Abstract
Ginsenoside Rh2, which is extracted from ginseng, exerts antitumor activity. Recent studies suggest that Rh2 may suppress the growth of colon cancer (CC) in vitro. However, the underlying mechanism remains unclear. In this study, we identified the relative levels of miR-150-3p in CC tissues and cells by a comprehensive strategy of data mining, computational biology, and real-time reverse transcription PCR (qRT-PCR) experiments. The regulatory effects of miR-150-3p/SRCIN1 on the proliferative and invasive abilities of CC cells are evaluated by CCK-8, EdU, wound healing, and transwell assays. Cell cycle- and apoptosis-related protein levels are assessed by western blot analysis. An in vivo tumor formation assay was conducted to explore the effects of miR-150-3p on tumor growth. Furthermore, bioinformatics and dual luciferase reporter assays are applied to determine the functional binding of miRNA to mRNA of the target gene. Finally, the relationship between Rh2 and miR-150-3p was further verified in SW620 and HCT-116 cells. miR-150-3p is downregulated in CC tissues and cell lines. Functional assays indicate that the upregulation of miR-150-3p inhibits tumor growth both in vivo and in vitro. In addition, SRCIN1 is upregulated in CC and predicts a poor prognosis, and it is the direct target for miR-150-3p. Moreover, the miR-150-3p mimic decreases Topflash/Fopflash-dependent luciferase activity, resulting in the inhibition of Wnt pathway activity. Rh2 can suppress the growth of CC by increasing miR-150-3p expression. Rh2 alleviates the accelerating effect on Wnt pathway activity, cell proliferation/migration, and colony formation caused by miR-150-3p inhibition. Rh2 inhibits the miR-150-3p/SRCIN1/Wnt axis to suppress colon cancer growth.
Keywords: ginsenoside Rh2; colon cancer; miR-150-3p, SRCIN1, Wnt pathway
Introduction
Colon cancer (CC) is one of the most prevalent cancers in the world in terms of its incidence and mortality rate [1], and as such, each year, more than one million new cases are diagnosed with CC, while approximately 600,000 CC patients die [2]. Similar to other cancer types, CC occurs as a result of various factors, such as the mutation of oncogenes and tumor suppressor genes, which activate and inactivate them, respectively [3]. Currently, resectable and advanced CC is mostly treated through radio- and chemotherapy [4], and additional efforts are being geared towards improving patient survival. However, for those who are in the later stages of CC, the prognosis remains poor. Therefore, to identify new and potentially more effective drug candidates, it is important to uncover the oncogenic mechanism of CC.
For thousands of years, ginseng has been traditionally applied as a Chinese herbal medicine [5], and its extract ginsenoside Rh2 was reported to exhibit potential antitumor activity against different types of cancer cell lines, such as the human breast cancer line MCF-7 and the sarcoma cell line SK-LMS-1 [6]. In particular, under hypoxic conditions, ginsenoside Rh2 was reported to prevent human lung cancer from migrating through the action of miRNA-491 [7], with similar effects observed against pancreatic cancer cells except that in this case the effects are mediated by matrix metalloproteinases (MMP)-2 and MMP-9 [8]. Despite the potential of ginsenoside Rh2 against tumors, its role in CC remains unknown.
MicroRNAs (miRNAs), which are small noncoding RNA molecules, control most of the signaling pathways of cells, including the development of both normal and cancer cells [ 9, 10] , and they prevent proteins from being translated by targeting mRNA and binding to their 3′ untranslated regions [11]. It was previously reported that miRNA-150-3p could potentially act as a diagnostic biomarker for CC, [12]. Indeed, when overexpressed, miRNA-150-3p downregulates SLCO4A1 expression, thereby repressing migration, invasion, sphere formation and tumorigenesis and increasing apoptosis in CC stem cells [13]. More recently, ginsenoside Rh2 was found to induce changes in miRNA in CC cells [14], with one example being the suppression of growth and metastasis of the cells through the action of miR-491 [15]. Interestingly, the ability of ginsenoside Rh2 to exert antitumor effects by inhibiting the activity of PBK/TOPK is specific to the human CC cell line HCT116 [16]; hence, ginsenoside Rh2 has the potential to be considered a promising drug for treating CC.
Cell proliferation, development and differentiation are regulated by the canonical Wnt signaling pathway [17], and the latter’s dysfunction has been linked to CC [18]. Indeed, mutation of the tumor suppressor adenomatous-polyposis coli (APC), an important feature of Wnt signaling, is found in 85% of colon cancer patients [19]. When Wnt is not activated, APC forms a protein complex with glycogen synthase kinase (GSK)-3β, casein kinase 1, axin and protein phosphatase 2A to carry out the degradation of β-catenin [20]. However, when APC is mutated, β-catenin is stabilized and translocated to the nucleus, where it complexes with T-cell factor/lymphoid enhancer factor (TCF/LEF) to trigger gene transcription [21]. Although the link between CC and mutations of the Wnt pathway has already been highlighted by TCGA [22], the underlying mechanism that activates a dysfunctional Wnt/β-catenin signaling pathway remains to be studied.
In this study, we demonstrated that the expression of miR-150-3p is negatively correlated with the proliferation, migration, and invasion abilities of CC cells in vitro. In addition, we also identified the SRC kinase signaling inhibitor 1 ( SRCIN1) gene as the target of miR-150-3p and the mediator of Wnt pathway activation. Finally, the antitumor function of ginsenoside Rh2 on CC was clarified to be relied on its regulation on miR-150-3p.
Materials and Methods
Data collection
Datasets from The Cancer Genome Atlas Colon Adenocarcinoma (TCGA-COAD, https://portal.gdc.cancer.gov/projects/TCGA-COAD) were used to obtain miRNA expression profiles. Using the edgeR package in R ( https://bioconductor.org/packages/release/bioc/html/edgeR.html), genes that are differentially expressed were analysed, with differential expression identified by selecting a P value of <0.05 and a |logFC| value of ≥ 1 as the thresholds. Data from the TCGA-COAD dataset were then used for correlational analyses, with statistically significant correlations indicated by P values<0.05.
Bioinformatics analysis
Interactions between miRNAs and mRNAs were predicted using miRDB ( http://mirdb.org/index.html) and TargetScan ( https://www.targetscan.org/vert_80/), with intersections between the results of the two software programs selected as the miRNAs’ possible targets. Correlational analyses of miRNAs-mRNAs were then carried out using Pearson’s correlation coefficient in the corrplot package in R ( https://www.rdocumentation.org/packages/corrplot/versions/0.92) before eventually visualizing the results using R’s ggpubr package. Significant correlations were identified from P values<0.05.
Prognostic analysis
The suitability of the SRCIN1 gene for predicting the progression-free survival (DFS), overall survival (OS) and specific survival (DSS) of CC patients was determined based on log-rank tests and Kaplan-Meier analyses using the survival package [23]. After determining the median expression level of SRCIN1, survival outcomes were compared between patients with high or low expression of the gene. The statistical significance of differences was determined at the 5% significance level.
Cell culture and treatment
Cell lines (colonic epithelial cells NCM460 and four human CC cells-HCT116, SW620, SW480, and CaCo-2) were obtained from the ATCC (Manassas, USA) and cultured in RPMI (Invitrogen) containing 1% penicillin/streptomycin mixture and 10% FBS (Gibco, Grand Island, USA) at 37°C under 5% CO 2. Following incubation, the cell lines HCT-116 and SW620 were subject to a 72-h treatment with different concentrations of ginsenoside Rh2 (0, 10 and 20 μM).
Cell transfection
miR-150-3p mimics (sense 5′-CUGGUACAGGCCUGGGGGA-3′, antisense 5′-CCCCAGGCCUGUACCAGUU-3′), miR-150-3p inhibitor (5′-UCCCCCAGGCCUGUACCAG-3′), and the negative controls (mimics NC: sense 5′-UUCUCCGAACGUGUCACGUTT-3′, antisense 5′-ACGUGACACGUUCGGAGAATT-3′; and inhibitor NC: 5′-GGCCUCACCGGGUGUAAAUCAG-3′) were obtained from RiboBio (Guangzhou, China). The SRCIN1 gene was then overexpressed by cloning its coding sequence into a pcDNA3.0 vector (Invitrogen, Carlsbad, USA). In brief, cells were cultured until a confluency of 80%–90%, and the transfection was performed. Cells were subsequently transfected using Lipofectamine 2000 (Thermo Fisher Scientific, Waltham, USA) according to the manufacturer’s instructions. The whole medium was changed after 6 h, and the cells were finally collected for further experiments after an additional incubation for 24–48 h. The expression level of miR-150-3p or SRCIN1 was determined by the RT-qPCR analysis.
Real-time reverse transcription PCR (qRT-PCR)
Total RNA was extracted with TRIzol reagent (Thermo Fisher Scientific) and reverse transcribed using the M-MLV First-Strand Synthesis reagent (Thermo Fisher Scientific) prior to qRT-PCR, performed on an iCycler IQ® RT-PCR Detection System (Bio-Rad Laboratories, Hercules, USA) using SYBR-Green Quantitative RT-qPCR kit (QR0100; Sigma-Aldrich, St Louis, USA). In this case, the PCR consisted of the following conditions: an initial denaturation for 2 min at 95°C, followed by 40 cycles, each with denaturation for 5 s at 95°C, annealing for 15 s at 60°C, and extension for 10 s at 72°C. The levels of SRCIN1 and miR-150-3p expression were finally quantified based on the 2 –ΔΔCq method using GAPDH and U6 respectively, as the internal controls. Primer sequences for this set of experiments are provided in Supplementary Table S1.
Cell viability assay
Cell viability was determined using the recommended protocol of the Cell Counting Kit-8 (CCK-8) system (Dojindo Laboratory, Kumamoto, Japan) [24]. Briefly, cells in logarithmic growth phase were collected 48 h after transfection and seeded at a density of 1×10 5 cells/well in a 96-well plate. At the designated time points (0, 24, 48, and 72 h), 10% CCK-8 reagent was added to the cells and incubated at 37°C for 2 h. The absorbance of each well was determined at 450 nm with a microplate reader (Bio-Rad Laboratories). Each group had six replicates, with all experiments carried out at least three times.
