Abstract
Neuroinflammation and immune dysregulation play a key role in Alzheimer’s disease (AD) and are also associated with severe Covid-19 and neurological symptoms. Also, genome-wide association studies found many risk single nucleotide polymorphisms (SNPs) for AD and Covid-19. However, our understanding of underlying gene regulatory mechanisms from risk SNPs to AD, Covid-19 and phenotypes is still limited. To this end, we performed an integrative multi-omics analysis to predict gene regulatory networks for major brain regions from population data in AD. Our networks linked transcription factors (TFs) to TF binding sites (TFBSs) on regulatory elements to target genes. Comparative network analyses revealed cross-region-conserved and region-specific regulatory networks, in which many immunological genes are present. Furthermore, we identified a list of AD–Covid genes using our networks involving known and Covid-19 genes. Our machine learning analysis prioritized 36 AD–Covid candidate genes for predicting Covid severity. Our independent validation analyses found that these genes outperform known genes for classifying Covid-19 severity and AD. Finally, we mapped genome-wide association study SNPs of AD and severe Covid that interrupt TFBSs on our regulatory networks, revealing potential mechanistic insights of those disease risk variants. Our analyses and results are open-source available, providing an AD–Covid functional genomic resource at the brain region level.
Introduction
Alzheimer’s Disease (AD), a neurodegenerative disease, affects over 50 million elders worldwide (1). Late-onset AD (LOAD) comprises > 97% of all AD cases, usually occurring after age 65 (2). AD patients experience phenotypic changes such as memory loss, cognitive decline, weak executive function (1) [e.g. poor Mini-Mental State Exam (MMSE) scores]. Many underlying molecular changes happen like an accumulation of amyloid-beta (Aβ) plaques and neurofibrillary tangles (NFTs), chronic neuroinflammation (may begin decades before clinical onset). Nonetheless, molecular mechanisms behind AD progression and phenotypes remain elusive. Misguided innate immunity may be a major culprit driving AD based on the neuroimmunomodulation theory of AD (3).
Molecular interconnections that exist between the central nervous system (CNS) and immune system (4) are also seen via the strong correlations between AD and the severity of Covid-19 infection (5). Covid-19, a robust marker for an overreactive immune system, can also mediate neuroinflammation (6). β-coronaviruses (like Covid-19) may attack the CNS, elevating AD dementia processes (7). Covid survivors have greater risk of neurological/psychiatric problems and brain fog [neuro-Covid (8) or long-Covid]; patients may have visible neuropathological abnormalities in brain structure (5) [e.g. Hippocampus atrophy (9)] similar to those found in AD patients (10). AD brains have high levels of circulating pro-inflammatory cytokines associated with activation of microglia [macrophage resident immune cells typically downregulated in healthy brains (4)]; these cytokines also contribute to the cytokine storm causing exaggerated inflammation characteristic of severe Covid (11). In fact, Covid patients experiencing delirium (symptom linked with high risk of AD) are at grave risk of death and typically sent to an Intensive Care Unit (ICU) (9). Elder patients (age > 65 years) are 70% likelier to be diagnosed with AD within a year of Covid infection (12). There is a 2-fold increased risk of Covid death in AD patients (13) and of higher severity of Covid for patients with APOE4 (E4 alleles for the key LOAD risk gene APOE) (14). Links among Covid, cognitive decline and neurodegenerative diseases like AD are puzzling and poorly understood (9). AD itself has over 34 canonical and intricately interconnected pathways, making that process daunting (15). Focusing on AD–Covid pathways may be a useful starting point of departure given their strong links. Thus, understanding genetic effects and underlying molecular mechanisms for shared AD–Covid paths may shed more insights on rogue immune responses not only in AD but also in severe and/or neuro Covid-19.
Several neuroimmunology pathways are shared by AD and Covid. One of them is the NF-κB (Nuclear Factor Kappa-light-chain-enhancer of activated B cells) pathway that is found in almost all cell-types and that regulates, inter alia, brain homeostasis (maintains synapse plasticity, learning, memory; moderates neuron survival/apoptosis) (16), innate immunity, inflammation (17). A prominent hypothesis believes AD may be caused by an impaired NF-κB pathway (16) with overactivated NF-κB transcription factors (TFs) like RELA and NFKB1. This may lead to more cytokines, neuroinflammation, oxidative stress complications, activated microglia, neuron death (16). NF-κB TFs are also involved in a positive feedback loop, activating pro-inflammatory cytokines in severe Covid (11). RELA, one of the most important TFs regulating Covid response (18), is associated with APOE4 (19). Gene regulatory networks (GRNs) can capture how these TFs regulate several genes of pro-inflammatory cytokines. Thus, to understand neuroimmunology in Covid-19 and AD better, it is important to analyze these underlying gene regulatory mechanisms.
Gene expression and regulation are a key mechanism leading to human diseases. Studies found differentially expressed genes (DEGs) in AD in various brain regions like the Hippocampus CA1 (Hippocampus), Lateral Temporal Lobe (LTL), Dorsolateral Prefrontal Cortex (DLPFC). Also, gene co-expression networks are widely used to identify co-expressed gene modules and link gene expression patterns to AD phenotypes (20). Genes in a module show similar expression dynamics across AD phenotypes, which denotes that they share certain molecular mechanisms that are dysregulated in AD (21). Nevertheless, understanding gene regulatory mechanisms controlling DEGs, co-expressed genes and modules for various AD phenotypes as they relate to the immune system is unclear. Gene expression and function are controlled by various regulatory factors working together in a GRN, like: TFs binding to TF binding sites (TFBSs) on regulatory elements (e.g. enhancers, promoters). However, our understanding of gene regulation in AD and in AD–Covid is still limited.
To address these issues, we performed an integrative analysis of multi-omics to reveal genes, functions and GRNs from AD and/or severe Covid-19 Single Nucleotide Polymorphisms (SNPs) to AD phenotypes for the three brain regions as mentioned above (Fig. 1, Materials and Methods). Given a brain region, we built a gene co-expression network using population gene expression data from an AD cohort and identified co-expressed genes and modules associated with AD phenotypes. We then integrated chromatin interaction data [e.g. High-throughput chromosome conformation capture (Hi-C)] and TF-gene expression relationships to predict TFs regulating co-expressed genes by binding to the regulatory elements that control these genes. Our machine learning (ML) analysis prioritized 36 AD–Covid candidate genes for predicting Covid severity and we evaluated further their ability to predict AD. Finally, we identified risk SNPs altering these TFBSs and analyzed their impact on our GRNs and AD phenotypes. We emphasized subnetworks and regulatory SNPs associated with our predicted AD–Covid genes. Thus, our analysis may provide deeper insights into molecular causes of neuroimmunology pertaining to AD, Covid-19 severity, neuro-Covid and AD–Covid.
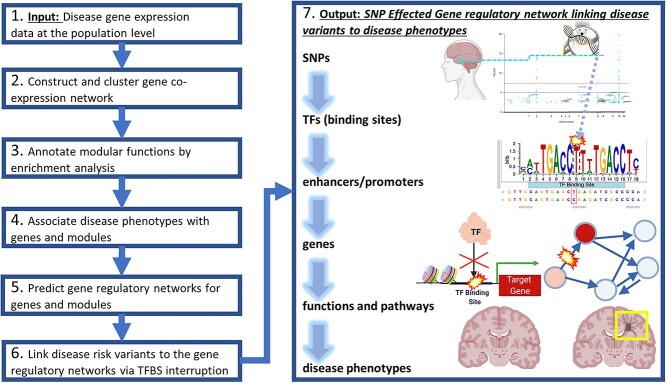
Figure 1.
Integrative analyses to predict GRNs from disease risk variants to phenotypes. Primarily, this analysis consists of seven major steps as a pipeline. First, it inputs the population gene expression data with phenotypic information (Step 1). Second, it uses that gene expression data to construct and cluster gene co-expression networks into gene modules (Step 2). Third, it performs enrichment analysis for these gene modules (Step 3). Fourth, it links genes and modules to various phenotypes from the input population (Step 4). Fifth, it predicts the Transcription Factors (TFs) and regulatory elements (e.g. TF binding sites along enhancers and/or promoters) that regulate genes and co-regulate modular genes as a gene regulatory network (GRN, Step5). Sixth, it further finds disease risk variants [e.g. Genome-Wide Association Studies (GWAS) Single Nucleotide Polymorphisms (SNPs)] that alter the binding sites of TFs from this GRN (Step 6). Seventh and finally, it outputs a SNP regulatory network (SNP-effected-GRN) linking functional non-coding disease risk variants to impacted TFs and enhancers/promoters to regulated genes and modules to enriched functions and pathways to disease phenotypes (Step 7). This network thus provides a deeper understanding of gene regulatory mechanisms in diseases. As a demo, in this paper, we applied this pipeline to AD population datasets from different brain regions. We predicted brain-specific GRNs for various AD phenotypes such as Alzheimer’s disease progression stages. Then, we built SNP-effected-GRNs by mapping single nucleotide polymorphisms (SNPs) from several AD GWAS datasets and a GWAS related to Covid-19 severity in Covid-positive individuals to these GRNs.
Results
Gene co-expression network analysis reveals gene expression dynamics for AD phenotypes across multiple brain regions
First, we applied our analysis to population gene expression datasets of three major brain regions relating to AD: Hippocampal CA1 (Hippocampus), LTL and DLPFC (Materials and Methods). We identified several gene co-expression modules showing specific gene expression dynamic changes for various AD phenotypes (Supplementary Material, Files S1–S3 for the Hippocampus, LTL and DLPFC, respectively), implying potential underlying gene regulatory mechanisms associated with the phenotypes. Given a brain region, we constructed and clustered a gene co-expression network to a set of gene co-expression modules. In a gene co-expression network for a brain region, nodes (or vertices) are genes and each edge represents that two respective genes have correlated gene expression profiles across the samples (i.e. co-expression). There are likely groups of co-expressed genes within the network that form densely connected sub-networks (gene modules). Genes in a module share similar gene expression dynamics in that respective brain region for the observed AD phenotypes. Modular eigengenes (MEs) represent expression dynamics for a gene module, using the first principal components of module gene expression matrices.
