Highlights
-
•
This work includes a ten-year FLUBV surveillance based on WGS.
-
•
Both FLUBV lineages were detected during the study period.
-
•
Phylogenetic analyses revealed that strains belonged to different genetic groups.
-
•
Intra-lineage reassortments in PB1, PB2, NA, and NS segments were detected.
-
•
An inter-lineage reassortment in NP was observed amongst B/Vic viruses from different seasons.
Keywords: Influenza B viruses, Reassortment event, Lineage, Molecular epidemiology
Abstract
Background
Influenza B viruses (FLUBV) have segmented genomes which enables the virus to evolve by segment reassortment. Since the divergence of both FLUBV lineages, B/Victoria/2/87 (FLUBV/VIC) and B/Yamagata/16/88 (FLUBV/YAM), PB2, PB1 and HA have kept the same ancestor, while some reassortment events in the other segments have been reported worldwide. The aim of the present study was to find out reassortment episodes in FLUBV strains detected in cases attended at Hospital Universitari Vall d'Hebron and Hospital de la Santa Creu i Sant Pau (Barcelona, Spain) from 2004 to 2015 seasons.
Methods
From October 2004 to May 2015, respiratory specimens were received from patients with respiratory tract infection suspicion. Influenza detection was carried out by either cell culture isolation, immunofluorescence or PCR-based assays. A RT-PCR was performed to distinguish both lineages by agarose gel electrophoresis. Whole genome amplification was performed using the universal primer set by Zhou et al. in 2012, and subsequently sequenced using Roche 454 GS Junior platform. Bioinformatic analysis was performed to characterise the sequences with B/Malaysia/2506/2007 and B/Florida/4/2006 corresponding sequences as reference of (B/VIC) and (B/YAM), respectively.
Results
A total of 118 FLUBV (75 FLUBV/VIC and 43 FLUBV/YAM), from 2004 to 2006, 2008–2011 and 2012–2015 seasons, were studied. The whole genome of 58 FLUBV/VIC and 42 FLUBV/YAM viruses was successfully amplified. Based on HA sequences, most FLUBV/VIC viruses (37; 64%) belonged to clade 1A (B/Brisbane/60/2008) except to 11 (19%), which fell within clade 1B (B/HongKong/514/2009) and 10 (17%) to B/Malaysia/2506/2004. Nine (20%) FLUBV/YAM viruses belonged to clade 2 (B/Massachusetts/02/2012), 18 (42%) to clade 3 (B/Phuket/3073/2013) and 15 (38%) fell within Florida/4/2006. Numerous intra-lineage reassortments in PB2, PB1, NA and NS were found in 2 2010–2011 viruses. An important inter-lineage reassortment event from 2008 to 2009 (11), 2010–2011 (26) and 2012–2013 (3) FLUBV/VIC (clade 1) strains to FLUBV/YAM (clade 3) was found, in addition to 1 reassortant NS in 2010–2011 B/VIC virus.
Conclusions
Intra- and inter-lineage reassortment episodes were revealed by WGS. While PB2-PB1-HA remained in complex, NP and NS reassortant viruses were found in both lineages. Despite reassorment events are not often, the characterisation only by HA and NA sequences might be underestimating their detection.
1. Background
Influenza viruses (FLUV), as causative agents of respiratory infections, contribute to morbidity and mortality in the community, mainly in patients at high-risk infections such as elderly, those with underlying comorbidities or pregnant women (Tewawong et al., 2017; Medina and García-Sastre 2011).
FLUV are single-stranded, negative-sense, segmented RNA viruses belonging to the Orthomyxoviridae family (Schrauwen et al., 2014; Bouvier and Palese 2008). Three FLUV types (A, B and C), which infect humans, can be described based on genetic and antigenic features and, amongst influenza B viruses (FLUBV), two lineages genetically and antigenically different can be distinguished, B/Victoria/2/87-like (FLUBV/VIC) and B/Yamagata/16/88-like (FLUBV/YAM) (Tewawong et al., 2015).
Both influenza A viruses (FLUAV) and FLUBV evolve through point mutations mainly in the surface glycoproteins (antigenic drift), haemagglutinin (HA) and neuraminidase (NA), to escape the existing human immunity and persist in the population, leading to the recurrent seasonal epidemics (Steinhauer 1999; Bedford et al., 2015). In addition, these viruses also evolve through the exchange of segments (antigenic shift) between clades, subtypes (FLUAV) or lineages (FLUBV) due to its segmented genome. These reassortment events can cause pandemics and FLUAV are considered the major threat over FLUBV owing to their large number of reservoirs compare to FLUBV (Steinhauer and Skehel 2002).
Since its divergence in 1983, both FLUBV lineages have co-circulated in the human population and reassortment episodes, despite being few, have been reported later than 1992 because of lineage mixing (Oong et al., 2017; Dudas et al., 2015). While most of FLUBV segments have suffered some reassortment events as the NA or the non-structural protein (NS), another ones like the polymerase subunits 1 (PB1) and 2 (PB2) or the HA have maintained separate FLUBV/VIC or FLUBV/YAM lineages (Dudas et al., 2015).