Colony formation assay
For this assay, 2 mL of medium supplemented with 10% FBS was added to 6-well plates before 600 cells were seeded into each well. Colonies formed after 14 days of culture were washed with PBS before cell fixation for 10 min with 4% paraformaldehyde. This was followed by a 40-min staining using Giemsa (Sigma-Aldrich), and after a second wash with PBS, visible colonies with diameters> 50 mm were counted.
EdU assay
To assess cell proliferation, an EdU assay kit (RiboBio) was used. For this purpose, after 1×10 6 transfected cells were seeded into each well of confocal plates, incubation was performed with 50 μM EdU buffer for 2 h at 37°C. After permeabilization with 0.1% Triton X-100 for 20 min, cells were subjected to fixation for half an hour using 4% formaldehyde. After addition of the EdU solution, the cell nuclei were stained with DAPI. Finally, an SP8 confocal microscope (Leica, Wetzlar, Germany) was used to measure the percentage of EdU-positive cells.
Wound healing assay
Forty-eight hours after transfection, cells were seeded into 6-well plates at 1×10 6 cells per well and cultured until the cell confluence was approximately 95%. Afterwards, a 200-μL pipette tip was used to make vertical linear scratches in the 6-well plate, and PBS was applied to remove the cell debris. After that, the cells were cultured in serum-free medium. Images of the wound gap were then captured under an IX71 inverted microscope (Olympus, Tokyo, Japan) after 0, 24, and 48 h. The differences in scratch healing among different groups were compared, and the healing rate was interpreted as the cell migration ability.
Transwell assay
For each group, Matrigel (BD Biosciences) was used to coat the membrane of the Transwell insert (Corning Incorporated, Corning, USA). Next, the transfected cells from all groups were collected and seeded in the upper chamber of the Transwell chamber, and 500 μL medium containing 10% FBS was added to the lower chamber of the 24-well plate. After being cultured for 24 h, the cells that did not penetrate the membrane in the Transwell chamber were gently removed, and the remaining cells were subjected to crystal violet staining prior to visualization under the IX71 inverted microscope. Finally, five visual fields were selected to calculate the average number of cells in each group. The number of cells passing through Matrigel was used as an indicator to judge the invasive ability of the cells. The migration assay was performed in accordance with the invasion assay except that the Transwell membrane was not precoated with Matrigel.
RNA immunoprecipitation (RIP) assay
RIP assay was performed using the Magna RIP RNA-Binding Protein Immunoprecipitation kit (Millipore, Billerica, USA). Briefly, cell lysate from 2×10 7 SW480 or HCT116 cells was incubated with magnetic beads to which human anti-Ago2 or normal mouse IgG (negative control) antibodies were conjugated (Millipore). Finally, the immunoprecipitated RNAs were subjected to qRT-PCR to confirm whether the bound targets were enriched, with the resulting products further subjected to 1% agarose gel electrophoresis.
Luciferase activity assay
The Topflash/Foplash assay was carried out to assess the activity of Wnt signaling, and for this purpose, Topflash and Fopflash plasmids were obtained from Addgene (Watertown, MA, USA). The former contained seven TCF/LEF binding sites, while the latter had six mutated sites located upstream of the luciferase reporter. Forty-eight hours after cotransfection using the plasmid pTK-renilla, the Topflash/Fopflash vector and 60 nM miRNA mimics or inhibitors, the intensity of luminescence was eventually recorded with the Dual-Glo luciferase assay kit (#E1910; Promega, Madison, USA).
For the miRNA binding luciferase activity assay, cotransfection of cells with NC mimic/miR-150-3p mimic and SRCIN1-WT/SRCIN1-MUT was achieved using Lipofectamine 2000 (Thermo Fisher Scientific). After 48 h, luciferase activity was then determined for lysed cotransfected cells using the Dual-Luciferase Reporter Assay System (Promega). Renilla luciferase units were used as the control to calculate the relative activities of the promoter.
Western blot analysis
After cell lysis with RIPA buffer (Cell Signaling Technology, Danvers, USA), the resulting proteins were quantified using the BCA Protein Assay kit (Cell Signaling Technology). Proteins (30 μg) from each sample were then separated by 12% SDS-PAGE prior to transfer onto PVDF membranes (Sangon, Shanghai, China). This was followed by membrane blocking using 5% nonfat milk (Sangon) and subsequent overnight incubation at 4°C with the following primary antibodies: SRCIN1 (ab122612, 1:1000, rabbit mAb; Abcam, Waltham, USA), PCNA (#13110, 1:1000, rabbit mAb; Cell Signaling Technology), Bax (#5023, 1:1000, rabbit mAb; Cell Signaling Technology), cleaved caspase3 (#9664, 1:1000, rabbit mAb; Cell Signaling Technology), Survivin (#2808, 1:1000, rabbit mAb; Cell Signaling Technology), Cyclin D1 (#55506, 1:1000, rabbit mAb; Cell Signaling Technology), MYC (#2276, 1:1000, rabbit mAb; Cell Signaling Technology), and β-catenin (#8480, 1:1000, rabbit mAb; Cell Signaling Technology). In this case, GAPDH (#5174, 1:1000, rabbit mAb; Cell Signaling Technology) was used as an internal control. After incubation, the secondary HRP-conjugated anti-rabbit IgG (#7074, 1:3000; Cell Signaling Technology) antibody was added and incubated for a 1 h at room temperature. Finally, SignalFire™ ECL Reagent (Cell Signaling Technology) was used to detect protein bands before analysing the results with ImageLab software v 4.1 (Bio-Rad). The ratio of the band intensity of the target protein to that of the internal reference was indicated as the relative expression of the gene.
In vivo tumor model
Four-week-old female BALB/c nude mice, obtained from Lanzhou University Laboratory Animal Center (Lanzhou, China), were kept under pathogen-free conditions in laminar airflow cabinets. After the mice were randomly assigned into three groups (miR-150-3p agomir group, NC agomir group and control group; five mice per group), the flanks of the animals were injected with 3×10 6 SW620 or HCT-116 cells. The tumor size was monitored every seven days. Tumor size was calculated as (length×width 2)/2. When the tumor reached a discernible size of 80 mm 3, drugs (miR-150-3p agomir, NC agomir or vehicle, 50 nM) were intraperitoneally administered every other day. After six weeks, the animals were sacrificed to collect the tumors for subsequent pathological examinations. Experiments involving animals were performed by following the Guidelines for the Care and Use of Laboratory Animals, with prior approval from the Ethics Committee of Lanzhou University Second Hospital.
Immunohistochemistry (IHC)
Both normal and colon cancer tissues collected from mice were fixed for 24 h using 4% paraformaldehyde. Then, immunohistochemical staining for SRCIN1, cleaved caspase 3, β-catenin and Ki-67 was performed by incubation with the following primary antibodies: SRCIN1 (ab5407, 1:200; Abcam), cleaved caspase 3 (ab32042, 1:100; Abcam), β-catenin (ab223075, 1:200; Abcam), and Ki-67 (ab15580, 1:200; Abcam). Finally, the IX71 inverted microscope (Olympus) was used to acquire images.
Statistical analysis
Data analysis was performed using Statistical Product and Service Solutions (SPSS) 19.0 (IBM Corp., Armonk, USA) and GraphPad Prism 7.0 (GraphPad, La Jolla, USA). The results were presented as the mean±standard deviation, with t test applied to check for significant differences between two groups. For the survival analysis, the Kaplan-Meier method and the log rank test were performed before comparing the curves. Correlational analyses were also undertaken to determine possible links between the expression levels of SRCIN1 and miR-150-3p based on Pearson’s correlation coefficient. P values<0.05 were indicative of statistically significant differences.
Results
miR-150-3p is downregulated in colon cancer
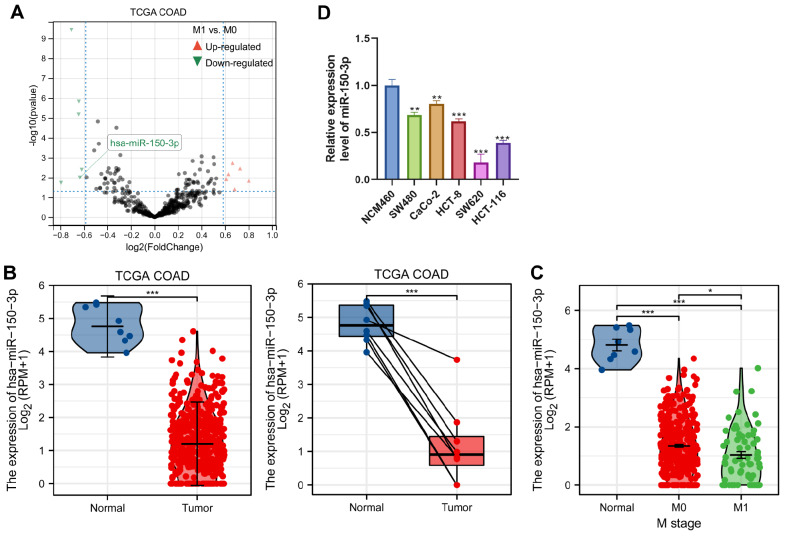
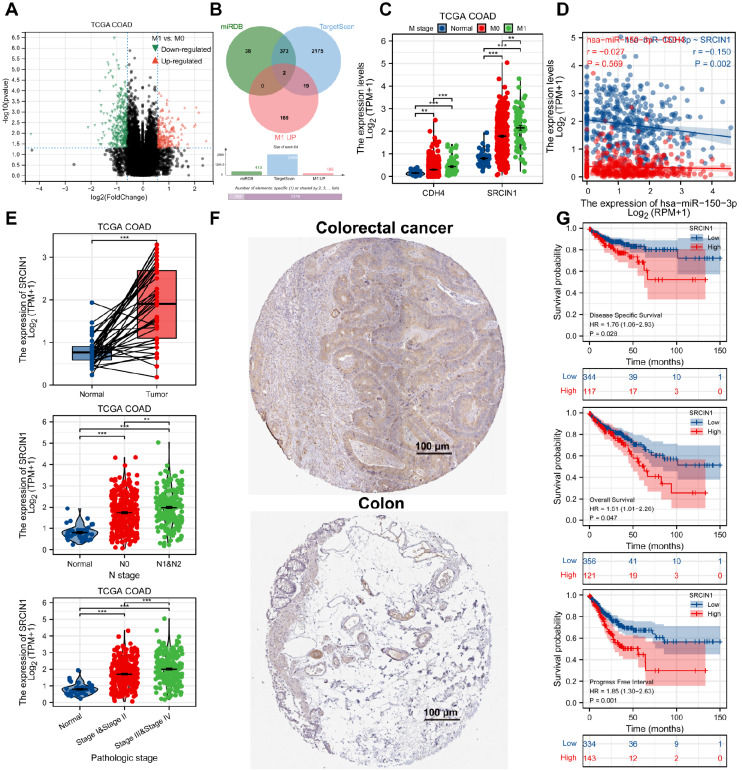
We first employed the TCGA database to analyse the expression of miR-150-3p in colon cancer. A volcano map ( Figure 1A) was drawn with differentially expressed genes. In addition, the expression of miR-150-3p was examined in colon cancer tissues using the TCGA dataset, and miR-150-3p expression level was demonstrated to be significantly decreased in colon carcinoma tissues compared with that in normal tissues ( Figure 1B). Moreover, low expression of miR-150-3p was significantly associated with the more invasive type ( Figure 1C). We also examined the level of miR-150-3p in SW480, CaCo-2, HCT-8, SW620, and HCT-116 cells as well as in the normal colon epithelial cell line NCM460. As shown in Figure 1D, miR-150-3p expression was downregulated in colon cancer cell lines compared with that in normal NCM460 cells. These results identified that miR-150-3p is downregulated in colon cancer.
