Abstract
Background
Despite current improvements in systemic cancer treatment, brain metastases (BM) remain incurable, and there is an unmet clinical need for effective targeted therapies.
Methods
Here, we sought common molecular events in brain metastatic disease. RNA sequencing of thirty human BM identified the upregulation of UBE2C, a gene that ensures the correct transition from metaphase to anaphase, across different primary tumor origins.
Results
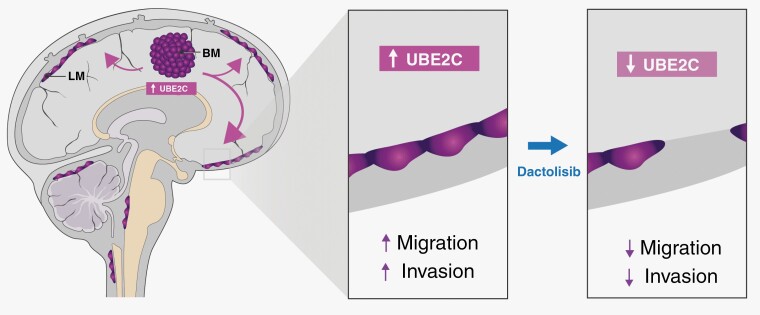
Tissue microarray analysis of an independent BM patient cohort revealed that high expression of UBE2C was associated with decreased survival. UBE2C-driven orthotopic mouse models developed extensive leptomeningeal dissemination, likely due to increased migration and invasion. Early cancer treatment with dactolisib (dual PI3K/mTOR inhibitor) prevented the development of UBE2C-induced leptomeningeal metastases.
Conclusions
Our findings reveal UBE2C as a key player in the development of metastatic brain disease and highlight PI3K/mTOR inhibition as a promising anticancer therapy to prevent late-stage metastatic brain cancer.
Keywords: brain metastases, leptomeningeal dissemination, UBE2C, PI3K/mTOR inhibition
Graphical Abstract
Graphical Abstract.
Key Points.
High levels of UBE2C in BM correlate with patients’ worse outcome.
UBE2C promotes leptomeningeal dissemination in vivo.
PI3K/mTOR inhibition prevents this late-stage complication of cancer.
Importance of the Study.
Brain metastases (BM) are a late-stage complication in cancer patients and remain an incurable disease. Using RNA sequencing and tissue microarrays in surgically resected BM from cancer patients with diverse primary cancers, we have identified UBE2C as a molecular marker of prognosis in patients with brain metastatic disease. High expression of UBE2C was associated with patients’ worse survival. We have shown that UBE2C increases migration and invasion and induces leptomeningeal dissemination in vivo. Importantly, early treatment of UBE2C-driven mouse and patient-derived xenografts with the dual PI3K/mTOR inhibitor dactolisib, prevented leptomeningeal dissemination, via downregulation of UBE2C.
The identification of UBE2C as a molecular marker of prognosis in brain metastatic disease across multiple cancer types and the ability to prevent leptomeningeal dissemination by UBE2C indirect targeting, open a new avenue for the designing of clinical trials toward the prevention of late-stage metastatic brain cancer.
Dissemination of cancer cells to the brain is a frequent and late-stage complication of many systemic cancers. Patients with brain metastatic disease have a poor prognosis, with a median survival between 3 and 11 months.1,2 Lung cancer (40%-60%), breast cancer (15%-30%), and melanoma (5%-15%) are the most common primary malignancies that disseminate to the brain.2–5 Autopsy studies showed that around 20%-26% of all cancer patients develop brain metastases (BM) but it is thought that the incidence may be higher and increasing.3,5,6 Advanced methods of BM diagnosis and more effective treatments for extracranial disease led to an improvement in patients’ survival, thus contributing to an increase in BM incidence. The standard of care treatment for BM includes surgical resection, whole-brain radiotherapy (WBRT), and stereotactic radiosurgery (SRS).7 Nonetheless, brain metastatic disease remains incurable.3 Furthermore, the most aggressive form of this disease is characterized by coating of the leptomeninges by cancer cells (leptomeningeal dissemination), and poor response to intensive chemotherapeutic regimens, including intrathecal therapy.8 Currently, molecular-targeted therapies in use for primary tumors lack good therapeutic responses in metastatic tumors located in the central nervous system (CNS). This might be explained either by the limited crossing of drugs through the blood-brain barrier (BBB)9 or by genetic differences between primary tumors and BM.10
Understanding the molecular mechanisms underlying the dissemination of cancer cells into the brain will open the possibility of identifying targets to be used in the development of novel therapies. Previous attempts to study the mechanisms of cancer dissemination to the brain focused mainly on analyzing genes differentially expressed between the most common primary tumors (lung and breast cancer) and their BM. The metastatic compartment often exhibits a distinct genetic profile from the primary tumor.10 Specific molecular targets have been found to play a role in the metastatic process to the brain, namely HER2, FOXC1, LEF1, and HOXB9 in lung cancer, COX2, HBEGF, and ST6GALNAC5 in breast cancer, and plasminogen activator (PA) inhibitory serpins in both these tumors.11–13
We hypothesized that brain metastatic tumors from diverse histological types share common genetic events which promote cancer cell dissemination and colonization of the brain, and these molecular alterations can be used as therapeutic targets. We performed RNA sequencing in a discovery cohort of thirty BM samples from patients diagnosed with various primary tumors and identified UBE2C as being differentially expressed. UBE2C is a ubiquitin-conjugating enzyme that functions together with anaphase-promoting complex/cyclosome (APC/C) involved in the cell cycle, ensuring a correct transition from metaphase to anaphase.14,15 UBE2C is present in the 20q13.1 locus, a region known to be amplified in a variety of malignancies, including gastro-esophageal carcinomas.16 High expression of UBE2C has been found in tumor tissues from different histological types, and it was associated with a worse prognosis in primary cancers.16,17 We analyzed an independent cohort of 89 BM from different primary origins and observed that patients with higher expression of UBE2C had a significantly worse prognosis. In orthotopic mouse models of BM, UBE2C promoted leptomeningeal dissemination and decreased survival, possibly due to an increase in cancer cell migration and invasion. Importantly, early treatment with a PI3K/mTOR inhibitor (dactolisib) prevented the UBE2C-induced leptomeningeal dissemination in these models, revealing a promising therapeutic avenue in advanced metastatic cancer prevention.
Materials and Methods
Key Resources
A full description of Materials and methods can be found in Supplementary Materials document.
Patients’ Samples
BM samples were collected in accordance with the Ethics board from Hospital de Santa Maria (Refª. Nº 367/18 and Refª. Nº 346/20) and a written informed consent was obtained from all patients, prior to study participation.
RNA Sequencing Analysis
The BM samples from diverse primary origins included in the study reflect the series of patients submitted to brain surgery at the Department of Neurosurgery (HSM-CHULN). Total RNA isolated from BM was processed using the TruSeq RNA Sample Preparation v2 kit (low-throughput protocol; Illumina, San Diego, CA, USA) to prepare the barcoded libraries. Libraries were validated and quantified using either DNA 1000 or high-sensitivity chips on a Bioanalyzer (Agilent, Santa Clara, CA, USA). 7.5 pM denatured libraries were input into cBot (Illumina), followed by deep sequencing using HiSeq 2500 (Illumina) for 101 cycles, with an additional seven cycles for index reading. For normal tissue samples, we downloaded the call sets from the ENCODE portal (https://www.encodeproject.org/) for tissues matching the tissue of origin of the BM.
Fastq files were imported into Partek Flow (Partek Incorporated, St. Louis, MO, USA). Quality analysis and quality control were performed on all reads to assess read quality and to determine the amount of trimming required (both ends: 13 bases 5ʹ and 1 base 3ʹ). Trimmed reads were aligned against the hg38 genome using the STAR v2.4.1d aligner. Unaligned reads were further processed using Bowtie 2 v2.2.5 aligner. Finally, aligned reads were combined before quantifying the expression against the ENSEMBL (release 84) database using the Partek Expectation-Maximization algorithm. Partek Flow default settings were used in all analyses. Files were then processed using Partek Genomic Suite (Partek Incorporated, St. Louis, MO, USA). Genes were filtered for expression values≤1 and 3 or more missing values, the remaining genes were then log2 transformed.
Microarray Datasets used in the Bioinformatic Analysis
Microarray datasets were also used in the analysis of RNA sequencing. GSE2109 and GSE7307 datasets were downloaded from GEO DataSets (https://www.ncbi.nlm.nih.gov/gds) and processed using Partek Genomic Suite. Files were imported into Partek Genomic Suites and normalized using the RMA method.
