Abstract
New approach methodologies (NAMs) that efficiently provide information about chemical hazard without using whole animals are needed to accelerate the pace of chemical safety assessments. Technological advancements in gene expression assays have made in vitro high-throughput transcriptomics (HTTr) a feasible option for NAMs-based hazard characterization of environmental chemicals. In the present study, we evaluated the Templated Oligo with Sequencing Readout (TempO-Seq) assay for HTTr concentration-response screening of a small set of chemicals in the human-derived MCF7 cell model. Our experimental design included a variety of reference samples and reference chemical treatments in order to objectively evaluate TempO-Seq assay performance. To facilitate analysis of these data, we developed a robust and scalable bioinformatics pipeline using open-source tools. We also developed a novel gene expression signature-based concentration-response modeling approach and compared the results to a previously implemented workflow for concentration-response analysis of transcriptomics data using BMDExpress. Analysis of reference samples and reference chemical treatments demonstrated highly reproducible differential gene expression signatures. In addition, we found that aggregating signals from individual genes into gene signatures prior to concentration-response modeling yielded in vitro transcriptional biological pathway altering concentrations (BPACs) that were closely aligned with previous ToxCast high-throughput screening (HTS) assays. Often these identified signatures were associated with the known molecular target of the chemicals in our test set as the most sensitive components of the overall transcriptional response. This work has resulted in a novel and scalable in vitro HTTr workflow that is suitable for high throughput hazard evaluation of environmental chemicals.
Keywords: transcriptomics, high-throughput screening, TempO-Seq, computational toxicology, toxicogenomics, omics research, high-throughput, in vitro models, human risk assessment, hazard assessment, gene expression, environmental chemicals, dose-response
Introduction
Current animal toxicity testing approaches only allow for a small fraction of the thousands of chemicals in U.S. commerce (USEPA 2020) to be thoroughly evaluated for human safety. New approach methodologies (NAMs) that can efficiently provide information about chemical hazard and risk without using whole animals are needed to accelerate the pace of chemical risk assessments (USEPA 2018). Formally, NAMs have been defined as any technology, methodology, approach or combination thereof that can be used to provide information on chemical hazard and risk that avoids the use of intact animals (USEPA 2018). This broad term can encompass many different types of in vitro bioactivity studies, in silico modeling of bioactivities and exposure predictions, cheminformatics and various combinations thereof. Recently, USEPA has made a commitment to phase out mammalian toxicity testing to the greatest extent possible by 2035 (Wheeler 2019), thereby increasing the impetus at USEPA for development and implementation of NAMs.
The USEPA has been evaluating high-throughput screening (HTS) and computational toxicology tools for over 10 years (Judson et al. 2010; Richard et al. 2016), resulting recently in the development of a NAM for screening of endocrine disrupting chemicals for use in a regulatory setting (USEPA 2016). Identifying and/or developing NAMs as effective replacements for in vivo toxicity testing is a significant challenge, but one that could be addressed by using a cross-disciplinary tiered toxicity testing approach as described in the USEPA Computational Toxicology Blueprint (i.e. EPA CompTox Blueprint) (Thomas et al. 2019). The success of this strategy depends on implementing “Tier 1” assays for hazard characterization. Toward this objective, the EPA CompTox Blueprint proposes the use of non-targeted high-throughput profiling (HTP) assays for initial characterization of the biological activity of environmental chemicals. Ideally, such profiling assays should be capable of being deployed in HTS format across multiple human-derived in vitro models while providing high content data that can be leveraged to identify potency thresholds for perturbation of cellular biology and predict putative mechanism of action. High-throughput transcriptomics (HTTr) using targeted RNA-Seq is one such assay that meets these criteria.
Gene expression profiling has long been considered an informative method for evaluating the biological activity and/or toxicity of chemicals (Joseph 2017). Past research focused on using gene expression data from in vivo animal studies to characterize the toxicity of environmental chemicals, to identify putative molecular mechanisms-of-action, and to define transcriptional points-of-departure (PODs) through the use of concentration-response modeling of multiplexed gene expression measurements (Blomme et al. 2009; Cui and Paules 2010; Farmahin et al. 2017; Harrill et al. 2019; Thomas et al. 2013). Such studies were necessarily low-throughput given the use of laboratory animals. Over the years, advances in transcriptomics research have included technological improvements in transcriptomics assay platforms (i.e. increased assay reproducibility and transcriptome coverage), the establishment of large-scale, open-access transcriptome profiling datasets housing both in vivo and in vitro chemical bioactivity data (Igarashi et al. 2015; Lamb et al. 2006; Svoboda et al. 2019), and development of many computational strategies for analyzing such data. However, research efforts relating to the latter two topics have primarily focused on mechanism-of-action characterization and chemical clustering/read-across (De Abrew et al. 2016). Concentration-response data and the analysis approaches needed for identifying biologic potency thresholds for environmental chemicals, referred to here as transcriptional biological pathway altering concentrations (BPACs) (Harrill et al. 2019; Judson et al. 2011) have been lacking. Fortunately, increasing efficiency and declining costs associated with generating whole transcriptome profiles have made in vitro HTTr screening in concentration-response mode a feasible option for NAMs-based hazard characterization of environmental chemicals.
In the present study, we evaluated the Templated Oligo with Sequencing Readout (TempO-Seq) assay (Yeakley et al. 2017) for HTTr screening of a small set of environmental chemicals. This assay requires small (picogram) amounts of input RNA, is scalable in terms of transcriptome coverage, is amenable to preparation of pooled sequencing libraries from many samples, yields sequencing reads of exactly 50 base pairs that can be rapidly aligned to generate gene counts, and is compatible with cell lysates prepared in multiwell (e.g. 384-well) format. This latter feature is important from an HTS perspective in that it eliminates the need for the time-consuming and costly process of RNA extraction. The version of the TempO-Seq assay we evaluated provides nearly whole transcriptome coverage (>20,000 genes), although lower coverage versions of the assay have been evaluated by other toxicology research groups (Grimm et al. 2019; Limonciel et al. 2018; Ramaiahgari et al. 2019). The MCF7 breast adenocarcinoma cell line was selected for use in the present study in order to anchor to the Connectivity Map (CMAP) database (Lamb et al. 2006), a large collection of whole transcriptome profiles from chemical-treated cells; MCF7 being the most highly represented. Our experimental design included a variety of reference samples and reference chemical treatments that facilitated objective evaluation of TempO-Seq assay performance. To facilitate analysis of these data (and data from potential future studies), we developed a robust and scalable bioinformatics pipeline using open-source tools and open-access data. Lastly, we developed a novel gene expression signature-based concentration-response modeling approach and compared the results to a previously proposed workflow for concentration-response analysis of transcriptomics data, as recommended by the National Toxicology Program (NTP 2018), and implemented as part of the BMDExpress software package (Phillips et al. 2019).
Analysis of reference samples and reference chemical treatments demonstrated highly reproducible differential gene expression signatures across assay plates. These signatures were associated with the known molecular targets and biological activity of the reference samples and treatments. For the in vitro HTTr data generated in the present study, large effect sizes (i.e. > 2-fold expression change) were rare at the individual gene level. However, we found that aggregating individual gene expression changes into signature scores prior to concentration-response modeling yielded transcriptional BPACs that were equivalent to, or in many cases, more sensitive than transcriptional BPACs determined using the previously proposed BMDExpress workflow. The transcriptional BPACs determined using the gene signature approach were closely aligned with in vitro BPACs determined from ToxCast HTS assays (Judson et al. 2010). The gene signature approach often identified signatures associated with the known molecular target of the test chemicals as the most sensitive drivers of the overall transcriptional response. In summary, we have developed a scalable experimental and bioinformatic workflow that can be used to conduct in vitro HTTr concentration-response screening of thousands of chemicals that may be found in the environment.
Methods
Materials
HTB-22™ MCF7 breast adenocarcinoma cells and dimethyl sulfoxide (DMSO) used for cryopreservation purposes were purchased from American Tissue Culture Collection (Manassas, VA). MediaTech™ Dulbecco’s Modified Eagles Medium (DMEM) with glucose (4.5 g/L), L-glutamine (584 mg/L) and sodium pyruvate (110 mg/L), rectangular 25 cm2 (T25), 75 cm2 (T75) 225 cm2 (T225) cell culture flasks with vented caps and barcoded 384-well optical imaging plates were purchased from Corning, Inc., (Corning, NY). Gibco™ Penicillin-Streptomycin-Glutamine (PSG), LifeTech™ TrypLE™ Select Enzyme, Invitrogen™ Countess® Cell Counting Chamber Slides, 0.4% Trypan Blue Stain, Applied Biosystems™ MicroAmp® Optical Adhesive Film, Nunc™ aluminum acrylate plate sealing tape, Hoechst 33342 trihydrochloride trihydrate (H-33342) 10 mg/mL aqueous solution, propidium iodide (PI) 1 mg/mL aqueous solution and CellEvent™ Caspase 3/7 Green Detection Reagent were purchased from ThermoFisher Scientific (Waltham, MA). Heat-Inactivated Fetal Bovine Serum (HI-FBS), 10X Phosphate Buffered Saline (PBS), and DMSO for test chemical solubilization and dilution were purchased from Millipore-Sigma (St. Louis, MO). Reference chemicals trichostatin A (TSA, catalog #: T8552), sirolimus (catalog #: R0395), genistein (catalog #: 345834), staurosporine (catalog #: S5921), ionomycin (catalog #: I9657), saccharin (catalog #: 240931) and sorbitol (catalog #: S1876) were also purchased from Millipore-Sigma. Echo® Qualified 384-Well Polypropylene Microplates (384PP) were purchased from Labcyte, Inc., (Sunnyvale, CA). Universal Human Reference RNA (UHRR, cataolog #: 636690) and Human Brain Reference RNA (HBRR, catalog #: 636658) were purchased from Takara-ClonTech (Mountain View, CA). For 1X PBS and paraformaldehyde working solutions, concentrated stocks were diluted in deionized water from a Dracor water purification system.
Chemical Selection
For this study, we selected a set of 44 chemicals (Table 1). For our screening design, this is the number of chemicals that would fit on a single 384 well plate in concentration-response format in addition to the reference chemical treatments, reference samples, and vehicle control wells. These chemicals were selected from three classes: those with effects on specific molecular targets expressed in MCF7 cells, chemicals causing broad cytotoxicity, and a set of herbicides that are active against targets that are not expressed in MCF7 cells (or not present in mammalian cells at all) but only exist in fungi or plants. In the first class, the six specific molecular targets tested for were estrogen receptor alpha (ESR1), androgen receptor (AR), peroxisome proliferator activating receptor alpha and gamma (PPARα and PPARγ, respectively), 3-Hydroxy-3-Methylglutaryl-CoA reductase (HMGCR), and thyroid hormone receptor (THRA). Relative baseline expression levels from the Human Protein Atlas (https://www.proteinatlas.org) (Uhlen et al. 2015) for expression of these molecular targets in MCF7 cells are as follows: ESR1 (24.3), HMGCR (10.4), THRA (3.2), PPARG (2.4), PPARA (1.7), AR (1.2). These are normalized expression levels (NX), where a value of 1.0 is the limit of detection. Several of the selected chemicals are known to be broadly cytotoxic through inhibition of sulfhydryl enzyme systems, disrupting mitochondrial electron transport or inhibiting protein synthesis (Kleinstreuer et al. 2014; Sipes et al. 2013). Our prior expectation was that the ESR1 agonists and antagonists should produce notable effects on gene expression in the MCF7 cell type. Antagonist activity should be seen because of the presence of estradiol in the culture media which causes the estrogen receptor (ER) pathway to be partially, but not fully active at baseline. In contrast, the herbicidal PPO inhibitors were expected to be largely inactive as they target enzymes that are only found in plants and have shown little activity in the broader ToxCast in vitro screening battery (Supplemental Material Table S1). For most of the target classes, we included more than one chemical to allow us to observe consistency in activity by target class. All literature references for the effects of the 44 chemicals used in this study are provided as Supplemental Material (Table S1).
Table 1:
Chemicals used in the study
| Name | CASRN | Target annotation |
|---|---|---|
| Cyproterone acetate | 427-51-0 | AR antagonist |
| Flutamide | 13311-84-7 | AR antagonist |
| Nilutamide | 63612-50-0 | AR antagonist |
| Vinclozolin | 50471-44-8 | AR antagonist |
| Amiodarone hydrochloride | 19774-82-4 | Blocks myocardial calcium, potassium and sodium channels |
| Cladribine | 4291-63-8 | DNA synthesis inhibitor |
| 4-Cumylphenol | 599-64-4 | ER agonist |
| 4-Nonylphenol, branched | 84852-15-3 | ER agonist |
| Bisphenol A | 80-05-7 | ER agonist |
| Bisphenol B | 77-40-7 | ER agonist |
| 4-Hydroxytamoxifen | 68392-35-8 | ER antagonist |
| Clomiphene citrate (1:1) | 50-41-9 | ER antagonist |
| Fulvestrant | 129453-61-8 | ER antagonist |
| Cyproconazole | 94361-06-5 | Ergosterol-biosynthesis inhibitor. Pan-cyp inhibitor |
| Imazalil | 35554-44-0 | Ergosterol-biosynthesis inhibitor. Pan-cyp inhibitor |
| Prochloraz | 67747-09-5 | Ergosterol-biosynthesis inhibitor. Pan-cyp inhibitor |
| Propiconazole | 60207-90-1 | Ergosterol-biosynthesis inhibitor. Pan-cyp inhibitor |
| Atrazine | 1912-24-9 | Herbicide, photosystem II inhibitor |
| Cyanazine | 21725-46-2 | Herbicide, photosystem II inhibitor |
| Simazine | 122-34-9 | Herbicide, photosystem II inhibitor |
| Butafenacil | 134605-64-4 | Herbicide, protoporphyrinogen oxidase (PPO) inhibition |
| Fomesafen | 72178-02-0 | Herbicide, protoporphyrinogen oxidase (PPO) inhibition |
| Lactofen | 77501-63-4 | Herbicide, protoporphyrinogen oxidase (PPO) inhibition |
| Lovastatin | 75330-75-5 | HMGCR inhibitor |
| Simvastatin | 79902-63-9 | HMGCR inhibitor |
| Maneb | 12427-38-2 | Inhibition of metal-dependent and sulfhydryl enzyme systems |
| Thiram | 137-26-8 | Inhibition of metal-dependent and sulfhydryl enzyme systems |
| Ziram | 137-30-4 | Inhibition of metal-dependent and sulfhydryl enzyme systems |
| Reserpine | 50-55-5 | inhibition of the ATP/Mg2+ pump |
| Rotenone | 83-79-4 | Mitochondria (complex I inhibitor) |
| Pyraclostrobin | 175013-18-0 | Mitochondria (complex III inhibitor) |
| Trifloxystrobin | 141517-21-7 | Mitochondria (complex III inhibitor) |
| Fenpyroximate (Z,E) | 111812-58-9 | Mitochondrial electron transport inhibitor |
| Clofibrate | 637-07-0 | PPARα agonist, upregulates extrahepatic lipoprotein lipase |
| Fenofibrate | 49562-28-9 | PPARα agonist, upregulates extrahepatic lipoprotein lipase |
| Farglitazar | 196808-45-4 | PPARα agonist |
| PFOA | 335-67-1 | PPARα, PPARα agonist |
| PFOS | 1763-23-1 | PPARα, PPARα agonist |
| Troglitazone | 97322-87-7 | PPARα, PPARα agonist |
| Cycloheximide | 66-81-9 | Protein synthesis inhibitor |
| Bifenthrin | 82657-04-3 | Sodium channel modulator |
| Cypermethrin | 52315-07-8 | Sodium channel modulator |
| Tetrac | 67-30-1 | T4 synthesis inhibitor |
| 3,5,3’-Triiodothyronine | 2/3/6893 | Thyroid hormone receptor agonist |
Cell Culture
Five vials of cryopreserved MCF7 (HTB-22™) cells were purchased from American Tissue Culture Collection, designated as passage 0 (P0) and stored in vapor phase liquid nitrogen prior to initial thawing and expansion. A passage 3 (P3) MCF7 cryostock was generated by thawing and pooling all five original source vials and culturing through three consecutive passages in complete growth medium (DMEM + 10% FBS + 1% PSG). At each passage, cells from different culture vessels were pooled prior to re-seeding for subsequent expansion. At P3, cells were cryopreserved at 4 million cells per mL in complete growth medium + 5% DMSO according to manufacturer’s protocol. MCF7 cultures used in all phases of this study were maintained in humidified incubators with 5% CO2 atmosphere and internal temperature of 37°C.
