Abstract
β-Glucuronidase (uidA) reporter gene fusions were constructed for the hrpZ, hrpL, and hrpS genes from the phytopathogen Pseudomonas syringae pv. maculicola strain ES4326. These reporters, as well as an avrRpt2-uidA fusion, were used to measure transcriptional activity in ES4326 and a ES4326 rpoN mutant. rpoN was required for the expression of avrRpt2, hrpZ, and hrpL in vitro in minimal media and in vivo when infiltrated into Arabidopsis thaliana leaves. In contrast, the expression of hrpS was essentially the same in wild-type and rpoN mutant strains. Constitutive expression of hrpL in an rpoN mutant restored hrpZ transcription to wild-type levels, restored the hypersensitive response when infiltrated into tobacco (Nicotiana tobacum), and partially restored the elicitation of virulence-related symptoms but not growth when infiltrated into Arabidopsis leaves. These data indicate that rpoN-mediated control of hrp gene expression acts at the level of hrpL and that in planta growth of P. syringae is not required for the elicitation of disease symptoms.
In gram-negative bacteria, transcriptional activation in response to external stimuli often involves the alternative sigma factor ς54 (1, 50). ς54, which is encoded by rpoN, works in conjunction with members of the NtrC superfamily of transcriptional activators (1, 50). Among the different enzymatic pathways under ς54 control are those responsible for nitrogen utilization, dicarboxylate transport, xylene degradation, and hydrogen utilization (8, 39, 47, 79).
In the case of some phytopathogenic bacterial species, rpoN has been implicated indirectly as a regulator of a large cluster of pathogenicity-related genes known as the hrp gene cluster (17, 27). For example, Pseudomonas syringae pv. syringae strain 61 contains a 25-kb hrp cluster consisting of 27 genes arranged as follows: hrpKL, hrpJ, hrcV, hrpQ, hrcN, hrpOP, hrcQRSTU, hrpVT, hrcC, hrpGF, hrpED, hrcJ, hrpBZA, and hrpSR (11, 33). The acronym hrp stands for hypersensitive response and pathogenicity. The genes within the hrp cluster are required not only for pathogenicity but also for elicitation of the plant resistance reaction known as the hypersensitive response (HR) (44, 45). The HR involves rapid, localized plant cell death, which is triggered by a gene-for-gene interaction between a particular avirulence (avr) gene in the pathogen and a corresponding resistance gene in the host (21, 56). The widely conserved members of the hrp cluster, designated hrc, show significant homology to members of the Yersinia type III secretory pathway and in the new simplified nomenclature bear the letter designation of the corresponding Yersinia gene (9). (The exception is hrcV, which is homologous to the Yersinia lcrD genes.)
Sequence analysis of the hrp cluster in P. syringae pv. phaseolicola suggested that ς54 would be required for hrp gene expression in conjunction with the hrpRS genes, which are required for expression of the remaining hrp genes in the cluster (17, 27). hrpR and hrpS encode proteins that contain the domain conserved among transcriptional activators such as NtrC, DctD, and NifA that work in concert with ς54 (29, 30; reviewed in references 1 and 50). Because of the HrpS-NtrC homology, it seemed likely that a P. syringae homologue of rpoN would be required for activation of the hrp gene cluster. Circumstantial evidence in support of this conclusion was provided by our findings presented in the accompanying study that the rpoN gene of P. syringae is required for pathogenesis and the HR (32).
The P. syringae hrpL gene product is related to the alternate sigma factor AlgU, which regulates genes involved in the biosynthesis and regulation of the exopolysaccharide alginate in P. aeruginosa (49, 80). The circuitry of hrp regulation in P. syringae appears to involve a transcriptional activation cascade in which HrpR activates hrpS, HrpS activates hrpL, and HrpL activates transcription of the remaining hrp genes (17, 26, 81), as well as avr genes that are responsible for eliciting the HR. One of these remaining hrp genes, hrpV, appears to negatively regulate hrp transcription and, while there is some evidence that hrpV functions upstream of hrpRS, the factor(s) involved in the regulation of hrpRS remain obscure (62).
Most of the remaining genes in the hrp cluster encode structural proteins that are involved in the synthesis or export of pathogenicity factors (78). As mentioned above, the nine hrc genes are homologous to members of the Yersinia type III secretory pathway that is responsible for translocating Yop proteins into host cells (18, 24, 53, 73). Similarly, hrp mutations block the export of PopA1 from P. solanacearum and HrpZ (harpin) and HrpW from P. syringae, of which the latter two elicit an HR-like response on incompatible and nonhost plants (2, 10, 11, 31). Given the role of the hrp cluster in protein secretion, the most likely cause for nonpathogenic and non-HR-eliciting phenotypes of hrp mutants is the inability to secrete a variety of virulence factors or elicitors. Recent studies suggest that several Avr proteins function directly inside of host plant cells in analogy with the type III-mediated transfer of virulence factor by Y. enterocolitica (23, 71, 75, 76).
avr genes are often coordinately regulated with hrp gene expression. A conserved sequence, GGAACCNA-N14-CCACNNA, which appears to be responsible for hrpL-dependent transcription, has been identified upstream of a variety of avr genes (81). In several cases, including avrRpt2 and avrRpm1, hrpL dependence has been confirmed experimentally (34, 35, 46, 63, 66, 70, 81). Thus, the loss of hrp by mutation of a putative hrp regulator such as rpoN should affect HR induction by blocking avr gene transcription and type III-mediated protein delivery.
As described above, the presence of the NtrC-like regulators HrpR and HrpS in P. syringae suggests a direct role for ς54 in hrp gene regulation. However, rpoN is often found to control multiple functions, including flagellar synthesis and nitrogen utilization (41, 50, 60). Moreover, P. syringae pathovars produce a number of toxins, including syringomycins, tabtoxin, phaseolotoxin, tagetitoxin, and coronatine (6), and we have found that rpoN is required for the expression of coronatine (32). Therefore, rpoN might be involved in the regulation of a number of virulence factors in addition to the hrp cluster, and our finding that rpoN mutants are nonpathogenic is not definitive evidence that rpoN regulates hrp gene expression.
To dissect the plant defense response using a molecular-genetic approach, our laboratory (13, 14, 16, 82) has established a model pathogenicity system that involves the infection of Arabidopsis thaliana with P. syringae pv. maculicola strain ES4326. Strain ES4326 proliferates extensively in Arabidopsis ecotype Columbia leaves and causes the development of water-soaked disease lesions (14, 16). In contrast, ES4326 carrying the avirulence gene avrRpt2 elicits a visible HR about 16 h after infiltration, and its multiplication in planta is 50- to 100-fold less than ES4326 (16). In the accompanying publication, we describe the isolation and characterization of an ES4326 rpoN mutant (32). We describe here experiments examining the effect of the rpoN mutant on hrp and avr gene regulation in ES4326. Our results indicate that neither hrp nor avr genes are transcribed in an rpoN mutant in vitro or during pathogenesis, that synthesis of the avrRpt2 gene product in the absence of hrp gene expression is insufficient to elicit an HR, that the coordinate rpoN regulation of hrp and avr genes acts through hrpL, that additional rpoN-regulated factors may be important for pathogenesis, and that the growth of ES4326 in Arabidopsis leaves is not required for the elicitation of disease symptoms.
MATERIALS AND METHODS
Strains and culture conditions.
Bacterial strains and plasmids used in this study are listed in Table 1. ES4326 and its derivatives were grown at 28°C in L broth (48), minimal M9 medium (48) or King's B (40) medium. Escherichia coli strains were grown at 37°C in L broth. Antibiotic concentrations for E. coli and P. syringae strains were as follows: streptomycin, 150 μg/ml; kanamycin, 25 μg/ml; tetracycline, 12 μg/ml; gentamicin, 20 μg/ml; spectinomycin, 20 μg/ml; and ampicillin, 100 μg/ml. Marker exchange of hrpZ::uidA was carried out by serially culturing ES4326 containing pHRPZgus without antibiotic selection, plating on medium containing kanamycin to select for the insertion, and screening on plates containing kanamycin and tetracycline for strains that had lost the vector (68).
TABLE 1.
