Abstract
A screen for nonsliding mutants of Mycobacterium smegmatis yielded 20 mutants with transposon insertions in the mps gene, which is involved in glycopeptidolipid biosynthesis. One mutant had an insertion in a gene predicted to encode a membrane transport protein. All mutants lacked glycopeptidolipids and were unable to form biofilms on polyvinyl chloride.
Recently, it has been shown that in spite of being nonflagellated microorganisms, mycobacteria can spread on the surface of solid growth medium by a sliding mechanism (9). This form of surface motility is produced by the expansive forces of the growing bacterial population, in combination with cell surface properties that favor reduced friction between the cells and the substrate, and it results in the slow movement of a uniform monolayer of cells as a unit (8). A time-lapse movie of sliding Mycobacterium smegmatis can be seen at http://gasp.med.harvard.edu/smegmatis/sliding.html. Both the fast-growing nonpathogenic M. smegmatis and the slow-growing opportunistic pathogen Mycobacterium avium are able to slide, and in both species this ability correlates with the presence of GPLs (9), a class of glycosylated peptidolipids present in the outermost layer of the cell envelope (11). However, no direct link between GPLs and sliding could be established, since the GPL-deficient strains in previous studies were not characterized genetically. Here we report the results of the first genetic analysis of sliding motility. Mutants unable to slide on motility plates lack GPLs and are also unable to form biofilms on polyvinyl chloride (PVC) plates.
Genetic screen for M. smegmatis nonsliding mutants.
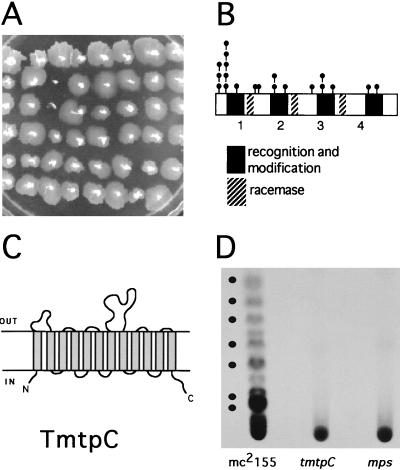
In order to gain more insight into the mechanism driving sliding motility in M. smegmatis mc2155, a screen was set up to look for genes that when disrupted would result in the inability to slide on the surface of plates containing 0.2% glucose–M63 salts medium solidified with 0.5% agarose (sliding medium). A mariner-based transposon (17) was used that contained a kanamycin resistance gene (kan) as a selectable marker. The temperature-sensitive plasmid pMycoMar, harboring the mariner-derived transposon, has been shown to work as an efficient transposon delivery system in M. smegmatis, resulting in random insertions in the genome (17). The temperature-sensitive mycobacterial replicon allows for the direct selection of transposition events on kanamycin-containing plates incubated at 40°C. A screen of 4,000 random transposon insertion mutants yielded a total of 21 nonsliding mutants that grew on the sliding medium but did not form the translucent halo surrounding the point of inoculation characteristic of wild-type mc2155 (Fig. 1A). All the nonsliding mutants had rough colony morphology and showed no detectable levels of GPLs by thin-layer chromatography (TLC) analysis (results not shown).
FIG. 1.
(A) Nonsliding motility screen. An M63 salts–0.2% glucose minimal medium plate containing 0.5% agarose as solidifying agent shows 48 mariner transposon mutants. Sliding motility results in the formation of a transparent halo surrounding the point of inoculation. On this plate, two nonsliding mutants are apparent. (B) Location of 20 nonsliding transposon insertion mutations within the mps locus. The predicted four modules (labeled 1, 2, 3, and 4) of the mycobacterial peptide synthetase (2) and the position of the mariner transposon insertions are indicated. (C) Schematic representation of the predicted secondary structure of the M. smegmatis TmtpC protein. The predicted amino acid sequence was analyzed using TopPred2 software (Stockholm University). (D) Autoradiogram of GPL profiles of pulse-labeled cultures of the mc2155, tmtpC, and mps strains. The positions of the GPL bands visible after spraying with 10% H2SO4 in ethanol are marked by dots.
Nonsliding mutants have transposon insertions in mps.
