Abstract
Autism is a multifaceted neurodevelopmental condition whose accurate diagnosis may be challenging because the associated symptoms and severity vary considerably. The wrong diagnosis can affect families and the educational system, raising the risk of depression, eating disorders, and self-harm. Recently, many works have proposed new methods for the diagnosis of autism based on machine learning and brain data. However, these works focus on only one pairwise statistical metric, ignoring the brain network organization. In this paper, we propose a method for the automatic diagnosis of autism based on functional brain imaging data recorded from 500 subjects, where 242 present autism spectrum disorder considering the regions of interest throughout Bootstrap Analysis of Stable Cluster map. Our method can distinguish the control group from autism spectrum disorder patients with high accuracy. Indeed the best performance provides an AUC near 1.0, which is higher than that found in the literature. We verify that the left ventral posterior cingulate cortex region is less connected to an area in the cerebellum of patients with this neurodevelopment disorder, which agrees with previous studies. The functional brain networks of autism spectrum disorder patients show more segregation, less distribution of information across the network, and less connectivity compared to the control cases. Our workflow provides medical interpretability and can be used on other fMRI and EEG data, including small data sets.
Subject terms: Computational biology and bioinformatics, Data mining, Data processing, Machine learning, Network topology, Programming language
Introduction
Autism is a multifactorial neurodevelopmental disorder with a complex genetic component1,2 and usually manifested since childhood (at least in the first three years of life) through deficits in social communication and restricted, repetitive patterns of behaviors or interests3. Because autism spectrum disorder (ASD) varies widely in symptoms and severity, an accurate diagnosis may be difficult. Indeed, there is no medical test to diagnose the disorder, such as a blood test. Diagnosis is based on observing the individual’s communication, social interaction, activities, and interests. This approach depends on experienced professionals, and an incorrect diagnosis can impact families and education, increasing the risk of depression, eating disorders, and self-harm4.
Furthermore, an autism misdiagnosis might occur because many other disorders have similar symptoms. In this way, it is essential to develop a quantitative and accurate method for autism diagnosis based on physical exams. This paper considers data from functional brain networks and machine learning algorithms to propose a computer-aid diagnostic methodology for autism.
Our approach is based on previous studies that suggested that autism is a manifestation of changes in the brain organization5. Abnormal neuronal connectivity has recently become the essential hypothesis for explaining the symptoms associated with autism6. By adopting the fMRI technique, Belmonte and Yurgelun-Todd7 demonstrated that the inputs of the autistic brain regions are cut off, with reduced activation and functional correlations with sensory areas. fMRI data from children with ASD8 suggest a strong parietal cortex activation responsible for visuospatial and sensory processing. In a resting state, regions of the medial prefrontal cortex related to the executive function comprised of skills that enable the individual to make decisions, pay attention, and differentiate conflicting thoughts are suppressed9. Apart from the medial prefrontal region, the rostral anterior cingulate cortex and the posterior cingulate cortex have also been investigated10. The function of the former includes memory recall and learning. In contrast, the posterior cingulate cortex is responsible for cognitive, emotional, and learning processes. Its metabolic activities during rest are deactivated during demanding cognitive tasks. According to Kennedy et al.10, the midline resting network of patients with ASD is less active than that of the control group, and task deactivation is insignificant. In structural terms, Keller et al.11 suggested the development of the brains of autistic children is atypical, showing an early overgrowth of white matter, followed by its reduction in adolescence and adulthood. Furthermore, Diffusion Tensor Imaging (DTI) results revealed the disorganization of white matter paths12.
These studies demonstrate that the structure of the brains of autistic people and healthy individuals differ. Therefore, we speculate that autism can be identified by reviewing information on brain anatomical organization. This data can be collected from electroencephalogram (EEG) or functional magnetic resonance imaging (fMRI) experiments. EEG is a relatively inexpensive method readily available in most contexts and has an excellent temporal resolution. Data from EEG has been used to enhance our understanding of human brain structural and functional networks13–15. On the other hand, fMRI has a low temporal resolution but a high spatial one, thus being well suited for analyses of spatial brain dynamics16,17. fMRI scans produce a set of three-dimensional images recorded over time and measure a signal (called BOLD signal (The decrease in the rate of deoxyhemoglobin can be detected with the increase of the NMR signal. This effect is called Blood Oxygenation Level-Dependent (BOLD))). The temporal evolution of the BOLD series is called the hemodynamic response function and is determined by the pixel intensity in fMRI images18,19. Each cube of an fMRI image, called a voxel, which anatomically maps a position in the brain, has a BOLD time series. Here, we consider the BOLD series to develop the classification method for autistic patients.
After mapping the brain, it is possible to classify people with ASD and typical development (TD) using machine learning methods. Machine learning (ML) techniques permit automatically extracting knowledge from a database. Previous studies have evaluated the effectiveness of machine learning in diagnosing ASD with supervised machine learning algorithms that distinguish between two classes, namely ASD and TD. Up to the present date, at least 45 articles have focused on supervised machine learning algorithms that aid in ASD diagnosis, where the most used ones are based on support vector machines (SVM)20 (see Table 1 for publications on the use of fMRI for distinguishing between ASD and TD).
Table 1.
Publications on using supervised ML algorithms on fMRI data for distinguishing ASD from TD patients. Based on20.
| Authors | Data size | ML methods | AUC | Accuracy | Recall | Precision |
|---|---|---|---|---|---|---|
| 21 | ASD ASD; TD (ABIDE) | Deep learning | – | 0.95 | 0.97 | – |
| 22 | ASD; TD (ABIDE) | Deep learning | 0.96 | 0.87 | 0.87 | – |
| 23 | ASD; TD (ABIDE) | RF | 0.91 | 0.89 | 0.93 | – |
| 24 | (ABIDE) | SVM | – | 0.80 | – | – |
| 25 | ASD; TD (ABIDE) | Lasso | 0.78 | 0.75 | 0.77 | 0.73 |
| 26 | ASD; TD (ABIDE) | Deep learning | – | 0.70 | 0.74 | 0.63 |
| 27 | ASD; TD | Deep belief network | 0.76 | – | – | – |
| 28 | ASD; TD | Linear SVM; ridge logistic regression | 0.73 | – | – | – |
Although ML has provided important advances in diagnosing autism, considerable challenges must be addressed. Many classification methods need to be more interpretable, which is disadvantageous, especially for understanding medical data29,30. Also, according to Table 125,28, small data sets are quite common31–34, which might cause unreliable results. To overcome the lack of interpretability, we can consider new techniques that have emerged in recent years towards facilitating the interpretation of machine learning results (e.g., SHapley Additive ExPlanations (SHAP) values35 identify the most important features for a model36–38). Moreover, to circumvent the use of small medical data, data augmentation techniques (e.g., sliding windows), which split data (e.g., time series from EEG and fMRI )39–41, might be adopted. However, one of their limitations is the loss of information during the splitting process, which the overlapping windows technique can solve. Part of the window information is repeated in each subsequent window and used for EEG42,43 and fMRI44,45 data. In this paper, we consider these methods to develop a new method for diagnosing autism that is interpretable and can be used in small data sets. In summary, our contributions are the following:
We design a method to classify fMRI time series using a connectivity matrix as input to the ML algorithm, which provides more accurate results than those reported in the literature.
Complex network measures characterize brain organization, quantifying the differences between ASD and TD patients. In addition, we use SHAP values for a biological interpretation of the connections between brain regions and their relation with ASD.
We adopt a sliding window data augmentation approach to increase the sample size by splitting the time series into smaller series with either mutually exclusive sections of the time series or overlapping sections of the sliding windows, in which portions of the sequence are repeated in multiple observations. This approach enables handling small medical data.
It is essential to point out that despite the extensive studies involving ML algorithms for the diagnosis of ASD (as mentioned in Table 1), previous works considered just one pairwise metric, i.e., Pearson correlation21,22,27. However, as verified in previous studies (e.g.46), correlation metrics are vital for diagnosing mental disorders. Therefore, we considered nine different pairwise metrics to find which best captures the ASD brain changes. Furthermore, unlike the studies in Table 1, we employed the SHAP (SHapley Additive exPlanations) values to identify the connections that differ in ASD and control patients. Moreover, we considered measures of complex networks to analyze how functional brain networks are modified in ASD. Thus, we proposed a more robust methodology that considers not just ML algorithms but also complex network measures while offering a medical interpretation of the results produced.
In the following sections, we describe the dataset, the methodology, and the results.
Data and data preprocessing
We consider the preprocessed version of the Autism Brain Imaging Data Exchange (ABIDE), which consists of 1112 datasets comprised of 539 ASD and 573 TD with 300s BOLD time series and provided by the Preprocessed Connectomes Project (PCP) dataset47. The PCP preprocessing pipeline includes cut time correction, motion correction, intensity normalization, and removal of artifacts such as breathing, heartbeat, and head motion. All data are properly anonymized in compliance with HIPAA requirements, and analyses are conducted following the University of Utah Institutional Review Board’s pre-approved protocols. All images were gathered with informed consent according to procedures established by human subjects research committees at each participating institution. The acquisition, informed consent, and site-specific protocols are described in detail at http://fcon1000.projects.nitrc.org/indi/abide/. Furthermore, it is available for use in Nilearn’s python package, a Python module for neuroimaging data. 242 ASD and 258 TD were used, and the preprocessed data were 0.5 Hz band-pass filtered since recent studies with fMRI have shown fluctuations may exist above that value48.
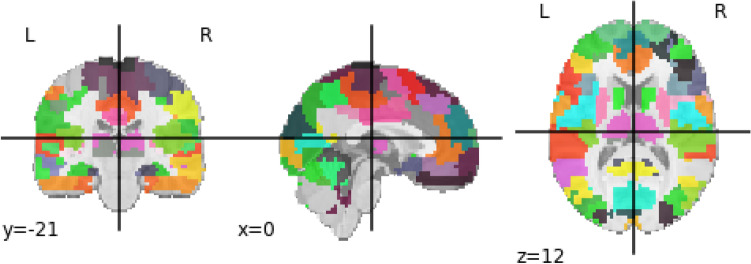
Brain regions of interest (ROI), rather than the entire BOLD time series obtained from each voxel of the brain image, are considered. A brain atlas containing these ROIs is used; therefore, only the BOLD time series voxels of this ROIS were adopted. Among the numerous predefined atlases, Bootstrap Analysis of Stable Clusters (BASC) was chosen since it was the map with the best performance for distinguishing ASD patients by deep learning model, according to22. It was proposed in49 and generated from group brain parcellation by BASC method, which is a k-means clustering-based algorithm that identifies brain networks with coherent activity in resting-state fMRI50. BASC map with a cluster number of 122 ROIs was used here (see Fig. 1). The preprocessed BOLD time series extracted for 122 regions can be found in the Supplementary Information.
Figure 1.
Figure developed using Python package Nilearn and containing BASC atlas with the 122 ROIs used in the present study.
A manual use of Yale BioImage Suite Package web application (Avaiable in https://bioimagesuiteweb.github.io/webapp/mni2tal.html) labelled the coordinates of each ROI for the identification of their names. After the extraction of the BOLD time series, the methodology described in “Section Methodology” was adopted.
