Abstract
Saccharomyces cerevisiae RAD53 (CHK2) and CHK1 control two parallel branches of the RAD9-mediated pathway for DNA damage-induced G2 arrest. Previous studies indicate that RAD9 is required for X-ray-associated sister chromatid exchange (SCE), suppresses homology-directed translocations, and is involved in pathways for double-strand break repair (DSB) repair that are different than those controlled by PDS1. We measured DNA damage-associated SCE in strains containing two tandem fragments of his3, his3-Δ5′ and his3-Δ3′::HOcs, and rates of spontaneous translocations in diploids containing GAL1::his3-Δ5′ and trp1::his3-Δ3′::HOcs. DNA damage-associated SCE was measured after log phase cells were exposed to methyl methanesulfonate (MMS), 4-nitroquinoline 1-oxide (4-NQO), UV, X rays and HO-induced DSBs. We observed that rad53 mutants were defective in MMS-, 4-NQO, X-ray-associated and HO-induced SCE but not in UV-associated SCE. Similar to rad9 pds1 double mutants, rad53 pds1 double mutants exhibited more X-ray sensitivity than the single mutants. rad53 sml1 diploid mutants exhibited a 10-fold higher rate of spontaneous translocations compared to the sml1 diploid mutants. chk1 mutants were not deficient in DNA damage-associated SCE after exposure to DNA damaging agents or after DSBs were generated at trp1::his3-Δ5′his3-Δ3′::HOcs. These data indicate that RAD53, not CHK1, is required for DSB-initiated SCE, and DNA damage-associated SCE after exposure to X-ray-mimetic and UV-mimetic chemicals.
Keywords: RAD53, Sister chromatid exchange, Saccharomyces cerevisiae, CHK1, Double-strand break repair
1. Introduction
Checkpoints play a critical role in maintaining genetic stability by ensuring that DNA repair mechanisms correct DNA damage before replication or segregation of damaged chromatids. In Saccharomyces cerevisiae (budding yeast), repair of double-strand breaks (DSBs) occurs by homologous recombination in which the undamaged sister chromatid is the preferred template for gap repair [1]. The G2 checkpoint mutant, rad9 [2], is defective in DSB-initiated SCE [3] and non-homologous end joining (NHEJ) [4]. rad9 diploids exhibit higher rates of homology-directed translocations and higher radiation-associated frequencies of chromosomal rearrangements, compared to wild-type [3]. Both genome instability [3] and NHEJ defects [5] in rad9 can be partially suppressed by imposing cell cycle delays, implying that G2 arrest suppresses the segregation of chromosomal fragments and facilitates DSB repair.
MEC1 (ATM/ATR) triggers two parallel RAD9-dependent pathways for signaling G2 arrest (for review, see [6]). After phosphorylation by Mec1, Rad9 associates with DSBs [7] and is required for the phosphorylation of Rad53 and Chk1. The CHK1-mediated pathway activates the securin Pds1; the RAD53 (CHK2)-mediated pathway inhibits the polo-like kinase Cdc5 and maintains Clb2/Cdc28 activity [8]. The CHK1 and RAD53 pathways converge by inhibiting cohesin degradation; Chk1 phosphorylates Pds1, thus inhibiting its ubiquitination by the Cdc20 anaphase promoting complex (APC), while Rad53 blocks the interaction of Pds1 and Cdc20 [9]. RAD53 is required for DNA repair throughout the cell cycle [10], while CHK1 is required for DNA repair only when DNA damage occurs in S phase [8]. Considering that both pds1 [11] and rad53 mutants are X-ray sensitive [12,13], it is possible that both CHK1 and CHK2 pathways contribute to recombinational repair of DSBs.
However, PDS1-mediated X-ray resistance is not CHK1-dependent since chk1 mutants are not X-ray sensitive [14]. Although the CHK1 pathway proceeds through PDS1 [15], Pds1 activation in S phase occurs by a mechanism that is CHK1-independent but MEC1-dependent [16]. CHK1, however, has been postulated to mediate homologous recombination in response to replication blocks in both mammalian cells [17] and in yeast [18]. Thus, it is important to determine whether the RAD9-dependent pathway that controls SCE is mediated by RAD53 or CHK1.
Mec1-dependent Rad53 phosphorylation occurs when Rad53 associates with Rad9 on DSBs [19]. Rad53 activation is required for DNA damage-induced transcription of DNA repair genes [20]. However, unlike RAD9, RAD53 is essential [21] due to its role in regulating ribonucleotide reductase (RNR) [22], and is activated by stalled replication forks [19,23]. The rad53 null mutant is viable when SML1, an inhibitor of RNR, is mutated or when RNR1 is over-expressed [22]. When deoxynucleotide concentration is reduced due to hydroxyurea (HU) exposure, RAD53 is required to keep stalled replication forks from irreversibly collapsing [24] and slows DNA replication by delaying the initiation of replication from late DNA origins [25]. The checkpoint signaling pathway that is initiated by replication fork stalling is mediated by Mrc1 and Tof1 [26,27]; however, a RAD9-mediated response may be initiated if nuclease digestion occurs at stalled forks [6]. Phosphorylated Rad53 triggers multiple changes through its activation of Dun1 and its association with Asf1 (for review, see [28]). asf1 exhibits higher rates of spontaneous SCE [29], while dun1 exhibits higher frequencies of UV-associated SCE [30]. Thus, Rad53 could participate in suppressing spontaneous SCE or facilitating DNA damage-associated SCE when both S phase and G2 checkpoints are triggered.
The RAD53 requirement for Rad55 phosphorylation [31] and the DNA damage-inducibility of RAD51 [32,33] further suggest that rad53 exhibits homologous recombination phenotypes. rad53 is defective in double-strand gap repair, as demonstrated by experiments in which a linear plasmid is introduced into yeast, and recombination occurs between the linear plasmid and the chromosome [34,35]. rad53 exhibits higher ratios of crossover events to non-crossover events in gap repair assays, suggesting that Rad53 is tilting the balance towards non-crossovers [35]. The exact molecular explanation for these observations is unknown. Cds1, the Schizosaccharomyces pompe Rad53 homologue, interacts with the Mus81 homologue [36], and is required for meiotic crossovers [37]. Evidence that such an interaction occurs in budding yeast is supported by a large scale protein screen [38]. These observations suggest that RAD53 facilitates DSB-repair in the genome so that crossover events are reduced.
Little is known concerning Rad53 function in regulating recombination between chromosomal sequences. Herein we determined the RAD53 and CHK1 requirement for spontaneous and DNA damage-associated SCE. rad53, but not chk1, mutants exhibited a two-fold higher rate of spontaneous SCE than wild-type. RAD53, but not CHK1, is required for SCE stimulated by a single HO endonuclease-induced DSB, and for DNA damage-associated SCE after exposure to X rays, 4-NQO and MMS. We thus suggest that RAD9-dependent DSB-initiated SCE pathway is mediated by RAD53.
2. Materials and methods
2.1. Media, chemicals, and yeast strains
Standard media, including YPD (yeast extract, peptone, dextrose), SC-HIS (synthetic complete lacking histidine), SC-TRP (synthetic complete lacking tryptophan), SC-URA (synthetic complete lacking uracil), and SC-LEU (synthetic media lacking leucine) have been previously described in Burke et al. [39]. YPD kanamycin is YPD supplemented with 200 μg/ml of kanamycin sulfate. YPD(HU) is YPD supplemented with 50 mM HU. Compounds used in this study, including methyl methanesulfonate (MMS) and 4-nitroquinoline 1-oxide (4-NQO), were purchased from Sigma or Aldrich Chemicals. 4-NQO was dissolved in dimethyl sulfoxide (DMSO).
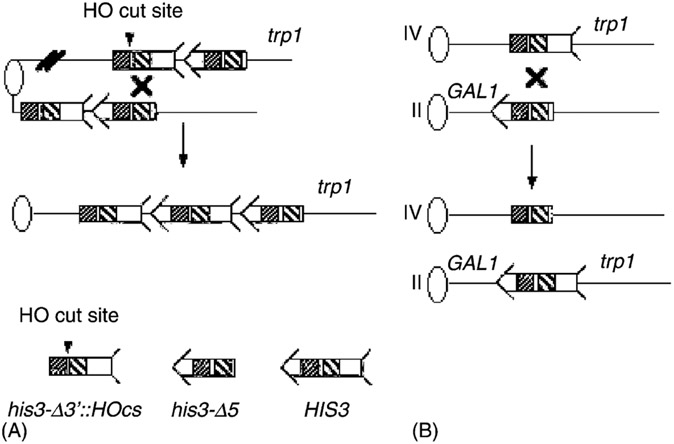
Relevant yeast strains are listed in Table 1. The strains used to measure homology-directed translocations and unequal SCE are derived from a S288c background, and their construction was previously described [3]. These strains contain two tandem truncated fragments of his3, his3-Δ3′ and his3-Δ5′ as illustrated (Fig. 1). The strains used to measure translocations contain his3 fragments on chromosomes II and IV, as described in Fasullo et al. [3]. Directed translocations were measured in diploids, which were derived from one haploid containing the his3 fragments and one haploid lacking the truncated fragments.