The DNA sequence of the transposon insertion site for 20 of the nonsliding mutants was obtained using the arbitrary PCR technique, as previously described (14). For the first round of amplification, the mariner primers used were 5′-GGGAATCATTTGAAGGTTGGT-3′ (for sequences 5′ of the insertion site) and 5′-GTCAATTCGAGCTCGGGTA-3′ (for sequences 3′ of the insertion site). For the second round of amplification, the mariner primer used for the 5′ insertion site reaction was 5′-TAGCGACGCCATCTATGTGTC-3′, and the one used for the 3′ insertion site was 5′-CTTGAAGGGAACTATGTTG-3′. The arbitrary PCR primers used were those described before (14). All 20 mutants contained transposon insertions in the same gene, mps, which was recently reported to encode a nonribosomal peptide synthetase in M. smegmatis involved in GPL biosynthesis (2). The location of the mariner transposon insertions in mps is shown in Fig. 1B. These results provide direct genetic evidence for the requirement of GPLs for sliding motility in mycobacteria.
Nonsliding mutant in a transport membrane protein homolog.
The arbitrary PCR method failed to provide the sequence of the transposon insertion site for one of the nonsliding mutants. Genomic mycobacterial DNA was isolated from this mutant, digested with XmaI, and cloned into pUC19. The kanamycin resistance gene in the mariner minitransposon was used to select for a DNA fragment that contained the transposon insertion. DNA sequencing revealed that the mariner transposon was inserted approximately in the middle of a 994-codon open reading frame encoding a putative transport membrane protein of 12 transmembrane segments with high similarity to the tmtpB and tmtpC genes of M. avium (GenBank accession no. AF143772) and the mmpL family in Mycobacterium tuberculosis (4) (Fig. 1C). The gene organization is highly conserved between M. smegmatis and M. avium (data not shown), with both organisms presenting genes encoding one small (tmtpA) and two large (tmtpB and tmtpC) putative transmembrane transport proteins in the same operon. The nonsliding mutant contains the mariner transposon inserted approximately in the middle of the coding region of the second large transport protein. We have therefore named the interrupted gene tmtpC. M. smegmatis TmtpC shows 64% identity and 78% similarity to M. avium TmtpC and 60% identity and 76% similarity to TmtpB. Interestingly, the tmtpC gene of M. avium is part of a locus involved in the synthesis of GPLs and in resistance to daunorubicin (GenBank accession no. AF143772). In addition, the mmpL genes have been suggested to be involved in lipid transport (19), and one of them, mmpL7, has been shown to be required for the transport of phthiocerol dimycocerosate to the cell wall of M. tuberculosis (5).
The sequence similarities and the phenotype of the tmtpC mutant in M. smegmatis (nonsliding, rough morphology, and lack of detectable GPLs by TLC analysis) suggest that the product of tmtpC is involved in the transport of GPLs across the cytoplasmic membrane. In an attempt to detect cytoplasmic GPLs that might be turned over rapidly due to the lack of transport to the envelope of the tmtpC mutant, we performed pulse-labeling experiments with 14C-amino acids. Briefly, 3-ml cultures of the strains grown in M63 salts–0.2% glucose were labeled with 150 μl of a mixture of 14C-labeled amino acids (Amersham). After labeling, cells were rapidly cooled on ice, collected by centrifugation, and immediately resuspended in 1 ml of a chloroform-methanol mixture (2:1). Extractions of GPLs and TLC analysis were performed as previously described (3). After a 5-min pulse, radioactively labeled GPLs were clearly detected in the parental strain, mc2155, but were completely absent in the tmtpC and mps mutants (Fig. 1D). The failure to detect any labeled products in the tmtpC mutant suggests that the TmtpC protein could play a role not only in the transport of GPLs to the envelope but also in the biosynthetic process.
Extracellular complementation of nonsliding mutants.
There are two possible mechanisms of GPL function in sliding. GPLs could remain associated with the outermost layer of the envelope or they could be secreted, lowering the surface tension of the water and creating a conditioning film for cells to slide on. The latter is the case with the serrawettings, which mediate surface spreading in Serratia (7, 10). In this organism, the presence of a surface-active exolipid can be detected by a drop collapsing test, and mutants deficient in serrawetting production can be complemented by the addition of surfactants to the growth medium, including not only serrawettings themselves but also other microbial surfactants. We have been unable to detect the presence of surfactants in the GPL-producing strains by the drop collapsing test. Also, attempts to complement the nonsliding phenotype by the addition of TLC-purified GPLs, surfactin (Sigma), and serrawettings W1 and W2 (provided by T. Matsuyama) to the sliding medium have failed.
