Hill slope nonlinear regression equation.
2.7. Binding assays for affinity and specificity evaluation
The same concentration (25 nM) of the aptamer and the oligonucleotide library was incubated separately with 0.67 µg of the 3ABC protein. The library used was the same that started the SELEX process. After incubation, the fraction of protein-bound oligonucleotides in each case was separated by filtration (as previously described for Kd determination) and quantified by real-time PCR using SYBR Green.
A specificity assay was also performed by incubating the aptamer (25 nM) separately with 0.67 µg of 3ABC and BSA (bovine serum albumin) proteins. The oligonucleotide fraction that bound to each of the proteins was separated and quantified as described before.
3. Results and discussion
The aptamer selection was performed by CE-SELEX. After protein incubation with the oligonucleotide library, capillary electrophoresis (CE) was used in the separation step. A representative CE electropherogram is shown in the Fig. 1. The separation conditions were standardized so that the peaks corresponding to the oligonucleotides that did not bind to the target and the 3ABC-aptamers complexes were eluted at different elution times. The collection window of 3ABC-aptamer complexes was 7.2 to 8.5 min. Five runs were performed to accumulate enough oligonucleotides for the next PCR amplification step. These runs were superimposed in Fig. 1.
Fig. 1.
Representative CE electropherogram of CE-SELEX separation step. The A region refers to oligonucleotides that did not bind to the target. The peak in the B corresponds to the 3ABC-aptamer complexes that were collected. The collection window was 7.2 to 8.5 min. Five runs, which were assigned by distinct color lines, were performed to accumulate enough DNA for PCR amplification. These runs were superimposed in the graph. AU = absorbance.
After five selection cycles (including incubation, separation and amplification steps) the obtained ssDNA pools were cloned and sequenced. The most prevalent sequences were chosen for further characterization. Among these, the FMDV1 (Fig. 2A) presented the best results in terms of affinity and specificity. Fig. 2B illustrates the most likely secondary structure of FMDV1 aptamer predicted by Mfold software.
Fig. 2.
(A) FMDV1 aptamer sequence with the conserved regions in bold. (B) Secondary structure of the FMDV1 aptamer predicted by Mfold (∆G = −10.42 kcal/mol).
To evaluate the increase in the affinity to the protein target obtained with the aptamer selection process, the 3ABC protein was incubated with the FMDV1 aptamer and the oligonucleotide library used as start material for selection. The DNA amount bound to the protein was negligible for the oligonucleotide library while it was considerably high for FMDV1 aptamer (Fig. 3), demonstrating the increment in affinity and the CE-SELEX efficiency. The aptamer quantification in this and next experiments was performed by qPCR (Peyrin, 2009; Yang and Bowser, 2013; Niazi et al., 2008; Hulme and Trevethick, 2010).
Fig. 3.
Evaluation of the increase in affinity to the 3ABC protein obtained with the aptamer selection process. The 3ABC protein (0.67 µg) was incubated separately with 25 nM of the FMDV1 aptamer and 25 nM of the oligonucleotide library used as start material for selection. Protein-bound DNA was quantified by qPCR. Statistical analyzes were performed using the GraphPad Prism 5 software (**p < 0.01).
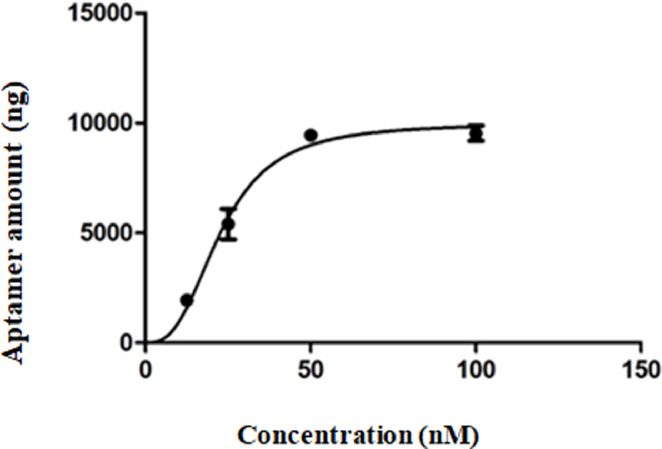
The aptamer affinity to the target is indicated by the dissociation constant in the equilibrium (Kd) which was determined by saturation assays. The binding to the target of an increasing series of FMDV1 aptamer concentrations was measured at equilibrium and the dissociation constant was obtained from a saturation-binding curve (Fig. 4). The FMDV1 aptamer showed high binding affinity to the 3ABC protein with Kd value in the nanomolar range: 22.69 ± 1.79 nM.
