Highlights
-
•
RO8191 strongly inhibits CHIKV replication in cells with defective type-1 IFN production.
-
•
RO8191 inhibited multiple stages of CHIKV replication cycle.
-
•
The highest antiviral activity was in the virucidal and post-entry stages.
-
•
Molecular docking analyses showed interactions between imidazonaphthyridine and CHIKV proteins.
-
•
ATR-FTIR analysis showed interactions mainly between RO8191 and CHIKV glycoproteins.
-
•
Our data suggest an in vitro antiviral activity of RO8191 against CHIKV.
Keywords: Imidazonaphthyridine, Antiviral activity, Chikungunya virus, Chikungunya Fever, Neglected tropical diseases, Type-1 IFN independent activity
Abstract
The Chikungunya virus (CHIKV) causes Chikungunya fever, a disease characterized by symptoms such as arthralgia/polyarthralgia. Currently, there are no antivirals approved against CHIKV, emphasizing the need to develop novel therapies. The imidazonaphthyridine compound (RO8191), an interferon-α (IFN-α) agonist, was reported as a potent inhibitor of HCV. Here RO8191 was investigated for its potential to inhibit CHIKV replication in vitro. RO8191 inhibited CHIKV infection in BHK-21 and Vero-E6 cells with a selectivity index (SI) of 12.3 and 37.3, respectively. Additionally, RO8191 was capable to protect cells against CHIKV infection, inhibit entry by virucidal activity, and strongly impair post-entry steps of viral replication. An effect of RO8191 on CHIKV replication was demonstrated in BHK-21 through type-1 IFN production mechanism and in Vero-E6 cells which has a defective type-1 IFN production, also suggesting a type-1 IFN independent mode of action. Molecular docking calculations demonstrated interactions of RO8191 with the CHIKV E proteins, corroborated by the ATR-FTIR assay, and with non-structural proteins, supported by the CHIKV-subgenomic replicon cells assay.
1. Introduction
Chikungunya fever is a viral disease with an acute phase characterized by dengue-like symptoms, such as high fever, nausea, severe arthralgia and polyarthralgia, and rashes, which can appear up to 12 weeks after the onset of viral infection (Kril et al., 2021; Silva et al., 2018; Thiberville et al., 2013). Aggravatingly, most CHIKV-infected patients develop chronic conditions of arthralgia and polyarthralgia, which persist for months to years, affecting the quality of life of infected people (Bedoui et al., 2021; Hibl et al., 2021; Kril et al., 2021; Schilte et al., 2013).
The Chikungunya virus (CHIKV) is the causative agent of Chikungunya fever, belonging to the Alphavirus genus within the Togaviridae family (Burt et al., 2017; Silva and Dermody, 2017). The viral particle consists of a positive single-stranded RNA genome (ssRNA+) of approximately 12 kb, protected by a protein capsid shell and a lipid envelope inserted with glycoproteins (Caglioti et al., 2013). The transmission of CHIKV occurs mainly through the bite of Aedes spp. Mosquitoes (Burt et al., 2017; Coffey et al., 2014), and since its first identification in Tanzania, East Africa, in 1952 (Schwartz and Albert, 2010), it has spread throughout the world, mainly affecting tropical and subtropical regions, such as the Americas (Pan American Health Organization, 2013).
Until December 2021, 131,630 cases of CHIKV fever were reported only in the Americas, of which about 127,487 cases and 11 deaths were confirmed in Brazil (Pan American Health Organization, 2021). Despite that, there are no approved vaccines or antiviral drugs by the Brazilian National Health Surveillance Agency (ANVISA) or the Food and Drug Administration of the United States of America (FDA) to manage Chikungunya fever (Subudhi et al., 2018). Therefore, the treatment of CHIKV infection is palliative, relying on non-steroidal anti-inflammatory and analgesics drugs, emphasizing the demand for antiviral development to treat this infection (Battisti et al., 2021; Tharmarajah et al., 2017).
The Pandemic Response Box (PRB), proposed by the Medicines for Malaria Venture (MMV) and the Drugs for Neglected Diseases initiative (DNDi), was designed with compounds that are highly active against malaria disease, and possess the potential to present biologic activities against other pathogens, being potentially applied in future outbreaks (Medicines for Malaria Venture and Drugs for Neglected Diseases initiative, n.d.; van Voorhis et al., 2016). Within the PRB, the imidazonaphthyridine (RO4948191 or RO8191) is a potent orally active interferon (IFN) agonist, binding directly to the IFNα/β receptor 2 (IFNAR2), triggering the JAK/STAT pathway, and consequently resulting in the activation and expression of IFN genes (Hwang et al., 2019; Konishi et al., 2012; Kota et al., 2018; Ying et al., 2021; Zeng et al., 2020). RO8191 was previously described as an inhibitor of Hepacivirus C (HCV) (S. Huang et al., 2014; Konishi et al., 2012; Wang et al., 2015), Hepatitis B virus (HBV) (Furutani et al., 2019; Takahashi et al., 2019), and Zika virus (ZIKV) (Fernandes et al., 2021) infections, and recently, as a synergetic molecule in antiviral treatments against CHIKV (Hwang et al., 2019). However, to the best of our knowledge, there is no description of RO8191 as an anti-CHIKV-specific inhibitor, as well as insights into its mechanism of action. Therefore, herein, RO8191 was evaluated as a potential inhibitor of CHIKV replication in vitro by the evaluation of its activity on several steps of the CHIKV replicative cycle in cells with production of type-1 IFN (BHK-21) and cells with defective production of type-1 IFN (Vero-E6) (Emeny and Morgan, 1979; Prescott et al., 2010), associated to bioinformatics, type-1 IFN quantification by QRT-PCR, and infrared spectroscopy analyzes that revealed the possible interactions between the RO8191 and CHIKV proteins.
2. Methods
2.1. Cells, virus and compound
Vero-E6 (isolated from the kidney of an African green monkey; ATCC CRL-1587) and BHK-21 cells (fibroblasts derived from Syrian golden hamster kidney; ATCC CCL-10) were maintained as previously described (Santos et al., 2021). Briefly, cells were maintained in Dulbecco's modified Eagle's medium (DMEM, SIGMA-ALDRICH), supplemented with 100 U/mL of penicillin (HYCLONE LABORATORIES), 100 mg/mL of streptomycin (HYCLONE LABORATORIES), 1% dilution of stock of non-essential amino acids (Hyclone Laboratories) and 1% of fetal bovine serum (FBS, HYCLONEN LABORATOIRES) in a humidified 5% CO2 incubator at 37 °C. Subgenomic replicon (SGR) harboring cells (BHK-CHIKV-NCT) (L Pohjala et al., 2011) were maintained under the same conditions of BHK-21 cells (ATCC CCL-10), except for the addition of G418 (SIGMA-ALDRICH) at 5 mg/mL.
The CHIKV expressing nanoluciferase reporter (CHIKV-nanoluc) (Matkovic et al., 2018; Leena Pohjala et al., 2011) (Fig. 1A) used for the antiviral assays is based on the CHIKV isolate LR2006OPY1 (East/Central/South African genotype). The CHIKV wild-type (CHIKVWT) used for infection assays, also from the genotype East/Central/South African genotype, was isolated and characterized by Nunes and coworkers (Nunes et al., 2015). CHIKV-nanoluc and CHIKVWT were produced, rescued, and titrated as previous described (Oliveira et al., 2020; Santos et al., 2021).
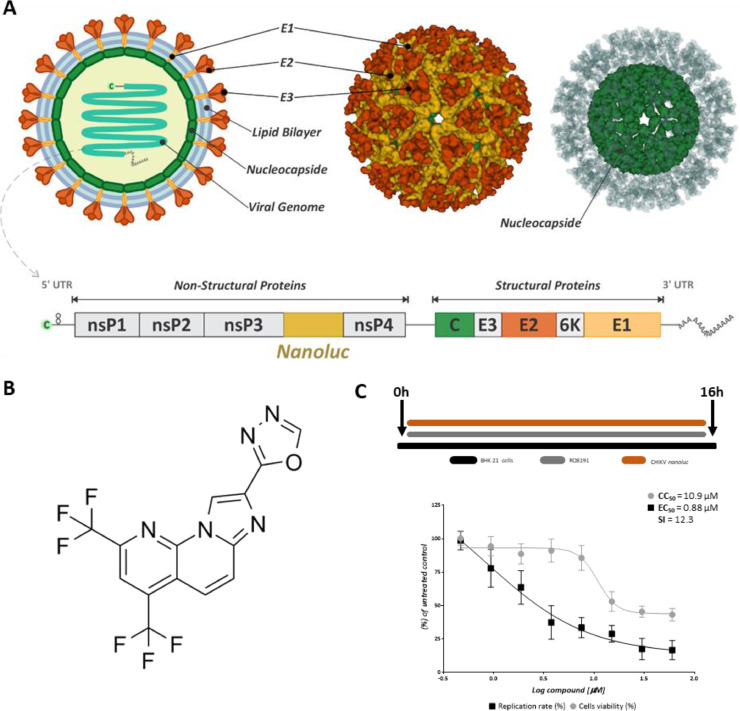
Fig. 1.
RO8191 activity on CHKV-nanoluc replication. A) Representative structure of CHIKV-nanoluc genome [PDB: 6NK5](Basore et al., 2019; Metz and Pijlman, 2016). B) Imidazonaphthyridine chemical structure (RO4948191 or RO8191) (CAS No.: 691,868–88–9; https://www.medchemexpress.com/ro8191.html). C) Representative scheme of the infection assays. BHK-21 cells were treated with two-fold serial dilutions of RO8191 at concentrations ranging from 0.46 to 60 µM. CHIKV-nanoluc replication was quantified by measuring nanoluciferase activity (indicated by a black square) and cell viability using MTT assay (indicated by a gray circle). Mean ± SD values from a minimum of three independent experiments, each measured in triplicate, are represented. All images were generated using GraphPad Prism 8 and GIMP 2.1v.
The imidazonaphthyridine compound (RO4948191 or RO8191) (Fig. 1B) was selected from PRB compounds (purity of > 90%) and was dissolved in 100% DMSO (v/v) to the concentration of 5 mM. The compound was diluted in the medium immediately before the assays (Fernandes et al., 2021).
2.2. Cell viability assay
Cell viability assays were performed as previously described (Oliveira et al., 2020; Santos et al., 2021), employing MTT [3-(4,5-dimethylthiazol-2-yl)−2,5-diphenyl tetrazolium bromide] (Sigma-Aldrich®) method. Briefly, BHK-21 and Vero-E6 cells were plated into 48-well plates at a density of 5 × 104 cells per well and incubated overnight at 37 °C and 5% CO2. Medium containing serial dilutions of RO8191 ranging from 60 µM to 0.46 µM was added to cells and incubated for 16 h. After this, the medium was replaced by MTT solution at 1 mg/ml, and cells were incubated for 30 min, after which the MTT solution was removed and replaced by 300 μL of DMSO (dimethyl sulfoxide) to solubilize the formazan crystals. The absorbance was measured at 490 nm on the Glomax microplate reader (Promega®). Cell viability was calculated according to the equation (T/C) × 100%, where T and C represent the mean optical density of the treated and untreated control groups, respectively. The cytotoxic concentration of 50% (CC50) was calculated using GraphPad Prism 8.
