Abstract
Saccharomyces cerevisiae rad9 checkpoint mutants exhibit pleiotropic phenotypes, including higher frequencies of chromosome loss, radiation sensitivity, and decreased induction of DNA damage-inducible genes. We had previously shown that rad9 mutants exhibit higher frequencies of DNA damage-associated translocations but lower frequencies of DNA damage-associated sister chromatid exchange (SCE), compared to wild type. Herein, we have shown that differences between the frequencies of DNA damage-associated recombination in the rad9 mutant and wild type depend on the identity and the concentration of the DNA damaging agent. Translocation and SCE frequencies were measured in strains containing truncated his3 fragments, located either on chromosomes II and IV, or located in tandem on chromosome IV, respectively. DNA damage-associated frequencies of translocations after exposure to hydrogen peroxide (H2O2), bleomycin, phleomycin, cisplatin, and camptothecin are higher in the rad9 diploid than in wild type. However, translocation frequencies after exposure to 4-nitroquinoline 1-oxide (4-NQO) and N-methyl-N′-nitro-N-nitrosoguanidine (MNNG) are similar in rad9 and wild-type strains. We suggest that the deficiency in triggering G2 arrest after exposure to specific DNA damaging agents results in the higher levels of DNA damage-associated translocations in rad9 mutants.
Keywords: Recombination, DNA damaging agent, Yeast, G2 checkpoint
1. Introduction
Cell cycle checkpoints may reduce chromosomal instability by allowing time for DNA repair before replication or segregation of chromatids. In Saccharomyces cerevisiae (yeast), four distinct cell cycle delays have been described: one G1/S phase, two S phase, and one G2/M phase [1]. Both single stranded DNA and stalled replication forks have been postulated to signal cell cycle checkpoints (for review, see [2]). Activation of one or more cell cycle checkpoints may occur after cells are exposed to specific DNA damaging agents. For example, alkylating agents that methylate in the DNA minor groove may block DNA polymerase progression, while methylation in the major groove may be repaired by enzymes which indirectly produce DNA strand breaks [3]. Thus, how DNA damage is processed and whether the damage is sufficient to block DNA replication may be key factors in triggering particular checkpoints.
RAD9 is required for cell cycle arrest in G2 after exposure to DNA damaging agents [4]. DNA damaging agents that trigger cell cycle arrest in G2 include UV and X rays [4], and chemical compounds that directly or indirectly cause double-strand breaks, such as hydrogen peroxide (H2O2) [5] and methyl methanesulfonate (MMS) [6]. Chemical compounds that cause intrastrand and interstrand cross-links, such as cisplatin [7], and topoisomerase I inhibitors, such as camptothecin [8], also trigger G2 arrest. However, other DNA damaging agents which cause bulky adducts, such as the UV-mimetic agent 4-Nitroquinoline 1-oxide (4-NQO), do not trigger G2 arrest in eukaryotic cells [9]. Thus, RAD9-mediated G2 arrest can be triggered both by radiomimetic chemical agents, as well as specific chemical compounds which indirectly cause DNA single or double-strand breaks.
RAD9-mediated G2 cell-cycle arrest allows more time for recombinational repair between sister chromatids, which are preferred substrates for X-ray-associated repair in yeast [10,11]. Consistent with this idea, rad9 mutants exhibit lower frequencies of X-ray-associated SCEs but higher frequencies of X-ray-associated translocations [12]. When rad9 cells are exposed to UV and MMS, there is only a minor decrease in frequencies of DNA damage-associated SCEs, compared to wild type [12]. These DNA damaging agents may generate a greater number of DNA lesions that trigger the MEC1-dependent S phase checkpoint to prevent replication fork collapse [13]. Thus, checkpoint mutants exposed to only a subset of DNA damaging agents exhibit higher frequencies of DNA damage-associated translocations and lower frequencies of DNA damage-associated SCE [12].
Besides its function to trigger cell cycle arrest in G2, RAD9 is required to prevent mitosis when DNA replication is incomplete [14] and delaying entry into S phase after exposure either to ionizing radiation, UV, or 4-NQO [15]. RAD9 is required to repair chromosome breaks that occur during S phase as a consequence of defective topoisomerase action [16] and in DNA damage tolerance mechanisms in excision repair mutants [17]. rad9 mutants are defective in maintaining large yeast artificial chromosomes (YACs) when such YACs contain only one replication origin [18]. Thus, RAD9 maintains genetic stability by triggering delays at other stages in the cell cycle.