Table 1.
Yeast strains
| Lab name | Genotype | Source (synonym) |
|---|---|---|
| YA190 | MATa sml1::URA3 rad53::HIS3 RAD5 | R. Rothstein (W2105) |
| YA191 | MATα ura3 his7 trp1 mec2-1::LEU2 | T. Weinert (mec2-1) |
| YA165 | MATα ura3-52 his3-Δ200 leu2-Δ1 trp1-Δ1 ADE2 | F. Winston (FY250) |
| YB300 | MATα ura3-52 his3-Δ200 ade2-101 leu2-Δ1 trp1-Δ63 Δcup1::ura3 pds1::LEU2 | This laboratory |
| Strains to measure SCEa | ||
| YB163 | MATa-in MATa-inc ura3-52 his3-Δ200 ade2-101 lys2-801; trp1-Δ1 trp1::[his3-Δ3′::HOcs, his3-Δ5′] | This laboratory |
| YB177 | MATa-in MATa-inc rad51::URA3 | rad51::URA3 disruption in YB163 |
| YB203 | MATa-inc ADE2 leu2-Δ1 | From cross of YB163 with YA165 |
| YB204 | MATα leu2-Δ1 | From cross of YB177 with YA165 |
| YB205 | MATα leu2-Δ1 rad51::URA3 | This laboratory |
| YB227 | MATa-inc ADE2 leu2-Δ1 sml1::KanMX | sml1::KanMX disruption in YB203 |
| YB228 | MATα leu2-Δ1 sml1::KanMX | sml1::KanMX disruption in YB204 |
| YB229 | MATα leu2-Δ1 sml1::KanMX rad53::LEU2 | rad53::LEU2 disruption in YB228 |
| YB230 | MATa-inc leu2-Δ1 mec2-1::LEU2 | From repeated cross of YA191 with YB163 |
| YB231 | MATa-inc leu2-Δ1 sml1::KanMX rad53::LEU2 | From cross of YB227 with YB229 |
| YB301 | MATα leu2-Δ1 pds1::LEU2 | This laboratory |
| YB335 | MATa-inc ADE2 leu2-Δ1 chk1::KanMX | chk1::KanMX disruption in YB203 |
| YB356 | MATa-inc leu2-Δ1 rad53::LEU2 pds1::LEU2; sml1::KanMX | From cross of YB231 × YB2301 |
| YB357 | MATa-inc leu2-Δ1 rad53::LEU2 rad51::URA3; sml1::KanMX | From cross of YB230 × YB205 |
| YB358 | MATa-inc leu2-Δ1 mec2-1::LEU2 pds1::LEU2 | From cross of YB230 × YB301 |
| YB359 | MATα leu2-Δ1 chk1::KanMX | From cross of FY250 × YB335 |
| YB360 | MATα leu2-Δ1 pds1::LEU2 chk1::KanMX | From cross of YB335 × YB301 |
| YB361 | MATα chk1::KanMX rad51::URA3 | From cross of YB335 × YB205 |
| Strains to measure translocations | ||
| YB318 | MATα ura3-52 his3-Δ200 ade2-n trp1-Δ1 gal3− leu2-3, 112 GAL1::his3-Δ 5′ trp1::his3-Δ3′::HOcs lys2− (leaky) | Derivative of YB109 |
| YB315 | MATa ura3-52 his3-Δ200 ade2-a lys2-801 trp1-Δ1 gal3− | Derived from YA102 |
| YB317 | MATa ura3-52 his3-Δ200 ade2-a lys2-801 trp1-Δ1 gal3− sml1::KanMX | sml1::KanMX disruption in YB315 |
| YB320 | MATα ura3-52 his3-Δ200 ade2-n trp1-Δ1 gal3− leu2-3, 112 GAL1::his3-Δ 5′ trp1::his3-Δ 3′::HOcs lys2− (leaky) sml1::KanMX | sml1::KanMX disruption in YB318 |
| YB353 | MATα ura3-52 his3-Δ200 ade2-n trp1-Δ1 gal3− leu2-3, 112 GAL1::his3-Δ 5′ trp1::his3-Δ 3′::HOcs lys2− (leaky) sml1::KanMX rad53::LEU2 | rad53::LEU2 disruption in YB317 |
| YB354 | MATα ura3-52 his3-Δ200 ade2-n trp1-Δ1 gal3− leu2-3, 112 GAL1::his3-Δ 5′ trp1::his3-Δ 3′::HOcs lys2− (leaky) sml1::KanMX rad53::LEU2 | rad53::LEU2 disruption in YB320 |
| YB348 | YB315 × YB318 | This laboratory |
| YB323 | YB317 × YB320 | This laboratory |
| YB355 | YB353 × YB354 | This laboratory |
If not indicated, genotype is the same as YB163; mating-type added for clarity.
Fig. 1.
Recombination assays used in this study. Recombination assays are shown for measuring unequal SCE and translocations. Ovals represent centromeres and lines represent chromosomes. For simplicity, the left arms of chromosomes are not included. An arrow and feathers together denote HIS3. As indicated on the bottom of the figure, the 5′ deletion lacks the feather and the 3′ deletion lacks the arrow. The two regions of the sequence identity shared by the his3 fragments are indicated by decorated boxes; dot-filled boxes indicate a region of 167 bp, and the diagonal line-filled boxes indicate a region of ~300 bp. The 117-bp HO cut site (HOcs), as indicated by an arrowhead, is located between these sequences within the his3-Δ3′ fragment. (A) To measure unequal SCE, the his3-truncated fragments are integrated into the trp1 locus on chromosome IV. The product of unequal SCE is a HIS3 gene flanked by the 5′ his3 truncated fragment and the 3′ truncated his3 fragment, respectively. (B) To measure homology-directed translocations, the his3-Δ5′ and his3-Δ3′::HOcs fragments are located on chromosomes II and IV, respectively. Recombination yields reciprocal translocations where HIS3 is linked to CEN2, and a novel his3 fragment, his3-Δ(5, 3′) is linked to CEN4.
We introduced rad53 (mec2-1), an allele that confers a checkpoint defect but not lethality [40], and rad53 null mutation into strains to measure SCE. We backcrossed mec2-1 mutant ten times to YB204 (Table 1) and screened for HU sensitive meiotic segregants. Since RAD53 is essential due to its regulation of ribonucleotide reductase [19], we first deleted SML1, a negative inhibitor of RNR1. The sml1::KanMX fragment was amplified by PCR using primers 5′CATATCGTTACTGTTTTGGAACATCGC3′ and 5′ATGAGTAGCAGCACGTTCCTT3′ (Resgen, Inc.) and introduced into the wild-type strains YB203 (MATa-inc) or YB204 (MATα) by selecting for kanamycin resistant transformants. To make a rad53 null mutant in the sml1 mutant background, BamHI-digested pWL22 was introduced into the YB228 strain (sml1::KanMX), selecting for Leu+ transformants. This plasmid contains a rad53 fragment that is deleted at both the 5′ and 3′ ends, so that upon integration at the Rad53 locus, two truncated gene fragments are generated [41]. We verified the HU sensitive phenotype of the Leu+ transformants, and the rad53 deficiency was confirmed by lack of complementation with known rad53 mutants YA190 and YA191 (Table 1).
We introduced the chk1::KanMX deletion into YB203 by one step gene disruption [42] to make YB335. The primers used to amplify the chk1::KanMX fragment were 5′CAACCTCAACCAAATACTATGTTCC3′ and 5′CTGTGGAAGAAAGAAGAAACTTGAG3′. This disruption was confirmed by PCR. Genetic crosses with YB335 and YB229, and YB335 and YB204 derived rad53 chk1 and chk1 pds1 double mutants, respectively.
