Highlights
-
•
A relatively low diversity of OsHV-1 genotypes was found in Australia.
-
•
Distinct OsHV-1 genotypes were geographically clustered in estuaries.
-
•
Australian OsHV-1 genotypes grouped in a globally distinct cluster.
-
•
Australian POMS outbreaks were not due to OsHV-1 microvariants.
Keywords: Ostreid herpesvirus, Variant, Diversity, Polymorphism, Phylogeny, Crassostrea gigas, Australia
Abstract
Since 2010, mass mortality events known as Pacific oyster mortality syndrome (POMS) have occurred in Crassostrea gigas in Australia associated with Ostreid herpesvirus 1. The virus was thought to be an OsHV-1 µVar or “microvariant”, i.e. one of the dominant variants associated with POMS in Europe, but there are few data to characterize the genotype in Australia. Consequently, the genetic identity and diversity of the virus was determined to understand the epidemiology of the disease in Australia. Samples were analysed from diseased C. gigas over five summer seasons between 2011 and 2016 in POMS-affected estuaries: Georges River in New South Wales (NSW), Hawkesbury River (NSW) and Pitt Water in Tasmania. Sequencing was attempted for six genomic regions. Numerous variants were identified among these regions (n = 100 isolates) while twelve variants were identified from concatenated nucleotide sequences (n = 61 isolates). Nucleotide diversity of the seven genotypes of C region among Australian isolates (Pi 0.99 × 10−3) was the lowest globally. All Australian isolates grouped in a cluster distinct from other OsHV-1 isolates worldwide. This is the first report that Australian outbreaks of POMS were associated with OsHV-1 distinct from OsHV-1 reference genotype, µVar and other microvariants from other countries. The findings illustrate that microvariants are not the only variants of OsHV-1 associated with mass mortality events in C. gigas. In addition, there was mutually exclusive spatial clustering of viral genomic and amino acid sequence variants between estuaries, and a possible association between genotype/amino acid sequence and the prevalence and severity of POMS, as this differed between these estuaries. The sequencing findings supported prior epidemiological evidence for environmental reservoirs of OsHV-1 for POMS outbreaks in Australia.
1. Introduction
Since late last century, the Pacific oyster, Crassostrea gigas, has been subject to recurrent disease events (Nicolas et al., 1992; Renault et al., 2000; Renault et al., 1994; Samain et al., 2007). However, in 2008 a marked increase in mass mortality outbreaks occurred in France (Cochennec-Laureau et al., 2010; Martenot et al., 2011; Segarra et al., 2010) and later in other areas in the world (Abbadi et al., 2018; Batista et al., 2015; Gittenberger et al., 2016; Hwang et al., 2013; Keeling et al., 2014; Mortensen et al., 2016; Peeler et al., 2012; Roque et al., 2012; Segarra et al., 2010). The disease was called Pacific oyster mortality syndrome (POMS) when it first appeared in Australia in 2010 in the Georges and Parramatta rivers, which are both major estuaries in Sydney (Jenkins et al., 2013; Paul-Pont et al., 2013a). POMS causes considerable economic losses in Australia, New Zealand and Europe (Fuhrmann et al., 2019). It is a multifactorial disease influenced by environmental conditions such as water temperature and host characteristics such as age and size (Hick et al., 2018; Petton et al., 2013) and although it has a proposed polymicrobial etiology involving bacteria (de Lorgeril et al., 2018; Friedman et al., 2005; Petton et al., 2021), outbreaks are induced by infection with Ostreid herpesvirus 1 microvariants (Batista et al., 2015; Gittenberger et al., 2016; Hwang et al., 2013; Jenkins et al., 2013; Keeling et al., 2014; Mortensen et al., 2016; Peeler et al., 2012; Renault et al., 2012; Roque et al., 2012). The disease typically occurs in summer and mortality can be reproduced experimentally by controlled exposure of oysters to the virus alone in a dose-responsive and temperature-dependent manner (de Kantzow et al., 2016; Paul-Pont et al., 2015).
Ostreid herpesvirus 1, OsHV-1, which is the only species included in the genus Ostreavirus, is a double-stranded DNA virus (Davison et al., 2009). The presence of the gene coding for the ATPase subunit of a terminase involved in the packaging of viral DNA inside the capsid, in addition to intranuclear replication, places OsHV-1 in the Malacoherpesviridae under the order Herpesvirales (Davison et al., 2009; Davison et al., 2005). Members of this taxon are widespread, both in terms of the range of species infected and also geographically. They have been detected in many hosts besides C. gigas including Crassostrea virginica (Farley et al., 1972), Ostrea angasi (Hine and Thorne, 1997), Ostrea edulis (Comps and Cochennec, 1993), Tiostrea chilensis (Hine et al., 1998), Pecten maximus (Arzul et al., 2001a), Ruditapes philippinarum (Renault et al., 2001), Ruditapes decusstaus (Renault and Arzul, 2001), Crassostrea angulata (Batista et al., 2015), Scapharca broughtonii (Bai et al., 2016; Xia et al., 2015), Carcinus maenas (Bookelaar et al., 2018), Chlamys farreri (Bai et al., 2019), Octopus vulgaris (Prado-Alvarez et al., 2021) and in numerous sites around the world including Europe, Asia, Oceania and the Americas (Abbadi et al., 2018; Batista et al., 2015; Burge et al., 2021; Gittenberger et al., 2016; Hwang et al., 2013; Jenkins et al., 2013; Keeling et al., 2014; Mortensen et al., 2016; Peeler et al., 2012; Roque et al., 2012; Segarra et al., 2010; Shimahara et al., 2012).
A specific microvariant, OsHV-1 µVar, was described in association with the large increase in mortality in C. gigas on the French coast in 2008 (Segarra et al., 2010), but the detection of this virus in a specimen collected before 2008 (Martenot et al., 2012) raises the question of its actual or exclusive involvement in the increasing global phenomenon of oyster mortality. OsHV-1 µVar was strictly characterised by mutations upstream of ORF 4 consisting mainly of 12 deletions in the microsatellite zone and a few mutations upstream and downstream which impact the amino acid sequence. In addition, at ORF 42-43 OsHV-1 µVar had a subtitution of a thymine instead of a cytosine downstream of the sequence without showing any impact on the amino acid sequence (Segarra et al., 2010). Following the publication of the OsHV-1µVar sequence from region C, partly located at ORF 4 (Segarra et al., 2010), several isolates were analysed and this revealed a range of « microvariants » close to the OsHV-1 µVar sequence (Burge et al., 2021; Burioli et al., 2018; Martenot et al., 2012; Morga et al., 2021). Data for numerous OsHV-1 genotypes characterised by sequencing of various coding and non-coding regions or whole genomes are available: in Europe, the reference genome isolated from C. gigas larvae (Davison et al., 2005), the variant OsHV-1var sampled from P. maximus (Arzul et al., 2001a) and the microvariant OsHV-1 µVar from C. gigas spat and juveniles (Abbadi et al., 2018; Burioli et al., 2017; Segarra et al., 2010); in China, Acute Viral Necrosis Virus (Ren et al., 2013), OsHV-1-SB isolated from S. broughtonii (Bai et al., 2016; Xia et al., 2015) and the variant isolated from C. farreri (Bai et al., 2019); in Sweden OsHV-1 SW6 found in O. edulis (Morga et al., 2021); and in many parts of the world, so called « microvariants » or µVars, which are similar to OsHV-1 µVar (Burge et al., 2021; Morga et al., 2021). Double-stranded DNA viruses are known to be genetically stable; DNA viruses in general have mutation rates of 10−8 to 10−6 substitutions per nucleotide site per cell infection, a lower rate than RNA viruses, due the viral DNA polymerase that exhibits proofreading activity (Peck and Lauring, 2018). However, Morga et al. (2021) recently reported a faster evolutionary rate than usual for OsHV-1. The emergence of variations may be due to selection pressure and the ability of the virus to adapt to its environment and host as reported in other members of the Herpesvirales (Renzette et al., 2013). The available data suggest that pathogenic subtypes of OsHV-1 known as microvariants possess genetic diversity beyond that defined for a single genotype by Segarra et al. (2010), but the extent of diversity and how this relates to their global emergence is unclear.
The first detection of OsHV-1 associated with mortality of C. gigas in Australia occured in 2010 but despite recurrent outbreaks there has been little characterisation of the genetic diversity of the viruses affecting C. gigas in Australia (Jenkins et al., 2013). Available data suggest that the genotype responsible for the index case in 2010 was close to OsHV-1 µVar but had some differences (Jenkins et al., 2013). Given that OsHV-1 genetic diversity could affect the gravity of the disease (Delmotte et al., 2022; Martenot et al., 2011; Morga et al., 2021), it is important to study the genetic composition of the Australian isolates to better understand epidemiological patterns in Australia. Detailed, long term, epidemiological studies in Australia revealed that POMS outbreaks recurred at the same sites over successive years (Whittington et al., 2019). The source of OsHV-1 for these disease events between 2011 and 2016 was environmental rather than spread through oyster farming activities (Whittington et al., 2018).
The aim of this report is to (i) study the identity and genetic diversity of OsHV-1 in archival samples collected during mortality events in Australia using six regions sequenced by Sanger technology, (ii) compare the Australian viruses with international isolates previously published, and iii) make inferences based on diversity and phylogeny about the source and transmission of the virus to inform management decisions for disease prevention and control.
2. Materials and methods
2.1. Oyster sample selection
Oyster samples were selected over the five summer seasons between 2011 and 2016 from the entire geographical distribution of POMS disease outbreaks in commercial aquaculture in Australia. Comprehensive data for each sample including location, age, size, mortality rate and viral load are provided in Supplementary data 1, linked by references to scientific publications on these populations. In summary, all samples were collected on oyster farms by researchers from the University of Sydney and all oysters in the final dataset were initially recruited as spat from an OsHV-1 free hatchery in Tasmania (Table 1). All samples were tested using OsHV-1 real-time PCR (Martenot et al., 2010) and positive samples containing the highest concentration of OsHV-1 DNA from among the samples collected that day were selected for sequencing. A total of 118 samples were selected and sequence was subsequently obtained from at least one region of the genome from 107 samples to which a unique “isolate ID” was then assigned. For seven of these (all wild C. gigas or Saccostrea commercialis from the Georges River in the 2013-2014 season) sequence was obtained only for Region 3 which had no variation (see below); these isolates were excluded from further analysis, leaving 100 isolates available for analysis, all being samples from farmed C. gigas (Supplementary data 1). Of the 100 samples of C. gigas, 38 were collected during periods of OsHV-1 infection in the Hawkesbury River estuary north of Sydney NSW (from Coba Bay n = 2; Kimmerikong Bay n = 5; Mooney Mooney n = 15; Mullet Creek n = 10; Porto Bay n = 6), 60 were collected in the Georges River estuary south of Sydney NSW (from Limekiln Bar n = 1; Neverfail Bay n = 2; Site A n = 13; Site B n = 10; Site C n = 25; Sylvania Waters n = 1; The Shed n = 8), while two were collected in Tasmania (from Pitt Water n = 2) (Fig. 1). Exact locations are illustrated in maps in Whittington et al. (2019) and in de Kantzow et al. (2017). Sampled oysters had an age range of 5.5 months to 17 months and a size range of 5mm to 110mm. They were all the subject of published studies in which details of the oyster populations are reported (de Kantzow et al., 2017; Hick et al., 2018; Paul-Pont et al., 2013a; Paul-Pont et al., 2013b; Paul-Pont et al., 2014; Whittington et al., 2015a; Whittington et al., 2015b; Whittington et al., 2019) (Supplementary data 1).
Table 1.