Figure 1 .
Identification of miR-150-3p expression via TCGA datasets
(A) Volcano plot chart showing the distribution of the differential miRNAs in TCGA-COAD according to different histological grades. Upregulated genes are red; downregulated genes are green. (B) The miR-150-3p mRNA expression level was significantly suppressed in COAD tissues compared with that in adjacent normal tissues in TCGA databases. (C) TCGA databases of miR-150-3p expression in COAD. The difference in the expression of miR-150-3p between tumor tissue and normal tissue was compared, and the expression of miR-150-3p was compared according to different histological grades and sample types. (D) qRT-PCR of miR-150-3plevels in colon cancer SW480, CaCo-2, HCT-8, SW620, and HCT-116 cell line and colonic epithelial NCM460 cell line. Experiments were repeated three times, and data are presented as the mean±standard deviation. * P<0.05, ** P<0.01, *** P<0.001.
Downregulation of miR-150-3p accelerates the proliferation, migration, and invasion abilities of colon cancer cells in vitro
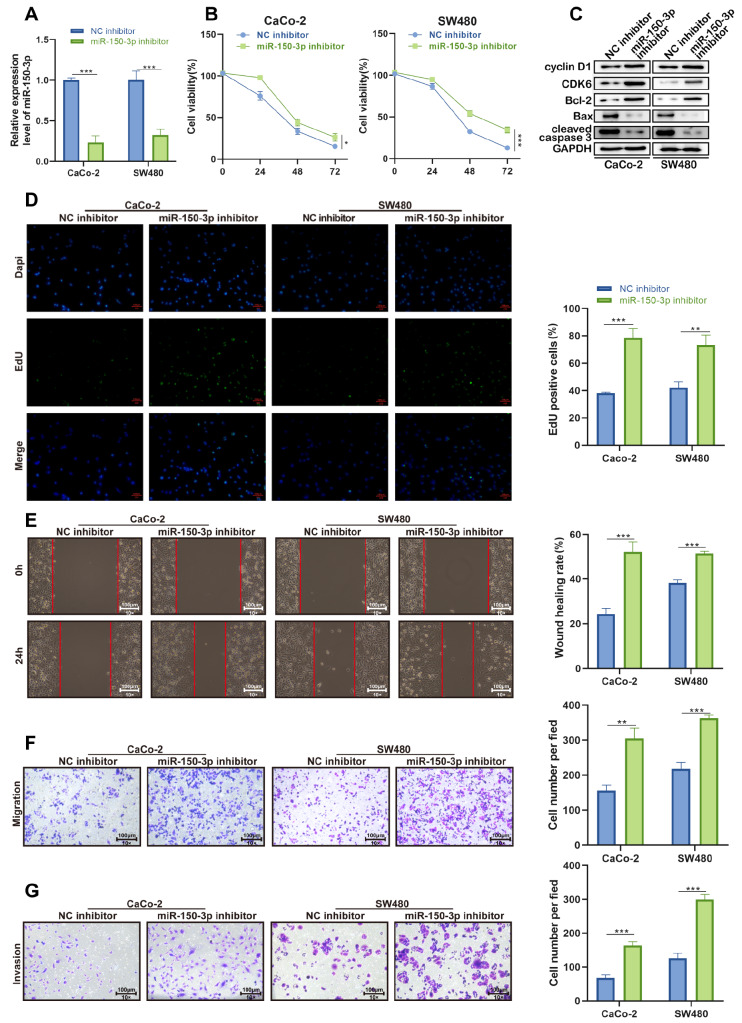
Next, functional assays were carried out, and miR-150-3p was knocked down for preparation. The efficiency of miR-150-3p interference was checked by qRT-PCR, and the data showed that miR-150-3p was noticeably downregulated in CaCo-2 and SW480 cells transfected with miR-150-3p inhibitor ( Figure 2A). Then, CCK-8 assay demonstrated that miR-150-3p silencing promoted the proliferation of CaCo-2 and SW480 cells ( Figure 2B). Afterwards, we assayed the expression levels of cell cycle- and apoptosis-related proteins by western blot analysis. miR-150-3p downregulation significantly increased the expressions of the G1/S-phase checkpoint proteins CyclinD1 and CDK6 as well as the oncogene Bcl-2. However, the expression levels of apoptosis-related proteins, including Bax and cleaved caspase3, were inhibited after miR-150-3p downregulation ( Figure 2C). Consistently, EdU assay demonstrated that downregulation of miR-150-3p promoted the proliferation of colon cancer cells ( Figure 2D). Moreover, wound healing analysis indicated that cell migration was enhanced by miR-150-3p deficiency ( Figure 2E,F). In addition, the number of invaded cells was increased by the knockdown of miR-150-3p ( Figure 2G). Collectively, these results indicated the potential anticancer ability of miR-150-3p in colon cancer in vitro.
Figure 2 .
Downregulated miR-150-3p facilitates cellular processes in colon cancer
(A) qRT-PCR of the interference efficiency of miR-150-3p in CaCo-2 and SW480 cell lines. (B) CCK-8 assay of cell proliferation in CaCo-2 and SW480 cells with silenced miR-150-3p. (C) Western blot analysis of cyclin D1, CDK6, Bcl-2, Bax, and cleaved caspase 3 expressions in CaCo-2 and SW480 cells with silenced miR-150-3p. (D) EdU assay of cell proliferation in miR-150-3p inhibitor-transfected cells. (E,F) Wound healing and transwell assays of cell migration ability in miR-150-3p inhibitor-transfected cells (magnification, ×100). Scale bar: 100 μm. (G) Transwell assay of cell invasive ability in CaCo-2 and SW480 cells with silenced miR-150-3p. Experiments were repeated three times, and data are presented as the mean±standard deviation. * P<0.05, ** P<0.01, *** P<0.001.
Overexpression of miR-150-3p inhibits the proliferation, migration, and invasion abilities of colon cancer cells in vitro
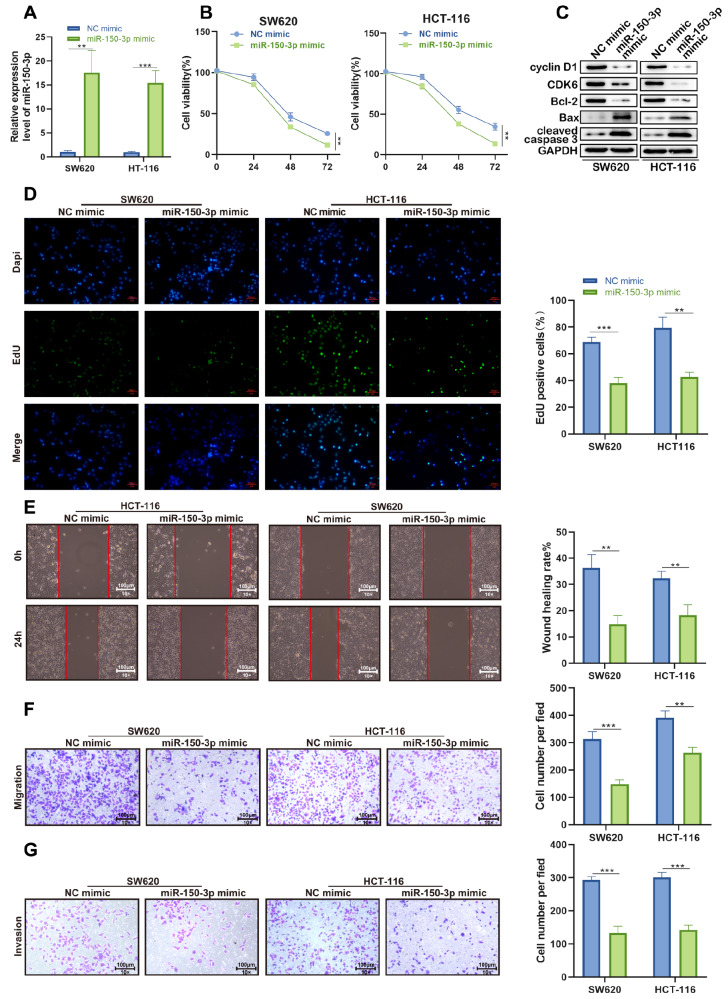
We next transfected colon cancer cells with miR-150-3p mimic and the negative control (NC mimic). RT-qPCR results showed that the transfection of miR-150-3p mimic induced a dramatic increase in the expression of miR-150-3p ( Figure 3A). The proliferation of SW620 and HCT-116 cells was significantly repressed in response to miR-150-3p overexpression by CCK-8 assay ( Figure 3B). In parallel, EdU assay showed significantly fewer positive cells after miR-150-3p overexpression ( Figure 3D). In addition, miR-150-3p overexpression significantly decreased the expressions of CyclinD1 and CDK6 as well as the oncogene Bcl-2, while the levels of Bax and cleaved caspase3 were pronouncedly increased in SW620 and HCT-116 cells after transfection with the miR-150-3p mimic ( Figure 3C). Consistent with the above findings, we observed that cell migration and invasion were suppressed in colon cancer cells transfected with miR-150-3p mimic compared with cells transfected with NC mimic, as revealed by wound healing and transwell assays ( Figure 3E,F). The abovementioned analyses suggested that miR-150-3p overexpression suppressed the malignant phenotypes of colon cancer cells.