In this manuscript, we have exposed cells to DNA damaging agents to identify compounds that stimulate more translocations but fewer SCEs in rad9 mutants, compared to wild type. In addition to DNA damaging agents that directly cause strand breaks, we also chose cross-linking and alkylating agents and agents that cause bulky adducts. Compared to wild type, we observed more DNA damage-associated translocations when rad9 cells were exposed to particular X-ray-mimetic agents, cisplatin or camptothecin but not to 4-NQO and MNNG.
2. Materials and methods
2.1. Strains and media
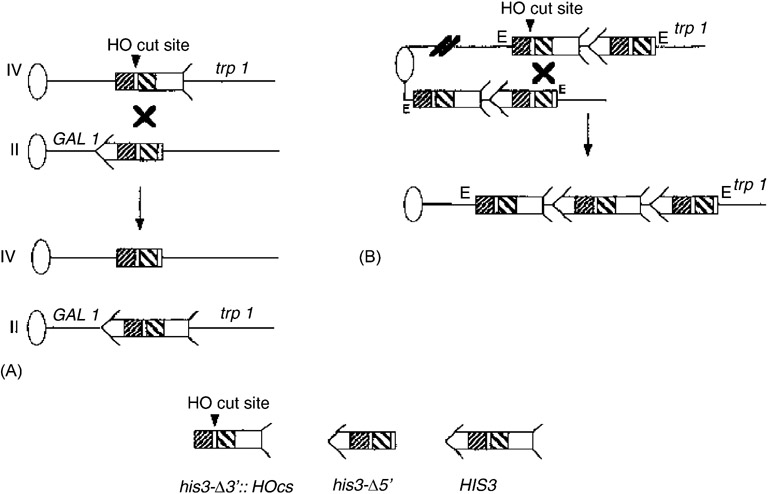
Standard media, including YPD (yeast extract, peptone, dextrose), synthetic complete lacking histidine (SC-HIS) have been previously described [19]. The strains used to measure homology-directed translocations and sister chromatid exchange (SCE) are listed in Table 1, and their construction was previously described [12]. These strains contain two truncated fragments of his3, his3-Δ3′ and his3-Δ5′ [12], as illustrated (Fig. 1). Directed translocations were measured in diploids, which were derived from one haploid containing the his3 fragments and one haploid lacking the truncated fragments.
Table 1.
Yeast strains
| Laboratory name | Genotypea | Source |
|---|---|---|
| YA102 | MATa ura3-52, his3–Δ200, ade2-101, 1ys2-801, trp1–Δ1, gal3− | M. Carlson |
| YB109 | MATα leu2-3,112,GAL1 :: his3-Δ5′,trp1 :: his3–Δ3’ :: HOcs, lys2− (leaky) | This laboratory |
| YB130 | MATa rad9 :: URA3 | This laboratory |
| YB132 | MATα leu2-3,112, GAL1 :: his3-Δ5′,trp1 :: his3-Δ3’ :: HOcs, lys2− (leaky), rad9 :: URA3 | This laboratory |
| YB110 | YB109 × YA102 | This laboratory |
| YB134 | YB130 × YB132 | This laboratory |
| YB163 | MATa-inc trp1 :: [his3-Δ3′ :: HOcs, his3-Δ5′] | This laboratory |
| YB146 | MATa trp1 :: [his3-Δ3′ :: HOcs, his3-Δ5′] | This laboratory |
| YB147 | MATa trp1 :: [his3-Δ3′ :: HOcs, his3-Δ5′] rad9 :: URA3 | This laboratory |
Genotype the same as YA102 unless indicated.
Fig. 1.
Recombination assays used in this study. Ovals represent centromeres and lines represent chromosomes. For simplicity, the left arms of chromosomes are not included. The position and orientation of the his3 recombinational substrates are shown as present in strains used to measure (A) reciprocal translocations and (B) unequal SCE. An “X” designates potential sites of crossovers, and the resulting chromosomal rearrangement is presented. An arrow and feathers denote HIS3. As indicated on the bottom of the figure, the 5′ deletion lacks the feathers and the 3′ deletion lacks the arrow. The two regions of sequence identity shared by the his3 fragments are indicated by decorated boxes; broadly spaced diagonal lines indicate a region of ~300 bp, and tightly spaced diagonal lines indicate a region of 167 bp.
2.2. DNA damaging agents
Compounds used in this study included methyl methanesulfonate (MMS), N-methyl-N′-nitro-N-nitrosoguanidine (MNNG), cisplatin, 4-nitroquinoline 1-oxide (4-NQO), bleomycin, phleomycin, H2O2, camptothecin. H2O2 (30% w/w) was purchased from Sigma. Chemicals were purchased from Sigma or Aldrich Chemicals. Chemicals were dissolved in either dimethyl sulfoxide (DMSO), dimethyl formamide (DMF) or water, depending on the specifications provided by the vendor.