Number of samples of OsHV-1 PCR-positive oysters from which nucleotide sequences were analysed according to the estuary and the season of collection. The number of sites sampled are in parentheses; sites differed between seasons.
| Estuary | 2011-2012 | 2012-2013 | 2013-2014 | 2014-2015 | 2015-2016 | Total |
|---|---|---|---|---|---|---|
| Georges River NSW | 19 (3) | 9 (3) | 14 (4) | 13 (3) | 5 (3) | 60 |
| Hawkesbury River NSW | 11 (3) | 15 (2) | 9 (2) | 3 (2) | 38 | |
| Pitt Water TAS | 2 (1) | 2 | ||||
| Total | 19 | 20 | 29 | 22 | 10 | 100 |
Fig. 1.
Sampling sites. George river estuary sites are located on the bottom map corresponding to the south of Sydney and Hawkesbury river estuary sites are located on the upper map corresponding to the north of Sydney.
2.2. Description of the selected regions
Six regions of the OsHV-1 genome were targeted for Sanger sequencing. Firstly, four regions were selected based on their discriminatory value for distinguishing microvariant genotypes from among other OsHV-1 variants. Region 1 and Region 2 targeted ORF 4 and ORF 43, respectively. They were selected in order to distinguish OsHV-1µVar (Segarra et al., 2010), OsHV-1µVarΔ9, OsHV-1µVarΔ 15 described in Europe (Martenot et al., 2012; Martenot et al., 2011) and other microvariants (Bai et al., 2016). Region 3 covered ORFs 35, 36, 37 and 38 and bounded a large deletion in OsHV-1µVar that induced the suppression of ORF 36 and 37 and disrupted ORF 38 (Burioli et al., 2017; Martenot et al., 2015; Martenot et al., 2013; Morga et al., 2021; Renault et al., 2012). Region 4 was selected for the high level of polymorphism suitable for distinguishing genotypes of OsHV-1 (Batista et al., 2015). It is located between ORF 49 and ORF 50 and is a non-coding site. Region 5 located on ORF 11, showed a large deletion of 1386bp in microvariants (Morga et al., 2021; Burioli et al., 2017; Martenot et al., 2013). Finally, Region 6, which could be involved in virulence mechanisms is a polymorphic region of interest; it targets ORF 88 coding for a transmembrane glycoprotein (Martenot et al., 2013). PCR and sequencing reaction primers used for each region are provided in Table 2 .
Table 2.
Regions of the OsHV-1 genome targeted for PCR amplification.
| Sequencing region | Amplification target |
Refs. | ||||
|---|---|---|---|---|---|---|
| Position a | Gene | Primer name and purposeb | Primer sequence (5’→3’) | Amplicon size (base pairs) | ||
| 1 | 178211 | ORF 4 | C2 (A, S) | CTCTTTACCATGAAGATACCCACC | 709 | (Renault and Arzul, 2001) |
| (C region) | 178919 | C6 (A, S) | GTGCACGGCTTACCATTTTT | |||
| C1585 (S) | GTATAAATAGGCGCG | This paper | ||||
| C1586 (S) | CGCGCCTATTTATAC | This paper | ||||
| C1587 (S) | CAGACGAGGTTAAC | This paper | ||||
| C1588 (S) | GTTAACCTCGTCTG | This paper | ||||
| C1593 (S) | GGAGCTGCGGCGCTATG | This paper | ||||
| 2 | 59950 | ORF 42-43 | IA1 (A, S) | CGCGGTTCATATCCAAAGTT | 607 | Segarra et al. (2010) |
| (A region) | 60557 | IA2 (A, S) | AATCCCCATGTTTCTTGCTG | |||
| C1589 (S) | CTTGCTCATCGTATTC | This paper | ||||
| 3 | 51979 | ORF 35 -36 -37 and 38 | Del 36-37F2 (A, S) | ATACGATGCGTCGGTAGAGC | 989, 384 or no amplification | (Renault et al., 2012) |
| 52968 | ||||||
| (605bpDel) | Del 36-37R (A, S) | CGAGAACCCCATTCCTGTAA | ||||
| C1590 (S) | CATGGTGATGAATGAAG | This paper | ||||
| C1591 (S) | CTTCATTCATCACCATG | This paper | ||||
| C1592 (S) | CATTCCTGTAAACAC | This paper | ||||
| 4 | 72414 | Between ORF49 and 50 | NC1 (A, S) | ACACCTAATGACCCCAAAGG | 506 | (Batista et al., 2015) |
| (NC1NC2 region) | 72919 | NC2 (A, S) | GACCAATCACCAGCTCAACA | |||
| 5 | 17402 | ORF 11 | ORF11For (A, S) | ACCACCGCGCCAAAATCTG | 2116 or 731 | (Martenot et al., 2013) |
| 19518 | ||||||
| ORF11Rev (A, S) | CGCTTCCTATCACCTTGTGG | |||||
| C1872 (S) | CTAGTCGTGCTCGTTCCTCTGC | This paper | ||||
| C1873 (S) | GGCAGAGATAGAACACAATG | This paper | ||||
| C1874 (S) | CATTGTGTTCTATCTCTGCC | This paper | ||||
| 6 | 133088 | ORF 88a | ORF88aFor (A,S) | CCCAGTCTATTATCCAGGTAC | 1020 | (Martenot et al., 2013) |
| 134107 | ||||||
| ORF88aRev (A,S) | ACCGTTCCTCAATCAGTCCC | |||||
Position on the OsHV-1 genome relative to the OsHV-1 reference genome, GenBank: AY509253
Purpose: A amplification, S sequencing
2.3. Sample processing
2.3.1. Extraction
Each sample was derived from whole animal or dissected mantle and gill tissues of one or more oysters, depending on their size (Supplementary data 1). The DNA extraction was performed according to previous studies (Evans et al., 2014; Whittington et al., 2019). Briefly, oysters were collected in the field and transported directly to the laboratory where they were frozen at −80°C until processed. A 400±100 mg sample was homogenised by bead beating using sterile stainless steel beads (Aussie Sapphires) and 1 mL distilled water and placed into a TissueLyser II machine (Qiagen) for 2 min at frequency 30, repeated once. All samples were clarified by centrifugation at 1340 g for 2 min in a microcentrifuge and supernatants were removed and stored in sterile tubes at −80°C. Nucleic acids were purified using a 5X MagMAXTM-96Viral RNA Isolation Kit (Ambion, Life TechnologiesTM, Mulgrave,Australia) and then MagMAXTM Express 96 magnetic particle processor (Applied BiosystemsTM, Life TechnologiesTM, Mulgrave, Australia) according to manufacturers’ instructions for a final volume of 50 µL using the AM1836 deep-well standard programme (Ambion, Life TechnologiesTM, Mulgrave, Australia). Purified nucleic acids were stored at −20°C.
2.3.2. PCR
Various PCR were perfomed in order to target the 6 regions described above in part 2.2. The primers are reported in Table 2. For Region 1 and Region 2, each reaction contained 5 µl DNA extract, 5µl 10X PCR buffer (66.6mM Tris-HCl,16.6mM (NH4)2SO4, 2.5mM MgCl2, 1.65mg/ml bovine serum albumin,10mM beta-mercaptoethanol), 10µl dNTP mix (1mM), 250nM each forward and reverse primer, 5U DNA polymerase (taq:pfu mix) and nuclease-free water to a final volume of 50µl. Thermocycling was performed (Corbett Research CGI960) according to the following conditions: 1 cycle at 94°C for 2min; 35 cycles consisted in 94°C for 1min, 50°C for 1 min and 72°C for 1min, and a final extension at 72°C for 5 min. For the remaining regions, each reaction contained 10µl 5X HiFi reaction buffer (Bioline) (containing 10mM Mg2+), 0.5µl dNTP mix (100mM), 1µl each forward and reverse primer (400nM), 1µl (2U) Velocity DNA polymerase (Bioline), 1.5µl DMSO (BIO-21098 -Bioline Aust Pty. Ltd), 5µl template DNA and nuclease-free water to a final volume of 50µl; thermocycling was performed according to the following conditions: 1 cycle at 98°C for 2min; 35 cycles consisted in 98°C for 30s, 62°C for 30s and 72°C for 30s, and a final extension at 72°C for 10 min.
The amplicons were visualised using 5µl of each amplification product on a 2% agarose gel, assessed against a molecular weight marker and visualised using RedSafe (iNtRON Biotechnology) on a GelDoc transilluminator (Biorad).
2.3.3. Sequencing
Amplified PCR product was purified by incubation with ExoSAP-IT/Cleansweep (ThermoFisher Scientific) then submitted for Sanger sequencing. For several samples where a single amplicon was not obtained, bands of the expected size were excised from the agarose gel and DNA was purified using the QIAquick Gel Extraction Kit (Qiagen). The amplicons were sent to the Australian Genome Research Facility or to the Monash Health Translation Precinct Medical Genomics Facility Australia for sequencing. Forward and reverse sequencing were performed in reactions with relevant PCR primers and internal primers (Table 2). Chromatograms were reviewed, analyzed and primer sequence was removed, using FinchTV (Geospiza).
2.4. DNA data processing
For each region targeted, nucleotide sequences were aligned with the reference genome NC_005881.2 and the two microvariant genomes KY242788 and KY271630, using MEGA10 (Kumar et al., 2018) with the MUSCLE algorithm. The sequences were trimmed and the chromatograms were again specifically checked for all mutations on forward and reverse strands. Base positions described in results are mapped to the reference genome NC_005881.2.
Phylogenetic trees were constructed using MEGA 10 with the Neighbour Joining method and 1000 replications of boostraps and the method based on Maximum Composite Likelihood was applied. Two trees were obtained: the first was based on the concatenated sequence of Regions 1, 2, 3, 4 and 6 (Region 5 was not included due to too few sequences - see results); the second tree was based on Region 1. The trees were unrooted because the ancestry of the reference genotype is uncertain. In addition, the use of another malacoherpesviridae, Haliotid herpesvirus, was impracticable because the ORFs had no sequence in common making alignment impossible. The unrooted tree allows the analysis of diversity but does not orient the genotypes temporally.
Nucleotide diversity (Pi) for Region 1 (C region) including the microsatellite region was calculated using DnaSP v6: DNA Sequence Polymorphism Analysis of Large Datasets (Rozas et al., 2017) and compared with the results of Mineur et al. (2015), after adding six sequences (JN800089, JN800075, JN800082, JN800083, JN800088 and JN800072) to be consistent with these authors. Briefly, the calculation was performed on the alignment; the indel polymorphism module with the “Multiallelic”gap option was used to consider all InDel events. Nucleotide diversity was listed according to geographic region. Pi values reported by Mineur et al. (2015) were corrected for frequency equal to one because multiple occurrences of an identical genotype may not be reported. To make the data comparable, Pi in the current study was not corrected because only one sequence per genotype was included in the calculation.
2.5. Amino acid sequence prediction
In order to estimate whether some putative mutations could induce a modification in the phenotype of the variant, a prediction of amino acid sequence was performed on coding sites in Regions 1, 2, 3, 5 and 6. The translations of the ORFs were performed with ORF Finder (https://www.ncbi.nlm.nih.gov/orffinder/) and the Bioedit sequence alignment editor. The translations were checked using Blastp (https://blast.ncbi.nlm.nih.gov/Blast.cgi?PAGE=Proteins). These predictions were aligned with the prediction of the reference genome NC_005881.2 and the two microvariant genomes KY242788 and KY271630 using Mega 10.
3. Results
3.1. DNA sequencing analysis
3.1.1. Description of the variants
All nucleotide sequences are available on the NCBI database and accession numbers are listed in Table 3. Results and metadata for each of the 100 samples is provided in Supplementary data 1. Sequence alignments are illustrated in Supplementary data 2.
Table 3.
