Abstract
Gene loss is a prevalent source of genetic variation in genome evolution. Calling loss events effectively and efficiently is a critical step for systematically characterizing their functional and phylogenetic profiles genome wide. Here, we developed a novel pipeline integrating orthologous inference and genome alignment. Interestingly, we identified 33 gene loss events that give rise to evolutionarily novel long noncoding RNAs (lncRNAs) that show distinct expression features and could be associated with various functions related to growth, development, immunity, and reproduction, suggesting loss relics as a potential source of functional lncRNAs in humans. Our data also demonstrated that the rates of protein gene loss are variable among different lineages with distinct functional biases.
Keywords: gene loss, long noncoding RNA, lncRNA origin, comparative genomics
Introduction
From bacteria to mammals, gene loss occurs in almost all kingdoms of life (Spanu et al. 2010; Kuraku and Kuratani 2011; McCutcheon and Moran 2011; Albalat and Cañestro 2016; Wang et al. 2018). Several studies have shown that the loss of protein-coding genes has led to significant phenotypic changes, such as the adoption of an herbivorous diet in pandas (Li et al. 2009) and the development of scales in pangolins (Meyer et al. 2013; Choo et al. 2016), as well as molecular adaptations, such as opsin adaptations in birds (Borges et al. 2015) and adaptation to aquatic ecosystems in cetaceans (Huelsmann et al. 2019). In humans, gene loss events have also been reported to cause not only improved disease resistance (Dean et al. 1996; Galvani and Novembre 2005; Wang et al. 2006; Shailendra 2009; Hedrick 2011; Okerblom et al. 2017) but also human-specific phenotypic changes (Stedman et al. 2004).
Efforts have been made to identify gene loss events genome widely in recent decades. Most of these studies have been based on directly searching for homologs of annotated source proteins in the target genome and may suffer from the identification of both false positives (due to out-paralogs, i.e., paralogs in different species derived from a more ancient shared duplication event) and false negatives (due to long-term divergence) (Wang et al. 2006; Zhu et al. 2007; Zhang et al. 2010; Zhao et al. 2015).
Here, we develop a novel pipeline that integrates orthologous inference and genome alignment called LOcal Sequence-based Tracing Functional Ortholog UNit Death, or LOST & FOUND. Applying the pipeline to mammals, we identified a number of previously missed gene loss events in the human genome after mouse–human divergence. Interestingly, we found that a set of “dead” protein-coding genes might undergo “rebirth” as functional long noncoding RNAs (lncRNAs), suggesting that relics of the lost protein could be the source of lncRNAs and thus perform important functions (Zhao et al. 2015; Hezroni et al. 2017). This highlights the importance of the systematic identification and annotation of gene loss events.
Results
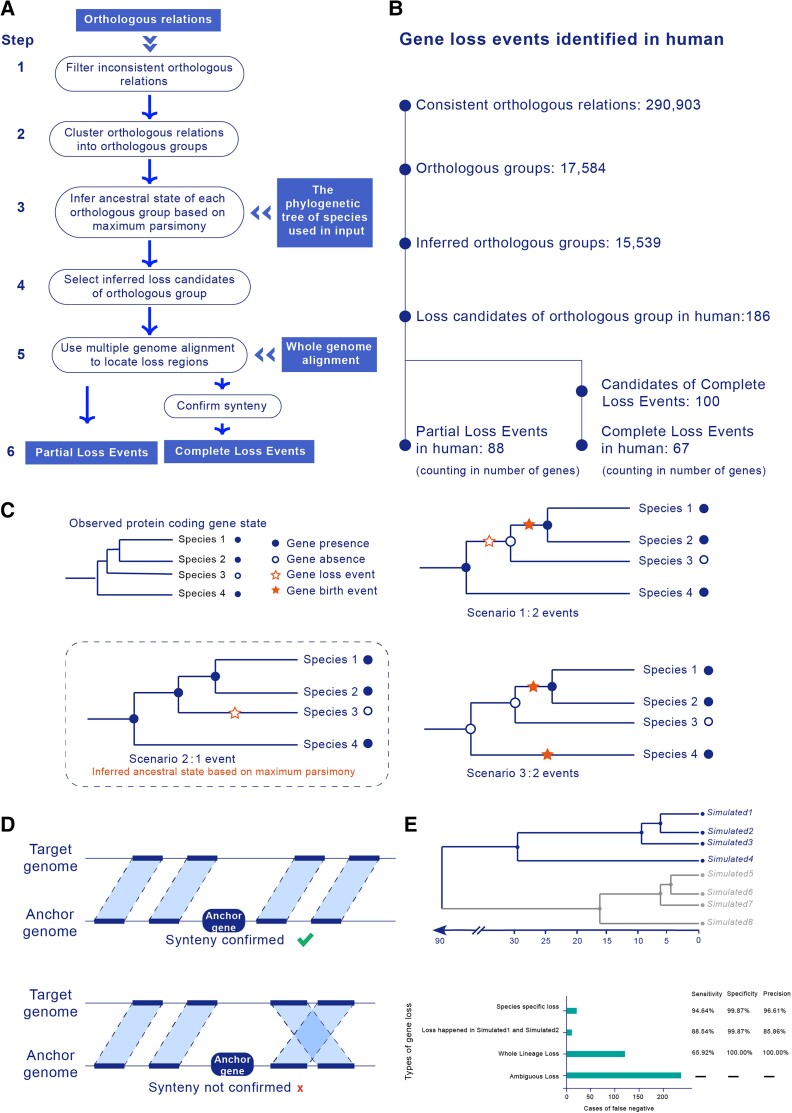
To effectively identify loss events across various time scales, we built a novel pipeline, LOST & FOUND (fig. 1A) (LOST & FOUND is available in https://github.com/gao-lab/LOST_and_FOUND), that combines existing annotations and genome alignment to map orthologous correspondence along the phylogenetic tree of species used in the input and infers ancestral state by minimizing the gene state turnover during divergence (i.e., following the maximum parsimony principle; fig. 1C). Here, we followed the broad definition of protein gene loss: a nonfunctionalization event occurring in a particular evolutionary lineage through either the complete deletion of the corresponding genomic locus (“Complete Loss”) or a loss-of-function mutation (“Partial Loss”) (Jensen 2001; Albalat and Cañestro 2016). For each candidate, the genome position of the anchor gene (i.e., the orthologous counterpart in the reference species) and the genome alignment are used for the back tracing of the loss relic or syntenic regions. Candidates whose relic or syntenic regions overlap with the assembly gap are excluded since we cannot determine whether the observation of such candidates is due to gene loss or simply the missing sequences of unassembled regions. Those candidates with relics are then classified as Partial Loss Events. For candidates without relics, nearby genome blocks are used to confirm synteny (fig. 1D). Those with synteny block pairs are then classified as Complete Loss Events.
Fig. 1.
Workflow and evaluation of LOST & FOUND. (A) Steps of the gene loss identification pipeline: (1) filter orthologous relationships, (2) cluster orthologous relation, (3) infer the ancestral state of the orthologous group, (4) select loss candidates, (5) locate loss regions by genome alignment, and (6) classify Complete Loss Events and Partial Loss Events. (B) Processes of human gene loss identification. The output counts of each step are listed. (C) Maximum parsimony principle for inferring the ancestral gene state. With a given observed gene state, there are various possible ancestral gene states. Using the maximum parsimony approach, the ancestral gene state that minimizes the number of changes required to generate the observed gene state is the inferred ancestral state. (D) Synteny confirmation for Complete Loss Events. Only when two upstream and downstream genomic blocks around the anchor gene are collinear is synteny confirmed. (E) Phylogenetic tree for genome simulation and pipeline performance for different types of gene loss. We simulated genome evolution based on this phylogenetic tree, and we curated the “ground truth” gene loss events in Simulated1. The genomes of Simulated5–8 were used as anchor species (i.e., species used as reference), whereas the genomes of Simulated1–4 were used as target species (i.e., query species and its sister used for better inference) for the pipeline evaluation. We examined the causes of the low sensitivity of our pipeline and showed that most of the false negatives were caused by ambiguous types of gene loss and the type of gene loss that simultaneously occurred in Simulated1, Simulated2, and Simulated3.