Figure 3 .
The effect of miR-150-3p upregulation on colon cancer cell biological behavior in vitro
(A) miR-150-3p expression was upregulated by the miR-150-3p mimic in CaCo-2 and SW480 cell lines. (B) CCK-8 assay showed that overexpression of miR-150-3p significantly decreased cell proliferation compared with NC mimic-transfected cells. (C) The protein levels of cell cycle- and apoptosis-related proteins in SW620 and HCT-116 cells after overexpression of the miR-150-3p mimic. (D) The numbers of Edu-positive SW620 and HCT-116 cells transfected with miR-150-3p mimic were significantly lower than in those cells transfected with NC mimic. (E,F) miR-150-3p overexpression impaired SW620 and HCT-116 cell migration, as measured by wound healing and transwell assays (magnification, ×100). Scale bar: 100 μm. (G) Transwell assay was used to determine cell invasion in SW620 and HCT-116 cells after miR-150-3p overexpression. Data are presented as the mean±SD of three independent experiments. * P<0.05, ** P<0.01, *** P<0.001.
Upregulation of miR-150-3p inhibits tumor growth in vivo
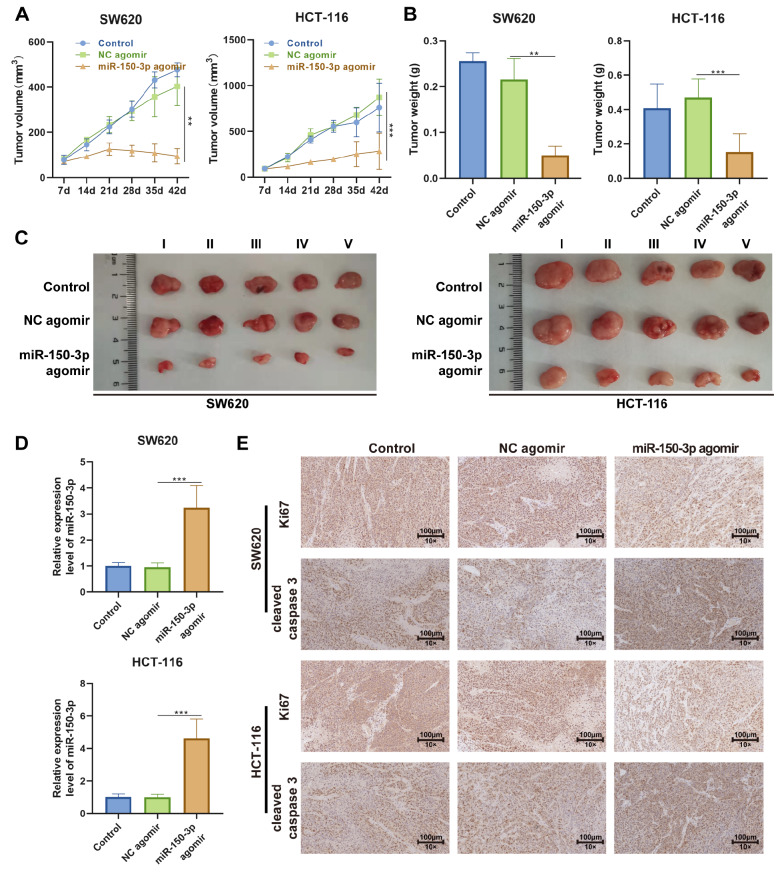
To validate the role of miR-150-3p in colon cancer in vivo, a xenograft tumor mouse model was constructed after subcutaneous injection of 3×10 6 SW620 and HCT-116 cells into the right posterior flank of nude mice. The tumor size was observed every seven days. When the tumor reached a discernible size of 80 mm 3, drugs were administered every other day. Forty-two days after inoculation, we excised the tumor from the sacrificed mouse and found that the tumor size and tumor weight of the miR-150-3p agomir group were much smaller than those of the NC agomir control group ( Figure 4B,C). The same result was observed in the tumor growth curve drawn from the estimated tumor size ( Figure 4A).
Figure 4 .
Upregulation of miR-150-3p inhibits tumor growth in vivo
SW620 and HCT-116 cells were implanted subcutaneously into immunocompromised mice. When the tumors reached 80 mm 3 in volume, the mice received intraperitoneal administration of miR-150-3p agomir or corresponding negative control every other day. The tumors were excised after 42 days of treatment for analysis. (A,B) Quantification of the growth curve and weight of tumors in each group. (C) Representative images of tumor formation in each group. (D) RT-qPCR analysis of miR-150-3p level in each tumor group. (E) Representative images of immunohistochemical staining of Ki67 and cleaved caspase 3 in tumor sections of each group. Each experiment was repeated three times. Data are presented as the mean±SD of three independent experiments. ** P<0.01, *** P<0.001.
RT-qPCR was performed to assess the expressions of miR-150-3p and SRCIN1. As shown in Figure 4D, the miR-150-3p agomir group had significantly increased expression level of miR-150-3p compared to the NC agomir group. To further elucidate the underlying mechanism of the anticancer effect on the tumor mass, we performed histological analysis of tumor sections. We stained the tumor sections with Ki67, a vital cellular marker for mitosis, and the results demonstrated that the miR-150-3p agomir was able to dramatically reduce the number of Ki67-positive cells ( Figure 4E). Meanwhile, IHC staining of cleaved caspase 3 was also performed to assess cell apoptosis in the xenograft tumor, which showed that cell apoptosis was increased in the miR-150-3p agomir group compared with that in the NC agomir group ( Figure 4E). These results strongly suggested that miR-150-3p exerted a suppressive effect on the growth of colon cancer.
SRCIN1 is upregulated in colon cancer and predicts a poor prognosis, and it is a direct target for miR-150-3p
For the identification of the underlying genes correlated with the poor prognosis and metastasis of colon cancer patients, the patients from the TCGA database were split into 2 groups based on M0 and M1 ( Figure 5A). Subsequently, to investigate the underlying mechanism of colon cancer inhibition modulated by miR-150-3p, we used the miRBase and TargetScan databases to predict the target genes of miR-150-3p, and the common genes of the two databases were considered potential target genes. Next, these 375 predicted genes overlapped with the 186 upregulated genes in the M1 subgroup of TCGA-COAD. The results indicated that SRCIN1 and CDH4 were closely correlated with miR-150-3p ( Figure 5B). Furthermore, the expressions of SRCIN1 and CDH4 were examined in colon cancer tissues using the TCGA-COAD dataset, and both SRCIN1 and CDH4 expression levels were demonstrated to be significantly increased in colon carcinoma tissues compared with those in normal tissues ( Figure 5C). The Pearson correlation test illustrated a negative correlation between the expression levels of miR-150-3p and SRCIN1 in colon cancer tissues ( Figure 5D). The SRCIN1 mRNA expression level was significantly increased in colon cancer tissues compared with that in adjacent normal tissues in TCGA databases. The SRCIN1 level was closely correlated with tumor pathological stage and lymphatic metastasis in colon cancer patients ( Figure 5E). Consistently, we detected the SRCIN1 level in colon cancer tissues as well, which was upregulated in colon cancer tissues compared with that in normal tissues ( Figure 5F). Furthermore, Kaplan-Meier analysis revealed that patients with high SRCIN1 expression displayed poorer survival, including DSS, OS, and PFS, than patients with low SRCIN1 expression ( Figure 5G). Collectively, these results identified that SRCIN1 is upregulated in colon cancer and predicts a poor prognosis, and it is a direct target for miR-150-3p.
Figure 5 .
SRCIN1 is upregulated in colon cancer and predicts a poor prognosis, and it is the direct target for miR-150-3p
(A) Volcano plot chart showing the distribution of the DEGs in TCGA-COAD datasets. Upregulated genes are red; downregulated genes are green. (B) Venn diagram of the mRNAs that may bind with miR-150-3p. (C) TCGA databases of CDH4 and SRCIN1 expression in COAD. The difference in the expression of CDH4 and SRCIN1 between tumor tissue and normal tissue was compared. (D) TCGA database of the correlation between SRCIN1 and miR-150-3p expression. (E) The SCRICN mRNA expression level was significantly increased in cancer tissues compared with that in adjacent normal tissues in TCGA databases. (F) Representative images of SRCIN1 IHC staining of 96 patients in colon cancer and normal tissues (×200, scale bar: 100 μm). (G) Kaplan-Meier analysis of the relationship between disease-specific survival (DSS), overall survival (OS), progression-free survival (PFS) and SRCIN1 expression in patients with colon cancer. The log-rank test was used to compare differences between the two groups. Data are presented as the mean±standard deviation. ** P<0.01, *** P<0.001.