2.3. Plate test assay for induced recombination in rad9 diploids
A single colony from the appropriate strain was inoculated in YPD medium and incubated at 30 °C with agitation. Approximately 107 cells were then plated on SC-HIS plates and the indicated amounts of the chemical were added in the center of the plate. The zone of toxicity is indicative of the lethality of the chemical, while a halo of induced His+ colonies is indicative of translocation events. Concentrations of solutions added to plates included H2O2 (30% w/w), 10.5 M MMS, 100 mM MNNG, 140 mM 4-NQO, 133 mM cisplatin, 10 mg/ml camptothecin, 3 units/ml bleomycin, 10 mM phleomycin.
2.4. Culturing and treatment of yeast
Protocols used to test the recombinogenicity of chemical DNA damaging agents have been described [20]. For measuring stimulation of SCE and translocations, cells were pre-incubated for 30 min in YPD after treatment with the DNA damaging agent, washed twice with sterile H2O, and then plated on selective medium (SC-HIS). We exposed cells to concentrations of chemicals that exhibit equivalent levels of lethality in both wild-type and rad9 strains (Table 2). Because spontaneous frequencies of translocations are significantly higher in rad9 mutants, we also measured net frequencies of DNA damage-associated recombination by subtracting the spontaneous frequency from the frequency obtained after chemical exposure, as done previously [12,21,22]. At least three independent experiments were done for each DNA damaging agent. The significance of the differences between rad9 mutants and RAD9+ strains was determined using the two-tailed paired sample t-test [23].
Table 2.
Stimulation of translocations by DNA damaging agents in RAD9 (YB110) and the rad9 :: URA3 diploid (YB134)
| Agent (concentration or dose) | Stimulation in
RAD9 (YB110)a |
Stimulation in
rad9− (YB134)a |
Ratioc | ||
|---|---|---|---|---|---|
| His+/CFU × 107 (survival%) | Netb | His+/CFU × 107 (survival%) | Netb | ||
| Spontaneous | 6 ± 2 | 0 | 23 ± 14 | 0 | NA |
| Alkylating agentsd | |||||
| MMS 0.1% (10.5 (mM)) | 62 ± 16(94) | 59 ± 13 | 248 ± 24(81) | 246 ± 21 | 4.2 |
| MNNG 50 (μM) | 100 ± 9 (88) | 92 ± 1 | 173 ± 46 (90) | 153 ± 38 | 1.7 |
| MNNG 100 (μM) | 250 ± 10 (85) | 248 ± 11 | 347 ± 40 (76) | 326 ± 37 | 1.3 |
| MNNG 200 (μM) | 345 ± 38 (53) | 338 ± 21 | 306 ± 62 (16) | 253 ± 43 | 0.7 |
| Bulky adducts | |||||
| 4-NQO 1 (μM) | 28 ± 9 (100) | 27 ± 14 | 43 ± 6 (100) | 33 ± 10 | 1.2 |
| 4-NQO 10 (μM) | 494 ± 163(67) | 484 ± 160 | 666 ± 91 (47) | 636 ± 96 | 1.3 |
| Cisplatin 1.3 (μM) | 122 ± 33 (100) | 120 ± 25 | 347 ± 71(87) | 330 ± 70 | 2.8 |
| Free radical agents | |||||
| Bleomycin 0.015 units | 60 ± 52 (94) | 54 ± 52 | 446 ± 130(86) | 403 ± 136 | 7.5 |
| Phleomycin 10 (μM) | 67 ± 31(93) | 59 ± 31 | 295 ± 117 (60) | 286 ± 116 | 4.8 |
| H2O2 2 (μM) | 219 ± 92 (76) | 212 ± 92 | 585 ± 259 (76) | 558 ± 260 | 2.6 |
| H2O2 4 (μM) | 346 ± 138(71) | 329 ± 192 | 533 ± 167 (54) | 523 ± 165 | 1.6 |
| TopoI inhibitor | |||||
| Camptothecin 50 (μg/ml) | 5 ± 5 (95) | <1 | 38 ± 14 (99) | 26 ± 14 | 5.2 |
| Camptothecin 100 (μg/ml) | 34 ± 7 (95) | 33 ± 7 | 163 ± 50 (85) | 153 ± 54 | 4.6 |
2.5. CHEF gels
Electrophoretic karyotypes of His+ recombinants were determined by pulse field electrophoresis using contour clamped gel electrophoresis (CHEF) to resolve chromosomal DNA. The running time was 26 h at 250 V (6 V/cm) and the pulse time was 90 s [22].