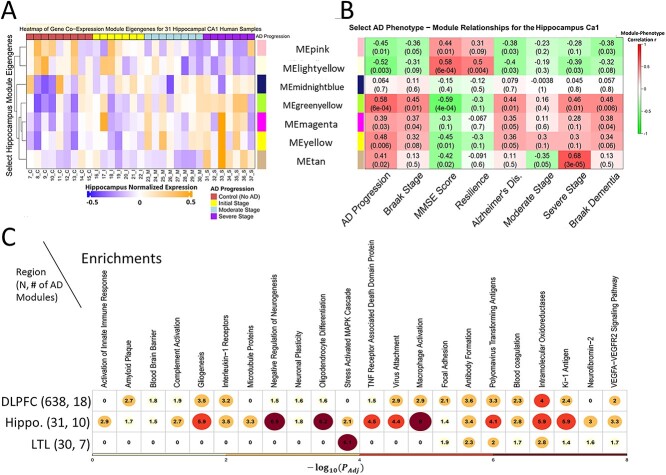
Hippocampus. Twenty-one of 30 gene modules (9,525 genes) are ‘phenotype-enriched’ as they are significantly positively associated with at least one key AD-related phenotype. Their MEs show specific expression dynamics (Fig. 2A: 7 select modules). Pink and lightyellow modules have high gene expression values for Controls and cluster together. On the other hand, greenyellow, yellow, tan and magenta modules cluster together given their high expression in AD. Next, we used expression dynamic patterns to link modules to phenotypes (Fig. 2B) by significant positive correlations. The tan module has the highest severe AD correlation (r = 0.68). The midnightblue module is significant for Braak 4 stage (mild dementia), the lightyellow module for cognitive resilience. The greenyellow module significantly correlates with AD, AD progression, moderate/severe AD, cognitive impairment, Braak 6 stage. LTL. Twenty-eight of 56 co-expression modules are phenotype enriched. We highlighted five MEs in Supplementary Material, Figure S1A. The sienna3 module has higher expression values for old and young Controls. Orange, magenta and yellow modules cluster together with high expression in AD. As shown in Supplementary Material, Figure S1B, the sienna3 module correlates positively with Controls (r = 0.63) and being asymptomatic for dementia or any other AD-related symptoms (r = 0.55). Yellow, orange and magenta modules associate with aging, AD/Braak progression, neuritic plaques; the orange module has r = 0.72 for AD/dementia. DLPFC. The sample size in the DLPFC, which is 20-fold larger relative to those of the other 2 regions, likely attributes to the comparatively lower module-phenotype correlations we observe in the DLPFC. Still, we see significantly correlated modules with select AD phenotypes and highlight 6 of 35 modules (all 35 are phenotype-enriched) (Supplementary Material, Fig. S1C and D, P < 0.05). The tan module is associated with the worst APOE genotype (E4/E4) and with the age for the diagnosis of AD; royalblue and green DLPFC modules correlate with severe AD based on the last MMSE score. In terms of better and healthier outcomes, the darkolivegreen module is significant for Controls, higher MMSE scores, cognitive resilience. Our gene modules across regions uncover gene expression dynamic patterns across phenotypes, suggesting that genes in a module are likely involved in similar functions and pathways. To understand this, we performed module enrichment analysis as follows.
Figure 2.
Gene co-expression modules significantly associated with AD phenotypes show specific expression dynamic patterns across phenotypes and enriched functions and pathways. The left and right heatmaps on the top row correspond to the same set of select gene modules for the Hippocampus (and the corresponding heatmaps for the LTL and DLPFC are in Supplementary Material, Figure S2). (A) Shows the module eigengenes (MEs) of seven gene co-expression modules in the Hippocampal CA1 region where rows: modules and columns: individual human samples. Red: high expression level. Blue: low expression level. On the left hand side of this heatmap is a dendrogram tree based on agglomerative hierarchical clustering so that similar modules (in terms of values for MEs) cluster close together. (B) Shows the correlation coefficients and P-values for the same 7 Hippocampal CA1 gene modules and various select AD phenotypes (Supplementary Materials contain additional phenotypes). Row: modules. Columns: AD phenotypes. Red: highly positive correlation (r > 0). Green: highly negative correlation (r < 0). (C) Shows select biological functions and pathways that are enriched for modules positively correlated (P < 0.05, r > 0) with AD across the three brain regions. Values in the cells, circle sizes and gradient color (yellow to red) correspond to the highest −log10(adjust P-value) enrichment value of any gene module associated with the phenotype of AD diagnosis (i.e. AD phenotype) from that given brain region. There are N = 31 post-mortem human samples in the Hippocampus, N = 30 in the Lateral Temporal Lobe (LTL) and N = 638 in the Dorsolateral Prefrontal Cortex (DLPFC).
Eigengenes and enrichments of co-expression modules reveal hub genes, gene functions and pathways in AD phenotypes
We performed gene set enrichment analyses (Materials andMethods) to understand better the biological functions, diseases, pathways, structures and other observed phenomena of our modules and link them to various AD phenotypes (Fig. 2). Healthy phenotypes are Control, cognitive resilience, protective APOE E2/E2 genotype. Our brain region module enrichments underscore the role of the immune system and neuroimmunology among other factors in AD progression and verify that the phenotype correlations we detected for our gene modules may indicate true biological signals.
In AD, the Hippocampus (Supplementary Material, Fig. S2A, Supplementary Material, File S1) has a major loss in volume, neurogenesis, memory, neuron density (22). Healthy gene modules are enriched with synaptic plasticity, dendrite development, calcium signaling. Perhaps resilient individuals are protected from microsatellite instability and amyloid accumulation. Age and AD progression modules are associated with abnormal innate immunity, Covid-19 spike protein, NF-κB pathway [overexpressed in AD (16)], activation of JNK and MAPK cascade [active in AD, involved in tau phosphorylation, neuroinflammation (23), synapse dysfunction, neuron death (70)]. Severe AD modules are associated with metabolic processes (24), immune memory, interferon signaling [high in AD mouse Hippocampus (25); this response to amyloid may activate microglia, initiating neuroinflammation and synapse loss in neurons (26)]. The LTL (Supplementary Material, Fig. S2B, Supplementary Material, File S2) is impacted early in AD (27). Control modules are enriched with Wnt signaling [inhibits tau protein hyperphosphorylation and production of amyloid-beta (Aβ) plaques (28)], whose dysregulation may lead to neurodegeneration. In AD and plaque modules, Nlp protein loss from Mitotic centrosomes is enriched [may cause microtubule instability, abnormal cell morphology, AD (29)]. We found cell-type and other pathway enrichments in AD progression phenotypes: astrocyte projection [glial cell type that is increasingly active near Aβ plaques in AD (30)], NF-κB activation, prion pathway [disruption may lead to Aβ plaques (31)]. Dramatic Histone H4 acetylation epigenetic losses on DNA regions near genes may decrease memory formation during aging and AD in the LTL (27). The DLPFC (Supplementary Material, Fi. S2C, Supplementary Material, File S3) works with the Hippocampus to mediate complex cognitive functions (32) and has plasticity deficits in AD (33). Microglia exclusively express AD genes like APOE (34). Here, APOE2 modules are associated with mitochondrial inheritance (P < 1e-16) and are shielded from neurotoxins, whereas APOE4 modules are enriched with Aβ response [may regulate microglia (88)], cognitive dysfunction. Promising associations (some with P < 1e-58) for APOE4 and AD-related modules support a crucial role of reactive microglia for AD disease progression. In AD, microglia may change shape, are more phagocytic, go awry and release pro-inflammatory cytokines, leading to Aβ and neurofibrillary tangles (NFTs) (35), synapse decline, neuroinflammation, neurodegeneration, cell death (34). Our results may shed light on links between APOE4 and neuroinflammation, with enrichments such as: autoimmune diseases (e.g. Wegener’s Granulomatosis), synapse pruning, astrocyte activation, microglia, abnormal innate immunity and cytokine levels [the DLPFC in AD patients typically has more pro-inflammatory cytokines like IL-1B, linked to Aβ plaques (35)]. Healthy modules are enriched with Electron Transport Chain [altered in AD (36)], neuron recognition, synapse plasticity, calcium ion regulated exocytosis. Finally, we compared three brain regions (Fig. 2C, Supplementary Material, Fig. S2D): Braak stage modules are enriched with Ki-1 antigen (tumor marker of activated immune cells regulating NF-κB and apoptosis), focal adhesion (plaques), VEGFA-VEGFR2 [altered levels in AD may impact microglia/neuron survival (37)]. Control and AD DLPFC/Hippocampus modules share neuroimmunomodulation. AD and Braak stage modules are enriched with blood–brain barrier (BBB), virus attachment, complement system (CS) activation [innate immune-mediated defense altered in AD (38)], oligodendrocyte differentiation [this change in this glial cell type is linked to neurodegeneration, Aβ accumulation (39)].
Prediction of brain region GRNs for AD phenotypes
To understand underlying molecular mechanisms regulating gene expression associated with various AD phenotypes, we predicted the GRNs for target genes (TGs) and gene modules of brain regions, especially using multi-omics data (Materials andMethods). Brain region GRNs link TFs and regulatory elements (e.g. enhancers or promoters) to TGs and co-expressed genes (e.g. from the same gene module). GRN edges can be activation or repression of TGs by TFs, which follow-ups can investigate. These GRNs can be further linked to AD phenotypes significantly associated with TGs and modules. We applied many popular approaches and public databases to predict networks and used their shared predictions to build our highly confident GRNs. We found: 1,043 candidate TFs in the Hippocampus, 1,580 in the LTL, (and 1,588 in the DLPFC), which we input into RTN, GENIE3 and TReNA Ensemble Solver for the Hippocampus and LTL, respectively. Supplementary Material, Table S1 shows statistics of TF–Regulatory Element–TG network nodes and edges. Supplementary Material, Files S4 and S5 contain our detailed final Hippocampus and LTL GRN edge lists. We found TFs significantly regulate 21 LTL gene co-expression modules and 21 Hippocampus gene modules; for example, in the Hippocampus, neurogenesis TF REST regulates a module of 883 genes, NFKB1 regulates one Control module, RELA regulates three gene modules (2 Control modules, 1 AD progression module).
