Abstract
The LasR-LasI and RhlR-RhlI quorum-sensing systems are global regulators of gene expression in the opportunistic pathogen Pseudomonas aeruginosa. Previous studies suggest that the RhlR-RhlI system activates expression of rpoS. We constructed merodiploid strains of P. aeruginosa containing the native rpoS gene and an rpoS-lacZ fusion. Studies of lacZ transcription in these strains indicated that rpoS was not regulated by RhlR-RhlI. We also generated an rpoS null mutant. This rpoS mutant showed elevated levels of rhlI (but not rhlR) transcription, elevated levels of the RhlI-generated acylhomoserine lactone quorum-sensing signal, and elevated levels of RhlR-RhlI-regulated gene transcription. These findings indicate that there is a relationship between RpoS and quorum sensing, but rather than the RhlR-RhlI system influencing the expression of rpoS, it appears that RpoS regulates rhlI.
Two acylhomoserine lactone (acyl-HSL) quorum-sensing signal systems are involved in transcriptional regulation of many genes in Pseudomonas aeruginosa. The two systems are the LasR-LasI and RhlR-RhlI (also termed the Vsm) regulatory circuits. LasI is required for synthesis of the signal N-(3-oxododecanoyl)-HSL (3OC12-HSL), and LasR is a transcriptional activator that responds to 3O-C12-HSL. The RhlI enzyme is responsible for synthesis of N-butyryl-HSL (C4-HSL), and RhlR is a transcriptional activator that responds to C4-HSL. The two quorum-sensing systems do not act independently of one another, but a hierarchy exists with LasR required for the activation of rhlR and to some extent rhlI (for reviews of quorum sensing see references 7, 8, and 24). Quorum sensing has been reported to control the expression of genes, including those coding for extracellular enzymes, secondary metabolites (e.g., pyocyanin and hydrogen cyanide), toxins, genes of unknown function, and the rpoS gene, which codes for a homolog of an Escherichia coli stationary-phase ς factor (30).
In E. coli, RpoS competes with the housekeeping sigma factor (ς70) for RNA polymerase, and RpoS enhances expression of more than 35 genes, many of which are involved in the general stress response (14, 18). P. aeruginosa rpoS has been cloned and sequenced (30). As with E. coli, the rpoS gene in P. aeruginosa is induced as cultures enter stationary phase (6). A P. aeruginosa rpoS null mutant shows a defect in the general stress response and a defect in the regulation of exoproducts, including pyocyanin and exotoxin A (12, 29).
The rhl quorum-sensing system has been reported to activate transcription of rpoS in P. aeruginosa (15). As an extension of an investigation of quorum-sensing-regulated genes in P. aeruginosa (31), we constructed an rpoS mutant. This mutant overproduced the RhlR-RhlI-controlled secondary metabolite pyocyanin. This finding prompted our study of the relationship between the RhlR-RhlI system and RpoS. The studies described here with the RpoS− mutant and with P. aeruginosa strains carrying an rpoS-lacZ allele led to the conclusion that RhlR-I does not regulate RpoS expression but that RpoS does influence the transcription of rhlI.