Methodology
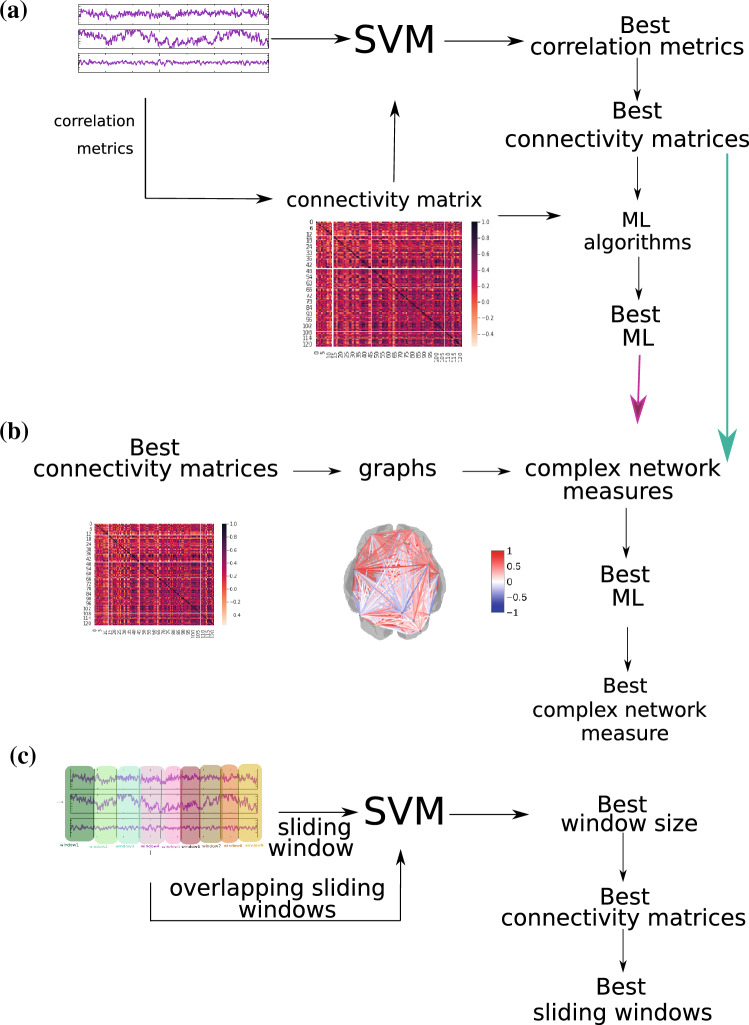
Figure 2 depicts the methodology workflow used and organized into three parts according to their aim, i.e., the finding of the best connectivity matrix (described in Fig. 2a and in “Section Connectivity matrix”), the best measures of complex networks (described in Fig. 2b and in “Section Complex network measures”), and the best sliding technique for differentiating ASD from TD patients (described in Fig. 2c and in “Section Sliding windows and overlapping sliding windows”). The python code with the methodology used in this work is available at: https://github.com/Carol180619/Paper-autism.git.
Figure 2.
The methodology used here for the diagnosis of autism. (a) methodology described in “Section Connectivity matrix”; (b) methodology reported in “Section Complex network measures”; (c) methodology described in “Section Sliding windows and overlapping sliding windows”.
Connectivity matrix
Once the time series for each of the 122 regions had been extracted, they were correlated according to Pearson Correlation (PC)51, Spearman Correlation (SC)52, Granger Causality (GC)53, Biweight Midcorrelation (BM)54, Sparce Canonical Correlation analysis (SCC)55, Graphical Lasso method (GL)56, Ledoit-Wolf shrinkage (LW)57, Mutual Information (MI)58, and Transfer Entropy (For the TE, MI, and GL metrics, a Min-max normalization and then a thresholding process was performed, with a value of 0.5, since these measures deal best with binary values) (TE)59. Finally, Fig. 3 displays the scheme to generate the connectivity matrices.
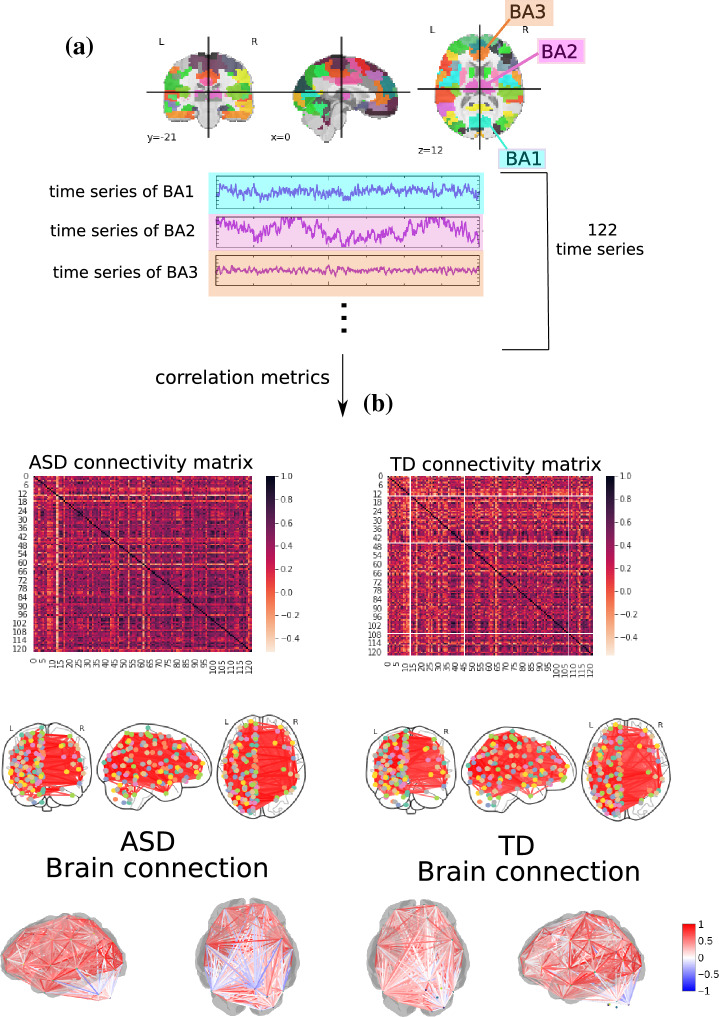
Figure 3.
Methodology to obtain the connectivity matrices. In (a), time series of 122 ROI is extracted from fMRI data with the use of BASC BOLD atlas (highlighted in blue, purple, and orange). The time series are correlated, (b), by pairwise statistical metrics (Pearson correlation was used in this example) towards forming the connectivity matrices, where each row and column correspond to one of the Brodmann areas for a patient with ASD for one with TD. The same highlighted matrices are arranged in a two-dimensional and three-dimensional brain schematic for better visualization.
Each matrix was reduced to the size of the vectors used as input to the ML algorithm. The support vector machine (SVM) algorithm60 was used to select the best methods to construct the correlation and connectivity matrices. We use this method because it has been considered in studies of ASD (see “Section Introduction”) and has a lower computational cost. The time series of each ROI was used for directly feeding SVM and finding the best connectivity metric that captured the brain changes due to ASD. It also checked whether the use of metrics was better than the direct use of time series - the one of better performance would be chosen. The results can be found in “Section Results related to the pairwise metrics”.
After the best brain connectivity metric had been determined, the following ML classifiers were used: Random Forest (RF)61, Naive Bayes (NB)62, Logistic regression (LG)63 with L-BFGS (Limited-memory Broyden Fletcher Goldfarb Shanno) solver64, Multilayer Perceptron (MLP)65, and tuned convolution neural network (called here tuned CNN) implemented in46. The SHAP value method was used for biological interpretation since it explains individual predictions of each attribute. The same sampling data set was used in all ML algorithms and split into training (train) and test sets, with of data comprising the test set. A k-fold cross-validation procedure was employed, with k = 10—this is a very used value for this method66–70). This procedure is used for model selection and hyper-parameter optimization. We considered the method called grid search, which was used for all ML algorithms except the untuned CNN model (since deep learning algorithms have a higher computational cost), as done in71–75. The hyper-parameter optimization values for each classifier model are provided in the “Appendix”. The standard performance metric accuracy76–80 was employed for evaluation. Due to the two-class (negative and positive) classification problem, other common metrics such as precision and recall were considered81–84. Precision (also called positive predictive value) corresponds to the hit rate in the negative class (here corresponding to the TD group), whereas recall (also called sensitivity) measures how well a classifier can predict positive examples (hit rate in the positive class), here related to ASD patients. F1 score72,85,86, another well-known measure, is the harmonic mean of recall and precision87. Regarding the visualization of the two latter measures, the Receiver Operating Characteristic (ROC) curve is a common method that displays the relation between the rate of true and false positives. The area below the curve, called Area Under ROC Curve (AUC), has been widely used in classification problems74,76,88,89. The AUC value ranges from 0 to 1- 1 corresponds to a classification result free of errors, and 0.5 indicates the classifier cannot distinguish the classes, as in a random choice. The micro average of the ROC curve, which computes the AUC metric independently for each class (it calculates AUC for healthy individuals, class zero, and separately calculates it for unhealthy ones, class one), was also considered. The average is computed considering the classes equally. The macro average was also employed in our evaluation - it does not consider the classes equally but aggregates their contributions separately and then calculates the average. The ML algorithms results can be found in “Section ML algorithms results”.
Complex network measures
A complex network (or a graph) was generated for each connectivity matrix to extract different measures. Towards inputting data into the ML algorithm, the complex network measures were stored in a matrix of attributes. Each column represents a complex network measure (feature), and each row denotes a subject. 2D matrices were generated for all subjects, as in90.
To describe the brain structure, the following complex network measures were calculated: assortativity coefficient91,92, betweenness centrality (BC)93, average shortest path length (APL)94, closeness centrality (CC)95, diameter96, hub score97, average degree of nearest neighbors98 (Knn), eigenvector centrality (EC)99, mean degree100, second moment of the degree distribution (SMD)101, entropy of the degree distributuion (ED)102, transitivity103,104, complexity, k-core105,106, eccentricity107, density108, and efficiency109.
Newly developed metrics (described in detail in90) reflecting the number of communities in a complex network were also applied. Community detection algorithms were also used in our study110–112. Since the community detection measures must be transformed into a single scalar value to be included in the matrix, community detection algorithms were applied to find the largest community. The average path length within the community was then calculated and received a single value as a result. The community detection algorithms used were the fastgreedy (FC)113, Infomap (IC)114, leading eigenvector (LC)115, label propagation (LPC)116, edge betweenness (EBC)117, spinglass (SPC)118, and multilevel community identification (MC)119. The abbreviations were extended with the letter “A” (for average path length) to indicate the approach (AFC, AIC, ALC, ALPC, AEBC, ASPC, and AMC).
These network measures were utilized to characterize the brain structure. Thus each observation (which represents the Patient’s brain network) is represented by a vector with these metrics. The results are provided in “Section Results for complex networks measures”.
Sliding windows and overlapping sliding windows
Due to the common issue of small datasets in neuroscience, the previously described methodology was expanded by a sliding window data augmentation approach. First, the sample size was increased by splitting each time series into smaller series. Such an increase can be achieved with either mutually exclusive sections of the time series or overlapping sections of the sliding windows, in which portions of the sequence are repeated in multiple observations.
A sample with 50 patients (25 ASD and 25 TD) was considered from the initial sample (242 ASD and 258 TD) for window sliding and overlapping windows sliding techniques evaluations. Then, the BOLD time series with 300 s were divided into windows of 15, 20, 30, 50, and 60 s and placed into the SVM to check the best way to split data. Then, with the best window size, it was also considered overlap sizes of 10%, 15%, 25%, 35%, 45%, and 55% of it. In other words, if the overlapping is 10%, each sliding window size has depicted a repetition of 10% of the previous window. This approach is used to avoid losing information when sliding.
The connection matrices are constructed using the best partitioning technique and the best correlation metric that fed the previously computed best classifier (see Fig. 2). The same sliding workflow was considered with samplings of 10, 20, 30, 50, 124, and 188 patients. The choice of such different sizes was based on previous neuroscience studies that used fMRI of similar sample sizes, respectively120–125.
Additionally to the performance metrics, a mean square error (MSE) was obtained for each sampling and each iteration of the k-fold cross-validation, resulting in an error vector. It was compared with the vector of the MSE obtained using the whole sample by statistical Student’s paired t-test126. The results are provided in “Section 3.4”.
Results
ML algorithms were applied for three different levels of data abstraction, namely (A) the connectivity matrix, (B) the matrix of attributes, whose elements are complex network measures calculated from (A), and (C) sliding data (see Fig. 2). In addition, the sliding window method was employed as an augmentation technique on small data samples to evaluate whether this methodology is advantageous when dealing with these data sets. We verify that all approaches automatically detected changes in the brain of ASD patients. The highest classification performance was obtained for the connectivity matrix with a 99% mean AUC (Table 2). Sections “Results related to the pairwise metrics”, “Results for complex networks measures”, and “Results from sliding windows and overlapping sliding windows” detail the results.
