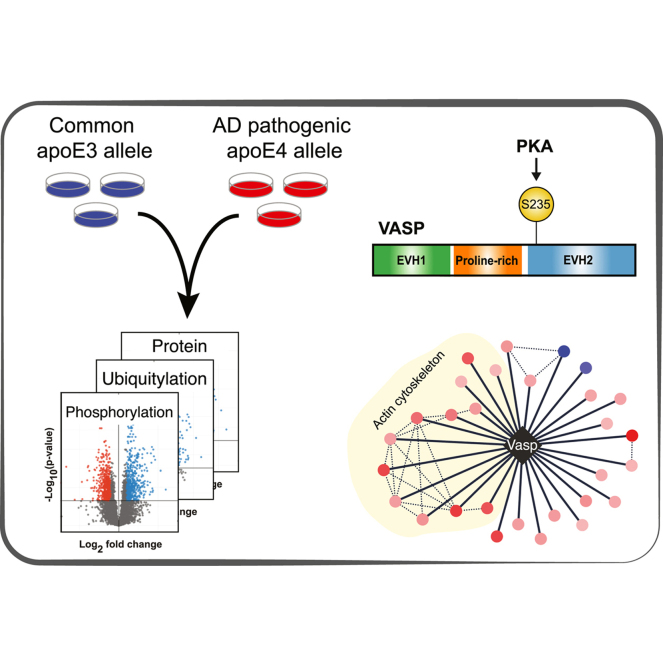
Abstract
Apolipoprotein (apo) E4 is the major genetic risk factor for Alzheimer’s disease. While neurons generally produce a minority of the apoE in the central nervous system, neuronal expression of apoE increases dramatically in response to stress and is sufficient to drive pathology. Currently, the molecular mechanisms of how apoE4 expression may regulate pathology are not fully understood. Here, we expand upon our previous studies measuring the impact of apoE4 on protein abundance to include the analysis of protein phosphorylation and ubiquitylation signaling in isogenic Neuro-2a cells expressing apoE3 or apoE4. ApoE4 expression resulted in a dramatic increase in vasodilator-stimulated phosphoprotein (VASP) S235 phosphorylation in a protein kinase A (PKA)-dependent manner. This phosphorylation disrupted VASP interactions with numerous actin cytoskeletal and microtubular proteins. Reduction of VASP S235 phosphorylation via PKA inhibition resulted in a significant increase in filopodia formation and neurite outgrowth in apoE4-expressing cells, exceeding levels observed in apoE3-expressing cells. Our results highlight the pronounced and diverse impact of apoE4 on multiple modes of protein regulation and identify protein targets to restore apoE4-related cytoskeletal defects.
Keywords: Alzheimer’s disease, Phosphorylation, Ubiquitylation, Protein-protein interaction, Kinase
Graphical Abstract
Highlights
-
•
Evaluation of PTMs regulated by the Alzheimer’s disease apoE4 pathogenic allele.
-
•
ApoE4 induces phosphorylation of VASP at position S235.
-
•
VASP S235 phosphorylation disrupts its interactions with cytoskeletal proteins.
-
•
PKA inhibition drives filopodia formation and neurite outgrowth in apoE4 cells.
In Brief
Apolipoprotein (apo) E4 is the major genetic risk factor for Alzheimer’s Disease (AD). Here, we measured the impact of apoE4 on protein phosphorylation and ubiquitylation. ApoE4 expression resulted in a dramatic increase in VASP S235 phosphorylation in a PKA-dependent manner resulting in the disruption of VASP interactions with numerous actin cytoskeletal and microtubular proteins. Reduction of VASP S235 phosphorylation via PKA inhibition resulted in a significant increase in neurite outgrowth in apoE4-expressing cells.
Apolipoprotein (apo) E is the major cholesterol transport protein in the brain and exists as three allelic variants, apoE2, apoE3, and apoE4, which each differ by single amino acid interchanges (1, 2, 3). The apoE4 variant is the major genetic risk factor for Alzheimer’s disease (AD) (4, 5, 6), occuring in 60 to 70% of all sporadic and familial AD cases (2, 4, 5, 6). ApoE4 lowers the age of onset and increases the risk of developing AD, and considerable evidence indicates that apoE4 has a fundamental role in AD neurodegeneration (2, 3, 7, 8, 9, 10, 11, 12, 13, 14, 15). However, the precise molecular mechanisms by which apoE4 drives pathology remain unclear (16, 17).
Current evidence suggests that neuronal expression of apoE4 is important to this process. Though neurons produce a minority of the apoE in the central nervous system (∼20%) (18), neuronal apoE expression levels increase dramatically in response to stress via a neuron-specific splicing mechanism (19). Furthermore, neuronal expression of apoE4 is sufficient to drive pathology (2, 10, 13, 18, 20), likely via an apoE4 gain-of-function mechanism as compared to apoE3 (21).
Numerous studies have shown that apoE4-driven neuropathology is associated with a variety of cellular phenotypes, including mitochondrial dysfunction (22, 23, 24), cytoskeletal defects, and altered microtubule structure (13). Remarkably, apoE4 expression increases phosphorylation of the microtubule-associated protein tau by activating the Erk pathway in the hippocampus of neuron-specific enolase (NSE)-apoE transgenic mice and in cultured neuronal cells (25). Furthermore, apoE4 induces neurofibrillary tangle (NFT) structures that include phosphorylated tau protein (26). Treatment of cultured neuronal cells with apoE4–lipid complexes impairs neurite outgrowth compared to apoE3–lipid complexes (27, 28, 29). Likewise, neuroblastoma cells stably expressing apoE4 display impaired neurite outgrowth; however, this can be restored by site-directed mutagenesis of apoE4 or by treatment with small-molecule structure correctors that convert the apoE4 structure to one that resembles apoE3 (30, 31).
Unbiased global proteomic analysis of protein abundance and posttranslational modifications by mass spectrometry (MS) represent powerful approaches to discover regulatory biological mechanisms. However, to date, efforts to identify the proteomic impact of apoE4 by MS have primarily been focused on specific organelles or subcompartments (32, 33) and have identified few differentially regulated proteins. Recently, we reported a global protein abundance analysis of Neuro-2a cells expressing apoE3 or apoE4 revealing hundreds of differentially regulated proteins and correlating these differences to apoE4-specific cellular processes (24). To further our understanding of the molecular mechanisms that may regulate apoE4-specific pathology, we have expanded upon our previous studies to include the analysis of protein phosphorylation and ubiquitylation signaling in isogenic Neuro-2a cells expressing human apoE3 or apoE4. The results highlight the diverse impact of apoE4 on multiple modes of protein regulation and identify a mechanism of apoE4-mediated cytoskeletal biology.
Experimental Procedures
Experimental Design and Statistical Rationale
Global proteomics analyses measuring protein abundances, phosphorylated peptides, and ubiquitylated peptides were each performed in biological triplicate for both apoE3 and apoE4 Neuro-2a cells. Each sample was analyzed on the mass spectrometer in technical duplicate. Affinity purification-mass spectrometry (APMS) experiments were performed in biological triplicate for each condition, with one replicate of data collected on the mass spectrometer. Label-free quantification and statistical analysis for both global proteomics and APMS data were performed using MSstats, and resulting log2-fold changes and adjusted p-values are reported (34).
Cell Culture
Mouse neuroblastoma Neuro-2a cell lines stably expressing full-length human apoE3 or apoE4 have been described previously (25). Cells were cultured in MEM containing 1× glutamax (Cat. #41090036, Gibco), 10% fetal bovine serum (Cat. #A3160402, Gibco), 1% nonessential amino acids (Cat. #11140050, Gibco), 1 mM sodium pyruvate (Cat. #11360-070, Gibco).
Cell Lysis and Digestion
Nearly confluent Neuro-2a apoE3 or apoE4 cells growing on 15-cm dishes were rinsed once with 10 ml ice-cold 1× PBS and scraped off the plate in 10 ml fresh ice-cold 1× PBS. Cells were pelleted at 4 °C for 10 min at 1500 rpm, the supernatant was decanted, and cells rinsed by resuspending in 10 ml fresh ice-cold 1× PBS and pelleting again at 4 °C for 10 min at 1500 rpm. Finally, cells were flash frozen on dry ice and stored at −80 °C until all replicates were collected. Three independent sets of cultures were harvested separately.
Cells were lysed by probe sonication on ice in a buffer composed of 8 M urea, 100 mM ammonium bicarbonate (pH 8.0), 150 mM NaCl, and cOmplete mini protease (Cat. #1183615300, Roche) and phosphatase inhibitors (PHOSS-RO, Roche). Protein disulfide bonds were reduced by the addition of 4 mM TCEP and incubated at 22 °C for 30 min. Cysteines were alkylated by the addition of 10 mM iodoacetamide and lysates were incubated in the dark at 22 °C for 30 min. Alkylation was quenched by the addition of 10 mM DTT and lysates were further incubated in the dark at 22 °C for 30 min. Next, proteins were digested overnight at 37 °C by the addition of trypsin (Promega) at a 1:100 enzyme:substrate ratio. Samples were acidified to pH 2 with TFA and desalted on C18 SepPak cartridges (Waters).
Phosphopeptide Enrichment
Immobilized metal affinity chromatography (IMAC) beads for phosphopeptide enrichment were prepared by stripping Ni-NTA Superflow beads (Qiagen) with 100 mM EDTA (pH 7.5), followed by loading of the beads with iron by incubation in 15 mM ferric chloride solution. The beads were then washed with HPLC grade water and 15 μl of beads were loaded into the top of a C18 microspin column. Approximately 1 mg of desalted peptides was resuspended in 75% acetonitrile (ACN), 0.15% TFA, and loaded onto the IMAC tips by pipetting and incubation for 4 min with intermittent mixing by pipetting. The lysate was then passed through the column, followed by four washes with 200 μl of 80% ACN, 0.1% TFA. The column was then equilibrated twice for 2 min with 200 μl of 0.5% formic acid (FA) with intermittent mixing by pipetting. Next, the bound phosphorylated peptides were eluted from the IMAC beads onto the C18 column by the addition of 200 μl of 500 mM potassium phosphate buffer (pH 7), which was incubated with the beads for 3 min with intermittent mixing by pipetting. The potassium phosphate elution was repeated once, and then the column was equilibrated twice for 15 s with 200 μl of 0.5% FA. Finally, the phosphopeptides were eluted from the C18 column twice by the addition of 75 μl of 50% ACN in 0.1% FA, and the eluted phosphopeptides were dried by vacuum centrifugation.