Fig. 4.
Dissociation constant (Kd) determination of FMDV1 aptamer. The saturation curve was obtained by plotting the aptamer concentration (nM) versus the total ssDNA (ng) bound to 3ABC protein. Protein-bound aptamer was quantified by qPCR. The Kd was calculated by non-linear regression analysis (One-Site Specific Binding with Hill slope software GraphPad Prism 5), resulting in Kd = 22.69 ± 1.79 nM.
The FMDV1 aptamer specificity was also assessed by comparing the aptamer binding to the 3ABC and BSA (control) proteins. The FMDV1 aptamer binding to 3ABC was significantly higher when compared with the BSA (Fig. 5), demonstrating its high specificity.
Fig. 5.
Specificity assay. Binding of FMDV1 aptamer to 3ABC and BSA proteins was performed by qPCR. The graph shows the ssDNA amount (ng) recovered after incubation of 25 nM of FMDV1 aptamer with 0.67 µg of each protein. Statistical analyzes were performed using the GraphPad Prism 5 software (**p < 0.01).
Aptamers has been obtained for components of FMDV. Two RNA aptamers were selected to RNA-dependent RNA polymerase (3Dpol) that were able to inhibit the enzyme activity in vitro (Ellingham et al., 2006; Forrest et al., 2004). These aptamers were useful in the study of virus replication with possible applications in diagnostics and therapeutics. Aptamers were also selected for a highly conserved peptide from the VP1 structural polypeptide and applied in the development of a competitive fluorescence resonance energy transfer (FRET)-based DNA aptamer test for FMD diagnosis (Bruno et al., 2008). The current work is the first to select an aptamer for 3ABC protein. The recombinant protein used as target for aptamer selection was obtained under mildly denaturing conditions and submitted to a refolding procedure (Clavijo et al., 2006). This way, FMDV1 aptamer is relevant in view of its application for FMD diagnosis by aptamer-based techniques, considering that it was selected for the native protein conformation. Although our aim was to obtain aptamers for diagnostic purposes, the FMDV1 potential to interfere with viral replication must also be investigated.
Aptamer-based biosensors, also called aptasensors use aptamers as bioreceptors. Aptasensors can overcome the lacking functional and storage stability of biosensors based on antibodies, since aptamers are chemically stable, cost effective, resistant to pH and temperature changes, and offer remarkable flexibility in the design of their structures. Aptasensors for hepatite C, H5N1 avian influenza and H1N1 viruses, among others, have been developed (Ozer et al., 2020). Aptasensors based in the FMDV1 aptamer would be very useful for FMD diagnosis and control due to the possibility of a rapid diagnosis in the point of care by using animal fluids samples. This evaluation will be the next stage of this study.
4. Conclusion
In the present work was obtained an aptamer with high affinity and specificity for the 3ABC protein of the foot-and-mouth disease virus, which presents potential for application in the FMD diagnosis by 3ABC detection.
Author statement
This study has not been published previously and it is not under consideration for publication elsewhere. The publication is approved by all authors and by the responsible authorities where the work was carried out. If accepted, it will not be published elsewhere in the same form, in English or in any other language, including electronically without the written consent of the copyright- holder.
CRediT authorship contribution statement
Camila Maria de Sousa Lacerda: Conceptualization, Methodology, Formal analysis, Investigation, Writing – original draft. Nathalie Bonatti Franco Almeida: Methodology, Formal analysis, Investigation. Viviane Cristina Fernandes dos Santos: Methodology, Formal analysis. Flávio Plentz: Formal analysis, Writing – review & editing, Funding acquisition, Resources, Supervision, Project administration. Antero Silva Ribeiro de Andrade: Conceptualization, Methodology, Formal analysis, Writing – original draft, Writing – review & editing, Funding acquisition, Resources, Supervision.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
The authors thank the Centro de Desenvolvimento da Tecnologia Nuclear/Comissão Nacional de Energia Nuclear (CDTN/CNEN) and Program for Technological Development in Tools for Health-PDTIS-FIOCRUZ for use of its facilities. This work was supported by Mediconchip Desenvolvimento Tecnológico and Banco Nacional de Desenvolvimento Econômico e Social (BNDES) (Project Contract No: 15.2.0163.1).