The bacterial strains and plasmids used in this study are described in Table 1. Unless otherwise specified, cultures were grown in Luria-Bertani (LB) broth or on LB agar (26) as described previously (28). For pyocyanin measurements, P. aeruginosa was grown in Pseudomonas broth. DNA manipulations involved standard methods (2), and DNA was sequenced at the University of Iowa DNA Core Facility. Southern blotting was performed using standard methods (2) with PCR-generated digoxigenin-11-dUTP-labeled DNA probes. Transformation of P. aeruginosa was done as follows. Cells from a mid-logarithmic-phase culture (optical density at 600 nm [OD600], 0.6 to 0.8) were harvested by centrifugation at 6,000 × g for 10 min at 4°C and suspended in cold 150 mM MgCl2 for 30 min. After another centrifugation, the cells were suspended for 10 min in 100 μl of cold 150 mM MgCl2 with 0.5 to 1 μg of plasmid DNA. The cells were heat shocked at 37°C for 3 min and then incubated on ice for 5 min, after which 1 ml of LB broth was added. This suspension was incubated with shaking at 37°C for 30 min and then cells were plated on selective agar.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics | Source or reference |
|---|---|---|
| P. aeruginosa | ||
| PAO1 | Wild type | 10 |
| PAO-MW20 | rpoS mutant of PAO1, Gmr | This study |
| PAO-R1 | ΔlasR derivative of PAO1, Tcr | 9 |
| PAO-MW1 | ΔlasI ΔrhlI derivative of PAO1, Hgr Tcr | 31 |
| PDO111 | rhlR::Tn501 derivative of PAO1, Hgr | 3 |
| E. coli | ||
| DH5α | F− φ80dlacZΔM15 Δ(lacZYA-argF)U169 endA1 recA1 hsdR17 deoR gyrA96 thi-1 relA1 supE44 | 26 |
| S17-1 | thi pro hsdR recA RP4-2 (Tet::Mu) (Km::Tn7) | 28 |
| Plasmids | ||
| pCR2.1 | TA cloning vector, Apr | Invitrogen |
| pEX1.8 | Broad-host-range expression vector, Apr | 23 |
| pSUP102 | pACYC184 carrying mobRP4, Cmr Tcr | 27 |
| pRK2013 | tra+ helper plasmid for triparental matings, Kmr | 5 |
| pHRP315 | Ω Smr/Spr cassette, Apr | 20 |
| pHRP309 | Promoterless lacZ, Gmr | 20 |
| pTL61T | lacZ transcriptional fusion vector, Apr | 17 |
| pBBR1MCS-5 | Broad-host-range vector, Gmr | 13 |
| pRPOS-1 | pCR2.1 containing a 2.8-kb rpoS fragment | This study |
| pRPOS-Gm | pRPOS-1 with aacC1 interrupting the rpoS gene, Gmr | This study |
| pMW102 | rpoS::aacC1 on pSUP102, mobilizable rpoS knockout plasmid, Gmr | This study |
| pMW105 | pEX1.8 containing ptac-rpoS, Apr | This study |
| pMW301 | Broad-host-range hcnAB-lacZ reporter, Apr Cbr | This study |
| pMW303 | Broad-host-range phzABC-lacZ reporter, Apr Cbr | This study |
| pMW304 | Broad-host-range rhlR-lacZ reporter, Gmr Spr | This study |
| pMW305 | Broad-host-range rhlI-lacZ reporter, Gmr Spr | This study |
| pTn5-Gm | pUTmini-Tn5 containing aacC1, Gmr | This study |
| pJBA24 | Cloning vector for creating reporter fusions on Tn5, Apr | 1 |
| pMRP24-RPOS | pJBA24 containing rpoS promoter, Apr | This study |
| pMW24-RPOS | rpoS promoter-lacZ fusion vector, Apr | This study |
| pUTminiTn5-Gm | Delivery plasmid for mini-Tn5-Gm, Apr Gmr | This study |
| pTn5-RPOS | pUTmini-Tn5 carrying rpoS-lacZ from pMW24-RpoS | This study |
To construct a P. aeruginosa rpoS mutant, we amplified a 2.8-kb P. aeruginosa chromosomal DNA fragment containing the rpoS gene by using the Expand long-template PCR system (Boehringer Mannheim). The rpoS fragment was cloned to form pRPOS-1 by using an Original TA cloning kit (Invitrogen, Carlsbad, Calif.). This plasmid was digested with HincII and ligated to a PCR-generated 1.1-kb aacC1 (encoding gentamicin acetyltransferase) fragment from pBBR1MCS-5 to generate pRPOS-Gm. A pRPOS-Gm EcoRI fragment containing the inactivated rpoS gene was cloned into EcoRI-digested pSUP102 to generate pMW102, which contains the aacC1 gene 266 bp downstream of the translational start codon of rpoS. The rpoS in pMW102 is flanked by 1.5 kb of P. aeruginosa DNA upstream and 1.3 kb downstream. With E. coli HB101(pRK2013) as a helper, we used triparental mating to mobilize pMW102 into P. aeruginosa PAO1 (5). We selected gentamicin-resistant colonies and identified a tetracycline-sensitive mutant, PAO-MW20. This mutant contained rpoS::aacC1 in place of rpoS, as shown by a Southern analysis with aacC1 and rpoS probes.
For complementation of PAO-MW20, a 1.2-kb chromosomal DNA fragment containing the rpoS gene from P. aeruginosa was created by PCR amplification using the Expand long-template PCR system, digested with EcoRI and HindIII, and ligated to EcoRI-HindIII-digested pEX1.8 to generate the ptac-rpoS plasmid pMW105. Mutations in rpoS were complemented by transformation of P. aeruginosa with pMW105.
