Abstract
We describe the use of a method called differential expression using customized amplification library (DECAL) to study the global changes in gene expression in iron-deficient versus iron-reconstituting cells of Synechocystis sp. strain PCC 6803. We identified a number of genes, such as isiA, idiA, psbA, cpcG, and slr0374, whose expression either increased or decreased in response to iron availability. Further analysis led to the identification of additional genes related to those identified by DECAL (e.g., psbC, psbO, psaA, apcABC, cpcBAC1C2D, and nblA) that were differentially regulated by iron availability. Expression of cpcG, psbC, psbO, psaA, apcABC, and cpcBAC1C2D increased, whereas that of isiA, idiA, nblA, psbA, and slr0374 decreased, in iron-reconstituting cells. S1 nuclease protection studies showed that increased transcript levels of psbA in iron-deficient cells was due to the increased expression of both psbA2 and psbA3 genes, although the steady-state level of psbA2 remained higher than that of psbA3.
Subtractive cDNA cloning, differential display, and DNA microarrays have been used to analyze global gene expression under different growth and environmental conditions (22, 40, 43, 46). These procedures are easier to perform in eukaryotes, due to the presence of a poly(A) tail on mRNA that permits easy separation from rRNA (3). Unfortunately, bacterial mRNA lacks this poly(A) tail and cannot be routinely extracted from total RNA. This problem also makes it difficult to use microarrays in bacteria, because of the high background that is observed when total RNA is used as a labeled probe (17). Moreover, the cost and/or commercial unavailability of DNA chips from prokaryotic organisms make their use largely beyond the financial resources of most within the scientific community. An alternative and cost-effective process to study the differential expression in prokaryotic organisms with small genomes may be available in the form of membranes on which are spotted arrays of Escherichia coli cells containing a library of clones. Nonetheless, an absolute requirement when using such membranes is the removal of the abundant rRNA from total RNA before labeling to avoid high background which may interfere with signal analysis. Recently, a new method called differential expression using customized amplification library (DECAL) has been used in global comparisons of gene expression in Mycobacterium tuberculosis (1). DECAL accomplishes this by first creating a customized amplification library (CAL) of genomic DNA that has been manipulated for optimal performance during analysis. The success of CAL depends on three factors: (i) physical removal of abundant sequences, i.e., rRNA genes; (ii) reduction in the complexity of the sequences and conversion of all DNA sequences to fragments of smaller and similar size; and (iii) selection of sequences that amplify efficiently.
In this study, we developed a DECAL for analysis of the global gene expression in the unicellular cyanobacterium Synechocystis sp. strain PCC 6803, an oxygenic photosynthetic organism. We chose Synechocystis sp. strain PCC 6803 as a model organism because it is transformable with external DNA, the frequency of homologous recombination is very high, thus facilitating genetic analysis, and it is capable of both photoheterotrophic and mixotrophic growth (45). Moreover, the complete sequence of Synechocystis sp. strain PCC 6803 has recently been determined (23). These characteristics have made Synechocystis sp. strain PCC 6803 a model organism for studies of photosynthetic processes and of modifications in response to external and internal stimuli. Such studies have wide application, because cyanobacteria are considered the progenitor of chloroplasts and because a number of biotic and abiotic stresses are known to directly affect photosynthesis and cyanobacterial growth (6, 14, 35, 37, 42).
To demonstrate proof of concept, we used iron-deficient growth of Synechocystis sp. strain PCC 6803 as a growth-limiting condition to analyze gene expression using DECAL. Iron deficiency is known to cause a variety of physiological and morphological changes in cyanobacteria, including loss of light-harvesting phycobilisomes (20), changes in the spectral characteristics of chlorophyll (Chl) within the thylakoids (20, 21, 31, 32), reduction in the number of thylakoids (41), and replacement of cofactors containing iron with noniron cofactors, such as ferredoxin with flavodoxin (24). A new Chl-binding protein, CPVI-4, which is similar to CP43 and encoded by isiA, associates with PSII (and possibly even with PSI) under iron-deficient conditions (31, 32). Similarly, synthesis of IdiA associated with the cytoplasmic side of the thylakoid membrane is greatly enhanced in iron-deficient cells (27). Despite these massive changes under iron-deficient conditions, cells continue to grow to high densities, although the growth rate is somewhat lower and the cells are smaller (41). Furthermore, these changes are reversible; after 24 h of iron addition, cells return to normal (41). In this work, we describe the information provided by the DECAL that can help us understand transcriptional regulation during reconstitution from iron starvation in Synechocystis sp. strain PCC 6803.
MATERIALS AND METHODS
Cyanobacterial strain and growth conditions.
Cultures of Synechocystis sp. strain PCC 6803 were grown phototrophically in liquid BG-11 medium at 30°C under a light intensity of 40 to 50 microeinsteins m−2 s−1 in the presence or absence of iron as described previously (41). Cells were subcultured in iron-deficient media at least two to three times prior to experimental use. Recovery of iron-deficient cultures was accomplished by addition of 6 mg of ferric ammonium citrate per liter of medium. Cells were harvested during recovery at the 0 h (iron-deficient culture) or at 3, 12, and 24 h after the addition of iron (reconstituting cultures).
Construction of a cosmid library.
A cosmid library of Synechocystis sp. strain PCC 6803 was constructed in pYUB328 (a kind gift of W. R. Jacobs) by using the double cosmid vector strategy as described previously (44). Genomic DNA from Synechocystis sp. strain PCC 6803 was isolated as described by Williams (45). Genomic DNA was partially restricted with Sau3AI, and fragments of 35 to 45 kb were isolated on a sucrose gradient as described previously (2). Arms of pYUB328 were prepared by digestion with XbaI, alkaline phosphatase, and then BamHI (4). These arms were ligated with genomic fragments at 1:10 molar ratio (insert to arms). The ligated DNA was in vitro packaged with GigaPack II Gold packaging mix (Stratagene); the resulting recombinant cosmids were transduced into E. coli XL1-Blue MRF′ and selected for ampicillin resistance on solid Luria-Bertani (LB) plates.
Construction of CAL.