GRNs and AD phenotypes associated with shared AD-Covid pathways
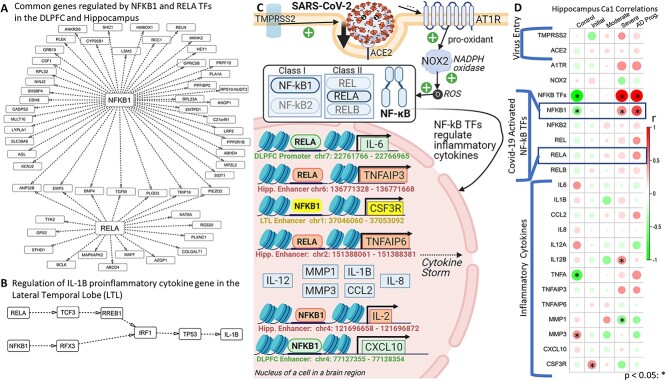
Rogue immune responses characterize AD and Covid-19. Our hypergeometric test of overlap between both AD and Covid-19 (SARS-CoV-2) Kyoto Encyclopedia of Genes and Genomes (KEGG) networks (P = 0.0034) was significant, suggesting that the shared AD–Covid mechanisms are important. We thus looked at these shared mechanisms implicated in adverse effects and inflammation in both diseases (16), like the NF-κB pathway. In mammals, the NF-κB TF family has five TFs: NFKB1 (or p105/p50 protein), NFKB2 (or p100/52 protein), REL (or c-Rel), RELA (or p65 protein) and RELB (proto-oncogene near APOE). Reactive Oxygen Species (ROS) activate RELA and NFKB1 TFs. Both TFs then transcribe pro-inflammatory cytokines (e.g. IL-6, IL-1B, TNF), reducing long-term potentiation (LTP) during AD (typically resulting in reduced strength of synaptic signal transmission between neurons, lower synaptic plasticity, memory loss and learning delays) and leading to exaggerated and potentially lethal immune responses in Covid [e.g. tissue injury, hypoxia (7), Acute Respiratory Distress Syndrome (ARDS) (40)] (Supplementary Material, Fig. S3). We found that gene expression levels of NF-κB TFs correlate positively with AD severity but negatively with controls in all three regions (Hippocampus: Supplementary Material, Fig. S4A). NFKB1 and RELB correlate negatively with controls in three regions, as do NFKB2 and RELA in the DLPFC and Hippocampus (Supplementary Material, Fig. S4B). All five TFs correlate positively with severe AD in the Hippocampus and two TFs correlate positively with AD in DLPFC (Supplementary Material, Fig. S4C). Upregulation of NF-κB TFs may be a key AD–Covid interplay as activation of these TFs is linked to greater inflammation in Covid and in AD (40). NFKB1 and RELA’s severe AD Hippocampus module has immune enrichments like PID IL-1 pathway, abnormal innate immunity, immunoglobulin level. We investigated our GRNs involving NF-κB TFs. Figure 3A shows shared target genes (TGs) for NFKB1 and/or RELA in the DLPFC and Hippocampus; seven TGs are regulated by both TFs in both regions, like ANP32B and EMP3. Figure 3B shows how RELA and NFKB1 indirectly regulate IL-1B in the LTL via TFs: TCF3, RFX3, RREB1, IRF1, TP53.
Figure 3.
Gene regulatory networks (GRNs) and phenotypes for Nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB), a shared pathway in both AD and Covid-19. (A) A subnetwork focusing on overlaps in the GRN between the DLPFC and Hippocampus, focusing on the target genes (TGs) regulated by NF-κB TFs: RELA (belongs to NF-κB class II) and NFKB1 (belongs to NF-κB class I). Here, only TF–TG links found in both brain regions are shown. (B) RELA and NFKB1 TFs regulate other TFs in a domino chain reaction that then regulate the pro-inflammatory cytokine IL-1B in the LTL. This illustrates the complexity of GRNs. For instance, RELA regulates TCF3, which then regulates RREB1, which then regulates IRF1, which then regulates TP53, which lastly regulates IL-1B. (C) The Covid-19 virus (SARS-CoV-2) spike protein enters and infects the cell. Gene regulation of pro-inflammatory cytokines by activated NF-κB TFs from our Hippocampal, LTL and/or DLPFC GRNs is linked with severe Covid-19 outcomes (e.g. cytokine storm and beyond). This visualization is adapted from the Covid-19 KEGG network (KEGG: hsa05171), focusing on the NF-κB pathway. Gray dashed arrows indicate regulation and black arrows indicate activation of cytokines by the respective TF. GRN edge lists in Supplementary Materials contain more examples. (D) Pearson correlations between AD phenotypes and expression levels of genes from (C) in the Hippocampus; the gene-phenotype correlations with P-value < 0.05 are denoted with an asterisk (*) on top.
We looked at the SARS-CoV-2 (Covid-19 KEGG: hsa05171) network to analyze how the NF-κB pathway and regulated cytokines may be associated with AD–Covid links and neuroinflammation (Fig. 3C). During Covid-19 infection, the SARS-CoV-2 Spike protein is primed by TMPRSS2, binds to the ACE2 [high expression in brain/macrophages (41)] receptor and interacts with AT1R (Angiotensin II Receptor Type 1) to enter and infect the cell (42). Neurons may be directly invaded by SARS-CoV-2 or by systemic infection compromising the blood-brain barrier (BBB, dysfunctional in AD), elevating brain levels of chemokines, Complement System (CS) factors and cytokines (increased in AD) (43) that damage neurons (9). TMPRSS2, ACE2 and AT1R expression levels correlate positively with severe/advanced AD in the Hippocampus (Fig. 3D). We used our GRNs to analyze how NFKB1 and RELA regulate genes of several pro-inflammatory cytokines that are involved in the severe Covid cytokine storm [associated with BBB dysfunction, anti-neuron antibodies, neuroinflammation, neurodegeneration (7), activation of microglia and astrocytes]. In the DLPFC, RELA binds to an enhancer of CXCL10, which has altered levels associated with immune dysfunction and inflammatory disease severity (44). NFKB1 binds to LTL enhancer of CSF3R, a regulator of neutrophil [innate immune system cells that change level and function in severe Covid (45)] and microglia maintenance (46). NFKB1 regulates IL-2 binding to an IL-2 enhancer on chromosome 4: 121,696,658–121,696,872. In AD brains, Aβ stimulation may activate NF-κB TFs to upregulate TNFa and IL-1B (regulates amyloid precursor protein (APP) synthesis) in microglia and astrocytes (16), likely triggering neuron death, cytokine cascade, more plaques, inflammation, tissue destruction (35,47). NFKB1 and RELA regulate TFs that further regulate inflammatory cytokines IL-1B, IL-12B, CCL2, MMP1/3, CLGN. In the Hippocampus, NFKB1 regulates SPI1 and BATF that then jointly regulate MMP1. RELA regulates TNFa-induced proteins TNFAIP3/6 (regulate long-term potentiation in AD) in the Hippocampus; IL-2 and TNFa are highly expressed in Covid patients with severe pneumonia who develop ARDS [needing to go to the ICU and receive emergency oxygen (40)] as well as in severe AD patients (as are cytokines CCL2, IL-1B, IL-12B) (Fig. 3D). RELA activates IL-12A/B (recruits Natural Killer cells (48)) and IL-1B via their respective enhancers and regulates IL-6 [induces C-Reactive Protein (CRP) synthesis that activates the CS (4)] by binding to an IL-6 promoter.
Activation of the CS is involved in an inflammatory feedback loop with neutrophil activation, resulting in tissue injury (49) in severe Covid. CS components like C1qrs activate microglia to the M1 state, releasing inflammatory mediators (50) causing Hippocampus atrophy (51) (these M1 microglia induce neurotoxicity; M2 microglia are instead anti-inflammatory and neuroprotective). We found that C1qrs correlates negatively with Control and Initial AD, but positively with AD severity in the Hippocampus (Supplementary Material, Fig. S5). APOE genotype is also associated with C1qrs expression in Covid patients (14) in the DLPFC (negative correlation with E2, positive with the APOE E4 allele). Indeed, many CS components correlate positively with AD progression. Immunoglobulin-G antibodies, whose responses to epitopes are key to Covid (52) immune response, correlate positively with moderate but not severe AD. Fibrinogen and the SELP protein changed from negative to positive associations from moderate to severe AD. Supplementary Material, Figure S6 shows correlations between other shared AD–Covid mechanisms and AD phenotypes (e.g. Hippocampus: Tumor Necrosis Factor Receptor (TNFR) with severe AD, IkappaB kinase (IKK) with cognitive impairment; LTL: IKK with neuritic plaques; DLPFC: c-Jun N-terminal kinases (JNKs) with cognitive resilience).
ML prediction of Covid-19 severity from AD-Covid-related GRNs
There are several other shared AD–Covid mechanisms. Supplementary Material, Files S6 and S7 have results for this section. We normalized gene expression data of a recent Covid-19 cohort (N = 50 Intensive Care Unit (ICU) vs. N = 50 non-ICU human samples) (108) and identified 5,085 differentially expressed genes (DEGs, 2,505 upregulated and 2,580 downregulated) for severe Covid (ICU). We looked at our three final brain region GRNs related to the 22 shared genes between AD and Covid KEGG networks, including TFs that regulate them and/or their TGs i.e. AD–Covid GRNs (Supplementary Material, Table S2). We then identified the AD–Covid lists from these three AD–Covid GRNs and filtered these gene lists down to only include the genes from the modules associated with AD phenotypes (21, 28, and 35 modules for the Hippocampus, LTL and DLPFC, respectively). Finally, a combined AD–Covid gene list had 2,153 genes (pooling our final AD–Covid genes lists from the Hippocampus: 1,146 genes, DLPFC: 895, LTL: 322) of which 733 are severe Covid DEGs. Covid severity correlates positively with many AD KEGG mechanisms and vice-versa (Supplementary Material, Figs S7 and S8). Seven DEGs are in all four gene lists (5 upregulated genes like SPI1, 2 downregulated genes: PIK3R3 and STAT2). AD–Covid genes strongly associate with Covid severity.