miR-150-3p regulates the inhibitory effect on colon cancer by functionally targeting SRCIN1
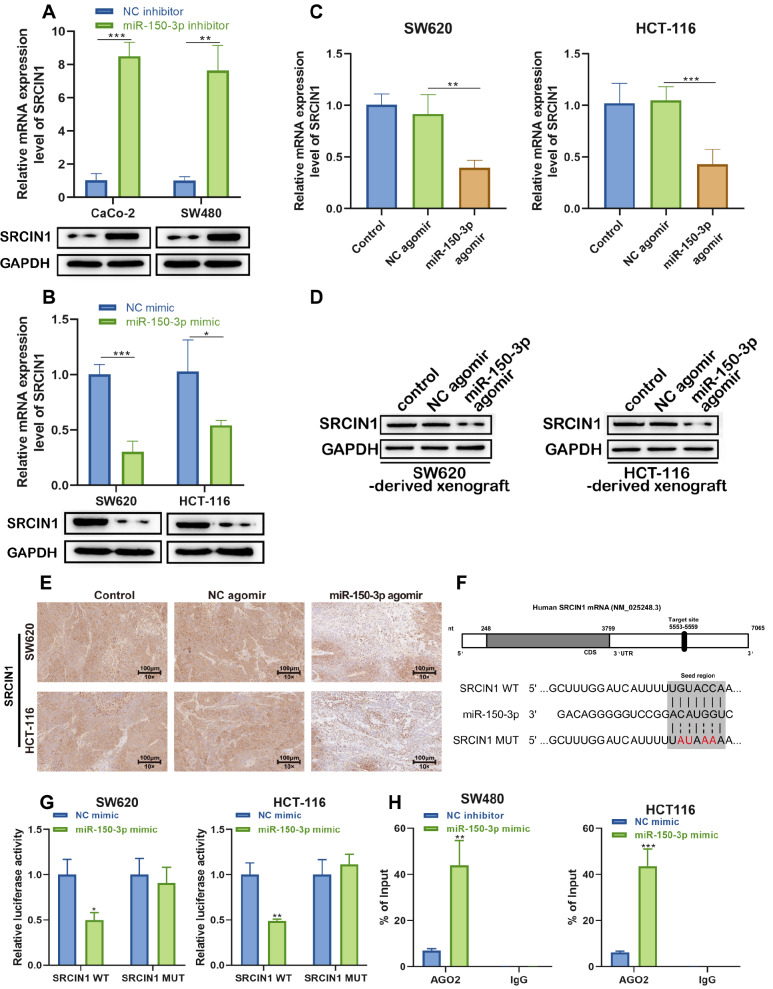
To test whether SRCIN1 has a binding site for miR-150-3p, the effect of miR-150-3p mimic or miR-150-3p inhibitor on SRCIN1 mRNA and protein expression levels in tumor cells was examined. As shown in Figure 6A,B, the protein and mRNA levels of SRCIN1 were downregulated by miR-150-3p mimics and upregulated by miR-150-3p inhibitor. In addition, to investigate whether miR-150-3p functionally targets SRCIN1 in the inhibition of colon cancer, we performed qRT-PCR, western blot, and immunohistochemical assays to detect the expression of SRCIN1 in xenografted tumors at both the mRNA and protein levels. As depicted in Figure 6C–E, the miR-150-3p agomir dramatically downregulated both the mRNA and protein levels of SRCIN1, but this effect was not observed in the NC agomir group.
Figure 6 .
miR-150-3p regulates the inhibitory effect on colon cancer by functionally targeting SRCIN1
(A) RT-qPCR and western blot analysis of SRCIN1 mRNA and protein levels in CaCo-2 and SW480 cells after treatment with miR-150-3p inhibitor. (B) RT-qPCR and western blot analysis of SRCIN1 mRNA and protein levels in SW620 and HCT-116 cells after treatment with the miR-150-3p mimic. (C) RT-qPCR of SRCIN1 mRNA in the control, NC agomir and miR-150-3p agomir groups. (D,E) Western blot analysis and immunohistochemical staining of SRCIN1 protein levels in the control, NC agomir and miR-150-3p agomir groups. (F) The binding sites between SRCIN1 and miR-150-3p. (G) Luciferase reporter assay of the binding between SRCIN1 and miR-150-3p. (H) Determination of the physical interaction between miR-150-3p and SRCIN1 by RNA immunoprecipitation experiments. Each experiment was repeated three times. Data are presented as the mean±standard deviation. * P<0.05, ** P<0.01, *** P<0.001.
Additionally, we found that SRCIN1 might be a target gene of miR-150-3p by using the online prediction software TargetScan ( http://www.targetscan.org/). We identified the potential binding site for miR-150-3p in the 3′UTR of SRCIN1 ( Figure 6F). To investigate whether miR-150-3p targets SRCIN1 directly, we constructed luciferase reporters containing a wild-type (WT) SRCIN1 3′UTR or a mutant SRCIN1 3′UTR sequence of the miR-150-3p binding site, and we found that miR-150-3p significantly suppressed the luciferase reporter activity of the WT miR-150-3p-3′UTR but not that of the mutant miR-150-3p-3′UTR ( Figure 6G). Moreover, RIP experiments showed that miR-150-3p coprecipitated with SRCIN1, confirming the miR-150-3p binds with the 3′UTR of SRCIN1 ( Figure 6H). On the basis of the above results, we speculated that SRCIN1 may be a target gene of miR-150-3p.
The SRCIN1/miR-150-3p axis regulates Wnt/β-catenin signaling
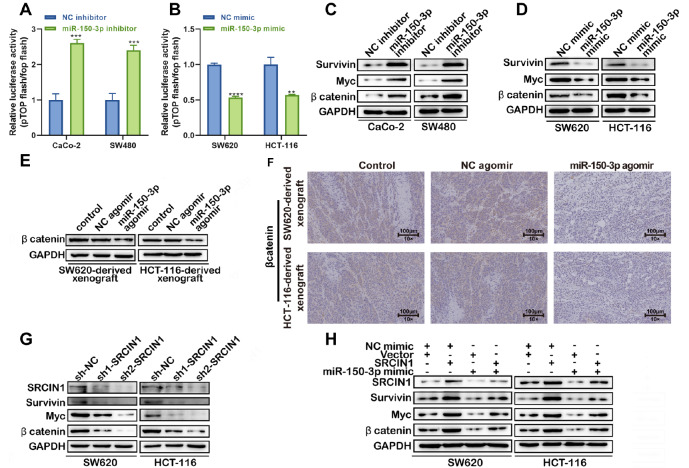
It is widely accepted that Wnt/β-catenin signaling is closely related to cancer progression, including colon cancer progression. To determine the relationship between miRNA-150-3p and Wnt signaling in colon cancer cells, a Topflash assay was performed. Inhibition of miRNA-150-3p resulted in increased relative TCF-dependent transcriptional activity, suggesting that miRNA-150-3p inhibits Wnt signaling ( Figure 7A), whereas the upregulation of miR-150-3p showed the opposite trend ( Figure 7B). Furthermore, western blot analysis also demonstrated that downregulation of miR-150-3p promoted the expressions of survivin, Myc, and β-catenin, but decreased expressions of Wnt signaling-related proteins with miR-150-3p overexpression ( Figure 7C,D). Moreover, western blot analysis and immunohistochemical staining of β-catenin were performed in SW620/HCT-116-derived xenografts. The results showed that the miR-150-3p agomir significantly inhibited the expression of β-catenin compared with the NC agomir ( Figure 7E,F). Thus, these data indicated that miR-150-3p agomir treatment may suppress Wnt signaling activation in vivo. In addition, we found that silencing of SRCIN1 exerted the opposite effect on miRNA-150-3p mimic transfection, which caused a significant decrease in the expressions survivin, Myc, and β-catenin in colon cancer cells ( Figure 7G).
Figure 7 .
The SRCIN1/miR-150-3p axis regulates Wnt/β-catenin signaling
(A,B) Luciferase reporter assay using pTopFlash and its mutant pFopFlash vectors was utilized to study β-catenin TCF/LEF promoter activity. (C,D) Downregulation of miR-150-3p resulted in elevated expression levels of survivin, Myc, and β-catenin in CaCo-2 and SW480 cells, while overexpression of miR-150-3p mimics led to a reduction in survivin, Myc, and β-catenin expression in SW620 and HCT-116 cells. Western blot analysis of survivin, Myc, and β-catenin was performed. (E,F) miR-150-3p agomir decreased the expression of β-catenin in SW620/HCT-116-derived xenografts as revealed by western blot analysis (E) and immunohistochemical staining (F). (G) Protein levels of SRCIN1, survivin, Myc, and β-catenin in SW620 and HCT-116 cells transfected with sh-NC, sh1-SRCIN1, or sh2-SRCIN1 were determined by western blot analysis. (H) Protein levels of SRCIN1, survivin, Myc, and β-catenin in SW620 and HCT-116 cells after transfection with miR-150-3p mimic or SRCIN1 overexpression vector alone or in combination were determined by western blot analysis. Data are presented as the mean±standard deviation. ** P<0.01, *** P<0.001.
In addition, SRCIN1 overexpression attenuated the miR-150-3p mimic-mediated downregulation of survivin, Myc, and β-catenin expression ( Figure 7H). Therefore, upregulation of SRCIN1 expression reversed the regulatory effects of the miR-150-3p mimic on Wnt/β-catenin signaling in colon cancer.
SRCIN1 reverses the regulatory effect of miR-150-3p on SW620 cells.
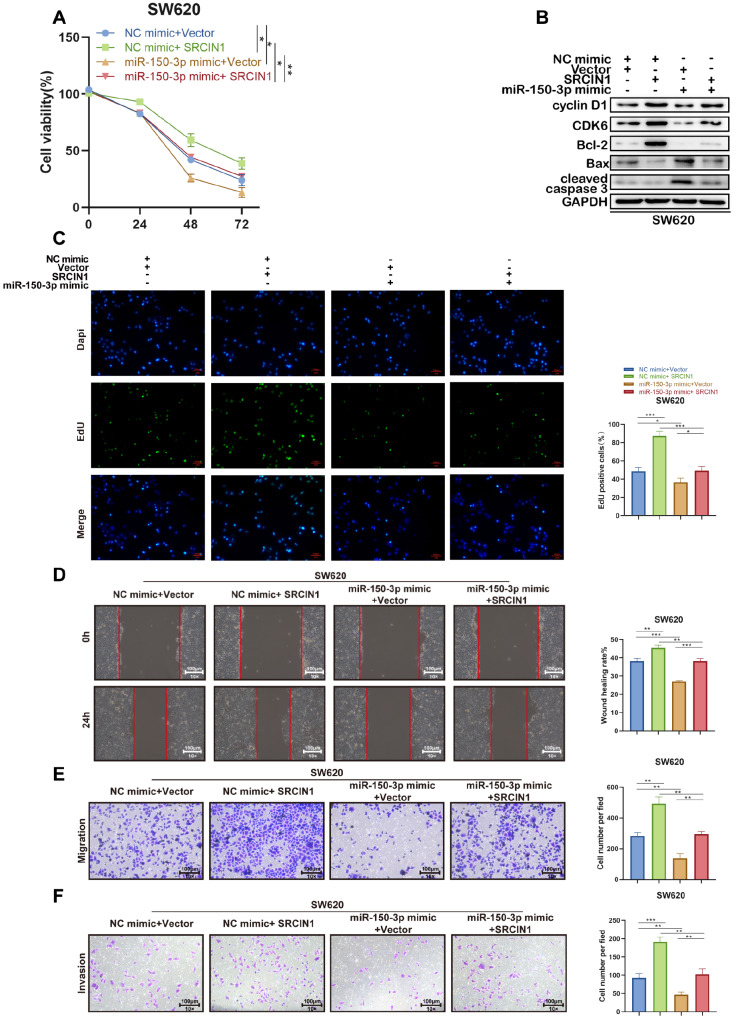
To further clarify the interaction between miR-150-3p and SRCIN1, a series of rescue experiments were conducted. Transfection of the miR-150-3p mimic reduced the viability of SW620 cells, which was partially reversed by cotransfection with the SRCIN1 overexpression vector ( Figure 8A). Furthermore, SRCIN1 overexpression attenuated the miR-150-3p mimic-mediated downregulation of Cyclin D1, CDK-6, and Bcl-2 expression and upregulation of Bax and cleaved caspase-3 expression ( Figure 8B). Meanwhile, the EdU assay showed that the miR-150-3p mimic-mediated inhibitory effect on the proliferation of SW620 cells was weakened after SRCIN1 was overexpressed Figure 8C). Similarly, the reduced migratory and invasive cell numbers in SW620 cells due to the transfection of miR-150-3p mimic were elevated after the overexpression of SRCIN1 ( Figure 8D–F). Thus, upregulation of SRCIN1 expression reversed the suppressive effects of the miR-150-3p mimic on the proliferative, migrative and invasive abilities of SW620 cells.