Footnotes
In present work, for the first time, an aptamer with high affinity and specificity for the 3ABC protein was obtained, which presents potential for applications that involves the 3ABC detection, mainly the FMD diagnosis.
References
- Arzt J., Belsham G.J., Lohse L., Bøtner A., Stenfeldt C. Transmission of foot-and-mouth disease from persistently infected carrier cattle to naive cattle via transfer of oropharyngeal fluid. mSphere. 2018;12 doi: 10.1128/mSphere.00365-18. e00365-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berezovski M., Drabovich A., Krylova S.M., Musheev M., Okhonin V., Krylov S.N. Nonequilibrium capillary electrophoresis of equilibrium mixtures: a universal tool for development of aptamers. J. Am. Chem. Soc. 2005;27:3165–3171. doi: 10.1021/ja042394q. [DOI] [PubMed] [Google Scholar]
- Berezovski M.V., Musheev M.U., Drabovich A.P., Jitkova J.V., Petrov A., Krylov S.N. Non-SELEX: selection of aptamers without intermediate amplification of candidate oligonucleotides. Nat. Protoc. 2006;1:1359–1369. doi: 10.1038/nprot.2006.200. [DOI] [PubMed] [Google Scholar]
- Bruno J.G., Carrillo M.P., Phillips T. Development of DNA aptamers to a foot-and-mouth disease peptide for competitive FRET-based detection. J. Biomol. Tech. 2008;19:109–115. [PMC free article] [PubMed] [Google Scholar]
- Clavijo A., Wright P., Kitching P. Developments in diagnostic techniques for differentiating infection from vaccination in foot-and-mouth disease. Vet. J. 2004;167(1):9–22. doi: 10.1016/s1090-0233(03)00087-x. [DOI] [PubMed] [Google Scholar]
- Clavijo A., Hole K., Li M., Collignon B. Simultaneous detection of antibodies to foot-and-mouth disease non-structural proteins 3ABC, 3D, 3A and 3B by a multiplexed Luminex assay to differentiate infected from vaccinated cattle. Vaccine. 2006;24:1693–1704. doi: 10.1016/j.vaccine.2005.09.057. [DOI] [PubMed] [Google Scholar]
- Drolet D.W., Moon-McDermott L., Romig T.S. An enzyme-linked oligonucleotide assay. Nat. Biotechnol. 1996;14:1021–1025. doi: 10.1038/nbt0896-1021. [DOI] [PubMed] [Google Scholar]
- Ellingham M., Bunka D.H.J., Rowlands D.J., Stonehouse N.J. Selection and characterization of RNA aptamers to the RNAdependent RNA polymerase from foot-and-mouth disease virus. RNA. 2006;12:1970–1979. doi: 10.1261/rna.161006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellington A.D., Szostak J.W. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- Fernández J., Agüero M., Romero L., Sánchez C., Belák S., Arias M., Sánchez-Vizcaíno J.M. Rapid and differential diagnosis of foot-and-mouth disease, swine vesicular disease, and vesicular stomatitis by a new multiplex RT-PCR assay. J. Virol. Methods. 2008;147:301–311. doi: 10.1016/j.jviromet.2007.09.010. [DOI] [PubMed] [Google Scholar]
- Forrest S., Lear Z., Herod M.R., Ryan M., Rowlands D.J., Stonehouse N.J. Inhibition of the foot-and-mouth disease virus subgenomic replicon by RNA aptamers. J. Gen. Virol. 2004;95:2649–2657. doi: 10.1099/vir.0.067751-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao T., Luo Y., Li W., Cao Y., Pei R. Progress in the isolation of aptamers to light-up dyes and the applications. Analyst. 2019;145:701–718. doi: 10.1039/c9an01825Ee. [DOI] [PubMed] [Google Scholar]
- Hulme E.C., Trevethick M.A. Ligand binding assays at equilibrium: validation and interpretation. Br. J. Pharmacol. 2010;161:1219–1237. doi: 10.1111/j.1476-5381.2009.00604.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutber A.M., Kitching R.P., Conway D.A. Control of foot-and-mouth disease through vaccination and the isolation of infected animals. Trop. Anim. Health Prod. 1998;30:217–227. doi: 10.3389/fvets.2020.00477. [DOI] [PubMed] [Google Scholar]