3. Results
3.1. Recombination assays
Our goal was to identify chemical DNA damaging agents that stimulate more translocations in rad9 mutants, compared to wild type. Considering that rad9 mutants are defective in X-ray-associated SCE [12], we then asked whether the frequencies of DNA damage-associated SCE were lower in rad9 mutants after exposure to these chemical agents. To measure frequencies of DNA damage-associated translocation and SCE, we select His+ prototrophs that result from recombination between the his3 fragments located on chromosomes II and IV or located in tandem at the trp1 locus, respectively. The rate of spontaneous translocations in the rad9 diploid YB134 is 2 × 10−7, which is fourfold higher than the rate in the wild-type diploid YB110 [12]. The rates of spontaneous SCE in rad9 haploid and diploid mutants and in wild-type haploid and diploid strains are the same, ~2 × 10−6 [12].
We employed a simple plate assay to detect recombinants that result from exposure to DNA damaging agents [20]. Because spontaneous rates of translocations are low [12,20], DNA damage-associated His+ recombinants containing translocations appear as halos after a chemical compound is diffused from the center of a SC-HIS plate inoculated with either wild-type or rad9 cells (Fig. 2). This assay indicates when there are more stimulated His+ recombinants per total number of inoculated cells after exposure to the DNA damaging agent. We observed a thicker halo of stimulated His+ recombinants and more spontaneous His+ recombinants after rad9 cells were exposed to MMS and H2O2, compared to wild type (Fig. 2). Other DNA damaging agents, including bleomycin, phleomycin, camptothecin and cisplatin, did not stimulate more His+ recombinants using this plate assay (data not shown). However, as shown in previous studies [20], particular DNA damaging agents stimulate more recombination in log phase cell cultures.
Fig. 2.
Plate assay demonstrating higher levels of DNA damage-associated translocations in a rad9 diploid compared to a wild-type diploid. (A) YB110 (wild-typediploid) + 0.2 μl of MMS. (B) YB110 (wild-typediploid) + 1 μl of H2O2. (C) YB134 (rad9mutant) + 0.2 μl of MMS. (D) YB134 (rad9mutant) + μl of H2O2.
3.2. Agents that directly cause DNA strand breaks stimulate more recombination in rad9 mutants
We had previously shown that rad9 cells exhibit higher frequencies of translocations after exposure to MMS [12], an agent known to promote DNA breaks. Actively growing log phase cultures of both wild-type and rad9 diploid strains were then exposed to other DNA damaging agents that create single and double-strand breaks. Exposure to 2 mM H2O2 increased translocation frequencies in both RAD9 and rad9 diploid strains by 31- and 22-fold, respectively; however, compared to wild type, there was a three-fold difference in the net frequency of DNA damage-associated translocations in the rad9 diploid compared to the RAD9 strain. Bleomycin and phleomycin exposure increased translocation frequencies 11- and 33-fold in the rad9 diploid, and we observed an eight and fivefold difference in the net translocation frequencies, respectively, in rad9 diploid compared to wild type. The higher frequencies of DNA damage-associated translocations obtained after exposure of rad9 cells to bleomycin, compared to the related compound phleomycin, may result from the higher number of double-strand breaks that are caused by bleomycin [24]. Thus, exposure to chemical agents that cause DNA strand breaks, including H2O2, bleomycin, and phleomycin, results in higher translocation frequencies in rad9 than in wild-type (Table 2).
3.3. Camptothecin and cross-linking agents that trigger G2 arrest stimulate more homology-directed translocations in rad9 mutants
DNA damaging agents, including cross-linking agents and topoisomerase inhibitors, do not directly induce single and double-strand breaks but can trigger G2 arrest in budding yeast. Exposure to 1.3 mM cisplatin increased translocation frequencies 81- and 22-fold in wild-type and rad9 strains, respectively; net frequencies were threefold higher in rad9 cells (Table 2). Net frequencies of camptothecin-associated translocations were fivefold higher in rad9 cells, compared to wild type, although we observed more modest increases in DNA damage-associated frequencies of translocations after camptothecin exposure compared to other compounds. These results indicate that frequencies of DNA damage-associated translocations may be higher in checkpoint mutants after exposure to compounds that do not directly induce strand breaks.