Strains and plasmids used
| Strain or plasmid | Relevant genotype, phenotype, or role in this study | Source or reference |
|---|---|---|
| Strains | ||
| E. coli | ||
| DH5a | F−lacZΔM15 endA1 recA1 hsdR17 supE44 thi-1 gyrA relA1 λ− | Bethesda Research Laboratories (28) |
| MM294 | Host of pRK2013 | G. Walker (4) |
| HB101 | Host of pHoGus and pSshe | 48 |
| MT609 | Recipient for cosmids with Tn3-uidA insertions | T. Finan (20) |
| P. syringae pv. phaseolicola LRG94 | hrpS::Tn3-spice | L. Rahme (64) |
| P. syringae pv. maculicola | ||
| ES4326 | Wild type | K. Davis (14) |
| ES4326 hrpZ::uidA | Contains chromosomal uidA translational fusion in hrpZ | This study |
| ES4326 rpoN::Kmr | Contains Kanr cassette inserted into rpoN gene | This study |
| P. syringae pv. tomato | ||
| DC3000 | Wild type | D. Cuppels (57) |
| DC3661 | Coronatine biosynthetic mutant | D. Cuppels (57) |
| MM1065 | Origin of avrRpt2 | M. Mindrinos (16) |
| Plasmids | ||
| pRK2013 | Kanr Tra+, mating helper | G. Ditta (15) |
| pLAFR3 | Tcr cosmid cloning vector | B. Staskawicz (72) |
| pBluescript SK(+) | Apr, cloning vector | Stratagene |
| pUC18 | Apr, plasmid cloning vector | Boehringer Mannheim |
| pRR54cos | Apr, cosmid cloning vector | L. Rahme (64) |
| pPL6 | Source of P. syringae pv. phaseolicola hrpL gene | L. Rahme (64) |
| pNN56 | Source of P. syringae pv. phaseolicola hrpS gene | L. Rahme (64) |
| pHoGus | Apr Kmr, source of Tn3-uidA | J. Glazebrook (22) |
| pSshe | Cmr, provides transposase in trans | J. Glazebrook (22) |
| pRAJ235 | Source of E. coli uidA gene | R. A. Jefferson (37) |
| pRSR0 | 1.4-kb SalI fragment containing avrRpt2 in pUC119 | R. Innes (35) |
| pLH12 | 1.4-kb SalI fragment containing avrRpt2 in pLAFR3 | R. Innes (77) |
| pAVR12 | avrRpt2-uidA translational fusion in pLAFR3 | This study |
| pAVRC | lacZ-avrRpt2 transcriptional fusion in pLAFR3 | This study |
| pEHS | ES4326 hrpS and hrpR in pBluescript SK(+) | This study |
| pEHSpuc | 2.0 HB SalI-ApaI fragment containing hrpS in pUC18 | This study |
| pEH10 | ES4326 hrp clone containing hrpS in pLAFR3 | This study |
| pEHL | ES4326 hrp clone containing hrpL in pRR54cos | This study |
| pEHL3 | 5.8-kb EcoRI fragment containing hrpL in pLAFR3 | This study |
| pHRPLC | lacZ-hrpL transcriptional fusion in pLAFR3 | This study |
| pHRPLgus | hrpL-uidA translational fusion in pLAFR3 | This study |
| pHRPZgus | hrpZ-uidA translational fusion in pLAFR3 | This study |
| pHRPSgus | hrpS-uidA translational fusion in pLAFR3 | This study |
| pXIVB4 | ES4326 hrp clone containing hrpL in pRR54cos | This study |
Bacterial genetics.
pLAFR3 derivatives were introduced into Pseudomonas strains via triparental matings with pRK2013 as the mobilizing plasmid (15).
Cloning of ES4326 hrpR, hrpS, hrpZ, and hrpL genes.
hrpS was cloned from a ES4326 genomic library constructed by Sau3A partial digestion of ES4326 genomic DNA (3). The partial digest was size selected for 5- to 10-kb fragments on a sucrose gradient (48) and ligated into the BamHI site of pBluescript. A plasmid carrying a 5.0-kb fragment containing hrpS, pEHS, was identified by colony hybridization using a randomly labeled probe (3) derived from cosmid pNN56 which contains hrpS from P. syringae pv. phaseolicola. Sequencing 500 bp of pEHS confirmed the presence of hrpR and a portion of hrpS on pEHS. A larger 24-kb clone, pEH10, containing the entire hrpS gene, was obtained by colony hybridization of an ES4326 genomic library constructed in pLAFR3 using probe containing a portion of hrpS generated from the 1.1-kb PvuII fragment of pEHS (Fig. 1). The presence of a functioning hrpS gene on pEH10 was confirmed by complementing the P. syringae pv. phaseolicola hrpS mutant LRG94 for disease symptoms on bean. Southern blot analysis delimited hrpS to a 2.8-kb KpnI fragment in pEH10. Sequencing the junction of a Tn3-uidA transposon insertion in pEH10 (see below) identified a homolog of the P. syringae pv. phaseolicola hrpZ gene on an adjacent 2.1-kb KpnI fragment.
FIG. 1.
Physical and genetic map of the P. syringae pathovar maculicola hrp cluster. Plasmid pEH10 was assayed for complementation of the P. syringae pv. phaseolicola hrpS mutant LRG94 as described in Materials and Methods. The regions of the hrp cluster contained in pEHL and pEH10 are shown. The fragment used to probe for overlap between the clones is hatched. The arrows represent the regions of the genes that have been sequenced and indicate the direction of transcription. Restriction enzymes: A, ApaI, B, BamHI, E, EcoRI, H, HindIII, K, KpnI, S, SalI.
A 36-kb clone, pEHL, containing hrpL was isolated from an ES4326 pRR54cos library by colony hybridization. This library was constructed similarly to the others except that 30- to 40-kb fragments were isolated. A hrpL probe was generated from a 2.4-kb EcoRI-SalI fragment from plasmid pPL6 containing hrpL from P. syringae pv. phaseolicola. A 5.8-kb EcoRI fragment identified as containing hrpL by Southern hybridization was subcloned into pLAFR3 to construct pEHL3 and sequenced.
RNA analysis.
RNA preparation and Northern blot analysis was carried out using standard techniques (3). An avrRpt2 probe was made by the Klenow random priming reaction using an internal fragment of avrRpt2 as the template (3). hrpL and hrpZ probes were made by PCR labeling using primers with the sequences 5′-CATACCCCCATTCAGGC-3′ and 5′-GTCTCAGTCTTAACAGC-3′, respectively (25, 67).
Construction of translational fusions.
A translational gene fusion that contains the amino-terminal half of avrRpt2 fused in frame to the entire E. coli uidA gene, including its ATG translation initiation codon, was constructed as follows. Plasmid pRAJ275, which contains the uidA gene, was digested with NcoI; the resulting 5′ overhangs were filled using Klenow polymerase (3), and the NcoI-cleaved DNA was digested with SalI. A 575-bp SalI-BalI fragment containing the promoter and the first 133 amino acids of avrRpt2 was purified from plasmid pRSR0 (35) and ligated to the NcoI-SalI-digested pRAJ275 DNA. The resulting plasmid containing the avrRpt2-uidA fusion was digested with HindIII and EcoRI, which cut within the polylinker and the 3′ end of the uidA gene, respectively. This 2.25-kb HindIII-EcoRI fragment was then cloned into pLAFR3 that had been linearized using the same two enzymes, resulting in the construction of pAVR12. ES4326 transconjugants carrying pAVR12 formed white colonies with blue centers on King's B or Luria-Bertani (LB) agar and dark blue colonies on minimal M9 medium containing indolyl-4-methylumbelliferyl-β-d-glucuronide (X-Gluc; Biosynth AG).
Translational gene fusions were made to hrpZ (pHRPZgus) and hrpL (pHRPLgus) by transposon mutagenesis using Tn3::uidA (22). For hrpZ, pEH10 was introduced by triparental mating into HB101 (pHoGus pSshe) which contains Tn3::uidA on pHoGus and the transposase gene on pSshe. The resulting strain was incubated overnight, mated with MT609, and then spread on LB agar containing tetracycline and kanamycin 6 h later. Because pHoGus is unable to replicate in MT609, Tcr Kmr exconjugants should only arise by a transposition event. Presumptive pEH10 Tn3::uidA isolates were analyzed by restriction mapping to locate the position of the uidA insert. A primer internal to the uidA gene (5′-GCAATTGCCCGGCTTTC-3′) was used to sequence the Tn3::uidA junctions. For hrpL, the 5.8-kb EcoRI hrpL subclone pEHL3 was mated into HB101 (pHoGus pSshe) and analyzed as described above for pEH10. An hrpS translational fusion to uidA was constructed by first subcloning a 2.0-kb SalI-KpnI fragment from pEH10 containing hrpS into pUC18 to produce pEHSpuc. Primers containing BspEI sites corresponding to the amino-terminal (5′-AGTTCTCCGGAGCATGCCTGCAGGTCG-3′) and carboxyl-terminal (5′-AGTTCTCCGGAGAGGCTGTAGCCGACG-3′) sequences of the uidA gene were used to generate PCR fragments containing uidA which were digested with BspEI and ligated into the BspEI site of pEHSpuc. The resulting fusion was digested with KpnI and HindIII which cut in the polylinker, the overhangs were filled using Klenow polymerase, and the fragment was ligated into the HindIII site of pLAFR3 (also filled using Klenow polymerase) to give pHRPSgus.
Construction of PlacZ-avrRpt2.
The 1.22-kb XmnI-PstI fragment of pRSR0 containing the P. syringae avrRpt2 gene and its putative Shine-Dalgarno sequence was cloned into the EcoRI (filled in with Klenow polymerase) and PstI sites of plasmid pLAFR3 to form pAVRC. Transcription originating at the lacZ promoter present in pLAFR3 (proximal to the EcoRI site) reads through into avrRpt2.
Construction of PlacZ-hrpL.
