Abstract
Reduction of the cobalt ion of cobalamin from the Co(III) to the Co(I) oxidation state is essential for the synthesis of adenosylcobalamin, the coenzymic form of this cofactor. A cob(II)alamin reductase activity in Salmonella enterica serovar Typhimurium LT2 was isolated to homogeneity. N-terminal analysis of the homogeneous protein identified NAD(P)H:flavin oxidoreductase (Fre) (EC 1.6.8.1) as the enzyme responsible for this activity. The fre gene was cloned, and the overexpressed protein, with a histidine tag at its N terminus, was purified to homogeneity by nickel affinity chromatography. His-tagged Fre reduced flavins (flavin mononucleotide [FMN] and flavin adenine dinucleotide [FAD]) and cob(III)alamin to cob(II)alamin very efficiently. Photochemically reduced FMN substituted for Fre in the reduction of cob(III)alamin to cob(II)alamin, indicating that the observed cobalamin reduction activity was not Fre dependent but FMNH2 dependent. Enzyme-independent reduction of cob(III)alamin to cob(II)alamin by FMNH2 occurred at a rate too fast to be measured. The thermodynamically unfavorable reduction of cob(II)alamin to cob(I)alamin was detectable by alkylation of the cob(I)alamin nucleophile with iodoacetate. Detection of the product, caboxymethylcob(III)alamin, depended on the presence of FMNH2 in the reaction mixture. FMNH2 failed to substitute for potassium borohydride in in vitro assays for corrinoid adenosylation catalyzed by the ATP:co(I)rrinoid adenosyltransferase (CobA) enzyme, even under conditions where Fre and NADH were present in the reaction mixture to ensure that FMN was always reduced. These results were interpreted to mean that Fre was not responsible for the generation of cob(I)alamin in vivo. Consistent with this idea, a fre mutant displayed wild-type cobalamin biosynthetic phenotypes. It is proposed that S. enterica serovar Typhimurium LT2 may not have a cob(III)alamin reductase enzyme and that, in vivo, nonadenosylated cobalamin and other corrinoids are maintained as co(II)rrinoids by reduced flavin nucleotides generated by Fre and other flavin oxidoreductases.
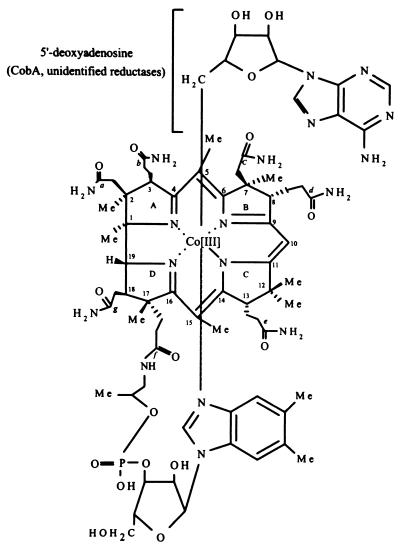
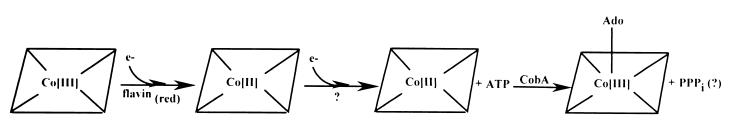
The biologically active, coenzymic form of cobalamin (Cbl) contains a 5′-deoxyadenosyl (Ado) group as the upper axial ligand (Fig. 1). Formation of the C-Co bond between the cobalt ion and the upper ligand requires that the cobalt ion be reduced to its +1 oxidation state. Reduction of Co(III) to Co(I) is thought to proceed in two consecutive one-electron reductions catalyzed by cob(III)alamin reductase (EC 1.6.99.8) and by cob(II)alamin reductase (EC 1.6.99.9) (36) (Fig. 2). The product of cob(II)alamin reductase, a Co(I) corrinoid, is the substrate for the ATP:co(I)rrinoid adenosyltransferase (CobA) (EC 2.5.1.17) enzyme that generates the C-Co bond. This bond is very labile and needs to be reformed to maintain activity. Hence, this branch of the Ado-Cbl biosynthetic pathway is essential for coenzyme recycling.
FIG. 1.
Structure of Ado-Cbl. In S. enterica the lower ligand base has been identified to be 5,6-dimethylbenzimidazole (16).
FIG. 2.
Proposed corrinoid adenosylation pathway in S. enterica serovar Typhimurium LT2. Ado, 5′-deoxyadenosine; e−, electron; flavin(red), reduced forms of flavin nucleotides; PPPi, tripolyphosphate (postulated to be the by-product of the CobA reaction in S. enterica (M. V. Fonseca and J. C. Escalante-Semerana, unpublished results).
In Salmonella enterica serovar Typhimurium LT2, the CobA enzyme responsible for the last step of the pathway has been isolated and partially characterized (30, 31). Although enzymic activities that can generate reduced corrinoids have been reported and in some cases isolated, the genes encoding the enzymes responsible for the reductive steps of the pathway have not been identified in any organism (2, 15, 36).
Reduction of cob(III)alamin by crude cell-free extracts of Clostridium tetanomorphum and Propionibacterium freundenreichii showed an absolute requirement for added flavin cofactors (3, 36). In these bacteria and in Pseudomonas denitrificans, NADH was the preferred source of electrons for the reaction (2). A system that included thiol compounds, a diaphorase enzyme, NADH, and flavin adenine dinucleotide (FAD) was used to reduce Cbl in P. freundenreichii (3, 15). Interestingly, this complex-reducing system was replaced by FADH2 when purified preparations of the adenosyltransferase from this organism were used. The cob(II)yrinic acid a,c-diamide reductase enzyme of P. denitrificans was isolated and shown to require the addition of flavin cofactors for activity (2). The purified enzyme was shown to reduce complete and incomplete Co(II) corrinoids. Here again, the gene encoding this activity was not identified.