2.3. Antiviral assays
To assess the antiviral activity of RO8191, BHK-21 and Vero-E6 cells were seeded at a density of 5 × 104 cells per well into 48-well plates 24 h prior the assays. Cells were infected with CHIKV-nanoluc at a multiplicity of infection (MOI) of 0.1 in the presence of the compound in two-fold serial dilutions ranging from 60 µM to 0.46 µM for BHK-21 cells and three-fold serial dilutions from 60 µM to 0.02 µM for Vero-E6 cells (Oliveira et al., 2020; Santos et al., 2021). Samples were harvested using Renilla luciferase lysis buffer (Promega®) at 16 h post-infection (h.p.i.), and virus replication levels were quantified by measuring nanoluciferase activity using the Renilla luciferase Assay System (Promega®). The effective concentration of 50% inhibition (EC50) was calculated using GraphPad Prism 8 software. The values of CC50 and EC50 were used to calculate the selectivity index (SI = CC50/ EC50) (Oliveira et al., 2020; Santos et al., 2021).
To analyze the CHIKV mRNA expression under the RO8191 treatment, an infection assay was performed using a CHIKVWT. To this, BHK-21 and Vero-E6 cells were seeded at 1 × 106 cells per well in a 6 wells plate. After 24 h, the cells were infected with CHIKVWT in an MOI of 0.1 PFU/well in the presence or absence of the RO8191 at 5 µM. After 16 h, the cell lysates (lysed with Passive Lysis buffer) and supernatant were collected in Trizol reagent and the RNA was extracted using phenol-chloroform extraction protocol (Tony et al., 2018). The cDNA was produced using the High-Capacity cDNA Reverse Transcription Kit following the manufacturer's instructions. The quantification was performed through qRT-PCR in the Applied Biosystems 7300 Real-Time PCR System, using the GoTaq® qPCR Master Mix A6002 (Promega®) following the manufacturer's instructions, and employing the primers to the region 3′-end genomic sequence of CHIKV genome, being the sequences forward (5′- TGYCTCTTAGGGGACACATATACCT −3′) and reverse (5′-TGYCTCTTAGGGGACACATATACCT-3′) (Simmons et al., 2016). Each PCR run contained two negative controls and a dilution series of CHIKVWT cDNA (6 to 6 × 106 copies/mL) derived from 1 mL of tittered CHIKVWT virus stock, which was used to generate the standard curve. Each sample was analyzed in triplicate.
All infection assays were performed at a BSL-2 laboratory under the authorization number CBQ: 163/02 and process SEI: 01,245.006267/2022–14 from the CTNBio - National Technical Commission for Biosecurity from Brazil.
2.4. Plaque reduction assay
A plaque reduction assay was performed using CHIKVWT. To this, BHK-21 and Vero-E6 cells were seeded at 1 × 105 cells per well in a 24 wells plate. After 24 h, the cells were infected with CHIKVWT in an MOI of 3 × 10−4 (30 PFU/well) for 1 h at 37 °C, in the presence or absence of the RO8191 at 5 µM. Then, medium containing carboxymethyl cellulose (CMC) at 2% with or without RO8191 at 5 µM was added to the cells for 48 h. After the treatment, the medium was removed, cells were fixed with 4% paraformaldehyde for 20 min and stained with violet crystal 0.5%. Plaque-forming units were counted and normalized according to the equation (T/C) × 100%, where T and C represent the mean PFU of the treated and untreated control groups, respectively.
2.5. Time-of-drug addition assay
For the time-of-drug addition assays, BHK-21 and Vero-E6 cells were seeded at a density of 5 × 104 cells per well into 48 well plates overnight and infected with CHIKV-nanonluc at a MOI of 0.1. The samples were harvested using Renilla luciferase lysis buffer (Promega®) 16 h after infection, and virus replication levels were quantified by measuring nanoluciferase activity using the Renilla luciferase Assay System (Promega®).
In the pre-treatment assay, cells were treated for 1 h at 37 °C with RO8191, washed 3 times with PBS or compound removal, and then infected with the virus for 1 h at 37 °C. Then, cells were washed again to remove the unbound virus and replaced with fresh medium for 16 h. For virus entry assays, cells were infected using medium containing RO8191 and virus for 1 h at 37 °C, extensively washed 3 times with PBS, and incubated with a fresh medium for 16 h. The virucidal activity was performed using the same protocol of virus entry assay, with exception that the inoculum containing compound and virus at MOI of 5 was incubated for an extra hour before being added to the cells. In the post-entry assay, cells were infected with CHIKV-nanoluc for 1 h, washed 3 times with PBS to remove unbound virus, and incubated with medium containing compound for 16 h at 37 °C.
2.6. Replication assay using BHK-CHIKV-NCT cells
BHK-CHIKV-NCT cells harboring CHIKV replicon that expresses the non-structural proteins of the virus, a selection marker (puromycin acetyltransferase, Pac), and reporter genes of Renilla luciferase and EGFP (L Pohjala et al., 2011), at a density of 1 × 104 cells per well in a 96-well plate and incubated overnight at 37 °C. Medium-containing serial dilutions of RO8191 at concentrations ranging from 60 µM to 0.46 µM was added to cells and incubated for 72 h. Samples were harvested using Renilla luciferase lysis buffer (Promega®), and the luminescence levels were quantified with the Renilla luciferase Assay System (Promega®). In parallel, cell viability values were analyzed by performing MTT assay. The effective concentration of 50% inhibition (EC50) was calculated using GraphPad Prism 8 software. The values of CC50 and EC50 were used to calculate the selectivity index (SI = CC50/ EC50).
2.7. mRNA quantification of type-1 IFN in BHK-21 cells
To quantify the type-1 IFN mRNA, BHK-21 cells were seeded at 1 × 106 cells per well in a 6 wells plate. After 24 h, the cells were infected with CHIKVWT in an MOI of 0.1 PFU/well in the presence or absence of the RO8191 at 5 µM. After 16 h.p.i., the cell lysates were collected using Trizol reagent and the RNA was extracted using phenol-chloroform extraction (Toni et al., 2018). The cDNA was produced using the High-Capacity cDNA Reverse Transcription Kit following the manufacturer's instructions. The quantification was performed through qRT-PCR, using the PowerUp SYBR® Green Master Mix (ThermoFisher®) and 10 ng of cDNA, following the manufacturer's instructions, in the StepOnePlus™ Real-Time PCR System. Primers for BHK-21-IFN-α and -β were designed and used in the amplification reactions (IFN-α forward: 5′- GCCCATAGGACAAGCAGCATC −3′; IFN-α reverse: 5′-GAGAGCAGGTTGACCAGTAGC-3′ - based on the sequence NCBI Accession: XM_040757440.1;_IFN-β forward: 5′-GAGGCCAGACAAAGCAGAAG-3′; IFN-β reverse 5′-TCTGAGGCAGAGAGGTCCAC-3′ – based on the sequence NCBI Accession: MW017682.1). GAPDH gene was used as endogenous control, using the designed primers forward (5′-GTCAAGGCTGAGAACGGGAAG-3′) and reverse (5′-CAACATACTCGGCACCAGC-3′), based on the sequence NCBI Accession: DQ403055.1. Amplification was performed following the conditions: incubation for 10 min at 95 °C, and then 40 cycles of 15 s at 95 °C and 1 min at ≥60 °C. The RT-qPCR 2 − C∆∆Ct method was employed to analyze the results (Livak and Schmittgen, 2001). Each sample was analyzed in duplicate.
2.8. Molecular docking assays
The compound RO8191 (PDB: 3R0L) and the CHIKV proteins were docked employing the GOLD program(Jones et al., 1997). GOLD performs a search for the best pose of the chosen molecule in the receptor-binding site using a genetic algorithm (GA) and the score ChemPL. The docking focused on the envelope glycoprotein complex (PDBid: 3N42), nsP1 (PDBid: 7DOP), nsP2 (PDBid:4ZTB), nsP3 (PDBid:6W8Z), and nsP4 were performed. For the nsP4, a database representative sequence of the nsP4 extracted from the virus polyprotein (uniport-id: Q8JUX6) was modeled using the RoseTTAFold (Minkyung et al., 2021) in Robetta online server (https://robetta.bakerlab.org/). The nsP4 tridimensional model was assessed using ERRAT (Colovos, 1993) Ramachandran Plot (Laskowski et al., 1993), and Verify 3D (Eisenberg et al., 1997) tools in SAVES v6.0 (https://saves.mbi.ucla.edu/). The nsP4 binding site was predicted using COACH (Yang et al., 2013) based on the RoseTTAFold structure prediction. COACH is a meta-server approach that combines multiple function annotation results to generate ligand binding site predictions. COACH results indicate a binding site similar to the site where the Uridine 5′-Triphosphate (UTP) interacts with the crystal structure of HCV ns5B polymerase (PDBid: 4RY5). The binding site was defined by the UTP position based on the 4RY5 structure and extrapolated to the modeled nsP4. The GA run parameters were maintained at default. The poses generated were then ranked and the solution with the best score was chosen. For the proteins nsP1–3 the interaction was analyzed using blind docking, while the envelop glycoprotein complex was directed to the seven probable ligand sites that were defined whit the support of Rashad and Keller (2013). The interaction on the best solution were analyzed using the program DS Visualizer (BIOVIA, Dassault Systèmes, Discovery Studio Visualizer, v 20.1, San Diego: Dassault Systèmes, 2020).
2.9. Infrared spectroscopy spectral assay through ATR-FTIR
Samples were recorded in a Fourier transform infrared connected to a micro-attenuated total reflectance spectrophotometer (ATR-FTIR Agilent Cary 630 FTIR, Agilent Technologies, Santa Clara, CA, USA), as previously described (Oliveira et al., 2020; Santos et al., 2021). The diamond unit in the ATR system performs an internal-reflection element to record the fingerprint infrared signature in 1800 cm−1 to 800 cm−1 regions. The samples were prepared as previously described (Oliveira et al., 2020; Santos et al., 2021), where RO8191 (50 µM) in PBS was mixed with CHIKV-nanoluc virions (1 × 106 PFU/mL) in PBS. A volume of 1.5 µL of each sample was inserted into the diamond unit and dehydrated for 6 min using airflow, forming a thin layer on the surface of the ATR crystal. The spectra were then recorded in triplicate (24 cm−1 resolution, 32 scans). All spectra were normalized by the vector method and corrected using the rubber-band baseline correction (Caixeta et al., 2020). Subsequently, the second derivative spectra were created based on raw data plotted in the Origin Pro 9.0 (OriginLab, Northampton, MA, USA) software, and corrected using the Savitzky-Golay algorithm with polynomial order 5 and 20 points of the window. To further elucidate the expression of the functional group evaluated we used the value of the valley heights (Oliveira et al., 2020; Santos et al., 2022, 2021).
2.10. dsRNA intercalation assay
A migration retardation assay was performed based on the previously described protocol (Campos et al., 2017; Krawczyk et al., 2009; Silva et al., 2019). The HCV JFH-1 3′ untranslated region (UTR) known for forming a dsRNA was amplified through PCR. The reaction product of 273 bp was purified by ReliaPrep™ DNA Clean-Up and Concentration System (Promega®) and used for in vitro transcription by the HiScribe™ T7 High Yield RNA Synthesis Kit (New England BioLabs®). The dsRNA molecule was obtained by complementary annealing, later incubated at 30 nM with compound at 5 µM for 45 min and analyzed in 1% agarose TAE 1x gel stained with ethidium bromide. The lack of band in the gel confirms the compound intercalation activity since it competes with ethidium bromide. Doxorubicin (100 µM) was used as a positive control of intercalation. The band quantification was performed using ImageJ.JS version 1.53j.