A PCR-based strategy using primers with engineered EcoRI sites was used to construct a translational fusion between hrpL and the lacZ promoter of cosmid pLAFR3. One primer (5′-AGTCAGAATTCCCAGAACCTTGTGATC-3′) corresponded to the amino-terminal end of hrpL starting with the third codon, and the other primer (5′-AGTTCGAATTCCTGTGTGGTTTCGGGC-3′) corresponded to hrpL sequence 27 bp downstream of the stop codon. The PCR product was digested with EcoRI and ligated to the EcoRI site of pLAFR3 to generate pHRPLC. The pHRPLC fusion was verified by sequencing the lacZ-hrpL junction.
Screen for hrp regulators.
ES4326 hrpZ::uidA was mutagenized using N-methyl-N′-nitro-N-nitrosoguanidine (NG) (52) by subculturing an overnight culture of ES4326 hrpZ::uidA in KB media, growing it to an optical density at 600 nm (OD600) of 1.0, resuspending four 5-ml samples in 4 ml of 0.1 M sodium citrate (pH 5.5), adding 0.2 ml of a 1-mg/ml solution of NG in 0.1 M sodium citrate (pH 5.5) to three of the cultures, and incubating the mixture in a 37°C water bath for 5, 15, and 30 min, respectively. Each sample was then pelleted, washed in 0.1 M phosphate buffer (pH 7.0), resuspended in 10 ml of KB, plated to determine the extent of killing, and grown overnight. The mutants were screened by plating 100 to 500 CFU on M9 plates containing 40 μg of 5-bromo-4-chloro-3-indolyl-β-d-glucuronide X-Gluc per ml.
β-Glucuronidase assays.
avr and hrp gene induction following transfer from rich medium to minimal medium was measured by growing cultures in King's B sucrose, in which hrp expression is suppressed, to late exponential phase (OD600 = 0.6 to 1.0), washing with M9 media, and resuspending the bacteria at an OD600 of 0.02 in M9 medium incubated at 28°C. At the indicated times, aliquots were pelleted in a microcentrifuge and resuspended in 10 mM MgSO4. Duplicate serial dilutions were then plated on LB media containing appropriate antibiotics, and the bacterial concentration was determined by CFU counting. Then, 100 μl of each sample was added to an equal volume of 2× extraction buffer (36). β-Glucuronidase assays were performed by adding 50 μl of sample to 500 μl of assay buffer containing 6.25 mM methylumbelliferyl-β-d-glucuronide as substrate (37). Fluorometric measurements were performed using a Hoefer Scientific Instruments fluorimeter with an excitation wavelength of 365 nm and an emission wavelength of 460 nm. Standard curves were obtained with 4-methyl-umbelliferone (Sigma). Enzymatic activity was calculated as rate of accumulation of 4-methylumbelliferone (nanomoles/minute) per bacterial cell and represents the average of two experiments with three replicates per experiment.
The accumulation of β-glucuronidase produced by bacteria in Arabidopsis leaves was determined after infiltrating the leaves with cultures grown in King's B to late exponential phase (OD600 = 0.6 to 1.0) that had been washed with 10 mM MgSO4 and resuspended in 10 mM MgSO4 at an OD600 of 0.2. At the indicated times, leaves were harvested, and six leaf punches obtained with a no. 2 cork borer were ground in 200 μl of 10 mM MgSO4 and then handled as described above. Values presented for glucuronidase activity represent the average of two or three experiments with three replicates per experiment.
Plant pathogenicity assays.
Arabidopsis ecotype Columbia was germinated, grown, and infiltrated with ES4326 strains at a titer of either 1 × 104 or 5 × 107 CFU/cm2 of leaf area as described earlier (74). Growth of P. syringae strains in leaves was measured by individually grinding four to six 0.2-cm2 leaf punches (excised with a no. 2 cork borer) in 10 mM MgSO4, plating appropriate dilutions on King's B medium (ES4326) containing appropriate antibiotics, and counting the CFU. Nicotiana tabacum (tobacco) cv. Xanthi was grown under greenhouse conditions, inoculated with ES4326 strains, and assayed for HR as previously described (72). Phaseolus vulgaris cv. Red Kidney (bean) was grown under greenhouse conditions and vacuum inoculated as described earlier (45).
RESULTS
Cloning of ES4326 hrpZ, hrpL, hrpS, and hrpR.
We cloned several genes that belong to the ES4326 hrp cluster which function at different levels in the hrp regulatory cascade. The ES4326 hrpRS genes were identified by colony hybridization using a heterologous hrpS probe from P. syringae pv. phaseolicola as described in Materials and Methods. Comparing the ES4326 sequences to hrpR and hrpS of P. syringae pv. syringae 61 revealed 83 and 85% DNA sequence identity, respectively. pEH10, a cosmid clone carrying ES4326 hrpS on a 24-kb insert complemented the inability of the P. syringae pv. phaseolicola hrpS mutant LRG94 to elicit disease symptoms on bean leaves, indicating that the ES4326 hrpS functions similarly to its counterpart in P. syringae pv. phaseolicola. As described in Materials and Methods, pEH10 was also shown to contain a presumptive hrpZ gene; a 90-bp sequence showed 82% identity to the P. syringae pv. syringae strain 61 hrpZ gene.
The ES426 hrpL gene was also identified by colony hybridization using a P. syringae pv. phaseolicola probe as described in Materials and Methods. A 5.8-kb EcoRI fragment containing hrpL was subcloned and sequenced, revealing a series of open reading frames corresponding to the P. syringae pv. syringae strain 61 hrpK, hrpL, hrpJ, hrcV, hrpJ3, and hrcN genes. The ES4326 and P. syringae pv. syringae strain 61 hrp genes are colinear and shared 73 to 89% DNA sequence identity, with hrpL being the most highly conserved.
Based on the structure of the hrp clusters in P. syringae pathovars phaseolicola and syringae, we expected that cosmid pEHL, which contains a 35-kb insert containing hrpL should overlap cosmid pEH10, which contains hrpS. Indeed, a 6-kb EcoRI fragment from pEH10 (that extends from the polylinker EcoRI site to the first EcoRI site; see hatched fragment in Fig. 1) hybridized to the 8-kb EcoRI fragment of pEHL. This allowed us to construct the restriction map of the ES4326 hrp cluster shown in Fig. 1.
Construction of uidA translational fusions.
Translational reporter fusions were constructed as described in Materials and Methods between the E. coli uidA structural gene for β-glucuronidase and the ES4326 hrpZ, hrpL, and hrpS genes, as well as the avirulence gene avrRpt2 from P. syringae pv. tomato. Plasmid pAVR12 contains 133 codons and 171 bp of upstream nontranslated sequence of avrRpt2 fused to the entire uidA gene, including the ATG translational initiation codon. Plasmid pHRPSgus contains 68 codons of hrpS and 1.7 kb of upstream sequence fused to the entire uidA gene. Plasmids pHRPZgus and pHRPLgus carry hrpZ-uidA and hrpL-uidA fusions at codons 135 and 25 of hrpZ and hrpL, respectively.
hrp and avrRpt2 transcription is rpoN dependent in vitro.
To test whether transcription of hrp and avr genes in ES4326 is rpoN dependent, we assayed expression of the hrpS-uidA, hrpL-uidA, hrpZ-uidA, and avrRpt2-uidA gene fusions in ES4326 rpoN::Kmr. Strains were grown overnight in KB media which suppresses hrp gene expression. The bacteria were then pelleted, washed, and resuspended in minimal M9 medium, a medium that derepresses hrp and avr gene expression (65). At various times, samples were taken and β-glucuronidase activity was determined, with zero time points taken in KB media before resuspension. The results, summarized in Fig. 2, show that the expression levels of hrpL-uidA, hrpZ-uidA, and avrRpt2-uidA fusions were at least 2 orders of magnitude lower in ES4326 rpoN::Kmr than in the wild type, whereas the hrpS-uidA fusion displayed the same low level of expression in both the wild type and in the rpoN::Kmr mutant. Interestingly, the timing of induction of hrpS-uidA was delayed in ES4326 rpoN::Kmr compared to the wild type, possibly due to the slower rate of growth of the rpoN mutant. It should be noted that we do not know what affect the Hrp and Avr protein regions in the fusion proteins have on the relative activity and stability of β-glucuronidase, and so expression data can only be compared for each fusion and not between different fusions.
FIG. 2.
hrpS, hrpZ, hrpL, and avrRpt2 transcriptional activity in ES4326 and ES4326 rpoN::Kmr following transfer from rich medium to minimal medium. ES4326 (□) and ES4326 rpoN::Kmr (◊) carrying fusions were shifted from rich (KB) to minimal (M9; pH 5.5) media at time zero, and the β-glucuronidase activity was measured at the indicated times as described in Materials and Methods. Glucuronidase activity is expressed as nanomoles per minute per cell. Time is given in hours after transfer to minimal medium. Reporter fusions: avrRpt2, avrRpt2-uidA; hrpZ, hrpZ-uidA; hrpL, hrpL-uidA; hrpS, hrpS-uidA.