Next generation sequencing (NGS) should help to recognise reassortant FLUBV strains through the study of the whole viral genome. The aim of this study was to find out reassortment events in FLUBV strains detected in cases attended at two hospitals in Barcelona (Catalonia, Spain) from 2004 to 2015 by whole-genome sequencing (WGS).
2. Materials and methods
From October 2004 (week 40/2004) to May 2015 (week 20/2015), upper (nasopharyngeal aspirates or swabs) and lower (bronchoalveolar lavages, bronchoaspirates and tracheal aspirates) respiratory tract specimens were collected for influenza laboratory-confirmation from patients with suspicion of acute respiratory tract infection who were attended at the Emergency Department or admitted to Hospital de la Santa Creu i de Sant Pau (HSCSP) and Hospital Universitari Vall d'Hebron (HUVH). Respiratory samples were processed within the first 24 h, being kept at 2–4 oC in several aliquots until its use. Based on medical records, demographic features (age and sex) and clinical data were retrospectively collected from FLUBV laboratory-confirmed cases.
2.1. Detection of FLUV from respiratory specimens
The detection of FLUV was carried out by either immunofluorescence (D3 Ultra 8TM DFA Respiratory Virus Screening & Identification Kit Diagnostic HYBRIDS, USA), PCR-based assays (Anyplex II RV16 Detection Kit Seegene, Korea and GeneXpert Flu, Cepheid, USA) or cell culture isolation by Madin-Darby canine kidney (MDCK) SIAT 1 cells (WHO, 2011). Prior to PCR-based assays, total nucleic acids were extracted using NucliSense easyMAG (bioMérieux, Marcy l'Etoile, France) according to the manufacturer's instructions, and kept frozen (−20 oC) until use.
2.2. Characterisation of FLUBV
A lineage reverse transcription RT-PCR (one-step RT-PCR system; Qiagen, Hilden, Germany) from an adapted protocol of the Centre for Control Disease and Prevention (CDC) was performed to initially distinguish both FLUBV lineages based on the HA sequence by agarose gel electrophoresis (WHO Chinese NIC, n.d.) (Fig. 1). The primer sequences were BVf (5′-ACATACCCTCGGCAAGAGTTTC-3′), BVr (5′-TGCTGTTTTGTTGTTGTCGTTTT-3′), BYf (5′-ACACCTTCTGCGAAAGCTTCA-3′) and BYr (5′-CATAGAGGTTCTTCATTTGGGTTT-3′), for FLUBV/VIC and FLUBV/YAM lineages, respectively.
Fig. 1.
Agarose gel electrophoresis of lineage RT-PCR.
2.3. Whole-genome amplification (WGA) and 454-pyrosequencing
The WGA was carried out using universal primer set and PCR protocol by Zhou and Wentworth (2012) with SuperScript III one-step RT-PCR kit (Invitrogen, USA) and subsequently sequenced with Roche 454 GS Junior platform. Briefly, full genome PCR amplicons were quantified using Quant-iT™ PicoGreen® Kit (Molecular Probes, Thermo Scientific™, USA) in an Infinite 200 PRO Tecan fluorimeter. A library for each patient was constructed by nebulisation and prepared using the GS DNA Rapid Library Preparation Kit (GS Junior Titanium Series, 454 Roche Life Sciences) according to manufacturer's recommendations. Each purified DNA was ligated to a multiplex identifier (MID). The resulting fragments were size-selected using Agencourt AMPure XP bead sizing (Beckman Coulter, USA), quantified again to make a dilution with the molecules needed to proceed with the emulsion PCR (emPCR) and then, ten samples per amplification were pooled. After emPCR, the emulsion was disrupted and the beads containing clonally amplified DNA were enriched. The beads were separated and deposited into individual picotiter-plate wells and combined with sequencing enzymes. Pyrosequencing was carried out on the picotiter-plate by successive flow of dNTPs (Deyde and Gubareva 2009; Margulies et al., 2005; Raca et al., 2010).
2.4. Bioinformatics analysis of the data
Sequencing data was obtained as standard flow gram format (SFF) files from the GS Run Processor after quality control filtering and read trimming using the manufacturer's default parameters (see 454 Sequencing System software manual version 2.5). The reads were trimmed using the MIDs for each sample, and mapped using the GS Reference Mapper with the reference sequences B/Malaysia/2506/2004 for FLUBV/VIC and B/Florida/4/2006 for FLUBV/YAM viruses.
2.5. Phylogenetic analysis and molecular characterisation
Phylogenetic analyses of the complete eight-segment large contigs were performed using the reference sequences by FLUBV lineage as recommended by season in the Northern Hemisphere by the European Centre for Disease Prevention and Control (ECDC). Nucleotide sequences were aligned using the MUSCLE algorithm, and the molecular evolutionary model of nucleotide substitutions were fitted to the multiple sequence alignments, both conducted in MEGA v11 (Tamura et al., 2011). The phylogenetic trees were constructed using a neighbor-joining distance method as implemented in MEGA v11 with the evolutionary model with the lowest Bayesian information criterion score (Tamura et al., 2011). The topological accuracy of the internal branch was evaluated by the bootstrap method (1000 replicates).