Extensive efforts to isolate mutants of S. enterica serovar Typhimurium LT2 defective in corrinoid reduction and adenosylation have failed to identify genetic loci other than cobA (9; M. V. Fonseca and J. C. Escalante-Semerena, unpublished results). Hence, we took a reverse genetics approach to the isolation of the Cbl reductase enzymes with the purpose of identifying the genes encoding these activities in this bacterium. In this paper, we show that homogeneous NAD(P)H:flavin oxidoreductase (Fre) (EC 1.6.8.1) enzyme drives the reduction of cob(III)alamin and cob(II)alamin by maintaining a pool of dihydroflavins (FMNH2, FADH2), not by directly using cob(III)alamin as the substrate. On the basis of this finding, we propose that S. enterica serovar Typhimurium LT2 may not have or need a cob(III)alamin reductase enzyme. We also show that reduction of cob(II)alamin to cob(I)alamin is detectable in vitro when FMNH2 is present in the reaction mixture. However, the amount of product formed under these conditions was insufficient for CobA enzyme function.
MATERIALS AND METHODS
In vitro activity assays. (i) Reduction of cob(III)alamin to cob(II)alamin.
The reaction mixture contained: N-(2-hydroxyethyl)piperazine-N′-(3-propanesulfonic acid) (HEPPS) buffer (pH 8.0, 0.2 μmol), flavin mononucleotide (FMN) (0.05 μmol), NADH (0.5 μmol), and cob(III)alamin (0.06 μmol). The final volume of the reaction mixture was 0.9 ml. Additional components of the reaction mixture were added using a Hamilton syringe (Hamilton Co., Reno, Nev.) previously flushed with anoxic water. The reaction mixture was incubated at 37°C for 2 min, the reaction was initiated by the addition of enzyme, and incubation was allowed to proceed for 30 min at 37°C under dim light. Cob(III)alamin reductase activity was determined by measuring the decrease in absorbance at 525 nm as a function of time, using the difference in the molar extinction coefficient (ɛ) between HOCbl and cob(II)alamin at that wavelength (Δɛ525 = 4.9 M−1 cm−1) (34). A unit of activity was defined as the amount of enzyme required to generate 1 nmol of cob(II)alamin per min.
(ii) Reduction of cob(II)alamin to cob(I)alamin.
The assay used to detect enzyme-dependent reduction of cob(II)alamin to cob(I)alamin was a modification of the method reported for the isolation of the cob(II)yrinic a,c-diamide reductase enzyme of P. denitrificans. This assay relied on the alkylation of the cob(I)alamin nucleophile (2). The reaction mixture contained the same buffer used to assay the reduction of cob(III)alamin to cob(II)alamin, with the FMN and NADH concentrations as described above. Iodoacetate (IA), (0.4 μmol) was added as a chemical trap for cob(I)alamin, and cob(II)alamin (0.06 μmol) substituted for cob(III)alamin as the substrate. Cob(II)alamin was generated by anoxic photolysis of methylcobalamin (CH3Cbl) (38). Briefly, a solution of CH3Cbl in the reaction buffer was degassed with oxygen-free N2, added to a methacrylate cuvette (Fisher, Itasca, Ill.) fitted with a red-butyl stopper, and flushed with oxygen-free N2 for 10 min. Photolysis of CH3Cbl was achieved by irradiating the sample for 5 min with a 150-W lamp placed ∼20 cm away from the cuvette. Conversion of cob(II)alamin to carboxymethyl (CMCbl) was monitored spectrophotometrically by the increase in absorbance at 525 nm as a function of time. This assay was used to monitor the purification of cob(II)alamin reductase activity.
(iii) Corrinoid adenosylation.
CobA assays were performed as described previously (31). To determine if S. enterica serovar Typhimurium LT2 cell-free extracts or the Fre protein had cob(II)alamin reductase activity, the corrinoid adenosylation assay was modified by substituting cell-free extracts or Fre, NADH, and FMN for potassium borohydride. A unit of CobA activity was defined as the amount of enzyme that generated 1 nmol of Ado-Cbl per min. Control experiments for the activity of CobA were performed with potassium borohydride as described previously (31).
(iv) Flavin reductase assays.
Flavin reductase activity was assayed by monitoring oxidation of NADH. Activity was assessed spectrophotometrically by the decrease in absorbance at 450 nm, as a function of time. One unit of activity was defined as the amount of enzyme required to reduce 1 nmol of FMN (ɛ450 = 12,200 M−1 cm−1) (8) per min. The assays contained FMN or FAD (0.05 μmol), NADH (0.5 μmol), and HEPPS buffer (pH 8.0, 0.18 mmol) in a final volume of 0.9 ml. Assays were performed under anoxic conditions to mimic conditions used in the cob(II)alamin reduction assays (see above).
(v) Photoreduction of FMN and chemical reduction of cob(III)alamin.
FMNH2 was generated by photoreduction of coenzyme F420 in the presence of potassium oxalate as a source of reducing equivalents (14, 21). Pyrex tubes (13 by 100 mm) were fitted with a red butyl stopper and degassed with oxygen-free N2 for 30 min. Three milliliters of an anoxic solution of 15 mM potassium oxalate in 0.2 M HEPPS buffer (pH 8.0) was anoxically transferred to the tube with a syringe previously flushed with anoxic buffer. This was followed by the addition of coenzyme F420 (0.005 μmol) and FMN (0.05 μmol). The solution was illuminated with a tungsten/halogen lamp for approximately 10 min. The headspace of the tube was constantly exchanged with oxygen-free N2 during the procedure. Spectra of the solution were recorded when bleaching of the flavin solution was observed. HOCbl (0.05 μmol) was anoxically added to the tube, and reduction of cob(III)alamin to cob(II)alamin was verified spectrophotometrically. After reduction of cob(III)alamin was deemed complete, IA (0.6 μmol) was added and alkylation of cob(I)alamin was monitored as described above.
