Abstract
Adipose plasticity is critical for metabolic homeostasis. Adipocyte transdifferentiation plays an important role in adipose plasticity, but the molecular mechanism of transdifferentiation remains incompletely understood. Here we show that the transcription factor FoxO1 regulates adipose transdifferentiation by mediating Tgfβ1 signaling pathway. Tgfβ1 treatment induced whitening phenotype in beige adipocytes, reducing UCP1 and mitochondrial capacity and enlarging lipid droplets. Deletion of adipose FoxO1 (adO1KO) dampened Tgfβ1 signaling by downregulating Tgfbr2 and Smad3 and induced browning of adipose tissue in mice, increasing UCP1 and mitochondrial content and activating metabolic pathways. Silencing FoxO1 also abolished the whitening effect of Tgfβ1 on beige adipocytes. The adO1KO mice exhibited a significantly higher energy expenditure, lower fat mass, and smaller adipocytes than the control mice. The browning phenotype in adO1KO mice was associated with an increased iron content in adipose tissue, concurrent with upregulation of proteins that facilitate iron uptake (DMT1 and TfR1) and iron import into mitochondria (Mfrn1). Analysis of hepatic and serum iron along with hepatic iron-regulatory proteins (ferritin and ferroportin) in the adO1KO mice revealed an adipose tissue-liver crosstalk that meets the increased iron requirement for adipose browning. The FoxO1-Tgfβ1 signaling cascade also underlay adipose browning induced by β3-AR agonist CL316243. Our study provides the first evidence of a FoxO1-Tgfβ1 axis in the regulation of adipose browning-whitening transdifferentiation and iron influx, which sheds light on the compromised adipose plasticity in conditions of dysregulated FoxO1 and Tgfβ1 signaling.
Graphical abstract
Highlights
-
•
Adipocyte transdifferentiation controls adipose plasticity and metabolic homeostasis.
-
•
Tgfβ1 downregulates iron metabolism and induces whitening phenotype in beige adipocytes.
-
•
Silencing FoxO1 dampens Tgfβ1 signaling, upregulating iron metabolism and inducing adipose browning.
-
•
FoxO1-Tgfβ1 signaling cascade underlies β3-AR agonist induced adipose browning.
-
•
Dysregulated FoxO1 and Tgfβ1 signaling may account for compromised adipose plasticity in obesity.
1. Introduction
Forkhead box O1 (FoxO1) is a transcription factor conserved across species, and it regulates genes involved in metabolism, cell cycle and differentiation, and tissue remodeling [[1], [2], [3], [4], [5]]. In mammals, activation of FoxO1 in the liver induces glucose production by upregulating gluconeogenic enzymes (e.g., glucose 6-phosphatase and phosphoenolpyruvate carboxykinase), which is associated with altered mitochondrial function [[6], [7], [8], [9], [10]]. Activation of FoxO1 in skeletal muscle regulates atrophy transcriptional program by cooperating with C/EBPδ and ATF4 during fasting [11], and mice overexpressing FoxO1 in skeletal muscle show less skeletal muscle mass, down-regulated slow twitch fiber genes, and impaired glycemic control [12]. In the pancreas, FoxO1 improves β-cell compensation under metabolic stress [13,14], and it regulates α-cell mass by controlling Arx expression through a Dnmt3a-mediated epigenetic mechanism [15]. A recent study shows that activation of hepatic FoxO1 suppresses Fgf21 secretion, whitening brown adipose tissue and impairing glucose metabolism [16]. A fine-tuned FoxO1 is thus critical for tissue function and metabolic homeostasis.
Adipose tissue has a high degree of plasticity through adipocyte differentiation, expansion, and transdifferentiation [17,18]. During adipocyte differentiation FoxO1 activity is fine tuned in the phases of clonal expansion, cell cycle arrest, and terminal differentiation [19,20]; overexpression of constitutively active FoxO1 or pharmacologic inhibition of FoxO1 suppresses adipogenesis, underscoring the important role of FoxO1 in adipocyte differentiation [19,21]. However, the in vivo role of FoxO1 in adipose biology remains incompletely understood. Activation of FoxO1 due to Pten overexpression resulted in adipose transdifferentiation (i.e., browning of white adipose tissue) and hyperactive brown adipose tissue in mice, concurrent with increased energy expenditure and improved glucose metabolism [22]. By contrast, activation of adipose FoxO1 due to Adipoq-Cre mediated conditional deletion of PDK1 in mice leads to less adipose tissue mass and impaired glucose tolerance [23]. In addition, use of aP2 promoter to overexpress a FoxO1 mutant (carboxyl terminal transactivation domain deficient) in the adipose tissue improves systemic insulin sensitivity and glucose metabolism in mice on high fat diet, which was associated with increased energy expenditure via activation of brown adipose tissue [24]. The discrepancy may arise in part from aP2 promoter mediated DNA recombination in non-adipose tissues and altered lineage plasticity during embryonic development [[25], [26], [27], [28], [29]]. Regardless, these studies suggest that FoxO1 is involved in adipose transdifferentiation and further studies are needed to elucidate the exact mechanism.
To address the question, we developed an inducible adipose FoxO1 knockout (adO1KO) mouse model by crossing adipoqCreERT2 mice [30] with FoxO1-floxed mice [6,31]. With this model, we achieved conditional deletion of FoxO1 in the adipose tissue in mouse adulthood to circumvent potential embryonic developmental issues, given that the lineage plasticity and adipose development can be compromised by factors dysregulating FoxO1 activity [3,28,29,32]. In addition, we used adiponectin (Adipoq) promoter to drive DNA recombination, which is more specific for adipocyte than aP2 promoter [[25], [26], [27]]. We found that deletion of adipose FoxO1 dampened Tgfb1-Smad3 signaling and mediated adipose browning-whitening transdifferentiation, concurrent with altered iron influx into adipocytes. This mechanism accounted at least in part for β3-adrenergic receptor (β3-AR) mediated adipose browning.
2. Results
2.1. Post-developmental deletion of adipose FoxO1 improved adipose and systemic metabolism
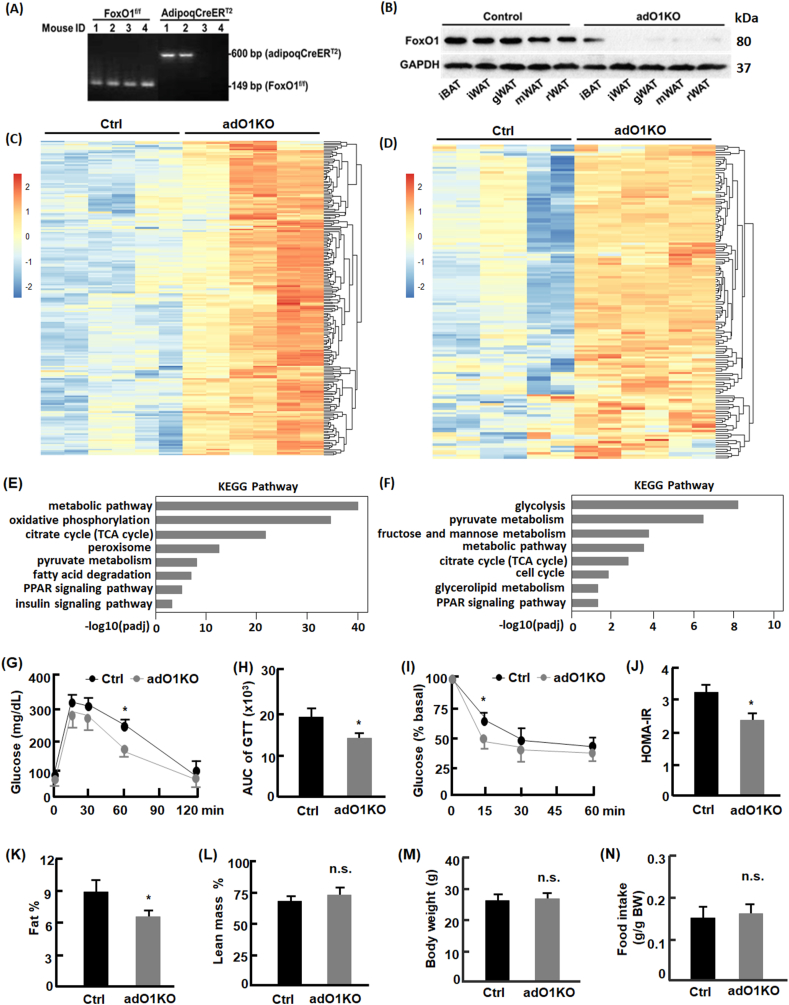
FoxO1flox/flox and adipoqCreERT2:FoxO1flox/flox mice breed at the expected frequency based on a Mendelian distribution, and newborn mice appear indistinguishable between FoxO1flox/flox and adipoqCreERT2:FoxO1flox/flox littermates. To trigger FoxO1 deletion from adipocytes in adult mice, we treated adipoqCreERT2:FoxO1flox/flox mice of 10–12 weeks old using tamoxifen that temporally activates the inducible DNA recombinase [30]. Detection of FoxO1 protein in different tissues using Western blotting analysis suggested that FoxO1 was effectively knocked out in white adipose tissue (Fig. 1, A-B). By contrast, deletion of FoxO1 in brown adipose tissue was mild, in line with the observation that adipoqCreERT2 mediated recombination in brown adipose tissue was about 15% [30]. FoxO1 expression was not affected in other tissues (Fig. 1s). Therefore, tamoxifen treated adipoqCreERT2:FoxO1flox/flox mice are an ideal conditional knockout model (adO1KO) for studying the role of FoxO1 in white adipose tissue. FoxO1flox/flox mice treated with tamoxifen were used as the control (Ctrl).
Fig. 1.
