Abstract
Ovarian cancer is the second leading cause of death in women with gynecological malignancy in China. Circular RNAs are a class of noncoding regulatory RNAs reported to be involved in cancer development and progression. Previous studies, including our own, have indicated that hsa_circ_0007444 is downregulated in ovarian cancer tissues. This study aims to elucidate the function and mechanism of hsa_circ_0007444 in ovarian cancer progression. The expression of hsa_circ_0007444 is determined by quantitative real-time PCR (qRT-PCR). Cell proliferation, invasion, migration and apoptosis are examined by cell counting-kit 8 (CCK-8), transwell and flow cytometry assays. Tumor growth and metastasis are assessed in vivo using Balb/c nude mouse xenograft model and tail vein injection model. And the mechanism of action of hsa_circ_0007444 is analysed by RNA-binding protein immunoprecipitation (RIP), luciferase reporter and rescue assays. hsa_circ_0007444 is downregulated in ovarian cancer tissues and cell lines compared with that in normal ovarian tissues and normal epithelial cell line. Gain- and loss-of-function results indicate that hsa_circ_0007444 inhibits cell proliferation, invasion, migration and increases cell apoptosis of ovarian cancer cells in vitro, and inhibits tumor growth and lung metastasis in vivo. Mechanistically, hsa_circ_0007444 can interact with AGO2 and sponge miR-23a-3p, thereby upregulating DICER1 expression, which is an important tumor suppressor in ovarian cancer. And miR-23a-3p mimics can rescue the inhibitory effect of hsa_circ_0007444 on ovarian cancer cell proliferation, invasion and migration. Therefore, hsa_circ_0007444 can inhibit ovarian cancer progression through the hsa_circ_0007444/miR-23a-3p/DICER1 axis.
Keywords: ovarian cancer, hsa_circ_0007444, miR-23a-3p, DICER1, progression
Introduction
Ovarian cancer is the third most commonly diagnosed gynecological cancer and the second leading cause of gynecological cancer death in China [1]. It is estimated that in 2022, there would be about 19,880 newly diagnosed cases of ovarian cancer and 12,810 ovarian cancer deaths in the United States [2]. Over 95% of ovarian malignancies are epithelial ovarian cancer [3]. Ovarian cancer which starts in the deep pelvic cavity is usually asymptomatic in the early stages of the disease. Symptoms like pelvic pain and abdominal swelling only become obvious in the late stages of ovarian cancer. As a result, about 70% of patients are diagnosed at stage III/IV and present with extensive metastasis. With such advanced disease, surgery combined with platinum-based chemotherapy is the standard treatment for ovarian cancer patients. However, due to the widespread metastasis and chemotherapy resistance in late-stage ovarian cancer, the 5-year survival rate of ovarian cancer patients is only about 46% [4]. Therefore, it is rather urgent to further understand the mechanisms of ovarian cancer development, progression and metastasis.
Circular RNAs (circRNAs) are a class of endogenous non-coding RNAs with covalently closed structure and widely present in eukaryotic cells [ 5, 6] . There are three main subtypes of circRNAs: exonic circRNAs (ecircRNAs) [7], which are mainly generated by back-splicing, predominantly cytoplasmic, may harbor microRNA (miRNA) response elements (MREs) and serve as miRNA sponges; circular intronic RNAs [8] (ciRNAs), which are formed by intron cyclization directly; exonic-intronic circRNAs [9] (EIciRNAs), which consist of both introns and exons. The circRNAs lack the 5’ cap and 3’ poly-A tail, and are resistant to RNA decay machineries, making circRNAs more stable than linear RNAs [5]. The rapid development of high-throughput sequencing has enabled the analysis and identification of new circRNAs and characterization of their functions in eukaryotic cells. Accumulating evidence has revealed that circRNAs perform several biological functions, such as acting as miRNA sponges and regulation of RNA splicing, chromatin remodeling, mRNA translation, and so on [ 5, 10, 11] . In recent years, numerous studies of circRNAs have indicated that they play crucial roles in the progression of cancers, including gastric cancer [12], lung cancer [13], liver cancer [14], breast cancer [15] and ovarian cancer [16].
miRNAs are a class of evolutionarily conserved, small, non-coding RNAs of 18 to 25 nucleotides, which regulate gene expression post-transcriptionally by promoting the degradation of target mRNAs or inhibiting the translation of target mRNAs [17]. circRNAs in the cytoplasm mainly act as competing endogenous RNAs (ceRNAs) to antagonize miRNA function. Emerging evidence indicates that the circRNA/miRNA/mRNA axis is critically important in various human cancers [5]. For example, hsa_circ_0051240 sponges miR-637 and thus induces KLK4 expression to promote proliferation, migration and invasion of ovarian cancer cells [18]. circPLEKHM3 can function as a tumor suppressor in ovarian cancer cells by targeting the miR-9/BRCA1/DNAJB6/KLF4/AKT1 axis [19]. circMUC16-mediated autophagy promotes epithelial ovarian cancer cell invasion and metastasis by sponging miR-199a-5p, thereby regulating the expressions of BECN1 and RUNX1 [20]. Therefore, circRNAs may represent novel molecules for understanding ovarian cancer pathogenesis and potential therapeutic target in ovarian cancer.
Through high-throughput sequencing and quantitative real-time polymerase chain reaction (qRT-PCR) validation, hsa_circ_0007444 was found to be downregulated in ovarian cancer tissues both in our previous studies and those of Lang’s group [ 21, 22] . In addition, the expression of hsa_circ_0007444 was also found to be negatively correlated with lymphatic metastasis of ovarian cancer patients [22].
In this study, we aim to investigate the role and mechanism of hsa_circ_0007444 in ovarian cancer growth and metastasis.
Materials and Methods
Sample collection
The tissue samples analysed in this study included 20 ovarian cancer tissues collected from ovarian cancer patients (43–77 years old) and 22 pathologically proven normal ovarian tissue obtained from patients (40–71 years old) with cervical cancer who underwent hysterectomy and ovary removal at the Women’s Hospital of Nanjing Medical University, Nanjing Maternity and Child Health Care Hospital between 2015 and 2017, without adjuvant chemotherapy or radiotherapy treatment before surgery. All samples were immediately frozen in liquid nitrogen and maintained at –80°C until further use. The protocol was reviewed and ethically approved by the Ethics Committee of Women’s Hospital of Nanjing Medical University (Nanjing Maternity and Child Health Care Hospital, Nanjing, China) (No. NFKSL-022), and all patients signed informed consent forms. And all experiments of this study were carried out in accordance with the approved guidelines and regulations.
Cell culture
The ovarian cancer cell lines SKOV3, A2780, and HO8910 were purchased from KeyGEN Biotech (Nanjing, China); 293T cells were purchased from the National Collection of Authenticated Cell Cultures (Shanghai, China); immortalized ovarian surface epithelial cells 80 (IOSE80) were a gift from Dr Li Jing in Nanjing Medical University. All cell lines were verified by STR profiling. SKOV3 cells were cultured in McCoy’s 5A medium (KeyGEN Biotech) supplemented with 10% fetal bovine serum (FBS; Thermo Fisher Scientific, Waltham, USA), 100 ng/mL streptomycin, and 100 U/mL penicillin (Thermo Fisher Scientific). A2780, HO8910, and 293T cells were cultured in high-glucose Dulbecco’s modified Eagle’s medium (DMEM; KeyGEN Biotech) supplemented with 10% FBS, 100 ng/mL streptomycin, and 100 U/mL penicillin. IOSE80 cells were cultured in RPMI-1640 supplemented with 10% FBS, 100 ng/mL streptomycin, and 100 U/mL penicillin. All cells were cultured at 37°C in an incubator with a humidified atmosphere containing 5% CO 2.
Agarose gel electrophoresis and Sanger sequencing
Electrophoresis was performed with a mixture of 10 μL PCR products and 2 μL loading buffer in a 1.5% agarose gel. The target gene products were recycled, purified before Sanger sequencing on an ABI3730 DNA Analyzer Instrument (Applied Biosystems, Foster city, USA). The sequencing results were analysed by Chromas system (Technelysium Pty, South Brisbane, Australia).
RNA isolation and quantitative real-time PCR (qRT-PCR)
Total RNA was extracted with TRIzol (Invitrogen, Carlsbad, USA). Total RNA (200 ng) was reversely transcribed into cDNA in a reaction volume of 20 μL using the Revert Aid First Strand cDNA Synthesis Kit (Thermo Fisher Scientific). Bulge_loop TM miRNA qRT-PCR Primer Sets (one RT primer and a pair of qPCR primers for each set) specific for miRNA, pri-miRNA and pre-miRNA were designed and synthesized by RiboBio (Guangzhou, China). cDNA amplification was performed and measured by using the AceQ Universal SYBR qPCR Master Mix (Vazyme Biotech, Nanjing, China) and the ABI Viia7 Real-Time PCR System (Applied Biosystems). The expression of mRNA and circRNA were normalized to β-actin, the expression of miRNA was normalized to U6 small nuclear RNA (U6 snRNA), and the relative expression was calculated by using the 2 –ΔΔCt method. All primers used are listed in Supplementary Table S1.
