Abstract
The Escherichia coli CheZ protein stimulates dephosphorylation of CheY, a response regulator in the chemotaxis signal transduction pathway, by an unknown mechanism. Genetic analysis of CheZ has lagged behind biochemical and biophysical characterization. To identify putative regions of functional importance in CheZ, we subjected cheZ to random mutagenesis and isolated 107 nonchemotactic CheZ mutants. Missense mutations clustered in six regions of cheZ, whereas nonsense and frameshift mutations were scattered reasonably uniformly across the gene. Intragenic complementation experiments showed restoration of swarming activity when compatible plasmids containing genes for the truncated CheZ1–189 peptide and either CheZA65V, CheZL90S, or CheZD143G were both present, implying the existence of at least two independent functional domains in each chain of the CheZ dimer. Six mutant CheZ proteins, one from each cluster of loss-of-function missense mutations, were purified and characterized biochemically. All of the tested mutant proteins were defective in their ability to dephosphorylate CheY-P, with activities ranging from 0.45 to 16% of that of wild-type CheZ. There was good correlation between the phosphatase activity of CheZ and the ability to form large chemically cross-linked complexes with CheY in the presence of the CheY phosphodonor acetyl phosphate. In consideration of both the genetic and biochemical data, the most severe functional impairments in this set of CheZ mutants seemed to be concentrated in regions which are located in a proposed large N-terminal domain of the CheZ protein.
Two-component regulatory systems allow bacteria to sense environmental changes and modify their behavior and gene expression patterns accordingly to optimally exploit available resources or survive adverse conditions (16, 31). This type of signal transduction pathway is minimally composed of a sensor kinase whose autophosphorylation state reflects environmental conditions and a response regulator whose activity is controlled by phosphoryl groups received from the sensor kinase. A fundamental prerequisite for any signal transduction pathway to provide current information is that signaling molecules must turn over on a time scale faster than that on which changes occur in the conditions being monitored. For two-component regulatory systems, there are four known classes of mechanisms by which response regulators may be dephosphorylated. First, response regulator phosphorylation occurs on aspartic acid residues and acyl phosphates are inherently labile to hydrolysis on the time scale of hours (20). Second, some response regulators possess intrinsic autodephosphorylation catalytic activities (13, 19, 26, 47). Third, some sensor kinases, either alone (1, 18) or in conjunction with ancillary proteins (19, 29), catalyze the removal of phosphoryl groups from response regulators in addition to their usual role of providing phosphoryl groups. Fourth, in some systems auxiliary proteins catalyze response regulator dephosphorylation (13, 30, 32).
The signal transduction pathway governing chemotaxis in Escherichia coli is one well-characterized example of a two-component regulatory system (43). This information-processing network enables E. coli to move in the direction of increasing attractant or decreasing repellant concentrations by controlling the phosphorylation state of the response regulator protein CheY. Transmembrane receptors detect environmental signals and suitably adjust the autophosphorylation rate of the sensor kinase CheA; CheA-P in turn acts as a phosphodonor to generate CheY-P from CheY (7, 15, 51). CheY-P loses its phosphoryl group via either an intrinsic autodephosphorylation capability or an accelerated dephosphorylation reaction in the presence of CheZ (13, 15). The balance between the rates of phosphorylation and dephosphorylation of CheY defines the intracellular concentration of CheY-P, which dictates the swimming behavior of the cell. CheY-P binds to the flagellar switch protein FliM (48, 49), which changes the direction of flagellar rotation from counterclockwise (CCW) to clockwise (CW) to produce the biased random walk which is necessary for chemotaxis. The phosphatase activity of CheZ, therefore, is critical in determining the intracellular concentration of CheY-P, and CheZ is essential for chemotaxis, as assayed by swarm formation in semisolid agar.
CheZ has been the subject of multiple investigations. The central portion of the CheZ amino acid sequence appears to be important for oligomerization (4), and the C-terminal portion binds to CheY-P (3, 27). Important questions concerning the mechanism and regulation of the CheZ-mediated dephosphorylation reaction remain. CheZ requires a Mg2+ cofactor, as does autodephosphorylation (23), but it is not known whether CheZ enhances the intrinsic autodephosphorylation activity of CheY or possesses an independent phosphatase activity. The possibility that CheZ may be regulated by other components of the chemotaxis pathway is attractive, as it could help account for the magnitude of signal amplification (or “gain”) observed in chemotaxis (9). Two interactions that affect CheZ activity have been reported, binding to a short version of CheA, CheAS (44, 45), and oligomerization with CheY-P (4–6), but the molecular basis for either potential regulatory mechanism has not been elucidated.
Most previously published studies of CheZ function have primarily employed biochemical or biophysical techniques. In order to address outstanding questions concerning the CheZ reaction mechanism, regulation, or even the potential existence of as yet unrecognized activities, a new experimental approach could be useful. CheZ does not have significant amino acid sequence similarity to any proteins currently in the sequence databases, so sequence comparison furnishes few clues concerning function. In this study, we pursued a genetic approach in which we isolated a large number of nonchemotactic CheZ mutants and mapped the corresponding cheZ mutations. The clustering of the resultant mutations suggested potential regions of functional importance in CheZ. The ability of specific pairs of different nonchemotactic cheZ mutations to complement one another and restore swarming behavior suggested the presence of at least two independent functional domains in CheZ. Finally, an investigation of the ability of selected mutant proteins to stimulate dephosphorylation of CheY and to undergo oligomerization gave information about specific regions or residues in CheZ which are important for function.
MATERIALS AND METHODS
Bacterial strains and plasmids.
KO642 is identical to RP1616, which carries cheZΔ6725 (22). The cheZΔ6725 strain KO642recA (9) and the mutD5 strain NR9458 (35) have been described previously.
Plasmid pKCB1 was constructed by the dut ung method of site-directed mutagenesis (21) to introduce an XhoI site at nucleotides 320 to 325 in the cheZ gene of the ptrpcheYZoripBR322 plasmid pRBB40 (8) without altering the amino acid sequence of CheZ. Plasmid pKCB2 was constructed by ligating the 2.0-kb EcoRI-BclI fragment carrying ptrpcheYcheZ from pKCB1 to the 3.5-kb EcoRI-BclI fragment of pACYC184 (10), transforming the resulting product into KO642, and selecting for Tetr. Derivatives of pKCB2 carrying cheZQ116(Oc), cheZQ142(Am), cheZQ190(Am), or cheZV205E alleles from mutant pKCB1 isolates were analogously constructed.
Isolation and identification of cheZ mutations.
Bacteria carrying the mutD5 allele exhibit mutation frequencies that are 50 to 100 times higher when the bacteria are grown in rich media than when they are grown in minimal media (11). Cells of NR9458 were therefore grown in minimal medium containing 1× M9 salts, 0.4% (wt/vol) glucose, 0.023% (wt/vol) proline, 1 μg of thiamine/ml, and 1 mM MgSO4, made competent by standard treatment with cold CaCl2, and transformed with pKCB1. After 1 h of growth at 37°C, the transformation cultures were diluted 10- or 100-fold in Luria-Bertani (LB) medium (1% [wt/vol] tryptone, 1% [wt/vol] NaCl, 0.5% [wt/vol] yeast extract) containing 100 μg of ampicillin/ml, and incubation continued overnight at 37°C. Mutagenized pKCB1 was isolated directly from NR9458 transformation cultures, transformed into KO642recA, and plated in Swarm Screening top agar (1% [wt/vol] tryptone, 0.5% [wt/vol] NaCl, 0.35% [wt/vol] Bacto agar) on Swarm Screening agar plates containing 100 μg of ampicillin/ml. After overnight incubation at 30°C, transformants were identified as deficient in swarming behavior by visual screening. Candidate mutants were single-colony purified, and their swarm phenotypes were confirmed by stabbing into motility (Mot) agar plates (1% [wt/vol] tryptone, 0.5% [wt/vol] NaCl, 0.3% [wt/vol] Bacto agar) with single colonies of KO642recA and KO642recA/pKCB1 as Che− and Che+ controls, respectively. Diameters of resultant swarms were measured after 9 h at 30°C. Plasmid DNA from candidate mutants (those with smaller swarms than the KO642recA/pKCB1-positive control) was isolated and retransformed into KO642recA. Ampr transformants were tested on Mot plates following single-colony purification to confirm plasmid linkage of the mutant phenotypes.
pKCB1 DNA from candidate mutants was subcloned to separate potential mutations in cheZ and cheY. Wild-type and mutant pKCB1s were double digested with EcoRI and BsmI, which cuts between codons 10 and 11 of cheZ. Following separation by electrophoresis on 1% (wt/vol) agarose gels, the two mutant pKCB1 fragments were each paired with the opposite wild-type fragment, ligated, and transformed into KO642recA. The presence of a mutation in the cheY or cheZ restriction fragment was then deduced from the swarm phenotypes on Mot plates of Ampr transformants in each pair of transformations. Confirmation that mutations responsible for reduced swarming were actually in the cheZ gene and not in the vector sequence was obtained in analogous subcloning experiments using other combinations of restriction enzymes. All cheZ mutations were mapped in this manner as being either between BsmI and XhoI sites in cheZ or between the XhoI site in cheZ and the StyI site in the partial flhB′ gene immediately 3′ of cheZ. We are aware of no reason to suspect that a mutation in flhB′ on pKCB1 could affect swimming behavior and therefore assume that mutations identified within cheZ by sequencing are responsible for the phenotypes produced by mutations mapping between XhoI and StyI sites.