For chemical screening, MCF7 cells were thawed and subjected to a uniform expansion protocol from P3 to P6 through increasingly larger sizes of tissue culture vessels (T25 to T75 to T225) prior to plating in 384-well format. The three cultures were initiated on consecutive days and subject to uniform handling procedures to ensure that dosing and sample collection also occurred on consecutive days. All chemical screening experiments were performed on P7 cells. Cell line authentication was performed on an aliquot of P6 cells using short tandem repeat (STR) profiling and comparison to the MCF7 (ATCC HTB-22) reference profile in accordance with the ANSI/ATCC ASN-0002–2011 method. STR profiling demonstrated a 100% match of the P6 profile to the reference profile (data not shown), thereby confirming that the cells used in this study were MCF7. The P6 cells were also negative for mycoplasma (data not shown) as assessed using the iNtRON Biotechnology eMYCO Plus kit.
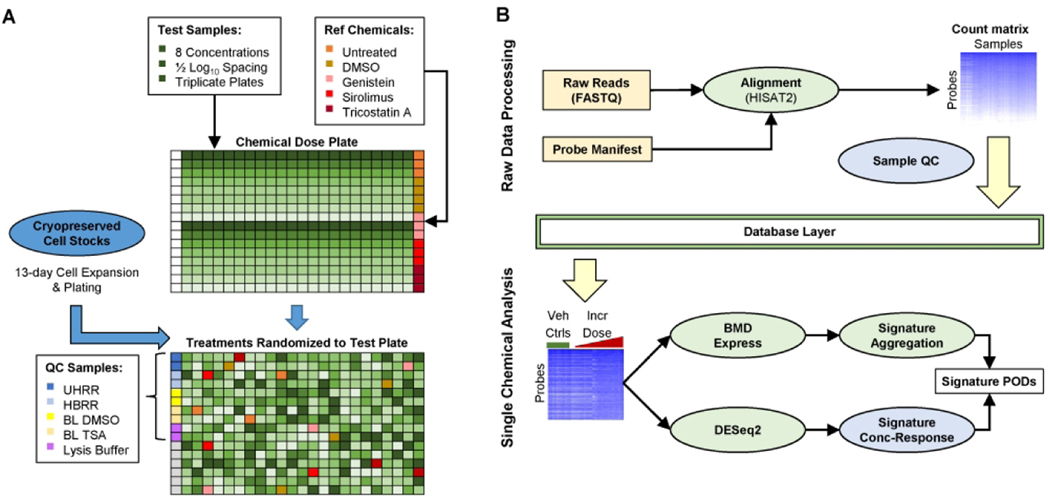
At the time of plating, cells were passaged and resuspended in complete growth media. A small aliquot (10–20 μL) of cell suspension was then labeled with Trypan Blue solution (1:1) and the number of live cells per volume of cell suspension determined using a Countess™ II Automated Cell Counter (ThermoFisher Scientific, Waltham, MA) according to manufacturer’s instructions. The concentration of the cell suspension was then adjusted to 250 cells/μL. The adjusted cell suspension was then dispensed into 384-well plates (40 μL/well) using a MultiFlo FX multi-mode dispenser equipped with a 10 μL dispensing cassette (BioTek, Winooski, VT). The total cell input at the time of seeding was 10,000 cells/well and vehicle control cultures reached a confluency of 50–60% by the time of sampling (data not shown). The screening experiment was performed using three independent cultures (i.e. biological replicates). Each biological replicate contained three assay plates of MCF7 cells used in: 1) the HTTr TempO-Seq assay, 2) the cell viability assay and 3) the apoptosis assay, respectively. The first column of each 384-well plate used in the HTTr TempO-Seq assay was left empty to accommodate addition of quality control samples prior to transcriptomics analysis (Figure 1). Cells were seeded in all wells of the cell viability and apoptosis assay plates. After seeding, cells were placed in an incubator and allowed a 24-hour recovery period prior to chemical treatments.
Figure 1. Overview of high-throughput transcriptomics workflow.
(A) Diagram of initial plating of test and reference chemicals on a TempO-Seq dose plate, followed by randomization of chemical exposures to test plates. The first column of each test plate is not loaded with cells and is reserved for dispensing of QC samples. Grey wells on the test plate are reserved for internal positive controls added and verified by the contractor. (B) Diagram of bioinformatics workflow. Light yellow boxes indicate raw data received from contractor after targeted RNA-Seq assay completion. Light green circles indicate steps performed using existing open-source methods. Light blue circles indicate novel methods developed as part of this work. Bioinformatic analysis is generally split into two phases. Raw data processing up to probe-level count matrix and samplelevel QC metrics is performed across entire data set. Subsequent analysis is performed separately for each chemical against plate-matched vehicle controls. Intermediate processing results are stored in a database layer to facilitate analysis.
Chemical Treatments
DMSO-solubilized chemical stock solutions were received frozen from the US EPA ToxCast chemical inventory management contractor (EvoTec, Princeton, NJ) and stored at −80°C prior to dose plate preparation. An eight-point dilution series (1/2 log10 spacing) of test chemicals was prepared in a LabCyte Echo-qualified 384-well polypropylene (384PP) plate at 200x the desired nominal test concentration for screening (0.03 – 100 μM). Singleton wells of dosing solutions for each test chemical concentration were arrayed in columns 2 through 23 of the dose plate (Figure 1A). Transcriptomics reference chemicals (genistein, trichostatin A, sirolimus) were solubilized as 200x dosing solutions corresponding to nominal test concentrations of 10, 1 and 0.1 μM, respectively. Triplicate wells of dosing solutions for each of the transcriptomic reference chemicals were added to the dose plate (column 24) along with quadruplicate wells of pure DMSO as a vehicle control. Reference treatments were included in the experiment in order to evaluate plate-to-plate reproducibility of transcriptional responses as measured using TempO-Seq, to confirm proper dispensing of test chemicals and optimize analysis methods by comparison with the Connectivity Map database (Lamb et al. 2006).
Cell viability positive control chemicals (staurosporine and ionomycin) were solubilized as 200x stocks corresponding to nominal test concentrations of 1 and 30 μM, respectively. Negative control (saccharin, sorbitol) chemicals were solubilized as 200x stocks corresponding to a nominal test concentration of 100 μM. Quadruplicate wells of cell viability positive and negative control chemicals were added to the dose plate (column 1). The dose plate was then sealed with a Nunc™ aluminum adhesive plate cover and stored at −80°C until use. On the first day of dosing, the dose plate was brought to room temperature inside a desiccator, centrifuged briefly to ensure even distribution of dose solutions in each well and unsealed. In between each dosing session (occuring on consecutive days), the dose plate was resealed with an aluminum cover and stored at room temperature, in the dark, within a desiccator to prevent hydration of DMSO test chemical solutions during the study.
At 24 h post-plating, 200 nL of 200x dosing solutions were transferred to the assay plates using a LabCyte Echo 550 acoustic dispenser. The final concentration of DMSO in all treatment, reference treatment and vehicle control wells was 0.5%. Well coordinates on each assay plate were uniquely randomized with respect to treatment so that any potential edge-well effects were distributed in an unbiased manner across all possible treatment conditions. For the HTTr TempO-Seq assay, only test chemicals, transcriptomic reference chemicals and vehicle control (i.e. DMSO) solutions were dispensed to columns 2 through 24 of assay plates. For the cell viability and apoptosis assays, test chemicals, transcriptomic reference chemicals, cell viability positive and negative control chemicals and vehicle control solutions were dispensed to columns 1 through 24 of the assay plates. All assay plates were placed back in an incubator (5% CO2, 37°C) immediately after dosing. All HTTr, cell viability and apoptosis experiments used a chemical exposure duration of 6 h prior to sampling as this was the most frequently used exposure duration for MCF7 cells in the CMAP database.
Cell Viability and Apoptosis Assays
The cell viability and apoptosis assays were conducted in a similar manner. For the cell viability assay, MCF7 cells were live-labeled with a combination of H-33342 and propidium iodide (PI) at nominal well concentrations of 8.1 and 3.75 μM, respectively. For the apoptosis assay, MCF7 cells were live-labeled with a combination of H-33342 and CellEvent™ Caspase 3/7 (Casp3/7) at nominal well concentrations of 8.1 and 5 μM, respectively. For both assays, labeling reagents were dispensed from 384PP plates to assay plates using a LabCyte Echo 550 acoustic dispenser. Following application of labeling reagents, assay plates were placed in a humidified incubator (5% CO2, 37°C) and incubated for 30 minutes. Plates were then fixed via direct addition of 12 μL of 16% paraformaldehyde solution followed by incubation at room temperature for 10 min, protected from light. Assay plates were then washed twice with 80 μL of 1X PBS and assay wells filled with 80 μL of 1X PBS as a storage buffer. Plates were sealed with optical adhesive film and stored at 4°C prior to imaging.
On the day of image acquisition, plates were removed from 4°C storage and equilibrated to room temperature, protected from light. Images were then acquired using a Cellomics™ ArrayScan XTI High-Content Screening system. For the cell viability assay, excitation/emission filters for H-33342 and propidium iodide image acquisition were 365/535 nm and 549/600 nm, respectively. For the apoptosis assay, image acquisition excitation/emission filters were 365/535 nm and 475/535 nm, respectively. For both assays, images were acquired using a 10X objective and 4 unique fields-of-view were imaged in each well. In each assay, nuclei were segmented in the H-33342 channel. Nuclei selection parameters were optimized to exclude border nuclei (i.e. instances where the entirety of the nuclei was not visible) and potential imaging artifacts (i.e. fibers, dust, etc.). For the cell viability assay, the mean pixel intensity of the PI channel was calculated within each valid nucleus. For the apoptosis assay, the mean pixel intensity of the Casp3/7 channel was calculated within each valid nucleus. Cell-level data for each plate was then exported for downstream analysis using the R statistical computing environment. Data were analyzed on a per plate basis. Each cell was identified as either PI-positive or CASP-3/7 positive if their mean intensity was above the 95th-percentile calculated for vehicle control wells on the same assay plate. The percentage of PI-positive or Casp3/7-positive cells were calculated for each assay well. Z’ values were calculated as described (Zhang et al. 1999) on a per plate basis using ionomycin and staurosporine positive control treatments and DMSO as the baseline treatment. Z’ values for negative control chemicals saccharin and sorbitol were also calculated using DMSO as the baseline treatment to demonstrate lack of effects on cell viability associated with the chemical dispensing procedure. Concentrations of test chemicals producing either ≥ 50% PI-positive or ≥ 50% Casp3/7-positive cells were flagged (Supplemental Table 2) and assay wells corresponding to those concentrations were not included in analysis of the TempO-Seq data.
HTTr Assay
Aqueous solutions (100 ng/μL) of Universal Human Reference RNA (UHRR) and Human Brain Reference RNA (HBRR) were prepared from source vials by dilution with RNase-free water. UHRR and HBRR solutions were then further diluted 1:1 in 2X BioSpyder lysis buffer, aliquoted and frozen at −80°C until use. Bulk lysates were prepared by treating MCF7 cells cultured in 384-well format with 0.5% DMSO (i.e. BLDMSO) or 1 μM trichostatin A (i.e. BLTSA) as described above and lysing the cells using the procedure described below. Lysates from wells receiving the same treatment were pooled in 50 mL conical tubes, aliquoted and frozen at −80°C until use. The RNA standards and the bulk lysates are collectively referred to as ‘reference samples’ throughout this study and were included in the experimental design in order to evaluate performance of the TempO-Seq assay across plates, absent of any influence of dosing or cell culture procedures.
Six hours after chemical treatment, HTTr assay plates were removed from the incubator and media in each assay well was drained to a residual volume of 10 μL using MultiFlo FX microdispenser equipped with a vacuum-driven aspiration manifold. To create cell lysates, 10 μL volumes of 2X BioSpyder lysis buffer were dispensed into each assay well using the same MultiFlo FX instrument equipped with 1 μL peristaltic pump dispensing cassette. Then, 20 μL volumes of UHRR, HBRR, BLDSMO, BLTSA and Lysis Buffer were manually dispensed into duplicate wells of column 1 of each HTTr assay plate as illustrated in Figure 1A. The plates were sealed with an adhesive aluminum plate cover and incubated at room temperature, protected from light, for 30 minutes. The plates were then stored at −80°C prior to shipment to BioSpyder, Inc.
Plates were shipped to BioSpyder, Inc. frozen (on dry ice) using overnight priority shipping. MCF7 cell lysates were then analyzed by BioSpyder using a custom-attenuated version of the TempO-Seq human whole transcriptome version 1 (hWTv1) assay (Yeakley et al. 2017), which includes 21,111 probes covering 19,287 genes (see Supplemental Material). Lysates were processed as described (House et al. 2017). In brief, 2 μL of each lysate was hybridized with 2 μL of detector oligos from the hWTv1 assay using the following thermal cycler protocol: 10 min at 70°C, followed by gradual decrease to 45°C over 49 min, terminating with 45°C incubation for 16–24 hours. Excess oligos were then removed via nuclease digestion (90 min at 37°C) and hybridized detector oligos were ligated (60 min at 37°) following respective additions of 24 μL TempO-Seq nuclease and ligation mixes. RNA/DNA duplexes were then heat-denatured and 10 μL of each ligation product was transferred to an amplification microplate containing 10 μL of PCR master mix per well. Ligation products were then uniquely labeled during product amplification (10 min at 37°C, 2 min at 95°C, 6 cycles of 95°C for 30 s, 54°C for 30 s, 72°C for 120 s, 16 cycles of 95°C for 30s and 72°C for 120s, followed by 72°C for 60 s) with well coordinate-specific “barcoded” primer pairs containing universal adaptors for sequencing. Samples were then pooled into a series of three sequencing libraries (1 for each plate). Pooled sequencing libraries were then distributed across multiple lanes of a HiSeq dual flow cell and analyzed on a HiSeq 2500 Ultra-High-Throughput Sequencing System (Illumina, San Diego, CA). The target depth for each test sample was 3 million sequenced reads.