Deletion of adipocyte FoxO1 activated metabolic pathways and improved systemic metabolic health. (A) Genotyping of FoxO1flox/flox mice and adipoqCreERT2:FoxO1flox/flox mice by PCR and DNA gel imaging. As floxed FoxO1 is located at 149 bp and adipoqCreERT2 at 600 bp, the images indicated that mice #1–2 were of adipoqCreERT2:FoxO1flox/flox and mice #3–4 of FoxO1flox/flox genotype. (B) Western blotting analysis of FoxO1 expression in control and adO1KO mice. Tamoxifen treated FoxO1flox/flox (control, Ctrl) showed normal expression of FoxO1, while tamoxifen-treated adipoqCreERT2:FoxO1flox/flox mice (adO1KO) showed drastic deletion of FoxO1 in white adipose tissue. iBAT, interscapular brown adipose tissues; iWAT inguinal white adipose tissue; gWAT, gonadal white adipose tissue; mWAT, mesenteric white adipose tissue; rWAT, retroperitoneal white adipose tissue. (C–D) adO1KO mice exhibited upregulation of metabolism-regulating genes in gonadal (C) and inguinal (D) adipose tissues. (E–F) KEGG pathway analysis of the upregulated genes in gonadal (E) and inguinal (F) adipose tissues. (G–H) Glucose tolerance test and the area under the curve (n = 6). (I) Insulin tolerance test (n = 8). (J) HOMA-IR indices of the control and adO1KO mice (n = 12). (K–M) Body composition analysis of adO1KO and control mice (n = 8). (N) Measurement of food intake (n = 6). *, p < 0.05; n.s., not significant.
RNA sequencing analysis revealed significant upregulation of an array of metabolism-related genes in both gonadal (Fig. 1, C) and inguinal adipose tissue from adO1KO mice versus the control mice (Fig. 1, D). The top ranked genes that underwent upregulation were associated with metabolic pathways, carbohydrate (e.g., glycolysis, fructose and mannose metabolism, pyruvate metabolism, and TCA cycle) and lipid metabolism (e.g., fatty acid degradation, PPAR signaling, and glyerolipid metabolism) according to the KEGG pathway analysis (Fig. 1, E-F). Other affected pathways include insulin signaling, peroxisome, and cell cycle. Further studies focused on gonadal adipose tissue given similar phenotype observed in gonadal and inguinal adipose tissue. Glucose tolerance test suggested that adO1KO mice had improved glucose clearance capacity in comparison to the control mice (Fig. 1, G-H). Insulin tolerance test revealed an enhancement of insulin sensitivity in the adO1KO mice, concurrent with a lower homeostasis model index of insulin resistance (HOMA-IR) (Fig. 1, I and J). In addition, fat mass in the adO1KO mice was significantly lower (by 2.7%, p < 0.05) than the control mice (Fig. 1, K), and lean mass was increased in adO1KO mice although the difference did not reach statistical significance (Fig. 1, L). Body weight and food intake were comparable between the control and adO1KO mice (Fig. 1, M − N). These data suggest that deletion of adipocyte FoxO1 activates metabolic pathways in adipose tissue and improves systemic metabolism.
2.2. Post-developmental deletion of adipose FoxO1 induced adipose browning
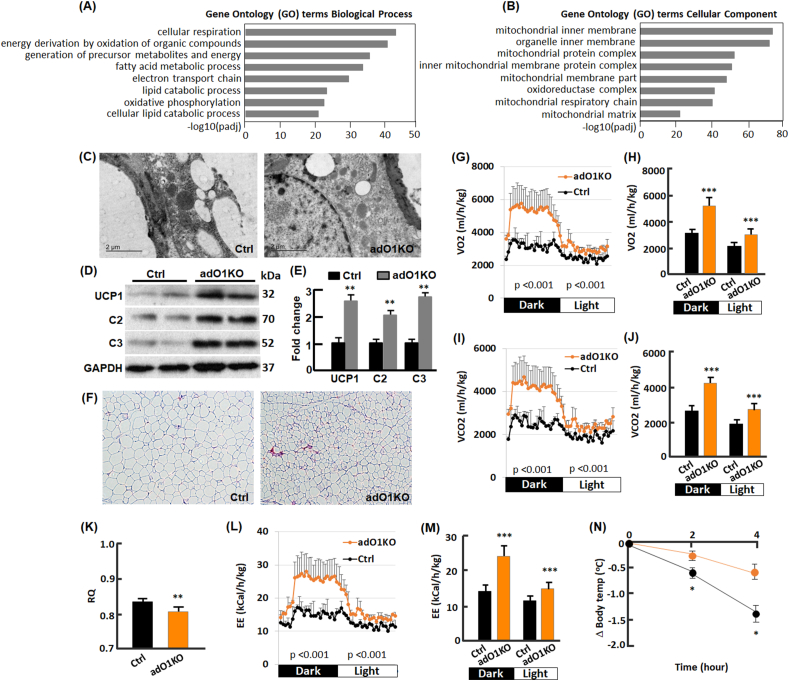
To further investigate the effects of post-developmental deletion of FoxO1 on adipose tissue, we conducted gene ontology (GO) enrichment analysis of the upregulated genes in adO1KO mice vs the control mice (Fig. 2, A-B). For biological processes, we observed a predominant GO enrichment of upregulated genes related to cellular respiration, energy metabolism, fatty acid/lipid metabolism, electron transport chain, and oxidative phosphorylation (Fig. 2, A). In line with this, GO enrichment underlines mitochondrial membrane, protein complex, respiration chain, and mitochondrial matrix as the top ranked cellular components with genes significantly upregulated (Fig. 2, B). Electron microscopic analysis of adipose tissues revealed an increase in mitochondrial content in adO1KO mice (Fig. 2, C), in parallel with upregulation of mitochondrial proteins analyzed by Western blotting (Fig. 2, D-E). In particular, we observed a significant increase in mitochondrial uncoupling protein 1 (UCP1; Fig. 2, D-E) and smaller adipocytes in adO1KO mice compared with the control mice (Fig. 2, F), the hallmarks of adipose browning [17,18]. Indeed, genes of UCP1 and Cidea (another browning marker [33]) were induced in adO1KO mice (Fig. 2s). These changes were associated with a drastic elevation in energy expenditure assessed by indirect calorimetry (Fig. 2, G-J). Respiratory quotient (RQ) was reduced by FoxO1 knockout (Fig. 2, K), indicative of increased fatty acid oxidation as a source of energy in adO1KO mice [34]. This is consistent with the later observation that inhibition of FoxO1 in beige cells promotes fatty acid oxidation (Fig. 4, J). The adO1KO mice showed greater energy expenditure (EE) than the control group (Fig. 2, L-M), concurrent with a stronger resistance to cold challenge (Fig. 2, N). Thus, post-developmental deletion of adipose FoxO1 induces browning of adipose tissue, increasing energy metabolism and thermogenic activities.
Fig. 2.
Deletion of adipocyte FoxO1 induced browning of white adipose tissue. (A) GO (biological process) analysis of the upregulated genes in gonadal adipose tissue. (B) GO (cellular component) analysis of the upregulated genes in gonadal adipose tissue. (C) Electron microscopic view of gonadal adipose tissue sections from control (Ctrl) and adO1KO mice. (D–E) Western blotting (D) and densitometric (E) analyses of mitochondrial proteins (n = 6). (F) H&E staining of gonadal adipose tissue sections (100x magnification). (G) Real time measurement of oxygen consumption with the TSE system (n = 7). (H) Average oxygen consumption in the dark and light cycles (n = 7). (I) Real time measurement of carbon dioxide production with the TSE system (n = 7). (J) Average carbon dioxide production in the dark and light cycles (n = 7). (K) Respiratory quotient (RQ) measured by the TSE system (n = 7). (L) Real time measurement of energy expenditure (EE) with the TSE system (n = 7). (M) Average energy expenditure (EE) in the dark and light cycles (n = 7). (N) Cold tolerance test (n = 5). *, p < 0.05; **, p < 0.01; ***, p < 0.001.
Fig. 4.
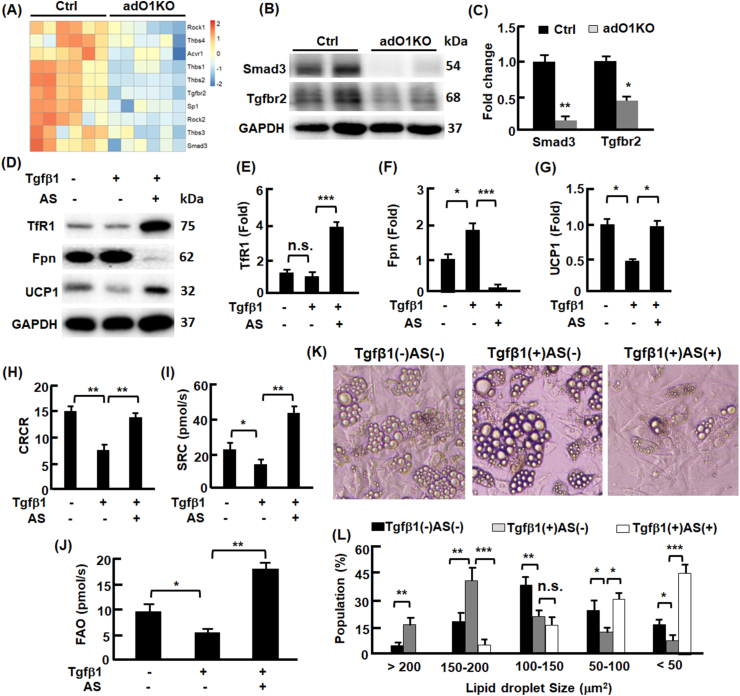
FoxO1-Tgfβ1 axis regulated adipocyte transdifferentiation. (A) adO1KO mice exhibited downregulation of genes involved in Tgfβ1 signaling pathway in gonadal adipose tissue. (B–C) The effects of FoxO1 deletion on the key proteins Tgfbr2 and Smad3 in Tgfβ1 signaling pathway in gonadal adipose tissue, analyzed by Western blotting (B) and densitometry (C), n = 6. (D–G) The effects of Tgfβ1 and FoxO1 inhibition on iron import (TfR1) and export (ferroportin, Fpn) proteins, and beige marker Ucp1 protein in C3H/10T1/2 adipocytes, evaluated by Western blotting (D) and densitometry (E–G), n = 3. (H–J) The effects of Tgfβ1 and FoxO1 inhibition on cellular respiration control ratio (panel H), mitochondrial spare respiration capacity (panel I), and fatty acid oxidation activities (panel J) in C3H/10T1/2 adipocytes, analyzed by high resolution respiratory, n = 4. (K) Representative images (200x magnification) of C3H/10T1/2 adipocytes without treatment, treated with Tgfβ1, and co-treated with Tgfβ1 and FoxO1 inhibitor AS. (L) The effects of Tgfβ1 and FoxO1 inhibition on the size and population of lipid droplets in C3H/10T1/2 adipocytes, n = 4. *, p < 0.05; **, p < 0.01; ***, p < 0.001; n.s., not significant.
