Summary
Human natural history and vaccine studies support a protective role of antibody dependent cellular cytotoxicity (ADCC) activity against many infectious diseases. One setting where this has consistently been observed is in HIV-1 vertical transmission, where passively acquired ADCC activity in HIV-exposed infants has correlated with reduced acquisition risk and reduced pathogenesis in HIV+ infants. However, the characteristics of HIV-specific antibodies comprising a maternal plasma ADCC response are not well understood. Here, we reconstructed monoclonal antibodies (mAbs) from memory B cells from late pregnancy in mother MG540, who did not transmit HIV to her infant despite several high-risk factors. Twenty mAbs representing 14 clonal families were reconstructed, which mediated ADCC and recognized multiple HIV Envelope epitopes. In experiments using Fc-defective variants, only combinations of several mAbs accounted for the majority of plasma ADCC of MG540 and her infant. We present these mAbs as evidence of a polyclonal repertoire with potent HIV-directed ADCC activity.
Subject areas: Immunology, Pediatrics, Virology
Graphical abstract
Highlights
-
•
ADCC-mediating antibodies were isolated from an HIV-infected non-transmitting mother
-
•
Multiple antibodies contributed to the plasma ADCC of this individual
-
•
The ADCC antibody repertoire in maternal and early infant plasma is similar
-
•
These findings suggest plasma ADCC in this mother is because of a polyclonal response
Immunology; Pediatrics; Virology
Introduction
Antibodies (Abs) prevent infection and modulate the immune response to pathogens via several mechanisms, preventing infection by neutralizing the virus and clearing infected cells through Fc-mediated effector functions such as antibody dependent cellular cytotoxicity (ADCC). Accumulating evidence, including numerous natural history studies and a vaccine clinical trial, implicates Abs that mediate ADCC in both reduced HIV-1 transmission and pathogenesis.1 Passive antibody studies in animal models using Abs where ADCC function was ablated have also suggested a role for Fc-mediated activity in protection.2,3 However, results in animal models are mixed, potentially because they are confounded by less efficient signaling of human antibodies in these models.4 Thus, human studies remain central to understanding the role of ADCC responses in HIV infection.
In early studies, ADCC responses have been associated with reduced viremia and slowed disease progression in individuals living with HIV.5 Similarly, elite controllers have been reported to have broader and higher magnitude ADCC than HIV progressors.6 Numerous correlative studies of the relationship between ADCC activity and disease outcome in chronic HIV infection have also suggested a beneficial role of ADCC Abs in reduced HIV pathogenesis.6,7,8,9,10,11,12,13,14 Recent studies from a multitude of transmission settings continue to support these early reports.15,16,17,18,19,20 A role for ADCC in an HIV vaccine setting remains controversial, perhaps in part because there is little or no efficacy associated with vaccine candidates tested to date. However, there was an association between a combination of low plasma IgA and high plasma ADCC with protection from HIV infection in the RV144 vaccine trial, which had an estimated efficacy of 31%.21,22 However, because a follow-up vaccine trial using a similar regimen in a distinct population, HVTN 702, did not demonstrate any efficacy, the role of ADCC in vaccine-mediated protection remains unclear.23
The one setting where there are consistent findings of a role for ADCC responses is in protection from vertical transmission of HIV. Vertical transmission is a unique setting through which the impact of circulating HIV-specific Abs on HIV acquisition and pathogenesis can be directly studied.24 Infants receive maternal HIV-specific Abs from their mothers during the third trimester of pregnancy via placental antibody transfer, and these Abs remain present in plasma for several months after birth.8,25,26 Within the setting of HIV vertical transmission, our group has shown in two different cohorts that ADCC mediated by passively transferred antibodies in infant plasma is correlated with improved survival of infants that acquire HIV.8,15 Moreover, passively acquired ADCC activity correlated with decreased HIV acquisition risk in a combined cohort analysis.15 More recently, Thomas et al. showed in a distinct cohort that HIV-exposed, uninfected infants had higher passively acquired autologous ADCC-mediating Abs than infants who acquired HIV during follow-up.16,27 There was also a correlation between passively acquired ADCC-mediating Abs and improved survival of infants that acquired HIV. Collectively, these studies demonstrated a correlation between passively acquired, Ab-mediated ADCC and improved clinical outcome in three different cohorts using three distinct assays. In each of these studies, there was no evidence for a role of neutralizing antibodies (nAbs) in protection from transmission or infant disease progression.8,16,28 In combination, these studies support a clinical benefit of pre-existing ADCC activity within the context of vertical transmission.
Despite the lack of a comprehensive approach to study ADCC antibodies, several major targets of ADCC mAbs have been identified in the various studies noted above. Although ADCC mAbs targeting nearly all major Env epitopes have been described, the most common targets of gp120-specific ADCC mAbs include variable loop 3 (V3) and CD4-inducible (CD4i) epitopes, which are only exposed on the Env trimer after CD4 engagement.29,30 Guan et al. further divided the CD4i epitopes into three major clusters (A, B, and C) based on competition experiments using prototypic mAbs A32 or C11, E51-M9, and 17b or 19e, respectively.31 Since then, additional CD4i mAbs have been described with epitopes that do not completely overlap with the prototypic cluster mAbs.31 Within gp41, early studies identified two distinct regions that are targeted by ADCC-capable, non-neutralizing Abs.32,33 Prior studies suggest that the majority of gp41-specific antibodies in HIV-infected individuals bind to two regions, the C-C loop and C-heptad repeat, also referred to as clusters I and II, respectively.33,34,35,36 Cluster I, a predominantly linear epitope, is targeted by prototypic mAbs 246-D and 50–69, whereas cluster II is discontinuous and is recognized by prototypic mAbs 98-6 and 167-7.
Even with increasing evidence that ADCC may play a protective role within the setting of vertical transmission, the specific characteristics of the individual Ab lineages that contribute to plasma ADCC are not well studied. In several instances, mAbs capable of mediating ADCC were identified incidentally using methods optimized to identify nAbs.37,38 Other reports have focused on the cumulative plasma ADCC response and/or on the Abs targeting a specific epitope.18,39,40,41 These efforts have been fruitful in identifying potent ADCC mAbs targeting distinct epitopes, but they fall short in failing to comprehensively evaluate plasma ADCC responses to both HIV Envelope (Env) subunits.29 Although gp120-specific ADCC has most consistently been associated with protection, ADCC directed against the gp41 subunit correlated with protection or reduced viremia in multiple animal studies.42,43 As such, few studies have taken a holistic approach to identify the lineages of mAbs that comprise plasma ADCC responses and how these Abs interact to promote potent cell killing.
In this study, we sought to identify the mAbs that contributed to the passively acquired plasma ADCC repertoire by sorting memory B cells circulating during late pregnancy, as the majority of placental Ab transfer occurs during the third trimester, thereby serving as a proxy identifying for passively acquired mAbs in an infant.25 We selected a mother, MG540, who was HIV seropositive at enrollment in the Nairobi Breastfeeding Trial (NBT), had a high viral load and did not transmit HIV to her infant, BG540.44 We identified 20 ADCC-capable mAbs representing 14 clonal families and targeting both gp120 and gp41 epitopes of Env. In competition ADCC experiments, combinations of mAbs recapitulated the majority of the plasma ADCC responses of MG540 and the passively acquired plasma ADCC of her infant, BG540. Single epitope-specific Abs that mediate ADCC did not individually contribute significantly to the overall plasma ADCC response, whereas combinations of isolated Abs targeting distinct epitopes largely recapitulated the plasma activity, which suggests that the MG540 plasma ADCC response is functionally polyclonal in nature.
Results
Supernatant screening and mAb reconstruction
HIV-1 non-transmitting mother MG540 was selected from the Nairobi Breastfeeding Trial (NBT), which was conducted before the availability of antiretroviral therapy for prevention of vertical transmission.44 We selected individual MG540 as a case where transmission did not occur despite the presence of known factors that correlate with transmission, including high plasma viral load (>105 copies mL−1 in late pregnancy) and high breastmilk viral levels (>104 copies mL−1 at delivery) during at least 10 months of breastfeeding.24,45,46 In addition, MG540 had gp120-specific plasma ADCC in the top quartile for the NBT cohort at pregnancy week 34 (P34).8 Her infant, BG540, born at gestational week 41, also had passively acquired plasma ADCC in the top quartile at week of life 0 (W0) compared to other infants in the cohort, a feature that is associated with reduced HIV acquisition risk and pathogenesis.15,16 Maternal monoclonal antibody (mAb) sequences present during late pregnancy, when the majority of placental antibody transfer occurs, were reconstructed by sorting memory B cells from an MG540 PBMC sample collected at P34, ∼7 weeks before the birth of BG540.25 This sample, stored since 1994, contained 10.8 x 106 live cells with 81% viability. Based on fluorescent antibody staining, 3.9% of single cells were B cells (CD19+ CD3− CD14− CD16−), and of these, 41.6% of cells were memory B cells (IgM− IgD−). For this study, we ultimately sorted 23,856 memory B cells into 11 x 384-well plates at a density of eight cells per well, which typically results in 1–2 viable cells per well, and cultured for 11 days to promote IgG expression.
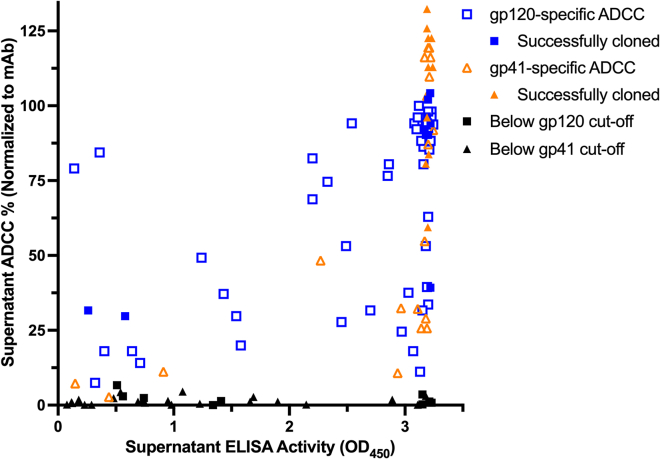
To select for memory B cells that expressed HIV-binding antibodies that also mediate ADCC, we developed a two-step protocol to screen first for antigen binding and then, within binding-positive wells, for ADCC activity. For binding assays, we included gp120 (clade A BG505), gp41 (clade C ZA1197), and Env trimer (BG505 T332N SOSIP) antigens to maximize detection of Env-specific antibodies. Screening well supernatants for HIV-specific IgG via ELISA identified 257 (8.23% of total) wells with activity >2x above the background. The highest activity subset (n = 192 wells) of these wells was subsequently screened for gp120-and gp41-specific ADCC activity via a multiplexed version of the RFADCC assay, which correlates well with clinical outcomes, and using the same antigens (Figure 1).6,8,15,47 Ninety-five of 192 wells (49.5%) demonstrated RFADCC activity >2x above background. Of these, 70% mediated gp120-specific ADCC, whereas 30% were active against gp41. We attempted mAb reconstruction for all 95 wells, yielding 20 pairs of heavy and light chain sequences (21% reconstruction success) and 17 unique mAbs representing 14 clonal families (Table 1). Identical heavy chain sequences were obtained for two distinct pairs of wells, which hereafter will only be referred to by a single identifier. Heavy and light chain nucleotide somatic hypermutation (SHM) ranged from 6.9–19.3% and 3.0–14.6% and the complementarity determining region three (CDR3) length ranged from 10 to 31 and 9–15 amino acids, respectively. All mAbs were initially tested to confirm HIV specificity via ELISA: eight families were gp120-specific and six were gp41-specific. All the gp120-specific families bound to both stabilized BG505 T332N (clade A) and CNE8 (CRF01) SOSIP trimers, whereas only one of the six gp41-specific families weakly bound to both trimers (Figure S1).
Figure 1.
HIV-binding and ADCC activity of primary B cell culture supernatants
Comparison of ELISA and RFADCC activity for B cell culture supernatants with HIV-specific ELISA activity (>2x background), where background-subtracted ELISA activity (x axis) is plotted against RFADCC activity normalized to positive control mAbs C11 (gp120) or 167-7 (gp41; y axis). Symbols indicate whether each well had gp120- (squares) or gp41-specific (triangles) RFADCC activity and whether a mAb was successfully cloned from the indicated well (closed symbols). MAb reconstruction was only attempted for wells with RFADCC activity 2-fold above the respective gp120-or gp41-specific background activities, which are distinct.
Table 1.