A CAL from Synechocystis sp. strain PCC 6803 was constructed essentially as described by Alland et al. (1), with certain modifications. A total of 1,080 cosmid clones were streaked on several solid LB plates containing 50 μg of ampicillin ml−1 using sterile toothpicks, and the toothpicks were subsequently transferred into liquid LB with 50 μg of ampicillin ml−1. LB plates were grown overnight at 37°C, and colonies were transferred onto a positively charged nylon membrane (Schleicher & Schuell). The lysis of colonies and DNA binding to membrane were performed as described previously (39). The liquid cultures were used to isolate cosmid DNA by the sodium dodecyl sulfate (SDS)-alkaline method. Identification of the clones containing rRNA genes was performed by colony hybridization and dot blotting. Cosmid DNA was mixed in a set of 10 and spotted onto the positively charged nylon membrane using a minifold apparatus (Schleicher & Schuell). PCR-amplified rRNA genes (5S, 16S, and 23S) were gel purified, radiolabeled with [α-32P]dCTP (Amersham Pharmacia Biotech) using a random primer Ready-to-Go labeling kit (Amersham Pharmacia Biotech); unincorporated radiolabeled nucleotides were removed by passing the solution through Probe Quant G-50 columns (Amersham Pharmacia Biotech) and used to probe the blots. The DNA from negative clones was pooled and digested individually with NotI and PacI. Insert DNA from NotI and PacI digest was purified from agarose gels with a QiaExII gel extraction kit (Qiagen). Approximately 1-μg fragments from NotI and PacI digest were mixed and separately digested with AluI and HaeIII and were then mixed and fractionated on a 2% NuSieve GTG low-melting-point agarose gel (FMC). Gel slices containing DNA fragments in the range of 400 to 1,500 bp were excised and stored at −20°C. Five microliters of gel slice solution was ligated with 1 μl of XhoI adapters (2 pmol μl−1). Ten microliters of the ligation mix was PCR amplified with 2 μl of 10 μM primers using TaqI DNA polymerase (Promega). The amplification cycle consisted of 3-min hot start followed by 10 cycles of PCR with 1-min segments of 94, 65, and 72°C. After the end of the 10th cycle, 4 U of fresh TaqI DNA polymerase was added and 27 additional cycles at 94°C for 1 min, 65°C for 2 min, and 72°C for 3 min were performed.
Total RNA extraction.
Total RNA from Synechocystis sp. strain PCC 6803 was isolated using the procedure described by Reddy et al. (34), with modifications (15). RNA was isolated using cells harvested from 1-liter cultures collected at 0, 3, 12, and 24 h after the addition of iron to the iron-deficient cells. At each time point, cells were mixed with 1/20 volume of stop solution (200 mM Tris-HCl [pH 8.0], 20 mM EDTA, 20 mM sodium azide) and 20 mM aurintricarboxylic acid pelleted, and stored at −80°C. Aurintricarboxylic acid was omitted from cells when the total RNA was used for enzymatic reactions.
Identification of differentially regulated genes.
One microgram of total RNA was treated with RQ1 RNase-free DNase (Promega) and reverse transcribed with 7.7 μg of biotin-labeled random hexamers and biotin-dATP (one-seventh of total dATP) using Superscript II (GIBCO-BRL) at 45°C for 1 h. Synthesis was terminated by heating the mix at 70°C for 15 min. The complementary RNA was removed by the addition of RNase H with incubation for 30 min at 37°C. Subsequently, 300 ng of CAL, 20 μg of salmon sperm DNA, and 20 μg of tRNA were added to the cDNA for a final volume of 150 μl. The mix was extracted with phenol-chloroform and ethanol precipitated overnight. The pellet was resuspended in 6 μl of 30 mM EPPS [N-(2-hydroxyethyl)piperazine-N′-3-propanesulfonic acid; Sigma], pH 8.0–3 mM EDTA, overlaid with oil, and denatured by heating at 99°C for 5 min followed by addition of 1.5 μl of 5 M NaCl preheated to 69°C. The sample was incubated at 69°C for 4 days followed by addition of 150 μl of incubation buffer (1× Tris-EDTA [pH 7.6], 1 M NaCl, 0.5% Tween 20) that had been preheated to 69°C. Fifty microliters of washed streptavidin-coated magnetic beads (Boehringer Mannheim) that had been preheated to 69°C was added to the mix and incubated at 55°C with occasional mixing for 60 min; the solution was then washed three times for 30 min each at room temperature and three times at 69°C with 0.1% SDS–0.2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) with slow continuous shaking. The sample was then washed with 2.5 mM EDTA and eluted by boiling in 50 μl of water. PCR was performed as for the CAL preparation, using 20 μl of eluted fragments in each sample.
Colony array hybridization.
The cosmid library arrays were prepared using a Bio-grid (BioRobotics) by double-spotting 1,080 cosmid clones into a 22- by 22-cm nylon membranes that rested on solid LB plates containing 50 μg of ampicillin ml−1. The biobank containing 1,080 clones was prepared manually by transferring 1,080 clones into three 384-well microtiter plates in LB containing 10% glycerol and 50 μg of ampicillin ml−1. The cells were stored at −80°C and regularly used for gridding. The colonies were spotted using a three-by-three double-offset pattern. The colonies were grown at 37°C to the optimum size. The lysis of colonies and DNA binding to membrane was done as described elsewhere (39). This yielded six identical replica blots containing 1,080 clones. The PCR-amplified fragments were labeled with [α-32P]dCTP (Amersham Pharmacia Biotech) for at least 6 h, using a random primer Ready-to-Go labeling kit (Amersham Pharmacia Biotech); unincorporated radiolabeled nucleotides were removed by passing the solution through Probe Quant G-50 columns (Amersham Pharmacia Biotech) and hybridized to cosmid library arrays for 16 to 18 h essentially as described elsewhere (39). The blots were washed twice at room temperature for 15 min each in 2× SSC–0.1% SDS and twice at 68°C for 30 min each in 0.1× SSC–0.1% SDS. Double-spotted colonies that showed different intensities with PCR probes were selected for further analysis.
Densitometry analysis.
Density measurements of the autoradiograms were performed with IPLab (Signal Analytics Corporation, Vienna, Va.) on a Macintosh G3 computer. A scanned image of the autoradiogram was segmented into 384 identical squares, and the density of each square was determined. The density value was inversely proportional to the intensity of spots and was divided into 256 segments. We standardized the autoradiograms by analyzing a series of spots with low density that did not change after the addition of iron. This led us to conclude that changes within ±10 density units should be considered identical. We observed 71 spots that differed by ±20 units and 36 spots that differed by more than ±50 units. We chose to analyze further a total of 14 spots: 8 with a density difference of ±50, 3 with a density difference of ∼±20, and 3 with a density difference of ∼±10.
Northern blots.
Five- to fifteen-microgram aliquots of total RNA isolated from cells taken at various time points were electrophoresed on 1% denaturing agarose gels. Each gel was soaked with 20 mM NaOH for 20 min and then transferred in 10× SSC for 45 min with slow shaking. RNA was capillary transferred onto positively charged nylon membranes (Schleicher & Schuell) for 2 to 6 h, using the procedure described in reference 11. RNA was fixed to the membrane by baking at 80°C for 1 to 2 h in a vacuum oven and stained with methylene blue to mark the position of the standard RNA markers and also to check the transfer. Prehybridization and hybridization were performed as described elsewhere (39). The membranes were washed twice at room temperature for 15 min each in 2× SSC–0.1% SDS and twice at 68°C for 15 min each in 0.1× SSC–0.1% SDS. Labeling and purification of various probes were done as described above.
S1 nuclease protection assay.
