Abstract
H-NS is an abundant nucleoid-associated protein involved in the maintenance of chromosomal architecture in bacteria. H-NS also has a role in silencing the expression of a variety of environmentally regulated genes during growth under nonpermissive conditions. In this study we demonstrate a role for H-NS in the negative modulation of expression of several genes within the ToxR virulence regulon of Vibrio cholerae. Deletion of hns resulted in high, nearly constitutive levels of expression of the genes encoding cholera toxin, toxin-coregulated pilus, and the ToxT virulence gene regulatory protein. For the cholera toxin- and ToxT-encoding genes, elevated expression in an hns mutant was found to occur in the absence of the cognate activator proteins, suggesting that H-NS functions directly at these promoters to decrease gene expression. Deletion analysis of the region upstream of toxT suggests that an extensive region located far upstream of the transcriptional start site is required for complete H-NS-mediated repression of gene expression. These data indicate that H-NS negatively influences multiple levels of gene expression within the V. cholerae virulence cascade and raise the possibility that the transcriptional activator proteins in the ToxR regulon function to counteract the repressive effects of H-NS at the various promoters as well as to recruit RNA polymerase.
Vibrio cholerae is the bacterial causative agent of the acute diarrheal disease called cholera. The organism is spread among individuals through the ingestion of contaminated water or food. In areas where cholera is endemic, the organism persists in an aquatic niche between periodic outbreaks. In the human host, V. cholerae pathogenesis involves the coordinated expression of a number of virulence factors, including cholera toxin (CT), which is directly responsible for the disease symptoms, and toxin-coregulated pilus (TCP), which is required for intestinal colonization. Expression of the genes that encode these virulence factors is regulated at the transcriptional level by a variety of parameters, such as osmolarity, temperature, pH, anaerobiosis, and chemoattractant amino acids (16, 30). Such regulation is thought to provide a mechanism by which the organism can induce the expression of virulence genes within the host and repress them under growth conditions where they are not required.
Transcriptional regulation of the genes encoding CT and TCP occurs via a cascade involving several activator proteins referred to collectively as the ToxR virulence regulon (for reviews, see references 12 and 45). The genes constituting the ToxR regulon are encoded both within the ancestral V. cholerae genome and on pathogenicity islands derived from the genomes of lysogenic bacteriophages. The tcp operon encodes the gene products required for formation of the TCP fiber and is located on the large TCP pathogenicity island also known as the Vibrio pathogenicity island (23, 24). The ctx operon encodes the CT subunits and is located within the genome of the CTX phage (55). This phage uses TCP as its receptor (55). Both the tcp and ctx operons are directly activated by ToxT, an AraC homolog that is encoded within the tcp operon and which regulates its own expression in addition to that of the other genes (4, 6, 59). Expression of toxT, in turn, is dependent on two cytoplasmic membrane protein pairs. The TcpP-TcpH protein pair is encoded by an operon located adjacent to the tcp operon on the pathogenicity island, and the ToxR-ToxS protein pair is encoded by an operon located elsewhere on the larger of the two V. cholerae chromosomes. Studies on the ToxR-ToxS protein pair have shown that ToxR directly binds DNA and activates transcription (28). Its stability in the membrane is enhanced by ToxS (10, 37). TcpP and TcpH are homologs of ToxR and ToxS, respectively, and are thought to function in a similar manner (18). No additional regulator of the toxRS operon is known, and its expression is constitutive over most growth conditions. In contrast, expression of the tcpPH operon is responsive to temperature and pH (5) and is dependent on at least two cytoplasmic activators, AphA and the LysR homolog AphB (25, 46). These two activators are encoded by unlinked genes that are not known to be associated with any pathogenicity islands. In addition, the cyclic AMP receptor protein (CRP) represses ToxR regulon gene expression at an early step in the pathway (44). This multitude of regulatory inputs provides a mechanism for virulence gene expression to respond to concurrent signals both within and outside the host.
It is becoming increasingly apparent that expression of many bacterial virulence gene regulons is controlled by overlapping regulatory systems encoded on pathogenicity islands, plasmids, and elsewhere within the genome. A protein that is broadly distributed within members of the family Enterobacteriaceae and has been demonstrated to have a role in modulating expression of virulence genes located on plasmids or pathogenicity islands is the histone-like nucleoid structuring protein H-NS. H-NS is a small, abundant protein that was first characterized with respect to its ability to mediate chromosomal DNA condensation (21, 54). H-NS is thought to influence expression of a myriad of seemingly unrelated genes by organizing promoter and regulatory regions into nucleoprotein complexes in response to environmental signals. Expression of genes that are influenced by H-NS is typically responsive to environmental parameters known to influence DNA topology, such as osmolarity, temperature, anaerobiosis, pH, and growth phase (2). H-NS preferentially binds to curved, AT-rich regions of DNA and favors the general consensus site 5′-TNTNAN-3′, where N is any nucleotide (39, 58). Examples of virulence genes best characterized with respect to the influence of H-NS on their expression include the fim, pap, and cfa genes of Escherichia coli (13, 17, 22) and several vir genes of Shigella flexneri (9, 14, 51). In most cases, H-NS modulates virulence gene expression in a negative manner, as evidenced by a large increase in expression under nonpermissive conditions and in the absence of appropriate activator proteins in hns mutant strains.
Changes in DNA topology have previously been shown to influence expression of some ToxR regulon genes (35). In this report we investigate the role of V. cholerae H-NS on the expression of the ToxR regulon. We have utilized the V. cholerae genome sequence to identify a gene encoding a protein with 41% identity to E. coli H-NS and have deleted the gene in various virulence gene promoter-lacZ fusion strains to determine the influence of H-NS at different levels in the virulence cascade. To further characterize the effect of H-NS at specific promoters, we have deleted genes encoding known activator proteins in various Δhns promoter-lacZ fusion strains. Finally, by using promoter deletions, we have determined that H-NS mediates repression over an extensive region upstream of the toxT promoter. These results indicate that H-NS influences multiple levels within the V. cholerae virulence cascade by repressing gene expression through a mechanism of transcriptional silencing.
