In most natural environments, association with a surface in a structure known as a biofilm is the prevailing microbial lifestyle. Surface association is an efficient means of lingering in a favorable microenvironment rather than being swept away by the current. Taken to the extreme, we may view the planktonic or free-swimming microbial phase primarily as a mechanism for translocation from one surface to another.
Genetic studies of single-species biofilms have shown that they form in multiple steps (46), require intercellular signalling (7), and demonstrate a profile of gene transcription that is distinct from that of planktonic cells (35). From this perspective, biofilm formation may be viewed as a developmental process that shares some of the features of other bacterial developmental processes such as sporulation of gram-positive bacteria (9), fruiting body formation in Myxococcus xanthus (33, 40, 44), and stalked-cell formation by Caulobacter crescentus (13, 19, 24, 37, 48). In natural environments, however, the biofilm is almost invariably a multispecies microbial community harboring bacteria that stay and leave with purpose, share their genetic material at high rates, and fill distinct niches within the biofilm. Thus, the natural biofilm is less like a highly developed organism and more like a complex, highly differentiated, multicultural community much like our own city.
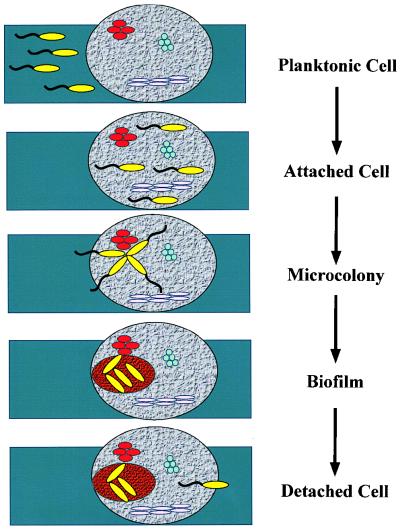
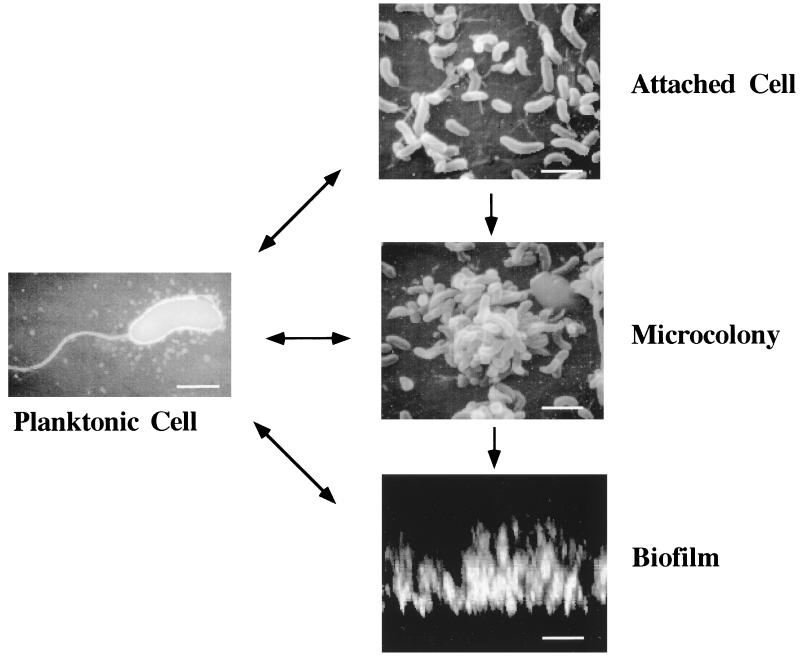
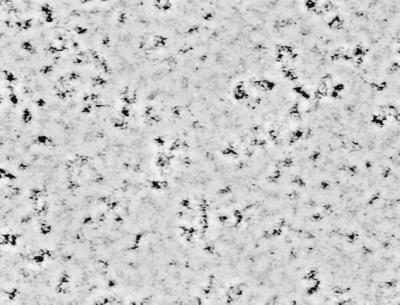
There are several steps that we must take to optimize our lives in a city. The first is to choose the city in which we will live, then we must select the neighborhood in the city that best suits our needs, and finally we must make our home amongst the homes of many others. Occasionally, when life in the city sours, we leave. The same steps occur in the formation of a bacterial biofilm (Fig. 1). First, the bacterium approaches the surface so closely that motility is slowed. The bacterium may then form a transient association with the surface and/or other microbes previously attached to the surface. This transient association allows it to search for a place to settle down. When the bacterium forms a stable association as a member of a microcolony, it has chosen the neighborhood in which to live. Finally, the buildings go up as a three-dimensional biofilm is erected. Occasionally, the biofilm-associated bacteria detach from the biofilm matrix. Micrographs of these steps in biofilm formation by a single bacterial species are shown in Fig. 2. Although these micrographs are static views of the steps in biofilm formation, a biofilm is not a motionless heap of cells. Figure 3 shows the first frame of a real time movie, accessible at http://gasp.med.harvard.edu/biofilms/jbmini/movie.html, that documents the activity in a mature biofilm. In this frame, the pillars of a mature biofilm are visible, distributed on top of a monolayer of surface-associated cells. The associated movie shows that, in addition to fixed cells, there are motile cells that maintain their association with the biofilm for long periods of time, swimming between pillars of biofilm-associated bacteria. The biofilm, therefore, demonstrates a level of activity similar to that of a bustling city.