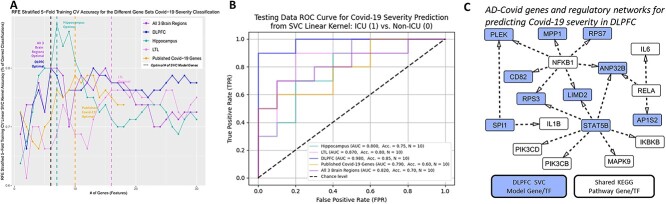
We applied support vector machine (SVM or SVC, Materials andMethods) models to predict the probability of severe Covid in Covid patients. Each model was trained using this normalized Covid gene expression data for a list of genes. We applied recursive feature elimination (RFE) cross validation (CV) on an SVM model for the 18 benchmark Covid genes [from previous studies (53–56)] (Fig. 4A); RFECV found 10 benchmark genes were optimal (highest 5-fold stratified CV accuracy on training data). We ran RFE on each of our five lists (benchmark and 4 AD–Covid lists) to select the top 10 optimal genes (predictive for Covid severity) for each list (on the basis of the training data), which we then used to build our benchmark model and four AD–Covid models, respectively. Forty-six genes were found across all five input lists (benchmark, combined, Hippocampus, LTL, DLPFC); the 36 genes from our four AD–Covid models are our AD–Covid genes (we found 0 overlaps with the 10 benchmark genes). Our four AD–Covid models outperformed the benchmark model on training data with higher average area under the receiver-operator characteristic curve (AUC) (Supplementary Material, Fig. S9) and accuracy (Supplementary Material, Table S3). Our models perform better than the benchmark model on test data (20 balanced samples) with higher AUC (except for the LTL model) and accuracy (Fig. 4B). Relative to the benchmark model, the DLPFC model (optimal; accuracy: 85%, AUC: 0.98) boosted accuracy by 25% and AUC by 0.19. Decision curve analysis (DCA, Supplementary Material, Supplementary Methods) found that our models generally have higher Net Benefits than the benchmark model across all probability thresholds (average Net Benefit increase of 0.153) and therefore have a greater clinical usability (Supplementary Material, Fig. S10). Hence, using our optimal AD–Covid model (for a given probability threshold) on average increases the number of truly severe Covid patients detected by approximately 153 per 1,000 Covid patients, without changing the number of non-severe patients who are needlessly sent to the ICU. Overall, our 36 genes have higher predictability for Covid severity than benchmark Covid genes on new Covid patient blood gene expression data. Our AD–Covid models may provide potential novel strategies to guide clinical decisions on sending Covid patients to the ICU or not.
Figure 4.
Predicting Covid-19 severity using AD–Covid GRNs. (A) Prediction accuracy of Covid-19 severity after selecting different numbers of genes from AD–Covid GRNs and recently found Covid-19 genes (benchmark genes). The accuracy was calculated on the basis of the support vector machine classification (SVM or SVC) model with 5-fold stratified cross-validation on 80 balanced training samples. The dashed lines correspond to the minimal numbers of select genes with the highest accuracy (i.e. optimal gene sets for predicting Covid-19 severity). (B) Receiver operating characteristic curves and corresponding AUC values for classifying Covid-19 severity in the test data of 20 balanced samples using the SVC machine learning models. (C) Subnetwork of the DLPFC GRN relating to the 10 AD–Covid DLPFC genes for predicting Covid-19 severity (N = 10) with the shared KEGG genes. Blue: genes/TFs found in the optimal DLPFC final model (which are also 10 of the 36 AD–Covid genes). White: 1 of the 22 shared KEGG genes (between AD and Covid KEGG networks). There is no overlap between both sets of genes.
We found that our 36 AD–Covid genes driving Covid severity may also drive neuroinflammation, which is predictive of AD. For this, we trained a logistic regression (LR) model to predict the probability of AD using Superior Frontal Gyrus (SFG) brain region gene expression data. Three AD–Covid genes (ANP32B, GPI, SPI1) are AMP-AD (57) nominated AD genes. GPI promotes neuron survival and immune-functions [e.g. serves as tumor-secreted cytokine (58)]. TF SPI1 regulates immune functions and microglia-mediated neurodegeneration in AD (59), and correlates strongly with AD/Braak Progression in the Hippocampus. We used 35 of those 36 genes (SMIM27 was missing) and added four binary features to control for the four cell-types (39 features in total). We compared our test performance for 24 samples: (12 AD, 12 Control) with that of a LR model using 597 AMP-AD (57) genes (601 features). Our AD–Covid LR model outperformed the AMP-AD LR model: AUC (0.583 vs. 0.569), accuracy (70.3% vs. 62.5%), DCA (29 optimal probability thresholds vs. 21) (Supplementary Material, Fig. S11).
Thus, our 36 AD–Covid genes are predictive of not only Covid severity but also of AD, as they performed better than their respective benchmark models and have promising clinical translational ability for predicting immune dysregulation, inflammation, AD and severe/neuro Covid. Gene ANP32B [enriched in extracellular vesicles in AD mice brain tissues (60)] strongly predicts Covid severity as it was found in DLPFC and Hippocampus models; ATM, EMP3 and LILRA6 were found in Hippocampus and combined models. Supplementary Material, Figure S12 reveals how 13 of these 36 genes are DEGs in excitatory (ExN) and/or inhibitory (InN) neurons for AD pathology overall and/or early AD pathology versus none using recent data (61); for instance, SPI1 is downregulated in ExN in both comparisons, whereas MYLIP is upregulated in InN for early AD (versus controls). We highlight the DLPFC GRN subnetwork for all 10 predictive genes directly regulating or regulated by at least 1 of the 22 shared KEGG genes (Fig. 4C, Hippocampus/LTL: Supplementary Material, Fig. S13). Our three brain region GRN subnetworks reveal TF–TG interactions that may predict pro-inflammatory cytokine levels and neuroinflammation. NFKB1 and RELA regulate several genes across all three regions, associated with immune dysregulation (able to predict Covid severity), like ANP32B in the DLPFC. SPI1 and NFKB1 target PLEK, whose expression is linked to synapse failure and cognitive dysfunction in AD (62). STAT5B regulates glucocorticoid receptor activity, which impacts the expression of pro- and anti-inflammatory genes (4). We found that STAT5B jointly regulates PI3K subunits PIK3CD and PIK3CB in the DLPFC (NFKB2 regulates PIK3R1 in the LTL); altered PI3K (shared AD–Covid mechanism) signaling may increase IRF5 activity (25) in AD (63) and in severe Covid. These 10 DLPFC SVC model-based AD–Covid genes are enriched (64) with immune system diseases like Hodgkin Lymphoma, T-cell Leukemia, Waldenstrom Macroglobulinemia.
Identification of disease risk variants for AD phenotypes via integration of genome-wide association studies (GWAS) and GRNs
It is crucial to understand how non-coding disease-associated SNPs [over 90% of risk SNPs (65)] affect gene regulatory mechanisms that eventually impact AD phenotypes. For this purpose, we looked at both AD SNPs and Covid-19 severity (on the basis of hospitalization status upon Covid infection) SNPs from recent GWAS. We did this for two main reasons. First, studies have found that even mild Covid-19 infection is associated with brain changes (66) and that severe Covid SNPs may contribute to cognitive dysfunction (9), thereby worsening AD phenotypes. Second, incorporating severe Covid SNPs may help us discover how Covid-related genetic risk variants are associated not only with Covid severity but also with AD and cognitive impairment (e.g. neuro-Covid), both of which are currently unknown (9). We mapped AD SNPs and Covid severity SNPs onto our final GRNs to see how these SNPs alter TFBSs on regulatory elements (enhancers and/or promoters) that regulate target genes (TGs) and gene modules. Furthermore, we linked these SNPs to AD phenotypes of corresponding TGs and modules for our three brain regions i.e. ‘brain-region SNP-effected-GRN for AD phenotypes’: SNP–TF–Regulatory Element–TG–Module–Phenotype. Thus, we could predict how AD and/or severe Covid SNPs impact TF regulation of TGs that belong to modules enriched with biological functions; TGs and modules may associate with AD phenotypes. Our SNP-effected-GRN predicted 144,098 total unique SNP–TF–TG relationships across the three regions (for 17,795 SNPs impacting TFBSs, 14 common AD–Covid SNPs); 6,245 SNP–TG relations had at least one validated blood/brain expression quantitative trait loci (eQTL) link. Supplementary Material, File S8 has metrics and our annotated SNP-effected-GRN. Below, we highlight some of the strong examples from our many SNP-effected-GRN predictions.
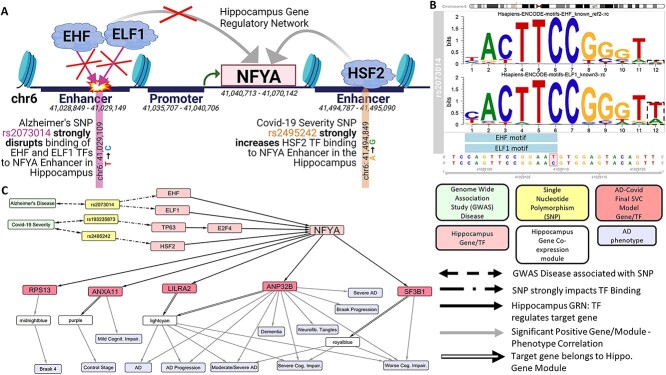
Our SNP-effected-GRN may predict how AD and/or severe Covid SNPs may alter the expression of TGs like our 36 AD–Covid genes. In Figure 5 we use our Hippocampus SNP-effected-GRN to focus on Nuclear Transcription Factor Y Subunit Alpha (NFYA), 1 of 525 common TGs dysregulated by AD SNPs and by severe Covid SNPs. Figure 5A shows many predicted NFYA enhancers. AD SNP rs2073014 strongly hinders EHF and ELF1 TFs [that both belong to the ETS TF family (67)] from binding to an NFYA enhancer. On the other hand, severe Covid SNP rs2495242 strongly increases HSF2 regulation of NFYA. SNP rs2073014 is a mutation that changes the DNA base from a T to a C at chromosome 6 position: 41,029,109, disrupting EHF and ELF1 motifs (Figure 5B); both motifs are significantly enriched in all single-cell assay for transposase-accessible chromatin (scATAC-seq) peaks (typically corresponding to enhancers) (67) of open chromatin in microglia. We found forty-eight prefrontal cortex (PFC) eQTL SNPs associated with reduced NFYA expression that correlate positively with rs2073014 on the basis of linkage disequilibrium (LD) analysis; this verifies that rs2073014 is associated with NFYA expression in the brain. Rs193235873, a Covid severity SNP, increases TP63 regulation of E2F4 (and TP63 significantly regulates E2F4’s Braak 6 stage module), which in turn regulates NFYA (Fig. 5C). NFYA is associated with AD (68), plays a key role in various cancers (69) and regulates 5 AD–Covid genes like LILRA2 and ANP32B (their shared module positively correlates with worse AD phenotypes). Supplementary Material, Figure S14a identifies cell-type eQTL SNPs impacting the regulation of ANXA11 (an AD-Covid gene) by five TFs, like NFKB1, in the Hippocampus. In particular, SNPs linked to our AD–Covid genes and correlated with AD phenotypes may help explain genetic mechanisms of critical illness, neuroimmunology and cognitive impairment in Covid (54) and in AD.
