Abstract
The minimal number of genes required for the formation of gas vesicles in halophilic archaea has been determined. Single genes of the 14 gvp genes present in the p-vac region on plasmid pHH1 of Halobacterium salinarum (p-gvpACNO and p-gvpDEFGHIJKLM) were deleted, and the remaining genes were tested for the formation of gas vesicles in Haloferax volcanii transformants. The deletion of six gvp genes (p-gvpCN, p-gvpDE, and p-gvpHI) still enabled the production of gas vesicles in H. volcanii. The gas vesicles formed in some of these gvp gene deletion transformants were altered in shape (ΔI, ΔC) or strength (ΔH) but still functioned as flotation devices. A minimal p-vac region (minvac) containing the eight remaining genes (gvpFGJKLM-gvpAO) was constructed and tested for gas vesicle formation in H. volcanii. The minvac transformants did not form gas vesicles; however, minvac/gvpJKLM double transformants contained gas vesicles seen as light refractile bodies by phase-contrast microscopy. Transcript analyses demonstrated that minvac transformants synthesized regular amounts of gvpA mRNA, but the transcripts derived from gvpFGJKLM were mainly short and encompassed only gvpFG(J), suggesting that the gvpJKLM genes were not sufficiently expressed. Since gvpAO and gvpFGJKLM are the only gvp genes present in minvac/JKLM transformants containing gas vesicles, these gvp genes represent the minimal set required for gas vesicle formation in halophilic archaea. Homologs of six of these gvp genes are found in Anabaena flos-aquae, and homologs of all eight minimal halobacterial gvp genes are present in Bacillus megaterium and in the genome of Streptomyces coelicolor.
Gas vesicles are formed by halophilic archaea, cyanobacteria, and some heterotrophic bacteria and allow these microorganisms to float at a favorable depth in their watery environment. These proteinaceous structures vary in length from 0.2 to 1.5 μm (for a review, see reference 39). The ribbed gas vesicle envelope exclusively consists of protein and appears to be watertight but is freely permeable to dissolved ambient gases. Electron micrographs indicate 4.6-nm-wide ribs arranged perpendicular to the long axis that are formed by a helix of low pitch and not by a stack of hoops (3, 30). The major constituent of the gas vesicle envelope is the hydrophobic 7- to 8-kDa protein GvpA (9, 38). Immunological studies revealed GvpC (20 to 42 kDa) as a second but minor protein constituent (8, 12, 15). In cyanobacteria, GvpC is located at the outer surface of the gas vesicle envelope and strengthens the entire structure (15, 22). In halophilic archaea, GvpC also appears to be responsible for a constant diameter of the cylindrical part of the gas vesicle (29).
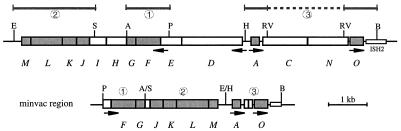
Genes encoding proteins involved in gas vesicle formation have been identified in a few species of cyanobacteria (Anabaena flos-aquae, Calothrix strain PCC 7601) and in halophilic archaea (Halobacterium salinarum, Haloferax mediterranei, and the haloalkaliphilic archaeon Natronobacterium vacuolatum) (6, 10, 14, 20, 21, 27), as well as in the soil bacterium Bacillus megaterium (26). In the case of halophilic archaea, 14 gvp genes cluster in an approximately 9-kb DNA region termed the vac region (10, 11, 19, 20). H. salinarum PHH1 harbors the two related vac regions p-vac (located on plasmid pHH1) and c-vac (located in the chromosome) (10, 18, 19). In both vac regions, the 14 gvp genes are identically arranged: gvpACNO form one cluster, and gvpDEFGHIJKLM are located upstream of gvpA and oriented in the opposite direction. Two promoters located in front of c-gvpA and c-gvpD drive the expression of these genes in the c-vac region, whereas four promoters could be identified in the p-vac region, resulting in p-gvpO and p-gvpFGHIJKLM transcripts in addition to the p-gvpA, p-gvpACNO, and p-gvpDE mRNAs (29). The expression of the p-vac region leads to predominantly spindle-shaped gas vesicles throughout growth, whereas the c-vac region is only expressed in p-vac deletion mutants, and then in the stationary growth phase only (9, 17). Fourteen gvp genes designated gvpAPQBRNFGLSKJTU are found in the gram-positive soil bacterium B. megaterium; the highly similar gvpA and gvpB genes of this gene cluster could encode the major gas vesicle structural protein GvpA (26). So far, the transcription of these gvp genes and gas vesicle formation have not been investigated; however, Escherichia coli transformants containing the gvpBRNFGLSKJTU genes of B. megaterium have been reported to form tiny gas vesicles (26).
The function of some of the halobacterial Gvp proteins has been determined by transformation experiments using halobacterial shuttle vectors conferring resistance to mevinolin (2, 25) or novobiocin (pMDS20) (16) and the gas vesicle-negative (Vac−) species Haloferax volcanii as the recipient strain. Also, an expression vector (pJAS35) is available which enables high-level expression of halobacterial reading frames under the control of the halobacterial ferredoxin (fdx) gene promoter (34). Such transformation experiments showed that (i) the entire p-vac region (construct M-O) leads to gas vesicle formation in H. volcanii (10), and (ii) the p-gvpACNO (A-O) and p-gvpDEFGHIJKLM (D-M) gene clusters present on different vector constructs (or A-O/F-M = ΔDE) allow the formation of spindle-shaped gas vesicles in H. volcanii transformants (28). The GvpD and GvpE proteins are involved in the regulation of gas vesicle formation: GvpE is a transcriptional activator required for gvpA promoter activity, whereas GvpD is involved in the repression of gas vesicle formation (11, 23, 24, 36).
Deletion studies have been carried out to determine the necessity of each gene found in the p-gvpACNO operon for gas vesicle formation (29). These experiments show that ΔA transformants (containing the entire p-vac region except for the gvpA gene) and ΔO transformants (p-vac region without gvpO) are Vac−, whereas ΔN (Vac+/−) and ΔC (Vac+) transformants produce gas vesicles. ΔC transformants contain many irregularly shaped gas vesicles with various diameters throughout a single gas vesicle. The finding that gas vesicles of ΔC+C transformants regain the wild-type shape implies that GvpC is involved in shape determination (29). A different approach has been employed by S. DasSarma's group, who inserted foreign DNA into various gvp genes present on endogenous plasmid pNRC100 and analyzed the effect in pNRC100-negative H. halobium mutant strain SD109 (7). This mutant still contains the c-vac region, although the cells appear to be devoid of gas vesicles. Since the two approaches revealed different results in 6 out of 14 gvp cases (including the data reported here), both methods will be discussed in more detail.