2.3. Adipose browning was associated with enhanced iron metabolism
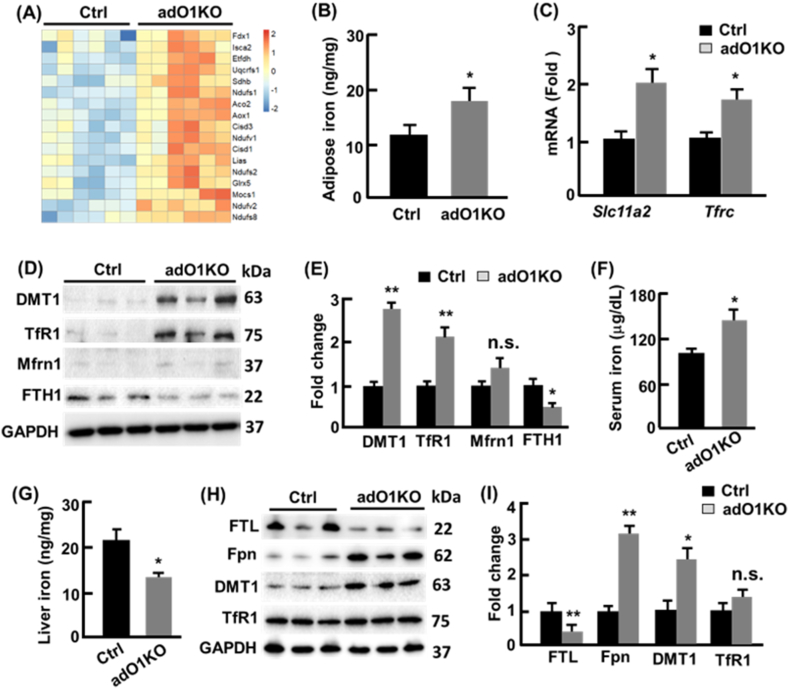
Mitochondria contain 20–50% of total cellular iron, largely in the form of iron-sulfur (Fe–S) clusters [35]. In line with the increased mitochondrial content in adO1KO mice, the genes encoding Fe–S binding proteins were significantly upregulated in the adipose tissues (Fig. 3, A), and non-heme iron was increased by 51% (p < 0.05; Fig. 3, B). Quantitative real time PCR analysis revealed 2-fold (p < 0.05) and 1.8-fold (p < 0.05) elevation in the expression of Slc11a2 (encoding divalent metal transporter 1, DMT1) and Tfrc (encoding transferrin receptor protein 1, TfR1), respectively (Fig. 3, C). Western blotting analysis suggested 2.7-fold and 2.1-fold increase in DMT1 and TfR1 proteins (Fig. 3, D-E). Mitoferrin 1 (Mfrn1), the mitochondrial metallochaperone that transports iron into mitochondria [35], was also upregulated but the change was not statistically significant (Fig. 3, D-E). Interestingly, the iron storage protein ferritin (FTH1) decreased by 55% (p < 0.05), suggesting that adipose browning imposes an increased iron demand and utilization thereby reducing iron storage (Fig. 3, D-E). Serum iron level was 1.47-fold higher in adO1KO mice than the control mice (Fig. 3, F), consistent with the increased adipose iron content in adO1KO mice (Fig. 3, B). By contrast, iron content in the liver was reduced by 36% (p < 0.05), concurrent with a 3.2-fold (p < 0.01) increase in hepatic ferroportin (Fpn) known to export iron into the circulation [35,36], and hepatic iron storing protein ferritin (FTL) level decreased by 63% (p < 0.01; Fig. 3, G-H). Downregulation of hepatic iron was associated with significantly increased DMT1 (Fig. 3, G-H). Iron content and iron-regulatory proteins in the spleen and small intestine were comparable in the control and adO1KO mice (data not shown). These findings suggest that cellular and mitochondrial iron transport in adipocytes was upregulated to accommodate the increased iron demand during adipose browning in adO1KO mice (i.e., an elevated iron influx into adipocytes) at the expense of hepatic iron storage.
Fig. 3.
Deletion of adipocyte FoxO1 altered iron metabolism in the adipose tissue and liver. (A) adO1KO mice showed upregulation of genes encoding Fe–S binding proteins in gonadal adipose tissue. (B) Non-heme iron content in gonadal adipose tissue (n = 6). (C) qPCR analysis of genes encoding DMT1 (Slc11a2) and TfR1 (Tfrc), n = 8. (D–E) Western blotting (D) and densitometric (E) analyses of iron regulating proteins (n = 6). (F) Serum iron in control (Ctrl) and adO1KO mice (n = 8–12). (G) Non-heme iron contents in the liver (n = 6). (H–I) Western blotting (H) and densitometric (I) analyses of iron-storing, exporting, and importing proteins (n = 6). *, p < 0.05; **, p < 0.01; n.s., not significant.
2.4. AdO1KO dampened Tgfβ1-Smad3 signaling for adipose browning
To explore the mechanism of adO1KO-induced adipose browning, we conducted pathway analysis of the RNA sequencing data and discovered that adO1KO mitigated the expression of critical genes involved in Tgfβ1 signaling pathway [37], including Tgfbr2 and Smad3 (Fig. 4, A). Consistently, Western blotting analysis revealed 89% and 62% downregulation of Smad3 and Tgfbr2 proteins in adO1KO mice, respectively (Fig. 4, B–C). These findings suggest that deletion of FoxO1 dampens Tgfβ1 signaling via Tgfbr2 and Smad3 during adipose browning. To determine whether Tgfβ1 signaling plays a role in white-beige adipocyte transdifferentiaiton, we treated beige cells (or brown-like adipocytes) with Tgfβ1, which led to 57% (p < 0.05) decrease in UCP1 protein, concurrent with moderate downregulation of TfR1 and upregulation of Fpn (Fig. 4, D-G), indicative of a whitening effect onbeige cells [17,18] and lowered iron demand. However, suppression of FoxO1 with the specific inhibitor AS1842856 (abbreviated as AS thereafter) reversed the effects of Tgfβ1 on UCP1, Tf1R, and Fpn (Fig. 4, D-G). High resolution respirometry (Oroboros, Austria) analysis of mitochondrial capacity in beige cells showed that Tgfβ1 treatment lowered cellular respiration control ratio (CRCR) by 49% (p < 0.01) and mitochondrial spare respiratory capacity (SRC) by 42% (p < 0.05) (Fig. 4, H–I). Mitochondrial fatty acid oxidation (FAO) activities were lower in Tgfβ1 treated beige cells in comparison to non-treated cells (Fig. 4, J). The Tgfβ1-induced changes in mitochondria were associated with increased population of large lipid droplets (62% lipid droplets with the area >150 μm2) in comparison to the control cells (75% lipid droplets with the area <150 μm2) (Fig. 4, K-L). Intriguingly, FoxO1 inhibitor AS reversed the effects of Tgfβ1 on FAO activities and reducing the size of lipid droplets (72% lipid droplets with the area <100 μm2) and lipid content in the cells (Fig. 4, J-L). Consistently, beige cells cotreated with Tgfβ1 and FoxO1 inhibitor AS showed normal expression of UCP1, concurrent with upregulated TfR1 and Fpn (Fig. 4, D-G), the proteins that facilitates iron metabolism by increasing iron uptake and retention [36,38]. Furthermore, Tgfβ1-induced reduction in CRCR and SRC was normalized or enhanced by AS (Fig. 4, H–I). Together, the results suggest that Tgfβ1 signaling promotes whitening of beige adipocytes, while silencing FoxO1 suppresses Tgfβ1 signaling and facilitates adipose browning, leading to a greater mitochondrial capacity and less lipid storage in the cells.