MATERIALS AND METHODS
Strains, media, and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. E. coli and V. cholerae strains were stored at −70°C in Luria-Bertani (LB) medium (27) containing 30% (vol/vol) glycerol. V. cholerae was grown in LB broth with a starting pH of either 6.5 or 8.5 at either 30 or 37°C. E. coli was grown in LB with a starting pH of 6.5 at 30°C. Antibiotic concentrations used in culture were as follows: ampicillin (Ap), 100 μg/ml; tetracycline (Tc), 15 μg/ml; gentamicin (Gm), 30 μg/ml; and streptomycin (Sm), 100 μg/ml generally or 1 mg/ml when selecting for loss of integrated plasmids from V. cholerae following standard allelic exchange. 5-Bromo-4-chloro-3-indolyl-β-d-galactoside (XGal) was used in LB agar at 40 μg/ml.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Characteristics | Reference or source |
|---|---|---|
| E. coli | ||
| MC4100 | F−araD139 Δ(argF-lac)U169 rpsL150 relA1 flbB5301 deoC1 ptsF25 rbsR Smr | 41 |
| DL1976 | MC4100 hns651 Tcr | 57 |
| RT4129 | MC4100 (λtoxT-lacZYA)4 (−656/+77) | This work |
| MBN199 | RT4129 hns651 Tcr | This work |
| DH92 | TE2680 (Kmr-ptoxT-lacZ) Kmr | 20 |
| RT4146 | MC4100 (Kmr-ptoxT-lacZ) Kmr (−172/+45) | This work |
| MBN201 | RT4146 hns-651 Tcr | This work |
| RT4317 | MC4100 (λtoxT-lacZYA)5 (−256/+37) | This work |
| MBN297 | RT4317 hns651 Tcr | This work |
| V. cholerae | ||
| O395 Sm | Classical Ogawa, Smr | 49 |
| CG842 | O395 Sm ΔlacZ | 8 |
| KSK218 | CG842 ctx-lacZ | 44 |
| MBN032 | CG842 (toxT-lacZ)1 | This work |
| MBN135 | CG842 tcpA-lacZ | This work |
| MBN019 | KSK218 ΔtoxT1 | This work |
| MBN142 | MBN135 ΔtoxT1 | This work |
| MBN147 | KSK218 Δhns1 | This work |
| MBN148 | MBN135 Δhns1 | This work |
| MBN153 | MBN019 Δhns1 | This work |
| MBN168 | MBN142 Δhns1 | This work |
| MBN170 | MBN032 Δhns1 | This work |
| MBN172 | MBN032 ΔtcpP1 | This work |
| MBN175 | MBN172 Δhns1 | This work |
| KSK236 | KSK218 toxR::pVM55 | 44 |
| MBN183 | MBN170 toxR::pVM55 | This work |
| MBN185 | MBN019 toxR::pVM55 | This work |
| MBN187 | MBN175 toxR::pVM55 | This work |
| MBN189 | MBN032 toxR::pVM55 | This work |
| MBN192 | MBN153 toxR::pVM55 | This work |
| MBN196 | MBN147 toxR::pVM55 | This work |
| MBN318 | MBN172 toxR::pVM55 | This work |
| Plasmids | ||
| pKAS32 | pGP704 rpsL Apr | 43 |
| pSAN9 | pKAS32 tcpF-tcpJ Apr | This work |
| pSAN10 | pSAN9 ΔtoxT Apr | This work |
| pMIN1 | pACYC184 Gmr cassette from pUCGM, Gmr Cmr | This work |
| pMIN3 | pSAN9 (toxT-lacZ)1 Apr | This work |
| pMIN26 | pKAS32, hns-flanking sequence, Apr | This work |
| pMIN27 | pKAS32, tcpP-flanking sequence, Apr | This work |
| pVM55 | pJM703.1::EcoRI-HpaI ‘toxR’ Apr | 30 |
| pRS415 | lacZYA transcriptional fusion vector | 42 |
| pJYT1 | pRS415::(toxT-lacZYA)4 | This work |
| pMIN38 | pRS415::(toxT-lacZYA)5 | This work |
| pTSK | pACYC184 toxR+ toxS+ Cmr | 4 |
Plasmid and strain construction.
Plasmid pMIN1 was constructed by cloning the HindIII-flanked Gmr gene from pUCGM (40) into the HindIII site of pACYC184 (7). This plasmid was used as a counterselection for conjugal matings between E. coli and V. cholerae recipient strains in this study. The plasmid was cured by overnight growth in LB broth without antibiotics.
The (toxT-lacZ)1 strain MBN032 was constructed by inserting an XhoI-SalI fragment containing a promoterless lacZ into the unique XhoI site within the toxT gene of pSAN9 to create pMIN3. Plasmid pSAN9 contains the V. cholerae tcpF, toxT, and tcpJ genes in pKAS32 (43). The orientation of the lacZ fragment was determined by PCR with a primer internal to lacZ, and the construct was used for allelic exchange with the O395 Smr ΔlacZ strain CG842. Proper integration of (toxT-lacZ)1 fusion in MBN032 was confirmed by PCR.
The V. cholerae Δhns1 mutation was constructed as follows. A sequence from the V. cholerae database identified as encoding an H-NS homolog by TBLASTN search (1) provided the basis for the design of oligonucleotide primers MN19 (5′-GATCGATCGCGGCCGCGAAGTTTTCGCCACTTGCCC-3′), MN20 (5′-GATCGATCGCGGCCGCTCTCGCTCAGGAAGACCACG-3′), MN21 (5′-GATCGGAATTCATGGCGCGATTGGCCGCTGC-3′), and MN22 (5′-GATCGTCTAGACCACGCCCTTGAGAAGCGGC-3′), which were used to amplify chromosomal sequences from O395 that flank the hns gene. The upstream 1,028-bp and downstream 835-bp fragments were inserted into the allelic exchange vector pKAS32, resulting in the Δhns1 plasmid pMIN26. The hns deletion was then introduced into the chromosome of various V. cholerae strains by allelic exchange. Constructs were verified by PCR.
Construction of the ΔtoxT1 mutation was accomplished by inverse PCR of pSAN9 using primers RT21 (5′-CCCAATCATTGCGTTCTACTCTGAAG-3′) and RT22 (5′-GAATATTTATTTATGTTGACAGGAGTTGCAG-3′). The resultant plasmid, pSAN10, lacks the toxT gene. Following allelic exchange with pSAN10, chromosomal deletions were confirmed by PCR.
The ΔtcpP1 mutation was constructed as follows. Two fragments generated by PCR amplification of O395 chromosomal DNA with primers TP-XBA (5′-GATCGTCTAGAAGATTTAGCAAGGTTACCGGG-3′), H-XS (5′-GATCGTCTAGAGAGCTCGAACATTAGGGTAAAGATGAAG-3′), TP-SPH (5′-GATCGGCATGCTTTCCCGATAACCTTTGGTGG-3′) and TP-BAM (5′-GATCGGGATCCAGTGATGCCGGCTAATTCATG-3′) were ligated into the multiple cloning site of pKAS32 to generate the ΔtcpP1 plasmid pMIN27. Introduction of ΔtcpP1 into the V. cholerae chromosome generates an 11-bp deletion 130 bp from the 3′ end of the gene and inserts 46 bp of noncoding polylinker sequence. The deletion was confirmed by PCR.
The toxR mutant strains were constructed via insertional inactivation of the chromosomal copy of toxR with pVM55 as previously described (30). All toxR::pVM55 strains were maintained in ampicillin. Correct chromosomal insertion of pVM55 was confirmed by observing the OmpU-OmpT switch by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) as described (30).
The E. coli (toxT-lacZYA)4 fusion (long) was constructed by amplifying the toxT promoter region from −656 to +77, with respect to the transcriptional start site, using primers RT19 (5′-AAAATCTAGATATGATATTGTGAATGTTGGTGGTG-3′) and RT45 (5′-AAAGGCCTAATCATTGCGTTCTACTCTGAAG-3′). The resulting fragment was cloned into the lacZ operon fusion vector pRS415 to generate pJYT1. The (toxT-lacZYA)4 fusion was recombined onto λRS45 (42) and integrated into the chromosome of MC4100 to create RT4129. Similarly, the (toxT-lacZYA)5 fusion (intermediate) was constructed with primers RT52 (5′-AAAAGAATTCAAGTGGTCAAATACTATGTTCTC-3′) and RT53 (5′-AAAAGGATCCGCAGAGAGCCATCCACGTA-3′) to amplify the toxT promoter from −256 to +37. This fragment was cloned into pRS415, generating pMIN38. This fusion was also recombined onto λRS45 and lysogenized into MC4100 to yield RT4317. The third toxT-lacZYA fusion (short), composed of the region from −172 to +45 of the toxT promoter, was previously constructed as a linearized plasmid integrant in the chromosome of E. coli strain DH92 (20). This fusion was transduced into MC4100 using P1vir, resulting in strain RT4146. The hns651 mutation from DL1976 was transduced into each of the fusion strains by P1vir, and plasmid pTSK was introduced by calcium chloride transformation.
β-Galactosidase assay.