Vector construction and cell transfection
Both the control and hsa_circ_0007444 overexpressing vector were constructed by Genomeditech (Shanghai, China). Briefly, after designing and synthesizing the hsa_circ_0007444 primers, PCR amplification was performed and the amplification products were cloned into hsa_circ_0007444 overexpressing vector GM-7183 or control vector PGMLV-6395. After separation of the PCR amplification products by agarose gel electrophoresis, the DNA fragments were recovered and purified by using a DNA Gel Extraction Kit (Axygen Biosciences, Union City, USA), and double restriction enzyme digestion. Eventually, the product was transferred to competent Escherichia coli cells, and the bacteria were evenly applied to a 10-cm Luria-Bertani (LB) culture plates with ampicillin, which selected for ampicillin resistant bacteria (successfully transfected bacteria). Bacteria were cultured in a humidified atmosphere at 37°C. After 12–18 h, a single bacterial colony was picked using a 10-μL pipette tip and inoculated into 15-mL sterilized ethylene-polypropylene (EP) tubes with LB culture medium for large scale production of the plasmids. The plasmids containing hsa_circ_0007444 were verified by Sanger sequencing. siRNAs including si-hsa_circ_0007444-1 (5′-ACACCAGGAAAGAAAAAAT-3′), si-hsa_circ_0007444-2 (5′-CCAGGAAAGAAAAAATGCC-3′); and siRNA-NC (siN0000001-1-5) were synthesized by RiboBio (Guangzhou, China). Ovarian cancer cells were seeded in 6-well plate. When the cell confluence reached 80%–90%, 1 μg of hsa_circ_0007444 overexpressing vector or control vector or 10 μL siRNA were transfected into cells using Lipofectamine 2000 (Invitrogen) following to instruction provided by the manufacturer.
Cell proliferation assay
Cell proliferation was measured using Cell Counting Kit-8 (CCK-8) kit (KeyGEN Biotech). Briefly, control, hsa_circ_0007444-overexpressing or hsa_circ_0007444-silenced SKOV3 and A2780 cells were seeded in 96-well plates at 2000 cells/well. After the cells were attached to the surface of the 96-well plate for 0, 24, 48 and 72 h, 10 μL of CCK-8 solution was added into each well and incubated at 37°C for 2 h, then the optical density (OD) of each well was measured at a wavelength of 450 nm on a Synergy H4 Hybrid Reader (BioTek Instruments, Shanghai, China).
Colony formation assay
About 3,000 ovarian cancer cells were seeded in 6-well plates. After incubation for 10 days, the colonies were gently washed twice with phosphate-buffered saline (PBS), fixed with 4% paraformaldehyde for 30 min and stained with 0.1% crystal violet (Sigma-Aldrich, St Louis, USA) for 15 min. Then, the number of colonies was counted and calculated.
Migration and invasion assays
The migration of SKOV3 and A2780 cells was analysed by wound-healing assay and transwell assay without using matrigel. In the wound-healing assay, SKOV3 and A2780 cells were seeded into 6-well plates. The cells were scratched linearly with a 200-μL pipette tip after the cell confluence reached 100%, washed three times with PBS and incubated in serum-free medium for 48 h. The images were recorded at 0, 24, and 48 h after scratching and the wound-healing area was measured by Image J (NIH, Bethesda, USA). In transwell migration assay, transwell chambers (Corning Co, Corning, USA) were placed in 24-well plates and 5×10 5 cells in 200 μL serum-free medium were loaded into the upper chamber of the transwell, while 600 μL of medium supplemented with 20% FBS was added into the lower chamber.
The invasion of SKOV3 and A2780 cells was evaluated by transwell invasion assay. Briefly, the upper chamber was coated with 60 μL Matrigel mix (1:7 dilution with serum-free medium) for 24 h at 4°C before the cells were seeded. Similar to the transwell migration assay, 200 μL of cell suspension (5×10 5 cells) in serum free medium were seeded in the upper chamber, while 600 μL of medium supplemented with 20% FBS was added into the lower chamber.
After 24–72 h of culture, the cells in the upper chamber were fixed with 4% paraformaldehyde for 1 h and stained with 0.1% crystal violet for 30 min. Then, the non-migrated or non-invaded cells on the upper surface were gently removed with a cotton swab. Images were captured under a microscope (Nikon, Tokyo, Japan). The migrated or invaded cells on the lower surface were lysed with RIPA lysis buffer and the protein concentration was determined by measuring the OD at 560 nm on a Synergy H4 Hybrid Reader (BioTek Instruments).
Flow cytometry analysis
Apoptosis of A2780 cells transfected with hsa_circ_0007444-overexpressing vector or control vector was assessed using an APC-Annexin V Apoptosis Detection kit (Thermo Fisher Scientific), while apoptosis in SKOV3 and A2780 cells transfected with siRNAs against hsa_circ_0007444 or control siRNA were assessed using a FITC-Annexin V Apoptosis Detection kit (Vazyme Biotech). After 24 h, apoptosis inducer from the Apoptosis Inducers Kit (Beyotime, Shanghai, China) was added to each well according to the manufacture’s protocol. After 48 h, the cells were suspended in Annexin V binding buffer and double-stained with fluorescein isothiocyanate (FITC) or allophycocyanin (APC) and propidium iodide (PI) for 25 min in the dark. Finally, cell apoptosis was measured with a BD FACS Calibur™ flow cytometer (BD Biosciences).
Nuclear and cytoplasmic RNA separation
The nuclear and cytoplasmic fractions of cells were isolated using the PARIS™ kit (Invitrogen). First, 1×10 7 ovarian cancer cells were washed twice with 10 mL of pre-cooled PBS. Then, the cells were digested with 1 mL trypsin and collected into 15-mL EP tube containing 3 mL of medium with 10% FBS to stop digestion. Cell pellets were re-suspended in 500 μL cell fraction buffer, incubated on ice for 10 min, and then centrifuged at 500 g and 4°C for 5 min to separate the nuclear and cytoplasmic cell fractions. Nuclear pellets were homogenized with 1 mL cell disruption buffer. Finally, the expression of reference genes or hsa_circ_0007444 in the fraction of nuclear and cytoplasmic were verified by qRT-PCR analysis. The nuclear RNA was normalized to 45S RNA, while cytoplasmic RNA was normalized to GAPDH.
RNA-binding protein immunoprecipitation assay
The RNA immunoprecipitation (RIP) assay was performed using the Magna RIP RNA-Binding Protein Immunoprecipitation Kit (Millipore, Billerica, USA) following the manufacturer’s instructions. Briefly, about 2×10 7 hsa_circ_0007444-overexpressing SKOV3 cells were dissolved in 210 μL cell lysis buffer. After 24 h, a 100-μL aliquot of cell lysis buffer was incubated with AGO2 RIP beads or IgG RIP beads and rotated overnight. RIP beads were pre-incubated with 5 μg AGO2 antibody (Abcam, Cambridge, UK) or 5 μg IgG antibody (Millipore). The next day, the immunoprecipitated RNA molecules were isolated from these mixtures following the RIP kit protocol. Eventually, the hsa_circ_0007444 level was measured by qRT-PCR analysis using the primers provided Supplementary Table S1.
RNA sequencing
Three replicates of SKOV3 cells transfected with control or si-hsa_circ_0007444-1 were collected, and total RNA was sequenced by Biotechnology Corporation (Shanghai, China). Briefly, total RNA was extracted using TRIzol (Invitrogen) according to standard protocols, followed by analysing the RNA quality with Agilent Bioanalyzer 2100 (Agilent technologies, Santa Clara, USA). An RNA-seq library was constructed using the RNA-seq Library Prep Kit (Vazyme), and then sequenced on the Illumina Hiseq 2000 platform (Illumina, San Diego, USA).
Dual luciferase reporter assay
The binding sites of miR-23a-3p in hsa_circ_0007444, and those of miR-23a-3p in DICER1 were predicted by STARBASE ( http://starbase.sysu.edu.cn/). The linear sequence of hsa_circ_0007444 cDNA or 3′UTR of DICER1 containing wild-type or mutant miR-23a-3p binding sites were subcloned into the pGL3-promoter vector (Promega, Madison, USA) by Nanjing Genebay Biotech (Nanjing, China). About 250 ng of pGL3-promoter plasmid with wild-type or mutant miR-23a-3p binding sites, 25 ng of pRL Renilla Luciferase Control Reporter Plasmid (Promega) and 50 nM miR-23a-3p mimics (5′-AUCACAUUGCCAGGGAUUUCC-3′) or miR-NC mimic (5′-UUUGUACUACACAAAAGUACUG-3′) synthesized by RiboBio were co-transfected into 293T cells using Lipofectamine 2000 (Invitrogen) in 24-well plates for 24 h. Finally, the luciferase activity was measured using a Dual-Luciferase Reporter Assay System (Promega).