For the construction of rhlI-lacZ and rhlR-lacZ transcriptional fusions, a two-step cloning procedure with pHRP315 and pHRP309 was used (20). A 588-bp PCR product beginning 435 bp upstream of the rhlR translational start site and a 445-bp PCR product beginning 250 bp upstream of the rhlI transcriptional start site were used as promoter fragments for making lacZ fusions. Each promoter fragment was directionally cloned downstream of the spectinomycin resistance cassette of pHRP315. The promoter fragments were then cloned upstream of the ′lacZ gene of pHRP309 to form the rhlI-lacZ and rhlR-lacZ transcriptional fusion plasmids pMW304 and pMW305. The hcnAB-lacZ and phzABC-lacZ fusion plasmids pMW301 and pMW303 were constructed as follows. Expand long-template PCR was used to amplify the chromosomal hcnAB-lacZ and phzABC-lacZ transcriptional fusions in P. aeruginosa qsc128 and qsc131 (31). The hcnAB-lacZ fragment begins 619 bp upstream of the qsc128 lacZ insertion, and the phzABC-lacZ fragment begins 886 bp upstream of the qsc131 lacZ insertion. The two PCR fragments were cloned into SalI-digested, end-polished pEX1.8 to form pMW301 (hcnAB-lacZ) and pMW303 (phzABC-lacZ). To minimize read-through from plasmid promoters, orientations were selected that placed the transcriptional terminator of pEX1.8 upstream of the promoter regions of the phz and hcn fusions. The addition of 3OC12-HSL and C4-HSL to the P. aeruginosa LasI− RhlI− mutant, PAO-MW1, containing pMW301 or pMW303 resulted in a >100-fold increase in β-galactosidase. This confirmed the presence of the quorum-sensing-controlled promoters on pMW301 and pMW303.
We constructed rpoS-lacZ reporters as follows. A 240-bp PCR-generated DNA fragment beginning 201 bp upstream of the rpoS transcriptional start site was ligated to KpnI-XbaI-digested pJBA24, forming pMRP24-RPOS. This rpoS plasmid was digested with SphI and HindIII and ligated to a 3.2-kb PCR-generated lacZ fragment from pTL61T to form pMW24-RPOS. NotI-digested pMW24-RPOS was ligated to NotI-digested pUTminiTn5-Gm to yield pTn5-RPOS. This plasmid is a mobilizable vector containing an rpoS-lacZ transcriptional fusion and a gentamicin resistance marker within mini-Tn5. The rpoS-lacZ fusion is flanked by transcriptional stops to minimize read-through from chromosomal promoters and is in the opposite orientation to the gentamicin resistance marker within the transposon. Introduction of pTn5-RpoS into P. aeruginosa was done by mating with E. coli S17-1 as described above. Five transconjugants that contained single Tn5 insertions at locations other than the chromosomal rpoS gene (as determined by a Southern blot analysis) that showed growth rates in LB broth that were indistinguishable from that of the wild-type strain (PAO1), and that showed a wild-type pyocyanin production phenotype (see below) were used for expression studies.
To analyze lacZ expression in reporter constructs, cultures were inoculated to an initial OD600 of 0.001 to 0.005, and β-galactosidase was measured at various time points as outlined by Miller (19). To monitor levels of acyl-HSLs, P. aeruginosa was grown in LB broth buffered with morpholinepropanesulfonic acid (MOPS) (250 mM, pH 7.0). Cells were removed from 5-ml samples by centrifugation at 6,000 × g for 15 min. The cell-free culture fluid was extracted three times with 5 ml of acidified ethyl acetate, and the extract was dried under a stream of nitrogen gas. The residue was dissolved in 500 μl of acidified ethyl acetate, and acyl-HSL levels were assessed by means of bioassays (21, 22) with synthetic 3OC12-HSL and C4-HSL as standards (Quorum Sciences, Inc., Coralville, Iowa). Pyocyanin was extracted from P. aeruginosa PAO1 and PAO-MW20 grown in Pseudomonas broth at 37°C, with shaking (starting OD600, 0.005), with chloroform. Pyocyanin in the chloroform extracts was measured spectrophotometrically at 520 nm according to the method of Essar et al. (4).