Tissue Microarrays (TMAs)
TMAs were kindly provided by the Neuropathology lab of HSM-CHULN. Protein levels were assessed by immunohistochemical (IHC) staining with UBE2C antibody (Boston Biochem, Cat# A650), ASF1B (Cell Signaling Technology, Cat# 2902), FoxM1 (Cell Signaling Technology, Cat# 5436)or Ki-67 (D2H10) (Cell Signaling Technology, Cat# 9027). For UBE2C, ASF1B and FoxM1 we used semi-quantitative scores of intensity (low or high staining intensity) and frequency (low: 0%-49% staining or high: 50%-100% staining), performed by two independent researchers and validated by a pathologist. For Ki67, an automated software was used to quantify the percentage of positive nuclei (ImmunoRatio).
Cell Culture
Human cell lines MDA-MB-231 (referred as MDA), A549 and HCT116 were maintained in the appropriate media. We induced brain tropism in these cell lines as previously described.12 All cell lines were genetically modified to overexpress UBE2C and MDA, additionally, for the KD of UBE2C.
MET-CF78 cells were derived from a patient with lung cancer BM (patient-derived cell culture), established in our laboratory as previously described.18
Cell Modulation
To achieve stable overexpression, the lentiviral vector was used for gene delivery. The plasmid for human UBE2C overexpression was made by subcloning the PCR-amplified UBE2C (IDT) fragment into the EcoRI and Mscl (NEB) site of LeGO-iV2.
The lentiviral vector pLV hU6-sgRNA hUBC-dCas9-KRAB-T2A-Puro (a gift from Charles Gersbach, Addgene plasmid # 71236) was used for stable KD. sgRNAs were obtained from IDT. After annealing, sgRNAs were ligated to BsmBI digested pLV hU6-sgRNA hUBc-dCas9-KRAB-T2a-Puro vector.
Target cells (MDA, A549, and HCT) were stably transduced and selected by flow cytometry using BD FACSAria III cell sorter or 1 µg/ml puromycin (InvivoGen, San Diego, USA, #ant-pr-1).
Immunoblotting
Whole-cell lysates were prepared as previously described19 and the immunoblotting was performed according to standard procedures. Two independent experiments were performed.
Cell Proliferation
CellTiter 96 Aqueous One Solution Reagent (MTS) was used as defined by the manufacturer’s protocol, for the time points of 0, 24, 48, 72, and 96 hours. Three independent experiments, with 3 technical replicates.
Colony Formation Assay
MDA or A549 were seeded onto 6 well plates, with 100 cells/well, and incubated at 37°C, 5% CO2. Cells were allowed to grow for two weeks. Pictures were analyzed using ColonyArea plugin for ImageJ.20
Migration
Seeding 2×104 cells in CIM-plate 16 and the impedance signals were recorded using the xCELLigence,21 for 72 hours.
Invasion
Seeding of 3×104 cells in matrigel-coated transwells for 30 h, at 37° C and 5%CO2.
In Vivo Orthotopic Xenografts
In accordance with Directive 2010/63/EU (transposed to Portuguese legislation through Decreto-Lei No. 113/2013, of August 7th), all animal procedures were approved by the institutional animal welfare body (ORBEA-iMM). NSG mice were acquired from Charles River Laboratories or in the NSG colony established in-house. Animals were subjected to procedures between the ages of 11 and 22 weeks old. Cancer cells were injected intracranially. MDA models were established by injecting 50,000 cells and 100,000 in MET-CF78 model. Histopathologic analysis was performed in CNS samples.
Mice In Vivo Imaging
Mice were injected with XenoLight D-Luciferin, Potassium Salt and imaged after 10 minutes in IVIS Lumina System, 5 minutes exposure.
Histopathological Analysis of Mouse Samples
H&E slides were blindly examined by two independent researchers and a specialized pathologist. Tumor score was defined as 0-no tumor, 1-mild, 2-moderate, 3-marked. Leptomeningeal dissemination score was 0-no neoplasic cell infiltration, 1-mild: minimal focal neoplasic cell infiltration in the meninges, 2-moderate: minimal multifocal neoplasic cell infiltration, 3-marked: high density multifocal neoplasic cell infiltration.
Drug Screening
A series of six-nine dilution steps of each inhibitor between 32.5 and 25,000 nM was tested. About 30 µl of cell suspension were seeded into each well of the drug library. After 72 hours of incubation at 37°C and 5% CO2, plates were analyzed using CellTiter-Glo reagent and read in the Spark MultiMode Plate reader.
In Vitro Drug Testing
In vitro drug assays were performed by seeding MDA, A549 (1000 cells), or MET-CF78 (500 cells) in 96-well plates and using dactolisib or Genz-644282 (25 nM, 100 nM, 250 nM, 500 nM, and 1 mM). Proliferation assays were performed as described above.
Drug Testing in Orthotopic BM Xenografts
NSG mice injected intracranially with MDA cancer cell line (5×104 cells/mouse) or MET-CF78 (1×105 cells/mouse) were treated with dual ATP-competitive PI3K and mTOR inhibitor dactolisib (30 mg/kg, based on the literature22), via oral gavage, daily from day 4 to day 14, for MDA, or from day 7 to day 18, with a wash-out period of 2 days after 5 days of treatment. Animals were randomized into treatment (dactolisib) and control (vehicle: N-Methyl-2-Pyrrolidone (NMP) and Polyethylene glycol 300 (PEG300), 10/90, v/v) groups. By the end of the treatment (day 15 for MDA cells, or 18 for MET-CF78 cells), CNS and organs (lungs, liver, spleen, and kidneys) were collected for histopathological analysis.
UBE2C IHC in Orthotopic BM Xenografts Samples
In mice injected with MDA and treated with dactolisib (or vehicle), UBE2C downregulation was analyzed by an in-house developed macro, using the ImageJ/Fiji macro to quantify the percentage of high UBE2C-staining in the tumor tissue (available at https://github.com/ClaraBarreto/UBE2C).
Statistical Analysis
For statistical differences using multiple testing, as in the RNA sequencing analysis, a Bonferroni adjusted p-value was used. Other statistical differences were determined using t-test (parametric) or Mann–Whitney tests (nonparametric) on GraphPad Prism v6.0 (GraphPad, California, USA, GraphPad Prism, RRID:SCR_002798), as stated in figure legends Differences were considered statistically significant for p≤0.05.
Results
UBE2C is Upregulated in Human BM
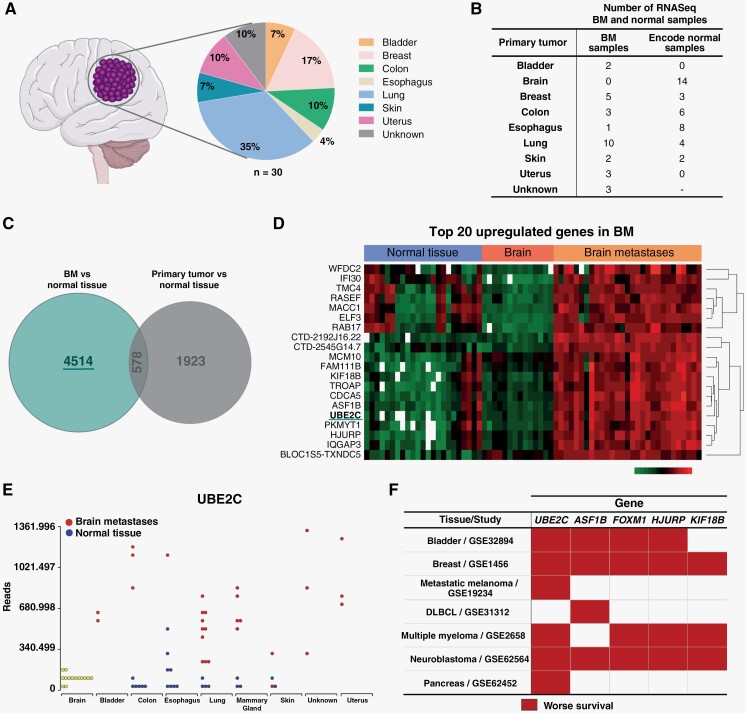
A discovery cohort from the neurosurgery department at Hospital de Santa Maria from Centro Hospitalar Universitário Lisboa Norte (HSM-CHULN) comprising thirty human BM from patients diagnosed with different primary tumor origins were analyzed using RNA sequencing. In this cohort, lung cancer was the most frequent primary tumor, followed by breast, uterus, and colon cancer (Figure 1A). Tumor samples were collected consecutively, and therefore, the representation of each primary tumor origin reflects the frequency in the neurosurgical series. The transcriptomic analysis included publicly available RNA sequencing datasets of normal tissues (Figure 1B) and microarray datasets of normal tissue and primary tumors (Supplementary Figure S1A), matching the BM cohort histological types. Genes upregulated exclusively in BM samples were identified when compared with normal tissue and primary tumors (n=4514 genes, Figure 1C). From this list, we selected the top 20 upregulated genes in BM based on p-value≤0.05 and positive or negative fold change≥1.7 (Figure 1D and Supplementary Figure S1B). The expression of these genes was then checked for directionality in all tumor types and the most promising genes were further investigated regarding their clinical relevance. Among the top 5 candidate genes (UBE2C, ASF1B, FOXM1, HJURP, and KIF18B), UBE2C was the most frequently associated with poor prognosis in publicly available clinical datasets from diverse cancer types, including metastatic melanoma (Figure 1E). UBE2C was found to be highly expressed in BM samples from diverse primary tumor origins when compared to normal tissues (Figure 1F).