β-Galactosidase activity was determined by the method of Miller (27) with the following modifications. In strains of the KSK218 (ctx-lacZ) background, cultures were assayed after 16 h of growth. Due to TCP-mediated bacterial autoagglutination of these strains, specific activity was calculated using the protein concentration determined by the bicinchoninic acid procedure (Pierce) rather than the optical density at 600 nm of the culture. For V. cholerae strains of the MBN032 background and E. coli strains, cultures were assayed during mid-log-phase growth.
SDS-PAGE and immunoblot.
Protein extracts from overnight cultures were prepared and analyzed by SDS–12.5% PAGE as described (49). Proteins were transferred to nitrocellulose and probed with anti-TcpA antibody (48) using the Renaissance Chemiluminescence Reagent Plus (NEN Life Science Products).
RESULTS
V. cholerae encodes an H-NS homolog.
A TBLASTN search (1) of the V. cholerae genome revealed the presence of a gene encoding an open reading frame with 41% identity and 51% similarity to the E. coli H-NS protein. The current genome sequence places the beginning of the hns gene at position 1221584 of the large chromosome. This gene has also recently been termed vicH by Bertin et al. (3). In order to determine any role of V. cholerae H-NS in the regulation of virulence gene expression, a deletion of the gene was constructed on a suicide plasmid that could be incorporated into reporter strains by allelic exchange. Introduction of the hns deletion into the genome of ctx-lacZ fusion strain KSK218 resulted in small, intensely blue colonies on agar containing XGal. The small colony size is consistent with the hns phenotype seen in other bacterial species, and the color suggested that a mutation in hns might cause an increase in ctx gene expression.
Deletion of hns results in high levels of ctx gene expression.
Regulation of expression from the ctx operon promoter is thought to be the last step in the ToxR virulence cascade. We therefore chose to first investigate any possible effect of H-NS at this promoter since it responds to many levels of input into the cascade. In V. cholerae O1 strains of the classical biotype, ctx transcription is optimally induced by growth in vitro at 30°C in LB with a starting pH of 6.5. Expression is reduced under growth conditions of increased temperature and starting pH, with growth at 37°C in LB with a starting pH of 8.5 used as a standard maximal repressing condition.
A comparison of ctx-lacZ expression between hns+ strain KSK218 and hns mutant strain MBN147 grown under various conditions revealed that the Δhns1 deletion resulted in derepression of ctx-lacZ expression under all growth conditions examined (Fig. 1A). Expression from the hns mutant strain at 30°C, regardless of pH, actually exceeded optimal wild-type expression at 30°C and pH 6.5. At 37°C and either pH, ctx expression in the hns mutant approached that observed for the hns+ strain grown under optimal expression conditions. At 30°C and pH 6.5, the finding that the level of expression in the hns mutant exceeded that of the wild type suggests that H-NS exerts a partial repressive effect even under inducing conditions. Together with the finding that loss of H-NS completely overrides the repressive effects of high pH, these results suggest that H-NS plays a role in silencing the ctx promoter under various environmental conditions. However, since Δhns1 does not permit maximal expression of ctx-lacZ at 37°C, this implies that repression by temperature may be influenced by other factors in addition to H-NS.
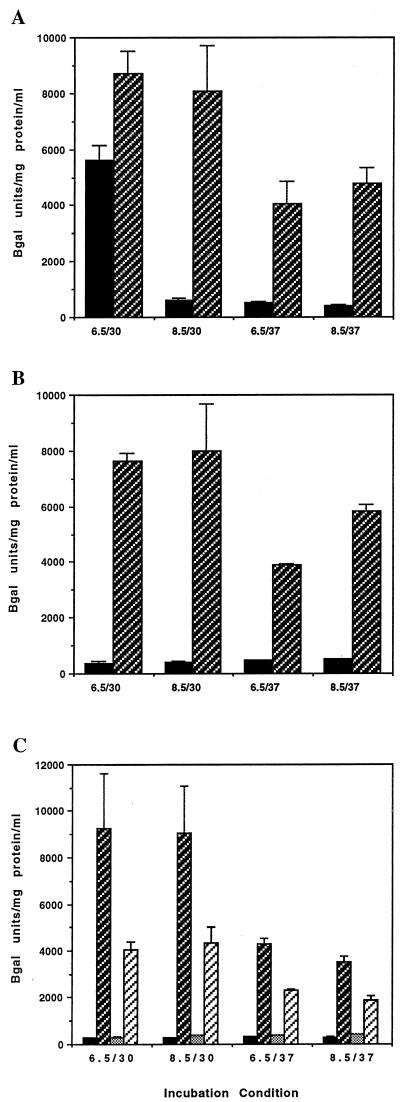
FIG. 1.
β-Galactosidase (Bgal) production in ctx-lacZ fusion strains. (A) Δhns1 mutation derepresses ctx expression under all conditions (pH/°C). Solid bars, KSK218 (hns+); hatched bars, MBN147 (Δhns1). (B) Loss of ctx expression in the ΔtoxT1 background is restored in the presence of the Δhns1 mutation. Solid bars, MBN019 (ΔtoxT1 hns+); hatched bars, MBN153 (ΔtoxT1 Δhns1). (C) Mutation of both toxR and toxT results in less ctx expression than for either mutation alone in the Δhns1 strain. Solid bars, KSK236 (toxT+ hns+ toxR); dark hatched bars, MBN196 (toxT+ Δhns1 toxR); shaded bars, MBN185 (ΔtoxT1 hns+ toxR); light hatched bars, MBN192 (ΔtoxT1 Δhns1 toxR). Error bars show standard deviations.
Expression of ctx no longer requires ToxT or ToxR in an hns mutant.
If H-NS exerts its influence directly at the ctx promoter, an hns deletion mutant might be derepressed for ctx expression even in the absence of the ToxT protein, which activates ctx expression by direct interaction at the ctx promoter (6). The dependence of ctx-lacZ expression on ToxT in LB (pH 6.5) at 30°C is readily apparent by comparing strain KSK218 (Fig. 1A) with the ΔtoxT1 strain MBN019 (Fig. 1B). Strikingly, deletion of hns in the ΔtoxT mutant strain MBN153 was found to restore high levels of ctx-lacZ expression under all conditions examined (Fig. 1B). The levels of ctx-lacZ expression in the ΔtoxT Δhns strain MBN153 were found to be essentially identical to those in the toxT+ Δhns strain MBN147 (Fig. 1A and B). These results suggest that H-NS mediates repression of gene expression directly at the ctx promoter in a manner that can be counteracted by the action of ToxT.
The expression of ctx is activated by ToxT, whereas the expression of ToxT is activated by the combined action of the ToxRS and TcpPH membrane protein pairs (18). Since ToxR is required to activate toxT expression, the effect of an hns mutation on ctx expression in a toxR mutant should be similar to that shown above for the toxT mutant. As shown in Fig. 1C, the normal dependence of ctx expression on ToxR, as evidenced in strain KSK236, was relieved by deletion of the hns gene in strain MBN196. As expected, the level of ctx-lacZ expression in strain MBN196 was found to be similar to that of the toxT hns double mutant MBN153 (Fig. 1B). However, an additional contribution of ToxR to ctx expression was suggested when the influence of H-NS on ctx expression was examined in a toxR toxT double mutant. As shown in Fig. 1C, the ΔtoxT toxR::pVM55 double mutant MBN185 was repressed for ctx-lacZ expression under all conditions examined, and transcription was restored upon introduction of the hns deletion into the double mutant MBN192. Surprisingly, expression did not increase to the levels achieved with either the ΔtoxT strain MBN153 (Fig. 1B) or the toxR::pVM55 strain MBN196 in the presence of the hns allele. If the only contribution of ToxR to ctx expression is to activate toxT expression, it would be expected that ctx-lacZ expression in the ΔtoxT Δhns strain MBN153 would be identical to that of the ΔtoxT Δhns toxR strain MBN192. The finding that the double toxT toxR mutant cannot achieve the same level of ctx expression as either mutation alone in the hns background suggests that either ToxT or ToxR can independently activate ctx expression in the absence of H-NS. This finding, although not expected in V. cholerae, is similar to previous results showing that either ToxR or ToxT can independently activate ctx expression in E. coli (11, 29).