To evaluate the performance of LOST & FOUND, we considered a stepwise genome evolution process across eight species with the same phylogenetic relationships (fig. 1E) as mouse (Mus musculus), rat (Rattus norvegicus), shrew mouse (Mus pahari), Ryukyu mouse (Mus caroli), macaque (Macaca mulatta), chimpanzee (Pan troglodytes), gorilla (Gorilla gorilla), and human (Homo sapiens; see Materials and Methods for more details). The genome evolution process was run based on EVOLVER (http://www.drive5.com/evolver/), a tool that simultaneously simulates the evolution of both annotations and sequences of a whole genome (Earl et al. 2014). With the synthetic data set, we systematically curated a set of “ground truth” gene loss events for benchmarking, showing 99.99% specificity and 66.23% sensitivity for the LOST & FOUND pipeline. After the examination of missed gene loss events, we found that the low sensitivity was mainly caused by two types of gene loss. Most of the false negatives (60.9%, 238 out of 391; fig. 1E) were due to Ambiguous Loss, a type of gene loss with ambiguous ancestral gene state inference (e.g., scenario in supplementary fig. S1, Supplementary Material online). Additionally, more than one-quarter of the false negatives (30.9%, 121 out of 391; fig. 1E) were caused by Whole Lineage Loss, a type of gene loss that occurred in multiple species along one entire lineage (i.e., loss took place simultaneously in Simulated1, Simulated2, and Simulated3), partly because the orthologous groups showing this type of gene loss present fewer orthologous relationships compared with other groups (and are thus more sensitive to annotation error of orthologs). The remaining 9.2% of false negatives suffered from unreliable orthologous relationships (e.g., the scenario in supplementary fig. S2, Supplementary Material online). Except for these two types of gene loss, LOST & FOUND showed high sensitivity as well as high specificity, especially for the species-specific type of gene loss. Overall, the high specificity of LOST & FOUND ensures that it will rarely produce false positives in gene loss identification.
We applied this pipeline to detect human gene loss after rodent and primate divergence. To improve the detection power (Thybert et al. 2018), we incorporated sister species in the rodent clade as reference species and the primate branch for orthologous inferences (fig. 3). With this tree and our pipeline, we ultimately identified 155 human gene loss events, 67 of which were Complete Loss Events, whereas 88 were Partial Loss Events (fig. 1B and supplementary data, Supplementary Material online). By using Exonerate (Slater and Birney 2005) to align the anchor genes and loss relics, we further validated these Partial Loss Events after their identification. Most of the Partial Losses suffered from disabling mutations (i.e., frameshifts or premature stop codons) or could not be completely aligned to the anchor gene (supplementary data, Supplementary Material online).
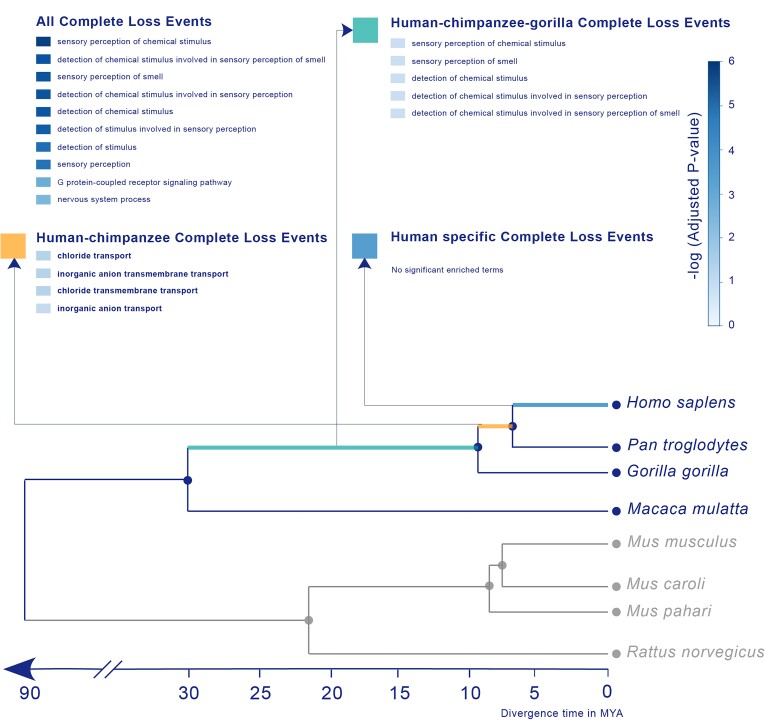
Fig. 3.
Functional bias of gene loss events in the human genome. The phylogenetic tree was used for human gene loss identification. Rodent species were used as anchors, whereas the primate species macaque, gorilla, and chimpanzee were used as sister species to trace human gene loss events. GO enrichment was performed for all loss events and for different loss events among different lineages.
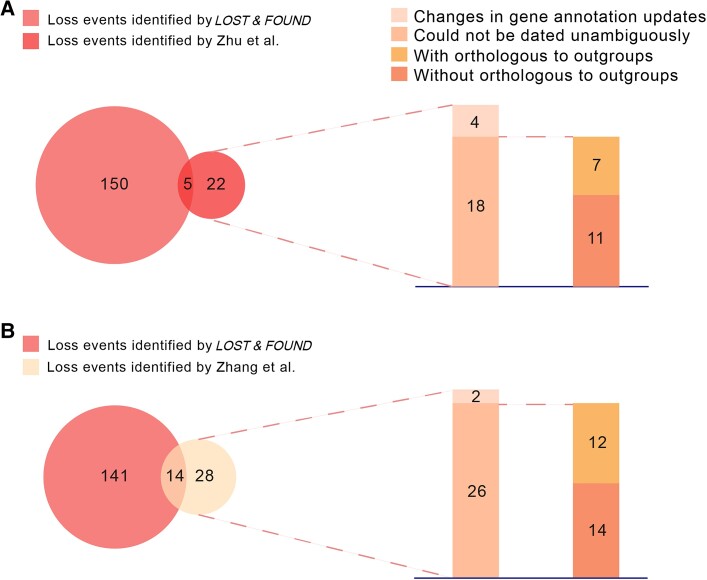
Notably, we found large differences when comparing our results with those of previous studies (fig. 2A and B) (Zhu et al. 2007; Zhang et al. 2010). Close inspection showed that 81.8% (18 out of 22; [Zhu et al. 2007]) and 92.8% (26 out of 28 [Zhang et al. 2010]) of “loss events” in the primate lineage excluded by our pipeline could not be dated unambiguously (supplementary fig. S3 and supplementary data, Supplementary Material online). In particular, half of these events (11 out of 18 [Zhu et al. 2007] and 14 out of 26 [Zhang et al. 2010]) showed no orthologs within the chicken and lizard genomes used as out-groups, suggesting that they may in fact represent gene origin events within the rodent lineage (fig. 2A and B and supplementary data, Supplementary Material online). The remaining inconsistent events (4 out of 22 [Zhu et al. 2007] and 2 out of 28 [Zhang et al. 2010]) were due to changes in gene annotation updates (fig. 2A and B and supplementary data, Supplementary Material online).
Fig. 2.
Comparison of gene loss events in humans with those identified in previous studies. (A) Comparison of our gene loss events with those reported by Zhu et al. (B) Comparison of our gene loss events with those reported by Zhang et al.