Table 2.
The table contains the summary of all results of the present work.
| Data abstraction level | Subset | AUC | Accuracy | F1 | Recall | Precision |
|---|---|---|---|---|---|---|
| Connectivity matrix | Train | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| Test | 0.99 | 0.99 | 0.99 | 0.99 | 0.99 | |
| Complex network | Train | 0.93 | 0.93 | 0.89 | 0.88 | 0.91 |
| Test | 0.98 | 0.98 | 0.98 | 0.98 | 0.98 | |
| Sliding data | Train | 0.84 | 0.84 | 0.84 | 0.84 | 0.84 |
| Test | 0.81 | 0.81 | 0.81 | 0.81 | 0.81 |
Classification using the data abstraction connectivity matrix best-captured brain changes due to ASD. The best performance is highlighted in bold.
Results related to the pairwise metrics
Table 3 contains the results for each connectivity matrix with different types of pairwise statistical metrics. SVM was used to detect the best one for capturing the brain changes due to ASD.
Table 3.
Results from different ML algorithms.
| Measures | Subset | AUC | Accuracy | F1 | Recall | Precision |
|---|---|---|---|---|---|---|
| Time series | Train | 0.49 | 0.51 | 0.00 | 0.27 | 0.35 |
| Test | 0.50 | 0.51 | 0.34 | 0.50 | 0.26 | |
| PC | Train | 0.67 | 0.67 | 0.69 | 0.67 | 0.66 |
| Test | 0.60 | 0.60 | 0.60 | 0.60 | 0.60 | |
| SC | Train | 0.98 | 0.98 | 0.97 | 0.98 | 0.98 |
| Test | 0.98 | 0.98 | 0.98 | 0.98 | 0.98 | |
| GC | Train | 0.51 | 0.52 | 0.00 | 0.32 | 0.37 |
| Test | 0.50 | 0.51 | 0.34 | 0.50 | 0.26 | |
| BM | Train | 0.75 | 0.75 | 0.72 | 0.75 | 0.75 |
| Test | 0.75 | 0.75 | 0.75 | 0.75 | 0.75 | |
| SCC | Train | 0.67 | 0.67 | 0.65 | 0.67 | 0.66 |
| Test | 0.62 | 0.62 | 0.62 | 0.62 | 0.62 | |
| GL | Train | 0.66 | 0.66 | 0.65 | 0.66 | 0.66 |
| Test | 0.57 | 0.58 | 0.57 | 0.57 | 0.57 | |
| LW | Train | 0.66 | 0.66 | 0.64 | 0.66 | 0.65 |
| Test | 0.58 | 0.58 | 0.58 | 0.58 | 0.58 | |
| MI | Train | 0.49 | 0.50 | 0.40 | 0.50 | 0.49 |
| Test | 0.49 | 0.49 | 0.49 | 0.49 | 0.49 | |
| TE | Train | 0.90 | 0.90 | 0.89 | 0.90 | 0.90 |
| Test | 0.91 | 0.91 | 0.91 | 0.91 | 0.91 |
The best MLs were RF and LR, whose performances are highlighted.
Significant values are in bold.
Spearman correlation coefficient (SC) achieved the best performance, followed by transfer entropy (TE). Finally, the best connectivity matrix was tested with the other ML algorithms to determine which best differentiated ASD patients from TD ones.
ML algorithms results
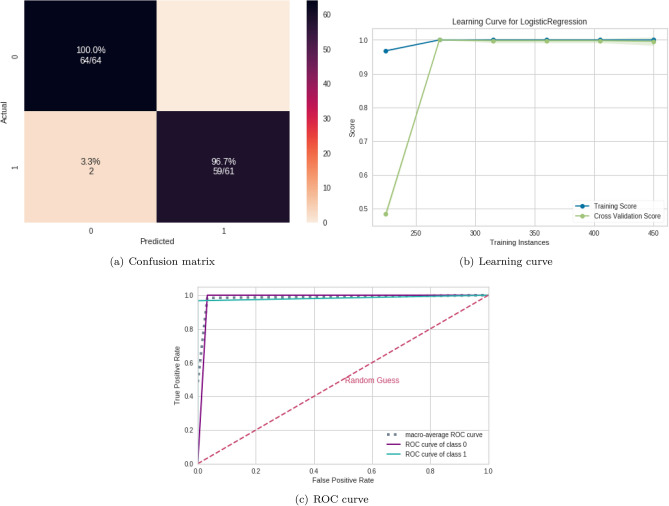
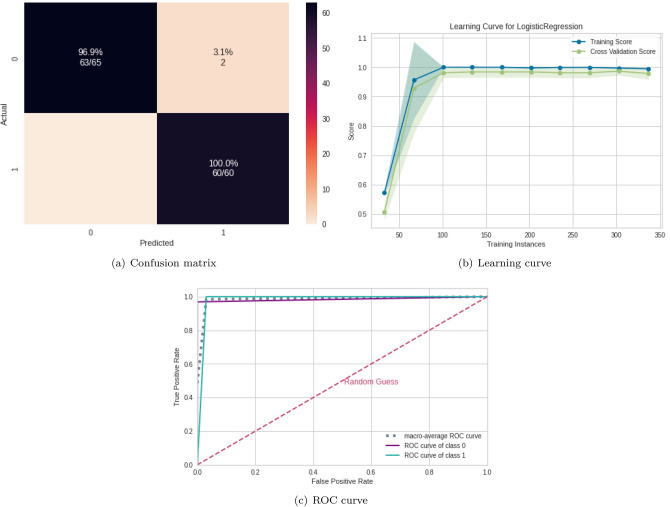
According to Table 4, the best classifiers are the random forest (RF) and logistic regression (LR). Since LR has a lower computational cost, it was chosen for the next steps. Its performance for the test set was equal to 0.99 for the mean AUC, precision, F1, recall, and accuracy. Figure 4 displays the confusion matrix (Fig. 4a), the learning curve (Fig. 4b), and the ROC curve (Fig. 4c), respectively.
Table 4.
Results from different ML algorithms.
| ML algorithm | Subset | AUC | Accuracy | F1 | Rec. | Pre. |
|---|---|---|---|---|---|---|
| SVM | Train | 0.99 | 0.99 | 0.99 | 0.99 | 0.99 |
| Test | 0.98 | 0.98 | 0.98 | 0.98 | 0.98 | |
| RF | Train | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| Test | 0.99 | 0.99 | 0.99 | 0.99 | 0.99 | |
| NB | Train | 1.00 | 1.00 | 0.99 | 1.00 | 0.99 |
| Test | 0.98 | 0.98 | 0.98 | 0.98 | 0.98 | |
| LR | Train | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| Test | 0.99 | 0.99 | 0.99 | 0.99 | 0.99 | |
| MLP | Train | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| Test | 0.98 | 0.99 | 0.99 | 0.99 | 0.99 | |
| Untuned CNN | Train | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| Test | 0.86 | 0.87 | 0.92 | 0.94 | 0.90 |
The best ML was RF and LR, whose performances are highlighted.
Significant values are in bold.
Figure 4.
ML results from connectivity matrices. (a) Confusion matrix indicating a TN rate (purple, according to the color bar) and a TP rate (blue, according to the color bar). (b) The learning curve for the training accuracy (blue) and for test accuracy (green). (c) ROC curve with classes 0 (TD) and 1 (ASD).
The learning curve evaluates the model’s predictability by varying the size of the training set38. The results show that the entire database is optional for achieving the highest validation accuracy. Regarding the classification model, TP (related to class 1) was higher than TN, showing that it better detects ASD patients (see confusion matrix in Fig. 4b).
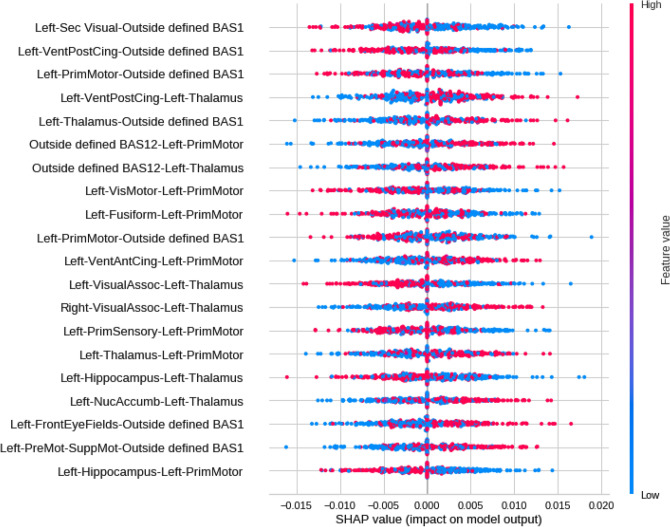
SHAP values were calculated to quantify the importance of brain connections for the logistic regression classifier (LR) (see Fig. 5 for the results). The area between regions Left-Sec Visual (visual cortex) and Outside defined BAS1 (area outside Brodmann’s map), identified as the cerebellum, was the most important connection. According to the data in Fig. 5, low correlation values (blue dots) for the connection (Left-Sec Visual and Outside defined BAS1) were essential for the detection of ASD patients, and high values of correlation (red dots) were important for the detection of TD ones. The second most crucial connection was detected between the Left ventral posterior cingulate cortex (Left-VentPostCing) and, again, the cerebellum (Outside defined BAS1). Figure 6 depicts the corresponding brain regions.
Figure 5.
Feature importance ranking for the LR classifier with brain regions in descending order. The connection between the Left-Sec Visual and Outside defined BAS1 regions is the most important for classifying ASD patients.
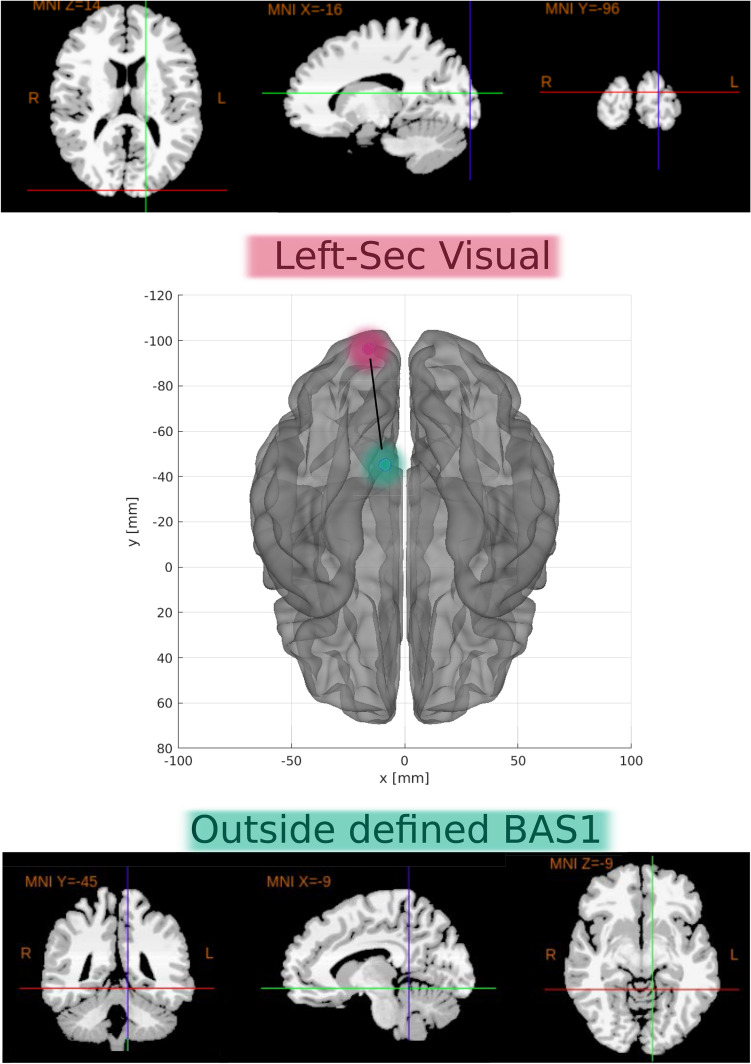
Figure 6.