Characteristics of reconstructed MG540 mAbs
| MG540 P34 Ab rearrangement characteristics |
SOSIP ELISA |
Epitope mapping |
RFADCC |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Clade A |
CRF01 |
|||||||||||||
| Family | ELISA Target | mAb ID | Heavy chain | CDRH3 length | VH SHM (%nt) | Light chain | CDRL3 length | VL SHM (%nt) | BG505.C2 T332N | CNE8 | PhIP-Seq | ELISA | Peak activity | EC50 |
| 1 | gp120 | MG540.7a | V1-69 D3-9 J3 | 26 | 12.7 | KV1-9 J5 | 9 | 5 | ++ | ++ | negative | C11 | 140 | 2.4 |
| 2 | MG540.16 | V5-51D2-15 J4 | 18 | 13.8 | LV3-1 J2 | 12 | 14 | +++ | ++ | V3 | NT | 135 | 9.9 | |
| MG540.67 | 18 | 12.7 | 12 | 13.4 | NT | NT | V3 | V3 | 133 | 8.0 | ||||
| MG540.90a | 18 | 14.3 | 12 | 14.6 | +++ | + | V3 | V3 | 141 | 6.2 | ||||
| 3 | MG540.17a | V3-21D3-9 J4 | 23 | 7.8 | KV1-5 J2 | 12 | 6.5 | ++ | – | negative | negative | 130 | 6.7 | |
| 4 | MG540.48 | V1-2 D3-10 J5 | 14 | 8.7 | LV8-61 J3 | 12 | 7.6 | ++ | ++ | negative | A32 | 146 | 3.7 | |
| 5 | MG540.50a | V1-69 D3-10 J5 | 18 | 10.3 | KV3-20 J4 | 11 | 5.5 | ++ | +++ | negative | A32 | 145 | 2.8 | |
| 6 | MG540.51 | V1-69 D3-16 J4 | 28 | 14.9 | KV3-20 J3 | 11 | 4.6 | ++ | ++ | negative | C11 | 133 | 3.8 | |
| 7 | MG540.82a | V1-69 D6-13 J4 | 10 | 13.6 | KV3-11 J5 | 12 | 6.2 | +++ | +++ | negative | 17b | 38.7 | 59 | |
| 8 | MG540.84 | V5-51D3-16 J4 | 19 | 9.9 | LV1-51 J2 | 15 | 8.6 | +++ | ++ | V3 | V3 | 130 | 11 | |
| 9 | gp41 | MG540.32 | V1-3 D2-15 J3 | 21 | 18.3 | KV3-15 J2 | 11 | 10.9 | + | – | IDE | NT | 141 | 33 |
| MG540.86a | 21 | 19.3 | 11 | 5.9 | + | + | IDE | IDE | 138 | 23 | ||||
| 10 | MG540.37a | V1-2 D2-21 J6 | 20 | 8.8 | KV4-1 K2 | 11 | 10 | – | – | negative | Cluster II | 134 | 47 | |
| 11 | MG540.56a | V4-39 D1-26 J4 | 15 | 14.3 | LV2-14 J3 | 11 | 8.3 | – | – | negative | Cluster II | 136 | 37 | |
| 12 | MG540.61 | V1-69 D4-17 J6 | 25 | 6.9 | KV2-28 J3 | 11 | 3 | – | – | negative | Cluster II | 158 | 56 | |
| 13 | MG540.63a | V3-64 D5-12 J6 | 22 | 13.9 | KV2-29 J4 | 11 | 7.7 | – | – | IDE | IDE | 122 | 19 | |
| 14 | MG540.88 | V3-30 D3-3 J6 | 31 | 14.8 | LV1-40 J3 | 13 | 11.7 | – | – | negative | Cluster II | 141 | 42 | |
Distinct, isolated MG540 P34 mAbs (rows) and their characteristics (columns) are reported. Clonally related mAbs are grouped into families. Antibodies are grouped by target, gp120 or gp41, as confirmed by ELISA (BG505 gp120 and ZA1197 gp41). Results of qualitative studies are also reported: ELISA activity against BG505 T332N and CNE8 SOSIP trimers, epitope mapping by PhIP-seq and ELISA, and RFADCC activity. RFADCC peak activity (% normalized to HIVIG) and EC50 (ng/mL) are averaged between two replicate experiments. See Figure S1 for representative SOSIP ELISA data and Table S1 for neutralization data.
NT, not tested; V3, variable loop 3; IDE, immunodominant epitope; +++, saturating ELISA activity at all/most concentrations; ++, activity above background for all concentrations; +, activity above background for some concentrations; -, no activity above background.
Indicate mAbs selected for GRLR variant generation.
ADCC activity of mAbs
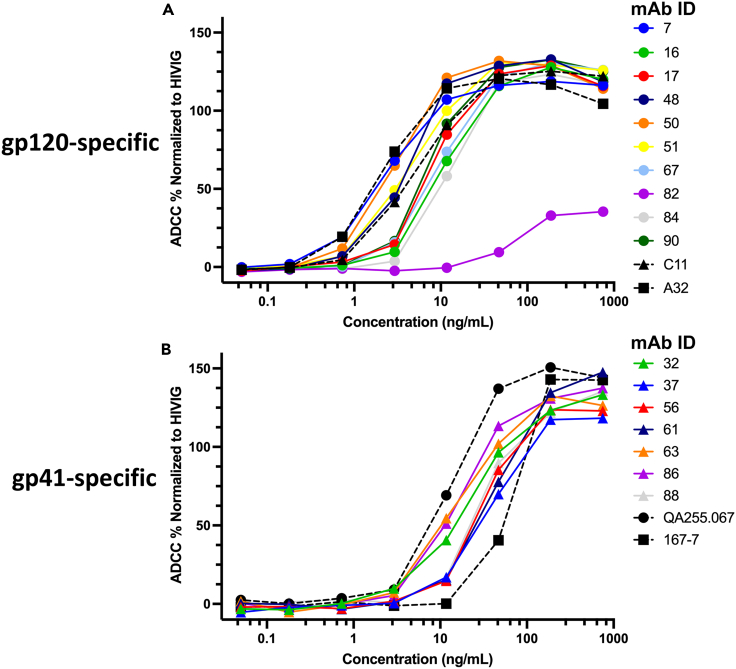
Reconstructed mAbs were next tested for ADCC activity using the RFADCC assay to identify ADCC capable antibodies for further characterization. We used this assay because RFADCC activity has been shown to correlate with infant outcomes.8,15 We used the same antigens as in the supernatant screening: BG505 gp120 and ZA1197 gp41. BG505 Env is a particularly relevant antigen because it is an early infection isolate from an infant in the NBT cohort and it is closely related to MG540 env sequences from P34 (∼88% pairwise nucleotide and 94% amino acid identity). Clade C ZA1197 is a transmitter-founder strain that has been used previously to measure ADCC.48,49 Compared to gp120, gp41 is more well-conserved, especially within regions targeted by ADCC antibodies.33 To determine the potency of each mAb, we tested serial dilutions of each antibody and quantified the effective concentration for 50% maximal killing (EC50) as well as peak (maximum) killing. Most of the gp120-specific mAbs potently mediated ADCC against BG505 gp120 coated target cells, with activity comparable to cluster A mAbs C11 and A32, which are prototype cluster A mAbs that targeted CD4i sites (Figure 2A).50,51 With the exception of less potent mAb MG540.82 (EC50 = 59 ng mL−1), the EC50 of the gp120-specific mAbs ranged from 2.4–11 ng mL−1, with several mAbs more potent than C11 (6.1 ng mL−1) but not A32 (1.7 ng mL−1). All gp120 mAbs exhibited similar maximum ADCC activity, again with the exception of mAb MG540.82, which mediated markedly lower ADCC (>3-fold) compared to the other mAbs. Similarly, the gp41-specific mAbs strongly mediated ADCC against ZA1197 gp41-coated cells (Figure 2B), with similar potency to the cluster I mAb QA255.067 and the cluster II mAb 167-7.33,48,49 The gp41-specific mAbs had EC50 values ranging from 19 to 98 ng mL−1, in line with those of QA255.067 (18 ng mL−1) and 167-7 (68 ng mL−1). The gp41-specific mAbs also mediated similar maximum ADCC to each other, apart from MG540.66, which is clonally related to more potent MG540.32 and 0.86 (family 9). The ADCC EC50 and peak activities of all mAbs are summarized in Table 1.
Figure 2.
RFADCC activity and potency of reconstructed MG540 mAbs
(A and B) Relative RFADCC activities of gp120-specific (A) and gp41-specific (B) mAbs per mAb concentration (ng/mL). Background-subtracted RFADCC activity of each mAb was normalized to that of positive control HIVIG. Normalization described in STAR Methods. Data are representative of two independent experiments. Gp120-specific positive control ADCC mAbs were C11 and A32 and gp41-specific positive controls were QA255.067 and 167-7. Background-subtracted activity of HIVIG in replicates ranged from 31 to 43% for gp120 and 21–28% for gp41.
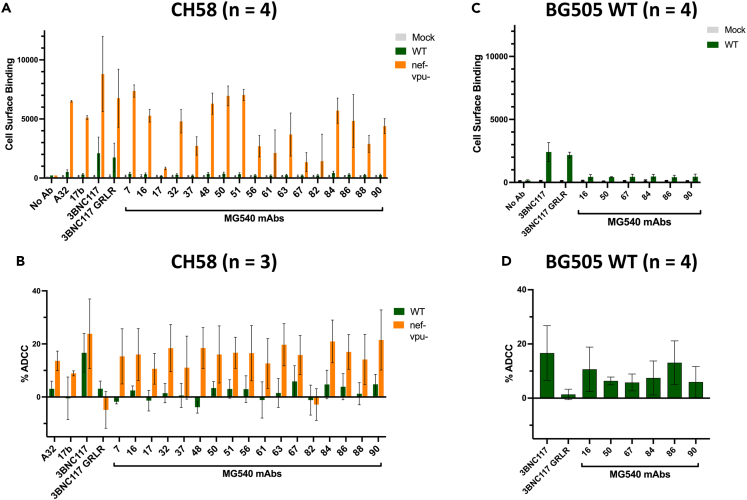
Although the RFADCC assay is a valuable measure of antibody effector function because of its correlation with clinical outcomes, there is not agreement in the field on whether it specifically detects ADCC or a combination of effector functions.47,52 More importantly, the use of gp120-or gp41-coated cells as targets in the RFADCC assays presents epitopes that are less accessible within the setting of HIV infection, largely because of CD4 downregulation by Nef and Vpu, as well as decreased surface Env expression.1 To examine the potential of MG540 mAbs to mediate ADCC of infected cells, we tested whether the mAbs could mediate ADCC against HIV-infected primary CD4+T cells (Figure 3). We compared the binding (Figure 3A) and ADCC (Figure 3B) mediated against cells infected with a wildtype or nef- vpu- CH58 (clade B) infectious molecular clone commonly used in this assay.53,54 Nearly all mAbs showed high levels of binding and ADCC against nef- vpu- CH58-infected cells, comparable to positive controls A32 and/or 17b, except for 0.82. A few showed (MG540.50, 0.51) even higher levels, comparable to a positive control that recognizes the CD4 binding site within the closed state of Env, 3BNC117 (Figures 3A and 3B, orange bars).55 By contrast, most mAbs showed markedly reduced binding to WT CH58-infected cells, in agreement with expected reduced epitope accessibility. Because several MG540 mAbs, 0.16, 0.50, 0.67, 0.84, 0.86, and 0.90, showed ADCC levels above the negative control 3BNC117 GRLR mAb, despite binding levels near background (Figures 3A and 3B, green bars), we tested this subset of mAbs for binding and ADCC against cells infected with BG505 T332N, the strain used in the RFADCC assay and more closely related to MG540 env sequences. These mAbs displayed binding levels (MFI: 416–466) ∼3-fold above the no Ab control (MFI: 148), albeit at ∼5-fold lower levels than 3BNC117 (MFI: 2208; Figure 3C). Similarly, there was measurable ADCC against BG505-infected cells, with activity ranging from 6 to 13%, compared to 3BNC117, which showed ∼15% ADCC (Figure 3D). This was consistently above the levels of the negative control (∼1%), suggesting that this subset of antibodies mediates low levels of ADCC against BG505-infected cells. Some mAbs mediated negative levels of ADCC in this assay, likely owing to killing of cells binding shed gp120.1
Figure 3.
Several MG540 mAbs bind and mediate ADCC against primary isolate infected CD4+T cells
(A–D) Surface antibody binding (A,C) and ADCC (B,D) of indicated mAbs was evaluated in CH58- (A,B) or BG505 T332N-infected (C,D) CD4+ T cells. Binding and ADCC were assessed in mock, WT, and nef-vpu- virus-infected cells depending on the assay and infecting variant, as indicated. Positive control mAbs included were A32, 17b, and 3BNC117. Negative controls include no antibody and a GRLR variant of 3BNC117. Mean and standard deviation are shown. Numbers of replicate experiments performed are indicated, except for MG540.50 which was tested in two replicate experiments in (C-D).
Defining MG540 mAb epitopes
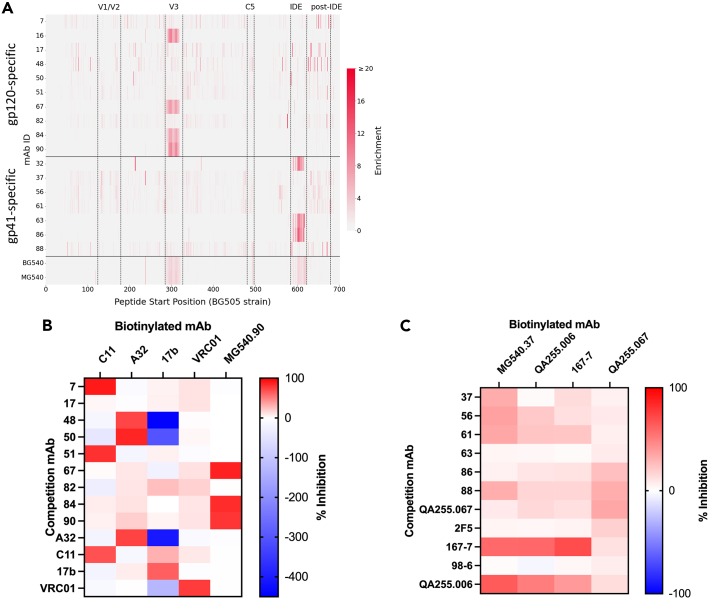
We mapped the epitope of each reconstructed mAb using a combination of phage display immunoprecipitation sequencing (PhIP-Seq) and competition ELISAs to identify both linear and conformational mAb epitopes. For PhIP-Seq, we used an HIV-specific phage display library expressing 38-mer Env peptides from six HIV strains. In some cases, Abs recognized well known linear epitopes in V3 region of gp120, centered on the GPxQ/R motif (mAbs MG540.16, 0.67, 0.84, 0.90), or the C-C loop (immunodominant epitope or IDE) of gp41 (mAbs MG540.32, 0.63, 0.66, 0.86; Figure 4A). Importantly, MG540 P34 and BG540 W0 plasma samples did not strongly enrich for responses for linear epitopes outside V3 or the IDE, suggesting that V3 and IDE-specific mAbs make up the majority of the linear epitope-directed plasma repertoire.
Figure 4.
Epitope mapping of MG540 mAbs
(A–C) Heatmap (A) showing the degree of enrichment of each peptide in red, with darker shades indicating higher enrichment (binding), as indicated by the scale on the right. Peptides are ordered by start position of the N-terminal amino acid, beginning at HXB2 amino acid 31 and spanning amino acid 704. MG540 mAbs are stratified by gp120 or gp41 specificity. Plasma samples are shown at the bottom. Dotted vertical lines indicate known linear Env epitopes, with the epitope indicated above. The Heatmaps (B,C) displaying the degree of competitive inhibition of biotinylated mAb ELISA activity using BG505 gp120- (B) or ZA1197 gp41-coated plates (C) are shown for a subset of mAbs. Competitor antibodies indicated in rows and biotinylated antibodies are listed as columns. Data are representative of two replicate experiments. Inhibition calculation is described in STAR Methods. See Figures S2 and S3 for additional epitope information and binding enhancement data.