Our pipeline also identified a number of loss events in humans that have been missed in previous studies. One major issue accounting for the disparity is that the genome alignment data that we employed enabled the more effective detection of gene loss events with large-scale genomic deletion (i.e., Complete Loss Event) relative to the plain Blast-based sequence comparison used previously (supplementary fig. S4, Supplementary Material online; 42.6%, 64 out of 150 [Zhu et al. 2007], and 43.2%, 61 out of 141 [Zhang et al. 2010]). Our Blast search experiment showed that compared with Partial Loss Events, Complete Loss Events are much more challenging for Blast searching (supplementary fig. S5, Supplementary Material online). Furthermore, we retained loss events detected in large families (21), such as olfactory or vomeronasal receptors (supplementary fig. S6, Supplementary Material online), which were filtered out in previous works (Zhu et al. 2007; Zhang et al. 2010).
We observed a statistically significant functional bias in genes hit by loss events. Complete Loss Events were enriched in sensory or stimulus detection (fig. 3 and supplementary data, Supplementary Material online), consistent with previous reports that most genes lost in humans are sensory related (Gilad et al. 2003; Young and Trask 2007; Kawamura and Melin 2017; Niimura et al. 2018; Qian et al. 2022). Complete Loss Events among different lineages also showed different patterns. Complete Loss Events occurring in the human–chimpanzee lineage were enriched in chloride transport–related processes, whereas those in the human–chimpanzee–gorilla lineage were enriched in sensory smell–related processes. In contrast, we did not find strong preferences among genes subject to Partial Loss. The anchors of the gene loss events were significantly enriched in multimember gene families compared with the anchors of known orthologous pairs (supplementary fig. S7, Supplementary Material online). Since the loss of genes belonging to multimember families can be compensated by their paralogs, this observation suggests that the loss of a gene is related to its dispensability (Albalat and Cañestro 2016).
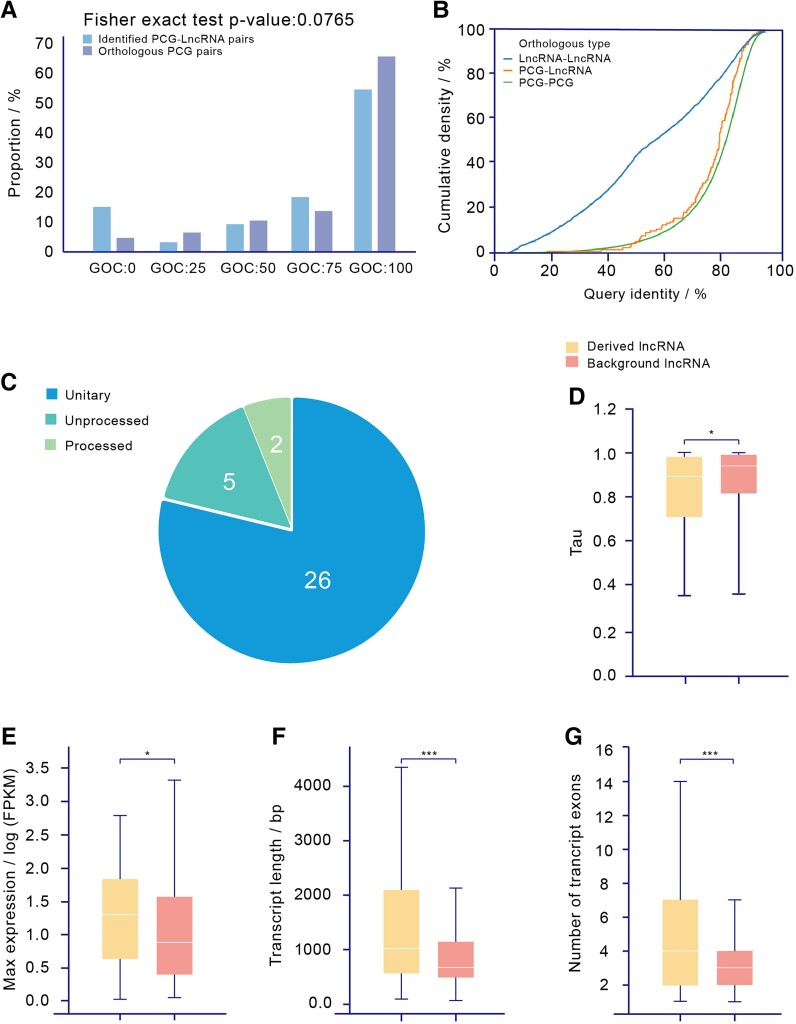
It has been suggested that lncRNA origins may be linked to the Partial Loss of former protein-coding genes (Duret et al. 2006; Hezroni et al. 2017; Liu et al. 2017). We compared the relic regions of all Partial Loss Events in the human genome and found that more than one-third (33 out of 88) of the loci overlapped with annotated lncRNAs (supplementary data, Supplementary Material online). Genomic synteny analysis (fig. 4A) and sequence comparison (fig. 4B) confirmed that these lncRNAs were derived from loss relics. Our results (fig. 4B) show that the cumulative distribution of the query identities between these lncRNAs and the anchor genes of their corresponding loss events are similar to those of protein-coding gene (PCG) orthologous pairs, whereas their query identities are significantly higher than those of lncRNA orthologous pairs.
Fig. 4.
Validation and characteristics of derived lncRNAs. (A) Distribution of GOC scores between anchor PCG–derived lncRNA pairs and annotated orthologous PCG pairs. To confirm that these lncRNAs are potentially derived from loss event relics, we first checked their synteny. We calculated the GOC scores between these lncRNAs and the anchor genes of their corresponding loss events and then compared them with the scores of existing orthologous protein-coding gene pairs. The distribution of GOC scores shows no significant differences (P = 7.65 * 10−2, Fisher's exact test). (B) Comparison of alignment query identity between PCG-derived lncRNA pairs, annotated orthologous PCG pairs, and annotated orthologous lncRNA pairs. We also considered the sequence similarity between these lncRNAs and their anchor genes. We collected a set of lncRNA orthologous pairs (Sarropoulos et al. 2019) and used them to compare the sequence similarities of different types of orthologs. (C) Classification of derived lncRNAs. (D) Comparison of tau between derived lncRNAs and background lncRNAs. (E) Comparison of maximum expression between derived lncRNAs and background lncRNAs. (F) Comparison of transcript length between derived lncRNAs and background lncRNAs. (G) Comparison of transcript exon numbers between derived lncRNAs and background lncRNAs.
Most of these derived lncRNAs (78.8%, 26 out of 33) were unitary (fig. 4C). Expression profiling across 28 tissues showed that 60 of 141 transcripts (for 20 out of 33 genes) of these lncRNAs were expressed in at least 1 tissue (Fragments Per Kilobase of transcript per Million mapped reads (FPKM) > 1; supplementary data, Supplementary Material online). In addition, most of these derived lncRNAs exhibited highly different expression patterns compared with their protein-coding ancestors (supplementary fig. S8, Supplementary Material online), suggesting that they are bona fide transcriptional units instead of transcriptional noise caused by partially degenerated promoters. Most of them were specifically expressed in one tissue, such as the testis or brain (supplementary fig. S9 and S10, Supplementary Material online), consistent with previous studies on tissue-specific lncRNAs. Coexpression analysis suggested statistically significant functional enrichment in reproduction and the immune response (supplementary data, Supplementary Material online), and further literature searches showed that several of the lncRNAs were involved in key biological processes such as cell proliferation and cell cycle regulation (table 1).
Table 1.