2.11. CHIKV nsP4 cloning, overexpression, and purification
The coding region of nsP4 was cloned into a pET-SUMO expression vector, generating the nsP4_pET-SUMO/LIC expression vector, as previously described (Freire et al., 2022). Rosetta (DE3) E. coli (Novagen) cells were transformed with nsP4_pET-SUMO/LIC and grown in TB medium, supplemented with 50 μM kanamycin and 34 μM chloramphenicol at 37 °C, until the OD600 reached 1.0. The protein expression was induced by adding 1 mM of Isopropyl β-d-1-thiogalactopyranoside (IPTG), at 18 °C for 16 h. Cells were harvested by centrifugation and cell pellets were resuspended in 50 mM Tris pH 8.0, 500 mM NaCl and 10% glycerol. Cells were lysed by sonication and cell debris was separated by centrifugation. The nsP4 was purified using an AKTA Purifier System (GE Healthcare). The first purification step was an affinity chromatography using a HisTrap HP 5.0 mL column (GE Healthcare). Concomitantly, the buffer was exchanged through dialysis and the His-tag-SUMO was cleavage by TEV protease during overnight at 4 °C. Another affinity chromatography was performed using the same system to collect the protein after cleavage. The protein was concentrated, and a final purification step was done through size-exclusion chromatography on a XK 26/1000 Superdex 75 column (GE Healthcare) pre-equilibrated in buffer 50 mM Tris pH 8.0, 200 mM NaCl and 10% glycerol. The final protein sample was analyzed in SDS-PAGE 12.5% to confirm its purity. Concentration was determined in a Nanodrop 1000 spectrophotometer.
2.12. MicroScale thermophoresis (MST)
Experiments were performed on a Monolith® NT.115 (Nanotemper technol-ogies), as previously described (Freire et al., 2022). The nsP4 was labelled on cysteine residues with NT-647-Maleimide dye (Nanotemper Technologies) as per manufacturer's instructions. The concentration of protein indicated for MicroScale Thermophoresis experiments was 25 nM and a serial dilution of the compound from 250 µM to 0.0076 µM (7.6 nM). The dissociation constant Kd was obtained by fitting the binding curve with the Hill function.
2.13. Statistical analysis
All experiments were performed in triplicates and all tests were performed a minimum of three times (events) to confirm the reproducibility of the results. GraphPad Prism 8 software was used to evaluate the statistical differences in the means of the readings using the student t-test for paired data or the Mann-Whitney for unpaired data. Values of p<0.01 were considered statistically significant.
3. Results
3.1. RO8191 is a potent inhibitor of CHIKV replication in BHK-21 cells
To assess the antiviral activity of RO8191 (Fig. 1B), a dose-response assay was carried out to determine the 50% effective concentration (EC50) and 50% cytotoxicity concentration (CC50) of this compound using previously described protocols (Oliveira et al., 2020; Santos et al., 2021). BHK-21 cells were treated with a two-fold serial dilution of RO8191 at concentrations ranging from 0.46 µM to 60 µM in the presence or absence of CHIKV-nanoluc (Fig. 1A) for 16 h (Fig. 1C). As a result, RO8191 demonstrated to possess CC50 of 10.9 µM and EC50 of 0.88 µM, with a selectivity index (SI) of 12.3 (Fig. 1D), emphasizing that this compound is a strong inhibitor of CHIKV replication in vitro. For further analysis, cells were treated with RO8191 at 5 µM, which significantly inhibited 95.7% of the CHIKV infection (cell viability > 100%).
3.2. RO8191 mainly inhibits post-entry stages of CHIKV replication
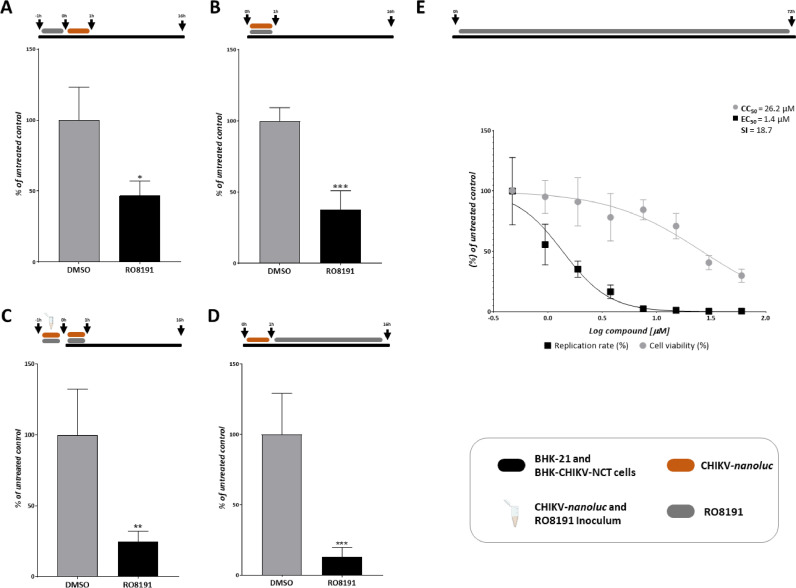
Since RO8191 compound exhibited a potent antiviral action against CHIKV, time-of-drug addition assay was carried out to investigate the effects of this molecule on different stages of viral replication (Santos et al., 2022, 2021). A protective assay was performed by pre-treating cells with RO8191 for 1 h at 37 °C, extensively washing cells with PBS, and infecting them with CHIKV-nanoluc for 1 h. Then, the virus-containing medium was removed, and a fresh medium was added (Fig. 2A). As a result, 16 h.p.i. RO8191 inhibited 52.9% of CHIKV infection (p = 0.04) (Fig. 2A), suggesting an effect of RO8191 as a protective molecule against CHIKV infection, providing an antiviral effect in host cells that blocked virus infection.
Fig. 2.
RO8191 activity on different stages of CHIKV-nanoluc replicative cycle in BHK-21 cells. A) BHK-21 cells were treated with RO8191 for 1 h, cells were washed with PBS to remove the compound, and were infected with CHIKV-nanoluc at MOI 0.1 for 1 h. Then, the medium was removed, cells were washed to remove unbound virus, and fresh medium was added. B) BHK-21 cells were infected with CHIKV-nanoluc (MOI 0.1) and simultaneously treated with RO8191 for 1 h. Then, cells were washed to completely remove the inoculum, and a fresh medium was added. C) RO8191 and CHIKV-nanoluc at MOI 5 were incubated for 1 h (representative inoculum). Then, the inoculum was added to the cells for an additional hour. Cells were washed to remove the inoculum and fresh medium was added. D) BHK-21 cells were infected with CHIKV-nanoluc (MOI 0.1) for 1 h, cells were washed with PBS to remove unbound virus, and were treated with RO8191. All infection assays were quantified 16 h.p.i through the measurement of luminescence levels. Schematic representation of each time-based assay as indicated by BHK-21 cells (black bars), RO8191 (gray bars), CHIKV-nanoluc (orange bars), and CHIKV-RO8191 inoculum (blue tube). E) Schematic representation of the time-based BHK-CHIKV cell assay as indicated by the BHK-CHIKV-NCT cells (black bars) and RO8191 (gray bars). BHK-CHIKV-NCT cells were incubated for 72 h with the RO8191 compound at concentrations ranging from 60 to 0.46 µM. Then, cells were washed with PBS and lysed with Passive Lysis Buffer to measure the activity of Renilla. The reduction of Renilla activity in BHK-CHIKV-NCT cells is shown by 50% of the highest concentrations (EC50) of RO8191 (black squares) and an effect in cell viability (CC50) using MTT assay (gray circles). Mean ± SD values of a minimum of three independent experiments, each measured in triplicate. (***) P<0.001, (**) P<0.01, and (*) P <0.05 are represented. Images were generated using GraphPad Prism 8 and GIMP 2.1v.
To further elucidate the activity of RO8191 in the early stages of CHIKV infection, virus and compound were simultaneously added to the cells for 1 h at 37 °C. Later, cells were washed with PBS and a fresh medium was added until 16 h.p.i. (Fig. 2B). As an outcome, the compound significantly inhibited 62.4% of CHIKV replication (p = 0.002), demonstrating the effect of RO8191 on viral entry into the cells (Fig. 2B). Additionally, we performed a virucidal assay by incubating the CHIKV particles with the compound for 1 h at 37 °C, and subsequently adding this inoculum to BHK-21 cells for an additional hour. Finally, the supernatant was removed, the cells were washed, and fresh medium was added (Fig. 2C). The results demonstrated that RO8191 possesses a virucidal activity inhibiting 75.4% of the CHIKV replication (p = 0.0024) (Fig. 2C), potentially by interacting with viral particles and blocking virus entry to the host cells.
To elucidate the post-entry effect of RO8191, the cells were infected with CHIKV-nanoluc for 1 h at 37 °C, then, the supernatant was removed, cells were washed with PBS, and medium with RO8191 was added (Fig. 2D). The results showed that the compound strongly inhibited CHIKV-nanoluc replication by 87% (p = 0.0004) (Fig. 2D).
Altogether, data obtained from the time-of-drug addition assay demonstrated that the main effect of RO8191 is on post-entry stages of virus replication (Fig. 2 and Supplementary Fig. 1A). Therefore, to further assess the RO8191 potential as a post-entry inhibitor, the effects of the compound on the replication stage of CHIKV replicative cycle, with no viral particles production and virion spread, were assessed through the employment of BHK-CHIKV-NCT cells, a subgenomic replicon system which expresses CHIKV non-structural proteins and the reporter genes of Renilla luciferase and EGFP (L Pohjala et al., 2011). As previously described, the measurement of the activity of these reporters allows the evaluation of the effect of compounds on replication complexes formed during the replication stage, as well as on the transcription and translation of subgenomic RNAs (L Pohjala et al., 2011; Santos et al., 2021). Thus, BHK-CHIKV-NCT cells were treated with two-fold serial dilution of the RO8191, at concentrations ranging from 0.46 to 60 µM for 72 h, and the expression of Renilla was subsequently measured (Fig. 2E). The results demonstrate that the compound strongly impaired viral replication, with an EC50 and CC50 of 1.4 µM and 26.2 µM, respectively, and SI of 18.7 (Fig. 2E), indicating a decrease in subgenomic RNA synthesis and/or translation, suggesting possible interactions of RO8191 with the virus nonstructural proteins or with the dsRNA intermediate of CHIKV replication.