2.2. Determining rates of spontaneous recombination and frequencies of DNA damage-associated recombination
The rates of spontaneous recombination were determined by the method of the median [43], as described by Esposito et al. [44]; eleven independent colonies were used for each rate calculation. We used the Mann–Whitney U-test [45] to determine the statistical significance of rate differences. The protocols used to measure DNA damage-associated recombination after exposure to MMS [46], 4-NQO [46], UV [47], and X rays [48] have been previously described. The X-ray radiation source was purchased from Faxitron, Inc. (Wheeling, IL), and the dose rate was 100 rad/min. A 254 nM germicidal lamp (2 J/m2/s) was used for UV irradiation. For measuring X-ray-associated SCE, Cells were incubated on ice during X-ray irradiation. To recover after radiation exposure, cells were incubated in YPD medium for 30 minutes at 30 °C. They were then washed with sterile H2O and plated directly onto selective medium (SC-HIS) and the appropriate serial dilution was plated on YPD medium to measure viability. Cells exposed to MMS or 4-NQO were not incubated in YPD after exposure and were simply washed and plated.
2.3. Measuring X-ray and UV sensitivity
Both wild-type and mutant strains were inoculated into 2 ml YPD and incubated at 23 or 30 °C, depending on the growth characteristics of the strain. Cells were grown to stationary phase. Appropriate serial dilutions were plated onto YPD medium and exposed to X rays or UV. Statistical significance of the differences between radiation sensitivities was determined by the two-tailed t-test. [45].
2.4. Plate test assay for induced recombination
A single colony from the appropriate strain was inoculated in YPD medium and incubated at 30 °C with agitation. Approximately 107 cells were then plated on SC-HIS plates and the indicated amounts of the chemical were added in the center of the plate. The zone of toxicity is indicative of the lethality of the chemical, while a halo of induced His+ colonies is indicative of SCE events.
2.5. Induction of HO endonuclease
The galactose-inducible HO gene present in plasmid pGHOT-GAL3 [3] was introduced into the designated strains by selecting for Trp+ transformants. Trp+ isolates were then cultured at 30 °C in liquid SC-TRP and diluted in YPL. At a density of approximately 107 cells/ml (mid log), glucose or galactose was added to a final concentration of 2%, to either repress or induce, respectively, the expression of HO endonuclease. After two hour incubation at 30 °C, cells were directly plated onto SC-HIS to measure SCE formation and onto YPD to measure viability. Colonies were replica plated onto SC-TRP to measure the stability of pGHOT-GAL3.
2.6. Single-strand annealing (SSA)
To measure SSA frequencies, we measured the frequencies of his3 deletions resulting from HO-induced DSBs targeted at trp1::his3Δ-5′his3Δ-3′::HOcs. SSA creates one single his3 fragment, his3-Δ(5′, 3′) that lacks the HO cut site and both 3′ and 5′ ends of his3; isolates containing this deletion can no longer generate His+ recombinants. YPD isolates were patched onto YPD and replica plated onto SC-HIS and SC-TRP to count colonies that papillated to His+ and retained pGHOT-GAL3, respectively. In 10 Trp+ colonies that did not papillate to His+, we confirmed the presence of his3-Δ(5′, 3′) by detecting a 1.4 kb PCR product using primers 5′CACGGCAGAGACCAATCAGTA3′ and 5′GCACTCCTGATTCCGCTAATA3′, as previously described [47].
3. Results
3.1. Spontaneous SCE frequencies are two-fold higher in rad53 but not chk1 mutants, compared to wild-type
The RAD9-dependent G2 checkpoint is achieved through two parallel pathways mediated by CHK1 and RAD53 [8]. Considering that rad9 mutants are defective in DSB-initiated SCE and exhibit higher rates of spontaneous translocations than wild-type [3], we investigated the role of RAD53 and CHK1 in spontaneous and DNA damage-associated SCE. The SCE recombination assay is shown in Fig. 1, consisting of tandem his3 fragments, his3-Δ5′ and his3-Δ3′::HOcs, located at the trp1 locus [3]; His+ recombinants can result from either sister chromatid gene conversion events or reciprocal exchange events. The strains used to measure translocation frequencies contains the same his3 fragments located on chromosomes II and IV, respectively (Fig. 1).
We measured rates of spontaneous SCE in rad53 (mec2-1) mutant [49], sml1::kanMX, rad53 sml1 null mutant and chk1::kanMXmutant strains (Table 2). The spontaneous SCE rates in chk1 and sml1 mutants were similar to the rate of spontaneous SCE in wild-type (P > 0.05). However, rad53 sml1 mutant exhibited a two-fold higher rate of spontaneous SCE than sml1 and wild-type (P < 0.05). Thus, G2 checkpoint mutants are not deficient in spontaneous SCE.
Table 2.
Rates of spontaneous SCEs in wild-type, chk1 and rad53 mutants
| Genotype (strain)a | Rate of recombination (× 106)b | Ratioc |
|---|---|---|
| RAD (YB163) | 1.2 ± 0.2 | 1 |
| sml1 (YB227) | 1.0 ± 0.3 | 0.8 |
| rad53 sml1 (YB229) | 2.2 ± 0.5 | 1.8 |
| rad53 (YB230) | 1.2 ± 0.2 | 1 |
| chk1 (YB335) | 1.4 ± 0.3 | 1.1 |
All strains are S288c(s) background. For full genotype, see Table 1.
Rate, number of events per cell division. n = 3.
Ratio, rate of SCE in mutant/rate of recombination in wild-type.
3.2. X-ray-associated SCE is decreased in rad53 but not chk1 mutants; however, UV-associated SCE is observed in rad53 mutants
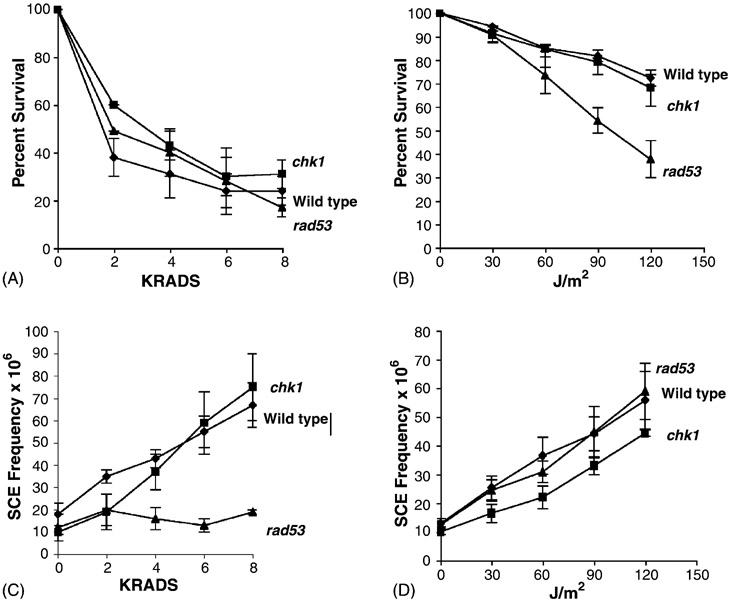
SCE frequencies increase after log phase rad9 cells are exposed to UV but not to X rays [3]. To measure radiation-associated SCE frequencies in rad53 and chk1 mutants, we exposed asynchronous log phase cultures of wild-type, rad53, and chk1 strains to X rays and UV (Fig. 2). Significant increases in SCE frequencies were observed after wild-type cells (YB163) were exposed to 4, 6, 8 krads of X rays, or to 60, 90, and 120 J/m2 of UV. We observed a maximum five-fold increase in SCE frequencies after 8 krads or 120 J/m2 exposures. Similar to wild-type, we observed significant increases in SCE frequencies after chk1 (YB335) was exposed to either X rays or UV; after 8 krads exposure, there was a seven-fold increase in SCE frequencies. There was no difference between the frequencies of UV-associated SCE in the wild-type and chk1 mutant (P > 0.05). No significant increase in SCE frequencies was observed after the rad53 mutant (mec2-1, YB230) or the rad53 null mutant (YB231) were exposed to X rays (Fig. 2). However, we observed a significant three-fold and four-fold increase in SCE frequencies after mec2-1 or the rad53 null mutant were exposed to UV, and this increase was similar to that observed in wild-type (P < 0.05). Thus, similar to rad9, rad53 mutants are deficient in X-ray-associated SCE but neither RAD9, CHK1 or RAD53 are required for UV-associated SCE.
Fig. 2.