GenBank accession numbers of the 29 DNA groups of the 6 regions analysed.
| Region | Gene | Sequence Group | GenBank Accession No. | |
|---|---|---|---|---|
| 1 | C region | ORF4 | G1 | ON953753 |
| G2 | ON953754 | |||
| G4 | ON953755 | |||
| G6 | ON953752 | |||
| G7 | ON953756 | |||
| 2 | A region | ORF 42-43 | G8 | ON953757 |
| G9 | ON953758 | |||
| 3 | 605 bpDel | ORF 35 to 38 | G10 | ON953759 |
| 4 | NC2 region | Non-coding, between ORF 49 and ORF 50 | G11 | ON954001 |
| G12 | ON954014 | |||
| G13 | ON954007 | |||
| G14 | ON954004 | |||
| G15 | ON954003 | |||
| G17 | ON954002 | |||
| G18 | ON954013 | |||
| G19 | ON954012 | |||
| G20 | ON954006 | |||
| G21 | ON954010 | |||
| G22 | ON954009 | |||
| G23 | ON954008 | |||
| G24 | ON954005 | |||
| G29 | ON954011 | |||
| 5 | ORF 11 gene | ORF 11 | G31 | ON953762 |
| G32 | ON953760 | |||
| G33 | ON953763 | |||
| G34 | ON953761 | |||
| 6 | ORF 88 | G35 | ON953765 | |
| G36 | ON953764 | |||
| G37 | ON953766 | |||
For Region 1, located at ORF 4, nucleotide sequence was obtained from 87 of 100 samples. None of the sequences were identical to the OsHV-1 reference sequence but they were partially similar to the microvariants, with the presence of most of the mutations except the two substitutions of adenine instead of guanine in the downstream region (bases 178,696 and 178,702). However, they all presented a novel substitution of an adenine instead of a guanine in a different position (base 178,705), also in the downstream region. Several mutations were observed allowing the identification of five different nucleotide sequences: G1, G2, G4, G6 and G7 (Fig. 1, Supplementary data 2). Eight of the sequences that grouped in G4 (Supplementary data 2) had 15 deletions (bases 178,558–178,572) in the microsatellite zone instead of the 12 deletions seen in microvariants (Burioli et al., 2017; Segarra et al., 2010), as was observed in OsHV-1µVarΔ 15 reported in Martenot et al. (2012).
For Region 2, situated at ORF 43, nucleotide sequence was obtained from 77 of 100 samples. They were all different from the reference sequence and the microvariants. They were separated into two groups, classified by one deletion of a thymine in the downstream region (base 60,504) (G8) for 36 samples and two deletions of two adenine in the upstream region (bases 60,059 - 60,060) for 41 samples (G9) (Fig. 2, Supplementary data 2).
In the third region coding for ORF 35 to ORF 38, nucleotide sequences were obtained from 96 of 100 samples and all were identical to the two microvariant sequences. The features included the large deletion zone of 606bp (bases 52,251–52,856) common to microvariants and the substitution of a cytosine instead of a thymine (base 52,885). They were classified into G10 (Fig. 3, Supplementary data 2).
Eighty-seven of 100 samples presented an amplification of the fourth region located on the non-coding site between ORF 49 and ORF 50. All the nucleotide sequences presented a common substitution of an adenine instead of a guanine in the upstream zone (base 72,489) and two deletions of two thymine next to the middle zone (bases 75,592–75,593). However there were many differences compared to the reference genotype and microvariants, consisting of 19 mutations over 408 nucleotides which were not observed in the microvariant genotypes. These mutations allowed the characterization of 14 groups (Fig. 4, Supplementary data 2), confirming the prior observations of high variability in this region (Batista et al., 2015). Four samples of a nucleotide sequence named G12 presented a thymine as well as a cytosine (Y) at the same position (base 72,536). The chromatograms of this and all the other regions of these four samples comprising G12 were unambiguous. All samples which contained the G12 variant were pools of 5 spat (Supplementary data 1).
For Region 5 sitting on ORF 11, nucleotide sequence was obtained from 42 of 100 samples. None had the large deletion observed in the microvariants. Even though the nucleotide sequences were close to the reference sequence, a few mutations were present allowing their classification into four groups (G31, G32, G33, G34) (Fig. 5, Supplementary data 2). All had a common substitution of a thymine instead of a guanine (base 18,621).
Of the 91 samples which amplified from Region 6 in the first part of ORF 88, 54 nucleotide sequences were similar to the reference sequence (G37). The others clustered into two groups, comprising the substitution of a cytosine instead of a guanine (base 133,574) in 36 samples (G36) and a substitution of a guanine instead of an adenine (base 133,574) in one sample (G35) (Fig. 6, Supplementary data 2).
Nucleotide sequences for Regions 1, 2, 3, 4 and 6 were obtained from 61 samples, which when concatenated led to the identification of 12 DNA variants (Table 4). V1 and V8 were the most numerous of these variants.
Table 4.
. Description of the 12 genotypic variants based on concatenated nucleotide sequences of Regions 1,2,3,4 and 6 for 61 isolates according to their group identification numbers for these regions.
| Variant | n | R1 | R2 | R3 | R4 | R6 |
|---|---|---|---|---|---|---|
| ORF 4 | ORF 43 | ORFs 35, 36, 37, 38 | uncoded site between ORF49 and 50 | ORF 88 | ||
| V1 | 32 | G7 | G8 | G10 | G11 | G36 |
| V2 | 3 | G1 | G9 | G10 | G11 | G37 |
| V3 | 1 | G7 | G9 | G10 | G20 | G37 |
| V4 | 1 | G7 | G9 | G10 | G21 | G37 |
| V5 | 1 | G7 | G9 | G10 | G29 | G37 |
| V6 | 2 | G7 | G8 | G10 | G11 | G37 |
| V7 | 5 | G7 | G9 | G10 | G13 | G37 |
| V8 | 10 | G7 | G9 | G10 | G11 | G37 |
| V9 | 2 | G4 | G9 | G10 | G12 | G37 |
| V10 | 2 | G7 | G9 | G10 | G14 | G37 |
| V11 | 1 | G7 | G9 | G10 | G15 | G37 |
| V12 | 1 | G7 | G9 | G10 | G17 | G37 |
3.1.2. Phylogenetic analyses
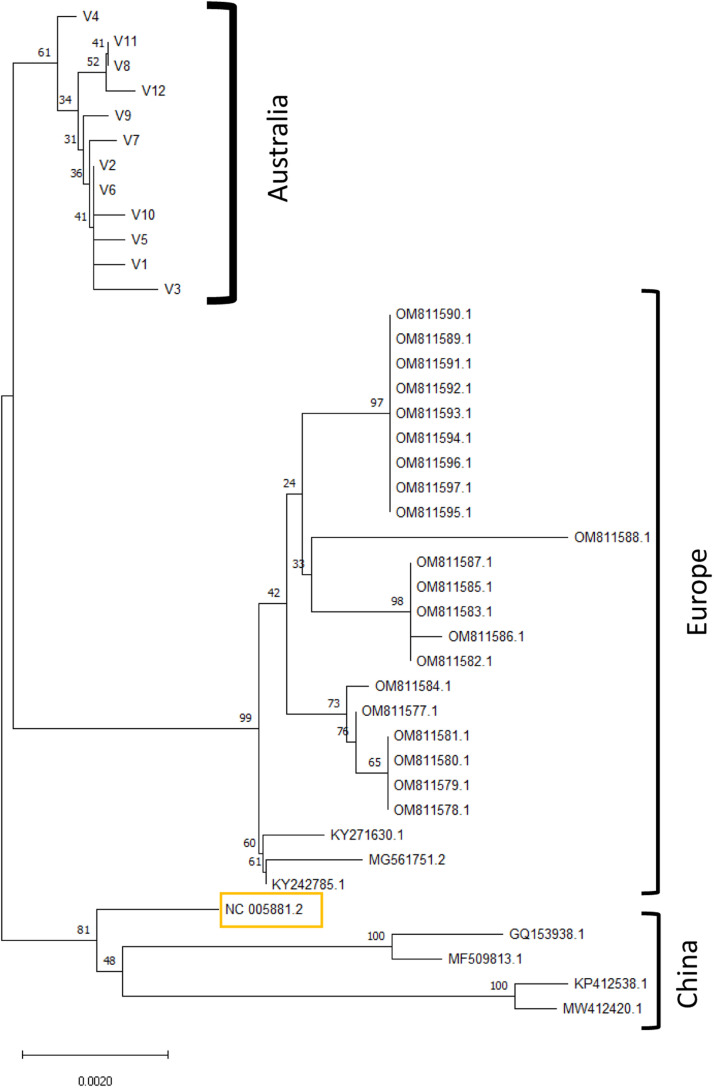
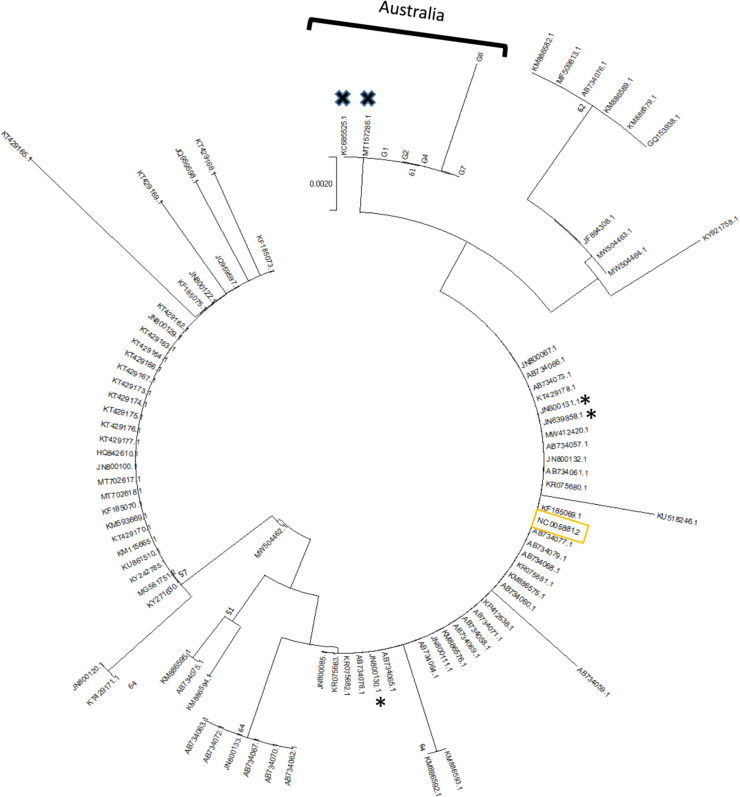
Firstly, a phylogenetic tree using Regions 1, 2, 3, 4 and 6 was constructed with the concatenated nucleotide sequences of the 12 variants and the 29 whole genomes present in NCBI. All Australian genotypes clustered in the same clade, distinct from isolates from Europe, China and the reference genotype and with a distinctly shorter branch length (Fig. 2). Secondly, a phylogenetic tree was constructed based only on the C region (Region 1), which has been well-studied internationally, in order to incorporate a wider range of isolates (Fig. 3). A single cluster distinguished all Australian genotypes from those from Europe, New Zealand, Japan and the Americas.
Fig. 2.
Distribution of the DNA variants according to the NCBI deposited nucleotide sequences. The unrooted phylogenetic tree was performed using the neighbour joining method and 1000 bootstrap replications were applied based on Method based on Maximum Composit Likehood. Regions 1,2,3,4 and 6 were concatenated on 61 Australian isolates showing 12 variants (V1, V2, V3, V4, V5, V6, V7, V8, V9, V10, V11, V12). Yellow rectangle is the reference nucleotide sequence of OsHV-1. Metadata for the nucleotide sequences of the current study that are included in the tree are in Supplementary data 1 while metadata for the nucleotide sequences that were already available in NCBI are in Table 1, Supplementary data 3.
Fig. 3.