To confirm the results obtained with the gene fusions, RNA blots containing total RNA isolated from ES4326, ES4326 rpoN::Kmr, ES4326 expressing avrRpt2 on plasmid pLH12, and ES4326 rpoN::kmr (pLH12) grown in M9 medium were probed with radiolabeled fragments containing avrRpt2. As shown in Fig. 3, the avrRpt2 transcript was detected only in the case of the wild-type strain carrying the avrRpt2 cosmid. The hrpL-uidA and hrpZ-uidA results were also confirmed by RNA blot analysis. Total RNA was isolated from ES4326 and ES4326 rpoN::Kmr grown in KB medium or after transfer to M9 minimal medium for 6 or 12 h. hrpL and hrpZ transcripts accumulated after transfer to minimal medium in ES4326 but not in ES4326 rpoN::Kmr (data not shown).
FIG. 3.
RNA blot analysis of avrRpt2 expression in P. syringae strains 6 h after transfer to minimal medium. Growth of bacteria and the RNA preparation was performed as described in Materials and Methods. A radiolabeled fragment encoding an internal portion of the avrRpt2 gene was used as a probe. Lanes: 1, ES4326(pLAFR3); 2, ES4326 rpoN (pLAFR3); 3, ES4326(pLH12); 4, ES4326 rpoN (pLH12).
rpoN is required for in planta induction of hrp and avr genes.
To determine whether hrp and avr gene regulation are substantially different in planta and in vitro, we monitored the expression of the hrpS-uidA, hrpL-uidA, hrpZ-uidA, and avrRpt2-uidA gene fusions in Arabidopsis leaves. As with the induction experiments in vitro, strains were grown overnight in KB medium but were then washed and resuspended in 10 mM MgSO4 before infiltration into Arabidopsis leaves. Figure 4, which presents β-glucuronidase activity data normalized to CFU, shows that avrRpt2-uidA, hrpZ-uidA, and hrpL-uidA were activated to significantly higher levels in wild-type ES4326 than in the ES4326 rpoN::Kmr mutant. In contrast to the in vitro experiment, some induction of the avrRpt2-uidA, hrpZ-uidA, and hrpL-uidA fusions appears to occur in the rpoN mutant, although the standard deviations in these strains were quite high and the β-glucuronidase activities were near the limits of detection. Another notable difference between the in vitro and in planta experiment was that hrpZ-uidA was induced to at least 10-fold-higher levels per cell in vitro than in planta, while hrpS-uidA was induced 10-fold higher in planta.
FIG. 4.
Time course of hrpS, hrpZ, hrpL, and avrRpt2 induction in ES4326 and ES4326 rpoN::Kmr following inoculation into Arabidopsis leaves. ES4326 (solid bars) and ES4326 rpoN::Kmr (cross-hatched bars) carrying hrpS, hrpZ, hrpL, and avrRpt2 fusions to uidA were inoculated into Arabidopsis leaves, and the glucuronidase activity was measured as described in Materials and Methods. Glucuronidase activity is reported as nanomoles per minute per cell. Time is given in hours after inoculation.
As was the case in vitro, hrpS-uidA activity in the strain ES4326 rpoN::Kmr mutant was comparable to wild-type levels at 6 h. Unexpectedly, however, as shown in Fig. 4, hrpS-uidA activity dropped sharply in the wild-type strain after 24 h, whereas activity dropped only slightly in ES4326 rpoN::Kmr. After 24 h, expression in the wild-type strain actually fell below the level in ES4326 rpoN::Kmr.
Constitutive expression of hrpL is sufficient to restore pathogenicity and the HR in rpoN mutants.
Because HrpL is sufficient to activate transcription of P. syringae pv. syringae hrp genes in E. coli (80), it appears likely that rpoN regulates hrp expression by activating the transcription of hrpL. If this were the case, constitutive expression of hrpL would be expected to suppress the nonpathogenic and non-HR-inducing phenotypes of an rpoN mutant. A fusion between the promoter region of lacZ and hrpL was constructed by cloning an engineered hrpL gene into the EcoRI site of pLAFR3 as described in Materials and Methods. Constitutive expression of hrpL (from plasmid pHRPLC) was confirmed by RNA blot analysis (data not shown). hrpZ was expressed at very high levels in ES4326 rpoN::Kmr (pHRPLC) even in hrp-suppressing KB media (data not shown). However, pHRPLC had no effect on other rpoN phenotypes, such as nonmotility and the inability to utilize nitrate as a sole nitrogen source (data not shown). As shown in the accompanying study (32), constitutive hrpL expression also did not restore expression of the phytotoxin coronatine, suggesting that ES4326 rpoN::Kmr (pHRPLC) phenotypes are most likely due to the specific activation of hrp and avrRpt2 genes. Interestingly, as shown in Fig. 5, ES4326 rpoN::Kmr carrying pHRPLC elicited a strong HR on tobacco leaves. ES4326 rpoN::Kmr (pHRPLC) also elicited water-soaked disease symptoms on Arabidopsis leaves, symptoms identical to the symptoms elicited by wild-type P. syringae (data not shown), when infiltrated at a titer of 5 × 107 CFU/ml. Lower doses of ES4326 rpoN::Kmr (pHRPLC) also elicited disease symptoms, though not to the same extent as the wild-type strain.
FIG. 5.
Constitutive expression of hrpL restores an HR when ES4326 rpoN is infiltrated into tobacco leaves. Tobacco leaves were infiltrated with bacterial suspensions (5 × 107 CFU/ml) as described in Materials and Methods, and symptoms were photographed 2 days postinoculation. Tobacco leaf sections: 1, ES4326 rpoN; 2, ES4326; 3, ES4326 rpoN (pHRPLC).
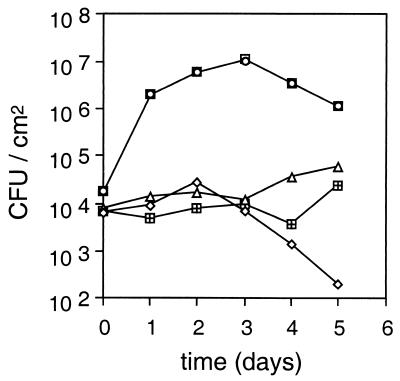
Despite the ability of strain ES4326 rpoN::Kmr (pHRPLC) to elicit disease symptoms, pHRPLC did not restore the ability of ES4326 rpoN::Kmr to grow in planta (Fig. 6). These data suggest that factors under rpoN control but not located in the hrp cluster may also be necessary for wild-type virulence in planta. However, there are two important caveats to this conclusion. First, ES4326 rpoN::Kmr (pHRPLC) grows even more slowly than the rpoN mutant itself, implying that constitutive expression of hrpL is deleterious. Second, the effect of pHRPLC on wild-type Psm ES4326 in planta could not be determined because the plasmid was rapidly lost from the wild-type strain upon infiltration (Fig. 6).
FIG. 6.
Constitutive expression of hrpL does not restore growth to strain ES4326 rpoN::Kmr in Arabidopsis leaves. Six-week-old Arabidopsis (Columbia) leaves were inoculated with bacterial suspension at 104 CFU/cm2, and bacterial titers were determined as described in Materials and Methods. Symbols: □, ES4326; ○, ES4326(pHRPLC) with no selection for pHRPLC; ◊, ES4326(pHRPLC) with antibiotic selection for pHRPLC; ▵, ES4326 rpoN::Kmr; ⊞, ES4326 rpoN::Kmr (pHRPLC).
Constitutive expression of avrRpt2 is not sufficient for the elicitation of the HR in an rpoN mutant.
We report in the accompanying study that an ES4326 rpoN mutant carrying the avirulence gene avrRpt2 failed to elicit an HR on A. thaliana (32). This is not surprising since avrRpt2 is expressed at very low levels in a rpoN mutant. Moreover, it is likely that AvrRpt2 is delivered directly into plant cells via the type III secretory system encoded by the hrp genes (58, 59, 61, 76). Therefore, we reasoned that constitutive avrRpt2 expression in ES4326 rpoN::Kmr would not be sufficient for elicitation of an HR. Indeed, ES4326 rpoN::Kmr containing plasmid pAVRC1 (which contains a lacZ-avrRpt2 fusion) did not elicit a detectable HR on tobacco or Arabidopsis leaves (data not shown).
Interestingly, ES4326(pAVRC1) did not elicit an HR in Arabidopsis at lower inoculation doses than ES4326 carrying pLH12, which contains avrRpt2 expressed from its own promoter. However, approximately 75% of the plants infiltrated with ES4326(pAVRC1) at a dose of 105 CFU/cm2 developed an HR 22 h after infiltration compared to only 12% of the plants inoculated with ES4326(pLH12). These data suggested that constitutive expression of avrRpt2 could potentially lead to enhanced resistance of the host. To investigate this latter point further, we determined the growth of ES4326(pLAFR3), ES4326(pLH12), and ES4326(pAVRC1) in Arabidopsis leaves. However, both pLH12 (avrRpt2) and pAVRC1 (lacZ-avrRpt2) resulted in the same decrease in growth compared to ES4326(pLAFR3) (data not shown).