The sequence of the entire cheZ gene in each mutant pKCB1 plasmid was determined at the University of North Carolina-Chapel Hill Automated DNA Sequencing Facility on a model 373A DNA sequencer (Applied Biosystems) using the Taq DyeDeoxy terminator cycle sequencing kit (Applied Biosystems).
Characterization of CheZE134K.
After all mutants had been isolated, DNA sequencing revealed that “wild-type” pKCB1 carried an inadvertent cheZE134K mutation. The same mutation is present in the two plasmid precursors of pKCB1, namely, pRBB40 (8) and the M. I. Simon laboratory isolate of pRL22 (25), but not in a pRL22 isolate from the A. J. Wolfe laboratory. Thus, unless stated otherwise, all cheZ alleles described in this report carry both the cheZE134K mutation, which for the sake of simplicity is not explicitly listed, and the given cheZ mutation. To maintain consistency, the designation wild type in this paper refers to the cheZE134K allele.
A version of pKCB1 carrying the genuine wild-type cheZ gene was created by site-specific mutagenesis, and the properties of CheZ and CheZE134K were compared both in vivo and in vitro. KO642recA/pKCB1 with Lys at position 134 is chemotactic and swarms on Mot plates at 80% of the rate of KO642recA/pKCB1 with Glu at position 134 (the true wild type). The in vitro Pi release assay described below showed that CheZE134K had a specific activity which was greater than 90% of that of the true wild-type CheZ. Our data are consistent with that of Sanna and Simon, who independently isolated cheZE134K and found that it caused a mild swarm defect and reduced CheY-P dephosphorylation activity about twofold (33).
Tethered-cell assay.
Flagellar rotational bias was determined using the tethered-cell assay as previously described (9).
Intragenic complementation.
For intragenic complementation experiments, competent KO642/pKCB2 cells carrying the appropriate cheZ allele were transformed with the desired mutant pKCB1 plasmids and plated on LB plates with 10 μg of tetracycline and 100 μg of ampicillin/ml. Transformants were single-colony purified, and their swarm phenotypes were determined by triplicate stabs into Mot agar plates. On each plate, KO642/pKCB1 was present as a positive control and KO642/pKCB2/pKCB1 with both plasmids carrying the cheZQ190(Am) mutation was present as a negative control. Diameters of resultant swarms were measured over the course of 14 h at 30°C. Swarm rates were determined and expressed as the rate relative to that of the positive control on the same plate, and the triplicate values were averaged for each test strain.
To rule out the possibility that swarming behavior in strains carrying two cheZ plasmids was a result of recombination rather than complementation, plasmid DNA was isolated from bacteria at the edge of swarms and transformed into KO642. Transformations were spread on LB plates containing 10 μg tetracycline/ml and on LB plates containing 100 μg of ampicillin/ml. Transformants were single-colony purified, stabbed into Mot plates to determine swarm phenotype, and patched onto LB plates containing both tetracycline and ampicillin (at the same concentrations specified above) to ascertain that transformants supporting swarming contained both plasmids.
Purification of CheZ and CheY.
Wild-type and mutant CheZ proteins were purified from the appropriate KO642recA/pKB1 strain as described previously (14), except that Whatman DE-52 was used for the ion-exchange step rather than MonoQ in order to increase the scale of the purification. For the DE-52 step, CheZ was eluted with a 200- by 200-ml gradient of 25 mM Tris (pH 7.5)–5 mM MgCl2–10% glycerol, with the limiting buffer containing 500 mM NaCl. CheY was purified from strain KO641recA/pRBB40 (8) as described previously (14). CheZL90S and CheZF117S exhibited some degradation during purification. However, full-length mutant proteins could be easily isolated in all cases.
Dephosphorylation assays.
The abilities of wild-type and mutant CheZ proteins to dephosphorylate CheY-P were compared using gel electrophoretic analysis of steady-state concentrations of [32P]CheY-P. In a 10-μl reaction mixture, 70 pmol of CheA, 70 pmol of CheY, and 3.5, 14, 70, or 350 pmol of wild-type or mutant CheZ were incubated in a solution containing 50 mM KCl, 10 mM MgCl2, and 50 mM Tris-HCl, pH 8.0. Reactions were initiated with the addition of [γ-32P]ATP to a final concentration of 0.3 mM ATP. After 2.5 min, the reactions were stopped with the addition of 2× sodium dodecyl sulfate (SDS) sample buffer (0.125 M Tris-Cl [pH 6.8], 4% SDS, 20% glycerol, 10% 2-mercaptoethanol), and the proteins were separated by SDS-polyacrylamide gel electrophoresis (PAGE) on 18% polyacrylamide gels, so that Pi and ATP migrated off the end of the gels. The gels were dried and subjected to analysis on a Molecular Dynamics Storm 860 phosphorimaging system.
The phosphatase activities of the CheZ variants were also assessed by measuring the rate of release of inorganic phosphate from reaction mixtures containing CheY-P and CheZ using the EnzChek phosphate assay kit (Molecular Probes). This is an enzyme-based spectroscopic assay in which Pi and 2-amino-6-mercapto-7-methylpurine riboside (MESG) act as substrates for phosphonucleoside phosphorylase to rapidly produce ribose-1-phosphate and 2-amino-6-mercapto-7-methylpurine with an accompanying change in extinction coefficient at 360 nm (46). The enzymatic conversion is rapid so that the kinetics reflect the rate of production of Pi in the system of interest. In the present application, CheY-P was produced in situ by mixing monophosphoimidazole (MPI) (39), CheY, and Mg2+, a necessary cofactor. A Beckman DU7500 microprocessor-controlled diode array spectrophotometer interfaced to a high-performance temperature controller was used for spectroscopic measurements. In a cuvette, MESG (200 μM), MPI (3 mM), and CheZ (0 to 12 μM) were mixed in a total volume of 440 μl containing 20 mM Tris, pH 7.5, and 10 mM MgCl2. While the absorbance at 360 nm was being monitored, nucleoside phosphorylase (0.45 U) was added. This resulted in an increase in absorbance (about 0.2 absorbance units) due to contaminating Pi in the MPI preparation, which stabilized within 30 s. To initiate the flow of phosphate through CheY and CheZ, CheY (6 μM) was added, and the rate of absorbance at 360 nm, reflecting the release of Pi from CheY, was recorded. The CheZ-dependent rate was the difference in rates observed in the presence and absence of CheZ. The temperature was maintained at 25°C. Rates calculated by the instrument software (absorbance per unit time) were converted to micromolar Pi per unit time by using a standard curve relating Pi concentration to absorbance at 360 nm, as described in the EnzChek protocol.
Peptide binding assay.
The association of CheY-P with peptides which correspond to the C-terminal 19 residues of CheZ (wild type or CheZV205E) was monitored by intrinsic protein fluorescence as described previously (27). The peptides were synthesized by the University of North Carolina PMBB Micro Protein Chemistry Facility and used as 5 mM stock solutions.
Protein cross-linking.
Chemical cross-linking of CheY and wild-type or mutant CheZ using 1-ethyl-3-(dimethylaminopropyl)carbodiimide hydrochloride and N-hydroxysuccinimide was carried out exactly as described previously (5). Samples were electrophoresed on SDS–10% polyacrylamide gels with the inclusion of Kaleidoscope prestained standards (Bio-Rad) and biotinylated ECL protein molecular weight markers (Amersham). The separated proteins were then transferred from gels to 0.45-μm-pore-size nitrocellulose (Optitran; Schleicher and Schuell). Blots were probed with a 1:400-diluted mouse monoclonal antibody against wild-type CheY (36), washed, and probed with a 1:10,000-diluted horseradish peroxidase-linked goat anti-mouse whole-immunoglobulin antibody (Sigma) and 1:1,500-diluted streptavidin-horseradish peroxidase (Amersham). The ECL Western blotting protocol (Amersham) was followed for the blocking, washing, and detection of blots. Following exposure to detection reagents, blots were exposed to Kodak X-OMAT scientific imaging film for 10 s.
