Abstract
Evolution by natural selection occurs in cultures of Escherichia coli maintained under carbon starvation stress. Mutants of increased fitness express a growth advantage in stationary phase (GASP) phenotype, enabling them to grow and displace the parent as the majority population. The first GASP mutation was identified as a loss-of-function allele of rpoS, encoding the stationary-phase global regulator, ςS (M. M. Zambrano, D. A. Siegele, M. A. Almirón, A. Tormo, and R. Kolter, Science 259:1757–1760, 1993). We now report that a second global regulator, Lrp, can also play a role in stationary-phase competition. We found that a mutant that took over an aged culture of an rpoS strain had acquired a GASP mutation in lrp. This GASP allele, lrp-1141, encodes a mutant protein lacking the critical glycine in the turn of the helix-turn-helix DNA-binding domain. The lrp-1141 allele behaves as a null mutation when in single copy and is dominant negative when overexpressed. Hence, the mutant protein appears to retain stability and the ability to dimerize but lacks DNA-binding activity. We also demonstrated that a lrp null allele generated by a transposon insertion has a fitness gain identical to that of the lrp-1141 allele, verifying that cells lacking Lrp activity have a competitive advantage during prolonged starvation. Finally, we tested by genetic analysis the hypothesis that the lrp-1141 GASP mutation confers a fitness gain by enhancing amino acid catabolism during carbon starvation. We found that while amino acid catabolism may play a role, it is not necessary for the lrp GASP phenotype, and hence the lrp GASP phenotype is due to more global physiological changes.
Natural microbial populations spend the majority of their lives under nutrient deprivation, due to intense competition for available resources (31, 32). The chemoorganotroph Escherichia coli can survive extended periods of carbon starvation; cells isolated from batch cultures starved for several years can still grow when supplied with exogenous carbon (11, 12). Prolonged starvation is a condition in which microbes such as E. coli undergo rapid evolution by natural selection: mutants with an increased fitness, termed the growth advantage in stationary phase (GASP) phenotype, grow and displace their wild-type parents as the majority (12, 41, 42, 43). The GASP phenomenon is continuous throughout the starvation period, as multiple rounds of population takeover have been observed (11, 43, 44). The first GASP mutation identified was an allele of rpoS (rpoS819) (43). Mutants with the rpoS819 allele are referred to as GI (GASPI) strains, as they have a GASP phenotype when competed against their wild-type (G0) parent (43, 44). Strain ZK1141, isolated from an aged culture of the rpoS819 GI strain (ZK819), expresses the GASP phenotype when competed against its GI parent and was thus designated a GII strain (41, 44). The GII GASP phenotype of ZK1141 is due to three mutations, designated sgaA, sgaB, and sgaC (44). In the work reported here, we identified the sgaB mutation as an allele of lrp, encoding the leucine-responsive regulatory protein, and demonstrate that a published lrp null allele also confers a GASP phenotype.
The sgaB GASP mutation is an allele of lrp.
Lrp is a dimeric DNA-binding protein that can act as either an activator or a repressor, depending on the promoter (6, 9, 33). Also depending on the promoter, Lrp's activity can be modulated positively or negatively by intracellular leucine levels. The Lrp regulon is extensive and includes many genes involved in amino acid metabolism and transport. In general, Lrp regulates amino acid metabolism by increasing anabolism and decreasing catabolism. Lrp appears to be a protein widely conserved in microbes, as lrp homologs have been detected in other proteobacteria such as Bradyrhizobium japonicum and Klebsiella aerogenes, in the gram-positive bacterium Bacillus subtilis, and even in the archaea Pyrococcus furiosus and Sulfolobus solfataricus (3, 7, 14, 21, 24).