Figure 1.
UBE2C is highly expressed in human BM. (A) Percentage of BM included in the RNA sequencing analysis, according to primary tumor origin (n=30, discovery cohort from HSM-CHULN). (B) Datasets included in the RNA sequencing (RNASeq) bioinformatic analysis, including data from normal tissue samples, from publicly available datasets. (C) Venn diagram analyzing the differentially expressed genes in BM and primary tumor samples, against genes from normal tissue. (D) Heatmap representing the top 20 upregulated genes in our discovery cohort when compared to publicly available data from normal brain and other normal tissues. (E) Comparison of RNA UBE2C levels in BM and normal tissues. (F) Top 5 upregulated genes in BM and their association with the overall survival (OS) of cancer patients, using datasets from several studies with publicly available RNAseq data: GSE32894: Hoglund, 201549; GSE1456: Bergh, 200550; GSE19234: Bhardwaj,51 2009; GSE31312: International DLBCL Rituximab-CHOP Consortium52; GSE2658: Hanamura, 200653; GSE62564: Tong, 201454; GSE62452: Hussain, 2016.55 DLBCL: Diffuse large B-cell lymphoma. Red color represents high gene expression significantly associated with decreased OS. The Figure was partly generated using Servier Medical Art, provided by Servier, licensed under a Creative Commons Attribution 3.0 unported license (smart.servier.com).
Patients with High Expression of UBE2C in BM have a Poorer Prognosis
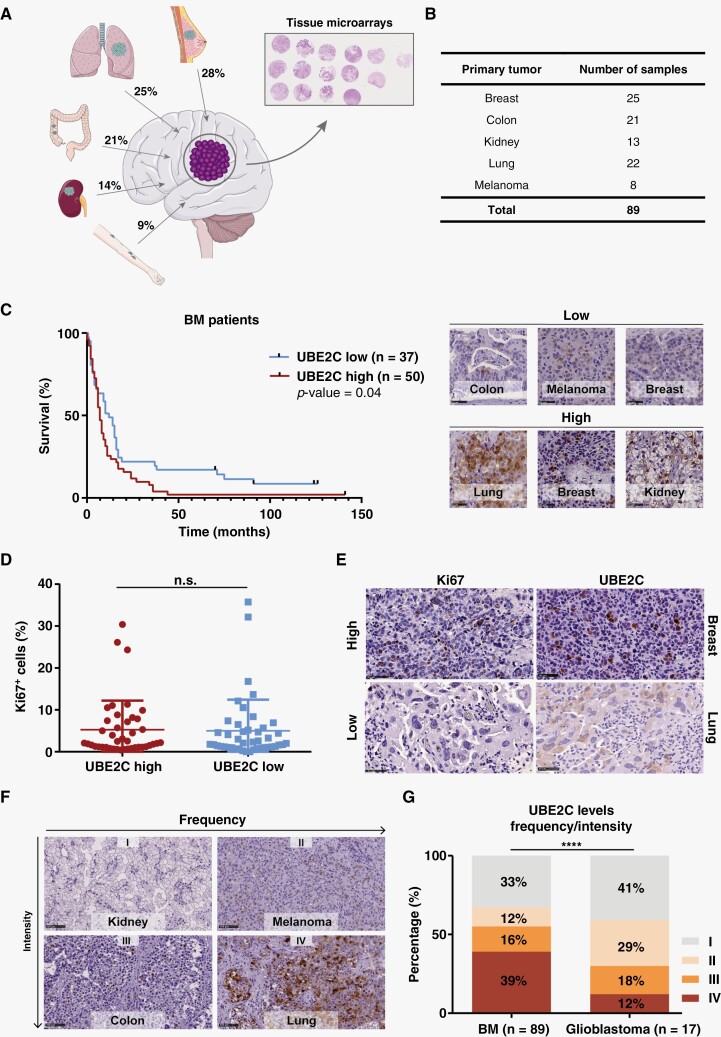
We validated the clinical relevance of UBE2C by immunohistochemistry (IHC) in tissue microarrays (TMAs), using an independent cohort of 89 patients with BM from different primary tumor origins (Figure 2A-B). Samples from patients with primary brain tumors (glioblastoma) were used as control. Patients had a median age of 62 years (28-90 years) (Supplementary Figure S2A), with a similar frequency of male and female patients (52% and 48%, respectively) (Supplementary Figure S2B). The median overall survival of these patients since the diagnosis of BM was 8 months (Supplementary Figure S2C), with different survival rates depending on the primary tumor origin (Supplementary Figure S2D-E). No differences were found in the survival of BM patients when comparing patients with high vs low expression levels (intensity of staining) of ASF1B (Supplementary Figure S2F) or FoxM1(Supplementary Figure S2G). In contrast, high expression of UBE2C (higher intensity of staining) was associated with a worse prognosis (median survival of 7 months), while patients with low UBE2C expression exhibited a median survival of 12 months (p-value=0.04) (Figure 2C). This finding was not due to a higher proliferative index in tumor cells (Figure 2D-E), nor dependent on the primary tumor type (Supplementary Figure S2H-L). Using a scoring system combining intensity and frequency of UBE2C staining (Figure 2F), we have observed that most BM samples (55%) have high levels of UBE2C (Score III and IV), in contrast with glioblastomas in which 70% of the samples have low expression of this protein (Figure 2G). This suggests that high UBE2C expression is a feature of brain metastatic tumors, but not of primary brain tumors, such as glioblastoma.
Figure 2.
High expression of UBE2C is associated with worse survival in patients with BM. (A) Percentage of BM by tumor type included in the tissue microarrays (TMA). (B) Number of BM samples by tumor origin in the TMA (independent validation cohort, N=89). (C) Kaplan-Meier analysis of patients’ survival according to UBE2C protein intensity levels and representative images of the IHC intensity score used (high and low). Differences were considered statistically significant for p-value≤0.05, according to the Log-rank (Mantel-Cox) test. (D) Comparison of UBE2C intensity and Ki67 staining in patients with BM. Data is expressed as median with interquartile range. Mann–Whitney test. (E) Representative images of IHC staining for UBE2C (scored as high or low) and Ki67 (assessed using ImageJ); Scale bar: 50 μm. (F) IHC score including intensity and frequency of the UBE2C staining in tumor tissue: I—low/low; II—low/high; III—high/low; III—high/high, respectively; Scale bar: 100 μm. (G) UBE2C expression was compared between BM and brain primary tumors (glioblastoma); ****p-value<0.0001, Chi-square (and Fisher’s exact) test. The Figure was partly generated using Servier Medical Art, provided by Servier, licensed under a Creative Commons Attribution 3.0 unported license (smart.servier.com).
UBE2C Promotes Cancer Cell Migration and Invasion In Vitro
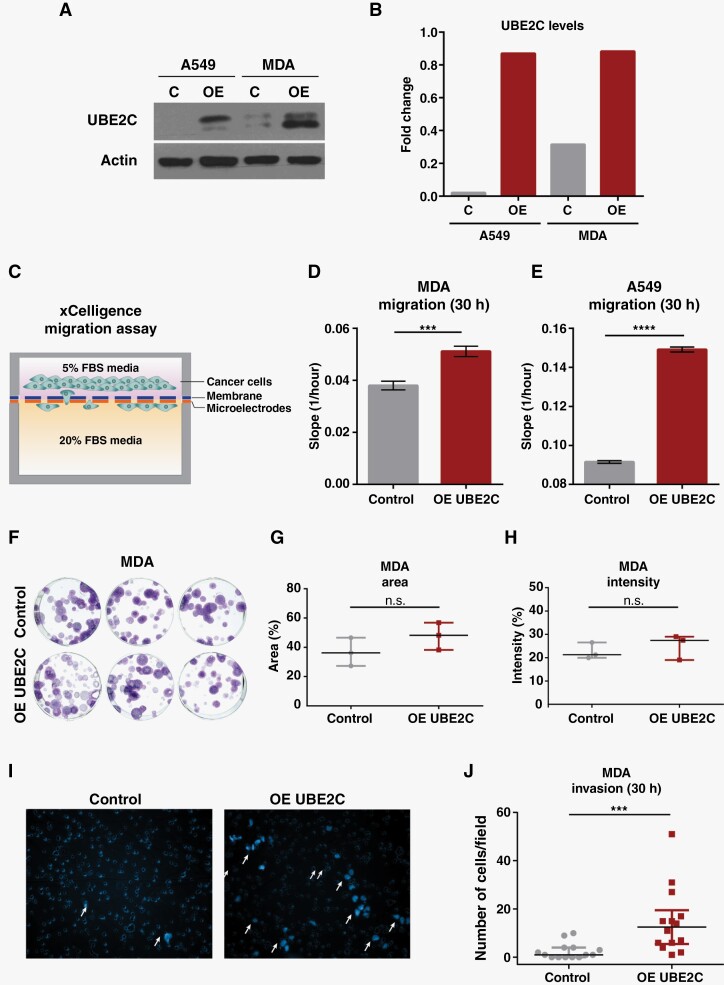
To validate and characterize the role of UBE2C in the context of brain metastatic disease we used two cancer cell lines, MDA (breast cancer) and A549 (lung cancer). Both cell lines were engineered to stably overexpress UBE2C (Figure 3A-B). To study the effect of UBE2C in cancer cell migration, we performed real-time cell analyses using the xCELLigence system (Figure 3C). The migration rate of MDA and A549 cells was significantly increased in UBE2C overexpressing cells (Figure 3D-E). This effect was not due to an increase in proliferation either in short-term (Supplementary Figure S3A-B) or long-term assays (Figure 3F-H and Supplementary Figure S3C-E), as assessed by MTS and colony formation assays, respectively. In addition, UBE2C increased the invasion ability of MDA cells in matrigel-coated transwells (Figure 3I-J).