Protein purification. (i) Purification of the cob(II)alamin reduction activity. (a) Mass culturing of S. enterica serovar Typhimurium LT2.
Cells of strain TR6583 (metE205 ara-9 cob+) were grown aerobically in Vogel-Bonner minimal medium (35) with the following additions: glucose (35 mM), cobinamide dicyanide (CN)2Cbi (20 nM), and 1,2-propanediol (12 mM). The latter was used to induce expression of the cob/pdu regulon under aerobic growth conditions (25). Cell mass was concentrated using a tangential flow cell concentrated using a tangential flow cell concentrator (Millipore, Bedford, Mass.), and cells were harvested by centrifugation at 10,000 × g for 10 min. Cell pellets (60 g [wet wt]) were resuspended in 130 ml of buffer A (50 mM Tris-Cl buffer [pH 8.0] at 4°C, 250 μM dithiothreitol) containing 16 μg of the protease inhibitor phenylmethylsulfonyl fluoride per ml.
(b) Generation of cell-free extracts.
Cell-free extracts were generated by passing the cell suspension three times through a French pressure cell at 1.4 × 108 kPa. The cell debris was removed by centrifugation at 40,000 × g for 2 h at 4°C.
(c) AS precipitation.
Precipitation of proteins with ammonium sulfate (AS) was performed at 4°C using finely ground Ultrapure AS (ICN Biochemicals, Aurora, Ohio). Protein pellets were obtained by centrifugation at 10,000 × g for 10 min at 4°C and resuspended in buffer A. Samples were desalted by dialysis at 4°C for use in subsequent steps. Cob(II)alamin reductase activity precipitated out of solution at between 30 and 45% saturation with AS.
(d) Anion-exchange chromatography.
The sample was loaded onto a DEAE-cellulose (Sigma) column (1.5 by 20 cm, 35-ml bed volume) equilibrated with buffer A. The column was developed at a flow rate of 35 ml per h. After the sample was loaded, the column was washed with 53 ml of buffer A and proteins bound to the resin were eluted with a 140-ml linear gradient of NaCl (0 to 0.5 M) in buffer A. Gradient elution was followed by a 53-ml wash with 1 M NaCl in buffer A.
(e) Affinity chromatography.
Fractions containing cob(II)alamin reductase activity from the anion-exchange chromatography step were pooled, concentrated using a Centriprep-10 concentrator (Amicon, Beverly, Mass.), and dialyzed against buffer A using a Microdialyzer (Pierce, Rockford, Ill.). The dialyzed sample was loaded onto an FMN-agarose (Sigma) column (0.8 by 4 cm, 2-ml bed volume). The column was washed with 3.0 ml of buffer A, followed by the stepwise elution of bound proteins with 20 μM, 50 μM, and 1 mM FMN in buffer A. Fractions were analyzed by alkylation assays and sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Fractions containing a single active protein, as judged by Coomassie blue staining, were pooled together, concentrated using a Centricon-10 microconcentrator (Amicon), and dialyzed against buffer A using a Microdialyzer (Pierce).
(ii) Purification of the CobA enzyme.
Purification of the CobA enzyme was performed as described previously, without modifications (31).
Protein techniques.
Protein concentration was determined by the method of Kunitz (17). Protein samples were analyzed by SDS-PAGE (18) and stained with Coomassie blue (26). The N-terminal sequence of purified protein samples was performed at the Protein/Nucleic Acid Shared Facility at the Medical College of Wisconsin (Milwaukee, Wis.) as described previously (33).
Genetic techniques.
Transductional crosses. Transductional crosses were performed using mutant P22 phage HT105/1 int-201 (27, 28) as described previously (4, 7).
Construction of an S. enterica fre mutant.
The fre::kan+ insertion in Escherichia coli strain LS1300 (a gift from C. DiRusso) was moved into the S. enterica chromosome as described previously (13).
Recombinant DNA techniques. (i) General.
All plasmid DNA isolations were performed using a QIAprep Spin Plasmid kit (Qiagen, Chatsworth, Calif.). Restriction enzymes and T4 DNA ligase were purchased from Promega (Madison, Wis.) and used according to the manufacturer's instructions. Vent exo mutant DNA polymerase was purchased from New England Biolabs (Beverly, Mass.). Purification of DNA fragments was performed using a QIAquick gel extraction kit. Pfu Turbo DNA polymerase was purchased from Stratagene (San Diego, Calif.). All primers used in this study were synthesized by Integrated DNA Technologies (Coralville, Iowa). DNA sequencing of PCR products and plasmids was performed at the Nucleic Acid and Protein Facility at the University of Wisconsin-Madison Biotechnology Center. PCR conditions and sequencing protocols used have been described elsewhere (32).
(ii) Cloning of the E. coli fre gene.
The E. coli fre gene was amplified from plasmid pCD140 using the primers FREPRIMER1 (5′-TTCAAAAATGGGGCTGGATG-3′) and (−)STRAND FRE (5′-CCTACGGTCGGACTATTTG-3′). The amplification profile was: 94°C for 5 min; 30 cycles of 94°C for 2 min, 45°C for 2 min, and 72°C for 1 min; 72°C for 10 min; and 4°C for 12 h. The amplified fragment was purified, cut with ClaI and BglII, and ligated into pSU21 (20) cut with ClaI and BamHI enzymes. The resulting plasmid was referred to as pFRE1.