The S1 nuclease protection assay was performed as described previously (2), with certain modifications. Sufficient primers specific to psbA2 and psbA3 were labeled with [γ-33P]ATP (Amersham Pharmacia Biotech) using T4 polynucleotide kinase (New England Biolabs). The reaction was terminated by heating at 75°C for 10 min and ethanol precipitated twice following the addition of tRNA and ammonium acetate. Twenty micrograms of total RNA was hybridized with excess primers at 51°C for 16 h. Nonhybridized nucleic acid was enzymatically removed by the addition of S1 nuclease (GIBCO-BRL). The mix was ethanol precipitated and loaded on an 8% polyacrylamide gel containing 6 M urea. After completion of the run, the gel was dried and exposed to the X-ray film.
DNA probes and primers.
The following probes were used: for psbA, a 0.6-kb BstEII fragment from Synechococcus sp. strain PCC 7942 (plasmid pSG200 [18]); for psbC, a 0.9-kb Bsu36I-EcoRI fragment from Synechocystis sp. strain PCC 6803 (plasmid pKW1344 [10]); for psbO, a 0.7-kb StuI-BstEII fragment from Synechocystis sp. strain PCC 6803 (plasmid pRB1 [7]); for psaA, a 2.8-kb EcoRI-BglII fragment from Synechococcus sp. strain PCC 7002 (plasmid pAQPR80 [9]); and for cpcBA, a 1.2-kb SmaI-XhoI fragment from Synechococcus sp. strain PCC 7002 (plasmid pAQPRI [9]).
The primers used were based on sequences available in Cyanobase and were synthesized at IDT (Coralville, Iowa). Primers used for rRNA were 5′-ACTTGGCATCGGACTATTGTGCCG-3′, 5′-ACGAGTGACCGTGTGCCTGTTGAA-3′, 5′-ATCGAGCTCCCATTGCTTGTAGGC-3′, and 5′-TTGATCCTGGCTCAGGATGAACGC-3′; for XhoI adapter sequence, 5′-CCTCTGAAGGTTCCAGAATCGATAGCTCGAGT-3′ (top strand) and 5′-P-ACTCGAGCTATCGATTCTGGAACCTTCAGAGGTTT-3′ (bottom strand); XhoI primer sequence, 5′-CCTCTGAAGGTTCCAGAATCGATAG-3′; for cpcG, 5′-GGCTCTGAAGAGAAGCCTGTTGTT-3′ and 5′-GGGCACAGAAGCTTCGATGTTGAT-3′; for slr0374, 5′-CAAGAAGAGCTGAGTGTACTGCTG-3′ and 5′-GAACTCCAGTCGCTGATATTCAGC-3′; for idiA, 5′-ATGACAACTAAGATTTCCCGGCGG-3′ and 5′-TGAATCGGGTTGGTAACGTCCCAA-3′; for T3 primer, 5′-AATTAACCCTCACTAAAGG-3′; for T7 primer, 5′-TAATACGACTCACTATAGGG-3′; for psbA2, 5′-CGCTGTTGGAGAGTCGTTGTCATTTGGT TATAAT TCCT TATGTAT T TGTCGATGT TCAGAT TGGAACTGACTAAACTTAGTC-3′; for psbA3, 5′-CGCTGTTGGAGAGTCGTTGTCATTTGGT TATAAT TCCT TATGTAT T TGTCAATGT TCAAAGGAT T TGGCCTCAAGCTC-3′, for apc, 5′-TTACGGGGGCAGTGTAATCAGG-3′ and 5′-TGGAGCAAAACG-GTTGGACG-3′; and for nblA, 5′-CCCAGAGCAACAACAAGAGTTACTG-3′ and 5′-CAGGTAAGATCAAGTTTGCGGC-3′.
RESULTS
Construction of CAL from Synechocystis sp. strain PCC 6803.
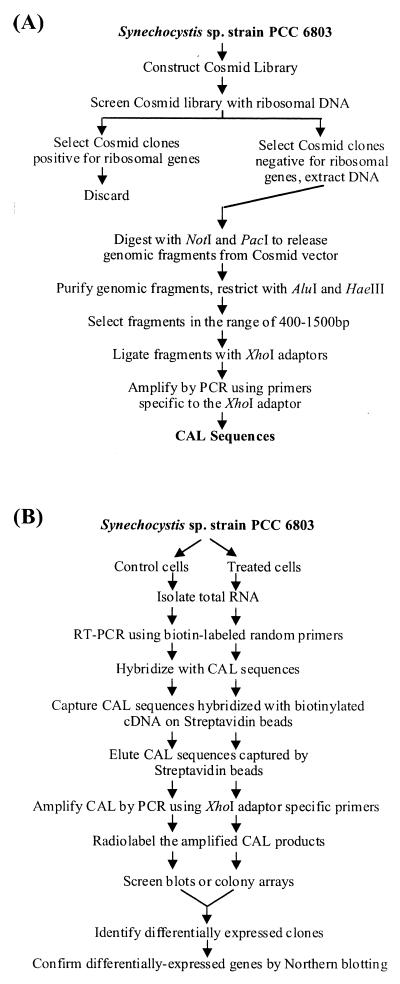
A CAL from Synechocystis sp. strain PCC 6803 was constructed as described by Alland et al. (1) (Fig. 1). Two restriction enzymes, NotI and PacI, were used to excise the genomic fragment from the pYUB328 vector. Both enzymes restrict Synechocystis sp. strain PCC 6803 DNA (12), and it was expected that this approach would minimize the loss of DNA fragments due to any internal site. The uniform size of fragments from the larger genomic fragments were generated following digestion with DraI and HaeIII. These selected fragments were PCR amplified a number of times to select the fragments, which are efficiently amplified. This step was necessary for maintaining the proportional amplification of mRNA in two different population of cells.
FIG. 1.
Flow chart showing various steps involved in construction of a DECAL for Synechocystis sp. strain PCC 6803. (A) Construction of CAL. A cosmid library carrying 35- to 45-kb genomic fragments of Synechocystis sp. strain PCC 6803 was constructed in pYUB328 and screened for clones containing ribosomal DNA sequences. Nonribosomal cosmids were pooled, restricted with suitable enzymes, and gel purified to generate smaller and similar-sized fragments. These fragments were ligated with adapters and PCR amplified to generate CAL sequences. (B) Identification of differentially regulated genes. Total RNA was isolated from Synechocystis sp. strain PCC 6803 cells that were treated under different conditions, reverse transcribed using biotin-labeled random hexamers, and hybridized to CAL sequences. CAL sequences representing cDNA in total RNA was eluted and amplified to generate PCR probes. These probes were radiolabeled and hybridized to replicate colony arrays of the cosmid library. Colonies showing differences in signal in the two arrays were selected, and differential gene expression was confirmed by Northern blot analysis.
Iron-starved cells.