Frequencies of DNA damage-associated SCE after exposure to X rays and UV in rad53 (YB231, black triangle), chk1(YB335, black square), and wild-type (YB163, black diamond) strains. Percent survival after exposure to X rays (A), and UV (B). Recombination frequencies after exposure to X rays (C), and UV (D).
3.3. rad53 but not chk1 mutants are defective in DSB-initiated SCE
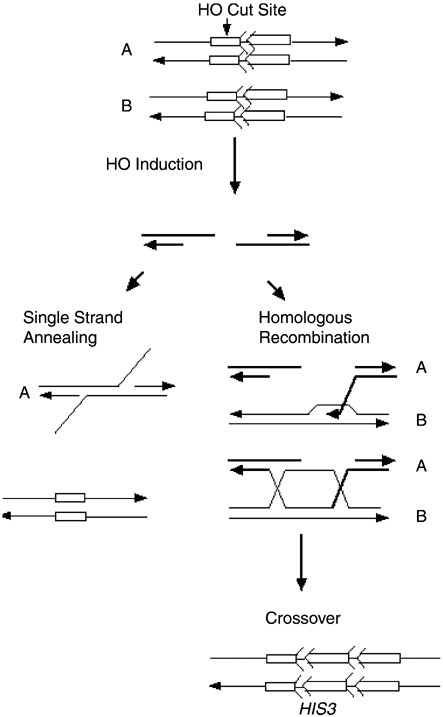
We had previously observed that SCE frequencies in rad9 mutants do not increase after X-ray exposure or when DSBs are directly targeted to trp1::his3-Δ5′ his3-Δ3’::HOcs [3]. We therefore determined whether HO-induced DSBs initiate SCE in rad53 and chk1 mutants. We introduced the galactose-inducible HO gene, present on pGHOT-GAL3, into wild-type, rad53 (mec2-1) rad53 null, sml1::kanMX, and chk1::kanMX strains. All strains contain the MATa-inc allele so that the MAT locus is not digested by HO endonuclease. The site-specific DSB at trp1::his3-Δ5′ his3-Δ3′::HOcs is repaired by SCE, SSA, or NHEJ (Fig. 3).
Fig. 3.
Double-strand break repair can occur either by SSA or unequal SCE. Symbols representing his3-Δ5′ and his3-Δ3′ are described in the legend to Fig. 1. “A” and “B” are two sister chromatids. The left panel shows repair of the HO endonuclease-generated DSB by the SSA pathway, which generates intrachromosomal deletions and a his3 fragment lacking both the 5′ and 3′ ends. Right panel of the figure shows repair of the HO endonuclease-induced DSB by homologous recombination. Unequal SCE generates HIS3 as shown at the bottom right. SCE events are measured by selecting for His+ recombinants while intrachromosomal deletions are screened, since the deletion renders cells unable to generate His+ recombinants.
We then induced HO endonuclease in asynchronous log phase cultures, and measured survival, SCE and deletion frequencies (Table 3). The pGHOT-GAL3 plasmid is retained equally well in all the strains. The percent viability after HO induction was 77–91% for all strains except for the rad53 and chk1 null mutants, which exhibited between 56 and 69% viability. After HO induction, the frequencies of SCE in wild-type and sml1 cells were increased ~13-fold, but only three-fold in either the rad53 (mec2-1) or the rad53 null mutant (Table 3). Interestingly, the highest SCE frequencies after HO induction were observed in the chk1 mutant. Because the rad53 sml1 mutant also exhibited higher levels of SCE before HO induction, we calculated net frequencies of recombination after HO induction, which were 9.3 × 10−5 for sml1 (n = 7) and 6.6 × 10−5 for rad53 sml1 (n = 6). Thus, although we could detect an increase in SCE frequencies following HO induction in the rad53 null mutant, it was consistently lower than in wild-type or in sml1 strains (P < 0.05) than in rad53 mutants.
Table 3.
Stimulation of SCE by HO-induced DSBs in rad53 and chk1 mutant strains
| Genotype (strain)a | % Viability after HO inductionb |
His+recombinants/Trp+CFU × 105 |
Fold increasee | Trp+CFU/total CFU (%) |
||
|---|---|---|---|---|---|---|
| Before HO inductionc |
After HO inductiond |
Before HO induction |
After HO induction |
|||
| RAD (YB163) | 84 ± 18 | 5.8 ± 1 | 76 ± 13 | 13 | 94 ± 3 | 91 ± 6 |
| sml1 (YB227) | 77 ± 22 | 7.9 ± 0.3 | 101 ± 33 | 13 | 93 ± 2 | 92 ± 1 |
| rad53 sml1 (YB231) | 69 ± 16 | 28 ± 14 | 93 ± 43 | 3.3 | 89 ± 6 | 87 ± 7 |
| rad53(YB230) | 91 ± 9 | 5.3 ± 0.6 | 17 ± 3 | 3.2 | 90 ± 1 | 91 ± 3 |
| rad53 pds1 (YB358) | 65 ± 8 | 4.7 ± 0.8 | 26 ± 9 | 5.5 | 84 ± 6 | 79 ± 4 |
| chk1 (YB335) | 56 ± 5 | 12 ± 9 | 161 ± 74 | 13 | 96 ± 2 | 95 ± 1 |
For complete genotype, see Table 1.
Trp+ CFU after HO induction/Trp+ CFU before HO induction × 100%.
His+ recombinants before HO induction/Trp+ CFU before HO induction.
His+ recombinants after HO induction/Trp+ CFU after HO induction.
His+ frequency after HO induction/His+ frequency before HO induction.
3.4. rad53 mutants exhibit higher frequency of DSB-initiated intrachromosomal deletions at trp1::his3-Δ5′ his3-Δ3′::HOcs than do chk1 mutants
SSA is the predominant DSB repair pathway at trp1::his3-Δ5′ his3-Δ3′::HOcs in wild-type strains [47,48], where it generates the fragment his3-Δ(5′,3′). Cells containing this his3 allele would not be able to generate His+ recombinants. Since rad53 and chk1 mutants exhibit lower viability after HO induction, compared to wild-type, we determined whether rad53 and chk1 mutants exhibited a lower frequency of intrachromosomal deletions by measuring the percentage of unselected Trp+ His− CFU arising after HO induction that can generate His+ recombinants. In wild-type, sml1, mec2-1, and the rad53 null mutant, 75–86% of unselected Trp+ His− colonies that appeared after HO induction did not generate His+ recombinants (Table 4). In contrast, only 60% of the chk1 unselected Trp+ His− colonies that appeared after HO induction did not generate His+ recombinants. Thus, SSA was stimulated as efficiently in rad53 and sml1 as in wild-type, but not as well in the chk1 mutant. Similar to rad9 mutants, rad53 mutants are not defective in SSA. We speculate that the higher frequencies of DSB-initiated SCE in chk1 may be due to a modest defect in SSA mechanisms.
Table 4.
Stimulation of intrachromosomal deletions by HO-induced DSBs in rad53 mutant strains
| Genotype (strain)a | His− CFU containing deletions (%)b |
Fold differencec |
|---|---|---|
| RAD (YB163) | 75 (124/165) | 1.0 |
| sml1::KanMX (YB227) | 76 (591/775) | 1.0 |
| rad53 sml1 (YB231) | 81 (244/301) | 1.1 |
| rad53 (YB230) | 86 (446/520) | 1.1 |
| chk1 (YB335) | 60 (304/504) | 0.8 |
For complete genotype, see Table 1.
Deletion events after HO induction/Total Trp+ CFU before HO induction × 100%.
% deletion in mutant strain/% deletion in wild-type strain.
3.5. rad53 mutants are defective in DNA damage-associated SCE after exposure to MMS and 4-NQO
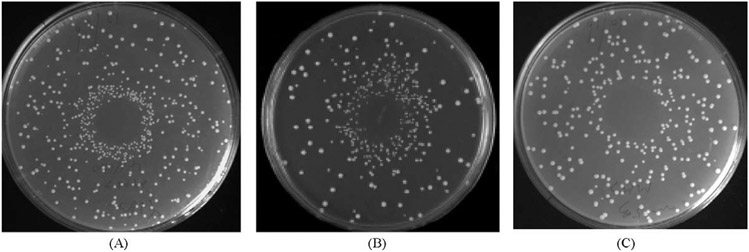
MMS and 4-NQO mimic the effects of ionizing radiation and UV, respectively, and are potent recombinagens that stimulate SCE [46]. In rad9 mutants, SCE frequencies increase after exposure to MMS and 4-NQO, similar to wild-type strains [50] but not after exposure to X rays [3]. We demonstrated that rad53 but not chk1 mutants were defective in MMS and 4-NQO-associated SCE using both a simple plate assay and a liquid assay. After cells were inoculated on SC-HIS plates and aliquots of MMS or 4-NQO were applied to the center of plates, we observed a dense halo of His+ recombinants on plates inoculated with wild-type and chk1 strains but not when plates were inoculated with either rad53 mutant (Fig. 4).