Ubiquitylated Peptide Enrichment
Approximately 1/8th of a tube of anti-PTMScan Ubiquitin Remnant Motif antibody beads (Cell Signaling Technologies) was used per sample. All steps were performed at 4 °C unless otherwise noted. The beads were first washed twice in IAP buffer (50 mM MOPS, 10 mM sodium phosphate buffer (pH 7.2), 50 mM NaCl), and then ∼5 mg of desalted peptides that had been resuspended in IAP was added to the beads. The samples were incubated with rotation for 90 min, and the beads were then washed twice with IAP buffer, followed by two washes with HPLC grade water. Next, peptides were eluted from the beads by the addition of 60 μl of 0.15% TFA to the beads for 10 min with gentle vortexing. The supernatant containing the ubiquitin-enriched peptide eluent was transferred to a new tube, and the elution was repeated by the addition of another 60 μl of 0.15% TFA. The second elution was combined with the first, desalted on C18 columns (Ultra Micro Spin Column, Silica C18, cat #SUM SS18V), and the desalted peptides were dried by vacuum centrifugation.
Mass Spectrometry Data Acquisition
Peptides were resuspended in 4% FA and 3% ACN and directly injected onto a 75 μm ID column (New Objective) packed with 25 cm of Reprosil C18 1.9 μm, 120 Å particles (Dr Maisch) for protein abundance, phosphorylation, and ubiquitylation experiments. For APMS experiments, peptides were resuspended in 0.1% FA and directly injected onto a PepSep column (Bruker) packed with 15 cm of Reprosil C18 1.5 μm, 120 Å particles (Dr Maisch). Peptides were eluted in positive ion mode into an Orbitrap Fusion Tribrid mass spectrometer (Thermo Fisher Scientific) with an ACN gradient delivered by an Easy1000 nLC system (protein abundance, phopshorylation, and ubiquitylation experiments) or an Orbitrap Fusion Lumos Tribrid mass spectrometer (Thermo Fisher Scientific) with an ACN gradient delivered by an Easy1200 nLC system (APMS experiments) (Thermo Fisher Scientific). All mobile phases contained 0.1% FA as buffer A. For protein abundance and phosphorylation experiments, the total MS acquisition time was 180 min, during which mobile phase buffer B (0.1% FA in 90% ACN) was ramped from 5% to 16% B over 114 min, followed by a ramp to 30% buffer B over a subsequent 50 min, and then a ramp to 95% B to wash the column. For the ubiquitylation experiment, the total MS acquisition time was 120 min, during which mobile phase buffer B (0.1% FA in 90% ACN) was ramped from 5% to 33% B over 105 min, followed by a ramp to 100% B to wash the column. For APMS experiments, the total MS acquisition time was 60 min, during which mobile phase buffer B (0.1% FA in 80% ACN) was ramped from 3% to 28% B over 44 min, followed by a ramp to 45% B over a subsequent 5 min, and lastly a ramp to 88% B to wash the column.
For protein abundance, phopshorylation, and ubiquitylation experiments, data was collected on an Orbitrap Fusion Tribrid Mass Spectrometer (Thermo Fisher Scientific). The ion transfer tube was set to 300 °C, the spray voltage was 1800 to 2000 V, MS1 scan was collected in the Orbitrap in profile mode with 120K resolution over a scan range of 400 to 1600 m/z, a 100 ms maximum injection time, 2e6 AGC target, S-Lens RF of 60. Charges states 2 to 7 were considered for MS2 scan selection using MIPS filtering, exclusion of undetermined charge states, a 5e3 minimum signal intensity, and a 20 s dynamic exclusion with a ±10 ppm window. MS2 scans were performed over the course of a 3 s cycle, a 1.6 m/z quadrupole isolation, 29% (ubiquitylation experiment) or 30% (protein abundance and phosphorylation experiments) normalized collision energy, HCD fragmentation, a 110 m/z first mass, ion trap detection with a rapid scan rate, 35 ms maximum injection time, 1e4 AGC target, centroid data type, and the allowance to injection ions for all available parallelizable time. For APMS experiments, data was collected on an Orbitrap Fusion Lumos Tribrid Mass Spectrometer (Thermo Fisher Scientific). The ion transfer tube was set to 275 °C, the spray voltage was 2000 V, MS1 scan was collected in the orbitrap in profile mode with 240K resolution over a scan range of 350 to 1350 m/z, a 50 ms maximum injection time, 1e6 AGC target, S-Lens RF of 30. Charges states 2 to 5 were considered for MS2 scan selection using MIPS filtering, advance peak determination, exclusion of undetermined charge states, a 5e3 minimum signal intensity, and a 20 s dynamic exclusion with a ±10 ppm window in which two occurrences were allowed. MS2 scans were performed over the course of a 1 s cycle, a 0.7 m/z quadrupole isolation, 32% normalized collision energy, HCD fragmentation, a 200 to 1200 m/z scan range, ion trap detection with a rapid scan rate, 18 ms maximum injection time, 3e4 AGC target, centroid data type.
Proteomics Data Analysis
All data were searched against the Mus musculus UniProt database canonical sequences (taxonomy ID 10090) downloaded on September second, 2021 and containing 17,085 sequences. Additionally, the human apoE3 and human apoE4 sequences were included. Additionally, APMS data was also searched against the Raph1 mouse sequence. Peptide and protein identification searches as well as label-free quantitation were performed using the MaxQuant data analysis algorithm (version 2.0.3.0) (35), and all peptide, protein, and modification site identifications were filtered to a 1% false-discovery rate. Default MaxQuant settings were used, with the exception that match between runs was enabled with a window of 2 min (protein abundance and phosphorylation experiments), 1 min (ubiquitylation experiment), or 0.7 min (APMS) for each data type respectively. Searches were performed with a precursor tolerance of 4.5 ppm and a fragment ion tolerance of 0.5 m/z, with trypsin/P specificity (cleaves after lysine and arginine also if a proline follows) allowing for up to two missed cleavages. For all datasets, a fixed modification of carbamidomethyl cysteine (57.02146 Da), a variable modification of methionine oxidation (15.99491 Da), and a variable modification protein N-terminal acetylation (42.01056 Da) were used. For phosphorylation experiments, an additional variable modification of phosphorylation on serine/threonine/tyrosine (79.96633 Da) was used, while for ubiquitylation experiments, an additional variable modification of di-glycine remnant on lysine (114.04293 Da) was applied. MS data files (raw and search results) and annotated spectra have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository with dataset identifier PXD041205 (36).
Label-free quantification and statistical analysis for both global proteomics and APMS data was performed using MSstats (34). Note, for consistent statistical testing across global proteomics data types, the previously published protein abundance analysis (24) was reanalyzed using a more recent version of the MSstats R-package (version 3.3.10), such that this same MSstats version was used for all global proteomics data types reported here. Enrichment analysis was performed using the enricher function from R packages clusterProfiler (version 3.99.0) (37). The Gene Ontology (GO) terms and annotations were obtained from the R annotation package org.Mm.eg.db (version 3.12.0). We selected the most significant GO term from among redundant GO terms, which were identified using a hierarchical tree based on distances (1-Jaccard Similarity Coefficients of shared genes in GO) between the significant terms and then cut the tree into clusters containing redundant terms (R functions hclust and cutree, cut height = 0.99). For tissue specificity, proteins with Log2FC(apoE4/apoE3) <−1 and adjusted p-value <0.05 were input into the “multiple proteins” function in STRING (38). For kinase activity analysis, kinase activities were estimated using known kinase-substrate relationships downloaded from the enzyme-substrate table of OmniPath (39) using the R package OmniPathR[2.0.0] on [01.13.2023]. Kinase-specific data was selected by requiring the modification type to match “phosphorylation” and the gene symbol to be annotated to GO term “GO:0016301 kinase activity” in the R annotation package org.Hs.eg.db [3.12.0].
For APMS experiments, interacting protein abundance values derived from MSstats (34) were visualized using Morpheus (https://software.broadinstitute.org/morpheus) and vasodilator-stimulated phosphoprotein (VASP) differential protein-protein interaction networks were visualized in Cytoscape (40) with edges between interacting proteins being supplemented from STRING (38).
siRNA Knockdowns
Predesigned silencer select siRNAs against APOE, VASP, and negative control were ordered from Thermo Fisher Scientific (Cat. #160728, Cat. #AM16708, Cat. #AM461, respectively). ApoE3 and apoE4 Neuro-2a cells were transfected with siRNA using Lipofectamine RNAiMax (Cat. #13778075, Thermo Fisher Scientific). Cells were seeded into 6-well plates and transfection protocol was followed according to manufacturer’s instructions (Cat. #13778075, Invitrogen).
Constructs
Human GFP-Vasp and mCherry-VASP was gifted by D. Mullins, University of California. Phospho-null mutation VASP S239A was generated using the QuikChange II site-directed mutagenesis kit (Cat. #200523, Agilent). Primers were designed using the QuikChange Primer Design Program and obtained from Integrated DNA Technologies IDT. VASP S239A Forward Primer: GGCCTCCTCCTGCTTGGCGACTTTCCTGAGTTTG, VASP S239A Reverse Primer: CAAACTCAGGAAAGTCGCCAAGCAGGAGGAGGCC. GFP-VASP and mCherry-VASP constructs were transfected using Polyjet transfection reagent (Cat. #SL100688, Signagen) following the manufacturer’s protocol.
Immunoblotting
Cells were lysed in ice-cold RIPA buffer supplemented with cOmplete mini protease inhibitor cocktail (Cat. #1183615300, Roche) and PhosSTOP (PHOSS-RO, Roche). Protein concentration was measured by a Pierce BCA protein assay kit (Cat. #A329645, Life Technologies), and 20 μg protein was loaded on a 4% to 12% acrylamide gel (Criterion TGX, Cat.#5671095, Bio-Rad). The samples were separated by SDS-PAGE and transferred with polyvinylidene difluoride membrane (Trans Blot Turbo, Cat. #1704157, Bio-Rad). Membranes are blocked in 5% milk with Tris-buffered saline with 0.1% Tween 20 detergent (TBST), or for phosphoproteins membranes were blocked 5% in BSA with TBST. Primary antibodies were prepared in 5% BSA-TBST and incubated overnight at 4 °C. The membranes were washed in TBST and incubated 2 h in species-specific horseradish peroxidase–conjugated secondary antibodies (1:3000–1:5000). Membranes were visualized using chemiluminescence substrate (Pierce ECL, Cat. #32209, Thermo Fisher Scientific) and imaged with ChemiDOC MP (Bio-Rad). Antibodies: VASP S239 1:1000 (ab194747, Abcam), VASP 1:1000 (13472-1-AP, Proteintech), apoE 1:2000 (178479, EMD Milipore), Actin 1:1000 (4970L, CST), Akt 1:1000 (9272S, CST), p-AKT T308 1:1000 (13038S, CST), p-AKT S473 1:1000 (4060S, CST).
