Abstract
Background and Aims
Smooth muscle cells (SMCs), interstitial cells of Cajal (ICCs), and platelet-derived growth factor receptor alpha (PDGFRα+) cells (PαCs) form a functional syncytium in the bowel known as the “SIP syncytium.” The SIP syncytium works in concert with the enteric nervous system (ENS) to coordinate bowel motility. However, our understanding of individual cell types that form this syncytium and how they interact with each other remains limited, with no prior single-cell RNAseq analyses focused on human SIP syncytium cells.
Methods
We analyzed single-nucleus RNA sequencing data from 10,749 human colon SIP syncytium cells (5572 SMC, 372 ICC, and 4805 PαC nuclei) derived from 15 individuals.
Results
Consistent with critical contractile and pacemaker functions and with known enteric nervous system interactions, SIP syncytium cell types express many ion channels, including mechanosensitive channels in ICCs and PαCs. PαCs also prominently express extracellular matrix–associated genes and the inhibitory neurotransmitter receptor for vasoactive intestinal peptide (VIPR2), a novel finding. We identified 2 PαC clusters that differ in the expression of many ion channels and transcriptional regulators. Interestingly, SIP syncytium cells co-express 6 transcription factors (FOS, MEIS1, MEIS2, PBX1, SCMH1, and ZBTB16) that may be part of a combinatorial signature that specifies these cells. Bowel region–specific differences in SIP syncytium gene expression may correlate with regional differences in function, with right (ascending) colon SMCs and PαCs expressing more transcriptional regulators and ion channels than SMCs and PαCs in left (sigmoid) colon.
Conclusion
These studies provide new insights into SIP syncytium biology that may be valuable for understanding bowel motility disorders and lead to future investigation of highlighted genes and pathways.
Keywords: SIP Syncytium, Smooth Muscle, Interstitial Cells of Cajal, PDGFRA, Single-Nucleus RNA Sequencing
Introduction
Human bowel digests food, absorbs nutrients, eliminates waste, and protects against luminal pathogens. This requires coordinated contraction and relaxation mediated by the “SIP syncytium” in conjunction with enteric, sympathetic, and parasympathetic nervous system, muscularis macrophages, and enteroendocrine cells. SIP syncytium cells form a functional unit composed of visceral smooth muscle cells (SMCs) that generate force, interstitial cells of Cajal (ICCs) that act as pacemakers, and platelet-derived growth factor receptor alpha (PDGFRα)-expressing cells (PαCs) that modulate smooth muscle contraction and relaxation.1, 2, 3 SMCs, ICCs, and PαCs are found in close proximity within the bowel wall and interact as functional units that collectively constitute the SIP syncytium.2
Visceral SMCs can produce tonic contractions to resist distension or phasic contractions to propel luminal contents.4, 5, 6, 7 Electrically coupled ICCs undergo spontaneous rhythmic depolarization and hyperpolarization (“slow waves”). Slow waves synchronize action potentials in SMCs to facilitate efficient propagation of muscle contractions.8 PαCs regulate smooth muscle contraction9,10 and mediate purinergic inhibitory signaling.11 SIP dysfunction can cause severe bowel dysmotility.3
Despite critical SIP roles in bowel motility, SMC, ICC, and PαC phenotypes and cellular interactions remain incompletely defined. We do not understand mechanisms that permit cross-talk between the SIP syncytium cell types or how these cell types may influence each other’s development. Furthermore, the pathways through which the SIP syncytium may respond to the variety of mechanical, chemical, and microbial stimuli in the bowel remain poorly understood. Few studies characterize gene expression for SMCs,12,13 ICCs,14,15 or PαCs,16 and all prior published data are from mice. We therefore reanalyzed our human colon single-nucleus RNAseq data17 to characterize 10,749 SIP syncytium cells. These results fit well with known physiology and provide new insight into SIP cell biology. We distinguished 2 PαCs subtypes. Both express many transcripts encoding for extracellular matrix (ECM) components and remodeling proteins. One subtype expresses primarily structural ECM genes and the other predominantly nonstructural ECM, plus ion channels and neurotransmitter receptors. Ion channels are abundant in ICCs and SMCs along with SMC contractile apparatus constituents. One novel finding is that ICCs and PαCs express mechanosensitive ion channels, suggesting direct responses to mechanical stimuli. PaCs also express VIPR2 neurotransmitter receptor, which is not previously reported. Six transcriptional regulators are relatively abundant in all SIP cells, suggesting this transcriptional network may define SIP cells. Gene expression differs in SMCs and PαCs from right compared with left colon. These analyses provide new insight into SIP functions and may facilitate new strategies to treat bowel motility disorders.
Methods
Human Tissue Collection and Single Nucleus Isolation
Data are from our recent manuscript.17 Deidentified colon was acquired with Institutional Review Board approval from Perelman School of Medicine at the University of Pennsylvania (IRB#804376) and Children’s Hospital of Philadelphia Institutional Review Board (IRB#13-010357) using the Abramson Cancer Center Tumor Bank.
Library Generation, Sequencing, and Data Processing
Aggregated data are identical to Gene Expression Omnibus series GSE156905: “human_aggregated_barcodes.tsv.gz,” “human_aggregated_genes.tsv.gz,” and “human_aggregated_matrix.mtx.gz.”
Analysis of Human Single-Nucleus RNA Sequencing Data
Using Seurat version 3.1.5,18,19 gene-barcode matrices were imported into R (RStudio Desktop version 1.2.5033, R version 3.6.2), filtered to remove low expressors or doublets (nGene = 200–5000) and mitochondrial contaminants (percent mitochondria <10%), normalized (Seurat default natural log-transformed RP10k normalization), and scaled to regress out variance due to differing percent mitochondrial RNA and number of unique molecular identified (UMI) RNA molecules per nucleus. Nuclei were clustered using the most statistically significant principal components up to the number where additional principal components contributed <5% of standard deviation and the principal components cumulatively contributed to 90% of the SD or when variation changed by <0.1% between consecutive principal components (17 principal components).20 After Uniform Manifold Approximation and Projection (UMAP) clustering, PαCs were identified based on PDGFRA expression. ICC cluster co-expressed KIT and ANO1. SMCs were defined by MYH11 expression. To evaluate if clustering was affected by sample origin, nuclei within UMAPs were color labeled. Sample #5035 was removed because this single sample significantly altered SIP clustering. After removing #5035, remaining data were renormalized and rescaled to regress out UMI and percent mitochondrial RNA and then clustered using 16 most statistically significant principal components.20 PαC#1 and PαC#2 were manually combined into a single cluster. “FindMarkers” was used to compare SMC, ICC, and PαC clusters to all other data set nuclei. For all analyses, only genes expressed by >10% of cells in a given cluster were included. Genes enriched by >0.25 loge (fold change of mean expression level) compared with cells in all other clusters were considered differentially expressed.
To identify genes differentially expressed between SIP clusters, data were reanalyzed using only this subset. Prior normalization and scaling were removed. Data were renormalized and rescaled to regress out UMI and percent mitochondrial RNA. Nuclei clustered using 10 most statistically significant principal components20 separated into SMC, ICC, and PαC clusters. Cell identity was confirmed using canonical markers for SMC (MYH11, ACTG2, ACTA2), ICC (KIT, ANO1), and PαC (KCNN3, PDGFRA). Differentially expressed genes were identified using “FindAllMarkers.” To identify gene expression differences by region, nuclei within each cluster were assigned identities (“right” or “left” colon). “FindMarkers()” was used to compare patterns within clusters. All P-values are adjusted based on Bonferroni correction. Adjusted P-value < .05 was considered significant. Differentially expressed gene lists are in Supplementary Data.
Data Visualization
Data were visualized using R. Analysis used GraphPad Prism version 9.3.1 for Windows (GraphPad Software, San Diego). Transcription factors were classified by “superclass” as outlined by Wigender et al.21 (http://tfclass.bioinf.med.uni-goettingen.de/).
Genes enriched by greater than 0.25 loge(fold change of mean expression level) were used for Metascape Gene Annotation and Analysis Resource for Gene Ontology and Gene Interaction Network Analysis.22
Access to Data
All authors had access to the study data and reviewed and approved the final article.
Results
Single-nucleus RNA sequencing data were from 16 individual human sigmoid (left) or ascending (right) colons and contain cells microdissected from perimyenteric plexus tissue (Table A1, and Wright and Schneider et al.17). Aggregated single-nucleus data were processed and analyzed using Seurat.19,23 After filtering, normalizing, and clustering, 14 distinct clusters were identified corresponding to 11 cell types (Figure A1A and Table A2), included 10,749 SIP nuclei (5572 SMC, 372 ICC, and 4805 PαC nuclei). Only nuclei expressing >200 unique genes (Figure A1B and C) with low mitochondrial RNA contamination (<10%; Figure A1D) were included. This resulted in a mean of 1138.21 unique genes and 1983.17 unique RNAs per nucleus (Figure A1B and C).