(iii) Site-directed mutagenesis.
Site-directed mutagenesis of the fre gene using the three-primer method was performed as described previously (22). An oligonucleotide with the sequence 5′-CGACAGAGAAAGCATATGACAAC-3′ was used to generate an NdeI site immediately 5′ of the fre coding sequence using plasmid pFRE1 DNA as the template. The −40 and reverse primers for M13mp19 were used as outer primers in the reaction. The PCR product was purified, cut with HindIII and XhoI, and ligated to plasmid pSU19 (20) cut with the same enzymes. The resulting plasmid was referred to as pFRE2.
(iv) Overproduction of His-tagged Fre enzyme.
Plasmid pFRE2 was cut with NdeI and XhoI, and the fragment containing fre was purified and ligated to the overexpression vector pET-15b cut with the same enzymes. The resulting plasmid was referred to as pFRE3. In this plasmid, expression of fre results in a Fre protein carrying an N-terminal histidine tag (His6Fre). His6Fre enzyme was overproduced from a 500-ml culture of strain JE4639 on Luria-Bertani broth containing 100 μg of ampicillin per ml. Cells were grown at 37°C with shaking to an A650 of ∼0.9. Isopropyl-1-thio-β-d-galactoside (IPTG) was added to a final concentration of 400 μM, and incubation was continued for 2 h. After incubation, cells were pelleted by centrifugation at 10,000 × g for 10 min.
(v) Purification of His6Fre protein.
Cell pellets were resuspended in 6 ml of binding buffer (5 mM imidazole in 20 mM Tris-Cl buffer [pH 7.9] containing 0.5 M NaCl). The cell suspension was broken by sonication using a 550 Sonic Dismembrator (Fisher) for 10 min (setting of 3, 50% duty). Cell-free extracts were generated by centrifugation at 40,000 × g for 2 h. His6Fre protein was purified from the cell-free extracts by nickel affinity chromatography on a His-bind resin (Novagen, Madison, Wis.) according to the manufacturer's instructions. After the column (1 by 10 cm, 3-ml bed volume) was loaded with cell-free extract, it was washed with 30 ml of binding buffer followed by 18 ml of the same buffer containing 0.06 M imidazole. Proteins bound to the resin were eluted with buffer containing 0.4 M imidazole. Fractions containing homogeneous His6Fre were pooled, dialyzed against 50 mM Tris-Cl (pH 8.0) at 4°C containing 1 mM EDTA, and concentrated using a Centricon microconcentrator (Amicon).
RESULTS
Isolation of cob(II)alamin reductase activity from cell-free extracts of S. enterica serovar Typhimurium LT2.
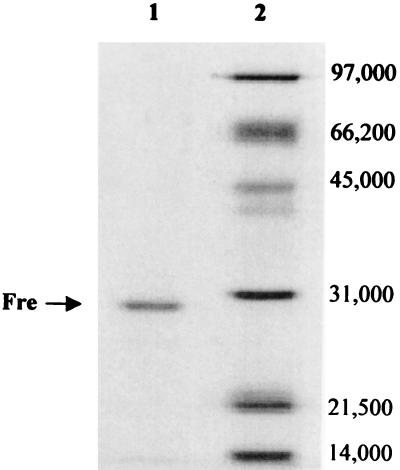
Initial approaches aimed at identifying the cob(II)alamin reductase enzyme of S. enterica demanded the involvement of the CobA enzyme under two different assay conditions. First, corrinoid adenosylation was assayed in a reaction mixture that contained cell-free extracts of the wild-type strain (i.e., the extracts contained chromosomal levels of the CobA enzyme), and cob(III)alamin or cob(II)alamin as the substrate. The second set of conditions were as described above, except that homogeneous CobA enzyme was added to the reaction mixture, with the cob(II)alamin reductase activity expected to be provided by cell-free extracts of the wild-type strain. Neither condition led to the synthesis of cob(I)alamin, the substrate for the adenosyltransferase enzyme (data not shown). To increase the sensitivity of the assay, the cob(I)alamin alkylation assay was used to detect cob(II)alamin reductase activity. Using this assay, a cob(II)alamin reductase activity dependent on FMN and NADH was detected in cell-free extracts of S. enterica. A protein with this activity was purified to homogeneity from cell extracts using conventional liquid chromatography procedures. The anion-exchange step proved very effective in separating a large number of contaminating proteins away from the cob(II)alamin reductase activity (data not shown). A small amount of enzyme (∼250 μg) was isolated from 60 g of cells (wet paste), indicating that this enzyme was not a very abundant protein. After the affinity chromatography step, some fractions containing this activity had a single protein of a molecular mass of approximately 29 kDa (Fig. 3). N-terminal sequence analysis of the first 15 amino acids of the purified protein yielded the sequence TTLSCKVTSVEAITD, a perfect match to the E. coli Fre (encoded by the fre gene) (11, 29). This finding was surprising because this enzyme was not expected to catalyze the reduction of cob(II)alamin to cob(I)alamin given the extremely low redox potential of the cob(II)alamin/cob(I)alamin couple (E0′ = −0.61V [1, 19]). Noteworthy is the fact that this reduction was monitored using in vitro alkylation assays. Under the conditions of the assay, reduction of cob(II)alamin was linear with time, but the rate of alkylation of cob(I)alamin did not change with increasing concentrations of Fre protein in the assay mixture (data not shown). These results suggested that the generation of cob(I)alamin was not enzymic. To test this possibility, FMNH2 was generated photochemically and was added to alkylation assay mixtures devoid of Fre protein and NADH. Complete reduction of cob(II)alamin was measured only when a 2:1 ratio of FMNH2 to cob(III)alamin was present in the reaction mixture.