Many of the mAbs directed to linear epitopes bound to peptides from multiple HIV strains (Figure S2). Among the V3-specific mAbs, MG540.84 bound to all strains tested except clade D QA013. Clonal family 2 (mAbs MG540.16, 0.67, 0.90) did not bind peptides from strains BF520 (clade A) and QA013. The strains differ in the residues upstream of the core binding motif: SIR/HI for B41, BG505, BL035, and ZA1197, GEHM for QA013, and SVHL for BF520, which may explain the observed differences in binding. These strain-specific results have similarly been reported for other V3 mAbs.56 MG540 and BG540 plasma also did not demonstrate detectable enrichment for V3 peptides from the QA013 strain, supporting strain-specificity of mAbs directed against this site.
Among the IDE-specific mAbs, MG540.63 and 0.86 displayed responses to peptides from all strains. By contrast, mAbs MG540.32 and 0.66, which are clonal to MG540.86, exhibited reduced responses to the clade A/D BL035 and QA013 IDE, with no binding by mAb MG540.32. Similarly, mAb MG540.32 bound to fewer epitopes overall within the IDE, which might suggest that it has a narrower epitope than the other IDE mAbs. At the center of the C-C loop, the BL035 and QA013 strains contain a negatively charged histidine at site 602 (HXB2 numbering), whereas all other strains utilize polar leucine or isoleucine residue. These data are in agreement with a study reporting that mutations at this site reduce mAb binding responses in PhIP-Seq.56
Because the remaining mAbs bound to gp120 or gp41 monomers but did not target linear epitopes represented by 38-mer peptides, we tested them for recognition of conformational epitopes targeted by previously described HIV-specific ADCC antibodies. As many potent ADCC mAbs target CD4-inducible (CD4i) regions of gp120, we focused on biotinylated variants of CD4i mAbs in competition ELISAs (Figure 4B). Competition was observed between five MG540 mAbs and CD4i mAbs: mAbs MG540.7 and 0.51 competed with C11, mAbs MG540.48 and 0.50 competed with A32, and MG540.82 competed with 17b. As expected based on the PhIP-Seq data, the V3-specific mAbs (MG540.67, 0.84, 0.90) competed with each other. Only one of the gp120-specific mAbs, MG540.17, did not exhibit clear competition with the biotinylated mAbs.
In the course of competition experiments, we noted that A32 and A32-like mAbs MG540.48 and 0.50 enhanced 17b ELISA activity, indicating that these mAbs may improve 17b epitope accessibility, similar to how stabilizing mutations or CD4-mimetics have been shown to enhance 17b affinity for Env expressed on HIV-infected cells.39,57,58 Although previous studies suggest enhancement of A32 binding to gp120 monomer by 17b, we did not observe enhanced binding mediated by 17b in initial competition experiments.59 By contrast, there was a modest increase in A32 binding to gp120 monomer coated wells by C11 and C11-like mAb MG540.51. When a lower concentration of biotinylated-A32 was used for competition, there was improved binding of A32 by these mAbs (Figure S3).
As PhIP-Seq identified the cluster I-specific mAbs (MG540.32, 0.63, 0.66, 0.86), we used mAbs specific for discontinuous cluster II, QA255.006 and 167-7, as competitors in a gp41 competition ELISA to determine if the remaining mAbs were directed against this region.33,48 Indeed, all four of the remaining mAbs, MG540.37, 0.56, 0.61, and 0.88, exhibited moderate competition of QA255.006 and/or 167-7 binding, indicating that they likely target cluster II (Figure 4C). As these results would suggest that the mAbs compete with each other, we also performed competition experiments using biotinylated mAb MG540.37. There was clear competition by mAbs MG540.37, 0.56, 0.61, 0.88, 167-7, and QA255.006 with MG540.37, which supported the cluster II specificity of these mAbs. Of note, mAb MG540.88 interestingly also competed with QA255.067, an IDE-specific mAb. MG540.37 was the only non-IDE mAb that did not compete with QA255.006, which suggests it may have a lower affinity or non-overlapping epitope within cluster II.
Altogether, mapping experiments identified the epitopes targeted by all of the isolated MG540 mAbs except for MG540.17. In total, the 14 clonal families recognized six non-overlapping Env regions, across both gp120 and gp41.
Neutralization activity of mAbs
Because ADCC-capable Abs that bind HIV Env can be neutralizing, we tested the ability of all mAbs to neutralize a small panel of cross-clade, heterologous viruses, many of which were only weakly neutralized by MG540 P34 plasma (Table S1). Most reconstructed mAbs were non-neutralizing. The two V3-specific families, 2 and 8, strongly neutralized the two tier 1 viruses tested, SF162 (clade B) and Q461.D1 (clade A), with IC50 values ranging from <0.625 to 5.4μg mL−1 but did not neutralize the tier 2 viruses tested.
Contribution of mAbs to overall plasma ADCC
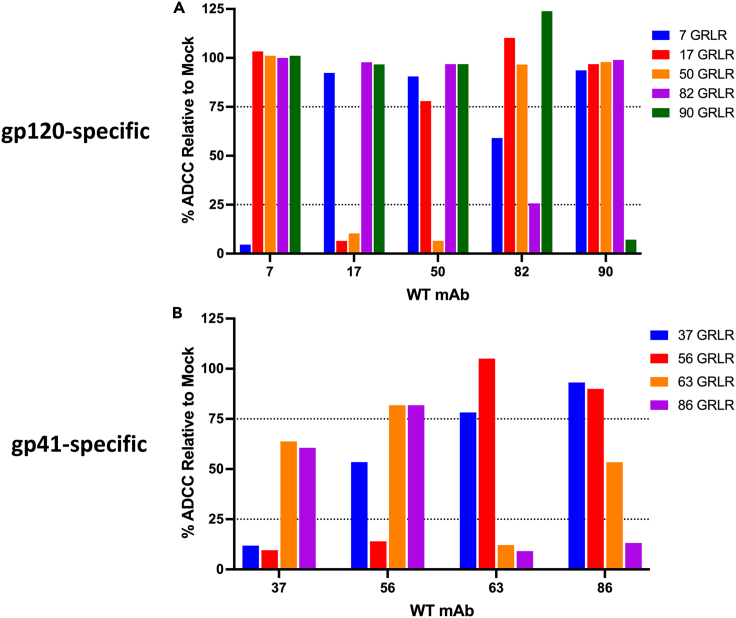
We performed competition experiments with the isolated MG540 mAbs to determine how they contribute to overall plasma ADCC. For this, we selected one mAb representing each epitope to distinguish between the epitopes targeted (∗indicated in Table 1) and generated GRLR variants, which introduces mutations that prevent binding to FcγRs and thus disrupt ADCC activity.60 For clonal families or mAbs targeting the same epitope, we selected the most potent mAb to reduce redundancy in evaluating the impact of each epitope on plasma ADCC, resulting in five distinct mAbs for gp120 and four for gp41.
We first validated the ability of serial dilutions of the GRLR variants to inhibit ADCC by its wildtype counterpart and used these results to define a concentration that resulted in at least 85% reduction in wildtype mAb ADCC for downstream studies (Figure S4). We next evaluated “cross-competition” between the GRLR variants and wildtype antibodies targeting distinct epitopes (Figure 5). As expected, the GRLR variants again potently reduced wildtype mAb ADCC (4–25% of mock activity), with little competition between most gp120 mAbs targeting distinct epitopes (which we define here as >75% of mock activity; maximum mock standard deviation = 4.5%). The exception is the GRLR variant of mAb MG540.50, which also strongly competed with mAb 0.17, although 0.17 GRLR did not compete with mAb 0.50 (Figure 5A). These results suggest there may be partial overlap between the epitope of mAbs MG540.17 and 0.50, which has been reported for other CD4i mAbs.31 However, we also cannot rule out steric hindrance of a binding site by the bound mAb that is not because of an overlapping epitope. As a result, we opted to include both MG540.17 and 0.50 in subsequent competition experiments.
Figure 5.
mAb GRLR variants exhibit cross-competition of ADCC activity
(A and B) Wildtype antibody ADCC of gp120- (A) or gp41-specific (B) mAbs in the presence of GRLR variants. mAb activity was normalized to that of the mock (no GRLR) mAb competition. WT mAbs were added to target cells at the respective EC95 concentration. Dashed lines indicate 25% and 75% of mock competition ADCC. GRLR mAbs were added at an empirically determined concentration, as indicated in Figure S4. Data are representative of two independent experiments. See Figures S5 and S6 for ADCC enhancement data.
For gp41, the GRLR mutants showed similar inhibition of wildtype mAbs as seen with the gp120 mAbs (86–88%). The Cluster I-specific gp41 mAbs, MG540.63 and 0.86, competed with each other, as did the cluster II mAbs (MG540.37 and −56), consistent with epitope mapping experiments (Figure 5B). The degree of competition was not equally bidirectional, with mAbs MG540.56 and 0.86 more potently inhibiting (<25% of mock) ADCC mediated by their respective gp41 cluster counterparts (<75% of mock), MG540.37 and 0.63. Owing to the high degree of non-reciprocal cross-competition between the gp41-specific mAbs, we opted to include all four mAbs in plasma competition experiments to maximize our chance of recapitulating the ADCC response.
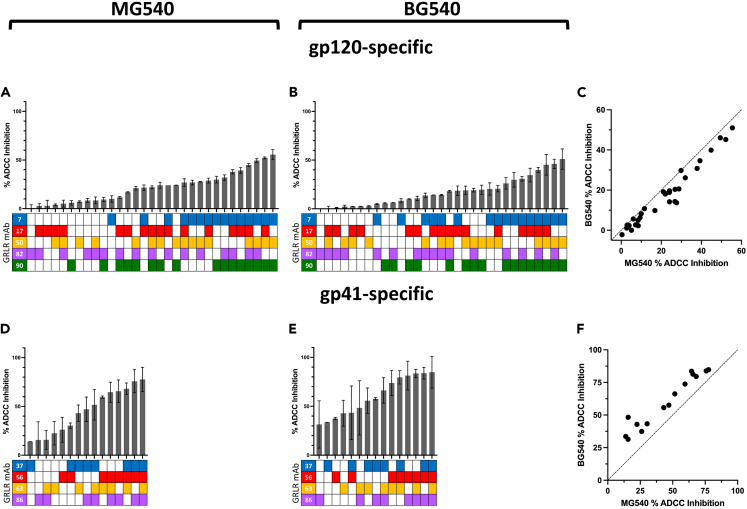
After validating the effectiveness of the GRL R variants to inhibit epitope-specific ADCC, we tested the ability of combinations of GRLR mAbs to inhibit either gp120-or gp41-specific plasma ADCC (Figure 6). The maximum inhibition by a single antibody (bars with only one shaded box in the lower chart) was only 10%, suggesting that none of the mAbs dominate the MG540 plasma ADCC response. Among the gp120-specific mAbs, pre-incubation with combination of 3–5 mAbs (indicated by shaded boxes in the lower chart) accounted for upwards of 55% of plasma ADCC, demonstrating that these mAbs collectively make up the majority of the gp120-specific plasma ADCC repertoire (Figure 6A). Overall, competitions involving more GRLR mAbs resulted in higher ADCC inhibition, although this trend was influenced by the specific mAbs present in each competition. In terms of epitope-specific contribution, combinations containing C11-like mAb MG540.7 tended to show the greatest ADCC inhibition. Conversely, combinations containing mAbs MG540.17 or 0.82 were somewhat lower ranking overall in terms of hierarchy for inhibition.
Figure 6.
Results of competition ADCC experiments with GRLR mAbs and MG540 P34 or BG540 W0 plasma
(A–F) Inhibition of MG540 P34 (A-B) or BG540 W0 (D-E) plasma gp120- (A-C) or gp41-specific (D-F) RFADCC activity by target cell pre-incubation with the indicated combinations of GRLR mAb variants. Below each bar, the shaded charts indicate which mAbs were present in the competition. Conditions are ordered by ascending degree of average plasma ADCC inhibition. Inhibition calculation is described in STAR Methods. Panels C and F show correlations between GRLR mAb plasma gp120- (C) or gp41-specific (F) ADCC competitions for MG540 and BG540. Mean ADCC inhibition values from maternal (A-B; MG540) and infant (D-E; BG540) plasma ADCC competition assays for gp120 (A,D) and gp41(B,E) were compared for all conditions, which includes single and combined mAbs. Dashed lines indicate line of identity. Mean and standard deviation from two replicate experiments are shown.
Compared to the gp120-specific antibodies, the gp41-targeting mAbs accounted for a greater proportion of total MG540 gp41-specific plasma ADCC (Figure 6D). Pre-incubation with all 4 mAbs led to a 78% reduction of gp41 plasma ADCC. Single mAb competitions resulted in 14–26% ADCC inhibition, with cluster II mAb MG540.56 having the greatest impact. Similarly, competitions containing mAb MG540.56 yielded more ADCC inhibition overall. Taken together, the plasma competition experiments indicate that the isolated mAbs largely account for MG540 plasma ADCC activity, with the gp41 mAbs resulting in near-complete abrogation of plasma ADCC.
We also examined whether the selected MG540 GRLR variants similarly recapitulated the passively acquired plasma ADCC of her infant, BG540. We tested the same combinations of GRLR MG540 mAbs for the ability to inhibit BG540 gp120-or gp41-specific ADCC using plasma from the first week of life. The GRLR mAbs recapitulated BG540 gp120-specific plasma ADCC to a strikingly similar extent (Figure 6B). Single mAb competitions reduced plasma ADCC by only up to 8%. A combination of three to five mAbs inhibited gp120 plasma ADCC by 30–51%, suggesting that the five GRLR mAbs recapitulate the BG540 plasma ADCC response to a similar degree as for MG540. As with MG540, C11-like mAb MG540.7 appeared to contribute the most to plasma ADCC, followed by V3-specific mAb MG540.90. Antibodies MG540.17 and 0.82 again did not appreciably reduce gp120-specific ADCC compared to competitions lacking these antibodies.
Similar to MG540, the gp41-specific GRLR variants recapitulated BG540 W0 plasma ADCC to a higher degree than for gp120-specific ADCC (Figure 6E). Combinations of two to four GRLR mAbs inhibited BG540 plasma ADCC by 81–85%. Single mAb competitions resulted in 31–43% ADCC inhibition, a higher proportion than observed for MG540 plasma. Competitions containing mAb MG540.56 again yielded greater ADCC inhibition, suggesting that contributes highly to passively acquired plasma ADCC activity in BG540.
In addition to recapitulating the level of ADCC activity mediated by plasma to a similar degree for MG540 and BG540, the contribution of each combination of GRLR mAbs was statistically significantly correlated across these individuals (Spearman r = 0.96 for gp120 and 0.96 for gp41). The gp120 competitions were slightly skewed toward having a higher impact on MG540 plasma ADCC (Figure 6C). Conversely, the gp41-specific competitions consistently had a greater impact on BG540 passively acquired plasma ADCC activity when compared to MG540 (Figure 6F). Altogether, these data suggest a high degree of concordance between the ADCC activity recapitulated by the GRLR mAbs for MG540 and BG540.