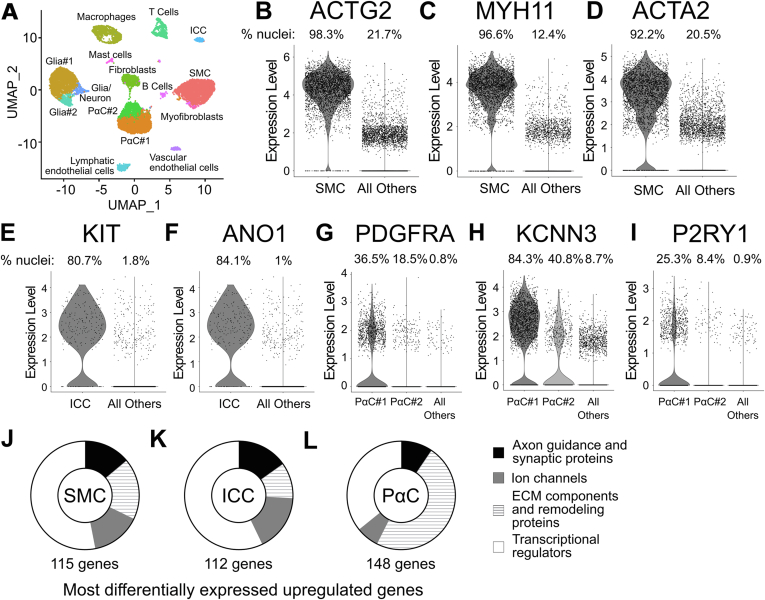
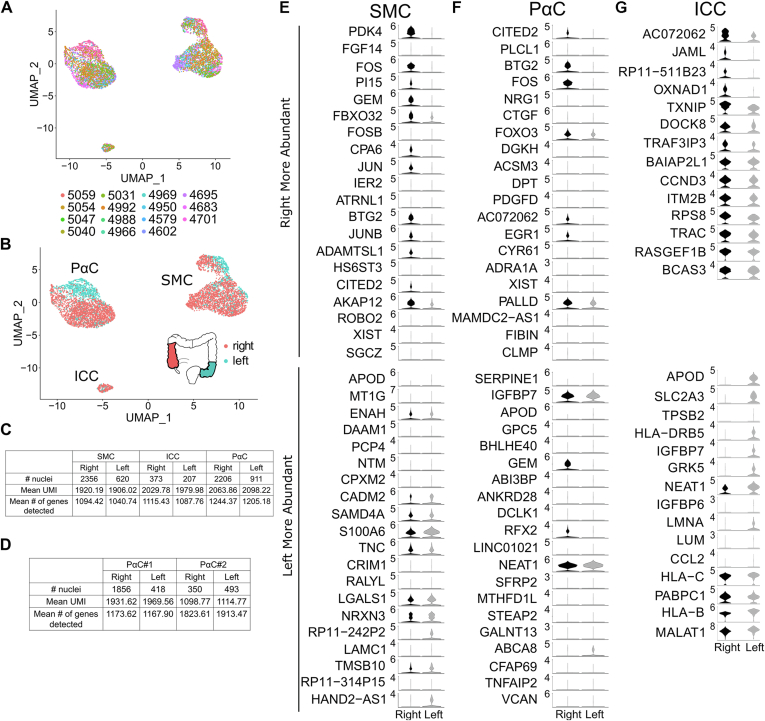
Unsupervised clustering of all nuclei yielded 2 SMC clusters (SMC#1 and SMC#2) that prominently express smooth muscle myosin heavy chain 11 (MYH11; Figure A1A and E), one PαC cluster, identified by PDGFRA (Figure A1F), and one ICC cluster expressing ANO1 and KIT (Figure A1G and H). Nuclei from individual samples were distributed across 14 clusters (Figure A1I). Initial analyses suggested sex (Figure A1J) and colon region (right/ascending colon vs left/sigmoid colon, Figure A1K) might determine SIP cell clustering, whereas there was no effect of age (Figure A1L). However, omitting a single sample (#5035) from a 24-year-old male with volvulus significantly changed SIP nuclei clustering (Figure A2A–C). Because volvulus causes ischemia and omitting other samples did not change clustering, we continued analyses excluding #5035 (final analyses included 2976 SMC, 233 ICC, and 3117 PαC). Without #5035, there is 1 ICC cluster, 2 distinct PαC clusters, and 1 SMC cluster (Figure 1A, Figure A2C). Excluding #5035 also minimized sex and colon region effects on unsupervised clustering patterns (Figure A2D and E).
Figure 1.
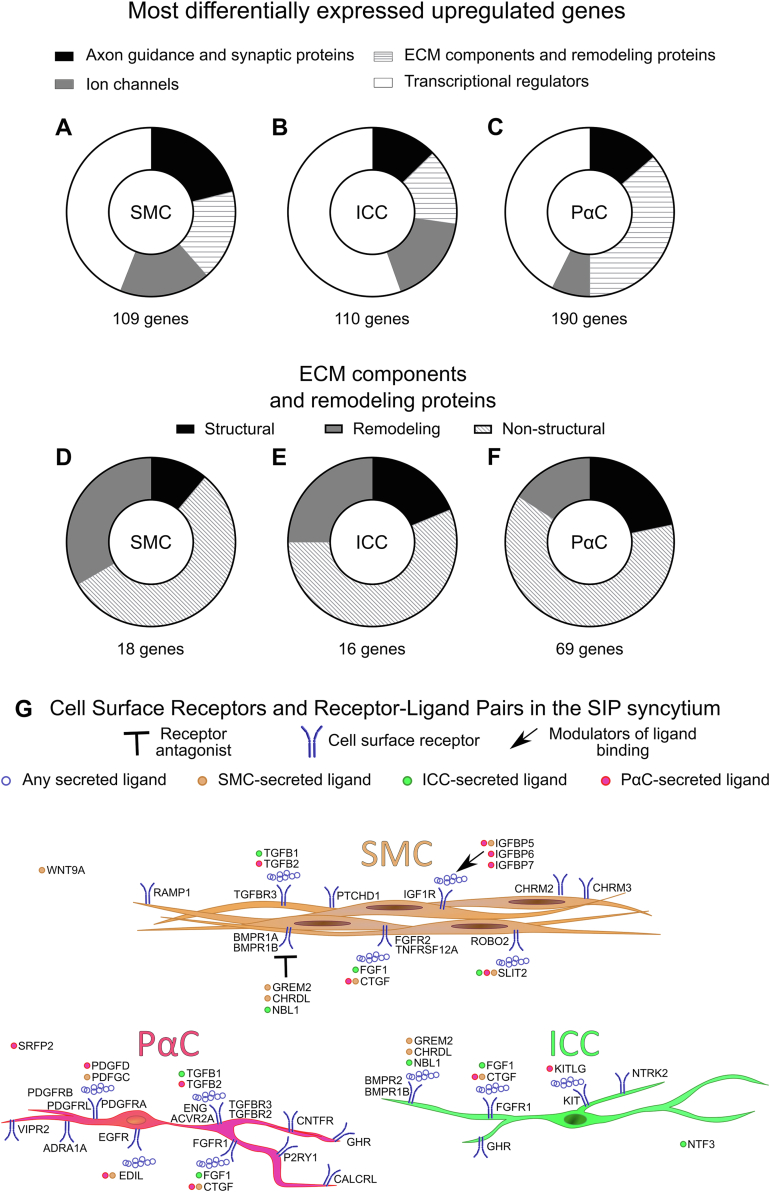
SMCs, ICCs, and PαCs have distinct gene expression profiles. (A) UMAP from 15 samples, excluding #5035. (B–D) SMCs express canonical markers ACTG2 (B), MYH11 (C), and ACTA2 (D). (E and F) ICC express KIT (E) and ANO1 (F). (G–I) PαC clusters express PDGFRA (G), KCNN3 (H), and P2YR1 (I). PαC markers were detected in more PαC#1 than PαC#2. (B–I) Expression level = loge(scaled RNA expression per nucleus). The numbers listed over each violin plot indicate the percentage of total nuclei per cluster expressing the gene. (J–L) Pie charts using only unique genes differentially enriched in SMC, ICC, and PαC compared with all other data set cells. Numbers indicate the number of genes in each pie chart.