FIG. 1.
A schematic representation of the steps a new bacterial species takes in forming a biofilm on a rock previously colonized with multiple species of bacteria. The yellow bacteria represent an aquatic species that swims towards the rock using polar flagella, forms random loose attachments to the rock, migrates over the surface to form a microcolony, and finally produces exopolysaccharide to form a three-dimensional biofilm. When environmental conditions become unfavorable, some of the bacteria may detach and swim away to find a surface in a more favorable environment.
FIG. 2.
A microscopic study of the steps in biofilm formation by V. cholerae. The planktonic bacterium was visualized by transmission electron microscopy (bar = 1 μM), the attached cells and microcolony were visualized by scanning electron microscopy (bar = 2 μM), and the biofilm micrograph represents a vertical section through a 20-μm biofilm taken by confocal scanning laser microscopy (bar = 10 μM).
FIG. 3.
The first frame in a movie taken of the activity in a mature V. cholerae biofilm. The dark collections of bacteria represent pillars in the biofilm, while a monolayer of cells is seen between the pillars. The corresponding movie, which demonstrates the activity associated with a mature biofilm, is accessible at http://gasp.med.harvard.edu/biofilms/jbmini/movie.html both in gray scale and in color to accentuate moving bacteria.
The genetic basis of the steps in biofilm formation has been investigated for a number of bacterial species, including Escherichia coli (34), Pseudomonas aeruginosa (31) and Vibrio cholerae (46). For these studies, a simple genetic screen was utilized in which random transposon mutants are grown in 96-well plates (5, 16, 32). After removal of the planktonic cells, the remaining biofilm-associated cells are stained with crystal violet. Those wells with no crystal violet staining correspond to mutants that are defective in biofilm formation. These genetic screens for biofilm-defective mutants have shown that the initial interaction with the surface is accelerated by force-generating organelles such as type IV pili and flagella. Once temporary contact with the surface is made, bacteria use either flagella or type IV pili to move along the surface in two dimensions until other bacteria are encountered and microcolonies are formed or enlarged (31, 34, 46). Finally, exopolysaccharide production is necessary to stabilize the pillars of the biofilm (46). Competition studies between wild-type V. cholerae and pilus or flagellar mutants show that these structures provide a great advantage in surface colonization (P. I. Watnick and R. Kolter, unpublished results). Thus, speed of attachment may be an important factor in garnering an apartment in the microbial city.
