Highlights
-
•
Four Tevenvirinae phages infecting Cronobacter and Enterobacter were isolated.
-
•
A point mutation in the Pet-CM3–4 phage adhesin influenced its host specificity.
-
•
Lipopolysaccharide was found to be receptor of both wild-type and mutated phage.
Keywords: Cronobacter, Enterobacter, Phage receptor, Phage adhesin, Long tail fiber
Abstract
Bacteria belonging to Cronobacter and Enterobacter genera are opportunistic pathogens responsible for infections in immunocompromised patients including neonates. Phage therapy offers a safe method for pathogen elimination, however, phages must be well characterized before application. In the present study we isolated four closely related bacteriophages from the subfamily Tevenvirinae infecting Cronobacter and Enterobacter strains. Bacteriophage Pet-CM3–4 which was isolated on C. malonaticus strain possessed broader host specificity than other three phages with primary Enterobacter hosts. Based on genome sequences all these phages have been assigned to the genus Karamvirus. We also studied factors influencing the host specificity of Pet-CM3–4 phage and its host range mutant Pet-CM3–1 and observed that a lysine to glutamine substitution in the long tail fiber adhesin was the reason of the Pet-CM3–1 reduced host specificity. By characterization of phage-resistant mutants from transposon library of C. malonaticus KMB-72 strain we identified that LPS is the receptor of both phages. C. malonaticus O:3 antigen is the receptor of Pet-CM3–1 phage and the Pet-CM3–4 phage binds to structures of the LPS core region. Obtained results will contribute to our understanding of biology and evolution of Tevenvirinae phages.
Abbreviations
- ANI
average nucleotide identity
- EOP
efficiency of plating
- LPS
lipopolysaccharide
- ECA
enterobacterial common antigen
1. Introduction
The genus Cronobacter belongs to the family Enterobacteriaceae and currently contains seven species (Iversen et al., 2007; Joseph et al., 2012). Members of the genus are opportunistic pathogens that can cause serious infections in neonates, including meningitis, necrotising enterocolitis and sepsis with low frequency, but high lethality rate (Holy and Forsythe, 2014). Cronobacter can also infect adults in particular the elderly and immunocompromised patients, but infections have generally milder manifestation compared to disease in new-borns (Alsonosi et al., 2015). Cronobacter is ubiquitous and has been isolated from various foods and environments (Turcovsky et al., 2011; Ueda, 2017). Main vehicle for Cronobacter transmission in neonatal infections is rehydrated powdered infant formula and bacterial contamination can be caused by its improper storage during the manufacture process (Henry and Fouladkhah, 2019).
Bacteria belonging to Enterobacter cloacae complex are common nosocomial pathogens capable of producing a wide variety of infections, such as pneumonia, urinary tract infections, and septicaemia. Spread of nosocomial multidrug resistant strains, including last-resort carbapenem resistant isolates, represent a major global threat (Annavajhala et al., 2019). Eighteen phylogenomic groups were described in E. cloacae complex, E. cloacae, E. hormaechei and related subspecies remain the most clinically relevant (Chavda et al., 2016).
One alternative to control bacterial pathogens is the application of bacteriophages due to their high specificity and efficiency. However, before application, it is necessary to obtain sufficient information about biocontrol phages to guarantee their safety and reliability (Moye et al., 2018; Ghosh et al., 2019; Altamirano and Barr, 2019). Presently, several Cronobacter and Enterobacter phages with sequenced genomes have been reported and their application for food decontamination was experimentally tested (Zuber et al., 2008; Kajsik et al., 2014; Endersen et al., 2017; Kajsik et al., 2019; Gibson et al., 2019).
Bacteriophages from the subfamily Tevenvirinae have been abundantly isolated on several bacterial hosts from Enterobacteriaceae family. They are a diverse group of lytic bacterial myoviruses that share genetic homologies and morphological similarities with the well-studied coliphage T4 (Petrov et al., 2010; Trojet et al., 2011; Grose and Casjens, 2014). Due to their high lytic potential, they are the accomplished candidates for the phage therapy (Gibson et al., 2019; Pham-Khanh et al., 2019).
Among other properties members of the Tevenvirinae subfamily differ in the host range (Gibson et al., 2019; Grose and Casjens, 2014). This is mainly determined by adsorption, the first step in the phage life cycle. The long tail fiber adhesins are the primary determinants of the host range in the Tevenvirinae phages and these proteins show high degree of variability between closely related phages. The dominant form of the phage adhesin in Tevenvirinae is a small protein gp38, but the distal tip of gp37 serves this purpose for T4 phage (Trojet et al., 2011; Suga et al., 2021). Several outer membrane proteins could be recognised as receptors by Tevenvirinae phages such as OmpA, OmpF, OmpC, Tsx, FadL and FhuA. Lipopolysaccharides (LPS) and bacterial capsule are also frequently used for attachment of various phages including Tevenvirinae members (Trojet et al., 2011; Wang et al., 2021; Gordillo Altamirano et al., 2021).
In this study, we describe four related Tevenvirinae bacteriophages infecting Cronobacter and Enterobacter strains, which were newly isolated from sewage. We studied factors influencing the host specificity, in particular we isolated phages with mutated tail fiber adhesins possessing narrowed host spectrum and showed that bacterial LPS is a probable phage receptor. These data are fundamental for the phage application in phage therapy and in food control.
2. Materials and methods
2.1. Bacterial strains and growth conditions
Cronobacter and Enterobacter strains from collections of Nottingham Trent University, Belgian Coordinated Collections of Microorganism or Czech Collection of Microorganism or isolated previously (Turcovsky et al., 2011; Vojkovska et al., 2016; Kadlicekova et al., 2018) were used and are listed in Table 1. Luria-Bertani (LB) broth and LB agar were used for bacterial cultivation.
Table 1.
Host range analysis of Pet-CM3–4 related phages.
| Strain | Pet-CM3–1 | Pet-CM3–4 | vKMB17 | vKMB19 | vKMB20 |
|---|---|---|---|---|---|
| C. condimenti KMB-130 | – | ++ | – | – | + (P) |
| C. dublinensis KMB-14 | – | + (P) | + (P) | – | + (P) |
| C. malonaticus KMB-17 | – | ++ (P) | – | – | – |
| C. malonaticus KMB-211 | – | + | – | – | – |
| C. malonaticus KMB-72 | ++ (P) | ++ (P) | – | ++ (P) | + (P) |
| C. muytjensii ATCC 51,329 | ++ (P) | ++ (P) | – | – | – |
| C. sakazakii BAA-894 | – | ++ (P) | – | – | – |
| C. sakazakii KMB-104 | – | ++ | – | – | – |
| C. sakazakii KMB-121 | – | ++ (P) | – | – | – |
| C. sakazakii ATCC 29,544 | – | ++ (P) | – | – | – |
| C. sakazakii KMB-203 | – | ++ | – | – | – |
| C. sakazakii KMB-204 | – | + | – | – | + (P) |
| C. turicensis KMB-131 | – | ++ | – | – | – |
| C. turicensis KMB-539 | – | ++ | – | – | + (P) |
| C. universalis KMB-126 | – | + (P) | – | – | – |
| E. asburiae KMB-219 | – | ++ | ++ (P) | – | ++ |
| E. asburiae KMB-220 | ++ (P) | ++ (P) | ++ (P) | ++ | ++ (P) |
| E. cloaceae KMB-221 | – | + (P) | – | – | – |
| E. cloaceae CCM 2320 | ++ (P) | ++ (P) | – | ++ (P) | – |
| E. hormaechei KMB-223 | – | ++ | – | – | – |
| E. hormaechei KMB-224 | – | ++ | – | – | + (P) |
| E. hormaechei KMB-243 | ++ (P) | ++ (P) | – | ++ (P) | – |
| E. hormaechei KMB-244 | ++ (P) | ++ (P) | – | ++ (P) | – |
| E. hormaechei KMB-245 | – | ++ | – | ++ | – |
| E. hormaechei KMB-246 | ++ (P) | ++ (P) | ++ (P) | ++ (P) | ++ (P) |
| E. hormaechei KMB-247 | ++ (P) | ++ (P) | – | ++ (P) | – |
| E. hormaechei KMB-261 | – | ++ | ++ (P) | – | ++ (P) |
| E. hormaechei KMB-265 | ++ (P) | + (P) | – | ++ (P) | – |
| E. hormaechei KMB-268 | – | ++ | – | – | – |
| E. hormaechei KMB-270 | – | ++ | – | – | – |
| E. hormaechei KMB-536 | ++ (P) | ++ (P) | – | ++ (P) | – |
| E. kobei CCM 1903 | ++ (P) | ++ (P) | ++ (P) | ++ (P) | ++ (P) |
| E. ludwigii KMB-686 | ++ (P) | ++ (P) | – | + (P) | – |
| E. ludwigii KMB-692 | ++ (P) | + (P) | – | ++ (P) | + (P) |
| E. ludwigii KMB-695 | ++ (P) | ++ (P) | + (P) | ++ (P) | + (P) |
| E. cloacae KMB-691 | ++ (P) | ++ | – | + (P) | – |
| E. cancerogenus CCM 2421 | – | – | ++ | ++ | ++ (P) |
| K. aerogenes CCM 2531 | – | – | – | – | – |
| P. gergoviae CCM 3459 | – | – | – | – | – |
Lysis in spots: ++: comparable to indicator strain (highlighted); +: more than 2 log reduced lysis compared with indicator strain; -: no lysis observed; (P): plaques observed.