Western blot analysis
About 2×10 7 ovarian cancer cells were lysed with RIPA lysis buffer (Beyotime), including 100 μL RIPA and 1 mM phenylmethanesulfonyl fluoride (PMSF), placed on ice for 30 min and then centrifuged for 30 min at 14,000 g and 4°C. The supernatant was collected and the protein concentration was measured using the bicinchoninic acid (BCA) Protein Assay Kit (Thermo Fisher Scientific). About 10 μg protein was separated by 8%–10% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE), and transferred to polyvinylidene fluoride (PVDF) membranes (Millipore). Subsequently, the membranes were blocked in 5% nonfat milk for 1 h, then incubated with specific primary antibodies overnight at 4°C. After being washed several times with Tris-buffered saline-tween 20 (TBST), membranes were incubated with the corresponding secondary antibody for 2 h. Finally, protein bands were imaged using the FluoroChem M imaging system (Protein Simple, San Jose, USA). The primary antibodies against β-actin (sc-81178; Santa Cruz Biotechnology, Santa Cruz, USA), α-tubulin (11224-1-AP; Proteintech, Rosemont, USA), Cleaved-Caspase 3 (9661; Cell Signaling Technology, Danvers, USA) and DICER1 (20567-1-AP; Proteintech) were used.
Lentivirus infection and stable cell line generation
Lentivirus packaging system was constructed using a lentivirus packaging kit (Genomeditech). Lentivirus vector and packaging vectors were co-transfected into HEK293T cells to generate lentivirus particles. Eventually, the supernatants containing lentivirus particles were collected and concentrated, and the titer of lentivirus was detected. The titer of hsa_circ_0007444-overexpressing lentivirus and control lentivirus was about 2×10 9 TU/mL and 1×10 9 TU/mL, respectively. Lentivirus was diluted to 1×10 7 TU/mL with complete culture medium. According to the multiplicity of infection (MOI) values (MOI=lentivirus concentration × lentivirus volume/cells number), 20 μL of diluted lentivirus and 2 μL of 1 mg/mL polybrene were added into one well of 6-well plates with about 5×10 4 cells. The cells were cultured in an incubator at 37°C for 48 h with a humidified atmosphere containing 5% CO 2. Stable cells lines were established after selection with 4 μg/mL puromycin for 48 h.
Animal experiment
Forty-six female BALB/c nude mice (4-6 weeks old, 13–18 g) used in this study were purchased from Gempharmatech (Nanjing, China). The mice were housed in a standard individually ventilated cage (IVC) system laboratory animal room. The animal experimental protocols were approved by Nanjing Medical University (IACUC-1601070), and all animal experiments were performed in accordance with Animal Research: Reporting of In Vivo Experiments (ARRIVE) guidelines.
Sixteen mice were randomly divided into two groups with 8 mice in each group, and then the control and hsa_circ_0007444-overexpressing SKOV3 cells (1×10 7 cells in 200 μL PBS) were subcutaneously injected into the left armpit of the mice. Mouse weight and tumor volume (volume (mm 3) = 0.5×length×width 2) were monitored every 2–3 days.
The other 30 mice were randomly divided into two groups with 15 mice in each group. One group was injected with control SKOV3 cells (2×10 6 cells in 200 μL PBS) and the other group was injected with hsa_circ_0007444-overexpressing SKOV3 cells (2×10 6 cells in 200 μL PBS) via the tail vein. Eight weeks later, mice were sacrificed, the lungs of each mouse were analysed, and the numbers of metastatic lesions in the lungs (diameter> 1 mm) were recorded. Lung metastatic nodules were analysed by hematoxylin and eosin (H&E) staining according to the protocols reported previously [23].
Immunohistochemistry analysis
Immunohistochemistry (IHC) analysis was performed using an IHC kit (PK10006; Proteintech) according to the manufacturer’s protocol. Control or hsa_circ_0007444-overexpressing tumor paraffin sections were stained with Ki67 (Proteintech).
Data retrieval and processing
The mRNA and miRNA expression data of The Cancer Genome Atlas (TCGA)-OV cohort were downloaded from the Genomic Data Commons Data Portal ( https://portal.gdc.cancer.gov/projects/TCGA-OV). The data from GEO dataset GSE65819 and GSE47841 were downloaded from NCBI ( https://www.ncbi.nlm.nih.gov/gds/?term=).
Statistical analysis
The statistical analysis of the results was performed using the SPSS 23.0 statistical analysis software (IBM, Armonk, USA) or prism (Graphpad, San Diego, USA). Data are expressed as the mean±SD. The differences between two groups were examined using the unpaired Student’s t-test or two-way ANOVA. One-way ANOVA followed by Turkey’s test was performed for comparison among more than two groups. Pearson correlation analysis was performed to confirm the correlation between miR-23a and DICER1 expression as well as between miR-23a and RHOBTB3 expression. The semi-quantification of protein expression was analysed by using the ImageJ software (NIH). P<0.05 was considered statistically significant.
Results
hsa_circ_0007444 is downregulated in ovarian cancer tissues and cell lines
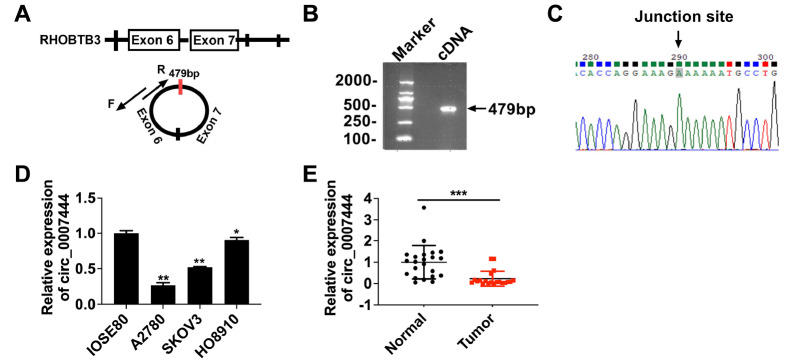
hsa_circ_0007444 is generated by circularization of back-spliced exons (exon 6 and exon 7) from the RHOBTB3 gene on chromosome 5 (from 95,091,099 to 95, 099,324,479 bp, hg19 genome build) ( Figure 1A). We designed divergent primers of hsa_circ_0007444, amplified hsa_circ_0007444 by using the cDNA template of A2780 cells ( Figure 1A,B), followed by Sanger sequencing ( Figure 1C) to verify its presence. Our previous study showed that hsa_circ_0007444 was significantly downregulated in ovarian cancer tissues compared with that in normal ovarian tissues [21]. In this study, we performed qRT-PCR analysis to further confirm the expression of hsa_circ_0007444 in normal ovarian tissues, normal ovarian epithelial cell line, ovarian cancer tissues and cell lines. Compared with that in normal ovarian epithelial cell line (IOSE80) or normal ovarian tissues, hsa_circ_0007444 was downregulated in ovarian cancer cell lines and ovarian cancer tissues ( Figure 1D,E). These results suggested that hsa_circ_0007444 might be involved in the progression of ovarian cancer.
Figure 1 .
hsa_circ_0007444 expression in ovarian cancer cell lines and tissues
(A) Schematic diagram of hsa_circ_0007444 generated by back-splicing from exon 7 to exon 6 of RHOBTB3. (B) The full length and circular structure of hsa_circ_0007444 in complementary DNA (cDNA) was detected by PCR using divergent primers. (C) Sanger sequencing revealed the back-splice junction site of hsa_circ_0007444. (D) qRT-PCR analysis of hsa_circ_0007444 expression in normal ovarian cells (IOSE80) and ovarian cancer cell lines (A2780, SKOV3, HO8910). (E) qRT-PCR analysis of hsa_circ_0007444 expression in normal ovarian tissues ( n=22) and ovarian cancer tissues ( n=20). * P<0.05, ** P<0.01, *** P<0.001.
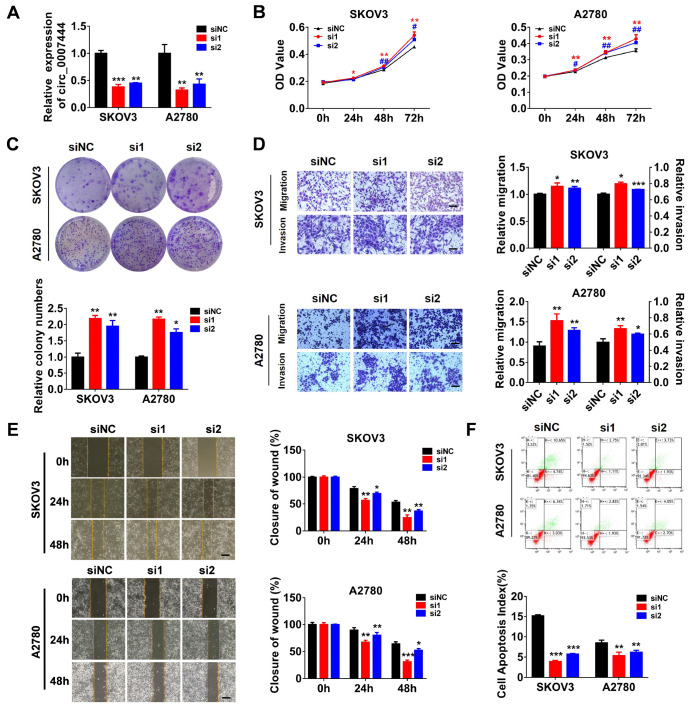
Silencing of hsa_circ_0007444 promotes ovarian cancer cell proliferation, invasion and migration and inhibits cell apoptosis in vitro
Considering its downregulation in ovarian cancer tissues and cell lines, we hypothesized that hsa_circ_0007444 might serve as a tumor suppressor in ovarian cancer. Two siRNAs were designed to target the back-splicing junction of hsa_circ_0007444, and qRT-PCR indicated that both siRNA can efficiently silence the expression of hsa_circ_0007444 without affecting the expression of RHOBTB3 mRNA ( Figure 2A and Supplementary Figure S1). Then, cell proliferation, migration, invasion, and apoptosis of the SKOV3 and A2780 ovarian cancer cells were analysed by CCK-8 assay, colony formation assay, transwell invasion and transwell migration assay, wound healing assay, and flow cytometry analysis. The results indicated that knockdown of hsa_circ_0007444 significantly promoted cell proliferation ( Figure 2B,C), invasion ( Figure 2D) and migration ( Figure 2D,E), and inhibited apoptosis ( Figure 2F) of both SKOV3 and A2780 cells.