3.3. RO8191 impairs CHIKV replication by dependent and independent of the type-1 IFN pathways
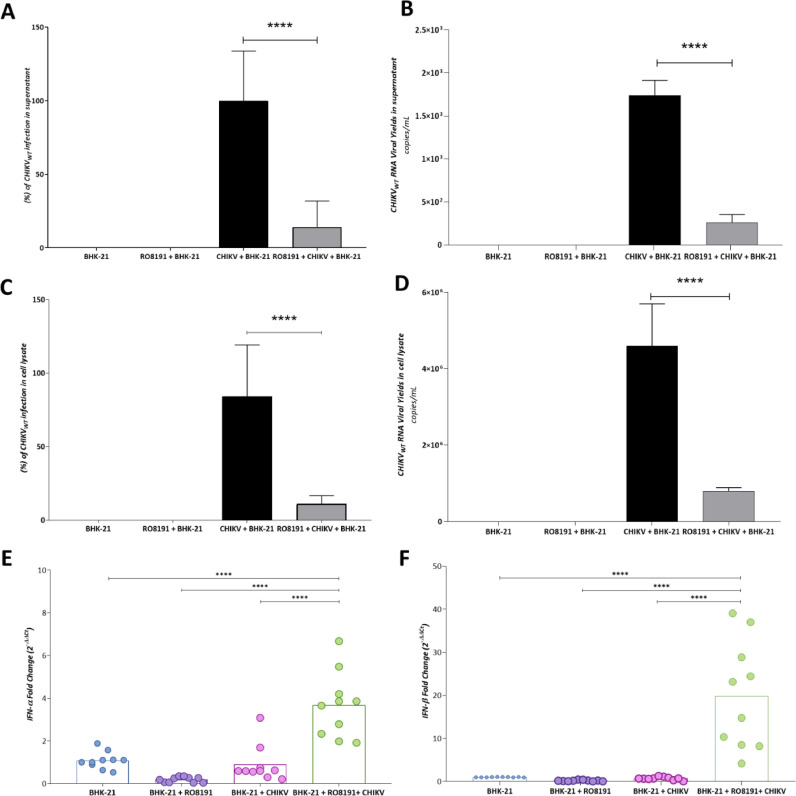
RO8191 was described as an agonist of type-1 IFN (Konishi et al., 2012), and BHK-21 cells possess the type-1 IFN pathway (MacDonald et al., 2007), which might suggest that the effect observed here is dependent on the intrinsic cellular immune response, and not only by the effects of the compound on CHIKV machinery. Therefore, an infection assay with BHK-21 cells and CHIKVWT was conducted in the presence or absence of RO8191 at 5 µM, and the levels of viral RNA, and IFN-α and -β were quantified by qRT-PCR. As an outcome, in the supernatant, CHIKVWT titers were reduced in 85.9% after treatment with RO8191 (Fig. 3A), representing by the infection decreased to 258.37 mRNA copies/mL under treatment, compared to 1.7 × 103 mRNA copies/mL of the untreated control (Fig. 3B). When samples from the cell lysates were analyzed, the same pattern was observed, showed by 89% of replication inhibition (Fig. 3C), observed by the viral RNA titers in the untreated control (4.5 × 106 mRNA copies/mL) and after treatment with RO8191 (7 × 105 mRNA copies/mL) (Fig. 3D).
Fig. 3.
BHK-21 cells express type-1 IFN (α/β) during RO8191 treatment. A) Percentage of CHIKVWT infection by measuring mRNA-CHIKVWT in supernatant of BHK-21 cells. B) CHIKVWT RNA titers by measuring mRNA-CHIKVWT in supernatant of BHK-21 cells. C) Percentage of CHIKVWT infection by measuring mRNA-CHIKVWT per mL in cell lysates of BHK-21 cells. D) CHIKVWT RNA titers by measuring mRNA-CHIKVWT per mL detected in cell lysates of BHK-21 cells. The CHIKV infection is presented in black bars while CHIKV in the presence of RO8191 treatment is demonstrated in gray bars. E) mRNA means of IFN-α expressed by BHK-21 during infection and/or treatment with RO8191. F) mRNA mean of IFN-β expressed by BHK-21 during infection and/or treatment with RO8191. BHK-21 cells were infected with CHIKVWT (MOI 0.1 PFU/well) in the presence or absence of RO8191 at 5 µM for 16 h. The supernatant and cell lysates were collected and processed to RNA extraction. cDNA was produced and mRNA levels was quantified by RT-qPCR. Mean values of three independent experiment each measured in triplicate, are represented. (****) P <0.0001. Images were generated using GraphPad Prism 8 and GIMP 2.1v.
Then, the type-1 IFN (α/β) expression was also quantified in the collected cell lysates (Fig. 3E-F). As a result, the BHK-21 cells did not express significant type-1 IFN levels (≥1 of fold change(-C∆∆Ct)) in the absence of stimulation with RO8191 treatment (Fig. 3E-F). On the other hand, the BHK-21 cells in the presence of CHIKV produced 0.9 of fold change(-C∆∆Ct) and 0.7 of fold change(-C∆∆Ct) of IFN-α (Fig. 3E) and -β (Fig. 3F), respectively. More interestingly, in the presence of both CHIKV and RO8191, the BHK-21 cells produced 3.6 of fold change(-C∆∆Ct) more of mRNA of IFN-α (Fig. 3E), and 19.8 of fold change(-C∆∆Ct) of IFN-β (Fig. 3F). Altogether, these data suggest that in BHK-21 cells, the molecule RO8191 is exerting its activity by dependent and independent of the type-1 IFN pathways.
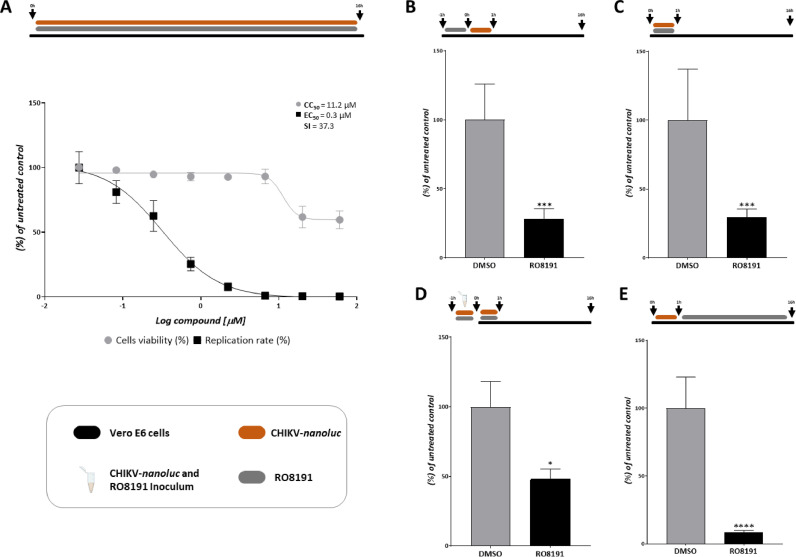
To further evaluate the RO8191 activity on CHIKV infection, Vero-E6 cells, which are cells with defective production of type-1 IFN (Emeny and Morgan, 1979; Prescott et al., 2010), were employed to perform the dose-response (Fig. 4A) and time-of-drug addition assay (Fig. 4B-E) assays on the same conditions used for BHK-21. The results demonstrated that RO8191 possesses CC50 and EC50 of 11.2 and 0.3 µM, respectively, with an SI of 37.3 (Fig. 4A). The time-of-drug addition assay demonstrated that RO8191 protected the cells from infection in 72% (p = 0.0005) (Fig. 4B), inhibited viral entry in 70.5% (p = 0.0008) (Fig. 4C), and presented virucidal activity in 51.8% (p = 0.0336) (Fig. 4D). The strongest effect observed using Vero-E6 cells was also in the post-entry stages, with inhibition of 91.6% of CHIKV replication (p<0.0001) (Fig. 4E and Supplementary Fig. 1B). Altogether, the data suggest an effect of RO8191 on the CHIKV replicative machinery, mainly on post-entry steps (Supplementary Fig. 1), however, in a type-1 IFN independent manner.
Fig. 4.
RO8191 activity against CHIKV-nanoluc infection in Vero-E6 cells. A) Representative scheme of the infection assays. Vero-E6 cells were treated with three-fold serial dilutions of RO8191 at concentrations ranging from 0.02 to 60 µM. CHIKV replication was quantified by measuring nanoluciferase activity (indicated by a black square) and cell viability using an MTT assay (indicated by a gray circle). Values of EC50, CC50, and SI were obtained from the treatment of Vero-E6 cells treated with RO8191. B) Vero-E6 cells were treated with RO8191 for 1 h, then the cells were washed with PBS and infected with CHIKV-nanoluc virus at MOI 0.1 for 1 h. The medium was removed, cells were washed with PBS and fresh medium was added. C) Vero-E6 cells were infected with CHIKV-nanoluc (MOI 0.1) and simultaneously treated with RO8191 for 1 h. Then, cells were washed, and fresh medium was added. D) RO891 and CHIKV-nanoluc at MOI 5 were incubated for 1 h, then the inoculum was added to cells for an extra 1 h, cells were washed, and fresh medium was added. E) Vero-E6 cells were infected with CHIKV-nanoluc (MOI 0.1) for 1 h, cells were washed with PBS and treated with RO8191. Schematic representation of each time-based assay as indicated by Vero-E6 cells (blue bars), RO8191 (gray bars), CHIKV-nanoluc (orange bars), and CHIKV-nanoluc/RO8191 inoculum (blue tube). Mean ± SD values of a minimum of three independent experiments, each measured in triplicate, are represented. (****) P <0.0001, (***) P <0.001 and (*) P <0.05. Images were generated using GraphPad Prism 8 and GIMP 2.1v.
The CHIKV RNA was also quantified in Vero-E6 cell lysates and supernatant and resulted in a significant decrease of mRNA-CHIKV expression (Supplementary Fig. 2). In the supernatant, 85.9% of CHIKVWT titers were observed (Supplementary Fig. 2A), showed by 2.1 × 105 mRNA copies/mL in the untreated controls, and 6.2 × 104 mRNA copies/mL after treatment with RO8191 (Supplementary Fig. 2B). In agreement, inhibition of 89% of CHIKVWT replication was seen in the cell lysates, since the viral RNA was 5 × 105 mRNA copies/mL and 1.1 × 105 mRNA copies/mL in the untreated control (Supplementary Fig. 2C) and RO8191 treatment (Supplementary Fig. 2D), respectively. It is important to emphasize that, since RO8191 protects cells against CHIKV infection and interferes with the entry and post-entry stages, the antiviral effects of this compound might also be due to an additional interference with host factors, other than type-1 IFN-related ones.
Finally, a plaque reduction assay employing the CHIKVWT was also performed to confirm the activity of RO8191 on the CHIKV replication cycle (Supplementary Fig. 3). RO8191 at 5 µM inhibited 88.1% and 86% of CHIKVWT replication in BHK-21 (Supplementary Fig. 3A) and Vero-E6 cells (Supplementary Fig. 3B), respectively. As expected, the levels of inhibition of RO8191 in both CHIKVWT and CHIKV-nanoluc were similar at the concentration tested.