Evidence exists that different genes are transcribed in the planktonic and biofilm-associated phases of the bacterial life cycle. This is again reminiscent of a developmental process. Prigent-Combaret et al. performed a screen for genes in E. coli that are differentially expressed in biofilm-associated cells, using a library of random insertion mutants generated with a MudX transposon carrying a promoterless lacZ gene (35). One interesting finding from this study is that flagellin synthesis is decreased in biofilm-associated cells, while production of colanic acid, an exopolysaccharide made by E. coli, is increased. The situation appears to be similar in P. aeruginosa. Alginate is an exopolysaccharide that is found in P. aeruginosa biofilms (14). Transcription of algC, a gene involved in the production of alginate, is increased approximately fourfold in biofilm-associated cells as compared with planktonic cells (6, 15). Furthermore, for many years, researchers have noted that pulmonary isolates of P. aeruginosa are mucoid due to production of copious amounts of alginate (14). Recently, Garrett and coworkers noted that flagella are absent from these mucoid isolates (15). In addition, they showed by mutational analysis that while alginate synthesis is positively regulated by the alternative sigma factor ς22, this sigma factor negatively regulates the synthesis of the flagellum. This suggests that when synthesis of the exopolysaccharide, alginate, is increased in biofilm-associated cells, flagellar synthesis decreases. Thus, to become a productive member of a biofilm community, the bacterium must differentiate into a biofilm-associated cell by repressing synthesis of the flagellum that might destabilize the biofilm and producing exopolysaccharide that will reinforce the biofilm structure.
Some genes may be expressed in response to a specific surface on which the bacterium has chosen to settle. For instance, chitin, a polymer of N-acetylglucosamine, is a component of crustacean and insect exoskeletons. Attachment to and degradation of chitin for use as a nutrient source is an important part of survival for many marine Vibrio species (3, 21). The structural genes that are important for attachment to chitin differ from those required for attachment to abiotic, nonnutritive surfaces such as plastic and glass (36, 45). Furthermore, although liquid medium that is rich in nutrients primes many bacteria for attachment to any local surface (32, 34, 45), the bacteria will attach to chitin, but not plastic or glass, even when surrounded by a nutrient-poor medium (45). In some marine bacteria, it has been shown that chitinase and chitin-binding genes are expressed selectively in the presence of chitin (29, 42). Thus, when the bathing medium is rich in nutrients, a bacterium will attach to any available surface, while in a nutrient-poor environment the bacterium will attach preferentially to a nutritive surface. This adaptation ensures that the bacterium will maximize access to nutrients in both nutrient-poor and nutrient-rich aqueous environments.
City dwellers distribute themselves geographically based on the neighbors and environment that best suits their needs and requirements. Chefs and grocers may settle together in the restaurant district, while musicians may settle near concert halls. The same is true for biofilm-associated cells. Specific coaggregation of oral bacteria is thought to determine the distribution of bacteria within multispecies dental biofilms known as plaque. These interactions are thought to be essential for successful plaque formation (22, 23, 47). Furthermore, the environment in a biofilm is not homogeneous. Microelectrode measurements have shown that the oxygen concentration and pH fall in a biofilm as the substratum is approached (30, 49). In single-species biofilms, the biofilm-associated bacteria alter gene expression to maximize survival in their particular microenvironment (20, 49). In mixed biofilms, which are more representative of biofilms occurring in nature, bacteria distribute themselves according to who can survive best in the particular microenvironment and also based on symbiotic relationships between the groups of bacteria (27, 28, 30). Thus, the bacteria in a multispecies biofilm are not randomly distributed but rather organized to best meet the needs of each.