2.2. Isolation of bacteriophages
Pet-CM3–4, vB-EclM_KMB17, vB-EclM_KMB19 and vB_EclM_KMB20 (in short Pet-CM3–4, vKMB17, vKMB19 and vKMB20) bacteriophages were isolated from wastewater on indicators stated in Table 1. 10 ml of wastewater was sterilized by 20 µm filtration and mixed with the same volume of two-fold concentrated LB medium and 200 µl overnight bacterial culture. Mixture was cultivated overnight at 37 °C with shaking. Phages were purified by three repeated isolations from single plaques on double agar and amplified in liquid cultures. Bacteriophage Pet-CM3–1 was unintentionally isolated as a spontaneous mutant of Pet-CM3–4 phage during cultivation on LB agar.
2.3. Determination of phage titer, growth curve and phage adsorption
The phage titer was determined by plaque assay as previously described (Kajsik et al., 2014). Briefly, 200 μl overnight bacterial culture supplemented with 10 µl 1 M CaCl2 and 10 µl 1 M MgCl2 was mixed with 5 ml top agar (0.2% peptone, 0.7% NaCl and 0.7% agar) and poured on LB agar plate. 10 μl phage lysate dilutions (102 −109 PFU/ml) were spotted onto the plate and incubated overnight. The phage host range was tested by plaque assay on strains listed in Table 1. EOP was calculated based by using references: C. malonaticus KMB-72 for Pet-CM3–1 and Pet-CM3–4 phages; E. asburiae KMB-220 for vKMB17 and vKMB20 phages; and E. hormaechei KMB-536 for vKMB19 phage.
The one step growth curve was measured by adding 100 μl 107 PFU/ml phage lysate to 10 ml exponentially grown bacterial culture. Phages were allowed to adsorb for 10 min at 37 °C. The mixture was then centrifuged, and the pellet was resuspended in 10 ml prewarmed LB. At this time a number of all phages (adsorbed and unadsorbed) was determined by double agar method. Then 1 ml samples were taken every 5 min into tube containing 50 µl chloroform, vortexed and the number of phages was counted by plaque assay.
The phage adsorption was measured in liquid cultures (Kajsik et al., 2014). 20 µl 108 PFU/ml phage suspension was added to 180 µl overnight bacterial culture and adsorbed 10 min at 37 °C. Subsequently, 10 μl sample was diluted into 1 ml SM buffer (100 mM NaCl; 8 mM MgSO4; 50 mM Tris–HCl, pH 7.5; 0.002% gelatine), adsorbed phages were removed by centrifugation and unadsorbed phages were counted by plaque assay. The measurements were repeated in triplicates. Significance of the results was determined by Student´s t-test.
Growth inhibition of Croobacter strains in the presence of bacteriophages was measured in 96-well microtiter plates by using 200 µl medium inoculated with diluted overnight bacterial culture (final OD600=0.1, approx. 107 CFU/ml) and a phage preparation (104 PFU/ml and 108 PFU/ml). Plates were incubated at 37 °C and the turbidity was measured every hour on Varioskan multimode microplate reader (Thermo Fisher, USA).
2.4. Isolation of DNA, genome sequencing and bioinformatics
Phage DNA was purified using a Phage DNA Isolation Kit (Norgen Biotek, Thorold, Ontario, Canada). A DNA fragment library was prepared using a Nextera kit (Illumina, San Diego, CA, USA). Paired-end sequencing with 2 × 150 bp reads was carried out on a MiSeq system (Illumina). De novo assembly into contigs was carried out on all reads using SPAdes (Bankevich et al., 2012). The genome was annotated using RAST (http://rast.nmpdr.org/) (Aziz et al., 2008) and PATRIC server (https://www.bv-brc.org/) (Wattam et al., 2017). The closest relatives of sequenced contigs were found using BLASTn to search the GenBank database and sequences were analysed manually using Geneious version 11.1.5 (Biomatters Ltd., Auckland, New Zealand). Phylogenomic tree was made in Victor (http://ggdc.dsmz.de/victor.php) (Meier-Kolthoff et al., 2017). The presence of resistance-encoding genes or virulence genes was detected using PATRIC server. Phage genomes were aligned and visualized in Easyfig 2.2.2 (Sullivan et al., 2011). Average Nucleotide Identity (ANI) was calculated by ANI calculator (https://www.ezbiocloud.net/tools/ani) (Yoon et al., 2017). Accession numbers of phage genomes are LT614807.1, OL849997, OL828290 and OL828291.
Bacterial DNA was isolated using a DNeasy Blood & Tissue Kit (Qiagen). The DNA library, next-generation sequencing and sequence analyses were performed as in the case of phage sequencing. The genomes were deposited into Cronobacter MLST database (https://pubmlst.org/organisms/cronobacter-spp) under accessions: C. malonaticus KMB-72 (ID:1763) and C. malonaticus KMB-33 (ID:3392).
2.5. Construction of C. malonaticus transposon library
A transposon mutagenesis library from the C. malonaticus KMB-72 strain was constructed by using the EZ-Tn5™<DHFR-1>Tnp Transposome™ Kit (epicentre, Illumina). Briefly, one microliter of transposon DNA was added to 50 μl of electrocompetent cells (efficiency >109 CFU/μg). After electroporation at 25 μF capacitance, 2,5 kV voltage and 200 Ω resistance, cells were recovered in LB medium at 37 °C with shaking at 250 rpm for 1 h. To select for transposon insertion clones, mixture was concentrated by centrifugation, spread onto LB agar plates containing 50 μg/ml trimethoprim, and incubated for 24 h at 37 °C. Grown colonies were washed down in 1 ml LB medium and the whole library was stored in 15% glycerol at −80 °C.
2.6. Selection of phage resistant mutants
100 µl of overnight culture after transposition (OD600 1) and 100 µl of either Pet-CM3–1 or Pet-CM3–4 phage (108 CFU/ml) was added to 800 µl LB medium. Mixture was incubated by shaking at 180 rpm and 37 °C. Samples were collected after 0, 1, 2, 3, 4, 5, 6, 8, 24 h, diluted and spread on selective LB medium containing trimethoprim. Colonies from the plate with the largest growth inhibition were selected and tested for the phage resistance.