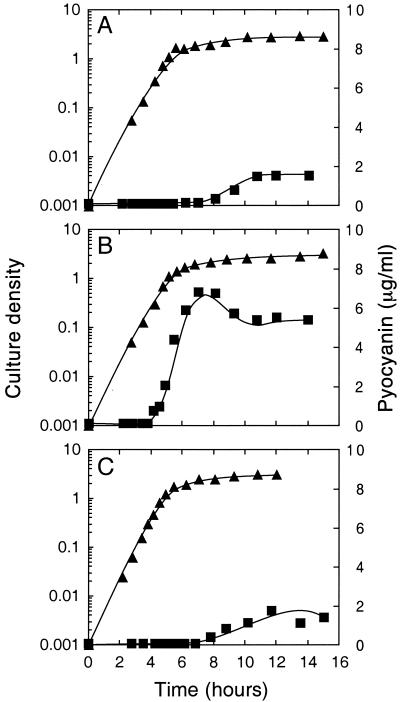
A recent report described a subset of quorum-sensing-controlled genes in P. aeruginosa that required acyl-HSLs for full activation, but even in the presence of acyl-HSLs, activation did not occur until the onset of stationary phase (31). Because previous investigations had indicated that the RhlR-RhlI quorum-sensing system activates rpoS (15), we hypothesized that the responses of genes that required both quorum sensing and stationary phase might be mediated by rpoS. To better understand the interactions between rpoS and quorum sensing in P. aeruginosa PAO1, an RpoS− mutant was constructed through allelic exchange of the wild-type rpoS with a plasmid-borne rpoS::aacC1 mutation. Cultures of the RpoS− mutant grown in LB broth or agar were obviously dark blue, whereas the parent strain, PAO1, was not. Because pyocyanin is a blue pigment, we measured the pyocyanin levels in cultures. Pyocyanin was detected earlier and accumulated to higher levels in the rpoS mutant than in the wild type (Fig. 1). In late-logarithmic-phase cultures, the RpoS− mutant produced approximately 10-fold more pyocyanin than did the wild type, PAO1. Complementation of the rpoS mutation in PAO-MW20 with pMW105 resulted in pyocyanin levels similar to those found in the parent (Fig. 1). These experiments indicate that rpoS negatively regulates the production or secretion of pyocyanin. Our results are consistent with a previous report of increased pyocyanin levels (twofold) in a P. aeruginosa rpoS mutant compared to the parent strain (29), although the closer examination of pyocyanin synthesis reported here shows that in logarithmic-phase cultures repression of pyocyanin synthesis by RpoS can be much greater than twofold.
FIG. 1.
Influence of RpoS on pyocyanin production. Growth was measured as OD600 (▴) and pyocyanin levels (■) are expressed as a function of time in P. aeruginosa PAO1 (A), the rpoS mutant strain PAO-MW20 (B), and PAO-MW20 with the ptac-rpoS plasmid pMW105 (C). For the latter (C), isopropyl-β-d-thiogalactopyranoside (IPTG) was added at a final concentration of 100 μM.
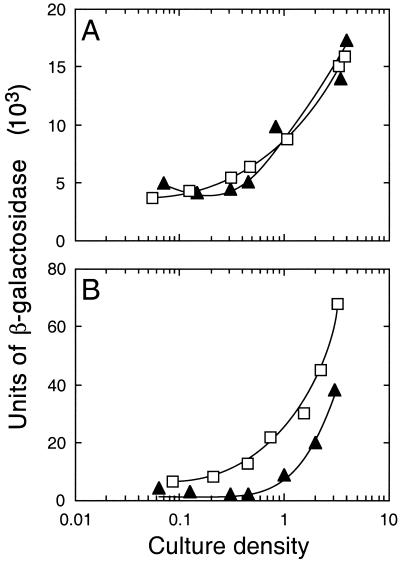
The production of pyocyanin is induced by the rhl quorum-sensing system in P. aeruginosa (16). Furthermore, the phzABCDEFG operon is required for pyocyanin production, and transcription of a phzABCDEFG operon-lacZ chromosomal fusion is increased more than 700-fold by the addition of 3OC12-HSL and C4-HSL to a LasI− RhlI− mutant (31). Thus, increased pyocyanin production in RpoS− mutants might be the result of enhanced transcription of rhlR or rhlI. To test this hypothesis, we measured β-galactosidase activity in P. aeruginosa carrying an rhlR-lacZ reporter on pMW304 or an rhlI-lacZ reporter on pMW305. Levels of β-galactosidase synthesis directed by the rhlR reporter on pMW304 were similar in the RpoS− mutant PAO-MW20 and the parent strain, PAO1 (Fig. 2A). However, β-galactosidase synthesis directed by the rhlI reporter on pMW305 occurred earlier during growth and reached a higher level in PAO-MW20 than it did in the parent (Fig. 2B). These experiments indicate that RpoS represses the transcription of rhlI (probably indirectly) but not of rhlR early in growth.