Enhancement between antibodies
There was some evidence to suggest enhancing interactions between antibodies in competition ADCC experiments. For example, there was an increase of 110–124% of mock ADCC activity for mAb MG540.82 when target cells were pre-incubated with the GRLR variants of mAb MG540.17 and 0.90 (Figure 5A). To explore this further, we tested wildtype mAbs at a lower EC50 concentration rather than saturating at EC95 concentrations. Indeed, there was stronger evidence for enhancement between multiple MG540 mAbs. This increased activity was most evident between MG540.82 and 0.90, which resulted in >260% ADCC relative to mock competition. The ADCC mediated by MG540.82 was also higher when cells were pre-incubated with MG540.17 (161% of mock) and 0.50 (131% of mock), although to a lesser extent (Figure S5). To probe these results further, we also tested for ADCC enhancement by the GRLR variants against BG505 T332N-infected CD4+T cells. Compared to MG540.82 alone (2% ADCC), co-incubation with GRLR variants of MG540.17, 0.50, and 0.90 resulted in a > 4-fold increase in ADCC (9–19%) against BG505 T332N-infected cells (Figure S6).
Discussion
In this study, we designed a two-step screening approach to identify and reconstruct ADCC-mediating HIV-specific mAbs from sorted memory B cells isolated from a non-transmitting mother living with HIV, MG540. Individual MG540 was selected because she did not transmit HIV to her infant despite a high viral load and nearly a year of breastfeeding in the absence of antiretroviral therapy, which was not available at the time. In total, we recovered 14 unique lineages, represented by 17 mAbs, which targeted a diversity of epitopes and together provide a thorough assessment of the ADCC-capable mAbs produced by circulating memory B cells during chronic HIV in this individual. These mAbs account for a large fraction of the late pregnancy plasma ADCC within MG540 as well as the passively acquired ADCC activity of her infant, BG540, with no gp120-specific mAb contributing more than 10%. These data suggest the polyclonal collection of MG540 mAbs were the dominant passively transferred ADCC antibodies circulating at the time of HIV exposure in BG540.
Although numerous studies have identified neutralizing Abs by culturing sorted B cells and testing the cell supernatants in neutralization assays, we opted to screen the supernatants via ELISA and the RFADCC assay to identify wells with B cells expressing ADCC-capable antibodies.61,62 Of the wells with HIV-specific ELISA activity, nearly half mediated ADCC against target cells coated with gp120 or gp41 monomer, likely reflecting the common elicitation of ADCC-capable antibodies in HIV infection.63,64
The mAbs reconstructed in this study recognized six non-overlapping epitopes among the gp120 and gp41 subunits of Env, including both CD4i (A32-, C11-, and 17b-like) and non-CD4i epitopes (V3, clusters I and II). One mAb, MG540.17, could not be mapped but based on non-reciprocal competition with a CD4i epitope targeting mAb, it may target a CD4i epitope that may partially overlap with that of A32. This finding is consistent with earlier studies describing ADCC mAbs that recognized CD4i epitopes that do not completely overlap with prototypic cluster A and C mAbs, thereby supporting the classification of more nuanced CD4i epitopes.31,65 The numerous epitopes targeted by the MG540 mAbs expand on previous attempts to characterize the ADCC repertoire of HIV-infected individuals by recovering a highly polyclonal collection of mAbs targeting diverse epitopes. Many such studies recovered mAbs targeting only one or two epitopes or only CD4i epitopes, which might indicate that ADCC is largely driven by antibodies targeting a single epitope.31,38,40,48,65 However, in several instances the ADCC-capable mAbs were identified by happenstance using approaches to screen for nAbs, and as such may have suffered from an inability to comprehensively detect broad lineages of nnAbs.
We selected the RFADCC assay for initial ADCC screening because activity in this assay has repeatedly been associated with positive clinical outcomes, including improved infant survival in vertical transmission studies.6,8,9,15,66 This assay also correlates well with other measures of ADCC and is tractable for high throughput screening.67,68,69 The ADCC-capable mAbs isolated in this study potently mediated ADCC against monomer coated cells in this assay. Many of the gp120 mAbs had an ADCC EC50 and peak activity comparable to prototype CD4i ADCC mAbs A32 and C11. The 17b-like mAb MG540.82 appears to be an outlier in that its lack of potency differs from previous studies suggesting most mAbs targeting CD4i epitopes are particularly potent mediators of ADCC.29 Regarding gp41 specificity, the mAbs we identified were noticeably less potent than the gp120-specific mAbs per the RFADCC assay.
Although RFADCC activity correlates with positive clinical outcomes, it may measure multiple effector functions and may not recapitulate the epitopes presented by HIV-infected CD4+T cells.70,71 In an assay that measures killing of infected cells, many of the MG540 mAbs also reproducibly mediated ADCC against nef- vpu- CH58-infected CD4+T cells. Surface antibody binding and ADCC activity was relatively diminished against WT CH58 infected cells, which suggests that some of the mAb epitope targets are less accessible on the surface of cells in which CD4 is downregulated, which is expected for CD4i antibodies. Nonetheless, ADCC activity was detected for six of the MG540 mAbs against both WT CH58- and BG505 T332N-infected cells. These findings provide evidence that the isolated mAbs likely mediate ADCC in vivo. This subset of six MG540 mAbs join those already described in the literature as capable of mediating ADCC against distinct HIV variants, including tier 2 viruses, even in the absence of neutralization capacity.8,48 It is also noteworthy that 8 of 14 MG540 antibodies used a VH1 heavy chain gene, which is consistent with a prior study and might indicate preferential induction of B cells using heavy chains from this family.65
An important goal of this study was to define the repertoire of antibodies that contributed to the ADCC activity of Abs passively transferred to BG540. Competition ADCC experiments indicated that combinations of the reconstructed mAbs recapitulated the majority of contemporaneous MG540 plasma ADCC. The same was true for BG540 using plasma from the first week of life, and the relative contribution of each antibody was highly correlated between MG540 and BG540. Although single MG540 mAbs did not contribute significantly to overall ADCC activity, mAbs targeting CD4i epitopes appeared to make the largest impact on overall gp120-specific plasma ADCC, in line with earlier studies.29 However, the limited contribution of single mAbs in this individual differs significantly from previous studies suggesting that the majority of plasma ADCC can be attributed to a single epitope, particularly those targeted by A32, C11, and 17b.65,72 By contrast, in this study, combinations of up to five MG540 mAbs targeting distinct epitopes blocked MG540 gp120-specific plasma ADCC by 55%, suggesting that the MG540 plasma ADCC repertoire is polyclonal. Given this study, which is the first to fully interrogate the ADCC repertoire in a non-transmitting mother and define a polyclonal response in this individual, future studies are needed to determine if this is more typical of non-transmitting mothers. The GRLR variants of the mAbs isolated here may be useful for this purpose.
Although representatives of the MG540 mAbs captured the majority of the plasma ADCC activities of MG540 and BG540, the tested mAbs did not achieve complete inhibition of plasma ADCC activity, particularly for gp120. For this there are two likely explanations: (1) It is possible that the full set of MG540 mAbs comprising the plasma ADCC repertoire were not recovered, and/or (2) the GRLR mAbs used in plasma competition ADCC experiments blocked epitope-specific ADCC, but at the same time also mediated enhancement of ADCC targeting distinct regions of Env, thereby confounding the perceived degree of ADCC inhibition. The latter possibility is supported in part by mAb competition experiments presented here that provide evidence for enhancing effects between some combinations of MG540 mAbs, but further work is needed to fully understand how mAbs synergize to potently mediate ADCC.
This detailed study focused on characterizing ADCC antibodies from a single individual to provide a deeper understanding of the qualities and synergy of ADCC-capable mAbs comprising a potent plasma ADCC response. We expanded on prior studies evaluating the contribution of mAbs targeting single epitopes to overall plasma ADCC by testing the ability of combinations of up to five mAbs isolated from this individual to recapitulate plasma ADCC. Notably, we present a case where ADCC responses appear to be highly polyclonal, which differs from prior studies indicating that plasma ADCC is driven by mAbs targeting a single epitope, particularly CD4i epitopes.65,72 We recognize that although the motivation for this study was a population-level association between passively acquired ADCC activity and infant outcomes, we cannot establish a detailed causal link to this association with a single case study. This detailed dissection of ADCC activity in a non-transmitting mother and her infant sets a direction for future studies to address whether polyclonality is a common feature of the passive ADCC repertoire in HIV-exposed, uninfected infants. If multiple antibodies are required for potent plasma ADCC, this would suggest that ADCC-directed vaccination and treatment efforts should consider methods to elicit or harness polyclonal responses, rather than focusing on a single epitope. Given recent studies implicating a protective role of effector functions other than ADCC, including ADCP, future studies to determine the broad range of effector functions mediated by the MG540 mAbs described here may also be informative of their overall functional activity.73,74 Finally, the tools developed here, including the supernatant screening approach and the GRLR mutants of the MG540 mAbs, could be used to further probe human plasma for ADCC responses directed against a variety of epitopes.
Limitations of the study
In this study, although we found a polyclonal collection of antibodies that largely recapitulated the plasma responses, we did not recover the entire gp120-specific ADCC activity of plasma antibodies. We also did not rule out that there is an antibody specificity, either among those identified or missed, that correlates with transmission at the population level. Rather, as noted above, this hypothesis-generating study of a single individual provides motivation for future work to examine if a polyclonal response is associated with lack of HIV vertical transmission in additional individuals. The RFADCC assay was used to screen for antibodies capable of ADCC because the measurements from this assay have been associated with infant clinical outcome in multiple cohorts.8,9,15 However, the underlying mechanistic activity captured in this assay has not been well defined. In addition, we used representative viral strains in this assay, rather than autologous virus because responses to those strains correlated with transmission risk at the population level. Studies of autologous responses could provide additional insights, particularly if escape from ADCC antibody activity contributes to selection during transmission, as it does with neutralizing antibodies.75
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Human monoclonal MG540 antibodies | This paper | Genbank OQ054484-OQ054517 |
| Polyclonal Anti-Human Immunodeficiency Virus Immune Globulin, Pooled Inactivated Human Sera (HIVIG) | HIV Reagent Program | Cat#ARP-3957 |
| Pierce High Sensitivity Streptavidin-HRP | Thermo Fisher | Cat#21130 |
| Anti-Human IgG (Fc specific)−Peroxidase antibody produced in goat | Sigma Aldrich | A0170-1ML |
| Monoclonal anti-Env gp41 QA255-067 | Williams et al., 201948 | N/A |
| Monoclonal anti-Env gp41 QA255-006 | Williams et al., 201948 | N/A |
| Monoclonal anti-Env gp41 167D | HIV Reagent Program | Cat#ARP-531 |
| Biological samples | ||
| Human PBMCs from individual MG540 | Prospective cohort of pregnant women living with HIV in Nairobi, Kenya, Nduati, 2000 | N/A |
| Plasma from individuals MG540 and BG540 | Prospective cohort of pregnant women living with HIV in Nairobi, Kenya, Nduati, 2000 | N/A |
| Human PBMCs from HIV-1 uninfected individuals | Bloodworks Northwest | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Q5 High-Fidelity MAster Mix | New England BioLabs | Cat#M0492S |
| FreeStyle MAX | Thermo Fisher | Cat#16447500 |
| Protein G agarose | Pierce | Cat#20397 |
| 293F FreeStyle Expression media | Invitrogen | Cat#12338-026 |
| BG505.W6M.ENV.B1 (BG505) gp120 | Cambridge Biologics | Cat#01-01-1028 |
| C.ZA.1197MB (ZA1197) gp41 ectodomain | Immune Technology Corp | Cat#IT-001-0052p |
| GibcoTM Roswell Park Memorial Institute 1640 medium (RPMI) | Thermo Fisher | Cat#11875-093 |
| Dynabeads™ Protein A for Immunoprecipitation | Thermo Fisher | Cat#10002D |
| Dynabeads™ Protein G for Immunoprecipitation | Thermo Fisher | Cat#10004D |
| Critical commercial assays | ||
| Vybrant CFDA SE Cell Tracker Kit | Invitrogen | Cat#V12883 |
| PKH26 Red Fluorescent Cell Linker Kit | Sigma Aldrich | Cat# MINI26-1KT |
| CellVue Claret Far Red Fluorescent Cell Linker Kit | Sigma Aldrich | MINCLARET-1KT |
| Deposited data | ||
| PhIP-Seq data for MG540 antibodies; individuals MG540 and BG540 | This paper | PRJNA870920 |
| Experimental models: Cell lines | ||
| Human: FreeStyle 293F | Invitrogen | Cat#R790-07; RRID:CVCL_D603 |
| Human: CEM.NKR | NIH HIV Reagent Program | Cat#ARP-458; RRID:CVCL_X622 |
| 3T3-msCD40L Cells | NIH HIV Reagent Program | Cat#ARP-12535; RRID:CVCL_1H10 |
| TZM-bl cells | NIH HIV Reagent Program | Cat#ARP-8129; RRID:CVCL_B478 |
| 293T cells | ATCC | Cat#CRL-3216; RRID:CVCL_0063 |
| Oligonucleotides | ||
| Primers for antibody gene amplification and sequencing | Vigdorovich, 2016 and Simonich, 2019 | N/A |
| Primers for PhIP-Seq Illumina sequencing library preparation | Stoddard, 2021 | N/A |
| Recombinant DNA | ||
| Human Igγ1 expression vector | Tiller, 2008 | Addgene: 80795 |
| Human Igκ expression vector | Tiller, 2008 | Addgene: 80796 |
| Human Igλ expression vector | Tiller, 2008 | Addgene: 99575 |
| Software and algorithms | ||
| Partis | Ralph, 2016 | https://github.com/psathyrella/partis |
| phippery | N/A | https://github.com/matsengrp/phippery |
| FlowJo v10 | TreeStar | RRID:SCR_008520 |
| Excel | Microsoft | RRID:SCR_016137 |
| Prism 9 | GraphPad | RRID:SCR_002798 |
| NumPy | NumPy | RRID:SCR_008633 |
| Seaborn | Michael Waskom | RRID:SCR_018132 |
| pandas | pandas | RRID:SCR_018214 |
| MatPlotLib | MatPlotLib | RRID:SCR_008624 |
| Geneious Prime v.2022.1.1 | Dotmatics | RRID:SCR_010519 |
| Other | ||
| BD LSRFortessa X-50 Flow Cytometer | BD Biosciences | N/A |
| 1-Step Ultra TMB-ELISA Substrate Solution | Thermo Fisher | Cat#34029 |
| Gal-Screen™ β-Galactosidase Reporter Gene Assay System for Mammalian Cells | Thermo Fisher | Cat#T1028 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Julie Overbaugh (joverbau@fredhutch.org).