HTTr data processing
Raw TempO-Seq data were provided by the vendor as individual FASTQ files for each sample well and were subsequently processed through a custom bioinformatics pipeline (Figure 1B). Each FASTQ file was aligned to the probe sequences in the hWTv1 assay (see Supplemental Material) using HISAT2 v2.1.0 (Kim et al. 2015; Kim et al. 2019) with spliced alignment disabled. Aligned reads in SAM format were processed with SAMtools v1.9 (Li et al. 2009) to compute the number of uniquely aligned reads for each probe. Probe counts and associated meta-data for each well were stored for analysis using MongoDB v3.6.14. Source code for all data processing steps are included in an open source package ‘httrpl’ (https://github.com/USEPA/httrpl_pilot). The probe counts for each sample are provided via FigShare (DOI: 10.23645/epacomptox.13368914, see Supplemental Material).
HTTr Quality Control
Extensive quality control (QC) criteria were developed to exclude probes and samples of low quality. The vendor updated the hWTv1 assay annotation to mask 151/21111 (0.7%) probes due to low quality as ascertained by correlation with RNA-Seq data (see Supplemental Material). All 21,111 probes in hTWv1 were used to align the raw data, but read counts for these 151 probes were excluded from all further analysis. Low quality samples were removed based on the QC criteria listed in Table 2. Sample level QC criteria were based on the cell viability results for each concentration of the test chemicals (Supplemental Table 2), and multiple well-level metrics computed from read mapping rate and count distribution across probes (Supplemental Table 3). The Gini coefficient (GiC) is a generalizable metric of overall inequality in any distribution, originally developed to measure income inequality but subsequently adapted to many other applications (van Mierlo et al. 2016) including biological data (Graczyk 2007). In this study, we computed a GiC for each sample based on the distribution of raw counts for all probes including those with 0 aligned reads. The thresholds for Fraction of Viable Cells (FrVC) and Fraction of Reads Uniquely Mapped to Probes (FMR) are simple majority cutoffs (majority of cells must be viable; majority of reads must uniquely map to probe sequences). The threshold for Number of Uniquely Mapped Reads (NMR) was set at 10% of target per-sample read depth. The thresholds for Number of Probes with at least 5 Reads (Ncov5), Number of Probes Capturing Top 80% of Signal (Nsig80), and GiC were set at approximately Tukey’s Outer Fence (Tukey 1977), defined as 3x the inter-quartile range (IQR), of the distribution for all samples cultured on each plate (test sample, vehicle control, and reference chemical treatments) excluding those with FrVC<0.5 (>50% cell death as noted above). All samples with QC flags were removed from further analysis except where otherwise noted. For each well corresponding to a vehicle control, reference sample, or reference chemical, we also computed the D-statistic (House et al. 2017), which is the average correlation against all other replicate wells of the same type.
Table 2.
Quality control metrics used for processing and analyzing the high-throughput transcriptomics data.
| Abbreviation | Description | Threshold | Additional Information |
|---|---|---|---|
| FrVC | Fraction of viable cells (PI-negative or Casp3/7-negative) | Reject < 50% | Highly cytotoxic conditions no longer represent molecular initiating event |
| NMR | Number of mapped reads, defined as sum of total read counts summed over all detected probes | Reject < 300,000 | Threshold =10% of target depth |
| FMR | Fraction of uniquely mapped reads | Reject < 50% | Majority of reads must align to a single probe sequence |
| Ncov5 | The number of probes with at least 5 uniquely mapped reads | Reject < 5,000 | Based on Tukey’s Outer Fence (3*IQR) of all viable samples cultured on each plate (test samples, vehicle controls, and reference chemical treatments) |
| Nsig80 | The number of probes capturing the top 80% of signal in a sample | Reject < 1,000 | |
| GiC | Gini coefficient computed for each sample based on the distribution of raw counts for all probes including those with 0 aligned reads | Reject > 0.95 |
Differential expression analysis
Differential expression analysis was performed independently for each reference chemical treatment against the matched DMSO controls. In addition, BLTSA samples were compared to BLDMSO samples. Two separate differential expression analyses, a plate-level analysis, and a plate-group analysis (specific to the reference chemical treatments only), were performed as follows. Read counts for all probes were tabulated for all relevant samples (all replicates of the reference chemical treatments and corresponding vehicle controls for each plate for the plate-level analysis or across all 3 plates for the plate-group analysis). Probes with mean read count < 5 across these subsets of samples were removed. Counts for the remaining samples and probes were modeled using DESeq2 v1.24 (Love et al. 2014). For the plate-level reproducibility analysis, individual probe counts were modeled as a function of treatment effect (i.e. counts ~ treatment). Size factors and dispersion were estimated using package defaults, and model-fitting was initially performed with ‘betaPrior=FALSE’. We then applied fold-change shrinkage to the treatment contrast (reference chemical vs DMSO control) to obtain moderated log2 fold-changes (L2FC) for each probe. Differential expression analysis of HBRR vs UHRR samples was also run, with the sample type in place of treatment effect.
For the combined analysis of all plates, counts were modeled by treating the experimental plate and treatment effect as independent factors (i.e. counts ~ plate + treatment). Size factors, dispersion estimates, and fold-change shrinkage were applied similarly to the plate-level analysis above except the moderated L2FC values were adjusted to remove any average plate effects. Additionally, we repeated these combined analyses without modeling plate effect (i.e. counts ~ treatment) and/or without applying the fold-change shrinkage step when computing moderated L2FC values, resulting in 4 possible DESeq2 configurations (+/− plate effect; +/− L2FC shrinkage).
Differential expression analysis of the 44 test chemicals was performed as described above, except that samples for all concentrations of the same test chemical (excluding samples with QC flags as noted above) were used together with all plate-matched DMSO wells to filter probes with mean count < 5 independently for each test chemical. The DESeq2 model used in this case treated plate effect and each concentration group as independent factors (i.e. counts ~ plate + concentration). Fold-change shrinkage was applied separately for each concentration vs DMSO controls to compute moderated L2FC values. Alternate configurations (without plate effect and/or without shrinkage) were not run for test chemicals. Differential expression p-values for each probe at each concentration were computed using the Wald test prior to fold-change shrinkage (DESeq2 default). The p-values were adjusted for multiple-testing within each concentration using the Benjamini-Hochberg step-up method, with the “independent filtering” option in DESeq2 turned off. We computed differentially expressed gene (DEG) accumulation scores from the DESeq2 results as follows: for each chemical, we first identified the sets of probes with adjusted p-value < 0.1 for each treatment concentration (c) vs DMSO. Second, the accumulated DEGs at each concentration (ci) were identified as the union of all significant probes for c ≤ ci. The accumulation score was defined as the number of accumulated DEGs for each concentration ci. Thus, for each chemical, the DEG accumulation score for the highest test concentration is equal to the size of the union of probes identified as differentially expressed in any pairwise comparison between a concentration and DMSO controls.
We summarized the DESeq2 differential expression analysis for each test chemical as a matrix in which the rows, columns and values correspond to the treatment concentrations, probes and L2FC values, respectively (which we will refer to as the probe level L2FC matrix). For each chemical, we then produced the gene level L2FC matrix (treatment conditions as rows; genes as columns) by aggregating the probe level L2FC values for each gene and using the highest magnitude fold change in either direction. All L2FC data for this study are included in the data release (see Supplemental Material). For comparison, we also computed “raw” L2FC values directly from the probe counts used as DESeq2 input, as follows: first, probe counts for each sample were converted to counts per million (CPM) and log2 transformed with a pseudo-count = 1. Next, we computed the mean log2(CPM) value for each treatment group (DMSO or individual chemical concentration). Raw L2FC values were then computed as the difference between mean log2(CPM) values for each treatment group vs the DMSO group.
Signature Gene Set Selection
A large collection of 37082 signatures, or gene sets (a list of Entrez gene symbols) was initially obtained from 4 major sources: Bioplanet (Huang et al. 2019) [ref v1.0, accessed 7/29/2019], CMAP (Subramanian et al. 2017), DisGeNET (Pinero et al. 2015), and MSigDB (Liberzon et al. 2015; Liberzon et al. 2011; Subramanian et al. 2005) [ref v6.2, accessed 3/8/2019]. CMAP signatures were created by taking the n most highly downregulated and the n most highly upregulated genes from each CMAP profile, where n=100, 200 or 300. For the current work only the 100 most down- and upregulated gene sets were used. For the current work, we included MSigDB (sub)collections C2 and H as these could be mapped to pathways that can be interpreted in meaningful ways with regards to molecular targets or processes. Signatures were annotated with a target name, indicating, for instance the molecular target that would elicit the response or the disease associated with the signature. The target names were derived from the signature names using an automated process and descriptions from the source database. The target names were then summarized into a curated set of target classes. For a few target names, such as PPAR, individual isoforms were summarized into a single target class (e.g. “PPAR”). We also created 1000 random signatures, which are random sets of genes with the same gene co-occurrence frequency and signature length distribution as the collection of real signatures. For the present analysis, we used a subset of 6586 signatures selected to cover all target classes matching the known target or mechanism of action of the 44 test chemicals (Table 1), plus the 1000 random signatures, for a total of 7586 signatures. Signatures can be either directional (as in CMAP) or nondirectional. Gene set enrichment analysis (see below) was performed separately for the up and down set of genes for directional gene signatures, and then the results were combined. The 7586 signatures used in this analysis include many up/down pairs, and when these are combined, there are 4431 final signatures covering at least 10 genes expressed in the current study. The complete collection of signature gene sets is included in the public data release containing the gene set catalog with all annotations, and an RData file containing the gene sets (see Supplemental Material).
Signature Concentration Response Modeling
We performed concentration-response modeling of signature-level enrichment scores for all test chemicals, starting from the gene-level L2FC matrix for all test chemicals, with rows corresponding to conditions (one row per chemical sample concentration combination) and columns corresponding to genes. For a gene to be retained for subsequent analyses, at least 95% of the conditions must contain data. For missing values, L2FC is set to zero. For each chemical-concentration-signature, a signature score is created. Here we use the single sample gene set enrichment analysis (ssGSEA) method (Barbie et al. 2009) as implemented in the GSVA R package (v1.32.0) to calculate the normalized enrichment score (NES). For directional signatures, NES scores are derived separately for the up- and down-regulated gene sets and the final signature score = signature score(up) – signature score(down). Signature scores can be positive or negative, but the distribution is expected to be zero centered. Henceforth, we use “signature scores” to refer to NES scores for directional and non-directional signatures. Note that for the single-concentration reference chemical treatments and bulk lysate samples, only the signature scores were used and no concentration-response modeling was performed.
For each combination of test chemical and signature, the concentration-response series of signature scores was fit to a set of models including constant, Hill, gain-loss (a rising Hill curve followed by a decreasing Hill curve), 2 polynomial models, a power model and 4 exponential models. These are ToxCast Pipeline (tcpl) (Filer et al. 2017) implementations of curve-fitting models included in BMDExpress (Phillips et al. 2019) except for the constant and gain-loss models which are specific to tcpl. Modeling was performed using the tcplfit2 R package (Sheffield et al. in press), which selects the model with the best (lowest) AIC value as the final curve-fit (Akaike 1974).
A key output from the tcplfit2 modeling is a “continuous hit call” for each concentration-response series. Whereas the original tcpl package provided “binary hit calls” to classify responses as either a hit (active) or a miss (inactive), continuous hit calls seek to quantify the strength of hits and identify borderline cases corresponding to low magnitude responses or highly variable (noisy) data. Calculation of the continuous hit call combines each of the following probabilistic criteria: 1) the probability that at least one median response at any test concentration is greater than the statistically defined-noise threshold (i.e. cutoff, described below), 2) the probability that the maximum absolute response (i.e. top) of the curve fit is above the noise threshold cutoff, and 3) the probability that the winning AIC is less than that of the constant model. This continuous hit call value falls between zero and one with higher values indicative of relative greater confidence in classifying a modeled endpoint as a hit. Details of the probabilistic calculation are provided with the tcplfit2 R package (Sheffield et al. in press).
A chemical-signature combination is considered active if a model other than the constant model had the minimum AIC. Additionally, the top must exceed a statistically defined noise threshold. To estimate noise, we first generated a set of randomized null L2FC data from the complete data set of 44 chemicals x 8 concentrations to generate a concentration-response data set for N (here N=1000) “random” chemicals. For each gene in each of the random chemicals, 8 L2FC values were generated from the distribution of L2FC values in the original data set for that gene, and these were assigned as the L2FC values for the 8 concentrations for that gene and that random chemical. Then, missing values were added in random locations of the random dataset to match the original fraction of missing values. Thus, the null data preserved the distributional properties of each gene, but any correlation between genes was broken. This null data set was then used to calculate null signature score distributions. The cutoff used to determine activity in the actual pathway data was set to the outer 95% confidence interval of this null distribution, corresponding to p=0.05.
For chemical-signature combinations with a model other than constant, a benchmark dose (BMD) was calculated as the potency estimate for the pathway. The BMD value is the concentration at which the winning model curve crosses the benchmark response level (BMR) which is set to 1.349 times the signature-specific noise level (Filipsson et al. 2003; Thomas et al. 2007; Yang et al. 2007). BMD bounds (i.e. BMDL and BMDU) were computed in accordance with the profile likelihood method (Banga et al. 2002). The transcriptomic BPAC based on signature scoring (BPACSig) analysis for each chemical was reported as the 5th lowest BMD value of active signatures that have a BMDU/BMDL ratio < 40 and a continuous hitcall >= 0.5.
Concentration-response Analysis with BMDExpress
For comparison, overall transcriptional BMD values were computed using the BMDExpress2 software (Phillips et al. 2019) based on a workflow described in the NTP approach to genomic dose-response modeling (NTP 2018). First, probe-level concentration response analysis was performed as follows. Probe counts for each chemical (subset to samples without QC flags, and probes with mean count > 5 as described for differential expression analysis above) were normalized to log2 counts per million (CPM) values using the sum of filtered probe counts as the sample depth and adding a pseudo-count of 1 before converting to log scale. For each test chemical, probe-level log2 CPM values were input to BMDExpress2 (Phillips et al. 2019) using the following parameters: pre-filtering was used to remove probes with fold-changes <2 at all concentrations; each pre-filtered probe was then fit to 8 different dose-response models (linear, poly2, power, Hill, exp2, exp3, exp4, and exp5); the best-fit model for each probe was selected based on the lowest Akaike information criterion (AIC); the benchmark response (BMR) was set to 1.349 x standard deviation of replicate samples, corresponding to 10% tail in a normal distribution; and Hill models with k parameter <1/3 the lowest positive dose were excluded from final model selection.