In order to detect amino acid substitutions in the different FLUBV segments, the translated amino acid sequences from aligned nucleotide sequences were related to reference sequences recommended by ECDC for influenza virus characterisation (B/Yamagata: B/Florida/04/2006, GISAID EPI134356; and B/Victoria: B/Malaysia/2506/2004, GISAID EPI117513).
2.6. Statistical analysis
Statistical analysis was performed using SPSS v22 (SPSS Inc., Chicago, IL, USA). Categorical variables were described through frequencies and proportions. Chi square test was calculated to assess associations between categorical variables. P values <0.05 were considered to be statistically significant.
3. Results
A total of 118 FLUBV (75 FLUBV/VIC and 43 FLUBV/YAM) strains from the 2005–2006, the 2008–2011 and the 2012–2015 seasons were studied. During the 2006–2008 and the 2011–2012 seasons, FLUBV strains could not be detected due to the low FLUBV circulation (Insituto Carlos III, n.d.). Whole genome was successfully amplified and sequenced from 58 (77%) FLUBV/VIC and 42 (98%) FLUBV/YAM viruses. Most of FLUBV laboratory-confirmed studied cases (98) were children under 5 years of age (40; 41%) and children between 5 and 14 years (35; 36%) with a 58:40 proportion of male and female (p = 0.375).
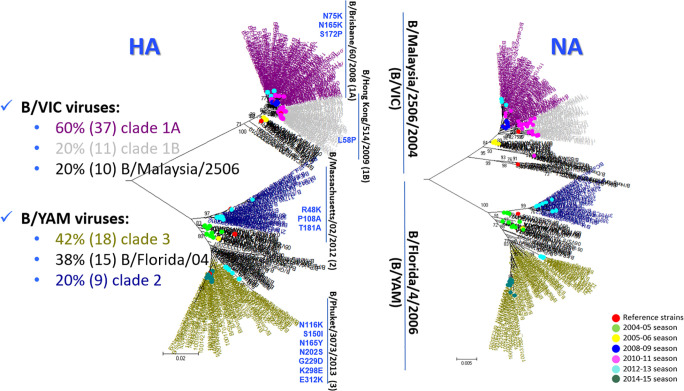
The phylogenetic analysis based on HA sequences revealed that most FLUBV/VIC viruses (37; 64%) belonged to clade 1A represented by B/Brisbane/60/2008 and the remaining sequences to clade 1B (11; 19%) represented by B/HongKong/514/2008 and B/Malaysia/2506/2004 (10; 17%). Most FLUBV/YAM strains belonged to clade 3 (18; 42%) represented by B/Phuket/3073/2013 and B/Florida/4/2006 (15; 38%) and the leftover sequences to clade 2 (9; 20%) represented by B/Massachusetts/02/2012. The comparison of HA and NA phylogenetic trees showed at first congruent classification between the different genetic groups as shown in Fig. 2. In addition, molecular characterisation of NA sequences revealed no mutations related to reduce sensibility to neuraminidase inhibitors (NAIs) (Francisco Pozo, 2013; Nguyen et al., 2012).
Fig. 2.
Phylogenetic trees based on the HA and NA coding-regions sequences of FLUBV strains using the Tamura 3-parameter method with gamma distribution 0.4. Sequences from the present study and the reference ones are labelled according to the legend on the right. The different coloured tree branches refer to the genetic groups of each lineage. Amino acid substitutions, relative to the reference sequences of every lineage, that define these genetic groups are labelled in blue.