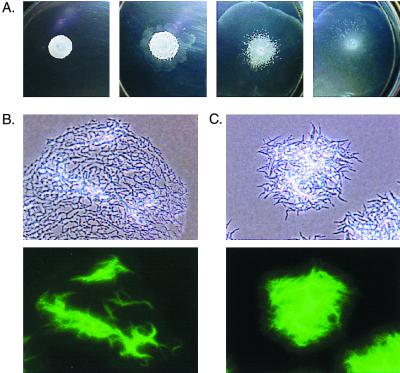
We also tried to complement the nonsliding mutants by growing them in sliding medium in the presence of mc2155. In order to clearly distinguish the mutant from the parental strain, we labeled the nonsliding mutants with green fluorescent protein (GFP) expressed from pGFP/O (9). Since the selectable marker in the plasmid (kan) is the same as the one present in the mariner transposon, we have used for this experiment the spontaneous rough mutant O2 (9), which is phenotypically indistinguishable from the nonsliding transposon mutants described in this work. Single cells of the GFP-labeled O2 strain were obtained by shaking a liquid culture with 1-mm sterile glass beads for 10 min and filtering it through Whatman no. 1 paper. A total of 15 μl of this preparation (containing approximately 106 CFU/ml) was plated on M63 salts–0.3% agarose plates with no added carbon source, either alone or with increasing amounts (2 × 10−2, 1 × 10−1, and 1 μl) of a saturated culture of mc2155. Plates were incubated at 37°C for 5 days. The results showed that the presence of increasing proportions of mc2155 cells in the inoculum changes the macroscopic appearance of the O2 strain, partially dispersing the compact cell clumps characteristic of nonsliding mutants (Fig. 2A). Microscopic analysis revealed that mc2155 can only partially complement the nonsliding phenotype of O2. Even when the wild type is at a >100-fold excess, the O2 cells present in the spreading halo still retain their characteristic clumped morphology (Fig. 2B). However, the clumps are not as compact as when O2 is grown by itself (Fig. 2C). Thus, although GPLs appeared to be released to some extent, the regularly spaced cell configuration characteristic of the sliding monolayer can be adopted only by the producing strain. These results, together with the lack of phenotypic suppression by purified surfactants, suggest that GPLs need to be located in their proper position in the cell envelope and that, therefore, their mechanism of action is different from that of secreted serrawettings (7, 10).
FIG. 2.
Macroscopic and microscopic analysis of mixtures of mc2155 and GFP-labeled cells of the nonsliding mutant O2. (A) Appearance of colonies grown on 0.3% agarose plates containing M63 salts with no added source of carbon for 5 days at 37°C. O2 cells were incubated alone (left) or in the presence of increasing amounts of mc2155 (see text for details). (B and C) Phase-contrast images (top) and fluorescent micrographs (bottom) of O2 cells incubated in the presence (B) or absence (C) of 1 μl of an M. smegmatis mc2155 culture.
Nonsliding mutants are defective in biofilm formation.
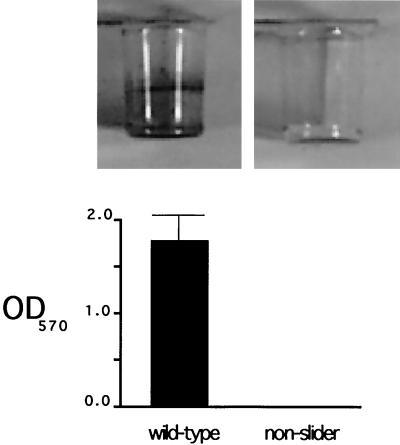
Surface-exposed molecules of the cell envelope have been shown to play crucial roles in the initial adhesion of bacteria to surfaces, as well as in the subsequent surface colonization during the process of biofilm development (12, 15, 20). Since the presence or absence of GPLs on the surface of M. smegmatis could affect the interactions of this organism with surfaces other than that of the growth plates, we decided to investigate the role of GPLs in biofilm formation. We adapted a previously described assay (13) to measure M. smegmatis biofilm formation on PVC plastic. Briefly, 100 μl of M63 2% glucose medium with 0.5% Casamino Acids was used per well in a 96-well PVC microtiter dish. The medium was inoculated with cells from a Luria-Bertani agar plate, and microtiter dishes were incubated with slight shaking (80 rpm) at room temperature for 48 h. The wells were rinsed twice with water, and 120 μl of a 1% cell-staining solution of crystal violet (CV) was added. Plates were incubated at room temperature for 30 min, rinsed with water three times, and scored for CV staining. Quantitation of biofilm formation was performed by extracting the biofilm-associated CV with 100% ethanol for 1 h and measuring the optical density at 570 nm. While mc2155 forms a clearly detectable biofilm under these conditions, no biofilm was detected in any of the nonsliding mutants (Fig. 3), indicating that GPLs are required for M. smegmatis biofilm formation on PVC plastic.