Mass spectrometry.
Electrospray ionization mass spectrometry was performed at the Duke University Medical Center Biomolecular Mass Spectrometry Laboratory. Data were collected using a Micromass (formerly Fisons)-VG Quattro BQ triple-quadrupole mass spectrometer equipped with a pneumatically assisted electrostatic ion source operating at atmospheric pressure and calibrated with horse heart myoglobin (16,951.48 amu).
RESULTS
Isolation of nonchemotactic CheZ mutants.
Random mutagenesis of cheZ was accomplished by passage of pKCB1, which carries cheYZ, through a strain defective in mismatch repair and subsequent transformation of mutagenized pKCB1 into KO642recA, which lacks cheZ. (Note that pKCB1 carries an inadvertent cheZ mutation, and therefore all mutant cheZ alleles described in this paper actually contain cheZE134K in addition to the reported mutation. See additional information in Materials and Methods.) Approximately 62,000 transformants were screened for a partial or complete loss of chemotactic ability using swarm assays; 130 transformants were found to carry mutations in pKCB1 that interfered with chemotaxis. The cheZ and cheY genes of pKCB1 from each of the mutants were subcloned and retested to determine which gene(s) harbored the mutation(s). Of the 130 pKCB1 mutants, 107 contained mutations only in cheZ, 10 had mutations only in cheY, and in 13 candidates the mutations could not easily be identified.
Each mutant cheZ gene was sequenced to determine the location of the mutation(s). One of the mutants had multiple cheZ mutations and was not considered further; the remaining 106 mutants possessed single mutations. There were 79 missense mutations comprising 48 different mutations at 37 different codons (Table 1). Fourteen nonsense mutations (at the codons for Lys7, Gln39, Gln77, Gln82 [twice], Trp94, Trp97 [twice], Gln116, Gln142 [twice], Gln147, and Gln190 [twice]) and 13 frameshift mutations (at the codons for Glu34, Glu46, Ala69, Lys88 [twice], Leu104, Ala105, Gln131, Ala138, Glu160, Asn170, Lys179, and Gly188) were isolated. Mutants carrying missense mutations exhibited a range of behavioral phenotypes (Table 1), whereas all nonsense and frameshift mutations resulted in complete loss of swarming ability and exclusively, CW flagellar rotation. A map (Fig. 1) displaying the locations types (missense, nonsense/frameshift), and phenotypes (loss or gain of function; partial or complete loss of swarming behavior) of cheZ mutations identified here and in other laboratories (4, 17, 33, 34, 40) has several striking features. Missense mutations seem to cluster in specific regions of cheZ, which are different for gain- and loss-of-function mutations. In contrast, nonsense and frameshift mutations appear to be distributed more randomly through the gene.
TABLE 1.
cheZ missense mutations
| Codon | Mutation
|
No. of isolates | Phenotype
|
||
|---|---|---|---|---|---|
| Wild-type amino acid | Mutant amino acid | Swarm diametera | CW flagellar rotationb | ||
| 47 | Ala | Asp | 1 | — | ++++ |
| 48 | Ile | Thr | 3 | 0.13 | + |
| 57 | Tyr | Cys | 2 | 0.25 | ++ |
| His | 3 | 0.25 | + | ||
| 58 | Val | Ala | 1 | 0.25 | +++ |
| 63 | Ala | Val | 1 | 0.17 | ++ |
| 64 | Gln | Pro | 2 | — | ++++ |
| 65 | Ala | Gly | 2 | 0.21 | + |
| Thr | 3 | — | +++ | ||
| Val | 3 | — | ++++ | ||
| 66 | Ala | Glu | 1 | — | ++++ |
| Thr | 4 | 0.21 | ++ | ||
| Val | 1 | 0.29 | +/− | ||
| 67 | Glu | Gly | 2 | 0.29 | + |
| Lys | 1 | 0.17 | +++ | ||
| 69 | Ala | Glu | 2 | — | ++++ |
| 70 | Leu | Pro | 1 | — | ++++ |
| 73 | Val | Ala | 3 | 0.17 | + |
| 74 | Glu | Lys | 2 | — | ++++ |
| 76 | Ser | Leu | 3 | 0.25 | + |
| 83 | Met | Thr | 1 | — | ++++ |
| 86 | Ser | Pro | 3 | 0.29 | + |
| 87 | Ala | Gly | 1 | — | ++++ |
| Val | 1 | — | ++++ | ||
| 90 | Leu | Ser | 1 | — | ++++ |
| 110 | Leu | Pro | 1 | — | ++++ |
| 114 | Thr | Ala | 1 | — | ++++ |
| 117 | Phe | Leu | 2 | 0.33 | + |
| Ser | 1 | — | ++++ | ||
| 124 | His | Tyr | 1 | — | +++ |
| 132 | Leu | Pro | 1 | — | ++++ |
| 137 | Met | Ile | 1 | 0.21 | + |
| Lys | 1 | — | ++++ | ||
| 138 | Ala | Val | 5 | — | ++++ |
| 139 | Gln | Arg | 1 | 0.08 | +++ |
| 140 | Asp | Asn | 1 | 0.25 | ++ |
| 141 | Phe | Leu | 1 | — | ++++ |
| 143 | Asp | Glu | 2 | — | +++ |
| Gly | 1 | — | ++++ | ||
| Val | 3 | — | ++++ | ||
| 145 | Thr | Pro | 1 | — | ++++ |
| 159 | Ile | Thr | 1 | 0.25 | + |
| 186 | Leu | Pro | 1 | — | +++ |
| 188 | Gly | Glu | 1 | — | ++++ |
| 189 | Pro | Leu | 1 | — | +++ |
| 204 | Gln | Arg | 1 | 0.33 | +/− |
| 205 | Val | Glu | 1 | — | ++++ |
| 206 | Asp | Gly | 1 | 0.50 | ++ |
Diameters of swarms on Mot plates, expressed as a fraction of the diameter of the swarm of the wild-type control. —, swarms with diameters identical to that of the negative control (2 mm).
Degree of CW flagellar rotation of tethered cells. ++++, exclusively CW rotation; +/−, CW rotation with frequent reversal of direction.
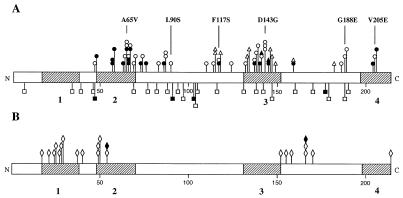
FIG. 1.
cheZ mutations resulting in a nonchemotactic phenotype. The CheZ protein is shown, with locations of amino acid substitutions resulting from mutations indicated. Cross-hatched regions, regions conserved among E. coli, Salmonella enterica serovar Typhimurium, P. aeruginosa, and P. putida with the following proposed functions: 1, unknown; 2, unknown; 3, oligomerization (4); 4, CheY binding (3). (A) Loss-of-function mutations, which result in an increase in the CW bias of flagellar rotation. Missense mutations are above the bar; nonsense and frameshift mutations are below. ○, complete loss-of-function missense mutations from this study; ●, partial loss-of-function missense mutations from this study; ▵ and ▴, missense mutations from reference 33 and from references 4 and 40, respectively; □ and ■, nonsense and frameshift mutations from this study and reference 40, respectively. The six mutant proteins chosen for further analysis are indicated. (B) Gain-of-function missense mutations, which result in a decrease in the CW bias of flagellar rotation, from references 33 and 34 (◊) and 17 and 40 (⧫).
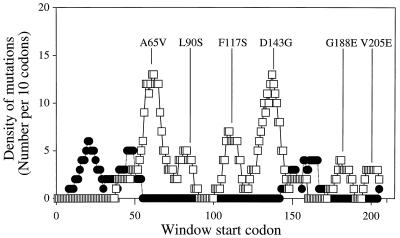
When the density of cheZ loss-of-function missense mutations was graphed against codon position, six peaks were evident (Fig. 2). These peaks presumably reveal functionally critical portions of the protein and are centered on residues 66, 86, 114, 141, 185, and 205. One mutant from each of the six clusters of cheZ loss-of-function mutations was selected for biochemical characterization. The six selected mutants were CheZA65V, CheZL90S, CheZF117S, CheZD143G, CheZG188E, and CheZV205E; the location of each is shown in Fig. 1 and 2. All of these mutations resulted in complete loss of swarming and each affects a residue strictly conserved in the four published CheZ protein sequences (12, 24, 28, 42). Western blot analysis showed that all six of the selected mutant strains expressed CheZ to levels similar to those for strains which contained wild-type cheZ (data not shown), suggesting that the phenotypes were due to alteration of function rather than changes in intracellular CheZ concentration.