Comparing Gene Expression Profiles of SIP Syncytium Cell Types
After omitting #5035, we reconfirmed cluster classification using ACTG2, MYH11, and ACTA2 in SMC (Figure 1B–D), KIT and ANO1 in ICC (Figure 1E and F), and PDGFRA, KCNN3, and P2RY1 (Figure 1G–I)2,16 in PαC clusters. To facilitate comparisons, we manually combined PαC#1 and PαC#2 into 1 cluster (PαC). We then identified messenger RNA (mRNA) relatively more abundant in SIP compared with other nuclei. From this enriched gene list, we identified mRNA differentially expressed between SMCs, ICCs, and PαCs. Based on known SIP functions, we focused on ECM components, ECM remodeling proteins, ion channels, axon guidance and synaptic proteins, and transcriptional regulators (Figure 1J–L). SMCs and ICCs express more mRNA for synapse-associated proteins, axon guidance proteins, ion channels and ion channel–associated proteins than PαCs (Figure 1J–L). PαCs express more ECM components and remodeling protein mRNA than SMCs or ICCs (Figure 1J–L).
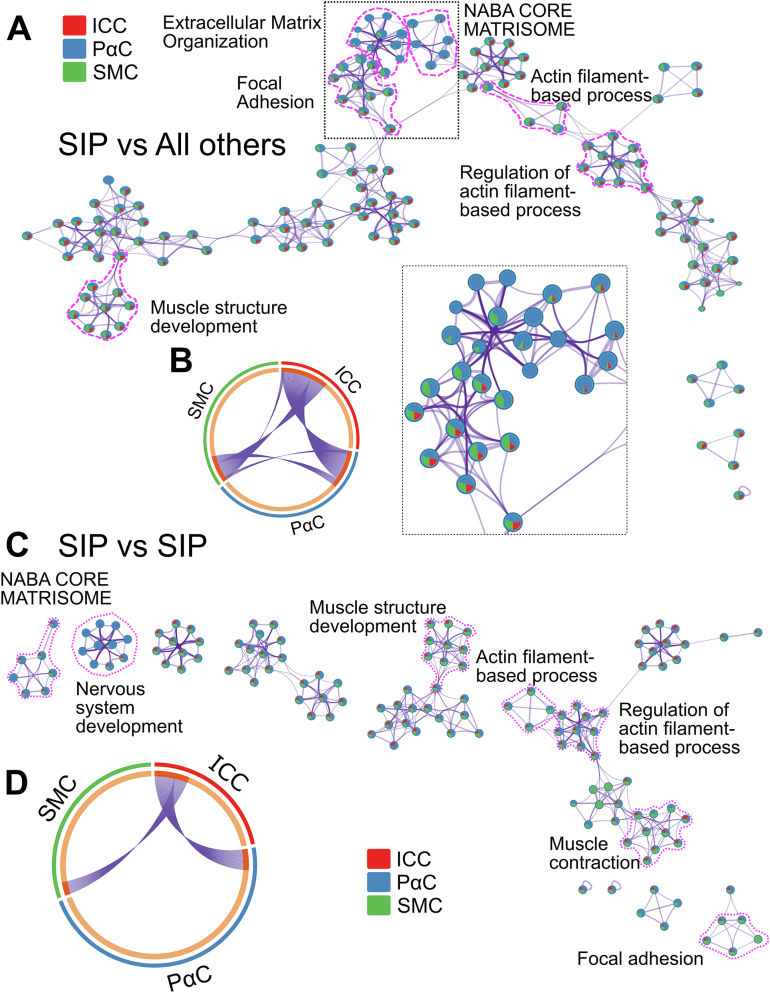
Network analysis for gene ontology (GO) term protein-protein interaction enrichment using Metascape22 (Figure 2) confirmed PαCs express more ECM components than all other cell types (Figure 2A). ICCs share many abundant, differentially expressed genes with SMCs and PαCs (Figure 2B). However, SMCs and PαCs share very few abundant, differentially expressed genes (Figure 2B). Significantly enriched protein-protein interaction networks by MCODE analysis show SMCs express networks involved in focal adhesions, axon and neuron projection guidance, transforming growth factor beta (TGFβ) signaling, and nonstructural ECM components (Figure A3A). ICCs expressed gene networks implicated in neurotransmission (Figure A3B). The most highly differentially expressed networks in PαCs are involved in ECM production, axon guidance, and nitrergic signaling (Figure A3C).
Figure 2.
Metascape protein-protein interaction network. (A and C) Network plot of highly enriched GO Terms. Each node is a pie chart representing percentage of GO Term–related genes differentially more abundant in each cell type. Similar GO Terms are connected by edges. (A) SIP syncytium compared with all other cells. (C) Each SIP cell type compared with other SIP cells. (B and D) Metascape Circos plots visualizing differentially enriched genes shared by cell types. Inner circle indicates genes differentially enriched in each cell type. Cell type unique genes are light orange. Shared genes are dark orange. (B) Genes enriched SIP compared with other cells. (D) Genes enriched in SIP compared with other SIP cells. (A and C) Selected regions are enlarged to show pie charts.
SMCs Express Genes That Facilitate Interactions With the Enteric Nervous System
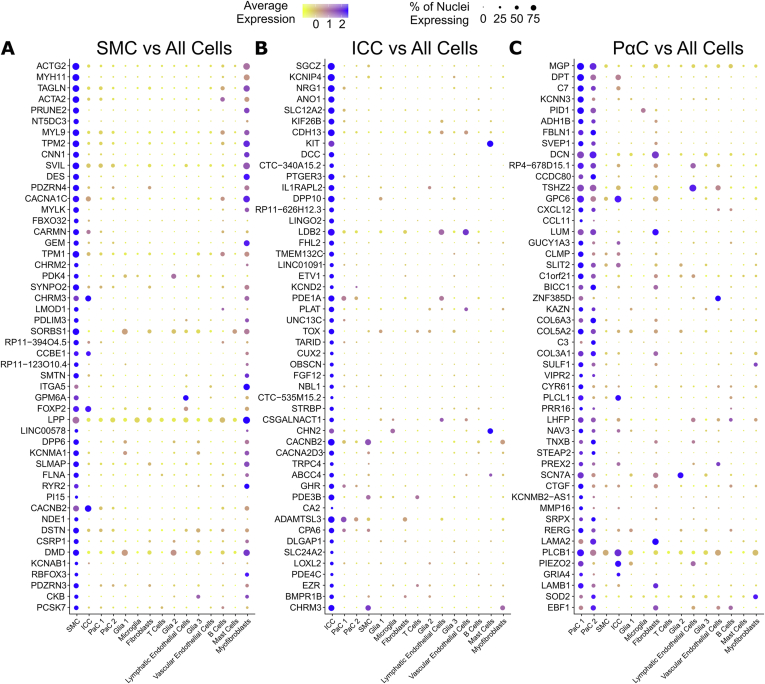
Genes most abundantly and differentially expressed in SMC are canonical smooth muscle genes (ACTG2, MYH11, TAGLN, ACTA2, MYL9, TPM2, CNN1) of the contractile apparatus. Several ion channels (CACNA1C, KCNMA1, RYR2, CACNB2, KCNAB1) and cholinergic G protein–coupled receptors (CHRM2, CHRM3) are much more abundant in SMC compared with all other cell types (Figure 3A, Figure A4A). SMC nuclei express many genes at relatively high levels that facilitate enteric nervous system (ENS) interactions. For example, SMCs express relatively high levels of GPM6A (Rank 31, Fold change 5.55), a glycoprotein involved in neuronal differentiation (24), SEMA3A (Rank 97, Fold change 2.79), an axon guidance molecule, NEGR1 (Rank 100, Fold change 2.76), a neuronal cell adhesion molecule, NLGN1 (Rank 113, Fold change 2.56), which promotes glutamatergic synapse formation, and NAV2 (Rank 128, Fold change 2.42), a neuron navigator. GPM6A expression has not been previously reported in SMCs.
Figure 3.
Most highly expressed genes differ for SMCs, ICCs, and PαCs. Dot plots showing top 50 most differentially expressed genes more abundant in (A) SMCs, (B) ICCs, and (C) PαCs compared with other data set cells. (A–C) Expression = loge(mean scaled RNA expression per cluster).
ICCs Express a Mechanosensitive Ion Channel, PIEZO2
ICC express relatively high levels of ion channels (KCNIP4, ANO1, KCND2, CACNB2, CACNA2D3, TRPC4) and cholinergic muscarinic receptor CHRM3 (Figure 3B, Figure A4B). Many highly differentially expressed genes relatively abundant in ICC were not previously reported in ICCs. This includes potassium voltage-gated channel interacting protein 4, KCNIP4 (Fold change 21.4), sarcoglycan zeta, SGCZ (Fold change 21.6), cadherin 13, CDH13 (Fold change 11.4), DCC Netrin 1 receptor, DCC (Fold change 10.4), and Piezo Type Mechanosensitive Ion Channel Component 2, PIEZO2 (Rank 61, Fold change 3.36).