GFP-VASP Affinity Purification MS
ApoE4 Neuro2a cells were seeded into a 15-cm dish. On the day of transfection, cells were transfected with Polyjet using 9 μg of GFP-VASP wild type (WT) and GFP-VASP S239A constructs according to the manufacturer's instructions (Cat. #SL100688, Signagen). After 8 h of transfection, cells were treated either with dimethyl sulfoxide (control) or with 15 μM H89, and cells were collected after 24 h of treatment.
Frozen cell pellets were thawed at room temperature and suspended in 0.5 ml lysis buffer [IP Buffer (50 mM Tris–HCl, pH 7.4 at 4 °C, 150 mM NaCl, 1 mM EDTA) supplemented with 0.5% Nonidet P 40 Substitute (NP40; Fluka Analytical) and cOmplete mini EDTA-free protease and PhosSTOP phosphatase inhibitor cocktails (Roche)]. Cell suspensions were immediately subjected to one freeze-thaw cycle (at least 20 min on dry ice and partial thawing at 37 °C) and then refrozen and kept at −80 °C until immunoprecipitation. Lysis was completed by partially thawing frozen samples at 37 °C and incubating on a tube rotator at 4 °C for 30 min. After pelleting debris at 13,000g, 4 °C for 15 min, 25 μl lysate was reserved before proceeding (below). A ThermoMixer C incubator was used to thaw samples at indicated temperatures with constant shaking at 1100 rpm for 1 to 2 min; otherwise, samples were kept cold.
For each equilibration/wash step (below), GFP-Trap magnetic agarose beads (20 μl in first plate only; Cat. #GTMA-10, Chromotek) and/or up to 1.0 ml buffer were pre-aliquoted into 96-well deep well plates, while 0.45 ml lysate was arrayed for each sample. All plates were kept on ice until loaded onto the KingFisher Flex (KFF) Purification System (Thermo Fisher Scientific) for automated processing as follows: GFP-Trap beads were equilibrated twice with IP wash buffer (IP buffer supplemented with 0.05% NP40) and incubated with cell lysates for 2 h. Protein-bound beads were washed three times with IP wash buffer and then once with IP buffer before elution. After transferring to 1.5-ml lobind tubes, beads were suspended with 50 μl 0.05% RapiGest in IP buffer and gently agitated on a vortex mixer at room temperature for 30 min to elute proteins and briefly rinsed in 25 μl IP buffer to recover residual proteins; both were combined for further processing (below). The KFF is operated in a cold room to maintain a 4 °C temperature during immunoprecipitation. Automated protocol steps were performed using the slow mix speed and the following mix times: 30 s for equilibration/wash steps, 2 h for binding, and 1 min for final bead release. Three 10 s bead collection times were used before bead transfer to the next plate.
Using a 96-well PCR plate, samples were incubated at 37 °C in a thermal cycler or at room temperature in a MixMate incubator with shaking at 1100 rpm for the following steps: denaturation and reduction at 37 °C for 30 min with 2 M urea and 1 mM DTT in 50 mM Tris–HCl pH 8.0, alkylation at room temperature for 45 min with 3 mM iodoacetamide, and quenching for 10 min with 3 mM DTT. Trypsin (0.5 μg/μl; Promega) was added twice (1.0 μl and 0.5 μl) and further incubated at 37 °C for 4 h and then another 2 h. Peptides were acidified with TFA (0.5% final, pH < 2.0) and desalted at room temperature using a BioPureSPE Mini 96-well plate (20 mg PROTO 300 C18; The Nest Group). Briefly, desalting columns were sequentially equilibrated with 100% methanol, 80% ACN, 0.1% TFA and 2% ACN, 0.1% TFA before passing acidified samples through columns twice and subsequently washed with 2% ACN, 0.1% TFA, and 0.1% FA. 0.2 to 0.3 ml of buffer was added at each step. Peptides were eluted twice with 50% ACN, 0.1% FA (0.12 ml total, divided) and dried under vacuum centrifugation (CentriVap Concentrator, Labconco). The desalting plate was centrifuged at 2000 rpm for 2 to 3 min. All buffers were prepared with HPLC or LC-MS grade reagents.
Immunofluorescence
Neuro-2a cells were cultured on coverslips. After one PBS wash, cells were fixed in 3.7% paraformaldehyde for 10 min. Then, cells were permeabilized with 0.5% TritonX-100 for 5 min, followed by the addition of 100 nM Phalloidin to the cells. Phalloidin staining protocol was followed according to manufacturer’s instructions (Cat. #PHDH1, Cytoskeleton Inc). For VASP staining, cells were blocked in 5% normal goat serum (NGS) for 15 min at room temperature. VASP antibody (13472-1-AP, Proteintech) was prepared in 5% NGS (diluted in 1:200) and incubated overnight at 4 °C. The next day, cells were washed with PBS and incubated with the secondary antibody (diluted in 1:500, 5% NGS) at room temperature for 45 min. The coverslips were mounted on glass slides using DAPI Fluoromount-G (Cat. #0100-20, Southernbiotech) and imaged using a Visitech iSIM superresolution module and Hamamatsu Quest camera on a Nikon Eclipse Ti-E with a 100x 1.45NA objective. Images were deconvolved using Microvolution, and neurite length was calculated using NeuronJ plugin in Fiji, as previously described (41). NeuronJ traces the neurite by finding two points, one at the start and one at the end and calculating the optimal intensity path. Neurite lengths were quantified using Prism graphpad with three independent biological replicates.
Conditioned Media Experiment
ApoE3 and apoE4 Neuro-2a cells were seeded in a T75 flask and after 72 h, media from either the apoE3 or apoE4 cells were collected from the T75 flask and briefly centrifuged to remove debris. Next, Neuro2a-apoE3 and Neuro2a-apoE4 cells were seeded into 6-well plates with a 50% density and to the cells was added either normal media or conditioned media (from apoE3 or apoE4 cells). Cells were incubated with the respective media for 3 days, then the cells were lysed, protein concentration was measured with BCA assay, and p-VASP levels were measured by immunoblotting.
Primary Neuron Lysates
Primary neuron cultures were prepared from embryonic day 18 to 20 or postnatal day 0 (P0) pups of three genotypes: (1) NSE promoter-driven APOE4-expressing (NSE-APOE4) transgenic mice, (2) NSE promoter-driven APOE3-expressing (NSE-APOE3) transgenic mice, and (3) WT C57BL/6 mice. Forty-eight well culture plates (Corning 3548) were coated with Poly-L-lysine (Sigma P4707) overnight and washed four times with water. The cortex and hippocampus were isolated, and the dissociated cells were plated at a density of 315,000 cells/cm2 (300,000 cells/well) in neurobasal medium (Gibco 21103-049) supplemented with B27 (Gibco 17504-044), 100 U/ml penicillin G, 100 μg/ml streptomycin (Gibco 15140-122), and 1% Glumax (Gibco 35050-061). Primary neurons were cultured in vitro for 7 days before treatment with 0.1% dimethyl sulfoxide three times during the following week. After 14 days, primary neurons were collected by adding 80 μl/well of low detergent buffer [50 mM Tris, pH 7.5, 150 mM NaCl, 1% NP40, 0.5% sodium deoxycholate, 0.1% SDS, one pellet of protease inhibitor (Roche 11836145001) per 25 ml], scratched off the plate, and homogenized with a P200 pipette tip. Lysates were transferred to labeled Eppendorf tubes and spun at 14,000 rpm for 5 min at room temperature. The neuron lysates were collected, stored at −20 °C, and analyzed in Western blot assays. Due to limited protein amounts, lysates from different cultures were randomly pooled to provide sufficient protein amounts. This resulted in a single lysate for apoE3 neurons and two lysates for apoE4 neurons.
Statistical Analysis
For nonproteomics data, a two-tailed t test was performed when two groups were compared, and one-way ANOVA or two-way ANOVA with repeated measures was performed when multiple groups were compared, according to different conditions. Where ANOVA tests revealed significant differences among means, a post hoc Sidak test was performed to identify significant differences. Statistical analyses were performed with Prism software (GraphPad Software, Inc). Differences were considered significant at ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, and ∗∗∗∗p < 0.0001. Analysis of filopodia and neurite length was performed with SuperPlots with data visualized in GraphPad Prism (42).
Results
PTM Networks are Highly Regulated by apoE4
Our previous work has shown that apoE allele status can significantly regulate the proteome, resulting in mitochondrial dysfunction and altered cellular metabolism (24). To further define the proteomic impact of apoE4, we analyzed differences for thousands of protein phosphorylation and ubiquitylation sites between isogenic mouse Neuro-2a cells expressing human apoE3 or apoE4 (Fig. 1 A-C and supplemental Table S1). These neuron-like cells have been extensively characterized and shown to recapitulate numerous phenotypes observed in AD patients, including increased amyloid β (10), tau hyperphosphorylation and NFTs (43), and mitochondrial dysfunction (24). Additionally, we found that apoE4 expression in Neuro-2a cells resulted in depletion of proteins normally expressed in the hippocampus (FDR = 4.5 × 10−3), a brain region that incurs neuronal loss in AD patients, further supporting the relevance of these cells as an AD model (supplemental Table S2).
Fig. 1.
Experimental overview. In all panels, differential regulation is defined as proteins or PTMs with adj.p < 0.05. A, ApoE3 and apoE4 Neuro-2a cells were subjected to proteomic analysis in biological triplicate, measuring protein abundance (data reanalyzed from our previous report (24)), phosphorylated peptides, and ubiquitylated peptides. B, total number of unique quantifications for each analysis method. C, fraction of differentially regulated qualifications for each analysis method. When a peptide or protein was only detected in 1 cell type, it was designated as E3-only or E4-only. D, volcano plots for quantified protein, phosphorylated peptides, and ubiquitylated peptides. Colored data points indicate analytes with adj.p < 0.05 and a log2 fold change >|1|. A selection of significantly regulated analytes are annotated with their gene name and the modified amino acid position. E, overlap of proteins or PTM-containing proteins from all quantified proteins/PTMs (top) or only from those proteins/PTMs that were differentially regulated (adj.p < 0.05) (bottom). F, structure of the Nfy trimeric protein complex bound to DNA (PDB: 4AWL). Nfyb is shown in light blue, with the position of K77 highlighted in dark blue. Nfya is shown in purple, and Nfyc is shown in gray. G, Gene Ontology enrichment analysis. Enrichment analysis was performed on proteins or PTMs with adj.p < 0.05. apo, apolipoprotein; PTM, posttranslational modification.