The probe-level best fit models were then aggregated to signature-level BMD values using the same collection of signature gene sets described above. Briefly, probe-level curve-fits were filtered to only those meeting the following criteria: best fit model produced convergent BMD, BMDL, and BMDU values; BMD < highest measured dose; BMDU:BMDL ratio < 40; and probe annotated as measuring a single gene. If multiple probes corresponding to the same gene had valid curve fits under this criterion, the gene-level BMD/BMDL/BMDU were taken as the average of all probes with valid curve fits. The signature-level BMD was computed as the median BMD for all associated genes passing the filters above. Only signatures containing at least 3 valid genes and 5% gene set coverage were retained for further analysis. The transcriptomic BPAC based on BMDExpress (BPACBMDX) analysis for each chemical was reported as the minimum signature-level BMD passing these filters. Root-Mean-Square Error (RMSE) and correlation coefficients between different BPAC derivation methods were computed on the log10 scale.
Results
We screened 44 chemicals in MCF7 cells in concentration-response and generated HTTr data using the TempO-Seq hWTv1 assay. First, we provide an outline of the quality of the HTTr data based on a set of QC metrics we developed for this platform. Second, we evaluate the reproducibility and mechanistic accuracy of the HTTr platform using inter-plate analysis of reference samples and comparison of reference chemical treatment effects with CMAP signatures, respectively. Third, we summarize the concentration-dependent HTTr responses for all 44 chemicals to stratify them in terms of their overall effect on the transcriptome. Fourth, we present a new gene signature-based concentration-response analysis that provides potency estimates for perturbation of cellular biology (i.e. BPACs).
A Robust, Scalable and Reproducible Workflow for High-Throughput Transcriptomics
In order to develop a screening platform that is scalable for high-throughput applications, we performed the TempO-Seq assay directly on cell lysates, bypassing the time-consuming and expensive task of RNA purification. The resulting trade-off is that some aspects of sample quality that would normally be assessed in low-throughput RNA-seq workflows (e.g. RNA integrity and concentration) can no longer be assessed prior to sequencing library preparation. Therefore, we developed a battery of QC metrics that can be used to identify and remove any low-quality TempO-Seq samples based on the resulting sequenced reads (Table 2).
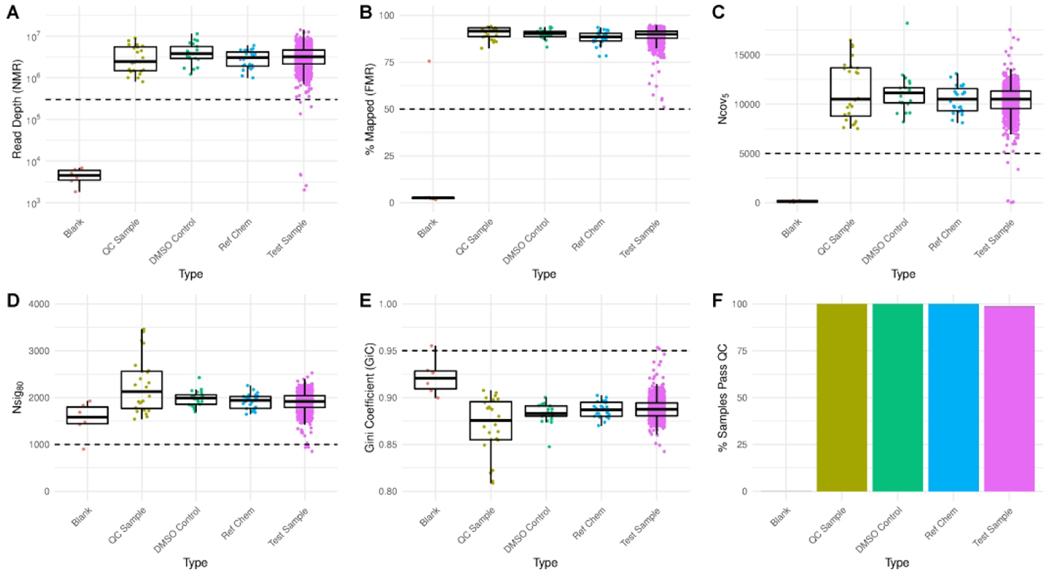
We first assessed metrics related to the sequencing and alignment of reads to probes. Median number of mapped reads (NMR) for all sample types was close to the target depth of 3 million reads per sample, although the dynamic range in depth is >10x, likely because measurement and adjustment of RNA concentrations prior to sample multiplexing was not performed (Figure 2A). Lysis buffer blanks (sequenced as negative controls) produced ~1,000x fewer aligned reads than most other samples. The median fraction of uniquely mapped reads (FMR) for all other sample types was >80% (Figure 2B), in line with previously published TempO-Seq results (Yeakley et al. 2017). As a final confirmation of reliable sequencing depth across samples, we assessed in each sample the number of probes meeting a minimum coverage threshold of 5 reads, a metric we denote as “Ncov5” (Figure 2C). We found that for most sample types in this study, the median Ncov5 value was ~10,000 probes. We applied the principle of Tukey’s Outer Fence (Tukey 1977) to determine a threshold for flagging samples with Ncov5 < 5,000 probes.
Figure 2. Quality assessment of high-throughput transcriptomics data.
(A-E) Distributions of all sample-level QC metrics, split by sample type. Dashed lines indicate thresholds for masking samples from further analysis. (F) Proportion of samples passing all QC thresholds by type. Blank = Lysis buffer negative controls containing no cellular material; QC Sample = samples prepared in larger batches and added to each plate prior to conduct of TempO-Seq assay; DMSO Control = vehicle control for all other wells; Ref Chem = single dose reference chemical treatments; Test Sample = wells treated with a test chemical.
We also assessed two distributional metrics designed to detect samples where low input RNA may have led to over-amplification and subsequent sequencing of PCR duplicates. The underlying principle here is that when library amplification is performed on a sample with low input, only a small number of hybridized probe molecules are present, and when the sample is sequenced deeply, these individual molecules may be sequenced and counted multiple times, resulting in a library with “lower complexity” compared to other samples (Adiconis et al. 2013). Again, due to the lack of RNA purification steps in our high-throughput procedure, we cannot identify and exclude low input samples prior to sequencing. We first defined the Nsig80 metric as the proportion of probes capturing the top 80% of read counts (Figure 2D). The median value for most sample types was ~2,000 probes, and again we used Tukey’s Outer Fence principle to determine a threshold of Nsig80 < 1,000 for flagging problematic samples by this metric. To account for the fact that Nsig80 is based on a single percentage of read counts in each sample, we also computed the Gini Coefficient (GiC), a generalizable measure of overall inequality (van Mierlo et al. 2016). We expect inequality in read counts across probes, owing to order of magnitude differences in expression across genes and the dynamic range of the TempO-Seq assay. However, samples that stand out as high outliers by this metric relative to other samples may also indicate potential problems with low input material. Therefore, we flagged any samples with a GiC > 0.95 based on the outer fence principle (Figure 2E).
In addition to identifying samples based on sequencing quality metrics, we flagged all 3 replicates of the highest concentration for two chemicals (6 samples total), clomiphene citrate (1:1) and 4-hydroxytamoxifen, as each of these treatment conditions produced >50% cell death on average in the complementary imaging plates used to assess overall cytotoxicity and apoptosis. Using ionomycin as the positive control, per plate Z’-values for the cell viability and apoptosis assays ranged from 0.78 to 0.93 and 0.33 to 0.91, respectively, indicating acceptable performance of each assay. In comparison, Z’-values using staurosporine were substantially lower for the cell viability (−3.78 to 0.53) and apoptosis (0.04 to 0.39) assays, indicating that 1 μM staurosporine is inappropriate for use as a positive control for these assays in MCF7 cells using a 6 hour exposure duration. Negative control chemicals saccharin and sorbitol (100 μM) did not produce any effects on cell viability or apoptosis in MCF7 cells.
Samples flagged for any of the reasons described here were removed from further analysis, resulting in the rejection of 13 individual samples treated with test chemicals, and 98.8% of all test samples passing QC (Figure 2F). All blank lysis buffer samples failed QC based on at least one flagging criteria, and 100% of all other control and reference samples types passed all QC criteria.
Evaluation of Assay Performance
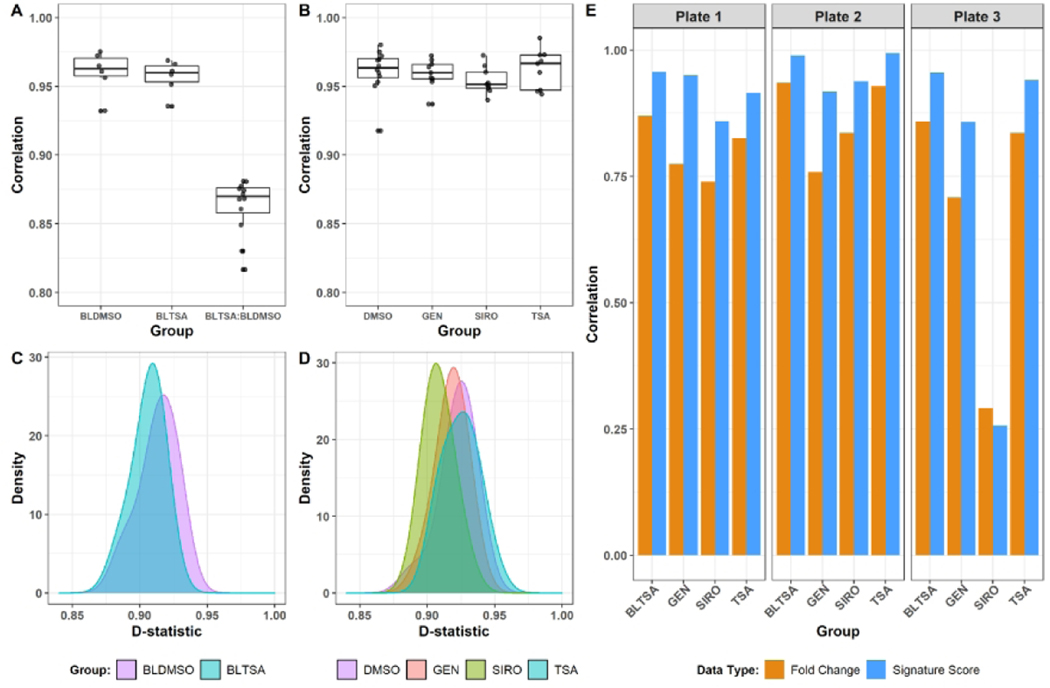
The reproducibility of the sequencing platform and experimental workflows associated with the HTTr assay were further evaluated by examining the bulk lysate reference samples and the single-concentration reference chemical treatments (genistein, sirolimus, and trichostatin A) throughout the bioinformatics pipeline. Plate-level reproducibility for each reference sample type was measured by the following metrics: the correlation of sample log2 CPM to the median log2 CPM of all replicates (Figure 3A–B), the D-statistic (Figure 3C–D) (House et al. 2017), the correlation of the DESeq2 moderated L2FCs for each plate compared to the median L2FC across plates (Figure 3E, orange bars), and the correlation of signature enrichment scores to the median signature scores (Figure 3E, blue bars). CPM correlations for replicate samples of the same type were all > 0.9 (Figure 3A, 3B). For comparison, CPM correlations between bulk lysate replicates from different treatments (e.g. BLTSA vs BLDMSO) were all lower than 0.9 (Figure 3A), demonstrating that the CPM correlation between replicates is higher than the background correlation between samples from the same cell type subject to different experimental treatments. The D-statistic has been previously used in outlier detection of vehicle control samples in TempO-Seq data, with three standard deviations below the median D-statistic previously proposed as a threshold for sample removal (House et al. 2017). Here, we applied the D-statistic to all reference and control sample types and saw that all samples passed this filtering criteria (Figure 3C, 3D), with the median D-statistic > 0.9 for all sample types reflecting high reproducibility between replicates. Lastly, we calculated the correlations of the DESeq2 moderated L2FCs and the signature enrichment scores for each reference treatment group and observed high correlations across all reference treatments except for the sirolimus replicates on plate 3 (Figure 3E). Correlations of the signature scores were higher compared to the probe-level L2FC correlations. We performed similar assessments on the UHRR and HBRR reference samples included on each plate and observed similar results with respect to overall reproducibility of the TempO-Seq assay and analysis workflow (Figure S1).
Figure 3. Reproducibility of high-throughput transcriptomics data.
(A-B) Pairwise correlations of log2 CPM values by treatment group. All correlations were calculated between individual samples of the same treatment group as indicated. The BLTSA:BLDMSO group shows the correlation between samples from different treatment conditions. (C-D) Density distribution of the D-statistic by treatment group. (E) Correlation of L2FC (orange bars) and ssGSEA signature scores (blue bars) in each of the three test plates to median L2FC and ssGSEA signature scores across all test plates. L2FC values and ssGSEA scores for each treatment group were calculated relative to the bulk lysate DMSO (for bulk lysate TSA treatments) or DMSO (for Genistein, Sirolimus, or Trichostatin A treatments) controls as described in the methods. BLDMSO = bulk lysate DMSO control; BLTSA = bulk lysate Trichostatin A; DMSO = DMSO control; GEN = Genistein; SIRO = Sirolimus; TSA = Trichostatin A.
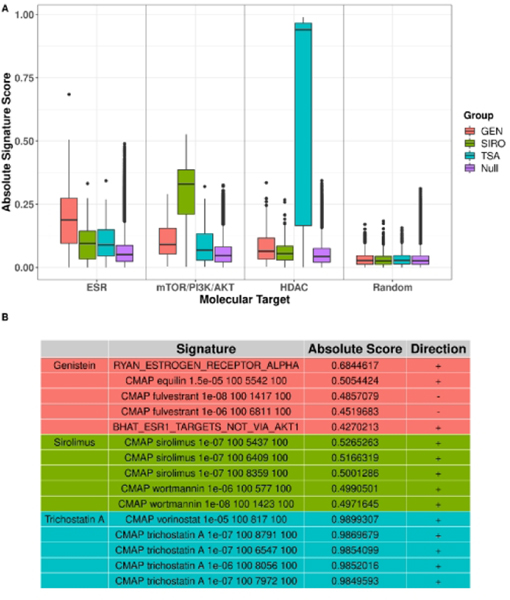
Next, we determined whether the signature scoring approach accurately identified the three reference chemicals genistein, sirolimus and trichostatin A as an estrogen receptor (ER) agonist, mammalian target of rapamycin (mTOR) inhibitor, and histone deacetylase (HDAC) inhibitor, respectively (references for mechanisms of reference chemicals are provided in Supplemental Material Table S1). This was accomplished by performing ssGSEA using gene-level DESeq2 moderated log2 fold changes computed across plates for each reference chemical treatment. ssGSEA is a modification of standard GSEA where scores are considered as the degree of enrichment of a given signature within an individual sample. Here, each sample corresponds to the rank-ordered log2 fold-changes for a single chemical treatment, and signature scores are calculated by integrating the difference between the Empirical Cumulative Distribution Functions of genes within a given signature and the genes not in the signature (Barbie et al. 2009). As a control, signature scores were also generated for the data set of 1,000 simulated chemicals derived from the null distribution (see methods). The absolute values of the signature scores were filtered to include only the gene sets associated with the ER, mTOR/PI3K/AKT, and HDAC target classes as well as the 1,000 randomly generated signatures.