The derived lncRNAs proved to be functional experimentally.
| Gene ID | Supported Studies | Phenotype |
|---|---|---|
| ENSG00000173209 | (Shalem et al. 2014) (Morgens et al. 2017) (Yilmaz et al. 2018) (Toledo et al. 2015) |
Cell proliferation |
| Response to toxin | ||
| Cell proliferation | ||
| Cell proliferation | ||
| ENSG00000197376 | (Shalem et al. 2014) | Response to chemicals |
| ENSG00000197847 | (Riba et al. 2017) (Wang et al. 2017) |
Regulation of signal transduction phenotype |
| Cell proliferation | ||
| ENSG00000261603 | (Riba et al. 2017) | Response to virus |
| ENSG00000088340 | (Yue et al. 2015) (Qiao and Li 2016) (Xia et al. 2015) (Sun et al. 2019) (Wu et al. 2017) |
Cell migration; cell invasion; cell proliferation; tumor-suppressive function; prognosis |
| Cell proliferation; cell cycle | ||
| Cell proliferation | ||
| Cell proliferation | ||
| Prognosis |
Notably, genetic analysis showed that these derived lncRNAs were more likely to be associated with growth and development processes (supplementary data, Supplementary Material online) and harbored more known cis-eQTL sites (P = 2.83 * 10−15, Mann–Whitney U test; supplementary fig. S11, Supplementary Material online) than other human lncRNAs. Moreover, we noted that these derived lncRNAs were highly and broadly expressed (P = 1.35 * 10−2 and P = 1.77 * 10−2, Mann–Whitney U test; fig. 4D and E and supplementary fig. S12, Supplementary Material online) and were significantly longer and had many more exons (P = 5.90 * 10−7 and P = 2.00 * 10−10, Mann–Whitney U test; fig. 4F and G) than other human lncRNAs. Such differences may be attributed to their protein-coding origin (i.e., as remnants of ancestral protein-coding genes). We also noted that these derived lncRNAs were more likely to be under positive selection than other lncRNAs in the human genome (P = 1.09 * 10−2, chi-square test; supplementary fig. S13, Supplementary Material online). Within-species tests of selective pressure, such as the Pi, Tajima's D, and derived allele frequency (DAF), did not show any differences between derived and nonderived lncRNAs, whereas between-species tests of Hudson–Kreitman–Aguadé (HKA) did show differences (supplementary fig. S13 and supplementary data, Supplementary Material online), suggesting that the positive selection on these lncRNAs was long term and that the sequences have now reached fixation. This indicates that these lncRNAs are under positive selection during speciation.
Discussion
Gene loss events can be inferred by the absence of orthologous genes (Jensen 2001; Albalat and Cañestro 2016). Briefly, when a gene is present in one species but not another species, it may indicate that either a gene gain event occurred in one species or a gene loss event occurred in another species. Using multiple orthologous relationships between species, our pipeline is able to trace the gene state of each common ancestral node backward and, thus, to distinguish whether the absence of a gene represents a loss event in the target species or a gain event in other lineages. For whole-genome alignment, the “correspondence” between genomes can be used to locate orthologous sequences where gene loss events could occur. This process involves intergenic regions, and the context of the query gene can also be considered under these circumstances, thus better supporting the identification of gene losses with large-scale deletion compared with canonical sequence-searching-based methodology (Zhu et al. 2007; Zhang et al. 2010; Zhao et al. 2015; Sharma, Hecker, et al. 2018). Our test shows that >96% (11,932 out of 12,349) of mouse and human orthologous pairs could be identified ab initio through multiple whole-genome alignments, further confirming its power.
It has long been proposed that Partial Loss Events may lead to the origination of novel lncRNAs (Duret et al. 2006; Ponting et al. 2009; Hezroni et al. 2017). Thus, investigating the function of these lncRNAs derived from gene loss is a viable method for conducting functional studies of gene loss. Our data suggested that more than one-third (33/88) of Partial Loss Events contribute to the origination of newly derived lncRNAs. More than half (17 out of 33) of these derived lncRNAs are under positive selection, and several have been shown to be functional experimentally (Oughtred et al. 2019; Zhao et al. 2020). For example, one of these lncRNAs, ENSG00000088340 (supplementary fig. S14, Supplementary Material online), has been associated with the progression of gastric cancer and colon cancer (Xia et al. 2015; Yue et al. 2015). ENSG00000088340 acts as a sponge of miR-106a-5p and thus suppresses oncogenesis. These results suggested that the loss of a protein-coding gene could lead to the “birth” of a novel-derived lncRNA that could further gain novel functionalities during follow-up evolution.
We noted that several identified derived lncRNAs (28 out of 33) overlap with introns of known protein-coding genes, which may lead to the speculation that the observed selection occurs because these lncRNAs are affected by the sequence constraint imposed by overlapping protein-coding genes. However, further inspection revealed no significant differences in the distribution of selection between intron-overlapping and nonoverlapping lncRNAs (14 out of 28 vs. 3 out of 5, P = 8.58 * 10−1, chi-square test).
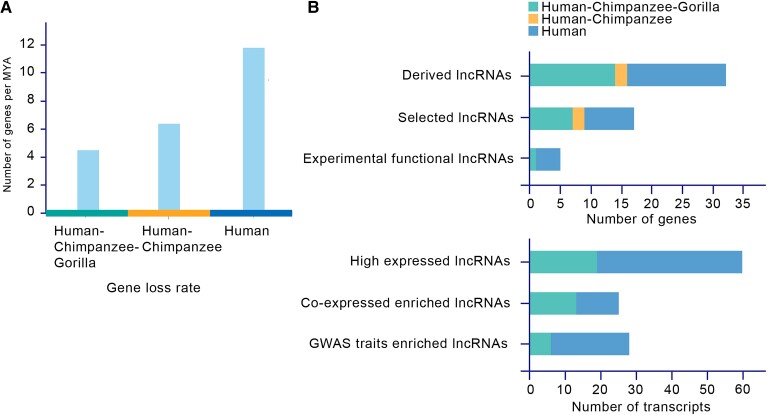
The peak gene loss rate (11.79 genes per MYA) is founded in the human lineage, whereas the gene loss rates in the human–chimpanzee lineage and human–chimpanzee–gorilla lineage are 6.36 and 4.46 genes per MYA, respectively. The gene loss rate in the human lineage is twice that in the other lineages, suggesting that gene loss is accelerated in the human lineage and that most human gene loss events are human-specific (fig. 5A). Even though the low sensitivity of LOST & FOUND for the type of gene loss that occurred in multiple species according to the test of the “ground truth” gene loss set was suggested to potentially lead to an underestimation of the gene loss rate by ∼40% in the human–chimpanzee–gorilla lineage, the gene loss rate in the human lineage was still much higher than the gene loss rate in the human–chimpanzee–gorilla lineage after putative correction (7.43 genes per MYA). In particular, the high specificity and high sensitivity of LOST & FOUND for the species-specific type of gene loss guarantee the loss rate observed in the human lineage. Intriguingly, functional gene losses (i.e., gene loss events related to the functional analysis candidates, such as gene loss events that give rise to lncRNAs or are related to highly expressed lncRNAs) showed a similar pattern. More than half of these functional gene losses were human-specific (fig. 5B). It has been reported that the human lineage is characterized by an accelerated evolutionary rate (λ) estimated by maximum likelihood based on gene family number analysis, and it has long been proposed that such drastic gene turnover helps shape the differences between modern humans and chimpanzees via the expansion of brain-related gene families (Hahn et al. 2007). Here, by focusing on the gene loss pathway, we also observed a consistent pattern along the human lineage; moreover, complementary to the expansion of functional gene families, we illustrated that the loss of certain genes could also contribute to modern human traits in multiple ways.
Fig. 5.
Gene loss rates and distribution of “functional gene loss”. (A) Gene loss rates among different lineages. The gene loss rate is highest in the human lineage. (B) Distribution of “functional gene loss”. Most of these “functional gene loss” events are human-specific.
LOST & FOUND takes advantage of whole-genome alignment and orthologous annotation. Sharma's pipeline (Sharma, Hecker, et al. 2018) also takes advantage of genome alignment. However, to rule out potential artifices, they implemented a series of “filters,” which could decrease sensitivity. For instance, the filter “mutation in several exons, <60% intact reading frame” would exclude one-third (11 out of 33) of derived lncRNA-related loss events identified by LOST & FOUND. Many of these “excluded derived lncRNAs” have vital functions (e.g., ENSG00000088340).