Figure 3.
Overexpression of UBE2C increases cancer cell migration and invasion. (A) Western Blot of UBE2C levels in MDA and A549 cell lines modulated for overexpression (OE) of UBE2C and (B) its densitometry analysis. (C) Schematic representation of migration assay using CIM-Plates in the xCELLigence system. Migration ability of (D) MDA and (E) A549 cells with OE of UBE2C was evaluated using the xCELLigence system at 30h and compared with control cells (empty vector); ****p-value<0.0001, ***p-value=0.001, unpaired t-test. Data represented as mean with SD. (F) Pictures of colony formation assay (CFA) performed in MDA OE UBE2C cells (100 cells/well) after 2 weeks, and (G) quantification of the area; p-value=0.23 and (H) intensity p-value=0.52 of the formed colonies. Mann-Whitney test. Data is presented as median and interquartile range. (I) Representative images and (J) quantification of the invasion capacity of MDA cells with OE UBE2C using matrigel-coated transwells at 30 h and compared with control cells (empty vector); ***p-value=0.0002, Mann-Whitney test. Data are presented as median with interquartile range.
UBE2C Drives Leptomeningeal Dissemination and Associates with Worse Survival In Vivo
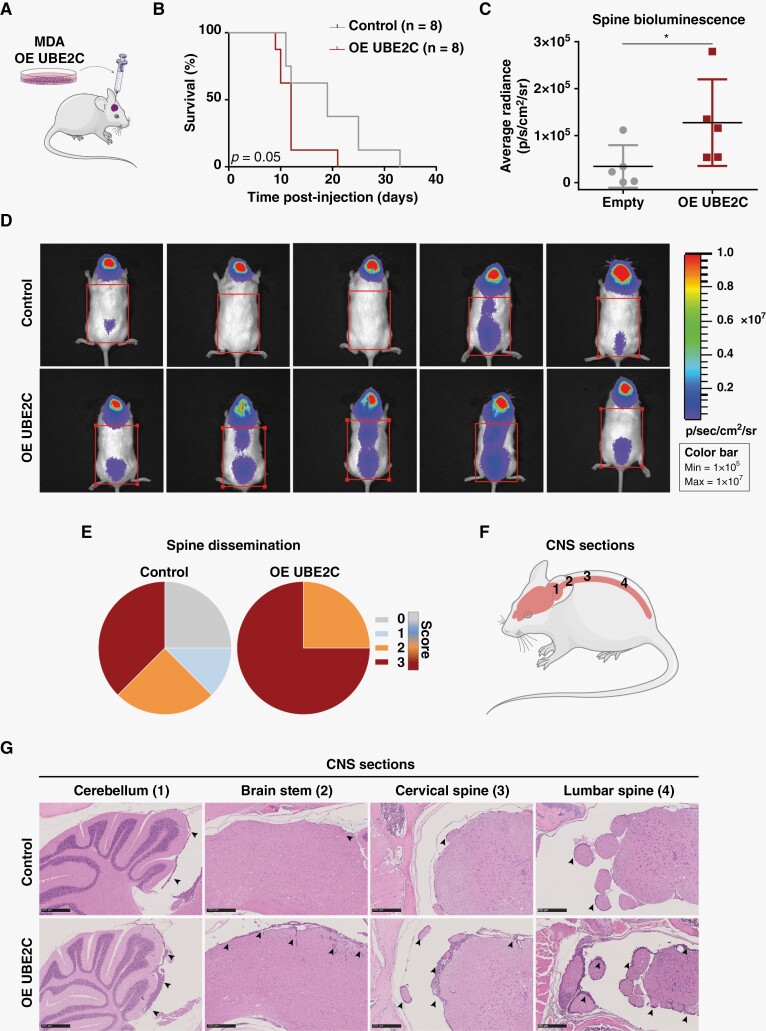
To assess the in vivo role of UBE2C in BM, we performed intracranial injections of MDA cells in NSG mice (Figure 4A). Animals injected with UBE2C-overexpressing cells showed decreased survival (Figure 4B, p-value=0.05), a phenotype associated with more aggressive disease, characterized by leptomeningeal dissemination. Although the levels of leptomeningeal dissemination in the brain were similarly high in both experimental groups (Supplementary Figure S4A), we observed a significant increase in spinal cord dissemination among animals with UBE2C-overexpressing cells, both in vivo (Figure 4C-D) and ex vivo (Figure 4E-G). We also established an MDA cell line with UBE2C knockdown (KD) using the CRISPR system (Supplementary Figure S4B). There were no differences in survival (Supplementary Figure S4C) or dissemination to the brain meninges (Supplementary Figure S4D) in orthotopic models transplanted with UBE2C KD or control cells. However, we found evidence of decreased dissemination of cancer cells with UBE2C silencing to the spinal cord meninges (Supplementary Figure S4E). We also observed that lung cancer sublines with tropism to the leptomeninges have increased levels of UBE2C in comparison with their parental counterparts (Supplementary Figure S4F-G). Thus, UBE2C drives a more aggressive disease phenotype with leptomeningeal dissemination and decreased survival in orthotopic models of BM.
Figure 4.
UBE2C decreases survival and mediates leptomeningeal dissemination driving an aggressive disease phenotype in vivo. (A) Intracranial injection of NSG mice (n=8/group) with MDA cell line, control and OE UBE2C; (B) and the survival analysis of both groups, *p-value=0.05, Kaplan-Meier test. (C) Representative images ofthewhole-body bioluminescence imaging of control and OE UBE2C mice at day 9 post-injection, and (D) quantification of the average radiance signal in the spine; *p-value=0.03, Mann-Whitney test. Data represented as median with interquartile range. (E) Spine leptomeningeal dissemination in animals injected with control and OE UBE2C cells. Histopathological score used to assess the leptomeningeal dissemination: 0- negative; 1- mild; 2- moderate; 3- marked. (F) CNS sections analyzed by histopathology, and (G) representative images of these sections with H&E staining (5x; scale bar: 250 μm) from mice injected with control and OE UBE2C cells. Black arrows indicate leptomeningeal dissemination. The Figure was partly generated using Servier Medical Art, provided by Servier, licensed under a Creative Commons Attribution 3.0 unported license (smart.servier.com).
Targeting the PI3K/mTOR Pathway In Vitro Decreases Cancer Cell Proliferation and Downregulates UBE2C
Due to the lack of effective treatment options for patients with BM and the inexistence of specific therapies targeting UBE2C, we performed a high-throughput drug screening to identify compounds with efficacy in treating UBE2C-driven BM. We used a chemical library of 650 compounds, FDA approved or in phase 3 or 4 clinical trials, to test cell lines with overexpression and knock-down of UBE2C (Supplementary Figure S5A). In this drug screening, we have selected two candidate compounds that targeted cells expressing high levels of UBE2C when compared to their counterparts, for further in vitro validation: dactolisib (PI3K/mTOR inhibitor) and Genz644282 (Topoisomerase I inhibitor) (Supplementary Figure S5B-C). Both compounds proved to be highly effective inhibiting cancer cell proliferation (Figure 5A-B and S5D-E).