2.7. Determination of transposon integration sites
The transposon insertion sites were identified by modified single-primer PCR, as described elsewhere (Alvarez-Ordonez et al., 2014). Single-primer PCR was performed using either TnPCR-F or TnPCR-R primers under the following conditions: 2 min at 94 °C; 30 cycles of 94 °C for 15 s, 60 °C for 30 s, 72 °C for 2 min; 30 cycles of 94 °C for 15 s, 40 °C for 30 s, 72 °C for 2 min; 30 cycles of 94 °C for 15 s, 60 °C for 30 s, 72 °C for 2 min; 7 min at 72 °C. PCR products were purified and Sanger sequenced using either DHFR-F or DHFR-R primers. Sequences were mapped to C. malonaticus KMB-72 genome and on GenBank database (NCBI). Alternatively, transposon integration site was determined by whole genome sequencing.
2.8. Construction of complementation plasmids
Construction of pBAD-waaL and pBAD-wzzE: The vector pBAD-gfp (Crameri et al., 1996) was used as a template in PCR with primers delGFP-F/delGFP-R and Phusion High-Fidelity DNA Polymerase. The PCR product was ligated, resulting in pBADdelGFP plasmid. The waaL and wzzE inserts were amplified from C. malonaticus KMB-72 genomic DNA by primers waaLF/waaLR and wzzEF/wzzER. PCR products were digested with EcoRI/KpnI and KpnI/SphI respectively and cloned into pBADdelGFP digested with the same enzymes. The ligation mixtures were transformed into E. coli DH5α.
Construction of pACYC-waa: the waa operon was amplified from C. malonaticus KMB-72 genomic DNA using primers IF-waa-117F and IF-waa-5250R. Vector DNA (origin, Cm cassette) was amplified from the pACYCDuet1 (Novagen) with primers Vec-pACYCf and Vec-pACYCr. The In-Fusion® HD Cloning Plus Kit (Takara) was used for direct joining of the resulting fragments into recombinant pACYC-waa plasmid.
All primers used in the study are listed in Table S1.
3. Results
3.1. Bacteriophage isolation and the host specificity
In this study, four bacteriophages were isolated from sewage. Pet-CM3–4 bacteriophage grown on C. malonaticus KMB-72 possessed the broadest host specificity. Three other related phages (vKMB17, vKMB19 and vKMB20) were isolated on Enterobacter strains. Pet-CM3–1 bacteriophage was unintentionally isolated during Pet-CM3–4 purification when the phage was purified from one plaque and the reduced host specificity of the preparation was later noticed.
The phage host specificity was tested on collection of 15 Cronobacter and 22 Enterobacter strains (Table 1). The zones of lysis were observed in all Cronobacter and 95% Enterobacter strains infected by Pet-CM3–4 phage, but single plaques were observed only in 53% Cronobacter and 59% Enterobacter strains. The vKMB17, vKMB19 and vKMB20 phages had substantially narrower host spectrum as lysis was present in 7–33% Cronobacter and 32–68% Enterobacter strains in tested set.
The mutant Pet-CM3–1 phage possessed narrowed host spectrum comparing with Pet-CM3–4 as only 13% Cronobacter and 59% E. cloaceae strains were infected by this phage (Table 1). We also observed faster formation of phage-resistant bacteria during Pet-CM3–1 infection of sensitive C. malonaticus KMB-72 strain compared to Pet-CM3–4 phage (Table S2, Fig. S1).
All phages formed small clear plaques on double agar plates (Fig. S2), Pet-CM3–1 mutant produced slightly bigger plaques then the other phages (Table S3). The one step growth curve of all phages showed similar paramethers, the life cycle was rather short (20 min latent period and 30–35 min whole cycle) with relatively small burst size of 17–61 phages per bacterial cell (Table S3, Fig. S3).
3.2. Genome sequence of the phages
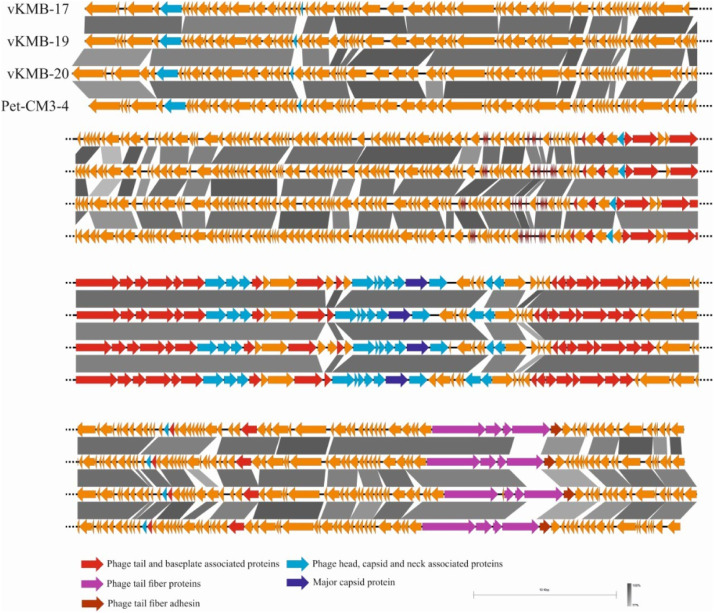
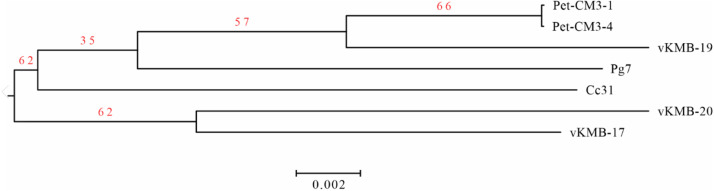
DNA sequence from the Pet-CM3–4 phage contained 172 060 bp and 39.8% GC pairs. Annotation detected 284 protein coding genes (orfs) and 19 tRNA genes. The vKMB17, vKMB19 and vKMB20 phages showed high DNA similarity and colinear genomes to Pet-CM3–4 (Fig. 1). We found two relative phage genomes deposited in GenBank database; CC31 (GU323318.1) and PG7 (KJ101592.1). The Pet-CM3–4 phage showed 83% and 89% similarity at DNA level and shared 252 (89%) and 262 (92%) common proteins with the CC31 and PG7 phages respectively. The vKMB17, vKMB19 and vKMB20 had average nucleotide similarity 82%, 94% and 79% to Pet-CM3–4 DNA and shared 87%, 94% and 85% ORFs whereas genes unique to particular phages encoded mostly hypothetical proteins (Fig. 1). Genome based tree separated phages into two clusters, the first one contained Pet-CM3–4 with vKMB19, the vKMB17 and vKMB20 phages created the second cluster and the reference phages were localized on separate branches (Fig. 2). According to these analyses the newly isolated phages have been assigned to the genus Karamvirus of the subfamily Tevenvirinae.
Fig. 1.
Genome comparison of Pet-CM3–4, vB-EclM_KMB17, vB-EclM_KMB19 and vB-EclM_KMB20 phages. Analysis and visualization were performed in Easyfig.
Fig. 2.
Phylogenomic tree of Pet-CM3–4 related phages. Phage genomes were compared by Genome-BLAST Distance Phylogeny (GBDP) method by Victor web resource (http://ggdc.dsmz.de/victor.php). The tree is based on amino acid comparison using the formula D4. Sequences of CC31(GU323318.1) and PG7 (KJ101592.1) phages were used as references.