Figure 5.
Select SNP regulatory networks (SNP-effected-GRNs) linking AD and Covid-19 severity risk variants (GWAS SNPs) to AD phenotypes. This example utilizes the SNP-effected-GRN in the Hippocampus. (A) This focuses on how an AD SNP rs2073014 disrupts binding of two TFs (EHF and ELF1) to an NFYA enhancer. Another Covid-19 severity SNP instead increases HSF2 TF binding and subsequent regulation (activation or repression is unknown) of NFYA. (B) This shows how AD SNP rs2073014 interrupts the binding sites of EHF and ELF1 TFs in the Hippocampus (on the basis of their respective sequence-specific motifs). (C) On the bottom right is a legend for this network. Bi-directional dashed arrows represent SNP association with AD and/or severe Covid-19 on the basis of summary statistics from recent GWAS. Arrows with dots in the middle represent that the SNP strongly impacts TF Binding (either increasing or disrupting it; details available in Supp. Files). The solid arrow represents that the TF regulates that given TG on the basis of the Hippocampus GRN. Grey arrows represent that the TG and/or TG module are statistically significantly positively correlated (P < 0.05, r > 0) with that given AD-related phenotype. In this figure, we analyze the role of one AD SNP and two Covid severity SNPs in eventually impacting the regulation of NFYA in the Hippocampus. For instance, SNP rs193235873 impacts the ability of TP63 to regulate E2F4 that then regulates NFYA. Furthermore, we show how NFYA regulates five AD–Covid genes (RPS13, ANXA11, LILRA2, ANP32B, SF3B1) and we highlight the Hippocampus gene modules for these 5 genes (using black arrows that contain a white rectangle). In addition, we link the gene modules and these five AD–Covid genes with their respective AD-related phenotypes. Thus, we predict how AD SNPs and Covid-19 severity SNPs may eventually impact the regulation of 5 of our 36 AD–Covid genes that are associated with various AD phenotypes.
We predicted five shared AD and severe Covid SNPs in IFNAR2 Hippocampus and/or LTL enhancers that may impact regulation of IFNAR2, a known Covid severity gene (54). These five SNPs are in LD with blood eQTL SNP rs7509997 that is strongly positively associated (P = 3.31e-49) with IFNAR2 expression. During AD, cytokine CSF3R’s overexpression in the LTL may be partly explained by SNPs like rs483341 that disrupt the ability of TF REST [protects neurons from Aβ-protein toxicity (70)] to bind to chromatin to repress its TGs (71) like CSF3R, leading to inflammation (72). Harmful AD SNP rs2564970 (P = 5.47e-08), which is four bases from a predicted CR1 Hippocampus enhancer (chromosome 1: 207,464,045 - 207,464,283), may strongly disrupt NFKB1 and RELA regulation of CR1 (Complement receptor type 1), a major AD gene associated with the CS. Moreover, our SNP-effected-GRN predicts previously unknown SNPs and specific TFs associated with NF-κB TF activation in AD, which may make NF-κB TFs neuroprotective or neurotoxic. We predict that harmful Covid severity SNP rs2736322 disrupts RREB1’s ability to bind to an FAM167A LTL enhancer and subsequently regulate FAM167A, a TG which correlates positively with AD and Braak stages and belongs to an AD LTL module. This may explain this SNP’s negative eQTL relationship with FAM167A expression across various brain cell-types. Furthermore, we have Supplementary Material, Figures S14 and S15 that elaborate on the following select stories: AD SNPs on regulatory elements may dysregulate Hippocampus expression of three AD–Covid genes: EMP3, LILRA2, SPI1 (Supplementary Material, Fig. S14b). Harmful AD SNP rs754366 (has a positive PFC eQTL link to APOC2 expression) may increase SPI1 binding to an APOC2 DLPFC enhancer where it activates APOC2 (Supplementary Material, Fig. S14c). Non-coding AD SNPs in microglia scATAC-seq peaks impact regulation of KCNN4 (Potassium Calcium-Activated Channel Subfamily N Member 4), an AD risk microglia gene with previously no known mutations (71) associated with its expression (Supplementary Material, Fig. S15). The above stories and more (in Supplementary Material) underscore the importance of our work and findings. Moreover, our SNP-effected-GRN may help explain GRN mechanisms behind several causal blood/brain eQTL links.
Discussion
In this paper, we performed an integrative multi-omics study to predict AD GRNs along with gene co-expression modules for three major brain regions. Using these networks and modules, we further linked a number of AD–Covid genes that improve AD and severe Covid predictions, and also revealed the regulatory mechanisms of genome-wide association study (GWAS) SNPs of AD and of severe Covid-19.
Brain regions are composed of varied cell types that may impact co-expression networks and gene regulation; for example, AD patients may have fewer neurons and more immune cells. Many human brain cell-type GRNs were predicted from recent single-cell sequencing data (e.g. scRNA-seq, scATAC-seq), which enable studying cell-type functional genomics and GRNs (73). We validated that many phenotype-associated SNPs are located on regulatory elements with cell-type epigenomic activities. An integrative analysis of cell-type GRNs can also be performed to understand regulatory mechanisms for GWAS risk variants for refined AD phenotypes [e.g. cerebrospinal fluid, psychotic symptoms (46)]. Using pooled cell types in the SFG to predict AD may have confounding factors as a human sample could have many corresponding cell-type samples. That the Covid transcriptomic data was from blood samples may present limitations as AD–Covid GRNs use transcriptomic data from brain tissues. Still, researchers found immune dysregulation in both AD brain and blood samples (74). Elevated pro-inflammatory molecules in Covid patients can compromise the blood-brain barrier (the BBB breaks down in AD), enter the brain and encounter astrocytes and microglia (both cell-types malfunction in AD); such patients are more susceptible to severe Covid and further neurological damage. Thus, Covid-19 patient blood gene expression data may predict future Central Nervous System (CNS) invasion and neuroinflammation. Our findings support further research into understanding better the causal links between AD and Covid (35) (e.g. it is unknown if Covid triggers new development of AD or accelerates AD progression). Treatments and drug development can perhaps be targeted at AD–Covid pathways to alleviate patient suffering i.e. care providers may suppress Interferon response or use acetylcholinesterase inhibitors (current AD treatment strategy) (25) to stimulate the cholinergic anti-inflammatory pathway in Covid patients. Such treatments may reduce the overall risk of cognitive decline in Covid survivors (9). Currently, research in AD–Covid, long- or neuro-Covid and AD neuroimmunology is nascent and more data is being generated, which may be used to eventually expand on our current work (especially given sample size limitations of our data).
Many large scientific consortia generate matched multi-omics data of individuals like AMP-AD (57), PsychENCODE (75), TCGA (76). We can extend our machine learning (ML) analysis to predict personalized phenotypes and prioritize phenotype-specific functional genomics and GRNs in diseases from this data. We found TFs regulate many gene modules and link to AD phenotypes, suggesting possible collinearity driven by TF regulations across phenotypes. Emerging ML approaches like neural networks may decouple phenotypic collinearity, uncovering phenotypic-specific TFs. Studies emphasize systems biology and ML approaches (like ours) to identify biomarkers for neuroinflammation (46) in AD, Covid-associated cognitive impairment (9) (e.g. neuro-Covid or long-Covid) and Covid severity. Neuroimmunology research is discovering the role of dysregulated immune responses in other complex neurologic diseases like Schizophrenia (SCZ), Amyotrophic Lateral Sclerosis, Myasthenia Gravis, Parkinson’s disease, Multiple Sclerosis (77). For instance, overactivated NF-κB signaling is also found in Post-Traumatic Stress and Bipolar Disorders (4). Covid-19 may similarly be used to understand the role of misguided immunity in SCZ, as SCZ is an autoimmune disease (excess pruning of synapses by microglia) and the second largest risk factor for Covid-19 death after age (78). Overall, we hope that our approach can be applied to help understand molecular mechanisms in other diseases by uncovering the association of orphaned GWAS loci in non-coding DNA regions with disease phenotypes and by using closely related diseases to help reveal additional mechanisms at play.
Materials and Methods
The pipeline of our integrative analysis for predicting gene regulatory mechanisms from AD and/or severe Covid-19 risk variants to AD phenotypes
Our analysis can be summarized as a pipeline to predict SNP-effected-GRNs (linking SNPs to GRNs) from disease risk variants to phenotypes (Fig. 1). The SNP-effected-GRNs for specific phenotypes link disease risk SNPs, non-coding regulatory elements and TFs to genes and genome functions, providing comprehensive mechanistic insights on gene regulation associated with disease phenotypes. Specifically, the pipeline includes the following steps. Here, our analysis is open-source available at https://github.com/daifengwanglab/ADSNPheno. We used human reference genome: hg19 (GRCh37: Genome Reference Consortium Human Build 37) for our genomic coordinates and our following analysis.
Step 1: Input population gene expression data of individuals and clinical information on AD phenotypes such as Braak staging and progression.
Step 2: Input data are used to construct a gene co-expression network linking all possible gene pairs. Network edge weights are correlations of gene expression profiles across input samples. The network is clustered further into gene co-expression modules. Genes in a module are likely to share similar functions and be co-regulated by specific regulatory mechanisms.
Step 3: Annotate gene co-expression modular functions and biological pathways by enrichment analyses of genes in the given module (using various biological resources).
Step 4: Associate modules and genes with AD phenotypes of the input samples, revealing potential driver genes (e.g. hubs) and modules for these phenotypes.