Fig. 4.
DNA damage-associated SCE after exposure to MMS is demonstrated by a simple plate assay. MMS stimulates SCE in wild-type and chk1, but less so in rad53 strains. One microliter of 10 mM MMS was applied to the center of each SC-HIS plate containing a lawn (107 cells) of the designated strain and. (A) Wild-type (YB163). (B) chk1 (YB335). (C) rad53 (YB229). Similar results were observed when 4-NQO was applied to the center of the plates.
We also measured SCE frequencies in asynchronous log phase cultures of wild-type, sml1, mec2-1 and rad53 null mutants after exposure to 4-NQO and MMS (Table 5). After exposure to 10 μM 4-NQO, SCE frequencies increased 11-fold and 32-fold in wild-type and sml1::kanMX strains, respectively, while SCE frequencies only increased three and four-fold in rad53 null mutant and mec2-1 mutant, respectively. Since rad53 mutants are hypersensitive to 4-NQO, we also measured SCE frequencies after exposure to 2 μM 4-NQO, which yields similar survival frequencies for wild-type and rad53 mutants. Wild-type and sml1::KanMX strains exhibited five-fold and 10-fold higher SCE frequencies after 2 μM 4-NQO exposure, whereas both rad53 mutants exhibited only a two-fold increase in SCE frequencies.
Table 5.
Stimulation of SCE by MMS and 4-NQO in wild-type, sml1, rad53 and chk1 strains
| Genotype (strain)a | His+/CFU × 106 (avg) after 4-NQO exposure (survival %) |
His+/CFU × 106 (avg) after MMS exposure(survival%) |
||||||
|---|---|---|---|---|---|---|---|---|
| Controlb | 2 μM | 10 μM | Ratioc | Controlb | 2 mM | 10 mM | Ratioc | |
| RAD (YB163) | 15 ± 4 | 74 ± 20 (71) | 164 ± 48 (30) | 5, 11 | 15 ± 5 | 59 ± 12 (92) | 118 ± 27 (60) | 4, 8 |
| sml1 (YB227) | 10 ± 1 | 96 ± 20 (69) | 323 ± 18 (9) | 10, 32 | 10 ± 1 | 51 ± 1 (100) | 99 ± 5 (70) | 5, 10 |
| rad53 sml1 (YB229) | 23 ± 2 | 53 ± 16 (51) | 79 ± 30 (2) | 2, 3 | 21 ± 2 | 54 ± 4 (75) | 42 ± 5 (24) | 3, 2 |
| rad53 (YB230) | 16 ± 7 | 28 ± 14 (61) | 73 ± 20 (7) | 2, 4 | 24 ± 10 | 37 ± 7 (70) | 81 ± 15 (8) | 2, 3 |
| chk1(YB335) | 19 ± 1 | 130 ± 42 (52) | 400 ± 190 (20) | 7, 21 | 19 ± 1 | 62 ± 17 (71) | 163 ± 15 (71) | 3, 8 |
For complete phenotype, see Table 1.
Frequency is events/viable cells; n = 3.
His+ frequency w/agents/spontaneous His+ frequency; first and second numbers indicate ratios for lower and higher concentrations, respectively.
After exposure to 10 mM MMS, we observed a maximum increase in SCE frequencies of three-fold in rad53 mutants, whereas there was an eight- and 10-fold increase in wild-type and sml1, respectively. To determine whether the rad53 deficiency in MMS-associated recombination resulted from MMS toxicity in rad53, we measured MMS-associated SCE frequencies after 2 mM MMS exposure. To compensate for the higher spontaneous SCE frequencies observed in rad53 sml1 mutants, we calculated net frequencies of recombination. The net frequency in rad53 sml1 was (2.9 ± 0.2) × 10−5, and significantly different (P = 0.04) from the net frequency in sml1, which was (4.1 ± 0.3) × 10−5. These results indicate that rad53 mutants are deficient in MMS and 4-NQO-associated SCE, and thus exhibit different SCE phenotypes from those observed in rad9 mutants [50].
3.6. rad53 diploid mutant exhibits a higher rate of spontaneous translocations than the sml1 or wild-type diploid
Considering that both rad9 and rad53 mutants were defective in DSB-initiated SCE, we determined whether rad53 diploid mutants would exhibit higher rates of spontaneous translocations and higher frequencies of X-ray-associated translocations, as observed in rad9 diploids [3]. We compared the spontaneous rates of wild-type (YB348), sml1 (YB3223), and sml1 rad53 (YB360) diploids. The rates of spontaneous translocations in wild-type and sml1 diploids were (3.0 ± 0.8) × 10−8 and (2.3 ± 0.3) × 10−8, respectively, while the rate in rad53 sml1 diploid was (2.6 ± 1.4) × 10−7, approximately 10-fold higher. The frequencies of X-ray-associated translocations were modestly two-fold higher in rad53 sml1 mutants compared to sml1 mutants after exposure to 4 krads X rays (P = 0.04), but not significantly different after exposure to 6 and 8 krads of X rays (P > 0.05). These data demonstrate that similar to rad9 mutants, rad53 mutants exhibit higher rates of spontaneous translocations.
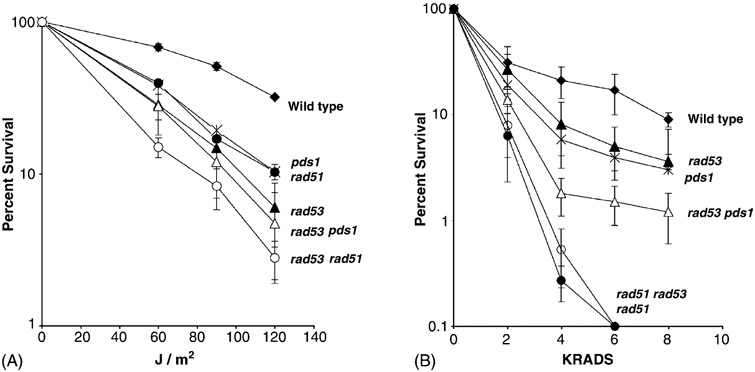
3.7. Radiation sensitivity and recombination phenotypes of rad53 rad51 and rad53 pds1 mutants
Considering that DSB-initiated SCE but not UV-associated SCE required RAD53, we determined the radiation sensitivity and recombination phenotypes of double rad53 pds1 and rad53 rad51 double mutants. Since pds1 mutants are temperature sensitive, pds1 single and double mutants were incubated at room temperature (23 °C) after radiation exposure to compare radiation-associated phenotypes. The pds1 rad53 (YB356) double mutant was more X-ray sensitive than the single mutants after exposure to 4, 6, and 8 krads (Fig. 5). The pds1 rad53 double mutant exhibited similar frequencies of SCE after HO induction as the rad53 mutant (Table 3). The rad53 rad51 double mutant (YB357) was not more X-ray sensitive than the rad51 mutant (YB177), and did not exhibit UV-associated SCE, consistent with previous studies that rad51 mutants are defective inUV-associated SCE. Thus, similar to rad9 pds1 mutants, the rad53 pds1 double mutant is more sensitive to X rays compared to the single mutants. This is consistent with the idea that both RAD9 and RAD53 are required for DSB-initiated SCE, while PDS1 is not.
Fig. 5.
Radiation resistance of rad53 (YB231, black triangle), pds1 (YB300, star), rad53 pds1 (YB356, open triangle), rad51 (YB177, black circle), rad53 rad51 (YB357, open circle) and wild-type (YB163, black diamond) cells after exposure to UV (left, A) and X rays (right, B). Percent survival is plotted against X-ray (KRADS) and UV (J/m2) dose.