FIG. 3.
Denaturing gel electrophoresis of purified Fre protein. Lane 1, purified Fre from FMN affinity chromatography; lane 2, molecular mass standards (numbers at right are in daltons).
FMNH2 did not substitute for potassium borohydride in the reaction catalyzed by the CobA enzyme.
CobA-dependent corrinoid adenosylation assays were performed to assess whether Fre generated the cob(I)alamin substrate for CobA. No CobA activity was detected under the conditions routinely used to assay this enzyme. Reaction mixtures containing varying amounts of Fre (6 to 12 μg), NADH, and FMN failed to substitute for potassium borohydride in the reduction of cob(II)alamin to cob(I)alamin. Control experiments where cob(I)alamin was generated with potassium borohydride (31) indicated that the CobA enzyme used in these experiments was active.
Fre protein is required for reduction of cob(III)alamin to cob(II)alamin.
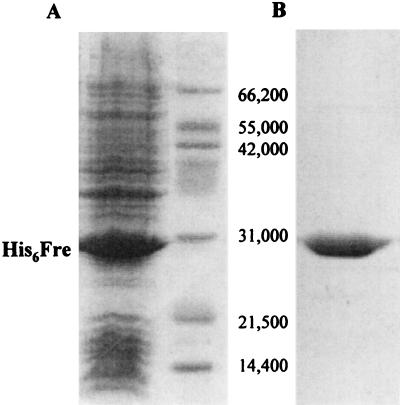
Reduction of cob(III)alamin by homogeneous His6Fre enzyme (Fig. 4) was tested in vitro by measuring the rate of reduction of cob(III)alamin to cob(II)alamin (i) under conditions that assumed cob(III)alamin served as substrate for Fre and (ii) under conditions where flavin reduction was allowed to proceed to completion before the addition of cob(III)alamin to the reaction mixture. Under the conditions where cob(III)alamin was assumed to serve as substrate for Fre, the rate of cob(III)alamin reduction correlated well with the rate of flavin reduction. Under these conditions, cob(III)alamin and FMN reduction was linear when the amount of Fre in the reaction mixture was between 0.2 and 0.45 μg of protein. Within this range of enzyme, FMN was reduced at a rate of 2.4 to 7.8 nmol of FMNH2 per min. Within the same range of Fre concentration, cob(III)alamin was reduced at a rate of 2.7 to 9.2 nmol per min. In contrast, the rate of reduction of cob(III)alamin to cob(II)alamin (Fig. 5) was too fast to be measured when cob(III)alamin was added after FMN or FAD was converted to FMNH2 or FADH2 (Fig. 5 shows the reaction with FADH2). These results suggested that reduction of cob(III)alamin to cob(II)alamin was not enzymic, but chemical. Thus, it was inferred that Fre was indirectly driving the reduction of cob(III)alamin to cob(II)alamin via either FMNH2 or FADH2 and that flavin reduction was the rate-limiting reaction for cob(III)alamin reduction. When cob(III)alamin was added to a solution of photochemically reduced FMNH2, cob(III)alamin reduction was again too fast to be measured, confirming that this reaction was dependent on FMNH2 and that cob(III)alamin was not used as a substrate by Fre.
FIG. 4.
(A) Denaturing gel electrophoresis of purified His6Fre enzyme. Lane 1, Overexpression of His6Fre enzyme in E. coli BL21(λDE3) cell-free extracts after IPTG induction; lane 2, molecular weight standards (numbers at right are masses [in daltons]). (B) Purified His6Fre enzyme after nickel affinity chromatography.
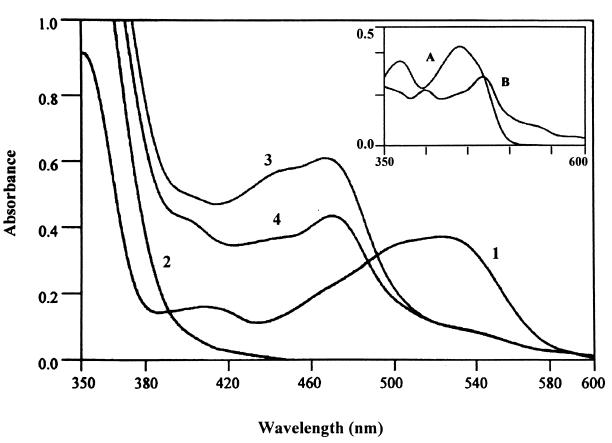
FIG. 5.
Nonenzymic, FADH2-dependent reduction of cob(III)alamin to cob(II)alamin. UV visible spectra, labeled as follows: 1, cob(III)alamin alone; 2, FADH2; 3, spectrum of the solution containing FADH2 obtained immediately after the addition of cob(III)alamin to it; 4, same as spectrum 3, but the spectrum was obtained 2 min after the addition of cob(III)alamin. The inset shows overlaid spectra of FAD (labeled A) and authentic cob(II)alamin (labeled B).
Phenotypes of an S. enterica fre mutant.
The phenotypes of an S. enterica fre mutant were assessed by their ability to synthesize methionine aerobically when provided with nonadenosylated cobinamide, or anaerobically, by demanding de novo Cbl synthesis (9). Under aerobic conditions, a cobA mutant (lacks adenosyltransferase activity) cannot synthesize Cbl (and thus is a methionine auxotroph) unless adenosylcobinamide is provided in the medium (9). Anaerobically, a cobA mutant is defective in corrin ring biosynthesis and is corrected by addition of Cbl to the medium (9). If the Fre enzyme were part of the corrinoid adenosylation pathway in this bacterium, a fre mutant would be predicted to have a CobA-like phenotype. The fre mutant did not display a CobA-like phenotype, i.e., it synthesized adenosylcobalamin de novo under anaerobic conditions and from nonadenosylated cobinamide, indicating that alternative means of achieving cob(II)alamin reduction exist in this bacterium.