The isolated phages contained no genes encoding for antibiotic resistance and virulece factors which could potentially enhance fitness of its bacterial hosts. In accordance with Tevenvirinae strictly virulent life mode, no integrases and lysogeny modules were present.
3.3. Tail fiber adhesins of Pet-CM3–4 phage
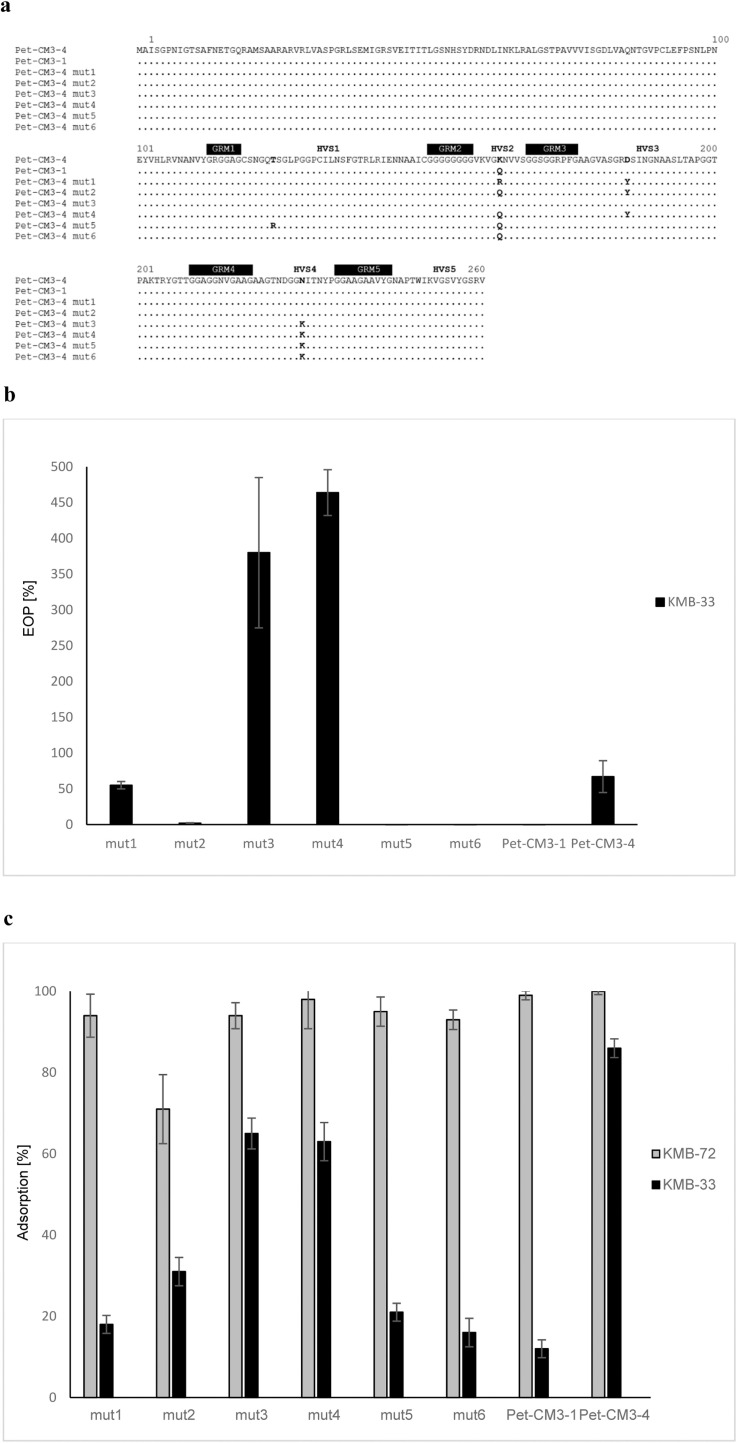
In the next part of the study we compared genomes of Pet-CM3–4 and its host range mutant Pet-CM3–1 in greater detail. Six missense mutations were revealed localized in gp6 (hypothetical protein), gp50 (recombination-related endonuclease), gp165 (baseplate wedge subunit and tail pin), gp167 (short tail fiber protein), gp171 (proximal tail sheath stabilization protein) and gp264 (tail fiber adhesin). Based on gene functions we proposed that the K163Q substitution in gp264 gene encoding for the long tail fiber adhesin was a source of the reduced Pet-CM3–1 host specificity.
To confirm this hypothesis, we compared infection of Pet-CM3–1 and Pet-CM3–4 phages on two strains; C. malonaticus KMB-72 which was used for the phage amplification and C. malonaticus KMB-33 which was sensitive to Pet-CM3–4 phage but resistant to Pet-CM3–1 infection when measured on double agar plates. Both strains possessed quite similar genomes with approximately 91% homologous proteins and 98.22% ANI value, but differed in their serotype.
We observed that overnight incubation of the Pet-CM3–1 with C. malonaticus KMB-33 in liquid medium repeatedly produced a small fraction of phages which were able to infect this strain. We isolated six phage mutants and sequenced their gp264 genes. We detected 1–3 amino acid substitutions in hypervariable regions of gp264 in each mutant (Fig. 3a). All mutants together with Pet-CM3–4 and Pet-CM3–1 phages were able to infect C. malonaticus KMB-72, but they differed in their ability to form plaques on C. malonaticus KMB-33; three possessed similar or higher EOP (efficiency of plating) values as Pet-CM3–4, one showed reduced level of EOP and two mutants did not form any plaques on C. malonaticus KMB-33 (Fig. 3b). Mutants differing in gp264 sequence displayed various adsorption to the host cells which mostly correlated with the plaque formation ability. A higher adsorption to C. malonaticus KMB-72 (71–100%) comparing to C. malonaticus KMB-33 (12–86%) was observed in all tested phages and the original Pet-CM3–4 phage possessed highest adsorption to both tested strains (Fig. 3c).
Fig. 3.
Comparison of Pet-CM3–4 and its gp264 long tail fiber adhesin mutants. Comparison of gp264 sequences (GRM – glycine-rich motifs, HVS – hypervariable sequences) (a), EOP of mutant phages on C. malonaticus KMB-33 comparing to C. malonaticus KMB-72 (b), Adsorption of mutant phages on C. malonaticus KMB-72 and C. malonaticus KMB-33 (c).