FIG. 2.
Influence of rpoS on rhlR and rhlI transcription in P. aeruginosa. (A) P. aeruginosa containing rhlR-lacZ plasmid pMW304; (B) P. aeruginosa containing rhlI-lacZ plasmid pMW305. Levels of β-galactosidase in the wild-type PAO1 (▴) and the rpoS mutant MW20 (□) are shown. Culture density is measured as OD600.
The repression of rhlI by RpoS appears to be relatively mild, but small changes in rhlI expression may have a significant impact on C4-HSL levels in early logarithmic phase, where C4-HSL levels are normally low. Thus, we monitored acyl-HSL synthesis during culture growth (Table 2). As assessed with an Rhl bioassay (22), C4-HSL levels in the rpoS mutant were elevated compared to those of the parent. As assessed with a Las bioassay (21), levels of 3OC12-HSL were unaffected by the RpoS mutation. In early-logarithmic-phase cultures of the RpoS− mutant, we detected sufficient levels of C4-HSL for induction of Rhl quorum-sensing-regulated genes, but in early-logarithmic-phase wild-type cultures there was no detectable C4-HSL. This suggests that the increase in pyocyanin production that results from an RpoS mutation is due to increased levels of C4-HSL, which are the result of enhanced transcription of rhlI.
TABLE 2.
Acyl-HSL concentrations in cultures of a P. aeruginosa RpoS− mutant (PAO-MW20) and the wild type, PAO1
| Culture density (OD600) | 3OC12-HSL (μM)a
|
C4-HSL (μM)a
|
||
|---|---|---|---|---|
| PAO1 | MW20 | PAO1 | MW20 | |
| 0.25 | 0.5 ± 0.2 | 0.7 ± 0.1 | <0.1 | 0.4 ± 0.1 |
| 1.5 | 3.1 ± 0.4 | 2.8 ± 0.3 | 0.5 ± 0.1 | 1.9 ± 0.1 |
| 3.0 | 4.4 ± 0.7 | 4.0 ± 0.6 | 3.9 ± 0.3 | 8.4 ± 0.7 |
| 4.0 | 3.8 ± 0.4 | 3.5 ± 0.5 | 12.8 ± 0.7 | 19.9 ± 1.1 |
Values are means ± 1 standard deviation.
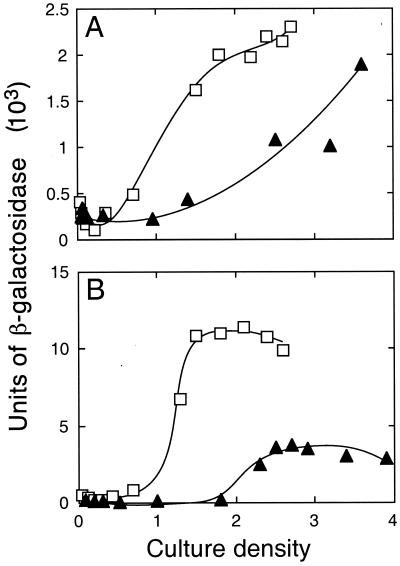
To establish that the increased pyocyanin levels in P. aeruginosa PAO-MW20 were due to increased transcription of the quorum-sensing-regulated phz genes, we compared phzC-lacZ transcription in the rpoS mutant to that in the parent (Fig. 3). Induction of phzC-lacZ occurred earlier in the rpoS mutant and coincided with the increased levels of C4-HSL (Fig. 2). To determine if other genes that are regulated by quorum sensing are affected by RpoS, we monitored expression of an hcnAB-lacZ transcriptional fusion. The hcn operon is involved in the production of the secondary metabolite hydrogen cyanide, and it is activated by the rhl quorum-sensing system (31). As with phzC-lacZ, transcription of hcnB-lacZ occurred earlier and was increased in the RpoS− mutant, compared to the parent (Fig. 3).