Screen to find additional hrp regulatory factors.
The results described above show that rpoN gene is required for the transcriptional activation of the hrpL, hrpZ, and avrRpt2 genes but that hrpS transcription is rpoN independent. Assuming that rpoN itself is not transcriptionally regulated under hrp-inducing conditions in vitro or in planta and that HrpS plus RpoN activate hrpL, these results suggest that HrpS plus RpoN activity is regulated by environmental factors. In turn, this line of reasoning suggests that there could be previously unknown genes outside of the hrp cluster that play a role in hrp gene regulation. In this section we describe a series of experiments that were undertaken to isolate such mutants.
The hrpZ-uidA fusion described above was transferred from pHRPZgus to the ES4326 chromosome to produce ES4326 hrpZ::uidA by marker exchange. ES4326 hrpZ::uidA was then mutagenized using nitrosoguanidine and plated onto M9 medium containing X-Gluc. Among approximately 14,000 colonies screened, 297 exhibited reduced or no blue color and were tested further.
Among the 297 putative mutants, we were particularly interested in identifying mutations that mapped outside of the hrp gene cluster. We first screened the 297 mutants for putative hrpR, hrpS, or hrpZ::uidA mutants by complementation with pEH10 or pHRPZgus. All but five of the mutants were complemented by one of these two plasmids. The five remaining mutants were each mated to a strain ES4326 chromosomal library in pRR54cos, and the exconjugants were plated on M9 X-Gluc plates to isolate plasmids that restored β-glucuronidase activity. Two such plasmids, pEHL and pXIVB4, restored activity to all five of the mutants. These clones were isolated and shown by DNA blot analysis to contain hrpL. The simplest interpretation of these data is that no hrp regulators outside of the hrp cluster were identified in the screen.
DISCUSSION
Circuitry of hrp regulation.
We have shown in the accompanying study that ES4326 rpoN is nonpathogenic and that ES4326 rpoN carrying avrRpt2 fails to elicit an HR on Arabidopsis (32). In the experiments reported here we sought to determine whether the alternative sigma factor ς54 encoded by rpoN is required for the expression of hrp and avr genes, which would explain the nonpathogenic and non-HR-inducing phenotypes of the ES4326 rpoN mutant. The rational for investigating whether rpoN regulates hrp expression is that hrpS and hrpR encode proteins that are homologous to the NtrC group of transcriptional activators that work in concert with ς54 to activate target gene expression (26, 27, 54).
In the case of P. syringae pv. phaseolicola, it was shown that HrpR is required for hrpS transcription and that hrpS mutants fail to activate transcription of the remaining hrp and hrc genes in the cluster (26, 27). While hrpRS transcription has been shown to be strongly affected by plant signals (65), it is not clear from previous work whether rpoN is regulated at the transcriptional level or whether the activity of the HrpR or HrpS is environmentally regulated.
Expression studies, conducted both in vitro and in planta, determined that hrpL, hrpZ, and avrRpt2 expression is significantly higher in wild-type ES4326 than in an rpoN mutant. Curiously, the hrpZ-uidA fusion was activated at least 10-fold higher in vitro than in planta. In contrast to the other hrp genes, the activation of hrpS transcription does not appear to be rpoN-dependent. A hrpS-uidA fusion was expressed at the same levels in vitro in both wild-type and rpoN mutant cells. In planta, hrpS-uidA expression per cell was about 10-fold higher than in vitro, a finding similar to what has been reported previously for P. syringae pv. phaseolicola (65). Despite some inconsistencies, these data shown that the loss of pathogenicity and HR phenotypes of the rpoN mutant is most easily explained by the failure to activate hrp and avr transcription in the rpoN mutant.
In general, our data are consistent with previously proposed models of hrp regulation in P. syringae in which HrpS plus RpoN (ς54) activate hrpL and in which HrpL activates avr genes and the remaining genes in the hrp cluster (63, 80, 81). To further test this model, we placed hrpL expression under control of the E. coli lacZ promoter. Constitutive expression of hrpL not only restored hrpZ transcription to ES4326 rpoN::Kmr but also restored the elicitation of an HR on tobacco and disease symptoms in Arabidopsis. These data suggest that rpoN does not play a significant role in hrp gene expression downstream of HrpL. Moreover, constitutive expression of hrpL did not result in complementation of other rpoN phenotypes, including lack of motility, slow growth, and the inability to utilize nitrate as a sole nitrogen source. The results suggest that the role of rpoN in eliciting disease symptoms and the HR, at least in Arabidopsis and tobacco plants, respectively, is primarily limited to the activation of hrpL. On the other hand, as discussed further below, we report in the accompanying study that rpoN is also required for the hrp-independent synthesis of the phytotoxin coronatine, and it is possible that the failure to activate the coronatine pathway contributes to the loss of pathogenicity phenotype of the rpoN mutant (32).
hrpS expression and the role of HrpS versus HrpR in the regulation of the hrp cluster.
In vitro, the final induction level of the hrpS-uidA fusion was similar in the wild type and the rpoN mutant; however, hrpS-uidA induction was delayed in ES4326 rpoN::Kmr, possibly due to the relatively slow growth of the rpoN mutant. In planta, hrpS-uidA induction was initially the same in the wild-type and rpoN mutant. Interestingly, hrpS-uidA activity dropped dramatically in the wild-type strain 24 h after infection but only declined moderately in the mutant. One explanation for these data is that the lack of growth of ES4326 rpoN::Kmr in planta and its failure to induce host defense responses (32) might mimic the early stages of infection at which hrpS-uidA activity is normally high. Preston et al. have reported that hrpV acts as a negative regulator of hrp transcription, possibly upstream of hrpS (62). Thus, hrpS downregulation could be the result of feedback inhibition by hrpV, an effect similar to the feedback regulation of the type III secretion system of Yersinia pestis (43). This seems unlikely though, as we would not then have expected to see the downregulation of the hrpS-uidA fusion in the rpoN mutant in vitro (see Fig. 2).
Another aspect of hrp gene regulation that needs further study is the relationship between HrpR and HrpS. The hrpS gene is downstream of hrpR, but they do not appear to constitute an operon (26). In both P. syringae pv. phaseolicola and ES4326, hrpS transcription must initiate within the hrpR gene and in P. syringae pv. phaseolicola, hrpR is required for hrpS transcription (26). However, because hrpS expression is not significantly reduced in rpoN mutants, either HrpR activity is rpoN independent or hrpS transcription is not dependent on HrpR in P. syringae pv. maculicola.
It is conceivable that in ES4326 hrpR is transcriptionally regulated by upstream factors and that HrpR plus RpoN activate hrpL. We have not specifically tested ES4326 hrpS or hrpR mutants to determine if they have the same phenotype as P. syringae pv. phaseolicola hrpS mutants.
rpoN- and hrpL-regulated virulence factors.
One unexpected aspect of our studies with constitutive hrpL expression is that hrpL restored both disease and HR symptoms to ES4326 rpoN::Kmr but did not restore growth in planta. This is a complicated result, and the failure of ES4326 rpoN::Kmr (pHRPLC) to grow in planta needs to be discussed separately from the fact that it elicited host symptoms. One simple explanation for the failure to grow relates to the highly pleiotropic nature of the rpoN mutation. For example, the rpoN mutant might be unable to metabolize a major nutrient source in the plant, irrespective of hrpL expression. Another explanation for the failure to grow in planta could be related to the fact that HrpL is an alternative sigma factor (80, 81) and that constitutive expression could interfere with a variety bacterial functions not related to pathogenicity. Indeed, ES4326 strains expressing hrpL constitutively grew more slowly in vitro, whereas in planta the plasmid carrying the hrpL construct was rapidly lost from a wild-type strain.
A more interesting possibility for the failure of ES4326 rpoN::Kmr (pHRPLC) to grow in planta is that important virulence factors required for in planta growth may lie outside of the hrp regulation pathway but under rpoN control. We show in the accompanying study that the phytotoxin coronatine is one such factor, being absent in rpoN mutants and not restored by constitutive hrpL expression (32). Coronatine is a non-host-specific toxin that primarily elicits chlorosis but is also known to induce hypertrophy and inhibit root elongation (7, 19, 69). Studies of coronatine-defective mutants of P. syringae pv. tomato have shown that the toxin normally contributes to lesion expansion, chlorosis, and bacterial multiplication in Arabidopsis (55). While the absence of coronatine alone seems insufficient to explain the failure of ES4326 rpoN::Kmr (pHRPLC) to grow in planta, coronatine may be one of several virulence factors under rpoN control but not hrpL control that contribute to pathogenicity.
In any case, our hrpL overexpression studies have shown that there is not a necessary linkage between symptom development in Arabidopsis and growth of P. syringae pv. maculicola in planta but that hrpL-dependent factors are required for the elicitation of these symptoms. This is consistent with the result reported in the accompanying study (32) that ES4326 rpoN::Kmr (pHRPLC) elicits defense gene induction, whereas ES4326 rpoN::Kmr does not when infiltrated into Arabidopsis leaves. An important future experiment is to determine whether the symptoms elicited by ES4326 rpoN::Kmr (pHRPLC) involve necrosis or programmed cell death (PCD) or both. One intriguing possibility is that hrpL-dependent factors are involved in eliciting PCD in virulent interactions that cause disease as well as in avirulent interactions that result in an HR.