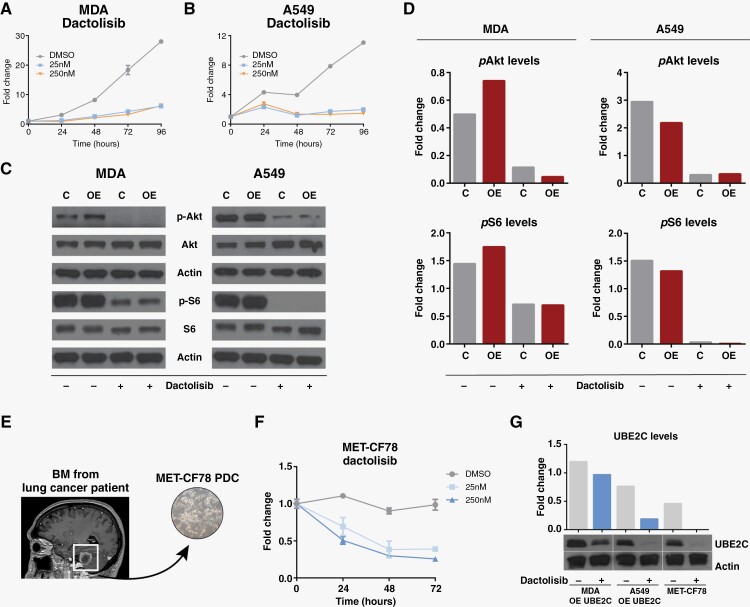
Figure 5.
Downregulation of PI3K/mTOR pathway inhibits cancer cell proliferation and decreases UBE2C levels. MTS assays using 25 nM and 250nM concentrations of dactolisib to assess proliferation in (A) MDA and (B) A459 cell lines with OE of UBE2C. (C) Evaluation of the effect of dactolisib in PI3K/mTOR pathway (p-Akt and p-S6 levels) by western blot using MDA and A549 cells with UBE2C overexpression (OE) in comparison with control cells (D). (E) MET-CF78 PDC derived from a BM of a lung cancer patient. (F) Inhibition of proliferation by dactolisib in patient-derived cultures from a lung cancer BM (MET-CF78). (G) UBE2C levels assessed by western blot in cancer cell lines treated with dactolisib (250 nM) for 24 h and its densitometry analysis.
Single-cell RNA sequencing data analysis of BM from patients with different primary tumors23 showed that metastatic tumor cells with high UBE2C expression levels (Supplementary Figure S5F) also presented high levels of MTOR1 (Supplementary Figure S5G), with a positive correlation between the mRNA levels of both genes (Supplementary Figure S5H). In addition, the PI3K/mTOR pathway has been implicated in brain metastatic cancer.10 Therefore, we decided to further evaluate the efficacy of inhibition of PI3K/mTOR signaling and examine its interplay with UBE2C. Dactolisib effectively inhibited the PI3K/mTOR pathway in breast and lung cancer cell lines in vitro, through the downregulation of pAkt and pS6, respectively (Figure 5C-D). Furthermore, we have also used a patient-derived culture (PDC), MET-CF78, isolated from a BM-derived from a lung cancer patient18 with constitutive expression of UBE2C (Figure 5E). We observed inhibition of MET-CF78 proliferation by dactolisib in a dose-dependent manner (Figure 5F). Interestingly, the targeting of PI3K/mTOR signaling pathway by dactolisib decreased the UBE2C levels in all cancer cell lines (Figure 5G), regardless of their different levels of PI3K activation (Supplementary Figure S5I).
Dactolisib Prevents Leptomeningeal Dissemination In Vivo
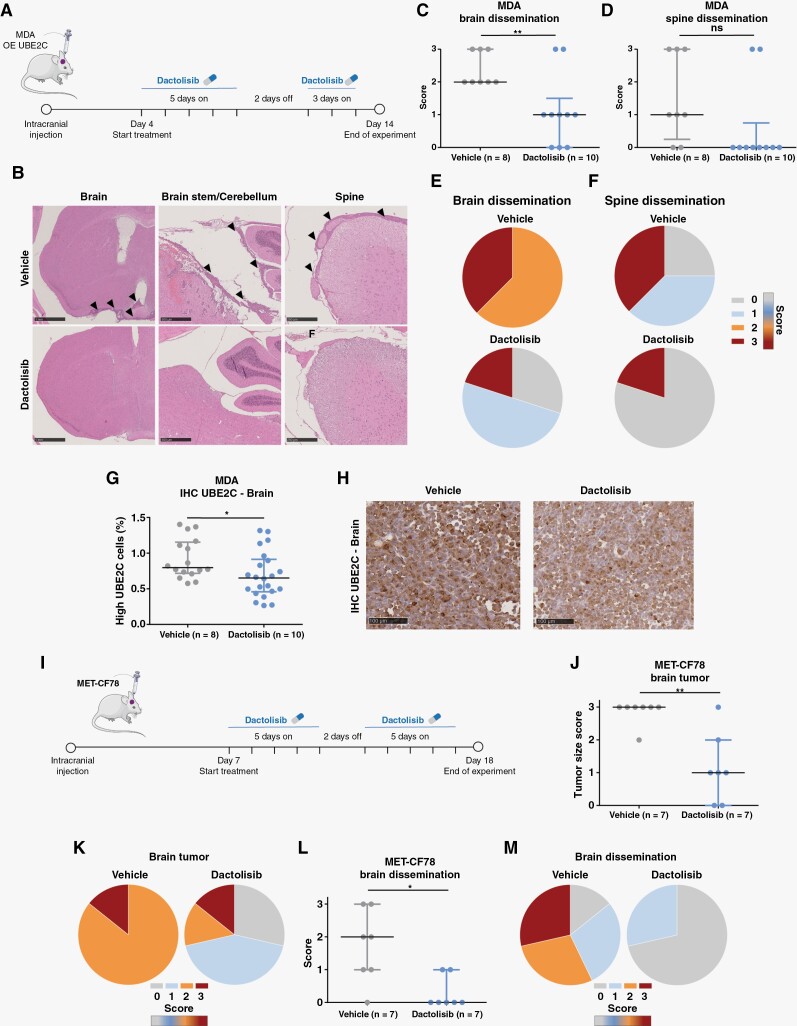
We then asked whether the aggressive metastatic phenotype induced by UBE2C in vivo could be prevented by early treatment with dactolisib, a compound known to cross the BBB.24 A preclinical therapeutic protocol was designed to early treat UBE2C-driven orthotopic mouse models of BM with dactolisib by oral gavage. Treatment was initiated on day 4 post-injection, and it was given daily for 10 days, with a wash-out period of two days (Figure 6A). There were no differences in weight between dactolisib-treated and untreated animals (Supplementary Figure S6A). Strikingly, although dactolisib was not able to significantly reduce brain tumor size (Supplementary Figure S6B), it was effective in preventing leptomeningeal dissemination both in the brain and spine (Figure 6C-F), reversing the aggressive phenotype induced by UBE2C-overexpressing cancer cells. Dactolisib-treated brain tumors exhibited a significant decrease in the number of cancer cells expressing high levels of UBE2C (Figure 6G-H).
Figure 6.
PI3K/mTOR dual inhibitor dactolisib prevents leptomeningeal dissemination in UBE2C-drivenmouse models of BM. (A) Treatment protocol in orthotopic xenografts using breast cancer cells (MDA) with overexpression of UBE2C. Animals were treated daily from day 4 to day 14 post-injection with dactolisib (30 mg/kg; n=10) or vehicle (NMP and PEG300, 10%-90%, v-v; n=8), with a wash-out period of 2 days. (B) Representative images of leptomeningeal dissemination in the control group and absence of dissemination in the dactolisib-treated group. Brain (scale: 1 mm), brain stem/cerebellum (scale: 500 μm), and spine (scale: 250 μm). (C) Histopathological scoring of leptomeningeal dissemination in the brain (**p-value=0.0083, t-test, data represented as mean and SD) and (D) in the spine (p-value=0.07, Mann-Whitney test, data represented as median and interquartile range). (E) Percentage of brain dissemination in vehicle (2: 62,5%, 3: 37,5%) and treatment (0: 30%, 1: 50%, 3: 20%) groups. (F) Percentage of spine dissemination in vehicle (0: 25%, 1: 37,5%, 3: 37,5%) and treatment (0: 80%, 3: 20%) groups. (G) Expression of UBE2C by IHC staining, in brain tumor samples upon treatment with dactolisib. *p-value=0.03, t-test. Data represented as mean with SD. (H) Representative images of UBE2C staining in brain tumor sections. (I) Treatment protocol in patient-derived xenografts of lung cancer BM (MET-CF78) (n=7/group). Animals were treated daily from day 7 to day 17 post-injection with dactolisib or vehicle, with a wash-out period of 2 days; Scale bar: 100 μm. (J) Histopathological scoring of the brain tumor size (0: no tumor; 1: minimal to mild; 2: moderate; 3: marked); **p-value=0.009; Mann-Whitney test. Data represented as median with interquartile range, and (K) brain tumor size score in vehicle (2: 14,286%, 3: 85,714%) and dactolisib (0: 28,571%, 1: 42,857%, 2: 14,286%, 3: 14,286%) treated mice. (L) Histopathological scoring of brain dissemination as described above; *p-value=0.026, Mann-Whitney test; data represented as median with interquartile range, and (M) percentage of brain dissemination in vehicle (0: 14,286%, 1: 28,571%, 2: 28,571%, 3: 28,571%) and treatment (0: 81,429%, 1: 28,571%) groups. Score used to assess the leptomeningeal dissemination: 0- negative; 1- mild; 2- moderate; 3- marked. The Figure was partly generated using Servier Medical Art, provided by Servier, licensed under a Creative Commons Attribution 3.0 unported license (smart.servier.com).