While there was a high degree of overlap in the proteins detected across data types, we observed apoE allele regulation to impact highly unique subsets of the proteome across the multiple modes of regulation (Fig. 1, D and E), as has been observed in the analysis of AD patient brain tissue (44). From the ubiquitylation data, we found several components of the proteasome to have lower ubiquitylation levels in apoE4 cells (e.g. Psmg2, Psma1, and Psma2). The most significantly regulated ubiquitylation detected was K77 on the nuclear transcription factor Y subunit beta (Nfyb) protein. Nfyb functions as part of a trimeric protein complex, along with subunits alpha and gamma, to bind to CCAAT motifs in the promoter regions of DNA. Interestingly, our previous publication reporting only the protein abundance data found that Nfy was predicted to regulate the differentially expressed proteins observed between apoE3 and apoE4 Neuro-2a cells (24). We evaluated the positioning of this ubiquitylation site within the structure of the trimeric Nfy protein complex (PDB: 4AWL) (45) and found that the Nfyb K77 side chain fits within the DNA minor groove, such that ubiquitylation of this residue would likely inhibit binding of this complex to DNA and consequently its Nfy transcriptional activity (Fig. 1F).
GO analysis of the datasets revealed a modest set of enriched terms, with cytoskeletal protein binding being among the most highly enriched terms among both protein abundance and phosphorylation sites that were found to be lower in apoE4 cells (Fig. 1G and supplemental Table S2). Indeed, in the case of phosphorylation sites, many of the most significantly regulated sites in our dataset, both higher and lower in apoE4 cells, are associated with cytoskeletal regulation (e.g., Map1b, Mapt, Flna, Flnc, Myo18a, and Vasp).
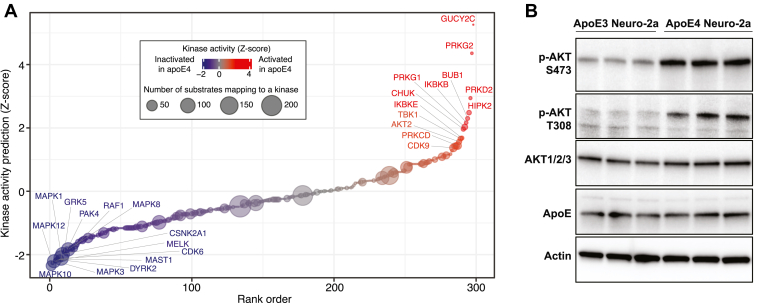
To better enable biological interpretation of regulated phosphorylation sites, they can be mapped to known upstream kinases. Here, we mapped our detected phosphorylation sites from mouse to their orthologous human phosphorylation sites, mapped these sites to kinases, and then predicted kinase activities based on the differential regulation of these phosphorylation sites (Fig. 2A and supplemental Table S2) (46). Evaluation of kinase activities from our dataset predicted strong inactivation of many components of the MAPK pathway in apoE4 cells, with activation of many kinases (GUCY2C, PRKD2, PRKG2, PRKG1) being driven by the strong increase in VASP S235 phosphorylation. To test the accuracy of these predictions, we selected one kinase, AKT2, for which we found nine substrate phosphorylation sites that predict an increase in AKT2 kinase activity in apoE4 Neuro-2a cells. In support of these predictions, we tested apoE3 and apoE4 lysates with antibodies that recognize different AKT phosphorylation sites and found that AKT phosphorylation was highly increased in apoE4 cells at two sites known to regulate its kinase activity (T308/T309/T305 and S473/S474/S472 on AKT1/2/3, respectively, Fig. 2B).
Fig. 2.
Analysis of kinase activities.A, kinase activity prediction analysis. Dot size is proportional to the number of substrates detected in the phosphoproteomics experiments, and dot color represents the predicted level of inactivation or activation of each kinase. B, Western blot measuring AKT phosphorylation at S473/S474/S472 and T308/T309/T305 on AKT1/2/3, AKT1/2/3 total protein, and apoE protein levels in Neuro-2a apoE3 or apoE4 cell lysates. Actin serves as a loading control. apo, apolipoprotein.
The Cytoskeletal Protein VASP is Phosphorylated at S235 in an apoE4-Dependent Manner
We hypothesized that the enrichment of cytoskeletal proteins in both protein abundance and phosphorylation analyses may provide mechanistic insights into the established cytoskeletal defects previously observed in apoE4 cells. Intriguingly, the most significantly regulated phosphorylation site in our data was VASP S235, which was 14-fold higher in apoE4 cells (Fig. 1D) and has previously been detected in apoE4-carrier AD patient brains (47).
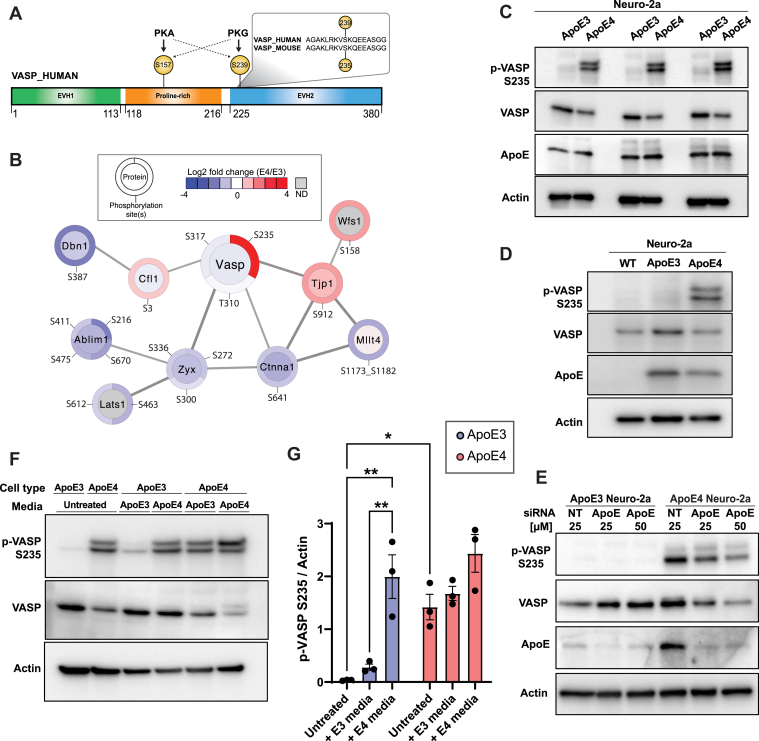
VASP is a member of the conserved ENA/VASP family of proteins regulating diverse cellular processes such as cell adhesion, neurite initiation, axon outgrowth, and actin cytoskeletal dynamics (48). VASP is composed of three conserved domains: an N-terminal Ena/VASP homology-1 (EVH-1) domain, a central poly-proline rich domain, and a C-terminal EVH-2 domain (49, 50) (Fig. 3A). While EVH-1 interacts with proteins such as zyxin and vinculin, which are important for cytoskeletal regulation, the protein rich domain mediates the interaction with profilin–actin complexes to enhance actin polymerization (50, 51). Beyond VASP itself, we found that many proteins related to VASP biology also had differentially regulated phosphorylation sites in apoE4-expressing cells, including zyxin (Zyx), drebrin (Dbn1), actin-binding LIM1 protein (Ablim1), and Cofilin-1 (Cfn1) proteins (Fig. 3B). Many of these had previously been linked to AD pathogenesis or nervous system development, including Dbn1 (52), Cfn1 (53), and Zyx (54, 55, 56, 57). In line with this, our global proteomic analysis showed the abundance levels of these four proteins to be reduced in Neuro-2a apoE4 cells (Fig. 3B).
Fig. 3.
VASP is phosphorylated at S235 in an apoE4-dependent manner.A, VASP protein domain organization: EVH1: Ens/VASP homology 1, proline rich region; EVH2: Ena/VASP homology 2. VASP proteins are the substrates of PKA or PKG. VASP S235 (mouse) and VASP S239 (human) are conserved across species. B, quantification of selected phosphorylation sites and protein abundance differences for proteins known to interact with VASP based on the STRING database. Protein abundance quantification is displayed in the interior circle. Phosphorylation site amino acid position and quantification annotations are displayed on the border. C, phosphorylation of VASP at S235 in apoE4 Neuro-2a cells validation by immunoblotting (n = 3 biological replicates), with actin as a loading control. D, VASP phosphorylation at S235 in Neuro-2a cells in the absence of apoE (WT) or different apoE alleles. Actin serves as a loading control. E, Western blot measuring VASP S235 phosphorylation, VASP protein, and apoE protein upon knockdown of apoE with the increasing concentrations of siRNA in apoE3 Neuro-2a and apoE4 Neuro-2a cells. NT indicates a nontargeting siRNA as a negative control. Actin serves as a loading control. F, Western blot of VASP S235 phosphorylation and VASP total protein in Neuro-2a apoE3 or apoE4 cells cultured with normal media (untreated) or with conditioned media from Neuro-2a apoE3 or apoE4 cells. Actin serves as a loading control. G, quantification of VASP S235 phosphorylation, relative to actin, for biological replicates of the experiments illustrated in panel F. Data are mean ± s.d. from n = 3 biological replicates per group ∗p < 0.05; ∗∗p < 0.01. apo, apolipoprotein; EVH, Ena/VASP homology; ND, not detected; VASP, vasodilator-stimulated phosphoprotein.
We validated the increase in VASP S235 phosphorylation in apoE4 cells by immunoblotting and found that it occurred without a concomitant increase in VASP protein abundance (Fig. 3C). This increase in phosphorylation was unique to apoE4-expressing cells, as VASP S235 phosphorylation was not detectable in WT Neuro-2a cells that lack exogenous human apoE expression (Fig. 3D). Furthermore, to evaluate if this phosphorylation site is present in apoE4 cells beyond Neuro-2a, we measured VASP S235 phosphorylation from primary neurons derived from either WT mice or knock-in mice with neuron-specific expression of either human apoE3 or apoE4. Here, we observed this phosphorylation site in two pools of apoE4 primary neuron lysates, as well as to a lower extent in WT mouse lysates (supplemental Fig. S1).
To further confirm the role of apoE4 in promoting VASP S235 phosphorylation, we performed siRNA knockdown of apoE and observed up to a 50% reduction in this phosphorylation site in apoE4 cells (Fig. 3E and supplemental Fig. S2). Lastly, while apoE can escape the secretory pathway to directly modulate intracellular biology, it is predominantly a secreted protein. Thus, we evaluated if conditioned media, which contains both apoE and a variety of other secreted factors, can regulate VASP S235 phosphorylation. ApoE3 or apoE4 Neuro-2a cells were cultured for 3 days using either normal media or conditioned media from either apo3 or apoE4 Neuro-2a cells. We found that conditioned media from apoE4 Neuro-2a cells significantly increased VASP S235 phosphorylation levels in apoE3 cells (Fig. 3, F and G). These results suggest that a secreted factor from apoE4 cells (potentially apoE itself) is responsible for initiating the cellular signaling cascade that induces VASP S235 phosphorylation.