Materials availability
The heavy and light chain sequences of the MG540 antibodies are available on GenBank (see key resources table). The Env peptide phage display library used for epitope mapping studies is available upon request. Additional materials or reagents generated in this study are available from the lead contact upon request, although a materials transfer agreement may be required.
Experimental model and subject details
Human plasma and PBMC samples
Plasma and peripheral blood mononuclear cell (PBMC) samples were collected from mother MG540, an individual living with HIV who enrolled in the Nairobi Breastfeeding Trial (NBT), or her infant, BG540.44 At the time of PBMC and plasma sampling, individual MG540 was a 32-year-old pregnant, adult, and married cisgender female living with HIV at pregnancy week 34. At the time of plasma sampling (delivery), BG540 was a newborn male of Kenyan ancestry. Additional socioeconomic data was not collected as part of the original study. As the NBT was carried out before the availability of antiretroviral therapy (ART), NBT participants did not receive ART. The infecting virus of MG540 was clade A based on available envelope sequences.76 Approval to conduct this study was provided by Kenyatta National Hospital - University of Nairobi Ethics and Research Committee, the Fred Hutchinson Cancer Research Center Institutional Review Board, and/or the University of Washington Institutional Review Board. Study participants provided written informed consent prior to enrollment for use of their data and samples for future studies.
Cell lines
For RFADCC: CEM.NKR cells (RRID:CVCL_X622; originally derived from female human T-lymphoblastoid cells) were obtained from NIH HIV Reagent Program (HRP; Germantown, MD, catalog #ARP-458) and grown at 37°C in RPMI 1640 media with added penicillin (100 U/mL), streptomycin (100 μg/mL), amphotericin B (250 ng/mL), L-glutamine (2mM), and fetal bovine serum (10%; all from Thermo Fisher Scientific).
For antibody production: FreeStyle 293F cells (Thermo Fisher Scientific, Waltham, MA, catalog #R790-07; RRID:CVCL_D603; originally derived from female fetal cells) were obtained from Invitrogen and grown at 37°C in Freestyle 293 Expression Media (Thermo Fisher Scientific) according to the manufacturer’s instructions.
For pseudovirus production: 293T cells (RRID:CVCL_0063; transformed cell line originally derived from female human embryonic kidney cells) were obtained from ATCC (Manassas, VA, catalog #CRL-3216) and grown at 37°C in DMEM media with added penicillin (100 U/mL), streptomycin 100 μg/mL, Amphotericin B (250 ng/mL), and 10% fetal bovine serum (all from Thermo Fisher Scientific).
For neutralization assays: TZM-bl cells were obtained from the NIH HRP(catalog #ARP-8129; RRID:CVCL_B478; originally derived from female cancerous human cervical tissue) and grown at 37°C in DMEM media with added penicillin (100 U/mL), streptomycin 100 μg/mL, Amphotericin B (250 ng/mL), and 10% fetal bovine serum (all from Thermo Fisher Scientific).
3T3-msCD40L were obtained from the NIH HRP (catalog # ARP-12535; RRID:CVCL_1H10; originally derived from spontaneous immortalized male Mus musculus cells). All cell lines were not further authenticated in our hands.
Method details
B cell sorting
An MG540 PBMC sample from pregnancy week 34 (P34) was thawed as previously described after storage in liquid nitrogen.77 Cells were stained on ice for 30mins using a cocktail of anti-CD19-BV510, anti-IgD-FITC, anti-IgM-FITC, anti-CD3-BV711, anti-CD14-BV711, and anti-CD16-BV711. Cells were then washed once and resuspended in fluorescence-activated cell sorting (FACS) wash (1x PBS, 2% FBS) prior to loading onto a BD FACS Aria II cell sorter. Memory B cells were identified and sorted as CD3− CD14− CD16− CD19+ IgD− IgM−. B cells were sorted into B cell medium (Iscove’s modified Dulbecco’s medium [GIBCO, subsidiary of Thermo Fisher Scientific], 10% heat-inactivated low-IgG FBS [Life Technologies, subsidiary of Thermo Fisher Scientific], 5 ml GlutaMAX [Life Technologies], 1 ml MycoZap plus PR [Lonza, Basel, Switzerland]). Immediately following sorting, memory B cells were plated at 8 B cells in 55 μL per well into 11 x 384-well plates in B cell medium supplemented with 100 U mL−1 interleukin-2 (IL-2; Roche, Basel Switzerland), 50 ng mL−1 IL-21 (Invitrogen), and 1 x 105 cells mL−1 irradiated 3T3-CD40L feeder cells (ARP Cat #12535; RRID:CVCL_1H10) using a Tecan automated liquid handling system. Cultured B cells were incubated for 11 days at 37°C in a 5% CO2 incubator based on the protocol by Huang et al.61 At day 10 of incubation, IgG was detected by ELISA in 53% of a random sample of wells at >10ng mL−1 and 51% > 100ng mL−1.
B cell culture harvest and supernatant ELISA
On day 11, B cells culture supernatants were aspirated and divided among two additional 384-well plates (20 μL per duplicate) by a Tecan automated liquid handling system. The B cells remained in their original plates, were resuspended in 20 μL RNA storage buffer (15 mM Tris pH 8 and 10 U murine RNAse inhibitor, New England Biolabs [NEB], Ipswich, MA), and were frozen at -80°C until needed. An ELISA to detect HIV specificity in cell supernatants was adapted from a previously described protocol, as follows.40 Maxisorp 384-well plates (Nunc, subsidiary of Thermo Fisher Scientific) were coated with 1μg mL−1 BG505.W6M.ENV.B1 (BG505) gp120 (Cambridge Bio, Brookline, MA, Genbank #ABA61515.1), 0.5μg mL−1 C.ZA.1197MB gp41 ectodomain (Immune Tech, New York, NY, Genbank #AYA63234.1), and 1μg mL−1 BG505.SOSIP.664 T332N (from BG505.W6.C2 Env, Genbank #DQ208458.1; kindly provided by Kelly Lee) in 1x PBS overnight at 4°C. Plates were washed four times between steps with PBS-0.05% Tween 20 (Thermo Fisher Scientific; PBST) wash buffer. All samples/reagents used in subsequent steps were diluted in B cell media (as above). Plates were blocked with B cell media for 1 hour at 37°C. B cell supernatants were then applied and incubated at 37°C for two hours. Next, Goat anti-human IgG-HRP (Sigma-Aldrich, St. Louis, MO) was applied at 1:2500 dilution for 1 hour at 37°C. After a final wash, 1-Step TMB-Ultra (Thermo Fisher Scientific) was added to the wells. The reaction was stopped after 15 min using 1M H2SO4 (Sigma-Aldrich). Absorbance was measured at 450nm optical density. Positive ELISA activity was defined was an absorbance greater than twice the background activity of wells incubated with B cell media alone.
Rapid and fluorometric ADCC assay
The rapid and fluorometric ADCC (RFADCC) assay was carried out as described previously.48,78 Briefly, CEM.NKR cells were stained with PKH26 cell linker dye (Sigma-Aldrich) and CFSE cytosolic dye (Vybrant CFDA-SE Cell Tracer Kit, Invitrogen). Double-stained cells were then coated with gp120 or gp41 at a ratio of 1.5μg antigen per 100,000 cells for 1 hour at room temperature. Coated cells were washed and a total of 5,000 target cells were added to wells containing 100μL of plasma diluted at 1:5000 in RPMI media or control mAbs at 100 – 500 ng mL−1. After 15 min, PBMCs from HIV negative donors were added at a ratio of 50:1 effector to target cells. RFADCC activity was allowed to occur for four hours at 37°C before cells were washed and fixed in 2% paraformaldehyde in PBS (Santa Cruz Biotechnology, Dallas, TX). Data were acquired via flow cytometry (BD Symphony or Fortessa X50; BD Biosciences, Franklin Lakes, NJ). PKH and CFSE were detected in the PE and FITC channels, respectively. Data were analyzed using FlowJo (v.10.1, Treestar). ADCC was defined as the percentage of PKH+, CFSE- cells out of total PKH+ cells after subtracting detected “background” activity mediated against uncoated target cells, which was standardized to be 3-5% during analysis in FlowJo software. ADCC activity was normalized to that mediated by Anti-HIV Immune Globulin (HIVIG, NIH ARP, Catalog #3957). For serial dilution experiments, gp120-specific and gp41-specific mAbs were tested at 0.05 – 750 ng mL−1and 0.15 – 2500 ng mL−1, respectively. The concentration at which 50% of maximal ADCC activity was detected (the 50% effective concentration; EC50) was determined for each mAb by fitting a sigmoidal dose-response curve to the data and interpolating.
Multiplex supernatant RFADCC assay
The RFADCC assay described above was adapted for use with B cell culture supernatants and multiple antigens. To simultaneously detect ADCC mediated against gp120 and gp41 coated cells, CEM.NKR cells were first labelled with CFSE to reduce batch effects. CFSE-labelled cells were then labelled with PKH26 or CellVue Claret cell linker dye (Sigma-Aldrich), which has similar properties to PKH26 and is spectrally compatible with both PKH26 and CFSE.79,80 PKH-26- and CFSE-labelled cells were coated with ZA1197 gp41, whereas CellVue Claret- and CFSE-labelled cells were coated with BG505 gp120. The remaining protocol steps of the protocol were followed as described above. CellVue Claret was detected in the APC channel. Gp41-specific ADCC was defined as the percentage of PKH+ CFSE- CellVue Claret- cells after subtracting background activity. Gp120-specific ADCC was defined as the percentage of CellVue Claret+ CFSE- PKH- cells after subtracting background activity. Positive ADCC was defined as activity greater than twice the background.
Reconstruction of antibodies
Wells demonstrating HIV-specific ELISA activity and gp120-or gp41-specific RFADCC activity were selected for antibody gene amplification and cloning. Plates containing frozen B cells for wells of interest were thawed on ice. Reverse transcription-PCR amplification of IgG heavy and light chain variable regions was performed as previously described, followed by nested chain-specific amplification PCRs.61 Two distinct primer sets were used to minimize PCR bias and maximize recovery of HIV-specific antibodies.81,82 Functional sequences were identified from second round amplified sequences using IMGT V-QUEST.83 Where sufficient sequencing for the 5’ constant region was available, we determined IgG subclass; all heavy chain sequences (8 of 17) were identified as IgG1 subclass. Amplified variable region sequences were cloned into corresponding IgG1, IgK, or IgL expression vectors.61,84 The Freestyle MAX system (Thermo Fisher Scientific) was used to co-transfect paired heavy and light chain expression plasmids for sequences isolated from the same well, and IgG was purified as described.40 Where multiple possible heavy and light chains pairs were present, all possible combinations were generated. To produce G236R/L328R (GRLR) FcγR null binding variants of select mAbs, heavy chain variable regions were cloned into an IgG1 expression vector containing the G236R/L328R substitutions.
Sequence analysis and clonal inference
Second round nested PCR sequences for heavy and light chains were grouped into clonal families (stemming from a single rearrangement event) and annotated with the B cell immunoglobulin analysis program, partis, using the paired clustering option (https://github.com/psathyrella/partis and https://arxiv.org/abs/2203.11367). This uses a Hidden Markov Model, together with a variety of more heuristic, but faster, steps, to first group together sequences from among each single (heavy or light) chain. It then incorporates pairing information by splitting apart such single-chain clusters whose partner clusters are incompatible.
Pseudovirus production and neutralization assays
TZM-bl-based neutralization assays and production of pseudoviruses carrying the Q23Δenv backbone were performed as previously described.85 Pseudoviruses were generated by co-transfecting plasmids containing env chimeras with an env-deficient sub-type A proviral plasmid (Q23ΔEnv) at a 1:2 mass ratio into 3 x 106 293T cells plated in a T-75 flask 24 hours before transfection. Four micrograms of total DNA was mixed with 12μL of Fugene6 transfection reagent (Roche). Forty-eight hours post-transfection, the supernatant was collected, sterile filtered, and then used to infected TZM-bl cells in the presence of DEAE-dextran (10μg mL−1). Neutralization assays were carried out by adding 500 infectious pseudovirus particles in a volume of 25 μL to an equal volume of serially diluted plasma or antibody sample at 37°C for 60 mins. TZM-bl cells (1 x 104) in 100 μL DMEM were then added to wells. Forty-eight hours post-infection, luminescence levels were measured using the Gal-Screen system (Thermo Fisher Scientific). Percent neutralization was calculated as the reduction in luminescence of each pseudovirus incubated in the presence of plasma or antibody compared to cells and virus alone. For MG540 plasma, the NT50 represents the reciprocal dilution of plasma that results in 50% reduction of virus infectivity. For mAbs, the IC50 represents the final concentration in μg mL−1which led to 50% neutralization of the indicated virus. Reported NT50 and IC50 values are the average of two experiments each performed in technical duplicate.
Phage display immunoprecipitation sequencing
To determine the targets of mAbs with linear epitopes, we used phage display immunoprecipitation sequencing (PhIP-Seq), as previously described.48,86,87 An oligonucleotide pool was generated for the env ectodomain and transmembrane domain (HXB2 AA 30-704) from six HIV-1 strains on GenBank: 9032.08_A1 (EU576114; B41), BF520.W14M.C2 (KX168094; BF520), BG505.W6.C2 (DQ208458; BG505), BL035.W6M.C1 (DQ208480; BL035), QA013.70I.H1 (FJ866134; QA013), and ZA1197.MB (AY463234; ZA1197). The BF520, BG505, and BL035 strains were isolated early in infection from infants enrolled in the NBT cohort. Oligonucleotides encoded 38 amino acid tiles, with 36 amino acid overlaps between adjacent tiles. Amplified phage at 2 x 105-fold representation were incubated with each sample mAb (10ng total at 1μg mL-1) in technical duplicate. After overnight incubation, phage were immunoprecipitated using protein A- and-G-coated beads (Pierce, a subsidiary of Thermo Fisher Scientific). Samples were then prepared for multiplexed sequencing on an Illumina MiSeq or HiSeq 2500 with 125bp single end reads using the rapid run setting, as described.88 Demultiplexing, read alignment, and enrichment calculations were performed as described.88 Demultiplexing was performed using the Illumina MiSeq reporter software. Index creation and alignment were performed with Bowtie 1.3, with up to 2 mismatches allowed after trimming 11 bases from the 3’ end of each 125bp read to match the 114bp oligonucleotides encoding the peptides in the library. We used Samtools to produce counts for each peptide counts using the idxstats module. Enrichment was calculated as the fold change in frequency of a given peptide within each sample compared to the library only control. See ‘data and code availability’ for more information.