2.5. FoxO1-Smad3 cascade was suppressed in β3-AR mediated adipose browning
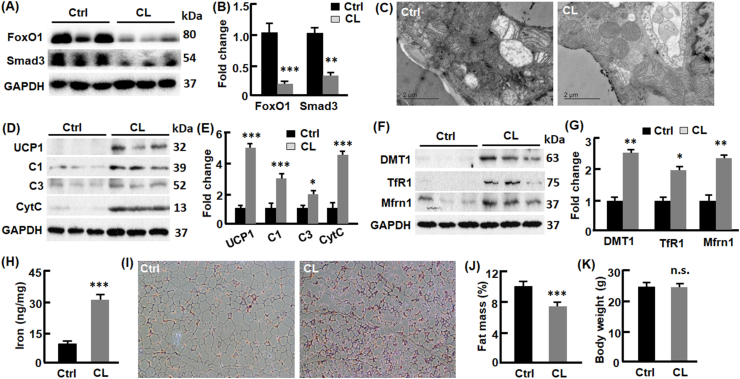
Activation of β3-adrenoceptor (β3-AR) induces adipose browning [39,40]. Using β3-AR agonist CL316243 (abbreviated as CL thereafter) to treat C57BL6/J mice, we found that CL downregulated adipose FoxO1 by 84% (p < 0.001) and Smad3 by 58% (p < 0.01) (Fig. 5, A-B). The downregulation of FoxO1- Smad3 cascade was associated with increased mitochondrial content in adipose tissue examined by electron microscope (Fig. 5, C). UCP1 was significantly upregulated by CL treatment (4.9-fold, p < 0.001), concurrent with elevation of other mitochondrial proteins such as complexes I and III as well as cytochrome C (Fig. 5, D-E). Consistently, mRNA levels of of UCP1 and Cidea were upregulated (Fig. 2s). As observed in adO1KO mice, the CL-treated mice showed higher DMT1, TfR1, and Mfrn1, the proteins that promote iron uptake and import into mitochondria (Fig. 5, F-G). Consistently, adipose iron level was increased significantly compared with the C57BL6/J mice treated with saline (31 vs 12 ng/mg, p < 0.001; Fig. 5, H). H&E staining reveals that CL treatment led to a higher population of smaller adipocytes (Fig. 5, I). Fat mass in the CL-treated mice was reduced significantly compared with the control mice (6.9% vs 10.1%, p < 0.001) although the body weight was comparable (Fig. 5, J-K). Overall, suppression of the FoxO1-Smad3 cascade by β3-AR agonist CL316243 was associated with adipose browning and increased iron metabolism.
Fig. 5.
CL downregulated FoxO1 and induced browning of white adipose tissue in C57BL6/J mice. (A–B) CL treatment downregulated FoxO1 and Smad3 in gonadal adipose tissue, evaluated by Western blotting (A) and densitometry (B), n = 6–8. (C) CL-treated mice show an increased mitochondrial content in gonadal adipose tissue, assessed by electron microscopy. (D–E) CL treatment upregulated mitochondrial proteins, evaluated by Western blotting (D) and densitometry (E), n = 6. (F–G) CL upregulated proteins importing iron into cells (DMT1 and TfR1) and mitochondria (Mfrn1), evaluated by Western blotting (F) and densitometry (G), n = 6. (H) Measurement of non-heme iron in gonadal adipose tissue (n = 6–8). (I) H&E staining of white adipose tissue (100x magnification). (J–K) Body composition analysis by NMR (n = 8–12). In all cases, mice treated with saline were used as the control (Ctrl) group. *, p < 0.05; **, p < 0.01; ***, p < 0.001; n.s., not significant.
3. Discussion
The plasticity of adipose tissue regulates its size, metabolism and function, and adaptative responses to physiological cues [17,41]. Transdifferentiation from white adipocytes to thermogenic beige adipocytes, or vice versa, represents an important mechanism of adipose plasticity [42]. In this study we discovered that FoxO1 mediates adipose transdifferentiation via Smad3 signaling (Fig. 1, Fig. 2, 4). Post-developmental deletion of FoxO1 downregulated Tgfbr2 and Smad3, associated with hallmarks of browning of white adipose tissue (e.g., upregulated UCP1 and smaller adipocyte size) and improved glucose and energy metabolism (Fig. 1, Fig. 2). In addition, treatment of beige adipocytes with Tgfβ1 induced whitening phenotype, while silencing FoxO1 pharmacologically with AS reversed the effect of Tgfβ1 (Fig. 4). Previous studies have shown that obese individuals have lower brown adipose mass or less pronounced response to browning stimuli than lean healthy controls [[43], [44], [45]]. The differences between lean and obese subjects may arise from dysregulated FoxO1 and Smad3 signaling, given that obesity activates FoxO1 through insulin resistance [3,46] and increases Tgfβ1 level [47,48]. Our findings shed light on the mechanism of FoxO1 regulating adipose transdifferentiation (Fig. 6), and add to the notion that silencing Tgfβ1-Smad3 signaling favors adipose browning [48,49]. Importantly, activation of β3-AR deactivates the FoxO1-Smad3 cascade, and deletion of adipose FoxO1 recapitulated the β3-AR agonist (CL) induced browning of adipose tissue (Fig. 4, Fig. 5). Thus, silencing FoxO1-Smad3 cascade may underlie β3-AR mediated adipose browning (Fig. 6). To this end, it was shown that obese subject had low or no response to β-AR agonists that cause adipose browning in healthy control [50,51], possibly because of activated FoxO1 by insulin resistance [3,46] and elevated Tgfβ1 [47,48]. Therefore, a fine-tuned FoxO1/Tgfβ1-Smad3 activity is critical to maintain browning plasticity.
Fig. 6.
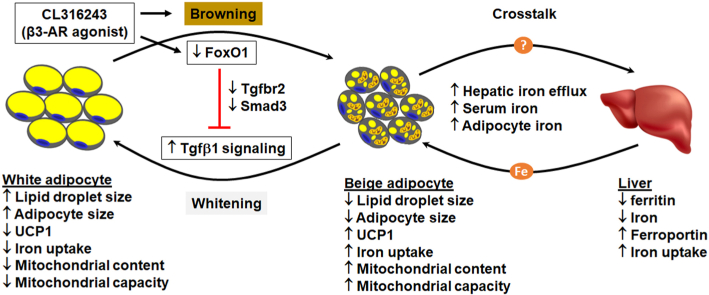
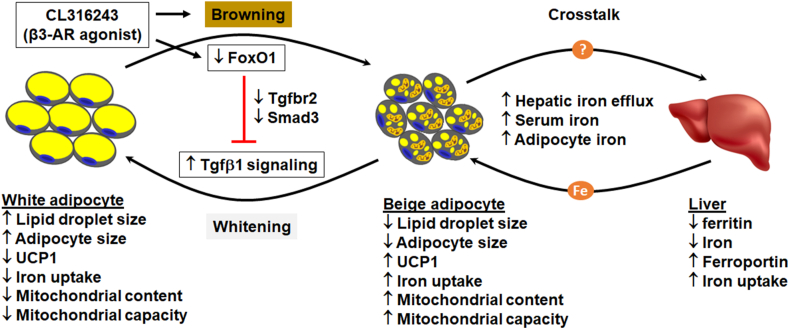
The proposed mechanism by which FoxO1-Tgfβ1 cascade mediates adipose transdifferentiation. Tgfβ1 signaling promotes whitening of beige adipocytes, while silencing FoxO1 blocks Tgfβ1 pathway by downregulating Tgfbr2 and Smad3, preventing Tgfβ1-induced whitening but favoring browning of white adipose tissue. The FoxO1-Tgfβ1 mechanism seems to be shared by adipose browning induced by CL, a classic browning agent. The question mark indicates a to-be-defined mechanism through which adipose tissue cross-talks with the liver, thereby triggering an increase in iron influx into the adipocytes in response to browning-imposed demand of iron.
Iron plays key roles in numerous enzymatic reactions, particularly in the mitochondria that are highly redox active and rich in heme-containing enzymes and Fe–S cluster proteins [35]. Iron deficiency dampens mitochondrial development and adipose browning and lowers body temperature in mice exposed to cold [52]. In line with increased mitochondrial content and UCP1 that promotes energy dissipation and thermogenesis in adO1KO mice (Fig. 1, Fig. 2), we found that adipose iron was significantly increased during adipose browning (Fig. 3, Fig. 5). Accordingly, the proteins and related genes that regulate cellular iron uptake (DMT1 and TfR1) and mitochondrial iron import (Mfrn1) were upregulated (Fig. 3, Fig. 5). The adO1KO mice showed lower iron content in the liver (the primary organ of body iron storage) but a higher iron content in the serum (Fig. 3, F-G), implying an increased iron flux from the liver to adipose tissue. Indeed, the iron exporter Fpn was significantly upregulated in the liver, concurrent downregulation of ferritin (the iron storage protein) in the liver from adO1KO mice (Fig. 3, H). Interestingly, ferritin level in the adipose tissue was also downregulated significantly regardless of elevation of DMT1 and TfR1 (Fig. 3, D-E). The reciprocal regulation of ferritin vs DMT1 and TfR1 in adO1KO adipose tissue suggests that the increased iron requirement for adipose browning may result in a secondary iron deficiency [53], which is known to upregulate DMT1 and downregulate ferritin via the IRP/IRE system [35,36]. Indeed, previous studies have reported iron deficiency secondary to increased iron requirements [54,55]. For instance, high-oxygen-affinity hemoglobin variants increased red cell mass requirement, which induced a compensatory erythrocytosis and secondary iron deficiency [56,57]. Thus, conditional deletion of adipose FoxO1 induces a crosstalk between the liver and adipose tissue to meet the increased iron requirement for adipose browning (Fig. 6). In the liver, FoxO1 was previously shown to regulate iron through heme metabolism [1,6,7]. By upregulating heme oxygenase 1 (heme catabolism) and down-regulating Fxn and Urod (heme anabolism), FoxO1 disrupts the integrity of mitochondrial electron transport chain and causes metabolic disorder in the liver [6,7], whereas silencing FoxO1 restores hepatic mitochondria and metabolic homeostasis [6,7,10]. This study unravels a novel role of FoxO1 in mediating the crosstalk of iron metabolism between adipose tissue and the liver.
Tgfbr2 is required for canonical TGFβ1 signaling to funnel to Smad3 in the regulation of target genes [37]. Our data reveals new target genes (e.g., Tgfbr2 and Smad3) by which FoxO1 mediates Tgfβ1 signaling pathway. Deletion of adipocyte FoxO1 downregulates Tgfbr2 and Smad3 genes, concurrent with pronounced decreases in Tgfbr2 and Smad3 proteins (Fig. 4, A-C). In keratinocyte FoxO1 transactivates Tgfβ1 by binding to the promoter of Tgfβ1 gene, which promotes keratinocyte transition to a wound-healing phenotype [58,59]. In chondrocyte, however, FoxO1 acts as a downstream target and exclusively induced by Tgfβ1 in a TGF-β activated kinase 1-dependent manner [60]. TGFβ1-SMAD signaling increases FoxO1 expression and activity during chondrogenic differentiation, and FoxO1 inhibition suppresses chondrocyte differentiation [61]. In liver cells, TGFβ1-Smad3 signaling stimulates gluconeogenesis by suppressing LKB1-AMPK pathway and promoting nuclear translocation of FoxO1, which in turn induces gluconeogenic enzymes [62]. These findings support the notion that FoxO1 as a transcription factor regulates cell proliferation, differentiation, and metabolism, in a cell type- and tissue-dependent manner [1,2,4].