Expression of tcpA is increased in parallel to ctx expression in an hns mutant.
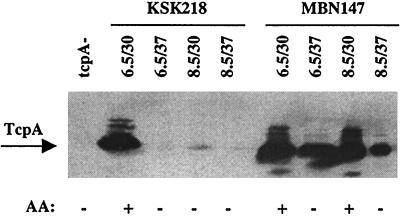
To discern whether promoters within the ToxR virulence cascade in addition to ctx might be affected by H-NS, we determined the influence of the hns mutation on tcpA expression. Protein extracts from overnight cultures of either KSK218 (hns+) or MBN147 (Δhns1) grown under the four conditions were subjected to SDS-PAGE and Western immunoblot analysis with anti-TcpA antibody. As expected, significant TcpA production was detected from strain KSK218 only after growth in the optimal inducing conditions of pH 6.5 and 30°C (Fig. 2). However, the presence of the hns mutation in strain MBN147 led to TcpA production under all four growth conditions. The trend in TcpA production appeared to essentially parallel the level of ctx transcription under the various growth conditions (Fig. 1A). A second measure of TcpA expression is the bacterial autoagglutination that occurs when large amounts of TCP are present on the bacterial surface. Autoagglutination of overnight cultures was evident for the wild-type strain grown at pH 6.5 and 30°C and for the hns mutant grown at pH 6.5 and 30°C or pH 8.5 and 30°C. This pattern was consistent with the Western blot analysis, showing that the highest levels of TcpA expression occurred under these conditions. These results indicate that H-NS acts to negatively influence tcpA expression, either by acting at the tcpA promoter or by influencing prior steps within the regulatory cascade.
FIG. 2.
TcpA production and autoagglutination (AA) by ctx-lacZ fusion strain KSK218 and its hns derivative MBN147. Strains were cultured under the conditions indicated (pH/°C) for each lane, and 15 μg of total protein extract was subjected to Western blot analysis with anti-TcpA. The tcpA mutant control strain was grown at pH 6.5 and 30°C.
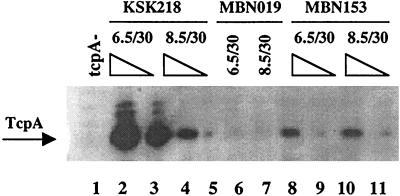
In order to determine whether H-NS acts directly at the tcpA promoter, we examined the effect of the hns mutation on the level of TcpA produced in a toxT mutant background. As shown in Fig. 3, the hns+ ctx-lacZ strain KSK218 produced high levels of TcpA at pH 6.5 and 30°C and less at pH 8.5 and 30°C, whereas no TcpA was detected from the ΔtoxT ctx-lacZ strain MBN019 for either growth condition. In contrast, the ΔtoxT Δhns ctx-lacZ strain MBN153 produced TcpA under both growth conditions, albeit only at a level comparable to that expressed by a wild-type strain grown under the semirepressive condition of LB at pH 8.5 and 30°C. The expression of TcpA in the absence of toxT suggests that H-NS directly affects the tcpA promoter. However, since TcpA production was not restored to wild-type levels, the effect of H-NS at the tcpA promoter appears to be smaller than at the ctx promoter, where wild-type levels were achieved. Interestingly, the low level of expression appears to be constitutive with respect to pH (Fig. 3, lanes 8 to 11).
FIG. 3.
Western immunoblot with anti-TcpA in toxT and hns mutant strains. Protein extracts were prepared from strains grown under the indicated conditions (pH/°C) and either 15 μg of total protein extract or a subsequent 1:5 dilution was loaded onto the gel. Lane 1, ΔtcpA control at pH 6.5 and 30°C; lanes 2 and 3, KSK218 (toxT+ hns+); lanes 4 and 5, KSK218; lanes 6 and 7 MBN019 (ΔtoxT1 hns+); lanes 8 and 9, MBN153 (ΔtoxT1 Δhns1); lanes 10 and 11, MBN153.
H-NS represses toxT expression.
The high level of TcpA produced under the nonpermissive condition of pH 8.5 and 30°C in the toxT+ Δhns strain MBN147 compared to that of the ΔtoxT Δhns strain MBN153 (Fig. 2 and 3) suggested that H-NS might directly influence toxT expression in addition to its role on expression from the ctx and tcpA promoters. To investigate this further, we used a series of toxT-lacZ fusion strains to directly examine the role of H-NS on toxT expression. Introduction of a toxT-lacZ transcriptional fusion into a wild-type strain (MBN032) revealed that toxT is most highly expressed during the logarithmic stage of growth (data not shown) at pH 6.5 and 30°C. Expression was found to be slightly attenuated at 37°C and greatly reduced by pH 8.5 at either incubation temperature (Fig. 4A). Thus, toxT expression responds most significantly to changes in pH but is also affected by temperature. Notably, this regulation mimics that previously determined for tcpP, which encodes an activator of toxT expression (5, 46).
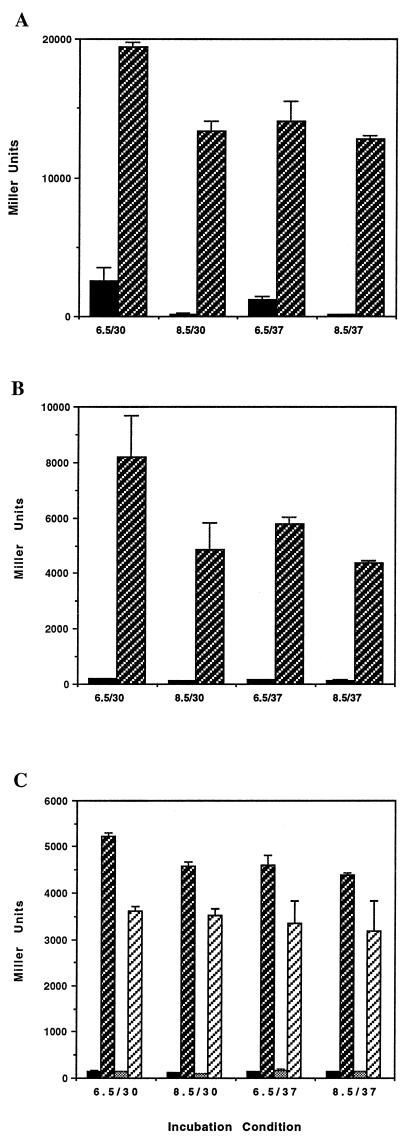
FIG. 4.
β-Galactosidase production by toxT-lacZ fusion strains. (A) The Δhns1 mutation derepresses toxT expression under all conditions (pH/°C). Solid bars, MBN032 (hns+); hatched bars, MBN170 (Δhns1). (B) The Δhns1 mutation permits toxT expression in a strain that lacks ToxR. Solid bars, MBN189 (hns+ toxR); hatched bars, MBN183 (Δhns1 toxR). (C) Neither ToxR nor TcpP is required to activate toxT in the absence of hns. Solid bars, MBN172 (hns+ ΔtcpP1); dark hatched bars, MBN175 (Δhns1 ΔtcpP1); shaded bars, MBN318 (hns+ ΔtcpP1 toxR); light hatched bars, MBN187 (Δhns1 ΔtcpP1 toxR). Error bars show standard deviations.