The most important connection found. Two-dimensional schematic (ventral-axis), where the connection between the Left-Sec Visual region (visual cortex, highlighted in pink) and Outsides BAS1 (cerebellum, highlighted in green) is observed in the central region. The brain plot was developed by the Braph tool127, and each region was plotted using the Brodmann map from Yale BioImage Suite Package (upper and lower regions in the Figure).
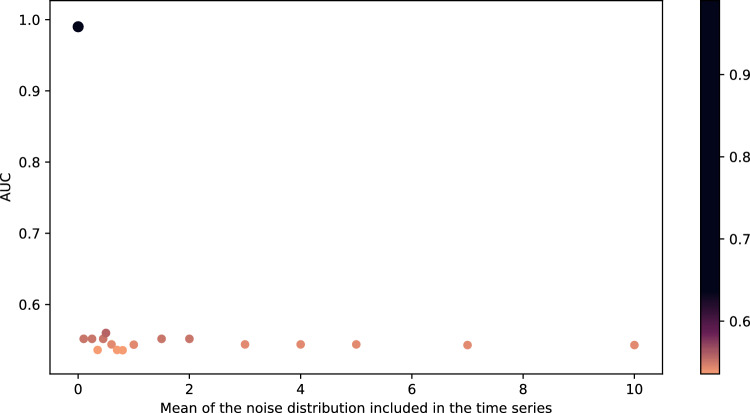
Since LG was the algorithm that provided the best performance, it was used in the following subsections. Furthermore, since the results were close to 100%, noises were inserted into the ASD and TD time series for further testing the model in this study. Such noises were generated by a normal distribution with a standard deviation equal to 0.1 and mean on the interval [0, 10]. After introducing the noises, Spearman’s correlation was used to generate the connectivity matrices from the time series. The results of the average AUC calculated on the test set are shown in Fig. 7. According to Fig. 7, the AUC according to the noise follows approximately a decreasing logarithmic function.
Figure 7.
The mean AUC of the test was obtained with the insertion of noise generated by a normal distribution with 0.1 standard deviation and 0–10 mean range.
Results for complex networks measures
The performance of the test sample considering the complex network yields a mean AUC equal to 0.98, 0.98 for precision, 0.98 for F1 score, 0.98 for recall, and 0.99 for accuracy. Confusion matrix Fig. 8, learning curve Fig. 8, and ROC curve Fig. 8 are shown in Fig. 8. Furthermore, according to Fig. 8, the whole dataset was unnecessary because the best result could be reached with only 100 train instances.
Figure 8.
ML results from complex network measures. (a) Confusion matrix indicating a TN rate (blue, according to the color bar) and a TP rate (purple, according to the color bar). (b) The learning curve for the training accuracy (blue) and for test accuracy (green). (c) ROC curve with classes 0 (TD) and 1 (ASD).
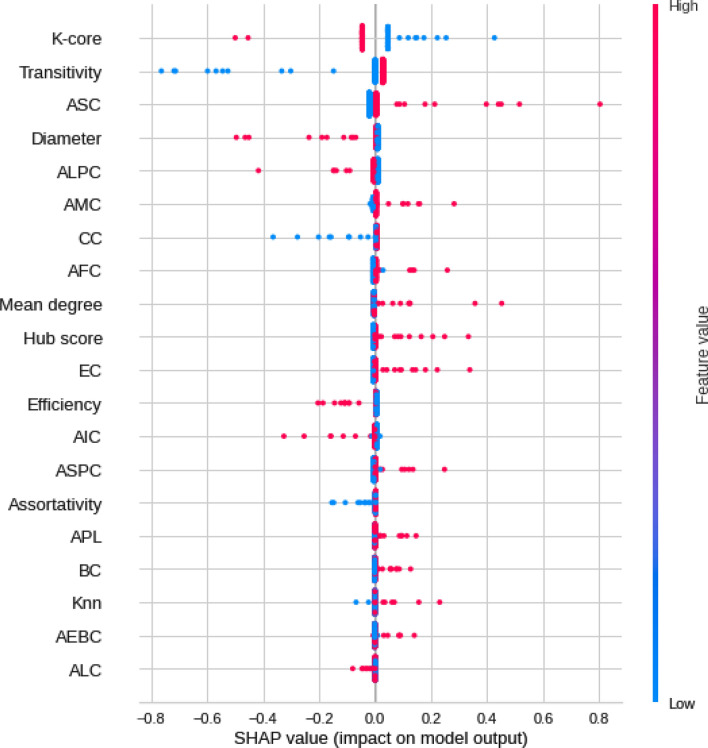
According to the SHAP values in Fig. 9, the most crucial measure for the model was the k-core, followed by the AEBC, introduced in90. High k-core values (pink dots) indicate their importance for the detection of TD, and low ones (blue dots) are important for the detection of ASD (Fig. 9). Low AEBC values (blue dots) indicate its importance for the detection of ASD, and high ones (pink dots) suggest its importance for the detection of TD. Higher values of efficiency were associated with TD patients; higher values of transitivity were associated with ASD, and low values indicated TD. Remarkably, the seven measures introduced in90 appeared in the ranking of best ones.
Figure 9.
Feature importance ranking for LR classifier with features in a descending order. K-core measure is the most important for the classification of ASD patients, followed by the AEBC measure.
Results from sliding windows and overlapping sliding windows
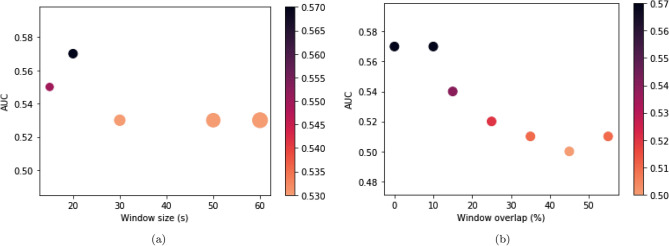
In this section, since two data augmentation techniques have been considered, a sample with 50 patients (25 ASD and 25 TD) was considered from the initial sample (242 ASD and 258 TD). Figure 10a shows the performance of SVM fed by time series divided into different window sizes. The best performance was achieved with a window size of 20 s. Figure 10b shows the best performance obtained with no overlapping or with a 10% of the time window size. Consequently, 10% overlapping was considered for the next step to avoid loss of information in the sliding process.
Figure 10.
Results of sliding and overlapping window sizes. (a) Mean AUC test obtained for the different window sizes. The width of the points in the graph corresponds to the window size variation, and their colors are indicated in the color bar. (b) The mean AUC test was obtained for the different overlapping sizes corresponding to the percentage of the window size. The colors of the points in the graph are depicted in the color bar.
The sliding process was used with different sample sizes, and the results are shown in Table 5.
Table 5.
Performance of the LR algorithm with the use of the sliding process and a varied number of samples of TD and ASC patients.
| Data abstraction level | Subset | AUC | Acccuracy | F1 score | Recall | Precision | T-test |
|---|---|---|---|---|---|---|---|
| Sample 10 | Train | 0.79 | 0.80 | 0.80 | 0.77 | 0.81 | 7.25 e-07 Threshold p-value |
| Test | 0.80 | 0.80 | 0.80 | 0.80 | 0.80 | ||
| Sample 20 | Train | 0.80 | 0.80 | 0.80 | 0.77 | 0.80 | 7.11 e-05 Reject null hypothesis |
| Test | 0.80 | 0.80 | 0.80 | 0.80 | 0.80 | ||
| Sample 30 | Train | 0.84 | 0.84 | 0.84 | 0.84 | 0.84 | 3.14 e-06 Reject null hypothesis |
| Test | 0.81 | 0.81 | 0.81 | 0.81 | 0.81 | ||
| Sample 50 | Train | 0.75 | 0.75 | 0.75 | 0.76 | 0.75 | 1.24 e-08 |
| Test | 0.75 | 0.75 | 0.75 | 0.75 | 0.75 | ||
| Sample 124 | Train | 0.70 | 0.70 | 0.70 | 0.72 | 0.70 | 2.77e-09 |
| Test | 0.73 | 0.73 | 0.73 | 0.73 | 0.73 | ||
| Sample 188 | Train | 0.70 | 0.70 | 0.70 | 0.70 | 0.70 | 7.60 e-12 |
| Test | 0.74 | 0.74 | 0.74 | 0.73 | 0.74 |
The best performance is highlighted in bold and was achieved with 30 patients.
Paired Student’s t-test (here called t-test) was also calculated between the sample performance and the performance for the whole data set. The null hypothesis is that the performances were statistically different. Therefore, a sample size of only ten patients was taken as a basis for comparison, given the premise that their performance should be statistically different when the entire database is considered for such a small sample size. Only samples for which the null hypothesis could be rejected (p-value greater than or equal to the baseline value for comparison) were considered, i.e., 10 and 20 patients. In other words, the performance of those two sizes showed no statistically significant differences from the data set but very similar results (Table 5). In other words, the performance of these two sample sizes showed no statistically significant differences from the data set but very similar results.
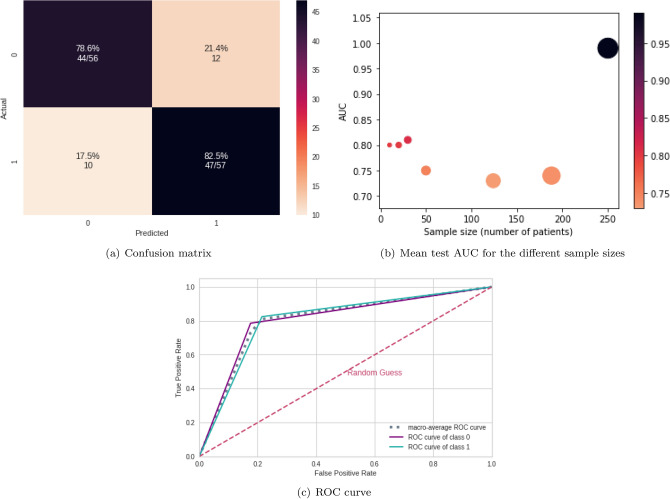
Figure 11 shows the confusion matrix (Fig. 11a) for the sample size of 30 patients, the mean AUC test for each sample size (Fig. 11b), and the ROC curve for the sample size of 30 patients (Fig. 11c). According to Fig. 11b, ASD and TD patients were differentiated even with different sample sizes, with above 79% AUC and accuracy.
Figure 11.
Results from connectivity matrices and ML. (a) Confusion matrix indicating an true negative rate (blue, according to the color bar) and a true positive rate (purple, according to the color bar). (b) Mean test AUC was obtained for the different sample sizes; the width of the points in the graph corresponds to the window size variation, and their colors are depicted in the color bar. (c) ROC curve with classes 0 (TD) and 1(ASD).
Discussion
The results from using the abstraction levels of the connectivity matrix and complex network data were superior to those reported in the literature (see Table 1). Therefore, the workflow developed here is more effective for detecting ASD patients with above 95% mean accuracy and mean AUC, and SC was the measure that best-captured brain changes in the patients (as an example, it is more robust for non-linear correlations than PC128). Since the Pearson correlation coefficient (PC) was ineffective in discriminating between the two classes (ASD and control subjects), we can conclude that brain changes due to ASD have a non-linear nature. Also, LG provides the best results, being the most suitable machine learning model, with lower computational cost than other ML algorithms used here (such as untuned CNN). Furthermore, we obtained better precision and recall compared with the studies presented in Table 1. A higher precision indicates that our model can better infer that an element belongs to class one (with ASD). In contrast, a higher recall implies that more elements with class one (with ASD) are captured. Furthermore, we obtained better precision and recall compared with the studies presented in Table 1. A higher precision indicates that our model can better infer that an element belongs to class one (with ASD). In contrast, a higher recall measure indicates very few false negatives, in our case, elements from class one, with ASD, and the model classifies as TD, which is helpful for medical data for correct diagnosing.