3.4. Bacterial receptors of Pet-CM3–1 and Pet-CM3–4 phages
3.4.1. Selection of phage resistant mutants
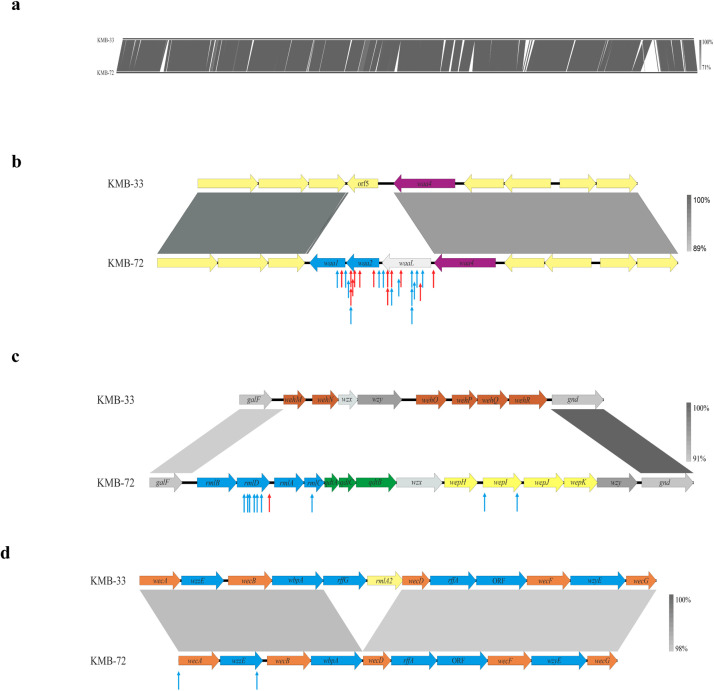
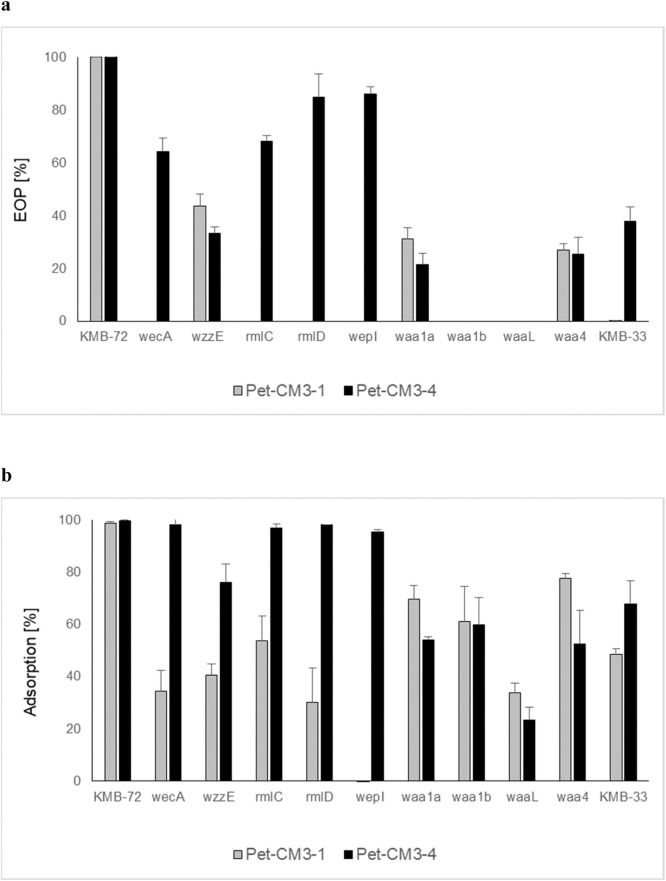
The bacterial receptors involved in adsorbtion of Pet-CM3–1 and Pet-CM3–4 phages were analyzed by using a transposon mutant library of the C. malonaticus KMB-72 strain. Aproximately 8000 clones with randomly inserted EZ-Tn5™ transposon were prepared and phage-resistant mutants were selected from pooled library in liquid media after Pet-CM3–4 or Pet-CM3–1 challenge. Next, the transposon integration sites were determined in randomly selected clones. We observed high frequency of the transposition into an operon consisting of four genes encoding for putative glycosyltransferases and O-antigen ligase involved in LPS biosynthesis (designated waa operon). Overall 27 mutants selected under both Pet-CM3–4 and Pet-CM3–1 selection contained transposon at this site. Glycosyltransferase waa2 and O-antigen ligase waaL were preferential transposon integration sites in this region (Fig. 4b). Eleven mutants contained transposon localized in rfb operon encoding synthesis of O-antigen polysaccharide, ten from these mutants were isolated under Pet-CM3–1 selection. Transposons were inserted in l-rhamnose biosynthesis (rmlD, rmlC) and in rhamnosyltransferase (wepI, Fig. 4c) genes. Two clones selected under Pet-CM3–1 phage infection contained transposon integrated in enterobacterial common antigen (ECA) gene cluster, namely wecA gene encoding for the undecaprenyl-phosphate-GlcNAc-phosphate transferase and wzzE gene encoding for the putative regulator of polysaccharide length (Fig. 4d). The remaining twelve integration sites were localized in several transcription regulators, transporters and membrane associated genes as well as genes encoding for metabolic enzymes.
Fig. 4.
Comparison of C. malonaticus KMB-72 and C. malonaticus KMB-33 genomes and transposon insertion sites in C. malonaticus KMB-72 mutants isolated under phage infection. Comparison of whole KMB-72 and KMB-33 genomes (a), comparison of waa operon encoding for two glycosyltransferases and O-antigen ligase in C. malonaticus KMB-72 with corresponding region of C. malonaticus KMB-33 (b), comparison of the rfb gene cluster (c) and comparison of the enterobacterial common antigen operon (d). Transposon sites of mutants selected under Pet-CM3–4 (red arrows) and under Pet-CM3–1 (blue arrows) phages are shown.
3.4.2. Characterization of phage resistant mutants
Selected mutants with inactivated waa, rfb or ECA genes were characterized by EOP and adsorption assays. We observed that the majority of mutants were still sensitive to Pet-CM3–4 infection although with slighty decreased EOP. The only two strains resistant to Pet-CM3–4 phage contained mutations localized in waa operon. In adition to waa mutants Pet-CM3–1 infection was also protected by transposon insertion in the rfb operon and wecA gene from ECA operon (Fig. 5a). Importance of rfb cluster for Pet-CM3–1 infection was supported by inability of this phage to infect C. malonaticus KMB-33 strain which had serotype CMa-O:2 comparing to CMa-O:3 serotype of C. malonaticus KMB-72.
Fig. 5.
EOP (a) and phage adsorption (b) in C. malonaticus KMB-72 transposon mutants.
In accordance with the decreased EOP, Pet-CM3–4 and Pet-CM3–1 phages showed significantly reduced adsorbtion to transposon mutants: Pet-CM3–4 to wzzE and waa genes and Pet-CM3–1 to all tested mutants (Fig. 5b).
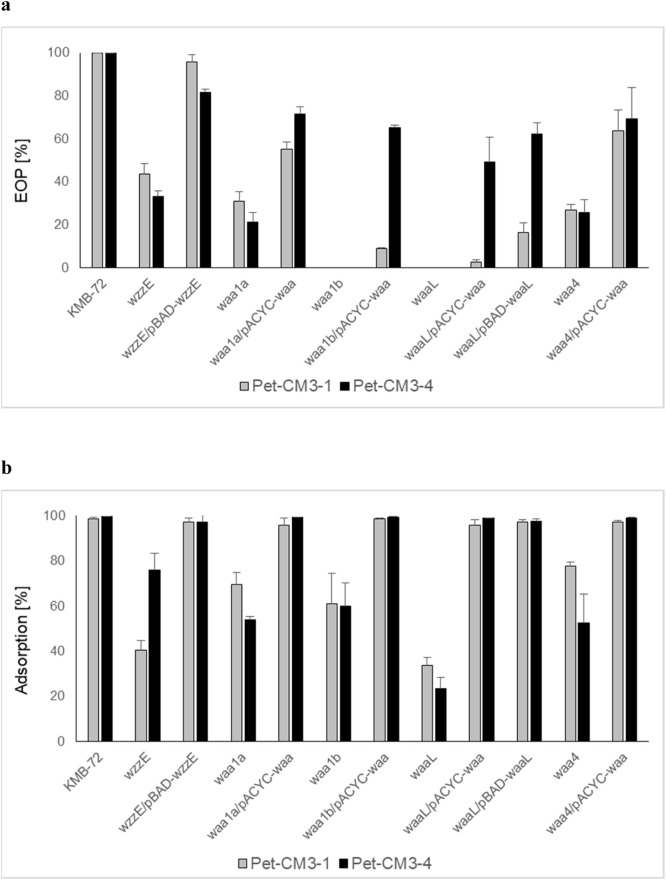
We used plasmid complementation to confirm the function of transposon inactivated genes in the phage sensitivity. We observed increased EOP values in complementation mutants, though their EOP reached only 2–70% values of the wilde type strain (Fig. 6a). The almost complete restoring of phage adsorption was detected in waa mutants after complementation of the waa operon present on pACYC-waa plasmid (Fig. 6b). Similar results were also obtained by complementation of waaL mutant with the plasmid encoding waaL gene. Good complementation of adsorption as well as EOP was reached by wzzE mutant complemented with a wzzE plasmid (Fig. 6b).