Step 5: Predict gene regulatory networks (GRNs) for genes and gene modules. We apply multiple computational methods to predict GRNs that link TFs to non-coding regulatory elements (e.g. enhancers, promoters) to genes and modules, providing regulatory mechanistic insights on AD genes and modules.
Step 6: Link disease risk variants [e.g. Single Nucleotide Polymorphisms (SNPs)] to the gene regulatory network. Our pipeline identifies functional AD and/or severe Covid risk SNPs that alter (increase or decrease) TF binding to TF binding sites (TFBSs) in regulatory elements in the GRN i.e. regulatory SNPs. We can then connect these non-coding regulatory SNPs to genes and modules and then to AD phenotypes and biological enrichments.
Step 7: Output a SNP-effected-GRN that links AD and/or severe Covid risk SNPs, non-coding regulatory elements, TFs to genes and their gene modules, genome functions (via module enrichment analysis in Step 3) for AD phenotypes in the input data. This network has SNP-Regulatory Element-TF-gene-module-phenotype links.
Population gene expression data and data processing in Alzheimer’s
We applied this pipelined analysis to post-mortem human AD population gene expression data for three major AD brain regions: Hippocampal CA1 (Hippocampus), LTL, DLPFC. We removed lowly expressed genes (zero variance; relative weights below 0.1), using the goodSamplesGenes() function in the weighted gene co-expression network analysis (WGCNA) (79) package in R. There are 12,183 shared genes across these three regions, including non-coding genes. We did not pre-adjust gene expression data using covariates (e.g. patient metadata) as those are used downstream as phenotypes. We performed feature engineering to create additional phenotypes for the human samples. We processed the data as follows, striving to meet quality control standards.
Hippocampus: We used microarray gene expression dataset (GSE1297) (80), which had total RNA expression values for 22 283 HG-U133 Affymetrix Human Genome U133 Plus 2.0 Microarray Identifier probes for 31 individual samples [9 control (no AD) and 22 samples in various AD stages: 7 initial, 8 moderate, 7 severe]. We used GEOquery (81), hgu133a.db (82), hgu133acdf (83) and Affy (84) R packages to download raw gene expression data and perform Robust Multichip Average (RMA) normalization (85) to account for background and technical variations among the samples. We mapped microarray probes to genes, averaging values that mapped to the same gene Entrez ID and removing unmapped probes. We applied a log2(x + 1) transform to the gene expression data (the x) and then standardized that data by R’s (86) scale() function. The final Hippocampus gene expression data has 13,073 genes for these 31 samples.
LTL: We used normalized bulk RNA-Seq dataset (GSE159699) (27) with total RNA expression values for 27,130 different genes for 30 individual samples. This group of individual samples includes 18 control samples [8 young (below age 60), 10 old (above age 60)] and 12 old samples with advanced AD. We applied a log2(x + 1) transformation to the data. The final LTL gene expression data has 25,292 genes for these 30 samples.
DLPFC: We used FPKM (Fragments per kilobase of exon per million mapped fragments) gene expression data from the ROSMAP Study on synapse.org (ID: syn3219045) (87). We found that 638 out of 640 individual RNA-Seq samples have mapped phenotypes. For instance, for the final consensus cognitive diagnosis (cogdx) phenotype on cognitive impairment: we have 201 samples in Group 1 (none), 168 in Groups 2–3 (mild), 269 in Groups 4–6 (AD/other dementia). We applied a log2(x + 1) data transform and then standardized the data with R’s scale() function (86). The final DLPFC gene expression data has 26,014 genes for these 638 samples.
Regulatory elements and chromatin interactions in human brain regions
Epigenomic data has identified a variety of regulatory elements like enhancers and promoters. Chromatin interaction data (e.g. Hi-C) further revealed interactions among enhancers and gene promoters. Thus, we integrated recent published epigenomic and chromatin interaction data to link enhancers to genes (via promoters). For the Hippocampus, we obtained enhancers and promoters from Brain Open Chromatin Atlas (88) and promoter-based interactions from GSE86189 (89). R package TxDb.Hsapiens.UCSC.hg19.knownGene (90) retrieved promoter start and stop positions of genes in the LTL and DLPFC, using a short ultra-conserved promoter length of 5,000 base pairs upstream of the protein-coding start site on the DNA (91). GSE130746 (27) H3K27ac (DNA Histone H3 protein acetylation of the lysine residue that is found at the N-terminal position 27 for H3) data (used for the LTL) has information on the gene, distance from the histone H3K27ac epigenetic mark to that gene’s Transcription Start Site (TSS), enhancer start and end positions; our final LTL enhancers were at least 1 kilobase pair (kbp) away from the TSS. We used PsychENCODE (75) enhancers and interacting enhancer-promoter pairs for the DLPFC.
Gene co-expression network analysis
We applied WGCNA (79) to population gene expression data to construct and cluster gene co-expression networks into gene co-expression modules (minimum module size = 30 genes; no modules were merged). Then, we applied an additional K-Means clustering step on the basis of code (92) and methodology that has been previously utilized and proven to improve conventional WGCNA module assignments and functional enrichments, in applications like finding brain-specific cell-type marker enrichments (93). This step utilizes modular eigengenes (MEs) from WGCNA modules as initial centroids, initial gene assignments from WGCNA and computable distance between the genes and MEs for K-Means to re-assign genes to optimal modules (retaining the number of modules originally detected by WGCNA) across the iterations (till convergence). In total, we obtained 30 gene co-expression modules for the Hippocampus (13,073 genes), 56 for the LTL (25,292 genes), 35 for the DLPFC (26,014 genes).
Enrichment analyses of gene co-expression modules
Co-expressed genes in the same module are highly likely to be involved in similar functions and pathways. Enrichment analysis has thus been widely used to identify such functions and pathways in a gene module. P-values for enrichments were adjusted using the Benjamini–Hochberg (B-H) correction procedure (for multiple hypothesis testing) and enriched terms with adjust P-value < 0.05 were selected. Given a group of genes (e.g. from a module) for each brain region, we performed enrichment analysis using multiple tools and their respective gene databases [e.g. Metascape (64), g:Profiler (94), WGCNA (79), rentrez (95), Bader Laboratory (96), Maayan Laboratory (97), ABAEnrichment (98), Psygenet2r (99), TissueEnrich (100), ClusterProfiler (101,102), CellMarker (103)]. Since we used multiple tools for enrichment analysis, a gene module could have many −log10(adjust P) enrichment values for a given enriched term; in that case, we used the highest enrichment value for that term for the module. To visualize enriched terms for a phenotype in a brain region, we averaged non-zero −log10(adjust P) values for only the gene modules that are significantly positively correlated (Pearson r > 0, P < 0.05) with that phenotype.
Association of genes and modules with AD phenotypes
We further associated genes and modules with these key AD developmental phenotypes, including: AD stages and progression (moderate stage, severe stage, AD Progression), healthy/resilient (Control individuals or resilient individuals with better cognitive abilities despite advanced AD pathology), APOE genotype [E4/E4 is a huge AD risk factor, while E2/E2 is protective (104)], Braak staging [stages from 1 to 6, with 6 linked to severe neuropathological damage and spread of neurofibrillary tangles across the brain], neuritic plaque accumulation (by CERAD score), cognitive impairment level. We associated gene co-expression modules with all possible AD phenotypes from the input data, by computing the pairwise correlations of each modular eigengene (ME) with each phenotype. WGCNA’s MEs are the first principal components of modular gene expression; an ME is a vector representing gene expression levels of input samples and is the likeliest gene expression pattern of the genes in that module. We used WGCNA’s moduleTraitCor() and moduleTraitPvalue() functions to correlate these MEs with phenotypes, finding the most significantly positively associated phenotypes for our gene modules for our analysis (P-value < 0.05, positive correlation r). Our modules of interest i.e. ‘phenotype-enriched modules’ are positively correlated with at least one AD-related phenotype (including the control stage phenotype since such modules may typically be down-regulated in expression during AD progression). We performed similar analysis (that we used for the MEs) for each of our genes using the expression data for that given gene to find significant phenotypes positively associated with that gene in that brain region. We used gene co-expression networks to examine the relationship between genes and AD phenotypes and identify potential driver (hub) genes for modules (based on the degree of connectivity for each gene in its respective gene co-expression module).
Prediction of GRNs from multi-omics
GRNs, a key molecular mechanism, fundamentally control gene transcription and expression. Co-expressed genes are likely co-regulated by similar GRNs. Thus, our analysis integrates multiple methods to predict GRNs from gene expression data. We predicted GRNs in brain regions using not only gene expression data but also chromatin interaction data to link TFs to regulatory elements to target genes (TGs)/modules. For our full DLPFC GRN, we used the published PsychENCODE GRN (Elastic Net regression weight cutoff: 0.1) filtered for genes in the DLPFC gene expression data (105). Our full GRNs linked TFs to regulatory elements (enhancers/promoters; chromosome #: regulatory region start—end) to TGs.
We used these four steps to construct our full Hippocampus GRN and full LTL GRN. First, we identified regulatory elements (e.g. enhancers, promoters) that potentially interact using recent chromatin interaction data (Hi-C) and Step 1 of the scGRNom pipeline (106). Second, we infer TFBSs on the basis of consensus binding site sequences on interacting enhancers and promoters by TFBSTools (107) and motifmatchr (108) using Step 2 of the scGRNom pipeline (106). We generate a chromatin interaction-based reference network linking TFs to regulatory elements (by TFBSs) to TGs (by interactions). Third, using gene expression data for a given brain region, we predicted all possible TF–TG pairs (or TF-modules) with strong expression relationships by applying three widely used tools: RTN (109), TreNA Ensemble Solver (110), GENIE3 (111) (and TF-gene-module pairs by RTN). Thus, we created a gene expression-based network by combining TF–TG pairs found by at least two of these three tools. Fourth and finally, we mapped the gene expression-based network TF–TG pairs to the TF-TG pairs in the chromatin interaction-based reference network. The full GRN (for the Hippocampus or the LTL) thus contains TF–TG pairs found in both the chromatin interaction and the gene expression-based networks (see Supplementary Materials, Supplementary Methods for more details).