The most important connection in the first five significant correlations was observed between Left-Sec Visual (visual cortex) and cerebellum (Outside defined BAS1) regions. Low correlation values (blue dots) were important for detecting ASD patients, whereas high values (red dots) indicated TD. The second most crucial connection was established between the Left- VentPostCing and, again, the cerebellum (Outside de- fined BAS1) regions. Finally, the cerebellum (Outside defined BAS1), Left-Thalamus, and Left-Prim Motor appeared in several primary connections. Notably, Left-Thalamus has been reported in other studies associated with ASD129,130.
Left-Sec Visual (visual cortex) is a part of the cerebral cortex that processes visual information, and a lower connection to the cerebellum (Outside defined BAS1) is more associated with ASD.
Left-VentPostCing corresponds to the upper part of the limbic system, i.e., part of the brain involved in behavioral and emotional responses. According to the literature, reductions in the functional connectivity of that brain area are expected in ASD patients131, which is consistent with our results since the region is less connected to the Outside BAS1 in ASD patients.
The brain region changes addressed elsewhere have been reported in the ASD literature. For example, both hyper- and hypo-connectivity were observed in ASD through stepwise functional connectivity in the resting state132. In the same study, hypoconnectivity was related to the parietal and frontal regions of the attention networks, whereas hyperconnectivity was observed for the default mode network in the visual cortex region. The authors in133 claimed ASD patients have higher activity in the occipital cortex bilaterally and in the Anterior Cingulate Cortex (ACC) but lower activation in the frontal gyri in comparison with a control group during automatic identification of visual changes. However131, reported reduced functional connectivity in ACC in ASD patients. The low correlation observed between the posterior cingulate region and the cerebellum (Left-VentPostCing vs. Outside BAS1) found in our study seems to point to a dysfunction, i.e., an alteration in functional communication in ASD. Such a correlation differs from the findings for ASD reported by other researchers, who have pointed to the anterior cingulate as one of the altered brain regions in ASD131,133,134 and found cortical thinning for ASD in the right ACC. Such results have led us to hypothesize that the ACC and other cingulate regions are implicated in ASD. Moreover, our attention has been drawn to the cingulate region and its relationships with other brain regions. The hypothesis can be reinforced by the findings of135, who reported abnormal functional connectivity between the posterior cingulate cortex and the ventromedial prefrontal cortex for ASD, with hypoconnectivity. Other studies have shown ASD patients have altered intra- and inter-network connectivity among the cerebellum, visual networks, and the sensory-motor region. According to136, the connectivity among those regions is related to problems in sensory and visual motor integration present in ASD. Such findings have corroborated our results of a low correlation between visual cortex regions and the cerebellum (first correlation of highest importance) and a correlation between the left primary motor region and the cerebellum (third correlation of highest importance). The cerebellum is associated with motor functions such as balance maintenance, executive control of movements, and cognitive, behavioral, and language functions137–141. Studies with fMRI have pointed to structural and functional changes in several cerebellum regions related to ASD. Lesions in the cerebellum compromise the cognitive, perceptual, and motor functioning of those systems142. Stoodley143 claimed abnormalities in the different cerebellar regions would produce behavioral symptoms associated with the functional breakdown of specific cerebrocerebellar circuits, thus compromising the acquisition of certain skills. Moreover, such long-term changes would significantly impact behavior, language, and social cognition, hence dysfunctions in behaviors associated with ASD, dyslexia, and Attention- Deficit/Hyperactivity Disorder (ADHD).
Our study’s third most important correlation was between Left-PrimMotor and the cerebellum. The motor cortex is also associated with alterations in ASD patients. Nebel et al.144 reported a delayed functional specialization within the motor cortex and alterations in both size and segregation of the primary motor cortex and that the functional sub-networks of the motor control system might be altered in autism. Mostofsky et al.145 observed a low motor ability in ASD related to increased white matter volume in the left hemisphere’s primary motor and premotor regions. We found a low correlation between Left-PrimMotor and the cerebellum for ASD, two important regions for motor control and skill, balance, and executive control of movements. Such a low correlation may cause problems in overall motor performance, thus interfering with socialization, which is commonly observed in ASD.
Regarding complex network measures, the most important measure for the model was the k-core, followed by the AEBC. K-Core decomposes the graph for finding important highly and mutually interconnected areas146,147. The k-core average was used for the calculation, which provides the degree of the subgraph in which all nodes have the same degree value, and helps identify small contiguous core areas in a network. High k-core values (pink dots) indicate its importance for the detection of TD, whereas low ones (blue dots) suggest ASD patients (Fig. 9, hence a weaker network connection among them. In contrast, EBC measures the average size of the largest community found by the edges betweenness method. For AEBC, low scores (blue dots) were important for detecting ASD, and high scores (pink dots) were important for detecting TD. Therefore, smaller communities can be detected by the presence of ASD. Higher values of efficiency were associated with TD patients and greater integration of networks and distributions of information in them. Therefore, the distribution of information in the functional networks of ASD patients is worse than that in TD. Concerning transitivity, a segregation network measure of the propensity of nodes to be grouped, higher values were associated with ASD, and low values indicated TD and the presence of more isolated communities clustered together.
The sliding process effectively differentiated TD from ASD patients since 30 patients achieved a 0.81 AUC and 0.81 mean accuracy. A statistical comparison between the sliding process and complete data showed no significant differences. Despite a lower performance with the use of the entire database, the technique could distinguish between ASD and TD patients with a significantly reduced amount of data, proving attractive for few data regarding ASD, as in25,28 (see Table 1). Furthermore, compared with some studies in Table 124–28, our model, using these data augmentation techniques in a smaller amount of data, performed better in terms of AUC, accuracy, recall, and precision.
Conclusions and future work
The workflow developed with the use of fMRI data could distinguish TD from ASD patients with both accuracy and AUC above 81%. The best pairwise statistical metric that captured brain changes due to ASD was SC, and the best-performing machine learning model was LG. According to the metric and the algorithm, the three most important brain connections with low values were established among Left-Sec Visual (visual cortex), Left-VentPostCing, and Left-PrimMotor with Outside defined BAS1 in ASD.
The functional connectivity of the Left-VentPostCing Posterior cingulate cortex is known to be reduced in ASD patients, which is consistent with our findings since this region is less connected to the cerebellum (Outside BAS1 region) in patients with ASD. Regarding complex networks, the brain networks of ASD patients showed more segregation, a weaker distribution of information across the network, and less connectivity. The sliding process employed effectively differentiated TD from ASD patients since a sample with 30 patients achieved 0.81 mean AUC and mean accuracy. A statistical comparison between the sliding process and complete data showed no significant differences. Therefore, the methodology is appropriate for cases of data of a small sample size.
Future studies may involve the application of the methodology to other fMRI data, as in148 for schizophrenia and fMRI data from ADHD-200 Global Competition. It can also be adopted with EEG data from patients with dystonia149. Other methodologies, such as the transfer learning method150, may be applied to small databases for comparison purposes.
Supplementary Information
Acknowledgements
F.A.R. is indebted to CNPq (Grant 309266/2019- 0) and FAPESP (Grant 19/23293-0) for the financial for the financial support provided to this research. T.G.L.O.T acknowledges FAPESB (Grant Number 307/2020 - Cota 2020; BOL0202/2020) for the financial support. A.M.P. is indebted to FAPESP (Grant 2019/22277-0) for the financial support. K.R. acknowledges FAPESP Grant 2019/26595-7. C.T. gratefully acknowledges financial support from the Zentrum für Wisschenschaftliche Services und Transfer (ZeWiS) Aschaffenburg, Germany. C.L.A acknowledges Angela Cristina Pregnolato Giampedro for revising the manuscript.
Abbreviations
- ABIDE
Brain Imaging Data Exchange
- APL
Average shortest path length
- ASD
Autism spectrum disorder
- AUC
Area Under ROC Curve
- BASC
Bootstrap Analysis of Stable Clusters
- BC
Betweenness centrality
- BM
Biweight Midcorrelation
- BOLD
Blood Oxygenation Level Dependent
- CC
Closeness centrality
- DTI
Diffusion Tensor Imaging
- EBC
Edge betweenness community detection
- EC
Eigenvector centrality
- ED
Entropy of the degree distributuion
- EEG
Electroencephalogram
- FC
Fastgreedy community detection
- fMRI
Functional magnetic resonance imaging
- GC
Granger Causality
- GL
Graphical Lasso method
- IC
Infomap community detection
- Knn
Average degree of nearest neighbors
- L-BFGS
Limited-memory Broyden Fletcher Goldfarb Shanno
- LC
Leading eigenvector community detection
- LG
Logistic regression
- LPC
Label propagation community detection
- LW
Ledoit-Wolf shrinkage
- MC
Multilevel community detection
- MI
Mutual Information
- ML
Machine learning
- MLP
Multilayer Perceptron
- MSE
Mean Square Error
- NB
Naive Bayes
- PC
Pearson Correlation
- PCP
Preprocessed Connectomes Project
- RF
Random Forest
- ROC
Receiver Operating Characteristic
- ROI
Brain regions of interest
- SC
Spearman Correlation
- SCC
Canonical Correlation analysis
- SHAP
SHapley Additive ExPlanations
- SMD
Second moment of the degree distribution
- SPC
Spinglass community detection
- SVM
Support vector machines
- TD
Typical development
- TE
Transfer Entropy
- Tuned CNN
Tuned convolution neural network
Appendix
Grid search hyperparameter tuning
See Table 6.
Table 6.
Hyperparameters for each classifier using Grid search optimizer.
| Classifier | Hyperparameters and description | Values |
|---|---|---|
| RF | max_depth: Maximum depth of the tree | [1,2,5,10,20,80] |
| max_features: Number of features to be considered toward a best split | [2, 3,5,10] | |
| min_samples_leaf : Minimum number of samples required to be at a leaf node | [1,2,3, 4, 5] | |
| min_samples_split: Minimum number of samples for the split of an internal node | [1,2,8, 10, 12,20] | |
| n_estimators: Number of trees in the forest | [1,2,3,5,10, 30,50,100, 200, 300,500] | |
| SVM | Kernel: Specifies the kernel type to be used in the algorithm | [rbf, linear] |
| Gamma: Kernel coefficient | [1e-3, 1e-4] | |
| C: Regularization parameter | [1, 10, 100, 1000] | |
| NB | Var_smoothin:Portion of the largest variance of all features that is added to variances for calculation stability | range 1e-09 to 1 |
| MLP | Activation: Activation function for the hidden layer | [identity, logistic, tanh, relu] |
| Solver: Solver for weight optimization | [lbfgs, sgd, adam] | |
| Alpha: L2 penalty (regularization term) parameter | [0.0001,1e-5,0.01,0.001] | |
| Batch_size: Size of minibatches for stochastic optimizers | [1000,5000] | |
| Learning_rate: Learning rate schedule for weight updates | [constant, invscaling, adaptive] | |
| Learning_rate_init: The initial learning rate used | [0.001,0.01,0.1,0.2,0.3] | |
| LR | C: Each value in Cs describes the inverse of r egularization strength | range 0.001 to 1000 |
| Penalty: Specifies the norm of the penalty | [l1, l2] |
Author contributions
Conceptualization: C.L.A, F.A.R. Data curation: C.L.A, F.A.R., T.G.L.O.T. Formal analysis: C.L.A, F.A.R., T.G.L.O.T, P.C.A, A.M.P, K.R, C.T, J.A.M.P. Investigation: C.L.A, F.A.R., T.G.L.O.T, P.C.A, A.M.P, K.R, C.T, J.A.M.P. Methodology: C.L.A Validation: C.L.A, T.G.L.O.T., K.R, F.A.R. Visualization: C.L.A Writing—original draft: C.L.A, F.A.R., T.G.L.O.T, P.C.A, A.M.P, K.R, C.T, J.A.M.P.
Funding
Open Access funding enabled and organized by Projekt DEAL.
Data availability
All data generated or analyzed during this study are included in this published article (and its Supplementary files).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-023-34650-6.