Gp120, gp41, and SOSIP ELISAs
ELISAs using plates coated with BG505 gp120, ZA1197 gp41, BG505 T332N SOSIP, and CNE8 SOSIP were performed similarly to the supernatant ELISAs described above. 384-well MaxiSorp plates (Nunc) were coated with single gp120 or SOSIP antigens at 1μg mL−1 or gp41 protein at 0.5μg mL−1. All subsequent steps were performed using 10% non-fat milk (RPI, Mt. Prospect, IL) in PBST. mAbs were tested in serial dilutions at 31.25 – 1000ng mL−1.
Competition ELISA
To determine the epitope of mAbs that did not map to a linear epitope via PhIP-Seq, we used antibodies with known epitopes in a competition binding assay, as previously described.40,89 Briefly, 384-well MaxiSorp plates were coated with either BG505 gp120 or ZA1197 gp41, as detailed above. Plates were then blocked with 3% BSA in PBST. Serial dilutions of antibodies of interested were added at 9.76 ng mL−1 to 10μg mL−1 and incubated at 37°C for 15 mins. Biotinylated (BT) antibodies were then added without washing and samples were incubated at 37°C for 45 mins. Next, plates were incubated with Pierce High Sensitivity Streptavidin-HRP (Thermo Fisher Scientific) at 1:10000 dilution for 1 hour at 37°C. After a final wash, 1-Step TMB-Ultra (Thermo Fisher Scientific) was added to the wells. The reaction was stopped 1-10 minutes later using 1M H2SO4 (Sigma-Aldrich). Absorbance was measured at 450nm optical density. Percent inhibition was calculated as: 100 ∗ [(AUC in presence of mAb of interest)/(scaled average absorbance of no mAb control and flu-specific mAb FI6v3)]. Antibodies were biotinylated using the EZ-Link Sulfo-NHS Biotinylation Kit (Thermo Fisher Scientific), according to the manufacturer’s instructions. Empirically determined optimal concentrations of the biotinylated mAbs were: 100ng mL−1 (MG540.90, 167-7, QA255.067), 250ng mL−1 (MG540.37, QA255.006), 500ng mL−1 (VRC01), and 750ng mL−1 (A32, C11, 17b).
Flow cytometry-based analysis of cell surface staining and ADCC
Cell surface mAb binding and ADCC responses mediated by mAbs (5μg mL−1) were measured at 48h post-infection as previously described.90,91 Cells infected with HIV-1 primary isolates (CH58 WT, CH58 nef- vpu-, or BG505 T332N) were stained intracellularly for HIV-1 p24, using the Cytofix/Cytoperm fixation/permeabilization kit (BD Biosciences, Franklin Lakes, NJ) and a fluorescent anti-p24 mAb (phycoerythrin [PE]-conjugated anti-p24, clone KC57; Beckman Coulter/Immunotech) at a 1:100 dilution. BG505 T332N infected cells were also stained with anti-CD4-FITC mAb (FITC anti-human CD4 Antibody, Biolegend, San Diego, CA) at a 1:100 dilution, and p24+CD4-low cells were chosen for the analysis. Infected cells were identified by gating the living cell population on the basis of the viability dye staining. The percentage of cytotoxicity was calculated using the following formula:[(% of p24+ cells in targets plus effectors) – (%p24+ cells in targets plus effectors plus Abs)]/(% of p24+ cells in targets). The target cells and effector cells without antibody were used to establish the ADCC threshold. For cell surface antibody staining, primary CD4+T cells were incubated with antibody samples at 5μg mL−1 for 30mins at 37°C. Cells were then washed with PBS and stained with goat anti-human (AlexaFluor 647; Jackson ImmunoResearch, West Grove, PA) secondary antibody for 20 min in PBS. A32, 17b, and 3BNC117 were used as positive controls. Background levels of binding were measured on mock infected cells. A GRLR variant of 3BNC117 was used as a negative control for the ADCC assay.
Competition ADCC assay
The competition ADCC assay was performed using a modified version of the RFADCC assay, as previously described.92 Target cells were coated with either BG505 gp120 or ZA1197 gp41, as detailed above. Coated target cells were added to wells containing 50 μL total of a GRLR variant of a competitor antibody or an equivalent volume of media.60 The solution was mixed by pipetting up and down repeatedly and the cells were incubated for 15mins at RT in the dark, allowing for competitor antibodies to bind. Next, 50 μL of plasma at a previously optimized dilution (1:5000 for gp120 and 1:2000 for gp41) or an indicated mAb at the indicated effective concentration (EC) was added to the wells. Wells were mixed again and incubated for 15mins at RT in the dark before adding effector cells. The concentration of GRLR mAb added was empirically determined via serial dilution by testing for complete abrogation of wildtype antibody ADCC when run at the EC95 concentration. GRLR variants were added at the following concentrations: 12.5μg mL−1 (MG540.7, −17, −50, −63, −82, and −90), 25μg mL-1 (MG540.37, -56, and -86). For wells containing multiple GRLR variants, each GRLR mAb was added at a higher working concentration so that the final concentration in 50μL remained the same optimal final concentration above. Percent ADCC inhibition was calculated as: 100 ∗ [(plasma ADCC in the presence of the indicated GRLR mAbs)/(plasma ADCC in the absence of GRLR mAbs)]
Quantification and statistical analysis
Raw data was processed in Microsoft Excel or GraphPad Prism v9. Graphs were generated using GraphPad Prism. PhiP-Seq heatmaps were generated with the NumPy, pandas, Matplotlib, and seaborn Python packages. Except where noted, all data represent the mean of two biological replicates, with two technical replicates in each experiment. Data were normalized and/or averaged across replicates using Microsoft Excel or GraphPad Prism. Enrichment was calculated as previously described for PhIP-Seq data.56 Additional details can be found in the figure legends.
Acknowledgments
We thank the participants and staff of the Nairobi Breastfeeding Trial. This work was supported by NIH R01 AI076105 to J.O., R01 AI146028 to F.M., and by training award T32 AI083203 and NIH F30 AI165112 to Z.Y. Dr. Matsen is an Investigator of the Howard Hughes Medical Institute. Scientific Computing Infrastructure at Fred Hutch funded by ORIP grant S10OD028685. A.F. is supported a Canada Research Chair on Retroviral Entry no. RCHS0235 950-232424. We thank Halima Medjahed for assistance processing PBMCs for the HIV-infected cell ADCC experiments. The following reagents was obtained through the NIH HIV Reagent Program, Division of AIDS, NIAID, NIH: Polyclonal Anti-Human Immunodeficiency Virus Immune Globulin, Pooled Inactivated Human Sera, ARP-3957, contributed by NABI and National Heart Lung and Blood Institute (Dr. Luiz Barbosa); Anti-Human Immunodeficiency Virus 1 (HIV-1) gp41 Monoclonal Antibody (167-D IV; “167-7”), ARP-11681, contributed by Dr. Susan Zolla-Pazner; CEM.NKR Cells, ARP-458, contributed by Dr. Peter Cresswell. 3BNC117 was kindly provided by Dr. Michel Nussenzweig. Graphical abstract created with Biorender.com.
Author contributions
Conceptualization, J.O. and Z.Y.; Methodology, Z.Y., S.D., L.D., D.R., and K.S.; Software, D.R., K.S., and F.M.; Validation, Z.Y., S.D., D.R., and K.S.; Formal analysis, Z.Y., L.D., V.C., S.D., K.S., D.R., and L.M.; Investigation, Z.Y., V.C., and S.D.; Resources, R.N., A.F., F.M., and J.O.; Data curation, Z.Y., V.C., S.D., D.R., K.S., and L.M.; Writing – original draft preparation, Z.Y. and J.O.; Writing – review and editing, Z.Y., L.D., A.F., F.M., and J.O.; Visualization, Z.Y. and K.S.; Supervision, Z.Y., A.F., F.M., and J.O.; Project administration, Z.Y. and J.O.; Funding acquisition, J.O.; Comments on the manuscript, all authors.
Declaration of interests
The authors declare no competing interests.
Inclusion and diversity
One or more of the authors of this paper self-identifies as a gender minority in their field of research. One or more of the authors of this paper self-identifies as a member of the LGBTQIA+ community. While citing references scientifically relevant for this work, we also actively worked to promote gender balance in our reference list.
Published: April 26, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2023.106762.
Supplemental information
Data and code availability
-
•
The PhIP-Seq sequencing dataset generated during this study is publicly available in the Short Read Archive, with the BioProject ID listed in the key resources table. MG540 antibody sequence accession numbers are listed in the key resources table. Pre-processed enrichment data for PhIP-Seq experiments will be shared by the lead contact upon request.
-
•
This paper does not report original code. The python API “phippery” was used to process the demultiplexed sequencing data, align reads to the reference oligonucleotide sequences, and calculate enrichment. “phippery” is available at (https://github.com/matsengrp/phippery). Clonal family inference was performed with the partis software package (available at https://github.com/psathyrella/partis/) paired clustering method, treating wells as droplets.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.Forthal D.N., Finzi A. Antibody-dependent cellular cytotoxicity in HIV infection. AIDS. 2018;32:2439–2451. doi: 10.1097/QAD.0000000000002011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Parsons M.S., Chung A.W., Kent S.J. Importance of Fc-mediated functions of anti-HIV-1 broadly neutralizing antibodies. Retrovirology. 2018;15:58. doi: 10.1186/s12977-018-0438-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brady J.M., Phelps M., MacDonald S.W., Lam E.C., Nitido A., Parsons D., Boutros C.L., Deal C.E., Garcia-Beltran W.F., Tanno S., et al. Antibody-mediated prevention of vaginal HIV transmission is dictated by IgG subclass in humanized mice. Sci. Transl. Med. 2022;14:eabn9662. doi: 10.1126/scitranslmed.abn9662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Crowley A.R., Ackerman M.E. Mind the gap: how interspecies variability in IgG and its receptors may complicate comparisons of human and non-human primate effector function. Front. Immunol. 2019;10:697. doi: 10.3389/fimmu.2019.00697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lewis G.K. Role of Fc-mediated antibody function in protective immunity against HIV-1. Immunology. 2014;142:46–57. doi: 10.1111/imm.12232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Madhavi V., Wren L.H., Center R.J., Gonelli C., Winnall W.R., Parsons M.S., Kramski M., Kent S.J., Stratov I. Breadth of HIV-1 Env-specific antibody-dependent cellular cytotoxicity: relevance to global HIV vaccine design. AIDS. 2014;28:1859–1870. doi: 10.1097/QAD.0000000000000310. [DOI] [PubMed] [Google Scholar]
- 7.Wren L.H., Chung A.W., Isitman G., Kelleher A.D., Parsons M.S., Amin J., Cooper D.A., Stratov I., Navis M., Kent S.J., Stratov I., ADCC study collaboration investigators Specific antibody-dependent cellular cytotoxicity responses associated with slow progression of HIV infection. Immunology. 2013;138:116–123. doi: 10.1111/imm.12016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Milligan C., Richardson B.A., John-Stewart G., Nduati R., Overbaugh J. Passively acquired antibody-dependent cellular cytotoxicity (ADCC) activity in HIV-infected infants is associated with reduced mortality. Cell Host Microbe. 2015;17:500–506. doi: 10.1016/j.chom.2015.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mabuka J., Nduati R., Odem-Davis K., Peterson D., Overbaugh J. HIV-specific antibodies capable of ADCC are common in breastmilk and are associated with reduced risk of transmission in women with high viral loads. PLoS Pathog. 2012;8:e1002739. doi: 10.1371/journal.ppat.1002739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tranchat C., Van de Perre P., Simonon-Sorel A., Karita E., Benchaïb M., Lepage P., Desgranges C., Boyer V., Trépo C. Maternal humoral factors associated with perinatal human lmmunodeficiency virus type-l transmission in a cohort from kigali, Rwanda, 1988-1994. J. Infect. 1999;39:213–220. doi: 10.1016/s0163-4453(99)90052-x. [DOI] [PubMed] [Google Scholar]
- 11.Ljunggren K., Moschese V., Broliden P.-A., Giaquinto C., Quinti I., Fenyö E.M., Wahren B., Rossi P., Jondal M. Antibodies mediating cellular cytotoxicity and neutralization correlate with a better clinical stage in children born to human immunodeficiency virus-infected mothers. J. Infect. Dis. 1990;161:198–202. doi: 10.1093/infdis/161.2.198. [DOI] [PubMed] [Google Scholar]
- 12.Broliden K., Sievers E., Tovo P.A., Moschese V., Scarlatti G., Broliden P.A., Fundaro C., Rossi P. Antibody-dependent cellular cytotoxicity and neutralizing activity in sera of HIV-1-infected mothers and their children. Clin. Exp. Immunol. 1993;93:56–64. doi: 10.1111/j.1365-2249.1993.tb06497.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baum L.L., Cassutt K.J., Knigge K., Khattri R., Margolick J., Rinaldo C., Kleeberger C.A., Nishanian P., Henrard D.R., Phair J. HIV-1 gp120-specific antibody-dependent cell-mediated cytotoxicity correlates with rate of disease progression. J. Immunol. 1996;157:2168–2173. doi: 10.4049/jimmunol.157.5.2168. [DOI] [PubMed] [Google Scholar]