FIG. 3.
Expression of hcnAB-lacZ (pMW301) (A) and phzABC-lacZ (pMW303) (B) in P. aeruginosa PAO1 (▴) and PAO-MW20 (□).
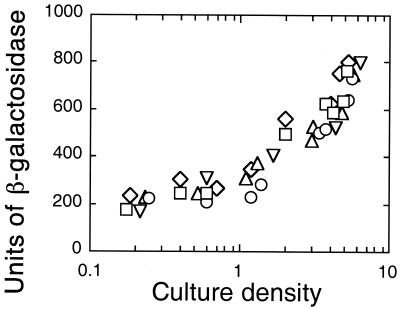
Expression of rpoS in P. aeruginosa has been shown by RNA analysis and immunodetection of the RpoS protein to be growth phase regulated (6, 30). As in E. coli, transcription of rpoS and the amount of RpoS in cells increases at the onset of stationary phase. It has also been reported that P. aeruginosa rpoS is activated by RhlR-RhlI (15). This suggests that there is a regulatory loop in which RhlR-RhlI activates rpoS and the rpoS product, in turn, represses rhlI. To begin an investigation of this possible loop, we constructed rpoS-lacZ merodiploids in the P. aeruginosa parent (PAO1), in a LasR− mutant (PAO-R1), in an RhlR− mutant (PDO111), and in a LasI− RhlI− mutant (PAO-MW1), as described above. As expected, in the parent, expression of the rpoS-lacZ fusion was growth phase dependent. However, contrary to published reports (15), the pattern of expression in the quorum-sensing mutants was indistinguishable from the pattern in the wild type. Furthermore, the addition of 3OC12-HSL and C4-HSL to the LasI− RhlI− mutant did not affect rpoS expression (Fig. 4). These results indicate that the RhlR-RhlI quorum-sensing system does not regulate rpoS transcription under the conditions of our experiment.
FIG. 4.
Transcription of chromosomal rpoS::lacZ in P. aeruginosa PAO1 (▵), the LasR− mutant PAO-R1 (□), the RhlR− mutant PDO111 (▿), the LasI− RhlI− double mutant PAO-MW1 (○), and PAO-MW1 in medium containing 3OC12-HSL (2 μM) and C4-HSL (5 μM) (◊). Five rpoS-lacZ transconjugants were analyzed, and the results were indistinguishable from those shown.
Our results indicate that RpoS functions to repress rhlI, and this repression manifests itself in early logarithmic phase. Expression of RpoS-controlled genes during exponential growth is known to occur in E. coli, where RpoS-dependent expression of xthA (25) and katG (11) occurs in the early logarithmic phase of growth. In the case described here, RpoS functions in early-logarithmic-phase repression of rhlI transcription. It is not an unreasonable hypothesis that RpoS functions to repress all early C4-HSL-regulated genes. Suh et al. (29) reported that an rpoS mutant caused more tissue damage in a rat chronic-lung-infection model than did the wild type and suggested that this could have resulted from elevated levels of pyocyanin in the mutant. Although our data are not inconsistent with this suggestion, it is possible that other genes regulated by C4-HSL might encode virulence factors that contribute to lung damage. In fact, hydrogen cyanide production could easily contribute to tissue damage.
In summary, we have investigated the influence of quorum sensing on rpoS expression in P. aeruginosa. Our data indicate that rpoS transcription is not regulated by quorum sensing. This is in contrast to a previous report that RhlR-RhlI is required for the growth phase-dependent expression of rpoS (15). In our study, quorum-sensing mutants as well as the wild type showed induction of the rpoS-lacZ reporter upon entry into stationary phase. Also, the addition of acyl-HSL signals to a quorum-sensing signal production mutant carrying rpoS-lacZ did not affect expression of β-galactosidase (Fig. 4). The differences between our results and those reported previously could be explained by the copy number of the reporters (single copy in our study versus multicopy), differences in P. aeruginosa strains, or experimental conditions. Regardless of the explanation for the conflicting results, we report a novel relationship between rpoS and quorum sensing in P. aeruginosa involving the repression of rhlI transcription by rpoS.