Villagers establish zoning laws and regulate settlement through communication with each other. Bacteria also communicate with each other. Intercellular communication between bacteria is generally carried out by bacterial products that are able to diffuse away from one cell and enter another cell. It is difficult to envision this as an effective means of communication between planktonic bacteria in natural, aquatic environments, since molecules are likely to be carried off in the aqueous phase with a very small probability of reaching neighboring bacteria. Rather, this method of intercellular signaling seems ideally suited for bacteria in a diffusion-limited environment such as the biofilm. Production of the quorum-sensing molecules known as acyl-homoserine lactones (acyl-HSLs) has been demonstrated in both natural and cultured biofilms (1, 7, 26, 41). The importance of acyl-HSLs in single-species biofilms has been clearly demonstrated. In P. aeruginosa, acyl-HSLs are responsible for defining the separations between bacterial pillars in the three-dimensional structure of the biofilm (7). P. aeruginosa mutants that do not produce acyl-HSL form biofilms in which the cells are closely packed together and are easily disrupted by sodium dodecyl sulfate. Acyl-HSLs are also mediators of surface attachment in Pseudomonas fluorescens (1). Extracellular signals, therefore, enforce the zoning laws in single-species biofilms.
Although little is known of the role of intercellular signaling in multispecies biofilms, we suspect it may differ significantly from that observed in single-species biofilms. We expect these signals to be especially important in favorable environments where surfaces are heavily colonized and competition for attachment to the surface is fierce. We define these signals broadly as any actively or passively transported bacterial products that alter the state of neighboring microbes. These might include bacterial metabolites, acyl-HSLs, secreted proteins, genetic material such as DNA or RNA, or as yet undiscovered bacterial products. These signals might alter the distribution of specific bacterial species in the biofilm, alter protein expression in neighboring cells, introduce new genetic traits into neighboring cells, or lure and incorporate bacteria into the biofilm for subsequent consumption. The last function of intercellular communication in multi-species biofilms is both fascinating and as yet uncharted. There are, however, laboratory models of lethal interspecies bacterial communication (38, 39). M. xanthus, for instance, is known to prey on E. coli. On soft agar plates, E. coli moves towards M. xanthus. Its chemotaxis machinery is required for this directed movement. The hypothesis is that M. xanthus secretes a signal that lures E. coli to its death (39). The bacteriocins are another example of cell-cell signals that result in lethal interspecies interactions. These are bacterially derived antibacterial proteins that act against closely related species (38). In fact, mathematical models predict that bacteriocin production would be most advantageous in a spatially structured environment such as a biofilm (10, 12), suggesting that these secreted proteins may have evolved specifically for the biofilm environment. The impact of intercellular communication on multispecies biofilms is potentially far reaching, and we predict that intercellular signalling, whether beneficial or detrimental to the recipient, will be a critical factor in the diversity and distribution of bacteria in a biofilm.
The thick biofilm is like a densely settled area. The buildings are back to back, and they are filled with people. It is difficult to imagine how bacteria can divide in such an environment. Thus, zero population growth may be the norm because the spatial constraints are such that cell division is impeded by surrounding exopolysaccharide. Such a situation may be akin to that of the polymer-encased bacteria that are used for biocatalytic engineering applications (25). Although these bacteria do not divide, they are viable and culturable once freed from the plastic encasement. Thus, one of the dictates of planktonic bacterial life, that consumed nutrients are funneled into procreation, may not apply to biofilm-associated cells. One possibility is that cell division is infrequent in a mature biofilm, and instead excess energy is used to make exopolysaccharide, an edible scaffold, that the cell can digest and use in time of need. As an example of this, production of an exopolysaccharide lyase has been shown for P. fluorescens under starvation conditions (1). This enzyme degrades the biofilm-associated exopolysaccharide for consumption and frees cells from the biofilm scaffold to seek more favorable environments. Both these functions seem adaptive during times of starvation.
One advantage of biofilm living is the ability to acquire transmissible, genetic elements at accelerated rates. There are many reports of accelerated rates of conjugation in bacterial biofilms (2, 18). This suggests that evolution by horizontal transfer of genetic material may occur rapidly in a biofilm, making it the perfect milieu for emergence of new pathogens by acquisition of antibiotic resistance, virulence factors, and environmental survival capabilities.