Failure to find upstream regulatory mutants.
Considering the relatively large number of mutants screened and because putative mutations were obtained in each of the known hrp regulatory loci, hrpR, hrpS, and hrpL, it seems likely that the screen for hrp regulatory mutants was saturated. However, there is a potential problem with the screen design that relates to the trans-complementation method used to identify, and thus remove for further consideration, hrpR, hrpS, and hrpZ::uidA mutants. It is conceivable that overexpression of hrpR and hrpS from pEH10 or pHRPZgus compensated for mutations in upstream regulatory factors or titrated out a factor that interacts directly with HrpS or HrpR to downregulate its activity.
It is possible that hrp gene regulation is mediated by a factor that interacts with HrpR and/or HrpS and/or RpoN. Another possibility is that rpoN expression itself is transcriptionally regulated. Regulation of rpoN transcription has been reported in Bradyrhizobium japonicum (42) and Rhodobacter capsulatus (12). In P. aeruginosa (38) and Klebsiella pneumonia (51), genes found downstream of rpoN appear to modify RpoN activity. Unfortunately, mutations that affect rpoN transcription or activity would probably have a slow-growth phenotype similar to rpoN mutants and would probably not have been identified in our screen.
Some regulatory genes such as the P. aeruginosa fur gene, which encodes an iron-responsive protein, are essential for bacterial survival (5). However, mutagenesis using NG should produce mutants that retain partial function, resulting in an intermediate phenotype allowing identification of such essential genes (52). We did not identify such mutants, though the screen might not have been sufficiently sensitive to detect small changes in activity.
hrpV has been reported to be a negative regulator of hrp expression, and there is some evidence that hrpV functions upstream of hrpRS (62). It is not likely, however, that we would have detected mutations in negative regulators because the screen was designed specifically to identify positive regulators outside of the hrp cluster. The high level of hrpZ-uidA expression in wild-type cells on the minimal media used in the screen most likely obscured the recovery of mutants with enhanced levels of expression. In any case, it is likely that hrpV mutants would have been complemented by the plasmids that we used to screen against hrpRS and hrpL mutants. Taken together, our results suggest that upstream regulators of hrp are either negative regulators, essential genes, or regulators of ς54 activity or transcription.
ACKNOWLEDGMENTS
We thank Carol Bender for editing the manuscript and Laurence Rahme for helpful discussions.
This work was supported by NIH grant GM48707 awarded to F.M.A.
REFERENCES
- 1.Albright L M, Huala E, Ausubel F M. Prokaryotic signal transduction mediated by sensor and regulator protein pairs. Annu Rev Genet. 1989;23:311–336. doi: 10.1146/annurev.ge.23.120189.001523. [DOI] [PubMed] [Google Scholar]
- 2.Arlat M, Van Gijsegem F, Huet J C, Pernollet J C, Boucher C A. PopA1, a protein which induces a hypersensitivity-like response on specific Petunia genotypes, is secreted via the Hrp pathway of Pseudomonas solanacearum. EMBO J. 1994;13:543–553. doi: 10.1002/j.1460-2075.1994.tb06292.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons; 2000. [Google Scholar]
- 4.Backman K, Ptashne M. Maximizing gene expression on a plasmid using recombination in vitro. Cell. 1978;13:65–71. doi: 10.1016/0092-8674(78)90138-1. [DOI] [PubMed] [Google Scholar]
- 5.Barton H A, Johnson Z, Cox C D, Vasil A I, Vasil M L. Ferric uptake regulator mutants of Pseudomonas aeruginosa with distinct alterations in the iron-dependent repression of exotoxin A and siderophores in aerobic and microaerobic environments. Mol Microbiol. 1996;21:1001–1017. doi: 10.1046/j.1365-2958.1996.381426.x. [DOI] [PubMed] [Google Scholar]
- 6.Bender C, Alarcon-Chaidez F, Gross D. Pseudomonas syringae phytotoxins: mode of action, regulation, and biosynthesis by peptide and polyketide synthetases. Microbiol Mol Biol Rev. 1999;63:266–292. doi: 10.1128/mmbr.63.2.266-292.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bender C L, Stone H E, Sims J J, Cooksey D A. Reduced pathogen fitness of Pseudomonas syringae pv. tomato Tn5 mutants defective in coronatine production. Physiol Mol Plant Pathol. 1987;30:273–283. [Google Scholar]
- 8.Black L K, Maier R J. IHF- and RpoN-dependent regulation of hydrogenase expression in Bradyrhizobium japonicum. Mol Microbiol. 1995;16:405–413. doi: 10.1111/j.1365-2958.1995.tb02406.x. [DOI] [PubMed] [Google Scholar]
- 9.Bogdanove A J, Beer S V, Bonas U, Boucher C A, Collmer A, Coplin D L, Cornelis G R, Huang H-C, Hutcheson S W, Panopoulos N J, Van Gijsegem F. Unified nomenclature for broadly conserved hrp genes of phytopathogenic bacteria. Mol Microbiol. 1996;20:681–683. doi: 10.1046/j.1365-2958.1996.5731077.x. [DOI] [PubMed] [Google Scholar]
- 10.Charkowski A, Alfano J, Preston G, Yuan J, He S, Collmer A. The Pseudomonas syringae pv. tomato HrpW protein has domains similar to haprins and pectate lyases and can elicit the plant hypersensitive response and bind to pectate. J Bacteriol. 1998;180:5211–5217. doi: 10.1128/jb.180.19.5211-5217.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Charkowski A, Huang H, Collmer A. Altered localization of HrpZ in Pseudomonas syringae pv. syringae hrp mutants suggests that different components of the type III secretion pathway control protein translocation across the inner and outer membranes of gram-negative bacteria. J Bacteriol. 1997;179:3966–3974. doi: 10.1128/jb.179.12.3866-3874.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cullen P J, Foster-Hartnett D, Gabbert K K, Kranz R G. Structure and expression of the alternative sigma factor, RpoN, in Rhodobacter capsulatus; physiological relevance of an autoactivated nifU2-rpoN superoperon. Mol Microbiol. 1994;11:51–65. doi: 10.1111/j.1365-2958.1994.tb00289.x. [DOI] [PubMed] [Google Scholar]
- 13.Davis K R, Ausubel F M. Characterization of elicitor-induced defense responses in suspension-cultured cells of Arabidopsis. Mol Plant-Microbe Interact. 1989;2:363–368. [Google Scholar]
- 14.Davis K R, Schott E, Ausubel F M. Virulence of selected phytopathogenic pseudomonads in Arabidopsis thaliana. Mol Plant-Microbe Interact. 1991;4:477–488. [Google Scholar]
- 15.Ditta G, Stanfield S, Corbin D, Helinski D R. Broad-host-range DNA cloning system for gram-negative bacteria: construction of a gene bank of Rhizobium meliloti. Proc Natl Acad Sci USA. 1980;27:7347–7451. doi: 10.1073/pnas.77.12.7347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dong X, Mindrinos M, Davis K R, Ausubel F M. Induction of Arabidopsis thaliana defense genes by virulent and avirulent Pseudomonas syringae strains and by a cloned avirulence gene. Plant Cell. 1991;3:61–72. doi: 10.1105/tpc.3.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fellay R, Rahme L G, Mindrinos M N, Frederick R D, Pisi A, Panopoulos N J. Genes and signals controlling the Pseudomonas syringae pv. phaseolicola-plant interaction. In: Hennecke H, Verma D P S, editors. Molecular genetics of plant-microbe interactions. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1991. pp. 45–52. [Google Scholar]
- 18.Fenselau S, Balbo I, Bonas U. Determinants of pathogenicity in Xanthomonas campestris pv. vesicatoria are related to proteins involved in secretion in bacterial pathogens of animals. Mol Plant-Microbe Interact. 1992;5:390–396. doi: 10.1094/mpmi-5-390. [DOI] [PubMed] [Google Scholar]
- 19.Feys B J F, Benedetti C E, Penfold C N, Turner J G. Arabidopsis mutants selected for resistance to the phytotoxin coronatine are male sterile, insensitive to methyl jasmonate, and resistant to a bacterial pathogen. Plant Cell. 1994;6:751–759. doi: 10.1105/tpc.6.5.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Finan T M, Kunkel B, de Vos G F, Signer E R. A second symbiotic megaplasmid in Rhizobium meliloti carrying exopolysaccharide and thiamine synthesis genes. J Bacteriol. 1986;167:66–72. doi: 10.1128/jb.167.1.66-72.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Flor H H. Current status of the gene-for-gene concept. Annu Rev Phytopathol. 1971;9:275–296. [Google Scholar]