Figure 8 .
SRCIN1 reverses the regulatory effect of miR-150-3p on SW620 cells
SW620 cells were transfected with NC mimic+Vector, NC mimic+SRCIN1 overexpression vector, miR-150-3p mimic+Vector, or miR-150-3p mimic+SRCIN1 overexpression vector. (A) Viability of SW620 cells at 24, 48, and 72 h. (B) Protein levels of cyclin D1, CDK6, Bcl-2, Bax and cleaved caspase 3 in SW620 cells with the indicated transfection were determined by western blot analysis. GAPDH was used as a loading control. (C) EdU assay was used to detect SW620 cell proliferation. (D) Would healing assay was used to detect SW620 cell migration. (E,F) Transwell assay was used to detect SW620 cell migration and invasion. Data are presented as the mean±standard deviation. ** P<0.01, *** P<0.001.
Ginsenoside Rh2 suppresses the growth of colon cancer cells via miR-150-3p in vitro
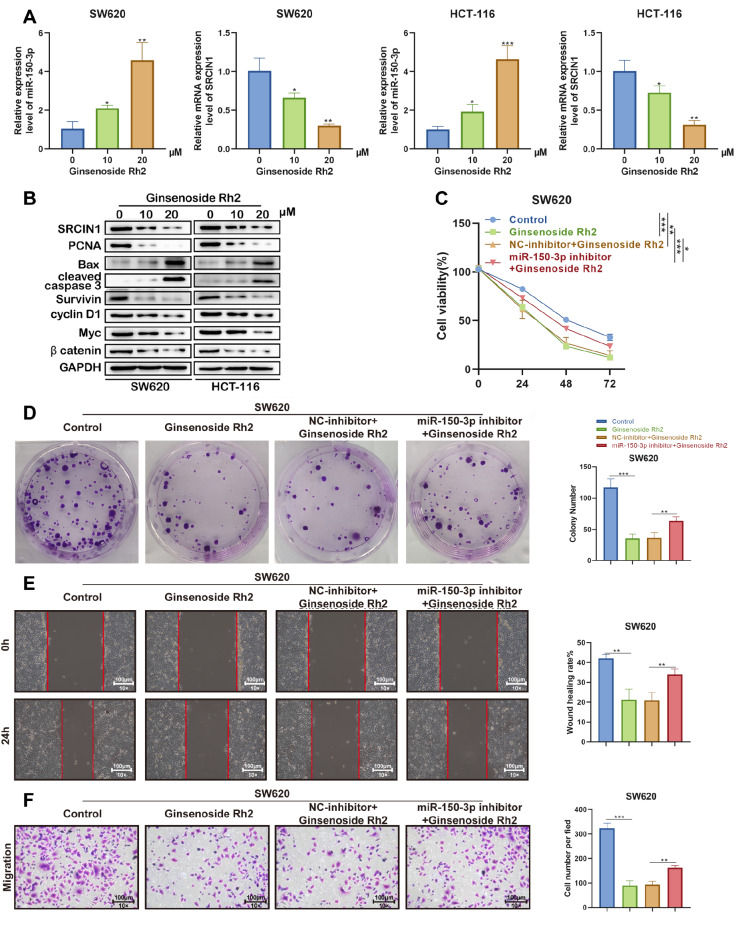
Next, we examined the expression levels of miR-150-3p and SRCIN1 in SW620 and HCT-116 cells treated with ginsenoside Rh2 (0, 10, 20 μM). The results showed that the expression of miR-150-3p was increased, while SRCIN1 was decreased in a dose-dependent manner ( Figure 9A). Moreover, ginsenoside Rh2 induced the expressions of the proapoptotic genes Bax and cleaved caspase-3 and decreased the expressions of PCNA, survivin, cyclin D1, Myc and β-catenin, as shown in Figure 9B, suggesting that ginsenoside Rh2 remarkably inhibited Wnt signaling and induced the apoptosis of colon cancer cells in a dose-depende nt manner in vitro.
Figure 9 .
Ginsenoside Rh2 suppresses the growth of colon cancer via miR-150-3p in vitro
(A,B) SW620 and SW620 cells were treated with or without 10 and 20 μM Rh2 for 18 h in DMEM/F12 medium with 10% FBS. (A) miR-150-3p and SRCIN1 relative expression levels were quantified by qRT-PCR in SW620 and HCT-116 cells, respectively. (B) Western blot analysis was used to analyse the expressions of SRCIN1, Bax, cleaved caspase-3, PCNA, survivin, cyclin D1, Myc, β-catenin and GAPDH. (C) CCK-8 assay was used to determine cell viability. (D) Colony formation ability was determined by colony formation assay. (E) A wound healing assay was performed to determine cell migration. (F) A Matrigel-coated transwell assay was performed to determine cell invasion.
To verify whether the impact of ginsenoside Rh2 on cell activity relies on the regulation of miR-150-3p, we used ginsenoside Rh2, ginsenoside Rh2 along with miR-150-3p or NC inhibitor to intervene with SW620 cells. According to the CCK-8 assay, treatment with ginsenoside Rh2 reduced the viability of SW620 cells, which was partially reversed by transfection with the miR-150-3p inhibitor ( Figure 9C). Moreover, colony formation assay showed that the ginsenoside Rh2-mediated inhibitory effect on the proliferation of SW620 cells was weakened after downregulation of miR-150-3p expression ( Figure 9D).
In addition to the inhibitory effect of ginsenoside Rh2 on cancer cell proliferation, we also evaluated the impact of ginsenoside Rh2 on the invasion and migration activities of SW620 cells. According to the assay results, under the treatment with ginsenoside Rh2, the invasion and migration activities of cancer cells were significantly inhibited. However, the reduced migratory and invasive cell numbers in SW620 cells caused by treatment with ginsenoside Rh2 were elevated after the downregulation of miR-150-3p expression ( Figure 9E,F). Thus, ginsenoside Rh2 suppresses the growth of colon cancer via miR-150-3p.
Discussion
This study showed downregulation of miR-150-3p expression in both colon cancer tissues and cell lines, and functional assays demonstrated its ability to promote tumor growth in vivo and in vitro. In addition, we found that SRCIN1 expression is upregulated in colon cancer, which is predictive of a poor prognosis, and SRCIN1 is targeted directly by miR-150-3p. Moreover, the miR-150-3p mimic reduced Topflash/Fopflash-dependent luciferase activity, resulting in the inhibition of Wnt pathway activity. Finally, we identified a novel ginsenoside Rh2-regulated miRNA and demonstrated that colon cancer cell growth is dependent on ginsenoside Rh2 in association with the miR-150-3p/SRCIN1/Wnt pathway.
miR-150-3p has been found to block the expressions of several oncogenic factors, including SP1 and SPOCK1, in glioma and prostate cancer cells and thus has the potential to inhibit tumor growth [ 25, 26] . miR-150-3p levels are also reduced in various cancer types, including lung squamous cell carcinoma [27], hepatocellular carcinoma [28], and breast cancer [29]. Here, using bioinformatics prediction and experimental verification, we showed that miR-150-3p binds to SRCIN1 and negatively regulates its expression in colon cancer cells. SRCIN1 is widely expressed in normal tissues and negatively regulates SRC activity, leading to reduced cell spreading and migration, and plays a role in Ca 2+-dependent exocytosis [30]. Its effects on SRC inactivation and tumor suppression have been observed in a variety of cancer types [ 31– 34] . Cao et al. [35] observed that miR-150 inhibited the translation of SRCIN1 to promote cell proliferation and migration in lung cancer. Sharma et al. [36] found that SRCIN1 blocked both growth and invasion in tumor cells through inhibition of SRC or E-cadherin/EGFR signaling. Moreover, SRCIN1 level was also negatively correlated with malignancy in breast cancer. It was also found by Damiano et al. that SRCIN1 prevented breast cancer cell invasion through the inhibition of cortactin, leading to reduced motility. However, a previous study reported the opposite effect of SRCIN1 on tumors in colorectal cancer, showing that SRCIN1 suppression facilitated the differentiation and tumorigenesis of colorectal cancer [37]. Consistent with this study, our data suggested the carcinogenicity potential of SRCIN1 in colon cancer.