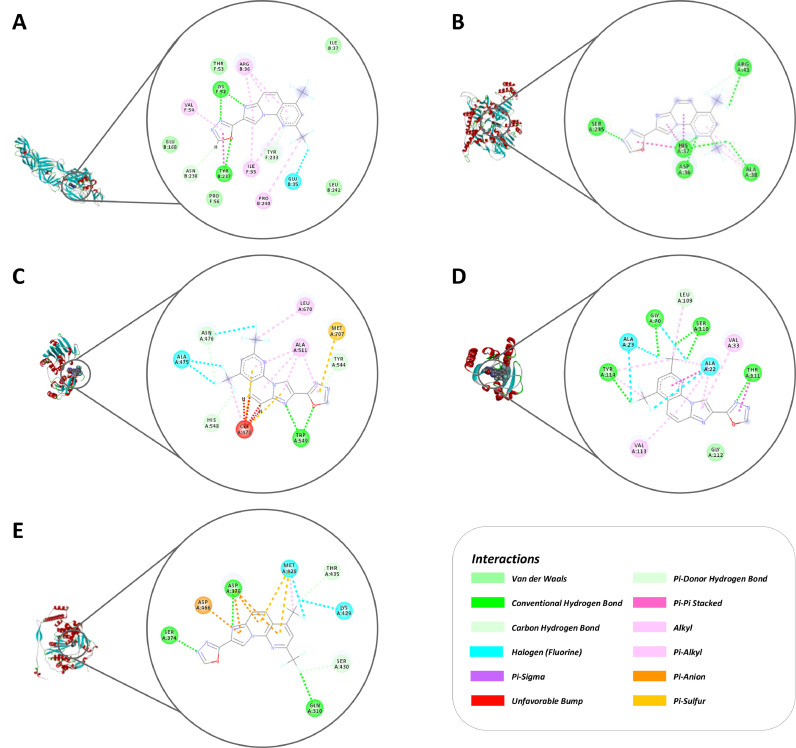
3.4. Molecular docking suggests the interaction of RO8191 with CHIKV glycoproteins and nonstructural proteins
Considering the effects of RO8191 on CHIKV replication stages, molecular docking data were generated by GOLD and suggested interesting interactions between the compound and CHIKV proteins. Based on the observed in vitro virucidal effect, interactions between CHIKV glycoproteins and the compounds were observed mainly in the domain II of viral protein E1, with docking ChemPLP scores ranging between 47.37 and 57.12, and in the domain A-B of the viral protein E2 and domain B of viral protein E3, with score 45.26 (Table 1, Fig. 5A). Alkyl and Pi-Alkyl interactions were identified between the compound and the residues VAL F:54, ARG B:36, ILE F:55, and PRO B:240 on the domain II; a Halogen interaction with residue GLU B:35; two conventional hydrogen bonds, one at residue LYS F:52 and other with Pi-Pi Stacked at TYR B:237 residues. The carbon-hydrogen bonds with the residue ASN B:238 and TYR F:233; and Van der Waals interaction at GLU B:168, PRO F:56, LEU B:241, THR F:53, and ILE B:37, were seen between RO8191 and CHIKV glycoproteins (Fig. 5A).
Table 1.
Localization and ChemPLP values showed between RO8191 and CHIKV proteins by molecular docking calculations.
| ChemPLP | Coordinates (x, y, z) | Volume (Å3) | Localization | ||
| Glycoproteins | Site 1 | 50.41 | −15.687, 2.019, −19.939 | 651.375 | Between II domain of E1 and C of E2. |
| Site 2 | 57.12 | −33.937, −18.731, −31.939 | 357.375 | Between II domain of E1 and beta-sheet of E2. | |
| Site 3 | 47.37 | −33.437, −6.731, −33.189 | 156.125 | Adjacent to site 2. | |
| Site 4 | 45.26 | −42.937, −28.731, −22.939 | 183.875 | Behind the fusion loop, between B domain of E3, and E2 domain B, and A domain of E2. | |
| Site 5 | −34.73 | −44.437, −14.731, −23.439 | 124 | Between beta-sheet of E2 and of E3. | |
| Site 6 | * | −16.187, −18.231, −36.439 | 20.5 | Inside the E3 cavity. | |
| Site 7 | 10.30 | −59.187, −15.731,−26.189 | 22.5 | Replacing the furin loop. | |
| nsP1 | 50.11 | Catalytic amino acid H37 | |||
| nsP2 | 35.26 | Catalytic amino acid H548 | |||
| nsP3 | 65.04 | ADP-ribose | |||
| nsP4 | 65.17 | Amino acid site ASP466 | |||
*There was no result on docking. Parameter based on Rashad and Keller (2013).
Fig. 5.
2D and 3D interactions between RO8191 and CHIKV proteins showed by molecular docking analysis. A) RO8181 interacts with CHIKV glycoproteins mainly through Hydrogen Bond (green lines) and alkyl ligands (pink lines). B) RO8191 showed interactions with nsP1 protein mainly through Conventional Hydrogen Bonds (green circles) interactions. C) RO8191 interacts with nsP2 mainly through Alkyl (pink lines) and Hydrogen Bonds (green lines) interactions. D) RO8191 interacts with nsP3 mainly through Hydrogens Bonds ligands and interactions (green lines and circles). E) RO8191 interactions with nsP4 mainly through Conventional Hydrogens Bonds (green lines) and Halogen (blue lines) ligands. RO8191 (PDB: 3R0L) and the CHIKV proteins (glycoprotein complex [PDBid: 3N42], nsP1 [PDBid: 7DOP], nsP2 [PDBid:4ZTB], nsP3 [PDBid:6W8Z], and database representative sequence of the nsP4 extracted from the virus polyprotein [uniport-id: Q8JUX6]) were docked employing the GOLD program site using a genetic algorithm (GA) and the score ChemPL. Images were generated using GraphPad Prism 8 and GIMP 2.1v.
The molecular docking performed with CHIKV nonstructural proteins (Fig. 5B-E) demonstrated the ChemPLP scores of 50.11, 35.26, 65.04, and 65.17 for nsP1, nsP2, nsP3, and nsP4, respectively (Table 1). The higher scores were observed for nsP3 and nsP4 (Table 1), in which at the nsP3 ADP-Ribose site the residues VAL A:33 and VAL A:113 interacted by alkyl and pi-alky bonds; the residues ALA A:23 and ALA A:22 by alkyl, amide-pi stacked, and halogen interactions; LEU A:109 by alkyl carbon-hydrogen bonds interaction; SER A:110 and GLY A:70 by halogen bond, TYR A:114 alkyl bond, and THR A:111 by amide-pi stacked and conventional hydrogen bonds, whereas the residue GLY A:112 interacted by van der Walls bond (Table 1, Fig. 5D). Additionally, nsP4 interacted with RO8191 through a pi-anion bond at ASP A:466, two halogen bonds at LYS A:429 and MET A:428; carbon-hydrogen bonds at THR A:435 and SER A:430; three conventional hydrogen bonds at GLN A:310, SER A:374, and ASP A:376 (with Pi-Anion ligands) (Table 1, Fig. 5E). Altogether, these data corroborate the in vitro effects shown by the compound.
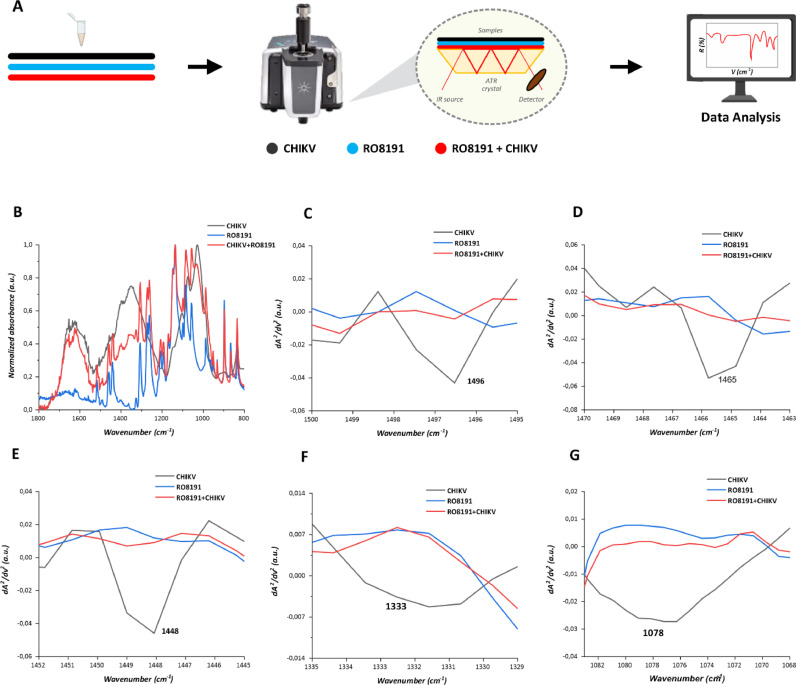
3.5. RO8191 induces molecular changes into CHIKV glycoproteins
A scheme of the ATR-FTIR technology based on infrared analysis to evaluate samples of CHIKV virions, RO8191, and CHIKV virions plus RO8191 is represented in Fig. 6A. We found a minimum of five molecular changes in CHIKV virions after incubation with RO8191 using ATR-FTIR analyses. The samples RO8191 at 50 µM, CHIKV-nanoluc virions (1 × 106 PFU/mL), and the mixture RO8191 (50 µM) and CHIKV-nanoluc virions were recorded in ATR-FTIR and the representative infrared spectra are shown in Fig. 6B. The biofingerprint in the range of 1800–800 cm−1 are capable to indicate absorption bands of glycoproteins, proteins, lipids, and RNA of CHIKV, and it may be exploited to suggest the interaction between the virus and RO8191. The second derivative spectrum is capable to identify the accurate spatial distribution of each wavenumber referring to each biochemical component (Kohler et al., 2007; Rieppo et al., 2012). In this context, infrared spectra can also detect binding among different functional groups of materials and biological samples (Haris, 2013). The second derivative function spectrum is capable to isolate and determine the suitable spatial distribution of each wavenumber related to the unique molecular component in each sample (Kohler et al., 2007; Rieppo et al., 2012). In this context, infrared spectra can also indicate molecular interactions between different functional groups of substances with biological samples (Haris, 2013). We highlighted the functional groups when a vibrational mode was present in the second derivative spectrum of CHIKV, and it was totally absent (above zero) after incubation of CHIKV with RO8191 (Fig. 6C–G). As an outcome, the binding interactions between CHIKV and RO8191 were suggested in five vibrational modes at 1496 cm−1, 1465 cm−1, 1448 cm−1, 1333 cm−1, and 1078 cm−1. The vibrational modes at 1496 cm−1 correspond to the in-plane CH bending vibrations (Fig. 6C). The vibrational mode at 1465 cm−1 represents CH2 scissoring mode of the acyl chain in lipids (Fig. 6D). The vibrational modes at 1448 cm−1 can be assigned to the asymmetric CH3 bending in methyl groups of proteins (Fig. 6E). The vibrational mode at 1333 cm−1 represents CH vibrations in polysaccharides (Fig. 6F), and 1078 cm−1 can be assigned to symmetric vibrations of PO2−in RNA (Fig. 6G). Altogether, these interactions displayed in infrared spectroscopy analyses are in accordance with the molecular docking analysis described above.
Fig. 6.
Infrared spectroscopy indicates molecular interactions between CHIKV virions with RO8191. A) Representative scheme of the ATR-FTIR technology with CHIKV virions (black line), RO8191 (blue line), and CHIKV virion plus incubation with RO8191 (red line). B) The representative infrared average spectrum of normalized mean spectra of CHIKV virion (black line), RO8191 (blue line), and CHIKV virion plus RO8191 (red line) employing ATR-FTIR platform from 1800 to 800 cm−1. C-F) The representative mean spectra of second derivative analysis to the vibrational modes at 1496 cm−1 (C), 1465 cm−1 (D), 1448 cm−1 (E), 1333 cm−1 (F), and 1078 cm−1 (G). Images were generated using GraphPad Prism 8 and GIMP 2.1v.