Incorporation of the hns mutation into the toxT-lacZ strain MBN170 resulted in significant derepression of expression under all conditions (Fig. 4A). As was observed for ctx expression, the most significant derepression occurred during growth under suboptimal expression conditions. At pH 6.5 and 30°C, expression was increased 8-fold in the hns mutant, whereas at pH 8.5 and 37°C, there was an 87-fold increase in expression. Interestingly, the trend of slight attenuation by temperature and a greater repressive influence of high pH was still observed. It is noteworthy that an insertion in hns was one of several mutations previously reported by Häse and Mekalanos (19) to increase expression of toxT.
Similar to the manner in which we examined the role of H-NS at the ctx promoter, we determined whether the presence of the activators of toxT expression, ToxR and TcpP, were required for derepression in the absence of hns. In the hns+ toxR strain MBN189, the loss of toxR abolished transcription of toxT-lacZ, as expected (Fig. 4B). However, in the absence of hns (strain MBN183), toxR was found not to be necessary for toxT expression (Fig. 4B). Similar results were found with respect to the requirement of TcpP for toxT expression. Deletion of tcpP in MBN172 significantly reduced toxT expression (Fig. 4C), whereas the level of β-galactosidase in the ΔtcpP Δhns toxT-lacZ strain MBN175 was similar to that of the wild type (Fig. 4C). Interestingly, the level of toxT-lacZ expression in the tcpP toxR double mutant strain in the hns mutant background (MBN187) was not as high as in the presence of either activator mutation alone (Fig. 4B and C). As in the case of the ctx promoter, this suggests that in the absence of H-NS, each activator can contribute independently to increase toxT expression. Of further interest was the trend of the toxR mutants to retain the characteristic regulatory pattern of toxT expression conferred by pH and temperature (Fig. 4B), whereas this fluctuation was absent in the tcpP mutant backgrounds (Fig. 4C). This is consistent with the nearly constitutive expression of ToxR for all of the growth conditions used in this assay (34) versus the regulated expression of TcpP under these conditions (5).
H-NS functions at a region upstream of the toxT promoter.
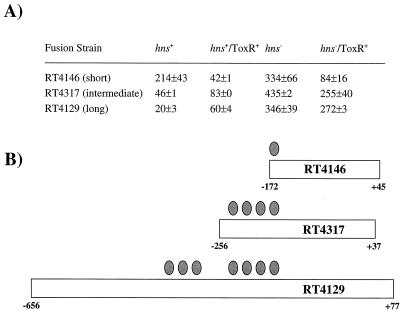
It has previously been shown that expression of a toxT-lacZ transcriptional fusion encompassing the region from −172 to +45 of the ToxR-dependent toxT transcriptional start site has a high basal level of activity that is actually repressed rather than activated by ToxR expressed in E. coli (20). The findings described above showing that H-NS exerts a repressive activity on toxT expression suggested that the high-level constitutive expression observed for this fusion construct might be due to deletion of sequences required for H-NS interaction near the toxT promoter. To examine this possibility, we constructed a series of toxT fusions with various lengths of upstream DNA and determined their expression in various toxR and hns backgrounds (Fig. 5). These included the original fusion transduced into the chromosome of strain MC4100 (short fusion strain RT4146), as well as two additional transcriptional fusions containing more extensive upstream regions spanning positions −256 to +37 (intermediate fusion strain RT4317) or −656 to +77 (long fusion strain RT4129). The latter two fusions were constructed as lambda lysogens of MC4100. These two fusions yielded substantially lower units of basal activity in the absence of ToxR than the original short fusion and, unlike the short fusion, were activated by ToxR (Fig. 5A).
FIG. 5.
Differential responses of toxT-lac fusions containing various amounts of the promoter region to H-NS and ToxR. (A) β-Galactosidase activity (Miller units, means ± standard deviations) from E. coli strains that are either hns+ or hns651 and carry a chromosomal copy of each fusion construct. ToxR is supplied from plasmid pTSK. (B) Schematic representation of the extent of the toxT promoter present in each fusion construct. Each oval represents an approximately twofold relative influence contributed by H-NS to decrease the basal level of β-galactosidase activity.
To determine whether H-NS contributed to the reduced basal level of expression from the fusions containing more extensive upstream regions, the hns651 mutation was transduced into the fusion backgrounds, and the effect on transcription was assessed by β-galactosidase assay. The hns651 mutation was found to elevate the level of transcription of these fusions to an approximately equivalent level at pH 6.5 and 30°C (Fig. 5A). The shortest fusion showed a 1.6-fold increase in basal expression in the hns strain, whereas the intermediate and long fusions showed 9-fold and 14-fold increases, respectively. These results suggest that the fusions with the more extensive regions upstream of the promoter are more strongly repressed by H-NS. Interestingly, although the intermediate and long fusions were moderately activated by ToxR in an hns+ background, they were repressed by ToxR in an hns mutant background. The repression of toxT expression by ToxR was similar to that seen for the short fusion regardless of hns background. This suggests a role for H-NS in the normal regulation of toxT expression and that this regulation is lacking in the short fusion strain. A model accounting for the influence of H-NS on the toxT promoter regions from each of the three fusion lengths is shown in Fig. 5B. Each oval represents an approximately twofold repression by H-NS on toxT expression. These data are consistent with a mechanism by which H-NS represses transcription by interactions at regions located at significant distances upstream of the RNA polymerase binding site. This type of repression by H-NS has been termed transcriptional silencing (17).
DISCUSSION
Regulation of the expression of the genes that encode the major virulence determinants of V. cholerae, CT and TCP, involves a complex interplay between regulators encoded within the ancestral genome and those encoded within the TCP pathogenicity island. The regulators encoded within the TCP island include the AraC homolog ToxT, which activates the expression of the ctx and tcp genes, and the transmembrane protein pair TcpP-TcpH, which activate the expression of toxT. Other virulence gene regulators that are not exclusively associated with pathogenicity islands and that likely participate in additional regulatory networks within the cell include the transmembrane protein pair ToxR-ToxS and the cytoplasmic proteins AphA and AphB. ToxR-ToxS is present in many Vibrio species and, in addition to its role in toxT activation, is required for expression of genes encoding homologs of the V. cholerae OmpU protein (56). AphA and AphB are required for expression of the tcpPH operon (25, 46), but other potential roles for these proteins have not yet been elucidated. In addition, gene expression within the V. cholerae ToxR virulence regulon is negatively influenced by the cyclic AMP-cyclic AMP receptor protein complex (44). The mechanism for this negative regulation within the virulence cascade is still under investigation, but it is known that this regulatory system influences the expression of multiple genes that affect cellular physiology in response to carbon source and perhaps additional growth parameters. In the present study we have determined that another protein with global effects, the H-NS protein, which influences chromatin structure and gene expression in response to numerous growth parameters, has a major negative influence at multiple levels of expression within the ToxR virulence gene regulon.
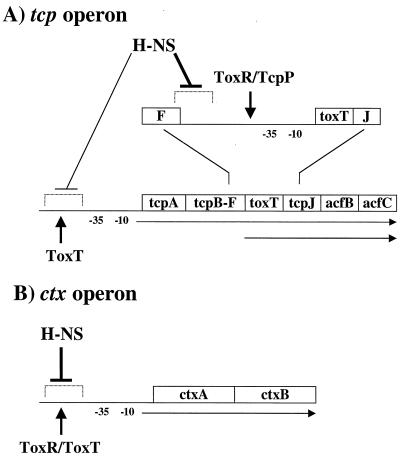
As depicted in Fig. 6, the studies reported here indicate that H-NS influences ToxR regulon gene expression by exerting a negative effect on at least three promoters, toxT, tcpA, and ctx. In the absence of H-NS, expression from each of these promoters was increased dramatically under noninducing conditions and, to a smaller degree, under inducing conditions. These results suggest that H-NS plays a role in repressing ToxR regulon gene expression under environmental conditions not normally permissive for its expression and that it represses expression even under normally inducing conditions. The observation that an hns mutation derepresses the expression of toxT, ctx, and tcpA under several environmental conditions even in the absence of their cognate activator proteins suggests that H-NS is directly influencing these promoters.