- 22.Glazebrook J, Walker G C. A novel exopolysaccharide can function in place of the calcofluor-binding exopolysaccharide in nodulation of alfalfa by Rhizobium meliloti. Cell. 1989;56:661–672. doi: 10.1016/0092-8674(89)90588-6. [DOI] [PubMed] [Google Scholar]
- 23.Gopalan S, Bauer D W, Alfano J R, Loniello A O, He S Y, Collmer A. Expression of the Pseudomonas syringae avirulence protein AvrB in plant cells alleviates its dependence on the hypersensitive response and pathogenicity (Hrp) secretion system in eliciting genotype-specific hypersensitive cell death. Plant Cell. 1996;8:1095–1105. doi: 10.1105/tpc.8.7.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gough C L, Genin S, Zischek C, Boucher C A. hrp genes of Pseudomonas solanacearum are homologous to pathogenicity determinants of animal pathogenic bacteria and are conserved among plant pathogenic bacteria. Mol Plant-Microbe Interact. 1992;5:384–389. doi: 10.1094/mpmi-5-384. [DOI] [PubMed] [Google Scholar]
- 25.Greenberg J T, Guo A, Klessig D F, Ausubel F M. Programmed cell death in plants: a pathogen-triggered response activated coordinately with multiple defense functions. Cell. 1994;77:551–563. doi: 10.1016/0092-8674(94)90217-8. [DOI] [PubMed] [Google Scholar]
- 26.Grimm C, Aufsatz W, Panopoulos N J. The hrpRS locus of Pseudomonas syringae pv. phaseolicola constitutes a complex regulatory unit. Mol Microbiol. 1995;15:155–165. doi: 10.1111/j.1365-2958.1995.tb02230.x. [DOI] [PubMed] [Google Scholar]
- 27.Grimm C G, Panopoulos N J. The predicted protein product of a pathogenicity locus from Pseudomonas syringae pv. phaseolicola is homologous to a highly conserved domain of several prokaryotic regulatory proteins. J Bacteriol. 1989;171:5031–5038. doi: 10.1128/jb.171.9.5031-5038.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hanahan D, Meselson M. Plasmid screening at high colony density. Methods Enzymol. 1983;100:333–342. doi: 10.1016/0076-6879(83)00066-x. [DOI] [PubMed] [Google Scholar]
- 29.Haula E, Ausubel F M. The central domain of Rhizobium meliloti NifA is sufficient to activate transcription from the R. meliloti nifH promoter. J Bacteriol. 1989;171:3354–3365. doi: 10.1128/jb.171.6.3354-3365.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Haula E, Moon A L, Ausubel F M. The central domain of Rhizobium leguminosarum DctD functions independently to activate transcription. J Bacteriol. 1992;174:1428–1431. doi: 10.1128/jb.174.4.1428-1431.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.He S Y, Huang H-C, Collmer A. Pseudomonas syringae pv. syringae HarpinPss: a protein that is secreted via the Hrp pathway and elicits the hypersensitive response in plants. Cell. 1993;73:1255–1266. doi: 10.1016/0092-8674(93)90354-s. [DOI] [PubMed] [Google Scholar]
- 32.Hendrickson E, Guevara P, Peñaloza-Vàzquez A, Shao J, Bender C L. Virulence of the phytopathogen Pseudomonas syringae pv. maculicola is rpoN dependent. J Bacteriol. 2000;182:3498–3507. doi: 10.1128/jb.182.12.3498-3507.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang H-C, Lin R-H, Chang C-J, Collmer A, Deng W-L. The complete hrp gene cluster of Pseudomonas syringae pv. syringae 61 includes two blocks of genes required for HarpinPss secretion that are arranged colinearly with Yersinia ysc homologs. Mol Plant-Microbe Interact. 1995;8:733–746. doi: 10.1094/mpmi-8-0733. [DOI] [PubMed] [Google Scholar]
- 34.Huynh T V, Dahlbeck D, Staskawicz B J. Bacterial blight of soybean: regulation of a pathogen gene determining host cultivar specificity. Science. 1989;245:1374–1377. doi: 10.1126/science.2781284. [DOI] [PubMed] [Google Scholar]
- 35.Innes R, Bent A, Kunkel B, Bisgrove S, Staskawicz B. Molecular analysis of avirulence gene avrRpt2 and identification of a putative regulatory sequence common to all known Pseudomonas syringae avirulence genes. J Bacteriol. 1993;175:4859–4869. doi: 10.1128/jb.175.15.4859-4869.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jefferson R A. Assaying chimeric genes in plants: the GUS gene fusion system. Plant Mol Biol Reporter. 1987;5:387–405. [Google Scholar]
- 37.Jefferson R A, Kavanagh T A, Bevan M W. GUS fusions: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 1987;13:3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jin S, Ishimoto K, Lory S. Nucleotide sequence of the rpoN gene and characterization of two downstream open reading frames in Pseudomonas aeruginosa. J Bacteriol. 1994;176:1316–1322. doi: 10.1128/jb.176.5.1316-1322.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kessler B, Marqués S, Köhler T, Ramos J L, Timmis K N, de Lorenzo V. Cross talk between catabolic pathways in Pseudomonas putida: XylS-dependent and -independent activation of the TOL meta operon requires the same cis-acting sequences within the Pm promoter. J Bacteriol. 1994;176:5578–5582. doi: 10.1128/jb.176.17.5578-5582.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.King E O, Ward M K, Raney D E. Two simple media for the demonstration of phycocyanin and fluorescin. J Lab Clin Med. 1954;44:301–307. [PubMed] [Google Scholar]
- 41.Köhler T, Michéa-Hamzehpour M, Henze U, Gotoh N, Curty L K, Pechère J-C. Characterization of MexE-MexF-OprN, a positively regulated multidrug efflux system of Pseudomonas aeruginosa. Mol Microbiol. 1997;23:345–354. doi: 10.1046/j.1365-2958.1997.2281594.x. [DOI] [PubMed] [Google Scholar]
- 42.Kullik I, Fritsche S, Knobel H, Sanjuan J, Hennecke H, Fischer H-M. Bradyrhizobium japonicum has two differentially regulated, functional homologs of the ς54 gene (rpoN) J Bacteriol. 1991;173:1125–1138. doi: 10.1128/jb.173.3.1125-1138.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee C A. Type III secretion systems: machines to deliver bacterial proteins into eukaryotic cells? Trends Microbiol. 1997;5:148–156. doi: 10.1016/S0966-842X(97)01029-9. [DOI] [PubMed] [Google Scholar]
- 44.Lindgren P B, Panopoulos N J, Staskawicz B J, Dahlbeck D. Genes required for pathogenicity and hypersensitivity are conserved and interchangeable among pathovars of Pseudomonas syringae. Mol Gen Genet. 1988;211:499–506. [Google Scholar]
- 45.Lindgren P B, Peet R C, Panopoulos N J. Gene cluster of Pseudomonas syringae pv. “phaseolicola” controls pathogenicity on bean and hypersensivity on non-host plants. J Bacteriol. 1986;168:512–522. doi: 10.1128/jb.168.2.512-522.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lorang J M, Keen N T. Characterization of avrE from Pseudomonas syringae pv. tomato: a hrp-linked avirulence locus consisting of at least two transcriptional units. Mol Plant-Microbe Interact. 1995;8:49–57. doi: 10.1094/mpmi-8-0049. [DOI] [PubMed] [Google Scholar]
- 47.Macaluso A, Best E A, Bender R A. Role of the nac gene product in the nitrogen regulation of some NTR-regulated operons of Klebsiella aerogenes. J Bacteriol. 1990;172:7249–7255. doi: 10.1128/jb.172.12.7249-7255.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1982. [Google Scholar]
- 49.Martin D W, Holloway B W, Deretic V. Characterization of a locus determining the mucoid status of Pseudomonas aeruginosa AlgU shows sequence similarities with a Bacillus sigma factor. J Bacteriol. 1993;175:1153–1164. doi: 10.1128/jb.175.4.1153-1164.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Merrick M J. In a class of its own—the RNA polymerase sigma factor ς54 (ςN) Mol Microbiol. 1993;10:903–909. doi: 10.1111/j.1365-2958.1993.tb00961.x. [DOI] [PubMed] [Google Scholar]
- 51.Merrick M J, Coppard J R. Mutations in genes downstream of the rpoN gene (encoding ς54) of Klebsiella pneumoniae affect repression from ς54-dependent promoters. Mol Microbiol. 1989;3:1765–1775. doi: 10.1111/j.1365-2958.1989.tb00162.x. [DOI] [PubMed] [Google Scholar]
- 52.Miller J H. Experiments in molecular genetics. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 53.Miller W, Mindrinos M N, Rahme L G, Frederick R D, Grimm C, Gressman R, Kyriakides X, Kokkinidis M, Panopoulos N J. Pseudomonas syringae pv. phaseolicola-plant interactions: host-pathogen signalling through cascade control of hrp gene expression. In: Nester E W, Verma D P S, editors. Advances in molecular genetics of plant-microbe interactions. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1993. pp. 267–274. [Google Scholar]