Fig. 6.
Complementation assay of EOP (a) and phage adsorption (b) in C. malonaticus KMB-72 transposon mutants.
4. Discussion
Infections caused by Enterobacter and Cronobacter strains pose serious health risks for immunocompromised individuals including neonates. Bacteriophages offer a safe approach for phage therapy and for eliminating pathogens in food. However, before being applied, selected bacteriophages must be characterized in detail to ensure safe and reliable effects (Moye et al., 2018; Ghosh et al., 2019).
In the present study, we characterized four closely related bacteriophages infecting Enterobacter and Cronobacter. The phages belonged to the subfamily Tevenvirinae which is extremely widespread group currently containing eleven genera and its members differ significantly in their host range (Grose and Casjens, 2014; Adriaenssens et al., 2018). The newly isolated phages showed high genome similarity to Escherichia phage CC31 and Enterobacter phage PG7, two previously described phages of Karamvirus genus (Petrov et al., 2010; Grose and Casjens, 2014).
We observed that the isolated phages greatly differed in their host specificity as phages primarily isolated on Enterobacter hosts (vKMB17, vKMB19 and vKMB20) showed narrower host range comparing to the Pet-CM3–4 phage which was isolated on a C. malonaticus strain (Table 1). However, Pet-CM3–1 phage, a spontaneous mutant of Pet-CM3–4 possessing reduced host specificity especially against Cronobacter and in a lesser extent also against Enterobacter was isolated during laboratory cultivations.
By comparative genome analysis of Pet-CM3–1 and Pet-CM3–4 phages six missense mutations were found, four of them were localized in tail structures. Adhesin of Tevenvirinae phages is localized on the distal tip of long tail fibers and it is encoded by C-terminal part of gp37 gene in T4 phage but in adjacent gp38 gene in T2 phage, this organization is present also in Pet-CM3–4 (Trojet et al., 2011; Bartual et al., 2010). Many studies proved that minor mutations in variable regions of adhesins changed the host range of Tevenvirinae phages by altered adsorption (Trojet et al., 2011; Chen et al., 2017; Pham-Khanh et al., 2019, Suga et al., 2021; Salem et al., 2021). Therefore, we proposed that lysine to glutamine substitution in the long tail fiber adhesin was the reason of the Pet-CM3–1 reduced host specificity. This observation was confirmed by significantly reduced adsorption of Pet-CM3–1 to C. malonaticus KMB-33 and by presence of adhesin mutations in phages with reverted ability to infect this strain (Fig. 3). Positively charged amino acid residues in hypervariable regions are probably important for adhesion to C. malonaticus KMB-33 as mutants containing lysine or arginine at position 163 and one mutant with lysine at site 229 showed the highest adsorption to this strain. Our results correspond to observations in other studies that single amino acid mutations in hypervariable regions of adhesins of Tevenvirinae phages are able to substantially change their host specificity (Trojet et al., 2011; Chen et al., 2017; Suga et al., 2021). It was also shown that hypervariable regions on S16 phage adhesin have a net preference for polar and aromatic amino acids, possibly involved in making high-affinity contacts with the bacterial receptor (Dunne et al., 2018).
In the next part of the study we focused on identification of bacterial receptors of Pet-CM3–1 and Pet-CM3–4 phages by characterization of phage-resistant mutants from transposon library of C. malonaticus KMB-72. Several outer membrane proteins and LPS were described as bacterial receptors of Tevenvirinae phages and some phages recognize more than one host receptor, such as T4 that recognizes both OmpC and LPS (Trojet et al., 2011). In our study, most mutants selected under phage presence had transposon localized into waa operon containing four genes encoding for two glycosyltransferases, O-antigen ligase and lipooligosaccharide phosphoetanolamine transferase (Fig. 4b). Genes waa1, waa2 and waaL were absent from C. malonaticus KMB-33 and were present in only 18% Cronobacter genomes in Cronobacter MLST database belonging to several species and serotypes. O-antigen ligase catalyses formation of glycosidic bond between undecaprenyl diphosphate linked O-antigen and teminal saccharide of LPS core oligosaccharide, deletion of this gene leads to formation of rough strains (Ruan et al., 2012). Accordingly to this function we observed absence of O-antigen repeats in LPS from waaL mutant on SDS-PAGE (data not shown). Four mutants with transposon localized in this region were analyzed in detail, all of them adsorbed phages weaker than the wild type strain and two do not form plaques on double agar plates (Fig. 5). LPS core oligosaccharide was detected as receptor also for three Yersinia Tevenvirinae phages (Salem et al., 2021).
Another large group of transposon mutants had interruptions in rfb gene cluster encoding for synthesis of O-antigen polysaccharide belonging to serotype of C. malonaticus O:3 (Sun et al., 2012). Selected phage-resistant mutants had insertions in l-rhamnose biosynthesis (rmlD, rmlC) and in rhamnosyltransferase (wepI, Fig. 4c) genes. By comparing host range, we observed that Pet-CM3–1 phage infect only C. malonaticus O:3 but not C. malonaticus O:2 strains and mutations in rhamnose biosynthesis genes protect host strains from infection. Therefore it is probable that Pet-CM3–1 phage uses rhamnose residues of C. malonaticus O:3 antigen as the attachment sites. On the other hand, Pet-CM3–4 phage infected strains belonging to different serotypes and rfb mutants of C. malonaticus KMB-72, therefore O-antigen is not the receptor of this phage. Rhamnose synthesis cluster is present in eleven of 30 Enterobacter rfb operons described recently including two of the most frequent serotypes which cover around 50% Enterobacter strains (Li et al., 2020), this explains relatively high infectivity of Pet-CM3–1 in these bacteria.
Two transposon mutants contained transposon inserted in ECA gene cluster encoding for an common enterobacterial surface antigen (Fig. 4d). The wecA gene encodes for transferase which catalyze the synthesis of undecaprenyl-N-acetyl-glucosaminyl diphosphate, the first step of biosynthesis in many O-antigens containing GlcNAc (Lehrer et al., 2007). The other gene, wzzE, encodes for regulator of polysaccharide length (Ogrodzki and Forsythe, 2015). Bacteria mutated in these genes possessed reduced phage sensitivity, wecA mutant was resistant only to Pet-CM3–1, but inactivation of wzzE caused increased resitance to both phages (Fig. 5). Based on all our analyses we can conclude that LPS is the major receptor of both phages. C. malonaticus O:3 antigen is the receptor of Pet-CM3–1 phage and the Pet-CM3–4 phage binds to structures of the LPS core region. These results contribute to our understanding of the biology and evolution of Tevenvirinae phages infecting Enterobacter and Cronobacter.
5. Conclusion
Four closely related bacteriophages from the subfamily Tevenvirinae infecting Cronobacter and Enterobacter strains were studied and factors influencing their host specificity were characterized. A single point mutation in the long tail fiber adhesin was observed as the source of substantial lowering of the phage host spectrum as the mutant phage Pet-CM3–1 was able to recognize only one specific O-antigen in comparison with the wild type phage Pet-CM3–4 which bound into the LPS core region present in broader group of Cronobacter and Enterobacter strains.