LOST & FOUND could be further improved. In particular, its sensitivity is clearly not high enough, suggesting that there are still a number of missed gene loss events. The constraint of maximum parsimony helps guarantee the high specificity of our pipeline. However, its limitation related to certain types of gene loss leaves the states of numerous orthologous groups undetermined and therefore causes false negatives. Consider Ambiguous Loss in the evaluation for instance, which cannot be identified through maximum parsimony and is the major cause of the low sensitivity of our pipeline, accounting for 22.0% (238 out of 1,081) of cases in the evaluation. During the identification of human gene loss, 185 candidates with ambiguous ancestral gene states were excluded, similar to the situation for Ambiguous Loss. Certain gene losses with ambiguous ancestral gene states might remain unidentified and need to be identified based on improved methodology or other evidence. It should also be mentioned that the evaluation of our pipeline was based on the phylogeny of primates and rodents and both the sensitivity and specificity might vary when it is applied to different phylogenies. In particular, the current analysis of derived lncRNAs depends on public genomic curation, which is actively evolving and may suffer from outdated annotation or misannotation. In addition, considering universality and applicability, we did not include complicated steps such as reannotating orthologs and relied on the existing annotations of genome or orthologs. When LOST & FOUND is applied in other species, especially for those species with poor gene annotation, further inspection should be performed since an unannotated gene might be misidentified as a loss.
Although several recent studies have demonstrated that gene loss is an essential part of the evolutionary landscape and may contribute to adaptive phenotype changes (Olson 1999; Jebb and Hiller 2018; Sharma, Lehmann, et al. 2018; Hecker et al. 2019), characterizing gene loss events effectively and efficiently remains a major challenge. Here, we present a novel pipeline for gene loss identification that not only combines the advantages of both orthologous inference and genome alignment data and thus detects gene loss more accurately but is also convenient to use and can be introduced in other studies. Using this pipeline, we systematically annotated human gene loss events. Most of the Complete Loss Events observed in humans were related to sensory smell–associated processes, whereas there were no significantly enriched processes related to Partial Loss Events. Such differences between Complete Loss Events and Partial Loss Events might indicate that different genes suffer from different loss processes, in that olfactory receptors in tandem regions tend to suffer from large-scale deletion. Most importantly, based on the loss events we identified, we discovered a set of lncRNAs derived from lost genes. These sets of lncRNAs are completely different from other lncRNAs. Most of them are under selection and show high, broad expression. They are also more likely to be involved in growth, development, immunity, and reproduction. The idea that the loss of a gene could generate a functional lncRNA extends our understanding of the effect of gene loss.
Materials and Methods
Genome-Wide Identification of Gene Loss Events
First, our pipeline, LOST & FOUND (fig. 1A), requires orthologous pairs between anchor species (i.e., species used as references) and target species (i.e., the query species and its sister used for better inference) as well as their multiple genome alignments from Ensembl as input. Because the “many to many” or “one to many” types of orthologous pairs show inconsistency between different Ensembl versions, the orthologous pairs are first filtered. Using the mean value of query identity (i.e., the percentage of the query sequence that matches the target sequence) and target identity (i.e., the percentage of the target sequence that matches the query sequence) between orthologous pairs as indicator, we found that at ∼55% (supplementary fig. S15, Supplementary Material online; 58%) identity, consistent and inconsistent orthologous pairs could be well classified. Therefore, we use this as a threshold to filter orthologous pairs. Then, these orthologous pairs are clustered as orthologous groups, each of which reflects the presence or absence of the gene. To avoid false positives, only orthologous groups with genes present in all anchor species are considered for further identification. Then, in each orthologous group, maximum parsimony (fig. 1C) is performed. The gene states, absence or presence of the orthologs, are mapped across the phylogeny for each orthologous group. Then, the gene states of the internal nodes (representing the ancestral gene state) are inferred from leaf to root across the whole phylogeny. The most likely ancestral gene state is the state that minimizes gene state turnover during divergence. For instance, the gene state of an internal node is inferred as “present” when both of its child node gene states are present, because the “present” state of this node requires zero gene state turnover for its child nodes. Those orthologous groups showing gene presence in the ancestor but absence in the query species are then selected as loss candidates. For each loss candidate, we use multiple genome alignment to locate the loss region in the query species based on the anchor gene (i.e., the gene in anchor species within an orthologous group or orthologous pairs). For each candidate, the coordinate of the anchor gene is used as the anchor of the genome alignment to retrieve homologous genomic regions in the query species. After retrieving the homologous genomic region, a merging process is performed. If the homologous genomic blocks are located on the same chromosome and the gap between the blocks is below a certain threshold (default = 500,000 bp, i.e., approximately ten times the average length of a human protein-coding gene), the blocks are merged into a complete region representing the homologous genomic region in the target species of the loss candidate. If the homologous genomic blocks are located on different chromosomes or the gap between the blocks is above the threshold (default = 500,000 bp), the blocks are separated into different regions. Those candidates with loss regions are classified as Partial Loss Event candidates, which indicates that the ancestral gene may have experienced mutations and lost protein-coding potential, whereas the relic gene region has been retained. For candidates for which the loss region cannot be located through genome alignment and their anchor genes, the nearby genome alignment blocks around the anchor genes are used for synteny confirmation. For each anchor gene of the candidate, two genome alignment blocks upstream and downstream are fetched, and their aligned regions in the target species are then obtained. If the order of these blocks shows conservation between query and target species, the synteny is confirmed, and the candidate is classified as a Complete Loss Event candidate (fig. 1D), which indicates that the ancestral gene region may have experienced whole-gene deletion and been removed. After obtaining the relic/syntenic regions of Partial/Complete Loss Event candidates, these regions will be compared with the assembly gap regions of query species. Those candidates whose relic/syntenic regions do not overlap with the assembly gap will eventually be identified as Partial/Complete Loss Events.
Simulation of Genome Evolution and Evaluation in LOST & FOUND
As in the Alignathon project (Earl et al. 2014), the simulation of genome evolution was performed using EVOLVER (http://www.drive5.com/evolver/). EVOLVER simulates genome evolution by first proposing mutations at randomly selected loci and then calling acceptance/rejection for the proposed mutations based on the base-wise “accept probability.” Accept probability is determined based on annotation. For instance, bases with more constraints or more conservation, such as those in genic regions, would be assigned lower accept probabilities than others. Since EVOLVER can only perform one cycle of evolution simulation at a time, we also used the evolverSimControl (https://github.com/dentearl/evolverSimControl) and evolverInfileGeneration (https://github.com/dentearl/evolverInfileGeneration/) tools to run the mammalian phylogeny simulation. The specific model parameter file required by EVOLVER is the same as in the Alignathon project (Earl et al. 2014).
We first initiated the simulation using the whole human genome, hg19/GRCh37, including the complete chromosome sequences and the annotations from the UCSC Genome Browser tracks mgcGenes, knownGene, knownGeneOld5, cpgIslandExt, and ensGene. All of these input data can be obtained through the evolverInfileGeneration tool. According to Alignathon (Earl et al. 2014), the distance of the simulator was set to 1.0 neutral substitutions per site for the simulation of the most recent vertebrate common ancestor, which experienced ∼500 million years of evolutionary time. Here, we set the distance to 0.18 neutral substitutions per site for the simulation of the most recent common ancestor of rodents and primates, which experienced ∼90 million years of evolutionary time. After acquiring the simulated genome of the most recent common ancestor, we used it as the input for mammalian phylogeny simulation. The mammalian phylogeny simulation was run based on the tree (in Newick format) ((((Simulated1: 6.5, Simulated2:6.5)S1-S2:2.0, Simulated3:8.5)S1-S2-S3:20.5, Simulated4:29)S1-S2-S3-S4:58, (((Simulated5:4.5, Simulated6:4.5)S5-S6:2.0, Simulated7:6.5)S5-S6-S7:9.5, Simulated8:16)S5-S6-S7-S8:71).