In this study, we examined the necessity of genes located in the p-gvpFGHIJKLM cluster for gas vesicle formation. Single gvp genes were deleted, and the expression of the remaining gvp genes was analyzed in H. volcanii transformants. ΔH and ΔI transformants still produced gas vesicles, whereas all other deletion variants were gas vesicle free. Together with the results obtained earlier (28, 29), these experiments show that 6 of the 14 gvp genes can be deleted without affecting the ability of the cell to form gas vesicles. We also investigated whether the eight genes gvpA, gvpO, and gvpFGJKLM represent the minimal number of gvp genes sufficient for gas vesicle formation in H. volcanii.
MATERIALS AND METHODS
Strains and growth conditions.
E. coli strains DH5α (13) and GM1674 (dam−) (31) harboring plasmid constructs were cultured at 37°C in LB broth (37) containing ampicillin at 100 μg/ml. H. volcanii WFD11 (lacking endogenous plasmid pHV2) (5) was grown in rich medium containing, per liter, 175 g of NaCl, 37 g of MgSO4 · 7H2O, 3.7 g of KCl, 5 g of Bacto Tryptone, 3 g of Bacto Yeast Extract, 25 ml of 1 M Tris/HCl (pH 7.2), 5 ml of 10% CaCl2 · 2H2O, and 100 μl of 100 μM MnCl2. Halobacterial transformants were selected on agar plates containing novobiocin at 0.2 μg/ml and/or mevinolin or lovastatin at 6 μg/ml. For inspection of the gas vesicle phenotype or isolation of total protein, transformants were grown in the medium described above, except that the NaCl concentration was raised to 206 g/liter to enhance gas vesicle formation. Lovastatin and mevinolin were a generous gift from Merck, Sharp and Dohme GmbH, Munich, Germany.
Constructs used for transformation of H. volcanii and transformation procedure.
The subfragments of the p-vac region used for the construction of plasmids are listed in Table 1. The protruding ends of DNA fragments E-O, F-O, and G-O were blunt ended by T4 polymerase and ligated to the blunt-ended BamHI site of halobacterial vector pWL102, whereas BglII fragment D-O was directly ligated to the BamHI site of pWL102 (25). The L1/2-O/pWL102 construct (Table 1) was used to yield the H-O, I-O, J-O, K-O, and L-O fragments. This construct was cut at the single NheI site located 36 bp upstream of the p-gvpH stop codon and at the single XbaI site located in the pWL102 sequence downstream of the truncated p-gvpL codon, resulting in a deletion of 36 bp of p-gvpH and all of p-gvpIJKL1/2. The remaining NheI/XbaI fragment, H9/10-O/pWL102 [Δgvp(H)IJKL1/2], was ligated to various fragments amplified by PCR. PCRs were performed with the p-vac region as the template and oligonucleotide CTCGAAATTCGGCTAGCACGAACA (positions 2746 to 2723 of the p-vac region, containing the NheI site [underlined] within p-gvpH) as primer 1 and the respective oligonucleotide containing an XbaI site (as listed in Table 1) as primer 2. The PCR products started at the NheI site in p-gvpH and contained 36 bp derived from the 3′ end of p-gvpH [= p-gvp(H)], p-gvp(H)I, p-gvp(H)IJ, p-gvp(H)IJK, or p-gvp(H)JKL. These fragments were cleaved with NheI/XbaI and ligated to NheI/XbaI fragment H9/10-O/pWL102, resulting in constructs H-O through L-O.
TABLE 1.
Constructs used in this study
| Construct | Restriction site(s) | Position(s) in p-vac sequencea | Size(s) (bp) | Second oligonucleotide used for PCRb or reference |
|---|---|---|---|---|
| D-O | BglII-BglII | 4633–9751 | 5,119 | |
| E-O | StyI-SspI | 4150–9909 | 5,760 | |
| F-O | NspI-SspI | 3494–9909 | 6,416 | |
| G-O | BstEII-SspI | 3122–9909 | 6,788 | |
| H-O | SspI | 2622–9909 | 7,288 | CCGGGCTTTCTAGAGGGTGATCTGCG |
| I-O | SspI | 2226–9909 | 7,684 | CCAGCATCTAGACGAGGTCGCCC |
| J-O | SspI | 1909–9909 | 8,001 | CGTCGTCTAGAGCTAGTTCCATC |
| K-O | SspI | 1505–9909 | 8,405 | CCACGCAGTCTAGATAGCGGCCG |
| L-O | SspI | 721–9909 | 9,189 | TCGCGTGTGTCTAGACTTTTGTTGGC |
| L1/2-O | XmnI-SspI | 1061–9909 | 8,849 | |
| M-O | MroI-SspI | 1–9909 | 9,909 | 28 |
| D-M | EcoRI-XcmI | 292–6515 | 6,224 | 28 |
| E-M | 364–4752 | 4,389 | ATCAGCCATGGAGAGTCGATCCCC | |
| F-M | 364–4158 | 3,795 | GTCAATCACCATGGCAAGGAAGAG | |
| G-M | 364–3529 | 3,166 | TTCGTGGACATTCCCATGGGCGCGGAACAG | |
| H-M | 364–3259 | 2,896 | CAGATGCGTGACCCCATGGAGG | |
| I-M | 364–2717 | 2,354 | ACATCGGCCATGGATGGAGG | |
| J-M | 364–2289 | 1,926 | GCGTCCGGGAGTCCCATGGGGACGAGGTGA | |
| K-M | 364–1940 | 1,577 | CCACGTCAGCCCATGGGACCAAATGAG | |
| L-M | 364–1623 | 1,260 | GACACCGCCATGGCACGGCTCGCC | |
| M-M | 364–775 | 412 | GCCGTGGCCATGGTACACGTTCGC | |
| ΔA | MroI-SspI, ΔXcmI-StuI | 1–9909, Δ6516–6744 | 9,909, Δ229 | 28 |
| ΔC | MroI-SspI, ΔScaI-KpnI | 1–9909, Δ6872–7716 | 9,909, Δ845 | 29 |
The p-vac sequence accession numbers in the EMBL database are X55648 (p-gvpD-M) and X64729 (p-gvpACNO). The positions refer to a combined sequence using an adenosine 486 nucleotides downstream of the p-gvpM stop codon (located within an MroI site) as arbitrary position 1.
Restriction sites are underlined (TCTAGA, XbaI; CCATGG, NcoI).
The p-vac subfragments E-M, F-M, G-M, H-M, I-M, J-M, K-M, and M-M (Table 1) were produced by PCR using oligonucleotide GTCTGGACGCTACCATGGCCTCTCGTTCCC (located downstream of p-gvpM at positions 351 to 380; the NcoI site is underlined) as primer 1 and the respective oligonucleotide complementary to different p-vac sequences as primer 2 (Table 1). Both primers contain an NcoI site, and the PCR products were ligated to pJAS35 cut with NcoI. The orientation of the DNA insert relative to the fdx promoter was determined by KpnI cleavage. The distance between the NcoI site and the start codon of the first gvp reading frame varied between 14 and 43 nucleotides, leading to short mRNA leader sequences. The expression of functional gvp gene products (especially in the case of the first gvp gene) was proven by the formation of gas vesicles in the respective control transformants.