Cyanobacteria, such as Synechococcus sp. strain PCC 7942, respond to iron deficiency with distinct morphological and physiological changes (36, 37, 41). We used iron-deficient growth and subsequent iron reconstitution to demonstrate proof of concept for DECAL, because iron deficiency leads to robust physiological alterations with continued cell growth. Major changes under iron-deficient growth include pigment changes, such as lowered Chl and phycobilisome composition, which result in a very yellowish culture. Such growth also leads to a shift in the Chl absorption peak to about 671 nm; this is due to the presence of a new Chl-binding protein (CP43′) which is encoded by the isiA gene (7, 25, 31–33). The readdition of iron leads to a reconstitution of normal cellular physiology and morphology within 12 to 24 h, a process which is easily identified by the regreening of the culture (41). Thus, analysis of iron deficiency and iron reconstitution provides specific well-characterized landmarks as well as an opportunity to discover the identity of many other genes that are involved in the significant cellular perturbations caused by iron-deficient growth.
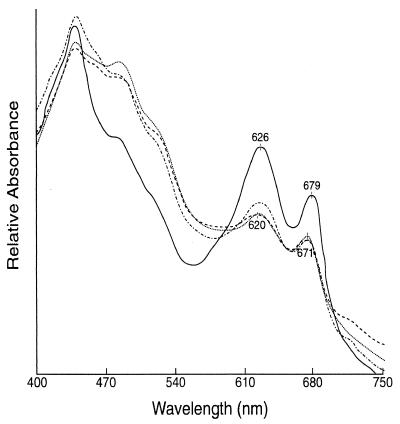
Therefore, we used DECAL to examine transcriptional changes during the first 24 h of reconstitution from iron deficiency. The pigment analysis of iron-starved Synechocystis sp. strain PCC 6803 cells showed decreased Chl and phycocyanin, and the PC/Chl ratio decreased as reported earlier for Synechococcus sp. strain PCC 7942 (36, 37). Addition of iron led to gradual accumulation of pigments, and by 24 h, the pigments levels were very similar to that found in normal cells (data not shown). Figure 2 shows the room temperature absorption spectra of Synechocystis sp. strain PCC 6803 cells that were collected at different time periods after the addition of iron to iron-starved cells. As reported earlier for Synechococcus sp. strain PCC 7942 and shown in Fig. 2 for Synechocystis sp. strain PCC 6803, the Chl absorption peak was blue shifted by 8 nm in iron-deficient cells relative to the peak in iron-sufficient cells. Addition of iron led to a gradual shift of the Chl absorption peak, which was restored to 679 nm by 24 h.
FIG. 2.
Absorption spectra of the iron-deficient Synechocystis sp. strain PCC 6803 cultures (····), and those collected at 3 (––––), 12 (—·—·—·—), and 24 (————) h after the addition of iron.
Identification of differentially regulated genes.
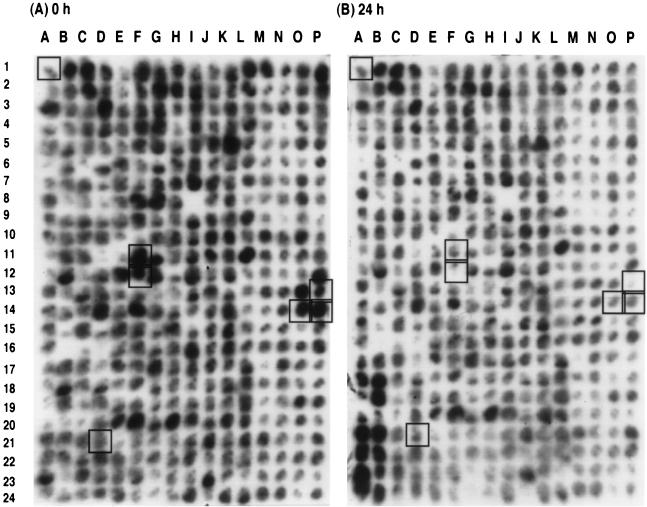
The differentially regulated genes in Synechocystis sp. strain PCC 6803 during iron starvation and iron reconstitution were identified by examining the differential hybridization patterns of cosmid library arrays with PCR probes prepared by the CAL library (Fig. 3). As indicated in Materials and Methods, we chose 11 spots which showed significant changes in iron-starving cells versus iron-reconstituting cells, plus 3 which showed virtually no change. Because of our interest in the physiological changes that occur during iron-deficient growth, this first group emphasized spots that decreased in density after the addition of iron. Cosmid clones representing these spots were further analyzed by dot blot and Northern blot analysis. Northern blot analysis revealed that six of the eight spots with density differences of ∼±50 units (D21, F11, F12, O14, P13, and P14) demonstrated differential expression, whereas only 1 of the 6 spots within ±20 units demonstrated differential expression. However, one of these (B15) included a cosmid containing cpcG. A1 is an example of a control spot with a density difference of ±10 that showed no change in transcription upon further analysis. These results provide some idea of the sensitivity of the Synechocystis sp. strain PCC 6803 DECAL library. We have an additional ∼25 spots with density differences in excess of 50 density units and an additional ∼40 spots with density differences of ±20 from this iron deficiency/iron reconstitution DECAL.
FIG. 3.
Cosmid arrays hybridized with radioactively labeled CAL sequences selected after hybridization with single-stranded cDNA prepared from iron-deficient cells (A) and 24 h after the addition of iron (B). The boxed spots represent cosmids that demonstrated major (D21, F11, F12, O14, P13, and P14) or minor (A1) density differences between the two conditions and that were analyzed further.
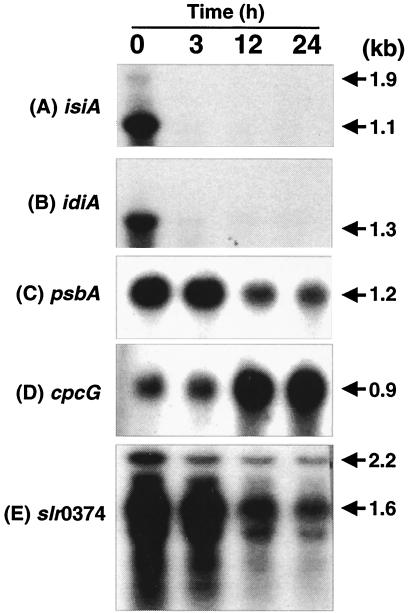
The genes in the seven clones that showed differential regulation were identified by sequencing the DNA from both ends using T3 and T7 primers. These sequences were then used to search for homologous regions in Cyanobase. The specific homologous region was mapped, and all of the genes present in this region were identified. The possible open reading frames (ORFs) were selected based on transcript size showing differential regulation on Northern blots. The primers specific to various ORFs were designed based on sequences available in Cyanobase, and the amplified fragment was used to hybridize the blots containing total RNA isolated from iron-deficient cultures and those at 3, 12, and 24 h during recovery. Using this strategy, we identified two clones containing the isiA gene and two clones with the psbA gene; the other three clones contained cpcG, idiA, and slr0374 (Fig. 4). Regulation of isiA and idiA in response to iron deficiency has been previously characterized (7, 25, 27). As shown in Fig. 4, the steady-state levels of both genes were high in iron-deficient cells and the addition of iron led to rapid loss of transcripts.