DISCUSSION
Fre is not responsible for the reduction of cob(II)alamin to cob(I)alamin.
We have demonstrated that Fre can promote the disproportionation reaction of cob(II)alamin to cob(I)alamin (39) by always maintaining FMN in a reduced state. This effect of Fre on cob(II)alamin reduction is indirect, because photochemically reduced FMN substituted for Fre and NADH in this reaction. On the basis of this result, we conclude that reduction of the cob(II)rrinoid via Fre does not require binding of this substrate to the Fre enzyme. Even though dihydroflavins have been alluded to be active in corrinoid reduction (3, 15), to the best of our knowledge, these observations were not published. Identification of bona fide cob(II)alamin reductase (EC 1.6.99.9) enzymes must demand in vitro coupling with the CobA enzyme. The lack of coupling between the Fre and CobA enzymes suggested that the level of cob(I)-alamin generated by FMNH2 oxidation was either below the Km of CobA for this substrate (5.2 μM [31]) or it was reoxidized to cob(II)alamin before binding to CobA. The results of alkylation assays were misleading due to the reactivity of the cob(I)alamin nucleophile and the IA electrophile.
Does S. enterica have cob(III)alamin reductase enzyme activity?
Reduction of cob(III)alamin to cob(II)alamin by FMNH2 suggested that flavin reductase enzymes such as Fre drive the reduction of cob(III)alamin to cob(II)alamin by maintaining a pool of dihydrofalvins. The existence of FMNH2 and FADH2 effectively bypasses the need for a specific reductase. Organisms like E. coli (and probably S. enterica) and luminous bacteria have more than one in vitro-detectable flavin reductase enzyme activity (5, 6, 10, 40, 41). Such a redundancy of function makes it likely that cob(II)alamin is the stable, intracellular form of this coenzyme, which would also explain why mutant strains defective in cob(III)alamin reductase function have not been isolated. Alternatively, reduction of cob(III)alamin to cob(I)alamin in S. enterica may be catalyzed by a single enzyme performing two one-electron reductions. In contrast with S. enterica, mammalian cells contain two different forms of cob(III)alamin reductase (24). Surprisingly, in this system, nonenzymatic reduction of cob(III)alamin by reducing substrates or low-molecular-weight factors has not been observed. Cob(III)alamin reductase has also been isolated and characterized in the protozoan Euglena gracilis (37). The E. gracilis and mammalian enzymes do not require FAD or FMN for the reduction of cob(III)alamin. The E. gracilis enzyme contains FAD or FMN as its prosthetic group (37).
Another plausible explanation for the lack of success in isolating Cbl reductase mutants could be that one of the redundant functions is required for de novo synthesis of the corrin ring and another is required for assimilation of exogenous corrinoids. A fre mutant was able to synthesize Ado-Cbl de novo anaerobically, and it assimilated exogenous corrinoids both aerobically and anaerobically, suggesting that Fre was not solely responsible for the reduction of the corrin ring.
Why have S. enterica mutants defective in cob(II)alamin reduction not been isolated?
Genetic approaches that led to the isolation of CobA mutants did not result in isolation of mutants defective in corrinoid reduction (9). Exhaustive mutant searches in an fre mutant background failed to isolate strains with a CobA-like phenotype (adenosylcobinamide auxotroph) with lesions in genes other than cobA (data not shown). The difficulty in the identification of the gene encoding the cob(II)alamin reductase enzyme would be explained if cob(II)alamin reduction in S. enterica were catalyzed by an essential enzyme, by more than two enzymes (i.e., redundancy), or by an enzyme that may be required for functions other than cob(II)rrinoid reduction.
Database searches for putative cobalamin reductases from known bacterial and archael genome sequences based on proximity to cobalamin biosynthetic genes have not been productive. Because the genes encoding previously isolated cob(II)alamin reductase activities in P. denitrificans, C. tetanomorphum, and P. freundenreichii were not been identified, it is not possible to identify homologous proteins in S. enterica based on sequence homology at this point.
Given the extreme reactivity of the co(I)rrinoid nucleophile, it is likely that during the synthesis of adenosylated corrinoids, reduction of co(II)rrinoids occurs on the adenosyltransferase enzyme once it has complexed with its substrate and ATP. Precedent for this idea has been provided through in vitro studies of activation of the cob(II)alamin form of methionine synthase (MetH) in E. coli (1, 23). In vitro, the reduction of cob(II)alamin bound to the methionine synthase MetH enzyme is catalyzed by reduced flavodoxin (FldA) (14, 23). Interactions between the FldA and MetH proteins change the coordination geometry of the cobalt from five-coordinate geometry to four-coordinate geometry due to the dissociation of residue His759 of MetH, which replaces the lower ligand base 5,6-dimethylbenzimidazole when Cbl is bound by the enzyme. The association of FldA and MetH proteins is specific and depends on pH and ionic strength (14).
Because the fldA gene is essential (12), the Cob phenotype associated with a lack of this function is unknown. If FldA indeed has a role in corrinoid reduction for adenosylation, this would imply some differences in the in vitro requirements of the previously identified reducing systems in other organisms, since all these systems required the addition of flavins for activity. The possible involvement of FldA in corrinoid reduction and adenosylation in S. enterica is currently under investigation in our laboratory.
ACKNOWLEDGMENTS
This work was supported by NIH grant GM40313 and by an Aid-to-Education grant from DuPont to J.C.E.-S. M.V.F. is the recipient of MARC predoctoral fellowship GM17528.