Author contributions statement
Lucia Oravcova, Veronika Kadličekova, Elham Ozaee: Phage isolation and host specificity characterization. Sulafa Elnwrani: Determination of the phage growth curves. Michal Andrezal and Michal Kajsik: Phage sequencing and genome analyses. Juraj Bugala: Recombinant construction. Hana Drahovska: Conceptualization, manuscript preparation.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper
Acknowledgments
This work was supported by the Research and Development Operational Programme for the project “Sustainable smart farming systems taking into account the future challenges” (SmartFarm, ITMS 313011W112) funded by the European Regional Development Fund and by the Operational Program Integrated Infrastructure for the project: “Research and development of the applicability of autonomous flying vehicles in the fight against the pandemic caused by COVID-19” (project no. 313011ATR9) funded by the European Regional Development Fund.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.virusres.2022.199025.
Appendix. Supplementary materials
References
- Adriaenssens E.M., et al. Taxonomy of prokaryotic viruses: 2017 update from the ICTV Bacterial and Archaeal Viruses Subcommittee. Arch. Virol. 2018;163:1125–1129. doi: 10.1007/s00705-018-3723-z. [DOI] [PubMed] [Google Scholar]
- Alsonosi A., Hariri S., Kajsik M., Orieskova M., Hanulik V., Roderova M., Petrzelova J., Kollarova H., Drahovska H., Forsythe S., Holy O. The speciation and genotyping of Cronobacter isolates from hospitalised patients. Eur. J. Clin. Microbiol. Infect. Dis. 2015;34:1979–1988. doi: 10.1007/s10096-015-2440-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altamirano F.L.G., Barr J.J. Phage therapy in the postantibiotic era. Clin. Microbiol. Rev. 2019;32 doi: 10.1128/CMR.00066-18. e00066-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez-Ordonez A., Begley M., Clifford T., Deasy T., Collins B., Hill C. Transposon mutagenesis reveals genes involved in osmotic stress and drying in Cronobacter sakazakii. Food Res. Int. 2014;55:45–54. doi: 10.1016/j.foodres.2013.10.037. [DOI] [Google Scholar]
- Annavajhala M.K., Gomez-Simmonds A., Uhlemann A.-.C. Multidrug-resistant Enterobacter cloacae complex emerging as a global, diversifying threat. Front Microbiol. 2019;10:44. doi: 10.3389/fmicb.2019.00044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aziz R.K., Bartels D., Best A., DeJongh M., Disz T., Edwards R.A., Formsma K., Gerdes S., Glass E.M., Kubal M., Meyer F., Olsen G.J., Olson R., Osterman A.L., Overbeek R.A., McNeil L.K., Paarmann D., Paczian T., Parrello B., Pusch G.D., Reich C., Stevens R., Vassieva O., Vonstein V., Wilke A., Zagnitko O. The RAST Server: rapid annotations using subsystems technology. BMC Genomics. 2008;9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankevich A., Nurk S., Antipov D., Gurevich A.A., Dvorkin M., Kulikov A.S., Lesin V.M., Nikolenko S.I., Pham S., Prjibelski A.D., Pyshkin A.V., Sirotkin A.V., Vyahhi N., Tesler G., Alekseyev M.A., Pevzner P.A. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartual S.G., Otero J.M., Garcia-Doval C., Llamas-Saiz A.L., Kahn R., Fox G.C., Van Raaij M.J. Structure of the bacteriophage T4 long tail fiber receptor-binding tip. The Proceedings of the National Academy of Sciences. 2010;107:20287–20292. doi: 10.1073/pnas.1011218107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavda K.D., Chen L., Fouts D.E., Sutton G., Brinkac L., Jenkin S.G., Bonomo R.A., Adams M.D., Kreiswirth B.N. Comprehensive genome analysis of carbapenemase-producing Enterobacter spp.: new insights into phylogeny, population structure, and resistance mechanisms. MBio. 2016;7 doi: 10.1128/mBio.02093-16. e02093-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M., Zhang L., Abdelgader S.A., Yu L., Xu J., Yao H., Lu C., Zhang W. Alterations in gp37 expand the host range of a T4-like phage. Appl. Environ. Microbiol. 2017;83 doi: 10.1128/AEM.01576-17. e01576-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crameri A., Whitehorn E.A., Tate E., Stemmer W.P.C. Improved green fluorescent protein by molecular evolution using DNA shuffling. Nat. Biotechnol. 1996;14 doi: 10.1038/nbt0396-315. 315-x1. [DOI] [PubMed] [Google Scholar]
- Dunne M., Denyes J.M., Arndt H., Loessner M.J., Leiman P.G., Klumpp J. Salmonella phage S16 tail fiber adhesin features a rare polyglycine rich domain for host recognition. Structure. 2018;26:1573–1582. doi: 10.1016/j.str.2018.07.017. [DOI] [PubMed] [Google Scholar]
- Endersen L., Buttimer C., Nevin E., Coffey A., Neve H., Oliveira H., Lavigne R., O'Mahony J. Investigating the biocontrol and anti-biofilm potential of a three phage cocktail against Cronobacter sakazakii in different brands of infant formula. Int. J. Food Microbiol. 2017;253:1–11. doi: 10.1016/j.ijfoodmicro.2017.04.009. [DOI] [PubMed] [Google Scholar]
- Ghosh C., Sarkar P., Issa R., Haldar J. Alternatives to conventional antibiotics in the era of antimicrobial resistance. Trends Microbiol. 2019;27(323–338):2019. doi: 10.1016/j.tim.2018.12.010. [DOI] [PubMed] [Google Scholar]
- Gibson S.B., Green S.I., Liu C.G., Salazar K.C., Clark J.R., Terwilliger A.L., Kaplan H.B., Maresso A.W., Trautner B.W., Ramig R.F. Constructing and characterizing bacteriophage libraries for phage therapy of human infections. Front Microbiol. 2019;10:2537. doi: 10.3389/fmicb.2019.02537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordillo Altamirano F., Forsyth J.H., Patwa R., Kostoulias X., Trim M., Subedi D., Archer S.K., Morris F.C., Oliveira C., Kielty L., Korneev D., O'Bryan M.K., Lithgow T.J., Peleg A.Y., Barr J.J. Bacteriophage-resistant Acinetobacter baumannii are resensitized to antimicrobials. Nature Microbiology. 2021;6:157–161. doi: 10.1038/s41564-020-00830-7. [DOI] [PubMed] [Google Scholar]
- Grose J.H., Casjens S.R. Understanding the enormous diversity of bacteriophages: the tailed phages that infect the bacterial family Enterobacteriaceae. Virology. 2014;468:421–443. doi: 10.1016/j.virol.2014.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry M., Fouladkhah A. Outbreak history, biofilm formation, and preventive measures for control of Cronobacter sakazakii in infant formula and infant care settings. Microorganisms. 2019;7:77. doi: 10.3390/microorganisms7030077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holy O., Forsythe S. Cronobacter spp. as emerging causes of healthcare-associated infection. J. Hosp. Infect. 2014;86:169–177. doi: 10.1016/j.jhin.2013.09.011. [DOI] [PubMed] [Google Scholar]
- Iversen C., Lehner A., Mullane N., Bidlas E., Cleenwerck I., Marugg J., Fanning S., Stephan R., Joosten H.C. The taxonomy of Enterobacter sakazakii: proposal of a new genus Cronobacter gen. nov. and descriptions of Cronobacter sakazakii comb. nov. Cronobacter sakazakii subsp. sakazakii, comb. nov., Cronobacter sakazakii subsp. malonaticus subsp. nov., Cronobacter turicensis sp. nov., Cronobacter muytjensii sp. nov., Cronobacter dublinensis sp. nov. and Cronobacter genomospecies 1. BMC Evol. Biol. 2007;7:64. doi: 10.1186/1471-2148-7-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph S., Cetinkaya E., Drahovska H., Levican A., Figueras M.J., Forsythe S.J. Cronobacter condimenti sp. nov., isolated from spiced meat, and Cronobacter universalis sp. nov., a species designation for Cronobacter sp. genomospecies 1, recovered from a leg infection, water and food ingredients. Int. J. Syst. Evol. Microbiol. 2012;62:1277–1283. doi: 10.1099/ijs.0.032292-0. [DOI] [PubMed] [Google Scholar]