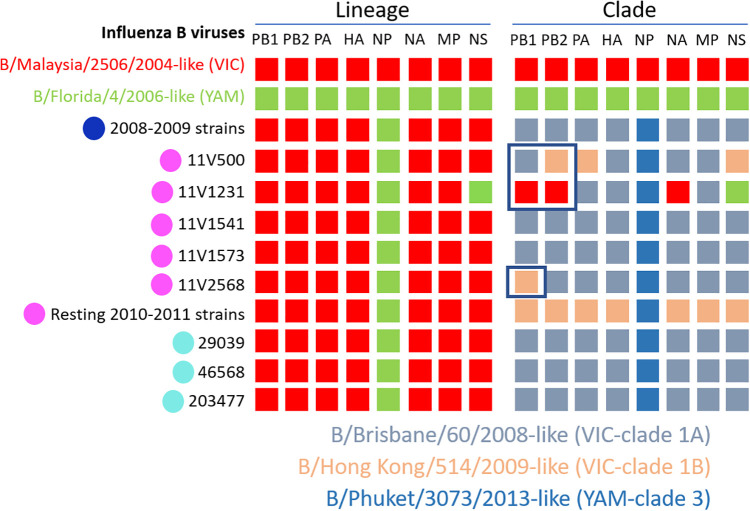
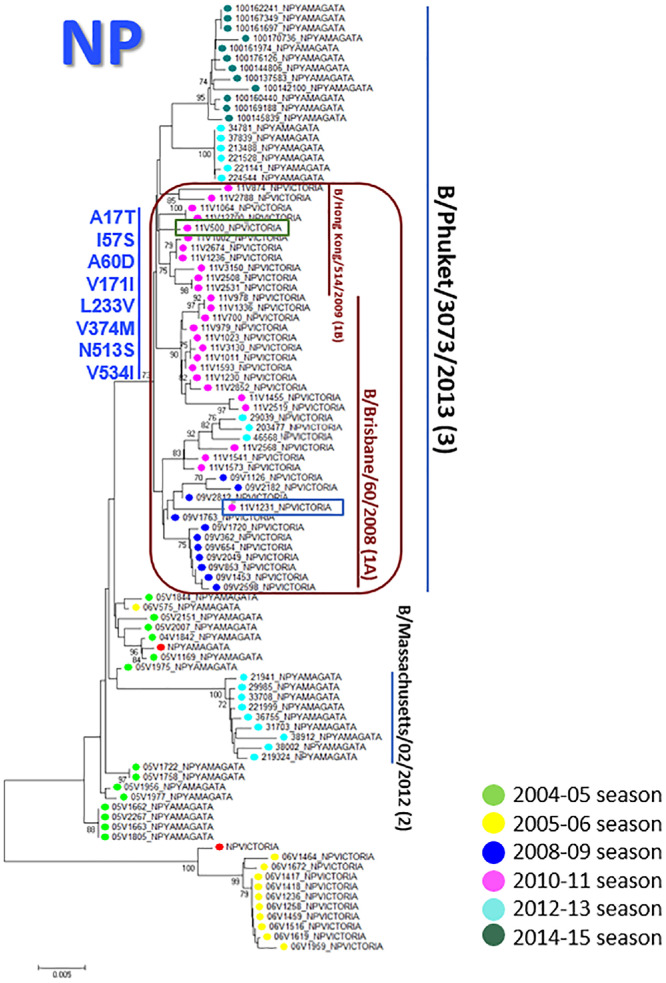
Several intra- and inter-lineage reassortment events were found by phylogenetic analysis of each segment as summarised in Fig. 3. Firstly, corresponding classification was observed amongst HA and NA glycoproteins except to one reassortant 2010–2011 FLUBV/VIC strain (11V1231), which showed genetic similarities to clade 1A in HA, but did not show any in NA (Supplementary Fig. 1). Regarding the polymerase subunits, the same genetic groups were maintained as observed in HA and NA, but three 2010–2011 FLUBV/VIC viruses (11V500, 11V1231 and 11V2568) shifted between clades. While 11V1231 did not show genetic similarities with clade 1 neither in PB1 nor in PB2, it fell within clade 1A in polymerase subunit A (PA). On the other hand, 11V500 fell within clade 1A in PB1, and shifted to clade 1B in the other subunits as it also happened to 11V2568, which fell within clade 1B in PB1 and changed to clade 1A in the other segments (Supplementary Fig. 2). Phylogenetic analysis of the nucleoprotein (NP) revealed an important inter-lineage reassortment event from 2008 to 2009 (11), 2010–2011 (26) and 2012–2013 (3) FLUBV/VIC (clade 1) strains to FLUBV/YAM (clade 3) as shown in Figs. 3 and 4. The molecular characterisation of these strains revealed the acquisition of characteristic FLUBV/YAM amino acid changes like A17T, A60D or V171I. Amongst the matrix protein (MP), the same genetic groups were kept as seen in HA, but in the NS, 11V500 shifted to clade 1B and 11V1231 reassorted to FLUBV/YAM gaining the typical FLUBV/YAM amino acid changes as N4D or E138D (Supplementary Fig. 3) and other mutations (I98V, S109P, T115S, N152S and V263M).
Fig. 3.
Summary of the reassortment events observed in the present study at the lineage and clade level.
Fig. 4.
Phylogenetic tree based on the NP coding-region sequences of FLUBV strains. Sequences from the present study and the reference ones are labelled according to the legend The reassortment event is red squared. Amino acid substitutions, relative to the reference sequence of B/YAM lineage, that define the genetic group B/Phuket/3073/2013, are labelled in blue.
Regarding the clinical data, hospitalisation, ICU-admission and mortality rates are summarised in Table 1. The main clinical symptoms of patients carrying reassortant strains (40/98; 41%) were fever (21; 52%) and respiratory disease (18; 45%), such as upper (laryngitis or pharyngitis) or lower (bronchitis) respiratory tract infections. No differences amongst hospitalisation, ICU-admission or mortality rates were found between reassortant and non-reassortant strains.
Table 1.
Hospitalisation, ICU-admission and mortality data from studied patients carrying or not reassortant strains.
| Patients carrying reassortant strains | Yes (%) | No (%) | |
|---|---|---|---|
| Hospitalisation | Total: 22 | p-value | |
| Yes | 6 (15) | 16 (28) | 0,296 |
| No | 33 (82) | 40 (69) | |
| ICU-admission | Total: 7 | ||
| Yes | 1 (3) | 6 (10) | 0,297 |
| No | 38 (95) | 50 (86) | |
| Mortality | Total: 5 | ||
| Yes | 0 (0) | 5 (9) | 0,081 |
| No | 40 (100) | 53 (91) | |
| TOTAL | 40 (41) | 58 (59) | 98 |
Percentages for Hospitalisation, ICU-admision and Mortality were calculated vertically, depending on the total patients carrying or not reassortant strains.