As shown in Figure 4A, the median signature score for each reference chemical was greatest for its corresponding target class. Specifically, the median signature score for ER was greater for genistein compared to trichostatin A, sirolimus, and the simulated chemicals. Similarly, sirolimus had the highest scores for signatures annotated as targeting mTOR/PI3K/AKT, and trichostatin A had the highest scores for signatures annotated as targeting HDAC. Importantly, all three reference chemicals and the simulated chemicals had relatively small and similar signature score distributions for the set of 1,000 randomly generated signatures as compared to the annotated signatures. These trends were robust to the specific parameters used in our differential expression analysis, as we observed similar results for different parameter choices (Figure S2). The specificity of the signature scores for correctly identifying the molecular target was also evident when we reviewed the top 5 signatures ranked by absolute signature score for each reference chemical (Figure 4B). For genistein, the top 5 signatures included biomarker signatures for ER (Ryan et al. 2016) as well as CMAP chemical treatment signatures associated with ER activity. The top 5 signatures for sirolimus included CMAP signatures for sirolimus as well as wortmannin, which is a known inhibitor of PI3K (Arcaro and Wymann 1993). The top signatures for trichostatin A included CMAP signatures for trichostatin A, as well as vorinostat, which are both hydroxamate-based HDAC inhibitors (Xu et al. 2007).
Figure 4. Signature set enrichment of reference chemical treatments.
(A) Distributions of absolute ssGSEA signature scores, calculated from the DESeq2 moderated log2 fold changes across the three test plates, for specific molecular target signatures across each treatment group. (B) Table of the top 5 highest ranked signatures by absolute signature score for Genistein (red), Sirolimus (green), or Trichostatin A (blue). GEN = Genistein; SIRO = Sirolimus; TSA = Trichostatin A; NULL = 1000 simulated chemicals derived from the null distribution (see methods).
Transcriptional Perturbations in Response to Chemical Treatment Reflect Mechanism of Action
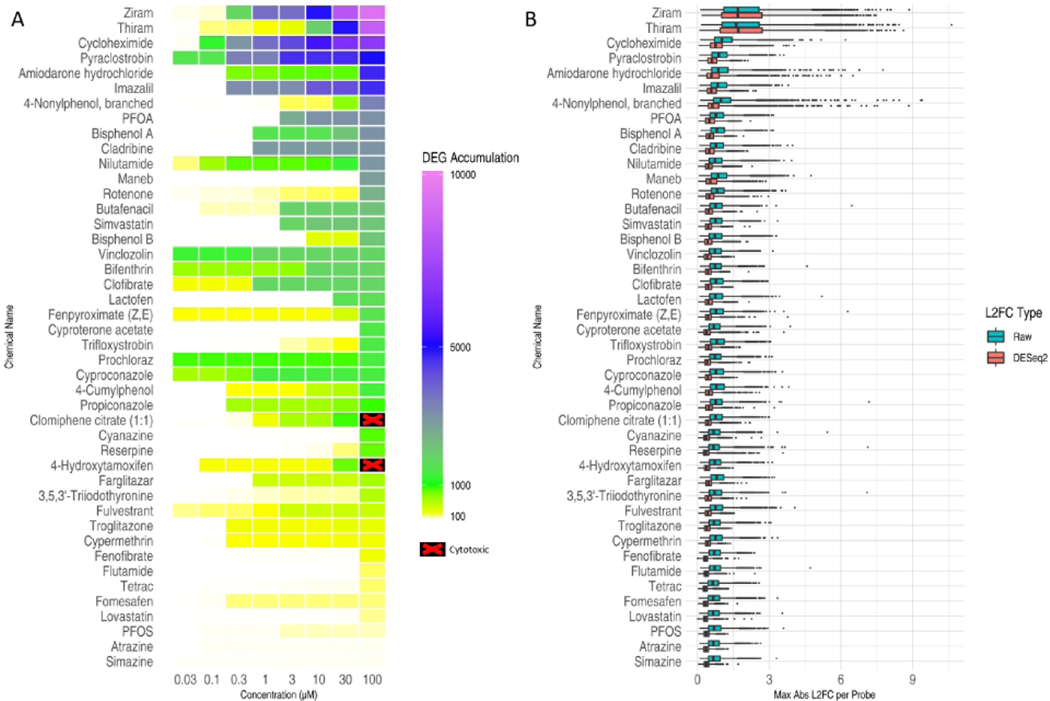
We first assessed overall transcriptional perturbation resulting from each test chemical by: 1) computing the accumulation of differentially expressed genes (DEGs) for each combination of chemical and concentration based on DESeq2 analysis p-values from pairwise comparisons (Figure 5A) and 2) plotting the distribution of absolute maximum L2FC and DESeq2 moderated L2FC observed at any test concentration of a chemical (Figure 5B). Accumulation scores across the chemicals suggest a diversity in both chemical potency and specificity of transcriptional response. Chemicals such as ziram and cycloheximide showed relatively steady escalation in transcriptional perturbation through the assayed concentrations, with a DEG accumulation score in excess of 1,000 and 10,000 at 1 and 10 μM, respectively. On the other hand, chemicals such as 4-nonylphenol and maneb achieved an accumulation score in excess of 1,000 only at the highest concentration assayed. In contrast, a subset of chemicals, such as simazine, appear relatively transcriptionally inert in MCF7 cells with a DEG accumulation score less than 10 even at the highest concentration. Overall, the magnitude of gene expression changes observed across the chemical set was small, with few genes exceeding a L2FC of ≥2-fold at any test concentration. Exceptions include chemicals with the highest DEG accumulation scores (ziram, thiram, cycloheximide, pyraclostrobin, amiodarone hydrochloride, and 4-nonylphenol, branched) where gene expression changes of ≥2-fold were observed with greater frequency. Of note, some chemicals whose known molecular targets are highly expressed in MCF7 cells (e.g. 4-hydroxytamoxifen, clomiphene citrate (1:1), 4-cumylphenol which target ESR1; lovastatin, simvastatin which target HMGCR) did not produce large-magnitude changes in expression for the majority of genes at 6 hours post treatment.
Figure 5. Transcriptional perturbations produced by chemical treatments.
(A) DEG accumulation scores for each test chemical (rows) at each concentration (columns). The color of each cell indicates the number of probes that were significantly differentially expressed at or below the corresponding concentration. Chemical concentrations that were masked from transcriptomic analysis due to cytotoxicity are shown as black with a red ‘X’ on the heatmap. (B) Distribution of gene response effect sizes. For each chemical, the maximum absolute L2FC value was computed for each probe across all concentrations and the distribution for all probes is represented as a boxplot. Blue boxplots indicate the distribution using “raw” L2FCs computed directly from mean log2(CPM) values. Red boxplots indicate the distributions of moderated L2FCs returned by DESeq2 analysis of raw counts across all plates.
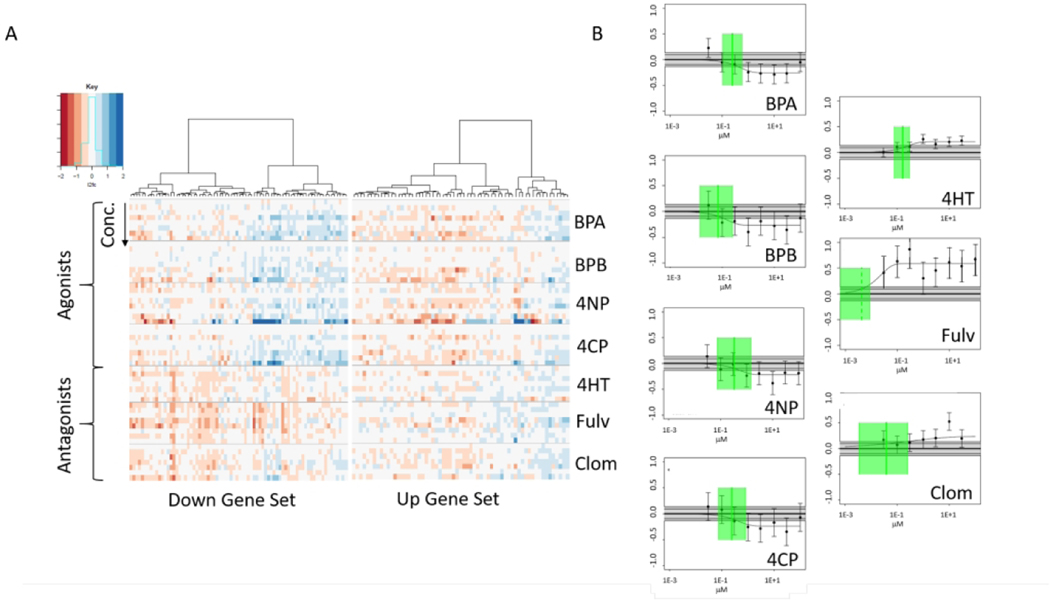
Cellular responses to environmental challenges (such as chemical exposures) involve the coordinated regulation of ensembles of transcripts to facilitate compensatory alterations in cellular function (Gaiteri et al. 2014; Subramanian et al. 2005; van Dam et al. 2018). Therefore, to better understand transcriptomic responses to chemical challenge at a mechanistic level, we used a signature-matching approach to pair gene signatures from the CMAP database to gene expression profiles generated in this experiment. The result of matching a gene signature for fulvestrant, as derived from the CMAP database, against the L2FC data for a subset of estrogenic and antiestrogenic chemicals are shown in Figure 6. The heatmap in Figure 6A is an illustrative example of the L2FC data and Figure 6B illustrates signature-level concentration-response results. This example uses one CMAP signature for a fulvestrant treatment (1E-8 M in MCF7 cells) represented by the 100 most up-regulated and 100 most down-regulated genes. Fulvestrant is an estrogen receptor (ER) antagonist, so one would expect other ER antagonists to have similar responses, and agonists to have opposite responses, at least in some respects. The heatmap of the CMAP downregulated genes (Figure 6A, left) shows that the antagonists 4-hydroxytamoxifen (4HT) and clomiphene citrate (1:1) (Clom) produce similar responses to fulvestrant (Fulv) among this set of genes. In addition, the 4 ER agonists (bisphenol A (BPA), bisphenol B (BPB), 4-nonylphenol (4NP) and 4-cumylphenol (4CP)) show a pattern of upregulation for the same genes that are downregulated with the antagonists. The CMAP fulvestrant upregulated genes (Figure 6A, right) shows less differentiation between the agonists and antagonists. Figure 6B shows signature-level concentration-response plots for the ER agonists and antagonists using the CMAP MCF7 fulvestrant 1E-8 M signature. Clear concentration-dependent enrichment scores for this signature and responses in opposite directions for agonists vs antagonists were observed. An important point is that the signal to noise is relatively low for all the chemicals except fulvestrant.
Figure 6. Concentration-dependent transcriptomic perturbations of estrogen receptor target genes.
(A) Heatmaps showing the log2-fold change (log2fc) values for genes in an example signature (CMAP fulvestrant 1e−08 1417 100. The naming convention for the CMAP signatures include the chemical name, the concentration in molar units, a sequential index and the number of genes.). The left-hand heatmap shows the most significantly down-regulated 100 genes and the right-hand panel shows the 100 most significantly up-regulated genes for this fulvestrant CMAP sample. Each horizontal block shows the results for the 8 separate concentrations of a given chemical where concentration increases from top to bottom. The first 4 chemicals are ER agonists and the final 3 are ER antagonists. Chemical abbreviations are: BPA: bisphenol A, BPB: bisphenol B, 4NP: 4-nonylphenol, branched, 4CP: 4-cumylphenol, 4hT: 4-hydroxytamoxifen, Fulv: fulvestrant, Clom: clomiphene citrate (1:1). (B) Signature-level concentration-response data for the same chemicals for this signature. Each panel shows the data points with 95% confidence intervals based on the fitting model error estimate, the winning concentration-response curve, the noise band (gray band spanning zero), the BMD (green vertical line) and its95% confidence interval (green box).The Y-axis is in arbitrary response units. Note that the response for the agonists is negative and for the antagonists is positive. All chemicals except 4-cumylphenol have a continuous hitcall > 0.9.
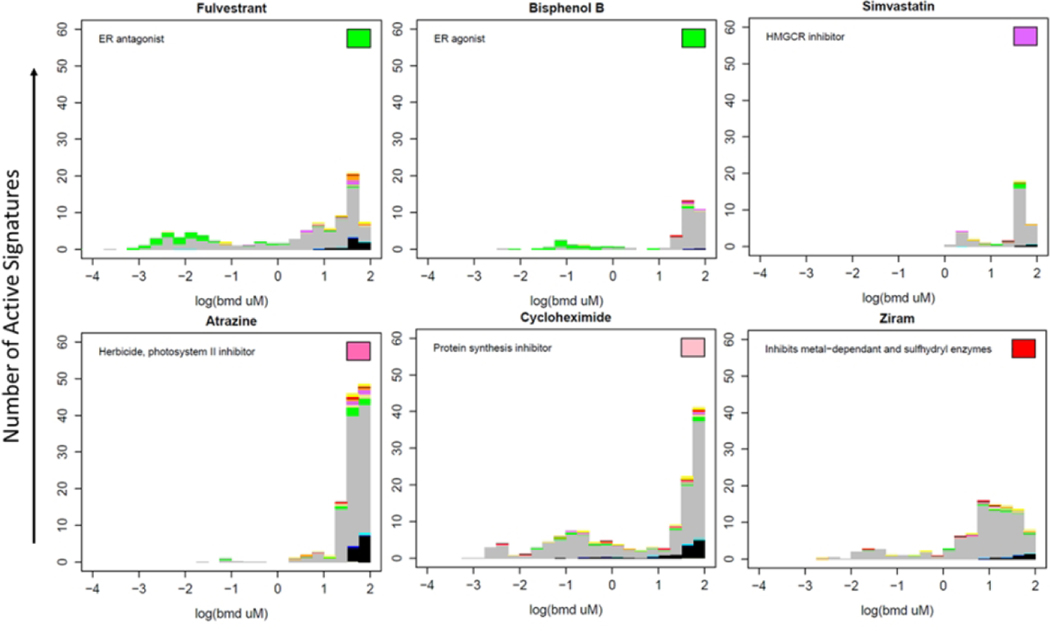
The concentration-dependent activity of six exemplar chemicals across thousands of signatures is illustrated in Figure 7. Each chemical was evaluated against 7,586 signatures, resulting in calls of active or inactive, and a potency (BMD) if active for each chemical × signature pair. Each histogram in Figure 7 shows the distribution of BMDs of the active signatures for a particular chemical. A selected set of signature target classes are indicated by colors other than gray, described in the figure legend. The color box in the top right hand of each panel indicates the target class of the chemical if there is a known specific human target. The six chemicals shown here illustrate several features seen across the larger chemical set. For instance, fulvestrant and bisphenol B are both active against the estrogen receptor (ER), indicated by green. The most potent signatures for these chemicals are associated with ER activity indicated by the green color in the histogram at low concentrations. Note that the color scheme does not distinguish agonist from antagonist mode. Simvastatin, an HMGCR inhibitor (indicated by purple), has among its most potent activities an HMGCR-related signature. Almost all chemicals, including those illustrated in Figure 7, show a typical large burst of activity at high concentrations which appears to be non-specific to any molecular target. A pair of highly toxic chemicals, cycloheximide and ziram show an extended tail of activity to lower concentrations without an obvious single target class. These chemicals also showed the broadest perturbation of the transcriptome in terms of the number of genes significantly differentially expressed at any test concentration (Figure 5). In all cases, the activity of the chemicals against the random gene signatures largely occurs at concentrations above 10 μM, in the non-specific burst region. Atrazine and simvastatin show the most typical behavior of having significant activity in the high concentration region and little to no activity below 1 μM.