The minimal p-vac region (minvac) was constructed in the following way. SmaI/Asp718-digested pUC18 was ligated with a PshAI/Asp718 DNA fragment (fragment 1 [see Fig. 4]) containing p-gvpFG (= FG × pUC). This construct was linearized by Asp718, blunt ended, recut with EcoRI, and ligated to an ScaI/EcoRI fragment (fragment 2 [see Fig. 4]) harboring p-gvpJKLM, resulting in construct FG-JKLM × pUC. A second construct containing the HindIII/BglII p-gvpACNO fragment (fragment 3 [see Fig. 4]) cloned in the HindIII/BamHI sites of pUC18 was cleaved with EcoRV to delete major portions of the p-gvpCN genes (= AO × pUC). Linearized AO × pUC was religated and linearized again at the HindIII site located upstream of p-gvpA (see Fig. 4). The HindIII site was blunt ended and used to insert the blunt-ended HindIII/EcoRI fragment containing p-gvpFGJKLM of FGJKLM × pUC. The resulting plasmid (= minvac × pUC) was analyzed by restriction digestion for the orientation of the p-gvp genes, which was determined as gvpFGJKLM-gvpAO. The minvac × pUC construct was cleaved with PvuII to yield the insert, which was then cloned in the BamHI-cleaved, blunt-ended pWL102 vector. The construction of the M-O, D-M, A-O, and ΔA fragments was described by Offner and Pfeifer (28), and the construction of ΔC, ACO, and ACN was described by Offner et al. (29). Prior to the transformation of H. volcanii, each construct was passaged through E. coli dam mutant strain GM1674 to avoid a halobacterial restriction barrier (16). Transformation was performed as described earlier (33). The presence of the desired plasmids in H. volcanii transformants was determined by Southern analyses of total DNA using a p-vac-region-specific, digoxigenin (DIG)-labeled DNA probe (DIG labeling kit of Boehringer Mannheim).
FIG. 4.
Genetic map of the p-vac region and strategy for the construction of the minvac region. The 14 gvp genes of the p-vac region are represented by boxes labeled A and C through O. Essential gvp genes are represented by grey boxes, and nonessential genes are represented by white boxes. Arrows indicate the locations of the endogenous promoters. Restriction sites used to obtain p-vac subfragments 1 to 3, used for the construction of the minvac plasmid, are shown at the top as follows: A, Asp718; B, BglII; E, EcoRI; H, HindIII; P, PshAI; RV, EcoRV; S, ScaI. The respective subfragments are shown as bars. For further explanations, see Materials and Methods. The minvac construct at the bottom acquired deletions of gvpDE, gvpHI, and gvpCN. ISH2, halobacterial insertion element.
RNA isolation and Northern analyses.
Total RNA was isolated as described by Chomczynski and Sacchi (4). For Northern analyses, 10 μg of each RNA was electrophoretically separated on denaturing, formaldehyde-containing 1.2% (wt/vol) agarose gels (1). Strand-specific RNA probes were synthesized using the following fragments cloned in pBluescript as the template: the 515-bp XhoI-Asp718 fragment containing the 3′ part of p-gvpF and p-gvpG (probe FG; Fig. 1), the 1,258-bp NcoI p-gvpLM fragment (probe LM), and the 476-bp HindIII-ScaI fragment containing p-gvpA (probe A). The RNA probes were synthesized using the DIG RNA labeling kit obtained from Boehringer Mannheim.
FIG. 1.
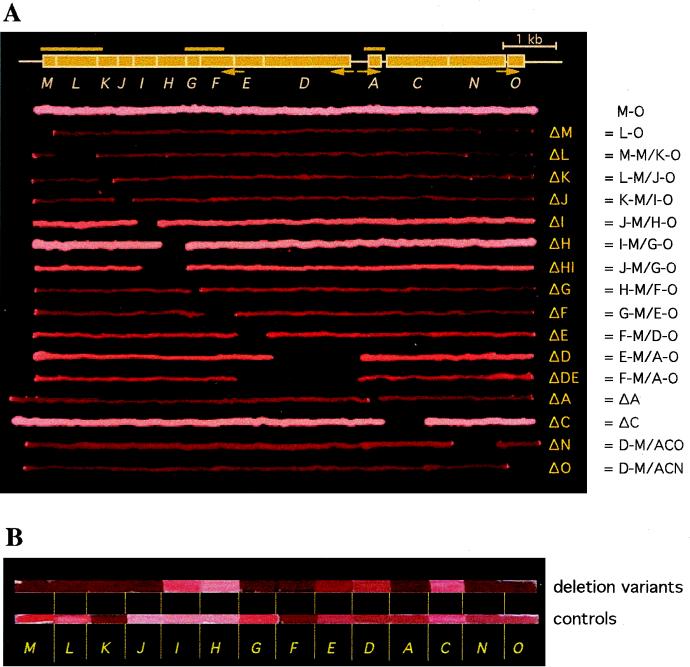
The p-vac region and phenotypes of various H. volcanii transformants. (A) The 14 gvp genes constituting the p-vac region are shown as boxes (A and C through O). The four endogenous p-vac promoters are indicated by arrows underneath. The bars above represent the A, FG, and LM probes used for Northern analyses. The lines underneath represent colonies grown according to the gvp fragment(s) present in the cells. The M-O (entire p-vac region), L-O (ΔM), ΔA, and ΔC transformants harbor the p-vac region on a single construct (plus pJAS35 as a second construct); all other transformants contain the p-vac region on two constructs. Vac+ colonies appear turbid and white; orange, turbid colonies possess fewer gas vesicles, and dark red translucent colonies are Vac−. The Δ designations to the right of the plate indicate the deleted gvp genes. The constructs used for transformation are shown further to the right. The construction of transformants M-O, ΔA, ΔC, ΔN, and ΔO has been described previously (28, 29). (B) Various gene deletion transformants (top) in comparison to the respective control transformants (bottom). The colonies are arranged according to the respective gene deletion given underneath (i.e., M = ΔM transformant on top and cΔM control transformant underneath).
Isolation of gas vesicles and electron microscopy.
H. volcanii transformants were grown on agar plates for 1 to 2 weeks. Cells were scraped from the agar and lysed in 10 mM Tris/HCl (pH 7.2) with 1 to 2 μl of DNase I (1 mg/ml). Gas vesicles were collected in small, narrow tubes (diameter, 4 mm) by centrifugation in an Eppendorf centrifuge for 20 min at 1,000 to 2,000 rpm to improve flotation and washed three times with 10 mM Tris/HCl (pH 7.2). For negative staining, a drop of the gas vesicle preparation was placed onto a carbon-coated copper grid and removed after 2 min with a pipette, and the grid was air dried. Gas vesicles were treated for 1 min with a solution of 1% uranyl acetate and 0.01% glucose in water, briefly rinsed with a drop of water, and then air dried. The specimens were examined with a ZEISS EM 912 transmission electron microscope operated with the OMEGA energy filter in the zero-loss mode.