With the simulated leaf genomes and their annotations, we labeled a set of orthologous groups as the “ground truth” gene loss in Simulated1. We inferred the orthologous relationships between the simulated genomes for those genes that share the exact same coding sequences (CDS) structure with the same ancestral gene. Genes that have different CDS structures from their ancestral gene due to CDS deletion, creation, or movement were inferred as “gene change.” The change in a gene during evolution could be the result of either new gene birth or previous gene death. Therefore, those orthologous groups consisting of “gene change” in the Simulated1 genome while containing orthologous genes in more than half of the simulated genomes as well as the Simulated5-8 genomes were classified into the “ground truth” gene loss set. The “gene change” in Simulated1 suggested that they could represent a loss in the Simulated1 lineage or birth in other lineages, whereas the orthologous genes existing in more than half of the simulated genomes as well as Simulated5-8 genomes confirmed that these genes previously existed in the anchor lineage and, thus, represent the gene loss in Simulated1.
After acquiring the “ground truth” gene loss set, we used it to test LOST & FOUND. In LOST & FOUND, we used the Ensembl orthologs as raw input. According to the Quest for Orthologs consortium, a community that contributes to providing the gold standard for orthologous annotation benchmarks and has already evaluated tens of public orthologous inference methods, the true positive rate and positive predictive value of Ensembl orthologs are 91.54% and 98.87%, respectively (Nevers et al. 2022). To eliminate the bias of using simulated orthologous relationships as input, we manually “corrupted” these simulated orthologous relationships by introducing ∼8.5% (≈100 – 91.54% of the true-positive rate) false-negative orthologs and ∼1.5% (≈100 – 98.87% of the positive predictive value) false-positive predictive orthologs. Then, we used these “corrupted” orthologs as the input to run LOST & FOUND and compared the identified loss events with the “ground truth” gene loss set.
Identification and Classification of Gene Loss Events in the Human–Mouse Clade
In this research, we considered the rodent branch as the anchor species set for tracing gene loss in the primate branch (target species set). The rodent branch consisted of mouse (M. musculus), rat (R. norvegicus), shrew mouse (M. pahari), and Ryukyu mouse (M. caroli), whereas the primate branch consisted of human (H. sapiens), chimpanzee (P. troglodytes), gorilla (G. gorilla), and macaque (M. mulatta). The original orthologous data set was the collection of anchor–target (one anchor species to one target species) species orthologous relationships, which includes all combinations of orthologous relationships between one rodent species and one primate species. The original orthologous relationships were obtained from Ensembl BioMart 94 (http://www.ensembl.org/biomart/martview/). Then, the “one to many” and “many to many” types of orthologous relationships with <55% mean values of query identity and target identity were filtered out due to the inconsistent orthologous annotations in different Ensembl versions, which might indicate unreliable annotated orthologous relationships. Since the rodent branch was considered the anchor, only those orthologous groups with genes present in all species of the rodent branch were retained for further analysis. Then, based on the maximum parsimony, the ancestral state of these groups was inferred, and the groups inferred as human gene losses were selected as gene loss candidates based on the 26 eutherian mammal alignments obtained from Ensembl Compara 94. Next, the assembly gap of the human genome was obtained through the UCSC Gap Locations track (http://genome.ucsc.edu). With the genome alignment, assembly gap regions, and loss candidates, Partial Loss Events and Complete Loss Events in humans were then identified based on our pipeline. Exonerate Ver 2.2.0 was used to validate the Partial Loss regions after identification. We chose the longest transcript of each anchor gene and aligned it to the corresponding loss relic region. Exonerate was employed using the coding2genome model with the best parameter set as 1.
Comparison of Gene Loss Events
The gene loss events reported in Zhang and Zhu were retrieved from their studies as gene symbols and then manually searched through Ensembl 94 and converted to Ensembl Gene IDs for comparison. Some of the gene symbols (8 out of 35 [Zhu et al. 2007] and 5 out of 47 [Zhang et al. 2010]) could not be identified through Ensembl 94 and were deprecated. The out-group species that we used to confirm the ancestral states of those inconsistent loss events were chicken and lizard. Orthologous pairs from chicken and lizard were obtained through Ensembl BioMart 94.
Evaluation of Gene Loss Rates, Gene Family Members, and Functional Patterns
Each gene loss event belongs to an orthologous group. In each group, based on the maximum parsimony, the gene state of each ancestral node can be inferred, and then, the loss node/lineage of gene loss can be determined. Combined with the divergence time acquired via TimeTree, the gene loss rate can be calculated by dividing the number of gene losses in each lineage by the divergence time of that lineage. Additionally, with the loss node of the loss events, Gene Ontology (GO) analysis for gene loss events among different lineages can be performed. GO analysis was performed based on the anchor protein-coding genes of loss events in mouse. The GO annotations were acquired through (http://geneontology.org/), and the enrichment analysis was performed based on AnnoLnc2 (Ke et al. 2020). Gene family member analysis was based on paralog annotation with data obtained from Ensembl BioMart (http://www.ensembl.org/biomart/martview/), archive version 94. If the paralog search for a gene identified any paralogs, the gene was considered to belong to the multimember family.
Identification and Confirmation of Gene Loss–Derived lncRNAs
For the Partial Loss Events that we identified, the relic region was also identified. The lncRNA annotation was acquired through Ensembl Perl API 94 (https://www.ensembl.org/). By using the Partial Loss region as a slice, the lncRNAs that overlap with these regions can be acquired. These lncRNAs are the derived lncRNAs. The Gene Order Conservation (GOC) scores of the annotated orthologous protein-coding genes were also acquired directly through BioMart (http://www.ensembl.org/biomart/martview/) 94. We calculated the GOC scores of the derived lncRNAs based on the Ensembl definition. The gene sequences of the derived lncRNAs and the annotated orthologous protein-coding genes were also acquired through the Ensembl Perl API, and global alignment was performed through EMBOSS.
Functional Annotation of Derived lncRNAs
The literature search for the derived lncRNAs was based on the LncTarD (Zhao et al. 2020) and BioGrid (Oughtred et al. 2019) databases. Genome-wide association studies (GWAS) traits were acquired through AnnoLnc2 (Ke et al. 2020). For all the lncRNAs, we searched their GWAS traits, and for each GWAS trait, we calculated the derived lncRNA numbers and background lncRNA numbers within it. Then, for each trait, we performed a chi-square test and checked which traits were enriched with the derived lncRNAs. The P value adjustment was performed through the Benjamini–Hochberg procedure. Weighted gene coexpression network analysis was performed using the WGCNA R package (Zhang and Horvath 2005). The expression data for the derived lncRNAs and protein-coding genes were acquired through AnnoLnc2 (Ke et al. 2020).
Transcript Structure, Expression, and Selection Comparison between Derived lncRNAs and Other lncRNAs
All of the lncRNA annotations and their length and exon numbers were determined based on Ensembl 94 and obtained from the Ensembl Perl API (https://www.ensembl.org/). The set of all annotated lncRNAs excluding the derived lncRNAs was considered the background lncRNAs. The lncRNA expression data were acquired from AnnoLnc2 (Ke et al. 2020). The selection analysis was based on sliding window analysis. The variant data were obtained from the 1000 Genomes Project (Siva 2008). We used 10 k, 20 k, and 50 k windows to scan the whole human genome, and Pi and Tajima's D values were calculated through VCFtools (Danecek et al. 2011). The DAF was calculated with our own script based on its definition. The HKA test was performed based on the 1000 Genomes Project (Siva 2008) and human–chimpanzee genome alignment data. The polymorphic sites and divergence sites in each window were counted with our own script and then preprocessed to the input format required by HKAdirect (Esteve-Codina et al. 2013). Then, the HKA of each window was calculated through HKAdirect (Esteve-Codina et al. 2013).