The sgaB locus was mapped roughly to min 20 on the E. coli chromosome (44), and further analysis by P1 transduction (29) showed that the sgaB locus was closely (89%) linked to the cydC locus (Table 1) at min 20.0. The lrp locus mapped nearby at min 20.1, and like sgaB mutants (44), lrp null mutants are more sensitive than the wild type to l-serine during growth on glucose (2). For these reasons, we considered lrp a candidate for sgaB and determined the sequence of the lrp allele of ZK1141 and the alleles of its parents, ZK819 (GI) and ZK126 (G0). The lrp genes (including 69 upstream bases that contains the minimal promoter [25]) were PCR amplified, and both strands were sequenced. Both alleles from the parental strains ZK126 and ZK819 were identical to the published wild-type allele (Genbank accession number M35869) (40). However, we found a single lesion in the coding region of the lrp gene of ZK1141: an in-frame, 3-bp deletion of the bases 5′-GGA-3′, resulting in a protein that lacks the Gly-39 residue of wild-type Lrp (40).
TABLE 1.
E. coli recipient and donor strains used in this study
| Strain | Genotype; phenotype | Source or reference |
|---|---|---|
| Recipients | ||
| ZK126 | W3110 tna2 ΔlacU169; G0 | 8 |
| ZK819 | ZK126 rpoS819 rpsL; Smr, GI | 43 |
| ZK1141 | ZK819 lrp-1141 sgaA sgaC; GII | 41 |
| ZK2552 | ZK819 ilvGMEDA+; Valr | 44 |
| ZK2553 | ZK819 bgl+; Bgl+ | 44 |
| Donors | ||
| ZK1240 | ZK126 cydC(surB1)::miniTn10Kmr | 35 |
| ZK2621 | ZK126 sdaA94::Tn10Cmr | This studya |
| BE2 | W3110 lrp-35::Tn10 | R. G. Matthews (10) |
| BE3479 | PS2209 gltB(psiQ32)::lacZ (Mu d1-1734) | R. G. Matthews (10) |
| CAG18467 | MG1655 zfd-1::Tn10 | 36 |
| CAG18493 | MG1655 zbi-29::Tn10 | 36 |
| DL39G | λ−aspC13 fnr-25 glyA42::Tn5 ilvE12 tyrB507 | E. coli Genetic Stock Center, Yale University |
| EC1051 | dadA279::Mu (Aprlac) araD139 ΔlacU169 met1 thi trp strA | M. Freundlich (28) |
The sdaA94::Tn10Cmr insertion mutation was isolated by random insertional mutagenesis of strain ZK126 with λNK1324 (23) and screening among the Cmr mutants for those that could not grow on serine as described elsewhere (37). The Tn10Cmr insertion of this mutant was mapped by arbitrary PCR (5, 44) to the middle of sdaA (nucleotide 654 of 1365). Mutants with this allele showed only background level of L-SD activity (1.4 ± 7.4 versus 121 ± 21 mg of pyruvate formed min−1 OD600−1 for ZK126 [18]).
Platko and Calvo (34) identified a putative helix-turn-helix (HTH) DNA-binding motif in the N-terminal region of Lrp (34). Most HTH proteins identified thus far have a glycine at position 9 in the HTH domain (4, 16), corresponding to the Gly-39 of Lrp absent in Lrp-1141. This glycine residue is thought to play a critical role in creating the turn structure, as its lack of a β carbon reduces any steric interference in creating the left-handed α helix of the turn (4, 16, 30). However, in some HTH proteins other amino acids can be substituted for glycine at position 9 of the motif and still allow proper folding (16, 17). To our knowledge this is the first characterization of an HTH protein deleted for the glycine at position 9. We predicted that the mutant protein has a misfolded DNA-binding motif and would behave as a loss-of-function protein.
The lrp-1141 GASP allele is a loss-of-function allele.
To determine if the lrp-1141 allele was a loss-of-function mutation, we compared several of its phenotypes with those of the wild-type allele and a published null allele, lrp-35 (Table 1). All three alleles were assayed in the ZK126 (G0) genetic background. To control for possible effects of expression from the tetR or tetA gene of the mini-Tn10 insertion of the lrp-35 allele (22, 23), we introduced into the lrp+ and lrp-1141 strains by P1 transduction a mini-Tn10 transposon linked (22%) to the lrp locus. This mini-Tn10 insertion did not affect the outcome of the assays described below, nor did it affect the GASP phenotype of the GI lrp-1141 strain (data not shown).