Figure 2 .
Down-regulation of hsa_circ_0007444 increases the proliferation, migration and invasion and promotes apoptosis of ovarian cancer cells
(A) hsa_circ_0007444 levels in ovarian cancer cells transfected with siRNA-NC, si-hsa_circ_0007444-1 or si-hsa_circ_0007444-2 were measured by qRT-PCR. The interfering efficiency of si-hsa_circ_0007444-1 or si-hsa_circ_0007444-2 in SKOV3 cells was 0.621±0.047 and 0.552±0.011, respectively. And the interfering efficiency of si-hsa_circ_0007444-1 or si-hsa_circ_0007444-2 in A2780 cells was 0.679±0.040 and 0.571±0.097, respectively. (B) Evaluation of the cell viability by CCK-8 assay in SKOV3 and A2780 cells transfected with siRNA-NC, si-hsa_circ_0007444-1 or si-hsa_circ_0007444-2. (C) Colony formation assay of SKOV3 and A2780 cells transfected with siRNA-NC, si-hsa_circ_0007444-1 or si-hsa_circ_0007444-2. The colony numbers were quantified by using image J. (D) Transwell migration and invasion assay was used to determine the migratory and invasive ability of SKOV3 and A2780 cells transfected with siRNA-NC, si-hsa_circ_0007444-1 or si-hsa_circ_0007444-2. The protein concentrations of the migrated and invaded cells were quantified by BCA assay. Scale bar: 100 μm. (E) Wound healing assay was used to verify the migratory ability of SKOV3 and A2780 cells transfected with siRNA-NC, si-hsa_circ_0007444-1 or si-hsa_circ_0007444-2. The wound healing area was determined by Image J. Scale bar: 200 μm. (F) The apoptosis rates of SKOV3 and A2780 cells were measured by flow cytometry analysis after transfection with siRNA-NC, si-hsa_circ_0007444-1 or si-hsa_circ_0007444-2. All experiments were performed either in duplicate or triplicate and repeated at least two times independently. The significance levels were evaluated by student’s t-test. * P<0.05, ** P<0.01, *** P<0.001.
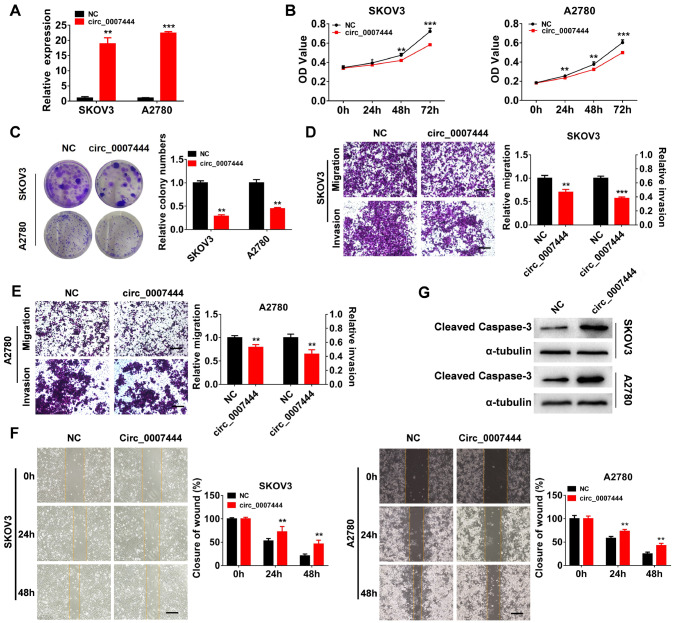
Overexpression of hsa_circ_0007444 significantly suppresses ovarian cancer cell proliferation, invasion and migration and promotes cell apoptosis in vitro
To further confirm the function of hsa_circ_0007444 in ovarian cancer, we transfected SKOV3 and A2780 cells with control or hsa_circ_0007444-overexpressing vector, qRT-PCR analysis revealed that hsa_circ_0007444 was significantly upregulated after transfecting the hsa_circ_0007444-overexpressing vector into SKOV3 and A2780 cells ( Figure 3A). As a result, overexpression of hsa_circ_0007444 significantly inhibited cell proliferation ( Figure 3B,C), migration and invasion ( Figure 3D–F). Additionally, overexpression of hsa_circ_0007444 increased the expression of cleaved caspase-3 in both SKOV3 and A2780 cells, indicating that upregulation of hsa_circ_0007444 enhances cell apoptosis ( Figure 3G and Supplementary Figure S2A). Flow cytometry analysis verified the increased apoptosis in hsa_circ_0007444-overexpressing ovarian cancer cells ( Supplementary Figure S2B,C).
Figure 3 .
Overexpression of hsa_circ_0007444 hinders the proliferation, migration and invasion of ovarian cancer cells
(A) The relative abundance of hsa_circ_0007444 in ovarian cancer cells transfected with hsa_circ_0007444-overexpressing and control plasmids was determined by qRT-PCR. The overexpression efficiency is 18.896±1.990 in SKOV3 cells, and 22.409±0.474 in A2780 cells. (B) Proliferation of SKOV3 and A2780 cells transfected with hsa_circ_0007444-overexrpressing and control vector was evaluated using the CCK-8 kit. (C) Colony formation assay was used to evaluate the proliferation capability of SKOV3 and A2780 cells transfected with hsa_circ_0007444-overexpressing or control vector. The colony numbers were quantified by using image J. (D,E) Migration and invasion abilities of SKOV3 and A2780 cells transfected with hsa_circ_0007444-overexpressing and control vector were measured by transwell migration and invasion assay. The protein concentration of the migrated and invaded cells was measured by BCA assay. Scale bar: 200 μm. (F) Wound healing assay was used to verify the migratory ability of SKOV3 and A2780 cells transfected with hsa_circ_0007444-overexpressing and control vector. The wound healing area was determined by Image J. Scale bar: 200 μm. (G) Western blot analysis of cleaved caspase-3 in SKOV3 and A2780 cells transfected with hsa_circ_0007444-overexpressing and control vector. All experiments were performed either in duplicate or triplicate and repeated at least two times independently. The significance levels were evaluated by Student’s t-test. * P<0.05, ** P<0.01, *** P<0.001.