3.6. Effects of RO8191 on viral dsRNA and CHIKV nsP4
The replication of ssRNA+ viruses is dependent on the dsRNA, a viral replicating intermediator (Rampersad and Tennant, 2018), and the viral RNA-dependent RNA polymerase (Rupp et al., 2015). Since RO8191 displayed activity in post-entry stages of infection and on subgenomic RNA synthesis and/or translation, and molecular docking calculations suggested interactions between RO8191 and CHIKV, interaction assays employing synthetic dsRNA and the CHIKV nsP4 were performed.
Employing the 3′ UTR region of JFH-1 HCV as a template, an amplicon flanked by a T7 promoter was produced and used for in vitro transcription, synthesizing a dsRNA molecule of 273 bp (Campos et al., 2017; Krawczyk et al., 2009; Silva et al., 2019). Then, RO8191 at 5 µM was incubated with 30 nM of dsRNA and loaded in an agarose gel for electrophoresis. The results showed that RO8191 did not intercalate with the dsRNA (90.6% of band density) compared with the band of the sample treated with doxorubicin, the positive control of dsRNA interaction (1.3% of band density), which does not appear in the gel (Supplementary Fig. 4).
The purified CHIKV nsP4 was obtained by cloning the nsP4 genomic region into pET-SUMO (nsP4_pET-SUMO/LIC) expression vector and transforming Rosetta (DE3) E. coli (Novagen) cells, as previously described (Freire et al., 2022). The nsP4 was purified using an AKTA Purifier System (GE Healthcare) and concentration was determined using spectrophotometry. The CHIKV nsP4 size is 54 kDa, and its presence has been verified and confirmed in all purification steps, as shown in SDS-PAGE gels (Supplementary Fig. 5). The assessment of nsP4 binding affinity was performed by MicroScale Thermophoresis (MST) on a Monolith® NT.115 (Nanotemper technol-ogies). The CHIKV nsP4 was labeled on cysteine residues with NT-647-Maleimide dye (Nanotemper Technologies) and the protein concentration was 25 nM. In addition, to estimate the interaction between RO8191 and CHIKV nsP4, we used a dissociation constant Kd, obtained by fitting the binding curve with the Hill function, and serial dilutions of the compound from 250 µM to 0.0076 µM. The results obtained for RO8191 are shown in Supplementary Fig. 6, where it is possible to observe the presence of the unbound states well defined, but not a bound state, suggesting that the occurrence of interaction between the protein and the compound is on a major scale concentration, approximated kd ± ∆kd=>207 ± 10 µM. With these results is possible to conclude that CHIKV nsP4 presented a low interaction with RO8191.
4. Discussion
CHIKV has shown an evident evasion of the first immune responses during infection (Tanabe et al., 2018), and also a high percentage of the infected people develop a chronic condition (Amaral et al., 2020; McCarthy et al., 2018). It highlights the clinical importance of chikungunya fever and the urgency to develop effective treatments. The data presented here suggest that RO8191 has strong anti-CHIKV activity in vitro, potentially through several antiviral mechanisms, making it an interesting promising treatment. Additionally, in vivo studies have shown the pharmacological potential of RO8191 as a small molecule that can be administered orally in a more controlled, simple, rapid, and economical way (Ishibashi et al., 2019; Konishi et al., 2012). RO8191 was described as a possible enhancer drug to be employed in combined therapy against CHIKV (Hwang et al., 2019), however, to the best of our knowledge, there is no description of the effect of the monotherapy with this compound on the CHIKV replicative cycle, as well as data on its mode of action. Data obtained here emphasized and supported the anti-CHIKV activity of RO8191 in vitro and provided insights into the mechanisms of action of this drug. Therefore, our results present RO8191 as a promising drug for the treatment of chikungunya fever.
RO8191 was previously described with antiviral activity by activating the type-I IFN pathway by specifically binding the IFNAR receptor (IFNAR2) subunit 2, resulting in a similar response to the interferon-alpha (IFN-α) and JAK/STAT signaling pathway (Furutani et al., 2019; Konishi et al., 2012; Takahashi et al., 2019). This was confirmed by the quantification of mRNA-type-1 IFN through RT-qPCR during CHIKV infection in the presence or absence of RO8191 treatment, demonstrating that BHK-21 cells express type 1 IFN (α/β) in response to the CHIKV infection, and it is increased in the presence of RO8191 as an agonist. Additionally, our data also demonstrate that RO8191 has anti-CHIKV activity in cells defective of type 1 interferon (α/β) production, an interesting, novel, and unreported activity for RO8191, likely suggesting multiple mechanisms of action for this compound. RO8191 exerts strong activity toward CHIKV infection in BHK-21 and Vero-E6 cells, emphasized by the SI of 12.3 and 37.3, respectively. It is important to mention that the antiviral effect shown in CHIKV-nanoluc was very similar to observed in CHIKVWT, which supports the use of CHIKV-nanoluc throughout our research. Also, RO8191 previously showed anti-ZIKV activity in infected Vero-E6 cells, with a strong antiviral effect (SI of 121.4) (Fernandes et al., 2021). Other authors described the effect of RO8191 impairing Hepatitis C and B virus infection (Furutani et al., 2019; Huang et al., 2014b; Konishi et al., 2012; Takahashi et al., 2019; Wang et al., 2015), among them, Konishi and coworkers demonstrated RO8191 activity against HCV with an EC50 of 0.2 µM (Konishi et al., 2012), as well as its synthetic analogs with an improved anti-HCV entry action (Wang et al., 2015). Additionally, Takahashi and collaborators described that a RO8191 analog (CDM-3008), with strong activity against HCV and HBV, impaired 92.1% and 90.1% of infection, respectively (Takahashi et al., 2019). These works corroborate our data, and altogether demonstrate a strong antiviral activity against important clinical viruses, suggesting a broad-spectrum activity for RO8191. Moreover, RO8191 exerts a potential antiviral effect mediated by distinct mechanisms of action.
Another point observed here is the potent effect of RO8191 in different cell lineages, inhibiting over 50% of viral replication at all stages, with the strongest effect observed in post-entry stages of the CHIKV replicative cycle. Even though RO8191 was previously described as an effective agonist of interferon signaling (Furutani et al., 2021; Ishibashi et al., 2019; Kitamura et al., 2022; Zeng et al., 2020), when the antiviral assays were performed in Vero-E6 cells, which do not naturally express type-1 IFN genes (Emeny and Morgan, 1979; Prescott et al., 2010), the compound still triggered a strong anti-CHIKV activity, probably through an independent of IFN-α production mechanism of action. The effect observed here is in agreement with Konishi and collaborators who demonstrated that RO8191 inhibited HCV virus through a similar mechanism to IFN-α (Konishi et al., 2012). Interestingly, Wang and coworkers demonstrated the activity of RO8191 analogs impairing HCV entry by interacting with the virions (Wang et al., 2015), which agrees with our results, and with the antiviral data in cells defective of type-1 IFN production. It might explain and emphasizes the potential of this molecule as antiviral through a different mechanism of action, being supported by the bioinformatics data that demonstrated strong interactions with the subunit E1, also corroborated by the ATR-IFTR results, which indicated interactions between RO8191 and viral glycoproteins and lipids from CHIKV virions.
The RO8191 post-entry activity was demonstrated to be the strongest effect in the time-of-drug addition assays, supported by the potent effect on the CHIKV subgenomic expressing cells. Additionally, the in-silico data showed the highest docking values for nsP3 and nsP4, suggesting that RO8191 might be also interacting and interfering with the nsP4 polymerase activity and/or with the ADP-ribose site of nsP3, both representing interesting antiviral targets for antiviral discovery against CHIKV (Chaudhary and Sehgal, 2021; Shimizu et al., 2020). Also, to the best of our knowledge, the inhibition of the nsP3 and/or nsP4 activities has not been described for RO8191 yet. It is important to highlight that our data did show an interaction between RO8191 and CHIKV nsP4, suggesting a low interference with nsP4. However, further analyses are needed to investigate the interference of RO8191 with the ADP-ribose site of nsP3 or with other viral and cellular proteins, which may potentially disrupt the CHIKV replication. Finally, ATR-FTIR assay demonstrated changes in RNA spectra, which agrees with the significant reduction of viral RNA in the CHIKVWT assays in BHK-21 and Vero-E6 cells.
5. Conclusion
Altogether, the data presented here support RO8191 as a potent CHIKV inhibitor, mainly by affecting post-entry stages of viral replication, but also through type-1 IFN dependent and independent pathways of intrinsic cellular response. The combined mechanisms of action exhibited by RO8191 highlight its potential to be further studied for its effects against CHIKV replication in vivo and present this compound as an interesting alternative for antiviral development against arboviruses. Therefore, this data might be useful for further approaches against CHIKV and provide a potential treatment for Chikungunya fever.
Credit author statement
Uriel Enrique Aquino Ruiz (Conceptualization, Methodology, Validation, Formal analysis, Investigation, Writing Original Draft); Igor Andrade Santos (Conceptualization, Methodology, Validation, Formal analysis, Investigation, Writing - Original Draft, Writing - Review and Editing); Victória Riquena Grosche (Methodology, Formal analysis, Investigation, Writing - Original Draft); Rafaela Sachetto Fernandes (Resources, Methodology, Formal analysis, Investigation, Writing – Review and Editing); Andre Schutzer de Godoy (Resources, Methodology, Formal analysis, Investigation, Writing – Review and Editing); Jhoan David Aguillón Torres (Methodology, Formal analysis, Investigation, Writing - Review and Editing); Marjorie Caroline Liberato Cavalcanti Freire (Methodology, Formal analysis, Investigation, Writing - Review and Editing); Nathalya Cristina de Moraes Roso Mesquita (Methodology, Formal analysis, Investigation, Writing - Review and Editing); Marco Guevara-Vega (Methodology, Formal analysis, Investigation, Writing - Original Draft, Writing - Review and Editing); Nilson Nicolau-Junior (Methodology, Software, Formal analysis, Investigation, Writing - Review and Editing); Robinson Sabino-Silva (Resources, Conceptualization, Methodology, Validation, Formal analysis, Investigation, Writing - Original Draft, Supervision); Tiago Wilson Patriarca Mineo(Conceptualization, Methodology, Validation, Formal analysis, Investigation, Writing - Review and Editing, Supervision); Glaucius Oliva (Resources, Conceptualization, Methodology, Validation, Formal analysis, Investigation, Writing - Review and Editing, Supervision); Ana Carolina Gomes Jardim (Resources, Conceptualization, Methodology, Validation, Formal analysis, Investigation, Writing - Original Draft, Writing - Review and Editing, Project Administration, Supervision),
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We thank Andres Merits (Institute of Technology, University of Tartu, Tartu, Estonia) for the provision of the CHIKV expressing-nanoluciferase and Paula Rahal from the Institute of Biosciences, Humanities and Exact Sciences, São Paulo State University, São José do Rio Preto, Sao Paulo, Brazil for provision of CHIKV wild-type. IAS thanks Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), scholarship #142495/2020–4 and CAPES.Print scholarship # 88887.700246/2022–00. ACGJ and RSS are grateful to Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES)-Brasil-Prevention and Combat of Outbreaks, Endemics, Epidemics and Pandemics-Finance Code #88881.506794/2020–01. ACGJ is also grateful to FAPEMIG (Minas Gerais Research Foundation APQ-01487–22) and CAPES-Finance Code 001. This work was also supported by Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP), CEPID grant 2013/07600–3 to GO, grant 2018/05130–3 to RSF, and 2016/19712–9 to ASG, and Coordenaçao de Aperfeiçoamento de Pessoal de Nível Superior (grant 88887.516153/2020–00) to ASG. We are grateful to the Medicine for Malaria Ventures (MMV, www.mmv.org) and the Drugs for Neglected Diseases initiative (DNDi, www.dndi.org) for their support, for the design of the Pandemic Response Box and for supplying the compounds. MarvinSketch was used to draw the chemicals, MarvinSketch version 20.17.0, ChemAxon (https://www.chemaxon.com).