4. Discussion
The limited number of FLUBV reservoirs causes that these viruses evolve through punctual mutations and, to a lesser degree, by the exchange of segments, compared to FLUAV. However, since the divergence of both FLUBV/VIC and FLUBV/YAM in 1983, several reassortment events have been reported between clades and lineages (Dudas et al., 2015; Oong et al., 2015, 2017), so these episodes might play a role in determining the evolutionary changes in both lineages. While PB1, PB2 and HA segments have kept the same ancestor (FLUBV/VIC or FLUBV/YAM) later than the lineage divergence, the other segments have suffered several reassortment episodes like PA, NP, NA and MP amongst FLUBV/YAM and NS amongst FLUBV/VIC viruses (Dudas et al., 2015). In order to detect these reassortment events, WGS is the ideal technique to study the whole viral genome such as its genetic features or the exchange of segments to have a better understanding of the genetic characteristics. In addition, the usage of universal primers avoids the selection of viral variants, and thus, the results are more reliable to further studies. Herein, the detection of different inter- and intra-lineage reassortment events in FLUBV strains from 2004 to 2015 seasons in Catalonia by WGS are described.
FLUBV, like FLUAV, have a variable and unpredictable circulation, with the shift of one lineage to another or their co-circulation season after season. In this study, the different FLUBV clades from both lineages are represented, which improves the study of reassortment episodes amongst all these strains. Dudas et al. (2015) reported in 2014 that PB1, PB2 and HA were the only segments to maintain separate FLUBV/VIC or FLUBV/YAM lineages in an almost 20-year study. They observed that the PB1-PB2-HA complex remained as a pure lineage, in comparison with the other segments. This could be caused because of the lack of reassortment between these segments due to their linkage disequilibrium since having a pure lineage of this complex might be beneficial for the whole viral fitness (Dudas et al., 2015). Nevertheless, in the present study, reassortment episodes of PB1-PB2-HA complex were observed (Fig. 3), in particular, amongst PB1 and PB2 segments, though they were detected only intra-lineages rather than inter-lineages. Thereby, it is worth to highlight these events due to the possible emergence of reassortant strains which might carry different lineage segments, as the appearance of A(H1)pdm09 amongst FLUAV (Garten et al., 2009; Fowlkes et al., 2011; Antón et al., 2012).
FLUBV/VIC and FLUBV/YAM lineages have suffered numerous reassortment events in the past as other authors reported (Kim et al., 2016; Dudas et al., 2015; Tewawong et al., 2015, 2017). Differences in phylogenetic groups between HA and NA are well known and have been deeply studied compared to the remaining segments due to their importance in the worldwide influenza surveillance (Antón et al., 2016). Thus, one of the reasons to carry out this work by WGS was to study the whole viral genome and detect reassortment episodes in the other segments, which just with the study of HA and NA would not be possible. The FLUBV/YAM NP has reassorted three times since 1995, in which the last one was in 2008 (Dudas et al., 2015). In this study, the NP segment from the 2008–2009, 2010–2011 and 2012–2013 FLUBV/VIC strains reassorted to FLUBV/YAM lineage (Fig. 4) and it corresponded to what Dudas et al. reported in 2014. These strains belonged initially to clade 1A and 1B from FLUBV/VIC lineage and then, these viruses gained distinctive amino acid changes related to clade 3 from FLUBV/YAM lineage. However, no relevant clinical differences could be distinguished between the patients carrying reassortant strains with those who did not carry.
As well as the reassortant NP segment, the NS segment from one 2010–2011 FLUBV/VIC strain shifted to FLUBV/YAM lineage in this study, acquiring those amino acid changes characteristic from that lineage. In addition, it gained another amino acid substitution different from the other strains (I98V, S109P, T115S, N152S and V263M), but these amino acids were from the same group, according to the side chain, with minor differences. Thus, these mutations might be compensating the reassortment into a different lineage. Though, this is a singular case (11V1231), which, as observed in Fig. 3, has shifted either between clades amongst the polymerase subunits, HA, NA and MP segments or between lineages in the NP and NS segments. However, regarding its clinical data, no proper differences could be found, and the patient was a four-year-old child, who did not have any comorbidities, only fever as the unique symptom.
Influenza surveillance is based on the molecular characterisation of only the HA and NA proteins, without taking into account the other segments, which could be misleading the emergence of novel strains carrying reassortant segments. The active implementation of the WGS in the influenza surveillance network would improve the detection of these strains and would give a wider view of what is really circulating worldwide. Although no clinical differences could be found amongst the patients carrying reassortant strains, further studies concerning its possible relationship are needed.