Acknowledgments
This research was supported by a grant from the National Institutes of Health (GM59026). M.R.P. was supported by a National Institutes of Health Postdoctoral Fellowship (GM 18740-01AI), and M.W. has been supported by a National Science Foundation Research Training Grant (DBI9602247) and by a Public Health Service Training Grant (732 GM8365).
REFERENCES
- 1.Andersen J B, Sternberg C, Poulsen L K, Bjorn S P, Givskov M, Molin S. New unstable variants of green fluorescent protein for studies of transient gene expression in bacteria. Appl Environ Microbiol. 1998;64:2240–2246. doi: 10.1128/aem.64.6.2240-2246.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ausubel F, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Short protocols in molecular biology. 3rd ed. New York, N.Y: John Wiley & Sons, Inc.; 1997. [Google Scholar]
- 3.Brint J M, Ohman D E. Synthesis of multiple exoproducts in Pseudomonas aeruginosa is under control of RhlR-RhlI, another set of regulators in strain PAO1 with homology to the autoinducer-responsive LuxR-LuxI family. J Bacteriol. 1995;177:7155–7163. doi: 10.1128/jb.177.24.7155-7163.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Essar D W, Eberly L, Hadero A, Crawford I P. Identification and characterization of genes for a second anthranilate synthase in Pseudomonas aeruginosa: interchangeability of the two anthranilate synthases and evolutionary implications. J Bacteriol. 1990;172:884–900. doi: 10.1128/jb.172.2.884-900.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Figurski D H, Helinski D R. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc Natl Acad Sci USA. 1979;76:1648–1652. doi: 10.1073/pnas.76.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fujita M, Tanaka K, Takahashi H, Amemura A. Transcription of the principal sigma-factor genes, rpoD and rpoS, in Pseudomonas aeruginosa is controlled according to the growth phase. Mol Microbiol. 1994;13:1071–1077. doi: 10.1111/j.1365-2958.1994.tb00498.x. [DOI] [PubMed] [Google Scholar]
- 7.Fuqua C, Greenberg E P. Self perception in bacteria: quorum sensing with acylated homoserine lactones. Curr Opin Microbiol. 1998;1:183–189. doi: 10.1016/s1369-5274(98)80009-x. [DOI] [PubMed] [Google Scholar]
- 8.Fuqua W C, Winans S C, Greenberg E P. Census and consensus in bacterial ecosystems: the LuxR-LuxI family of quorum-sensing transcriptional regulators. Annu Rev Microbiol. 1996;50:727–751. doi: 10.1146/annurev.micro.50.1.727. [DOI] [PubMed] [Google Scholar]
- 9.Gambello M J, Iglewski B H. Cloning and characterization of the Pseudomonas aeruginosa lasR gene, a transcriptional activator of elastase expression. J Bacteriol. 1991;173:3000–3009. doi: 10.1128/jb.173.9.3000-3009.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Holloway B W, Krishnapillai V, Morgan A F. Chromosomal genetics of Pseudomonas. Microbiol Rev. 1979;43:73–102. doi: 10.1128/mr.43.1.73-102.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ivanova A, Miller C, Glinsky G, Eisenstark A. Role of rpoS (katF) in oxyR-independent regulation of hydroperoxidase I in Escherichia coli. Mol Microbiol. 1994;12:571–578. doi: 10.1111/j.1365-2958.1994.tb01043.x. [DOI] [PubMed] [Google Scholar]
- 12.Jorgensen F, Bally M, Chapon-Herve V, Michel G, Lazdunski A, Williams P, Stewart G S. RpoS-dependent stress tolerance in Pseudomonas aeruginosa. Microbiology. 1999;145:835–844. doi: 10.1099/13500872-145-4-835. [DOI] [PubMed] [Google Scholar]
- 13.Kovach M E, Elzer P H, Hill D S, Robertson G T, Farris M A, Roop II R M, Peterson K M. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene. 1995;166:175–176. doi: 10.1016/0378-1119(95)00584-1. [DOI] [PubMed] [Google Scholar]
- 14.Lange R, Hengge-Aronis R. Identification of a central regulator of stationary-phase gene expression in Escherichia coli. Mol Microbiol. 1991;5:49–59. doi: 10.1111/j.1365-2958.1991.tb01825.x. [DOI] [PubMed] [Google Scholar]