RESULTS
Deletion of single gvp genes and effect on gas vesicle formation in H. volcanii transformants.
The deletion of a specific gvp gene in the p-gvpDEFGHIJKLM cluster was achieved by the complementation of two subfragments present on different vector plasmids, except for the deletion of gvpM. The ΔM transformant harbored a single p-vac construct (L-O) without the gvpM gene together with the “empty” pJAS35 expression vector. Each construct produced was designated according to the gvp gene located near the boundary of the subfragment (Table 1; Fig. 1A). Four of these fragments (D-O, E-O, F-O, and G-O) were obtained by restriction endonuclease digestions, whereas all other subfragments were, at least in part, amplified by PCR. Subfragments D-O through L-O were inserted into pWL102, and the various gvp genes were expressed under the control of the endogenous promoters. In contrast, constructs E-M through M-M contained gvp reading frames inserted into pJAS35, where they are expressed under ferredoxin (fdx) promoter control (Table 1).
Various combinations of two of these constructs were used to transform H. volcanii. Figure 1A shows the series of transformants as colony streaks according to the p-vac subfragment(s) present in the cells. For completeness, all transformants with gvp gene deletions are included; the deletion of genes found within the p-gvpACNO cluster has already been described (29). Gas vesicle-producing (Vac+) colonies appear pink-white and turbid, whereas Vac− colonies are red and translucent. Gas vesicles seen by phase-contrast microscopy appear as light refractile bodies inside the cells. M-O transformants contain the entire p-vac region and showed the expected pink-white and turbid Vac+ phenotype (28). In these transformants, gas vesicle formation became visible 2 days after colony formation. The ΔC (29) and ΔH transformants were also Vac+; the latter one started to produce gas vesicles significantly earlier than ΔC and M-O. The ΔI, ΔHI, ΔE, ΔD, and ΔDE transformants formed turbid colonies, but the amount of gas vesicles (also seen as light refractile bodies in the cells) was lower compared to the M-O wild type (Fig. 1A and data not shown). Minor amounts of gas vesicles were seen in ΔN transformants when the cells were inspected by phase-contrast microscopy (29). The lack of any of the gvpM, gvpL, gvpK, gvpJ, gvpG, gvpF, gvpA, or gvpO genes yielded red translucent colonies (Fig. 1A), and no light refractile bodies were detected in any of the cells inspected by phase-contrast microscopy. The results implied that these eight genes are essential for gas vesicle formation whereas the six genes gvpDE, gvpHI, and gvpCN are not required.
To demonstrate that each of the constructs was expressed and produced functional Gvp proteins, the respective control transformants were prepared. Each control transformant contained the gvp gene missing in the gene deletion transformant as the first reading frame in the subfragment cloned in pJAS35. Figure 1B shows the colony phenotype of the gene deletion transformants in comparison to that of the respective control transformants. One would expect each control transformant to express a Vac+ phenotype similar to that of the wild type; however, the phenotype indicated some variations. A wild-type Vac+ phenotype (pink-white colonies and many light refractile bodies inside the cells) was observed with the cΔC, cΔG, cΔH, cΔI, cΔL, and cΔM transformants, while cΔJ indicated an overproducer phenotype. The cΔA, cΔD, cΔE, cΔN, and cΔO transformants contained somewhat fewer gas vesicles (Fig. 1B, bottom line), and only minor amounts were observed in cΔF and cΔK, as verified by phase-contrast microscopy. The ΔI and cΔJ transformants contained extremely long gas vesicles (see below). These results demonstrated that the gvp genes present in these control transformants were expressed and that the gene products were functional in gas vesicle formation.
Northern analyses to investigate transcription from the gvpF-M cluster.
To analyze the gvp mRNA levels in the transformants, total RNA was isolated and used for Northern analyses. H. volcanii transformants with the wild-type p-vac region produce the 4-kb p-gvpF-M mRNA during exponential growth (29). Since parts of the p-gvpFGHIJKLM gene cluster were present on different vector constructs in the other transformants, the mRNAs derived from this region were investigated in more detail.
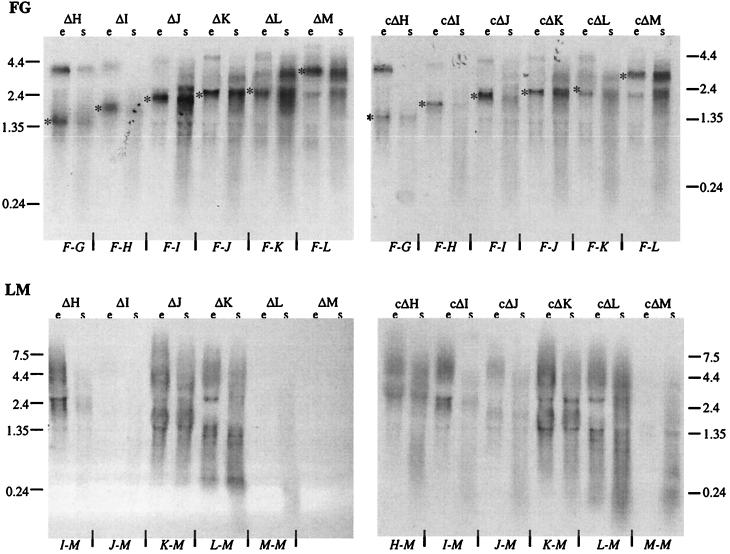
The FG probe (Fig. 1) was used to monitor mRNAs starting at the pF promoter present in the pWL102 constructs, whereas the LM probe was used to determine transcripts derived from the respective pJAS35 constructs in each transformant. Using the FG probe for Northern analyses, mRNAs of the expected length were detected in all gene deletion and control transformants (Fig. 2). The LM probe detected transcripts starting at the fdx promoter in the pJAS35 construct. This promoter usually leads to a large amount of mRNA during the exponential growth phase and a smaller amount during the stationary growth phase (34). Using the LM probe, such a transcript pattern was seen in transformants harboring the H-M, I-M, K-M, and L-M constructs whereas the amount of mRNA was smaller in transformants containing the J-M construct (ΔI and cΔJ) (Fig. 2). This was surprising, since the cΔJ transformant showed overproduction of gas vesicles. The M-M construct was expressed predominantly during the stationary growth phase in both cases (ΔL and cΔM) (Fig. 2). Transformants containing the M-M construct by itself showed the same pattern of gvpM mRNA production throughout growth (data not shown). In summary, these analyses demonstrated that each fragment was transcribed and that the observed variations in transcription did not reflect the different gas vesicle phenotypes observed.
FIG. 2.
Northern analyses of gene deletion and control transformants to investigate the various p-gvpF-M transcripts. RNA samples were derived from the exponential (e) and stationary (s) growth phases. A 10-μg sample of total RNA was applied to each slot. Transcripts starting at the pF promoter were visualized using the FG probe (top), whereas the LM probe was used to detect transcripts starting at the fdx promoter in pJSA35 (bottom). The construct responsible for mRNA hybridization is indicated at the bottom of each gel. For the FG probe, the expected mRNA is indicated by an asterisk. The values on the side of each gel are the sizes (in kilobases) of the RNA markers.