Distribution of the DNA variants according to nucleotide sequences deposited in NCBI. The unrooted phylogenetic tree was performed using the neighbour joining method and 1000 bootstrap replications were applied based on maximum composite likelihood. Region 1 from 87 Australian isolates was used, showing 5 groups (G1, G2, G4, G6, G7). Crosses indicate Australian nucleotide sequences extant in NCBI; asterisks indicate the New Zealand nucleotide sequences. Yellow rectangle is the reference nucleotide sequence of OsHV-1. Metadata for the nucleotide sequences of the current study that are included in the tree are in Supplementary data 1 while metadata for the nucleotide sequences that were already available in NCBI are in Table 2, Supplementary data 3.
3.1.3. Diversity
The number of genotypes available for analysis had almost doubled from 48 to 95 since the study of Mineur et al in 2015 (Table 5). The Australian isolates had the lowest genetic diversity overall (Pi 0.99 × 10−3) when compared to viruses from other regions of the world where diversity ranged from 5.31 × 10−3 to 7.61 × 10−3 (Table 5).
Table 5.
Nucleotide diversity in the C region (including the microsatellite region) of OsHV-1 from different geographical regions. The data are based on sequences listed in Table 2 of Supplementary data 3 as well as sequences from the current study. Six sequences were added to be consistent with Mineur et al (2015) (see materials and methods). NW Pacific includes isolates from China, Japan and Korea; NE Pacific includes Mexico and USA; Europe includes France, Ireland, Italy, Portugal and Spain; Australasia includes Australia and New Zealand.
| Current study |
Mineur et al (2015) |
||||
|---|---|---|---|---|---|
| Geographic region | No. of genotypes | Nucleotide diversity (Pi) | No. of genotypes | Nucleotide diversity (Pi)** | |
| All | 95 | 7.46 × 10−3 | 48 | 14.97 × 10−3 | |
| NW Pacific | 45 | 6.45 × 10−3 | 27 | 9.34 × 10−3 | |
| Australasia | 10 | 1.49 × 10−3 | 3 | 2.09 × 10−3 | |
| Australia | 7 | 0.99 × 10−3 | nr | nr | |
| NE Pacific | 5 | 6.24 × 10−3 | 2 | 6.27 × 10−3 | |
| Europe all years | 41 | 7.61 × 10−3 | 16 | 15.30 × 10−3 | |
| *Europe < 2008 | 9 | 3.43 × 10−3 | 10 | 4.67 × 10−3 | |
| Europe ≥ 2008 | 32 | 2.07 × 10−3 | 6 | 3.45 × 10−3 | |
| France | 27 | 5.31 × 10−3 | nr | nr | |
*One sequence (AY459363) with a high frequency of undetermined nucleotides (“N”) was excluded from the current study
**corrected for Hd=1, see materials and methods
nr, no result
3.1.3. Influencing factors
The distribution among the sites over time of the 12 DNA variants identified from concatenated nucleotide sequences is illustrated in Table 6 while similar data for all genomic regions is presented in Table 1, Supplementary data 2.
Table 6.
Distribution of 12 genomic variants of OsHV-1 among 61 isolates in the Georges and Hawkesbury River estuaries according to site and date of collection over five summer seasons between 2011-2012 and 2015–2016.
| 2011-2012 | 2012-2013 | 2013-2014 | 2014-2015 | 2015-2016 | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Isolate | Site | Date collected | Variant | Isolate | Site | Date collected | Variant | Isolate | Site | Date collected | Variant | Isolate | Site | Date collected | Variant | Isolate | Site | Date collected | Variant | |
| Georges River | ||||||||||||||||||||
| 13 | Site C | 16-11-11 | V9 | 19 | Site A | 07-11-12 | V4 | 65 | The Shed | 28-01-14 | V7 | 102 | Neverfail | 16-12-14 | V10 | 108 | Site C | 09-11-15 | V8 | |
| 14 | Site C | 16-11-11 | V9 | 26 | Site B | 28-11-12 | V5 | 66 | The Shed | 28-01-14 | V7 | 103 | Neverfail | 16-12-14 | V10 | 109 | Site C | 09-11-15 | V8 | |
| 4 | Site A | 24-11-11 | V8 | 67 | The Shed | 28-01-14 | V7 | 104 | Site B | 27-1-15 | V11 | |||||||||
| 9 | Site B | 10-02-12 | V3 | 68 | The Shed | 28-01-14 | V7 | 105 | Site B | 27-1-15 | V12 | |||||||||
| 69 | The Shed | 28-01-14 | V7 | 79 | Site C | 27-1-15 | V8 | |||||||||||||
| 70 | Site C | 25-02-14 | V2 | 80 | Site C | 27-1-15 | V8 | |||||||||||||
| 71 | Site C | 25-02-14 | V2 | 81 | Site C | 27-1-15 | V8 | |||||||||||||
| 73 | Site C | 25-02-14 | V2 | 82 | Site C | 27-1-15 | V8 | |||||||||||||
| 84 | Site C | 27-1-15 | V8 | |||||||||||||||||
| 85 | Site C | 27-1-15 | V8 | |||||||||||||||||
| 106 | Site B | 24-2-15 | V8 | |||||||||||||||||
| Hawkesbury River | 31 | Mullet | 21-01-13 | V1 | 42 | Kimmerikong | 31-12-13 | V1 | 74 | Mooney | 18-11-14 | V1 | 114 | Mullet | 26-10-15 | V1 | ||||
| 32 | Mullet | 21-01-13 | V1 | 43 | Kimmerikong | 31-12-13 | V1 | 75 | Mooney | 18-11-14 | V1 | |||||||||
| 33 | Mullet | 21-01-13 | V1 | 44 | Kimmerikong | 31-12-13 | V1 | 76 | Mooney | 18-11-14 | V1 | |||||||||
| 34 | Mullet | 21-01-13 | V1 | 46 | Kimmerikong | 31-12-13 | V6 | 77 | Mooney | 18-11-14 | V1 | |||||||||
| 36 | Mullet | 21-01-13 | V1 | 48 | Mooney | 25-01-14 | V1 | 78 | Mooney | 18-11-14 | V1 | |||||||||
| 37 | Mullet | 21-01-13 | V1 | 49 | Mooney | 25-01-14 | V1 | 110 | Porto | 28-11-14 | V1 | |||||||||
| 38 | Mullet | 21-01-13 | V1 | 50 | Mooney | 25-01-14 | V1 | 111 | Porto | 28-11-14 | V1 | |||||||||
| 41 | Coba | 29-01-13 | V1 | 51 | Mooney | 25-01-14 | V1 | 112 | Porto | 28-11-14 | V1 | |||||||||
| 39 | Porto | 15-02-13 | V1 | 52 | Mooney | 25-01-14 | V1 | 113 | Porto | 28-11-14 | V1 | |||||||||
| 40 | Porto | 15-02-13 | V1 | 53 | Mooney | 25-01-14 | V1 | |||||||||||||
| 54 | Mooney | 25-01-14 | V1 | |||||||||||||||||
| 55 | Mooney | 25-01-14 | V1 | |||||||||||||||||
| 56 | Mooney | 25-01-14 | V1 | |||||||||||||||||
| 57 | Mooney | 25-01-14 | V6 | |||||||||||||||||
Considering the concatenated nucleotide sequences, up to four variants were found in one summer season in the Georges River, compared to only one or two variants in the Hawkesbury River. Nine of the 12 variants occurred at only one site and in only one summer season. The variants V1 and V6 were present only in the Hawkesbury River; V1 was present at most sites and in each of the four summers, while V6 was found only at Kimmerikong Bay and Mooney Mooney in summer 2013–2014. The other variants were confined to the Georges River where V8 was identified at various sites within Woolooware Bay in three summer seasons. The other variants in the Georges River were detected only at a single site and a single sampling date.
More than one variant was detected over time at some sites; for example, five were seen at Site B in the Georges River while two were seen at both Kimmerikong Bay and Mooney Mooney in the Hawkesbury river (Table 7). More than one variant was found in the same summer season at some sites; for example, in 2014–2015 V8, V11 and V12 were found at Site B, while in 2013-2014 both V1 and V6 were found at Kimmerikong Bay and at Mooney Mooney. Furthermore, there were also several instances where different variants were detected in the same population of oysters at the same time: V11 and V12 were both present at Site B on 27/1/15; V1 and V6 were both present at Kimmerong Bay and Mooney Mooney on 13/12/13 and also on 25/1/14.
Table 7.
Distribution of 12 DNA variants and protein prediction profiles among 61 samples from ten different sites in the Georges and Hawkesbury river estuaries between 2011 and 2016.
| Estuary | Site | No. samples | DNA variant | Predicted protein |
|---|---|---|---|---|
| Hawkesbury river (n = 34) | Coba Bay | 1 | V1 | ACH |
| Kimmerikong Bay | 4 | V1 (n = 3) V6 (n = 1) | ACH | |
| Mooney Mooney | 15 | V1 (n = 14) V6 (n = 1) | ACH | |
| Mullet Creek | 8 | V1 | ACH | |
| Porto Bay | 6 | V1 | ACH | |
| George river (n = 27) | Neverfail | 2 | V10 | ADH |
| Site A | 2 | V4 V8 | ADH | |
| Site B | 5 | V3 V5 V8 V11 V12 | ADH | |
| Site C | 13 | V8 (n = 8) V9 (n = 2) V2 (n = 3) | ADH | |
| The shed | 5 | V7 | ADH |
There were similar observations for Region 4 (Fig. 4, Supplementary data 2), which had the most variability; 12 of the 14 variants were found in only one season and at only one site. Three of five variants in Region 1 and two of four variants in Region 5 were also seen in only one season and at only one site.
Two isolates from Tasmania were not incorporated into the phylogenetic analysis because of incomplete sequencing of regions from these samples. However one of these samples was the only isolate classified as G6 in Region 1 (Table 1 and Fig. 1, Supplementary data 2); there were no sequencing data from this region of the other sample from Tasmania. The oysters sampled in New South Wales had been supplied from at least nine different production batches (i.e. different broodstock spawnings) from one hatchery in Tasmania. Oysters grown from five of these batches yielded more than one variant, and in one case, five different OsHV-1 variants, which has important implications when assessing the sources of the virus (see discussion).
3.2. Amino acid sequence prediction of variants
3.2.1. Description of the predicted amino acid sequences
Predicted amino acid sequences were aligned with the reference and microvariant sequences. All alignments are illustrated in Supplementary data 2. Regions 3 and 4 were not included in this analysis because Region 3 had a large deletion of ORF 36 and 37, inducing overlap of ORF 35 and 38 and creating uncertainty about whether a longer ORF could be translated, while Region 4 was a non-coding area.
For Region 1, two amino acid sequences were identified for ORF 4 among the five DNA sequences described above in part 3.1.1. They were both different to the reference genotype and the microvariants. The first group was named <<A>> and contained G1, G2, G4 and G7; it had an asparagine (N) instead of an aspartic acid (D). The second group named <<B>> contained G6 and had in addition a leucine (L) instead of a serine (S) (Fig. 7, Supplementary data 2).
For Region 2, the two DNA groups characterised for ORF 43 presented two different amino acid sequences. The first was named <<C>> and contained G8. It had a similar amino acid sequence to the reference genotype and the microvariant. The second was named <<D>>. It contained G9 and was characterised by three successive mutations of a serine (S), lysine (K) and a deletion instead of a phenylalanine (F), a glutamic acid (E) and an arginine (R) (Fig. 8, Supplementary data 2).
Region 5 located on ORF 11 had four DNA groups which translated into 3 amino acid sequences. One was similar to OsHV-1 reference genotype, and was named « E ». The second was named « F » and had a mutation of an asparagine (N) instead of a lysine (K). The third was named « G » and had 4 substitutions of a lysine (K) (Fig. 9, Supplementary data 2).
Region 6 coding for the first part of ORF 88 presented two amino acid sequences named « H » containing G36 and G37 and « I » containing G35. <<H>> had a similar sequence to the reference genotype and one of the microvariants, KY242785, while <<I>> had one mutation of a glutamic acid (E) instead of lysine (K) (Fig. 10, Supplementary data 2).