3.4. DNA damage-associated SCE is decreased in rad9 mutants after exposure to a subset of DNA damaging agents
To determine whether higher frequencies of DNA damage-associated translocations correlated with lower frequencies of DNA damage-associated SCE, we measured SCE frequencies in both wild-type (YB163 and YB146) and rad9 (YB147) strains after exposure to H2O2, camptothecin, phleomycin, cisplatin and 4-NQO. After exposure to phleomycin and camptothecin, we observed significant twofold increases in SCE frequencies in wild type (P < 0.05) but not in the rad9 strain (P > 0.05) (Table 3). SCE frequencies after exposure to bleomycin and cisplatin were 4.7 × 10−5 (average, n = 3) and 3.2 × 10−5 (average, n = 3) but not higher than wild type (YB147), 2.4 × 10−5 (P > 0.05). We observed a fourfold stimulation of recombination in both wild-type and rad9 cells after exposure to H2O2 (Table 3) and 4-NQO (data not shown). Thus, differences in SCE recombination frequencies in wild type and rad9 mutants after exposure to DNA damaging agents was only observed for a subset of DNA damaging agents.
Table 3.
Stimulation of SCE by H2O2, camptothecin, and phleomycin in RAD9 and rad9 :: URA3 strains
| Chemical agent | Stimulation in
RAD9 (YB163)a |
Stimulation in
rad9 :: URA3 (YB147) |
||
|---|---|---|---|---|
| His+/CFU × 106 (survival %) | Fold increaseb | His+/CFU × 106 (survival %) | Fold increaseb | |
| Spontaneous | 16 ± 2 (100) | 1 | 15 ± 2 (100) | 1 |
| 0.04% H2O2 | 59 ± 12 (40) | 3.7 | 60 ± 20 (35) | 4 |
| 50 μg/ml camptothecin | 32 ± 7 (88) | 2 | 20 ± 3 (91) | 1.3 |
| 10 μM phleomycin | 33 ± 6 (41) | 2.1 | 17 ± 3 (45) | 1.1 |
For complete genotype, see Table 1.
His+ frequency with agent/spontaneous His+ frequency.
3.5. RAD9 checkpoint is less efficient in suppressing 4-NQO and MMNG-associated recombination
Several DNA damaging agents create particular DNA adducts that require replication to initiate recombination, while other DNA damaging agents more frequently form DNA adducts in replicating regions. For example, recombinogenicity of 4-NQO is enhanced when cells undergo replication [25] and DNA adducts formed after MNNG exposure occur most frequently in active replicating regions [26]. We observed a 43- and 23-fold increase in translocation frequencies after wild-type and rad9 cells were exposed to 4-NQO, respectively (Table 2). We observed a 52- and 21-fold increase in translocation frequencies after wild-type and rad9 cells were exposed to MNNG, respectively (Table 2). Net DNA damage-associated frequencies after exposure to either MNNG or 4-NQO were less than twofold higher in rad9. This data suggest that RAD9 poorly suppresses recombination that is either stimulated by MMNG or 4-NQO.
3.6. Electrophoretic karyotypes of DNA damage-associated His+ recombinants
We previously observed that most spontaneous and radiation-associated His+ recombinants from wild type contain reciprocal translocations, CEN2 :: IV and CEN4 :: II, while most His+ recombinants from rad9 contain one non-reciprocal translocation, CEN2 :: IV [12]. We therefore characterized the electrophoretic karyotypes of DNA damage-associated translocations that occurred after exposure to H2O2 and camptothecin, DNA damaging agents that do not directly stimulate double-strand breaks (Fig. 3). We observed that four out of four H2O2-associated His+ recombinants from the rad9 strain contain only one non-reciprocal translocation, CEN2 :: IV After exposure to camptothecin, we characterized the electrophoretic karyotype of six His+ recombinants from the rad9 strain and three from wild type. Of the His+ recombinants from the rad9 strain, we observed that four contain non-reciprocal translocations and two contain reciprocal translocations. We observed that one His+ recombinant contained reciprocal translocations and two contained non-reciprocal translocations from the wild-type strain. This data suggest that DNA damage caused by camptothecin stimulates both reciprocal and non-reciprocal translocations.
Fig. 3.
Electrophoretic karyotype of camptothecin-associated His+ recombinants generated in the rad9 diploid YB134. (A) YB134 His−-parent. (B) His+ recombinant containing a non-reciprocal translocations. (C) His+ recombinant containing a reciprocal translocation. (D) His+ recombinant containing a reciprocal translocation. The positions of chromosomes II, IV, and translocations CEN2 :: IV and CEN4 :: V are indicated by the arrow.
4. Discussion
S. cerevisiae rad9 mutants exhibit complex phenotypes, including radiation sensitivity, enhanced frequencies of chromosome loss, and failure to arrest the cell cycle in G2 [15,17,18,27]. Here we report that, depending on the chemical compound, RAD9 can partially suppress the frequencies of DNA damage-associated translocations and increase the frequencies of DNA damage-associated SCE. We observed higher frequencies of translocations in rad9 mutants after exposure to DNA damaging agents that are known to specifically trigger cell cycle arrest in the G2 phase, and these agents include bleomycin, phleomycin, H2O2, cisplatin and camptothecin. Since frequencies of DNA damage-associated SCE were the same in wild type and rad9 after exposure to specific DNA damaging agents, we speculate that S phase checkpoints are important in enhancing recombinational repair. Because rad9 and wild-type strains did not exhibit different recombination phenotypes after exposure to MNNG and 4-NQO but rad9 mutants are hypersensitive to both MNNG and 4-NQO, we speculate that RAD9 participates in additional DNA repair pathways to confer resistance both to alkylating compounds and compounds that cause bulky adducts.