FIG. 2.
The numbers of unique cheZ mutations from this and previous studies (4, 17, 33, 34) falling within consecutive windows of 10 codons were determined. The first window included codons 1 to 10, and the final window included codons 205 to 214, for a total of 205 windows. The location of each mutant chosen for biochemical testing is designated as the window in which the mutation falls at the central codon. □, loss-of-function mutations; ●, gain-of-function mutations.
Chemotaxis depends on the balance between tumbling and smooth-swimming behavior, so either gain-of-function mutations, resulting in increased CCW flagellar rotation and increased smooth-swimming behavior, or loss-of-function mutations, resulting in increased CW flagellar rotation and increased tumbling behavior, could result in a loss of chemotaxis. To distinguish between these possibilities, the tethered-cell assay was used to determine the direction of flagellar rotation in each mutant. Each mutant showed some degree of increased CW bias of flagellar rotation and therefore increased tumbling behavior, indicating that all mutants isolated in this study were loss-of-function mutants. The degree of swarming capability retained by each mutant generally correlated inversely to the degree of increased CW bias. Mutants showing partial swarming had less CW biases than mutants showing a complete loss in swarming behavior, consistent with previous descriptions of bacterial migration in semisolid agar (50).
Intragenic complementation of cheZ mutations.
CheZ is a dimer (5). We subcloned several cheZ mutations from pKCB1 into compatible plasmid pKCB2 and determined whether coexpression of various pairs of cheZ mutations could restore swarming activity to the ΔcheZ strain KO642. The presence of CheZ1–189 (the product of cheZQ190(Am), i.e., an amber mutation in the cheZ codon for Gln190) significantly enhanced swarm formation by strains also expressing CheZA65V, CheZL90S, or CheZD143G but not CheZF117S or CheZG188E (Table 2). These intragenic complementation results are consistent with the presence of at least two independent functional domains in CheZ that can be supplied by different subunits of a heterodimer. To further explore the boundaries of such putative domains, all 15 possible combinations of cheZA65V, cheZL90S, cheZF117S, cheZD143G, or cheZG188E on pKCB1 and cheZQ116(Oc), cheZQ142(Am), or cheZV205E on pKCB2 were tested in KO642. However, none gave swarms that were distinguishable from those observed with KO642 alone (data not shown).
TABLE 2.
Intragenic complementation of cheZ mutations
| cheZ allele on pKCB1 | Swarm ratea of:
|
|
|---|---|---|
| KO642/pKCB1 | KO642/pKCB1/pKCB2. cheZQ190(Am) | |
| Wild type | 1.0 | 0.92 |
| cheZA65V | 0.03 | 0.21 |
| cheZL90S | 0.03 | 0.23 |
| cheZF117S | 0.02 | 0.05 |
| cheZD143G | 0.03 | 0.38 |
| cheZG188E | 0.08 | 0.04 |
| cheZQ190(Am) | 0.02 | 0.02 |
Rates of swarm formation on Mot plates, expressed as a fraction of the swarm rate of K0642/pKCB1.
It was possible that restoration of the swarming observed in the Table 2 experiment was a result not of complementation but rather of genetic recombination between the two mutant copies of cheZ to recreate wild-type CheZ. Two tests performed with bacteria isolated from the outer portions of swarms of KO642/pKCB1/pKCB2 strains, where recombinants might be expected, are inconsistent with recombination. First, both plasmids were necessary to observe swarming. All isolates that produced swarms on Mot plates following growth on media selecting for only one of the two plasmids actually carried both plasmid-encoded antibiotic resistances (data not shown). Conversely, those isolates that did not support swarming following growth in the presence of only one antibiotic were sensitive to the other antibiotic. Second, the flagellar rotation patterns of bacteria isolated from the edges of swarms were similar to the flagellar rotation patterns of the original transformants and different from that supported by wild-type cheZ. Finally, the fact that some pairs of cheZ alleles produced swarms whereas others did not, in a pattern that was not correlated with the physical distance between mutations, is also inconsistent with recombination.
Dephosphorylation-stimulating activity of mutant CheZ proteins.
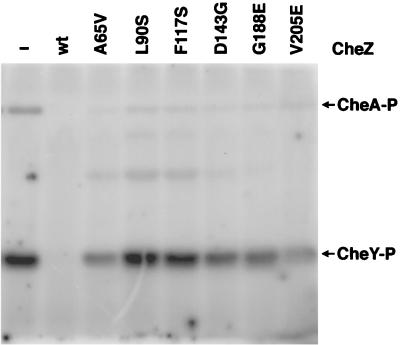
Each of the six chosen mutant CheZ proteins was purified and tested for the ability to dephosphorylate wild-type CheY-P in two different in vitro assays. In the first assay, equimolar amounts of CheA and CheY were incubated for 2.5 min with [γ-32P]ATP and quantities of wild-type or mutant CheZ proteins which spanned a 100-fold concentration range. The reactions were quenched, the products were separated by electrophoresis, and the amount of CheY-P present was assessed by phosphorimaging. At a molar ratio of 1 CheA:1 CheY:0.05 CheZ, the amount of CheY-P formed was reduced approximately fourfold by the presence of wild-type CheZ but was not significantly affected by any of the mutant CheZ proteins (data not shown). At a molar ratio of 1 CheA:1 CheY:0.2 CheZ, no CheY-P was detected in the presence of wild-type CheZ, whereas the mutant CheZ proteins still had no significant effect (data not shown). At a molar ratio of 1 CheA:1 CheY:1 CheZ, the mutant CheZ proteins reduced the amount of CheY-P by at most twofold compared to the control reaction lacking CheZ (data not shown). Even at a molar ratio of 1 CheA:1 CheY:5 CheZ, detectable amounts of CheY-P were found in the presence of each of the six mutant CheZ proteins (Fig. 3). The dramatic differences in the sensitivity of CheY-P to the presence of wild-type or mutant CheZ proteins in this crude assay suggest that each of the mutant CheZ proteins is profoundly deficient in the ability to stimulate dephosphorylation of CheY-P.
FIG. 3.
Ability of mutant CheZ proteins to reduce the concentration of [32P]CheY-P. Wild-type CheA, wild-type CheY, and indicated (none [−], wild-type [wt], or mutant) CheZ proteins in a 1:1:5 molar ratio were incubated with [γ-32P]ATP for 2.5 min. Reaction products were separated by SDS-PAGE and visualized with a phosphorimager.
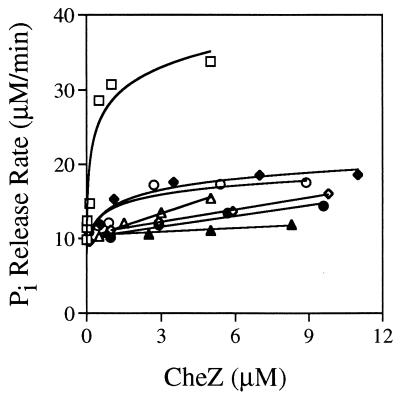
Given that the CheZ and CheA binding surfaces on CheY are believed to overlap (52) and given the 5 CheZ:1 CheY molar ratio used, the mutant CheZ proteins could conceivably reduce the amount of CheY-P observed in Fig. 3 by interfering with phosphotransfer from CheA to CheY rather than by stimulating dephosphorylation of CheY-P. To more accurately quantitate the dephosphorylation-stimulating abilities of the mutant CheZ proteins, we developed a new assay which measured the steady-state rate of Pi release instead of the amount of CheY-P at a single time point. In this assay, CheY-P is continuously generated by autophosphorylation with a large molar excess of MPI and Pi is detected spectrophotometrically following a rapid reaction with commercially available assay reagents. The addition of increasing amounts of wild-type or mutant CheZ proteins increases the steady-state rate of Pi release (Fig. 4). The upper limit of CheZ activity reflects the maximal rate of CheY-P formation through autophosphorylation (i.e., CheY-P is dephosphorylated as fast as it can be made). Note that all assays project back to a common Pi release rate at zero CheZ concentration, which reflects both the autophosphorylation and autodephosphorylation rates of CheY. Specific activities of the various CheZ proteins were determined by measuring the concentration dependence of the CheZ-stimulated component of the Pi release rate in the linear portion of the curves, i.e., at low CheZ concentrations. In decreasing order, the mutant CheZ proteins had the following activities in comparison to that of wild-type CheZ: CheZG188E, 16%; CheZV205E, 6.8%; CheZF117S, 3.0%; CheZA65V, 1.5%; CheZL90S, 1.3%; CheZD143G, 0.45%. CheZF117S and CheZA65V were previously reported to have ∼40 and ∼25% of the dephosphorylating activity of CheZE134K, respectively, on the basis of a different assay (33). The truncated CheZ1–189 protein did not exhibit detectable activity in the Pi release assay (data not shown).