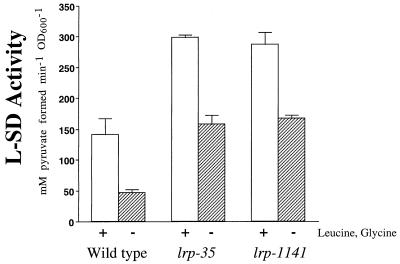
Strains with lrp null alleles are reported to grow more slowly than wild type on glucose and have an increased sensitivity to serine during growth on glucose (2). We confirmed these results and observed that the lrp-1141 mutant behaves identically to the lrp-35 null mutant in both assays. As a more sensitive assay for Lrp activity, we measured l-serine deaminase (L-SD) activity in these strains (18). The major L-SD of E. coli is encoded by sdaA, whose expression is repressed directly by Lrp and activated by leucine and glycine; the leucine activation is due in part to relief of Lrp-mediated repression (26, 37). We assayed L-SD activity as previously described (18) for the wild type and the two lrp mutants grown in the presence or absence of the L-SD inducers glycine and leucine. In both conditions, the lrp-1141 mutant showed L-SD activities (milligrams of pyruvate formed per minute per unit of optical density at 600 nm [OD600]) indistinguishable from the lrp null allele activity (Fig. 1). Similar results were observed in assays of an Lrp-activated gene, gltB (see below). These results indicate that lrp-1141 is a null allele, as it shows no residual repression or activation activity at these promoters.
FIG. 1.
The lrp-1141 mutant has the same L-SD activity as the lrp-35 mutant. L-SD activity was assayed in mid-exponential-phase cells (OD600 of 0.3) grown in M63 glucose (0.2%)–valine (0.005%)–isoleucine (0.005%) medium, with (open bars) or without (hatched bars) the L-SD inducers glycine (0.0075%) and leucine (0.005%). Bars indicate standard deviations (n ≥ 4).
The 2-fold-higher L-SD activity in the lrp mutants grown in the presence of leucine and glycine is interesting, considering previous reports demonstrating that leucine alone increased L-SD activity only 1.2-fold in lrp mutants (26). As glycine and leucine increase L-SD activity in an additive manner (18), these results indicate that unlike leucine, glycine may induce L-SD in an Lrp-independent manner. Alternatively, the different findings may be due differences in strain backgrounds.
The lrp-1141 allele is dominant negative when overexpressed.
Mutations in the DNA-binding domain of lrp that disrupt the DNA-binding activity but do not affect its stability or its ability to dimerize act in a dominant-negative manner, as their protein products are stable and can form heterodimers with the wild-type Lrp monomers that cannot bind DNA (34). To determine if the lrp-1141 allele is dominant negative, we cloned both the mutant and wild-type genes, including 288 bases of upstream sequence, and introduced these clones by transformation into strains with different chromosomal lrp alleles. The lrp+ and lrp-1141 alleles of ZK126 and ZK1141 were amplified by PCR with primers engineered with 5′ ends recognizable by the restriction enzyme EcoRI, and these EcoRI-digested PCR products were ligated into the low-copy-number plasmid pSU2718 (27), creating plasmids pEZ1 (lrp+) and pEZ2 (lrp-1141). We sequenced both strands of the cloned inserts and confirmed that they were the correct alleles. For both constructs, pEZ1 and pEZ2, the lrp gene was inserted in an orientation opposite that of the lac promoter of the pSU2718 vector.
To determine the dominant or recessive nature of the lrp-1141 allele, three assays were performed on the strains carrying various combinations of lrp alleles on the chromosome (single copy) and on a plasmid (multicopy). We assayed two growth phenotypes, colony size and serine sensitivity, on M63 glucose plates as described above. We also assayed transcription of the gltBDF operon, encoding glutamate synthase, which is positively regulated by Lrp (10). The gltB::lacZ transcriptional fusion (Table 1) was crossed into our strains by P1 transduction. The results of all three assays were the same: when the lrp-1141 allele is in multicopy, it is dominant to the single-copy wild-type allele (Table 2). These results indicate that the Lrp-1141 protein is stable and can form inactive heterodimers with the wild-type Lrp monomers. However, the wild-type allele in multicopy is dominant to the lrp-1141 allele in single copy. Overexpression of the wild-type allele most likely restores Lrp-dependent activity by increasing the total concentration of wild-type monomers such that, in addition to the inactive heterodimers, a sufficient amount of active wild-type homodimers form.