In summary, several intra- and inter-lineage reassortment episodes were herein revealed by WGS and phylogenetic analysis. PB1-PB2-HA remained in complex within lineages as reported before, although some inter-clade reassortment events were detected in three strains. In addition, an important reassortment in the NP and another in the NS segments were found in both lineages. Therefore, surveillance must be continued due to the continue emergence of reassortant FLUBV, although they are not often, but with the implementation of WGS.
Ethical approval
Institutional Review Board approval (PR(AG)329/2014) was obtained from the Vall d'Hebron University Hospital (HUVH) Clinical Research Ethics Committee.
CRediT authorship contribution statement
Cristina Andrés: Conceptualization, Visualization, Formal analysis, Methodology, Writing – original draft, Writing – review & editing. Margarita del Cuerpo: Conceptualization, Visualization, Formal analysis, Methodology, Writing – original draft, Writing – review & editing. Núria Rabella: Writing – review & editing, Conceptualization, Visualization, Writing – review & editing. Maria Piñana: Writing – review & editing. Manuel Jesús Iglesias-Cabezas: Writing – review & editing. Alejandra González-Sánchez: Writing – review & editing. Juliana Esperalba: Writing – review & editing. Ariadna Rando: Writing – review & editing. Maria Carmen Martín: Methodology. Francisco Fuentes: Methodology. Susana Rubio: Methodology. Narcís Saubi: Writing – review & editing. Tomàs Pumarola: Conceptualization, Visualization, Writing – review & editing. Andrés Antón: Conceptualization, Visualization, Writing – review & editing.
Declaration of Competing Interest
The authors declare no conflicts of interest.
Acknowledgements
This study was supported by the Spanish Ministry of Economy and Competitiveness, Fondo de Investigación Sanitaria (Grants FIS PI08/0118, FIS PI11/01864, FIS PI14/01838) del Instituto de Salud Carlos III, co-financed by the European Development Regional Fund (ERDF) "A way to achieve Europe", Spanish Network for Research in Infectious Diseases (REIPI RD16/0016/0003), and by CIBER -Consorcio Centro de Investigación Biomédica en Red- (CB 2021) del Instituto de Salud Carlos III, Ministerio de Ciencia e Innovación and Unión Europea – NextGenerationEU.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.virusres.2023.199089.
Appendix. Supplementary materials
Supplementary Figure 1: Phylogenetic trees based on the HA and NA coding-regions sequences of FLUBV strains. Sequences from the present study and the reference ones are labelled according to the legend. The squares refer to the reassortant strains.
Supplementary Figure 2: Phylogenetic trees based on the polymerase subunits sequences of FLUBV strains. Sequences from the present study are labelled according to the legend. The squares refer to the reassortant strains.
Supplementary Figure 3: Phylogenetic trees based on the MP and NS coding-regions sequences of FLUBV strains. Sequences from the present study are labelled according to the legend. The squares refer to the reassortant strains. Regarding the NS segment, the amino acid substitutions acquired by the reassortant strain, relative to the reference sequence of B/YAM lineage, are labelled in blue.
Data availability
Data will be made available on request.
References
- (WHO), World Health Organisation . Manual For the Laboratory Diagnosis and Virological Surveillance of Influenza. WHO Library Cataloguing-in-Publication Data; 2011. WHO global influenza surveillance network. [Google Scholar]
- Antón A., Marcos M.A., Torner N., Isanta R., Camps M., Martínez A., Domínguez A., Jané M., Jiménez de Anta M.T., Pumarola T. Virological surveillance of influenza and other respiratory viruses during six consecutive seasons from 2006 to 2012 in Catalonia, Spain. Clin. Microbiol. Infect. 2016;22(6):564. doi: 10.1016/j.cmi.2016.02.007. https://www.ncbi.nlm.nih.gov/pubmed/26939538 e1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antón A., Pozo F., Niubó J., Casas I., Pumarola T. Influenza A(H1N1)pdm09 virus: viral characteristics and genetic evolution. Enferm. Infecc. Microbiol. Clin. 2012;30(Suppl 4):10–17. doi: 10.1016/S0213-005X(12)70099-X. http://www.ncbi.nlm.nih.gov/pubmed/23116787 [DOI] [PubMed] [Google Scholar]