In the patient-derived xenograft of the lung cancer BM (MET-CF78), early preclinical treatment with oral dactolisib (Figure 6I) had no differences in spine dissemination (Supplementary Figure S6C) and there was no impact of the treatment in the weight of the animals (Supplementary Figure S6D). However, dactolisib significantly reduced both brain tumor size (Figure 6J-K) and leptomeningeal dissemination to the brain (Figure 6L-M).
To validate if the effect of dactolisib in vivo was, at least in part, UBE2C-dependent, we developed orthotopic mouse models using MDA cells with KD of UBE2C (n=7 mice/group). There were no differences in the weight of treated animals compared to the untreated (Supplementary Figure S6E). In UBE2C-depleted tumors, dactolisib did not significantly impact brain tumor size (Supplementary Figure S6F) or CNS dissemination (Supplementary Figure S6G-H).
Discussion
Despite significant therapeutic advances in the treatment of primary cancers, BM remain a major clinical hurdle. We began addressing this challenge by interrogating BM from diverse primary cancers using RNA sequencing coupled with functional approaches, in both human and mouse models. Focusing on BM from diverse primary tumors, we were able to identify common genetic events leading to cancer cell dissemination and/or colonization into the brain. Simultaneously, these molecular targets constitute novel targets for therapy.
We identified UBE2C as a differentially expressed gene in human BM and showed that high levels of UBE2C are associated with shorter survival in cancer patients with brain metastatic disease. UBE2C was first described by Okamoto as an oncogene, overexpressed in primary tumors and cancer cell lines, when compared to normal tissues.17 It has also been shown that UBE2C correlates with higher tumor grades. In lung cancer patients, UBE2C was associated with poorer survival25 and with tumor progression, as a consequence of autophagy inhibition and increased tumor invasiveness.26 In breast cancer, UBE2C was identified as a prognostic marker27 and associated with cancer grade progression28 and drug resistance.29 Similar observations were made in colon, ovarian, and bladder cancers, among other types of tumors.30–33 Interestingly, UBE2C was associated with BM in a recent publication, where single-cell RNA sequencing of BM from multiple cancers led to the identification of two subgroups, being UBE2C one of the signature genes in the proliferative group.23
UBE2C has been described as a promoter of migration and invasion of tumor cells in endometrial cancer models, inducing epithelial to mesenchymal transition (EMT), via downregulation of p53 levels.34 Similar effects were also observed in gastric35 and hepatocellular cancers.36 We observed that orthotopic mouse models injected with cancer cells overexpressing UBE2C developed a more aggressive disease phenotype, with leptomeningeal dissemination in the brain and spinal cord. This phenomenon may be explained by the increase in cancer cell migration and invasion induced by overexpression of this gene, as observed in vitro. One possible explanation for the increased colonization of the leptomeninges, particularly in the spinal cord, in a poor microenvironment such as the cerebrospinal fluid (CSF), is that cancer cells with high levels of UBE2C have higher glycolytic activity, further promoting their migration and invasion abilities even in such environmental conditions.37,38 In fact, a metabolic diagnostic tool based on nuclear magnetic resonance (NMR) analysis of patients with leptomeningeal carcinomatosis showed increased lactate levels, a product of glycolysis.39
In cancer patients, leptomeningeal dissemination can occur in approximately 5%-15% of the cases,8 having a dismal prognosis with a median survival between 2 and 4 months.8,40–42 Very little is known about the molecular biology of this advanced stage of metastatic cancer, making it exceedingly difficult to treat. Aggressive therapies including intrathecal chemotherapy and whole-brain radiation have failed to alter the natural history of leptomeningeal dissemination. Recent phase 2 studies have reported the clinical benefit of patients treated with EGRF inhibitors and immunotherapy,43,44 although the duration of responses in leptomeningeal disease was limited in time.
Since late-stage CNS dissemination seems very difficult to target, the best chance for improved survival in these patients would be to prevent cancer cells from disseminating into the leptomeninges. Our observations revealed that early treatment with oral dactolisib (a dual PI3K/mTOR inhibitor) prevented the development of leptomeningeal dissemination in orthotopic mouse models driven by UBE2C. This effect of dactolisib might be UBE2C-dependent because, in UBE2C-depleted tumors, treatment did not reduce brain tumor size or CNS dissemination.
Moreover, dactolisib effectively reduced dissemination and brain tumor size in a patient-derived xenograft from a lung cancer BM. Dactolisib brain bioavailability and clinical effect have been previously reported, even at lower doses.45,46 In both in vivo models, the dual PI3K/mTOR inhibitor was well tolerated and did not induce toxicity. Interestingly, mutations in PI3K/Akt/mTOR signaling have been found in BM, but not in primary tumors.10 Also, activation of this signaling pathway has already been associated with leptomeningeal dissemination in melanoma patients.47 We have shown that PI3K/mTOR pathway inhibition with dactolisib decreased cancer migration and invasion, and downregulated UBE2C both in vitro and in vivo. The connection between PI3K/Akt/mTOR pathway and UBE2C has been previously reported in gastric cancer cells, where UBE2C led to the activation of AURKA and consequent EMT by the decrease in p-AKT1 levels.35 Furthermore, the mTOR inhibitor CCI-779 decreased the levels of UBE2C in castration-resistant prostate cancer, by disrupting its transcription and blocking UBE2C-dependent invasion.48 We believe dactolisib targets key steps in the metastatic cascade (mobilization of cancer cells from the primary tumor and invasion) and, therefore, may be considered as a novel therapeutic strategy to prevent brain metastatic disease.
Further studies are warranted to better explore the mechanisms of UBE2C-driven dissemination to the brain and to dissect the cellular interplay between cancer, stroma, and immune cells, mediated by UBE2C. Moreover, the limited size of our BM patients’ cohort and the number of models used for validation (breast and lung cancer) do not allow the generalization of our results for each primary cancer type. The comparison of UBE2C expression levels in BM with matched primary tumors or extracranial metastases would also add layers of information on UBE2C specificity. Additional functional studies should be performed to validate if the leptomeningeal dissemination driven by high levels of UBE2C is a general feature of cancers from different primary origins, using other BM cancer models, besides breast and lung cancer.
This notwithstanding, we have shown that UBE2C is a relevant player in brain metastatic disease, especially relevant for leptomeningeal dissemination, and that it can be used as a prognostic marker in cancer patients with this condition. Furthermore, we demonstrated that UBE2C-induced leptomeningeal dissemination can be prevented in vivo by targeting the PI3K/mTOR pathway. The results from our study may prompt the advancement of PI3K/mTOR inhibitors into clinical trials for patients with advanced metastatic cancer of the CNS from multiple primary origins.
Supplementary Material
Acknowledgments
The authors would like to acknowledge the patients who kindly provided the tumor specimens included in the RNA sequencing and TMA analysis for this research, and the Biobanco-iMM CAML who enabled the process of tumor specimen collection, processing, and storage. Finally, the authors thank Pedro Pereira, the Histology Service of Comparative Pathology Unit, the Bioimaging Unit, and the Rodent Facility from Instituto de Medicina Molecular João Lobo Antunes for technical assistance.
Contributor Information
Eunice Paisana, Instituto de Medicina Molecular João Lobo Antunes, Faculdade de Medicina da Universidade de Lisboa; Av. Prof. Egas Moniz, 1649-028, Lisboa, Portugal.
Rita Cascão, Instituto de Medicina Molecular João Lobo Antunes, Faculdade de Medicina da Universidade de Lisboa; Av. Prof. Egas Moniz, 1649-028, Lisboa, Portugal.
Carlos Custódia, Instituto de Medicina Molecular João Lobo Antunes, Faculdade de Medicina da Universidade de Lisboa; Av. Prof. Egas Moniz, 1649-028, Lisboa, Portugal.
Nan Qin, Department of Pediatric Oncology, Hematology and Clinical Immunology, Heinrich Heine University Düsseldorf, Medical Faculty, and University Hospital Düsseldorf; Moorenstraße 5, 40225 Düsseldorf, Germany; German Cancer Consortium (DKTK), Partner Site Essen/Düsseldorf, Düsseldorf, Germany; Moorenstraße 5, 40225 Düsseldorf, Germany.