FIG. 6.
Model for H-NS-mediated negative modulation of virulence gene expression. (A) H-NS negatively affects two steps in tcp operon expression. A major effect occurs at the ToxR/TcpP-dependent transcriptional start site upstream of toxT. The major region required for this activity is located upstream of position −172 with respect to the toxT mRNA start site. The negative effect of H-NS at the toxT promoter is overcome by ToxR and TcpP. H-NS also exerts a minor effect at the tcp operon promoter located upstream of tcpA. This effect is overcome by ToxT. (B) H-NS also exerts a major negative effect at the ctx promoter. This effect is counteracted by ToxT, with a possible additional contribution from ToxR.
H-NS appears to exert its largest repressive effect on the toxT promoter. Expression of toxT is activated by ToxR-ToxS together with TcpP-TcpH (18). In the absence of H-NS, expression of toxT was close to or greater than wild-type levels under inducing conditions in the absence of either ToxR, TcpP, or both. This was the case even under normally repressive environmental conditions. These findings are further supported by the recent report of an insertion mutation in hns that significantly increased toxT transcription (19). Analysis of E. coli toxT-lacZ fusions suggests that H-NS exerts its negative effects on transcription by influencing the promoter over an extensive region from −172 to beyond −256 with respect to the start of transcription (Fig. 5 and 6). The region of the promoter spanning from −114 to −73 has been shown to be required for the interaction of ToxR with DNA (20). The location of the TcpP binding site has not yet been reported but TcpP appears to influence transcription downstream of −172 (31). Previous experiments with toxT-lacZ fusions carrying DNA only to −172 (short) have indicated that toxT is actually repressed by ToxR (20). This paradoxical finding may be explained by the results shown here, that upon increasing the length of the upstream region in these fusions, to −256 or to −656, the basal level of expression is decreased by H-NS such that it becomes activated by ToxR.
H-NS also has a significant effect on expression of the ctx promoter. This promoter is directly activated by ToxT in V. cholerae (6). Although at least some influence on the ctx promoter in the toxT+ background may be the result of increased expression of the toxT promoter, as discussed above, deletion of hns resulted in expression from ctx that was close to or greater than wild-type expression in the absence of ToxT. This result suggests that H-NS also influences the ctx promoter directly. Interestingly, the level of ctx expression in the hns mutant lacking both ToxT and ToxR is lower than the level of expression in an hns mutant lacking only ToxT. This result suggests that ToxR directly influences the ctx promoter in the absence of hns. It has previously been shown in E. coli that either ToxR or ToxT is capable of activating ctx expression (11, 28). Genetic footprint analysis indicates that ToxR interacts at two positions, one at −69 to −57 and the other at −47 to −39 (36). Although a direct role for ToxR at the ctx promoter in V. cholerae has not been demonstrated in vitro, recent studies indicate that ToxR and ToxT have a dual role at the ctx promoter in vivo (26).
H-NS has a more moderate effect at the tcpA promoter than at the toxT and ctx promoters. In a toxT mutant background, the absence of H-NS restored expression to wild-type levels at pH 8.5 and 30°C but not at pH 6.5 and 30°C. ToxT is the only known activator that functions at the tcpA promoter, but the cyclic AMP-cyclic AMP receptor protein complex has been implicated in exerting a negative influence at the tcpA promoter. Further investigation of how these factors interact to influence expression from the tcpA promoter is under way.
The results presented here indicate that in V. cholerae, H-NS affects both the expression of a positive transcriptional regulator, ToxT, and the expression of the target genes of the regulator, tcpA and ctx. This is similar to the situation observed in the VirF-VirB regulatory cascade of S. flexneri (38). The toxT, tcpA, and ctx promoters possess characteristics that have been correlated with H-NS binding. The high AT content of these promoters likely promotes local curvature within these regions. Molecular models for transcriptional silencing by H-NS vary with respect to the position at which the protein interacts with the DNA. For genes in which the H-NS binding sites overlap the promoter elements directly, H-NS is proposed to reduce transcription by preventing the binding of RNA polymerase at the promoter (53). Repression of E. coli rrnB (50) and S. flexneri virB (51) is thought to occur in this manner. Alternatively, many H-NS binding sites have been found to lie outside the immediate promoter region. This appears to be the situation with toxT. In the case of Salmonella enterica serovar Typhimurium proU expression, the H-NS binding site is a curved region 200 bp downstream from the transcriptional start site (33). For proU and other genes where H-NS binds outside of the promoter elements, H-NS-DNA binding causes a change in DNA topology that leads to a subsequent influence on gene expression (33). In such a case, H-NS binding could generate locally constrained supercoiling that specifically silences the promoter (52). Finally, it has been suggested that H-NS can repress transcription by decreasing the rate of open complex formation at the promoter (47).
The mechanisms by which activator proteins function to counteract H-NS-mediated modulation of gene expression are not well understood. Atlung and Ingmer (2) have suggested that H-NS generally functions as an activator antagonist at genes for which expression is repressed by H-NS and specifically induced by positive regulators. Examples include the cfaAB, pap, and coo genes of E. coli that are repressed by H-NS and activated by CfaD, PapB, and Rns, respectively (15, 22, 32). Although these activators are dispensable in the absence of H-NS, CfaD and Rns are able to further increase transcription of their target genes in hns mutant strains (22, 32). A similar situation occurs at the toxT and ctx promoters. At least one of the functions of such activators appears to be to counteract H-NS repression of their respective target genes. Whether these activators interact directly with H-NS to displace it from the DNA is not known. Other transcriptional activators that function at H-NS-repressed promoters are still required to activate gene expression in the absence of H-NS. For example, expression of the Shigella virB gene is repressed by H-NS but still requires VirF for activation in hns mutants (51). This appears to be the case for the tcpA promoter, since ToxT is still required for maximal expression in the absence of H-NS. Further analysis of the molecular interactions between H-NS and the regulatory proteins TcpP, ToxR, and ToxT at the various promoters within the ToxR regulon will provide insights into the mechanisms by which this protein influences virulence gene expression.
ACKNOWLEDGMENTS
We thank Cori Sandoe and Jennifer Thibert for plasmids, Christine White-Ziegler and Victor DiRita for strains, and Tom Kirn for assistance with genome analysis.