In line with the notion that silencing Smad3 promotes adipose browning [48], our data show that ablation of FoxO1 dampened Smad3 and induced browning phenotype in white adipose tissue of adO1KO mice (Fig. 1, Fig. 2). In addition, we found that TGFβ1 signaling resulted in whitening of beige adipocytes, while inhibition of FoxO1 reversed the whitening effect (Fig. 4). The mechanism of FoxO1-TGFβ1 axis in adipose transdifferentiation remains to be investigated. Of note, recent studies suggest that TGFβ1 signaling negatively regulate cyclooxygenase-2 (Cox-2), a rate-limiting enzyme in the synthesis of prostaglandin that facilitates the recruitment and activation of beige adipocytes [49]. TGFβ1 may induce hepatocyte-derived exosomal miRNA let-7b-5p, which suppresses sympathetic stimulation and recruitment of beige adipocytes by downregulating β3-AR [63].
Taken together, our study identifies the FoxO1/Tgfβ1-Smad3 axis as an important mechanism of adipose transdifferentiation (Fig. 6). Silencing FoxO1 abrogates the whitening effects of Tgfβ1 on beige adipocytes. Post-developmental deletion of FoxO1 results in browning of white adipose tissue in mice. Adipose browning due to FoxO1 ablation is associated with increased iron influx into adipocytes, during which a crosstalk is initiated between the liver and adipose tissue to meet the increased requirement of iron. The FoxO1-Smad3 cascade appears to underlie β3-AR agonist CL induced browning of adipose tissue (Fig. 6). Further studies are warranted to delineate the molecular mechanisms that regulate the liver-adipose tissue crosstalk in iron metabolism.
4. Materials and methods
4.1. Mice The FoxO1flox/flox and adipoqCreERT2 mice were described previously [6,9,30,31]
To obtain adipoqCreERT2: FoxO1flox/flox mice, we crossed male adipoqCreERT2 mice with female FoxO1flox/flox mice. Genotyping was conducted as described [6,9,30]. To induce the deletion of FoxO1, adult mice (10–12 week old) were treated with tamoxifen (50 mg/kg body weight) by I.P. injection once a day for 5 consecutive days [64,65]. C57BL6/J mice (10–12 weeks old) mice were injected (I.P.) with CL (1 mg/kg body weight) or an equivalent volume of sterile saline once per day for five consecutive days [66]. All the mice were housed in plastic cages on a 12-h (7:00 a.m.–7:00 p.m.) light–dark photocycle, with free access to water and regular chow diet. Body composition analysis and glucose tolerance tests were conducted at the age of 16 weeks old, followed by tissue collection for molecular analysis. All the procedures followed the NIH guideline and were approved by the Institutional Animal Care and Use Committees at the University of Florida and Virginia Tech.
4.2. Indirect calorimetry
Whole body energy expenditure was analyzed by indirect calorimetry with a TSE PhenoMaster system where mice were housed individually with free access to food and water [67]. Mice were allowed 3 days to acclimate to the new housing conditions before the measurement of O2 consumption (ml kg−1 h−1) and CO2 production (ml kg−1 h−1). Energy expenditure (EE) was calculated using the Weir equation: EE = 3.941 × VO2 + 1.106 × VCO2 and normalized to body weight.
4.3. Histological analysis
Adipose tissues were fixed with 10% formalin. The fixed tissues were embedded in paraffin and 7 μm sections were stained with hematoxylin and eosin. Images were taken at 20 × magnification with a Nikon Eclipse Ts2 microscope (Melville, NY, USA).
4.4. Transmission electron microscopy (TEM)
TEM was conducted as described previously with some modifications [6,68]. Specifically, adipose tissues were cut into small pieces (∼1 mm3) and fixed in 2.5% glutaraldehyde in sodium cacodylate buffer and post fixed in 1% osmium tetroxide and embedded in an Epon-Araldite mixture. Ultrathin (80 nm) sections were obtained with an Ultracut E ultramicrotome and post stained with 3% uranyl acetate, followed by 0.4% lead citrate. The ultrathin sections were examined with a JEOL 1200EX transmission electron microscope and images were recorded digitally.
4.5. Cell culture and treatment
C3H/10T1/2 cells (CCL-226, ATCC) were cultured and maintained in Dulbecco's modified Eagle's medium–10% FBS as described previously [69]. To induce beige differentiation, cells were maintained for 3 days in medium supplemented with 1 μg/ml insulin, 0.5 mM 3-isobutyl-1-methylxanthine, 2 μg/ml dexamethasone, 5 μM rosiglitazone and then switched to medium supplemented with 1 μg/ml insulin for 6 days. The beige adipocytes were then treated for 5 days with Tgfβ1 (5 ng/ml), AS1842856 (1 μM), or combination of Tgfβ1 (5 ng/ml) and AS1842856 (1 μM) as described previously [19,[70], [71], [72], [73]]. Images of the cells were captured with a Nikon Eclipse Ts2 microscope (Melville, NY, USA), and the size and number of lipid droplets were analyzed with the NIH ImageJ software (Bethesda, MD, USA) as described previously [73,74].
4.6. Respirometry
Oxygen consumption of beige adipocytes was measured with high resolution respirometry (Oxygraph-2k, Oroboros). Adipocytes (7 × 104) were suspended in 2.1 mL mitochondrial respiration medium (Oxygraph-2k, Oroboros). Basal cellular respiration was recorded after the injection of pyruvate (5 mM), and proton leak respiration was assessed after injection of oligomycin (10 ng/mL). Maximal cellular respiration rates were measured by titration of carbonyl cyanide 4-(trifluoromethoxy) phenylhydrazone (FCCP, 0.5-μM steps). Nonmitochondrial oxygen consumption was determined after injection of antimycin A (2.5 μM), and it was subtracted from all other respiratory states. Spare respiratory capacity (SRC) was calculated by subtracting basal from maximal oxygen consumption rates [75,76]. Cellular respiratory control ratio (CRCR) was calculated as the ratio of maximal to leak oxygen consumption [75,76]. Fatty acid oxidation (FAO) activities were examined by measuring oxygen consumption in the presence of octanoyl-carnitine (0.5 mM) and malate (2 mM) as the substrates [77].
4.7. Cold tolerance test
The mice were maintained in a climate chamber (Columbus Instruments, Ohio) at 5 °C with free access to water but not food. The rectal temperature was measured with a TH5 thermometer (Physitemp, Clifton, NJ) at the indicated time after exposure to the cold.
4.8. GTT, ITT and HOMA-IR
Glucose tolerance test (GTT) and insulin tolerance test (ITT) were performed as described previously [78]. For GTT, the mice were fasted overnight (∼14 h), and the animals were injected intraperitoneally (i.p.) with d-glucose (2 g/kg of body weight), and blood glucose was measured at the indicated time points with Contour Blood Glucose Meters and Test Strips. For ITT, mice were fasted for 4–6 h, followed by i.p. injection of diluted insulin (1 U/kg Humulin R; Lilly), and blood glucose was measured at indicated time points. For homeostasis model index of insulin resistance (HOMA-IR), ultra-sensitive mouse insulin ELISA kits were used to measure insulin in plasma from overnight-fasted mice according to the manufacturer (Crystal Chem, Elk Grove Village, IL); glucose was measured with Contour Blood Glucose Meters and Test Strips for overnight-fasted mice. HOMA-IR were calculated as follows [79]: HOMA-IR = insulin (mU/L) × glucose (mg/dL)/405, derived from HOMA-IR = insulin (mU/L) × glucose (mmol/L)/22.5.
4.9. Iron assay
Total non-heme iron was analyzed with Iron Assay Kits (Cat # MAK025) from Sigma according to the manufacturer's instructions [80,81].
4.10. Western blotting
The procedure was performed as described [20,[82], [83], [84]]. Specifically, tissue and cell lysates were prepared with PLC lysis buffer (30 mM Hepes, pH 7.5, 150 mM NaCl, 10% glycerol, 1% Triton X-100, 1.5 mM MgCl2, 1 mM EGTA, 10 mM NaPPi, 100 mM NaF, 1 mM Na3VO4) supplemented with protease inhibitor cocktail (Roche), and 1 mM PMSF. Total protein concentrations of the lysates were determined using a DC protein assay kits (Bio-Rad). Antibody (catalog number) information: GAPDH (MA5-15738) and β-actin (MA5-15739) antibodies were purchased from Pierce (Rockford, IL, USA); FoxO1 antibody (9454s) from Cell Signaling Technology (Beverly, MA, USA); C1 (A21344), C2 (A11142) and C3 (A21362) antibodies from Invitrogen; cytochrome C antibody (K257-100-5) from Biovision; antibodies against DMT1 (bs-3577R-TR) and TfR1 (bs-0988R) from Bioss Antibodies; Ferroportin antibody (NBP1-21503) from Novus Biologicals; antibodies against UCP1 (23673-1-AP), FTL (10727-1-AP), Tgfbr2 (20000-1-AP), and Mitoferrin 1 (26469-1-AP) from Proteintech; Smad3 (SC-101154) from Santa Cruz; FTH antibody (SAB2108662-100UL) from Sigma (Billerica, MA, USA).