The amino acid prediction of the 12 DNA variants identified among 61 samples resulted in two profiles based on ORF 4, ORF 43 and ORF88: ACH (n = 34) and ADH (n = 27) (Table 8). Region 5 (ORF11) was not included because there were only 42 sequences available for it.
Table 8.
Protein profiles of the 12 DNA variants identified among 61 samples from ten different sites in the Georges and Hawkesbury river estuaries between 2011 and 2016.
| Variant | n | ORF 4 | ORF 43 | ORF 88 |
|---|---|---|---|---|
| V1 | 32 | A | C | H |
| V2 | 3 | A | D | H |
| V3 | 1 | A | D | H |
| V4 | 1 | A | D | H |
| V5 | 1 | A | D | H |
| V6 | 2 | A | C | H |
| V7 | 5 | A | D | H |
| V8 | 10 | A | D | H |
| V9 | 2 | A | D | H |
| V10 | 2 | A | D | H |
| V11 | 1 | A | D | H |
| V12 | 1 | A | D | H |
3.2.2. Influencing factors
The amino acid profile of isolates was different between the two estuaries in NSW (Tables 6 and 7). The ACH profile was found only in the Hawkesbury River, an estuary located 40 km north of Sydney while the ADH profile was found only in the Georges River, an estuary located 20 km south of Sydney.
4. Discussion
4.1. Australian OsHV-1 isolates have lower diversity than OsHV-1 isolates globally
It is difficult to compare genomic diversity between published studies of OsHV-1 due to differences in the genomic regions sequenced, sample sizes, time span, geographic extent, host species and disease state (healthy, sick, or dead). However, in this study we report diversity of Ostreid herpes virus1 in estuaries near Sydney Australia that may be lower than in other geographic regions of the world.
Regions of the OsHV-1 genome under selection pressure and other regions which accumulate a high number of substitutions have been reported (Morga et al., 2021). Selection pressure is less on a non-coding area and mutations acquired over time are less likely to be eliminated. The diversity revealed in this study is mainly explained by inclusion of non-coding Region 4 between ORF 49 and ORF 50, which is a polymorphic area (Batista et al., 2015). In this region alone, 14 groups were identified among the 87 samples (one variant for every 6.2 isolates). Similarly, Batista et al. (2015) identified one variant for every 6.0 isolates examined in a study of 18 isolates from mostly diseased C. gigas in Portugal.
C region, corresponding to Region 1 in the present study, is the most commonly analysed part of the OsHV-1 genome. With the typical primers C2/C6 used for amplification there is representation of part of a non-coding region and part of ORF 4 (Batista et al., 2015). Nucleotide diversity in the C region was calculated formally and compared to data published in 2015 (Mineur et al., 2015). There were more sequences in public databases, but the nucleotide diversity had not changed markedly except for Europe where it appeared to have decreased (from 15.3 × 10−3 to 7.6 × 10−3). The Australian isolates had the lowest genetic diversity (Pi 0.99 × 10−3) overall. Globally, 48 different genotypes had been identified in C region (including the microsatellite region) by 2015 (Mineur et al., 2015). Expressed in simple terms of the frequency of genotypes among isolates examined, in Australia seven genotypes have been identified among 89 samples in diseased C. gigas (five from 87 isolates in the present study and two existing in NCBI), which equates to one variant for every 12.7 isolates. In contrast, in a study covering a wide area of coastal Japan, 23 genotypes were found among 123 samples from healthy Crassostrea species (one variant for every 5.4 isolates) (Shimahara et al., 2012); in both countries samples were collected over about 5 years. The genetic diversity among diseased oysters in Australia was clearly much less than that in healthy oysters in Japan. The data from Japan in the north west Pacific region likely represent the diverse environmental reservoir of viral types detectable in healthy shellfish, while the low diversity in Australia may represent selected environmental strains of OsHV-1 capable of causing disease in farmed oysters under certain environmental conditions, that then became locally dominant. Relatively low diversity was seen in Europe after 2008 when OsHV-1 µVar became the most common strain in diseased oysters (Table 5). In Europe there had been spread of the virus with movements of live oysters for farming, for example from France to Ireland and Jersey (Peeler et al., 2012). Within France, OsHV-1 µVar was dispersed widely with commercial movements of live oysters from Marennes-Oléron to farming areas on both the Atlantic and Mediterranean coasts (Delmotte et al., 2022). Thus, in addition to the health/disease condition of oysters and their geographic locations, the observed regional differences in the diversity of OsHV-1 genotypes depends on epidemiological differences between the sources and modes of spread of the viruses. All these factors will need to be considered in future studies of the diversity of OsHV-1.
In Region 4 we observed a double nucleotide (thymine and cytosine) in four individual samples for which remaining nucleotide sequences were unambiguous (G12 – Fig. 4, and Supplementary data 2) and otherwise matched the nucleotide sequence of G18 which had a cytosine at this position (Fig. 4, Supplementary data 2). Our first hypothesis is that there has been a recent mutation at this position to explain why we found two different copies of this part of the genome, one being the ancestral form and the other being the mutated one. This is feasible due to the persistence of the herpesvirus in surviving oyster hosts (Evans et al., 2017b), because gene exchange in herpesviruses can occur by recombination (Thiry et al., 2005) and because Rosani and Venier (2017) showed the presence of DNA recombination-initiating promoter binding in Malacoherpesviridae. The four samples with this mutation were from oysters in close proximity to one another and they could have been infected almost simultaneously from a host in which the mutation first occured, resulting in a pool of two variants in each of the four samples. This is consistent with Martenot et al (2011) who reported various genotypes in the same batch of oysters and Morga et al (2021) who reported various genotypes in the same individual. A second possibility would be that there were two otherwise identical but long-standing OsHV-1 variants in each of the four samples, due to different individuals containing a different genotype of OsHV-1, because each sample came from a pool of five individual oysters. While this does increase the chance of mixing several variants into one DNA extract, in no other sample did we find a group with a thymine at this position. The least likely explanation because the non-coding area between ORF 49 and ORF 50 is present as a single copy in the complete OsHV-1 genomes available on NCBI, is the existence of a double copy of this region.
The microsatellite zone of the C region of OsHV-1 is polymorphic because of repeated trinucleotides which allow replication mistakes, as observed by many authors (Martenot et al., 2012; Martenot et al., 2011; Mineur et al., 2015; Renault et al., 2014). The range in the number of ACT trinucleotide repeats across all genotypes globally was reported to be 3 - 13 (Mineur et al., 2015), corresponding to a range of 3–10 trinucleotide deletions (9–30 base deletions). However, we observed only two profiles among a large number of isolates, either 12 or 15 base deletions. This is consistent with the only other Australian sequences deposited in NCBI; these came from the first episode of mortality in 2010 and had 12 base deletions.
4.2. Australian isolates differed from OsHV-1 isolates globally
Concatenated nucleotide sequences showed a distinct cluster including all the Australian isolates, different from all other genotypes reported worldwide and supported by the distinct distribution with a shorter branch size for the Australian genotypes (Fig. 2). Although the bootstrap value was only 63, this could be explained by the presence of Region 3 located between ORF 35 and 38 including a large deletion area and region 5 located on ORF 11, with a 462bp deletion only in the microvariant. These differences are calculated as multiple mutation events when it is more likely that each corresponds to a single mutation event. Burioli et al. (2016) studied the same disturbed region and discussed the need for sophisticated algorithms for alignment and phylogenetical analysis, adapted to take into account DNA-virus evolution analysis, in order to obtain accurate results with the correct phylogenetic distance. The Australian clade was also confirmed by the inclusion of the only two NCBI sequences from Australia, KC685525.1 and MT157286.1 (Burge et al., 2020; Jenkins et al., 2013) in the C region tree (Fig. 3). This is consistent with the phylogeny reported in Mineur et al. (2015), which was performed on the same locus with another algorithm, median joining genotype, and showed three closely related genotypes from temperate Australasia. In contrast, the phylogenetic tree presented in Burge et al. (2021) with the neigbour joining method showed that the Australian genotype was close to the Japanese, American, Korean and French microvariants. However, unlike the current phylogeny, the tree described in Burge et al. (2021) is not based on evolutionary distance and contains too few samples - a single genotype from Australia - which is not sufficient to construct a cluster. The Australian viruses appear to be genetically different across the genomic regions analyzed and this raises the question about their true phylogenetic origins. Sanger sequencing of a limited number of genomic regions may bias analyses compared to whole genome analysis, which will be indispensable for determining clades of OsHV-1 and should be the subject of future studies.
4.3. Australian isolates were not OsHV-1 reference strain, OsHV-1 µVar or microvariants
Neither OsHV-1 reference genotype nor OsHV-1 µVar were identified among the Australian samples. Currently OsHV-1 “microvariants” are described as being close to OsHV-1 µVar as defined by Segarra et al. (2010) but not identical to it (Burge et al., 2021; Morga et al., 2021). In all of the Australian isolates the two regions which characterise OsHV-1 µVar according to Segarra et al. (2010), ORF 4 and ORF 43, had dissimilar nucleotide sequences. None of these sequences had the large deletion in ORF 11 reported in the microvariant sequences (Burioli et al., 2017; Martenot et al., 2013; Morga et al., 2021) and presented a similar nucleotide sequence to the OsHV-1 reference virus on ORF 11 except for one point mutation. Moreover, half the samples from Australia did not present an amplification product from this region. Since there was enough biological material in the same samples to perform sequencing of the other regions, we suggest that there were changes at the primer binding sites, which others have also mentioned to be a likely reason for this problem (Arzul et al., 2001b; Martenot et al., 2011); PCR-sequencing of the other regions was not uniformly successful either, consistent with many other studies of OsHV-1.
The variant of OsHV-1 known as µVar is thought to be responsible for the sudden escalation of mass mortality events in C. gigas in France from 2008, and then later in other European countries. However, µVar and the more recently described “microvariants” are clearly not the only variants of OsHV-1 associated with mass mortality events in C. gigas. The disease outbreaks in Australia since 2010 have not been caused by these types of OsHV-1 but have been just as sudden in onset, recurrent and economically devastating for the oyster industry as those in France (Fuhrmann et al., 2019). Therefore further research is required to understand the virulence determinants of OsHV-1 and how these are coded in the genome.
4.4. Internal comparison of Australian isolates
The two estuaries studied in New South Wales had different OsHV-1 profiles. At the Georges River sites, ten variants were identified over five summer seasons while at the Hawkesbury River sites only two variants were found. Delmotte et al. (2022) showed a great diversity of variants in areas with high oyster farming activity. However, the causes of the different patterns of distribution of variants here are unknown since these two estuaries in Australia had equivalent, intense oyster farming activity using similar genetic stock of C. gigas from the same OsHV-1 free hatchery in Tasmania. As a consequence of the two distributions of DNA variants, the amino acid sequences of the viruses present in the Hawkesbury and Georges river estuaries were different: the ACH profile in the Hawkesbury River and ADH in the Georges River. This distinct pattern has a precedent in Europe, but with far greater geographic separation than the 60 km that exists between these two estuaries. Based on RNA-Seq SNP analysis, Delmotte et al. (2020) reported two genetically different OsHV-1 populations in POMS outbreaks in C. gigas on the Atlantic and Mediterranean coasts of France; they had amino acid sequence differences in viral membrane proteins, which may affect virulence (Delmotte et al., 2020).
Due to the selection of samples based on high viral load, which in general is correlated with mortality (Oden et al., 2011), it was not appropriate to compare the OsHV-1 isolates in this study according to their degree of replication fitness or observed mortalities. However, in the oyster populations from which the samples were collected there were differences in the patterns of mortality: spat in the Georges River estuary were 6.4 times more likely to have mortality due to OsHV-1 than those in the Hawkesbury River estuary (Whittington et al., 2019). Subclinical infection of oysters with OsHV-1 was also more likely in the Georges River (Evans et al., 2017b; Whittington et al., 2019). It is possible that the mutually exclusive genotypic variants of OsHV-1 identified between the two estuaries differed in virulence. However, other factors could also be involved. While oyster genotype was similar between the two rivers because of the common origin of spat, there could be a wide range of environmental differences. These could include differences in the co-occurrence of reservoirs/hosts, paticulate vectors, filter feeding species and seaweeds that could all influence the severity of POMS (Dugeny et al., 2022; Evans et al., 2017c; Pernet et al., 2021; Whittington et al., 2018).