The data supporting these conclusions was obtained using both a simple plate assay and a quantitative liquid assay. The plate assay provides a quick method to identify a subset of chemical compounds that stimulate more translocations in rad9 mutants. However, the liquid assay provides a more accurate measurement of the translocation frequencies after exposure to particular DNA damaging agents, which are active in log phase cells or in the presence of oxygen. Although a subset of DNA damage-associated translocation frequencies are higher in rad9 mutants than in wild type, the fold induction (ratio of the DNA damage-associated translocation frequency to the spontaneous frequencies) may be higher in wild type than in the rad9 mutant because the spontaneous translocation frequencies are lower in the wild-type strain.
We previously documented that frequencies of X-ray-associated translocations are increased but frequencies of X-ray-associated SCE are decreased in rad9 mutants [12]. Particular DNA damaging agents that mimic the effects of ionizing radiation, such as phleomycin, thus stimulate more translocations but fewer SCEs in rad9 mutants. We suggest that RAD9 may confer resistance to several radiomimetic compounds by simply triggering a G2 arrest and allowing more time for sister chromatid gap repair.
Higher frequencies of DNA damage-associated translocations in rad9 cells after camptothecin and cisplatin exposure support ideas that both camptothecin [28] and cisplatin [7] create DNA adducts that indirectly generate recombinogenic double-strand breaks. Cisplatin-induced interstrand crosslinks creates replication-dependent double-strand breaks that trigger G2 arrest [29]. Modest stimulation of SCE after cisplatin exposure could result from the low percentage of interstrand crosslinks among total number of DNA adducts [29] and the relatively high frequencies of spontaneous SCE.
Double-strand breaks resulting from camptothecin exposure occur when the DNA replication apparatus encounters trapped topoisomerase I intermediates, leading to replication fork collapse [16]. His+ recombinants resulting from either unequal SCE or translocations after camptothecin exposure could result from break-induced replication (BIR) mechanisms [30,31], and RAD9-mediated G2 arrest may provide time for unequal SCE. Non-reciprocal translocations are postulated to occur by BIR [22]. However, we detected both reciprocal and non-reciprocal translocations among camptothecin-associated His+ recombinants. Considering that we exposed asynchronous log phase cell cultures to camptothecin, it is possible that camptothecin-associated DNA damage occurred after DNA replication was completed and gap repair mechanisms initiated recombination resulting in reciprocal translocations.
We observed that in rad9 mutants, higher frequencies of DNA damage-associated translocations and lower frequencies of DNA damage-associated SCE do not correlate after exposure to particular DNA damaging agents, such as H2O2. Frequencies of MMS and UV-associated SCE are also similar in wild type and rad9 mutants [12]. Both UV and MMS create DNA adducts that impede DNA replication fork progression and trigger S phase checkpoints [32], although longer MMS-induced S phase has been also attributed to the inhibition of replication initiation at late replication origins [13]. Thus, it would be interesting to determine whether DNA damage-associated SCE was deficient in mutants defective in S phase checkpoints.
We found that RAD9 does not function to suppress DNA damage-associated translocations after exposure to either 4-NQO or MNNG. Consistent with previous literature [26,33], we speculate that these compounds are recombinogenic in S phase, and RAD9-independent S phase checkpoints regulate DNA damage-associated recombination. The RAD9 requirement for efficient UV excision repair [34] and for the G1 cell cycle delay [15] after exposure to DNA damaging agents may explain the sensitivity of rad9 mutants to the UV-mimetic agent 4-NQO and to particular alkylating agents.
Our results are interesting to compare with those from a related study concerning the types of DNA damaging agents that stimulate gross chromosomal rearrangements [35]. The DNA damage-associated frequencies of translocations are higher than frequencies of gross chromosomal rearrangements, possibly reflecting the greater efficiency of homologous recombination, compared to non-homologous recombination. DNA damage resulting from X-rays and bleomycin exposure, generate more gross chromosomal rearrangements than DNA damaging agents that do not directly cause double-strand breaks. However, our results indicate that a subset of alkylating agents and bulky adducts are potent stimulators of chromosomal rearrangements directed by homologous recombination. Although there are several reasons that may account for these differences, it is interesting to note that the RAD9 checkpoint suppresses directed translocations; whereas, the S phase checkpoint is important in suppressing gross chromosomal rearrangements [36]. Thus, it will be interesting to extend these studies and determine whether RAD9-independent S phase checkpoints also suppress homology-directed translocations.