FIG. 4.
Northern blot confirmation of genes that were differentially expressed in iron-deficient versus iron-sufficient conditions. Total RNA was isolated from iron-starved cells (0 h) and from cells harvested at 3, 12, and 24 h after the addition of iron. Total RNA (10 μg/lane) was separated on a 1% denaturing agarose gel, capillary transferred, and hybridized with corresponding radiolabeled probes (see Materials and Methods). (A) isiA; (B) idiA; (C) psbA; (D) cpcG; (E) slr0374.
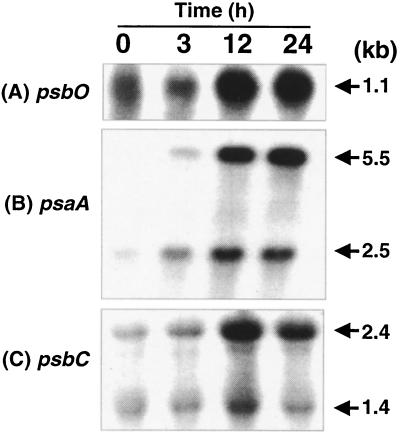
Two clones showing differential regulation contained the psbA gene. As shown in Fig. 4, the steady-state level of the psbA transcript gradually decreased with increasing time periods after the addition of iron. The high psbA transcript level in iron-starved cells was surprising, since it has been shown that Synechocystis sp. strain PCC 6803 cells growing in iron-limiting conditions have decreased levels of PSII and PSI centers (36, 37). Therefore, we expected that steady-state levels of transcripts coding for photosystem proteins would be minimal in iron-deficient cells, with rapid accumulation following addition of iron. Indeed, the transcript levels of psaA, psbC, and psbO, coding for the reaction center protein of PSI, a Chl-binding protein of PSII, and the Mn-stabilizing protein of the oxygen-evolving complex, respectively, were low in iron-starved cells and increased following the addition of iron (Fig. 5).
FIG. 5.
Steady-state transcript levels of psbO (A), psaA (B), and psbC (C) genes in iron-starved and iron-reconstituted cells. Experimental conditions are as described in the legend to Fig. 3; sources of gene probes are detailed in Materials and Methods.
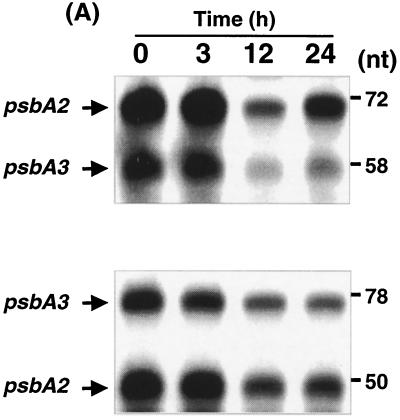
In Synechocystis sp. strain PCC 6803, there are three genes for psbA: psbA1, psbA2, and psbA3 (28). psbA1 is a cryptic gene and does not code for a functional protein, whereas psbA2 and psbA3 are highly homologous and both code for a functional D1 protein (38). Recently, Mate et al. (26) showed the differential regulation of psbA genes in Synechocystis sp. strain PCC 6803 in response to UV-B. Thus, it is possible that the increase in the psbA transcript levels in iron-deficient cells was due to differential expression from the two psbA genes. To decipher the gene-specific regulation by iron, we performed an S1 nuclease protection assay using a synthetic oligonucleotide specific to either psbA2 or psbA3. As shown in Fig. 6, iron starvation led to the accumulation of transcripts originating from both psbA2 and psbA3. Addition of iron to the iron-starved cells had little effect during the first 3 h after iron addition (Fig. 6). However, steady-state transcript levels of the psbA genes decreased after further recovery toward iron sufficiency. It is interesting that whereas transcription of psbA3 continued to decrease until 24 h, the transcription of psbA2 increased between 12 and 24 h after iron addition.
FIG. 6.
S1 nuclease protection assays of psbA2 (A) and psbA3 (B) genes in iron-starved and iron-reconstituted Synechocystis sp. strain PCC 6803 cells. Primers specific to psbA2 and psbA3 were hybridized (at 51°C for 16 h) to 20 μg of total RNA from iron-starved cells or cells collected at 3, 12, and 24 h after the addition of iron. Nonhybridized primers were digested with S1 nuclease, and remaining sample was fractionated on an 8% polyacrylamide gel in the presence of 8 M urea, vacuum dried, and exposed to X-ray films. Lengths of the gene-specific protected fragments, 78 nucleotides (nt) for psbA3 and 50 nt for psbA2, were obtained using the primer homologous to psbA3. In the case of the primer specific to psbA2, gene-specific protected fragments of 72 nt for psbA2 and 58 nt for psbA3 were obtained.
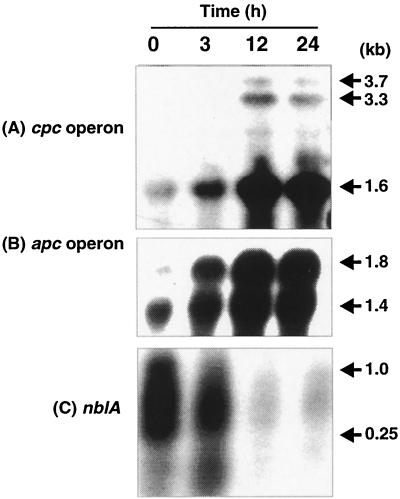
Another gene differentially regulated in response to iron availability was identified as cpcG (Fig. 4B). cpcG encodes for a 30-kDa linker polypeptide that serves in the attachment of phycocyanin hexamers to the phycobilisome core (19). The steady-state level of the cpcG transcript was very low in iron-deficient cells, and iron addition led to the accumulation of transcript. Following this observation, Northern blot analysis was performed to determine the expression of genes coding for allophycocyanin and phycocyanin subunits. Analysis of transcript originating from allophycocyanin operon (slr2067, slr1986, and ssr3383) revealed two transcripts of 1.4 and 1.8 kb, both of which were differentially regulated by iron starvation (Fig. 7A). Similarly, analysis of the phycocyanin operon (sll1577, sll1578, sll1579, sll1580, and ssl3093) revealed three transcripts of 3.7, 3.3, and 1.6 kb which were differentially regulated in response to iron availability (Fig. 7B). The steady-state level of the transcripts originating from phycocyanin and allophycocyanin operons followed kinetics similar to that of cpcG.
FIG. 7.