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.virusres.2022.199029.
Appendix. Supplementary materials
Data Availability
Data will be made available on request.
References
- Amaral J.K., Bilsborrow J.B., Schoen R.T. Chronic chikungunya arthritis and rheumatoid arthritis: what they have in common. Am. J. Med. 2020;133:e91–e97. doi: 10.1016/J.AMJMED.2019.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basore K., Kim A.S., Nelson C.A., Zhang R., Smith B.K., Uranga C., Vang L., Cheng M., Gross M.L., Smith J., Diamond M.S., Fremont D.H. Cryo-EM structure of chikungunya virus in complex with the Mxra8 receptor. Cell. 2019;177 doi: 10.1016/J.CELL.2019.04.006. 1725-1737.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battisti, V., Urban, E., Langer, T., 2021. Antivirals against the Chikungunya Virus. Viruses 13, 1307. https://doi.org/10.3390/v13071307. [DOI] [PMC free article] [PubMed]
- Bedoui Y., Septembre-Malaterre A., Giry C., Jaffar-Bandjee M.C., Selambarom J., Guiraud P., Gasque P. Robust cox-2-mediated prostaglandin response may drive arthralgia and bone destruction in patients with chronic inflammation post-Chikungunya. PLoS Negl Trop Dis. 2021;15 doi: 10.1371/journal.pntd.0009115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burt F.J., Chen W., Miner J.J., Lenschow D.J., Merits A., Schnettler E., Kohl A., Rudd P.A., Taylor A., Herrero L.J., Zaid A., Ng L.F.P., Mahalingam S. Chikungunya virus: an update on the biology and pathogenesis of this emerging pathogen. Lancet Infect. Dis. 2017;17:e107–e117. doi: 10.1016/S1473-3099(16)30385-1. [DOI] [PubMed] [Google Scholar]
- Caglioti C., Lalle E., Castilletti C., Carletti F., Capobianchi M.R., Bordi L. Chikungunya virus infection: an overview. New Microbiol. 2013;36:211–227. [PubMed] [Google Scholar]
- Caixeta D.C., Aguiar E.M.G., Cardoso-Sousa L., Coelho L.M.D., Oliveira S.W., Espindola F.S., Raniero L., Crosara K.T.B., Baker M.J., Siqueira W.L., Sabino-Silva R. Salivary molecular spectroscopy: a sustainable, rapid and non-invasive monitoring tool for diabetes mellitus during insulin treatment. PLoS One. 2020;15 doi: 10.1371/journal.pone.0223461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos G.R.F., Bittar C., Jardim A.C.G., Shimizu J.F., Batista M.N., Paganini E.R., Assis L.R.de, Bartlett C., Harris M., Bolzani V.da S., Regasini L.O., Rahal P. Hepatitis C virus in vitro replication is efficiently inhibited by acridone Fac4. J. Gen. Virol. 2017;98:1693–1701. doi: 10.1099/jgv.0.000808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhary M., Sehgal D. In silico identification of natural antiviral compounds as a potential inhibitor of chikungunya virus non-structural protein 3 macrodomain. J. Biomol. Struct. Dyn. 2021:1–11. doi: 10.1080/07391102.2021.1960195. [DOI] [PubMed] [Google Scholar]
- Coffey L.L., Failloux A.-.B., Weaver S.C. Chikungunya Virus–Vector Interactions. Viruses. 2014;6:4628–4663. doi: 10.3390/v6114628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colovos C. Verification of protein structures: patterns of nonbonded atomic interactions. Mol. Biol. 1993;2:1511–1519. doi: 10.1002/pro.5560020916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg D., Lüthy R., Bowie J.U. VERIFY3D: assessment of protein models with three-dimensional profiles. Methods Enzymol. 1997;277:396–404. doi: 10.1016/S0076-6879(97)77022-8. [DOI] [PubMed] [Google Scholar]
- Emeny J.M., Morgan M.J. Regulation of the interferon system: evidence that vero cells have a genetic defect in interferon production. J. Gen. Virol. 1979;43:247–252. doi: 10.1099/0022-1317-43-1-247. [DOI] [PubMed] [Google Scholar]
- Fernandes R.S., de Godoy A.S., Santos I.A., Noske G.D., de Oliveira K.I.Z., Gawriljuk V.O., Gomes Jardim A.C., Oliva G. Discovery of an imidazonaphthyridine and a riminophenazine as potent anti-Zika virus agents through a replicon-based high-throughput screening. Virus Res. 2021;299 doi: 10.1016/J.VIRUSRES.2021.198388. [DOI] [PubMed] [Google Scholar]
- Freire M.C.L.C., Basso L.G.M., Mendes L.F.S., Mesquita N.C.M.R., Mottin M., Fernandes R.S., Policastro L.R., Godoy A.S., Santos I.A., Ruiz U.E.A., Caruso I.P., Sousa B.K.P., Jardim A.C.G., Almeida F.C.L., Gil L.H.V.G., Andrade C.H., Oliva G. Characterization of the RNA-dependent RNA polymerase from Chikungunya virus and discovery of a novel ligand as a potential drug candidate. Sci. Rep. 2022;12:10601. doi: 10.1038/S41598-022-14790-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furutani Y., Toguchi M., Higuchi S., Yanaka K., Gailhouste L., Qin X.-.Y., Masaki T., Ochi S., Matsuura T. Establishment of a Rapid Detection System for ISG20-Dependent SARS-CoV-2 Subreplicon RNA Degradation Induced by Interferon-α. Int. J. Mol. Sci. 2021;22:11641. doi: 10.3390/ijms222111641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furutani Y., Toguchi M., Shiozaki-Sato Y., Qin X.-.Y., Ebisui E., Higuchi S., Sudoh M., Suzuki H., Takahashi N., Watashi K., Wakita T., Kakeya H., Kojima S. An interferon-like small chemical compound CDM-3008 suppresses hepatitis B virus through induction of interferon-stimulated genes. PLoS One. 2019;14 doi: 10.1371/journal.pone.0216139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haris P.I. Probing protein–protein interaction in biomembranes using Fourier transform infrared spectroscopy. Biochim. Biophys. Acta Biomembr. 2013;1828:2265–2271. doi: 10.1016/j.bbamem.2013.04.008. [DOI] [PubMed] [Google Scholar]
- Hibl B.M., Dailey Garnes N.J.M., Kneubehl A.R., Vogt M.B., Spencer Clinton J.L., Rico-Hesse R.R. Mosquito-bite infection of humanized mice with chikungunya virus produces systemic disease with long-term effects. PLoS Negl. Trop. Dis. 2021;15 doi: 10.1371/journal.pntd.0009427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S., Qing J., Wang S., Wang H., Zhang L., Tang Y. Design and synthesis of imidazo[1,2-α][1,8]naphthyridine derivatives as anti-HCV agents via direct C–H arylation. Org. Biomol. Chem. 2014;12:2344–2348. doi: 10.1039/C3OB42525H. [DOI] [PubMed] [Google Scholar]
- Huang S., Qing J., Wang S., Wang H., Zhang L., Tang Y. Design and synthesis of imidazo[1,2-α][1,8]naphthyridine derivatives as anti-HCV agents via direct C–H arylation. Org. Biomol. Chem. 2014;12:2344–2348. doi: 10.1039/C3OB42525H. [DOI] [PubMed] [Google Scholar]
- Hwang J., Wang Y., Fikrig E. Inhibition of chikungunya virus replication in primary human fibroblasts by liver X receptor agonist. Antimicrob. Agents Chemother. 2019 doi: 10.1128/AAC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishibashi D., Homma T., Nakagaki T., Fuse T., Sano K., Satoh K., Mori T., Atarashi R., Nishida N. Type I interferon protects neurons from prions in in vivo models. Brain. 2019;142:1035–1050. doi: 10.1093/brain/awz016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones G., Willett P., Glen R.C., Leach a R., Taylor R. Development and validation of a genetic algorithm for flexible docking. J. Mol. Biol. 1997;267:727–748. doi: 10.1006/jmbi.1996.0897. [DOI] [PubMed] [Google Scholar]
- Kitamura H., Tanigawa T., Kuzumoto T., Nadatani Y., Otani K., Fukunaga S., Hosomi S., Tanaka F., Kamata N., Nagami Y., Taira K., Uematsu S., Watanabe T., Fujiwara Y. Interferon-α exerts proinflammatory properties in experimental radiation-induced esophagitis: possible involvement of plasmacytoid dendritic cells. Life Sci. 2022;289 doi: 10.1016/J.LFS.2021.120215. [DOI] [PubMed] [Google Scholar]
- Kohler A., Bertrand D., Martens H., Hannesson K., Kirschner C., Ofstad R. Multivariate image analysis of a set of FTIR microspectroscopy images of aged bovine muscle tissue combining image and design information. Anal. Bioanal. Chem. 2007;389:1143–1153. doi: 10.1007/s00216-007-1414-9. [DOI] [PubMed] [Google Scholar]
- Konishi H., Okamoto K., Ohmori Y., Yoshino H., Ohmori H., Ashihara M., Hirata Y., Ohta A., Sakamoto H., Hada N., Katsume A., Kohara M., Morikawa K., Tsukuda T., Shimma N., Foster G.R., Alazawi W., Aoki Y., Arisawa M., Sudoh M. An orally available, small-molecule interferon inhibits viral replication. Sci. Rep. 2012;2:259. doi: 10.1038/srep00259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kota S.K., Roening C., Patel N., Kota S.B., Baron R. PRMT5 inhibition promotes osteogenic differentiation of mesenchymal stromal cells and represses basal interferon stimulated gene expression. Bone. 2018;117:37–46. doi: 10.1016/J.BONE.2018.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krawczyk M., Wasowska-Lukawska M., Oszczapowicz I., Boguszewska-Chachulska A.M. Amidinoanthracyclines - a new group of potential anti-hepatitis C virus compounds. Biol. Chem. 2009;390:351–360. doi: 10.1515/BC.2009.040. [DOI] [PubMed] [Google Scholar]
- Kril V., Aïqui-Reboul-Paviet O., Briant L., Amara A. New insights into chikungunya virus infection and pathogenesis. Annu. Rev. Virol. 2021 doi: 10.1146/annurev-virology-091919. [DOI] [PubMed] [Google Scholar]
- Laskowski R.A., Macarthur M.W., Moss D.S., Thornton J.M. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Cryst. 1993;26:283–291. [Google Scholar]
- Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- MacDonald M.R., Machlin E.S., Albin O.R., Levy D.E. The zinc finger antiviral protein acts synergistically with an interferon-induced factor for maximal activity against alphaviruses. J. Virol. 2007;81:13509–13518. doi: 10.1128/jvi.00402-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matkovic R., Bernard E., Fontanel S., Eldin P., Chazal N., Hassan Hersi D., Merits A., Péloponèse J.-.M., Briant L. The host DHX9 DExH-box helicase is recruited to chikungunya virus replication complexes for optimal genomic RNA translation. J. Virol. 2018;93:1–17. doi: 10.1128/JVI.01764-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy M.K., Davenport B.J.J., Morrison T.E. In: Chikungunya Virus. Heise M., editor. Springer International Publishing; Cham: 2018. Chronic chikungunya virus disease; pp. 55–80. [DOI] [Google Scholar]
- Medicines for Malaria Venture, 2022 Drugs for neglected diseases initiative, n.d. pandemic response box [WWW Document]. URL https://www.mmv.org/mmv-open/pandemic-response-box (accessed 3.28.22).