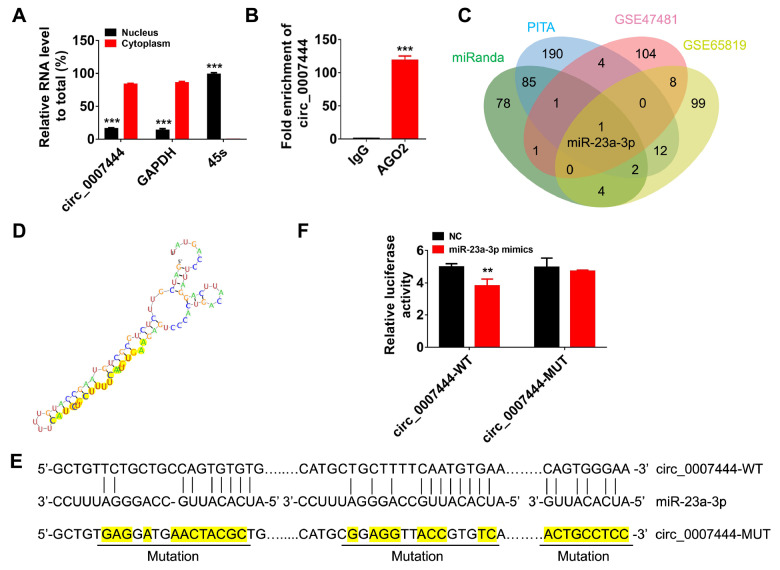
hsa_circ_0007444 sponges miR-23a-3p in ovarian cancer cells
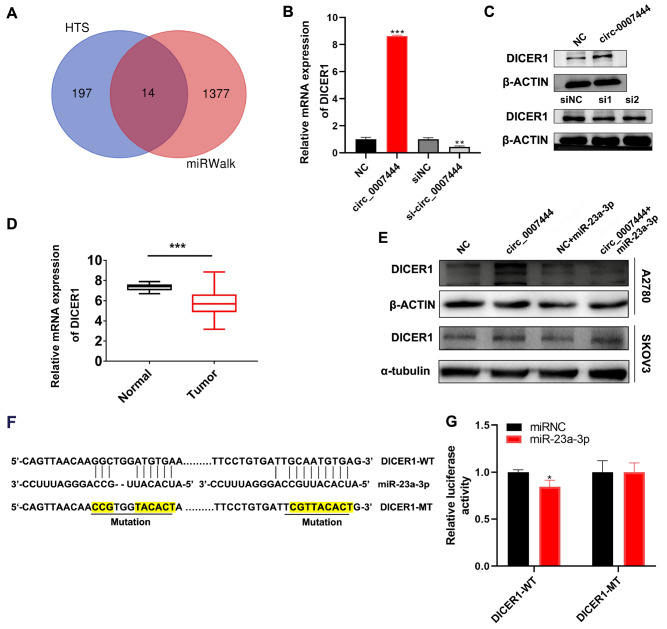
The potential mechanism of circRNAs’ functions is mainly related to their subcellular location [5]. Accordingly, we first analysed the hsa_circ_0007444 expression in the fraction of nuclear and cytoplasmic compartment of SKOV3 cells, and our results revealed that hsa_circ_0007444 was most abundant in the cytoplasm ( Figure 4A). CircRNAs in the cytoplasm mainly functions as competing endogenous RNAs (ceRNAs), thereby inhibiting the role of miRNAs-regulated RNA-induced silencing complex (RISC) which degrades targeted mRNAs [ 5, 24, 25] . AGO2 is the catalytic engine of RISC [26]. Thus, we predicted the binding ability of hsa_circ_0007444 with AGO2 using the Circinteractome database [27], which showed that there are 8 AGO2 binding sites in the sequence of hsa_circ_0007444 ( Supplementary Table 2). AGO2 RIP-qPCR analysis was performed to confirm the interaction between AGO2 and hsa_circ_0007444, and the results indicated that hsa_circ_0007444 could directly bind to AGO2 ( Figure 4B). Prediction of the miRNAs that may bind with hsa_circ_0007444 by miRanda and PITA bioinformatics tools revealed 172 and 295 miRNA targets, respectively, which were predicted to have at least two binding sites in hsa_circ_0007444 ( Supplementary Table 3 and 4). Since hsa_circ_0007444 mainly acts as a tumor suppressor in ovarian cancer, miRNAs sponged by hsa_circ_0007444 might be oncogenic miRNAs and upregulated in ovarian cancer tissues. Then we analysed the expressions of miRNAs in GEO datasets (GSE47841 and GSE65819) and found 119 and 126 upregulated miRNAs in ovarian cancer tissues, respectively. Further analysis of the above results showed that only miR-23a-3p, which was predicted to have at least two binding sites in hsa_circ_0007444 by both miRanda and PITA, was up-regulated in ovarian cancer patients as compared with that in normal ovarian epithelial (GSE47841) or fallopian tube (GSE65819) ( Figure 4C). Then the binding sites of miR-23a-3p in hsa_circ_0007444 was predicted by RNAhybrid [28] ( Figure 4D). Then we constructed the luciferase reporter vectors containing mutant or wild-type miR-23a-3p binding sequences ( Figure 4E). As expected, transfection of miR-23a-3p mimics decreased the luciferase activity of the luciferase reporter containing wild-type miR-23a-3p binding sites and the suppressing effect was concentration dependent ( Supplementary Figure S3A), but it did not affect the luciferase activity of the luciferase reporter containing the mutant miR-23a-3p binding sites, suggesting that miR-23a-3p can directly target hsa_circ_0007444 ( Figure 4F).
Figure 4 .
hsa_circ_0007444 targets miR-23a-3p
(A) qRT-PCR analysis of hsa_circ_0007444 expression in the nuclear and cytoplasmic fraction of SKOV3 cells. The nuclear RNA was normalized to 45S RNA, while cytoplasmic RNA was normalized to GAPDH. (B) RIP-qPCR assay was used to measure the amount of hsa_circ_0007444 pulled down by anti-AGO2 and IgG antibodies in SKOV3 cells. (C) The miRNAs that may interact with hsa_circ_0007444 predicted by miRanda and PITA were intersected with differentially expressed miRNAs with |fold change|>1 and P<0.05 in the GEO dataset GSE47841 and GSE65819. The Venn diagram shows the number of overlapping miRNAs. (D) The RNA binding structure between hsa_circ_0007444 and miR-23a-3p was predicted by RNAhybrid. The sequence in yellow indicates the complementary part of circ_0007444 with miR-23a-3p and the vertical bars between nucleotides show the pairing sites. (E) The binding sites between hsa_circ_00074444 and miR-23a-3p predicted by StarBase v.3.0, and the binding sites that were mutated in hsa_circ_0007444-MUT vector were highlighted. (F) The relative luciferase activity of 293T cells co-transfected with hsa_circ_0007444-WT+miR-23a-3p mimics or hsa_circ_0007444-WT+miR-NC or hsa_circ_0007444-MUT+miR-23a-3p mimics or hsa_circ_0007444-MUT+miR-NC were detected by dual-luciferase reporter assay. * P<0.05, ** P<0.01, *** P<0.001.
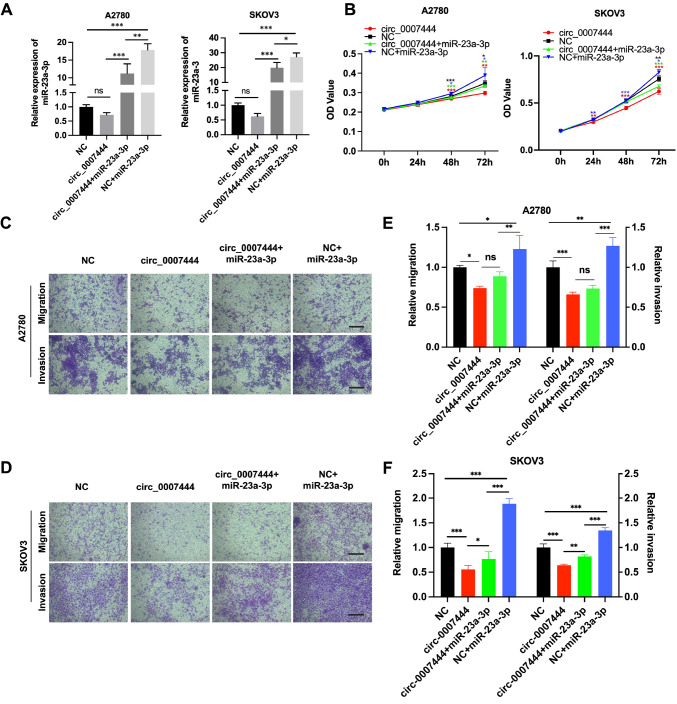
The inhibitory effect of hsa_circ_0007444 on ovarian cancer cell proliferation, migration and invasion could be reversed by miR-23a-3p
Previous studies have shown that miR-23a-3p acts as an oncogene in various cancers such as liver, colorectal and pancreatic cancers [ 29– 31] , and it is upregulated in ovarian cancer tissues [32]. To investigate whether hsa_circ_0007444 exerts its anti-cancer effects by sponging miR-23a-3p, we conducted rescue experiments in ovarian cancer cells. Our results showed that miR-23a-3p expression was increased by about 27- fold and 17- fold respectively in SKOV3 and A2780 cells transfected with miR-23a-3p mimics compared to that in ovarian cancer cells transfected with miR-NC mimic, and slightly decreased in hsa_circ_0007444-overexpressing ovarian cancer cells. Meanwhile, compared with that in ovarian cancer cells transfected with miR-23a-3p alone, miR-23a-3p was significantly reduced in the cells transfected with both hsa_circ_0007444-overexpressing vector and miR-23a-3p mimics ( Figure 5A). The RNA level of pri-miR-23a and pre-miR-23a were not affected by hsa_circ_0007444, excluding the transcriptional regulation of hsa_circ_0007444 on miR-23a ( Supplementary Figure S3B). The decrease of miR-23a-3p in hsa_circ_0007444-overexpressing cells is possibly caused by target-directed miRNA degradation as reported previously [33]. As a result, transfection of miR-23a-3p mimics alone significantly promoted ovarian cancer cell proliferation ( Figure 5B; A2780 48 h: P<0.001, A2780 72 h: P<0.05, SKOV3 48 h: ns, SKOV3 72 h: P<0.01), invasion and migration ( Figure 5C–F), and transfection of miR-23a-3p mimics into hsa_circ_0007444-overexpressing ovarian cancer cells recovered the inhibitory effects of hsa_circ_0007444 on cell proliferation ( Figure 5B), migration, and invasion ( Figure 5C–F) in SKOV3 and A2780 cells.