To determine whether rad53 pds1 or rad53 rad51 double mutants were more UV sensitive than single mutants, we measured percent survival after cells were exposed to 60, 90, and 120 J/m2 UV. rad53 rad51, but not rad53 pds1 double mutants, were more UV sensitive than the single mutants (P < 0.05). Although rad53 mutants exhibit UV-associated SCE after irradiated cells were incubated at 30 °C, we found that neither mec2-1 or mec2-1 pds1 double mutant exhibited higher frequencies of SCE after cells were exposed 60, 90, and 120 J/m2 UV and incubated at room temperature (23 °C). In contrast, we observed a three-fold increase in SCE frequencies after wild-type cells were exposed to 120 J/m2 UV and incubated at room temperature. These results suggest that RAD53 and PDS1 are involved in similar pathways for UV repair, and that RAD51 confers UV resistance in rad53 mutants.
4. Discussion
RAD9-dependent G2 arrest is mediated by CHK1 and RAD53 (CHK2) [8]. We had previously observed that rad9 mutants are defective in DSB-initiated SCE and suppress homology-directed translocations [3], and wished to clarify whether CHK1 or RAD53 is required for DNA damage-associated SCE. Our results indicate that rad53 mutants but not chk1 mutants were defective in DNA damage-associated SCE after exposure to radiomimetic DNA damaging agents, X rays, and after a single HO-induced DSB. Neither rad53 or chk1 mutants were defective in UV-associated SCE. Similar to rad9 pds1 double mutants, rad53 pds1 double mutants exhibit more X-ray sensitivity than corresponding single mutants. These results lead to the following conclusions. First, RAD9-dependent pathway for DSB-initiated SCE does not require CHK1, but likely involves RAD53. Second, rad53 mutants are defective in more types of DNA damage-associated SCE than rad9, indicating both S phase and G2 signaling is important in triggering SCE after exposure to specific DNA damaging agents. Third, rad53 mutants, similar to rad9 mutants, exhibit higher rates of spontaneous translocations, suggesting that defective DNA damage-associated SCE correlates with higher levels of ectopic recombination.
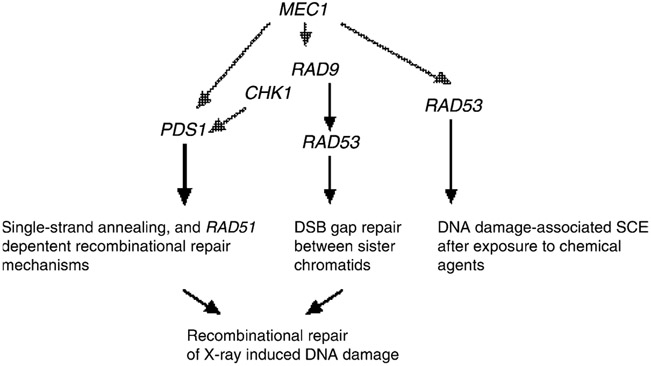
Considering that RAD51 is epistatic to RAD53, RAD9 and PDS1 for X-ray resistance, we propose a model (Fig. 6) where different RAD51-dependent recombinational repair mechanisms may be triggered by PDS1, RAD9, or RAD53. We suggest that RAD53 is involved in sister chromatid gap repair mechanisms, which are either directly initiated by DSBs or indirectly initiated by DNA adducts. The RAD53-dependent RAD9 pathway is involved in DSB-initiated SCE, a RAD9-independent RAD9 pathway is required for DNA damage-associated SCE after exposure to chemical agents that may trigger replication blocks, such as MMS. The RAD51-dependent recombination mechanism by which PDS1 confers X-ray resistance has not been defined [11]; however, PDS1 does function in RAD51-independent SSA mechanisms [11] Since mec1 mutants fail to activate either Pds1 or, Rad53 and exhibit both SSA [51] and gap repair deficiencies [35], it is possible that the PDS1 and RAD53-mediated recombination pathways are directly activated by MEC1. However, observations that TEL1 also participates in signaling DNA damage suggest that there are MEC1-independent pathways for activating the RAD53 and PDS1-mediated recombination pathways [52].
Fig. 6.
Model of checkpoint control of recombinational repair between sister chromatids. Sensor and transducer genes are shown with arrows pointing to the appropriate repair pathway. Black arrows indicate that the gene is required for the particular type of recombination event. The dotted arrows indicate known interactions but not established recombination pathways; MEC1 is the hypothetical checkpoint gene that is required for both PDS1 and RAD53-mediated pathways. DSB repair that occurs by sister chromatid recombination proceeds by a RAD9 and RAD53-dependent pathway, while PDS1 is involved in other types of recombinational repair, such as SSA. RAD9-independent SCE resulting from exposure to chemical agents is likely initiated by multiple lesions. All DNA damage-associated SCE events require RAD51.
Our studies were based on using both a rad53 null mutant and a rad53 point mutation, mec2-1. Since the rad53 null mutant requires over-expression of RNR1 or deletion of Sml1 for viability [19], we also used mec2-1 [49] that is defective in checkpoint function but not in essential function. The mec2-1 and the mec1 sml1 null mutants have similar DNA damage-associated SCE phenotypes, indicating that abolishing the RAD53 checkpoint function is sufficient to confer the defect in DNA damage-associated SCE phenotypes. However, we observed a modest two-fold increase in rates of spontaneous SCE and higher frequencies of SCE after HO induction in the rad53 sml1 null mutant, compared to mec2-1. We do not understand the higher frequencies of DSB-initiated SCE events obtained in the rad53 null mutant, compared to mec2-1, considering that SCE frequencies in both mutants do not increase after X-ray exposure. However, RAD53 is required for efficient NHEJ [53], and ku70 mutants, deficient in NHEJ, exhibit higher frequencies of DSB-initiated SCE, compared to wild-type [54]. Rad53 interacts with other proteins that suppress spontaneous SCE, such as Asf1 [55]. Thus, it is important to determine whether Rad53 interacts with Asf1 in mec2-1, and whether the mec2-1mutant is proficient at NHEJ.
Although it is feasible that the RAD53 pathway for DSB-initiated SCE is mediated by RAD9-dependent G2 arrest, additional RAD53 checkpoint functions may contribute to DSB-initiated SCE. DNA damage-associated SCE is RAD55-dependent [47] and RAD53 is required for the efficient phosphorylation and activation of Rad55 [31]. Mutations that abolish Rad55 phosphorylation have been shown to confer X-ray sensitivity, suggesting a defect in X-ray-associated SCE [36]. Third, RAD53 reduces histone accumulation [56], facilitating DNA repair; DNA repair deficiencies in checkpoint mutants can be suppressed by inhibiting histone deacetylation [57]. Thus, it would be interesting to determine whether rad53 hypomorphic mutants that are defective in a subset of these phenotypes still exhibit similar SCE phenotypes to the rad53 null mutant.
The RAD53 dependence of MMS and 4-NQO damage-associated SCE likely results from RAD53 checkpoint functions not shared with RAD9. 4-NQO and MMS-associated SCE may proceed through MRC1, which is postulated to provide an adaptor function when replication forks stall [26]. Alternatively, RAD53 may be required to directly interact with proteins that are speculated to bind to and cleave collapsed replication forks, such as Mus81 [36]. These results indicate that S phase-specific checkpoint responses may also be important in triggering DNA damage-associated SCE.
Interestingly, RAD53 is not required for UV-associated SCE. Multiple pathways for UV-associated recombination have been described in Escherichia coli [58], and similar pathways have been suggested in yeast [59]. The temperature dependence for RAD53-independent recombination, may result from the requirement of Rad53 for phosphorylation of recombination proteins, such as Rad55, which exhibit temperature dependence [60] and may be required for Rad53-independent recombination. Genetic differences between 4-NQO-associated SCE and UV-associated SCE may result from the free radical properties of 4-NQO, which account for the different sensitivities of DNA repair mutants to 4-NQO and UVC [61]. Considering 4-NQO resistance also requires genes in the RAD52 epistasis group, we suggest that 4-NQO adducts may stimulate SCE by forming DSBs.
Although we did not observe any SCE phenotypes in chk1 mutants, CHK1 may still be required for some types of DNA damage-associated SCE when the DSB is generated in S phase. For example, DNA damage caused by cross-linking agents, such as cisplatin, may initiate break-induced replication in S phase [62]. In Schizosaccharomyces pombe, CHK1 is required for recombinational repair in response to DNA interstrand cross-links [63]. Thus, further studies are needed to determine whether CHK1 is required for cross-link repair in budding yeast and whether a homologous recombination phenotype can be ascribed directly to chk1.
In summary, we have shown that RAD53 (CHK2), but not CHK1, is required for DSB-initiated SCE. CHK2 and CHK1 have different roles in different eukaryotic organisms (for review, see [64]). Thus, it would be interesting if similar SCE phenotypes were observed in other eukaryotic chk1 and chk2 mutants.