Daniel Picard, Department of Pediatric Oncology, Hematology and Clinical Immunology, Heinrich Heine University Düsseldorf, Medical Faculty, and University Hospital Düsseldorf; Moorenstraße 5, 40225 Düsseldorf, Germany; German Cancer Consortium (DKTK), Partner Site Essen/Düsseldorf, Düsseldorf, Germany; Moorenstraße 5, 40225 Düsseldorf, Germany.
David Pauck, Department of Pediatric Oncology, Hematology and Clinical Immunology, Heinrich Heine University Düsseldorf, Medical Faculty, and University Hospital Düsseldorf; Moorenstraße 5, 40225 Düsseldorf, Germany; German Cancer Consortium (DKTK), Partner Site Essen/Düsseldorf, Düsseldorf, Germany; Moorenstraße 5, 40225 Düsseldorf, Germany.
Tânia Carvalho, Instituto de Medicina Molecular João Lobo Antunes, Faculdade de Medicina da Universidade de Lisboa; Av. Prof. Egas Moniz, 1649-028, Lisboa, Portugal.
Pedro Ruivo, Instituto de Medicina Molecular João Lobo Antunes, Faculdade de Medicina da Universidade de Lisboa; Av. Prof. Egas Moniz, 1649-028, Lisboa, Portugal.
Clara Barreto, Instituto de Medicina Molecular João Lobo Antunes, Faculdade de Medicina da Universidade de Lisboa; Av. Prof. Egas Moniz, 1649-028, Lisboa, Portugal.
Delfim Doutel, Anatomic Pathology Department, Instituto Português de Oncologia Francisco Gentil, R. Prof. Lima Basto, 1099-023, Lisboa, Portugal.
José Cabeçadas, Anatomic Pathology Department, Instituto Português de Oncologia Francisco Gentil, R. Prof. Lima Basto, 1099-023, Lisboa, Portugal.
Rafael Roque, Neurology Department, Laboratory of Neuropathology, Hospital de Santa Maria, Centro Hospitalar Universitário Lisboa Norte (CHULN), Av. Prof. Egas Moniz, 1649-028, Lisboa, Portugal.
José Pimentel, Neurology Department, Laboratory of Neuropathology, Hospital de Santa Maria, Centro Hospitalar Universitário Lisboa Norte (CHULN), Av. Prof. Egas Moniz, 1649-028, Lisboa, Portugal.
José Miguéns, Department of Neurosurgery, Hospital de Santa Maria, Centro Hospitalar Universitário Lisboa Norte (CHULN), Av. Prof. Egas Moniz, 1649-028, Lisboa, Portugal.
Marc Remke, Department of Pediatric Oncology, Hematology and Clinical Immunology, Heinrich Heine University Düsseldorf, Medical Faculty, and University Hospital Düsseldorf; Moorenstraße 5, 40225 Düsseldorf, Germany; German Cancer Consortium (DKTK), Partner Site Essen/Düsseldorf, Düsseldorf, Germany; Moorenstraße 5, 40225 Düsseldorf, Germany.
João T Barata, Instituto de Medicina Molecular João Lobo Antunes, Faculdade de Medicina da Universidade de Lisboa; Av. Prof. Egas Moniz, 1649-028, Lisboa, Portugal.
Claudia C Faria, Instituto de Medicina Molecular João Lobo Antunes, Faculdade de Medicina da Universidade de Lisboa; Av. Prof. Egas Moniz, 1649-028, Lisboa, Portugal; Department of Neurosurgery, Hospital de Santa Maria, Centro Hospitalar Universitário Lisboa Norte (CHULN), Av. Prof. Egas Moniz, 1649-028, Lisboa, Portugal.
Funding
Fundação para a Ciência e a Tecnologia (PTDC/MED-ONC/32222/2017, PD/BD/128288/2017 to EP, SFRH/BD/140299/2018 to CC); Millennium bcp; private donations.
Authorship Statement
Study design: EP, RC, JTB and CCF. Study conduct: EP, RC, CC, NQ, DPicard, DPauck, TC, PR, RR, CF, JP and CCF. Data collection: EP, RC, CC, NQ, DPicard, DPauck, DD and JC. Data analysis: EP, RC, CC, NQ, DPicard, DPauck, TC, PR, CB, and CCF. Data interpretation: EP, RC, DPicard, TC, PR, JTB and CCF. Drafting manuscript: EP, RC and CCF. Revising manuscript content: EP, RC, CC, NQ, DPicard, DPauck, TC, PR, CB, DD, JC, RR, JP, JM, MR, JTB and CCF. All authors approved the final version of this manuscript and take responsibility for the integrity of the data analysis.
Conflict of Interest StatementCCF is Medical Director at TargTex—Targeted Therapeutics. The other authors declare no potential conflicts of interest.
References
- 1. Kocher M, Soffietti R, Abacioglu U, et al. Adjuvant whole-brain radiotherapy versus observation after radiosurgery or surgical resection of one to three cerebral metastases: results of the EORTC 22952-26001 study. J Clin Oncol. 2011;29(2):134–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Berghoff AS, Schur S, Füreder LM, et al. Descriptive statistical analysis of a real life cohort of 2419 patients with brain metastases of solid cancers. ESMO Open. 2016;1(2):e000024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Achrol AS, Rennert RC, Anders C, et al. Brain metastases. Nat Rev Dis Primers. 2019;5(1):5. [DOI] [PubMed] [Google Scholar]
- 4. Nayak L, Lee EQ, Wen PY.. Epidemiology of brain metastases. Curr Oncol Rep. 2012;14(1):48–54. [DOI] [PubMed] [Google Scholar]
- 5. Barnholtz-Sloan JS, Sloan AE, Davis FG, et al. Incidence proportions of brain metastases in patients diagnosed (1973 to 2001) in the Metropolitan Detroit Cancer Surveillance System. J Clin Oncol. 2004;22(14):2865–2872. [DOI] [PubMed] [Google Scholar]
- 6. Smedby KE, Brandt L, Bäcklund ML, Blomqvist P.. Brain metastases admissions in Sweden between 1987 and 2006. Br J Cancer. 2009;101(11):1919–1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Rosenfelder, N,Brada, M.. Integrated treatment of brain metastases. Curr Opin Oncol. 2019;31(6):501–507. [DOI] [PubMed] [Google Scholar]
- 8. Beauchesne P. Intrathecal chemotherapy for treatment of leptomeningeal dissemination of metastatic tumours. Lancet Oncol. 2010;11(9):871–879. [DOI] [PubMed] [Google Scholar]
- 9. Fortin D. The blood-brain barrier: its influence in the treatment of brain tumors metastases. Curr Cancer Drug Targets. 2012;12(3):247–259. [DOI] [PubMed] [Google Scholar]
- 10. Brastianos PK, Carter SL, Santagata S, et al. Genomic characterization of brain metastases reveals branched evolution and potential therapeutic targets. Cancer Discov. 2015;5(11):1164–1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Eichler AF, Chung E, Kodack DP, et al. The biology of brain metastases-translation to new therapies. Nat Rev Clin Oncol. 2011;8(6):344–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bos PD, Zhang XH-F, Nadal C, et al. Genes that mediate breast cancer metastasis to the brain. Nature. 2009;459(7249):1005–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Valiente M, Obenauf AC, Jin X, et al. Serpins promote cancer cell survival and vascular co-option in brain metastasis. Cell. 2014;156(5):1002–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Townsley FM, Aristarkhov A, Beck S, Hershko A, Ruderman JV.. Dominant-negative cyclin-selective ubiquitin carrier protein E2-C/UbcH10 blocks cells in metaphase. Proc Natl Acad Sci U S A. 1997;94(6):2362–2367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Reddy SK, Rape M, Margansky WA, Kirschner MW.. Ubiquitination by the anaphase-promoting complex drives spindle checkpoint inactivation. Nature. 2007;446(7138):921–925. [DOI] [PubMed] [Google Scholar]
- 16. Wagner KW, Sapinoso LM, El-Rifai W, et al. Overexpression, genomic amplification and therapeutic potential of inhibiting the UbcH10 ubiquitin conjugase in human carcinomas of diverse anatomic origin. Oncogene. 2004;23(39):6621–6629. [DOI] [PubMed] [Google Scholar]
- 17. Okamoto Y, Ozaki T, Miyazaki K, et al. UbcH10 is the cancer-related E2 ubiquitin-conjugating enzyme. Cancer Res. 2003;63(14):4167–4173. [PubMed] [Google Scholar]