Figure 5 .
miR-23a-3p rescues the inhibitory effect of hsa_circ_0007444 on ovarian cancer cell proliferation, invasion and migration
(A) The relative expression of miR-23a-3p in SKOV3 and A2780 cells transfected with control vector, hsa_circ_0007444-overexpressing vector, hsa_circ_007444-overexpressing vector+miR-23a-3p mimics or control vector+miR-23a-3p mimics was measured by qRT-PCR. (B) The proliferation of SKOV3 and A2780 cells was assessed by CCK-8 assay (* P<0.05, ** P<0.01, *** P<0.001; *: NC versus circ_0007444, *: circ_0007444 versus circ_0007444 + miR-23a-3p, *: circ_0007444+miR-23a-3p versus NC+miR-23a-3p, *: NC versus NC+miR-23a-3p). (C,D) The representative images of migrated and invaded cells in transwell migration assay and transwell invasion assay. Scale bar: 200 μm. (E,F) Quantitative results of the migrated and invaded cells in transwell assay. All the experiments were performed in triplicates and repeated independently at least three times. The significance levels were evaluated using one way ANOVA followed by Tukey’s test. ns, not significant.
hsa_circ_0007444 upregulates DICER1 expression by sponging miR-23a-3p
The prediction of the potential target genes of miR-23a-3p by miRwalk revealed 1391 target genes ( Supplementary Table 5). Meanwhile, RNA-sequencing analysis of control and hsa_circ_0007444-knockdown SKOV3 cells found a total of 211 downregulated mRNAs ( Supplementary Table 6). As shown in the RNA-sequencing results, 14 of the potential target genes predicted by miRwalk were significantly downregulated in hsa_circ_0007444-silenced ovarian cancer cells ( Figure 6A). As shown in Supplementary Figure S4A,B and Figure 6B, qRT-PCR analysis indicated that only 5 predicted target genes ( KLF6, TRMT9B, ZNF107, ZNF512, and DICER1) were significantly downregulated in hsa_circ_0007444-silenced SKOV3 cells, whereas only DICER1 was significantly upregulated in the hsa_circ_0007444-overexpressing SKOV3 cells. Western blot analysis confirmed the upregulation of DICER1 in the hsa_circ_0007444-overexpressing SKOV3 cells and downregulation of DICER1 in the hsa_circ_0007444-silenced SKOV3 cells ( Figure 6C). Recent studies showed that DICER1 was downregulated in various tumors and associated with a poor prognosis [ 34, 35] . And it has been reported that DICER1 expression is decreased in ovarian cancer tissues and downregulation of DICER1 promotes ovarian cancer progression [36]. In our study, DICER1 expression was also significantly downregulated in ovarian cancer tissues compared to that in normal ovarian tissues ( Figure 6D). Transfection of miR-23a-3p mimics attenuated the upregulation of DICER1 induced by overexpression of hsa_circ_0007444 ( Figure 6E and Supplementary Figure S4C,D). In addition, the luciferase reporter activity was significantly decreased after co-transfection of miR-23a-3p mimics and the DICER1-3′UTR-WT luciferase reporter in ovarian cancer cells and this effect was enhanced after increasing the concentration of miR-23a-3p ( Figure 6F,G and Supplementary Figure S4E), indicating that DICER1 is a potential downstream target of miR-23a-3p. Importantly, we found that the expression level of miR-23a-3p was negatively correlated with the abundance of DICER1 mRNA in ovarian cancer tissues, but there was no relationship between miR-23a-3p and RHOBTB3 expression in ovarian cancer tissues ( Supplementary Figure S4F,G). Meanwhile, transfection of miR-23a-3p mimics did not alter the expression of RHOBTB3 in either SKOV3 cells or A2780 cells ( Supplementary Figure S4H). These results revealed that hsa_circ_0007444 could act as a miR-23a-3p sponge to regulate DICER1 expression.
Figure 6 .
hsa_circ_0007444 upregulates DICER1 expression by sponging miR-23a-3p
(A) Overlapping data from high-throughput sequencing and miRWalk database showed that 14 genes that may bind to miR-23a-3p were also down-regulated in hsa_circ_0007444-silenced ovarian cancer cells. (B) qRT-PCR analysis of DICER1 expression in SKOV3 cells transfected with control vector, hsa_circ_0007444-overexpressing vector, siRNA-NC or si-hsa_circ_0007444. (C) Western blot analysis of DICER1 protein expression in SKOV3 transfected with control vector, hsa_circ_0007444-overexpressing vector, siRNA-NC, si-hsa_circ_0007444-1 or si-hsa_circ_0007444-2. (D) qRT-PCR analysis of DICER1 expression in normal ovarian tissues ( n=22) and ovarian cancer tissues ( n=20). (E) Western blot analysis of DICER1 in SKOV3 and A2780 transfected with control vector, hsa_circ_0007444-overexpressing vector, control vector + miR-23a-3p mimics, or hsa_circ_0007444-overexpressing vector+miR-23a-3p mimics. (F) The putative binding sites between miR-23a-3p and 3′UTR of DICER1 were predicted by StarBase v.3.0, and the binding sites that were mutated in DICER1-MT vector were highlighted. (G) The relative luciferase activity of 293T cells co-transfected with DICER1-WT or DICER1-MUT and miR-23a-3p or control mimics was assessed by dual-luciferase reporter assay. * P<0.05, ** P<0.01, *** P<0.001.
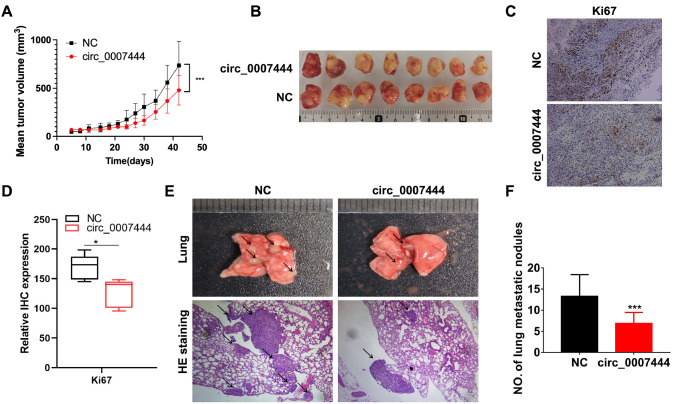
Overexpression of hsa_circ_0007444 suppresses ovarian cancer growth and lung metastasis in vivo
To further confirm the tumor-suppressing role of hsa_circ_0007444 in ovarian cancer in vivo. We first established a SKOV3 cell line stably overexpressing hsa_circ_0007444 by infecting SKOV3 cells with hsa_circ_0007444-overexpressing lentivirus. Subsequently, the hsa_circ_0007444-overexpressing or control SKOV3 cells were subcutaneously injected into BALB/c nude female mice ( n=8 in each group), and the tumor volume was measured every two to three days. The results showed that the tumors growing in mice injected with the hsa_circ_0007444-overexpressing SKOV3 cells was significantly slower than that in mice injected with control SKOV3 cells ( Figure 7A). As a result, the tumor volumes were smaller in hsa_circ_0007444-overexpressing group than in the control group ( Figure 7A,B). IHC analysis showed that the expression of Ki67 in hsa_circ_0007444-overexpressing group was decreased compared with that in the control group ( Figure 7C,D), indicating that hsa_circ_0007444 can inhibit the growth of ovarian cancer in vivo.
Figure 7 .
Overexpression of hsa_circ_0007444 suppresses the growth and lung metastasis of ovarian cancer growth in vivo
(A) Tumor volumes in xenografts of control and hsa_circ_0007444-overexpressing group at different time points ( n=8 in each group). Statistical significance was evaluated using two-way ANOVA. (B) Representative images of the xenograft tumors in A. (C) Representative immunohistochemical results of Ki67 in the xenograft tumor in mice injected with control or hsa_circ_0007444-overexpressing SKOV3 cells. Scale bar: 100 μm. (D) Quantitative analysis of the ki67 expression in the xenograft tumors. Statistical significance was evaluated using unpaired Student’s t test. (E) Representative images of lungs and H&E staining results in mice that were injected with control or hsa_circ_0007444-overexpressing SKOV3 cells via the tail vein. Scale bar: 200 μm. The arrow indicates the tumor metastasis nodules in the lung. (F) Quantitative analysis of the lung metastasis nodules. Statistical significance was evaluated using unpaired Student’s t test. * P<0.05, *** P<0.001.
To investigate the role of hsa_circ_0007444 in tumor metastasis, hsa_circ_0007444-overexpressing and control SKOV3 cells were injected into the BALB/c nude female mice via the tail vein ( n=15 in each group). Eight weeks later, the mice were sacrificed and the lungs of these mice were examined, which showed that the mice in hsa_circ_0007444-overexpressing group had less lung metastatic nodules than the control group mice ( Figure 7E,F). HE staining of the lungs of these mice showed the same result ( Figure 7E). These results further confirmed that hsa_circ_0007444 may serve as a tumor suppressor in ovarian cancer progression.
Discussion
Our previous study identified five circRNAs, including hsa_circ_0007444 (circRHOBTB3), that were significantly downregulated in ovarian cancer tissues compared with those in normal ovarian tissues [21]. Notably, it has been reported that hsa_circ_0007444 expression was negatively correlated with lymphatic metastasis in ovarian cancer patients [22]. Accordingly, in this study, we further investigated the role and mechanism of hsa_circ_0007444 in ovarian cancer growth and metastasis.
Consistent with the results reported by Deng’s, Sun’s and Wu’s group, which indicated that hsa_circ_0007444 inhibited gastric cancer growth by sponging miR-654-3p [37] and ovarian cancer progression by sponging miR-570-3p or regulating the PTEN/PI3K/AKT signalling pathway [ 38, 39] , our study also demonstrated the tumor-suppressing function of hsa_circ_0007444 in ovarian cancer growth and metastasis both in vitro and in vivo. Both our group and Sun’s group demonstrated that hsa_circ_0007444 could inhibit ovarian tumor growth in vivo. By using the tail vein injection model, we showed that overexpression of hsa_circ_0007444 also significantly impaired the lung metastasis of ovarian cancer cells in vivo, which is consistent with the negative correlation between hsa_circ_0007444 expression and lymphatic metastasis, further indicating that hsa_circ_0007444 may become the potential therapeutic target of ovarian cancer.