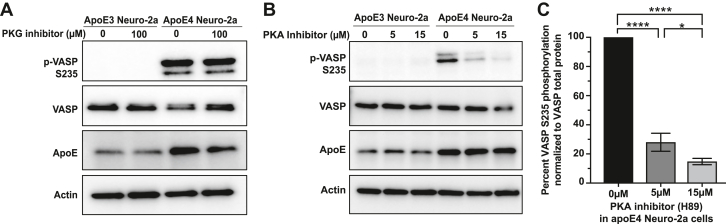
VASP S235 Phosphorylation is Mediated by PKA
Phosphorylation of VASP at serine 235 (mouse), or the orthologous serine 239 (human), is known to impair F-actin accumulation, cell adhesion, and filopodia formation (58) (Fig. 3A). The serine threonine kinases protein kinase G (PKG) and protein kinase A (PKA) can phosphorylate VASP at S235. PKG is the preferred kinase for this site (59) and consequently was predicted to be activated in apoE4 cells (Fig. 2A), while PKA has been shown to preferentially phosphorylate VASP S157 (Fig. 3A) (48). In order to understand which protein kinase is responsible for VASP S235 phosphorylation in apoE4 Neuro-2a cells, we first treated cells with two different PGK inhibitors RRKAARE (Fig. 4A) or KT5823 (supplemental Fig. S3); however no decrease in phosphorylation was observed. In contrast, treatment of cells with two different PKA inhibitors, H89 (Fig. 4, B and C) or KT5720 (supplemental Fig. S3), both dramatically reduced VASP S235 phosphorylation. These results suggest that PKA is responsible for the majority of VASP S235 phosphorylation and is the preferential kinase for this site in apoE4 Neuro-2a cells.
Fig. 4.
VASP S235 is phosphorylated by PKA. Immunoblots of VASP S235 phosphorylation in apoE3 and apoE4 Neuro-2a cells upon kinase inhibition. A, 24 h inhibition with the PKG inhibitor, RKRARKE. B, 24 h inhibition with increasing concentrations of the PKA inhibitor H89. Actin serves as a loading control. C, quantification of VASP S235 phosphorylation levels from panel B. Data are mean ± s.d. from n = 3 biological replicates per group ∗p < 0.05; ∗∗∗∗p < 0.0001. apo, apolipoprotein; VASP, vasodilator-stimulated phosphoprotein.
VASP Interactions With Actin Cytoskeletal Proteins are Regulated by PKA
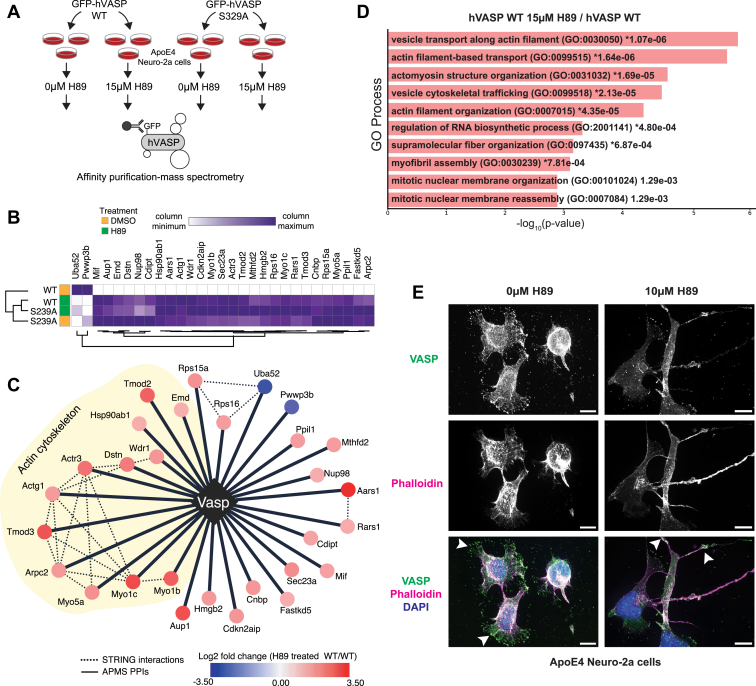
To investigate the role of VASP S235 phosphorylation in apoE4 Neuro-2a cells, we performed APMS to identify how VASP protein-protein interactions (PPIs) are regulated by its phosphorylation status. We expressed WT human VASP, which was highly phosphorylated at the orthologous S239 position in apoE4 Neuro-2a cells (supplemental Fig. S4), similar to what we observed for the endogenous mouse VASP S235 phosphorylation. Separately, we expressed human VASP with an S239A mutation (hVASP S239A) which prohibits phosphorylation at this position. APMS experiments were performed in the presence or absence of PKA inhibition by H89 (Fig. 5A).
Fig. 5.
Phosphorylated VASP and phospho-null VASP protein–interacting networks.A, GFP-tagged human VASP (hVASP) WT and hVASP S239A are expressed in apoE4 Neuro-2a cells in triplicate in the absence or presence of 15 μM H89. GFP is used as a tag to pull down VASP protein–interacting partners which were identified using MS. B, heat map represents scaled protein abundance values for a selection of interacting proteins specifically regulated by hVASP S239 phosphorylation. C, comparison of hVASP WT with H89-treated hVASP WT protein interaction network for proteins displayed from panel B. (Red, increase; blue, decrease). D, bar plot of Gene Ontology analysis of proteins differentially purified from hVASP expressing apoE4 Neuro-2a cells in the absence or presence of H89. Proteins in Gene ontology analysis using EnrichR were determined by a p < 0.05 and a log2 fold change >1 and sorted with p-value ranking. E, localization of VASP in apoE4 Neuro-2a cells in the absence or presence of 10 μM H89 for 24 h. Phalloidin staining is for F-actin. DAPI: nuclear staining. The scale bar is 10 μm. apo, apolipoprotein; hVASP, human VASP; MS, mass spectrometry; VASP, vasodilator-stimulated phosphoprotein.
To prioritize PPIs dependent on hVASP S239 phosphorylation status, we filtered the APMS data for proteins with differential binding (>2-fold) between WT hVASP and S239A hVASP, as well as differential binding upon dephosphorylation of WT S239 by H89 treatment. Additionally, to control for H89 effects on hVASP PPIs that may not be related to its S239 phosphorylation status, we further filtered PPIs to those that were relatively unchanged (<2-fold) in S239A mutant cells upon H89 treatment. These criteria resulted in a set of 29 proteins whose interactions were consistent across all conditions except when hVASP was in a highly phosphorylated state at position S239 (i.e., WT hVASP) (Fig. 5B and supplemental Table S3). Blocking hVASP S239 phosphorylation increased PPIs with components of the actin cytoskeleton (Fig. 5C). For instance, hVASP interactions with microfilament motor activity proteins Myo1b, Myo1c, and Myo5a were increased following PKA inhibition in apoE4 Neuro-2a cells. Myo1b is involved in neurite outgrowth, cell migration, axon formation, and growth cone dynamics (60). Actin nucleating and branching protein Arp2/3 and its component Actr3, as well as actin protein Actg1, also preferentially interacted with hVASP upon S239 dephosphorylation by H89 (Fig. 5C). In line with this, GO analysis of changes in this protein interaction network showed an enrichment of pathways associated with actin filament transport, trafficking, and reorganization (Fig. 5D).
We next asked whether the observed changes in PPIs were influenced by VASP subcellular localization in apoE4 Neuro-2a cells, comparing VASP and F-actin subcellular localization patterns in apoE4 Neuro-2a cells in the absence or presence of H89 (Fig. 5E). When VASP S235 was phosphorylated, it was predominantly localized at the leading edge and in the cytosol (Fig. 5E). However, upon dephosphorylation of S235 by H89 treatment, VASP became less cytosolic and was enriched as clusters in F-actin–containing neurites (Fig. 5E). Taken together with APMS data, these findings suggest VASP phosphorylation at S235 by PKA alters actin cytoskeleton protein interactions and subsequently VASP subcellular localization in apoE4 Neuro-2a cells.
Inhibition of PKA Promotes Filopodia Formation and Neurite Outgrowth in apoE4 Cells
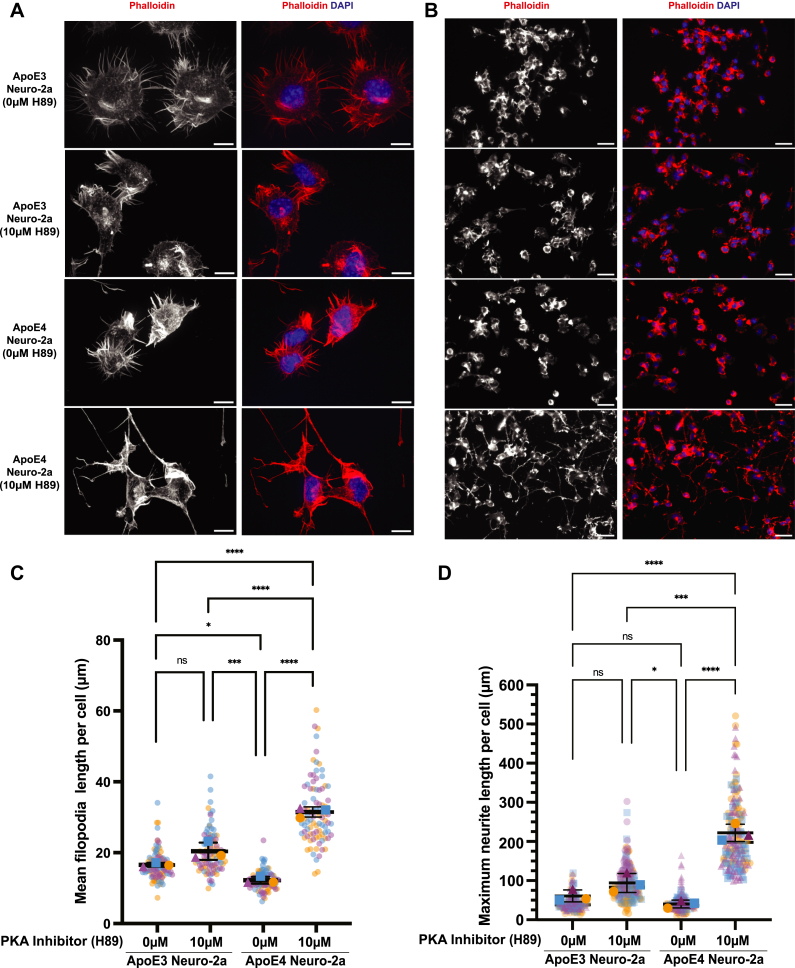
Previous studies have shown that apoE4 expression inhibits neurite outgrowth (28, 29), a process which is initiated by robust filopodia formation (61). The observation of long F-actin neurite structures upon PKA inhibition in apoE4 Neuro-2a cells led us to hypothesize that VASP S235 phosphorylation could be an apoE4 allele-specific mechanism regulating neurite outgrowth and synaptic remodeling through actin cytoskeleton-dependent processes. To test this, we used H89 to inhibit VASP S235 phosphorylation in both apoE3 and apoE4 Neuro-2a cells and quantified the impact on filopodia formation and neurite outgrowth. As expected, under normal conditions, apoE4 cells had moderately shorter filopodia than apoE3 cells. However, PKA inhibition resulted in a dramatic increase in the length of filopodia in apoE4 cells, some of which appeared to form neurites (Fig. 6, A and C). Next, we measured neurite outgrowth under these same conditions and observed that indeed, neurite outgrowth was significantly increased in H89-treated apoE4 cells compared with control apoE4 cells (Fig. 6, B and D). In contrast, H89 treatment cells did not significantly increase neurite outgrowth in apoE3 cells. Taken together, these results show that in apoE4 cells, depletion of VASP S235 phosphorylation by inhibition of PKA leads to extended filopodia length and subsequently neurite outgrowth.