FIG. 3.
Nonsliding mutants are defective in biofilm formation. Biofilm formation was assayed by CV staining of cultures incubated in M63 salts minimal medium supplemented with 2% glucose and 0.5% Casamino Acids, and CV quantitation was performed afterwards (see text for details). Results for M. smegmatis mc2155 and a nonslider transposon mutant are shown. OD570, optical density at 570 nm.
Model.
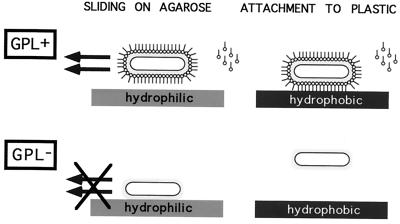
The experiments presented here demonstrate that GPLs are required for sliding motility on agarose plates, as well as for attachment and biofilm formation on PVC plastic. The following model could explain the role of these amphiphilic molecules in the interaction of M. smegmatis with both types of surfaces (Fig. 4). We propose that the GPLs, which are known to be present in the outermost layer of the envelope (11), are bound via the hydrophilic head to the cell capsule (composed mostly of polysaccharides [6]), thereby exposing the hydrophobic tails to the outside. This would result in a hydrophobic cell that cannot interact with the hydrophilic agarose surface. The resulting reduction of friction would lead to sliding on the surface of the agarose. Conversely, hydrophobic interactions between the exposed fatty acid tails of the GPLs and the hydrophobic surface of the PVC plastic would mediate attachment to plastic and biofilm formation. This type of interaction has been recognized to play a role in bacterial attachment to surfaces (16). In M. smegmatis mutants unable to synthesize or export GPLs (such as the mps and tmtpC mutants described in this work), the hydrophilic capsule would be exposed to the outside. There it could prevent attachment to the hydrophobic plastic surface and favor interaction with the agarose surface, restraining movement on the growth medium. The characteristic clumping of the nonsliding mutants could also be a consequence of increased exposure of the capsule polysaccharides, since there is some evidence indicating that sugars are involved in mycobacterial clumping (1). For the sake of simplicity, we depict GPLs in our model as a continuous layer on the cell surface. However, this is not likely to be the case. The cross section of the head group of GPLs is larger than that of the hydrocarbon tail, a typical feature of amphiphilic molecules that form micelles or fibrils, and indeed a discontinuous lamellar structure made up of GPL fibrils has been observed in M. avium (18). A discontinuous GPL distribution is also possible in the M. smegmatis capsule and might explain how spreading cells maintain close cell-to-cell proximity restricted to small surface areas, mainly around the cell poles (9). Although the details of their location in the cell surface remain to be defined, the work presented here provides direct evidence for the critical role of GPLs in sliding motility and attachment to surfaces.
FIG. 4.
Model for the action of GPLs in sliding motility and attachment to plastic. See text for details.
Nucleotide sequence accession number.
The sequence of the 994-codon open reading frame obtained in this study has been submitted to GenBank with accession no. AF271635.
Acknowledgments
We thank Eric Rubin for providing the pMycoMar shuttle vector and Tohey Matsuyama for providing purified serrawettings.
This work was supported by NIH grants GM58213 and GM55199 to R.K. and an NIH minority postdoctoral supplement to J.R.