Acknowledgements
This work was supported by grant CA70105 from the National Institutes of Health and from a grant from the March of Dimes. We thank T. Weinert for the mec2-1 strain and the pWL22 plasmid for disrupting RAD53. We thank Cinzia Cera for carefully reading the manuscript.
References
- [1].Kadyk LC, Hartwell LH, Sister chromatids are preferred over homologs as substrates for recombinational repair in Saccharomyces cerevisiae, Genetics 132 (1992) 387–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Weinert TA, Hartwell LH, The RAD9 gene controls the cell cycle response to DNA damage in Sacchromyces cerevisiae, Science 241 (1988) 317–322. [DOI] [PubMed] [Google Scholar]
- [3].Fasullo MT, Bennett T, AhChing P, Koudelik J, The Saccharomyces cerevisiae RAD9 checkpoint reduces the DNA damage-associated stimulation of directed reciprocal translocations, Mol. Cell Biol 18 (1998) 1190–2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Lewis LK, Kirchner JM, Resnick MA, Requirement for end-joining and checkpoint functions, but not RAD52-mediated recombination, after EcoRI endonuclease cleavage of Saccharomyces cerevisiae DNA, Mol. Cell Biol 18 (1998) 1891–1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].de la Torre-Ruiz M, Lowndes NF, The Saccharomyces cerevisiae DNA damage checkpoint is required for efficient repair of double strand breaks by non-homologous end joining, FEBS Lett. 467 (2000) 311–315. [DOI] [PubMed] [Google Scholar]
- [6].Nyberg K, Michelson R, Putnam C, Weinert TA, Toward maintaining the genome: DNA damage and replication checkpoints, Ann. Rev. Genet 36 (2002) 617–657. [DOI] [PubMed] [Google Scholar]
- [7].Naiki T, Wakayama T, Nakada D, Matsumoto K, Sugimoto K, Association of Rad9 with double-strand breaks through a Mec1-dependent mechanism, Mol Cell Biol. 24 (2004) 3277–3285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Sanchez Y, Bachant J, Wang H, Hu F, Liu D, Tetzlaff M, Elledge SJ, Control of the DNA damage checkpoint by Chk1 and Rad53 protein kinases through distinct mechanisms, Science 286 (1999) 1166–1171. [DOI] [PubMed] [Google Scholar]
- [9].Agarwal R, Tang Z, Yu H, Cohen-Fix O, Two distinct pathways for inhibiting Pds1 ubiquitination in response to DNA damage, J. Biol. Chem 278 (2003) 45027–45033. [DOI] [PubMed] [Google Scholar]
- [10].Longhese MP, Foiani M, Muzi-Falconi M, Lucchini G, Plevani P, DNA damage checkpoint in budding yeast, EMBO J. 17 (1998) 5525–5528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].DeMase D, Zeng L, Cera C, Fasullo M, The Saccharomyces cerevisiae PDS1 and RAD9 checkpoint genes control different DNA double-strand break repair pathways, DNA Repair (Amst) 4 (2005) 59–69. [DOI] [PubMed] [Google Scholar]
- [12].Game JC, Mortimer RK, A genetic study of X-ray sensitive mutants in yeast, Mutat. Res 24 (1974) 281–292. [DOI] [PubMed] [Google Scholar]
- [13].Moore CW, Responses of radiation-sensitive mutants of Saccharomyces cerevisiae to lethal effects of bleomycin, Mutat. Res 51 (1978) 165–180. [DOI] [PubMed] [Google Scholar]
- [14].Liu Y, Vidanes G, Lin YC, Mori S, Siede W, Characterization of a Saccharomyces cerevisiae homologue of Schizosaccharomyces pombe Chk1 involved in DNA-damage-induced M-phase arrest, Mol. Gen. Genet 262 (2000) 1132–1146. [DOI] [PubMed] [Google Scholar]
- [15].Wang H, Liu D, Wang Y, Qin J, Elledge SJ, Pds1 phosphorylation in response to DNA damage is essential for its DNA damage checkpoint function, Genes Dev. 15 (2001) 1361–1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Clarke DJ, Segal M, Jensen S, Reed SI, Mec1p regulates Pds1p levels in S phase: complex coordination of DNA replication and mitosis, Nat. Cell Biol 3 (2001) 619–627. [DOI] [PubMed] [Google Scholar]
- [17].Sorensen CS, Hansen LT, Dziegielewski J, Syljuasen RG, Lundin C, Bartek J, Helleday T, The cell-cycle checkpoint kinase Chk1 is required for mammalian homologous recombination repair, Nat. Cell Biol 7 (2005) 195–201. [DOI] [PubMed] [Google Scholar]
- [18].Schollaert KL, Poisson JM, Searle JS, Schwanekamp JA, Tomlinson CR, Sanche Y, A role for Saccharomyces cerevisiae Chk1p in the response to replication blocks, Mol Biol Cell. 15 (2004) 4051–4063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Sun Z, Fay DS, Marini F, Foiani M, Stern DF, Spk1/Rad53 is regulated by Mec1-dependent protein phosphorylation in DNA replication and damage checkpoint pathways, Genes Dev. 10 (1996) 395–406. [DOI] [PubMed] [Google Scholar]
- [20].Allen JB, Zhou Z, Siede W, Friedberg EC, Elledge SJ, The SAD1/RAD53 protein kinase controls multiple checkpoints and DNA damage-induced transcription in yeast, Genes Dev. 8 (1994) 2401–2415. [DOI] [PubMed] [Google Scholar]
- [21].Zheng P, Fay DS, Burton J, Xiao H, Pinkham JL, Stern DF, SPK1 is an essential S-phase-specific gene of Saccharomyces cerevisiae that encodes a nuclear serine/threonine/tyrosine kinase, Mol. Cell. Biol 13 (1993) 5829–5842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Zhao X, Muller EG, Rothstein R, A suppressor of two essential checkpoint genes identifies a novel protein that negatively affects dNTP pools, Mol. Cell 2 (1998) 329–340. [DOI] [PubMed] [Google Scholar]
- [23].Sanchez Y, Desany BA, Jones WJ, Liu Q, Wang B, Elledge SJ, Regulation of RAD53 by the ATM-like kinases MEC1 and TEL1 in yeast cell cycle checkpoint pathways 271 (1996) 357–360. [DOI] [PubMed] [Google Scholar]
- [24].Sogo JM, Lopes M, Foiani M, Fork reversal and ssDNA accumulation at stalled replication forks owing to checkpoint defects, Science 297 (2002) 599–602. [DOI] [PubMed] [Google Scholar]
- [25].Tercero JA, Diffley JF, Regulation of DNA replication fork progression through damaged DNA by the Mec1/Rad53 checkpoint, Nature 412 (2001) 553–557. [DOI] [PubMed] [Google Scholar]
- [26].Alcasabas AA, Osborn AJ, Bachant J, Hu F, Werler PJ, Bousset K, Furuya K, Diffley JF, Carr AM, Elledge SJ, Mrc1 transduces signals of DNA replication stress to activate Rad53, Nat. Cell Biol 3 (2001) 958–965. [DOI] [PubMed] [Google Scholar]
- [27].Katou Y, Kanoh Y, Bando M, Noguchi H, Tanaka H, Ashikari T, Sugimoto K, Shirahige KS, S-phase checkpoint proteins Tof1 and Mrc1 form a stable replication-pausing complex, Nature 424 (2003) 1078–1083. [DOI] [PubMed] [Google Scholar]
- [28].Koundrioukoff S, Polo S, Almouzni G, Interplay between chromatin and cell cycle checkpoints in the context of ATR/ATM-dependent checkpoints, DNA Repair (Amst) 3 (2004) 969–978. [DOI] [PubMed] [Google Scholar]
- [29].Prado F, Cortes-Ledesma F, Aguilera A, The absence of the yeast chromatin assembly factor Asf1 increases genomic instability and sister chromatid exchange, EMBO 5 (2004) 497–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Fasullo MT, Koudelik J, AhChing P, Giallanza P, Cera C, Radiosensitive and mitotic recombination phenotypes of the Saccharomyces cerevisiae dun1 mutant defective in DNA damage-inducible gene expression, Genetics 152 (1999) 909–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Bashkirov VI, King JS, Bashkirova EV, Schmuckli-Maurer J, Heyer WD, DNA repair protein Rad55 is a terminal substrate of the DNA damage checkpoints, Mol. Cell. Biol 20 (2000) 4393–4404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].De Sanctis V, Bertozzi C, Costanzo G, Di Mauro E, Negri R, Cell cycle arrest determines the intensity of the global transcriptional response of Saccharomyces cerevisiae to ionizing radiation, Radiat. Res 156 (2001) 379–387. [DOI] [PubMed] [Google Scholar]