To our knowledge there were no transfers of commercial oysters between the Georges and Hawkesbury Rivers after first detection of OsHV-1 in the Georges River late in 2010 due to government biosecurity directives (Paul-Pont et al., 2014). The only common factor between the two oyster production locations was the recruitment site of all batches of farmed C. gigas in this study: Tasmania. However, the current data indicate that the virus contamination did not come from this recruitment step but from the culture environment. There are three reasons for this: (i) consistent pre-shipment PCR testing of all batches of spat from the hatchery by the government laboratory in Tasmania and again upon arival in NSW by the researchers (all negative results) means that spat were specific-pathogen free (SPF); (ii) different genotypes of OsHV-1 were detected in the same batch of spat after deployment in NSW; (iii) a unique genotype (G6) of OsHV-1 was detected in Tasmania; G6 was not found among 85 samples from the Hawkesbury River or Georges River. Epidemiological observations alone had suggested that recurrent POMS outbreaks in Australia were derived from the environment and were not due to disease transfer with oysters during farming (Whittington et al., 2018), a feature which is now strongly supported by DNA sequence evidence. This scenario is in direct contrast to France where evidence points to a radiation of disease associated with commercial spat distribution (Delmotte et al., 2022). The time of first appearance of OsHV-1 in a putative environmental host in Australia cannot be determined, but it could have been a point source introduction to a naive population, then adaptation to local conditions. In fact there are historical records of a virus like Ostreid herpesvirus from Ostrea angasi in Western Australia (Hine and Thorne, 1997). These authors demonstrated herpes-like viral particles by electron microscopy, but did not have the molecular tools to identify the virus. In any case, this observation raises the possibility that OsHV-1 could be either indigenous to Australia or could have been introduced many decades ago and become endemic, and it could have mutated to form its own lineage over time, as is supported by the current phylogenetic analysis. Knowing that OsHV-1 µVar emerged around 2008 in Europe and then dispersed rapidly, the absence of OsHV-1 µVar in Australia suggests that there has been effective compliance with the strict quarantine laws that aim to prevent introduction of exotic pathogens into Australia.
Arzul et al. (2001a) speculated 20 years ago that pathogenic forms of OsHV-1 arose from possibly benign bivalve herpesviruses which could be transmitted between bivalve species and which would be favoured by modern shellfish farming practices (Arzul et al., 2001a; Arzul et al., 2001b). In Australia, there is evidence that invertebrate species in the Georges River besides C. gigas are exposed to OsHV-1 and may become infected, but their role in the epidemiology of POMS remains to be determined (Evans et al., 2017c). The environment is an important factor in the transmission cycle because OsHV-1 can be found in seawater and sediments (Evans et al., 2014; 2017) and is likely transported on suspended particles over 5µm (Evans et al., 2017a; Evans et al., 2017c; Paul-Pont et al., 2013b; Whittington et al., 2015b). Depending on conditions such as temperature, the virus is expected to persist in seawater and remain infectious for several days according to in vitro experimentation (Hick et al., 2016; Martenot et al., 2015). This gives the virus time to transit between hosts through the environment. Ostreid herpesvirus 1 can infect several different host species (Arzul et al., 2001a; Bai et al., 2016; Batista et al., 2015; Bookelaar et al., 2018; Comps and Cochennec, 1993; Evans et al., 2017c; Farley et al., 1972; Hine and Thorne, 1997; Hine et al., 1998; Prado-Alvarez et al., 2021; Renault and Arzul, 2001; Renault et al., 2001; Xia et al., 2015) and may persist in survivors of outbreaks or remain latent (Arzul et al., 2002; Evans et al., 2017b), which may explain why each environment has its own assemblage of viruses. While the molecular evidence strongly suggested that the virus contamination came from reservoirs in the local culture environment rather than a point source such as an oyster hatchery, the distribution of the variants across sites and time suggests that some reservoirs were persistent and widespread, while others were highly site and time specific and therefore possibly short-lived, or that transmission was infrequent. The reservoirs may be subject to local environmental influences which affect their distribution and abundance. Further field research is required to discover the environmental reservoir of OsHV-1 and the specific environmental factors that facilitate emergence of the virus. Climate change, short term climatic and water quality variations and intensification of farming are among the many factors to be considered.
5. Conclusion
Sequencing analyses of OsHV-1 isolates from estuaries near Sydney, Australia, revealed a low diversity of genotypes relative to other geographic regions, reflecting differences in sources and modes of spread of the viruses and the types of oyster populations (healthy, or diseased) that have been examined so far. Importantly, Australian POMS outbreaks were associated with a type of OsHV-1 distinct from others globally and were not a “microvariant” genotype. Elsewhere, since 2008 OsHV-1 microvariants have been the only variants associated with mass mortality events in C. gigas. Future whole genome sequencing investigations will allow more detailed classification of Australian isolates of OsHV-1. Based on both DNA and amino acid sequences, OsHV-1 variants were absolutely correlated to estuary. The C. gigas populations in the two estuaries near Sydney had different prevalence and severity of POMS, which suggests that the geographically mutally exclusive genotypes differed in virulence. The molecular evidence supports field epidemiological evidence that the disease-causing virus came from reservoirs in the local environment. The sequencing findings in this study help inform understanding of the various sources and modes of spread of OsHV-1 that have led to mass mortality events in farmed C. gigas globally.
List of supplemental data
Supplemental data 1
Table 1 Metadata for all samples
Table 2 Citations of scientific publications describing the populations of oysters from which samples were collected
Supplemental data 2
Table 1 Variants by genomic region and geographic location
Figs. 1 to 10 DNA and amino acid sequence alignments
Supplemental data 3
Genbank accession numbers of isolates included in phylogenetic trees
CRediT authorship contribution statement
Suzanne Trancart: Investigation, Data curation, Methodology, Visualization, Formal analysis, Writing – original draft, Writing – review & editing. Alison Tweedie: Investigation, Data curation, Writing – review & editing. Olivia Liu: Investigation, Writing – review & editing. Ika Paul-Pont: Conceptualization, Investigation, Writing – review & editing. Paul Hick: Investigation, Writing – review & editing. Maryline Houssin: Resources, Investigation, Supervision, Writing – review & editing. Richard J. Whittington: Funding acquisition, Project administration, Supervision, Resources, Conceptualization, Investigation, Data curation, Formal analysis, Writing – original draft, Writing – review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
Sample collection and sequencing was funded by Fisheries Research and Development Corporation Australia under projects 2011/053, 2012/032 and 2014/040 and The University of Sydney and LABÉO France. Sanger sequencing was undertaken by Australian Genome Research Facility and Monash Health Translation Precinct Medical Genomics Facility Australia. Bruce Allford, Len Drake, Bob Drake and Robert Hill are thanked for their invaluable assistance with field work. Shellfish Culture Tasmania kindly provided all batches of spat that were deployed and sampled in cited research trials. Vickie Patten and Ann-Michele Whittington provided skilled laboratory assistance with oyster processing.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.virusres.2022.198994.
Appendix. Supplementary materials
Data Availability
Comprehensive metadata for each sample are provided in Supplementary Data, linked to prior publications that contain more details. All sequences are available in GenBank.
References
- Abbadi M., Zamperin G., Gastaldelli M., Pascoli F., Rosani U., Milani A., Schivo A., Rossetti E., Turolla E., Gennari L., Toffan A., Arcangeli G., Venier P. Identification of a newly described OsHV-1 mu var from the North Adriatic Sea (Italy) J. Gen. Virol. 2018;99(5):693–703. doi: 10.1099/jgv.0.001042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arzul I., Nicolas J.L., Davison A.J., Renault T. French scallops: a new host for ostreid herpesvirus-1. Virology. 2001;290(2):342–349. doi: 10.1006/viro.2001.1186. [DOI] [PubMed] [Google Scholar]
- Arzul I., Renault T., Lipart C., Davison A.J. Evidence for interspecies transmission of oyster herpesvirus in marine bivalves. J. Gen. Virol. 2001;82:865–870. doi: 10.1099/0022-1317-82-4-865. [DOI] [PubMed] [Google Scholar]
- Arzul I., Renault T., Thébault A., Gérard A. Detection of oyster herpesvirus DNA and proteins in asymptomatic Crassostrea gigas adults. Virus Res. 2002;84(1-2):151–160. doi: 10.1016/s0168-1702(02)00007-2. [DOI] [PubMed] [Google Scholar]
- Bai C., Gao W., Wang C., Yu T., Zhang T., Qiu Z., Wang Q., Huang J. Identification and characterization of Ostreid herpesvirus 1 associated with massive mortalities of Scapharca broughtonii broodstocks in China. Dis. Aquat. Org. 2016;118(1):65–75. doi: 10.3354/dao02958. [DOI] [PubMed] [Google Scholar]
- Bai C.M., Morga B., Rosani U., Shi J., Li C., Xin L.S., Wang C.M. Long-range PCR and high-throughput sequencing of Ostreid herpesvirus 1 indicate high genetic diversity and complex evolution process. Virology. 2019;526:81–90. doi: 10.1016/j.virol.2018.09.026. [DOI] [PubMed] [Google Scholar]
- Batista F.M., López-Sanmartín M., Grade A., Morgado I., Valente M., Navas J.I., Power D.M., Ruano F. Sequence variation in Ostreid herpesvirus 1 microvar isolates detected in dying and asymptomatic Crassostrea angulata adults in the Iberian Peninsula: insights into viral origin and spread. Aquaculture. 2015;435(0):43–51. doi: 10.1016/j.aquaculture.2014.09.016. [DOI] [Google Scholar]
- Bookelaar B.E., O'Reilly A.J., Lynch S.A., Culloty S.C. Role of the intertidal predatory shore crab Carcinus maenas in transmission dynamics of Ostreid herpesvirus-1 microvariant. Dis. Aquat. Org. 2018;130(3):221–233. doi: 10.3354/dao03264. [DOI] [PubMed] [Google Scholar]
- Burge C.A., Friedman C.S., Kachmar M.L., Humphrey K.L., Moore J.D., Elston R.A. The first detection of a novel OsHV-1 microvariant in San Diego, California, USA. J. Invert. Pathol. 2021;184 doi: 10.1016/j.jip.2021.107636. [DOI] [PubMed] [Google Scholar]
- Burge C.A., Reecez K.S., Dhar A.K., Kirkland P., Morga B., Degremont L., Faury N., Wippel B.J.T., MacIntyre A., Friedman C.S. First comparison of French and Australian OsHV-1 mu vars by bath exposure. Dis. Aquat. Org. 2020;138:137–144. doi: 10.3354/dao03452. [DOI] [PubMed] [Google Scholar]
- Burioli E.A.V., Prearo M., Houssin M. Complete genome sequence of Ostreid herpesvirus type 1 microVar isolated during mortality events in the Pacific oyster Crassostrea gigas in France and Ireland. Virology. 2017;509:239–251. doi: 10.1016/j.virol.2017.06.027. [DOI] [PubMed] [Google Scholar]
- Burioli E.A., Prearo M., Riina M., Bona M., Fioravanti M., Arcangeli G., Houssin M. Ostreid herpesvirus type 1 genomic diversity in wild populations of Pacific oyster Crassostrea gigas from Italian coasts. J. Invert. Pathol. 2016;137:71–83. doi: 10.1016/j.jip.2016.05.004. [DOI] [PubMed] [Google Scholar]
- Burioli E.A.V., Varello K., Lavazza A., Bozzetta E., Prearo M., Houssin M. A novel divergent group of Ostreid herpesvirus 1 mu Var variants associated with a mortality event in Pacific oyster spat in Normandy (France) in 2016. J. Fish Dis. 2018;41(11):1759–1769. doi: 10.1111/jfd.12883. [DOI] [PubMed] [Google Scholar]
- Cochennec-Laureau, N., Baud, J., Bedier, E., Boudry, P., Huvet, A., Nicolas, J., Pepin, J., Petton, B., 2010. Bilan des « Journées Surmortalité des huîtres creuses, Crassostrea gigas » du Programme P7 « Aquaculture Durable » des 8 et 9 décembre 2009 (Review of "Oysters Crassostrea gigas mortality days" Program P7 "Sustainable Aquaculture" December 8–9, 2009), https://archimer.ifremer.fr/doc/00000/7393/.