TABLE 2.
Phenotypes of strains with various alleles of lrp on the chromosome and in the pSU2718 vectora
|
lrp allele
|
Phenotype on M63 glucose (0.2%)
|
gltB::lacZ activityc | ||
|---|---|---|---|---|
| Chromosome | Plasmid | Relative colony size | Growth on serineb | |
| + | None | ++ | r | 286 (10) |
| + | + | ++ | r | 297 (22) |
| + | lrp-1141 | + | s | 21.4 (1.0) |
| lrp-1141 | None | + | s | 13.6 (1.0) |
| lrp-1141 | + | ++ | r | 265 (24) |
| lrp-1141 | lrp-1141 | + | s | 25.2 (1.3) |
| lrp-35 | None | + | s | 27.1 (3.5) |
| lrp-35 | + | ++ | r | 296 (9) |
| lrp-35 | lrp-1141 | + | s | 27.3 (3.2) |
All assays included chloramphenicol (30 μg/ml) to ensure plasmid maintenance.
Determined as sensitive (s) or resistant (r) by filter disk method (44).
The lrp-35 null mutation also confers a GASP phenotype.
If lrp-1141 is a null allele, then the lrp-35 null allele should exhibit the same GASP phenotype as lrp-1141. We therefore determined the stationary-phase fitness of the lrp-35 null allele relative to the lrp+ allele by competition assays in the GI (rpoS819) background, as described previously (44). We eliminated any possible fitness losses of the lrp-35 strains due to expression from the mini-Tn10 insertion by competing the lrp-35 mutants against two different lrp+ strains carrying either the zbi-29::Tn10 or zfd-1::Tn10 intergenic insertion (Table 1). Competing strains carried one of two neutral markers to distinguish them: the ability to grow on β-glucosides (Bgl+) or the ability to grow on glucose in the presence of valine (Valr) (44). These markers were switched between strains to confirm that they did not affect fitness.
Unlike the lrp+ control strain (Fig. 2A), the lrp-1141 strain expresses a GASP phenotype when inoculated as a 1,000-fold minority into a culture of the lrp+ strain (Fig. 2B) (44). As predicted, the lrp-35 null mutant exhibited a GASP phenotype essentially identical to that of the lrp-1141 mutant (Fig. 2C). This result confirmed that loss-of-function mutations in lrp confer a GASP phenotype on E. coli. This indicates that the Lrp regulon is induced during prolonged carbon starvation, and that activity of this regulon under these conditions results in a fitness loss relative to those cells unable to express this regulon.
FIG. 2.
The lrp-35 mutation confers a competitive advantage to GI cells. Into a 1-day-old LB culture of a Valr GI mutant (●) was inoculated as a 1,000-fold minority a 1-day-old LB culture of a Bgl+ GI mutant (A; ○), GI lrp-1141 mutant (B; □), or GI lrp-35 mutant (C; ▵). Viable counts of the two competing populations were determined by titering the mixed culture onto M63-glucose-valine or M63-salicin plates. Asterisks indicate that viable counts fell below detectable levels (<103 CFU/ml). Similar patterns were observed in six replicate mixtures per strain pairing, including ones when the selectable markers were switched between strains. Data for panel B are from reference 44.
The lrp-1141 and lrp-35 alleles compete with equal fitness.
The possibility remained that there is some undetected activity of the Lrp-1141 protein that plays a role in GASP. In fact, lrp-1141 mutants, unlike lrp-35 null mutants, display mucoid growth on glucose at 30°C (44). However, the lrp-1141 mutants displayed no fitness advantage relative to the lrp-35 null mutants when competed in stationary phase (data not shown). Hence, the mucoidy phenotype of lrp-1141 plays no role in stationary-phase competitions, and we conclude that it is the loss-of-function nature of the lrp-1141 allele that is responsible for the GASP phenotype.