- 18. Faria CC, Cascão R, Custódia C, et al. Patient-derived models of brain metastases recapitulate human disseminated disease. Cell Rep Med. 2022;3(5):100623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Barata JT, Silva A, Brandao JG, et al. Activation of PI3K is indispensable for interleukin 7-mediated viability, proliferation, glucose use, and growth of T cell acute lymphoblastic leukemia cells. J Exp Med. 2004;200(5):659–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Guzman C, Bagga M, Kaur A, Westermarck J, Abankwa D.. ColonyArea: an ImageJ plugin to automatically quantify colony formation in clonogenic assays. PLoS One. 2014;9(3):e92444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Qin N, Paisana E, Langini M, et al. Intratumoral heterogeneity of MYC drives medulloblastoma metastasis and angiogenesis. Neuro Oncol. 2022;24(9):1509–1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Almeida ARM, Neto JL, Cachucho A, et al. Interleukin-7 receptor alpha mutational activation can initiate precursor B-cell acute lymphoblastic leukemia. Nat Commun. 2021;12(1):7268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gonzalez H, Mei W, Robles I, et al. Cellular architecture of human brain metastases. Cell. 2022;185(4):729–745.e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gil del Alcazar CR, Hardebeck MC, Mukherjee B, et al. Inhibition of DNA double-strand break repair by the dual PI3K/mTOR inhibitor NVP-BEZ235 as a strategy for radiosensitization of glioblastoma. Clin Cancer Res. 2014;20(5):1235–1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Perrotta I, Bruno L, Maltese L, et al. Immunohistochemical analysis of the ubiquitin-conjugating enzyme UbcH10 in lung cancer: a useful tool for diagnosis and therapy. J Histochem Cytochem. 2012;60(5):359–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Guo J, Wu Y, Du J, et al. Deregulation of UBE2C-mediated autophagy repression aggravates NSCLC progression. Oncogenesis. 2018;7(6):49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Loussouarn D, Campion L, Leclair F, et al. Validation of UBE2C protein as a prognostic marker in node-positive breast cancer. Br J Cancer. 2009;101(1):166–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jayanthi V, et al. Grade-specific diagnostic and prognostic biomarkers in breast cancer. Genomics. 2020;112(1):388–396. [DOI] [PubMed] [Google Scholar]
- 29. Wang C, Pan Y-H, Shan M, et al. Knockdown of UbcH10 enhances the chemosensitivity of dual drug resistant breast cancer cells to epirubicin and docetaxel. Int J Mol Sci . 2015;16(3):4698–4712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Presta I, et al. UbcH10 a Major Actor in Cancerogenesis and a Potential Tool for Diagnosis and Therapy. Int J Mol Sci . 2020;21(6):2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Fujita T, Ikeda H, Taira N, et al. Overexpression of UbcH10 alternates the cell cycle profile and accelerate the tumor proliferation in colon cancer. BMC Cancer.2009;9:87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Berlingieri MT, Pallante P, Guida M, et al. UbcH10 expression may be a useful tool in the prognosis of ovarian carcinomas. Oncogene. 2007;26(14):2136–2140. [DOI] [PubMed] [Google Scholar]
- 33. Morikawa T, Kawai T, Abe H, et al. UBE2C is a marker of unfavorable prognosis in bladder cancer after radical cystectomy. Int J Clin Exp Pathol.2013;6(7):1367–1374. [PMC free article] [PubMed] [Google Scholar]
- 34. Liu Y, Zhao R, Chi S, et al. UBE2C is upregulated by estrogen and promotes epithelial-mesenchymal transition via p53 in endometrial cancer. Mol Cancer Res. 2020;18(2):204–215. [DOI] [PubMed] [Google Scholar]
- 35. Wang R, Song Y, Liu X, et al. UBE2C induces EMT through Wnt/betacatenin and PI3K/Akt signaling pathways by regulating phosphorylation levels of Aurora-A. Int J Oncol. 2017;50(4):1116–1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Xiong Y, Lu J, Fang Q, et al. UBE2C functions as a potential oncogene by enhancing cell proliferation, migration, invasion, and drug resistance in hepatocellular carcinoma cells. Biosci Rep. 2019;39(4):BSR20182384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Cao JZ, Nie G, Hu H, et al. UBE2C promotes the progression of pancreatic cancer and glycolytic activity via EGFR stabilization-mediated PI3K-Akt pathway activation. J Gastrointest Oncol. 2022;13(3):1444–1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yang YF, Chang Y-C, Tsai K-W, Hung M-H, Kang B-H.. UBE2C triggers HIF-1alpha-glycolytic flux in head and neck squamous cell carcinoma. J Cell Mol Med. 2022;26(13):3716–3725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. An YJ, Cho HR, Kim TM, et al. An NMR metabolomics approach for the diagnosis of leptomeningeal carcinomatosis in lung adenocarcinoma cancer patients. Int J Cancer. 2015;136(1):162–171. [DOI] [PubMed] [Google Scholar]
- 40. Venur VA, Chukwueke UN, Lee EQ.. Advances in management of brain and leptomeningeal metastases. Curr Neurol Neurosci Rep. 2020;20(7):26. [DOI] [PubMed] [Google Scholar]
- 41. Scott BJ, Kesari S.. Leptomeningeal metastases in breast cancer. Am J Cancer Res.2013;3(2):117–126. [PMC free article] [PubMed] [Google Scholar]
- 42. Le Rhun E, Weller M, Brandsma D, et al. ; EANO Executive Board and ESMO Guidelines Committee. EANO-ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up of patients with leptomeningeal metastasis from solid tumours. Ann Oncol. 2017;28(suppl 4):iv84–iv99. [DOI] [PubMed] [Google Scholar]
- 43. Yang JCH, Kim S-W, Kim D-W, et al. Osimertinib in patients with epidermal growth factor receptor mutation-positive non-small-cell lung cancer and leptomeningeal metastases: The BLOOM Study. J Clin Oncol. 2020;38(6):538–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Brastianos PK, et al. Phase II study of pembrolizumab in leptomeningeal carcinomatosis. J Clin Oncol. 2018;36(suppl 15):2007.29763341 [Google Scholar]
- 45. Lin F, Chandrasekaran G, de Gooijer MC, Beijnen JH, van Tellingen O.. Determination of NVP-BEZ235, a dual PI3K and mTOR inhibitor, in human and mouse plasma and in mouse tissue homogenates by reversed-phase high-performance liquid chromatography with fluorescence detection. J Chromatogr B Analyt Technol Biomed Life Sci. 2012;901:9–17. [DOI] [PubMed] [Google Scholar]
- 46. Netland IA, Førde HE, Sleire L, et al. Dactolisib (NVP-BEZ235) toxicity in murine brain tumour models. BMC Cancer. 2016;16:657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Smalley I, Law V, Wyatt C, et al. Proteomic analysis of CSF from patients with leptomeningeal melanoma metastases identifies signatures associated with disease progression and therapeutic resistance. Clin Cancer Res. 2020;26(9):2163–2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wang H, Zhang C, Rorick A, et al. CCI-779 inhibits cell-cycle G2-M progression and invasion of castration-resistant prostate cancer via attenuation of UBE2C transcription and mRNA stability. Cancer Res. 2011;71(14):4866–4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Sjodahl G, Lauss M, Lövgren K, et al. A molecular taxonomy for urothelial carcinoma. Clin Cancer Res. 2012;18(12):3377–3386. [DOI] [PubMed] [Google Scholar]
- 50. Pawitan Y, Bjöhle J, Amler L, et al. Gene expression profiling spares early breast cancer patients from adjuvant therapy: derived and validated in two population-based cohorts. Breast Cancer Res. 2005;7(6):R953–R964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Bogunovic D, O'Neill DW, Belitskaya-Levy I, et al. Immune profile and mitotic index of metastatic melanoma lesions enhance clinical staging in predicting patient survival. Proc Natl Acad Sci U S A. 2009;148(48):20429–20434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Frei E, Visco C, Xu-Monette ZY, et al. Addition of rituximab to chemotherapy overcomes the negative prognostic impact of cyclin E expression in diffuse large B-cell lymphoma. J Clin Pathol. 2013;66(11):956–961. [DOI] [PubMed] [Google Scholar]
- 53. Hanamura I, Huang Y, Zhan F, Barlogie B, Shaughnessy J.. Prognostic value of cyclin D2 mRNA expression in newly diagnosed multiple myeloma treated with high-dose chemotherapy and tandem autologous stem cell transplantations. Leukemia. 2006;20(7):1288–1290. [DOI] [PubMed] [Google Scholar]
- 54. Su Z, Fang H, Hong H, et al. An investigation of biomarkers derived from legacy microarray data for their utility in the RNA-seq era. Genome Biol. 2014;15(12):523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Yang S, He P, Wang J, et al. A Novel MIF signaling pathway drives the malignant character of pancreatic cancer by targeting NR3C2. Cancer Res. 2016;76(13):3838–3850. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.