- Jamal S.M., Belsham G.J. Foot-and-mouth disease: past, present and future. Vet. Res. 2013;44:116. doi: 10.1186/1297-9716-44-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K.H., Zeng H. Aptamer-based ELISA assay for highly specific and sensitive detection of Zika NS1 protein. Anal. Chem. 2017;89:12743–12748. doi: 10.1021/acs.analchem.7b02862. [DOI] [PubMed] [Google Scholar]
- Mendosa S.D., Bowser M.T. In vitro selection of high-affinity DNA ligands for human IgE using capillary electrophoresis. Anal. Chem. 2004;76:5387–5392. doi: 10.1021/ac049857v. [DOI] [PubMed] [Google Scholar]
- Niazi J.H., Lee S.J., Kim Y.S., Gu M.B. ssDNA aptamers that selectively bind oxytetracycline. Bioorg. Med. Chem. 2008;16:1254–1261. doi: 10.1016/j.bmc.2007.10.073. [DOI] [PubMed] [Google Scholar]
- Ozer T., Geiss B.J., Henry C.S. Review—chemical and biological sensors for viral detection. J. Electrochem. Soc. 2020;167 doi: 10.1149/2.0232003jes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peyrin E. Nucleic acid aptamer molecular recognition principles and application in liquid chromatography and capillary electrophoresis. J. Sep. Sci., 2009;32:1531–1536. doi: 10.1002/jssc.200900061. [DOI] [PubMed] [Google Scholar]
- Ramos E., Piñeiro D., Soto M., Abanades D.R., Martín M.E., Salinas M., González V.M. A DNA aptamer population specifically detects Leishmania infantum H2A antigen. Lab. Invest. 2007;87:409–416. doi: 10.1038/labinvest.3700535. [DOI] [PubMed] [Google Scholar]
- Sariya L., Thangthumniyom N., Wajjwalku W., Chumsing W., Ramasoota P., Lekcharoensuk P. Expression of foot and mouth disease virus nonstructural polyprotein 3ABC with inactive 3Cpro in Escherichia coli. Protein Expr. Purif. 2011;80:17–21. doi: 10.1016/j.pep.2011.07.006. [DOI] [PubMed] [Google Scholar]
- Smith P.F., Howerth E.W., Carter D, Gray E.W., Noblet R., Berghaus R.D., Stallknecht D.E., Mead D. Host predilection and transmissibility of vesicular stomatitis New Jersey virus strains in domestic cattle (Bos taurus) and swine (Sus scrofa) BMC Vet. Res. 2012;8:183. doi: 10.1186/1746-6148-8-183. http://www.biomedcentral.com/1746-6148/8/183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song S., Wang L., Li J., Zhao J., Fan C. Aptamer-based biosensors. Trends Anal. Chem. 2008;27:108–117. doi: 10.1021/acs.analchem.7b02862. [DOI] [Google Scholar]
- Tuerk C., Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- Vidic J., Manzano M., Chang CM., JaffrezicRenault N. Advanced biosensors for detection of pathogens related to livestock and poultry. Vet. Res. 2017;48:11. doi: 10.1186/s13567-017-0418-5. Vidic et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T., Chen C., Larcher L.M., Barrero R, Veedu R.N. Three decades of nucleic acid aptamer technologies: Lessons learned, progress and opportunities on aptamer development. Biotechnol. Adv. 2019;37:28–50. doi: 10.1016/j.biotechadv.2018.11.001. [DOI] [PubMed] [Google Scholar]
- Wong C.L., Yong C.Y., Ong H.K., Ho K.L., Tan W.S. Advances in the diagnosis of foot-and-mouth disease. Front. Vet. Sci. 2020;7:477. doi: 10.3389/fvets.2020.00477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Bowser M.T. Capillary electrophoresis–SELEX selection of catalytic DNA aptamers for a small-molecule porphyrin target. Anal. Chem. 2013;85:1525–1530. doi: 10.1021/ac302721j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]