There are other advantages to living in a city. People live together because this is advantageous in times of adversity. Similarly, biofilm-associated cells are more resistant to many toxic substances such as antibiotics, chlorine, and detergents (4). There is evidence that decreased diffusion into the biofilm (8, 43), decreased bacterial growth rate in a biofilm (11), biofilm-specific substances such as exopolysaccharide (50), and the quorum-sensing specific effects (7, 17) may be reasons for this resistance. This property of biofilms, thus, is most likely multifactorial.
If the bacteria were unable to escape the biofilm, the biofilm would, like an old apartment building, become a death trap when the nutrient supply was exhausted, environmental conditions became unfavorable, or an unfriendly neighbor entered the community. Once the bacterium is encased in exopolysaccharide, however, abandoning the biofilm becomes a significant task. At such times, a polysaccharide lyase may provide the bacterium with an escape (1). This product hastens detachment of biofilm-associated cells. Thus, the cycle of attachment shown in Fig. 1 is completed.
We liken the multispecies bacterial biofilm to a city where bacteria settle selectively, limit settlements of new bacteria, store energy in exopolysaccharide, and transfer genetic material horizontally all for the good of the many. A genetic and biochemical understanding of the interactions between species in a biofilm, complex though they may be, is critical to our understanding of how the biofilm city functions and survives. We predict that in multiple-species biofilms many different types of soluble biofilm-specific signals will be discovered whose influence on dissimilar bacterial neighbors will be sometimes helpful and sometimes detrimental or even fatal. When conditions in the biofilm change, such interactions may determine which cells survive, which perish, and which move on. An understanding of the relationships among species in the biofilm city is essential to our appreciation of the benefits of biofilm-associated living.
REFERENCES
- 1.Allison D G, Ruiz B, SanJose C, Jaspe A, Gilbert P. Extracellular products as mediators of the formation and detachment of Pseudomonas fluorescens biofilms. FEMS Microbiol Lett. 1998;167:179–184. doi: 10.1111/j.1574-6968.1998.tb13225.x. [DOI] [PubMed] [Google Scholar]
- 2.Angles M L, Marshall K C, Goodman A E. Plasmid transfer between marine bacteria in the aqueous phase and biofilms in reactor microcosms. Appl Environ Microbiol. 1993;59:843–850. doi: 10.1128/aem.59.3.843-850.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Colwell R R, Spira W M. The ecology of Vibrio cholerae. In: Barua D, Greenough W B I, editors. Cholera. New York, N.Y: Plenum; 1992. pp. 107–127. [Google Scholar]
- 4.Costerton J W, Cheng K-J, Geesey G G, Ladd T I, Nickel J C, Dasgupta M, Marrie T J. Bacterial biofilms in nature and disease. Annu Rev Microbiol. 1987;41:435–464. doi: 10.1146/annurev.mi.41.100187.002251. [DOI] [PubMed] [Google Scholar]
- 5.Cowan M M, Fletcher M. Rapid screening methods for detection of bacterial mutants with altered adhesion abilities. J Microbiol Methods. 1987;7:241–249. [Google Scholar]
- 6.Davies D G, Chakrabarty A M, Geesey G G. Exopolysaccharide production in biofilms: substratum activation of alginate gene expression by Pseudomonas aeruginosa. Appl Environ Microbiol. 1993;59:1181–1186. doi: 10.1128/aem.59.4.1181-1186.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davies D G, Parsek M R, Pearson J P, Iglewski B H, Costerton J W, Greenberg E P. The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science. 1998;280:295–298. doi: 10.1126/science.280.5361.295. [DOI] [PubMed] [Google Scholar]