FIG. 4.
The concentration dependence of dephosphorylation-stimulating activity of various CheZ proteins was assessed using the EnzChek phosphate assay kit from Molecular Probes, which converts Pi to a product that can be measured spectrophotometrically at 360 nm. CheY-P was continuously generated from CheY and MPI, and the rate of Pi release was measured continuously in the presence of various concentrations of CheZ. The total concentration of CheY in the assay was 6 μM. Data for wild-type CheZ (□), CheZA65V (◊), CheZL90S (●), CheZF117S (▵), CheZD143G (▴), CheZG188E (⧫), and CheZV205E (○) are shown. Data points are the mean values of at least two independent assays.
Complex formation by mutant CheZ proteins.
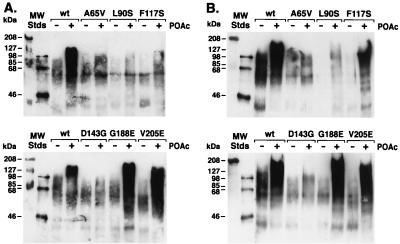
CheZ has been reported to form oligomers of composition ([CheZ]2 · CheY)4–5 in the presence of CheY-P (5), and oligomerization has been correlated with the dephosphorylation-stimulating activity of CheZ (4, 6). The phosphorylation-dependent ability of mutant CheZ proteins to promote the formation of large-molecular-weight complexes containing CheY was assessed in a cross-linking assay (5) (Fig. 5). The molecular weights of the products of cross-linking wild-type CheZ to CheY increased in the presence of the CheY phosphodonor acetyl phosphate. CheZG188E, CheZV205E, and CheZF117S formed complexes similar to those obtained with wild-type CheZ. CheZA65V, CheZL90S, and CheZD143G did not promote the formation of high-molecular-weight complexes. The prominent CheY-containing band at about 60 kDa observed with the latter set of mutant CheZ proteins and with all CheZ proteins in the absence of phosphorylation is consistent with crosslinking of a CheZ dimer to a CheY monomer (expected molecular mass, 62 kDa).
FIG. 5.
Cross-linking of CheY and CheZ proteins. Wild-type (wt) and mutant CheZ proteins were incubated with chemical cross-linking agents and CheY in the presence or absence of the phosphodonor acetyl phosphate (POAc) as previously described (5). Reaction products were separated by SDS-PAGE, transferred to a nitrocellulose membrane, probed with anti-CheY antibody, and detected by enhanced chemiluminescence. Note that for optimal resolution of high-molecular-weight complexes, monomeric CheY (14 kDa) has been electrophoresed off the end of the gels. Panels A and B display the results of duplicate experiments.
Binding of CheZ peptides to CheY-P.
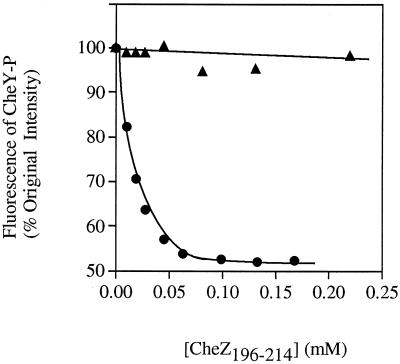
A peptide consisting of the 19 C-terminal amino acids of CheZ has been shown to bind CheY, and the affinity is increased significantly when CheY is phosphorylated (3, 27), implicating the C terminus of CheZ in binding interactions with CheY (3). Therefore it was notable that CheZV205E, which has an amino acid substitution in the putative CheY binding region, behaved similarly to wild-type CheZ in the cross-linking assay (Fig. 5). Presumably, the binding of CheZ to CheY-P is a prerequisite to forming the high-molecular-weight cross-linked products observed in this assay. To further explore the impact of the CheZV205E substitution on binding to CheY-P, we compared the interaction of CheY-P with peptides that correspond to the C-terminal 19 amino acids of wild-type and CheZV205E using fluorescence (Fig. 6) (27). Whereas the wild-type peptide produced the predicted fluorescence quench indicative of binding (Kd, ∼19 μM), the mutant peptide caused no significant change in fluorescence. Assuming that the absence of fluorescence change in the experiment using the mutant peptide was due to inability to bind (rather than binding which did not affect fluorescence), it appears that the single Val-to-Glu substitution severely affected the interaction of the peptide with CheY-P. Therefore, this mutation has a large impact on the binding between the CheZ peptide and CheY but does not appear to affect the interaction between full-length CheZ and CheY, as measured by the cross-linking assay.
FIG. 6.
Fluorescence detection of the interaction of CheZ peptides with CheY-P. Nineteen-amino-acid peptides corresponding to the C terminus of wild-type CheZ (AGVVASQDQVDDLLDSLGF) (●) or CheZV205E (AGVVASQDQEDDLLDSLGF) (▴) were added to CheY (20 μM) in the presence of phosphoramidate (100 mM) in 50 mM NaPi (pH 7.2)–20 mM MgCl2. The fluorescence intensity of CheY-P (λex = 285 nm; λem = 340 nm) was monitored after each addition, corrected for dilution due to volume changes, and expressed as the percentage of the fluorescence of CheY-P in the absence of added peptide.
CheZ degradation product.
A degraded form of CheZ is often observed during purification (42). When one of our preparations of CheZE134K exhibited degradation during storage, it was subjected to mass spectral analysis, which revealed two species. The mass of the major species was 23,988.9 ± 2.4 amu, in good agreement with the mass expected (23,989.18 amu) for CheZE134K methylated at the N terminus (38, 41). The mass of the minor species was 20,574.9 ± 2.0 amu, consistent with either N-methylated CheZE134K1–181 (expected mass, 20,575.48 amu) or CheZE134K17–198 (expected mass, 20,575.44 amu). CheZE134K1–181 is the more probable identification because CheZE134K17–198 would require multiple proteolytic events and no intermediate single-cleavage products (i.e., CheZE134K1–198 or CheZE134K17–214) were observed.
DISCUSSION
Mutagenesis.
The random mutagenesis used to generate cheZ mutations that do not support chemotaxis was designed to obtain as large a pool of mutant CheZ proteins as possible. The number of different cheZ mutations that potentially could be isolated by our method depends on the randomness of the mutagenesis itself, the degree of saturation of the search (related to how many candidate mutants were screened), and the particular screening assay employed to identify nonchemotactic mutants. Among the single mutations isolated were examples of 11 of the 12 possible single base substitutions (only G→C was not observed), but most were transition rather than transversion mutations (data not shown). A sense of the high degree of saturation of the mutagenesis comes from the fact that we obtained many mutations multiple times (Table 1) and also reisolated 4 of the 17 previously described loss-of-function cheZ missense mutations and 2 of the 3 previously described cheZ nonsense mutations. Gain-of-function mutations are expected to be much less common than loss-of-function mutations, and we did not isolate any using this screening method. Most of the previously described gain-of-function CheZ mutants were isolated using a screen designed specifically to identify such mutants (33). Although additional nonchemotactic CheZ mutants presumably remain to be discovered, this is by far the largest collection of cheZ mutations ever reported.
Distribution of mutations.
Several conclusions can be drawn from the observed distribution of cheZ mutations altering CheZ function. First, the nonrandom distribution of loss-of-function missense mutations, which seem to cluster in six distinct segments of the cheZ gene (Fig. 2), suggests that the missense mutations may define discrete regions in the CheZ protein critical for structure and/or function. Although loss-of-function CheZ mutants have been identified in other studies, (4, 33, 40), they have not been of sufficient quantity to elucidate the six clusters revealed here. In contrast to the missense mutations, the nonsense and frameshift mutations appear to be distributed fairly randomly through cheZ (Fig. 1), although the relatively small number of such mutations in our sample does not allow a firm conclusion. Nonsense and frameshift mutations would be expected to impair CheZ function regardless of their locations, so long as the C-terminal portion of the protein is essential for function, and the C-terminal region of CheZ has previously been implicated in CheY binding (3). The failure of the nonsense and frameshift mutations to cluster is consistent with a mutagenesis protocol that introduces lesions into the cheZ gene at random locations.