Inspection of the gas vesicles formed by transformants ΔH and ΔI and the respective control transformants.
The cells of Vac+ transformants ΔH and ΔI were inspected by phase-contrast microscopy. ΔH transformants contained light refractile bodies throughout growth. ΔI transformants indicated large, cylinder-shaped gas vesicles spanning the entire length of the cell and sometimes even altering the cell shape by pushing out lobes. A similar phenotype was observed with the cΔJ transformants.
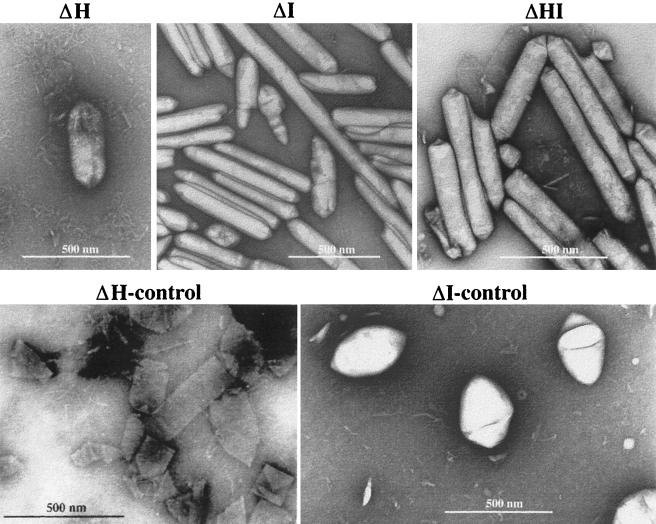
Gas vesicles were isolated from ΔH and ΔI (and ΔHI) transformants and also from the respective control transformants and investigated by electron microscopy (Fig. 3). Gas vesicles of the ΔI transformant were extremely long and cylinder shaped, with an average length of more than 0.6 μm (Fig. 3). Some even measured 2.7 μm in length and were thus longer than the average H. volcanii cell. The appearance of these extremely long gas vesicles explained the shape alterations of ΔI transformant cells observed in the phase-contrast microscope. The control transformant cΔI, however, contained spindle-shaped gas vesicles, as found in wild-type H. salinarum (Fig. 3). Many gas vesicles were obtained by flotation from the ΔH transformant; however, intact gas vesicles were rarely found after staining for electron microscopy (Fig. 3). They disintegrated into single ribs, demonstrating that the gas vesicles synthesized without GvpH were unstable during the staining procedure. Gas vesicles isolated from the control transformant cΔH were stable but appeared cylinder and not spindle shaped (Fig. 3). Gas vesicles isolated from the double deletion transformant ΔHI were stable and cylinder shaped.
FIG. 3.
Electron micrographs of isolated gas vesicles of ΔH, ΔI, ΔHI, and the respective control transformants.
Construction of the minvac region containing eight gvp genes.
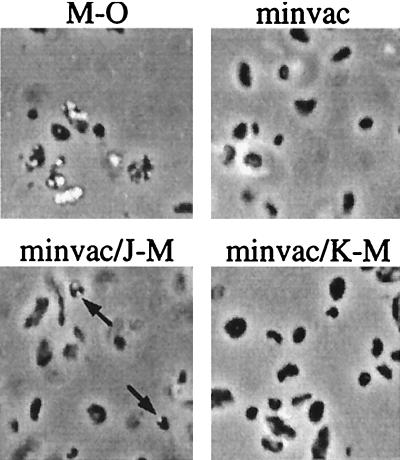
The results of the transformation experiments implied that the genes p-gvpFGJKLM and p-gvpAO constitute the minimal number of genes required for gas vesicle formation. In order to prove that these genes are really sufficient, a plasmid containing these eight gvp genes was constructed: the gvpDE and gvpHI genes were deleted from the p-gvpDEFGHIJKLM cluster, as were the gvpCN genes from the p-gvpACNO gene cluster. The resulting minimal p-vac (minvac) region contained the remaining gvp genes arranged consecutively under the control of the endogenous promoters pA, pO, and pF (Fig. 4). The H. volcanii transformants containing the minvac plasmid did not contain light refractile bodies and were Vac−, suggesting that the six gvp genes that were lacking (gvpCN, gvpDE, and gvpHI) could not be deleted all at once and that the remaining eight gvp genes were not sufficient for gas vesicle formation. To identify the missing gene(s) required for gas vesicle formation, double transformants were produced containing the minvac construct together with a second plasmid harboring an additional fragment of the p-vac region (A-O or E-M). The minvac/A-O transformants were Vac−, whereas the minvac/E-M transformants were Vac+, suggesting that the p-gvpHI genes of the p-gvpFGHIJKLM unit might be lacking. Double transformants containing minvac plus further deletions in E-M (H-M, I-M, J-M, or K-M) were produced, and the cells were inspected for gas vesicle formation. Light refractile bodies were found in the minvac/H-M and minvac/I-M transformants and even in the minvac/J-M transformants, although the gvp genes present on construct J-M were already present on the minvac plasmid (Fig. 5 and data not shown). The minvac/K-M transformants did not reveal light refractile bodies and were thus Vac−. These results suggested that the gvpHI genes are not required for gas vesicle formation and that the gvpJKLM genes present on the minvac construct (especially gvpJ) were not sufficiently expressed.
FIG. 5.
Phase-contrast microscopy of various H. volcanii transformants. Gas vesicles are visible as white bodies inside the cells. The constructs present in the cells are shown above the images. The M-O transformant contains the entire p-vac region and produces many light refractile bodies, whereas the minvac and minvac/K-M transformants are gas vesicle free. Gas vesicles found in minvac/J-M cells are indicated by arrows. Magnification, ×1,050.
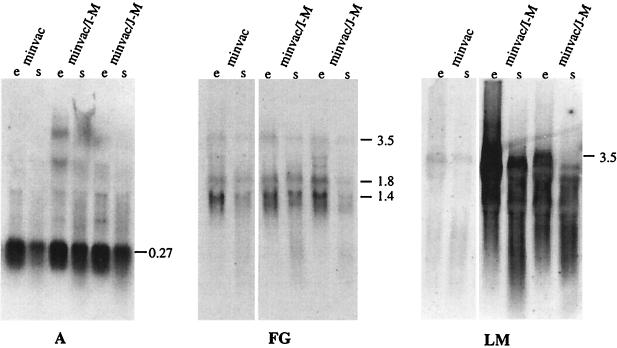
Northern analyses were performed to investigate the amount of gvp transcripts using the A, FG, and LM probes. The A probe detected a large amount of 0.27-kb gvpA mRNA in each transformant (Fig. 6). The FG probe used to detect the gvpFGJKLM mRNA indicated a small amount of a 3.5-kb transcript that could span the entire gvpFGJKLM region (Fig. 4). In addition to this transcript, larger amounts of 1.8- and 1.4-kb mRNAs were also detected, suggesting early termination or degradation of the 3.5-kb transcript (Fig. 6). The LM probe indicated the small amount of the 3.5-kb mRNA in minvac transformants. The double transformant minvac/I-M or minvac/J-M contained a large amount of I-M or J-M mRNA due to the expression of these genes under fdx promoter control in pJAS35 (Fig. 6). Thus, the lack of gas vesicles in minvac transformants might be due to the early termination (or processing) of the gvpFGJKLM mRNA.