- Bedford T., Riley S., Barr I.G., Broor S., Chadha M., Cox N.J., Daniels R.S., Gunasekaran C.P., Hurt A.C., Kelso A., Klimov A., Lewis N.S., Li X., McCauley J.W., Odagiri T., Potdar V., Rambaut A., Shu Y., Skepner E., Smith D.J., Suchard M.A., Tashiro M., Wang D., Xu X., Lemey P., Russell C.A. Global circulation patterns of seasonal influenza viruses vary with antigenic drift. Nature. 2015;523(7559):217–220. doi: 10.1038/nature14460. http://www.ncbi.nlm.nih.gov/pubmed/26053121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouvier N.M., Palese P. The biology of influenza viruses. Vaccine. 2008;26(Suppl 4):D49–D53. doi: 10.1016/j.vaccine.2008.07.039. http://www.ncbi.nlm.nih.gov/pubmed/19230160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deyde V.M., Gubareva L.V. Influenza genome analysis using pyrosequencing method: current applications for a moving target. Expert Rev. Mol. Diagn. 2009;9(5):493–509. doi: 10.1586/erm.09.21. http://www.ncbi.nlm.nih.gov/pubmed/19580433 [DOI] [PubMed] [Google Scholar]
- Dudas G., Bedford T., Lycett S., Rambaut A. Reassortment between influenza B lineages and the emergence of a coadapted PB1-PB2-HA gene complex. Mol. Biol. Evol. 2015;32(1):162–172. doi: 10.1093/molbev/msu287. https://www.ncbi.nlm.nih.gov/pubmed/25323575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowlkes A.L., Arguin P., Biggerstaff M.S., Gindler J., Blau D., Jain S., Dhara R., McLaughlin J., Turnipseed E., Meyer J.J., Louie J.K., Siniscalchi A., Hamilton J.J., Reeves A., Park S.Y., Richter D., Ritchey M.D., Cocoros N.M., Blythe D., Peters S., Lynfield R., Peterson L., Anderson J., Moore Z., Williams R., McHugh L., Cruz C., Waters C.L., Page S.L., McDonald C.K., Vandermeer M., Waller K., Bandy U., Jones T.F., Bullion L., Vernon V., Lofy K.H., Haupt T., Finelli L. Epidemiology of 2009 pandemic influenza A (H1N1) deaths in the United States, April-July 2009. Clin. Infect. Dis. 2011;52(Suppl 1):S60–S68. doi: 10.1093/cid/ciq022. https://www.ncbi.nlm.nih.gov/pubmed/21342901 [DOI] [PubMed] [Google Scholar]
- Pozo F., Lina B., Rebelo de Andrade H., Enouf V., Kossyvakis A., Broberg E., Daniels R., Lackenby A., Meijer A. Guidance for clinical and public healt laboratories testing influenza virus antiviral drug susceptibility in Europe. J. Clin. Virol. 2013:7. doi: 10.1016/j.jcv.2013.01.009. [DOI] [PubMed] [Google Scholar]
- Garten R.J., Davis C.T., Russell C.A., Shu B., Lindstrom S., Balish A., Sessions W.M., Xu X., Skepner E., Deyde V., Okomo-Adhiambo M., Gubareva L., Barnes J., Smith C.B., Emery S.L., Hillman M.J., Rivailler P., Smagala J., de Graaf M., Burke D.F., Fouchier R.A., Pappas C., Alpuche-Aranda C.M., López-Gatell H., Olivera H., López I., Myers C.A., Faix D., Blair P.J., Yu C., Keene K.M., Dotson P.D., Boxrud D., Sambol A.R., Abid S.H., St George K., Bannerman T., Moore A.L., Stringer D.J., Blevins P., Demmler-Harrison G.J., Ginsberg M., Kriner P., Waterman S., Smole S., Guevara H.F., Belongia E.A., Clark P.A., Beatrice S.T., Donis R., Katz J., Finelli L., Bridges C.B., Shaw M., Jernigan D.B., Uyeki T.M., Smith D.J., Klimov A.I., Cox N.J. Antigenic and genetic characteristics of swine-origin 2009 A(H1N1) influenza viruses circulating in humans. Science. 2009;325(5937):197–201. doi: 10.1126/science.1176225. http://www.ncbi.nlm.nih.gov/pubmed/19465683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.I., Lee I., Park S., Bae J.Y., Yoo K., Lemey P., Park M.S., Song J.W., Kee S.H., Song K.J. Reassortment compatibility between PB1, PB2, and HA genes of the two influenza B virus lineages in mammalian cells. Sci. Rep. 2016;6:27480. doi: 10.1038/srep27480. https://www.ncbi.nlm.nih.gov/pubmed/27270757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margulies M., Egholm M., Altman W.E., Attiya S., Bader J.S., Bemben L.A., Berka J., Braverman M.S., Chen Y.J., Chen Z., Dewell S.B., Du L., Fierro J.M., Gomes X.V., Godwin B.C., He W., Helgesen S., Ho C.H., Irzyk G.P., Jando S.C., Alenquer M.L., Jarvie T.P., Jirage K.B., Kim J.B., Knight J.R., Lanza J.R., Leamon J.H., Lefkowitz S.M., Lei M., Li J., Lohman K.L., Lu H., Makhijani V.B., McDade K.E., McKenna M.P., Myers E.W., Nickerson E., Nobile J.R., Plant R., Puc B.P., Ronan M.T., Roth G.T., Sarkis G.J., Simons J.F., Simpson J.W., Srinivasan M., Tartaro K.R., Tomasz A., Vogt K.A., Volkmer G.A., Wang S.H., Wang Y., Weiner M.P., Yu P., Begley R.F., Rothberg J.M. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437(7057):376–380. doi: 10.1038/nature03959. http://www.ncbi.nlm.nih.gov/pubmed/16056220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina R.A., García-Sastre A. Influenza A viruses: new research developments. Nat. Rev. Microbiol. 2011;9(8):590–603. doi: 10.1038/nrmicro2613. http://www.ncbi.nlm.nih.gov/pubmed/21747392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen H.T., Fry A.M., Gubareva L.V. Neuraminidase inhibitor resistance in influenza viruses and laboratory testing methods. Antivir. Ther. 2012;17(1 Pt B):159–173. doi: 10.3851/IMP2067. http://www.ncbi.nlm.nih.gov/pubmed/22311680 [DOI] [PubMed] [Google Scholar]