For each of the three brain regions, we built final GRNs by using our prior analysis (see earlier), which had assigned TGs to gene co-expression modules and associated the modules with AD phenotypes and biological enrichments. This prior analysis provided richer annotations for TGs in our full GRNs. Our final GRN for each brain region comprehensively linked TFs to non-coding regulatory elements to TGs and these TGs to gene modules to AD phenotypes/enrichments.
Identification of AD-Covid GRNs and genes using GRNs and gene modules
To investigate potential mechanistic interplays between AD and Covid-19, we compared AD (hsa05010) and Covid-19 (hsa05171) KEGG networks (112) and found AD–Covid mechanisms like: NF-κB, Inhibitor of Nuclear Factor Kappa B Kinase (IKK), c-Jun N-terminal Kinase (JNK), Interleukin-6 (IL-6), Phosphoinositide 3-Kinase (PI3K), Tumor Necrosis Factor alpha (TNFa), TNF Receptor (TNFR). We found a statistically significant overlap of 22 genes between both KEGG networks on the basis of a hypergeometric test (7,559 human genes in the KEGG universe, 384 human KEGG genes in AD, 232 human KEGG genes in Covid-19). The 22 shared KEGG genes correlate highly with AD phenotypes in different brain regions. This motivated us to find neuroimmunology genes in AD–Covid. We used the Pathview (113) tool to visualize correlations of KEGG network mechanisms with AD phenotypes. For each region, we constructed an AD–Covid gene list using its respective final GRN and gene co-expression modules as follows. First, we built an AD–Covid GRN: a subnetwork of the GRN with TFs regulating and/or TGs of the 22 shared KEGG genes such that each GRN edge contains at least 1 shared KEGG gene. Second, we filtered these AD–Covid GRNs to only include genes that belong to an ‘AD-phenotype enriched’ gene module. Hence, genes in our AD–Covid GRN (our AD–Covid genes) were either 1 of the 22 shared KEGG genes or directly linked to them by a GRN link. Moreover, the AD–Covid genes had altered expression dynamic patterns associated with AD. Thus, we built four AD–Covid gene lists: Hippocampus, LTL, DLPFC and a combined list of the three. These four lists were later used to predict Covid-19 severity (see next).
Gene expression analysis and ML prediction for Covid-19 severity from AD-Covid GRNs
To gauge the clinical performance of our AD–Covid genes in terms of predicting Covid severity (proxy for immune system dysregulation), we looked at recent population RNA-seq gene expression data of human Covid-19 blood samples (GSE157103) (114). We median normalized this data (19,472 genes) and applied differential expression analysis by DESeq2 (115) between 50 severe (Intensive Care Unit (ICU)) and 50 non-severe (non-ICU) Covid patients. Aside from applying differential expression analysis to find individual-associated genes, we performed machine learning (ML) analysis to determine if any of our four AD–Covid gene lists (from our AD–Covid GRNs) and the respective normalized blood gene expression data could predict the probability of severe Covid (being in the ICU) for Covid patients better than a benchmark list of Covid genes could. We used a SVM classifier model [linear kernel, balanced class weights, on the basis of Python’s Scikit-Learn (116) svm.SVC package] to output the predicted probabilities of severe Covid for Covid samples. We randomly partitioned our data using an 80–20 training–testing split with 80 samples (40 ICU, 40 non-ICU) in training data and held out 20 samples (10 ICU, 10 non-ICU) in test data. Stratified 5-Fold Cross Validation (CV) was used to calculate training classification accuracy; each fold held out 16 samples (8 from each class) for validation and trained an SVM model on the remaining 64 samples (32 from each class). Input data used to build each model was the median normalized Covid gene expression data for the respective selected genes (features) for the training samples. We did not use age and gender as predictors given their low correlations with Covid severity.
For our ML analysis, we gathered a benchmark list of 18 known and published Covid genes from the following four studies (53–56). A study (53) used U.K. Biobank GWAS and Covid mortality data to discover eight genes associated with high Covid mortality: DNAH7, CLUAP1, DES, SPEG, STXBP5, PCDH15, TOMM7, WSB1. Another study (54) has identified seven risk genes (OAS1, OAS2, OAS3, TYK2, DPP9, IFNAR2, CCR2) associated with life-threatening Covid outcomes (e.g. inflammatory organ damage). Numerous studies (55) implicate SNPs in ACE2 and TMPRSS2 as risk factors for Covid susceptibility. Another Covid severity study used a Random Forests ML model and has identified VEGF-D as the most predictive indicator (56). To build our benchmark model, we first performed RFE CV (RFECV) on a SVM model using these 18 benchmark genes to calculate the accuracy of adding a gene to the model and optimal number of genes to use i.e. smallest number of genes with the maximum stratified 5-fold CV training accuracy when classifying ICU versus non-ICU Covid patients. Second, we ran RFE on a SVM model with that optimal number of genes to select the predictive genes from the training data. Third, we used these selected benchmark genes to train another SVM model as our benchmark model. We fixed all models to use this same number of genes to help facilitate direct comparison of the predictive models. We performed the second and third steps instead on each of our respective input AD–Covid gene lists to build our four AD–Covid models. Thus, we built five models to predict Covid severity: benchmark, combined, Hippocampus, LTL, and DLPFC. We compared the prediction performance of each of our four AD–Covid models with that of the benchmark model using: accuracy, AUC and Decision Curve Analysis (DCA, Supplementary Materials, Supplementary Methods) on training and test (generalize potential clinical impact of models) data. For each model, we report training metrics by averaging values across all five stratified folds. We flagged ‘AD–Covid genes’ used in any of our four AD–Covid models (predictive for severe Covid) as potential candidate biomarkers for AD–Covid-neuroimmunology.
ML prediction for AD and Covid severity from AD–Covid genes
We analyzed the performance of our AD–Covid genes (those from among our 4 AD–Covid models) for predicting AD on a new human population cohort (GSE125050) (117) of 22 AD and 21 control postmortem SFG tissues in the frontal cortex (linked with AD pathology). That study isolated RNA-seq data for four brain cell types (neurons, astrocytes, endothelial cells, microglia) from SFG tissues. We pooled raw gene expression data for these four cell-types (62 control, 46 AD samples, Supplementary Material, Table S4) for our task. For each cell type, we held out three AD and three Control cell-type samples for testing (total: 12 AD, 12 Control). The remaining 84 samples were used to train a LR model (Python’s Scikit-Learn (116) Logistic Regression package, liblinear solver, balanced class weights) to predict AD or control for a given sample. Our features were pooled gene expression data values for AD–Covid genes and four dummy (0 or 1) features noting the cell-type for each sample. We built another LR model as a benchmark, only changing the gene features we used, which were now 597 AMP-AD (58) nominated genes for AD identified in the SFG gene expression data. The AMP-AD consortium (58) flagged these AMP-AD genes as good targets for AD treatment and/or prevention on the basis of computational analyses in previous studies of human samples. We kept the shared and common AMP-AD and AD-Covid genes as gene features to train both LR models. We compared the test performance of the optimal AD–Covid LR model and benchmark AMP-AD LR model to better quantify the effectiveness of our AD–Covid genes in predicting AD as well. Furthermore, we noted our AD–Covid genes that were differentially expressed genes (DEGs) in recent single-cell transcriptomic data (61) analysis for AD pathology versus controls in Excitatory (ExN) and/or Inhibitory (InN) neurons.
Linking GWAS SNPs for AD and for severe Covid-19 to gene regulatory elements
Genome-Wide Association Studies (GWAS) have identified genetic risk variants associated with diseases like AD. However, most AD SNPs lie on non-coding regions, hindering finding AD genes and understanding downstream disease functions. We consider SNPs with P < 5e-5 to include candidate disease SNPs via interrupting gene regulation at large (Supplementary Material, Table S5). We looked at summary statistics of 26,969 AD risk GWAS SNPs across five studies (118–122) and 1,642 SNPs from the seventh round of GWAS meta-analyses related to severity across all Covid-19 positive human populations (123): 16,512 hospitalized cases (severe) versus 71,321 not hospitalized controls (non-severe). We mapped SNPs to regulatory elements in the GRNs via altered TFBSs (Fig. 1 pipeline step 6). We overlapped 28,597 AD and/or severe Covid risk SNPs (14 common) with regulatory elements (enhancers or promoters) in our final GRNs. MotifbreakR (124) identified 24,576 SNPs altering TFBSs of 791 TFs. These regulatory SNPs either increase TF affinity for the TFBS (on the basis of TF sequence-specific motif) or interrupt and subsequently decrease TF binding to that regulatory region. We linked these SNPs to TGs from regulatory elements with altered TFBSs, adding a 10 kilobase (kbp) and 2 kbp buffer extension to the start and end positions of enhancers and promoters, respectively. Thus, we mapped our SNPs to our final GRNs. Our SNP regulatory network (SNP-effected-GRN: SNP effect on our final GRN) comprised our predicted SNP–Regulatory Element–TF–TG–Module–Phenotype links. We used expression quantitative trait loci (eQTL) data (associates SNP with changes in TG expression) from various sources [Supplementary Material, Table S6; tissues: brain (75,125,126)/blood (126); cell-types: brain (127,128)] to annotate SNP-effected-GRN links with this external SNP-TG validation as highly-confident; our SNP-effected-GRN may explain GRN mechanisms behind these causal eQTL links. Furthermore, we performed linkage disequilibrium (LD) via LDlink (129) (GRCh37 genome, all human populations) to correlate a pair of SNPs (on the same chromosome); linked SNPs have significantly correlated alleles and tend to be non-randomly inherited together in all populations. Supplementary Material, Supplementary Methods have more details and a framework for analyzing our SNP-effected-GRN.
Supplementary Material
Acknowledgements
The authors are grateful to Jonathan Edward Bryan for his assistance in tasks related to this project and Renu Poochie Khullar and Armaan Khullar for their helpful comments on the manuscript.
Conflict of Interest statement. None declared.
Contributor Information
Saniya Khullar, Department of Biostatistics and Medical Informatics, University of Wisconsin – Madison, Madison, WI 53076, USA; Waisman Center, University of Wisconsin – Madison, Madison, WI 53705, USA.
Daifeng Wang, Department of Biostatistics and Medical Informatics, University of Wisconsin – Madison, Madison, WI 53076, USA; Waisman Center, University of Wisconsin – Madison, Madison, WI 53705, USA; Department of Computer Sciences, University of Wisconsin – Madison, Madison, WI 53706, USA.