- 8.De Beer D, Srinivasan R, Stewart P S. Direct measurement of chlorine penetraion into biofilms during disinfection. Appl Environ Microbiol. 1994;60:4339–4344. doi: 10.1128/aem.60.12.4339-4344.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dunny G M, Leonard B A B. Cell-cell communication in gram-positive bacteria. Annu Rev Microbiol. 1997;51:527–564. doi: 10.1146/annurev.micro.51.1.527. [DOI] [PubMed] [Google Scholar]
- 10.Durrett R, Levin S. Allelopathy in spatially distributed populations. J Theor Biol. 1997;185:165–171. doi: 10.1006/jtbi.1996.0292. [DOI] [PubMed] [Google Scholar]
- 11.Evans D J, Brown M R W, Gilbert P. Susceptibility of bacterial biofilms to tobramycin: role of specific growth rate and phase in the division cycle. J Antimicrob Chemother. 1990;25:585–591. doi: 10.1093/jac/25.4.585. [DOI] [PubMed] [Google Scholar]
- 12.Frank S. Spatial polymorphism of bacteriocins and other allelopathic traits. Evol Ecol. 1994;8:369–386. [Google Scholar]
- 13.Fukuda A, Iba H, Okada Y. Stalkless mutants of Caulobacter crescentus. J Bacteriol. 1977;131:280–287. doi: 10.1128/jb.131.1.280-287.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gacesa P. Bacterial alginate biosynthesis—recent progress and future prospects. Microbiology. 1998;144:1133–1143. doi: 10.1099/00221287-144-5-1133. [DOI] [PubMed] [Google Scholar]
- 15.Garrett E S, Perlegas D, Wozniak D J. Negative control of flagellum synthesis in Pseudomonas aeruginosa is modulated by the alternative sigma factor AlgT (AlgU) J Bacteriol. 1999;181:7401–7404. doi: 10.1128/jb.181.23.7401-7404.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Genevaux P, Muller S, Bauda P. A rapid screening procedure to identify mini-Tn10 insertion mutants of Escherichia coli K-12 with altered adhesion properties. FEMS Microbiol Lett. 1996;142:27–30. doi: 10.1111/j.1574-6968.1996.tb08402.x. [DOI] [PubMed] [Google Scholar]
- 17.Hassett D J, Ma J F, Elkins J G, McDermott T R, Ochsner U A, West S E, Huang C T, Fredericks J, Burnett S, Stewart P S, McFeters G, Passador L, Iglewski B H. Quorum sensing in Pseudomonas aeruginosa controls expression of catalase and superoxide dismutase genes and mediates biofilm susceptibility to hydrogen peroxide. Mol Microbiol. 1999;34:1082–1093. doi: 10.1046/j.1365-2958.1999.01672.x. [DOI] [PubMed] [Google Scholar]
- 18.Hausner M, Wuertz S. High rates of conjugation in bacterial biofilms as determined by quantitative in situ analysis. Appl Environ Microbiol. 1999;65:3710–3713. doi: 10.1128/aem.65.8.3710-3713.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hecht G B, Newton A. Identification of a novel response regulator required for the swarmer-to-stalked-cell transition in Caulobacter crescentus. J Bacteriol. 1995;177:6223–6229. doi: 10.1128/jb.177.21.6223-6229.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang C T, Xu K D, McFeters G A, Stewart P S. Spatial patterns of alkaline phosphatase expression within bacterial colonies and biofilms in response to phosphate starvation. Appl Environ Microbiol. 1998;64:1526–1531. doi: 10.1128/aem.64.4.1526-1531.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Keyhani N O, Roseman S. The chitin catabolic cascade in the marine bacterium Vibrio furnissii. J Biol Chem. 1996;271:33414–33424. doi: 10.1074/jbc.271.52.33414. [DOI] [PubMed] [Google Scholar]