Both our group and Sun’s group found that hsa_circ_0007444 is mainly located in the cytoplasm of ovarian cancer cells. The results of our RIP-qPCR analysis also showed that hsa_circ_0007444 binds to AGO2 and may act as a miRNA sponge. Our further study indicated that hsa_circ_0007444 binds to miR-23a-3p and thus regulates the expression of miR-23a-3p and its target gene DICER1. DICER1 is the key enzyme in miRNA production and acts as a tumor suppressor in several cancers. Silencing of DICER1 can promote cancer cell invasion and metastasis and is associated with poor prognosis [35]. It has been reported that DICER1 is downregulated in ovarian cancer tissues, and downregulation of DICER1 can promote ovarian cancer metastasis [36]. In addition, several miRNAs were found to be downregulated in cancer due to the downregulation of DICER1. However, miR-23a was found to be upregulated in several cancers, such as ovarian cancer [40], nasopharyngeal carcinoma [41] and gastric cancer [42]. miRNAs can also act as the upstream regulators of DICER1, which promotes the degradation of DICER1 [ 34, 43] . Furthermore, despite having a reduced level of DICER1, Treg cells were reported to significantly overexpress several miRNAs, including miR-23a [44], which indicated that miR-23a might also act as the upstream regulator of DICER1. Our study first demonstrated the involvement of the hsa_circ_0007444/miR-23a-3p/DICER1 axis in ovarian cancer development.
Notably, previous studies have shown that miR-23a-3p can inhibit PTEN and thus elevate the expression of PD-L1 and promote the phosphorylation of AKT in macrophages [29]. In addition, miR-23a-3p was found to regulate the PTEN/PI3K/AKT signalling pathway in osteosarcoma cells, hepatic stellate cells, and hepatic fibrosis tissues [ 45, 46] . Hotspot mutations in DICER1 were found to result in the activation of PI3K/AKT/mTOR signalling by modulating specific miRNA population [47]. These results indicated that hsa_circ_0007444 may also regulate the PI3K/AKT pathway by directly sponging miR-23a-3p.
We also noticed that the circRNA could regulate the same process through different mechanisms. For example, Zeng et al. [48] reported that circHIPK3 promotes colorectal cancer growth and metastasis by sponging miR-7, while Yan et al. [49] showed that circHIPK3 promotes colorectal cancer cell proliferation and metastasis through the circHIPK3/miR-1207-5p/FMNL2 axis. Gao et al. [50] demonstrated that the circRNA MYLK promotes hepatocellular carcinoma progression by sponging miR-29a and thus regulates KMT5C. Li et al. [51] showed that circular RNA MYLK sponges miR-362-3p and regulates Rab23 expression to promote hepatocellular carcinoma progression. These results further demonstrated the multi-function of circRNAs in the disease regulatory network.
Finally, through bioinformatics analysis using available databases (riboCIRC: http://www.ribocirc.com/rna_calling.html, and circRNADb: http://reprod.njmu.edu.cn/circrnadb/circRNADb.php), we also found that hsa_circ_0007444 has the potential to encode a peptide. Therefore, further studies are needed to understand the function and mechanism of hsa_circ_0007444-encoded peptide in cancer and to develop new therapeutic drugs for the treatment of cancer.
In conclusion, we found that hsa_circ_0007444, which is downregulated in ovarian cancer tissues and cell lines, could interact with AGO2 to sponge miR-23a-3p and upregulate DICER1 expression, and its overexpression effectively impairs cell proliferation, invasion, migration and apoptosis in vitro, and inhibits tumor growth and lung metastasis in vivo. This newly discovered ceRNA network may offer a possible therapeutic target for the treatment of ovarian cancer.
Supporting information
Supplementary Data
Supplementary data is available at Acta Biochimica et Biphysica Sinica online.
COMPETING INTERESTS
The authors declare that they have no conflict of interest.
Funding Statement
This work was supported by the grants from the National Natural Science Foundation of China (No. 81902651) and the 333 Project of Jiangsu Province (to X.J.).
References
- 1.Zheng R, Zhang S, Zeng H, Wang S, Sun K, Chen R, Li L, Wei W, He J. Cancer incidence and mortality in China, 2016. J Nat Cancer Cent. 2022, 2: 1–9 . [DOI] [PMC free article] [PubMed]
- 2.Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer Statistics, 2022. CA Cancer J Clin. . 2022;72:7–33. doi: 10.3322/caac.21708. [DOI] [PubMed] [Google Scholar]
- 3.Lheureux S, Braunstein M, Oza A. Epithelial ovarian cancer: evolution of management in the era of precision medicine. CA Cancer J Clin. 2019, 69: 280–304 . [DOI] [PubMed]
- 4.Lheureux S, Gourley C, Vergote I, Oza AM. Epithelial ovarian cancer. Lancet. . 2019;393:1240–1253. doi: 10.1016/S0140-6736(18)32552-2. [DOI] [PubMed] [Google Scholar]
- 5.Li X, Yang L, Chen LL. The biogenesis, functions, and challenges of circular RNAs. Mol Cell. . 2018;71:428–442. doi: 10.1016/j.molcel.2018.06.034. [DOI] [PubMed] [Google Scholar]
- 6.Hsu MT, Coca-prados M. Electron microscopic evidence for the circular form of RNA in the cytoplasm of eukaryotic cells. Nature. . 1979;280:339–340. doi: 10.1038/280339a0. [DOI] [PubMed] [Google Scholar]
- 7.Chen I, Chen CY, Chuang TJ. Biogenesis, identification, and function of exonic circular RNAs. WIREs RNA. . 2015;6:563–579. doi: 10.1002/wrna.1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang Y, Zhang XO, Chen T, Xiang JF, Yin QF, Xing YH, Zhu S, et al. Circular intronic long noncoding RNAs. Mol Cell. . 2013;51:792–806. doi: 10.1016/j.molcel.2013.08.017. [DOI] [PubMed] [Google Scholar]
- 9.Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, Marzluff WF, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. . 2013;19:141–157. doi: 10.1261/rna.035667.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Qu S, Liu Z, Yang X, Zhou J, Yu H, Zhang R, Li H. The emerging functions and roles of circular RNAs in cancer. Cancer Lett. . 2018;414:301–309. doi: 10.1016/j.canlet.2017.11.022. [DOI] [PubMed] [Google Scholar]
- 11.Qu S, Yang X, Li X, Wang J, Gao Y, Shang R, Sun W, et al. Circular RNA: A new star of noncoding RNAs. Cancer Lett. . 2015;365:141–148. doi: 10.1016/j.canlet.2015.06.003. [DOI] [PubMed] [Google Scholar]
- 12.Shi P, Wan J, Song H, Ding X. The emerging role of circular RNAs in gastric cancer. Am J Cancer Res. 2018, 8: 1919–1932 . [PMC free article] [PubMed]
- 13.Hu W, Bi ZY, Chen ZL, Liu C, Li LL, Zhang F, Zhou Q, et al. Emerging landscape of circular RNAs in lung cancer. Cancer Lett. . 2018;427:18–27. doi: 10.1016/j.canlet.2018.04.006. [DOI] [PubMed] [Google Scholar]
- 14.Qin M, Liu G, Huo X, Tao X, Sun X, Ge Z, Yang J, et al. Hsa_circ_0001649: a circular RNA and potential novel biomarker for hepatocellular carcinoma. Cancer Biomark. . 2016;16:161–169. doi: 10.3233/CBM-150552. [DOI] [PubMed] [Google Scholar]
- 15.Wang X, Fang L. Advances in circular RNAs and their roles in breast Cancer. J Exp Clin Cancer Res. . 2018;37:206. doi: 10.1186/s13046-018-0870-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen Q, Zhang J, He Y, Wang Y. hsa_circ_0061140 knockdown reverses FOXM1-mediated cell growth and metastasis in ovarian cancer through miR-370 sponge activity. Mol Ther Nucleic Acids. . 2018;13:55–63. doi: 10.1016/j.omtn.2018.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 17.Ha M, Kim VN. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol. . 2014;15:509–524. doi: 10.1038/nrm3838. [DOI] [PubMed] [Google Scholar]
- 18.Zhang M, Xia B, Xu Y, Zhang Y, Xu J, Lou G. Circular RNA (hsa_circ_0051240) promotes cell proliferation, migration and invasion in ovarian cancer through miR-637/KLK4 axis. Artif Cells Nanomed Biotechnol. . 2019;47:1224–1233. doi: 10.1080/21691401.2019.1593999. [DOI] [PubMed] [Google Scholar]