PαCs Express High Levels of ECM Components and Mechanosensitive Ion Channels
Combined PαC cluster prominently expresses ion channels (KCNN3, SCN7A, PIEZO2, and GRIA4). PDGFRA was not in the top 50 most differentially expressed genes in PαC, but well-known PαC marker KCNN3 is the fourth-most differentially expressed gene in PαCs compared with all other cells. PαC highly differentially express neurotransmitter receptor VIPR2 (Fold change 4.32), not previously reported, and express relatively high levels of GUCY1A3 (Fold change 5.23), which is activated by nitric oxide (Figure 3C, Figure A4C). This suggests nitric oxide directly signals in human PαCs. PαC clusters express high levels of many genes encoding ECM components or ECM remodeling proteins (16 of the top 50 most differentially expressed genes; Figure 3C, Figure A4C). These include MGP, DPT, FBLN1, SVEP1, DCN, GPC6, LUM, KAZN, COL6A3, COL5A2, COL3A1, SULF1, TNXB, MMP16, LAMA2, and LAMB1. Furthermore, compared with SMC and ICC, PαC express high levels of structural ECM (collagens and versican), nonstructural ECM (such as fibronectin, laminin, proteoglycans),24 and ECM remodelers (such as matrix metalloproteinases; Figure 3C, Figures A4C and A5A–C). Relative proportions of structural and nonstructural ECM and ECM remodelers are shown in Figure A5D.
SIP Syncytium Cells Express Diverse ion Channels
All SIP express many ion channels and ion channel–associated proteins, including potassium, calcium, and sodium channels, and glutamate ionotropic receptors (Figure A6A–C). ICC and PαC differentially express relatively high levels of mechanosensitive channels (PIEZO2 in ICC and PαC, and TMEM150C in ICC; Figure A6B and C). Many axon guidance and synapse-associated proteins are enriched in SIP cells (Figure A7).
SIP Syncytium Shares a Unique Signature of 6 Transcriptional Regulators
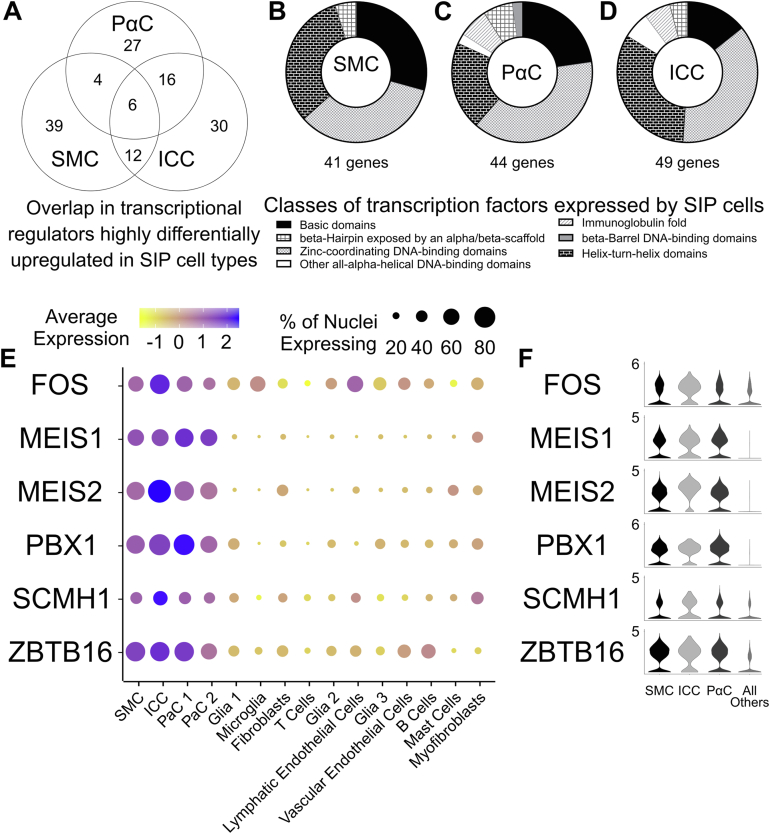
SMCs, ICCs, and PαCs each differentially express at relatively high levels many transcriptional regulators (epigenetic regulators, direct transcriptional activators, repressors; Figure A8A–C). Diverse transcription factor classes are expressed by all SIP cells (Figure 4A–D), with a subset expressed relatively abundantly by all 3 SIP syncytium cell types compared with all other data set clusters (Figure 4E and F). This shared subset includes FOS, MEIS1, MEIS2, PBX1, SCMH1, and ZBTB16.
Figure 4.
Six transcriptional regulators are shared among SIP cells. (A) Venn Diagram for transcriptional regulators more abundant in SIP syncytium than other cells. (B–D) Transcriptional regulator classes. (E) Dot plot shows relative expression of 6 transcriptional regulators co-expressed by all SIP cells. Colors indicate mean gene expression (blue = high, yellow = low). Dot size visualizes percentage of cells expressing genes. (F) Violin plots. (E and F) Expression = loge(mean scaled RNA expression per cluster).
Compared With Other SIP Clusters, PαCs Abundantly Express Transcriptional Regulators and ECM Components
The preceding analyses compared SIP with all other data set cells. This global comparison could mask subtle differences between SIP cell types. We therefore removed non-SIP clusters, renormalized and rescaled. We again identified many differentially expressed ion channels, ion channel–associated proteins, axon guidance and synaptic proteins, ECM components, and remodeling proteins (Figure 5A–C). Curiously, structural ECM mRNA was relatively more abundant in PαCs, whereas SMCs and ICCs expressed more ECM remodelers (Figure 5D–F).
Figure 5.
Relative abundance of the most differentially expressed SIP genes and receptor-ligand pairs. (A–F) Pie charts show the relative proportion of differentially expressed transcripts compared with other SIP cells. Numbers indicate genes in each pie chart. (G) Receptor-ligand pairs that may support cross-talk between SIP cells. Neurotransmitter receptors RAMP1, CALCRL, VIPR2, CHRM2, CHRM3, P2RY1, and ADRA1A are differentially expressed at high levels among SIP cells. Some ligands and corresponding receptors are produced in the same cell. Colored dots indicate ligand cells of origin.
Metascape network analysis comparing SIP to each other (Figure 2C) highlighted many of the same GO term networks identified when SIP were compared with all other cells (Figure 2A). SMC and ICC share differentially enriched transcripts. ICC and PαCs share a nonoverlapping set of differentially enriched mRNA. SMCs and PαCs had no differentially enriched transcripts in common when evaluating only SIP cells (Figure 2D). MCODE protein-protein interaction networks show axon guidance pathways enriched in SMCs compared with other SIP cells (Figure A9A). Additional ICC networks involved in neurotransmission were identified when non-SIP cells were omitted from analyses (Figure A9B). Surprisingly, no MCODE enrichment terms were identified for PαCs when we compared only to other SIP cells.
Limiting analyses to SIP cells identified many of the same differentially expressed synaptic proteins and axon guidance molecules (Figure A10), ion channels (Figure A11), ECM proteins (Figure A12), and transcriptional regulators (Figure A13), as when all cells were included. However, many additional genes differentially expressed among SIP cells were missed in prior analyses (Figures A5–A8). For example, 48% (30/63) of synaptic and axon guidance molecules differentially expressed between SIP syncytium cells (Figure A10D–F) are also likely expressed in enteric glia and other non-SIP cell types, as these genes were not identified in our initial analysis that included all cell types. In contrast, most ion channels and channel-associated proteins differentially expressed between SIP cells were identified in analyses that include non-SIP cells (Figure A11D–F). This suggests only a few ion channels differentially expressed between SIP cells are abundantly expressed in non-SIP cell types (6/51 [12%]; Figure A11). PαCs differentially express at high levels many more ECM components and ECM remodeling proteins compared with SMCs or ICCs (Figure A12). These observations highlight many genes differentially expressed among SIP cells not yet well studied in these cells.