Second, loss-of-function mutations and gain-of-function mutations cluster in different regions of cheZ (Fig. 2), a finding that extends previous findings (33). With the exception of a mutation at residue 214, gain-of-function mutations are found in two regions, between residues 17 and 54 and between residues 152 and 170. Sanna and Simon (33) suggested that these might define two regions of interaction with CheY. Our graphical analysis (Fig. 2) further suggests that the region between residues 17 and 54 may actually contain two clusters of gain-of-function mutants. The large number of loss-of-function mutations reported here, like those described by Sanna and Simon (33), are found almost exclusively in regions of cheZ separate from those regions defined by gain-of-function mutations.
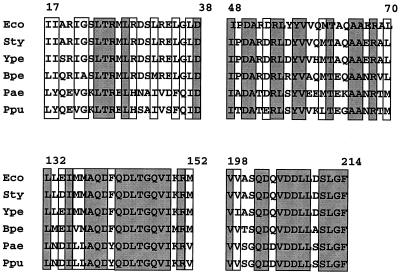
Third, several of the clusters defined in this study coincide with regions of CheZ which are highly conserved among sequenced cheZ genes of six species (Fig. 7). Notably, the two largest clusters (those at residues 132 to 149 and residues 57 to 76) correspond to regions of CheZ with extremely high conservation. Similarly, the C-terminal cluster (residues 204 to 206) is located in a region of nearly 100% amino acid sequence identity among the six species. Highly conserved regions of CheZ presumably suggest regions of functional or structural significance; clustering of mutations at these regions of the gene is consistent with their inferred importance. However, this was not the case with all of the clusters. The clusters corresponding to residues 83 to 90, residues 110 to 118, and residues 182 to 189 are in regions of relatively low sequence homology between the various CheZ proteins. No loss-of-function mutations were found in a fourth region of CheZ (residues 17 to 38), which possesses a high degree of sequence conservation (Fig. 7), but one cluster of gain-of-function mutations was located there (Fig. 2).
FIG. 7.
Sequence alignment of the four regions of CheZ proteins which display a high degree of sequence conservation. Amino acid sequences from E. coli (Eco), S. enterica serovar Typhimurium (Sty), P. aeruginosa (Pae), and P. putida (Ppu) were obtained from published reports (12, 24, 28, 42). The presence of a cheZ gene in the genomes of Bordetella pertussis (Bpe) and Yersinia pestis (Ype) was indicated by a TBlastn search for E. coli cheZ. The portions of those CheZ sequences which were not available from the TBlastn search results were obtained by manual translation of nucleotide sequences produced by the B. pertussis and Y. pestis sequencing groups at the Sangre Centre (ftp://ftp.sanger.as.uk/pub/). Note that the B. pertussis and Y. pestis sequences are tentative and await confirmation. Gray shading, amino acid sequence identity; no shading, conservation of the chemical nature of the side chain (acidic, basic, polar, or nonpolar).
The domain structure of CheZ.
Of the five cheZ missense mutations tested, three of the four located nearest the 5′ end of the gene complemented a nonsense mutation located at the 3′ end of the cheZ gene (Table 2). The simplest interpretation of this pattern of intragenic complementation is that CheZ has at least two independent functional domains that can be supplied by different subunits of a heterodimer. In this view, CheZ mutants CheZA65V, -L90S, and -D143G would have functional C-terminal but not N-terminal domains, whereas CheZG188E and CheZ1–189 would have functional N-terminal but not C-terminal domains. The inactivity of CheZ1–189 in the Pi release assay indicates that the N-terminal domain of CheZ is not sufficient to promote dephosphorylation of CheY-P. Several additional observations are consistent with division of CheZ into a large N-terminal domain and a small C-terminal domain separated by an exposed linker. A peptide consisting of the C-terminal 19 amino acids of CheZ binds CheY-P (3, 27), suggesting that a rather small portion of the C-terminal region is capable of folding into a functional structure. Furthermore, following removal of the C-terminal fragment, a large N-terminal fragment of CheZ is stable in the presence of trypsin (42). Lys195 is readily accessible to trypsin cleavage (3), and we have provided mass spectrometry evidence for cleavage of E. coli CheZ following Glu181. In addition to experimental evidence, sequence information is also consistent with a linker in this segment of CheZ. The region of CheZ from residues 168 to 187 (E. coli numbering) contains no residues conserved between the six species with known sequences (data not shown). Furthermore, the CheZ sequences from Pseudomonas aeruginosa (24) and Pseudomonas putida (12) each contain an insertion of 12 amino acids in this region relative to those of the other CheZ proteins (data not shown).
The model of two independent CheZ domains developed above predicts that cheZV205E, like cheZQ190(Am), should encode a mutant protein with a functional N-terminal domain and therefore should complement cheZ alleles (cheZA65V, cheZL90S, and cheZD143G) encoding mutant proteins with functional C-terminal domains, but this was not observed. Perhaps CheZV205E has a steric constraint to functional heterodimer formation that is absent in CheZ1–189. Tests for intragenic complementation employing shorter nonsense peptides truncated at residues 116 or 142 failed to reveal a subdivision of the large N-terminal CheZ fragment into smaller domains.
Strains lacking functional CheZ rotate their flagella CCW following exposure to positive chemotactic stimuli, but the latent time between stimulus and response is 10 times longer than in wild-type bacteria (37), presumably because destruction of CheY-P via autodephosphorylation is much slower than destruction via CheZ-mediated dephosphorylation. The inability to rapidly change the direction of flagellar rotation while swimming through a chemoeffector gradient renders bacteria with mutant cheZ nonchemotactic in most circumstances. Although the addition of CheZ1–189 permitted chemotactic swarm formation by several strains expressing mutant CheZ proteins (Table 2), CheZ1–189 had little apparent effect on nonstimulated flagellar rotational bias, which remained predominantly CW (data not shown). The CheZ heterodimers evidently provide sufficient dephosphorylating activity towards CheY-P to decrease the response time to positive stimuli and thus restore chemotaxis yet provide insufficient CheZ activity to dramatically affect the bias of nonstimulated cells, given the highly nonlinear relationship between CheY-P concentration and bias (2, 36).
Dephosphorylation and complex formation.
The order of ability of mutant CheZ proteins to stimulate dephosphorylation of CheY-P (Fig. 4) (the prefix CheZ is omitted), wild type > G188E and V205E > F117S > A65V and L90S > D143G, is the same as the order of ability to support formation of cross-linked complexes with large molecular weights (Fig. 5): wild type, G188E, and V205E > F117S > A65V, L90S, and D143G. These data substantially extend the correlation between these two assays previously reported for CheZ mutant proteins CheZF141I, -D143E, and -T145M (4). However, the ability of CheZG188E and CheZV205E to form complexes apparently indistinguishable from those formed by wild-type CheZ while possessing only ∼10% of the dephosphorylating activity of wild-type CheZ further suggests that the property of CheZ measured by the cross-linking assay is necessary but not sufficient for maximal dephosphorylation activity. Similarly, the inability of the peptide corresponding to the C terminus of CheZV205E to bind CheY-P demonstrates that this binding activity is not necessary for CheY-CheZ cross-linked complexes.
The nature of defects in the mutant CheZ proteins.
Chemoeffector-mediated regulation of CheZ activity has been proposed as one mechanism to account for amplification in the chemotaxis signal transduction pathway (9). Mutants defective in this hypothetical regulatory function would be unable to support chemotaxis but could retain dephosphorylation-stimulating activity. The cheZ mutations discovered in this work were isolated on the basis of impaired chemotaxis, but none of the six mutant CheZ proteins that were tested supported normal dephosphorylation of CheY-P and hence do not contain strictly regulatory defects.
Functional implications of clusters of cheZ mutations.
Taking into account phenotypic characterization of entire mutant sets as well as biochemical characterization of single mutant proteins, differentiations can be made regarding likely roles of the six regions delineated here in the structure and/or function of CheZ. One of the larger clusters, corresponding to residues 132 to 149 stands out in several respects as a possible region of high functional importance in CheZ activity. First, this cluster coincides with a stretch of highly conserved residues deduced from sequenced cheZ genes (Fig. 7). Second, most of the mutations in this cluster result in a complete loss of CheZ function (Table 1). Third, this region contains many polar and charged residues, which could be solvent exposed and which could actively participate in catalysis or binding. It is notable that our random mutagenesis generated three independent substitutions at Asp143, two of which gave no activity in vivo, and even the conservative glutamate substitution resulted in severe defects. Finally, biochemical analysis showed that the mutant chosen from this cluster, CheZD143G, was the least active of any of the tested mutant proteins (less than 1% of wild-type phosphatase activity) and did not form high-molecular-weight complexes in the presence of CheY-P and the cross-linker, as does wild-type CheZ. Therefore, this region appears to be a likely candidate to contain a residue or residues which are essential for CheZ activity. Three previously described mutants containing mutations within this region were shown to be defective in phosphatase activity and oligomerization (4).