We thank C. DiRusso (The Albany Medical College, Albany, N.Y.) for plasmids and strains. We thank H. Beinert (Institute for Enzyme Research, University of Wisconsin-Madison) for assistance in performing the photochemical reduction of FMN. We thank B. Mukhopadhyay and R. S. Wolfe (University of Illinois-Urbana-Champaign) for the gift of coenzyme F420.
REFERENCES
- 1.Banerjee R V, Harder S R, Ragsdale S W, Matthews R G. Mechanism of reductive activation of cobalamin-dependent methionine synthase: an electron paramagnetic resonance spectroelectrochemical study. Biochemistry. 1990;29:1129–1135. doi: 10.1021/bi00457a005. [DOI] [PubMed] [Google Scholar]
- 2.Blanche F, Maton L, Debussche L, Thibaut D. Purification and characterization of cob(II)yrinic acid a,c-diamide reductase from Pseudomonas denitrificans. J Bacteriol. 1992;174:7452–7454. doi: 10.1128/jb.174.22.7452-7454.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brady R O, Castanera E G, Barker H A. The enzymatic synthesis of cobamide coenzymes. J Biol Chem. 1962;237:2325–2332. [PubMed] [Google Scholar]
- 4.Chan R K, Botstein D, Watanabe T, Ogata Y. Specialized transduction of tetracycline resistance by phage P22 in Salmonella typhimurium. II. Properties of a high transducing lysate. Virology. 1972;50:883–898. doi: 10.1016/0042-6822(72)90442-4. [DOI] [PubMed] [Google Scholar]
- 5.Covès J, Fontecave M. Reduction and mobilization of iron by a NAD(P)H:flavin oxidoreductase from Escherichia coli. Eur J Biochem. 1993;211:635–641. doi: 10.1111/j.1432-1033.1993.tb17591.x. [DOI] [PubMed] [Google Scholar]
- 6.Covès J, Niviere V, Eschenbrenner M, Fontecave M. NADPH-sulfite reductase from Escherichia coli. A flavin reductase participating in the generation of the free radical of ribonucleotide reductase. J Biol Chem. 1993;268:18604–18609. [PubMed] [Google Scholar]
- 7.Davis R W, Botstein D, Roth J R. A manual for genetic engineering: advanced bacterial genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press, Cold Spring Harbor Laboratory Press; 1980. [Google Scholar]
- 8.Dawson R M C, Elliott D C, Elliott W H, Jones K M. Data for biochemical research. 3rd ed. Oxford, United Kingdom: Oxford University Press; 1986. [Google Scholar]
- 9.Escalante-Semerena J C, Suh S-J, Roth J R. cobA function is required for both de novo cobalamin biosynthesis and assimilation of exogenous corrinoids in Salmonella typhimurium. J Bacteriol. 1990;172:273–280. doi: 10.1128/jb.172.1.273-280.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fieschi F, Nivière V, Frier C, Décount J-L, Fontecave M. The mechanism and substrate specificity of the NADPH:flavin oxidoreductase from Escherichia coli. J Biol Chem. 1995;270:30392–30400. doi: 10.1074/jbc.270.51.30392. [DOI] [PubMed] [Google Scholar]
- 11.Fontecave M, Eliasson R, Reichard P. NAD(P)H:flavin oxidoreductase of Escherichia coli. A ferric iron reductase participating in the generation of the free radical of ribonucleotide reductase. J Biol Chem. 1987;262:12325–12331. [PubMed] [Google Scholar]
- 12.Gaudu P, Weiss B. Flavodoxin mutants of Escherichia coli K-12. J Bacteriol. 2000;182:1788–1793. doi: 10.1128/jb.182.7.1788-1793.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hamilton C M, Aldea M, Washburn B K, Babitzke P, Kushner S R. New method for generating deletions and gene replacements in Escherichia coli. J Bacteriol. 1989;171:4617–4622. doi: 10.1128/jb.171.9.4617-4622.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoover D M, Jarrett J T, Sands R H, Dunham W R, Ludwig M L, Matthews R G. Interactions of Escherichia coli cobalamin-dependent methionine synthase and its physiological partner flavodoxin: binding of flavodoxin leads to axial ligand dissociation from the cobalamion cofactor. Biochemistry. 1997;36:127–138. doi: 10.1021/bi961693s. [DOI] [PubMed] [Google Scholar]
- 15.Huennekens F M, Vitols K S, Fujii K, Jacobsen D W. Biosynthesis of cobalamin coenzymes. In: Dolphin D, editor. B12. Vol. 1. New York, N.Y: John Wiley & Sons, Inc.; 1982. pp. 145–168. [Google Scholar]
- 16.Johnson M G, Escalante-Semerena J C. Identification of 5,6-dimethylbenzimidazole as the Coα ligand of the cobamide synthesized by Salmonella typhimurium: nutritional characterization of mutants defective in biosynthesis of the imidazole ring. J Biol Chem. 1992;267:13302–13305. [PubMed] [Google Scholar]
- 17.Kunitz M J. Crystalline inorganic pyrophosphatase isolated from baker's yeast. J Gen Physiol. 1952;35:423–450. doi: 10.1085/jgp.35.3.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Laemmli U K. Cleavage and structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 19.Lexa D, Saveant J-M. The electrochemistry of vitamin B12. Acc Chem Res. 1983;16:235–243. [Google Scholar]
- 20.Martínez E, Bartolomé B, de la Cruz F. pACY184-derived cloning vectors containing the multiple cloning site and lacZα reporter gene of pUC8/9 and pUC18/19 plasmids. Gene. 1988;68:159–162. doi: 10.1016/0378-1119(88)90608-7. [DOI] [PubMed] [Google Scholar]