Since there was no detectable fitness difference between the lrp-1141 and insertion (null) allele, we found it surprising that the GASP allele we isolated from a starved culture was lrp-1141. Small, in-frame deletions such as lrp-1141 should be rare among the total array of spontaneous null mutations possible, as only one-third of all spontaneous deletions are in frame, and deletions themselves would occupy only a small percentage of the null mutations possible. At least two factors independent of relative fitness may have increased the likelihood of isolating a strain with such an unexpected GASP allele.
One possible factor is that lrp contains a potential deletion hot spot. The 3-bp deletion of the lrp-1141 allele overlaps the sequence 5′-GTGG-3′ of the wild-type allele, which has been identified as a hot spot for spontaneous mutation in E. coli: a high frequency of mutations was found at or in close proximity to this sequence in wild-type strains (13), and even higher frequencies were found in PolA− strains (13, 19). Among the classes of spontaneous mutations associated with this sequence was a high frequency of deletion mutations, many of which were small. It is therefore possible that this same (unknown) mechanism acted at the 5′-GTGG-3′ hot spot of lrp and facilitated the lrp-1141 deletion. If so, this may indicate a lowered level of DNA polymerase I activity during prolonged starvation, that is, that polA+ cells may be phenotypically PolA− in stationary phase.
A second possible factor leading to the selection of lrp-1141 is its dominant nature. While recessive mutations do not change the cell's physiology until the wild-type product disappears (by degradation or dilution resulting from cell division), dominant mutations can potentially change the cell's physiology as soon as its mutant product is made. Hence, dominant GASP mutations (dominant negative or gain of function) may predominate in starved populations because they can change the physiology of the cell more rapidly than recessive mutations. This may be of critical importance to starved cells, if dilution of wild-type protein by cell division is rare or absent and the cells have only a limited time to acquire GASP mutations before they are outcompeted for nutrients and die. Of particular relevance to this model is the finding that starved E. coli cells can have multiple copies of their chromosome (1), which would make it even more difficult for a recessive loss-of-function GASP phenotype to be expressed. It would therefore be of great interest to isolate and characterize the nature of lrp and other GASP mutations of other starvation survivors to determine the frequency of dominant versus recessive GASP alleles.
Mutations that disrupt alanine, serine, and threonine catabolism do not prevent the lrp GASP phenotype.
Previously, we demonstrated that the sgaB GASP allele (lrp-1141) conferred faster growth when serine, threonine, or alanine was used as the sole carbon and energy source (44). Lrp represses the genes encoding the primary catabolic enzymes for these amino acids: sdaA (L-SD), glyA (serine hydroxymethyltransferase), dadAX (d-alanine deaminase and alanine racemase), and kbl-tdh (2-amino-3-ketobutyrate coenzyme A lyase and threonine dehydrogenase) (28, 33). Thus, the most likely reason why lrp mutants grow more rapidly than the wild type on these amino acids is that the quantity of these enzymes in the wild type is growth rate limiting, and lrp mutants produce more of these enzymes.
We proposed a model that the increased catabolism of these amino acids is responsible for the GASP phenotype of the sgaB (lrp-1141) mutant (44). To address this model, we constructed sdaA, glyA, and dadA insertion mutants of the GI lrp-1141 strain by P1 transduction (Table 1) and assayed for a loss of the GASP phenotype. Significantly, GI lrp-1141 strains with mutations in any of the three genes, or any combination of the three, were still able to outcompete the GI strain. These results indicate that enhanced catabolism of alanine, serine, and threonine is dispensable for the lrp GASP phenotype. However, while not essential, these catabolic activities may still play a significant role stationary-phase competition, as sdaA mutants in the G0, GI, and GI lrp-1141 backgrounds were outcompeted by their respective sdaA+ parents (data not shown).