Fig. 6.
PKA inhibition increases filopodia and neurite length.A and B, phalloidin staining of apoE3 or apoE4 expressing Neuro-2a cells in the absence or presence of 10 μM PKA inhibitor (H89), DAPI: nuclear staining. Images in panel B are lower magnification to show more cells; scale bar is 10 μm (A) and 200 μm (B). C and D, measurement of mean of filopodia and maximum neurite length per cell using NeuronJ (41). Data represents n = 3 independent samples (biological replicates). Each small data point (purple, blue, yellow) represents the length per cell, color-coded by the sample; error bars are ±s.d. of the biological replicates. The triangle represents the average of purple data points, the square represents the average of blue data points, the sphere represents the average of yellow data points. p values were calculated using the means of the biological replicates (ns: non-significant, ∗p < 0.05; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001; with one-way ANOVA analysis with Tukey’s multiple comparison test). apo, apolipoprotein.
Discussion
Neurons can undergo dramatic morphological changes, forming abundant neurite extensions, axonal elongations, synaptic spines and synapses, all guided by cytoskeletal elements. Disruptions to these cytoskeletal processes are a common feature of AD neuropathogenesis, leading to abnormal actin-containing inclusions, NFT formation, and synaptic deficits (62). These phenotypes can be specifically regulated by apoE4 expression, which has been documented to result in cytoskeletal impairments via mechanisms that have remained unclear (27, 28, 29, 30, 31). The global phosphoproteomics analysis presented here provides insights into the molecular mechanisms of this process, implicating PKA and the actin-interacting protein VASP.
Our findings demonstrate that apoE4 expression results in differential phosphorylation resulting in rewiring of the phosphorylation signaling architecture. Our observations in Neuro-2a cells are supported by clinical data, in which phosphorylation signaling was found to readily distinguish AD patients from other conditions, including progressive supranuclear palsy, mild cognitive impairment, or individuals with high pathology but low cognitive impact (44). Additionally, several phosphoproteomics studies have demonstrated that PKA activity is upregulated in AD patient brains (44, 47). Human VASP phosphorylation at S239 (the orthologous site to mouse S235) has also been detected in a handful of patients carrying the apoE4 allele (47), however, likely due to the high density of lysine and arginine residues proximal to this phosphorylation site, insufficient detections of this site have been reported in clinical cohorts. Thus, further studies utilizing alternative proteolytic enzymes or detection methods would be required to statistically correlate this phosphorylation site with apoE4 allele status, AD diagnosis, or cytoskeletal dysfunction in patients.
Previous work has established a role for apoE4 in promoting PKA activity. For example, in smooth muscle cells and mouse embryonic fibroblasts, extracellular apoE4 can bind to LRP1, resulting in increased cAMP levels and consequently activation of PKA (63). Internalization of apoE4 through the LRP receptor has been shown to reduce neurite extension and branching in Neuro-2a cells (27). Furthermore, incubation of rat primary hippocampal neurons with extracellular apoE4 results in the stimulation of the cAMP response element-binding protein (64), a downstream PKA target. Our results build upon these observations and indicate that of the many potential downstream PKA targets, VASP S235 phosphorylation is dramatically upregulated in the apoE4-expressing cells. Interestingly, we also found that factors present in apoE4 conditioned media are sufficient to drive VASP S235 phosphorylation; however, it is unclear if this is regulated by secreted apoE4 or other factors present uniquely in the apoE4 media.
We found that PKA inhibition increased the interaction of VASP with proteins such as Myo1b, Myo5a, Tmod2, and Tmod3, which are involved in actin filament organization, vesicle transport along the actin filaments, and actomyosin structure organization. This suggests that apoE4-dependent cytoskeletal dysfunction through VASP phosphorylation at S235 could be reversed by PKA inhibition. There is evidence that in response to an increase in cAMP levels in neurons, PKA subunit C translocates from the cytosol to the plasma membrane to phosphorylate its substrates (65). As VASP is localized at the leading edge, phosphorylation of S235 by PKA may prevent VASP from being involved in filopodia formation and neurite initiation.
Filopodia are composed of VASP clusters, parallel filament actin bundles, and other actin-binding proteins such as lamellipodin (66). Together with microtubules, filopodia are required for neurite initiation and axon extension, which are essential processes for neuronal migration, synapse formation, and axon outgrowth and guidance (66, 67, 68). Accordingly, our abundance and phosphoproteomics data suggest that numerous proteins involved in actin cytoskeleton organization, actin filament-based processes, cell leading edge, postsynaptic actin cytoskeleton organization, and microtubule-based transportation are reduced in apoE4 Neuro-2a cells. In contrast, proteins involved in actin filament fragmentation, actin filament severing, and depolymerization processes are elevated in apoE4 Neuro-2a cells. Surprisingly, in contrast to our findings, previous studies have shown that neurite initiation and length are reduced upon PKA inhibition in zebrafish neurons and rat hippocampal neurons (69, 70), however, these studies did not evaluate the impact of apoE4 on these phenotypes.
In summary, our results suggest a mechanism where increased PKA activity and VASP S235 phosphorylation may limit cytoskeletal dynamics in apoE4 neurons and consequently limit the capacity for such neurons to engage in synaptic remodeling. Importantly, our results also indicate that this phenotype can be reversed upon PKA inhibition. Thus, further elucidation of the role of VASP and PKA in actin cytoskeletal reorganization may provide valuable insights into apoE4-dependent neuropathological mechanisms.
Data Availability
Mass spectrometry data files (raw and search results) have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository with dataset identifier PXD041205 (36).
Supplemental data
This article contains supplemental data.
Conflict of interest
The N. J. K. laboratory has received research support from Vir Biotechnology, F. Hoffmann-La Roche, and Rezo Therapeutics. N. J. K. has financially compensated consulting agreements with the Icahn School of Medicine at Mount Sinai, New York, Maze Therapeutics, Interline Therapeutics, Rezo Therapeutics, GEn1E Lifesciences, Inc, and Twist Bioscience Corp. He is on the Board of Directors of Rezo Therapeutics and is a shareholder in Tenaya Therapeutics, Maze Therapeutics, Rezo Therapeutics, and Interline Therapeutics. D. L. S. has a consulting agreement with Maze Therapeutics and Rezo Therapeutics. R. W. M. is the cofounder and shareholder of Escape Bio, Inc. R. W. M. is the CEO, CSO, and shareholder of the stem cell company GABAeron, Inc. A. d. C. is an employee of GABAeron, Inc.
Acknowledgments
We thank Yadong Huang, Kristen Skruber, Anke Meyer-Franke, and Michael McGregor for helpful discussions. We thank Einar Krogsaeter and Kliment Verba for assistance with figures.
Funding and additional information
This work was supported by an NIH grant R01AG059751 to R. W. M., N. J. K., and D. L. S., an NIH grant R35GM118119 to R. D. M., and funding from the Howard Hughes Medical Institute Investigator Program to R. D. M. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Author contributions
Z. C., N. J. K., R. W. M., and D. L. S. conceptualization; Z. C., S. J. L., G. M. J., B. W. N., A. L. O., J. R. J., and A. d. C. investigation; Z. C., S. J. L., Y. Z., B. J. P., and D. L. S. visualization; Z. C. validation; Z. C., Y. Z., B. J. P., and D. J.-M. formal analysis; Z. C. and D. L. S. writing–original draft; Z. C., M. E., R. W. M., and D. L. S. writing–review and editing; S. J. L., R. D. M., R.W. M., and D. L. S. resources; J. R. J. and D. L. S. methodology; J. R. J. software; J. R. J., N. J. K., R. W. M., and D. L. S. supervision; N. J. K., R. W. M., and D. L. S. funding acquisition; R. W. M. and D. L. S. project administration.
Footnotes
Present addresses for: David Jimenez-Morales, Department of Medicine, Division of Cardiovascular Medicine, Stanford University, Stanford, California, USA; Adam L. Orr, Helen and Robert Appel Alzheimer’s Disease Research Institute, Feil Family Brain and Mind Research Institute, Weill Cornell Medicine, New York, New York, USA; Jeffrey R. Johnson, Department of Microbiology, Icahn School of Medicine at Mount Sinai, New York, New York, USA.
Supplementary Data
Immunoblotting of VASP 235 phosphorylation, VASP total protein, apoE, and actin in mouse primary neurons expressing apoE3 or apoE4. Actin serves as the loading control. Due to limited protein amounts lysates from different cultures were randomly pooled to provide sufficient protein amounts. This resulted in a single lysate for apoE3 neurons, and two lysates for apoE4 neurons. The right panel is a shorter exposure of the blot.
Quantification of VASP S235 phosphorylation levels upon apoE siRNA knockdown. Values are normalized to VASP protein levels, (n = 2 biological replicates), ns:non-significant, ∗p < 0.05.
VASP S235 phosphorylation levels in apoE3 Neuro-2a and apoE4 Neuro-2a cells are determined by immunoblotting following treatment with PKA (KT5720) (1 μM and 3 μM) or PKG (KT5823) inhibitors. Cells were treated either with 1 μM and 3 μM of KT5720 or 0.2 μM and 2 μM of KT5823 for 24 h.