The mutant proteins tested from the cluster corresponding to residues 57 to 76 (CheZA65V), and the cluster corresponding to residues 83 to 90 (CheZL90S) also showed nearly complete loss of phosphatase and oligomerization abilities in biochemical assays, suggesting important roles in structure and/or function for these regions. However, unlike what was found for the cluster corresponding to residues 132 to 149, most of the mutations in the cluster corresponding to residues 57 to 76 resulted in only partial loss of function (intermediate swarming ability) (Table 1), and most of these substitutions occurred at nonpolar residues. Likewise, all of the mutations which resulted in full loss of activity for the cluster corresponding to residues 83 to 90 affected nonpolar residues. These patterns could suggest that these regions are critical for structural integrity and may not themselves participate directly in CheZ function. However, further study is required to better understand the role of these regions.
The small cluster of three CheZ mutations at the C terminus (residues 204 to 206) correlates with a region of CheZ which has been previously implicated in CheY binding (3). CheZV205E was the only mutant of this group to show a total loss of function on the swarm assay. The peptide which corresponded to this mutant showed complete loss of ability to bind CheY-P relative to the peptide with the wild-type sequence, demonstrating that Val205 plays a critical role in the binding of the peptide to CheY-P. In contrast, full-length CheZV205E had measurable phosphatase activity and formed cross-linked products to a similar extent as wild-type CheZ. Whereas the cross-linking experiment is unlikely to be capable of sensitively distinguishing intermediate levels of cross-linked products (resulting from intermediate levels of binding), the presence of cross-linked products for CheZV205E shows that this mutant can bind to CheY-P. That binding between CheY and CheZ can occur in the absence of the interactions involved in binding the CheZ peptide implies that the interface between the C-terminal peptides of CheZ and CheY-P likely represents only a subset of CheY-CheZ binding interactions. This conclusion is in agreement with McEvoy et al. (27), who noted that the difference in the affinities of CheY and CheY-P for full-length CheZ was greater than that for the CheZ196–214 peptide. Similarly, Sanna and Simon, based on the isolation of suppression mutations in cheZ that restored the ability to bind CheY23ND, have proposed that CheY binding regions on CheZ are located at the far N terminus and in the area of residues 150 to 170 (34).
The role of the region of CheZ delineated by the final small cluster of mutations, including cheZF117S, is uncertain. The amino acid sequence in this portion of CheZ is not well conserved between different bacterial species. CheZF117S has low but measurable phosphorylation and complex formation capabilities. Therefore this region may possess an indirect role in function, such as a structural role in aligning critical residues from other functional regions.
Due to the large number of mutants mapped in these studies, we have been able to identify regions of the primary structure of CheZ which have structural and/or functional importance. It will eventually be of great interest to locate the amino acid substitutions resulting from each cluster of mutations on the (currently unknown) three-dimensional structure of CheZ. Although some differentiations regarding the biochemical and genetic behavior of mutants in these clusters can now be made, one or more of these clusters likely combine to form single functional units in the folded protein so that it may not be possible to assign discrete functions to each region of primary sequence. In addition to the identification of important regions of CheZ, these studies have provided a physical resource of ∼50 unique missense mutants. Together, these will combine to shape further biochemical and biophysical studies of CheZ.
ACKNOWLEDGMENTS
We thank Roel Schaaper for the mutD strain, Yuval Blat and Michael Eisenbach for advice on the cross-linking assay, Birgit Scharf and Howard Berg for anti-CheY antibody, Bob Stevens for mass spectrometry, Megan McEvoy for sharing details of the peptide binding assay prior to publication, Karen Fahrner for guidance to unpublished cheZ sequences, and Jeryl Appleby, Ken Bott, Janne Cannon, Tom Kawula, and Martin Schuster for useful discussions.
This work was supported by Public Health Service grant GM-50860 from the National Institute of General Medical Sciences.
REFERENCES
- 1.Aiba H, Mizuno T, Mizushima S. Transfer of phosphoryl group between two regulatory proteins involved in osmoregulatory expression of the ompF and ompC genes in Escherichia coli. J Biol Chem. 1989;264:8563–8567. [PubMed] [Google Scholar]
- 2.Alon U, Surette M G, Aguera y Arcas B, Liu Y, Leibler S, Stock J B. Response regulator output in bacterial chemotaxis. EMBO J. 1998;17:4238–4248. doi: 10.1093/emboj/17.15.4238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blat Y, Eisenbach M. Conserved C-terminus of the phosphatase CheZ is a binding domain for the chemotactic response regulator CheY. Biochemistry. 1996;35:5679–5683. doi: 10.1021/bi9530447. [DOI] [PubMed] [Google Scholar]
- 4.Blat Y, Eisenbach M. Mutants with defective phosphatase activity show no phosphorylation-dependent oligomerization of CheZ. The phosphatase of bacterial chemotaxis. J Biol Chem. 1996;271:1232–1236. doi: 10.1074/jbc.271.2.1232. [DOI] [PubMed] [Google Scholar]
- 5.Blat Y, Eisenbach M. Oligomerization of the phosphatase CheZ upon interaction with the phosphorylated form of CheY. The signal protein of bacterial chemotaxis. J Biol Chem. 1996;271:1226–1231. doi: 10.1074/jbc.271.2.1226. [DOI] [PubMed] [Google Scholar]
- 6.Blat Y, Gillespie B, Bren A, Dahlquist F W, Eisenbach M. Regulation of phosphatase activity in bacterial chemotaxis. J Mol Biol. 1998;284:1191–1199. doi: 10.1006/jmbi.1998.2224. [DOI] [PubMed] [Google Scholar]
- 7.Borkovich K A, Simon M I. The dynamics of protein phosphorylation in bacterial chemotaxis. Cell. 1990;63:1339–1348. doi: 10.1016/0092-8674(90)90429-i. [DOI] [PubMed] [Google Scholar]
- 8.Bourret R B, Hess J F, Simon M I. Conserved aspartate residues and phosphorylation in signal transduction by the chemotaxis protein CheY. Proc Natl Acad Sci USA. 1990;87:41–45. doi: 10.1073/pnas.87.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bray D, Bourret R B, Simon M I. Computer simulation of the phosphorylation cascade controlling bacterial chemotaxis. Mol Biol Cell. 1993;41:469–482. doi: 10.1091/mbc.4.5.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chang A C Y, Cohen S N. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J Bacteriol. 1978;134:1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dengen G E, Cox E C. Conditional mutator gene in Escherichia coli: isolation, mapping, and effector studies. J Bacteriol. 1974;117:477–487. doi: 10.1128/jb.117.2.477-487.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ditty J L, Grimm A C, Harwood C S. Identification of a chemotaxis gene region from Pseudomonas putida. FEMS Microbiol Lett. 1998;159:267–273. doi: 10.1111/j.1574-6968.1998.tb12871.x. [DOI] [PubMed] [Google Scholar]
- 13.Hess J F, Bourret R B, Oosawa K, Matsumura P, Simon M I. Protein phosphorylation and bacterial chemotaxis. Cold Spring Harbor Symp Quant Biol. 1988;53:41–48. doi: 10.1101/sqb.1988.053.01.008. [DOI] [PubMed] [Google Scholar]
- 14.Hess J F, Bourret R B, Simon M I. Phosphorylation assays for proteins of the two-component regulatory system controlling chemotaxis in Escherichia coli. Methods Enzymol. 1991;200:188–204. doi: 10.1016/0076-6879(91)00139-n. [DOI] [PubMed] [Google Scholar]
- 15.Hess J F, Oosawa K, Matsumura P, Simon M I. Phosphorylation of three proteins in the signaling pathway of bacterial chemotaxis. Cell. 1988;53:79–87. doi: 10.1016/0092-8674(88)90489-8. [DOI] [PubMed] [Google Scholar]
- 16.Hoch J A, Silhavy T J, editors. Two-component signal transduction. Washington, D.C.: ASM Press; 1995. [Google Scholar]
- 17.Huang C, Stewart R C. CheZ mutants with enhanced ability to dephosphorylate CheY, the response regulator in bacterial chemotaxis. Biochim Biophys Acta. 1993;1202:297–304. doi: 10.1016/0167-4838(93)90019-n. [DOI] [PubMed] [Google Scholar]