It is of particular note that the two known GASP loci, lrp and rpoS, are both regulators of many genes. Mutations in regulators such as these make global shifts in metabolism and physiology, often with a coordinated effect (for instance, enhanced catabolism of multiple amino acids in lrp and rpoS mutants [44]). Therefore, while altering a single activity may increase fitness, altering many activities simultaneously by altering the function of a global regulator may result in an even higher fitness gain. Hence, beneficial mutations in global regulators may be selected over other mutations when bacteria are exposed to new environments. In support of this hypothesis are reports that clinical and soil isolates of E. coli and salmonellae have extensive allele variation of rpoS (15, 20, 38; E. R. Zinser and R. Kolter, unpublished data) indicating that there is considerable selective pressure acting on the rpoS locus in the natural world.
Acknowledgments
We thank M. Freundlich and R. Matthews for providing strains. We thank G. O'Toole for technical assistance. We thank S. E. Finkel for critical reading of the manuscript and members of the Kolter lab for helpful comments.
This work was supported by grants from the National Science Foundation (MCB9728936), the National Institutes of Health (GM55199), and the Department of Health and Human Services (ES07155-15) (E.R.Z.).
REFERENCES
- 1.Åkerlund T, Nordström K, Bernander R. Analysis of cell size and DNA content in exponentially growing and stationary-phase batch cultures of Escherichia coli. J Bacteriol. 1995;177:6791–6797. doi: 10.1128/jb.177.23.6791-6797.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ambartsoumian G, D'Ari R, Lin R T, Newman E B. Altered amino acid metabolism in lrp mutants of Escherichia coli K12 and their derivatives. Microbiology. 1994;140:1737–1744. doi: 10.1099/13500872-140-7-1737. [DOI] [PubMed] [Google Scholar]
- 3.Beloin C, Ayora S, Exley R, Hirschbein L, Ogasawara N, Kasahara Y, Alonso J C, Hegarat L. Characterization of an lrp-like (lrpC) gene from Bacillus subtilis. Mol Gen Genet. 1997;256:63–71. doi: 10.1007/s004380050546. [DOI] [PubMed] [Google Scholar]
- 4.Brennan R G, Matthews B W. The helix-turn-helix DNA binding motif. J Biol Chem. 1989;264:1903–1906. [PubMed] [Google Scholar]
- 5.Caetano-Annoles G. Amplifying DNA with arbitrary oligonucleotide primers. PCR Methods Appl. 1993;3:85–92. doi: 10.1101/gr.3.2.85. [DOI] [PubMed] [Google Scholar]
- 6.Calvo J M, Matthews R G. The leucine-responsive regulatory protein, a global regulator of metabolism in Escherichia coli. Microbiol Rev. 1994;58:466–490. doi: 10.1128/mr.58.3.466-490.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Charlier D, Roovers M, Thia-Toong T-L, Durbecq V, Glansdorff N. Cloning and identification of the Sulfolobus solfataricus lrp gene encoding an archaeal homologue of the eubacterial leucine-responsive global transcriptional regulator Lrp. Gene. 1997;201:63–68. doi: 10.1016/s0378-1119(97)00428-9. [DOI] [PubMed] [Google Scholar]
- 8.Connell N, Han Z, Moreno F, Kolter R. An E. coli promoter induced by the cessation of growth. Mol Microbiol. 1987;1:195–201. doi: 10.1111/j.1365-2958.1987.tb00512.x. [DOI] [PubMed] [Google Scholar]
- 9.Cui Y, Midkiff M A, Wang Q, Calvo J M. The leucine-responsive regulatory protein (Lrp) from Escherichia coli. J Biol Chem. 1996;271:6611–6617. doi: 10.1074/jbc.271.12.6611. [DOI] [PubMed] [Google Scholar]