- 14.Lambotte O., Ferrari G., Moog C., Yates N.L., Liao H.-X., Parks R.J., Hicks C.B., Owzar K., Tomaras G.D., Montefiori D.C., et al. Heterogeneous neutralizing antibody and antibody-dependent cell cytotoxicity responses in HIV-1 elite controllers. AIDS. 2009;23:897–906. doi: 10.1097/QAD.0b013e328329f97d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yaffe Z.A., Naiman N.E., Slyker J., Wines B.D., Richardson B.A., Hogarth P.M., Bosire R., Farquhar C., Ngacha D.M., Nduati R., et al. Improved HIV-positive infant survival is correlated with high levels of HIV-specific ADCC activity in multiple cohorts. Cell Rep. Med. 2021;2:100254. doi: 10.1016/j.xcrm.2021.100254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thomas A.S., Moreau Y., Jiang W., Isaac J.E., Ewing A., White L.F., Kourtis A.P., Sagar M. Pre-existing infant antibody-dependent cellular cytotoxicity associates with reduced HIV-1 acquisition and lower morbidity. Cell Rep. Med. 2021;2:100412. doi: 10.1016/j.xcrm.2021.100412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Curtis A.D., Saha P.T., Dennis M., Berendam S.J., Ramasubramanian P., Cross K.A., Alam S.M., Ferrari G., Kozlowski P.A., Fouda G.G., et al. Vaccine-Induced, high-magnitude HIV env-specific antibodies with fc-mediated effector functions are insufficient to protect infant rhesus macaques against oral SHIV infection. mSphere. 2022;7:e0083921. doi: 10.1128/msphere.00839-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mielke D., Bandawe G., Pollara J., Abrahams M.-R., Nyanhete T., Moore P.L., Thebus R., Yates N.L., Kappes J.C., Ochsenbauer C., et al. Antibody-dependent cellular cytotoxicity (ADCC)-Mediating antibodies constrain neutralizing antibody escape pathway. Front. Immunol. 2019;10:2875. doi: 10.3389/fimmu.2019.02875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen X., Lin M., Qian S., Zhang Z., Fu Y., Xu J., Han X., Ding H., Dong T., Shang H., Jiang Y. The early antibody-dependent cell-mediated cytotoxicity response is associated with lower viral set point in individuals with primary HIV infection. Front. Immunol. 2018;9:2322. doi: 10.3389/fimmu.2018.02322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ruiz M.J., Salido J., Abusamra L., Ghiglione Y., Cevallos C., Damilano G., Rodriguez A.M., Trifone C., Laufer N., Giavedoni L.D., et al. Evaluation of different parameters of humoral and cellular immune responses in HIV serodiscordant heterosexual couples: humoral response potentially implicated in modulating transmission rates. EBioMedicine. 2017;26:25–37. doi: 10.1016/j.ebiom.2017.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rerks-Ngarm S., Pitisuttithum P., Nitayaphan S., Kaewkungwal J., Chiu J., Paris R., Premsri N., Namwat C., de Souza M., Adams E., et al. Vaccination with ALVAC and AIDSVAX to prevent HIV-1 infection in Thailand. N. Engl. J. Med. 2009;361:2209–2220. doi: 10.1056/NEJMoa0908492. [DOI] [PubMed] [Google Scholar]
- 22.Haynes B.F., Gilbert P.B., McElrath M.J., Zolla-Pazner S., Tomaras G.D., Alam S.M., Evans D.T., Montefiori D.C., Karnasuta C., Sutthent R., et al. Immune-correlates analysis of an HIV-1 vaccine efficacy trial. N. Engl. J. Med. 2012;366:1275–1286. doi: 10.1056/NEJMoa1113425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gray G.E., Bekker L.-G., Laher F., Malahleha M., Allen M., Moodie Z., Grunenberg N., Huang Y., Grove D., Prigmore B., et al. Vaccine efficacy of ALVAC-HIV and bivalent subtype C gp120–MF59 in adults. N. Engl. J. Med. 2021;384:1089–1100. doi: 10.1056/NEJMoa2031499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Milligan C., Slyker J.A., Overbaugh J. Advances in Virus Research. Elsevier; 2018. The role of immune responses in HIV mother-to-child transmission; pp. 19–40. [DOI] [PubMed] [Google Scholar]
- 25.Parekh B.S., Shaffer N., Coughlin R., Hung C.-H., Krasinski K., Abrams E., Bamji M., Thomas P., Hutson D., Schochetman G., et al. Dynamics of maternal IgG antibody decay and HIV-specific antibody synthesis in infants born to seropositive mothers. AIDS Res. Hum. Retrovir. 1993;9:907–912. doi: 10.1089/aid.1993.9.907. [DOI] [PubMed] [Google Scholar]
- 26.Kourtis A.P., Bulterys M., Nesheim S.R., Lee F.K. Understanding the timing of HIV transmission from mother to infant. JAMA. 2001;285:709–712. doi: 10.1001/jama.285.6.709. [DOI] [PubMed] [Google Scholar]
- 27.Yaffe Z.A., Overbaugh J. HIV-1 protection: antibodies move in for the kill. Cell Rep. Med. 2021;2:100428. doi: 10.1016/j.xcrm.2021.100428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lynch J.B., Nduati R., Blish C.A., Richardson B.A., Mabuka J.M., Jalalian-Lechak Z., John-Stewart G., Overbaugh J. The breadth and potency of passively acquired human immunodeficiency virus type 1-specific neutralizing antibodies do not correlate with the risk of infant infection. J. Virol. 2011;85:5252–5261. doi: 10.1128/JVI.02216-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pollara J., Bonsignori M., Moody M.A., Pazgier M., Haynes B.F., Ferrari G. Epitope specificity of human immunodeficiency virus-1 antibody dependent cellular cytotoxicity [ADCC] responses. CHR. 2013;11:378–387. doi: 10.2174/1570162X113116660059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Butler A.L., Fischinger S., Alter G. The antibodiome—mapping the humoral immune response to HIV. Curr. HIV AIDS Rep. 2019;16:169–179. doi: 10.1007/s11904-019-00432-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guan Y., Pazgier M., Sajadi M.M., Kamin-Lewis R., Al-Darmarki S., Flinko R., Lovo E., Wu X., Robinson J.E., Seaman M.S., et al. Diverse specificity and effector function among human antibodies to HIV-1 envelope glycoprotein epitopes exposed by CD4 binding. Proc. Natl. Acad. Sci. USA. 2013;110:E69–E78. doi: 10.1073/pnas.1217609110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tyler D.S., Stanley S.D., Zolla-Pazner S., Gorny M.K., Shadduck P.P., Langlois A.J., Matthews T.J., Bolognesi D.P., Palker T.J., Weinhold K.J. Identification of sites within gp41 that serve as targets for antibody-dependent cellular cytotoxicity by using human monoclonal antibodies. J. Immunol. 1990;145:3276–3282. doi: 10.4049/jimmunol.145.10.3276. [DOI] [PubMed] [Google Scholar]
- 33.Xu J.-Y., Gorny M.K., Palker T., Karwowska S., Zolla-Pazner S. Epitope mapping of two immunodominant domains of gp4l, the transmembrane protein of human immunodeficiency virus type 1, using ten human monoclonal antibodies. J. Virol. 1991;65:4832–4838. doi: 10.1128/JVI.65.9.4832-4838.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gorny M.K., Zolla-Pazner S. Recognition by human monoclonal antibodies of free and complexed peptides representing the prefusogenic and fusogenic forms of human immunodeficiency virus type 1 gp41. J. Virol. 2000;74:6186–6192. doi: 10.1128/JVI.74.13.6186-6192.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Earl P.L., Broder C.C., Doms R.W., Moss B. Epitope map of human immunodeficiency virus type 1 gp41 derived from 47 monoclonal antibodies produced by immunization with oligomeric envelope protein. J. Virol. 1997;71:2674–2684. doi: 10.1128/jvi.71.4.2674-2684.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pietzsch J., Scheid J.F., Mouquet H., Seaman M.S., Broder C.C., Nussenzweig M.C. Anti-gp41 antibodies cloned from HIV-infected patients with broadly neutralizing serologic activity. J. Virol. 2010;84:5032–5042. doi: 10.1128/JVI.00154-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Simonich C., Shipley M.M., Doepker L., Gobillot T., Garrett M., Cale E.M., Hennessy B., Itell H., Chohan V., Doria-Rose N., et al. A diverse collection of B cells responded to HIV infection in infant BG505. Cell Rep. Med. 2021;2:100314. doi: 10.1016/j.xcrm.2021.100314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Doepker L.E., Simonich C.A., Ralph D., Shipley M.M., Garrett M., Gobillot T., Vigdorovich V., Sather D.N., Nduati R., Matsen F.A., Overbaugh J.M. Diversity and function of maternal HIV-1-Specific antibodies at the time of vertical transmission. J. Virol. 2020;94:e01594-19. doi: 10.1128/JVI.01594-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Richard J., Pacheco B., Gohain N., Veillette M., Ding S., Alsahafi N., Tolbert W.D., Prévost J., Chapleau J.-P., Coutu M., et al. Co-Receptor binding site antibodies enable CD4-mimetics to expose conserved anti-cluster A ADCC epitopes on HIV-1 envelope glycoproteins. EBioMedicine. 2016;12:208–218. doi: 10.1016/j.ebiom.2016.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Williams K.L., Cortez V., Dingens A.S., Gach J.S., Rainwater S., Weis J.F., Chen X., Spearman P., Forthal D.N., Overbaugh J. HIV-Specific CD4-induced antibodies mediate broad and potent antibody-dependent cellular cytotoxicity activity and are commonly detected in plasma from HIV-infected humans. EBioMedicine. 2015;2:1464–1477. doi: 10.1016/j.ebiom.2015.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Anand S.P., Prévost J., Baril S., Richard J., Medjahed H., Chapleau J.-P., Tolbert W.D., Kirk S., Smith A.B., Wines B.D., et al. Two families of env antibodies efficiently engage fc-gamma receptors and eliminate HIV-1-Infected cells. J. Virol. 2019;93:e01823-18. doi: 10.1128/JVI.01823-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bomsel M., Tudor D., Drillet A.-S., Alfsen A., Ganor Y., Roger M.-G., Mouz N., Amacker M., Chalifour A., Diomede L., et al. Immunization with HIV-1 gp41 subunit virosomes induces mucosal antibodies protecting nonhuman primates against vaginal SHIV challenges. Immunity. 2011;34:269–280. doi: 10.1016/j.immuni.2011.01.015. [DOI] [PubMed] [Google Scholar]
- 43.Moog C., Dereuddre-Bosquet N., Teillaud J.-L., Biedma M.E., Holl V., Van Ham G., Heyndrickx L., Van Dorsselaer A., Katinger D., Vcelar B., et al. Protective effect of vaginal application of neutralizing and nonneutralizing inhibitory antibodies against vaginal SHIV challenge in macaques. Mucosal Immunol. 2014;7:46–56. doi: 10.1038/mi.2013.23. [DOI] [PubMed] [Google Scholar]
- 44.Nduati R., John G., Mbori-Ngacha D., Richardson B., Overbaugh J., Mwatha A., Ndinya-Achola J., Bwayo J., Onyango F.E., Hughes J., Kreiss J. Effect of breastfeeding and formula feeding on transmission of HIV-1: a randomized clinical trial. Obstet. Gynecol. Surv. 2000;55:479–481. doi: 10.1097/00006254-200008000-00010. [DOI] [PubMed] [Google Scholar]
- 45.Milligan C., Omenda M.M., Chohan V., Odem-Davis K., Richardson B.A., Nduati R., Overbaugh J. Maternal neutralization-resistant virus variants do not predict infant HIV infection risk. mBio. 2016;7:e02221-15. doi: 10.1128/mBio.02221-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rousseau C.M., Nduati R.W., Richardson B.A., Steele M.S., John-Stewart G.C., Mbori-Ngacha D.A., Kreiss J.K., Overbaugh J. Longitudinal analysis of human immunodeficiency virus type 1 RNA in breast milk and of its relationship to infant infection and maternal disease. J. Infect. Dis. 2003;187:741–747. doi: 10.1086/374273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kramski M., Schorcht A., Johnston A.P.R., Lichtfuss G.F., Jegaskanda S., De Rose R., Stratov I., Kelleher A.D., French M.A., Center R.J., et al. Role of monocytes in mediating HIV-specific antibody-dependent cellular cytotoxicity. J. Immunol. Methods. 2012;384:51–61. doi: 10.1016/j.jim.2012.07.006. [DOI] [PubMed] [Google Scholar]
- 48.Williams K.L., Stumpf M., Naiman N.E., Ding S., Garrett M., Gobillot T., Vézina D., Dusenbury K., Ramadoss N.S., Basom R., et al. Identification of HIV gp41-specific antibodies that mediate killing of infected cells. PLoS Pathog. 2019;15:e1007572. doi: 10.1371/journal.ppat.1007572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Doepker L.E., Danon S., Harkins E., Ralph D.K., Yaffe Z., Garrett M.E., Dhar A., Wagner C., Stumpf M.M., Arenz D., et al. Development of antibody-dependent cell cytotoxicity function in HIV-1 antibodies. Elife. 2021;10:e63444. doi: 10.7554/eLife.63444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Moore J.P., McCutchan F.E., Poon S.W., Mascola J., Liu J., Cao Y., Ho D.D. Exploration of antigenic variation in gp120 from clades A through F of human immunodeficiency virus type 1 by using monoclonal antibodies. J. Virol. 1994;68:8350–8364. doi: 10.1128/jvi.68.12.8350-8364.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Moore J.P., Willey R.L., Lewis G.K., Robinson J., Sodroski J. Immunological evidence for interactions between the first, second, and fifth conserved domains of the gp120 surface glycoprotein of human immunodeficiency virus type 1. J. Virol. 1994;68:6836–6847. doi: 10.1128/jvi.68.11.6836-6847.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kramski M., Parsons M.S., Stratov I., Kent S.J. HIV-specific antibody immunity mediated through NK cells and monocytes. Curr. HIV Res. 2013;11:388–406. doi: 10.2174/1570162x113116660061. [DOI] [PubMed] [Google Scholar]