Northern blot analysis of total RNA from Synechocystis sp. strain PCC 6803 cells probed with DNA from the phycocyanin operon cpcBAC1C2D (A), the allophycocyanin operon apcABC (B), and the nblA gene (C). Experimental conditions are same as for Fig. 3; sources of gene probes are detailed in Materials and Methods.
Recently, nblA has been identified in Synechococcus sp. strain PCC 7942 as a gene involved in phycobilisome degradation (13). It has been shown that the nblA transcript is quite abundant during sulfur and nitrogen starvation in Synechococcus sp. strain PCC 7942 (13), conditions which led to the rapid loss of phycobilisomes. Since iron deficiency leads to the loss of phycobilisomes, we were interested in determining the iron regulation of nblA. Northern blot analysis revealed that nblA transcripts were present at very high levels in iron-starved cells and at low levels 3 h into reconstitution and were virtually absent at 12 and 24 h after the addition of iron (Fig. 7C). Similar to the case for Synechococcus sp. strain PCC 7942, transcripts originating from nblA in Synechocystis sp. strain PCC 6803 revealed a smear of transcripts ranging from 0.25 to 1.0 kb.
Another gene found to be differentially regulated during iron starvation was slr0374. This ORF, which is classified as a cell division cycle protein in Cyanobase, has homology with cell division proteins CDC48 and FtsH from a number of organisms (23). It also demonstrated homology with hypothetical chloroplast protein RF46 from Guillardia theta and hypothetical protein ycf46 from Odontella sinensis and Porphyra purpurea. Although its function in Synechocystis sp. strain PCC 6803 is not known, iron starvation led to an increase in the steady-state level of the slr0374 transcript. Reconstitution of iron-starved cells led to a decrease in the transcript; however, unlike transcripts of iron-regulated isiA and idiA genes, its transcript was present even after 24 h of iron addition. In addition, the blot probed with slr0374 showed a large smear indicative of multiple transcripts.
DISCUSSION
Cyanobacteria are found in virtually all terrestrial niches and face fluctuating chemical and physical parameters such as nutrient availability, light quality and quantity, and temperature. Like other bacteria, they have a plethora of regulatory systems that enable them to respond quickly to such environmental alterations (5), something they have done successfully for billions of years. These adaptive processes involve global changes in gene expression. This study had two objectives: to determine if DECAL was useful for the study of global gene expression in cyanobacteria, and to learn more about genes regulated by iron availability in this unicellular strain. We have demonstrated that our DECAL identified two genes, isiA and idiA, that had previously been shown to be regulated by iron availability (7, 25, 27). In addition, we identified a series of genes involved in photosynthesis or pigment synthesis that had not previously been identified as under iron regulation (psbA, cpcG, and slr0374). We conclude that DECAL can be used successfully to detect genes that are differentially regulated by environmental fluctuations, but that the sensitivity of the current library must be improved.
The differential regulation of the cpcG gene was consistent with the pigment and photosynthetic alterations found in iron-deficient cyanobacteria (20, 21, 31, 32, 41). cpcG is not the only gene which encodes a phycobilisome component that is regulated by iron, since transcripts originating from the apc and cpc operons were also differentially regulated by iron availability. Significantly, the kinetics of transcript accumulation of cpcG, apcABC, and cpcBAC1C2D during reconstitution were similar. In addition, the transcript level of nblA increased in the iron-starved cells. nblA has recently been identified as a gene whose product is involved in the degradation of phycobilisomes (13).
Although it was anticipated that psbA transcription would modulate during transition from growth in iron-deficient to iron-sufficient conditions, the exact expression pattern of the psbA genes was surprising. An increased transcript levels of psbA was observed in iron-deficient cells, whereas iron reconstitution led to a gradual decrease in the transcript level. Previous studies have shown that iron-deficient growth leads to a reduced PSI/PSII ratio and to induction of the isiA gene, which encodes a modified Chl-binding protein, CP43′ (7, 36, 37). Expression analysis of psbC, psbO, and psaA showed decreased transcript levels in iron-deficient cells, compared to an increase in the transcript level of all the three genes during iron reconstitution. The reason for such high levels of psbA transcription is not known, but the overall transcriptional regulation of psbA genes in cyanobacteria is complex. Several studies have suggested that the increase in the steady-state level of psbA transcripts under adverse conditions is due to increased transcription of the psbA3 gene (8, 26, 28, 29). Under normal growth conditions, psbA2 constitutes more than 90% of the psbA transcripts (29). The results with the S1 nuclease protection assay suggested that iron deficiency led to an increased transcription level of both genes in similar patterns. This was in contrast to the regulation of these genes by UV-B, which led to a 20- to 25-fold increase in transcript level of psbA3 but only a 2- to 3-fold increase in the level of psbA2 (26). The addition of iron to iron-deficient cells initially led to the loss of both transcripts at the same rate; however, the accumulation of psbA2 transcript increased between 12 to 24 h during reconstitution, whereas the accumulation of the psbA3 transcript continued to decrease. Finally, it is important to note that in iron-deficient cells, the steady-state level of the psbA2 transcripts was higher than that of psbA3.
This study also resulted in the identification of a previously uncharacterized gene, slr0374, as iron regulated. It is important to note that the transcript originating from slr0374 was abundant in iron-starved cells, suggesting that the gene product might be of greater importance during growth-limiting conditions. Another interesting finding related to slr0374 was the identification of multiple transcripts. Our preliminary data suggest that slr0374 may be part of an operon consisting of at least two additional genes, slr0373 and slr0376 (A. K. Singh and L. A. Sherman, unpublished data). Although the function of slr0374 is not known, analysis of its primary sequence based on amino acid sequence homology shows that it belongs to class of proteins with important cellular functions. A preliminary sequence analysis indicated that slr0374 may contain a leucine zipper and a Walker motif found in members of the AAA protein family, including a conserved second region of homology. AAA modules function as regulatory subunits in many complexes, including the 26S proteasome, in the assembly of various membrane-targeting protein complexes during membrane fusion, peroxisome biogenesis, assembly of mitochondrial membrane proteins, cell cycle control, mitotic spindle formation, cytoskeleton interactions, vesicle secretion, signal transduction, and transcription factors (30). We will determine if slr0374 is induced during alterations in other environmental parameters and whether it is a specific or a general stress response protein.