Figure 7. Assigning putative molecular targets based on connectivity with CMAP chemicals and signatures. BMD distribution histograms for example chemicals.
Each active pathway is represented by an element of the histogram at the corresponding BMD value. Colors in the bar chart indicate the signature target classes: green = estrogen, beige = thyroid, blue = CYP P450, tan = ion channel, purple = HMGCR/cholesterol, orange = mitochondria, red = cell stress, yellow = PPAR, black = random, gray = other. The color of the rectangle in the top right indicates the target class of the chemical. Of note are (1) the burst of activity at high concentrations; (2) most of the stress or random activity being observed at high concentrations in a burst; (3) the estrogenic chemicals showing estrogenic pathway activity at the lower concentrations (specificity). The number of signatures with continuous hitcalls >0.5 and number of signatures tested is listed in each panel.
Transcriptomic BPACs Recapitulate HTS Screening Results
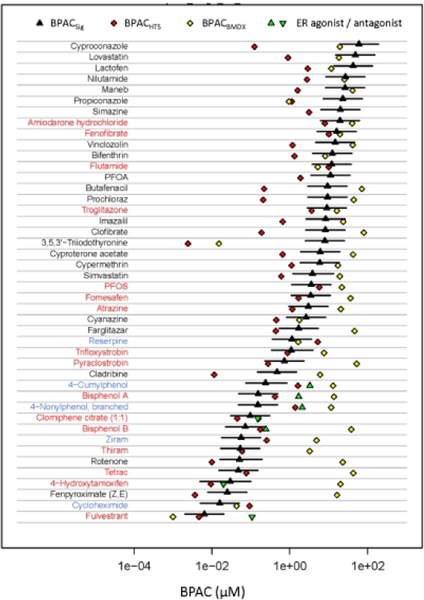
One key use of the HTTr data is defining an in vitro biological pathway-altering concentration (BPAC) for each chemical. The BPAC is a concentration below which there is little or no observed bioactivity. Figure 8 shows BPACSig as a black triangle, with confidence intervals being the lower and upper confidence bounds for the signature defining the BPACSig. The most sensitive signature potency from concentration-response modeling with BMDExpress was also used to define an in vitro BPAC (BPACBMDX, yellow diamonds) based on a workflow described in the NTP recommended approach to genomic dose-response modeling (see methods) (NTP 2018). We compared these results to previously derived in vitro BPACs from the ToxCast HTS data set. The 44 chemicals analyzed here were previously screened in up to 1045 HTS assays. The ToxCast BPAC (BPACHTS, indicated by a red diamond) is the lower 5th percentile of the active AC50 values for assays that passed a series of quality filters (Paul Friedman et al. 2020). The names of the chemicals are color-coded based on the comparison between BPACSig and BPACHTS: red indicates that the BPACHTS is within the BPACSig confidence intervals; black indicates that BPACHTS is more potent than BPACSig; and blue indicates that BPACSig is more potent than BPACHTS. For most test chemicals, the BPACBMDX values are above those from the other two methods, often by at least an order of magnitude.
Figure 8. Comparison of transcriptomic and HTS-derived BPACs.
The black triangle and confidence intervals correspond to BPACSig and associated upper and lower 95% confidence bounds of BPACSig respectively; yellow diamonds correspond to BPACBMDX; red diamonds correspond to BPACHTS; green up and down triangles indicate potencies from the EPA ER Pathway Model (https://www.epa.gov/endocrine-disruption/endocrine-disruptor-screening-programedsp-estrogen-receptor-bioactivity). The names of chemicals are colored red if the BPACHTS is within the BPACSig confidence limits; colored black if the BPACHTS is lower than BPACSig; and colored blue if the BPACHTS is above the BPACSig. The BPACHTS is the lower 5th percentile of the active AC50 values for assays that passed a series of quality filters (Paul Friedman et al. 2020).
Overall, BPACHTS and BPACSig are in better agreement (RMSE = 1.09; cor = 0.62) than BPACHTS and BPACBMDX (RMSE = 1.84; cor = 0.45). However, there are some notable chemicals for which BPACHTS is markedly more potent than BPACSig. In most of these cases BPACHTS is also more potent than BPACBMDX. The majority of these cases can be explained by the use of ToxCast assays for the specific target of the chemical, which is not active/expressed in MCF7 cells. There are twelve chemicals where the BPACHTS is more than 10x below the BPACSig as listed in Table 3. The most extreme case is 3,5,3’-triiodothyronine, the natural hormone T3, which is the ligand for the thyroid hormone receptor. The most potent activity of T3 is against the alpha and beta forms of the receptor. Although the alpha form is expressed in MCF7 cells, the baseline expression level is relatively low. There are several pan-cytochrome P450 (CYP) inhibitors (cyproconazole, butafenacil, prochloraz, imazalil and propiconazole) which are active against a set of cell-free enzyme activity assays for a variety of human and rat CYP targets. Cladribine is a DNA synthesis inhibitor whose most potent targets are DNA repair enzymes and oxidative stress pathways. The remaining chemicals are lovastatin, clofibrate, maneb, lactofen, and vinclozolin whose most potent assay targets in ToxCast are not related to the known targets of those chemicals.
Table 3:
Chemicals with BPACHTS 10x lower than BPACSig.
| Chemical | Ratio (BPACHTS / BPACSig) | Target | Potent ToxCast Targets | MCF7 Expression level (log2 CPM) |
|---|---|---|---|---|
| 3,5,3’-Triiodothyronine | 0.003 | Thyroid hormone receptor | Thyroid hormone receptors THRA, THRB | THRA=2.6 THRB=3.0 |
| Cyproconazole | 0.002 | Pan-cyp inhibitor | Cell-free assays for CYP2A1, CYP2C9, CYP2C13, CYP2C19 | CYP2A, CYP2B, CYP2C all <1 |
| Lovastatin | 0.02 | HMGCR | Multiple cell-free and cell based targets but not HMGCR | HMGCR=7.4 |
| Prochloraz | 0.02 | Pan-cyp inhibitor | Cell-free assays for CYP2A2, CYP2C19, CYP2B1, CYP2C11 | CYP2A, CYP2B, CYP2C all <1 |
| Clofibrate | 0.02 | PPARα | Activity against cell-free assays for protein phosphatases PTPN2, PTPN12, PTPRF | PPARA=2.6 |
| Butafenacil | 0.02 | Protoporphyrinogen oxidase (PPO) inhibition | Cell-free assays for CYP2A2, CYP2C19, CPP2B1, CYP2B6, CYP2C11 | CYP2A, CYP2B, CYP2C all <1 |
| Cladribine | 0.02 | DNA Synthesis inhibitor | DNA repair, oxidative stress assays | |
| Propiconazole | 0.05 | Pan-cyp inhibitor | Cell-free assays for CYP2A2, CYP2B1, CYP2C19, CYP2C11, CYP2C9 | CYP2A, CYP2B, CYP2C all <1 |
| Maneb | 0.06 | Inhibition of metal-dependent and sulfhydryl enzyme systems | Cell-free assays for IRAK4, PTPN4, PTPN9 | IRAK4=1.8, PTPN4=3.9, PTPN9=4.1 |
| Lactofen | 0.07 | Protoporphyrinogen oxidase (PPO) inhibition | Cell-based assays for HMGCS2, PPARα | PPARA=2.6, HMGCS2<1 |
| Imazalil | 0.08 | Pan-cyp inhibitor | Cell-free assays for PTPN11, CYP2B1, CYP2A2, CYP3A4, CYP2D2, CYP2C19 Cell-based assays for CYP1A2, CYP1A1 | CYP2A, CYP2B, CYP2C, CYP1A2, CYP2D, CYP3A all <1 CYP1A2=2.3, PTPN11=8.8 |
| Vinclozolin | 0.08 | Lipid Peroxidation | Cell-free and cell-based assays for AR Cell-free assay for TSPO Cell-based assays for CYP1A1, CYP1A2 | CYP1A2 <1 CYP1A2=2.3, TSPO=2.4, AR=3.8 |
Discussion
NAMs that inform chemical hazard and mechanism are needed to accelerate the pace of chemical risk assessments. We objectively evaluated the performance of the TempO-Seq human whole transcriptome assay for HTTr analysis (Harrill et al. 2019) in MCF7 cells using 44 ToxCast chemicals and 3 reference chemical treatments. We developed scalable and robust laboratory and bioinformatics workflows aimed at screening hundreds of environmental chemicals in concentration-response to provide two key pieces of information: 1) the potency threshold where chemicals perturb cellular biology as measured by changes in gene expression (i.e. BPACs) and 2) putative mechanisms of chemical toxicity. Overall, we found that the TempO-Seq human whole transcriptome assay was highly reproducible and gene signature scores used for concentration-response modeling yielded transcriptional BPACs that were closely aligned with in vitro BPACs determined from ToxCast HTS assays.
Key aspects of the TempO-Seq technology that enable whole transcriptome HTTr screening include compatibility with cell lysates without the need for additional RNA purification, and the ability to generate multiplexed sequencing libraries from picrogram amounts of RNA where each read can be tracked to an individual sample (Yeakley et al. 2017). The number of samples in this study (1,134) is small in comparison with traditional HTS screening (Pereira and Williams 2007; Szymanski et al. 2012) but quite large in comparison to the number of samples associated with even the most complex in vivo toxicology studies involving whole transcriptome profiling (Gong et al. 2014a; Waring et al. 2001). Unlike typical RNA-Seq workflows, our TempO-Seq assay implementation bypasses RNA purification, quantification and concentration adjustment steps to achieve high sample throughput. However, this also eliminates the option to exclude low quality samples prior to preparing pooled sequencing libraries. As such, our bioinformatics pipeline includes multiple QC filters to detect and remove data from low-quality samples in lieu of typical pre-sequencing quality checks. Several of these QC metrics evaluate the distribution of read counts in TempO-Seq data based on the observation that degraded or low-input RNA-Seq samples are often associated with characteristic changes in sample “complexity”: i.e. a lower number of detected genes and a higher number of duplicative reads (Adiconis et al. 2013). We observed a small number of samples with distributional metrics (NSig80, GiC) that differed substantially from most samples with 97% of samples being retained for analysis. We anticipate that these QC metrics can be extended to HTTr screens using cell types other than MCF7. However, differences in the number and relative abundance of genes expressed in diverse cell types, or changes in the probe sequences used in future versions of the TempO-Seq assay, may necessitate study-specific adjustment of QC thresholds. This factored into the decision to incorporate IQR-based QC thresholds for the NCov5, NSig80 and Gini metrics into the bioinformatics workflow as this calculation is simple to implement, robust, and flexible for different experimental contexts.
Important criteria for the incorporation of NAMs into chemical risk assessments include demonstrations of assay reproducibility and accuracy (Bal-Price et al. 2018) (A8382_Bal-Price; OECD 2014). In this experiment, an objective evaluation of the technical reproducibility of the TempO-Seq assay was performed using bulk lysate reference samples and the two reference RNA standards located in each assay plate. Unlike test samples, gene expression measurements in these samples would not be influenced by potential heterogeneity in the biological state of MCF7 cells across different cultures or the mechanics of chemical dispensing; rather variations would be strictly associated with conduct of the TempO-Seq assay. Using this approach, we observed that the technical reproducibility of the TempO-Seq assay was high, as assessed using correlation of L2FC values, correlation of signature scores and the D-statistic (Figure 3A, 3C, 3E; Figure S1) (House et al. 2017).
In addition to technical reproducibility, we evaluated the biological reproducibility of the MCF7 in vitro test system using reference chemical treatments. These reference treatments (i.e. trichostatin A, sirolimus, genistein) were selected based on the expression of molecular targets for each chemical in MCF7 cells (i.e. HDAC, mTOR/PI3K/AKT, ER) and the availability of transcriptomic signatures associated with these targets in the CMAP database and other public gene set collections. Variations in gene expression measurements observed in the reference treatment samples could be due to heterogeneity in independent MCF7 cell cultures, the mechanics of chemical dispensing, or technical variability in the TempO-Seq assay. We observed that the biological reproducibility of the MCF7 in vitro test system was high in most cases, as assessed using the same metrics employed for evaluating technical reproducibility (Figure 3B, 3D). This was notable given previously published reports of cellular and phenotypic heterogeneity observed in MCF7 cells from the same batch in response to chemical treatment (Kleensang et al. 2016). Responses to trichostatin A and genistein were highly correlated across all 3 plates. The reason for the comparatively lower correlations of L2FC values and ssGSEA signature scores for sirolimus in plate 3 as compared to across plate median values (Figure 3E) is unknown, but may be due to heterogeneity in the response of MCF7 cells to this particular chemical as compared to the culture batches on the other two plates or technical variability associated with the TempOSeq assay or sample preparation steps for these particular samples. In spite of this technical variability, the combined analysis of all three plates demonstrated clear enrichment for mTOR/PI3K/AKT signaling pathways in our test system in response to sirolimus (Figure 4A, 4B) regardless of whether plate effects were directly modeled by DESeq2 (Supplementary Figure 2), demonstrating that our workflow is robust to occasional variability on a single plate.
The availability of previously established transcriptomic signatures for the reference treatment chemicals allowed us to evaluate the accuracy of the TempO-Seq assay in terms of identifying ‘correct’ biological responses. Using the signature scoring approach, strong associations were observed between the gene expression profiles measured by TempO-Seq and gene expression signatures of chemicals known to affect the same molecular targets as the reference chemicals (Figure 4). For example, the top five high-scoring signatures matching genistein, a known ER agonist, consisted of estrogenic chemicals from the CMAP database, estrogen-related gene sets from MSigDB and a well-characterized gene expression biomarker for ERα (Ryan et al. 2016). Likewise, the top signature matches for sirolimus and trichostatin A were either the same chemicals or those with similar biological activity from the CMAP database. These trends were robust to specific parameters used in our differential expression analysis, as we observed similar results for different parameter choices (Figure S2). By comparison, absolute signature score distributions were lower for: 1) reference chemicals against signatures for mismatched targets, 2) null chemicals generated by randomly permuting the observed L2FC matrix, and 3) randomly generated gene signatures with the same gene number and co-occurrence frequency as the real gene signatures. In each case, the absolute signature scores for a reference treatment queried against its matching target gene signature subsets were markedly higher than all other query and subject combinations. Thus, the enriched biological signals observed in the MCF7 cells appear to be non-random in nature, and accurate with respect to identification of characteristic gene expression responses following molecular target activation.