4.11. qPCR
The procedure was performed as described [72]. Briefly, RNA was extracted with RNeasy Mini Kits (Qiagen, Germantown, MD, USA) according to the manufacturer's instruction. The RNA samples were used to synthesize cDNA by reverse transcription PCR using iScript™ cDNA Synthesis Kits (Bio-Rad, Hercules, CA, USA) according to the manufacturer's instruction. Gene expression was analyzed by quantitative real-time PCR on a CFX96 system (Bio-Rad, Hercules, CA, USA). The primers used in this study were 5′- CTG CTG AGC GAA GAT ACC AG-3′ (forward) and 5′- CTC AGG AGC TTA GGT CAG AAG-3′ (reverse) for Slc11a2; 5′- CCC ATG ACG TTG AAT TGA ACCT-3′ (forward) and 5′- GTA GTC TCC ACG AGC GGA ATA-3′ (reverse) for Tfrc; 5′- ACT GCC ACA CCT CCA GTC ATT -3′ (forward) and 5′- CTT TGC CTC ACT CAG GAT TGG -3′ (reverse) for UCP1; 5′- ATC ACA ACT GGC CTG GTT ACG -3′ (forward) and 5′- TAC TAC CCG GTG TCC ATT TCT -3′ (reverse) for Cidea; 5′- CTC TGG CTC CTA GCA CCA TGA AGA -3′ (forward) and 5′- GTA AAA CGC AGC TCA GTA ACA GTC CG -3′ (reverse) for ACTB as a reference gene.
4.12. RNASeq
Total RNA from adipose tissue was prepared with RNeasy Mini Kits (Qiagen), and mRNA was purified from total RNA using poly-T oligo-attached magnetic beads [85]. After library construction, diluting library to 1.5 ng/μl with the preliminary quantitative result by Qubit2.0 and detecting the insert size by Agilent 2100. Q-PCR was used to accurately quantify the library effective concentration (>2 nM), in order to ensure the library quality. Libraries were fed into Illumina machine (Illumina Platform PE150) after pooling according to activity and expected data volume. Raw image data file from high-throughput sequencing was transformed to Sequenced Reads (Raw Reads) by CASAVA base recognition (Base Calling). Raw reads were filtered to remove reads containing adaptors, or >10% bases that could not be determined, or when the Qscore of over 50% bases were ≤ 5. STAR was used to map the clean reads to the reference genome (Mus MusculusGRCm38/mm10). Differential expression analysis was performed with a DESeq2 R package and enrichment analysis with ClusterProfiler [85].
4.13. Statistical analysis
Measurements were duplicated or triplicated, with 6–12 mice included in each group. Data were presented as mean ± SD. Unless the use of female mice were specified, the animal studies were conducted on males. Differences between groups and treatments were validated by one-way analysis of variance or a two-sided t-test. A value of p < 0.05 was considered statistically significant.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was supported in part by the American Heart Association (18TPA34230082 to Z.C.), and the USDA National Institute of Food and Agriculture (1020373 to Z.C.).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2023.102727.
Appendix A. Supplementary data
The following is the Supplementary data to this article.
Data availability
Data will be made available on request.
References
- 1.Cheng Z. FoxO transcription factors in mitochondrial homeostasis. Biochem. J. 2022;479(4):525–536. doi: 10.1042/BCJ20210777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cheng Z. The FoxO-autophagy Axis in health and disease. Trends Endocrinol. Metabol.: TEM (Trends Endocrinol. Metab.) 2019;30(9):658–671. doi: 10.1016/j.tem.2019.07.009. [DOI] [PubMed] [Google Scholar]
- 3.Cheng Z. FoxO1: mute for a tuned metabolism? Trends Endocrinol. Metabol.: TEM (Trends Endocrinol. Metab.) 2015;26(7):402–403. doi: 10.1016/j.tem.2015.06.006. [DOI] [PubMed] [Google Scholar]
- 4.Cheng Z., White M.F. Targeting Forkhead box O1 from the concept to metabolic diseases: lessons from mouse models. Antioxidants Redox Signal. 2011;14(4):649–661. doi: 10.1089/ars.2010.3370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tao Z., Cheng Z. Hormonal regulation of metabolism-recent lessons learned from insulin and estrogen. Clin. Sci. (Lond.) 2023;137(6):415–434. doi: 10.1042/CS20210519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheng Z., Guo S., Copps K., Dong X., Kollipara R., Rodgers J.T., Depinho R.A., Puigserver P., White M.F. Foxo1 integrates insulin signaling with mitochondrial function in the liver. Nat. Med. 2009;15(11):1307–1311. doi: 10.1038/nm.2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang W.B., Yan H., Pan Q., Shen J.Z., Zhou F.H., Wu C.D., Sun Y.X., Guo S.D. Glucagon regulates hepatic mitochondrial function and biogenesis through FOXO1. J. Endocrinol. 2019;241(3):265–278. doi: 10.1530/JOE-19-0081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lu M., Wan M., Leavens K.F., Chu Q., Monks B.R., Fernandez S., Ahima R.S., Ueki K., Kahn C.R., Birnbaum M.J. Insulin regulates liver metabolism in vivo in the absence of hepatic Akt and Foxo1. Nat. Med. 2012;18(3):388–395. doi: 10.1038/nm.2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dong X.C., Copps K.D., Guo S., Li Y., Kollipara R., DePinho R.A., White M.F. Inactivation of hepatic Foxo1 by insulin signaling is required for adaptive nutrient homeostasis and endocrine growth regulation. Cell Metabol. 2008;8(1):65–76. doi: 10.1016/j.cmet.2008.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.O-Sullivan I., Zhang W., Wasserman D.H., Liew C.W., Liu J., Paik J., DePinho R.A., Stolz D.B., Kahn C.R., Schwartz M.W., Unterman T.G. FoxO1 integrates direct and indirect effects of insulin on hepatic glucose production and glucose utilization. Nat. Commun. 2015;6:7079. doi: 10.1038/ncomms8079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Oyabu M., Takigawa K., Mizutani S., Hatazawa Y., Fujita M., Ohira Y., Sugimoto T., Suzuki O., Tsuchiya K., Suganami T., Ogawa Y., Ishihara K., Miura S., Kamei Y. FOXO1 cooperates with C/EBP delta and ATF4 to regulate skeletal muscle atrophy transcriptional program during fasting. Faseb. J. 2022;36(2) doi: 10.1096/fj.202101385RR. [DOI] [PubMed] [Google Scholar]
- 12.Kamei Y., Miura S., Suzuki M., Kai Y., Mizukami J., Taniguchi T., Mochida K., Hata T., Matsuda J., Aburatani H., Nishino I., Ezaki O. Skeletal muscle FOXO1 (FKHR) transgenic mice have less skeletal muscle mass, down-regulated type I (slow twitch/red muscle) fiber genes, and impaired glycemic control. J. Biol. Chem. 2004;279(39):41114–41123. doi: 10.1074/jbc.M400674200. [DOI] [PubMed] [Google Scholar]
- 13.Kobayashi M., Kikuchi O., Sasaki T., Kim H.J., Yokota-Hashimoto H., Lee Y.S., Amano K., Kitazumi T., Susanti V.Y., Kitamura Y.I., Kitamura T. FoxO1 as a double-edged sword in the pancreas: analysis of pancreas-and beta-cell-specific FoxO1 knockout mice. Am J Physiol-Endoc M. 2012;302(5):E603–E613. doi: 10.1152/ajpendo.00469.2011. [DOI] [PubMed] [Google Scholar]
- 14.Zhang T., Kim D.H., Xiao X., Lee S., Gong Z., Muzumdar R., Calabuig-Navarro V., Yamauchi J., Harashima H., Wang R., Bottino R., Alvarez-Perez J.C., Garcia-Ocana A., Gittes G., Dong H.H. FoxO1 plays an important role in regulating beta-cell compensation for insulin resistance in male mice. Endocrinology. 2016;157(3):1055–1070. doi: 10.1210/en.2015-1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kodani N., Nakae J., Kobayashi M., Kikuchi O., Kitamura T., Itoh H. FCoR-Foxo1 Axis regulates alpha-cell mass through repression of Arx expression. iScience. 2020;23(1) doi: 10.1016/j.isci.2019.100798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stohr O., Tao R., Miao J., Copps K.D., White M.F. FoxO1 suppresses Fgf21 during hepatic insulin resistance to impair peripheral glucose utilization and acute cold tolerance. Cell Rep. 2021;34(12) doi: 10.1016/j.celrep.2021.108893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sakers A., De Siqueira M.K., Seale P., Villanueva C.J. Adipose-tissue plasticity in health and disease. Cell. 2022;185(3):419–446. doi: 10.1016/j.cell.2021.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ghaben A.L., Scherer P.E. Adipogenesis and metabolic health. Nat. Rev. Mol. Cell Biol. 2019;20(4):242–258. doi: 10.1038/s41580-018-0093-z. [DOI] [PubMed] [Google Scholar]
- 19.Zou P., Liu L., Zheng L., Stoneman R.E., Cho A., Emery A., Gilbert E.R., Cheng Z. Targeting FoxO1 with AS1842856 suppresses adipogenesis. Cell Cycle. 2014;13(23):3759–3767. doi: 10.4161/15384101.2014.965977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shi L., Tao Z., Cheng Z. Assessing the activity of transcription factor FoxO1. Methods Mol. Biol. 2023;2594:97–106. doi: 10.1007/978-1-0716-2815-7_8. [DOI] [PubMed] [Google Scholar]
- 21.Nakae J., Kitamura T., Kitamura Y., Biggs W.H., 3rd, Arden K.C., Accili D. The forkhead transcription factor Foxo1 regulates adipocyte differentiation. Dev. Cell. 2003;4(1):119–129. doi: 10.1016/s1534-5807(02)00401-x. [DOI] [PubMed] [Google Scholar]