- Comps M., Cochennec N. A Herpes-like virus from the European oyster Ostrea edulis L. J. Invert. Pathol. 1993;62(2):201–203. doi: 10.1006/jipa.1993.1098. [DOI] [Google Scholar]
- Davison A.J., Eberle R., Ehlers B., Hayward G.S., McGeoch D.J., Minson A.C., Pellett P.E., Roizman B., Studdert M.J., Thiry E. The order herpesvirales. Arch. Virol. 2009;154(1):171–177. doi: 10.1007/s00705-008-0278-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davison A.J., Trus B.L., Cheng N.Q., Steven A.C., Watson M.S., Cunningham C., Le Deuff R.M., Renault T. A novel class of herpesvirus with bivalve hosts. J. Gen. Virol. 2005;86:41–53. doi: 10.1099/vir.0.80382-0. [DOI] [PubMed] [Google Scholar]
- de Kantzow M., Hick P., Becker J., Whittington R. Effect of water temperature on mortality of Pacific oysters Crassostrea gigas associated with microvariant Ostreid herpesvirus 1 (OsHV-1 μVar) Aquac. Environ. Interact. 2016;8:419–428. [Google Scholar]
- de Kantzow M.C., Hick P.M., Dhand N.K., Whittington R.J. Risk factors for mortality during the first occurrence of Pacific Oyster Mortality Syndrome due to Ostreid herpesvirus - 1 in Tasmania, 2016. Aquaculture. 2017;468:328–336. doi: 10.1016/j.aquaculture.2016.10.025. [DOI] [Google Scholar]
- de Lorgeril J., Lucasson A., Petton B., Toulza E., Montagnani C., Clerissi C., Vidal-Dupiol J., Chaparro C., Galinier R., Escoubas J.M., Haffner P., Degremont L., Charriere G.M., Lafont M., Delort A., Vergnes A., Chiarello M., Faury N., Rubio T., Leroy M.A., Perignon A., Regler D., Morga B., Alunno-Bruscia M., Boudry P., Le Roux F., Destoumieux-Garzon D., Gueguen Y., Mitta G. Immune-suppression by OsHV-1 viral infection causes fatal bacteraemia in Pacific oysters. Nat. Commun. 2018;9:ARTN 4215. doi: 10.1038/s41467-018-06659-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delmotte J., Chaparro C., Galinier R., de Lorgeril J., Petton B., Stenger P.L., Vidal-Dupiol J., Destoumieux-Garzon D., Gueguen Y., Montagnani C., Escoubas J.M., Mitta G. Contribution of viral genomic diversity to oyster susceptibility in the pacific oyster mortality syndrome. Front. Microbiol. 2020;11:1579. doi: 10.3389/fmicb.2020.01579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delmotte J., Pelletier C., Morga B., Galinier R., Petton B., Lamy J.B., Kaltz O., Avarre J.C., Jacquot M., Montagnani C., Escoubas J.M. Genetic diversity and connectivity of the Ostreid herpesvirus 1 populations in France: a first attempt to phylogeographic inference for a marine mollusc disease. Virus Evol. 2022;8(1) doi: 10.1093/ve/veac039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dugeny E., de Lorgeril J., Petton B., Toulza E., Gueguen Y., Pernet F. Seaweeds influence oyster microbiota and disease susceptibility. J. Anim. Ecol. 2022;91(4):805–818. doi: 10.1111/1365-2656.13662. [DOI] [PubMed] [Google Scholar]
- Evans O., Hick P., Alford B., Whittington R.J. Transmission of Ostreid herpesvirus-1 microvariant in seawater: detection of viral DNA in seawater, filter retentates, filter membranes and sentinel Crassostrea gigas spat in upwellers. Aquaculture. 2017;473:456–467. doi: 10.1016/j.aquaculture.2017.03.021. [DOI] [Google Scholar]
- Evans O., Hick P., Whittington R.J. Detection of Ostreid herpesvirus-1 microvariants in healthy Crassostrea gigas following disease events and their possible role as reservoirs of infection. J. Invert. Pathol. 2017;148:20–33. doi: 10.1016/j.jip.2017.05.004. [DOI] [PubMed] [Google Scholar]
- Evans O., Paul-Pont I., Hick P., Whittington R.J. A simple centrifugation method for improving the detection of Ostreid herpesvirus-1 (OsHV-1) in natural seawater samples with an assessment of the potential for particulate attachment. J. Virol. Methods. 2014;210(0):59–66. doi: 10.1016/j.jviromet.2014.09.023. [DOI] [PubMed] [Google Scholar]
- Evans O., Paul-Pont I., Whittington R.J. Detection of Ostreid herpesvirus 1 microvariant DNA in aquatic invertebrate species, sediment and other samples collected from the Georges River estuary, New South Wales, Australia. Dis. Aquat. Org. 2017;122(3):247–255. doi: 10.3354/dao03078. [DOI] [PubMed] [Google Scholar]
- Farley C.A., Banfield W.G., Kasnic G., Jr., Foster W.S. Oyster Herpes-Type Virus. Science. 1972;178(4062):759–760. doi: 10.2307/1735848. [DOI] [PubMed] [Google Scholar]
- Friedman C., Estes R., Stokes N., Burge C., Hargove J., Barber B., Elston R., Burreson E., Reece K. Herpes virus in juvenile Pacific oysters Crassostrea gigas from Tomales Bay, California, coincides with summer mortality episodes. Dis. Aquat. Org. 2005;63(1):33–41. doi: 10.3354/dao063033. [DOI] [PubMed] [Google Scholar]
- Fuhrmann M., Castinel A., Cheslett D., Nozal D.F., Whittington R.J. The impacts of Ostreid herpesvirus 1 microvariants on Pacific oyster aquaculture in the Northern and Southern Hemispheres since 2008. Rev. Sci. Et Tech. Off. Int. Des. Epizoot. 2019;38(2):491–509. doi: 10.20506/rst.38.2.3000. [DOI] [PubMed] [Google Scholar]
- Gittenberger A., Voorbergen-Laarman M.A., Engelsma M.Y. Ostreid herpesvirus OsHV-1 muVar in Pacific oysters Crassostrea gigas (Thunberg 1793) of the Wadden Sea, a UNESCO world heritage site. J. Fish Dis. 2016;39(1):105–109. doi: 10.1111/jfd.12332. [DOI] [PubMed] [Google Scholar]
- Hick P., Evans O., Looi R., English C., Whittington R.J. Stability of Ostreid herpesvirus-1 (OsHV-1) and assessment of disinfection of seawater and oyster tissues using a bioassay. Aquaculture. 2016;450:412–421. doi: 10.1016/j.aquaculture.2015.08.025. [DOI] [Google Scholar]
- Hick P.M., Evans O., Rubio A., Dhand N.K., Whittington R.J. Both age and size influence susceptibility of Pacific oysters (Crassostrea gigas) to disease caused by Ostreid herpesvirus-1 (OsHV-1) in replicated field and laboratory experiments. Aquaculture. 2018;489:110–120. doi: 10.1016/j.aquaculture.2018.02.013. [DOI] [Google Scholar]
- Hine P.M., Thorne T. Replication of herpes-like viruses in haemocytes of adult flat oysters Ostrea angasi: an ultrastructural study. Dis. Aquat. Org. 1997;29(3):189–196. http://www.scopus.com/inward/record.url?eid=2-s2.0-0030953661&partnerID=40&md5=14baec6c1cd641cbd52daa52c1cadf8d [Google Scholar]
- Hine P.M., Wesney B., Besant P. Replication of a herpes-like virus in larvae of the flat oyster Tiostrea chilensis at ambient temperatures. Dis. Aquat. Org. 1998;32(3):161–171. doi: 10.3354/dao032161. [DOI] [Google Scholar]
- Hwang J.Y., Park J.J., Yu H.J., Hur Y.B., Arzul I., Couraleau Y., Park M.A. Ostreid herpesvirus 1 infection in farmed Pacific oyster larvae Crassostrea gigas (Thunberg) in Korea. J. Fish Dis. 2013;36(11):969–972. doi: 10.1111/Jfd.12093. [DOI] [PubMed] [Google Scholar]
- Jenkins C., Hick P., Gabor M., Spiers Z., Fell S.A., Gu X., Read A., Go J., Dove M., O'Connor W., Kirkland P.D., Frances J. Identification and characterisation of an Ostreid herpesvirus-1 microvariant (OsHV-1 micro-var) in Crassostrea gigas (Pacific oysters) in Australia. Dis. Aquat. Org. 2013;105(2):109–126. doi: 10.3354/dao02623. [DOI] [PubMed] [Google Scholar]
- Keeling S., Brosnahan C., Williams R., Gias E., Hannah M., Bueno R., McDonald W., Johnston C. New Zealand juvenile oyster mortality associated with Ostreid herpesvirus 1—an opportunistic longitudinal study. Dis. Aquat. Org. 2014;109(3):231–239. doi: 10.3354/dao02735. [DOI] [PubMed] [Google Scholar]
- Kumar S., Stecher G., Li M., Knyaz C., Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018;35(6):1547. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martenot C., Denechere L., Hubert P., Metayer L., Oden E., Trancart S., Travaille E., Houssin M. Virulence of Ostreid herpesvirus 1 mu Var in sea water at 16 degrees C and 25 degrees C. Aquaculture. 2015;439:1–6. doi: 10.1016/j.aquaculture.2015.01.012. [DOI] [Google Scholar]
- Martenot C., Fourour S., Oden E., Jouaux A., Travaillé E., Malas J.P., Houssin M. Detection of the OsHV-1 μVar in the Pacific oyster Crassostrea gigas before 2008 in France and description of two new microvariants of the Ostreid Herpesvirus 1 (OsHV-1) Aquaculture. 2012;338–341(0):293–296. doi: 10.1016/j.aquaculture.2011.12.030. [DOI] [Google Scholar]
- Martenot C., Oden E., Travaillé E., Malas J.-P., Houssin M. Detection of different variants of Ostreid Herpesvirus 1 in the Pacific oyster, Crassostrea gigas between 2008 and 2010. Virus Res. 2011;160(1–2):25–31. doi: 10.1016/j.virusres.2011.04.012. [DOI] [PubMed] [Google Scholar]
- Martenot C., Oden E., Travaillé E., Malas J.P., Houssin M. Comparison of two real-time PCR methods for detection of ostreid herpesvirus 1 in the Pacific oyster Crassostrea gigas. J. Virol. Methods. 2010;170(1-2):86–89. doi: 10.1016/j.jviromet.2010.09.003. [DOI] [PubMed] [Google Scholar]
- Martenot C., Travaille E., Lethuillier O., Lelong C., Houssin M. Genome exploration of six variants of the Ostreid Herpesvirus 1 and characterization of large deletion in OsHV-1muVar specimens. Virus Res. 2013;178(2):462–470. doi: 10.1016/j.virusres.2013.08.006. [DOI] [PubMed] [Google Scholar]
- Mineur F., Provan J., Arnott G. Phylogeographical analyses of shellfish viruses: inferring a geographical origin for ostreid herpesviruses OsHV-1 (Malacoherpesviridae) Mar. Biol. 2015;162(1):181–192. doi: 10.1007/s00227-014-2566-8. [DOI] [Google Scholar]
- Morga B., Jacquot M., Pelletier C., Chevignon G., Degremont L., Bietry A., Pepin J.F., Heurtebise S., Escoubas J.M., Bean T.P., Rosani U., Bai C.M., Renault T., Lamy J.B. Genomic diversity of the Ostreid Herpesvirus type 1 across time and location and among host species. Front. Microbiol. 2021;12 doi: 10.3389/fmicb.2021.711377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortensen S., Strand A., Bodvin T., Alfjorden A., Skar C.K., Jelmert A., Aspan A., Saelemyr L., Naustvoll L.J., Albretsen J. Summer mortalities and detection of Ostreid herpesvirus microvariant in Pacific oyster Crassostrea gigas in Sweden and Norway. Dis. Aquat. Org. 2016;117(3):171–176. doi: 10.3354/dao02944. [DOI] [PubMed] [Google Scholar]