- 19.Zhang L, Zhou Q, Qiu Q, Hou L, Wu M, Li J, Li X, et al. CircPLEKHM3 acts as a tumor suppressor through regulation of the miR-9/BRCA1/DNAJB6/KLF4/AKT1 axis in ovarian cancer. Mol Cancer. . 2019;18:144. doi: 10.1186/s12943-019-1080-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gan X, Zhu H, Jiang X, Obiegbusi SC, Yong M, Long X, Hu J. CircMUC16 promotes autophagy of epithelial ovarian cancer via interaction with ATG13 and miR-199a. Mol Cancer. . 2020;19:45. doi: 10.1186/s12943-020-01163-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Teng F, Xu J, Zhang M, Liu S, Gu Y, Zhang M, Wang X, et al. Comprehensive circular RNA expression profiles and the tumor-suppressive function of circHIPK3 in ovarian cancer. Int J Biochem Cell Biol. . 2019;112:8–17. doi: 10.1016/j.biocel.2019.04.011. [DOI] [PubMed] [Google Scholar]
- 22.Ning L, Long B, Zhang W, Yu M, Wang S, Cao D, Yang J, et al. Circular RNA profiling reveals circEXOC6B and circN4BP2L2 as novel prognostic biomarkers in epithelial ovarian cancer. Int J Oncol. . 2018;53:2637–2646. doi: 10.3892/ijo.2018.4566. [DOI] [PubMed] [Google Scholar]
- 23.Feldman AT, Wolfe D. Tissue processing and hematoxylin and eosin staining. Methods Mol Biol. 2014, 1180: 31–43 . [DOI] [PubMed]
- 24.Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. . 2011;146:353–358. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thomson DW, Dinger ME. Endogenous microRNA sponges: evidence and controversy. Nat Rev Genet. . 2016;17:272–283. doi: 10.1038/nrg.2016.20. [DOI] [PubMed] [Google Scholar]
- 26.Li X, Wang X, Cheng Z, Zhu Q. AGO2 and its partners: a silencing complex, a chromatin modulator, and new features. Crit Rev Biochem Mol Biol. . 2020;55:33–53. doi: 10.1080/10409238.2020.1738331. [DOI] [PubMed] [Google Scholar]
- 27.Dudekula DB, Panda AC, Grammatikakis I, De S, Abdelmohsen K, Gorospe M. CircInteractome: a web tool for exploring circular RNAs and their interacting proteins and microRNAs. RNA Biol. . 2016;13:34–42. doi: 10.1080/15476286.2015.1128065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rehmsmeier M, Steffen P, Höchsmann M, Giegerich R. Fast and effective prediction of microRNA/target duplexes. RNA. . 2004;10:1507–1517. doi: 10.1261/rna.5248604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu J, Fan L, Yu H, Zhang J, He Y, Feng D, Wang F, et al. Endoplasmic reticulum stress causes liver cancer cells to release exosomal miR-23a-3p and up-regulate programmed death ligand 1 expression in macrophages. Hepatology. . 2019;70:241–258. doi: 10.1002/hep.30607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang X, Gao X, Tian J, Zhang R, Qiao Y, Hua X, Shi G. LINC00261 inhibits progression of pancreatic cancer by down-regulating miR-23a-3p. Arch Biochem Biophys. . 2020;689:108469. doi: 10.1016/j.abb.2020.108469. [DOI] [PubMed] [Google Scholar]
- 31.Lee Y, Kim SJ, Choo J, Heo G, Yoo JW, Jung Y, Rhee SH, et al. miR-23a-3p is a key regulator of IL-17C-induced tumor angiogenesis in colorectal Cancer. Cells. . 2020;9:1363. doi: 10.3390/cells9061363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Prahm K, Høgdall C, Karlsen M, Christensen I, Novotny G, Høgdall E. Identification and validation of potential prognostic and predictive miRNAs of epithelial ovarian cancer. PLoS One. 2018, 13: e0207319 . [DOI] [PMC free article] [PubMed]
- 33.Han J, LaVigne CA, Jones BT, Zhang H, Gillett F, Mendell JT. A ubiquitin ligase mediates target-directed microRNA decay independently of tailing and trimming. Science. . 2020;370:eabc9846. doi: 10.1126/science.abc9546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rupaimoole R, Ivan C, Yang D, Gharpure KM, Wu SY, Pecot CV, Previs RA, et al. Hypoxia-upregulated microRNA-630 targets Dicer, leading to increased tumor progression. Oncogene. . 2016;35:4312–4320. doi: 10.1038/onc.2015.492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Foulkes WD, Priest JR, Duchaine TF. DICER1: mutations, microRNAs and mechanisms. Nat Rev Cancer. . 2014;14:662–672. doi: 10.1038/nrc3802. [DOI] [PubMed] [Google Scholar]
- 36.To SKY, Mak ASC, Eva Fung YM, Che CM, Li SS, Deng W, Ru B, et al. β-catenin downregulates Dicer to promote ovarian cancer metastasis. Oncogene. . 2017;36:5927–5938. doi: 10.1038/onc.2017.185. [DOI] [PubMed] [Google Scholar]
- 37.Deng G, Mou T, He J, Chen D, Lv D, Liu H, Yu J, et al. Circular RNA circRHOBTB3 acts as a sponge for miR-654-3p inhibiting gastric cancer growth. J Exp Clin Cancer Res. . 2020;39:1. doi: 10.1186/s13046-019-1487-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yalan S, Yanfang L, He C, Yujie T. Circular RNA circRHOBTB3 inhibits ovarian cancer progression through PI3K/AKT signaling pathway. Panminerva Med. . 2020 doi: 10.23736/S0031-0808.20.03957-9. [DOI] [Google Scholar]
- 39.Wu X, Liu D, Wang S, Liu J. Circ_0007444 inhibits the progression of ovarian cancer via mediating the miR-570-3p/PTEN axis. Onco Targets Ther. . 2021;14:97–110. doi: 10.2147/OTT.S266186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang Z, Wang X, Bai R, Liu W, Li X, Liu M, Tang H. miR-23a promotes IKKα expression but suppresses ST7L expression to contribute to the malignancy of epithelial ovarian cancer cells. Br J Cancer. . 2016;115:731–740. doi: 10.1038/bjc.2016.244. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 41.Bao L, You B, Shi S, Shan Y, Zhang Q, Yue H, Zhang J, et al. Metastasis-associated miR-23a from nasopharyngeal carcinoma-derived exosomes mediates angiogenesis by repressing a novel target gene TSGA10. Oncogene. . 2018;37:2873–2889. doi: 10.1038/s41388-018-0183-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hu X, Wang Y, Liang H, Fan Q, Zhu R, Cui J, Zhang W, et al. miR-23a/b promote tumor growth and suppress apoptosis by targeting PDCD4 in gastric cancer. Cell Death Dis. . 2017;8:e3059. doi: 10.1038/cddis.2017.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Martello G, Rosato A, Ferrari F, Manfrin A, Cordenonsi M, Dupont S, Enzo E, et al. A microRNA targeting dicer for metastasis control. Cell. . 2010;141:1195–1207. doi: 10.1016/j.cell.2010.05.017. [DOI] [PubMed] [Google Scholar]
- 44.Divekar AA, Dubey S, Gangalum PR, Singh RR. Dicer insufficiency and microRNA-155 overexpression in lupus regulatory T cells: an apparent paradox in the setting of an inflammatory milieu. J Immunol. . 2011;186:924–930. doi: 10.4049/jimmunol.1002218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dong Z, Li S, Wang X, Si L, Ma R, Bao L, Bo A. lncRNA GAS5 restrains CCl 4-induced hepatic fibrosis by targeting miR-23a through the PTEN/PI3K/Akt signaling pathway . Am J Physiol-Gastrointestinal Liver Physiol. . 2019;316:G539–G550. doi: 10.1152/ajpgi.00249.2018. [DOI] [PubMed] [Google Scholar]
- 46.Liu J, Chen M, Ma L, Dang X, Du G. LncRNA GAS5 Suppresses the Proliferation and Invasion of Osteosarcoma Cells via the miR-23a-3p/PTEN/PI3K/AKT Pathway. Cell Transplant. . 2020;29:096368972095309. doi: 10.1177/0963689720953093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Klein SD, Martinez-Agosto JA. Hotspot mutations in DICER1 causing GLOW syndrome-associated macrocephaly via modulation of specific microRNA populations result in the activation of PI3K/ATK/mTOR signaling. MicroRNA. . 2019;9:70–80. doi: 10.2174/2211536608666190624114424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zeng K, Chen X, Xu M, Liu X, Hu X, Xu T, Sun H, et al. CircHIPK3 promotes colorectal cancer growth and metastasis by sponging miR-7. Cell Death Dis. . 2018;9:417. doi: 10.1038/s41419-018-0454-8. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 49.Yan Y, Su M, Qin B. CircHIPK3 promotes colorectal cancer cells proliferation and metastasis via modulating of miR-1207-5p/FMNL2 signal. Biochem Biophys Res Commun. . 2020;524:839–846. doi: 10.1016/j.bbrc.2020.01.055. [DOI] [PubMed] [Google Scholar]
- 50.Gao J, Li E, Liu W, Yang Q, Xie C, Ai J, Zhou F, et al. Circular RNA MYLK promotes hepatocellular carcinoma progression through the miR29a/KMT5C signaling pathway. Onco Targets Ther. . 2020;13:8615–8627. doi: 10.2147/OTT.S258715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li Z, Hu Y, Zeng Q, Wang H, Yan J, Li H, Yu Z. Circular RNA MYLK promotes hepatocellular carcinoma progression by increasing Rab23 expression by sponging miR-362-3p. Cancer Cell Int. . 2019;19:211. doi: 10.1186/s12935-019-0926-7. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.