- 22.Ortega-Molina A., Efeyan A., Lopez-Guadamillas E., Munoz-Martin M., Gomez-Lopez G., Canamero M., Mulero F., Pastor J., Martinez S., Romanos E., Mar Gonzalez-Barroso M., Rial E., Valverde A.M., Bischoff J.R., Serrano M. Pten positively regulates brown adipose function, energy expenditure, and longevity. Cell Metabol. 2012;15(3):382–394. doi: 10.1016/j.cmet.2012.02.001. [DOI] [PubMed] [Google Scholar]
- 23.Hosooka T., Hosokawa Y., Matsugi K., Shinohara M., Senga Y., Tamori Y., Aoki C., Matsui S., Sasaki T., Kitamura T., Kuroda M., Sakaue H., Nomura K., Yoshino K., Nabatame Y., Itoh Y., Yamaguchi K., Hayashi Y., Nakae J., Accili D., Yokomizo T., Seino S., Kasuga M., Ogawa W. The PDK1-FoxO1 signaling in adipocytes controls systemic insulin sensitivity through the 5-lipoxygenase-leukotriene B4 axis. Proc. Natl. Acad. Sci. U. S. A. 2020;117(21):11674–11684. doi: 10.1073/pnas.1921015117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nakae J., Cao Y., Oki M., Orba Y., Sawa H., Kiyonari H., Iskandar K., Suga K., Lombes M., Hayashi Y. Forkhead transcription factor FoxO1 in adipose tissue regulates energy storage and expenditure. Diabetes. 2008;57(3):563–576. doi: 10.2337/db07-0698. [DOI] [PubMed] [Google Scholar]
- 25.Jeffery E., Berry R., Church C.D., Yu S., Shook B.A., Horsley V., Rosen E.D., Rodeheffer M.S. Characterization of Cre recombinase models for the study of adipose tissue. Adipocyte. 2014;3(3):206–211. doi: 10.4161/adip.29674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang Q.A., Scherer P.E., Gupta R.K. Improved methodologies for the study of adipose biology: insights gained and opportunities ahead. J. Lipid Res. 2014;55(4):605–624. doi: 10.1194/jlr.R046441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee K.Y., Russell S.J., Ussar S., Boucher J., Vernochet C., Mori M.A., Smyth G., Rourk M., Cederquist C., Rosen E.D., Kahn B.B., Kahn C.R. Lessons on conditional gene targeting in mouse adipose tissue. Diabetes. 2013;62(3):864–874. doi: 10.2337/db12-1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lynes M.D., Schulz T.J., Pan A.J., Tseng Y.H. Disruption of insulin signaling in Myf5-expressing progenitors leads to marked paucity of brown fat, but normal muscle development. Endocrinology. 2015 doi: 10.1210/en.2014-1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sanchez-Gurmaches J., Guertin D.A. Adipocytes arise from multiple lineages that are heterogeneously and dynamically distributed. Nat. Commun. 2014;5:4099. doi: 10.1038/ncomms5099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sassmann A., Offermanns S., Wettschureck N. Tamoxifen-inducible Cre-mediated recombination in adipocytes. Genesis. 2010;48(10):618–625. doi: 10.1002/dvg.20665. [DOI] [PubMed] [Google Scholar]
- 31.Paik J.H., Kollipara R., Chu G., Ji H., Xiao Y., Ding Z., Miao L., Tothova Z., Horner J.W., Carrasco D.R., Jiang S., Gilliland D.G., Chin L., Wong W.H., Castrillon D.H., DePinho R.A. FoxOs are lineage-restricted redundant tumor suppressors and regulate endothelial cell homeostasis. Cell. 2007;128(2):309–323. doi: 10.1016/j.cell.2006.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cheng Z., White M.F. The AKTion in non-canonical insulin signaling. Nat. Med. 2012;18(3):351–353. doi: 10.1038/nm.2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jash S., Banerjee S., Lee M.J., Farmer S.R., Puri V. CIDEA transcriptionally regulates UCP1 for britening and thermogenesis in human fat cells. iScience. 2019;20:73–89. doi: 10.1016/j.isci.2019.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Qian S.W., Tang Y., Li X., Liu Y., Zhang Y.Y., Huang H.Y., Xue R.D., Yu H.Y., Guo L., Gao H.D., Liu Y., Sun X., Li Y.M., Jia W.P., Tang Q.Q. BMP4-mediated brown fat-like changes in white adipose tissue alter glucose and energy homeostasis. Proc. Natl. Acad. Sci. U. S. A. 2013;110(9):E798–E807. doi: 10.1073/pnas.1215236110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ward D.M., Cloonan S.M. Mitochondrial iron in human health and disease. Annu. Rev. Physiol. 2019;81:453–482. doi: 10.1146/annurev-physiol-020518-114742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Muckenthaler M.U., Rivella S., Hentze M.W., Galy B. A red carpet for iron metabolism. Cell. 2017;168(3):344–361. doi: 10.1016/j.cell.2016.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Akhurst R.J., Hata A. Targeting the TGFbeta signalling pathway in disease. Nat. Rev. Drug Discov. 2012;11(10):790–811. doi: 10.1038/nrd3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Crielaard B.J., Lammers T., Rivella S. Targeting iron metabolism in drug discovery and delivery. Nat. Rev. Drug Discov. 2017;16(6):400–423. doi: 10.1038/nrd.2016.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kurylowicz A., Puzianowska-Kuznicka M. Induction of adipose tissue browning as a strategy to combat obesity. Int. J. Mol. Sci. 2020;21(17) doi: 10.3390/ijms21176241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bartelt A., Heeren J. Adipose tissue browning and metabolic health. Nat. Rev. Endocrinol. 2014;10(1):24–36. doi: 10.1038/nrendo.2013.204. [DOI] [PubMed] [Google Scholar]
- 41.Chouchani E.T., Kajimura S. Metabolic adaptation and maladaptation in adipose tissue. Nat Metab. 2019;1(2):189–200. doi: 10.1038/s42255-018-0021-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cinti S. Transdifferentiation properties of adipocytes in the adipose organ. Am J Physiol-Endoc M. 2009;297(5):E977–E986. doi: 10.1152/ajpendo.00183.2009. [DOI] [PubMed] [Google Scholar]
- 43.Kulterer O.C., Herz C.T., Prager M., Schmoeltzer C., Langer F.B., Prager G., Marculescu R., Kautzky-Willer A., Hacker M., Haug A.R., Kiefer F.W. Brown adipose tissue prevalence is lower in obesity but its metabolic activity is intact. Front. Endocrinol. 2022;13 doi: 10.3389/fendo.2022.858417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Orava J., Nuutila P., Noponen T., Parkkola R., Viljanen T., Enerback S., Rissanen A., Pietilainen K.H., Virtanen K.A. Blunted metabolic responses to cold and insulin stimulation in Brown adipose tissue of obese humans. Obesity. 2013;21(11):2279–2287. doi: 10.1002/oby.20456. [DOI] [PubMed] [Google Scholar]
- 45.Sidossis L., Kajimura S. Brown and beige fat in humans: thermogenic adipocytes that control energy and glucose homeostasis. J. Clin. Invest. 2015;125(2):478–486. doi: 10.1172/JCI78362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cheng Z., Tseng Y., White M.F. Insulin signaling meets mitochondria in metabolism. Trends Endocrinol. Metabol.: TEM (Trends Endocrinol. Metab.) 2010;21(10):589–598. doi: 10.1016/j.tem.2010.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Alessi M.C., Bastelica D., Morange P., Berthet B., Leduc I., Verdier M., Geel O., Juhan-Vague I. Plasminogen activator inhibitor 1, transforming growth factor-beta(1), and BMI are closely associated in human adipose tissue during morbid obesity. Diabetes. 2000;49(8):1374–1380. doi: 10.2337/diabetes.49.8.1374. [DOI] [PubMed] [Google Scholar]
- 48.Yadav H., Quijano C., Kamaraju A.K., Gavrilova O., Malek R., Chen W.P., Zerfas P., Duan Z.G., Wright E.C., Stuelten C., Sun P., Lonning S., Skarulis M., Sumner A.E., Finkel T., Rane S.G. Protection from obesity and diabetes by blockade of TGF-beta/smad3 signaling. Cell Metabol. 2011;14(1):67–79. doi: 10.1016/j.cmet.2011.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wankhade U.D., Lee J.H., Dagur P.K., Yadav H., Shen M., Chen W.P., Kulkarni A.B., Mccoy J.P., Finkel T., Cypess A.M., Rane S.G. TGF-beta receptor 1 regulates progenitors that promote browning of white fat. Mol. Metabol. 2018;16:160–171. doi: 10.1016/j.molmet.2018.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Carey A.L., Formosa M.F., Van Every B., Bertovic D., Eikelis N., Lambert G.W., Kalff V., Duffy S.J., Cherk M.H., Kingwell B.A. Ephedrine activates brown adipose tissue in lean but not obese humans. Diabetologia. 2013;56(1):147–155. doi: 10.1007/s00125-012-2748-1. [DOI] [PubMed] [Google Scholar]
- 51.Qiu Y., Sun L.Z., Hu X.L., Zhao X., Shi H.Y., Liu Z., Yin X. Compromised browning plasticity of primary subcutaneous adipocytes derived from overweight Chinese adults. Diabetol. Metab. Syndrome. 2020;12(1) doi: 10.1186/s13098-020-00599-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yook J.S., Thomas S.S., Toney A.M., You M., Kim Y.C., Liu Z., Lee J., Chung S. Dietary iron deficiency modulates adipocyte iron homeostasis, adaptive thermogenesis, and obesity in C57bl/6 mice. J. Nutr. 2021;151(10):2967–2975. doi: 10.1093/jn/nxab222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Camaschella C. Iron deficiency. Blood. 2019;133(1):30–39. doi: 10.1182/blood-2018-05-815944. [DOI] [PubMed] [Google Scholar]
- 54.Kumar A., Sharma E., Marley A., Samaan M.A., Brookes M.J. Iron deficiency anaemia: pathophysiology, assessment, practical management. Bmj Open Gastroenter. 2022;9(1) doi: 10.1136/bmjgast-2021-000759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Balendran S., Forsyth C. Non-anaemic iron deficiency. Aust. Prescr. 2021;44(6):193–196. doi: 10.18773/austprescr.2021.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wajcman H., Galacteros F. Abnormal hemoglobins with high oxygen affinity and erythrocytosis. Hematol. Cell Ther. 1996;38(4):305–312. doi: 10.1007/s00282-996-0305-4. [DOI] [PubMed] [Google Scholar]