- 22.Klier C M, Roble A G, Kolenbrander P E. Actinomyces serovar WVA963 coaggregation-defective mutant strain PK2407 secretes lactose-sensitive adhesin that binds to coaggregation partner Streptococcus oralis 34. Oral Microbiol Immunol. 1998;13:337–340. doi: 10.1111/j.1399-302x.1998.tb00688.x. [DOI] [PubMed] [Google Scholar]
- 23.Kolenbrander P E, Parrish K D, Andersen R N, Greenberg E P. Intergeneric coaggregation of oral Treponema spp. with Fusobacterium spp. and intrageneric coaggregation among Fusobacterium spp. Infect Immun. 1995;63:4584–4588. doi: 10.1128/iai.63.12.4584-4588.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Losick R, Shapiro L, editors. Microbial development. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1984. [Google Scholar]
- 25.Lyngberg O K, Thiagarajan V, Stemke D J, Schottel J L, Scriven L E, Flickinger M C. A patch coating method for preparing biocatalytic films of Escherichia coli. Biotechnol Bioeng. 1999;62:44–55. doi: 10.1002/(sici)1097-0290(19990105)62:1<44::aid-bit6>3.0.co;2-w. [DOI] [PubMed] [Google Scholar]
- 26.McLean R J, Whiteley M, Strickler D J, Fuqua W C. Evidence of autoinducer activity in naturally occurring biofilms. FEMS Microbiol Lett. 1997;154:259–263. doi: 10.1111/j.1574-6968.1997.tb12653.x. [DOI] [PubMed] [Google Scholar]
- 27.Moller S, Pedersen A R, Poulsen L K, Arin E, Molin S. Activity and three-dimensional distribution of toluene-degrading Pseudomonas putida in a multispecies biofilm assessed by quantitative in situ hybridization and scanning confocal laser microscopy. Appl Environ Microbiol. 1996;62:4632–4640. doi: 10.1128/aem.62.12.4632-4640.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moller S, Sternberg C, Andersen J B, Christensen B B, Ramos J L, Givskov M, Molin S. In situ gene expression in mixed-culture biofilms: evidence of metabolic interactions between community members. Appl Environ Microbiol. 1998;64:721–732. doi: 10.1128/aem.64.2.721-732.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Montgomery M T, Kirchman D L. Induction of chitin-binding proteins during the specific attachment of the marine bacterium Vibrio harveyi to chitin. Appl Environ Microbiol. 1994;60:4284–4288. doi: 10.1128/aem.60.12.4284-4288.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Okabe S, Satoh H, Watanabe Y. In situ analysis of nitrifying biofilms as determined by in situ hybridization and the use of microelectrodes. Appl Environ Microbiol. 1999;65:3182–3191. doi: 10.1128/aem.65.7.3182-3191.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.O'Toole G A, Kolter R. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol Microbiol. 1998;30:295–304. doi: 10.1046/j.1365-2958.1998.01062.x. [DOI] [PubMed] [Google Scholar]
- 32.O'Toole G A, Kolter R. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol Microbiol. 1998;28:449–461. doi: 10.1046/j.1365-2958.1998.00797.x. [DOI] [PubMed] [Google Scholar]
- 33.Plamann L, Li Y, Cantwell B, Mayor J. The Myxococcus xanthus asgA gene encodes a novel signal transduction protein required for multicellular development. J Bacteriol. 1995;177:2014–2020. doi: 10.1128/jb.177.8.2014-2020.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pratt L A, Kolter R. Genetic analysis of Escherichia coli biofilm formation: roles of flagella, motility, chemotaxis and type I pili. Mol Microbiol. 1998;30:285–293. doi: 10.1046/j.1365-2958.1998.01061.x. [DOI] [PubMed] [Google Scholar]