This work was supported by NIH grants AI39654 to R.K.T. and AI41558 to K.S. J.D.P. was supported by postdoctoral fellowship PF-4286 from the ACS.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Atlung T, Ingmer H. H-NS: a modulator of environmentally regulated gene expression. Mol Microbiol. 1997;24:7–17. doi: 10.1046/j.1365-2958.1997.3151679.x. [DOI] [PubMed] [Google Scholar]
- 3.Bertin P, Benhabiles N, Krin E, Laurent-Winter C, Tendeng C, Turlin E, Thomas A, Danchin A, Brasseur R. The structural and functional organization of H-NS-like proteins is evolutionarily conserved in gram-negative bacteria. Mol Microbiol. 1999;31:319–329. doi: 10.1046/j.1365-2958.1999.01176.x. [DOI] [PubMed] [Google Scholar]
- 4.Brown R C, Taylor R K. Organization of tcp, acf, and toxT genes within a ToxT-dependent operon. Mol Microbiol. 1995;16:425–439. doi: 10.1111/j.1365-2958.1995.tb02408.x. [DOI] [PubMed] [Google Scholar]
- 5.Carroll P A, Tashima K T, Rogers M B, DiRita V J, Calderwood S B. Phase variation in tcpH modulates expression of the ToxR regulon in Vibrio cholerae. Mol Microbiol. 1997;25:1099–1111. doi: 10.1046/j.1365-2958.1997.5371901.x. [DOI] [PubMed] [Google Scholar]
- 6.Champion G A, Neely M N, Brennan M A, DiRita V J. A branch in the ToxR regulatory cascade of Vibrio cholerae revealed by characterization of toxT mutant strains. Mol Microbiol. 1997;23:323–331. doi: 10.1046/j.1365-2958.1997.2191585.x. [DOI] [PubMed] [Google Scholar]
- 7.Chang A C, Cohen S N. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the p15A cryptic miniplasmid. J Bacteriol. 1978;134:1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chiang S L, Taylor R K, Koomey M, Mekalanos J J. Single amino acid substitutions in the N-terminus of Vibrio cholerae TcpA affect colonization, autoagglutination, and serum resistance. Mol Microbiol. 1995;17:1133–1142. doi: 10.1111/j.1365-2958.1995.mmi_17061133.x. [DOI] [PubMed] [Google Scholar]
- 9.Colonna B, Casalino M, Fradiani P A, Zagaglia C, Naitza S, Leoni L, Prosseda G, Coppo A, Ghelardini P, Nicoletti M. H-NS regulation of virulence gene expression in enteroinvasive Escherichia coli harboring the virulence plasmid integrated into the host chromosome. J Bacteriol. 1995;177:4703–4712. doi: 10.1128/jb.177.16.4703-4712.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.DiRita V J, Mekalanos J J. Periplasmic interaction between two membrane regulatory proteins, ToxR and ToxS, results in signal transduction and transcriptional activation. Cell. 1991;64:29–37. doi: 10.1016/0092-8674(91)90206-e. [DOI] [PubMed] [Google Scholar]
- 11.DiRita V J, Parsot C, Jander G, Mekalanos J J. Regulatory cascade controls virulence in Vibrio cholerae. Proc Natl Acad Sci USA. 1991;88:5403–5407. doi: 10.1073/pnas.88.12.5403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DiRita V J. Three-component regulatory system controlling virulence in Vibrio cholerae. In: Hoch J A, Silhavy T J, editors. Two-component signal transduction. Washington, D.C.: American Society for Microbiology; 1995. pp. 351–365. [Google Scholar]
- 13.Donato G M, Lelivelt M J, Kawula T H. Promoter-specific repression of fimB expression by the Escherichia coli nucleoid-associated protein H-NS. J Bacteriol. 1997;179:6618–6625. doi: 10.1128/jb.179.21.6618-6625.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dorman C J, Porter M E. The Shigella virulence gene regulatory cascade: a paradigm of bacterial gene control mechanisms. Mol Microbiol. 1998;29:677–684. doi: 10.1046/j.1365-2958.1998.00902.x. [DOI] [PubMed] [Google Scholar]
- 15.Forsman K, Sonden B, Goransson M, Uhlin B E. Antirepression function in Escherichia coli for the cAMP-cAMP receptor protein transcriptional activator. Proc Natl Acad Sci USA. 1992;89:9880–9884. doi: 10.1073/pnas.89.20.9880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gardel C L, Mekalanos J J. Regulation of cholera toxin by temperature, pH and osmolarity. Methods Enzymol. 1994;235:517–526. doi: 10.1016/0076-6879(94)35167-8. [DOI] [PubMed] [Google Scholar]
- 17.Göransson M, Sondén B, Nilsson P, Dagberg B, Forsman K, Emanuelsson K, Uhlin B E. Transcriptional silencing and thermoregulation of gene expression in Escherichia coli. Nature. 1990;344:682–685. doi: 10.1038/344682a0. [DOI] [PubMed] [Google Scholar]
- 18.Häse C C, Mekalanos J J. TcpP protein is a positive regulator of virulence gene expression in Vibrio cholerae. Proc Natl Acad Sci USA. 1998;95:730–734. doi: 10.1073/pnas.95.2.730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Häse C C, Mekalanos J J. Effects of changes in membrane sodium flux on virulence gene expression in Vibrio cholerae. Proc Natl Acad Sci USA. 1999;96:3183–3187. doi: 10.1073/pnas.96.6.3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Higgins D E, DiRita V J. Transcriptional control of toxT, a regulatory gene in the ToxR regulon of Vibrio cholerae. Mol Microbiol. 1994;14:17–29. doi: 10.1111/j.1365-2958.1994.tb01263.x. [DOI] [PubMed] [Google Scholar]
- 21.Hulton C S J, Seirafi A, Hinton J C D, Sidebotham J M, Waddell L, Pavitt G D, Owen-Hughes T, Spassky A, Buc H, Higgins C F. Histone-like protein H1 (H-NS), DNA supercoiling, and gene expression in bacteria. Cell. 1990;63:631–642. doi: 10.1016/0092-8674(90)90458-q. [DOI] [PubMed] [Google Scholar]
- 22.Jordi B J A M, Dagberg B, de Haan L A M, Hamers A M, van der Zeijst B A M, Gaastra W, Uhlin B E. The positive regulator CfaD overcomes the repression mediated by histone-like protein H-NS (H1) in the CFA/I fimbrial operon of Escherichia coli. EMBO J. 1992;11:2627–2632. doi: 10.1002/j.1460-2075.1992.tb05328.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Karaolis D K R, Johnson J A, Bailey C C, Boedeker E C, Kaper J B, Reeves P R. A Vibrio cholerae pathogenicity island associated with epidemic and pandemic strains. Proc Natl Acad Sci USA. 1998;95:3134–3139. doi: 10.1073/pnas.95.6.3134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Karaolis D K R, Somara S, Maneval D R, Jr, Johnson J A, Kaper J B. A bacteriophage encoding a pathogenicity island, a type-IV pilus and a phage receptor in cholera bacteria. Nature. 1999;399:375–379. doi: 10.1038/20715. [DOI] [PubMed] [Google Scholar]
- 25.Kovacikova G, Skorupski K. A Vibrio cholerae LysR homolog, AphB, cooperates with AphA at the tcpPH promoter to activate expression of the ToxR virulence cascade. J Bacteriol. 1999;181:4250–4256. doi: 10.1128/jb.181.14.4250-4256.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee S H, Hava D L, Waldor M K, Camilli A. Regulation and temporal expression patterns of Vibrio cholerae virulence genes during infection. Cell. 1999;99:625–634. doi: 10.1016/s0092-8674(00)81551-2. [DOI] [PubMed] [Google Scholar]
- 27.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 28.Miller V L, Taylor R K, Mekalanos J J. Cholera toxin transcriptional activator ToxR is a transmembrane DNA binding protein. Cell. 1987;48:271–279. doi: 10.1016/0092-8674(87)90430-2. [DOI] [PubMed] [Google Scholar]