SIP Cells Express Many Growth Factor and Neurotransmitter Receptors
SIP cells express at relatively high levels many cell surface receptors and ligands (Figure 5G). These include tyrosine kinase growth factor receptors FGFR2 and IGF1R preferentially expressed in SMCs, and FGFR1, NTRK2, and KIT preferentially expressed in ICCs. PαCs preferentially express PDGFRA (by definition) and FGFR1, GHR, and CNTFR. SIP cells differentially express cell surface receptors that regulate differentiation (TGFß receptors, bone morphogenetic protein receptors, Hedgehog receptor PTCHD1) and neurotransmitter receptors. In particular, vasoactive intestinal peptide receptor VIPR2, calcitonin receptor CALCRL, and adrenergic receptor ADRA1A are preferentially expressed in PαCs, whereas acetylcholine receptors CHRM2 and CHRM3 and calcitonin gene–related peptide (CGRP) receptor RAMP1 are preferentially expressed in SMCs. Interestingly, PαCs express KITLG, a trophic factor that activates KIT to support ICCs survival. Ligands for transforming growth factor receptors, fibroblast growth factor (FGF) receptors, and PDGF receptors are produced in many SIP cells and might work cell autonomously.
Two PαC Populations Differ in Ion Channels and Structural ECM mRNA Expression
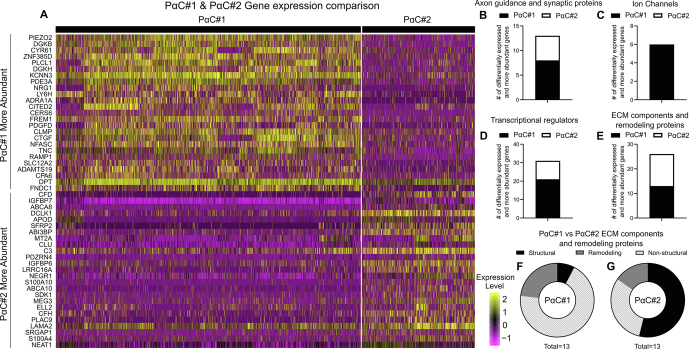
Given previous reports of distinct PαC subtypes16 and 2 PαC clusters in our UMAP (Figure 1A), we compared PαC#1 with PαC#2 (Figure 6A). PDGFRA expression was significantly increased in both PαC clusters compared with all other cells (Figure 1G). Both PαC clusters express KCNN3 (Figure 1H) and P2RY1 (Figure 1I) at higher proportions than other cells, but these transcripts were much more abundant in PαC#1 than PαC#2. Also, compared with PαC#2, PαC#1 had higher levels of mechanosensitive ion channel PIEZO2, neurotransmitter receptors ADRA1A and RAMP1 (Figure 6A), and more transcripts encoding axon guidance and synaptic proteins (Figure 6B and Figure A14A and B), ion channels and ion channel–associated proteins (Figure 6C and Figure A14C and D), and transcriptional regulators (Figure 6D and Figure A14E and F). PαC#2 express more structural ECM components (Figure 6E–G and Figure A14G and H).
Figure 6.
PαC clusters have distinct gene expression profiles. (A) Heatmap shows relative transcript abundance for 50 genes most differentially expressed between PαC#1 and PαC#2. Rows correspond to single genes. 3117 vertical lines each representing a single nucleus. Colors indicate relative expression = loge(mean scaled RNA expression per nucleus). (B–E) Number of genes most abundantly expressed in PαC#1 compared with PαC#2. (F and G) Numbers indicate genes represented.
PαC are Distinct From Fibroblasts
As PαCs were previously described as fibroblast-like cells,25 we compared PαC clusters against the fibroblasts (identified by MEOX2, COL1A2, COL1A1, FMO1, LSP1, and VIM) in our data set.26 Metascape network analysis showed most GO term enrichment for nervous system development genes came from PαC#1 (Figure A15A). PαC#1 and PαC#2 had few differentially expressed genes in common. PαC#2 and fibroblasts shared a larger proportion of differentially expressed genes. PαC#1 and fibroblasts had no differentially expressed genes in common (Figure A15B). Protein-protein MCODE Interaction Enrichment Analysis, which provides GO terms for differentially expressed genes when >3 encoded proteins interact with each other, showed most enriched interaction networks for PαC#1 involved ribosomal proteins (Figure A16A). Other enriched networks for PαC#1 included smooth muscle contraction and muscle development, purinergic and nitrergic signaling, and anti-inflammatory cytokine production. In contrast, while PαC#2 are enriched for the complement system in neuronal development and plasticity, most other enriched protein-protein interaction networks were ECM related (Figure A16B). Fibroblast cluster primarily showed enrichment of ECM-related networks, similar to PαC#2 (Figure A16C).
Right Colon SIP Express More Ion Channels and Transcriptional Regulators and Left Colon SIP Express More Axon Guidance and Synaptic Proteins
Because the colon has regional differences in motility and epithelial cell composition,27 we wondered if SIP cells gene expression differed between right (ascending) and left (sigmoid) colon. SIP cells organized into 3 distinct clusters (Figure 7A and B). Forced comparison between right and left colon showed SIP cells had similar mean UMIs and mean numbers of genes detected in each region (Figure 7C). Most nuclei corresponding to PαC#1 came from right colon, whereas right and left colon contributed similar PαC#2 numbers (Figure 7D). Most differentially expressed genes between the right and left colon are shown in Figure 7E–G. The number of significantly differentially expressed genes was limited in ICCs, perhaps because of low cell numbers (right colon: 373 nuclei, left colon: 207 nuclei; Figure 7G).
Figure 7.
SIP gene expression differs in right vs left colon. (A) SIP nuclei do not cluster by sample ID after removing non-SIP cells. (B) Color coding shows partial segregation of gene expression by bowel region. (C) Nuclei from the left or right colon express similar numbers of unique RNA (UMI) and unique genes. (D) PαC#1 and PαC#2 in the left and right colon express similar numbers of UMIs and unique genes. PαC#1 nuclei were 5.3-fold more abundant than PαC#2 nuclei in the right colon. Left colon had similar numbers of PαC#1 and PαC#2. (E–G) Violin plots of 40 most differentially expressed genes in right/ascending vs left/sigmoid colon. Expression level = loge(scaled RNA expression per nucleus).
Compared with left colon, right colon PαCs (Figure A17A and B) and SMCs (Figure A18A and B) had more axon guidance and synaptic function/maintenance mRNA, many more ion channels and ion channel–associated mRNA (PαC: Figure A17C and D; SMC: Figure A18C and D) and more transcriptional regulators (PαC: Figure A17E and F; SMC: Figure A18G and H). Only a few neurotransmitter receptors were differentially expressed between bowel regions. Most of these transcripts were more abundant in the right colon. For example, ADRA1A and CALCRL were more abundant in right colon PαCs. Muscarinic acetylcholine receptors CHRM2 and CHRM3 were more abundant in right colon SMCs. Only tachykinin receptor TACR2 is more abundant in left colon SMCs. While right and left colon PαCs and SMCs expressed similar numbers of ECM components and ECM remodeling proteins (PαC: Figure A17G and H; SMC: Figure A18E and F), right colon PαCs expressed fewer structural ECM components compared with left colon (Figure A17I and J). These regional differences in SMC and PαC gene expression might be linked to differences in bowel motility.
Discussion
We present the first analysis of adult human colon single-nucleus RNA sequencing focused on SIP syncytium, highlighting novel roles and interactions for SIP cells. For example, adult SMCs express many repulsive axon guidance molecules that could restrict axon entry into muscle layers to specific types of enteric neurons (eg, excitatory and inhibitory motor neurons). This is unexplored biology. ICCs and PαCs express mechanosensitive ion channels, suggesting they directly respond to mechanical forces prevalent in the bowel. There were 2 distinct PαCs populations. PαC#1 expresses numerous ion channels consistent with PαC roles in inhibitory neurotransmission. PαC#2 prominently expresses structural ECM genes. In contrast, PαC#1 expresses many nonstructural ECM genes and ECM remodeling proteins. Furthermore, SMCs and PαCs in the right colon express many more transcriptional regulators and ion channels than SMCs and PαCs in the left colon, suggesting regional differences in SIP function. Finally, all SIP cells share differentially enriched expression of at least 6 transcription factors not previously reported in this context. These proteins may form a transcriptional network to direct SIP cell differentiation and/or maintenance. These major findings reinforce and extend existing SIP syncytium literature.