- 21.Massey V, Hemmerich P. A photochemical procedure for reduction of oxidation-reduction proteins employing deazariboflavin as catalyst. J Biol Chem. 1977;252:5612–5614. [PubMed] [Google Scholar]
- 22.Michael S F. Mutagenesis by incorporation of a phosphorylated oligo during PCR amplification. BioTechniques. 1994;16:410–414. [PubMed] [Google Scholar]
- 23.Osborne C, Chen L-M, Matthews R G. Isolation, cloning, mapping, and nucleotide sequencing of the gene encoding flavodoxin in Escherichia coli. J Bacteriol. 1991;173:1729–1737. doi: 10.1128/jb.173.5.1729-1737.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pezacka E H. Identification and characterization of two enzymes involved in the intracellular metabolism of cobalamin. Cyanocobalamin β-ligand transferase and microsomal cob(III)alamin reductase. Biochim Biophys Acta. 1993;1157:167–177. doi: 10.1016/0304-4165(93)90061-c. [DOI] [PubMed] [Google Scholar]
- 25.Rondon M R, Escalante-Semerena J C. The poc locus is required for 1,2-propanediol-dependent transcription of the cobalamin biosynthetic (cob) and propanediol utilization (pdu) genes of Salmonella typhimurium. J Bacteriol. 1992;174:2267–2272. doi: 10.1128/jb.174.7.2267-2272.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sasse J. Detection of proteins. In: Ausubel F A, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. Vol. 1. New York, N.Y: Wiley Interscience; 1991. pp. 10.6.1–10.6.8. [Google Scholar]
- 27.Schmieger H. A method for detection of phage mutants with altered transduction ability. Mol Gen Genet. 1971;100:378–381. doi: 10.1007/BF00438281. [DOI] [PubMed] [Google Scholar]
- 28.Schmieger H, Bakhaus H. The origin of DNA in transducing particles of P22 mutants with increased transduction frequencies (HT-mutants) Mol Gen Genet. 1973;120:181–190. doi: 10.1007/BF00267246. [DOI] [PubMed] [Google Scholar]
- 29.Spyrou G, Haggård-Ljungquist E, Krook M, Jörnvall H, Nilsson E, Reichard P. Characterization of the flavin reductase gene (fre) of Escherichia coli and construction of a plasmid for overproduction of the enzyme. J Bacteriol. 1991;173:3673–3679. doi: 10.1128/jb.173.12.3673-3679.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Suh S-J. Ph.D. dissertation. University of Wisconsin, Madison; 1994. [Google Scholar]
- 31.Suh S-J, Escalante-Semerena J C. Purification and initial characterization of the ATP:corrinoid adenosyltransferase encoded by the cobA gene of Salmonella typhimurium. J Bacteriol. 1995;177:921–925. doi: 10.1128/jb.177.4.921-925.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thomas M G, O'Toole G A, Escalante-Semerena J C. Molecular characterization of eutF mutants of Salmonella typhimurium LT2 identifies eutF lesions as partial-loss-of-function tonB alleles. J Bacteriol. 1999;181:368–374. doi: 10.1128/jb.181.2.368-374.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Trzebiatowski J R, Escalante-Semerena J C. Purification and characterization of CobT, the nicotinate mononucleotide:5,6-dimethylbenzimidazole phosphoribosyltransferase enzyme from Salmonella typhimurium LT2. J Biol Chem. 1997;272:17662–17667. doi: 10.1074/jbc.272.28.17662. [DOI] [PubMed] [Google Scholar]
- 34.Vitols E, Walker G A, Huennekens F M. Enzymatic conversion of vitamin B12s to a cobamide coenzyme, α-(5,6-dimethylbenzimidazolyl)deoxy-adenosylcobamide (adenosyl-B12) J Biol Chem. 1966;241:1455–1461. [PubMed] [Google Scholar]
- 35.Vogel H J, Bonner D M. Acetylornithase of Escherichia coli: partial purification, and some properties. J Biol Chem. 1956;218:97–106. [PubMed] [Google Scholar]
- 36.Walker G A, Murphy S, Huennekens F M. Enzymatic conversion of vitamin B12a to adenosyl-B12: evidence for the existence of two separate reducing systems. Arch Biochem Biophys. 1969;134:95–102. doi: 10.1016/0003-9861(69)90255-0. [DOI] [PubMed] [Google Scholar]
- 37.Watanabe F, Oki Y, Nakano Y, Kitaoka S. Purification and characterization of aquacobalamin reductase (NADPH) from Euglena gracilis. J Biol Chem. 1987;262:11514–11518. [PubMed] [Google Scholar]
- 38.Yamada R, Shimizu S, Fukui S. Preparation of solid vitamin B12r by anaerobic photolysis of methylcobalamin. Methods Enzymol. 1971;18C:52–54. [Google Scholar]
- 39.Yamada R-H, Schimizu S, Fukui S. Disproportionation of vitamin B12r under various mild conditions. Biochemistry. 1968;7:1713–1719. doi: 10.1021/bi00845a014. [DOI] [PubMed] [Google Scholar]
- 40.Zenno S, Kobori T, Tanokura M, Saigo K. Conversion of NfsA, the major Escherichia coli nitroreductase, to a flavin reductase with an activity similar to that of Frp, a flavin reductase in Vibrio harveyi, by a single amino acid substitution. J Bacteriol. 1998;180:422–425. doi: 10.1128/jb.180.2.422-425.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zenno S, Saigo K, Kanoh H, Inouye S. Identification of the gene encoding the major NAD(P)H-flavin oxidoreductase of the bioluminescent bacterium Vibrio fischeri ATCC 7744. J Bacteriol. 1994;176:3536–3543. doi: 10.1128/jb.176.12.3536-3543.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]