Acknowledgements
This research was supported by grant CA70105 from the National Institutes of Health. We thank Cinzia Cera for carefully reading this manuscript.
References
- [1].Weinert T, DNA damage checkpoints update: getting molecular, Curr. Opin. Genet. Dev 8 (1988) 183–185. [DOI] [PubMed] [Google Scholar]
- [2].Nyberg K, Michelson R, Putnam C, Weinert TA, Toward maintaining the genome: DNA damage and replication checkpoints, Ann. Rev. Genet 36 (2002) 617–657. [DOI] [PubMed] [Google Scholar]
- [3].Friedberg EC, Walker GC, Siede W, DNA Repair and Mutagenesis, ASM Press, Washington DC, 1995. [Google Scholar]
- [4].Weinert TA, Hartwell LH, The RAD9 gene controls the cell cycle response to DNA damage in Sacchromyces cerevisiae, Science 241 (1988) 317–322. [DOI] [PubMed] [Google Scholar]
- [5].Flattery-O’Brien JA, W Dawes I, Hydrogen peroxide causes RAD9-dependent cell cycle arrest in G2 in Saccharomyces cerevisiae whereas menadione causes G1 arrest independent of RAD9 function, J. Biol. Chem 273 (1998) 8564–8571. [DOI] [PubMed] [Google Scholar]
- [6].Kupiec M, Simchen G, Arrest of the mitotic cell cycle and of meiosis in Saccharomyces cerevisiae by MMS, Mol. Gen. Genet 201 (1985) 558–564. [DOI] [PubMed] [Google Scholar]
- [7].Grossman KF, Brown J, Moses RE, Cisplatin DNA cross-links do not inhibit S-phase and cause only a G2/M arrest in Saccharomyces cerevisiae, Mutat. Res 434 (1999) 29–39. [DOI] [PubMed] [Google Scholar]
- [8].Levin NA, Bjornsti MA, Fink GR, A novel mutation in DNA topoisomerase I of yeast causes DNA damage and RAD9-dependent cell cycle arrest, Genetics 133 (1993) 799–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Levin P, Escalza P, Mateos S, Cortes F, Mitomycin C, 4-nitroquinoline-1-oxide and ethyl methanesulfonate induce long-lived lesions in DNA which result in SCEs during successive cell cycles in human lymphocytes, Mutat. Res 270 (1992) 177–183. [DOI] [PubMed] [Google Scholar]
- [10].Fabre F, Boulet A, Roman H, Gene conversion at different points in the mitotic cycle of Saccharomyces cerevisiae, Mol. Gen. Genet 195 (1984) 139–143. [DOI] [PubMed] [Google Scholar]
- [11].Kadyk LC, HartwelL LH, Sister chromatids are preferred over homologs as substrates for recombinational repair in Saccharomyces cerevisiae, Genetics 132 (1992) 387–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Fasullo M, Bennett T, AhChing P, Koudelik J, The Saccharomyces cerevisiae RAD9 checkpoint reduces the DNA damage-associated stimulation of directed translocations, Mol. Cell. Biol 18 (1998) 1190–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Tercero JA, Diffley JF, Regulation of DNA replication fork progression through damaged DNA by the Mec1/Rad53 checkoint, Nature 412 (2001) 553–557. [DOI] [PubMed] [Google Scholar]
- [14].Garvick B, Carson M, Hartwell L, Single-stranded DNA arising at telomeres in cdc13 mutants may constitute a specific signal for the RAD9 checkpoint, Mol Cell Biol. 15, 6128–6138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Siede W, Friedberg A, Friedberg E, RAD9-dependent G1 arrest defines a second checkpoint for damaged DNA in the cell cycle of Saccharomyces cerevisiae, Proc. Nat. Acad. Sci. USA 90 (1993) 7985–7989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Pouliot JJ, Robertson CA, Nash HA, Pathways for repair of topoisomerase I covalent complexes in Saccharomyces cerevisiae, Genes Cells 6 (2001) 677–687. [DOI] [PubMed] [Google Scholar]
- [17].Paulovich AG, Armour CD, Hartwell LH, The Saccharomyces cerevisiae RAD9, RAD17, RAD24 and MEC3 genes are required for tolerating irreparable, ultraviolet-induced DNA damage, Genetics 150 (1998) 75–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Van Brabant A, Buchanan C, Charboneau E, Fangman W, Brewer B, An origin-deficient yeast artificial chromosome triggers a cell cycle checkpoint, Mol. Cell 7 (2001) 705–713. [DOI] [PubMed] [Google Scholar]