- 54.Mindrinos M N, Rahme L G, Frederick R D, Hatziloukas E, Grimm C, Panopoulos N J. Pseudomonas biotransformations, pathogenesis and evolving biotechnology. Washington, D.C.: American Society for Microbiology; 1990. Structure, function, regulation, and evolution of genes involved in pathogenicity, the hypersensitive response, and phaseolotoxin immunity in the bean halo blight pathogen; pp. 74–81. [Google Scholar]
- 55.Mittal S, Davis K R. Role of the phytotoxin coronatine in the infection of Arabidopsis thaliana by Pseudomonas syringae pv. tomato. Mol Plant-Microbe Interact. 1995;8:165–171. doi: 10.1094/mpmi-8-0165. [DOI] [PubMed] [Google Scholar]
- 56.Mittler R, Lam E. Sacrifice in the face of foes: pathogen-induced programmed cell death in plants. Trends Microbiol. 1996;4:10–15. doi: 10.1016/0966-842x(96)81499-5. [DOI] [PubMed] [Google Scholar]
- 57.Moore R A, Starratt A N, Ma S-W, Morris V L, Cuppels D A. Identification of a chromosomal region required for biosynthesis of the phytotoxin coronatine by Pseudomonas syringae pv. tomato. Can J Microbiol. 1989;35:910–917. [Google Scholar]
- 58.Mudgett M, Staskawicz B. Characterization of the Pseudomonas syringae pv. tomato AvrRpt2 protein: demonstration of secretion and processing during bacterial pathogenesis. Mol Microbiol. 1999;32:927–41. doi: 10.1046/j.1365-2958.1999.01403.x. [DOI] [PubMed] [Google Scholar]
- 59.Mudgett M, Staskawicz B. Protein signaling via type III secretion pathways in phytopathogenic bacteria. Curr Opin Microbiol. 1998;1:109–114. doi: 10.1016/s1369-5274(98)80150-1. [DOI] [PubMed] [Google Scholar]
- 60.O'Toole R, Milton D L, Wolf Watz H. Chemotactic motility is required for invasion of the fish pathogen Vibrio anguillarum. Mol Microbiol. 1996;19:625–637. doi: 10.1046/j.1365-2958.1996.412927.x. [DOI] [PubMed] [Google Scholar]
- 61.Pirhonen M U, Lidell M C, Rowley D L, Lee S W, Jin S, Liang Y, Silverstone S, Keen N T, Hutcheson S W. Phenotypic expression of Pseudomonas syringae avr genes in E. coli is linked to the activities of the hrp-encoded secretion system. Mol Plant-Microbe Interact. 1996;9:252–260. doi: 10.1094/mpmi-9-0252. [DOI] [PubMed] [Google Scholar]
- 62.Preston G, Deng W, Huang H, Collmer A. Negative regulation of hrp genes in Pseudomonas syringae by HrpV. J Bacteriol. 1988;180:4532–4537. doi: 10.1128/jb.180.17.4532-4537.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Puri N, Jenner C, Bennett M, Stewart R, Mansfield J, Lyons N, Taylor J. Expression of avrPphB, an avirulence gene from Pseudomonas syringae pv. phaseolicola, and the delivery of signals causing the hypersensitive reaction in bean. Mol Plant-Microbe Interact. 1997;10:247–256. doi: 10.1094/MPMI.1997.10.2.247. [DOI] [PubMed] [Google Scholar]
- 64.Rahme L G, Mindrinos M N, Panopoulos N J. Genetic and transcriptional organization of the hrp cluster of Pseudomonas syringae pv. phaseolicola. J Bacteriol. 1991;173:575–586. doi: 10.1128/jb.173.2.575-586.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rahme L G, Mindrinos M N, Panopoulos N J. Plant and environmental sensory signals control the expression of hrp genes in Pseudomonas syringae pv. phaseolicola. J Bacteriol. 1992;174:3499–3507. doi: 10.1128/jb.174.11.3499-3507.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ritter C, Dangl J L. The avrRpm1 gene of Pseudomonas syringae pv. maculicola is required for virulence on Arabidopsis. Mol Plant-Microbe Interact. 1995;8:444–453. doi: 10.1094/mpmi-8-0444. [DOI] [PubMed] [Google Scholar]
- 67.Rogers E E, Ausubel F M. Arabidopsis enhanced disease susceptibility mutants exhibit enhanced susceptibility to several bacterial pathogens and alterations in PR-1 gene expression. Plant Cell. 1997;9:305–316. doi: 10.1105/tpc.9.3.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ruvkun G B, Ausubel F M. A general method for site directed mutagenesis in prokaryotes: construction of mutations in symbiotic nitrogen fixation genes of Rhizobium meliloti. Nature. 1981;289:85–88. doi: 10.1038/289085a0. [DOI] [PubMed] [Google Scholar]
- 69.Sakai R, Nishiyama K, Ichihara A, Shiraishi K, Sakamura S. The relation between bacterial toxic action and plant growth regulation. In: Daly J M, Uritani I, editors. Recognition and specificity in host-parasite interactions. Baltimore, Md: University Park Press; 1979. pp. 165–179. [Google Scholar]
- 70.Salmeron J M, Staskawicz B J. Molecular characterization and hrp dependence of the avirulence gene avrPto from Pseudomonas syringae pv. tomato. Mol Gen Genet. 1993;239:6–16. doi: 10.1007/BF00281595. [DOI] [PubMed] [Google Scholar]
- 71.Scofield S R, Tobias C M, Rathjen J P, Chang J H, Lavelle D T, Michelmore R W, Staskawicz B J. Molecular basis of gene-for-gene specificity in bacterial speck disease of tomato. Science. 1996;274:2063–2065. doi: 10.1126/science.274.5295.2063. [DOI] [PubMed] [Google Scholar]
- 72.Staskawicz B J, Dahlbeck D, Keen N T, Napoli C. Molecular characterization of cloned avirulence genes from race 0 and race 1 of Pseudomonas syringae pv. glycinea. J Bacteriol. 1987;169:5789–5794. doi: 10.1128/jb.169.12.5789-5794.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Straley S C, Plano G V, Skrzypek E, Haddix P L, Fields K A. Regulation by Ca2+ in the Yersinia low-Ca2+ response. Mol Microbiol. 1993;8:1005–1010. doi: 10.1111/j.1365-2958.1993.tb01644.x. [DOI] [PubMed] [Google Scholar]
- 74.Swanson J, Kearney B, Dahlbeck D, Staskawicz B J. Cloned avirulence gene of Xanthomonas campestris pv. vesicatoria complements spontaneous race change mutant. Mol Plant-Microbe Interact. 1988;1:5–9. [Google Scholar]
- 75.Tang X, Frederick R D, Zhou J, Halterman D A, Jia Y, Martin G B. Initiation of plant disease resistance by physical interaction of AvrPto and Pto kinase. Science. 1996;274:2060–2063. doi: 10.1126/science.274.5295.2060. [DOI] [PubMed] [Google Scholar]
- 76.van Dijk K, Fouts D E, Rehm A H, Hill A R, Collmer A, Alfano J R. The Avr (effector) proteins HrmA (HopPsyA) and AvrPto are secreted in culture from Pseudomonas syringae pathovars via the Hrp (type III) protein secretion system in a temperature- and pH-sensitive manner. J Bacteriol. 1999;181:4790–4797. doi: 10.1128/jb.181.16.4790-4797.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Whalen M C, Innes R W, Bent A F, Staskawicz B J. Identification of Pseudomonas syringae pathogens of Arabidopsis and a bacterial locus determining avirulence on both Arabidopsis and soybean. Plant Cell. 1991;3:49–59. doi: 10.1105/tpc.3.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Willis D K, Hrabak E M, Rich J J, Barta T M, Lindow S E, Panopoulos N J. Isolation and characterization of a Pseudomonas syringae pv. syringae mutant deficient in lesion formation on bean. Mol Plant-Microbe Interact. 1990;3:149–156. [Google Scholar]
- 79.Wu Z-L, Charles T C, Wang H, Nester E W. The ntrA gene of Agrobacterium tumefaciens: identification, cloning, and phenotype of a site-directed mutant. J Bacteriol. 1992;174:2720–2723. doi: 10.1128/jb.174.8.2720-2723.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Xiao Y, Heu S, Yi J, Lu Y, Hutcheson S W. Identification of a putative alternate sigma factor and characterization of a multicomponent regulatory cascade controlling the expression of Pseudomonas syringae pv. syringae Pss61 hrp and hrmA genes. J Bacteriol. 1994;176:1025–1036. doi: 10.1128/jb.176.4.1025-1036.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Xiao Y, Hutcheson S W. A single promoter sequence recognized by a newly identified alternate sigma factor directs expression of pathogenicity and host range determinants in Pseudomonas syringae. J Bacteriol. 1994;176:3089–3091. doi: 10.1128/jb.176.10.3089-3091.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yu G-L, Katagiri F, Ausubel F M. Arabidopsis mutations at the RPS2 locus result in loss of resistance to Pseudomonas syringae strains expressing the avirulence gene avrRpt2. Mol Plant-Microbe Interact. 1993;6:434–443. doi: 10.1094/mpmi-6-434. [DOI] [PubMed] [Google Scholar]