GFP and mcherry-tagged hVASP constructs were expressed in apoE3 and apoE4 Neuro-2a cells for 24 h. Endogenous VASP was knocked down by VASPsiRNA. hVASP phosphorylation at S239 was validated by immunoblotting. Endogenous VASP is shown with arrows and hVASP is shown as tagged in brackets. Actin serves a loading control.
References
- 1.Mahley R.W. Apolipoprotein E: cholesterol transport protein with expanding role in cell biology. Science. 1988;240:622–630. doi: 10.1126/science.3283935. [DOI] [PubMed] [Google Scholar]
- 2.Mahley R.W., Weisgraber K.H., Huang Y. Apolipoprotein E4: a causative factor and therapeutic target in neuropathology, including Alzheimer’s disease. Proc. Natl. Acad. Sci. U. S. A. 2006;103:5644–5651. doi: 10.1073/pnas.0600549103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mahley R.W., Weisgraber K.H., Huang Y. Apolipoprotein E: structure determines function, from atherosclerosis to Alzheimer’s disease to AIDS. J. Lipid Res. 2009;50 Suppl:S183–S188. doi: 10.1194/jlr.R800069-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Corder E.H., Saunders A.M., Strittmatter W.J., Schmechel D.E., Gaskell P.C., Small G.W., et al. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer’s disease in late onset families. Science. 1993;261:921–923. doi: 10.1126/science.8346443. [DOI] [PubMed] [Google Scholar]
- 5.Farrer L.A., Adrienne Cupples L., Haines J.L., Hyman B., Kukull W.A., Mayeux R., et al. Effects of age, sex, and ethnicity on the association between apolipoprotein E genotype and Alzheimer disease: a meta-analysis. JAMA. 1997;278:1349–1356. [PubMed] [Google Scholar]
- 6.Genin E., Hannequin D., Wallon D., Sleegers K., Hiltunen M., Combarros O., et al. APOE and Alzheimer disease: a major gene with semi-dominant inheritance. Mol. Psychiatry. 2011;16:903–907. doi: 10.1038/mp.2011.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Biundo F., Ishiwari K., Del Prete D., D’Adamio L. Interaction of ApoE3 and ApoE4 isoforms with an ITM2b/BRI2 mutation linked to the Alzheimer disease-like danish dementia: effects on learning and memory. Neurobiol. Learn. Mem. 2015;126:18–30. doi: 10.1016/j.nlm.2015.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bu G. Apolipoprotein E and its receptors in Alzheimer’s disease: pathways, pathogenesis and therapy. Nat. Rev. Neurosci. 2009;10:333–344. doi: 10.1038/nrn2620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Holtzman D.M., Herz J., Bu G. Apolipoprotein E and apolipoprotein E receptors: normal biology and roles in Alzheimer disease. Cold Spring Harb. Perspect. Med. 2012;2 doi: 10.1101/cshperspect.a006312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang Y., Mucke L. Alzheimer mechanisms and therapeutic strategies. Cell. 2012;148:1204–1222. doi: 10.1016/j.cell.2012.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim J., Basak J.M., Holtzman D.M. The role of apolipoprotein E in Alzheimer’s disease. Neuron. 2009;63:287–303. doi: 10.1016/j.neuron.2009.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu C.-C., Liu C.-C., Kanekiyo T., Xu H., Bu G. Apolipoprotein E and Alzheimer disease: risk, mechanisms and therapy. Nat. Rev. Neurol. 2013;9:106–118. doi: 10.1038/nrneurol.2012.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mahley R.W., Huang Y. Apolipoprotein e sets the stage: response to injury triggers neuropathology. Neuron. 2012;76:871–885. doi: 10.1016/j.neuron.2012.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mahley R.W., Huang Y. Small-molecule structure correctors target abnormal protein structure and function: structure corrector rescue of apolipoprotein E4-associated neuropathology. J. Med. Chem. 2012;55:8997–9008. doi: 10.1021/jm3008618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Spinney L. Alzheimer’s disease: the forgetting gene. Nature. 2014;510:26–28. doi: 10.1038/510026a. [DOI] [PubMed] [Google Scholar]
- 16.Koutsodendris N., Nelson M.R., Rao A., Huang Y. Apolipoprotein E and Alzheimer’s disease: findings, hypotheses, and potential mechanisms. Annu. Rev. Pathol. 2022;17:73–99. doi: 10.1146/annurev-pathmechdis-030421-112756. [DOI] [PubMed] [Google Scholar]
- 17.Martens Y.A., Zhao N., Liu C.-C., Kanekiyo T., Yang A.J., Goate A.M., et al. ApoE Cascade hypothesis in the pathogenesis of Alzheimer’s disease and related dementias. Neuron. 2022;110:1304–1317. doi: 10.1016/j.neuron.2022.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Knoferle J., Yoon S.Y., Walker D., Leung L., Gillespie A.K., Tong L.M., et al. Apolipoprotein E4 produced in GABAergic interneurons causes learning and memory deficits in mice. J. Neurosci. 2014;34:14069–14078. doi: 10.1523/JNEUROSCI.2281-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu Q., Walker D., Bernardo A., Brodbeck J., Balestra M.E., Huang Y. Intron-3 retention/splicing controls neuronal expression of apolipoprotein E in the CNS. J. Neurosci. 2008;28:1452–1459. doi: 10.1523/JNEUROSCI.3253-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tong L.M., Djukic B., Arnold C., Gillespie A.K., Yoon S.Y., Wang M.M., et al. Inhibitory interneuron progenitor transplantation restores normal learning and memory in ApoE4 knock-in mice without or with Aβ accumulation. J. Neurosci. 2014;34:9506–9515. doi: 10.1523/JNEUROSCI.0693-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang C., Najm R., Xu Q., Jeong D.-E., Walker D., Balestra M.E., et al. Gain of toxic apolipoprotein E4 effects in human iPSC-derived neurons is ameliorated by a small-molecule structure corrector. Nat. Med. 2018;24:647–657. doi: 10.1038/s41591-018-0004-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Parihar M.S., Brewer G.J. Mitoenergetic failure in Alzheimer disease. Am. J. Physiol. Cell Physiol. 2007;292:C8–23. doi: 10.1152/ajpcell.00232.2006. [DOI] [PubMed] [Google Scholar]
- 23.Atamna H., Frey W.H., 2nd Mechanisms of mitochondrial dysfunction and energy deficiency in Alzheimer’s disease. Mitochondrion. 2007;7:297–310. doi: 10.1016/j.mito.2007.06.001. [DOI] [PubMed] [Google Scholar]
- 24.Orr A.L., Kim C., Jimenez-Morales D., Newton B.W., Johnson J.R., Krogan N.J., et al. Neuronal apolipoprotein E4 expression results in proteome-wide alterations and compromises bioenergetic capacity by disrupting mitochondrial function. J. Alzheimers Dis. 2019;68:991–1011. doi: 10.3233/JAD-181184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harris F.M., Brecht W.J., Xu Q., Mahley R.W., Huang Y. Increased tau phosphorylation in apolipoprotein E4 transgenic mice is associated with activation of extracellular signal-regulated kinase: modulation by zinc. J. Biol. Chem. 2004;279:44795–44801. doi: 10.1074/jbc.M408127200. [DOI] [PubMed] [Google Scholar]
- 26.Harris F.M., Brecht W.J., Xu Q., Tesseur I., Kekonius L., Wyss-Coray T., et al. Carboxyl-terminal-truncated apolipoprotein E4 causes Alzheimer’s disease-like neurodegeneration and behavioral deficits in transgenic mice. Proc. Natl. Acad. Sci. U. S. A. 2003;100:10966–10971. doi: 10.1073/pnas.1434398100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bellosta S., Nathan B.P., Orth M., Dong L.M., Mahley R.W., Pitas R.E. Stable expression and secretion of apolipoproteins E3 and E4 in mouse neuroblastoma cells produces differential effects on neurite outgrowth. J. Biol. Chem. 1995;270:27063–27071. doi: 10.1074/jbc.270.45.27063. [DOI] [PubMed] [Google Scholar]
- 28.Nathan B.P., Bellosta S., Sanan D.A., Weisgraber K.H., Mahley R.W., Pitas R.E. Differential effects of apolipoproteins E3 and E4 on neuronal growth in vitro. Science. 1994;264:850–852. doi: 10.1126/science.8171342. [DOI] [PubMed] [Google Scholar]
- 29.Nathan B.P., Chang K.C., Bellosta S., Brisch E., Ge N., Mahley R.W., et al. The inhibitory effect of apolipoprotein E4 on neurite outgrowth is associated with microtubule depolymerization. J. Biol. Chem. 1995;270:19791–19799. doi: 10.1074/jbc.270.34.19791. [DOI] [PubMed] [Google Scholar]
- 30.Brodbeck J., McGuire J., Liu Z., Meyer-Franke A., Balestra M.E., Jeong D.-E., et al. Structure-dependent impairment of intracellular apolipoprotein E4 trafficking and its detrimental effects are rescued by small-molecule structure correctors. J. Biol. Chem. 2011;286:17217–17226. doi: 10.1074/jbc.M110.217380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen H.-K., Liu Z., Meyer-Franke A., Brodbeck J., Miranda R.D., McGuire J.G., et al. Small molecule structure correctors abolish detrimental effects of apolipoprotein E4 in cultured neurons. J. Biol. Chem. 2012;287:5253–5266. doi: 10.1074/jbc.M111.276162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.James R., Searcy J.L., Le Bihan T., Martin S.F., Gliddon C.M., Povey J., et al. Proteomic analysis of mitochondria in APOE transgenic mice and in response to an ischemic challenge. J. Cereb. Blood Flow Metab. 2012;32:164–176. doi: 10.1038/jcbfm.2011.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shi L., Du X., Zhou H., Tao C., Liu Y., Meng F., et al. Cumulative effects of the ApoE genotype and gender on the synaptic proteome and oxidative stress in the mouse brain. Int. J. Neuropsychopharmacol. 2014;17:1863–1879. doi: 10.1017/S1461145714000601. [DOI] [PubMed] [Google Scholar]
- 34.Choi M., Chang C.-Y., Clough T., Broudy D., Killeen T., MacLean B., et al. MSstats: an R package for statistical analysis of quantitative mass spectrometry-based proteomic experiments. Bioinformatics. 2014;30:2524–2526. doi: 10.1093/bioinformatics/btu305. [DOI] [PubMed] [Google Scholar]