- Nicolas J., Comps M., Cochennec N. Herpes-like virus infecting Pacific oyster larvae, C.gigas. Bull. Eur. Assoc. Fish Pathol. 1992;12(1):11–13. http://eafp.squarespace.com/bulletin-archive/1992-volume-12/issue-1/ [Google Scholar]
- Oden E., Martenot C., Berthaux M., Travaille E., Malas J.P., Houssin M. Quantification of ostreid herpesvirus 1 (OsHV-1) in Crassostrea gigas by real-time PCR: determination of a viral load threshold to prevent summer mortalities. Aquaculture. 2011;317(1-4):27–31. doi: 10.1016/j.aquaculture.2011.04.001. [DOI] [Google Scholar]
- Paul-Pont I., Dhand N.K., Whittington R.J. Influence of husbandry practices on OsHV-1 associated mortality of Pacific oysters Crassostrea gigas. Aquaculture. 2013;412:202–214. doi: 10.1016/j.aquaculture.2013.07.038. [DOI] [Google Scholar]
- Paul-Pont I., Dhand N.K., Whittington R.J. Spatial distribution of mortality in Pacific oysters Crassostrea gigas: reflection on mechanisms of OsHV-1 transmission. Dis. Aquat. Org. 2013;105(2):127–138. doi: 10.3354/dao02615. [DOI] [PubMed] [Google Scholar]
- Paul-Pont I., Evans O., Dhand N.K., Rubio A., Coad P., Whittington R.J. Descriptive epidemiology of mass mortality due to Ostreid herpesvirus-1 (OsHV-1) in commercially farmed Pacific oysters (Crassostrea gigas) in the Hawkesbury River estuary, Australia. Aquaculture. 2014;422–423:146–159. doi: 10.1016/j.aquaculture.2013.12.009. [DOI] [Google Scholar]
- Paul-Pont I., Evans O., Dhand N.K., Whittington R.J. Experimental infections of Pacific oyster Crassostrea gigas using the Australian Ostreid herpesvirus-1 (OsHV-1) uVar strain. Dis. Aquat. Org. 2015;113(2):137–147. doi: 10.3354/dao02826. http://www.int-res.com/abstracts/dao/v113/n2/p137-147/ [DOI] [PubMed] [Google Scholar]
- Peck K.M., Lauring A.S. Complexities of viral mutation rates. J. Virol. 2018;92(14) doi: 10.1128/jvi.01031-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peeler E.J., Allan Reese R., Cheslett D.L., Geoghegan F., Power A., Thrush M.A. Investigation of mortality in Pacific oysters associated with Ostreid herpesvirus-1 μVar in the Republic of Ireland in 2009. Prev. Vet. Med. 2012;105(1-2):136–143. doi: 10.1016/j.prevetmed.2012.02.001. [DOI] [PubMed] [Google Scholar]
- Pernet F., Lugue K., Petton B. Competition for food reduces disease susceptibility in a marine invertebrate. Ecosphere. 2021;12(4) doi: 10.1002/ecs2.3435. [DOI] [Google Scholar]
- Petton B., Destoumieux-Garzón D., Pernet F., Toulza E., de Lorgeril J., Degremont L., Mitta G. The pacific oyster mortality syndrome, a polymicrobial and multifactorial disease: state of knowledge and future directions. Front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.630343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petton B., Pernet F., Robert R., Boudry P. Temperature influence on pathogen transmission and subsequent mortalities in juvenile Pacific oysters Crassostrea gigas. Aquac. Environ. Interact. 2013;3(3):257–273. doi: 10.3354/Aei00070. [DOI] [Google Scholar]
- Prado-Alvarez M., Garcia-Fernandez P., Faury N., Azevedo C., Morga B., Gestal C. First detection of OsHV-1 in the cephalopod Octopus vulgaris. Is the octopus a dead-end for OsHV-1? J. Invert. Pathol. 2021;183 doi: 10.1016/j.jip.2021.107553. [DOI] [PubMed] [Google Scholar]
- Ren W., Chen H., Renault T., Cai Y., Bai C., Wang C., Huang J. Complete genome sequence of acute viral necrosis virus associated with massive mortality outbreaks in the Chinese scallop, Chlamys farreri. Virol J. 2013;10 doi: 10.1186/1743-422x-10-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renault T., Arzul I. Herpes-like virus infections in hatchery-reared bivalve larvae in Europe: specific viral DNA detection by PCR. J. Fish Dis. 2001;24(3):161–167. doi: 10.1046/j.1365-2761.2001.00282.x. [DOI] [Google Scholar]
- Renault T., Le Deuff R.M., Chollet B., Cochennec N., Gerard A. Concomitant herpes-like virus infections in hatchery-reared larvae and nursery-cultured spat Crassostrea gigas and Ostrea edulis. Dis. Aquat. Org. 2000;42(3):173–183. doi: 10.3354/dao042173. [DOI] [PubMed] [Google Scholar]
- Renault T., Le Deuff R.M., Cochennec N., Maffart P. Herpesviruses associated with mortalities among Pacific oyster, Crassostrea gigas, in France - comparative study. Rev. Med. Veter. 1994;145:735–742. [Google Scholar]
- Renault T., Lipart C., Arzul I. A herpes-like virus infecting Crassostrea gigas and Ruditapes philippinarum larvae in France. J. Fish Dis. 2001;24(6):369–376. doi: 10.1046/j.1365-2761.2001.00300.x. [DOI] [PubMed] [Google Scholar]
- Renault T., Moreau P., Faury N., Pepin J.-F., Segarra A., Webb S. Analysis of clinical Ostreid herpesvirus 1 (Malacoherpesviridae) specimens by sequencing amplified fragments from three Vvrus genome areas. J. Virol. 2012;86(10):5942–5947. doi: 10.1128/jvi.06534-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renault T., Tchaleu G., Faury N., Moreau P., Segarra A., Barbosa-Solomieu V., Lapegue S. Genotyping of a microsatellite locus to differentiate clinical Ostreid herpesvirus 1 specimens. Vet. Res. 2014;45(3):3. doi: 10.1186/1297-9716-45-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renzette N., Gibson L., Bhattacharjee B., Fisher D., Schleiss M.R., Jensen J.D., Kowalik T.F. Rapid intrahost evolution of human cytomegalovirus is shaped by demography and positive selection. PLos Genet. 2013;9(9) doi: 10.1371/journal.pgen.1003735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roque A., Carrasco N., Andree K.B., Lacuesta B., Elandaloussi L., Gairin I., Rodgers C.J., Furones M.D. First report of OsHV-1 microvar in Pacific oyster (Crassostrea gigas) cultured in Spain. Aquaculture. 2012;324:303–306. doi: 10.1016/j.aquaculture.2011.10.018. [DOI] [Google Scholar]
- Rosani U., Venier P. Oyster RNA-seq data support the development of Malacoherpesviridae genomics. Front. Microbiol. 2017;8 doi: 10.3389/fmicb.2017.01515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozas J., Ferrer-Mata A., Sanchez-DelBarrio J.C., Guirao-Rico S., Librado P., Ramos-Onsins S.E., Sanchez-Gracia A. DnaSP v6: DNA sequence polymorphism analysis of large datasets. Mol. Biol. Evol. 2017;34:3299–3302. doi: 10.1093/molbev/msx248. [DOI] [PubMed] [Google Scholar]
- Samain J.F., Degremont L., Soletchnik P., Haure J., Bedier E., Ropert M., Moal J., Huvet A., Bacca H., Van Wormhoudt A., Delaporte M., Costil K., Pouvreau S., Lambert C., Boulo V., Soudant P., Nicolas J.L., Le Roux F., Renault T., Gagnaire B., Geret F., Boutet I., Burgeot T., Boudry P. Genetically based resistance to summer mortality in the Pacific oyster (Crassostrea gigas) and its relationship with physiological, immunological characteristics and infection processes. Aquaculture. 2007;268(1-4):227–243. doi: 10.1016/J.Aquaculture.2007.04.044. [DOI] [Google Scholar]
- Segarra A., Pépin J.F., Arzul I., Morga B., Faury N., Renault T. Detection and description of a particular Ostreid herpesvirus 1 genotype associated with massive mortality outbreaks of Pacific oysters, Crassostrea gigas, in France in 2008. Virus Res. 2010;153(1):92–99. doi: 10.1016/j.virusres.2010.07.011. [DOI] [PubMed] [Google Scholar]
- Shimahara Y., Kurita J., Kiryu I., Nishioka T., Yuasa K., Kawana M., Kamaishi T., Oseko N. Surveillance of Type 1 Ostreid herpesvirus (OsHV-1) variants in Japan. Fish Pathol. 2012;47(4):129–136. doi: 10.3147/jsfp.47.129. [DOI] [Google Scholar]
- Thiry E., Meurens F., Muylkens B., McVoy M., Gogev S., Thiry J., Vanderplasschen A., Epstein A., Keil G., Schynts F. Recombination in alphaherpesviruses. Rev. Med. Virol. 2005;15(2):89–103. doi: 10.1002/rmv.451. [DOI] [PubMed] [Google Scholar]
- Whittington R., Dhand N., Evans O., Paul-Pont I. Further observations on the influence of husbandry practices on OsHV-1 μVar mortality in Pacific oysters Crassostrea gigas: age, cultivation structures and growing height. Aquaculture. 2015;438:82–97. doi: 10.1016/j.aquaculture.2014.12.040. [DOI] [Google Scholar]
- Whittington R., Hick P., Evans O., Rubio A., Alford B., Dhand N., Paul-Pont I. Protection of Pacific oyster (Crassostrea gigas) spat from mortality due to Ostreid herpesvirus-1 OsHV-1 µVar) using simple treatments of incoming seawater in land-based upwellers. Aquaculture. 2015;437:10–20. doi: 10.1016/j.aquaculture.2014.11.016. [DOI] [Google Scholar]
- Whittington R.J., Liu O., Hick P.M., Dhand N., Rubio A. Long-term temporal and spatial patterns of Ostreid herpesvirus 1 (OsHV-1) infection and mortality in sentinel Pacific oyster spat (Crassostrea gigas) inform farm management. Aquaculture. 2019;513 doi: 10.1016/j.aquaculture.2019.734395. [DOI] [Google Scholar]
- Whittington R.J., Paul-Pont I., Evans O., Hick P., Dhand N.K. Counting the dead to determine the source and transmission of the marine herpesvirus OsHV-1 in Crassostrea gigas. Vet. Res. 2018;49(1):34. doi: 10.1186/s13567-018-0529-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia J., Bai C., Wang C., Song X., Huang J. Complete genome sequence of Ostreid herpesvirus-1 associated with mortalities of Scapharca broughtonii broodstocks. Virol. J. 2015;12 doi: 10.1186/s12985-015-0334-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Comprehensive metadata for each sample are provided in Supplementary Data, linked to prior publications that contain more details. All sequences are available in GenBank.