Wnt has a strong association with cancer, including both the development and progression of tumors [38]. The majority of colon tumors have a high level of Wnt pathway activity. The 2012 report from the TCGA consortium estimated that up to 92% of sporadic colorectal cancers contain at least one alteration in a known Wnt regulator [39]. More importantly, an increasing amount of evidence supports that Wnt signaling plays a crucial role in the growth and invasion of colon tumors [ 40, 41] . It has been reported that the attenuation of Wnt/β-catenin signaling by diverse genes or small molecules may contribute to a decrease in colon cell proliferation as well as migration [42]. Several miRNAs, including miR-150, are documented to modulate Wnt signaling by influencing the expressions of Wnt-associated proteins [43], which suggests that the suppressive effect of miR-150-5p on colon cancer cells may be mediated by the Wnt/β-catenin pathway. For instance, miR-150-5p has been demonstrated to suppress the stem cell-like characteristics of glioma cells by restraining the Wnt/β-catenin pathway [44]. The negative regulatory role of miR-150-5p in the Wnt/β-catenin pathway has also been reported in laboratory research on thyroid cancer [45]. Notably, a recent study reported that miR-150 could serve as a tumor suppressor in colorectal cancer by inactivating the Wnt/β-catenin pathway [46]. Despite this evidence, the relationship between miR-150-3p and the Wnt/β-catenin signaling pathway is far from clear. In this study, we found that overexpression of miR-150-3p led to suppression of Wnt/β-catenin signaling in colon cancer in vitro and in vivo, which is in line with a previous report [46]. SRCIN1, the identified target gene of miR-150-3p in this study, has been demonstrated to activate Wnt/β-catenin signaling, thereby contributing to the carcinogenesis and metastasis of colorectal cancer [37]. Similarly, our data showed that silencing of SRCIN1 resulted in significant suppression of Wnt/β-catenin signaling in colon cancer cells and vice versa, verifying that SRCIN1 serves as an activator of Wnt/β-catenin signaling. The possible mechanism may be that SRCIN1 could upregulate downstream genes of β-catenin by affecting the location and function of the TCF/β-catenin complex [37]. To clarify whether the effect of miR-150-3p on Wnt/β-catenin signaling relies on SRCIN1, the SRCIN1 overexpression vector was cotransfected into miR-150-3p-overexpressing colon cancer cells. The results showed that the suppression of Wnt/β-catenin signaling induced by the miR-150-3p mimic was rescued by overexpressing SRCIN1 in colon cancer cells. Taken together, our study revealed that miR-150-3p inhibits the Wnt/β-catenin pathway by targeting SRCIN1.
As one of the saponins in ginseng, ginsenoside Rh2 is a relatively small, nontoxic molecule with a variety of advantageous properties, including anti-inflammatory, antioxidant, and antitumour actions [ 47, 48] . Ginsenoside Rh2 is documented to promote apoptosis, block proliferation, regulate the immune response, and inhibit both angiogenesis and metastasis in human tumor cells [ 49, 50] . It has also been found that ginsenoside Rh2 is effective for treating ulcerative colitis by downregulating STAT3/miR-214 levels [51]. Ginsenoside Rh2 could upregulate miR-419 to inhibit hypoxia-induced lung adenocarcinoma cell migration via modulation of MMP9 [7]. Our study showed that the expression of miR-150-3p was elevated as the dose of ginsenoside Rh2 increased in colon cancer cells, revealing the regulatory effect of ginsenoside Rh2 on miR-150-3p in colon cancer. Notably, functional experiments also indicated that the suppressive effect of ginsenoside Rh2 on colon cancer cell proliferation, migration, and invasion was restrained by silencing of miR-150-3p. Together with the current findings, this suggests that ginsenoside Rh2 upregulates the expression of miRNA-150-3p to suppress SRCIN1/Wnt, thereby restraining the proliferation and migration of colon cancer cells.
Nevertheless, our present study is preliminary in nature, since the precise mechanism by which ginsenoside Rh2 regulates miR-150-3p expression remains unknown. DNA hypermethylation is associated with the aberrant expression pattern of miRNAs. A previous study revealed that ginsenoside Rg3 could regulate miR-603 level by antagonizing DNMT3A-mediated DNA methylation in the miR-603 precursor gene promoter, thereby suppressing the malignancy of ovarian cancer cells [52]. In addition, ginsenoside Rh2 suppresses the expression of C3orf67-AS1 by promoter methylation to exert an antiproliferative effect on breast cancer cells [53]. Hence, it is possible that ginsenoside Rh2 influences miR-150-3p expression by affecting transcription, for instance, by hypermethylation of the promoter region, or posttranscriptional activity, for instance, directly by altering miRNA processing or indirectly by cytokines or hormones. In the future, we will further focus on how miR-150-3p is regulated by ginsenoside Rh2.
This appears to be the first study demonstrating the influence of miR-150-3p/SRCIN1/Wnt on colon cancer cell growth and its regulation by ginsenoside Rh2. We hope that this study will provide a foundation for further studies on the potential use of ginsenoside Rh2 in the treatment of colon cancer.
Supporting information
Supplementary Data
Supplementary data is available at Acta Biochimica et Biphysica Sinica online.
COMPETING INTERESTS
The authors declare that they have no conflict of interest.
Funding Statement
This work was supported by the grants from the Science and Technique Innovation Project of Lanzhou University Second Hospital (No. CY2018-MS14) and Doctoral Students Training Research Fund of Lanzhou University Second Hospital (No. YJS-BD-32).
References
- 1.Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2022. CA Cancer J Clin. . 2022;72:7–33. doi: 10.3322/caac.21708. [DOI] [PubMed] [Google Scholar]
- 2.Baidoun F. Colorectal cancer epidemiology: recent trends and impact on outcomes. Curr Drug Targets. . 2021;22:998–1009. doi: 10.2174/18735592MTEx9NTk2y. [DOI] [PubMed] [Google Scholar]
- 3.Haraldsdottir S, Einarsdottir HM, Smaradottir A, Gunnlaugsson A, Halfdanarson TR. [Colorectal cancer - review]. Laeknabladid 2014, 100: 75-82. [DOI] [PubMed]
- 4.Biller LH, Schrag D. Diagnosis and treatment of metastatic colorectal cancer. JAMA. . 2021;325:669–685. doi: 10.1001/jama.2021.0106. [DOI] [PubMed] [Google Scholar]
- 5.Lee IS, Kang KS, Kim SY. Panax ginseng pharmacopuncture: current status of the research and future challenges. Biomolecules. . 2019;10:33. doi: 10.3390/biom10010033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hu C, Yang L, Wang Y, Zhou S, Luo J, Gu Y. Ginsenoside Rh2 reduces m6A RNA methylation in cancer via the KIF26B-SRF positive feedback loop. J Ginseng Res. . 2021;45:734–743. doi: 10.1016/j.jgr.2021.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen Y, Zhang Y, Song W, Zhang Y, Dong X, Tan M. Ginsenoside Rh2 inhibits migration of lung cancer cells under hypoxia via mir-491. ACAMC. . 2019;19:1633–1641. doi: 10.2174/1871520619666190704165205. [DOI] [PubMed] [Google Scholar]
- 8.Tang XP. Effects of ginsenoside Rh2 on growth and migration of pancreatic cancer cells. World J Gastroenterol. . 2013;19:1582–1592. doi: 10.3748/wjg.v19.i10.1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shi Y, Liu Z, Lin Q, Luo Q, Cen Y, Li J, Fang X, et al. MiRNAs and cancer: key link in diagnosis and therapy. Genes. . 2021;12:1289. doi: 10.3390/genes12081289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Saliminejad K, Khorram Khorshid HR, Soleymani Fard S, Ghaffari SH. An overview of microRNAs: Biology, functions, therapeutics, and analysis methods. J Cell Physiol. . 2019;234:5451–5465. doi: 10.1002/jcp.27486. [DOI] [PubMed] [Google Scholar]
- 11.Song P, Yang F, Jin H, Wang X. The regulation of protein translation and its implications for cancer. Sig Transduct Target Ther. . 2021;6:68. doi: 10.1038/s41392-020-00444-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roman-Canal B, Tarragona J, Moiola CP, Gatius S, Bonnin S, Ruiz-Miró M, Sierra JE, et al. EV-associated miRNAs from peritoneal lavage as potential diagnostic biomarkers in colorectal cancer. J Transl Med. . 2019;17:208. doi: 10.1186/s12967-019-1954-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu K, Xu T, Song X, Shen J, Zheng S, Zhang L, Tao G, et al. LncRNA SLCO4A1-AS1 modulates colon cancer stem cell properties by binding to miR-150-3p and positively regulating SLCO4A1. Lab Invest. . 2021;101:908–920. doi: 10.1038/s41374-021-00577-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ma J, Gao G, Lu H, Fang D, Li L, Wei G, Chen A, et al. Reversal effect of ginsenoside Rh2 on oxaliplatin‑resistant colon cancer cells and its mechanism. Exp Ther Med. . 2019;18:630–636. doi: 10.3892/etm.2019.7604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wei W, Guo Q, Guo C, Cui X, Ma X, Shen X, Luo Y. Ginsenoside Rh2 Suppresses Metastasis and Growth of Colon Cancer via miR-491. J Oncol. 2021, 2021: 6815713 . [DOI] [PMC free article] [PubMed]
- 16.Yang J, Yuan D, Xing T, Su H, Zhang S, Wen J, Bai Q, et al. Ginsenoside Rh2 inhibiting HCT116 colon cancer cell proliferation through blocking PDZ-binding kinase/T-LAK cell-originated protein kinase. J Ginseng Res. . 2016;40:400–408. doi: 10.1016/j.jgr.2016.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Steinhart Z, Angers S. Wnt signaling in development and tissue homeostasis. Development. . 2018;145:dev146589. doi: 10.1242/dev.146589. [DOI] [PubMed] [Google Scholar]