- [19].Sherman F, Fink GR, Hicks JB, Appendix A, in: Methods in Yeast Genetics, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, 1986, pp. 163–168. [Google Scholar]
- [20].Fasullo M, Dave P, Rothstein R, DNA-damaging agents stimulate the formation of directed reciprocal translocations in Saccharomyces cerevisiae, Mutat. Res 314 (1994) 121–133. [DOI] [PubMed] [Google Scholar]
- [21].Fasullo M, P Dave, Mating-type regulates the DNA damage-associated stimulation of reciprocal translocation events in Saccharomyces cerevisiae, Mol. Gen. Genet 243 (1994) 63–70. [DOI] [PubMed] [Google Scholar]
- [22].Fasullo M, Giallanza P, Dong Z, Cera C, Bennett T, Saccharomyces cervisiae rad51 mutants are defective in DNA damage-associated sister chromatid exchanges but exhibit increased rates of homology directed translocations, Genetics 158, 959–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Zar JH, Biostatistical Analysis, Prentice Hall, Englewood Cliffs, NJ, 1996. [Google Scholar]
- [24].McKoy JF, P. Pleninger L Wall A Pramanik M Martinez BW Morre, Genetic changes and bioassays in bleomycin- and phleomycin-treated cells, and their relationship to chromosomal breaks, Mutat. Res 336 (1995) 19–27. [DOI] [PubMed] [Google Scholar]
- [25].Galli A, Schiestl RH, Cell division transforms mutagenic lesions into deletion-recombinogenic lesions in yeast cells, Mutat. Res 429 (1999) 13–26. [DOI] [PubMed] [Google Scholar]
- [26].Burke W, Fangman WL, Temporal order in yeast chromosome replication, Cell 5 (1975) 263–269. [DOI] [PubMed] [Google Scholar]
- [27].Aboussekhra A, Vialard JE, Morison D, Torr-Ruiz MA, Cernakova L, Fabre F, Lowndes N, The novel role for the budding yeast RAD9 checkpoint gene in DNA damage-dependent transcription, EMBO J. 14 (1996) 3912–3922. [PMC free article] [PubMed] [Google Scholar]
- [28].Nitiss J, Wang J, DNA topoisomerase-targeting antitumor drugs can be studied in yeast, Proc. Natl. Acad. Sci. USA 85 (1998) 7501–7505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Grossman KF, Ward A, Moses RE, Saccharomyces cerevisiae. Lacking Snm1, Rev3, or Rad51 have a normal S-phase but arrest permanently in G2 after cisplatin treatment, Mutat. Res 461 (2000) 1–13. [DOI] [PubMed] [Google Scholar]
- [30].Malkova A, Ivanov EL, Haber JE, Double-strand break repair in the absence of RAD51 in yeast: a possible role for break-induced DNA replication, Proc. Natl. Acad. Sci. USA 93 (1996) 7131–7136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Haber JE, DNA recombination: the replication connection, Trends Biochem. Sci 24 (1999) 271–275. [DOI] [PubMed] [Google Scholar]
- [32].Foiani M, Gerrari M, Liberi G, Lopes M, Lucca C, Marini F, Pelllicioli A, Muzi Falconi M, P. Plevani S S-phase DNA damage checkpoint in budding yeast, J. Biol. Chem 379 (1999) 1019–1023. [DOI] [PubMed] [Google Scholar]
- [33].Romotar R, Belanger E, Brodeur I, Masson JY, Drobetsky EA, A yeast homolgue of the human phosphotyrozyl phosphatase activator PTPA is implicated in protection against oxidative DNA damage induced by the model carcinogen 4-nitroquinoline 1-oxide, J. Biol. Chem 273 (1998) 21489–21496. [DOI] [PubMed] [Google Scholar]
- [34].Al-Mograbi NM, Al-Sharif IS, Aboussekhra A, The Saccahromyces cerevisiae RAD9 cell cycle checkpoint gene is required for optimal repair of UV-induced pyrimidine dimers in both G(1) and G(2)/M phases of the cell cycle, Nucleic Acids Res. 29 (2001) 2020–2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Myung K, Kolodner R, Induction of genome instability by DNA damage in Saccharomyces cerevisiae, DNA Rep. 2 (2003) 243–258. [DOI] [PubMed] [Google Scholar]
- [36].Kolodner R, Putnam C, Myung K, Maintenance of genome stability in Saccharomyces cerevisiae, Science 297 (2002) 552–557. [DOI] [PubMed] [Google Scholar]