- 10.Ernsting B R, Denninger J W, Blumenthal R M, Matthews R G. Regulation of the gltBDF operon of Escherichia coli: how is a leucine-insensitive operon regulated by the leucine-responsive regulatory protein? J Bacteriol. 1993;175:7160–7169. doi: 10.1128/jb.175.22.7160-7169.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Finkel S E, Kolter R. Evolution of microbial diversity during prolonged starvation. Proc Natl Acad Sci USA. 1999;96:4023–4027. doi: 10.1073/pnas.96.7.4023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Finkel S E, Zinser E, Gupta S, Kolter R. Life and death in stationary phase. In: Busby S J W, Thomas C M, Brown N L, editors. Molecular microbiology. Berlin, Germany: Springer-Verlag; 1997. pp. 3–16. [Google Scholar]
- 13.Fix D F, Burns P A, Glickman B W. DNA sequence analysis of spontaneous mutation in a PolA1 strain of Escherichia coli indicates sequence-specific effects. Mol Gen Genet. 1987;207:267–272. doi: 10.1007/BF00331588. [DOI] [PubMed] [Google Scholar]
- 14.Friedberg D, Platko J V, Tyler B, Calvo J M. The amino acid sequence of Lrp is highly conserved in four enteric microorganisms. J Bacteriol. 1995;177:1624–1626. doi: 10.1128/jb.177.6.1624-1626.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gupta S. Mutations that confer a competitive advantage during starvation. M.A. thesis. Cambridge, Mass: Harvard University; 1997. [Google Scholar]
- 16.Harrison S C, Aggarwal A K. DNA recognition by proteins with the helix-turn-helix motif. Annu Rev Biochem. 1990;59:933–969. doi: 10.1146/annurev.bi.59.070190.004441. [DOI] [PubMed] [Google Scholar]
- 17.Hochschild A, Irwin N, Ptashne M. Repressor structure and the mechanism of positive control. Cell. 1983;32:319–325. doi: 10.1016/0092-8674(83)90451-8. [DOI] [PubMed] [Google Scholar]
- 18.Isenberg S, Newman E B. Studies on l-serine deaminase in Escherichia coli K-12. J Bacteriol. 1974;118:53–58. doi: 10.1128/jb.118.1.53-58.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jankovic M, Kostic T, Savic D J. DNA sequence analysis of spontaneous histidine mutations in a polA1 strain of Escherichia coli K12 suggests a specific role of the GTGG sequence. Mol Gen Genet. 1990;223:481–486. doi: 10.1007/BF00264457. [DOI] [PubMed] [Google Scholar]
- 20.Jordan S J, Dodd C E R, Stewart G S A B. Use of single-strand conformation polymorphism analysis to examine the variability of the rpoS sequence in environmental isolates of salmonellae. Appl Environ Microbiol. 1999;65:3582–3587. doi: 10.1128/aem.65.8.3582-3587.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.King N D, O'Brian M R. Identification of the lrp gene in Bradyrhizobium japonicum and its role in regulation of δ-aminolevulinic acid uptake. J Bacteriol. 1997;179:1828–1831. doi: 10.1128/jb.179.5.1828-1831.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kleckner N. Transposon Tn10. In: Berg D E, Howe M M, editors. Mobile DNA. Washington, D.C.: ASM Press; 1989. pp. 227–268. [Google Scholar]
- 23.Kleckner N, Bender J, Gottesman S. Uses of transposons with emphasis on Tn10. Methods Enzymol. 1991;204:139–180. doi: 10.1016/0076-6879(91)04009-d. [DOI] [PubMed] [Google Scholar]
- 24.Kyrpides N C, Ouzounis C A. The eubacterial transcription activator Lrp is present in the archaeon Pyrococcus furiousus. Trends Biochem Sci. 1995;20:140–141. doi: 10.1016/s0968-0004(00)88988-4. [DOI] [PubMed] [Google Scholar]
- 25.Landgraf J R, Wu J, Calvo J M. Effects of nutrition and growth rate on Lrp levels in Escherichia coli. J Bacteriol. 1996;178:6930–6936. doi: 10.1128/jb.178.23.6930-6936.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin R, D'Ari R, Newman E B. The leucine regulon of Escherichia coli K-12: a mutation in rblA alters expression of l-leucine-dependent metabolic operons. J Bacteriol. 1990;172:4529–4535. doi: 10.1128/jb.172.8.4529-4535.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martinez E, Bartolome B, De la Cruz F. pACYC184-derived cloning vectors containing the multiple cloning site and lacZα reporter gene of pUC8/9 and pUC18/19 plasmids. Gene. 1988;68:159–162. doi: 10.1016/0378-1119(88)90608-7. [DOI] [PubMed] [Google Scholar]