- 18.Wang J, Cai H, Liu Q, Xia Y, Xing LK, Zuo Q, Zhang Y, et al. Cinobufacini inhibits colon cancer invasion and metastasis via suppressing Wnt/β-catenin signaling pathway and eMT. Am J Chin Med. . 2020;48:703–718. doi: 10.1142/S0192415X20500354. [DOI] [PubMed] [Google Scholar]
- 19.Phull MS, Jadav SS, Gundla R, Mainkar PS. A perspective on medicinal chemistry approaches towards adenomatous polyposis coli and Wnt signal based colorectal cancer inhibitors. Eur J Medicinal Chem. . 2021;212:113149. doi: 10.1016/j.ejmech.2020.113149. [DOI] [PubMed] [Google Scholar]
- 20.Cheng X, Xu X, Chen D, Zhao F, Wang W. Therapeutic potential of targeting the Wnt/β-catenin signaling pathway in colorectal cancer. Biomed Pharmacother. . 2019;110:473–481. doi: 10.1016/j.biopha.2018.11.082. [DOI] [PubMed] [Google Scholar]
- 21.Schweigert A, Fischer C, Mayr D, von Schweinitz D, Kappler R, Hubertus J. Activation of the Wnt/β-catenin pathway is common in wilms tumor, but rarely through β-catenin mutation and APC promoter methylation. Pediatr Surg Int. . 2016;32:1141–1146. doi: 10.1007/s00383-016-3970-6. [DOI] [PubMed] [Google Scholar]
- 22.Liu Z, Yang C, Li X, Luo W, Roy B, Xiong T, Zhang X, et al. The landscape of somatic mutation in sporadic Chinese colorectal cancer. Oncotarget. . 2018;9:27412–27422. doi: 10.18632/oncotarget.25287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aalen OO. Further results on the non-parametric linear regression model in survival analysis. Statist Med. . 1993;12:1569–1588. doi: 10.1002/sim.4780121705. [DOI] [PubMed] [Google Scholar]
- 24.Li N, Zhou P, Zheng J, Deng J, Wu H, Li W, Li F, et al. A polymorphism rs12325489C>t in the lincrna-enst00000515084 exon was found to modulate breast cancer risk via gwas-based association analyses. PLoS ONE. . 2014;9:e98251. doi: 10.1371/journal.pone.0098251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tan Z, Jia J, Jiang Y. MiR-150-3p targets SP1 and suppresses the growth of glioma cells . Biosci Rep. . 2018;38:BSR20180019. doi: 10.1042/BSR20180019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Okato A, Arai T, Kojima S, Koshizuka K, Osako Y, Idichi T, Kurozumi A, et al. Dual strands of pre-miR-150 (miR-150-5p and miR-150-3p) act as antitumor miRNAs targeting SPOCK1 in naïve and castration-resistant prostate cancer. Int J Oncol. . 2017;51:245–256. doi: 10.3892/ijo.2017.4008. [DOI] [PubMed] [Google Scholar]
- 27.Mizuno K, Tanigawa K, Misono S, Suetsugu T, Sanada H, Uchida A, Kawano M, et al. Regulation of oncogenic targets by tumor-suppressive miR-150-3p in lung squamous cell carcinoma. Biomedicines. . 2021;9:1883. doi: 10.3390/biomedicines9121883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yugawa K, Yoshizumi T, Mano Y, Itoh S, Harada N, Ikegami T, Kohashi K, et al. Cancer-associated fibroblasts promote hepatocellular carcinoma progression through downregulation of exosomal miR-150-3p. Eur J Surg Oncol. . 2021;47:384–393. doi: 10.1016/j.ejso.2020.08.002. [DOI] [PubMed] [Google Scholar]
- 29.Shinden Y, Hirashima T, Nohata N, Toda H, Okada R, Asai S, Tanaka T, et al. Molecular pathogenesis of breast cancer: impact of miR-99a-5p and miR-99a-3p regulation on oncogenic genes. J Hum Genet. . 2021;66:519–534. doi: 10.1038/s10038-020-00865-y. [DOI] [PubMed] [Google Scholar]
- 30.Grasso S, Cangelosi D, Chapelle J, Alzona M, Centonze G, Lamolinara A, Salemme V, et al. The SRCIN1/p140Cap adaptor protein negatively regulates the aggressiveness of neuroblastoma. Cell Death Differ. . 2020;27:790–807. doi: 10.1038/s41418-019-0386-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Damiano L, Le Dévédec SE, Di Stefano P, Repetto D, Lalai R, Truong H, Xiong JL, et al. p140Cap suppresses the invasive properties of highly metastatic MTLn3-EGFR cells via impaired cortactin phosphorylation. Oncogene. . 2012;31:624–633. doi: 10.1038/onc.2011.257. [DOI] [PubMed] [Google Scholar]
- 32.Damiano L, Di Stefano P, Camacho Leal MP, Barba M, Mainiero F, Cabodi S, Tordella L, et al. p140Cap dual regulation of E-cadherin/EGFR cross-talk and Ras signalling in tumour cell scatter and proliferation. Oncogene. . 2010;29:3677–3690. doi: 10.1038/onc.2010.128. [DOI] [PubMed] [Google Scholar]
- 33.Dong J, Li B, Lin D, Lu D, Liu C, Lu X, Tang X, et al. LincRNA00494 suppresses non-small cell lung cancer cell proliferation by regulating SRCIN1 expression as a ceRNA. Front Oncol. . 2020;10:79. doi: 10.3389/fonc.2020.00079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guo L, Zhu Y, Li L, Zhou S, Yin G, Yu G, Cui H. Breast cancer cell‐derived exosomal miR‐20a‐5p promotes the proliferation and differentiation of osteoclasts by targeting SRCIN1. Cancer Med. . 2019;8:5687–5701. doi: 10.1002/cam4.2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cao M, Hou D, Liang H, Gong F, Wang Y, Yan X, Jiang X, et al. miR-150 promotes the proliferation and migration of lung cancer cells by targeting SRC kinase signalling inhibitor 1. Eur J Cancer. . 2014;50:1013–1024. doi: 10.1016/j.ejca.2013.12.024. [DOI] [PubMed] [Google Scholar]
- 36.Sharma N, Repetto D, Aramu S, Grasso S, Russo I, Fiorentino A, Mello-Grand M et al. Identification of two regions in the p140Cap adaptor protein that retain the ability to suppress tumor cell properties. Am J Cancer Res. 2013, 3: 290–301 . [PMC free article] [PubMed]
- 37.Zhang M, Ma F, Xie R, Wu Y, Wu M, Zhang P, Peng Y, et al. Overexpression of Srcin1 contributes to the growth and metastasis of colorectal cancer. Int J Oncol. . 2017;50:1555–1566. doi: 10.3892/ijo.2017.3952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Patel S, Alam A, Pant R, Chattopadhyay S. Wnt signaling and its significance within the tumor microenvironment: novel therapeutic insights. Front Immunol. . 2019;10:2872. doi: 10.3389/fimmu.2019.02872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Muzny DM, Bainbridge MN, Chang K, Dinh HH, Drummond JA, Fowler G, Kovar CL, et al. Comprehensive molecular characterization of human colon and rectal cancer. Nature. . 2012;487:330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schneikert J, Behrens J. The canonical Wnt signalling pathway and its APC partner in colon cancer development. Gut. . 2007;56:417–425. doi: 10.1136/gut.2006.093310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Caspi M, Wittenstein A, Kazelnik M, Shor-Nareznoy Y, Rosin-Arbesfeld R. Therapeutic targeting of the oncogenic Wnt signaling pathway for treating colorectal cancer and other colonic disorders. Adv Drug Deliver Rev. . 2021;169:118–136. doi: 10.1016/j.addr.2020.12.010. [DOI] [PubMed] [Google Scholar]
- 42.Birkenkamp-Demtroder K, Maghnouj A, Mansilla F, Thorsen K, Andersen CL, Øster B, Hahn S, et al. Repression of KIAA1199 attenuates Wnt-signalling and decreases the proliferation of colon cancer cells. Br J Cancer. . 2011;105:552–561. doi: 10.1038/bjc.2011.268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Peng Y, Zhang X, Lin H, Deng S, Qin Y, He J, Hu F, et al. Dual activation of Hedgehog and Wnt/β-catenin signaling pathway caused by downregulation of SUFU targeted by miRNA-150 in human gastric cancer. Aging. . 2021;13:10749–10769. doi: 10.18632/aging.202895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tian W, Zhu W, Jiang J. miR‐150‐5p suppresses the stem cell‐like characteristics of glioma cells by targeting the Wnt/β‐catenin signaling pathway. Cell Biol Int. . 2020;44:1156–1167. doi: 10.1002/cbin.11314. [DOI] [PubMed] [Google Scholar]
- 45.Bai D, Sun H, Wang X, Lou H, Zhang J, Wang X, Jiang L. MiR-150 inhibits cell growth in vitro and in vivo by restraining the RAB11A/WNT/β-catenin pathway in thyroid cancer. Med Sci Monit. . 2017;23:4885–4894. doi: 10.12659/MSM.906997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cui X, Jiang X, Wei C, Xing Y, Tong G. Astragaloside IV suppresses development of hepatocellular carcinoma by regulating miR-150-5p/β-catenin axis. Environ Toxicol Pharmacol. . 2020;78:103397. doi: 10.1016/j.etap.2020.103397. [DOI] [PubMed] [Google Scholar]
- 47.Li X, Chu S, Lin M, Gao Y, Liu Y, Yang S, Zhou X, et al. Anticancer property of ginsenoside Rh2 from ginseng. Eur J Medicinal Chem. . 2020;203:112627. doi: 10.1016/j.ejmech.2020.112627. [DOI] [PubMed] [Google Scholar]
- 48.Zhang H, Park S, Huang H, Kim E, Yi J, Choi SK, Ryoo Z, et al. Anticancer effects and potential mechanisms of ginsenoside Rh2 in various cancer types (Review) Oncol Rep. . 2021;45:33. doi: 10.3892/or.2021.7984. [DOI] [PubMed] [Google Scholar]
- 49.Huang Y, Huang H, Han Z, Li W, Mai Z, Yuan R. Ginsenoside Rh2 inhibits angiogenesis in prostate cancer by targeting CNNM1. J Nanosci Nanotechnol. . 2019;19:1942–1950. doi: 10.1166/jnn.2019.16404. [DOI] [PubMed] [Google Scholar]
- 50.Guan N, Huo X, Zhang Z, Zhang S, Luo J, Guo W. Ginsenoside Rh2 inhibits metastasis of glioblastoma multiforme through Akt-regulated MMP13. Tumor Biol. . 2015;36:6789–6795. doi: 10.1007/s13277-015-3387-1. [DOI] [PubMed] [Google Scholar]
- 51.Chen X, Xu T, Lv X, Zhang J, Liu S. Ginsenoside Rh2 alleviates ulcerative colitis by regulating the STAT3/miR-214 signaling pathway. J EthnoPharmacol. . 2021;274:113997. doi: 10.1016/j.jep.2021.113997. [DOI] [PubMed] [Google Scholar]
- 52.Lu J, Wang L, Chen W, Wang Y, Zhen S, Chen H, Cheng J, et al. miR-603 targeted hexokinase-2 to inhibit the malignancy of ovarian cancer cells. Arch Biochem Biophys. . 2019;661:1–9. doi: 10.1016/j.abb.2018.10.014. [DOI] [PubMed] [Google Scholar]
- 53.Jeong D, Ham J, Park S, Kim HW, Kim H, Ji HW, Kim SJ. Ginsenoside Rh2 suppresses breast cancer cell proliferation by epigenetically regulating the long noncoding RNA C3orf67-AS1. Am J Chin Med. . 2019;47:1643–1658. doi: 10.1142/S0192415X19500848. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.