- Oong X.Y., Ng K.T., Lam T.T., Pang Y.K., Chan K.G., Hanafi N.S., Kamarulzaman A., Tee K.K. Epidemiological and evolutionary dynamics of influenza B viruses in Malaysia, 2012-2014. PLoS One. 2015;10(8) doi: 10.1371/journal.pone.0136254. https://www.ncbi.nlm.nih.gov/pubmed/26313754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oong X.Y., Ng K.T., Tan J.L., Chan K.G., Kamarulzaman A., Chan Y.F., Sam I.C., Tee K.K. Whole-genome phylogenetic analysis of Influenza B/Phuket/3073/2013-like viruses and unique reassortants detected in Malaysia between 2012 and 2014. PLoS One. 2017;12(1) doi: 10.1371/journal.pone.0170610. https://www.ncbi.nlm.nih.gov/pubmed/28129386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO Collaborating Centre for Reference and Research on Influenza Chinese National Influenza Centre. National Institute for Viral Disease Control and. 155 Changbai Roadm changping district. 102206 Beijing China: Chinese centre for disease control and prevention.
- Instituto Carlos III, "Sistema de Vigilancia de la Gripe en España." Available at: http://vgripe.isciii.es/gripe/inicio.do.
- Raca G., Jackson C., Warman B., Bair T., Schimmenti L.A. Next generation sequencing in research and diagnostics of ocular birth defects. Mol. Genet. Metab. 2010;100(2):184–192. doi: 10.1016/j.ymgme.2010.03.004. http://www.ncbi.nlm.nih.gov/pubmed/20359920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrauwen E.J., de Graaf M., Herfst S., Rimmelzwaan G.F., Osterhaus A.D., Fouchier R.A. Determinants of virulence of influenza A virus. Eur. J. Clin. Microbiol. Infect. Dis. 2014;33(4):479–490. doi: 10.1007/s10096-013-1984-8. http://www.ncbi.nlm.nih.gov/pubmed/24078062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinhauer D.A. Role of hemagglutinin cleavage for the pathogenicity of influenza virus. Virology. 1999;258(1):1–20. doi: 10.1006/viro.1999.9716. http://www.ncbi.nlm.nih.gov/pubmed/10329563 [DOI] [PubMed] [Google Scholar]
- Steinhauer D.A., Skehel J.J. Genetics of influenza viruses. Annu. Rev. Genet. 2002;36:305–332. doi: 10.1146/annurev.genet.36.052402.152757. http://www.ncbi.nlm.nih.gov/pubmed/12429695 [DOI] [PubMed] [Google Scholar]
- Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011;28(10):2731–2739. doi: 10.1093/molbev/msr121. http://www.ncbi.nlm.nih.gov/pubmed/21546353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tewawong N., Suntronwong N., Korkong S., Theamboonlers A., Vongpunsawad S., Poovorawan Y. Evidence for influenza B virus lineage shifts and reassortants circulating in Thailand in 2014-2016. Infect. Genet. Evol. 2017;47:35–40. doi: 10.1016/j.meegid.2016.11.010. https://www.ncbi.nlm.nih.gov/pubmed/27845268 [DOI] [PubMed] [Google Scholar]
- Tewawong N., Suwannakarn K., Prachayangprecha S., Korkong S., Vichiwattana P., Vongpunsawad S., Poovorawan Y. Molecular epidemiology and phylogenetic analyses of influenza B virus in Thailand during 2010 to 2014. PLoS One. 2015;10(1) doi: 10.1371/journal.pone.0116302. https://www.ncbi.nlm.nih.gov/pubmed/25602617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou B., Wentworth D.E. Influenza A virus molecular virology techniques. Methods Mol. Biol. 2012;865:175–192. doi: 10.1007/978-1-61779-621-0_11. http://www.ncbi.nlm.nih.gov/pubmed/22528160 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1: Phylogenetic trees based on the HA and NA coding-regions sequences of FLUBV strains. Sequences from the present study and the reference ones are labelled according to the legend. The squares refer to the reassortant strains.
Supplementary Figure 2: Phylogenetic trees based on the polymerase subunits sequences of FLUBV strains. Sequences from the present study are labelled according to the legend. The squares refer to the reassortant strains.
Supplementary Figure 3: Phylogenetic trees based on the MP and NS coding-regions sequences of FLUBV strains. Sequences from the present study are labelled according to the legend. The squares refer to the reassortant strains. Regarding the NS segment, the amino acid substitutions acquired by the reassortant strain, relative to the reference sequence of B/YAM lineage, are labelled in blue.
Data Availability Statement
Data will be made available on request.