- 15.Latifi A, Foglino M, Tanaka K, Williams P, Lazdunski A. A hierarchical quorum-sensing cascade in Pseudomonas aeruginosa links the transcriptional activators LasR and RhlR to expression of the stationary-phase sigma factor RpoS. Mol Microbiol. 1996;21:1137–1146. doi: 10.1046/j.1365-2958.1996.00063.x. [DOI] [PubMed] [Google Scholar]
- 16.Latifi A, Winson K M, Foglino M, Bycroft B W, Stewart G S A B, Lazdunski A, Williams P. Multiple homologues of LuxR and LuxI control expression of virulence determinants and secondary metabolites through quorum sensing in Pseudomonas aeruginosa PAO1. Mol Microbiol. 1995;17:333–344. doi: 10.1111/j.1365-2958.1995.mmi_17020333.x. [DOI] [PubMed] [Google Scholar]
- 17.Linn T, St. Pierre R. Improved vector system for constructing transcriptional fusions that ensures independent translation of lacZ. J Bacteriol. 1990;172:1077–1084. doi: 10.1128/jb.172.2.1077-1084.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Loewen P C, Hengge-Aronis R. The role of the sigma factor sigma S (KatF) in bacterial global regulation. Annu Rev Microbiol. 1994;48:53–80. doi: 10.1146/annurev.mi.48.100194.000413. [DOI] [PubMed] [Google Scholar]
- 19.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. pp. 352–355. [Google Scholar]
- 20.Parales R E, Harwood C S. Construction and use of a new broad-host-range lacZ transcriptional fusion vector, pHRP309, for gram− bacteria. Gene. 1993;133:23–30. doi: 10.1016/0378-1119(93)90220-w. [DOI] [PubMed] [Google Scholar]
- 21.Pearson J P, Gray K M, Passador L, Tucker K D, Eberhard A, Iglewski B H, Greenberg E P. Structure of the autoinducer required for expression of Pseudomonas aeruginosa virulence genes. Proc Natl Acad Sci USA. 1994;91:197–201. doi: 10.1073/pnas.91.1.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pearson J P, Passador L, Iglewski B H, Greenberg E P. A second N-acylhomoserine lactone signal produced by Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 1995;92:1490–1494. doi: 10.1073/pnas.92.5.1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pearson J P, Pesci E C, Iglewski B H. Roles of Pseudomonas aeruginosa las and rhl quorum-sensing systems in control of elastase and rhamnolipid biosynthesis genes. J Bacteriol. 1997;179:5756–5767. doi: 10.1128/jb.179.18.5756-5767.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pesci E C, Iglewski B H. The chain of command in Pseudomonas quorum sensing. Trends Microbiol. 1997;5:132–135. doi: 10.1016/S0966-842X(97)01008-1. [DOI] [PubMed] [Google Scholar]
- 25.Sak B D, Eisenstark A, Touati D. Exonuclease III and the catalase hydroperoxidase II in Escherichia coli are both regulated by the katF gene product. Proc Natl Acad Sci USA. 1989;86:3271–3275. doi: 10.1073/pnas.86.9.3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 27.Simon R, O'Connell M, Labes M, Puhler A. Plasmid vectors for the genetic analysis and manipulation of Rhizobia and other Gram-negative bacteria. Methods Enzymol. 1986;118:640–659. doi: 10.1016/0076-6879(86)18106-7. [DOI] [PubMed] [Google Scholar]
- 28.Simon R, Priefer U, Puhler A. A broad-host-range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram-negative bacteria. Bio/Technology. 1983;1:37–45. [Google Scholar]
- 29.Suh S-J, Silo-Suh L, Woods D E, Hassett D J, West S E H, Ohman D E. Effect of rpoS mutation on the stress response and expression of virulence factors in Pseudomonas aeruginosa. J Bacteriol. 1999;181:3890–3897. doi: 10.1128/jb.181.13.3890-3897.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tanaka K, Takahashi H. Cloning, analysis and expression of an rpoS homologue gene from Pseudomonas aeruginosa PAO1. Gene. 1994;150:81–85. doi: 10.1016/0378-1119(94)90862-1. [DOI] [PubMed] [Google Scholar]
- 31.Whiteley M, Lee K M, Greenberg E P. Identification of genes controlled by quorum sensing in Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 1999;96:13904–13909. doi: 10.1073/pnas.96.24.13904. [DOI] [PMC free article] [PubMed] [Google Scholar]