- 53.Salazar-Gonzalez J.F., Salazar M.G., Keele B.F., Learn G.H., Giorgi E.E., Li H., Decker J.M., Wang S., Baalwa J., Kraus M.H., et al. Genetic identity, biological phenotype, and evolutionary pathways of transmitted/founder viruses in acute and early HIV-1 infection. J. Exp. Med. 2009;206:1273–1289. doi: 10.1084/jem.20090378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Prévost J., Richard J., Medjahed H., Alexander A., Jones J., Kappes J.C., Ochsenbauer C., Finzi A. Incomplete downregulation of CD4 expression affects HIV-1 env conformation and antibody-dependent cellular cytotoxicity responses. J. Virol. 2018;92:e00484-18. doi: 10.1128/JVI.00484-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Scheid J.F., Mouquet H., Ueberheide B., Diskin R., Klein F., Oliveira T.Y.K., Pietzsch J., Fenyo D., Abadir A., Velinzon K., et al. Sequence and structural convergence of broad and potent HIV antibodies that mimic CD4 binding. Science. 2011;333:1633–1637. doi: 10.1126/science.1207227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Garrett M.E., Itell H.L., Crawford K.H.D., Basom R., Bloom J.D., Overbaugh J. Phage-DMS: a comprehensive method for fine mapping of antibody epitopes. iScience. 2020;23:101622. doi: 10.1016/j.isci.2020.101622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dey B., Svehla K., Xu L., Wycuff D., Zhou T., Voss G., Phogat A., Chakrabarti B.K., Li Y., Shaw G., et al. Structure-based stabilization of HIV-1 gp120 enhances humoral immune responses to the induced Co-receptor binding site. PLoS Pathog. 2009;5:e1000445. doi: 10.1371/journal.ppat.1000445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Richard J., Veillette M., Brassard N., Iyer S.S., Roger M., Martin L., Pazgier M., Schön A., Freire E., Routy J.-P., et al. CD4 mimetics sensitize HIV-1-infected cells to ADCC. Proc. Natl. Acad. Sci. USA. 2015;112:E2687–E2694. doi: 10.1073/pnas.1506755112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liao H.-X., Alam S.M., Mascola J.R., Robinson J., Ma B., Montefiori D.C., Rhein M., Sutherland L.L., Scearce R., Haynes B.F. Immunogenicity of constrained monoclonal antibody A32-human immunodeficiency virus (HIV) env gp120 complexes compared to that of recombinant HIV type 1 gp120 envelope glycoproteins. J. Virol. 2004;78:5270–5278. doi: 10.1128/JVI.78.10.5270-5278.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Horton H.M., Bernett M.J., Peipp M., Pong E., Karki S., Chu S.Y., Richards J.O., Chen H., Repp R., Desjarlais J.R., Zhukovsky E.A. Fc-engineered anti-CD40 antibody enhances multiple effector functions and exhibits potent in vitro and in vivo antitumor activity against hematologic malignancies. Blood. 2010;116:3004–3012. doi: 10.1182/blood-2010-01-265280. [DOI] [PubMed] [Google Scholar]
- 61.Huang J., Doria-Rose N.A., Longo N.S., Laub L., Lin C.-L., Turk E., Kang B.H., Migueles S.A., Bailer R.T., Mascola J.R., Connors M. Isolation of human monoclonal antibodies from peripheral blood B cells. Nat. Protoc. 2013;8:1907–1915. doi: 10.1038/nprot.2013.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Doria-Rose N., Doria-Rose N., Bailer R., Louder M., Lin C.-L., Turk E., Laub L., Longo N., Connors M., Mascola J. High throughput HIV-1 microneutralization assay. Protocol Exchange. 2013 doi: 10.1038/protex.2013.069. [DOI] [Google Scholar]
- 63.Ljunggren K., Broliden P.A., Morfeldt-Månson L., Jondal M., Wahren B. IgG subclass response to HIV in relation to antibody-dependent cellular cytotoxicity at different clinical stages. Clin. Exp. Immunol. 1988;73:343–347. [PMC free article] [PubMed] [Google Scholar]
- 64.Tyler D.S., Stanley S.D., Nastala C.A., Austin A.A., Bartlett J.A., Stine K.C., Lyerly H.K., Bolognesi D.P., Weinhold K.J. Alterations in antibody-dependent cellular cytotoxicity during the course of HIV-1 infection. Humoral and cellular defects. J. Immunol. 1990;144:3375–3384. [PubMed] [Google Scholar]
- 65.Bonsignori M., Pollara J., Moody M.A., Alpert M.D., Chen X., Hwang K.-K., Gilbert P.B., Huang Y., Gurley T.C., Kozink D.M., et al. Antibody-dependent cellular cytotoxicity-mediating antibodies from an HIV-1 vaccine efficacy trial target multiple epitopes and preferentially use the VH1 gene family. J. Virol. 2012;86:11521–11532. doi: 10.1128/JVI.01023-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dhande J., Angadi M., Murugavel K.G., Poongulali S., Nandagopal P., Vignesh R., Ghate M., Kulkarni S., Thakar M. The anti–HIV-1 ADCC-mediating antibodies from cervicovaginal secretions of HIV-infected women have an ability to mediate lysing of autologous CD4+ HIV-infected cells. J Acquir Immune Defic Syndr. 2018;79:277–282. doi: 10.1097/QAI.0000000000001788. [DOI] [PubMed] [Google Scholar]
- 67.McLean M.R., Madhavi V., Wines B.D., Hogarth P.M., Chung A.W., Kent S.J. Dimeric fcγ receptor enzyme-linked immunosorbent assay to study HIV-specific antibodies: a New look into breadth of fcγ receptor antibodies induced by the RV144 vaccine trial. J. Inf. 2017;199:816–826. doi: 10.4049/jimmunol.1602161. [DOI] [PubMed] [Google Scholar]
- 68.Huang Y., Ferrari G., Alter G., Forthal D.N., Kappes J.C., Lewis G.K., Love J.C., Borate B., Harris L., Greene K., et al. Diversity of antiviral IgG effector activities observed in HIV-infected and vaccinated subjects. J. Inf. 2016;197:4603–4612. doi: 10.4049/jimmunol.1601197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cheng H.D., Dowell K.G., Bailey-Kellogg C., Goods B.A., Love J.C., Ferrari G., Alter G., Gach J., Forthal D.N., Lewis G.K., et al. Diverse antiviral IgG effector activities are predicted by unique biophysical antibody features. Retrovirology. 2021;18:35. doi: 10.1186/s12977-021-00579-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Alrubayyi A., Schuetz A., Lal K.G., Jongrakthaitae S., Paolino K.M., Ake J.A., Robb M.L., de Souza M.S., Michael N.L., Paquin-Proulx D., Eller M.A. A flow cytometry based assay that simultaneously measures cytotoxicity and monocyte mediated antibody dependent effector activity. J. Immunol. Methods. 2018;462:74–82. doi: 10.1016/j.jim.2018.08.012. [DOI] [PubMed] [Google Scholar]
- 71.Lewis G.K., Ackerman M.E., Scarlatti G., Moog C., Robert-Guroff M., Kent S.J., Overbaugh J., Reeves R.K., Ferrari G., Thyagarajan B. Knowns and unknowns of assaying antibody-dependent cell-mediated cytotoxicity against HIV-1. Front. Immunol. 2019;10:1025. doi: 10.3389/fimmu.2019.01025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ferrari G., Pollara J., Kozink D., Harms T., Drinker M., Freel S., Moody M.A., Alam S.M., Tomaras G.D., Ochsenbauer C., et al. An HIV-1 gp120 envelope human monoclonal antibody that recognizes a C1 conformational epitope mediates potent antibody-dependent cellular cytotoxicity (ADCC) activity and defines a common ADCC epitope in human HIV-1 serum. J. Virol. 2011;85:7029–7036. doi: 10.1128/JVI.00171-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hioe C.E., Li G., Liu X., Tsahouridis O., He X., Funaki M., Klingler J., Tang A.F., Feyznezhad R., Heindel D.W., et al. Non-neutralizing antibodies targeting the immunogenic regions of HIV-1 envelope reduce mucosal infection and virus burden in humanized mice. PLoS Pathog. 2022;18:e1010183. doi: 10.1371/journal.ppat.1010183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Spencer D.A., Goldberg B.S., Pandey S., Ordonez T., Dufloo J., Barnette P., Sutton W.F., Henderson H., Agnor R., Gao L., et al. Phagocytosis by an HIV antibody is associated with reduced viremia irrespective of enhanced complement lysis. Nat. Commun. 2022;13:662. doi: 10.1038/s41467-022-28250-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wu X., Parast A.B., Richardson B.A., Nduati R., John-Stewart G., Mbori-Ngacha D., Rainwater S.M.J., Overbaugh J. Neutralization escape variants of human immunodeficiency virus type 1 are transmitted from mother to infant. J. Virol. 2006;80:835–844. doi: 10.1128/JVI.80.2.835–844.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Neilson J.R., John G.C., Carr J.K., Lewis P., Kreiss J.K., Jackson S., Nduati R.W., Mbori-Ngacha D., Panteleeff D.D., Bodrug S., et al. Subtypes of human immunodeficiency virus type 1 and disease stage among women in Nairobi, Kenya. J. Virol. 1999;73:4393–4403. doi: 10.1128/JVI.73.5.4393-4403.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Simonich C.A., Williams K.L., Verkerke H.P., Williams J.A., Nduati R., Lee K.K., Overbaugh J. HIV-1 neutralizing antibodies with limited hypermutation from an infant. Cell. 2016;166:77–87. doi: 10.1016/j.cell.2016.05.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gómez-Román V.R., Florese R.H., Patterson L.J., Peng B., Venzon D., Aldrich K., Robert-Guroff M. A simplified method for the rapid fluorometric assessment of antibody-dependent cell-mediated cytotoxicity. J. Immunol. Methods. 2006;308:53–67. doi: 10.1016/j.jim.2005.09.018. [DOI] [PubMed] [Google Scholar]
- 79.Wallace P.K., Tario J.D., Fisher J.L., Wallace S.S., Ernstoff M.S., Muirhead K.A. Tracking antigen-driven responses by flow cytometry: monitoring proliferation by dye dilution: tracking Cell Proliferation. Cytometry. 2008;73:1019–1034. doi: 10.1002/cyto.a.20619. [DOI] [PubMed] [Google Scholar]
- 80.Tario J.D., Gray B.D., Wallace S.S., Muirhead K.A., Ohlsson-Wilhelm B.M., Wallace P.K. Novel lipophilic tracking dyes for monitoring cell proliferation. Immunol. Invest. 2007;36:861–885. doi: 10.1080/08820130701712933. [DOI] [PubMed] [Google Scholar]
- 81.Doria-Rose N.A., Bhiman J.N., Roark R.S., Schramm C.A., Gorman J., Chuang G.-Y., Pancera M., Cale E.M., Ernandes M.J., Louder M.K., et al. New member of the V1V2-directed CAP256-VRC26 lineage that shows increased breadth and exceptional potency. J. Virol. 2016;90:76–91. doi: 10.1128/JVI.01791-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Liao H.-X., Levesque M.C., Nagel A., Dixon A., Zhang R., Walter E., Parks R., Whitesides J., Marshall D.J., Hwang K.-K., et al. High-throughput isolation of immunoglobulin genes from single human B cells and expression as monoclonal antibodies. J. Virol. Methods. 2009;158:171–179. doi: 10.1016/j.jviromet.2009.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lefranc M.-P., Giudicelli V., Ginestoux C., Jabado-Michaloud J., Folch G., Bellahcene F., Wu Y., Gemrot E., Brochet X., Lane J., et al. IMGT(R), the international ImMunoGeneTics information system(R) Nucleic Acids Res. 2009;37:D1006–D1012. doi: 10.1093/nar/gkn838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tiller T., Meffre E., Yurasov S., Tsuiji M., Nussenzweig M.C., Wardemann H. Efficient generation of monoclonal antibodies from single human B cells by single cell RT-PCR and expression vector cloning. J. Immunol. Methods. 2008;329:112–124. doi: 10.1016/j.jim.2007.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Goo L., Milligan C., Simonich C.A., Nduati R., Overbaugh J. Neutralizing antibody escape during HIV-1 mother-to-child transmission involves conformational masking of distal epitopes in envelope. J. Virol. 2012;86:9566–9582. doi: 10.1128/JVI.00953-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Xu G.J., Kula T., Xu Q., Li M.Z., Vernon S.D., Ndung’u T., Ruxrungtham K., Sanchez J., Brander C., Chung R.T., et al. Comprehensive serological profiling of human populations using a synthetic human virome. Science. 2015;348:aaa0698. doi: 10.1126/science.aaa0698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Stoddard C.I., Galloway J., Chu H.Y., Shipley M.M., Sung K., Itell H.L., Wolf C.R., Logue J.K., Magedson A., Garrett M.E., et al. Epitope profiling reveals binding signatures of SARS-CoV-2 immune response in natural infection and cross-reactivity with endemic human CoVs. Cell Rep. 2021;35:109164. doi: 10.1016/j.celrep.2021.109164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Garrett M.E., Galloway J.G., Wolf C., Logue J.K., Franko N., Chu H.Y., Matsen F.A., Overbaugh J.M. Comprehensive characterization of the antibody responses to SARS-CoV-2 Spike protein finds additional vaccine-induced epitopes beyond those for mild infection. Elife. 2022;11:e73490. doi: 10.7554/eLife.73490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sok D., van Gils M.J., Pauthner M., Julien J.-P., Saye-Francisco K.L., Hsueh J., Briney B., Lee J.H., Le K.M., Lee P.S., et al. Recombinant HIV envelope trimer selects for quaternary-dependent antibodies targeting the trimer apex. Proc. Natl. Acad. Sci. USA. 2014;111:17624–17629. doi: 10.1073/pnas.1415789111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Richard J., Veillette M., Batraville L.-A., Coutu M., Chapleau J.-P., Bonsignori M., Bernard N., Tremblay C., Roger M., Kaufmann D.E., Finzi A. Flow cytometry-based assay to study HIV-1 gp120 specific antibody-dependent cellular cytotoxicity responses. J. Virol. Methods. 2014;208:107–114. doi: 10.1016/j.jviromet.2014.08.003. [DOI] [PubMed] [Google Scholar]
- 91.Veillette M., Désormeaux A., Medjahed H., Gharsallah N.-E., Coutu M., Baalwa J., Guan Y., Lewis G., Ferrari G., Hahn B.H., et al. Interaction with cellular CD4 exposes HIV-1 envelope epitopes targeted by antibody-dependent cell-mediated cytotoxicity. J. Virol. 2014;88:2633–2644. doi: 10.1128/JVI.03230-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Naiman N.E., Slyker J., Richardson B.A., John-Stewart G., Nduati R., Overbaugh J.M. Antibody-dependent cellular cytotoxicity targeting CD4-inducible epitopes predicts mortality in HIV-infected infants. EBioMedicine. 2019;47:257–268. doi: 10.1016/j.ebiom.2019.08.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
The PhIP-Seq sequencing dataset generated during this study is publicly available in the Short Read Archive, with the BioProject ID listed in the key resources table. MG540 antibody sequence accession numbers are listed in the key resources table. Pre-processed enrichment data for PhIP-Seq experiments will be shared by the lead contact upon request.
-
•
This paper does not report original code. The python API “phippery” was used to process the demultiplexed sequencing data, align reads to the reference oligonucleotide sequences, and calculate enrichment. “phippery” is available at (https://github.com/matsengrp/phippery). Clonal family inference was performed with the partis software package (available at https://github.com/psathyrella/partis/) paired clustering method, treating wells as droplets.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.