FIG. 6.
Northern analyses of minvac and minvac double transformants to detect gvpA and gvpF-M mRNA. RNA samples were derived from the exponential (e) and stationary (s) growth phases. A 10-μg sample of total RNA was applied to each slot. Transcripts starting at the pA promoter were visualized using the A probe (A), and transcripts derived from the pF promoter were visualized using an FG probe (FG), whereas the LM probe also detected transcripts starting at the fdx promoter in pJSA35 in the cases of the I-M and J-M constructs (LM). The values on the right side of each gel are the sizes (in kilobases) of the hybridizing mRNAs.
DISCUSSION
The results presented here suggest that 8 out of 14 gvp genes found in the vac regions of halophilic archaea are sufficient for gas vesicle formation: gvpA (encoding the major gas vesicle structural protein); gvpO, whose function is unknown; and gvpFGJKLM. In contrast to the deletion analysis described here, DasSarma's group reported that gas vesicle formation in H. salinarum (formerly H. halobium) requires at least 10 out of 13 gvp genes (gvpO was not recognized at that time; reference 7). That group inserted foreign DNA into various gvp genes derived from pNRC100 and analyzed the effect on H. salinarum mutant strain SD109 lacking the pNRC100-encoded vac region. One reason for the discrepancies in 6 out of 14 cases could be the use of different recipient strains for the transformation experiments. We chose H. volcanii as the recipient because this strain is easy to transform, grows faster than H. salinarum, and—most importantly—offers a clean genetic background for the functional analysis of gvp genes. The expression of various gvp genes in this recipient strain appears to be similar to that found for H. salinarum or H. mediterranei. H. salinarum strains harboring plasmid deletion variants lacking the p-vac region (such as H. salinarum PHH4 or SD109; reference 7) are deficient in the plasmid vac region but still contain the c-vac region.
Another difference is the mutation method used for the analyses. Insertion of foreign DNA into a gvp gene can cause a polar mutation affecting not only the expression of the gvp gene investigated but also that of gvp genes located farther downstream, especially when they are cotranscribed. Without the determination of the expression of these gvp genes, the results obtained are not sufficient to define the function of a single gvp gene. Another problem is encountered when the integration site is close to the 5′ or 3′ terminus of the gvp gene and the insert does not destroy the gene function at the protein level. In all such cases, the presence or absence of the respective gvp gene products (mRNAs or proteins) should be determined; however, this has not been done (7). In contrast, deletion of a single gvp gene within the vac region clearly destroys its function and if the control transformant containing the respective gvp gene produces gas vesicles, the phenotype of the gvpΔX transformants provides a good indication of the effect of the mutation. The deletion of the gvp gene of interest is, however, often achieved by the complementation of gvp genes present on two vector plasmids. In such cases, gvp genes are expressed either by their endogenous promoter(s) or under ferredoxin (fdx) promoter control (28, 29, and this report). In such double transformants, gas vesicle synthesis could be affected by an imbalance of Gvp proteins due to different plasmid copy numbers or the higher fdx promoter activity compared to the normally regulated gvp gene expression. However, gvp gene function can still be determined by this method, especially when the respective control transformant indicates normal gas vesicle formation. The differences obtained with mutations in the gvpACNO gene cluster have already been discussed (29, 35).
The analyses of the gvpFGHIJKLM genes presented in this report indicated that deletion of the gene gvpF, gvpJ, gvpK, or gvpL in the p-vac region resulted in Vac− transformants, whereas the respective control transformants produced gas vesicles. Similar results have been obtained by insertional mutation of these genes (7). Contradictory results were observed with the gvpG, gvpH, gvpI, and gvpM genes. Deletion of p-gvpG or p-gvpM (and also mc-gvpM; reference 11) resulted in Vac− transformants, whereas gas vesicle production was still observed when each of these gvp genes was mutated by a respective insertion (7). However, the integration site of the insertion in the gvpM gene is very close to the 3′ end and the absence of GvpM has not been checked in these transformants. An insert in gvpH results in transformants containing minor amounts of gas vesicles (7), but the ΔH transformants described in this report produced large amounts of gas vesicles that could be isolated by flotation but were unstable in electron microscopy. These results imply that the presence of GvpH is important for the formation of stable gas vesicles. Transformants carrying an insert in the gvpI gene are Vac− (7), whereas the ΔI transformants described here contained extremely long, cylinder-shaped gas vesicles, and the respective ΔI/I control transformant harbored large numbers of spindle-shaped gas vesicles. All of these discrepancies indicate that careful analyses are required to unequivocally determine a gvp gene function.
Our deletion analyses suggested that the p-gvpC, p-gvpDE, p-gvpHI, and p-gvpN genes are not essential for gas vesicle formation and that the remaining eight genes (gvpAO and gvpFGJKLM) consequently represent the minimal number of genes required for their assembly. In order to test whether these eight genes are indeed sufficient, the minvac construct containing these genes was used to transform H. volcanii. The minvac construct was initially inadequate for the formation of gas vesicles; however, the addition of extra copies of the gvpJKLM genes resulted in transformants containing gas vesicles whereas extra copies of gvpKLM revealed Vac− transformants, suggesting that the lack of gas vesicles in minvac transformants is mainly caused by the lack of sufficient GvpJ protein. Northern analyses demonstrated that the J-M genes on the original minvac construct were insufficiently expressed due to early termination of transcription. Nevertheless, since only the gvpAO and gvpFGJKLM genes were present in the minvac/JKLM transformants, these eight gvp genes represent the minimal number of genes required for gas vesicle formation. Thus, it is possible to delete six gvp genes simultaneously without losing the ability to synthesize gas vesicles. Similar experiments have not been done for the system used by DasSarma et al. (7).
The essential gene products determined in this study involve all of the small Gvp proteins with molecular masses of 8 to 12.7 kDa (GvpA, GvpG, GvpJ, GvpK, and GvpM) that also exhibit hydrophobic stretches in their amino acid sequences (10, 28). Surprisingly, GvpJ and GvpM show more than 60% sequence similarity to the GvpA protein, suggesting that they are structural components of the gas vesicle (20, 32). However, the amino acid composition of isolated gas vesicles exclusively reflects that of the GvpA protein and does not indicate a recognizable proportion of other proteins (9); thus, GvpM and GvpJ may only be required in early stages of gas vesicle assembly, being replaced by GvpA later on. Neither a GvpA-specific antiserum (12) nor an anti-gas vesicle serum (8) reacts with other Gvp proteins in a gas vesicle preparation. For some of the products encoded by the gvpFGHIJKLM gene cluster, chaperone functions have been suggested; they might keep the highly hydrophobic GvpA protein in a conformation that is required for the assembly process or even comprise an incorporation mechanism.