- 18.Igo M M, Ninfa A J, Stock J B, Silhavy T J. Phosphorylation and dephosphorylation of a bacterial activator by a transmembrane receptor. Genes Dev. 1989;3:1725–1734. doi: 10.1101/gad.3.11.1725. [DOI] [PubMed] [Google Scholar]
- 19.Keener J, Kustu S. Protein kinase and phosphoprotein phosphatase activities of nitrogen regulatory proteins NTRB and NTRC of enteric bacteria: roles of the conserved amino-terminal domain of NTRC. Proc Natl Acad Sci USA. 1988;85:4976–4980. doi: 10.1073/pnas.85.14.4976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koshland D E., Jr Effect of catalysts on the hydrolysis of acetyl phosphate: nucleophilic displacement mechanisms in enzymatic reactions. J Am Chem Soc. 1952;74:2286–2292. [Google Scholar]
- 21.Kunkel T A, Roberts J D, Zakour R A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–383. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- 22.Liu J D, Parkinson J S. Role of CheW protein in coupling membrane receptors to the intracellular signaling system of bacterial chemotaxis. Proc Natl Acad Sci USA. 1989;86:8703–8707. doi: 10.1073/pnas.86.22.8703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lukat G S, Stock A M, Stock J B. Divalent metal ion binding to the CheY protein and its significance to phosphotransfer in bacterial chemotaxis. Biochemistry. 1990;29:5436–5442. doi: 10.1021/bi00475a004. [DOI] [PubMed] [Google Scholar]
- 24.Masduki A, Nakamura J, Ohga T, Umezaki R, Kato J, Ohtake H. Isolation and characterization of chemotaxis mutants and genes of Pseudomonas aeruginosa. J Bacteriol. 1995;177:948–952. doi: 10.1128/jb.177.4.948-952.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Matsumura P, Rydel J J, Linzmeier R, Vacante D. Overexpression and sequence of the Escherichia coli cheY gene and biochemical activities of the CheY protein. J Bacteriol. 1984;160:36–41. doi: 10.1128/jb.160.1.36-41.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McCleary W R. The activation of PhoB by acetylphosphate. Mol Microbiol. 1996;20:1155–1163. doi: 10.1111/j.1365-2958.1996.tb02636.x. [DOI] [PubMed] [Google Scholar]
- 27.McEvoy M M, Bren A, Eisenbach M, Dahlquist F W. Identification of the binding interfaces on CheY for two of its targets, the phosphatase CheZ and the flagellar switch protein, FliM. J Mol Biol. 1999;289:1423–1433. doi: 10.1006/jmbi.1999.2830. [DOI] [PubMed] [Google Scholar]
- 28.Mutoh N, Simon M I. Nucleotide sequence corresponding to five chemotaxis genes in Escherichia coli. J Bacteriol. 1986;165:161–166. doi: 10.1128/jb.165.1.161-166.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ninfa A J, Magasanik B. Covalent modification of the glnG product, NRI, by the glnL product, NRII, regulates the transcription of the glnALG operon in Escherichia coli. Proc Natl Acad Sci USA. 1986;83:5909–5913. doi: 10.1073/pnas.83.16.5909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ohlsen K L, Grimsley J K, Hoch J A. Deactivation of the sporulation transcription factor SpoOA by the SpoOE protein phosphatase. Proc Natl Acad Sci USA. 1994;91:1756–1760. doi: 10.1073/pnas.91.5.1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Parkinson J S. Signal transduction schemes of bacteria. Cell. 1993;73:857–871. doi: 10.1016/0092-8674(93)90267-t. [DOI] [PubMed] [Google Scholar]
- 32.Perego M, Hanstein C, Welsh K M, Djavakhishvili T, Glaser P, Hoch J A. Multiple protein aspartate phosphatases provide a mechanism for the integration of diverse signals in the control of development in Bacillus subtilis. Cell. 1994;79:1047–1055. doi: 10.1016/0092-8674(94)90035-3. [DOI] [PubMed] [Google Scholar]
- 33.Sanna M G, Simon M I. In vivo and in vitro characterization of Escherichia coli protein CheZ gain- and loss-of-function mutants. J Bacteriol. 1996;178:6275–6280. doi: 10.1128/jb.178.21.6275-6280.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sanna M G, Simon M I. Isolation and in vitro characterization of CheZ suppressors for the Escherichia coli chemotactic response regulator mutant CheYN23D. J Biol Chem. 1996;271:7357–7361. doi: 10.1074/jbc.271.13.7357. [DOI] [PubMed] [Google Scholar]
- 35.Schaaper R M, Cornacchio R. An Escherichia coli dnaE mutation with suppressor activity toward mutator mutD5. J Bacteriol. 1992;174:1974–1982. doi: 10.1128/jb.174.6.1974-1982.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Scharf B E, Fahrner K A, Turner L, Berg H C. Control of direction of flagellar rotation in bacterial chemotaxis. Proc Natl Acad Sci USA. 1998;95:201–206. doi: 10.1073/pnas.95.1.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Segall J E, Manson M D, Berg H C. Signal processing times in bacterial chemotaxis. Nature. 1982;296:855–857. doi: 10.1038/296855a0. [DOI] [PubMed] [Google Scholar]
- 38.Silverman M, Simon M. Chemotaxis in Escherichia coli: methylation of che gene products. Proc Natl Acad Sci USA. 1977;74:3317–3321. doi: 10.1073/pnas.74.8.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Silversmith R E, Appleby J L, Bourret R B. The catalytic mechanism of phosphorylation and dephosphorylation of CheY: kinetic characterization of imidazole-phosphates as phosphodonors and the role of acid catalysis. Biochemistry. 1997;36:14965–14974. doi: 10.1021/bi9715573. [DOI] [PubMed] [Google Scholar]
- 40.Sockett H, Yamaguchi S, Kihara M, Irikura V M, MacNab R. Molecular analysis of the flagellar switch protein FliM of Salmonella typhimurium. J Bacteriol. 1992;174:793–806. doi: 10.1128/jb.174.3.793-806.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stock A M, Schaeffer E, Koshland D E, Jr, Stock J. A second type of methylation reaction in bacterial chemotaxis. J Biol Chem. 1987;262:8011–8014. [PubMed] [Google Scholar]
- 42.Stock A M, Stock J B. Purification and characterization of the CheZ protein of bacterial chemotaxis. J Bacteriol. 1987;169:3301–3311. doi: 10.1128/jb.169.7.3301-3311.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stock J B, Surette M. Chemotaxis. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. Washington, D.C.: American Society for Microbiology; 1996. pp. 1103–1129. [Google Scholar]
- 44.Wang H, Matsumura P. Characterization of the CheAS/CheZ complex: a specific interaction resulting in enhanced dephosphorylating activity on CheY-phosphate. Mol Microbiol. 1996;19:695–703. doi: 10.1046/j.1365-2958.1996.393934.x. [DOI] [PubMed] [Google Scholar]
- 45.Wang H, Matsumura P. Phosphorylating and dephosphorylating complexes in bacterial chemotaxis. J Bacteriol. 1997;179:287–289. doi: 10.1128/jb.179.1.287-289.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Webb M R. A continuous spectrophotometric assay for inorganic phosphate and for measuring phosphate release kinetics in biological systems. Proc Natl Acad Sci USA. 1992;89:4884–4887. doi: 10.1073/pnas.89.11.4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Weiss V, Magasanik B. Phosphorylation of nitrogen regulator I (NRI) of Escherichia coli. Proc Natl Acad Sci USA. 1988;85:8919–8923. doi: 10.1073/pnas.85.23.8919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Welch M, Oosawa K, Aizawa S-I, Eisenbach M. Effects of phosphorylation, Mg2+, and conformation of the chemotaxis protein CheY on its binding to the flagellar switch protein FliM. Biochemistry. 1994;33:10270–10276. doi: 10.1021/bi00200a031. [DOI] [PubMed] [Google Scholar]
- 49.Welch M, Oosawa K, Aizawa S-I, Eisenbach M. Phosphorylation-dependent binding of a signal molecule to the flagellar switch of bacteria. Proc Natl Acad Sci USA. 1993;90:8787–8793. doi: 10.1073/pnas.90.19.8787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wolfe A J, Berg H C. Migration of bacteria in semisolid agar. Proc Natl Acad Sci USA. 1989;86:6973–6977. doi: 10.1073/pnas.86.18.6973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wylie D, Stock A, Wong C-Y, Stock J. Sensory transduction in bacterial chemotaxis involves phosphotransfer between Che proteins. Biochem Biophys Res Commun. 1988;151:891–896. doi: 10.1016/s0006-291x(88)80365-6. [DOI] [PubMed] [Google Scholar]
- 52.Zhu X, Volz K, Matsumura P. The CheZ-binding surface of CheY overlaps the CheA- and FliM-binding surfaces. J Biol Chem. 1997;272:23758–23764. doi: 10.1074/jbc.272.38.23758. [DOI] [PubMed] [Google Scholar]