In this study, we compared two different approaches for deriving transcriptional BPACs. The first was an established workflow for concentration-response modeling of gene expression data based on probe-level curve fitting and mapping to gene sets (Harrill et al. 2019; NTP 2018) using the BMDExpress software package (Phillips et al. 2019). The second was a novel workflow that aggregates gene level information into signature scores (Barbie et al. 2009) prior to concentration-response modeling with an updated version of the tcpl R-package; tcplfit2 (Sheffield et al. in press). When comparing BPACs from the two approaches, we noted many instances where BPACBMDX was notably higher than BPACSig. We hypothesize that these potency differences may be driven by two factors: 1) the low magnitude of gene expression changes observed for a majority of chemicals in our test set (Figure 5B) and 2) the manner in which the respective methods take into account subtle changes in gene expression that may be associated with true biological signal. The low magnitude gene expression changes we observed in MCF7 cells, including responses to known estrogenic and anti-estrogenic chemicals, are consistent with previous transcriptomics studies in this cell type (Gong et al. 2014b; Lecomte et al. 2019; Ryan et al. 2016; Stanislawska-Sachadyn et al. 2015) and thus appear to be an inherent feature of this in vitro model. The BMDExpress approach models the response for each probe independently. A set of filtering criteria is then applied to remove probes where the response does not surpass a certain threshold, where the BMD is not within the bounds of the tested concentration range or where the BMD is associated with a large degree of uncertainty. Any individual probe with a fit not meeting these criteria would not be used in the signature mapping step used to define the BPACBMDX. In contrast the signature modeling approach does not involve probe/gene level curve-fitting, but instead aggregates signals from multiple genes into a signature score prior to concentration-response modeling. Thus, low-magnitude gene responses that are coordinately regulated are included in determination of BPACSig and uncertainty in the potency estimate is calculated at the signature as opposed to the gene level.
The behavior of coordinately expressed genes has received much attention in the biological sciences (Gaiteri et al. 2014; Singh et al. 2018; van Dam et al. 2018). However, to our knowledge, methods for addressing this behavior in the context of concentration-response modeling and next generation risk assessment have rarely been explored. In an in vivo toxicity study in multiple tissues, Dean et al. implemented a modified BMDExpress workflow where a GSEA scoring method that takes into account expression of all genes (with no pre-filtering) was used to exclude gene sets where there was no evidence of a coordinated transcriptional response from calculation of the overall transcriptional POD (Dean et al. 2017). They observed that transcriptional PODs calculated in this manner were within one order of magnitude of apical PODs. Further, in an in vivo study in the mouse liver, Parfett et al. observed that significant changes in coregulated gene sets as assessed via concentration-response modeling of a ‘cumulative expression difference’ score could be observed at lower doses / test concentrations than statistically significant changes in individual genes (Parfett et al. 2013). This is consistent with the trend we observe in the present study of BPACBMDX generally being less sensitive than BPACSig. The present data support that aggregating signal prior to concentration-response modeling can provide more conservative estimates of chemical bioactivity as compared to mapping statistically significant changes in individual genes to signatures. It should be noted that our observations regarding the comparison of BPACs derived from different methods is specific to the current study. More extensive comparison of methods for BPAC determination using a larger number of chemicals, test conditions and cell lines would be required to make definite conclusions regarding performance of the modeling approaches.
Aggregation of signal into gene signatures prior to concentration-response modeling yielded potency estimates for bioactivity that were well-aligned with those derived from the ToxCast suite of HTS assays (i.e. BPACHTS). In contrast, BPACBMDX was often greater than BPACHTS, in some cases by several orders of magnitude. This suggests that that BPACSig values are more reflective of the in vitro biological activity of the test chemicals as compared to BPACBMDX, if using the ToxCast assay suite as the benchmark. For example, we were able to observe signatures associated with the known molecular targets of some of the test chemicals, particularly estrogens, as being among the most sensitive using the signature scoring method (e.g. fulvestrant, bisphenol B, Figure 7). The estrogen response signatures were affected prior to the onset of what appears to be non-specific effects across many different signature types at higher test concentrations. By design, some of the chemicals in our test set have primary molecular targets that are either not expressed in MCF7 cells or not expressed in humans at all (Table 1). Therefore, transcriptional perturbations associated with activity at a specific molecular target was not expected for every chemical. Rather, we hypothesized that some chemicals would produce transcriptional effects associated with promiscuous bioactivity and general cell stress. This appears to be the case, as the distribution of signature BMDs for each of the test chemicals was skewed to the right, regardless of the presence or absence of mechanistically-relevant pathway hits at lower test concentrations. For example, the signature response of atrazine (Figure 7) appears to be largely associated with transcriptional perturbations of many different signature types at the upper end of the tested concentration range. The increased frequency of signature BMDs in the upper end of the tested concentration range is reminiscent of the cytotoxicity burst that is observed in ToxCast HTS screening data as test concentrations increase (Judson et al. 2016). The fact that randomly constructed signatures (Figure 7, black) are also being activated at concentrations greater than ~10 μM support that these transcriptional responses may be associated with non-specific bioactivity or cellular stress. However, we contend that in the absence of evidence of a clear, molecular target-driven transcriptional response, transcriptional changes associated with this non-specific burst are relevant for establishing BPACs for use in NAMs-based chemical risk assessment.
For chemicals where BPACHTS was lower than either BPACBMDX or BPACSig, the difference was attributable to the nature of the response within the ToxCast assay suite. Among the most sensitive HTS hits for cyproconazole, propiconazole, butafenacil and prochloraz were cell-free assays for cytochrome P450 inhibition (CYP2A1, CYP2B, CYP2C9, CYP2C1, CYP2C19), a type of biological activity unlikely to be associated with transcriptional changes in MCF7 cells that lack basal expression of these enzymes. For other chemicals whose targets are expressed in MCF7 cells (i.e. clofibrate, PFOA, lovastatin), the difference between BPACHTS and BPACSig or BPACBMDX may be due to the hypothesized increased sensitivity of cell-free assay systems as compared to cell-based assay systems, or an insufficient amount of time (6 hours) for transcriptional effects associated with the annotated bioactivity of the test chemical to manifest.
Of note, in this work we have used concentration-response modeling of TempO-Seq data in an objective way to identify the threshold where perturbations in gene expression begin to occur. In the context of this study, we make no presumptions regarding whether or not changes in gene expression observed for any given chemical are adverse, adaptive or benign in nature. A previous study by Paul-Friedman et al. concluded that in vitro bioactivity estimates derived from ToxCast assays (e.g. BPACHTS) could be used as a reasonable lower bound estimate of in vivo adverse effect levels (Paul Friedman et al. 2020). In the previous study of 448 substances, 89% of substances had administered equivalent doses (AEDs) that were less than traditional PODs determined from an extensive database of mammalian adverse effect values. Given that in the present study, BPACSig values were well-aligned with BPACHTS values, our results suggest that BPACSig would likewise provide reasonable lower bound estimates of in vivo adverse effect values. However, additional studies are needed to rigorously test this hypothesis given that our present study evaluates only a small number of chemicals (n = 44) in a single human-derived cell type at a single time point. In vitro to in vivo extrapolation (IVIVE) and comparison to in vivo effect values as described in Paul-Friedman et al. is of interest in the regulatory community for tiering and prioritization of large collections of chemicals and will be an important topic of study when larger scale HTTr data sets have been generated.
In summary, we have established robust experimental workflows for HTTr screening with a low fail rate at the individual sample level and a flexible and scalable open-source bioinformatics pipeline for sequence alignment, count generation, and QC flagging that can be applied to future HTTr screening studies. The use of reference samples and reference chemical treatments to evaluate the technical reproducibility of the transcriptomic assay and the biological reproducibility of the culture model can be carried forward to future large-scale HTTr screening studies. We also conclude that the signature modeling approach combined with curated and annotated signature collections can be used to identify putative molecular targets underlying transcriptional bioactivity and provide potency estimates for perturbation of cellular biology in the absence of large changes in gene expression. The present proof-of-concept study uses MCF7 cells and a single exposure duration. However, no single in vitro cell model can capture the diversity of biological responses that may occur in humans following exposure to environmental chemicals. Likewise, no single snapshot in time is likely to provide a complete characterization of the biological response of a cell to chemical exposure. In the future, we intend to apply the laboratory and bioinformatics workflows developed here to multiple, complementary human-derived cell types in order to increase the number of biological targets, pathways and temporal responses evaluated beyond those that are present in the MCF7 cell model or described in the present manuscript (Thomas et al. 2019). The experimental and bioinformatic workflows described here can be used across diverse cell models and will increase the pace of hazard evaluation for thousands of chemicals that may be found in the environmental and inform next generation risk assessments.
Supplementary Material
Acknowledgements
The authors would like to thank Dr. Matthew Martin and Dr. Agnes Karmaus for contributions during the scoping and planning phases of this work. The authors would also like to thank Daniel Hallinger, Terri Fairly, Sandra Roberts and David Murphy for operations support activities during the conduct of this research. The authors would also like to thank colleagues at BioSpyder, Inc. Joanne Yeakley, Bruce Seligmann, Pete Shepherd, Joel McComb, Milos Babic, Kyle LeBlanc and Garrett McComb for conduct of the TempO-Seq assays and informative discussions regarding the technology. The authors would also like to thank Drs. Leah Wehmas, Chris Corton, Scott Auerbach, John Cowden, Kimberly Slentz-Kesler and Maureen Gwinn for their insightful comments during review of this manuscript.
Funding
Funding for this work was provided by the US EPA Office of Research and Development.
List of abbreviations
- 384PP
384-Well Polypropylene Microplates
- 4NP
4-Nonylphenol
- 4CP
4-Cumylphenol
- AIC
Akaike Information Criterion
- AR
androgen receptor
- BPA
Bisphenol A
- BPB
Bisphenol B
- BLDMSO
Bulk Lysate DMSO-treated
- BLTSA
Bulk Lysate TSA-treated
- BMC
Benchmark Concentration
- BMD
Benchmark Dose
- BMDL
BMD Lower Bound
- BMDU
BMD Upper Bound
- BMR
Benchmark Response
- BMDX
BMDExpress
- BPAC
Biological Pathway Altering Concentration
- Clom
Clomiphene Citrate (1:1)
- CMAP
Connectivity Map
- CPM
Counts per Million
- DEG
Differentially Expressed Gene
- DMEM
Dulbecco’s Modified Eagles Medium
- DMSO
Dimethyl Sulfoxide
- ER
estrogen receptor
- ESR1
Estrogen receptor alpha
- FMR
Fraction of Reads Uniquely Mapped to Probes
- FrVC
Fraction of Viable Cells
- Fulv
Fulvestrant
- GiC
Gini Coefficient
- GSEA
Gene Set Enrichment Analysis
- HBRR
Human Brain Reference RNA
- HDAC
Histone Deacetylase
- HI-FBS
Heat-Inactivated Fetal Bovine Serum
- HMGCR
3-Hydroxy-3-Methylglutaryl-CoA Reductase
- HTP
High-Throughput Profiling
- HTS
High-Throughput Screening
- HTTr
High-throughput transcriptomics
- hWTv1
Human Whole Transcriptome version 1
- IQR
Inter-Quartile Range
- L2FC
Log2 Fold-Change
- mTOR
Mammalian Target of Rapamycin
- NAMs
New approach methodologies
- Ncov5
Number of Probes with at least 5 Reads
- NMR
Number of Uniquely Mapped Reads
- Nsig80
Number of Probes Capturing Top 80% of Signal
- NTP
National Toxicology Program
- NX
Normalized Expression Levels
- PBS
Phosphate Buffered Saline
- PI
Propidium Iodide
- POD
Point of departure
- PPARα
peroxisome proliferator activating receptor alpha
- PPARγ
peroxisome proliferator activating receptor gamma
- PSG
Penicillin-Streptomycin-Glutamine
- QC
Quality Control
- ssGSEA
Single-Sample Gene Set Enrichment Analysis
- tcpl
ToxCast Pipeline
- TempO-Seq
Templated Oligo with Sequencing Readout
- THRA
Thyroid hormone receptor
- TSA
Trichostatin A
- UHRR
Universal Human Reference RNA
Footnotes
Competing financial interests: The authors declare they have no actual or potential competing financial interests
Disclaimer: The views expressed in this article are those of the authors and do not necessarily represent the views or policies of the U.S. Environmental Protection Agency.
Supplementary Information
The source code used to conduct analyses in this work can be found https://github.com/USEPA/httrpl_pilot. Raw read data for all samples (FASTQ format) and probe set manifest can be found under GEO data series GSE162855. Other data files described in the Supplementary Material are located on FigShare (DOI: 10.23645/epacomptox.13368914).
Conflict of Interest
The authors declare no conflict of interest. This manuscript has been reviewed by the Center for Computational Toxicology and Exposure, Office of Research and Development, U.S. Environmental Protection Agency, and approved for publication. Approval does not signify that the contents reflect the view of the Agency, nor does mention of trade names or commercial products constitute endorsement or recommendation for use.