References
- 1.Lord C, Brugha TS, Charman T, Cusack J, Dumas G, Frazier T, Jones EJ, Jones RM, Pickles A, State MW, et al. Autism spectrum disorder. Nat. Rev. Dis. Primers. 2020;6:1. doi: 10.1038/s41572-019-0138-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Al-Beltagi M. Autism medical comorbidities. World J. Clin. Pediatrics. 2021;10:15. doi: 10.5409/wjcp.v10.i3.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.A. P. Association et al., American psychiatric association: Diagnosti c and statistical manual of mental disorders. Arlington (2013)
- 4.Hosozawa M, Sacker A, Cable N. Timing of diagnosis, depression and self-harm in adolescents with autism spectrum disorder. Autism. 2021;25:70. doi: 10.1177/1362361320945540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beaudet AL. Autism: Highly heritable but not inherited. Nat. Med. 2007;13:534. doi: 10.1038/nm0507-534. [DOI] [PubMed] [Google Scholar]
- 6.Belmonte MK, Allen G, Beckel-Mitchener A, Boulanger LM, Carper RA, Webb SJ. Autism and abnormal development of brain connectivity. J. Neurosci. 2004;24:9228. doi: 10.1523/JNEUROSCI.3340-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Belmonte MK, Yurgelun-Todd DA. Functional anatomy of impaired selective attention and compensatory processing in autism. Cogn. Brain Res. 2003;17:651. doi: 10.1016/S0926-6410(03)00189-7. [DOI] [PubMed] [Google Scholar]
- 8.DeRamus TP, Black BS, Pennick MR, Kana RK. Enhanced parietal cortex activation during location detection in children with autism. J. Neurodev. Disord. 2014;6:1. doi: 10.1186/1866-1955-6-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Euston DR, Gruber AJ, McNaughton BL. The role of medial prefrontal cortex in memory and decision making. Neuron. 2012;76:1057. doi: 10.1016/j.neuron.2012.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kennedy DP, Redcay E, Courchesne E. Failing to deactivate: Resting functional abnormalities in autism. Proc. Natl. Acad. Sci. 2006;103:8275. doi: 10.1073/pnas.0600674103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Keller TA, Kana RK, Just MA. A developmental study of the structural integrity of white matter in autism. NeuroReport. 2007;18:23. doi: 10.1097/01.wnr.0000239965.21685.99. [DOI] [PubMed] [Google Scholar]
- 12.Aoki Y, Abe O, Nippashi Y, Yamasue H. Comparison of white matter integrity between autism spectrum disorder subjects and typically developing individuals: A meta-analysis of diffusion tensor imaging tractography studies. Mol. Autism. 2013;4:1. doi: 10.1186/2040-2392-4-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.De Vico Fallani F, Rodrigues FA, da Fontoura Costa L, Astolfi L, Cincotti F, Mattia D, Salinari S, Babiloni F. Multiple pathways analysis of brain functional networks from EEG signals: An application to real data. Brain Topogr. 2011;23:344. doi: 10.1007/s10548-010-0152-z. [DOI] [PubMed] [Google Scholar]
- 14.Alves CL, Pineda AM, Roster K, Thielemann C, Rodrigues FA. EEG functional connectivity and deep learning for automatic diagnosis of brain disorders: Alzheimer’s disease and schizophrenia. J. Phys. Complex. 2022;3:025001. doi: 10.1088/2632-072X/ac5f8d. [DOI] [Google Scholar]
- 15.Pineda, A. M. & Rodrigues, F. A. Complex networks to differentiate elderly and young people. In Annual International Conference on Information Management and Big Data 435–444 (Springer, 2020)
- 16.Menon V, Crottaz-Herbette S. Combined EEG and FMRI studies of human brain function. Int. Rev. Neurobiol. 2005;66:291. doi: 10.1016/S0074-7742(05)66010-2. [DOI] [PubMed] [Google Scholar]
- 17.Formisano E, Kim D-S, Di Salle F, Van de Moortele P-F, Ugurbil K, Goebel R. Mirror-symmetric tonotopic maps in human primary auditory cortex. Neuron. 2003;40:859. doi: 10.1016/S0896-6273(03)00669-X. [DOI] [PubMed] [Google Scholar]
- 18.Sturzbecher, M. J. Detecção e caracterização da resposta hemodinâmica pelo desenvolvimento de novos métodos de processamento de imagens funcionais por ressonância magnética, Ph.D. thesis, Universidade de São Paulo (2006)
- 19.Biswal B, Zerrin Yetkin F, Haughton VM, Hyde JS. Functional connectivity in the motor cortex of resting human brain using echo-planar mri. Magn. Reson. Med. 1995;34:537. doi: 10.1002/mrm.1910340409. [DOI] [PubMed] [Google Scholar]
- 20.Hyde KK, Novack MN, LaHaye N, Parlett-Pelleriti C, Anden R, Dixon DR, Linstead E. Applications of supervised machine learning in autism spectrum disorder research: A review. Rev. J. Autism Dev. Disord. 2019;6:128. doi: 10.1007/s40489-019-00158-x. [DOI] [Google Scholar]
- 21.Al-Hiyali MI, Yahya N, Faye I, Al-Quraishi MS, Al-Ezzi A. Principal subspace of dynamic functional connectivity for diagnosis of autism spectrum disorder. Appl. Sci. 2022;12:9339. doi: 10.3390/app12189339. [DOI] [Google Scholar]
- 22.Subah FZ, Deb K, Dhar PK, Koshiba T. A deep learning approach to predict autism spectrum disorder using multisite resting-state FMRI. Appl. Sci. 2021;11:3636. doi: 10.3390/app11083636. [DOI] [Google Scholar]
- 23.Chen CP, Keown CL, Jahedi A, Nair A, Pflieger ME, Bailey BA, Müller R-A. Diagnostic classification of intrinsic functional connectivity highlights somatosensory, default mode, and visual regions in autism. NeuroImage Clin. 2015;8:238. doi: 10.1016/j.nicl.2015.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nunes AS, Vakorin VA, Kozhemiako N, Peatfield N, Ribary U, Doesburg SM. Atypical age-related changes in cortical thickness in autism spectrum disorder. Sci. Rep. 2020;10:1. doi: 10.1038/s41598-020-67507-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yamagata B, Itahashi T, Fujino J, Ohta H, Nakamura M, Kato N, Mimura M, Hashimoto R-I, Aoki Y. Machine learning approach to identify a resting-state functional connectivity pattern serving as an endophenotype of autism spectrum disorder. Brain Imaging Behav. 2019;13:1689. doi: 10.1007/s11682-018-9973-2. [DOI] [PubMed] [Google Scholar]
- 26.Devi, B., Kumar, S., Shankar, V. G. et al. Anadata: A novel approach for data analytics using random forest tree and SVM. In Computing, Communication and Signal Processing 511–521 (Springer, 2019)
- 27.Huang Z-A, Zhu Z, Yau CH, Tan KC. Identifying autism spectrum disorder from resting-state FMRI using deep belief network. IEEE Trans. Neural Netw. Learn. Syst. 2020;32:2847. doi: 10.1109/TNNLS.2020.3007943. [DOI] [PubMed] [Google Scholar]
- 28.McBride JC, Zhao X, Munro NB, Jicha GA, Schmitt FA, Kryscio RJ, Smith CD, Jiang Y. Sugihara causality analysis of scalp EEG for detection of early Alzheimer’s disease. NeuroImage Clin. 2015;7:258. doi: 10.1016/j.nicl.2014.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rudin C. Stop explaining black box machine learning models for high stakes decisions and use interpretable models instead. Nat. Mach. Intell. 2019;1:206. doi: 10.1038/s42256-019-0048-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ekanayake, I., Meddage, D. & Rathnayake, U. A novel approach to explain the black-box nature of machine learning in compressive strength predictions of concrete using shapley additive explanations (shap). Case Stud. Constr. Mater. e01059 (2022).
- 31.Steyerberg EW, Eijkemans MJ, Harrell FE, Jr, Habbema JDF. Prognostic modelling with logistic regression analysis: A comparison of selection and estimation methods in small data sets. Stat. Med. 2000;19:1059. doi: 10.1002/(SICI)1097-0258(20000430)19:8<1059::AID-SIM412>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 32.Ferguson AR, Nielson JL, Cragin MH, Bandrowski AE, Martone ME. Big data from small data: Data-sharing in the ‘long tail’ of neuroscience. Nat. Neurosci. 2014;17:1442. doi: 10.1038/nn.3838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bae H-J, Kim C-W, Kim N, Park B, Kim N, Seo JB, Lee SM. A perlin noise-based augmentation strategy for deep learning with small data samples of HRCT images. Sci. Rep. 2018;8:1. doi: 10.1038/s41598-018-36047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.D’souza RN, Huang P-Y, Yeh F-C. Structural analysis and optimization of convolutional neural networks with a small sample size. Sci. Rep. 2020;10:1. doi: 10.1038/s41598-020-57866-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lundberg, S. M. & Lee, S.-I. A unified approach to interpreting model predictions. In Proceedings of the 31st International Conference on Neural Information Processing Systems 4768–4777 (2017)
- 36.Bowen, D. & Ungar, L. Generalized shap: Generating multiple types of explanations in machine learning. arXiv preprint arXiv:2006.07155 (2020)
- 37.Rodríguez-Pérez R, Bajorath J. Interpretation of compound activity predictions from complex machine learning models using local approximations and shapley values. J. Med. Chem. 2019;63:8761. doi: 10.1021/acs.jmedchem.9b01101. [DOI] [PubMed] [Google Scholar]
- 38.Spadon G, de Carvalho AC, Rodrigues-Jr JF, Alves LG. Reconstructing commuters network using machine learning and urban indicators. Sci. Rep. 2019;9:1. doi: 10.1038/s41598-019-48295-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lashgari E, Liang D, Maoz U. Data augmentation for deep-learning-based electroencephalography. J. Neurosci. Methods. 2020;346:108885. doi: 10.1016/j.jneumeth.2020.108885. [DOI] [PubMed] [Google Scholar]
- 40.Qiang N, Dong Q, Liang H, Ge B, Zhang S, Sun Y, Zhang C, Zhang W, Gao J, Liu T. Modeling and augmenting of FMRI data using deep recurrent variational auto-encoder. J. Neural Eng. 2021;18:0460b6. doi: 10.1088/1741-2552/ac1179. [DOI] [PubMed] [Google Scholar]
- 41.Luo Y, Zhu L-Z, Wan Z-Y, Lu B-L. Data augmentation for enhancing EEG-based emotion recognition with deep generative models. J. Neural Eng. 2020;17:056021. doi: 10.1088/1741-2552/abb580. [DOI] [PubMed] [Google Scholar]
- 42.Chang C, Liu Z, Chen MC, Liu X, Duyn JH. EEG correlates of time-varying bold functional connectivity. Neuroimage. 2013;72:227. doi: 10.1016/j.neuroimage.2013.01.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li Y, Yang H, Li J, Chen D, Du M. EEG-based intention recognition with deep recurrent-convolution neural network: Performance and channel selection by grad-cam. Neurocomputing. 2020;415:225. doi: 10.1016/j.neucom.2020.07.072. [DOI] [Google Scholar]
- 44.Chang C, Metzger CD, Glover GH, Duyn JH, Heinze H-J, Walter M. Association between heart rate variability and fluctuations in resting-state functional connectivity. Neuroimage. 2013;68:93. doi: 10.1016/j.neuroimage.2012.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jie B, Liu M, Lian C, Shi F, Shen D. Designing weighted correlation kernels in convolutional neural networks for functional connectivity based brain disease diagnosis. Med. Image Anal. 2020;63:101709. doi: 10.1016/j.media.2020.101709. [DOI] [PubMed] [Google Scholar]