Supplementary Material
Acknowledgments
The authors thank Drs. Zemin Zhang, Cheng Li, Letian Tao, Jian Lu, and Liping Wei at Peking University for their helpful comments and suggestions during the study. This work was supported by funds from the National Key Research and Development Program of China (2021YFC2502000, 2016YFC0901603) as well as the State Key Laboratory of Protein and Plant Gene Research, the Beijing Advanced Innovation Center for Genomics (ICG) at Peking University, and the Shaw Foundation Hong Kong Limited. The research of G.G. was supported in part by the National Program for Support of Top-notch Young Professionals. Part of the analysis was performed on the Computing Platform of the Center for Life Sciences of Peking University and supported by the High-Performance Computing Platform of Peking University.
Contributor Information
Zheng-Yang Wen, State Key Laboratory of Protein and Plant Gene Research, School of Life Sciences, Biomedical Pioneering Innovative Center (BIOPIC) & Beijing Advanced Innovation Center for Genomics (ICG), Center for Bioinformatics (CBI), Peking University, Beijing, China.
Yu-Jian Kang, State Key Laboratory of Protein and Plant Gene Research, School of Life Sciences, Biomedical Pioneering Innovative Center (BIOPIC) & Beijing Advanced Innovation Center for Genomics (ICG), Center for Bioinformatics (CBI), Peking University, Beijing, China.
Lan Ke, State Key Laboratory of Protein and Plant Gene Research, School of Life Sciences, Biomedical Pioneering Innovative Center (BIOPIC) & Beijing Advanced Innovation Center for Genomics (ICG), Center for Bioinformatics (CBI), Peking University, Beijing, China.
De-Chang Yang, State Key Laboratory of Protein and Plant Gene Research, School of Life Sciences, Biomedical Pioneering Innovative Center (BIOPIC) & Beijing Advanced Innovation Center for Genomics (ICG), Center for Bioinformatics (CBI), Peking University, Beijing, China.
Ge Gao, State Key Laboratory of Protein and Plant Gene Research, School of Life Sciences, Biomedical Pioneering Innovative Center (BIOPIC) & Beijing Advanced Innovation Center for Genomics (ICG), Center for Bioinformatics (CBI), Peking University, Beijing, China.
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Author Contributions
G.G. conceived the study and supervised the research; Z.Y.W. contributed to the computational framework and data curation of the pipeline; Z.Y.W., Y.J.K., L.K., and D.C.Y. contributed to the genomic analysis and data collection; and Z.Y.W. and G.G. wrote the manuscript with comments and input from all coauthors.
Conflict of interest statement. The authors declare no competing interests.
References
- Albalat R, Cañestro C. 2016. Evolution by gene loss. Nat Rev Genet. 17:379–391. [DOI] [PubMed] [Google Scholar]
- Borges R, Khan I, Johnson WE, Gilbert MTP, Zhang G, Jarvis ED, O’Brien SJ, Antunes A. 2015. Gene loss, adaptive evolution and the co-evolution of plumage coloration genes with opsins in birds. BMC Genomics 16:751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choo SW, Rayko M, Tan TK, Hari R, Komissarov A, Wee WY, Yurchenko AA, Kliver S, Tamazian G, Antunes A, et al. 2016. Pangolin genomes and the evolution of mammalian scales and immunity. Genome Res. 26:1312–1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA, Handsaker RE, Lunter G, Marth GT, Sherry ST, et al. 2011. The variant call format and VCFtools. Bioinformatics 27:2156–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean M, Carrington M, Goedert J, OBrien SJ. 1996. Genetic restriction of HIV-1 infection and progression to AIDS by a deletion allele of the CKR5 structural gene. Science 274:1069–1069. [DOI] [PubMed] [Google Scholar]
- Duret L, Chureau C, Samain S, Weissenbach J, Avner P. 2006. The Xist RNA gene evolved in eutherians by pseudogenization of a protein-coding gene. Science 312:1653–1655. [DOI] [PubMed] [Google Scholar]
- Earl D, Nguyen N, Hickey G, Harris RS, Fitzgerald S, Beal K, Seledtsov I, Molodtsov V, Raney BJ, Clawson H, et al. 2014. Alignathon: a competitive assessment of whole-genome alignment methods. Genome Res. 24:2077–2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteve-Codina A, Paudel Y, Ferretti L, Raineri E, Megens H-J, Silió L, Rodríguez MC, Groenen MAM, Ramos-Onsins SE, Pérez-Enciso M. 2013. Dissecting structural and nucleotide genome-wide variation in inbred Iberian pigs. BMC Genomics 14:148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galvani AP, Novembre J. 2005. The evolutionary history of the CCR5-Δ32 HIV-resistance mutation. Microbes Infect. 7:302–309. [DOI] [PubMed] [Google Scholar]
- Gilad Y, Man O, Paabo S, Lancet D. 2003. Human specific loss of olfactory receptor genes. Proc Natl Acad Sci U S A. 100:3324–3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn MW, Demuth JP, Han S-G. 2007. Accelerated rate of gene gain and loss in primates. Genetics 177:1941–1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecker N, Sharma V, Hiller M. 2019. Convergent gene losses illuminate metabolic and physiological changes in herbivores and carnivores. Proc Natl Acad Sci U S A. 116:3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedrick PW. 2011. Population genetics of malaria resistance in humans. Heredity (Edinb). 107:283–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hezroni H, Ben-Tov Perry R, Meir Z, Housman G, Lubelsky Y, Ulitsky I. 2017. A subset of conserved mammalian long non-coding RNAs are fossils of ancestral protein-coding genes. Genome Biol. 18:162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huelsmann M, Hecker N, Springer MS, Gatesy J, Sharma V, Hiller M. 2019. Genes lost during the transition from land to water in cetaceans highlight genomic changes associated with aquatic adaptations. Sci Adv. 5:eaaw6671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jebb D, Hiller M. 2018. Recurrent loss of HMGCS2 shows that ketogenesis is not essential for the evolution of large mammalian brains. eLife 7:e38906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen RA. 2001. Orthologs and paralogs - we need to get it right. Genome Biol. 2:INTERACTIONS1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamura S, Melin AD. 2017. Evolution of genes for color vision and the chemical senses in primates. In: Saitou N, editor. Evolution of the human genome I: the genome and genes. Tokyo: Springer Japan. p. 181–216. [Google Scholar]
- Ke L, Yang D-C, Wang Y, Ding Y, Gao G. 2020. Annolnc2: the one-stop portal to systematically annotate novel lncRNAs for human and mouse. Nucleic Acids Res. 48:W230–W238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuraku S, Kuratani S. 2011. Genome-wide detection of gene extinction in early mammalian evolution. Genome Biol Evol. 3:1449–1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Fan W, Tian G, Zhu H, He L, Cai J, Huang Q, Cai Q, Li B, Bai Y, et al. 2009. The sequence and de novo assembly of the giant panda genome. Nature 463:311–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W-H, Tsai ZT-Y, Tsai H-K. 2017. Comparative genomic analyses highlight the contribution of pseudogenized protein-coding genes to human lincRNAs. BMC Genomics 18:786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCutcheon JP, Moran NA. 2011. Extreme genome reduction in symbiotic bacteria. Nat Rev Microbiol. 10:13–26. [DOI] [PubMed] [Google Scholar]