In summary, this work has enabled us to determine the sensitivity and utility of a DECAL for analyses of differential gene expression in the cyanobacterium Synechocystis sp. strain PCC 6803. The results indicated that spots demonstrating the largest density difference had a high probability of containing clones with genes that are transcriptionally regulated by iron concentration. We have an additional ∼25 spots which reproducibly show density difference of ±50 units plus another 40 with medium density differences. Nonetheless, some of these spots, as well as those we chose as controls, indicated no change in expression upon further study. This could be for a number of reasons, the most important of which is the use of cosmids carrying large fragments (35 to 45 kb) of DNA. Thus, it is possible that a spot contains a clone with some genes that were up-regulated and some that were down-regulated. It is also possible that some clones may contain genes with highly abundant transcripts, which might mask the difference in the signal pattern caused by differentially regulated genes present in the same fragment. Nonetheless, once the DECAL has been constructed, it represents a relatively simple method that can generate important information on differential gene expression. We will extend this work by constructing a plasmid library array containing 6,000 clones with ∼2-kb fragments, and this will be used in conjunction with the DECAL. We are also involved with the production of a complete genome microarray containing all 3,168 genes of Synechocystis sp. strain PCC 6803. Nonetheless, these microarrays will require a great deal of fine tuning and many duplicates before they are considered reliable. The use of a rapid and inexpensive DECAL library will still remain of use as an adjunct to such high-resolution microarrays.
ACKNOWLEDGMENTS
We thank Torin Weisbrod and Bill Jacobs for pYUB328, Kim Hirsch for technical help, and Hong Li for discussions and for comments on the manuscript.
This research was supported by grant DE-FG02-99ER20342 from the Department of Energy.
REFERENCES
- 1.Alland D, Kramnik I, Weisbrod T R, Otsubo L, Cerny R, Miller L P, Jacobs W R J, Bloom B R. Identification of differentially expressed mRNA in prokaryotic organisms by customized amplification libraries (DECAL): the effect of isoniazid on gene expression in Mycobacterium tuberculosis. Proc Natl Acad Sci USA. 1998;95:13227–13232. doi: 10.1073/pnas.95.22.13227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: Greene Publishing and Wiley-Interscience; 1999. [Google Scholar]
- 3.Aviv H, Leder P. Purification of biologically active globin messenger RNA by chromatography on oligothymidylic acid-cellulose. Proc Natl Acad Sci USA. 1972;69:1408–1412. doi: 10.1073/pnas.69.6.1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Balasubramanian V, Pavelka M S, Jr, Bardarov S S, Martin J, Weisbrod T R, McAdam R A, Bloom B R, Jacobs W R J. Allelic exchange in Mycobacterium tuberculosis with long linear recombination substrates. J Bacteriol. 1996;178:273–279. doi: 10.1128/jb.178.1.273-279.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Behrenfeld M J, Kolber Z S. Widespread iron limitation of phytoplankton in the South Pacific Ocean. Science. 1999;283:840–843. doi: 10.1126/science.283.5403.840. [DOI] [PubMed] [Google Scholar]
- 6.Bhaya D, Watanabe N, Ogawa T, Grossman A R. The role of an alternative sigma factor in motility and pili formation in the cyanobacterium Synechocystis sp. strain PCC 6803. Proc Natl Acad Sci USA. 1999;96:3188–3193. doi: 10.1073/pnas.96.6.3188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burnap R L, Troyan T, Sherman L A. The highly abundant chlorophyll-protein complex of iron deficient Synechococcus sp. PCC7942 (CP43′) is encoded by the isiA gene. Plant Physiol. 1993;103:893–902. doi: 10.1104/pp.103.3.893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Campbell D, Eriksson M-J, Oquist G, Gustafsson P, Clarke A K. The cyanobacterium Synechococcus resists UV-B by exchanging photosystem II reaction-center D1 proteins. Proc Natl Acad Sci USA. 1998;95:364–369. doi: 10.1073/pnas.95.1.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cantrell A, Bryant D A. Molecular cloning and nucleotide sequence of the psaA and psaB genes of the cyanobacterium Synechococcus sp. PCC 7002. Plant Mol Biol. 1987;9:453–468. doi: 10.1007/BF00015877. [DOI] [PubMed] [Google Scholar]
- 10.Chisholm D, Williams J G K. Nucleotide sequence of psbC, the gene encoding the CP-43 chlorophyll a-binding protein of Photosystem II, in the cyanobacterium Synechocystis 6803. Plant Mol Biol. 1988;10:293–301. doi: 10.1007/BF00029879. [DOI] [PubMed] [Google Scholar]
- 11.Chomczynski P, Mackey K. One-hour downward capillary blotting of RNA at neutral pH. Anal Biochem. 1994;221:303–305. doi: 10.1006/abio.1994.1416. [DOI] [PubMed] [Google Scholar]
- 12.Churin Y N, Shalak I N, Borner T, Shestakov S V. Physical and genetic map of the chromosome of the unicellular cyanobacterium Synechocystis sp. strain PCC 6803. J Bacteriol. 1995;177:3337–3343. doi: 10.1128/jb.177.11.3337-3343.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Collier J L, Grossman A R. A small polypeptide triggers complete degradation of light-harvesting phycobiliproteins in nutrient-deprived cyanobacteria. EMBO J. 1994;13:1039–1047. doi: 10.1002/j.1460-2075.1994.tb06352.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Collier J L, Herbert S K, Fork D C, Grossman A R. Changes in the cyanobacterial photosynthetic apparatus in response to macronutrient deprivation. Photosyn Res. 1994;42:173–183. doi: 10.1007/BF00018260. [DOI] [PubMed] [Google Scholar]
- 15.Colon-Lopez M S, Sherman D M, Sherman L A. Transcriptional and translational regulation of nitrogenase in light-dark- and continuous-light-grown cultures of the unicellular cyanobacterium Cyanothece sp. strain ATCC 51142. J Bacteriol. 1997;179:4319–4327. doi: 10.1128/jb.179.13.4319-4327.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Conway G. A novel gene expressed during zebrafish gastrulation identified by differential RNA display. Mech Dev. 1995;52:383–391. doi: 10.1016/0925-4773(95)00416-x. [DOI] [PubMed] [Google Scholar]
- 17.de Saizieu A, Certa U, Warrington J, Gray C, Keck W, Mous J. Bacterial transcript imaging by hybridization of total RNA to oligonucleotide arrays. Nat Biotechnol. 1998;16:45–48. doi: 10.1038/nbt0198-45. [DOI] [PubMed] [Google Scholar]