- [33].Cohen Y, Dardalhon M, Averbeck D, Homologous recombination is essential for RAD51 up-regulation in Saccharomyces cerevisiae following DNA crosslinking damage, Nucl. Acids Res 30 (2002) 1224–1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Bartsch S, Kang LE, Symington LS, RAD51 is required for the repair of plasmid double-stranded DNA gaps from either plasmid or chromosomal templates, Mol. Cell Biol 20 (2000) 1194–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Haghnazari E, Heyer WD, The DNA damage checkpoint pathways exert multiple controls on the efficiency and outcome of the repair of a double-stranded DNA gap, Nucl. Acids Res 32 (2004) 4257–4268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Boddy MN, Lopez-Girona A, Shanahan P, Interthal H, Heyer WD, Russell P, Damage tolerance protein Mus81 associates with the FHA1 domain of checkpoint kinase Cds1, Mol. Cell Biol 20 (2000) 8758–8766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Smith GR, Boddy MN, Shanahan P, Russell P, Fission yeast Mus81.Eme1 Holliday junction resolvase is required for meiotic crossing over but not for gene conversion, Genetics 165 (2003) 2289–2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Ho Y, Gruhler A, Heilbut A, Bader GD, Moore L, Adams SL, Millar A, Taylor P, Bennett K, Boutilier K, Yang L, Wolting C, Donaldson I, Schandorff S, Shewnarane J, Vo M, Taggart J, Goudreault M, Muskat B, Alfarano C, Dewar D, Lin Z, Michalickova K, Willems AR, Sassi H, Nielsen PA, Rasmussen KJ, Andersen JR, Johansen LE, Hansen LH, Jespersen H, Podtelejnikov A, Nielsen E, Crawford J, Poulsen V, Sorensen BD, Matthiesen J, Hendrickson RC, Gleeson F, Pawson T, Moran MF, Durocher D, Mann M, Hogue CW, Figeys D, Tyers M, Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry, Nature 415 (2002) 180–183. [DOI] [PubMed] [Google Scholar]
- [39].Burke D, Dawson D, Stearns T, Methods in yeast genetics. A Cold Spring Harbor Laboratory Course Manual, Cold Spring Harbor Press, New York, 2000. [Google Scholar]
- [40].Weinert TA, Kiser GL, Hartwell LH, Mitotic checkpoint genes in budding yeast and the dependence of mitosis on DNA replication and repair, Genes Dev. 15 (1994) 652–665. [DOI] [PubMed] [Google Scholar]
- [41].Shortle D, Haber JE, Botstein D, Lethal disruption of the yeast actin gene by integrative DNA transformation, Science 217 (1982) 371–373. [DOI] [PubMed] [Google Scholar]
- [42].Rothstein RJ, One-step gene disruption in yeast, Methods Enzymol. 101 (1983) 202–211. [DOI] [PubMed] [Google Scholar]
- [43].Lea DE, Coulson CA, The distribution of the numbers of mutants in bacterial populations, J. Genet 49 (1949) 264–284. [DOI] [PubMed] [Google Scholar]
- [44].Esposito MS, Maleas DT, Bjornstad KA, Bruschi CV, Simultaneous detection of changes in chromosome number, gene conversion and intergenic recombination during mitosis of Saccharomyces cerevisiae, Curr. Genet 6 (1982) 5–11. [DOI] [PubMed] [Google Scholar]
- [45].Zar JH, Biostatistical Analysis, Prentice Hall, Englewood Cliffs, NJ, 1996. [Google Scholar]
- [46].Fasullo MT, Dave P, Rothstein R, DNA-damaging agents stimulate the formation of directed reciprocal translocations in Saccharomyces cerevisiae, Mutat. Res 314 (1994) 121–133. [DOI] [PubMed] [Google Scholar]
- [47].Dong Z, Fasullo M, Multiple recombination pathways for sister chromatid exchange in Saccharomyces cerevisiae: role of RAD1 and the RAD52 epistasis group genes, Nucl. Acids Res 31 (2003) 2576–2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Fasullo MT, Giallanza P, Bennett T, Cera C, Dong Z, Saccharomyces cerevisiae rad51 mutants are defective in DNA damage-stimulated sister chromatid exchange but exhibit increase rates of homology-directed translocations, Genetics 158 (2001) 959–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Kim S, Weinert TA, Characterization of the checkpoint gene RAD53/MEC2 in Saccharomyces cerevisiae, Yeast 13 (1997) 735–745. [DOI] [PubMed] [Google Scholar]
- [50].Fasullo M, Zeng L, Giallanza P, Enhanced stimulation of chromosomal translocations by radiomimetic DNA damaging agents and camptothecin in Saccharomyces cerevisiae rad9 checkpoint mutants, Mutat. Res 547 (2004) 123–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Kaye JA, Melo JA, Cheung SK, Vaze MB, Haber JE, Toczyski DP, DNA breaks promote genomic instability by impeding proper chromosome segregation, Curr. Biol 14 (2004) 2096–2106. [DOI] [PubMed] [Google Scholar]
- [52].Lisby M, Barlow JH, Burgess RC, Rothstein R, Choreography of the DNA damage response: spatiotemporal relationships among checkpoint and repair proteins, Cell 118 (2004) 699–713. [DOI] [PubMed] [Google Scholar]
- [53].de la Torre-Ruiz M, Lowndes NF, The Saccharomyces cerevisiae DNA damage checkpoint is required for efficient repair of double strand breaks by non-homologous end joining, FEBS Lett. 467 (2000) 311–315. [DOI] [PubMed] [Google Scholar]
- [54].Fasullo M, St. Amour C, Zeng L, Enhanced stimulation of chromosomal translocations and sister chromatid exchanges by either HO-induced double-strand breaks or X rays in Saccharomyces cerevisiae yku70 mutants, Mutat. Res 578 (2005) 158–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Hu F, Alcasabas AA, Elledge SJ, Asf1 links Rad53 to control of chromatin assembly, Genes Dev. 15 (2001) 1061–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Gunjan A, Verreault A, A Rad53 kinase-dependent surveillance mechanism that regulates histone protein levels in S. cerevisiae, Cell 115 (2003) 537–549. [DOI] [PubMed] [Google Scholar]
- [57].Scott KL, Plon SE, Loss of Sin3/Rpd3 histone deacetylase restores the DNA damage response in checkpoint-deficient strains of Saccharomyces cerevisiae, Mol. Cell Biol 13 (2003) 4522–4531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Courcelle J, Hanawalt PC, RecA-dependent recovery of arrested DNA replication forks, Annu. Rev. Genet 37 (2003) 611–646. [DOI] [PubMed] [Google Scholar]
- [59].Kadyk LC, Hartwell LH, Replication-dependent sister chromatid recombination in rad1 mutants of Saccharomyces cerevisiae, Genetics 133 (1993) 469–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Lovett ST, Mortimer RK, Characterization of null mutants of the RAD55 gene of Saccharomyces cerevisiae: effects of temperature, osmotic strength and mating type, Genetics 116 (1987) 547–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Ramotar D, Belanger E, Brodeur I, Masson JY, Drobetsky EA, A yeast homologue of the human phosphotyrosyl phosphatase activator PTPA is implicated in protection against oxidative DNA damage induced by the model carcinogen 4-nitroquinoline 1-oxide, J. Biol. Chem 273 (1998) 21489–21496. [DOI] [PubMed] [Google Scholar]
- [62].Sancar A, Lindsey-Boltz LA, Unsal-Kacmaz K, Linn S, Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints, Ann. Rev. Biochem 73 (2004) 39–85. [DOI] [PubMed] [Google Scholar]
- [63].Lambert S, Mason SJ, Barber LJ, Hartley JA, Pearce JA, Carr AM, McHugh PJ, Schizosaccharomyces pombe checkpoint response to DNA interstrand cross-links, Mol. Cell Biol 23 (2003) 4728–4737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Chen Y, Sanchez Y, Chk1 in the DNA damage response: conserved roles from yeasts to mammals, DNA Repair 3 (2004) 1025–1032. [DOI] [PubMed] [Google Scholar]