References
- Adiconis X, Borges-Rivera D, Satija R, DeLuca DS, Busby MA, Berlin AM, Sivachenko A, Thompson DA, Wysoker A, Fennell T et al. 2013. Comparative analysis of rna sequencing methods for degraded or low-input samples. Nat Methods. 10(7):623–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akaike H 1974. New look at statistical-model identification. Ieee T Automat Contr. Ac19(6):716–723. [Google Scholar]
- Arcaro A, Wymann MP. 1993. Wortmannin is a potent phosphatidylinositol 3-kinase inhibitor: The role of phosphatidylinositol 3,4,5-trisphosphate in neutrophil responses. Biochem J. 296 ( Pt 2):297–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bal-Price A, Hogberg HT, Crofton KM, Daneshian M, FitzGerald RE, Fritsche E, Heinonen T, Hougaard Bennekou S, Klima S, Piersma AH et al. 2018. Recommendation on test readiness criteria for new approach methods in toxicology: Exemplified for developmental neurotoxicity. ALTEX. 35(3):306–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banga S, Patil GP, Taillie C. 2002. Direct calculation of likelihood-based benchmark dose levels for quantitative responses. Environ Ecol Stat. 9(3):295–315. [Google Scholar]
- Barbie DA, Tamayo P, Boehm JS, Kim SY, Moody SE, Dunn IF, Schinzel AC, Sandy P, Meylan E, Scholl C et al. 2009. Systematic rna interference reveals that oncogenic kras-driven cancers require tbk1. Nature. 462(7269):108–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blomme EA, Yang Y, Waring JF. 2009. Use of toxicogenomics to understand mechanisms of drug-induced hepatotoxicity during drug discovery and development. Toxicol Lett. 186(1):22–31. [DOI] [PubMed] [Google Scholar]
- Cui Y, Paules RS. 2010. Use of transcriptomics in understanding mechanisms of drug-induced toxicity. Pharmacogenomics. 11(4):573–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Abrew KN, Kainkaryam RM, Shan YK, Overmann GJ, Settivari RS, Wang X, Xu J, Adams RL, Tiesman JP, Carney EW et al. 2016. Grouping 34 chemicals based on mode of action using connectivity mapping. Toxicol Sci. 151(2):447–461. [DOI] [PubMed] [Google Scholar]
- Dean JL, Zhao QJ, Lambert JC, Hawkins BS, Thomas RS, Wesselkamper SC. 2017. Editor’s highlight: Application of gene set enrichment analysis for identification of chemically induced, biologically relevant transcriptomic networks and potential utilization in human health risk assessment. Toxicol Sci. 157(1):85–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farmahin R, Williams A, Kuo B, Chepelev NL, Thomas RS, Barton-Maclaren TS, Curran IH, Nong A, Wade MG, Yauk CL. 2017. Recommended approaches in the application of toxicogenomics to derive points of departure for chemical risk assessment. Arch Toxicol. 91(5):2045–2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filer DL, Kothiya P, Setzer RW, Judson RS, Martin MT. 2017. Tcpl: The toxcast pipeline for highthroughput screening data. Bioinformatics. 33(4):618–620. [DOI] [PubMed] [Google Scholar]
- Filipsson AF, Sand S, Nilsson J, Victorin K. 2003. The benchmark dose method--review of available models, and recommendations for application in health risk assessment. Crit Rev Toxicol. 33(5):505–542. [PubMed] [Google Scholar]
- Gaiteri C, Ding Y, French B, Tseng GC, Sibille E. 2014. Beyond modules and hubs: The potential of gene coexpression networks for investigating molecular mechanisms of complex brain disorders. Genes Brain Behav. 13(1):13–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong B, Wang C, Su Z, Hong H, Thierry-Mieg J, Thierry-Mieg D, Shi L, Auerbach SS, Tong W, Xu J. 2014a. Transcriptomic profiling of rat liver samples in a comprehensive study design by rna-seq. Sci Data. 1:140021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong P, Madak-Erdogan Z, Li J, Cheng J, Greenlief CM, Helferich W, Katzenellenbogen JA, Katzenellenbogen BS. 2014b. Transcriptomic analysis identifies gene networks regulated by estrogen receptor alpha (eralpha) and erbeta that control distinct effects of different botanical estrogens. Nucl Recept Signal. 12:e001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graczyk PP. 2007. Gini coefficient: A new way to express selectivity of kinase inhibitors against a family of kinases. J Med Chem. 50(23):5773–5779. [DOI] [PubMed] [Google Scholar]
- Grimm FA, House JS, Wilson MR, Sirenko O, Iwata Y, Wright FA, Ball N, Rusyn I. 2019. Multi-dimensional in vitro bioactivity profiling for grouping of glycol ethers. Regul Toxicol Pharmacol. 101:91–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrill J, Shah I, Setzer RW, Haggard D, Auerbach S, Judson R, Thomas RS. 2019. Considerations for strategic use of high-throughput transcriptomics chemical screening data in regulatory decisions. Curr Opin Toxicol. 15:64–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- House JS, Grimm FA, Jima DD, Zhou YH, Rusyn I, Wright FA. 2017. A pipeline for high-throughput concentration response modeling of gene expression for toxicogenomics. Front Genet. 8:168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang R, Grishagin I, Wang Y, Zhao T, Greene J, Obenauer JC, Ngan D, Nguyen DT, Guha R, Jadhav A et al. 2019. The ncats bioplanet - an integrated platform for exploring the universe of cellular signaling pathways for toxicology, systems biology, and chemical genomics. Front Pharmacol. 10:445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igarashi Y, Nakatsu N, Yamashita T, Ono A, Ohno Y, Urushidani T, Yamada H. 2015. Open tg-gates: A large-scale toxicogenomics database. Nucleic Acids Res. 43(Database issue):D921–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph P 2017. Transcriptomics in toxicology. Food Chem Toxicol. 109(Pt 1):650–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Judson R, Houck K, Martin M, Richard AM, Knudsen TB, Shah I, Little S, Wambaugh J, Setzer RW, Kothiya P et al. 2016. Analysis of the effects of cell stress and cytotoxicity on in vitro assay activity across a diverse chemical and assay space. Toxicol Sci. 153(2):409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Judson RS, Houck KA, Kavlock RJ, Knudsen TB, Martin MT, Mortensen HM, Reif DM, Rotroff DM, Shah I, Richard AM et al. 2010. In vitro screening of environmental chemicals for targeted testing prioritization: The toxcast project. Environ Health Perspect. 118(4):485–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Judson RS, Kavlock RJ, Setzer RW, Hubal EA, Martin MT, Knudsen TB, Houck KA, Thomas RS, Wetmore BA, Dix DJ. 2011. Estimating toxicity-related biological pathway altering doses for high-throughput chemical risk assessment. Chem Res Toxicol. 24(4):451–462. [DOI] [PubMed] [Google Scholar]
- Kim D, Langmead B, Salzberg SL. 2015. Hisat: A fast spliced aligner with low memory requirements. Nat Methods. 12(4):357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Paggi JM, Park C, Bennett C, Salzberg SL. 2019. Graph-based genome alignment and genotyping with hisat2 and hisat-genotype. Nat Biotechnol. 37(8):907–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleensang A, Vantangoli MM, Odwin-DaCosta S, Andersen ME, Boekelheide K, Bouhifd M, Fornace AJ Jr, Li HH, Livi CB, Madnick S et al. 2016. Genetic variability in a frozen batch of mcf-7 cells invisible in routine authentication affecting cell function. Scientific Reports. 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinstreuer NC, Yang J, Berg EL, Knudsen TB, Richard AM, Martin MT, Reif DM, Judson RS, Polokoff M, Dix DJ et al. 2014. Phenotypic screening of the toxcast chemical library to classify toxic and therapeutic mechanisms. Nat Biotechnol. 32(6):583–591. [DOI] [PubMed] [Google Scholar]
- Lamb J, Crawford ED, Peck D, Modell JW, Blat IC, Wrobel MJ, Lerner J, Brunet JP, Subramanian A, Ross KN et al. 2006. The connectivity map: Using gene-expression signatures to connect small molecules, genes, and disease. Science. 313(5795):1929–1935. [DOI] [PubMed] [Google Scholar]
- Lecomte S, Demay F, Pham TH, Moulis S, Efstathiou T, Chalmel F, Pakdel F. 2019. Deciphering the molecular mechanisms sustaining the estrogenic activity of the two major dietary compounds zearalenone and apigenin in er-positive breast cancer cell lines. Nutrients. 11(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, Genome Project Data Processing S. 2009. The sequence alignment/map format and samtools. Bioinformatics. 25(16):2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberzon A, Birger C, Thorvaldsdottir H, Ghandi M, Mesirov JP, Tamayo P. 2015. The molecular signatures database (msigdb) hallmark gene set collection. Cell Syst. 1(6):417–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberzon A, Subramanian A, Pinchback R, Thorvaldsdottir H, Tamayo P, Mesirov JP. 2011. Molecular signatures database (msigdb) 3.0. Bioinformatics. 27(12):1739–1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limonciel A, Ates G, Carta G, Wilmes A, Watzele M, Shepard PJ, VanSteenhouse HC, Seligmann B, Yeakley JM, van de Water B et al. 2018. Comparison of base-line and chemical-induced transcriptomic responses in heparg and rptec/tert1 cells using tempo-seq. Arch Toxicol. 92(8):2517–2531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for rna-seq data with deseq2. Genome Biol. 15(12):550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NTP. 2018. Ntp research report on national toxicology program approach to genomic doseresponse modeling: Research report 5. Durham (NC). [PubMed] [Google Scholar]
- Parfett C, Williams A, Zheng JL, Zhou G. 2013. Gene batteries and synexpression groups applied in a multivariate statistical approach to dose-response analysis of toxicogenomic data. Regul Toxicol Pharmacol. 67(1):63–74. [DOI] [PubMed] [Google Scholar]
- Paul Friedman K, Gagne M, Loo LH, Karamertzanis P, Netzeva T, Sobanski T, Franzosa JA, Richard AM, Lougee RR, Gissi A et al. 2020. Utility of in vitro bioactivity as a lower bound estimate of in vivo adverse effect levels and in risk-based prioritization. Toxicol Sci. 173(1):202–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira DA, Williams JA. 2007. Origin and evolution of high throughput screening. Br J Pharmacol. 152(1):53–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips JR, Svoboda DL, Tandon A, Patel S, Sedykh A, Mav D, Kuo B, Yauk CL, Yang L, Thomas RS et al. 2019. Bmdexpress 2: Enhanced transcriptomic dose-response analysis workflow. Bioinformatics. 35(10):1780–1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinero J, Queralt-Rosinach N, Bravo A, Deu-Pons J, Bauer-Mehren A, Baron M, Sanz F, Furlong LI. 2015. Disgenet: A discovery platform for the dynamical exploration of human diseases and their genes. Database (Oxford). 2015:bav028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramaiahgari SC, Auerbach SS, Saddler TO, Rice JR, Dunlap PE, Sipes NS, DeVito MJ, Shah RR, Bushel PR, Merrick BA et al. 2019. The power of resolution: Contextualized understanding of biological responses to liver injury chemicals using high-throughput transcriptomics and benchmark concentration modeling. Toxicological Sciences. 169(2):553–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard AM, Judson RS, Houck KA, Grulke CM, Volarath P, Thillainadarajah I, Yang C, Rathman J, Martin MT, Wambaugh JF et al. 2016. Toxcast chemical landscape: Paving the road to 21st century toxicology. Chem Res Toxicol. 29(8):1225–1251. [DOI] [PubMed] [Google Scholar]
- Ryan N, Chorley B, Tice RR, Judson R, Corton JC. 2016. Moving toward integrating gene expression profiling into high-throughput testing: A gene expression biomarker accurately predicts estrogen receptor alpha modulation in a microarray compendium. Toxicol Sci. 151(1):88–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheffield T, Brown J, Paul Friedman K, Judson R. in press. Tcplfit2: An r-language general purpose concentration-response modeling package. Bioinformatics. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh AJ, Ramsey SA, Filtz TM, Kioussi C. 2018. Differential gene regulatory networks in development and disease. Cell Mol Life Sci. 75(6):1013–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB. 2013. Profiling 976 toxcast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 26(6):878–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanislawska-Sachadyn A, Sachadyn P, Limon J. 2015. Transcriptomic effects of estrogen starvation and induction in the mcf7 cells. The meta-analysis of microarray results. Curr Pharm Biotechnol. 17(2):161–172. [DOI] [PubMed] [Google Scholar]
- Subramanian A, Narayan R, Corsello SM, Peck DD, Natoli TE, Lu X, Gould J, Davis JF, Tubelli AA, Asiedu JK et al. 2017. A next generation connectivity map: L1000 platform and the first 1,000,000 profiles. Cell. 171(6):1437–1452 e1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES et al. 2005. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 102(43):15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svoboda DL, Saddler T, Auerbach SS. 2019. An overview of national toxicology program’s toxicogenomic applications: Drugmatrix and toxfx. Chall Adv Comput Che. 30:141–157. [Google Scholar]
- Szymanski P, Markowicz M, Mikiciuk-Olasik E. 2012. Adaptation of high-throughput screening in drug discovery-toxicological screening tests. Int J Mol Sci. 13(1):427–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas RS, Allen BC, Nong A, Yang L, Bermudez E, Clewell HJ 3rd, Andersen ME. 2007. A method to integrate benchmark dose estimates with genomic data to assess the functional effects of chemical exposure. Toxicol Sci. 98(1):240–248. [DOI] [PubMed] [Google Scholar]
- Thomas RS, Bahadori T, Buckley TJ, Cowden J, Deisenroth C, Dionisio KL, Frithsen JB, Grulke CM, Gwinn MR, Harrill JA et al. 2019. The next generation blueprint of computational toxicology at the u.S. Environmental protection agency. Toxicol Sci. 169(2):317–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas RS, Wesselkamper SC, Wang NC, Zhao QJ, Petersen DD, Lambert JC, Cote I, Yang L, Healy E, Black MB et al. 2013. Temporal concordance between apical and transcriptional points of departure for chemical risk assessment. Toxicol Sci. 134(1):180–194. [DOI] [PubMed] [Google Scholar]
- Tukey JW. 1977. Exploratory data analysis. Reading (MA): Addison-Wesley. [Google Scholar]
- Uhlen M, Fagerberg L, Hallstrom BM, Lindskog C, Oksvold P, Mardinoglu A, Sivertsson A, Kampf C, Sjostedt E, Asplund A et al. 2015. Proteomics. Tissue-based map of the human proteome. Science. 347(6220):1260419. [DOI] [PubMed] [Google Scholar]
- Endocrine disruptor screening program tier 1 battery of assays. 2016. United States Environmental Protection Agency; [accessed 2020 April 27]. https://www.epa.gov/endocrine-disruption/endocrine-disruptor-screening-program-tier-1-battery-assays. [Google Scholar]
- USEPA. 2018. Strategic plan to promote the development and implementation of alternative test methods within the tsca program. Washington (DC): Office of Chemical Safety and Pollution Prevention. [Google Scholar]
- Tsca chemical substance inventory. 2020. United States Environmental Protection Agency; [accessed 2020 April 27]. https://www.epa.gov/tsca-inventory.
- van Dam S, Vosa U, van der Graaf A, Franke L, de Magalhaes JP. 2018. Gene co-expression analysis for functional classification and gene-disease predictions. Brief Bioinform. 19(4):575–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Mierlo T, Hyatt D, Ching AT. 2016. Employing the gini coefficient to measure participation inequality in treatment-focused digital health social networks. Netw Model Anal Health Inform Bioinform. 5(1):32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waring JF, Jolly RA, Ciurlionis R, Lum PY, Praestgaard JT, Morfitt DC, Buratto B, Roberts C, Schadt E, Ulrich RG. 2001. Clustering of hepatotoxins based on mechanism of toxicity using gene expression profiles. Toxicol Appl Pharmacol. 175(1):28–42. [DOI] [PubMed] [Google Scholar]
- Wheeler A 2019. Memorandum from administrator wheeler. Directive to prioritize efforts to reduce animal testing. Washington (DC: ): United States Environmental Protection Agency. [Google Scholar]
- Xu WS, Parmigiani RB, Marks PA. 2007. Histone deacetylase inhibitors: Molecular mechanisms of action. Oncogene. 26(37):5541–5552. [DOI] [PubMed] [Google Scholar]
- Yang L, Allen BC, Thomas RS. 2007. Bmdexpress: A software tool for the benchmark dose analyses of genomic data. BMC Genomics. 8:387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeakley JM, Shepard PJ, Goyena DE, VanSteenhouse HC, McComb JD, Seligmann BE. 2017. A trichostatin a expression signature identified by tempo-seq targeted whole transcriptome profiling. PLoS One. 12(5):e0178302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang JH, Chung TD, Oldenburg KR. 1999. A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J Biomol Screen. 4(2):67–73. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.