- 29.Miller V L, Mekalanos J J. Synthesis of cholera toxin is positively regulated at the transcriptional level by toxR. Proc Natl Acad Sci USA. 1984;81:3471–3475. doi: 10.1073/pnas.81.11.3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miller V L, Mekalanos J J. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J Bacteriol. 1988;170:2575–2583. doi: 10.1128/jb.170.6.2575-2583.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Murley Y M, Carroll P A, Skorupski K, Taylor R K, Calderwood S B. Differential transcription of the tcpPH operon confers biotype-specific control of the Vibrio cholerae ToxR virulence regulon. Infect Immun. 1999;67:5117–5123. doi: 10.1128/iai.67.10.5117-5123.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Murphree D, Froehlich B, Scott J R. Transcriptional control of genes encoding CS1 pili: negative regulation by a silencer and positive regulation by Rns. J Bacteriol. 1997;179:5736–5743. doi: 10.1128/jb.179.18.5736-5743.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Owen-Hughes T A, Pavitt G D, Santos D S, Sidebotham J M, Hulton C S J, Hinton J C D, Higgins C F. The chromatin-associated protein H-NS interacts with curved DNA to influence DNA topology and gene expression. Cell. 1992;71:255–265. doi: 10.1016/0092-8674(92)90354-f. [DOI] [PubMed] [Google Scholar]
- 34.Parsot C, Mekalanos J J. Expression of ToxR, the transcriptional activator of the virulence factors in Vibrio cholerae, is modulated by the heat shock response. Proc Natl Acad Sci USA. 1990;87:9898–9902. doi: 10.1073/pnas.87.24.9898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Parsot C, Mekalanos J J. Structural analysis of the acfA and acfD genes of Vibrio cholerae: effects of DNA topology and transcriptional activators on expression. J Bacteriol. 1992;174:5211–5218. doi: 10.1128/jb.174.16.5211-5218.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pfau J D, Taylor R K. Genetic footprint of the ToxR-binding site in the promoter for cholera toxin. Mol Microbiol. 1996;20:213–222. doi: 10.1111/j.1365-2958.1996.tb02502.x. [DOI] [PubMed] [Google Scholar]
- 37.Pfau J D, Taylor R K. Mutations in toxR and toxS that separate transcriptional activation from DNA binding at the cholera toxin gene promoter. J Bacteriol. 1998;180:4724–4733. doi: 10.1128/jb.180.17.4724-4733.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Prosseda G, Fradiani P A, Di Lorenzo M, Falconi M, Micheli G, Casalino M, Nicoletti M, Colonna B. A role for H-NS in the regulation of the virF gene of Shigella and enteroinvasive Escherichia coli. Res Microbiol. 1998;149:15–25. doi: 10.1016/s0923-2508(97)83619-4. [DOI] [PubMed] [Google Scholar]
- 39.Rimsky S, Spassky A. Sequence determinants for H1 binding on Escherichia coli lac and gal promoters. Biochemistry. 1990;29:3765–3771. doi: 10.1021/bi00467a024. [DOI] [PubMed] [Google Scholar]
- 40.Schweizer H P. Small broad-host-range gentamicin resistance gene cassettes for site-specific insertion and deletion mutagenesis. BioTechniques. 1993;15:831–834. [PubMed] [Google Scholar]
- 41.Silhavy T J, Berman M L, Enquist L W. Experiments with gene fusions. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1984. [Google Scholar]
- 42.Simons R W, Houman F, Kleckner N. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene. 1987;53:85–96. doi: 10.1016/0378-1119(87)90095-3. [DOI] [PubMed] [Google Scholar]
- 43.Skorupski K, Taylor R K. Positive selection vectors for allelic exchange. Gene. 1996;169:47–52. doi: 10.1016/0378-1119(95)00793-8. [DOI] [PubMed] [Google Scholar]
- 44.Skorupski K, Taylor R K. Cyclic AMP and its receptor protein negatively regulate the coordinate expression of cholera toxin and toxin-coregulated pilus in Vibrio cholerae. Proc Natl Acad Sci USA. 1997;94:265–270. doi: 10.1073/pnas.94.1.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Skorupski K, Taylor R K. Control of the ToxR virulence regulon in Vibrio cholerae by environmental stimuli. Mol Microbiol. 1997;25:1003–1009. doi: 10.1046/j.1365-2958.1997.5481909.x. [DOI] [PubMed] [Google Scholar]
- 46.Skorupski K, Taylor R K. A new level in the Vibrio cholerae ToxR virulence cascade: AphA is required for transcriptional activation of the tcpPH operon. Mol Microbiol. 1999;31:763–771. doi: 10.1046/j.1365-2958.1999.01215.x. [DOI] [PubMed] [Google Scholar]
- 47.Spassky A, Rimsky S, Garreau H, Buc H. H1a, an E. coli DNA-binding protein which accumulates in stationary phase, strongly compacts DNA in vitro. Nucleic Acids Res. 1984;12:5321–5340. doi: 10.1093/nar/12.13.5321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sun D J, Sayer M, Kovari I, Sumrada R A, Taylor R K. Localization of protective epitopes within the pilin subunit of the Vibrio cholerae toxin-coregulated pilus. Infect Immun. 1991;59:114–118. doi: 10.1128/iai.59.1.114-118.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Taylor R K, Miller V L, Furlong D B, Mekalanos J J. Use of phoA gene fusions to identify a pilus colonization factor coordinately regulated with cholera toxin. Proc Natl Acad Sci USA. 1987;84:2833–2837. doi: 10.1073/pnas.84.9.2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tippner D, Afflerbach H, Bradaczek C, Wagner R. Evidence for a regulatory function of the histone-like Escherichia coli protein H-NS in ribosomal RNA synthesis. Mol Microbiol. 1994;11:589–604. doi: 10.1111/j.1365-2958.1994.tb00339.x. [DOI] [PubMed] [Google Scholar]
- 51.Tobe T, Yoshikawa M, Mizuno T, Sasakawa C. Transcriptional control of the invasion regulatory gene virB of Shigella flexneri: activation by VirF and repression by H-NS. J Bacteriol. 1993;175:6142–6149. doi: 10.1128/jb.175.19.6142-6149.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tupper A E, Owen-Hughes T A, Ussery D W, Santos D S, Ferguson D J P, Sidebotham J M, Hinton J C D, Higgins C F. The chromatin-associated protein H-NS alters DNA topology in vitro. EMBO J. 1994;13:258–268. doi: 10.1002/j.1460-2075.1994.tb06256.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ueguchi C, Minuzo T. The Escherichia coli nucleoid protein H-NS functions directly as a transcriptional repressor. EMBO J. 1993;12:1039–1046. doi: 10.1002/j.1460-2075.1993.tb05745.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Varshavsky A J, Nedospasov S A, Bakayev V V, Bakayeva T G, Georgiev G P. Histone-like proteins in the purified Escherichia coli deoxyribonucleoprotein. Nucleic Acids Res. 1977;4:2725–2745. doi: 10.1093/nar/4.8.2725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Waldor M K, Mekalanos J J. Lysogenic conversion by a filamentous phage encoding cholera toxin. Science. 1996;272:1910–1914. doi: 10.1126/science.272.5270.1910. [DOI] [PubMed] [Google Scholar]
- 56.Welch T J, Bartlett D H. Identification of a regulatory protein required for pressure-responsive gene expression in the deep-sea bacterium Photobacterium species strain SS9. Mol Microbiol. 1998;27:977–985. doi: 10.1046/j.1365-2958.1998.00742.x. [DOI] [PubMed] [Google Scholar]
- 57.White-Ziegler C A, Hill M L A, Braaten B A, van der Woude M W, Low D A. Thermoregulation of Escherichia coli pap transcription: H-NS is a temperature-dependent DNA methylation blocking factor. Mol Microbiol. 1998;28:1121–1137. doi: 10.1046/j.1365-2958.1998.00872.x. [DOI] [PubMed] [Google Scholar]
- 58.Yamada H, Muramatsu S, Mizuno T. An Escherichia coli protein that preferentially binds to sharply curved DNA. J Biochem. 1990;108:420–425. doi: 10.1093/oxfordjournals.jbchem.a123216. [DOI] [PubMed] [Google Scholar]
- 59.Yu R R, DiRita V J. Analysis of an autoregulatory loop controlling ToxT, cholera toxin, and toxin-coregulated pilus production in Vibrio cholerae. J Bacteriol. 1999;181:2584–2592. doi: 10.1128/jb.181.8.2584-2592.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]