Our initial strategy resulted in 2 SMC clusters, 1 ICC cluster, and 1 PαC cluster. Eliminating 1 sample (#5035) condensed SMC into a single cluster and split PαC into 2 clusters. #5035 was from a young adult with sigmoid volvulus, an uncommon condition that typically occurs when sigmoid colon is massively dilated. Massive dilation may occur because of distal obstruction or dysfunction in the dilated bowel. We do not know if this tissue was ischemic. We therefore omitted #5035 because our goal was to define normal SIP biology. Interestingly, 2 human PαC clusters delineated after removing #5035 may correspond to 2 murine PαC subpopulations distinguished by high or low PDGFRA expression (PDGFRAhigh vs PDGFRAlow).16 In mice, these cell types respond differently to partial intestinal obstruction. As the muscle layer increases in size, PDGFRAlow proliferate and PDGFRAhigh hypertrophy. PDGFRAlow cells have relatively high expression of Cacna1g,16 but we did not detect CACNA1G in human PαC clusters, possibly due to the limited read depth or species differences. For example, our human data show THBS4 at relatively high levels in PαC (3.16-fold higher than all other cells), but Thbs4 was recently described as an ICC-specific marker in mice.14
Our human data show fewer PαC#2 nuclei had detectable PDGFRA compared with PαC#1. This suggests human PαC#2 may correspond to murine28 PDGFRAlow, but this needs further validation. Nonetheless, divergent gene expression in PαC#1 vs PαC#2 is interesting in light of limited understanding of PαC function. PαCs have been called “fibroblast-like”25 and have roles in inhibitory neurotransmission within the SIP syncytium.1,2 Our data suggest these attributes might be distinct functions of PαC#2 and PαC#1, respectively, because PαC#2 express higher levels of structural ECM components and PαC#1 express higher levels of ion channels, channel-associated proteins, and neurotransmitter receptors. PαC#1 thus appear more electrically and transcriptionally active than PαC#2. If human PαC#2 are similar to murine PDGFRAlow, then partial intestinal obstruction may cause an increase in ECM-producing PαC#2 with less expansion of PαC#1, a testable hypothesis.
New Insights
We were not surprised to see prominent expression of ion channels, ion channel–associated proteins, axon guidance molecules, and synaptic proteins because all SIP cells are innervated by neurons and electrically active.1,3,29 We were surprised to see mechanosensitive ion channels (PIEZO2 and TMEM150C) in ICCs and PαCs suggesting ICCs and PαC directly detect mechanical force and could modulate smooth muscle contraction without input from enteric primary sensory neurons. Our data also show for the first time high expression of vasoactive intestinal peptide receptor 2 (VIPR2) in PαC (Figures 3C and 5G, Figure A4C), which suggests VIP could directly modulate human PαC. In addition, we detected relatively high levels of CGRP co-receptors CALCRL in PαCs and RAMP1 in SMCs. β-CGRP, a neurotransmitter expressed by enteric neuron subtypes such as intrinsic primary afferent neurons,30,31 has been implicated in murine intestinal peristaltic reflex.32,33 Expression of CGRP receptors in SMCs and PαCs may explain why CGRP induces smooth muscle relaxation even when neurotransmission is blocked with tetrodotoxin.34, 35, 36 This may have clinical relevance since recently approved anti-CGRP migraine therapeutics Erenumab and Fremanezumab can cause prominent gastrointestinal symptoms, including constipation, diarrhea, abdominal pain, and nausea.37, 38, 39 Our data suggest SIP cells may respond to neurotransmitters in ways that remain to be delineated by functional experiments. Expression of cell surface receptors and ligands for several growth factors, differentiation factors, and axon guidance molecules suggests significant cross-talk between the SIP syncytium cell types.
Another interesting aspect of SIP syncytium cell biology is that murine ICC and SMC share a common embryonic KIT+ precursor at least in longitudinal muscle.40, 41, 42 This common precursor expresses PDGFRα and PDGFRß receptors, and during fetal development, ligands PDGF-A and PDGF-B are expressed in circular muscle SMC and in enteric neurons, respectively. Blocking PDGFR with chemical antagonist AG1295 suppresses SMC differentiation in longitudinal muscle and appears to induce ICC formation.43 The origin of mature PαC remains unclear, but it is tempting to speculate that PDGFR-expressing mesenchymal cells are common precursors for all SIP cell types at least during fetal development.16,43 However, this has not been experimentally confirmed. The widespread PDGFRA expression in mesodermal and ectodermal derivatives during embryonic organogenesis44 and in adult mouse, where PDGFRA is expressed in most but not all fibroblasts,45 suggests we need to delineate bowel PαC subpopulations based on additional cell type–specific markers.
Supporting their close developmental relationship, all SIP syncytium cells express high levels of 6 transcriptional regulators relative to other cells (MEIS1, MEIS2, PBX1, FOS, ZBTB16, and SCMH1; Figure 4E and F). Finding MEIS1, MEIS2, and PBX1 in the same cells is not surprising. MEIS1 and MEIS2 orthologs regulate PBX1 translocation to the nucleus and MEIS1 and PBX1 heterodimerize on DNA to control gene expression, including a set of HOX genes.46 The immediate early gene FOS mediates TGF-β signaling, and TGF-β receptors are expressed by all SIP cells in our data set (Figure 4E and F). TGF-β signaling is important for differentiation of ICC and vascular smooth muscle.47,48 ZBTB16/PLZF regulates cellular responsiveness to FGF signaling49 and was recently described as a likely causative genomic locus in a rat model of myocardial hypertrophy, fibrosis, and hypertension.50 FGF receptors are expressed by all SIP cells in our data set (Figure 5G). SCMH1 is a component of the polycomb repressive complex 1 that regulates the expression of many genes through chromatin modifications.51 These differentially expressed factors may be part of a shared SIP-specific transcriptional regulatory network. Supporting this hypothesis, when only SIP cells are compared against each other, these transcriptional regulators are either no longer detected as differentially expressed (MEIS1, SCMH1, and ZBTB16) or only upregulated in a single SIP syncytium cell type (FOS, MEIS2, and PBX1).
A curious observation is that SMCs and PαCs in ascending/right colon express more transcriptional regulators, ion channels (Figure 7E and F, Figures A17 and A18), and neurotransmitter receptors than the same cell types in sigmoid/left colon. This correlates with 5.3-fold more PαC#1 compared with PαC#2 in our right colon data set and a ratio of 0.85:1 for PαC#1:PαC#2 in the left colon (Figure 7D). Increased right colon transcriptional, electrical, and chemical complexity in SIP syncytium parallels the observation that ENS circuits are also more complex in proximal (right) compared with distal (left) colon.27 In contrast, the greater number of structural ECM genes in left colon PαCs compared with right colon PαCs may correlate with thicker muscularis layer and denser ECM in sigmoid colon. To the best of our knowledge, this is the first report of regional differences in gene expression for colon SMCs and PαCs.
Systems-level analysis22 to identify unexpected protein networks or functions using Metascape protein-protein MCODE Interaction Enrichment Analysis (Figures A3 and A9) highlighted additional aspects of SIP biology. As expected, SMC GO terms include “Smooth Muscle Contraction” and “Muscle Contraction” based on many contractile apparatus proteins. Compared with other data set cells, SMCs are enriched in focal adhesion–associated proteins, proteins that metabolize heparan sulfate and chondroitin sulfate proteoglycans, and axon guidance molecules. GO terms for ICC include “Purine metabolism,” driven in part by GUCY1A1 and GUCY1B1. These soluble guanylate cyclase subunits make cyclic guanosine monophosphate in response to nitric oxide. ICC also express phosphodiesterases that degrade cGMP as reported.52 ICC GO terms highlight interactions of ICC with neurons (“Neuronal System,” “Protein-Protein Interaction at Synapses”) when ICCs are compared with all other cells and highlight responses to monoamines/catecholamines when ICC are compared with other SIP cells. GO terms for PαC vs all other cells highlight many ECM components (collagens, fibrillin, tenascin C, laminins, fibulins), collagen-specific endoplasmic reticulum chaperone SERPINIH1, and TIMP1 that prevents ECM degradation, as well as proteins that permit direct responses to nitric oxide, ciliary neurotrophic factor, leukemia inhibitory factor, interleukin 6, and oncostatin M.53 interleukin 6 and oncostatin M have potent pro-inflammatory roles in Crohn’s disease54,55 where striking proliferation and hypertrophy of bowel muscle cause fibrostenosing strictures.53 This suggests thickening of muscularis propria in the setting of inflammation may at least in part be attributed to the proliferation of PαC already known to proliferate in some contexts.16
Conclusion
This study has limitations. Cells analyzed were microdissected from near myenteric plexus. We did not capture distinct types of ICC found distant from myenteric plexus.15 Single-nucleus RNAseq data provide low read depth, so many genes were only detected in a small proportion of nuclei in a given cluster. Analyses were not followed by protein-level validation. However, RNAseq data correlates well with known RNA and protein data from SIP syncytium. Furthermore, ENS gene expression patterns in this same data set were extensively validated and correlate well with the prior ENS literature.17 Despite limitations, these analyses provide deep insight into SIP syncytium biology, revealing previously unrecognized gene expression patterns. This first-of-its-kind analysis of human colon data should facilitate fascinating hypotheses that guide future studies and has relevance for bowel motility disorders, bowel inflammation, and mechanical obstruction.