- 28.Mathew E, Zhi J, Freundlich M. Lrp is a direct repressor of the dad operon in Escherichia coli. J Bacteriol. 1996;178:7234–7240. doi: 10.1128/jb.178.24.7234-7240.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miller J H. A short course in bacterial genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Press; 1992. [Google Scholar]
- 30.Mondragón A, Subbiah S, Almo S C, Drottar M, Harrison S C. Structure of the amino-terminal domain of phage 434 repressor at 2.0 Å resolution. J Mol Biol. 1989;205:189–200. doi: 10.1016/0022-2836(89)90375-6. [DOI] [PubMed] [Google Scholar]
- 31.Morita R Y. Bioavailability of energy and its relationship to growth and starvation survival in nature. Can J Microbiol. 1988;34:436–441. [Google Scholar]
- 32.Morita R Y. Bioavailability of energy and the starvation state. In: Kjelleberg S, editor. Starvation in bacteria. New York, N.Y: Plenum Press; 1993. pp. 1–23. [Google Scholar]
- 33.Newman E B, Lin R T, D'Ari R. The leucine/Lrp regulon. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 1513–1525. [Google Scholar]
- 34.Platko J V, Calvo J M. Mutations affecting the ability of Escherichia coli Lrp to bind DNA, activate transcription, or respond to leucine. J Bacteriol. 1993;175:1110–1117. doi: 10.1128/jb.175.4.1110-1117.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Siegele D A, Kolter R. Isolation and characterization of an Escherichia coli mutant defective in resuming growth after starvation. Genes Dev. 1993;7:2629–2640. doi: 10.1101/gad.7.12b.2629. [DOI] [PubMed] [Google Scholar]
- 36.Singer M, Baker T A, Schnitzler G, Deischel S M, Goel M, Dove W, Jaacks K J, Grossman A D, Erickson J W, Gross C A. A collection of strains containing genetically linked alternating antibiotic resistance elements for genetic mapping of Escherichia coli. Microbiol Rev. 1989;53:1–24. doi: 10.1128/mr.53.1.1-24.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Su H, Lang B F, Newman E B. l-Serine degradation in Escherichia coli K-12: cloning and sequencing of the sdaA gene. J Bacteriol. 1989;171:5095–5102. doi: 10.1128/jb.171.9.5095-5102.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Waterman S R, Small P L C. Characterization of the acid resistance phenotype and rpoS alleles of Shiga-like toxin-producing Escherichia coli. Infect Immun. 1996;64:2808–2811. doi: 10.1128/iai.64.7.2808-2811.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wild J, Obrepalska B. Regulation of expression of the dadA gene encoding d-amino acid dehydrogenase in Escherichia coli: analysis of dadA-lac fusions and direction of dadA transcription. Mol Gen Genet. 1982;186:405–410. doi: 10.1007/BF00729461. [DOI] [PubMed] [Google Scholar]
- 40.Willins D A, Ryan C W, Platko J V, Calvo J M. Characterization of Lrp, an Escherichia coli regulatory protein that mediates a global response to leucine. J Biol Chem. 1991;266:10768–10774. [PubMed] [Google Scholar]
- 41.Zambrano M M, Kolter R. Escherichia coli mutants lacking NADH dehydrogenase-I have a competitive disadvantage in stationary phase. J Bacteriol. 1993;175:5642–5647. doi: 10.1128/jb.175.17.5642-5647.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zambrano M M, Kolter R. GASPing for life in stationary phase. Cell. 1996;86:181–184. doi: 10.1016/s0092-8674(00)80089-6. [DOI] [PubMed] [Google Scholar]
- 43.Zambrano M M, Siegele D A, Almirón M, Tormo A, Kolter R. Microbial competition: Escherichia coli mutants that take over stationary phase cultures. Science. 1993;259:1757–1760. doi: 10.1126/science.7681219. [DOI] [PubMed] [Google Scholar]
- 44.Zinser E R, Kolter R. Mutations enhancing amino acid catabolism confer a growth advantage in stationary phase. J Bacteriol. 1999;181:5800–5807. doi: 10.1128/jb.181.18.5800-5807.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]