- Kadlicekova V., Kajsik M., Soltys K., Szemes T., Slobodnikova L., Janosikova L., Hubenakova Z., Ogrodzki P., Forsythe S., Turna J., Drahovska H. Characterisation of Cronobacter strains isolated from hospitalised adult patients. Antonie Van Leeuwenhoek. 2018;111:1073–1085. doi: 10.1007/s10482-017-1008-2. [DOI] [PubMed] [Google Scholar]
- Kajsik M., Oslanecova L., Szemes T., Hyblova M., Bilkova A., Drahovska H., Turna J. Characterization and genome sequence of Dev2, a new T7-like bacteriophage infecting Cronobacter turicensis. Arch. Virol. 2014;159:3013–3019. doi: 10.1007/s00705-014-2173-5. [DOI] [PubMed] [Google Scholar]
- Kajsik M., Bugala J., Kadlicekova V., Szemes T., Turna J., Drahovska H. Characterization of Dev-CD-23823 and Dev-CT57, new Autographivirinae bacteriophages infecting Cronobacter spp. Arch. Virol. 2019;164:1383–1391. doi: 10.1007/s00705-019-04202-3. [DOI] [PubMed] [Google Scholar]
- Lehrer J., Vigeant K.A., Tatar L.D., Valvano M.A. Functional characterization and membrane topology of Escherichia coli WecA, a sugar-phosphate transferase initiating the biosynthesis of enterobacterial common antigen and O-antigen lipopolysaccharide. J. Bacteriol. 2007;189:2618–2628. doi: 10.1128/JB.01905-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Huang J., Wang X., Xu C., Han T., Guo X. Genetic characterization of the O-Antigen and development of a molecular serotyping scheme for Enterobacter cloacae. Front Microbiol. 2020;11:727. doi: 10.3389/fmicb.2020.00727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier-Kolthoff J.P., Göker M. VICTOR: genome-based phylogeny and classification of prokaryotic viruses. Bioinformatics. 2017;33:3396–3404. doi: 10.1093/bioinformatics/btx440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moye Z.D., Woolston J., Sulakvelidze A. Bacteriophage applications for food production and processing. Viruses. 2018;10:205. doi: 10.3390/v10040205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogrodzki P., Forsythe S. Capsular profiling of the Cronobacter genus and the association of specific Cronobacter sakazakii and C. malonaticus capsule types with neonatal meningitis and necrotizing enterocolitis. BMC Genomics. 2015;16:758. doi: 10.1186/s12864-015-1960-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrov V.M., Ratnayaka S., Nolan J.M., Miller E.S., Karam J.D. Genomes of the T4-related bacteriophages as windows on microbial genome evolution. Virol. J. 2010;7:292. doi: 10.1186/1743-422X-7-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham-Khanh N.H., Sunahara H., Yamadeya H., Sakai M., Nakayama T., Yamamoto H., Truong Thi Bich V., Miyanaga K., Kamei K. Isolation, characterisation and complete genome sequence of a Tequatrovirus phage, Escherichia phage KIT03, which simultaneously infects Escherichia coli O157:H7 and Salmonella enterica. Curr. Microbiol. 2019;76:1130–1137. doi: 10.1007/s00284-019-01738-0. [DOI] [PubMed] [Google Scholar]
- Ruan X., Loyola D.E., Marolda C.L., Perez-Donoso J.M., Valvano M.A. The WaaL O-antigen lipopolysaccharide ligase has features in common with metal ion-independent inverting glycosyltransferases. Glycobiology. 2012;22:288–299. doi: 10.1093/glycob/cwr150. [DOI] [PubMed] [Google Scholar]
- Salem M., Pajunen M.I., Jun J.W., Skurnik M. T4-like bacteriophages isolated from pig stools infect Yersinia pseudotuberculosis and Yersinia pestis using LPS and OmpF as receptors. Viruses. 2021;13:296. doi: 10.3390/v13020296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suga A., Kawaguchi M., Yonesaki T., Otsuka Y. Manipulating interactions between T4 phage long tail fibers and Escherichia coli receptors. Appl. Environ. Microbiol. 2021;87:1–17. doi: 10.1128/AEM.00423-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan M.J., Petty N.K., Beatson S.A. Easyfig: a genome comparison visualizer. Bioinformatics. 2011;27:btr039. doi: 10.1093/bioinformatics/btr039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., Wang M., Wang Q., Cao B., He X., Li K., Feng L., Wang L. Genetic analysis of the Cronobacter sakazakii O4 to O7 O-antigen gene clusters and development of a PCR assay for identification of all C. sakazakii O serotypes. Appl. Environ. Microbiol. 2012;78:3966–3974. doi: 10.1128/AEM.07825-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trojet S.N., Caumont-Sarcos A., Perrody E., Comeau A.M., Krisch H.M. The gp38 adhesins of the T4 superfamily: a complex modular determinant of the phage's host specificity. Genome Biol Evol. 2011;3:674–686. doi: 10.1093/gbe/evr059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turcovsky I., Kunikova K., Drahovska H., Kaclikova E. Biochemical and molecular characterization of Cronobacter spp. (formerly Enterobacter sakazakii) isolated from foods. Antonie Van Leeuwenhoek. 2011;99:257–269. doi: 10.1007/s10482-010-9484-7. [DOI] [PubMed] [Google Scholar]
- Ueda S. Occurrence of Cronobacter spp. in dried foods, fresh vegetables and soil. Biocontrol Sci. 2017;22:55–59. doi: 10.4265/bio.22.55. [DOI] [PubMed] [Google Scholar]
- Vojkovska H., Karpiskova R., Orieskova M., Drahovska H. Characterization of Cronobacter spp. isolated from food of plant origin and environmental samples collected from farms and from supermarkets in the Czech Republic. Int. J. Food Microbiol. 2016;217:130–136. doi: 10.1016/j.ijfoodmicro.2015.10.017. [DOI] [PubMed] [Google Scholar]
- Wang X., Loh B., Gordillo Altamirano F., Yu Y., Hua X., Leptihn S. Colistin-phage combinations decrease antibiotic resistance in Acinetobacter baumannii via changes in envelope architecture. Emerging Microbes and Infections. 2021;10:2205–2219. doi: 10.1080/22221751.2021.2002671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wattam A.R., Davis J.J., Assaf R., Boisvert S., Brettin T., Bun C., Conrad N., Dietrich E.M., Disz T., Gabbard J.L., Gerdes S., Henry C.S., Kenyon R.W., Machi D., Mao C., Nordberg E.K., Olsen G.J., Murphy-Olson D.E., Olson R., Overbeek R., Parrello B., Pusch G.D., Shukla M., Vonstein V., Warren A., Xia F., Yo H., Stevens R.L. Improvements to PATRIC, the all-bacterial bioinformatics database and analysis resource center. Nucleic Acid Research. 2017;45:D535–D542. doi: 10.1093/nar/gkw1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon S.H., Ha S.M., Lim J., Kwon S., Chun J. A large-scale evaluation of algorithms to calculate average nucleotide identity. Antonie Van Leeuwenhoek. 2017;110:1281–1286. doi: 10.1007/s10482-017-0844-4. [DOI] [PubMed] [Google Scholar]
- Zuber S., Boissin-Delaporte C., Michot L., Iversen C., Diep B., Brüssow H., Breeuwer P. Decreasing Enterobacter sakazakii (Cronobacter spp.) food contamination level with bacteriophages: prospects and problems. Microb. Biotechnol. 2008;1:532–543. doi: 10.1111/j.1751-7915.2008.00058.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.