- 18.Golden S S, Brusslan J, Haselkorn R. Expression of a family of psbA genes encoding a photosystem II polypeptide in the cyanobacterium Anacystis nidulans R2. EMBO J. 1986;5:2789–2798. doi: 10.1002/j.1460-2075.1986.tb04569.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grossman A R, Schaefer M R, Chiang G G, Collier J L. The phycobilisome, a light-harvesting complex responsive to environmental conditions. Microbiol Rev. 1993;57:725–749. doi: 10.1128/mr.57.3.725-749.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guikema J A, Sherman L A. Organization and function of chlorophyll in membranes of cyanobacteria during iron starvation. Plant Physiol. 1983;73:250–256. doi: 10.1104/pp.73.2.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guikema J A, Sherman L A. Influence of iron deprivation on the membrane composition of Anacystis nidulans. Plant Physiol. 1984;74:90–95. doi: 10.1104/pp.74.1.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gupta R, Thomas P, Beddington R S P, Rigby P W J. Isolation of developmentally regulated genes by differential display screening of cDNA libraries. Nucleic Acids Res. 1998;26:4538–4539. doi: 10.1093/nar/26.19.4538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaneko T, Sato S, Kotani H, Tanaka A, Asamizu E, Nakamura Y, Miyajima N, Hirosawa M, Sugiura M, Sasamoto S, Kimura T, Hosouchi T, Matsuno A, Muraki A, Nakazaki N, Naruo K, Okumura S, Shimpo S, Takeuchi C, Wada T, Watanabe A, Yamada M, Yasuda M, Tabata S. Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC6803. II. Sequence determination of the entire genome and assignment of potential protein-coding regions. DNA Res. 1996;3:185–209. doi: 10.1093/dnares/3.3.185. [DOI] [PubMed] [Google Scholar]
- 24.Laudenbach D E, Reith M E, Straus N A. Isolation, sequence analysis, and transcriptional studies of the flavodoxin gene from Anacystis nidulans R2. J Bacteriol. 1988;170:258–265. doi: 10.1128/jb.170.1.258-265.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Laudenbach D E, Straus N A. Characterization of a cyanobacterial iron stress-induced gene similar to psbC. J Bacteriol. 1988;170:5018–5026. doi: 10.1128/jb.170.11.5018-5026.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mate Z, Sass L, Szekeres M, Vass I, Nagy F. UV-B-induced differential transcription of psbA genes encoding the D1 protein of photosystem II in the cyanobacterium Synechocystis 6803. J Biol Chem. 1998;273:17439–17444. doi: 10.1074/jbc.273.28.17439. [DOI] [PubMed] [Google Scholar]
- 27.Michel K-P, Thole H H, Pistorius E K. IdiA, a 34 kDa protein in the cyanobacteria Synechococcus sp. strains PCC 6301 and PCC 7942, is required for growth under iron and manganese limitations. Microbiology. 1996;142:2635–2645. doi: 10.1099/00221287-142-9-2635. [DOI] [PubMed] [Google Scholar]
- 28.Mohamed A, Jansson C. Influence of light on accumulation of photosynthesis-specific transcripts in the cyanobacterium Synechocystis 6803. Plant Mol Biol. 1989;13:693–700. doi: 10.1007/BF00016024. [DOI] [PubMed] [Google Scholar]
- 29.Mohamed A, Eriksson J, Osiewacz M D, Jansson C. Differential expression of the psbA genes in the cyanobacterium Synechocystis 6803. Mol Gen Genet. 1993;238:161–168. doi: 10.1007/BF00279543. [DOI] [PubMed] [Google Scholar]
- 30.Neuwald A F, Aravind L, Spouge J L, Koonin E V. AAA+: a class of chaperone-like ATPases associated with the assembly, operation, and disassembly of protein complexes. Genome Res. 1999;9:27–43. [PubMed] [Google Scholar]
- 31.Pakrasi H B, Goldenberg A, Sherman L A. Membrane development in the cyanobacterium, Anacystis nidulans, during recovery from iron starvation. Plant Physiol. 1985;79:290–295. doi: 10.1104/pp.79.1.290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pakrasi H B, Riethman H C, Sherman L A. Organization of pigment proteins in the photosystem II complex of the cyanobacterium Anacystis nidulans R2. Proc Natl Acad Sci USA. 1985;82:6903–6907. doi: 10.1073/pnas.82.20.6903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Park Y I, Sandstrom S, Gustafsson P, Oquist G. Expression of the isiA gene is essential for the survival of the cyanobacterium Synechococcus sp. PCC 7942 by protecting photosystem II from excess light under iron limitation. Mol Microbiol. 1999;32:123–129. doi: 10.1046/j.1365-2958.1999.01332.x. [DOI] [PubMed] [Google Scholar]
- 34.Reddy K J, Webb R, Sherman L A. Bacterial RNA isolation with one hour centrifugation in a table-top ultracentrifuge. BioTechniques. 1990;8:250–251. [PubMed] [Google Scholar]
- 35.Reumann S, Keegstra K. The endosymbiotic origin of the protein import machinery of chloroplastic envelope membranes. Trends Plant Sci. 1999;4:302–307. doi: 10.1016/s1360-1385(99)01449-1. [DOI] [PubMed] [Google Scholar]
- 36.Riethman H C, Sherman L A. Purification and characterization of an iron stress-induced chlorophyll-protein from the cyanobacterium Anacystis nidulans R2. Biochim Biophys Acta. 1988;935:141–151. doi: 10.1016/0005-2728(88)90211-3. [DOI] [PubMed] [Google Scholar]
- 37.Riethman H C, Bullerjahn G, Reddy K J, Sherman L A. Regulation of cyanobacterial pigment-protein composition and organization by environmental factors. Photosyn Res. 1988;18:133–161. doi: 10.1007/BF00042982. [DOI] [PubMed] [Google Scholar]
- 38.Salih G F, Jansson C. Activation of the silent psbA1 gene in the cyanobacterium Synechocystis sp strain 6803 produces a novel and functional D1 protein. Plant Cell. 1997;9:869–878. doi: 10.1105/tpc.9.6.869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 40.Schena M, Shalon D, Davis R W, Brown P O. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science. 1995;270:467–470. doi: 10.1126/science.270.5235.467. [DOI] [PubMed] [Google Scholar]
- 41.Sherman D M, Sherman L A. Effect of iron deficiency and iron restoration on ultrastructure of Anacystis nidulans. J Bacteriol. 1983;156:393–401. doi: 10.1128/jb.156.1.393-401.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sherman L A, Meunier P, Colon-Lopez M S. Diurnal rhythms in metabolism: a day in the life of a unicellular, diazotrophic cyanobacterium. Photosyn Res. 1999;58:25–42. [Google Scholar]
- 43.Vernon D M, Bohnert H J. A novel methyl transferase induced by osmotic stress in the facultative halophyte Mesembryanthemum crystallinum. EMBO J. 1992;11:2077–2085. doi: 10.1002/j.1460-2075.1992.tb05266.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wahl G M, Lewis K A, Ruiz J C, Rothenberg B, Zhao J, Evans G A. Cosmid vectors for rapid genomic walking, restriction mapping and gene transfer. Proc Natl Acad Sci USA. 1987;84:2160–2164. doi: 10.1073/pnas.84.8.2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Williams J G K. Construction of specific mutations in photosystem II photosynthetic reaction center by genetic engineering methods in Synechocystis 6803. Methods Enzymol. 1988;167:766–778. [Google Scholar]
- 46.Zhao N, Hashida H, Takahashi N, Misumi Y, Sakaki Y. High-density cDNA filter analysis: a novel approach for large-scale quantitative analysis of gene expression. Gene. 1995;156:207–213. doi: 10.1016/0378-1119(95)00023-y. [DOI] [PubMed] [Google Scholar]