Comparison of the minvac region to gvp gene clusters found in bacteria.
With the recent finding of gas vesicle genes in B. megaterium (26) and also in Streptomyces coelicolor (cosmid 1E6; EMBL database) it is interesting to compare these eight essential gvp genes of halophilic archaea with gas vesicle genes identified in bacteria. In the cyanobacterium A. flos-aquae, six homologs to essential archaeal gvp genes have been identified, including multiple copies of gvpA and the genes gvpC, gvpN, and gvpJKL (22). According to our studies, homologs to gvpFG, gvpM, and gvpO are still lacking (Table 2). Since neither transcript analyses nor transformation experiments have been done, it is not clear whether the gvp gene cluster of Anabaena is complete.
TABLE 2.
Comparison of gvp loci identified in archaea and bacteria
The gram-positive bacterium B. megaterium contains a gvp gene cluster consisting of 14 genes (gvpAPQ-gvpBRNFGLSKJTU) (26). This gvp gene cluster has been used to transform E. coli, leading to the formation of tiny gas vesicles (average length, 40 nm). The first three genes (gvpAPQ) of this gvp cluster can be deleted without disturbing the ability of E. coli transformants to synthesize gas vesicles (26). The product of the gvpB gene presumably constitutes the major gas vesicle structural protein. The product of the second gene of this cluster, GvpR, exhibits 44% similarity to the GvpO protein assigned to S. coelicolor and 39% similarity to the GvpO protein of halophilic archaea. Thus, GvpR might be a more distant homolog of GvpO (Table 2). The gvpNFGLKJ genes are homologs to essential genes determined in this study. The gvpS gene product shows similarity to GvpA and GvpJ and could be a more distant homolog of the GvpM protein of halophilic archaea. A phylogenetic tree constructed with the sequences of GvpA, GvpJ, and GvpS indicates that all three sequences cluster together (data not shown). The gvpTU genes located at the end of this gene cluster have no archaeal homolog. Thus, among the 14 gvp genes found in B. megaterium, all eight essential gvp genes determined for halophilic archaea are present (Table 2).
During the genome sequencing project of the gram-positive bacterium S. coelicolor, eight gvp genes were found in cosmid 1E6; these genes are arranged as a gvpOAFGxxJLSK cluster (with xx representing two hypothetical protein genes; EMBL gene sequence data bank). The gvpS gene product is highly similar to the GvpS protein of B. megaterium and most likely a more distant homolog of GvpM. The arrangement of certain gvp genes (i.e., gvpLSK) is the same in both gram-positive bacteria but differs from the arrangement of the homologous genes (gvpKLM) in halophilic archaea. Strikingly, the eight gvp genes found in S. coelicolor exactly match the gvp genes in minvac that are required for gas vesicle formation in H. salinarum (Table 2). The expression of gvp genes in the gram-positive bacteria has not been investigated. It might be interesting to see whether, where, and when S. coelicolor produces gas vesicles and what functions these flotation devices might have in an organism that forms mycelia and does not usually exist in an aquatic environment. The possession of genes encoding gas vesicles is obviously more widely distributed than currently thought, and they occur in archaea as well as in gram-positive and gram-negative bacteria.
ACKNOWLEDGMENTS
This work was supported by grant Pf 165/6-3 obtained from the Deutsche Forschungsgemeinschaft.
We thank Christa Schleper, Jobst Gmeiner, and Kathryn Nixdorff for critical reading of the manuscript. The technical assistance of Ilka Dürr with electron miroscopy of gas vesicles is gratefully acknowledged. Lovastatin and mevinolin were a generous gift of Merck, Sharp and Dohme GmbH, Munich, Germany.
REFERENCES
- 1.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. Vol. 1. New York, N.Y: Greene Publishing Associates and Wiley-Interscience; 1988. [Google Scholar]
- 2.Blaseio U, Pfeifer F. Transformation of Halobacterium halobium: development of vectors and investigation of gas vesicle synthesis. Proc Natl Acad Sci USA. 1990;87:6772–6776. doi: 10.1073/pnas.87.17.6772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blaurock A E, Walsby A E. Crystalline structure of the gas vesicle wall from Anabaena flos-aquae. J Mol Biol. 1976;105:183–199. doi: 10.1016/0022-2836(76)90106-6. [DOI] [PubMed] [Google Scholar]
- 4.Chomczynski P, Sacchi N. Single step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 5.Cline S W, Schalkwyk L, Doolittle W F. Transformation of the archaebacterium Halobacterium volcanii with genomic DNA. J Bacteriol. 1989;171:4987–4991. doi: 10.1128/jb.171.9.4987-4991.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Damerval T, Houmard J, Guglielmi G, Csiszar K, Tandeau de Marsac N. A developmentally regulated gvpABC operon is involved in the formation of gas vesicles in the cyanobacterium Calothrix 7601. Gene. 1987;54:83–92. doi: 10.1016/0378-1119(87)90350-7. [DOI] [PubMed] [Google Scholar]
- 7.DasSarma S, Arora P, Lin F, Molinari E, Yin L. Wild-type gas vesicle formation requires at least ten genes in the gvp gene cluster of Halobacterium halobium plasmid pNRC100. J Bacteriol. 1994;176:7646–7652. doi: 10.1128/jb.176.24.7646-7652.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Englert C, Pfeifer F. Analysis of gas-vesicle gene expression in Haloferax mediterranei reveals that GvpA and GvpC are both gas-vesicle structural proteins. J Biol Chem. 1993;268:9329–9336. [PubMed] [Google Scholar]
- 9.Englert C, Horne M, Pfeifer F. Expression of the major gas vesicle protein in the halophilic archaebacterium Haloferax mediterranei is modulated by salt. Mol Gen Genet. 1990;222:225–232. doi: 10.1007/BF00633822. [DOI] [PubMed] [Google Scholar]
- 10.Englert C, Krüger K, Offner S, Pfeifer F. Three different but related gene clusters encoding gas vesicles in halophilic archaea. J Mol Biol. 1992;227:586–592. doi: 10.1016/0022-2836(92)90914-6. [DOI] [PubMed] [Google Scholar]
- 11.Englert C, Wanner G, Pfeifer F. Functional analysis of the gas-vesicle gene cluster of the halophilic archaeon Haloferax mediterranei defines the vac-region boundary and suggests a regulatory role for the gvpD gene or its product. Mol Microbiol. 1992;6:3543–3550. doi: 10.1111/j.1365-2958.1992.tb01789.x. [DOI] [PubMed] [Google Scholar]