- 35.Cox J., Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 36.Vizcaíno J.A., Csordas A., Del-Toro N., Dianes J.A., Griss J., Lavidas I., et al. 2016 Update of the PRIDE database and its related tools. Nucleic Acids Res. 2016;44 doi: 10.1093/nar/gkw880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yu G., Wang L.-G., Han Y., He Q.-Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Szklarczyk D., Gable A.L., Nastou K.C., Lyon D., Kirsch R., Pyysalo S., et al. The STRING database in 2021: customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021;49:D605–D612. doi: 10.1093/nar/gkaa1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Türei D., Korcsmáros T., Saez-Rodriguez J. OmniPath: guidelines and gateway for literature-curated signaling pathway resources. Nat. Methods. 2016;13:966–967. doi: 10.1038/nmeth.4077. [DOI] [PubMed] [Google Scholar]
- 40.Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Meijering E., Jacob M., Sarria J.-C.F., Steiner P., Hirling H., Unser M. Design and validation of a tool for neurite tracing and analysis in fluorescence microscopy images. Cytometry A. 2004;58:167–176. doi: 10.1002/cyto.a.20022. [DOI] [PubMed] [Google Scholar]
- 42.Lord S.J., Velle K.B., Mullins R.D., Fritz-Laylin L.K. SuperPlots: communicating reproducibility and variability in cell biology. J. Cell Biol. 2020;219 doi: 10.1083/jcb.202001064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huang Y., Liu X.Q., Wyss-Coray T., Brecht W.J., Sanan D.A., Mahley R.W. Apolipoprotein E fragments present in Alzheimer’s disease brains induce neurofibrillary tangle-like intracellular inclusions in neurons. Proc. Natl. Acad. Sci. U. S. A. 2001;98:8838–8843. doi: 10.1073/pnas.151254698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bai B., Wang X., Li Y., Chen P.-C., Yu K., Dey K.K., et al. Deep multilayer brain proteomics identifies molecular networks in Alzheimer’s disease progression. Neuron. 2020;105:975–991.e7. doi: 10.1016/j.neuron.2019.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nardini M., Gnesutta N., Donati G., Gatta R., Forni C., Fossati A., et al. Sequence-specific transcription factor NF-Y displays histone-like DNA binding and H2B-like ubiquitination. Cell. 2013;152:132–143. doi: 10.1016/j.cell.2012.11.047. [DOI] [PubMed] [Google Scholar]
- 46.Ochoa D., Jonikas M., Lawrence R.T., El Debs B., Selkrig J., Typas A., et al. An atlas of human kinase regulation. Mol. Syst. Biol. 2016;12:888. doi: 10.15252/msb.20167295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Morshed N., Lee M., Rodriguez F.H., Lauffenburger D.A., Mastroeni D., White F. Quantitative phosphoproteomics uncovers dysregulated kinase networks in Alzheimer’s disease. Nat. Aging. 2021;1:550–565. doi: 10.1038/s43587-021-00071-1. [DOI] [PubMed] [Google Scholar]
- 48.Döppler H., Storz P. Regulation of VASP by phosphorylation: consequences for cell migration. Cell Adh. Migr. 2013;7:482–486. doi: 10.4161/cam.27351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kwiatkowski A.V., Gertler F.B., Loureiro J.J. Function and regulation of Ena/VASP proteins. Trends Cell Biol. 2003;13:386–392. doi: 10.1016/s0962-8924(03)00130-2. [DOI] [PubMed] [Google Scholar]
- 50.Reinhard M., Jarchau T., Walter U. Actin-based motility: stop and go with Ena/VASP proteins. Trends Biochem. Sci. 2001;26:243–249. doi: 10.1016/s0968-0004(00)01785-0. [DOI] [PubMed] [Google Scholar]
- 51.Ferron F., Rebowski G., Lee S.H., Dominguez R. Structural basis for the recruitment of profilin-actin complexes during filament elongation by Ena/VASP. EMBO J. 2007;26:4597–4606. doi: 10.1038/sj.emboj.7601874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Harigaya Y., Shoji M., Shirao T., Hirai S. Disappearance of actin-binding protein, drebrin, from hippocampal synapses in Alzheimer’s disease. J. Neurosci. Res. 1996;43:87–92. doi: 10.1002/jnr.490430111. [DOI] [PubMed] [Google Scholar]
- 53.Flynn K.C., Pak C.W., Shaw A.E., Bradke F., Bamburg J.R. Growth cone-like waves transport actin and promote axonogenesis and neurite branching. Dev. Neurobiol. 2009;69:761–779. doi: 10.1002/dneu.20734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Luo S., Schaefer A.M., Dour S., Nonet M.L. The conserved LIM domain-containing focal adhesion protein ZYX-1 regulates synapse maintenance in Caenorhabditis elegans. Development. 2014;141:3922–3933. doi: 10.1242/dev.108217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gomez T.M., Roche F.K., Letourneau P.C. Chick sensory neuronal growth cones distinguish fibronectin from laminin by making substratum contacts that resemble focal contacts. J. Neurobiol. 1996;29:18–34. doi: 10.1002/(SICI)1097-4695(199601)29:1<18::AID-NEU2>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 56.Hoffman L.M., Jensen C.C., Kloeker S., Wang C.-L.A., Yoshigi M., Beckerle M.C. Genetic ablation of zyxin causes Mena/VASP mislocalization, increased motility, and deficits in actin remodeling. J. Cell Biol. 2006;172:771–782. doi: 10.1083/jcb.200512115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Legerstee K., Geverts B., Slotman J.A., Houtsmuller A.B. Dynamics and distribution of paxillin, vinculin, zyxin and VASP depend on focal adhesion location and orientation. Sci. Rep. 2019;9 doi: 10.1038/s41598-019-46905-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Benz P.M., Blume C., Seifert S., Wilhelm S., Waschke J., Schuh K., et al. Differential VASP phosphorylation controls remodeling of the actin cytoskeleton. J. Cell Sci. 2009;122:3954–3965. doi: 10.1242/jcs.044537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Butt E., Abel K., Krieger M., Palm D., Hoppe V., Hoppe J., et al. cAMP-and cGMP-dependent protein kinase phosphorylation sites of the focal adhesion vasodilator-stimulated phosphoprotein (VASP) in vitro and in intact human platelets. J. Biol. Chem. 1994;269:14509–14517. [PubMed] [Google Scholar]
- 60.Iuliano O., Yoshimura A., Prospéri M.-T., Martin R., Knölker H.-J., Coudrier E. Myosin 1b promotes axon formation by regulating actin wave propagation and growth cone dynamics. J. Cell Biol. 2018;217:2033–2046. doi: 10.1083/jcb.201703205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dent E.W., Kwiatkowski A.V., Mebane L.M., Philippar U., Barzik M., Rubinson D.A., et al. Filopodia are required for cortical neurite initiation. Nat. Cell Biol. 2007;9:1347–1359. doi: 10.1038/ncb1654. [DOI] [PubMed] [Google Scholar]
- 62.Bamburg J.R., Bloom G.S. Cytoskeletal pathologies of Alzheimer disease. Cell Motil. Cytoskeleton. 2009;66:635–649. doi: 10.1002/cm.20388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhu Y., Hui D.Y. Apolipoprotein E binding to low density lipoprotein receptor-related protein-1 inhibits cell migration via activation of cAMP-dependent protein kinase A. J. Biol. Chem. 2003;278:36257–36263. doi: 10.1074/jbc.M303171200. [DOI] [PubMed] [Google Scholar]
- 64.Ohkubo N., Mitsuda N., Tamatani M., Yamaguchi A., Lee Y.D., Ogihara T., et al. Apolipoprotein E4 stimulates cAMP response element-binding protein transcriptional activity through the extracellular signal-regulated kinase pathway. J. Biol. Chem. 2001;276:3046–3053. doi: 10.1074/jbc.M005070200. [DOI] [PubMed] [Google Scholar]
- 65.Tillo S.E., Xiong W.-H., Takahashi M., Miao S., Andrade A.L., Fortin D.A., et al. Liberated PKA catalytic subunits associate with the membrane via myristoylation to preferentially phosphorylate membrane substrates. Cell Rep. 2017;19:617–629. doi: 10.1016/j.celrep.2017.03.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Drees F., Gertler F.B. Ena/VASP: proteins at the tip of the nervous system. Curr. Opin. Neurobiol. 2008;18:53–59. doi: 10.1016/j.conb.2008.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kwiatkowski A.V., Rubinson D.A., Dent E.W., Edward van Veen J., Leslie J.D., Zhang J., et al. Ena/VASP is required for neuritogenesis in the developing cortex. Neuron. 2007;56:441–455. doi: 10.1016/j.neuron.2007.09.008. [DOI] [PubMed] [Google Scholar]
- 68.Lin Y.-L., Lei Y.-T., Hong C.-J., Hsueh Y.-P. Syndecan-2 induces filopodia and dendritic spine formation via the neurofibromin-PKA-Ena/VASP pathway. J. Cell Biol. 2007;177:829–841. doi: 10.1083/jcb.200608121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kao H.-T., Song H.-J., Porton B., Ming G.-L., Hoh J., Abraham M., et al. A protein kinase A-dependent molecular switch in synapsins regulates neurite outgrowth. Nat. Neurosci. 2002;5:431–437. doi: 10.1038/nn840. [DOI] [PubMed] [Google Scholar]
- 70.Guo B., Qi M., Huang S., Zhuo R., Zhang W., Zhang Y., et al. Cadherin-12 regulates neurite outgrowth through the PKA/Rac1/Cdc42 pathway in cortical neurons. Front. Cell Dev. Biol. 2021;9 doi: 10.3389/fcell.2021.768970. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Immunoblotting of VASP 235 phosphorylation, VASP total protein, apoE, and actin in mouse primary neurons expressing apoE3 or apoE4. Actin serves as the loading control. Due to limited protein amounts lysates from different cultures were randomly pooled to provide sufficient protein amounts. This resulted in a single lysate for apoE3 neurons, and two lysates for apoE4 neurons. The right panel is a shorter exposure of the blot.
Quantification of VASP S235 phosphorylation levels upon apoE siRNA knockdown. Values are normalized to VASP protein levels, (n = 2 biological replicates), ns:non-significant, ∗p < 0.05.
VASP S235 phosphorylation levels in apoE3 Neuro-2a and apoE4 Neuro-2a cells are determined by immunoblotting following treatment with PKA (KT5720) (1 μM and 3 μM) or PKG (KT5823) inhibitors. Cells were treated either with 1 μM and 3 μM of KT5720 or 0.2 μM and 2 μM of KT5823 for 24 h.
GFP and mcherry-tagged hVASP constructs were expressed in apoE3 and apoE4 Neuro-2a cells for 24 h. Endogenous VASP was knocked down by VASPsiRNA. hVASP phosphorylation at S239 was validated by immunoblotting. Endogenous VASP is shown with arrows and hVASP is shown as tagged in brackets. Actin serves a loading control.
Data Availability Statement
Mass spectrometry data files (raw and search results) have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository with dataset identifier PXD041205 (36).