REFERENCES
- 1.Anton V P, Rouge P, Daffe M. Identification of the sugars involved in mycobacterial cell aggregation. FEMS Microbiol Lett. 1996;144:167–170. doi: 10.1111/j.1574-6968.1996.tb08525.x. [DOI] [PubMed] [Google Scholar]
- 2.Billman-Jacobe H, McConville M J, Haites R E, Kovacevic S, Coppel R C. Identification of a peptide synthetase involved in the biosynthesis of glycopeptidolipids of Mycobacterium smegmatis. Mol Microbiol. 1999;33:1244–1253. doi: 10.1046/j.1365-2958.1999.01572.x. [DOI] [PubMed] [Google Scholar]
- 3.Brennan P J, Souhrada M, Ullom B, McClatchy J K, Goren M B. Identification of atypical mycobacteria by thin-layer chromatography of their surface antigens. J Clin Microbiol. 1978;8:374–379. doi: 10.1128/jcm.8.4.374-379.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cole S T, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon S V, Eiglmeier K, Gas S, Barry III C E, Tekaia F, Badcock K, Basham D, Brown D, Chillingworth T, Connor R, Davies R, Devlin K, Feltwell T, Gentles S, Hamlin N, Holroyd S, Hornsby T, Jagels K, Barrell B G, et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 5.Cox J S, Chen B, McNeil M, Jacobs W R., Jr Complex lipid determines tissue-specific replication of Mycobacterium tuberculosis in mice. Nature. 1999;402:79–83. doi: 10.1038/47042. [DOI] [PubMed] [Google Scholar]
- 6.Daffe M, Draper P. The envelope layers of mycobacteria with reference to their pathogenicity. Adv Microb Physiol. 1998;39:131–203. doi: 10.1016/s0065-2911(08)60016-8. [DOI] [PubMed] [Google Scholar]
- 7.Eberl L, Molin S, Givskov M. Surface motility of Serratia liquefaciens MG1. J Bacteriol. 1999;181:1703–1712. doi: 10.1128/jb.181.6.1703-1712.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Henrichsen J. Bacterial surface translocation: a survey and a classification. Bacteriol Rev. 1972;36:478–503. doi: 10.1128/br.36.4.478-503.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martínez A, Torello S, Kolter R. Sliding motility in mycobacteria. J Bacteriol. 1999;181:7331–7338. doi: 10.1128/jb.181.23.7331-7338.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Matsuyama T, Kaneda K, Nakagawa Y, Isa K, Hara-Hotta H, Yano I. A novel extracellular cyclic lipopeptide which promotes flagellum-dependent and -independent spreading growth of Serratia marcescens. J Bacteriol. 1992;174:1769–1776. doi: 10.1128/jb.174.6.1769-1776.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ortalo-Magné A, Lemassu A, Lanéelle M-A, Bardou F, Silve G, Gounon P, Marchal G, Daffé M. Identification of the surface-exposed lipids on the cell envelopes of Mycobacterium tuberculosis and other mycobacterial species. J Bacteriol. 1996;178:456–461. doi: 10.1128/jb.178.2.456-461.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.O'Toole G A, Kolter R. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol Microbiol. 1998;30:295–304. doi: 10.1046/j.1365-2958.1998.01062.x. [DOI] [PubMed] [Google Scholar]
- 13.O'Toole G A, Kolter R. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol Microbiol. 1998;28:449–461. doi: 10.1046/j.1365-2958.1998.00797.x. [DOI] [PubMed] [Google Scholar]
- 14.O'Toole G A, Pratt P A, Watnick P I, Newman D K, Weaver V B, Kolter R. Genetic approaches to study biofilms. Methods Enzymol. 1999;310:91–109. doi: 10.1016/s0076-6879(99)10008-9. [DOI] [PubMed] [Google Scholar]
- 15.Pratt L A, Kolter R. Genetic analysis of Escherichia coli biofilm formation: roles of flagella, motility, chemotaxis and type I pili. Mol Microbiol. 1998;30:285–293. doi: 10.1046/j.1365-2958.1998.01061.x. [DOI] [PubMed] [Google Scholar]
- 16.Rosenberg M, Kjelleberg S. Hydrophobic interactions in bacterial adhesion. Adv Microb Ecol. 1986;9:353–393. [Google Scholar]
- 17.Rubin E, Akerley B J, Novik V N, Lampe D J, Husson R N, Mekalanos J J. In vivo transposition of mariner-based elements in enteric bacteria and mycobacteria. Proc Natl Acad Sci USA. 1999;96:1645–1650. doi: 10.1073/pnas.96.4.1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rulong S, Aguas A P, daSilva P P, Silva M T. Intramacrophagic Mycobacterium avium bacilli are coated by a multiple lamellar structure: freeze fracture analysis of infected mouse liver. Infect Immun. 1991;59:3895–3902. doi: 10.1128/iai.59.11.3895-3902.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tekaia F, Gordon S V, Garnier T, Brosch R, Barrell B G, Cole S T. Analysis of the proteome of Mycobacterium tuberculosis in silico. Tuber Lung Dis. 1999;79:329–342. doi: 10.1054/tuld.1999.0220. [DOI] [PubMed] [Google Scholar]
- 20.Watnick P I, Fullner K J, Kolter R. A role for the mannose-sensitive hemagglutinin in biofilm formation by Vibrio cholerae El Tor. J Bacteriol. 1999;181:3606–3609. doi: 10.1128/jb.181.11.3606-3609.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]