- 46.Alves, C. L. Diagnóstico de doenças mentais baseado em mineração de dados e redes complexas. Ph.D. thesis, Universidade de São Paulo
- 47.Nielsen JA, Zielinski BA, Fletcher PT, Alexander AL, Lange N, Bigler ED, Lainhart JE, Anderson JS. Multisite functional connectivity MRI classification of autism: Abide results. Front. Hum. Neurosci. 2013;7:599. doi: 10.3389/fnhum.2013.00599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Trapp C, Vakamudi K, Posse S. On the detection of high frequency correlations in resting state FMRI. Neuroimage. 2018;164:202. doi: 10.1016/j.neuroimage.2017.01.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bellec P, Rosa-Neto P, Lyttelton OC, Benali H, Evans AC. Multi-level bootstrap analysis of stable clusters in resting-state FMRI. Neuroimage. 2010;51:1126. doi: 10.1016/j.neuroimage.2010.02.082. [DOI] [PubMed] [Google Scholar]
- 50.Yang X, Zhang N, Schrader P. A study of brain networks for autism spectrum disorder classification using resting-state functional connectivity. Mach. Learn. Appl. 2022;8:100290. [Google Scholar]
- 51.Benesty, J., Chen, J., Huang, Y. & Cohen, I. Pearson correlation coefficient. In Noise Reduction in Speech Processing 1–4 (Springer, 2009)
- 52.Lubinski D. Introduction to the special section on cognitive abilities: 100 years after spearman’s (1904) general intelligence’,objectively determined and measured. J. Pers. Soc. Psychol. 2004;86:96. doi: 10.1037/0022-3514.86.1.96. [DOI] [PubMed] [Google Scholar]
- 53.Granger CW. Investigating causal relations by econometric models and cross-spectral methods. Econom. J. Econom. Soc. 1969;37:424–438. [Google Scholar]
- 54.Wilcox RR. Introduction to Robust Estimation and Hypothesis Testing. New York: Academic press; 2011. [Google Scholar]
- 55.Hardoon DR, Shawe-Taylor J. Sparse canonical correlation analysis. Mach. Learn. 2011;83:331. doi: 10.1007/s10994-010-5222-7. [DOI] [Google Scholar]
- 56.Sojoudi S. Equivalence of graphical lasso and thresholding for sparse graphs. J. Mach. Learn. Res. 2016;17:3943. [Google Scholar]
- 57.Ledoit O, Wolf M. Nonlinear shrinkage estimation of large-dimensional covariance matrices. Ann. Stat. 2012;40:1024. doi: 10.1214/12-AOS989. [DOI] [Google Scholar]
- 58.Kraskov A, Stögbauer H, Grassberger P. Estimating mutual information. Phys. Rev. E. 2004;69:066138. doi: 10.1103/PhysRevE.69.066138. [DOI] [PubMed] [Google Scholar]
- 59.Schreiber T. Measuring information transfer. Phys. Rev. Lett. 2000;85:461. doi: 10.1103/PhysRevLett.85.461. [DOI] [PubMed] [Google Scholar]
- 60.Bottou L, Lin C-J. Support vector machine solvers. Large Scale Kernel Mach. 2007;3:301. [Google Scholar]
- 61.Breiman L. Random forests. Mach. Learn. 2001;45:5. doi: 10.1023/A:1010933404324. [DOI] [Google Scholar]
- 62.Friedman N, Geiger D, Goldszmidt M. Bayesian network classifiers. Mach. Learn. 1997;29:131. doi: 10.1023/A:1007465528199. [DOI] [Google Scholar]
- 63.Tolles J, Meurer WJ. Logistic regression: Relating patient characteristics to outcomes. JAMA. 2016;316:533. doi: 10.1001/jama.2016.7653. [DOI] [PubMed] [Google Scholar]
- 64.Najafabadi MM, Khoshgoftaar TM, Villanustre F, Holt J. Large-scale distributed l-BFGS. J. Big Data. 2017;4:1. doi: 10.1186/s40537-017-0084-5. [DOI] [Google Scholar]
- 65.Hinton G, Rumelhart D, Williams R. Learning internal representations by error propagation. Parallel Distrib. Process. 1986;1:318. [Google Scholar]
- 66.Berrar, D. Cross-validation (2019).
- 67.Bengio Y, Grandvalet Y. No unbiased estimator of the variance of k-fold cross-validation. J. Mach. Learn. Res. 2004;5:1089. [Google Scholar]
- 68.Shah AA, Khan YD. Identification of 4-carboxyglutamate residue sites based on position based statistical feature and multiple classification. Sci. Rep. 2020;10:1. doi: 10.1038/s41598-020-73107-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kawamoto T, Kabashima Y. Cross-validation estimate of the number of clusters in a network. Sci. Rep. 2017;7:1. doi: 10.1038/s41598-017-03623-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chan J, Rea T, Gollakota S, Sunshine JE. Contactless cardiac arrest detection using smart devices. NPJ Digital Med. 2019;2:1. doi: 10.1038/s41746-019-0128-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sato M, Morimoto K, Kajihara S, Tateishi R, Shiina S, Koike K, Yatomi Y. Machine-learning approach for the development of a novel predictive model for the diagnosis of hepatocellular carcinoma. Sci. Rep. 2019;9:1. doi: 10.1038/s41598-019-44022-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhong Z, Yuan X, Liu S, Yang Y, Liu F. Machine learning prediction models for prognosis of critically ill patients after open-heart surgery. Sci. Rep. 2021;11:1. doi: 10.1038/s41598-021-83020-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Arcadu F, Benmansour F, Maunz A, Willis J, Haskova Z, Prunotto M. Author correction: Deep learning algorithm predicts diabetic retinopathy progression in individual patients. NPJ Digital Med. 2020;3:1. doi: 10.1038/s41746-020-00365-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Krittanawong C, Virk HUH, Kumar A, Aydar M, Wang Z, Stewart MP, Halperin JL. Machine learning and deep learning to predict mortality in patients with spontaneous coronary artery dissection. Sci. Rep. 2021;11:1. doi: 10.1038/s41598-021-88172-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rashidi HH, Sen S, Palmieri TL, Blackmon T, Wajda J, Tran NK. Early recognition of burn-and trauma-related acute kidney injury: A pilot comparison of machine learning techniques. Sci. Rep. 2020;10:1. doi: 10.1038/s41598-019-57083-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mincholé A, Rodriguez B. Artificial intelligence for the electrocardiogram. Nat. Med. 2019;25:22. doi: 10.1038/s41591-018-0306-1. [DOI] [PubMed] [Google Scholar]
- 77.Tolkach Y, Dohmgörgen T, Toma M, Kristiansen G. High-accuracy prostate cancer pathology using deep learning. Nat. Mach. Intell. 2020;2:411. doi: 10.1038/s42256-020-0200-7. [DOI] [Google Scholar]
- 78.Dukart J, Weis S, Genon S, Eickhoff SB. Towards increasing the clinical applicability of machine learning biomarkers in psychiatry. Nat. Hum. Behav. 2021;5:431. doi: 10.1038/s41562-021-01085-w. [DOI] [PubMed] [Google Scholar]
- 79.Li RC, Asch SM, Shah NH. Developing a delivery science for artificial intelligence in healthcare. NPJ Digital Med. 2020;3:1. doi: 10.1038/s41746-020-00318-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Park Y, Kellis M. Deep learning for regulatory genomics. Nat. Biotechnol. 2015;33:825. doi: 10.1038/nbt.3313. [DOI] [PubMed] [Google Scholar]
- 81.Ito Y, Unagami M, Yamabe F, Mitsui Y, Nakajima K, Nagao K, Kobayashi H. A method for utilizing automated machine learning for histopathological classification of testis based on johnsen scores. Sci. Rep. 2021;11:1. doi: 10.1038/s41598-021-89369-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kim J, Lee J, Park E, Han J. A deep learning model for detecting mental illness from user content on social media. Sci. Rep. 2020;10:1. doi: 10.1038/s41598-020-68764-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Li Y, Nowak CM, Pham U, Nguyen K, Bleris L. Cell morphology-based machine learning models for human cell state classification. NPJ Syst. Biol. Appl. 2021;7:1. doi: 10.1038/s41540-021-00180-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yu X, Pang W, Xu Q, Liang M. Mammographic image classification with deep fusion learning. Sci. Rep. 2020;10:1. doi: 10.1038/s41598-020-71431-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Berryman S, Matthews K, Lee JH, Duffy SP, Ma H. Image-based phenotyping of disaggregated cells using deep learning. Commun. Biol. 2020;3:1. doi: 10.1038/s42003-020-01399-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yang S, Kweon J, Roh J-H, Lee J-H, Kang H, Park L-J, Kim DJ, Yang H, Hur J, Kang D-Y, et al. Deep learning segmentation of major vessels in X-ray coronary angiography. Sci. Rep. 2019;9:1. doi: 10.1038/s41598-019-53254-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hannun AY, Rajpurkar P, Haghpanahi M, Tison GH, Bourn C, Turakhia MP, Ng AY. Cardiologist-level arrhythmia detection and classification in ambulatory electrocardiograms using a deep neural network. Nat. Med. 2019;25:65. doi: 10.1038/s41591-018-0268-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Bracher-Smith M, Crawford K, Escott-Price V. Machine learning for genetic prediction of psychiatric disorders: A systematic review. Mol. Psychiatry. 2021;26:70. doi: 10.1038/s41380-020-0825-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Patel D, Kher V, Desai B, Lei X, Cen S, Nanda N, Gholamrezanezhad A, Duddalwar V, Varghese B, Oberai AA. Machine learning based predictors for Covid-19 disease severity. Sci. Rep. 2021;11:1. doi: 10.1038/s41598-021-83967-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Alves, C. L., Cury, R. G., Roster, K., Pineda, A. M., Rodrigues, F. A., Thielemann, C. & Ciba, M. Application of machine learning and complex network measures to an EEG dataset from ayahuasca experiments. medRxiv (2022) [DOI] [PMC free article] [PubMed]
- 91.Newman ME. The structure and function of complex networks. SIAM Rev. 2003;45:167. doi: 10.1137/S003614450342480. [DOI] [Google Scholar]
- 92.Newman ME. Assortative mixing in networks. Phys. Rev. Lett. 2002;89:208701. doi: 10.1103/PhysRevLett.89.208701. [DOI] [PubMed] [Google Scholar]
- 93.Freeman LC. A set of measures of centrality based on betweenness. Sociometry. 1977;40:35. doi: 10.2307/3033543. [DOI] [Google Scholar]
- 94.Albert R, Barabási A-L. Statistical mechanics of complex networks. Rev. Mod. Phys. 2002;74:47. doi: 10.1103/RevModPhys.74.47. [DOI] [Google Scholar]
- 95.Freeman LC. Centrality in social networks conceptual clarification. Soc. Netw. 1978;1:215. doi: 10.1016/0378-8733(78)90021-7. [DOI] [Google Scholar]
- 96.Albert R, Jeong H, Barabási A-L. Diameter of the world-wide web. Nature. 1999;401:130. doi: 10.1038/43601. [DOI] [Google Scholar]
- 97.Kleinberg JM. Hubs, authorities, and communities. ACM Comput. Surv. (CSUR) 1999;31:5. doi: 10.1145/345966.345982. [DOI] [Google Scholar]
- 98.Eppstein D, Paterson MS, Yao FF. On nearest-neighbor graphs. Discrete Comput. Geometry. 1997;17:263. doi: 10.1007/PL00009293. [DOI] [Google Scholar]
- 99.Bonacich P. Power and centrality: A family of measures. Am. J. Sociol. 1987;92:1170. doi: 10.1086/228631. [DOI] [Google Scholar]
- 100.Doyle J, Graver J. Mean distance in a graph. Discrete Math. 1977;17:147. doi: 10.1016/0012-365X(77)90144-3. [DOI] [Google Scholar]
- 101.Snijders TA. The degree variance: An index of graph heterogeneity. Soc. Netw. 1981;3:163. doi: 10.1016/0378-8733(81)90014-9. [DOI] [Google Scholar]
- 102.Dehmer M, Mowshowitz A. A history of graph entropy measures. Inf. Sci. 2011;181:57. doi: 10.1016/j.ins.2010.08.041. [DOI] [Google Scholar]