Acknowledgments
Authors' Contributions:
Sabine Schneider and Sohaib K. Hashmi contributed to conceptualization and methodology. Sabine Schneider, Sohaib K. Hashmi, A. Josephine Thrasher, and Deepika R. Kothakapa contributed to investigation; A. Josephine Thrasher, Deepika R. Kothakapa, Sabine Schneider, and Sohaib K. Hashmi contributed to formal analysis. A. Josephine Thrasher and Deepika R. Kothakapa contributed to data curation. Sabine Schneider and Sohaib K. Hashmi wrote the article. Sabine Schneider, Sohaib K. Hashmi,. A. Josephine Thrasher, Deepika R. Kothakapa, Christina M. Wright, and Robert O. Heuckeroth reviewed and edited the article. Sabine Schneider, Christina M. Wright, and Robert O. Heuckeroth contributed to resources. Sabine Schneider and Sohaib K. Hashmi contributed to supervision. Robert O. Heuckeroth contributed to funding acquisition.
Footnotes
Conflicts of Interest: This author discloses the following: R.O.H. was a consultant for BlueRock Therapeutics and served on a Scientific Advisory Panel for Takeda. The remaining authors disclose no conflicts.
Funding: This study was funded by NIHF30DK118827 (S.K.H.), NIHR01 DK128282 (R.O.H.), NIHR01 R01DK122798 (R.O.H.), the Irma and Norman Braman Endowment (R.O.H.), the Suzi and Scott Lustgarten Center Endowment (R.O.H.), The Children’s Hospital of Philadelphia Frontier Program for Precision Diagnosis and Therapy for Pediatric Motility Disorders (R.O.H.), and The Children’s Hospital of Philadelphia Research Institute (R.O.H.). The study sponsors had no role in study design, collection, analysis, or interpretation of data.
Ethical Statement: The corresponding author, on behalf of all authors, jointly and severally, certifies that their institution has approved the protocol for any investigation involving humans or animals and that all experimentation was conducted in conformity with ethical and humane principles of research.
Data Transparency Statement: Full data sets are deposited in Gene Expression Omnibus: Gene Expression Omnibus accession number GSE156905. All lists of differentially expressed genes generated in this analysis can be accessed as supplementary data files with this article.
Reporting Guidelines: Not applicable.
Material associated with this article can be found in the online version at https://doi.org/10.1016/j.gastha.2022.12.004.
Supplementary Materials
References
- 1.Blair P.J., Rhee P.-L., Sanders K.M., et al. The significance of interstitial cells in neurogastroenterology. J Neurogastroenterol Motil. 2014;20:294–317. doi: 10.5056/jnm14060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sanders K.M., Ward S.M., Koh S.D. Interstitial cells: regulators of smooth muscle function. Physiol Rev. 2014;94:859–907. doi: 10.1152/physrev.00037.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schneider S., Wright C.M., Heuckeroth R.O. Unexpected roles for the Second Brain: enteric nervous system as Master regulator of bowel function. Annu Rev Physiol. 2018;81:235–259. doi: 10.1146/annurev-physiol-021317-121515. [DOI] [PubMed] [Google Scholar]
- 4.Bitar K.N. Function of gastrointestinal smooth muscle: from signaling to contractile proteins. Am J Med. 2003;115(Suppl 3A):15S–23S. doi: 10.1016/s0002-9343(03)00189-x. [DOI] [PubMed] [Google Scholar]
- 5.Bitar K.N., Gilmont R.R., Raghavan S., et al. In: Physiology of the gastrointestinal tract. Fifth Ed. Johnson L.R., Ghishan F.K., Kaunitz J.D., et al., editors. Academic Press; Boston: 2012. Chapter 17 - cellular physiology of gastrointestinal smooth muscle; pp. 489–509. [Google Scholar]
- 6.Gabella G. Cells of visceral smooth muscles. J Smooth Muscle Res. 2012;48:65–95. doi: 10.1540/jsmr.48.65. [DOI] [PubMed] [Google Scholar]
- 7.Gabella G. Development of visceral smooth muscle. Results Probl Cell Differ. 2002;38:1–37. doi: 10.1007/978-3-540-45686-5_1. [DOI] [PubMed] [Google Scholar]
- 8.Maeda H., Yamagata A., Nishikawa S., et al. Requirement of c-kit for development of intestinal pacemaker system. Development. 1992;116:369–375. doi: 10.1242/dev.116.2.369. [DOI] [PubMed] [Google Scholar]
- 9.Lee H., Koh B.H., Peri L.E., et al. Purinergic inhibitory regulation of murine detrusor muscles mediated by PDGFRα+ interstitial cells. J Physiol. 2014;592:1283–1293. doi: 10.1113/jphysiol.2013.267989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baker S.A., Hennig G.W., Ward S.M., et al. Temporal sequence of activation of cells involved in purinergic neurotransmission in the colon. J Physiol. 2015;593:1945–1963. doi: 10.1113/jphysiol.2014.287599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kurahashi M., Mutafova-Yambolieva V., Koh S.D., et al. Platelet-derived growth factor receptor-α-positive cells and not smooth muscle cells mediate purinergic hyperpolarization in murine colonic muscles. Am J Physiol Cell Physiol. 2014;307:C561–C570. doi: 10.1152/ajpcell.00080.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chi J.-T., Rodriguez E.H., Wang Z., et al. Gene expression programs of human smooth muscle cells: tissue-specific differentiation and prognostic significance in breast cancers. Plos Genet. 2007;3:1770–1784. doi: 10.1371/journal.pgen.0030164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee M.Y., Park C., Berent R.M., et al. Smooth muscle cell genome Browser: Enabling the identification of novel Serum response factor target genes. PLoS One. 2015;10:e0133751. doi: 10.1371/journal.pone.0133751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee M.Y., Ha S.E., Park C., et al. Transcriptome of interstitial cells of Cajal reveals unique and selective gene signatures. PLoS One. 2017;12:e0176031. doi: 10.1371/journal.pone.0176031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen H., Ordög T., Chen J., et al. Differential gene expression in functional classes of interstitial cells of Cajal in murine small intestine. Physiol Genomics. 2007;31:492–509. doi: 10.1152/physiolgenomics.00113.2007. [DOI] [PubMed] [Google Scholar]
- 16.Ha S.E., Lee M.Y., Kurahashi M., et al. Transcriptome analysis of PDGFRα+ cells identifies T-type Ca2+ channel CACNA1G as a new pathological marker for PDGFRα+ cell hyperplasia. PLoS One. 2017;12:e0182265. doi: 10.1371/journal.pone.0182265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wright C.M., Schneider S., Smith-Edwards K.M., et al. scRNA-sequencing reveals new enteric nervous system roles for GDNF, NRTN, and TBX3. Cell Mol Gastroenterol Hepatol. 2021;11:1548–1592.e1. doi: 10.1016/j.jcmgh.2020.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Butler A., Hoffman P., Smibert P., et al. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol. 2018;36:411–420. doi: 10.1038/nbt.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stuart T., Butler A., Hoffman P., et al. Comprehensive integration of single-cell data. Cell. 2019;177:1888–1902.e21. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Piper M., Pantano L., Mistry M., et al. Single-cell RNA-seq: clustering analysis. In-depth-NGS-Data-Analysis-Course 2018. https://hbctraining.github.io/In-depth-NGS-Data-Analysis-Course/sessionIV/lessons/SC_clustering_analysis.html Available at:
- 21.Wingender E., Schoeps T., Haubrock M., et al. TFClass: expanding the classification of human transcription factors to their mammalian orthologs. Nucleic Acids Res. 2018;46:D343–D347. doi: 10.1093/nar/gkx987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou Y., Zhou B., Pache L., et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun. 2019;10:1523. doi: 10.1038/s41467-019-09234-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Satija R., Farrell J.A., Gennert D., et al. Spatial reconstruction of single-cell gene expression data. Nat Biotechnol. 2015;33:495–502. doi: 10.1038/nbt.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kular J.K., Basu S., Sharma R.I. The extracellular matrix: Structure, composition, age-related differences, tools for analysis and applications for tissue engineering. J Tissue Eng. 2014;5 doi: 10.1177/2041731414557112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Baker S.A., Hennig G.W., Salter A.K., et al. Distribution and Ca(2+) signalling of fibroblast-like (PDGFR(+)) cells in the murine gastric fundus. J Physiol. 2013;591:6193–6208. doi: 10.1113/jphysiol.2013.264747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tabib T., Morse C., Wang T., et al. SFRP2/DPP4 and FMO1/LSP1 define major fibroblast populations in human Skin. J Invest Dermatol. 2018;138:802–810. doi: 10.1016/j.jid.2017.09.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li Z., Hao M.M., Van den Haute C., et al. Regional complexity in enteric neuron wiring reflects diversity of motility patterns in the mouse large intestine. Elife. 2019;8:e42914. doi: 10.7554/eLife.42914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Breland A., Ha S.E., Jorgensen B.G., et al. Smooth Muscle Transcriptome Browser: offering genome-wide references and expression profiles of transcripts expressed in intestinal SMC, ICC, and PDGFRα+ cells. Sci Rep. 2019;9:387. doi: 10.1038/s41598-018-36607-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sanders K.M., Koh S.D., Ro S., et al. Regulation of gastrointestinal motility--insights from smooth muscle biology. Nat Rev Gastroenterol Hepatol. 2012;9:633–645. doi: 10.1038/nrgastro.2012.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Furness J.B., Robbins H.L., Xiao J., et al. Projections and chemistry of Dogiel type II neurons in the mouse colon. Cell Tissue Res. 2004;317:1–12. doi: 10.1007/s00441-004-0895-5. [DOI] [PubMed] [Google Scholar]
- 31.Nurgali K., Stebbing M.J., Furness J.B. Correlation of electrophysiological and morphological characteristics of enteric neurons in the mouse colon. J Comp Neurol. 2004;468:112–124. doi: 10.1002/cne.10948. [DOI] [PubMed] [Google Scholar]
- 32.Grider J.R. Neurotransmitters mediating the intestinal peristaltic reflex in the mouse. J Pharmacol Exp Ther. 2003;307:460–467. doi: 10.1124/jpet.103.053512. [DOI] [PubMed] [Google Scholar]
- 33.Bornstein J.C., Costa M., Grider J.R. Enteric motor and interneuronal circuits controlling motility. Neurogastroenterol Motil. 2004;16:34–38. doi: 10.1111/j.1743-3150.2004.00472.x. [DOI] [PubMed] [Google Scholar]
- 34.Holzer P., Barthó L., Matusák O., et al. Calcitonin gene-related peptide action on intestinal circular muscle. Am J Physiol. 1989;256:G546–G552. doi: 10.1152/ajpgi.1989.256.3.G546. [DOI] [PubMed] [Google Scholar]
- 35.Barthó L., Lembeck F., Holzer P. Calcitonin gene-related peptide is a potent relaxant of intestinal muscle. Eur J Pharmacol. 1987;135:449–451. doi: 10.1016/0014-2999(87)90699-6. [DOI] [PubMed] [Google Scholar]
- 36.Barthó L. Calcitonin gene-related peptide and capsaicin inhibit the circular muscle of the Guinea-pig ileum. Regul Pept. 1991;35:43–48. doi: 10.1016/0167-0115(91)90252-c. [DOI] [PubMed] [Google Scholar]
- 37.Falkenberg K., Bjerg H.R., Olesen J. Two-hour CGRP infusion causes gastrointestinal Hyperactivity: possible relevance for CGRP antibody treatment. Headache. 2020;60:929–937. doi: 10.1111/head.13795. [DOI] [PubMed] [Google Scholar]
- 38.Holzer P., Holzer-Petsche U. Constipation caused by anti-calcitonin gene-related peptide migraine therapeutics explained by antagonism of calcitonin gene-related peptide's motor-Stimulating and prosecretory function in the intestine. Front Physiol. 2021;12:820006. doi: 10.3389/fphys.2021.820006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Haanes K.A., Edvinsson L., Sams A. Understanding side-effects of anti-CGRP and anti-CGRP receptor antibodies. J Headache Pain. 2020;21:26. doi: 10.1186/s10194-020-01097-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Klüppel M., Huizinga J.D., Malysz J., et al. Developmental origin and Kit-dependent development of the interstitial cells of cajal in the mammalian small intestine. Dev Dyn. 1998;211:60–71. doi: 10.1002/(SICI)1097-0177(199801)211:1<60::AID-AJA6>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 41.Torihashi S., Nishi K., Tokutomi Y., et al. Blockade of kit signaling induces transdifferentiation of interstitial cells of cajal to a smooth muscle phenotype. Gastroenterology. 1999;117:140–148. doi: 10.1016/s0016-5085(99)70560-3. [DOI] [PubMed] [Google Scholar]
- 42.Torihashi S., Ward S.M., Sanders K.M. Development of c-Kit-positive cells and the onset of electrical rhythmicity in murine small intestine. Gastroenterology. 1997;112:144–155. doi: 10.1016/s0016-5085(97)70229-4. [DOI] [PubMed] [Google Scholar]
- 43.Kurahashi M., Niwa Y., Cheng J., et al. Platelet-derived growth factor signals play critical roles in differentiation of longitudinal smooth muscle cells in mouse embryonic gut. Neurogastroenterol Motil. 2008;20:521–531. doi: 10.1111/j.1365-2982.2007.01055.x. [DOI] [PubMed] [Google Scholar]
- 44.Takakura N., Yoshida H., Ogura Y., et al. PDGFR alpha expression during mouse embryogenesis: immunolocalization analyzed by whole-mount immunohistostaining using the monoclonal anti-mouse PDGFR alpha antibody APA5. J Histochem Cytochem. 1997;45:883–893. doi: 10.1177/002215549704500613. [DOI] [PubMed] [Google Scholar]
- 45.Muhl L., Genové G., Leptidis S., et al. Single-cell analysis uncovers fibroblast heterogeneity and criteria for fibroblast and mural cell identification and discrimination. Nat Commun. 2020;11:3953. doi: 10.1038/s41467-020-17740-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Safgren S.L., Olson R.J., Pinto E Vairo F., et al. De novo PBX1 variant in a patient with glaucoma, kidney anomalies, and developmental delay: an expansion of the CAKUTHED phenotype. Am J Med Genet A. 2022;188:919–925. doi: 10.1002/ajmg.a.62576. [DOI] [PubMed] [Google Scholar]
- 47.Vetuschi A., Sferra R., Latella G., et al. Smad3-null mice lack interstitial cells of Cajal in the colonic wall. Eur J Clin Invest. 2006;36:41–48. doi: 10.1111/j.1365-2362.2006.01593.x. [DOI] [PubMed] [Google Scholar]
- 48.Guo X., Chen S.-Y. Transforming growth factor-β and smooth muscle differentiation. World J Biol Chem. 2012;3:41–52. doi: 10.4331/wjbc.v3.i3.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gaber Z.B., Butler S.J., Novitch B.G. PLZF regulates fibroblast growth factor responsiveness and maintenance of neural progenitors. PLoS Biol. 2013;11 doi: 10.1371/journal.pbio.1001676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liška F., Mancini M., Krupková M., et al. Plzf as a candidate gene predisposing the spontaneously hypertensive rat to hypertension, left ventricular hypertrophy, and interstitial fibrosis. Am J Hypertens. 2014;27:99–106. doi: 10.1093/ajh/hpt156. [DOI] [PubMed] [Google Scholar]
- 51.Takada Y., Isono K.-I., Shinga J., et al. Mammalian Polycomb Scmh1 mediates exclusion of Polycomb complexes from the XY body in the pachytene spermatocytes. Development. 2007;134:579–590. doi: 10.1242/dev.02747. [DOI] [PubMed] [Google Scholar]
- 52.Baker S.A., Drumm B.T., Cobine C.A., et al. Inhibitory neural regulation of the Ca 2+ transients in intramuscular interstitial cells of cajal in the small intestine. Front Physiol. 2018;9:328. doi: 10.3389/fphys.2018.00328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen W., Lu C., Hirota C., et al. Smooth muscle hyperplasia/hypertrophy is the most prominent histological change in Crohn's fibrostenosing bowel strictures: a semiquantitative analysis by using a novel histological grading scheme. J Crohns Colitis. 2017;11:92–104. doi: 10.1093/ecco-jcc/jjw126. [DOI] [PubMed] [Google Scholar]
- 54.Danese S., Vermeire S., Hellstern P., et al. Randomised trial and open-label extension study of an anti-interleukin-6 antibody in Crohn's disease (ANDANTE I and II) Gut. 2019;68:40–48. doi: 10.1136/gutjnl-2017-314562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.West N.R., Hegazy A.N., Owens B.M.J., et al. Oncostatin M drives intestinal inflammation and predicts response to tumor necrosis factor-neutralizing therapy in patients with inflammatory bowel disease. Nat Med. 2017;23:579–589. doi: 10.1038/nm.4307. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.