- 57.Pavic M., Francina A., Durand D.V., Rousset H. Polycythaemia and iron deficiency. Lancet. 2003;362(9396):1624. doi: 10.1016/S0140-6736(03)14796-4. [DOI] [PubMed] [Google Scholar]
- 58.Zhang C.Y., Ponugoti B., Tian C., Xu F.X., Tarapore R., Batres A., Alsadun S., Lim J., Dong G.Y., Graves D.T. FOXO1 differentially regulates both normal and diabetic wound healing. JCB (J. Cell Biol.) 2015;209(2):289–303. doi: 10.1083/jcb.201409032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ponugoti B., Xu F.X., Zhang C.Y., Tian C., Pacios S., Graves D.T. FOXO1 promotes wound healing through the up-regulation of TGF-beta 1 and prevention of oxidative stress. JCB (J. Cell Biol.) 2013;203(2):327–343. doi: 10.1083/jcb.201305074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang C.C., Shen J., Ying J., Xiao D., O'Keefe R.J. FoxO1 is a crucial mediator of TGF-beta/TAK1 signaling and protects against osteoarthritis by maintaining articular cartilage homeostasis. Proc. Natl. Acad. Sci. U. S. A. 2020;117(48):30488–30497. doi: 10.1073/pnas.2017056117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kurakazu I., Akasaki Y., Hayashida M., Tsushima H., Goto N., Sueishi T., Toya M., Kuwahara M., Okazaki K., Duffy T., Lotz M.K., Nakashima Y. FOXO1 transcription factor regulates chondrogenic differentiation through transforming growth factor beta 1 signaling. J. Biol. Chem. 2019;294(46):17555–17569. doi: 10.1074/jbc.RA119.009409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yadav H., Devalaraja S., Chung S.T., Rane S.G. TGF-1/Smad3 pathway targets PP2A-AMPK-FoxO1 signaling to regulate hepatic gluconeogenesis. J. Biol. Chem. 2017;292(8):3420–3432. doi: 10.1074/jbc.M116.764910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhao J., Hu L., Gui W., Xiao L., Wang W., Xia J., Fan H., Li Z., Zhu Q., Hou X., Chu H., Seki E., Yang L. Hepatocyte TGF-beta signaling inhibiting WAT browning to promote NAFLD and obesity is associated with let-7b-5p. Hepatol Commun. 2022;6(6):1301–1321. doi: 10.1002/hep4.1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Imai T., Jiang M., Chambon P., Metzger D. Impaired adipogenesis and lipolysis in the mouse upon selective ablation of the retinoid X receptor alpha mediated by a tamoxifen-inducible chimeric Cre recombinase (Cre-ERT2) in adipocytes. Proc. Natl. Acad. Sci. U. S. A. 2001;98(1):224–228. doi: 10.1073/pnas.011528898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Liu L., Zou P., Zheng L., Linarelli L.E., Amarell S., Passaro A., Liu D., Cheng Z. Tamoxifen reduces fat mass by boosting reactive oxygen species. Cell Death Dis. 2015;6:e1586. doi: 10.1038/cddis.2014.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Buzelle S.L., MacPherson R.E., Peppler W.T., Castellani L., Wright D.C. The contribution of IL-6 to beta 3 adrenergic receptor mediated adipose tissue remodeling. Physiological reports. 2015;3(2) doi: 10.14814/phy2.12312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Brierley D.I., Holt M.K., Singh A., de Araujo A., McDougle M., Vergara M., Afaghani M.H., Lee S.J., Scott K., Maske C., Langhans W., Krause E., de Kloet A., Gribble F.M., Reimann F., Rinaman L., de Lartigue G., Trapp S. Central and peripheral GLP-1 systems independently suppress eating. Nat Metab. 2021;3(2):258–273. doi: 10.1038/s42255-021-00344-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sidossis L.S., Porter C., Saraf M.K., Borsheim E., Radhakrishnan R.S., Chao T., Ali A., Chondronikola M., Mlcak R., Finnerty C.C., Hawkins H.K., Toliver-Kinsky T., Herndon D.N. Browning of subcutaneous white adipose tissue in humans after severe adrenergic stress. Cell Metabol. 2015;22(2):219–227. doi: 10.1016/j.cmet.2015.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ferrari A., Longo R., Fiorino E., Silva R., Mitro N., Cermenati G., Gilardi F., Desvergne B., Andolfo A., Magagnotti C., Caruso D., De Fabiani E., Hiebert S.W., Crestani M. HDAC3 is a molecular brake of the metabolic switch supporting white adipose tissue browning. Nat. Commun. 2017;8 doi: 10.1038/s41467-017-00182-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Liguori T.T.A., Liguori G.R., Moreira L.F.P., Harmsen M.C. Fibroblast growth factor-2, but not the adipose tissue-derived stromal cells secretome, inhibits TGF-beta1-induced differentiation of human cardiac fibroblasts into myofibroblasts. Sci. Rep. 2018;8(1) doi: 10.1038/s41598-018-34747-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang W., Chun H., Baek J., Sadik J.E., Shirazyan A., Razavi P., Lopez N., Lyons K.M. The TGFbeta type I receptor TGFbetaRI functions as an inhibitor of BMP signaling in cartilage. Proc. Natl. Acad. Sci. U. S. A. 2019;116(31):15570–15579. doi: 10.1073/pnas.1902927116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Liu L., Tao Z., Zheng L.D., Brooke J.P., Smith C.M., Liu D., Long Y.C., Cheng Z. FoxO1 interacts with transcription factor EB and differentially regulates mitochondrial uncoupling proteins via autophagy in adipocytes. Cell death discovery. 2016;2 doi: 10.1038/cddiscovery.2016.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Liu L., Zheng L.D., Zou P., Brooke J., Smith C., Long Y.C., Almeida F.A., Liu D., Cheng Z. FoxO1 antagonist suppresses autophagy and lipid droplet growth in adipocytes. Cell Cycle. 2016;15(15):2033–2041. doi: 10.1080/15384101.2016.1192732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Deutsch M.J., Schriever S.C., Roscher A.A., Ensenauer R. Digital image analysis approach for lipid droplet size quantitation of Oil Red O-stained cultured cells. Anal. Biochem. 2014;445:87–89. doi: 10.1016/j.ab.2013.10.001. [DOI] [PubMed] [Google Scholar]
- 75.Schottl T., Kappler L., Braun K., Fromme T., Klingenspor M. Limited mitochondrial capacity of visceral versus subcutaneous white adipocytes in male C57BL/6N mice. Endocrinology. 2015;156(3):923–933. doi: 10.1210/en.2014-1689. [DOI] [PubMed] [Google Scholar]
- 76.Brand M.D., Nicholls D.G. Assessing mitochondrial dysfunction in cells. Biochem. J. 2011;435(2):297–312. doi: 10.1042/BJ20110162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gonzalez-Armenta J.L., Gao Z.R., Appt S.E., Vitolins M.Z., Michalson K.T., Register T.C., Shively C.A., Molina A.J.A. Skeletal muscle mitochondrial respiration is elevated in female cynomolgus macaques fed a western compared with a mediterranean diet. J. Nutr. 2019;149(9):1493–1502. doi: 10.1093/jn/nxz092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Guo S., Copps K.D., Dong X., Park S., Cheng Z., Pocai A., Rossetti L., Sajan M., Farese R.V., White M.F. The Irs1 branch of the insulin signaling cascade plays a dominant role in hepatic nutrient homeostasis. Mol. Cell Biol. 2009;29(18):5070–5083. doi: 10.1128/MCB.00138-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mather K. Surrogate measures of insulin resistance: of rats, mice, and men. Am. J. Physiol. Endocrinol. Metab. 2009;296(2):E398–E399. doi: 10.1152/ajpendo.90889.2008. [DOI] [PubMed] [Google Scholar]
- 80.Liu X.Y., Wei D.G., Li R.S. Capsaicin induces ferroptosis of NSCLC by regulating SLC7A11/GPX4 signaling in vitro. Sci. Rep. 2022;12(1) doi: 10.1038/s41598-022-16372-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ma S., Dubin A.E., Zhang Y., Mousavi S.A.R., Wang Y., Coombs A.M., Loud M., Andolfo I., Patapoutian A. A role of PIEZO1 in iron metabolism in mice and humans. Cell. 2021;184(4):969–982 e13. doi: 10.1016/j.cell.2021.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tao Z., Shi L., Parke J., Zheng L., Gu W., Dong X.C., Liu D., Wang Z., Olumi A.F., Cheng Z. Sirt1 coordinates with ERalpha to regulate autophagy and adiposity. Cell death discovery. 2021;7(1):53. doi: 10.1038/s41420-021-00438-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tao Z., Liu L., Zheng L.D., Cheng Z. Autophagy in adipocyte differentiation. Methods Mol. Biol. 2019;1854:45–53. doi: 10.1007/7651_2017_65. [DOI] [PubMed] [Google Scholar]
- 84.Tao Z., Zheng L.D., Smith C., Luo J., Robinson A., Almeida F.A., Wang Z., Olumi A.F., Liu D., Cheng Z. Estradiol signaling mediates gender difference in visceral adiposity via autophagy. Cell Death Dis. 2018;9(3):309. doi: 10.1038/s41419-018-0372-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wang W., Yan T.Q., Guo W., Niu J.F., Zhao Z.Q., Sun K.K., Zhang H.L., Yu Y.Y., Ren T.T. Constitutive GLI1 expression in chondrosarcoma is regulated by major vault protein via mTOR/S6K1 signaling cascade. Cell Death Differ. 2021;28(7):2221–2237. doi: 10.1038/s41418-021-00749-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.