- 35.Prigent-Combaret C, Vidal O, Dorel C, Lejeune P. Abiotic surface sensing and biofilm-dependent regulation of gene expression in Escherichia coli. J Bacteriol. 1999;181:5993–6002. doi: 10.1128/jb.181.19.5993-6002.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pruzzo C, Crippa A, Bertone S, Pane L, Carli A. Attachment of Vibrio alginolyticus to chitin mediated by chitin-binding proteins. Microbiology. 1996;142:2181–2186. doi: 10.1099/13500872-142-8-2181. [DOI] [PubMed] [Google Scholar]
- 37.Quon K C, Marczynski G T, Shapiro L. Cell cycle control by an essential bacterial two-component signal transduction protein. Cell. 1996;84:83–93. doi: 10.1016/s0092-8674(00)80995-2. [DOI] [PubMed] [Google Scholar]
- 38.Riley M A. Molecular mechanisms of bacteriocin evolution. Annu Rev Genet. 1998;32:255–278. doi: 10.1146/annurev.genet.32.1.255. [DOI] [PubMed] [Google Scholar]
- 39.Shi W, Zusman D R. Fatal attraction. Nature. 1993;366:414–415. doi: 10.1038/366414a0. [DOI] [PubMed] [Google Scholar]
- 40.Shimkets L J. Intercellular signaling during fruiting-body development of Myxococcus xanthus. Annu Rev Microbiol. 1999;53:525–549. doi: 10.1146/annurev.micro.53.1.525. [DOI] [PubMed] [Google Scholar]
- 41.Stickler D J, Morris N S, McLean R J, Fuqua C. Biofilms on indwelling urethral catheters produce quorum-sensing signal molecules in situ and in vitro. Appl Environ Microbiol. 1998;64:3486–3490. doi: 10.1128/aem.64.9.3486-3490.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stretton S, Techkarnjanaruk S, McLennan A M, Goodman A E. Use of green fluorescent protein to tag and investigate gene expression in marine bacteria. Appl Environ Microbiol. 1998;64:2554–2559. doi: 10.1128/aem.64.7.2554-2559.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Suci P A, Mittelman M W, Yu F P, Geesey G G. Investigation of ciprofloxacin penetration into Pseudomonas aeruginosa biofilms. Antimicrob Agents Chemother. 1994;38:2125–2133. doi: 10.1128/aac.38.9.2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wall D, Kaiser D. Type IV pili and cell motility. Mol Microbiol. 1999;32:1–10. doi: 10.1046/j.1365-2958.1999.01339.x. [DOI] [PubMed] [Google Scholar]
- 45.Watnick P I, Fullner K J, Kolter R. A role for the mannose-sensitive hemagglutinin in biofilm formation by Vibrio cholerae El Tor. J Bacteriol. 1999;181:3606–3609. doi: 10.1128/jb.181.11.3606-3609.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Watnick P I, Kolter R. Steps in the development of a Vibrio cholerae biofilm. Mol Microbiol. 1999;34:586–595. doi: 10.1046/j.1365-2958.1999.01624.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Whittaker C J, Klier C M, Kolenbrander P E. Mechanisms of adhesion by oral bacteria. Annu Rev Microbiol. 1996;50:513–552. doi: 10.1146/annurev.micro.50.1.513. [DOI] [PubMed] [Google Scholar]
- 48.Wu J, Newton A. Regulation of the Caulobacter flagellar gene hierarchy; not just for motility. Mol Microbiol. 1997;24:233–239. doi: 10.1046/j.1365-2958.1997.3281691.x. [DOI] [PubMed] [Google Scholar]
- 49.Xu K D, Stewart P S, Xia F, Huang C T, McFeters G A. Spatial physiological heterogeneity in Pseudomonas aeruginosa biofilm is determined by oxygen availability. Appl Environ Microbiol. 1998;64:4035–4039. doi: 10.1128/aem.64.10.4035-4039.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yildiz F H, Schoolnik G K. Vibrio cholerae O1 El Tor: identification of a gene cluster required for the rugose colony type, exopolysaccharide production, chlorine resistance, and biofilm formation. Proc Natl Acad Sci USA. 1999;96:4028–4033. doi: 10.1073/pnas.96.7.4028. [DOI] [PMC free article] [PubMed] [Google Scholar]