- 12.Halladay J T, Jones J G, Lin F, MacDonald A B, DasSarma S. The rightward gas vesicle operon in Halobacterium plasmid pNRC100: identification of the gvpA and gvpC gene products by use of antibody probes and genetic analysis of the region downstream of gvpC. J Bacteriol. 1993;175:684–692. doi: 10.1128/jb.175.3.684-692.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 14.Hayes P K, Powell R S. The gvpA/C cluster of Anabaena flos-aquae has multiple copies of a gene encoding GvpA. Arch Microbiol. 1995;164:50–57. doi: 10.1007/BF02568734. [DOI] [PubMed] [Google Scholar]
- 15.Hayes P K, Buchholz B, Walsby A E. Gas vesicles are strengthened by the outer-surface protein, GvpC. Arch Microbiol. 1992;157:229–234. doi: 10.1007/BF00245155. [DOI] [PubMed] [Google Scholar]
- 16.Holmes M L, Nuttall S D, Dyall-Smith M. Construction and use of halobacterial shuttle vectors and further studies on Haloferax DNA gyrase. J Bacteriol. 1991;12:3807–3813. doi: 10.1128/jb.173.12.3807-3813.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Horne M, Pfeifer F. Expression of two gas vacuole protein genes in Halobacterium halobium. Mol Gen Genet. 1989;218:437–444. doi: 10.1007/BF00332407. [DOI] [PubMed] [Google Scholar]
- 18.Horne M, Englert C, Pfeifer F. Two genes encoding gas vacuole proteins in Halobacterium halobium. Mol Gen Genet. 1988;213:459–464. doi: 10.1007/BF00339616. [DOI] [PubMed] [Google Scholar]
- 19.Horne M, Englert C, Wimmer C, Pfeifer F. A DNA region of 9 kbp contains all genes necessary for gas vesicle synthesis in halophilic archaebacteria. Mol Microbiol. 1991;5:1159–1174. doi: 10.1111/j.1365-2958.1991.tb01889.x. [DOI] [PubMed] [Google Scholar]
- 20.Jones J G, Young D C, DasSarma S. Structure and organization of the gas-vesicle gene cluster on the Halobacterium halobium plasmid pNRC100. Gene. 1991;102:117–122. doi: 10.1016/0378-1119(91)90549-q. [DOI] [PubMed] [Google Scholar]
- 21.Kinsman R, Hayes P K. Genes encoding proteins homologous to halobacterial Gvps N, J, K, F and L are located downstream of gvpC in the cyanobacterium Anabaena flos-aquae. DNA Sequence. 1997;7:97–106. doi: 10.3109/10425179709020156. [DOI] [PubMed] [Google Scholar]
- 22.Kinsman R, Walsby A E, Hayes P K. GvpCs with reduced numbers of repeating sequence elements bind to and strengthen cyanobacterial gas vesicles. Mol Microbiol. 1995;17:147–154. doi: 10.1111/j.1365-2958.1995.mmi_17010147.x. [DOI] [PubMed] [Google Scholar]
- 23.Krüger K, Pfeifer F. Transcript analysis of the c-vac region and differential synthesis of the two regulatory gas vesicle proteins GvpD and GvpE in Halobacterium salinarium PHH4. J Bacteriol. 1996;178:4012–4019. doi: 10.1128/jb.178.14.4012-4019.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krüger K, Hermann T, Armbruster V, Pfeifer F. The transcriptional activator GvpE for the halobacterial gas vesicle genes resembles a basic region leucine-zipper regulatory protein. J Mol Biol. 1998;279:761–771. doi: 10.1006/jmbi.1998.1795. [DOI] [PubMed] [Google Scholar]
- 25.Lam W L, Doolittle W F. Shuttle vectors for the archaebacterium Halobacterium volcanii. Proc Natl Acad Sci USA. 1989;86:5478–5482. doi: 10.1073/pnas.86.14.5478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li N, Cannon M. Gas vesicle genes identified in Bacillus megaterium and functional expression in Escherichia coli. J Bacteriol. 1998;180:2450–2458. doi: 10.1128/jb.180.9.2450-2458.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mayr A, Pfeifer F. The characterization of the nv-gvpACNOFGH gene cluster involved in gas vesicle formation in Natronobacterium vacuolatum. Arch Microbiol. 1997;168:24–32. doi: 10.1007/s002030050465. [DOI] [PubMed] [Google Scholar]
- 28.Offner S, Pfeifer F. Complementation studies with the gas vesicle-encoding p-vac region of Halobacterium salinarium PHH1 reveal a regulatory role for the p-gvpDE genes. Mol Microbiol. 1995;16:9–19. doi: 10.1111/j.1365-2958.1995.tb02387.x. [DOI] [PubMed] [Google Scholar]
- 29.Offner S, Wanner G, Pfeifer F. Functional studies of the gvpACNO operon of Halobacterium salinarium reveal that the GvpC protein shapes gas vesicles. J Bacteriol. 1996;178:2071–2078. doi: 10.1128/jb.178.7.2071-2078.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Offner S, Ziese U, Wanner G, Typke D, Pfeifer F. Structural characteristics of halobacterial gas vesicles. Microbiology. 1998;144:1331–1342. doi: 10.1099/00221287-144-5-1331. [DOI] [PubMed] [Google Scholar]
- 31.Palmer B, Marinus M. The dam and dcm strains of Escherichia coli—a review. Gene. 1994;143:1–12. doi: 10.1016/0378-1119(94)90597-5. [DOI] [PubMed] [Google Scholar]
- 32.Pfeifer F, Englert C. Function and biosynthesis of gas vesicles in halophilic archaea. J Bioenerg Biomembr. 1992;24:577–585. doi: 10.1007/BF00762350. [DOI] [PubMed] [Google Scholar]
- 33.Pfeifer F, Ghahraman P. Plasmid pHH1 of Halobacterium salinarium: characterization of the replicon region, the gas-vesicle gene cluster and insertion elements. Mol Gen Genet. 1993;238:193–200. doi: 10.1007/BF00279547. [DOI] [PubMed] [Google Scholar]
- 34.Pfeifer F, Offner S, Krüger K, Ghahraman P, Englert C. Transformation of halophilic archaea and investigation of gas-vesicle synthesis. Syst Appl Microbiol. 1994;16:569–577. [Google Scholar]
- 35.Pfeifer F, Krüger K, Röder R, Mayr A, Ziesche S, Offner S. Gas vesicle formation in halophilic archaea. Arch Microbiol. 1997;167:259–268. doi: 10.1007/s002030050441. [DOI] [PubMed] [Google Scholar]
- 36.Röder R, Pfeifer F. Influence of salt on the transcription of the gas-vesicle genes of Haloferax mediterranei and identification of the endogenous transcriptional activator gene. Microbiology. 1996;142:1715–1723. doi: 10.1099/13500872-142-7-1715. [DOI] [PubMed] [Google Scholar]
- 37.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 38.Walker J E, Hayes P K, Walsby A E. Homology of gas vesicle proteins in cyanobacteria and halobacteria. J Gen Microbiol. 1984;130:2709–2715. [Google Scholar]
- 39.Walsby A E. Gas vesicles. Microbiol Rev. 1994;58:94–144. doi: 10.1128/mr.58.1.94-144.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]