- Metz S.W., Pijlman G.P. In: Chikungunya Virus. Okeoma C.M., editor. Springer International Publishing; Cham: 2016. Function of Chikungunya Virus Structural Proteins; pp. 63–74. [DOI] [Google Scholar]
- Minkyung B., Frank D., Ivan A. Accurate prediction of protein structures and interactions using a three-track neural network. Science. 2021;373:871–876. doi: 10.1126/science.abj8754. 1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunes M.R.T., Faria N.R., de Vasconcelos J.M., Golding N., Kraemer M.U., de Oliveira L.F., Azevedo R. Emergence and potential for spread of Chikungunya virus in Brazil. BMC Med. 2015;13:102. doi: 10.1186/s12916-015-0348-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira D.M.de, Santos I.de A., Martins D.O.S., Gonçalves Y.G., Cardoso-Sousa L., Sabino-Silva R., von Poelhsitz G., Franca E.de F., Nicolau-Junior N., Pacca C.C., Merits A., Harris M., Jardim A.C.G. Organometallic complex strongly impairs chikungunya virus entry to the host cells. Front. Microbiol. 2020;11 doi: 10.3389/fmicb.2020.608924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan American Health Organization, 2021. Epidemiological update dengue, chikungunya and zika in the context of COVID-19 [WWW Document]. URL https://iris.paho.org/handle/10665.2/55639 (accessed 3.28.22).
- Pan American Health Organization, 2013. Epidemiological Alert, Chikungunya Fever [WWW Document]. URL https://iris.paho.org/handle/10665.2/50703 (accessed 3.28.22).
- Pohjala L., Utt A., Varjak M., Lulla A., Merits A. Inhibitors of alphavirus entry and replication identified with a stable chikungunya replicon cell line and virus-based assays. PLoS One. 2011;6:28923. doi: 10.1371/journal.pone.0028923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohjala Leena, Utt A., Varjak M., Lulla A., Merits A., Ahola T., Tammela P. Inhibitors of alphavirus entry and replication identified with a stable Chikungunya replicon cell line and virus-based assays. PLoS One. 2011;6:e28923. doi: 10.1371/journal.pone.0028923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prescott J., Hall P., Acuna-Retamar M., Ye C., Wathelet M.G., Ebihara H., Feldmann H., Hjelle B. New World Hantaviruses Activate IFNλ Production in Type I IFN-Deficient Vero E6 Cells. PLoS One. 2010;5:e11159. doi: 10.1371/journal.pone.0011159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rampersad S., Tennant P. Viruses. Elsevier; 2018. Replication and Expression Strategies of Viruses; pp. 55–82. [DOI] [Google Scholar]
- Rashad A.A., Keller P.A. Structure based design towards the identification of novel binding sites and inhibitors for the chikungunya virus envelope proteins. J. Mol. Graph. Model. 2013;44:241–252. doi: 10.1016/j.jmgm.2013.07.001. [DOI] [PubMed] [Google Scholar]
- Rieppo L., Saarakkala S., Närhi T., Helminen H.J., Jurvelin J.S., Rieppo J. Application of second derivative spectroscopy for increasing molecular specificity of fourier transform infrared spectroscopic imaging of articular cartilage. Osteoarthritis Cartilage. 2012;20:451–459. doi: 10.1016/J.JOCA.2012.01.010. [DOI] [PubMed] [Google Scholar]
- Rupp J.C., Sokoloski K.J., Gebhart N.N., Hardy R.W. Alphavirus RNA synthesis and non-structural protein functions. J. Gen. Virol. 2015;96:2483–2500. doi: 10.1099/jgv.0.000249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos I.A., Pereira A.K., dos S., Guevara-Vega M., de Paiva R.E.F., Sabino-Silva R., Bergamini F.R.G., Corbi P.P., Jardim A.C.G. Repurposing potential of rimantadine hydrochloride and development of a promising platinum(II)-rimantadine metallodrug for the treatment of Chikungunya virus infection. Acta Trop. 2022;227 doi: 10.1016/J.ACTATROPICA.2021.106300. [DOI] [PubMed] [Google Scholar]
- Santos I.A., Shimizu J.F., de Oliveira D.M., Martins D.O.S., Cardoso-Sousa L., Cintra A.C.O., Aquino V.H., Sampaio S.V., Nicolau-Junior N., Sabino-Silva R., Merits A., Harris M., Jardim A.C.G. Chikungunya virus entry is strongly inhibited by phospholipase A2 isolated from the venom of Crotalus durissus terrificus. Sci. Rep. 2021;11:8717. doi: 10.1038/s41598-021-88039-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilte C., Staikovsky F., Couderc T., Madec Y., Carpentier F., Kassab S., Albert M.L., Lecuit M., Michault A. Correction: chikungunya Virus-associated Long-term Arthralgia: a 36-month Prospective Longitudinal Study. PLoS Negl. Trop. Dis. 2013;7 doi: 10.1371/annotation/850ee20f-2641-46ac-b0c6-ef4ae79b6de6. null. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz O., Albert M.L. Biology and pathogenesis of chikungunya virus. Nat. Rev. Microbiol. 2010;8:491–500. doi: 10.1038/nrmicro2368. [DOI] [PubMed] [Google Scholar]
- Shimizu J.F., Martins D.O.S., McPhillie M.J., Roberts G.C., Zothner C., Merits A., Harris M., Jardim A.C.G. Is the ADP ribose site of the Chikungunya virus NSP3 Macro domain a target for antiviral approaches? Acta Trop. 2020;207 doi: 10.1016/J.ACTATROPICA.2020.105490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva N.M.da, Teixeira R.A.G., Cardoso C.G., Siqueira Junior J.B., Coelho G.E., de Oliveira E.S.F. Chikungunya surveillance in Brazil: challenges in the context of Public Health. Epidemiol. Health Serv.: J. Sistema Unico de Saude do Brasil. 2018;27 doi: 10.5123/S1679-49742018000300003. [DOI] [PubMed] [Google Scholar]
- Silva L.A., Dermody T.S. Chikungunya virus: epidemiology, replication, disease mechanisms, and prospective intervention strategies. J. Clin. Invest. 2017;127:737–749. doi: 10.1172/JCI84417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva S., Shimizu J.F., Oliveira D.M.de, Assis L.R.de, Bittar C., Mottin M., Sousa B.K.de P., Mesquita N.C.de M.R., Regasini L.O., Rahal P., Oliva G., Perryman A.L., Ekins S., Andrade C.H., Goulart L.R., Sabino-Silva R., Merits A., Harris M., Jardim A.C.G. A diarylamine derived from anthranilic acid inhibits ZIKV replication. Sci. Rep. 2019;9:17703. doi: 10.1038/s41598-019-54169-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons M., Myers T., Guevara C., Jungkind D., Williams M., Houng H.-.S. Development and validation of a quantitative, one-step, multiplex, real-time reverse transcriptase PCR assay for detection of dengue and chikungunya viruses. J. Clin. Microbiol. 2016;54:1766–1773. doi: 10.1128/JCM.00299-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subudhi B., Chattopadhyay S., Mishra P., Kumar A. Current Strategies for Inhibition of Chikungunya Infection. Viruses. 2018;10:235. doi: 10.3390/v10050235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi N., Hayashi K., Nakagawa Y., Furutani Y., Toguchi M., Shiozaki-Sato Y., Sudoh M., Kojima S., Kakeya H. Development of an anti-hepatitis B virus (HBV) agent through the structure-activity relationship of the interferon-like small compound CDM-3008. Bioorg. Med. Chem. 2019;27:470–478. doi: 10.1016/J.BMC.2018.11.039. [DOI] [PubMed] [Google Scholar]
- Tanabe I.S.B., Tanabe E.L.L., Santos E.C., Martins W.v., Araújo I.M.T.C., Cavalcante M.C.A., Lima A.R.v., Câmara N.O.S., Anderson L., Yunusov D., Bassi Ê.J. Cellular and Molecular Immune Response to Chikungunya Virus Infection. Front. Cell Infect. Microbiol. 2018;8:345. doi: 10.3389/fcimb.2018.00345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tharmarajah, K., Mahalingam, S., Zaid, A., 2017. Chikungunya: vaccines and therapeutics. F1000Res 6, 2114. https://doi.org/10.12688/f1000research.12461.1. [DOI] [PMC free article] [PubMed]
- Thiberville S.D., Moyen N., Dupuis-Maguiraga L., Nougairede A., Gould E.A., Roques P., de Lamballerie X. Chikungunya fever: epidemiology, clinical syndrome, pathogenesis and therapy. Antiviral Res. 2013;99:345–370. doi: 10.1016/J.ANTIVIRAL.2013.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Voorhis W.C., Adams J.H., Adelfio R., Ahyong V., Akabas M.H., Alano P., Alday A. Open Source Drug Discovery with the Malaria Box Compound Collection for Neglected Diseases and Beyond. PLoS Pathog. 2016;12 doi: 10.1371/journal.ppat.1005763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Wang S., Cheng L., Chen L., Wang Yongguang, Qing J., Huang S., Wang Yuanhao, Lei X., Wu Y., Ma Z., Zhang L., Tang Y. Discovery of Imidazo[1,2-α][1,8]naphthyridine Derivatives as Potential HCV Entry Inhibitor. ACS Med. Chem. Lett. 2015;6:977–981. doi: 10.1021/acsmedchemlett.5b00159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Roy A., Zhang Y. Protein-ligand binding site recognition using complementary binding-specific substructure comparison and sequence profile alignment. Bioinformatics. 2013;29:2588–2595. doi: 10.1093/bioinformatics/btt447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying Z.H., Li H.M., Yu W.Y., Yu C.H. Iridin prevented against lipopolysaccharide-induced inflammatory responses of macrophages via inactivation of pkm2-mediated glycolytic pathways. J. Inflamm. Res. 2021;14:341–354. doi: 10.2147/JIR.S292244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng J., Wu D., Hu H., Young J.A.T., Yan Z., Gao L. Activation of the Liver X Receptor Pathway Inhibits HBV Replication in Primary Human Hepatocytes. Hepatology. 2020;72 doi: 10.1002/hep.31217/suppinfo. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.