- Meyer W, Liumsiricharoen M, Suprasert A, Fleischer LG, Hewicker-Trautwein M. 2013. Immunohistochemical demonstration of keratins in the epidermal layers of the Malayan pangolin (Manis javanica), with remarks on the evolution of the integumental scale armour. Eur J Histochem. 57:e27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgens DW, Wainberg M, Boyle EA, Ursu O, Araya CL, Tsui CK, Haney MS, Hess GT, Han K, Jeng EE, et al. 2017. Genome-scale measurement of off-target activity using Cas9 toxicity in high-throughput screens. Nat Commun. 8:15178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nevers Y, Jones TEM, Jyothi D, Yates B, Ferret M, Portell-Silva L, Codo L, Cosentino S, Marcet-Houben M, Vlasova A, et al. 2022. The quest for orthologs orthology benchmark service in 2022. Nucleic Acids Res. 50:W623–W632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niimura Y, Matsui A, Touhara K. 2018. Acceleration of olfactory receptor gene loss in primate evolution: possible link to anatomical change in sensory systems and dietary transition. Mol Biol Evol. 35:1437–1450. [DOI] [PubMed] [Google Scholar]
- Okerblom JJ, Schwarz F, Olson J, Fletes W, Ali SR, Martin PT, Glass CK, Nizet V, Varki A. 2017. Loss of CMAH during human evolution primed the monocyte-macrophage lineage toward a more inflammatory and phagocytic state. J Immunol. 198:2366–2373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson MV. 1999. When less is more: gene loss as an engine of evolutionary change. Am J Hum Genet. 64:18–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oughtred R, Stark C, Breitkreutz B-J, Rust J, Boucher L, Chang C, Kolas N, O’Donnell L, Leung G, McAdam R, et al. 2019. The BioGRID interaction database: 2019 update. Nucleic Acids Res. 47:D529–D541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponting CP, Oliver PL, Reik W. 2009. Evolution and functions of long noncoding RNAs. Cell 136:629–641. [DOI] [PubMed] [Google Scholar]
- Qian SH, Chen L, Xiong Y-L, Chen Z-X. 2022. Evolution and function of developmentally dynamic pseudogenes in mammals. Genome Biol. 23:235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao Q, Li H. 2016. LncRNA FER1L4 suppresses cancer cell proliferation and cycle by regulating PTEN expression in endometrial carcinoma. Biochem Biophys Res Commun. 478:507–512. [DOI] [PubMed] [Google Scholar]
- Riba A, Emmenlauer M, Chen A, Sigoillot F, Cong F, Dehio C, Jenkins J, Zavolan M. 2017. Explicit modeling of siRNA-dependent on- and off-target repression improves the interpretation of screening results. Cell Syst. 4:182–193. [DOI] [PubMed] [Google Scholar]
- Sarropoulos I, Marin R, Cardoso-Moreira M, Kaessmann H. 2019. Developmental dynamics of lncRNAs across mammalian organs and species. Nature 571:510–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shailendra KS. 2009. Controversial role of smallpox on historical positive selection at the CCR5 chemokine gene (CCR5-Δ32). J Infect Dev Ctries. 3:324–326. [DOI] [PubMed] [Google Scholar]
- Shalem O, Sanjana Neville E, Hartenian E, Shi X, Scott David A, Mikkelsen Tarjei S, Heckl D, Ebert Benjamin L, Root David E, Doench John G, et al. 2014. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 343:84–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma V, Hecker N, Roscito JG, Foerster L, Langer BE, Hiller M. 2018. A genomics approach reveals insights into the importance of gene losses for mammalian adaptations. Nat Commun. 9:1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma V, Lehmann T, Stuckas H, Funke L, Hiller M. 2018. Loss of RXFP2 and INSL3 genes in Afrotheria shows that testicular descent is the ancestral condition in placental mammals. PLoS Biol. 16:e2005293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siva N. 2008. 1000 Genomes project. Nat Biotechnol. 26:256–256. [DOI] [PubMed] [Google Scholar]
- Slater GSC, Birney E. 2005. Automated generation of heuristics for biological sequence comparison. BMC Bioinf. 6:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spanu PD, Abbott JC, Amselem J, Burgis TA, Soanes DM, Stuber K, Ver Loren van Themaat E, Brown JK, Butcher SA, Gurr SJ, et al. 2010. Genome expansion and gene loss in powdery mildew fungi reveal tradeoffs in extreme parasitism. Science 330:1543–1546. [DOI] [PubMed] [Google Scholar]
- Stedman HH, Kozyak BW, Nelson A, Thesier DM, Su LT, Low DW, Bridges CR, Shrager JB, Minugh-Purvis N, Mitchell MA. 2004. Myosin gene mutation correlates with anatomical changes in the human lineage. Nature 428:415–418. [DOI] [PubMed] [Google Scholar]
- Sun X, Zheng G, Li C, Liu C. 2019. Long non–coding RNA Fer–1–like family member 4 suppresses hepatocellular carcinoma cell proliferation by regulating PTEN in vitro and in vivo. Mol Med Rep. 19:685–692. [DOI] [PubMed] [Google Scholar]
- Thybert D, Roller M, Navarro FCP, Fiddes I, Streeter I, Feig C, Martin-Galvez D, Kolmogorov M, Janoušek V, Akanni W, et al. 2018. Repeat associated mechanisms of genome evolution and function revealed by the Mus caroli and Mus pahari genomes. Genome Res. 28:448–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo CM, Ding Y, Hoellerbauer P, Davis Ryan J, Basom R, Girard Emily J, Lee E, Corrin P, Hart T, Bolouri H, et al. 2015. Genome-wide CRISPR-Cas9 screens reveal loss of redundancy between PKMYT1 and WEE1 in glioblastoma stem-like cells. Cell Rep. 13:2425–2439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Grus WE, Zhang J. 2006. Gene losses during human origins. PLoS Biol. 4:e52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Moore BM, Panchy NL, Meng F, Lehti-Shiu MD, Shiu S-H. 2018. Factors influencing gene family size variation among related species in a plant family. Genome Biol Evol. 10:2596–2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Yu H, Hughes NW, Liu B, Kendirli A, Klein K, Chen WW, Lander ES, Sabatini DM. 2017. Gene essentiality profiling reveals gene networks and synthetic lethal interactions with oncogenic Ras. Cell 168:890–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Huang J, Wang W, Xu J, Yin M, Cheng N, Yin J. 2017. Long non-coding RNA Fer-1-like protein 4 acts as a tumor suppressor via miR-106a-5p and predicts good prognosis in hepatocellular carcinoma. Cancer Biomark. 20:55–65. [DOI] [PubMed] [Google Scholar]
- Xia T, Chen S, Jiang Z, Shao Y, Jiang X, Li P, Xiao B, Guo J. 2015. Long noncoding RNA FER1L4 suppresses cancer cell growth by acting as a competing endogenous RNA and regulating PTEN expression. Sci Rep. 5:13445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yilmaz A, Peretz M, Aharony A, Sagi I, Benvenisty N. 2018. Defining essential genes for human pluripotent stem cells by CRISPR–Cas9 screening in haploid cells. Nat Cell Biol. 20:610–619. [DOI] [PubMed] [Google Scholar]
- Young JM, Trask BJ. 2007. V2R gene families degenerated in primates, dog and cow, but expanded in opossum. Trends Genet. 23:212–215. [DOI] [PubMed] [Google Scholar]
- Yue B, Sun B, Liu C, Zhao S, Zhang D, Yu F, Yan D. 2015. Long non-coding RNA Fer-1-like protein 4 suppresses oncogenesis and exhibits prognostic value by associating with miR-106a-5p in colon cancer. Cancer Sci. 106:1323–1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang ZD, Frankish A, Hunt T, Harrow J, Gerstein M. 2010. Identification and analysis of unitary pseudogenes: historic and contemporary gene losses in humans and other primates. Genome Biol. 11:R26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Horvath S. 2005. A general framework for weighted gene co-expression network analysis. Stat Appl Genet Mol Biol. 4:17. [DOI] [PubMed] [Google Scholar]
- Zhao H, Shi J, Zhang Y, Xie A, Yu L, Zhang C, Lei J, Xu H, Leng Z, Li T, et al. 2020. LncTarD: a manually-curated database of experimentally-supported functional lncRNA–target regulations in human diseases. Nucleic Acids Res. 48:D118–D126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Tang L, Li Z, Jin J, Luo J, Gao G. 2015. Identification and analysis of unitary loss of long-established protein-coding genes in Poaceae shows evidences for biased gene loss and putatively functional transcription of relics. BMC Evol Biol. 15:66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Sanborn JZ, Diekhans M, Lowe CB, Pringle TH, Haussler D. 2007. Comparative genomics search for losses of long-established genes on the human lineage. PLoS Comput Biol. 3:e247. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.