- 103.Watts DJ, Strogatz SH. Collective dynamics of ‘small-world’ networks. Nature. 1998;393:440. doi: 10.1038/30918. [DOI] [PubMed] [Google Scholar]
- 104.Newman ME, Watts DJ, Strogatz SH. Random graph models of social networks. Proc. Natl. Acad. Sci. 2002;99:2566. doi: 10.1073/pnas.012582999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Seidman SB. Network structure and minimum degree. Soc. Netw. 1983;5:269. doi: 10.1016/0378-8733(83)90028-X. [DOI] [Google Scholar]
- 106.Newman M. Networks: An Introduction. Oxford: Oxford University Press; 2010. [Google Scholar]
- 107.Hage P, Harary F. Eccentricity and centrality in networks. Soc. Netw. 1995;17:57. doi: 10.1016/0378-8733(94)00248-9. [DOI] [Google Scholar]
- 108.Anderson BS, Butts C, Carley K. The interaction of size and density with graph-level indices. Soc. Netw. 1999;21:239. doi: 10.1016/S0378-8733(99)00011-8. [DOI] [Google Scholar]
- 109.Latora V, Marchiori M. Economic small-world behavior in weighted networks. Eur. Phys. J. B Condensed Matter Complex Syst. 2003;32:249. doi: 10.1140/epjb/e2003-00095-5. [DOI] [Google Scholar]
- 110.Newman ME. Communities, modules and large-scale structure in networks. Nat. Phys. 2012;8:25. doi: 10.1038/nphys2162. [DOI] [Google Scholar]
- 111.Kim J, Lee J-G. Community detection in multi-layer graphs: A survey. ACM SIGMOD Rec. 2015;44:37. doi: 10.1145/2854006.2854013. [DOI] [Google Scholar]
- 112.Zhao X, Liang J, Wang J. A community detection algorithm based on graph compression for large-scale social networks. Inf. Sci. 2021;551:358. doi: 10.1016/j.ins.2020.10.057. [DOI] [Google Scholar]
- 113.Clauset A, Newman ME, Moore C. Finding community structure in very large networks. Phys. Rev. E. 2004;70:066111. doi: 10.1103/PhysRevE.70.066111. [DOI] [PubMed] [Google Scholar]
- 114.Rosvall M, Axelsson D, Bergstrom CT. The map equation. Eur. Phys. J. Spec. Topics. 2009;178:13. doi: 10.1140/epjst/e2010-01179-1. [DOI] [Google Scholar]
- 115.Newman ME. Finding community structure in networks using the eigenvectors of matrices. Phys. Rev. E. 2006;74:036104. doi: 10.1103/PhysRevE.74.036104. [DOI] [PubMed] [Google Scholar]
- 116.Raghavan UN, Albert R, Kumara S. Near linear time algorithm to detect community structures in large-scale networks. Phys. Rev. E. 2007;76:036106. doi: 10.1103/PhysRevE.76.036106. [DOI] [PubMed] [Google Scholar]
- 117.Girvan M, Newman ME. Community structure in social and biological networks. Proc. Natl. Acad. Sci. 2002;99:7821. doi: 10.1073/pnas.122653799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Reichardt J, Bornholdt S. Statistical mechanics of community detection. Phys. Rev. E. 2006;74:016110. doi: 10.1103/PhysRevE.74.016110. [DOI] [PubMed] [Google Scholar]
- 119.Blondel VD, Guillaume J-L, Lambiotte R, Lefebvre E. Fast unfolding of communities in large networks. J. Stat. Mech. Theory Exp. 2008;2008:P10008. doi: 10.1088/1742-5468/2008/10/P10008. [DOI] [Google Scholar]
- 120.Hajebrahimi F, Velioglu HA, Bayraktaroglu Z, Helvaci Yilmaz N, Hanoglu L. Clinical evaluation and resting state FMRI analysis of virtual reality based training in Parkinson’s disease through a randomized controlled trial. Sci. Rep. 2022;12:1. doi: 10.1038/s41598-022-12061-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Liu J, Chen H, Gao X, Cui M, Ma L, Zheng X, Guan B, Ma X. Surgical treatment of diffuse and multi-lobes involved glioma with the assistance of a multimodal technique. Sci. Rep. 2022;12:1. doi: 10.1038/s41598-022-07287-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Perovnik M, Tomše P, Jamšek J, Emeršič A, Tang C, Eidelberg D, Trošt M. Identification and validation of Alzheimer’s disease-related metabolic brain pattern in biomarker confirmed Alzheimer’s dementia patients. Sci. Rep. 2022;12:1. doi: 10.1038/s41598-022-15667-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Ashar YK, Gordon A, Schubiner H, Uipi C, Knight K, Anderson Z, Carlisle J, Polisky L, Geuter S, Flood TF, et al. Effect of pain reprocessing therapy vs placebo and usual care for patients with chronic back pain: A randomized clinical trial. JAMA Psychiat. 2022;79:13. doi: 10.1001/jamapsychiatry.2021.2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Hack LM, Zhang X, Williams LM. Striato-cortical neuroimaging markers in the reward network distinguish melancholic depression and response to treatment: An ispot-d report. Biol. Psychiat. 2021;89:S270. doi: 10.1016/j.biopsych.2021.02.676. [DOI] [Google Scholar]
- 125.Polli A, Weis L, Biundo R, Thacker M, Turolla A, Koutsikos K, Chaudhuri KR, Antonini A. Anatomical and functional correlates of persistent pain in Parkinson’s disease. Mov. Disord. 2016;31:1854. doi: 10.1002/mds.26826. [DOI] [PubMed] [Google Scholar]
- 126.William S. The probable error of a mean. Biometrika. 1908;6:1. doi: 10.2307/2331554. [DOI] [Google Scholar]
- 127.Mijalkov M, Kakaei E, Pereira JB, Westman E, Volpe G, Initiative ADN. BRAPH: A graph theory software for the analysis of brain connectivity. PLoS ONE. 2017;12:e0178798. doi: 10.1371/journal.pone.0178798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Wang Y, Li Y, Cao H, Xiong M, Shugart YY, Jin L. Efficient test for nonlinear dependence of two continuous variables. BMC Bioinform. 2015;16:1. doi: 10.1186/1471-2105-16-S12-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.McGrath J, Johnson K, O’Hanlon E, Garavan H, Leemans A, Gallagher L. Abnormal functional connectivity during visuospatial processing is associated with disrupted organisation of white matter in autism. Front. Hum. Neurosci. 2013;7:434. doi: 10.3389/fnhum.2013.00434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Alaerts K, Woolley DG, Steyaert J, Di Martino A, Swinnen SP, Wenderoth N. Underconnectivity of the superior temporal sulcus predicts emotion recognition deficits in autism. Soc. Cognit. Affect. Neurosci. 2014;9:1589. doi: 10.1093/scan/nst156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Leech R, Sharp DJ. The role of the posterior cingulate cortex in cognition and disease. Brain. 2014;137:12. doi: 10.1093/brain/awt162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Martínez K, Martínez-García M, Marcos-Vidal L, Janssen J, Castellanos FX, Pretus C, Villarroya Ó, Pina-Camacho L, Díaz-Caneja CM, Parellada M, et al. Sensory-to-cognitive systems integration is associated with clinical severity in autism spectrum disorder. J. Am. Acad. Child Adolescent Psychiatry. 2020;59:422. doi: 10.1016/j.jaac.2019.05.033. [DOI] [PubMed] [Google Scholar]
- 133.Clery H, Andersson F, Bonnet-Brilhault F, Philippe A, Wicker B, Gomot M. FMRI investigation of visual change detection in adults with autism. NeuroImage Clin. 2013;2:303. doi: 10.1016/j.nicl.2013.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Laidi C, Boisgontier J, de Pierrefeu A, Duchesnay E, Hotier S, d’Albis M-A, Delorme R, Bolognani F, Czech C, Bouquet C, et al. Decreased cortical thickness in the anterior cingulate cortex in adults with autism. J. Autism Dev. Disord. 2019;49:1402. doi: 10.1007/s10803-018-3807-3. [DOI] [PubMed] [Google Scholar]
- 135.Lau WK, Leung M-K, Zhang R. Hypofunctional connectivity between the posterior cingulate cortex and ventromedial prefrontal cortex in autism: Evidence from coordinate-based imaging meta-analysis. Prog. Neuropsychopharmacol. Biol. Psychiatry. 2020;103:109986. doi: 10.1016/j.pnpbp.2020.109986. [DOI] [PubMed] [Google Scholar]
- 136.Oldehinkel M, Mennes M, Marquand A, Charman T, Tillmann J, Ecker C, Dell’Acqua F, Brandeis D, Banaschewski T, Baumeister S, et al. Altered connectivity between cerebellum, visual, and sensory-motor networks in autism spectrum disorder: Results from the eu-aims longitudinal european autism project. Biol. Psychiatry Cognit. Neurosci. Neuroimaging. 2019;4:260. doi: 10.1016/j.bpsc.2018.11.010. [DOI] [PubMed] [Google Scholar]
- 137.Amore G, Spoto G, Ieni A, Vetri L, Quatrosi G, Di Rosa G, Nicotera AG. A focus on the cerebellum: From embryogenesis to an age-related clinical perspective. Front. Syst. Neurosci. 2021;15:646052. doi: 10.3389/fnsys.2021.646052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Mariën P, Borgatti R. Language and the cerebellum. Handb. Clin. Neurol. 2018;154:181. doi: 10.1016/B978-0-444-63956-1.00011-4. [DOI] [PubMed] [Google Scholar]
- 139.Jeremy D, Schmahmann J. The cerebellum and cognition. Neurosci. Lett. 2019;688:62. doi: 10.1016/j.neulet.2018.07.005. [DOI] [PubMed] [Google Scholar]
- 140.Wang SS-H, Kloth AD, Badura A. The cerebellum, sensitive periods, and autism. Neuron. 2014;83:518. doi: 10.1016/j.neuron.2014.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Van Overwalle F, Manto M, Cattaneo Z, Clausi S, Ferrari C, Gabrieli JD, Guell X, Heleven E, Lupo M, Ma Q, et al. Consensus paper: Cerebellum and social cognition. Cerebellum. 2020;19:833. doi: 10.1007/s12311-020-01155-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Delgado-García J. Estructura y función del cerebelo. Rev. Neurol. 2001;33:635. [PubMed] [Google Scholar]
- 143.Stoodley CJ. The cerebellum and neurodevelopmental disorders. Cerebellum. 2016;15:34. doi: 10.1007/s12311-015-0715-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Nebel MB, Joel SE, Muschelli J, Barber AD, Caffo BS, Pekar JJ, Mostofsky SH. Disruption of functional organization within the primary motor cortex in children with autism. Hum. Brain Mapp. 2014;35:567. doi: 10.1002/hbm.22188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Mostofsky SH, Burgess MP, Gidley Larson JC. Increased motor cortex white matter volume predicts motor impairment in autism. Brain. 2007;130:2117. doi: 10.1093/brain/awm129. [DOI] [PubMed] [Google Scholar]
- 146.Daianu, M. et al. Breakdown of brain connectivity between normal aging and Alzheimer’s disease: A structural k-core network analysis. Brain connectivity3, 407 (2013). [DOI] [PMC free article] [PubMed]
- 147.Hagmann P, Cammoun L, Gigandet X, Meuli R, Honey CJ, Wedeen VJ, Sporns O. Mapping the structural core of human cerebral cortex. PLoS Biol. 2008;6:e159. doi: 10.1371/journal.pbio.0060159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Bellec, P. Cobre preprocessed with NIAK 0.17-lightweight release. 10, m9 (2016)
- 149.Baltazar CA, Machado BS, De Faria DD, Paulo AJM, Silva SMCA, Ferraz HB, de Carvalho Aguiar P. Brain connectivity in patients with dystonia during motor tasks. J. Neural Eng. 2020;17:056039. doi: 10.1088/1741-2552/abbbd6. [DOI] [PubMed] [Google Scholar]
- 150.Wan Z, Yang R, Huang M, Zeng N, Liu X. A review on transfer learning in EEG signal analysis. Neurocomputing. 2021;421:1. doi: 10.1016/j.neucom.2020.09.017. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article (and its Supplementary files).