Funding
National Institutes of Health, R01AG067025, RF1MH128695, R03NS123969, R21NS127432, R21NS128761 and U01MH116492 to D.W. and P50HD105353 to Waisman Center. S.K. was supported by an NLM training grant to the Computation and Informatics in Biology and Medicine Training Program (NLM 5T15LM007359).
Authors’ Contributions
D.W. conceived and designed the study. S.K. and D.W. analyzed the data and wrote the manuscript.
Ethics approval and consent to participate
Not applicable. Public data was utilized for this analysis as well as approved ROSMAP data for the DLPFC.
Consent for publication
All authors read and approved the final manuscript.
Data availability
Our analysis (codes and data including diagnosis information) is open-source available at https://github.com/daifengwanglab/ADSNPheno and our functional genomics resource for AD is available at https://adsnpheno.shinyapps.io/AlzheimersDisease_SNPheno.
References
- 1. Alzheimer’s Statistics . Alzheimers.nethttps://www.alzheimers.net/resources/alzheimers-statistics.
- 2. Rabinovici, G.D. (2019) Late-onset Alzheimer disease. Continuum (Minneap Minn), 25, 14–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Maccioni, R.B., González, A., Andrade, V., Cortés, N., Tapia, J.P. and Guzmán-Martínez, L. (2018) Alzheimer’s disease in the perspective of neuroimmunology. Open Neurol J, 12, 50–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zass, L.J., Hart, S.A., Seedata, S., Hemmings, S.M.J. and Malan-Muller, S. (2017) Neuroinflammatory genes associated with post-traumatic stress disorder: implications for comorbidity. Psychiatry Genetics, 27, 1–16. [DOI] [PubMed] [Google Scholar]
- 5. COVID Appears to Raise Risk for Alzheimer’s Disease. https://www.usnews.com/news/health-news/articles/2022-09-16/covid-appears-to-raise-risk-for-alzheimers-disease.
- 6. Amruta, N., Chastain, W.H., Paz, M., Solch, R.J., Murray-Brown, I.C., Befeler, J.B., Gressett, T.E., Longo, M.T., Engler-Chiurazzi, E.B. and Bix, G. (2021) SARS-CoV-2 mediated neuroinflammation and the impact of COVID-19 in neurological disorders. Cytokine Growth Factor Rev., 58, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. de Erausquin, G.A., Snyder, H., Carrillo, M., Hosseini, A.A., Brugha, T.S. and Seshadri, S. (2021) The chronic neuropsychiatric sequelae of COVID-19: the need for a prospective study of viral impact on brain functioning. Alzheimers Dement., 17, 1056–1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Heming, M., Li, X., Räuber, S., Mausberg, A.K., Börsch, A.-L., Hartlehnert, M., Singhal, A., Lu, I.-N., Fleischer, M., Szepanowski, F.et al. (2021) Immunity, 54, 164–175.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gordon, M.N., Heneka, M.T., Le Page, L.M., Limberger, C., Morgan, D., Tenner, A.J., Terrando, N., Willette, A.A., Willette, S.A. (2022) Impact of COVID-19 on the Onset and Progression of Alzheimer’s Disease and Related Dementias: A Roadmap for Future Research - Gordon - 2022 - Alzheimer’s & Dementia - Wiley Online Library. 10.1002/alz.12488. [DOI] [PMC free article] [PubMed]
- 10. Reiken, S., Sittenfeld, L., Dridi, H., Liu, Y., Xiaoping, L. and Marks, A.R. (2022) Alzheimer’s-like signaling in brains of COVID-19 patients - Reiken - 2022 - Alzheimer’s & Dementia - Wiley Online Library. 10.1002/alz.12558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Su, C.-M., Wang, L. and Yoo, D. (2021) Activation of NF-κB and induction of proinflammatory cytokine expressions mediated by ORF7a protein of SARS-CoV-2. Sci. Rep., 11, 13464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wang, L., Davis, P.B., Volkow, N.D., Berger, N.A., Kaelber, D.C. and Xu, R. (2022) Association of COVID-19 with new-onset Alzheimer’s disease. J. Alzheimers Dis., 89, 411–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Anderson, P. (2021) COVID-19 tied to acceleration of Alzheimer’s pathology. Medscape. AAIC. http://www.medscape.com/viewarticle/955755. [Google Scholar]
- 14. Inal, J. (2020) Biological factors linking ApoE ε4 variant and severe COVID-19. Curr. Atheroscler. Rep., 22, 70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mizuno, S., Iijima, R., Ogishima, S., Kikuchi, M., Matsuoka, Y., Ghosh, S., Miyamoto, T., Miyashita, A., Kuwano, R. and Tanaka, H. (2012) AlzPathway: a comprehensive map of signaling pathways of Alzheimer’s disease. BMC Syst. Biol., 6, 52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jha, N.K., Jha, S.K., Kar, R., Nand, P., Swati, K. and Goswami, V.K. (2019) Nuclear factor-kappa β as a therapeutic target for Alzheimer’s disease. J. Neurochem., 150, 113–137. [DOI] [PubMed] [Google Scholar]
- 17. What is NF-κB pathway? MBInfo (2018) https://www.mechanobio.info/what-is-mechanosignaling/signaling-pathways/what-is-the-nf-%ce%bab-pathway/.
- 18. Fagone, P., Ciurleo, R., Lombardo, S.D., Iacobello, C., Palermo, C.I., Shoenfeld, Y., Bendtzen, K., Bramanti, P. and Nicoletti, F. (2020) Transcriptional landscape of SARS-CoV-2 infection dismantles pathogenic pathways activated by the virus, proposes unique sex-specific differences and predicts tailored therapeutic strategies. Autoimmun. Rev., 19, 102571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Xiong, N., Schiller, M.R., Li, J., Chen, X. and Lin, Z. (2021) Severe COVID-19 in Alzheimer’s disease: APOE4’s fault again? Alzheimers Res. Ther., 13, 111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Morabito, S., Miyoshi, E., Michael, N. and Swarup, V. (2020) Integrative genomics approach identifies conserved transcriptomic networks in Alzheimer’s disease. Hum. Mol. Genet., 29, 2899–2919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wan, Q., Tang, J., Han, Y. and Wang, D. (2018) Co-expression modules construction by WGCNA and identify potential prognostic markers of uveal melanoma. Exp. Eye Res., 166, 13–20. [DOI] [PubMed] [Google Scholar]
- 22. Bartsch, T., Dohring, J., Rohr, A., Jansen, O. and Deuschl, G. (2011) CA1 neurons in the human hippocampus are critical for autobiographical memory, mental time travel, and autonoetic consciousness. Proc. Natl. Acad. Sci. U S A, 108, 17562–17567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lee, J.K. and Kim, N.-J. (2017) Recent advances in the inhibition of p38 MAPK as a potential strategy for the treatment of Alzheimer’s disease. Molecules, 22(8), 1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mapstone, M., Gross, T.J., Macciardi, F., Cheema, A.K., Petersen, M., Head, E., Handen, B.L., Klunk, W.E., Christian, B.T., Silverman, W.et al. (2020) Metabolic correlates of prevalent mild cognitive impairment and Alzheimer’s disease in adults with Down syndrome. Alzheimers Dement. (Amst), 12, e12028. 10.1002/dad2.12028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Naughton, S.X., Raval, U. and Pasinetti, G.M. (2020) Potential novel role of COVID-19 in Alzheimer’s disease and preventative mitigation strategies. J. Alzheimers Dis., 76, 21–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Roy, E.R., Wang, B., Wan, Y., Chiu, G., Cole, A., Yin, Z., Propson, N.E., Xu, Y., Jankowsky, J.L. and Liu, Z. (2020) et al., JCI - type I interferon response drives neuroinflammation and synapse loss in Alzheimer disease. J. Clin. Invest., 130, 1912–1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Nativio, R., Lan, Y., Donahue, G., Sidoli, S., Berson, A., Srinivasan, A.R., Shcherbakova, O., Amlie-Wolf, A., Nie, J., Cui, X.et al. (2020) An integrated multi-omics approach identifies epigenetic alterations associated with Alzheimer’s disease. Nat. Genet., 52, 1024–1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Inestrosa, N.C. and Varela-Nallar, L. (2014) Wnt signaling in the nervous system and in Alzheimer’s disease. J. Mol. Cell Biol., 6, 64–74. [DOI] [PubMed] [Google Scholar]
- 29. Dubey, J., Ratnakaran, N. and Koushika, S.P. (2015) Neurodegeneration and microtubule dynamics: death by a thousand cuts. Front Cell Neurosci., 9, 343. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4563776/. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Vasile, F., Dossi, E. and Rouach, N. (2017) Human astrocytes: structure and functions in the healthy brain. Brain Struct. Funct., 222, 2017–2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kellett, K.A.B. and Hooper, N.M.. 2009. Prion protein and Alzheimer disease. Prion 3, 190–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Brinton, R.D., Gore, A.C., Schmidt, P.J. and Morrison, J.H. (2009) 68 - reproductive aging of females: neural systems. In Pfaff, D.W., Arnold, A.P., Etgen, A.M., Fahrbach, S.E. and Rubin, R.T. (eds), Hormones, Brain and Behavior (Second Edition). Academic Press, San Diego, pp. 2199–2224. [Google Scholar]
- 33. Kumar, S., Iwata, Y., Zomorrodi, R., Blumberger, D.M., Fischer, C.E., Daskalakis, Z.J., Mulsant, B.H., Pollock, B.G., Graff-Guerrero, A. and Rajji, T.K. (2020) Dorsolateral prefrontal cortex metabolites and their relationship with plasticity in Alzheimer’s disease. Alzheimers Dement., 16(Suppl. 4), 1, e045879. [Google Scholar]
- 34. Hemonnot, A.-L., Hua, J., Ulmann, L. and Hirbec, H. (2019) Microglia in Alzheimer disease: well-known targets and new opportunities. Frontiers in Aging Neuroscience, 11. 10.3389/fnagi.2019.00233/full. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kinney, J.W., Bemiller, S.M., Murtishaw, A.S., Leisgang, A.M., Salazar, A.M. and Lamb, B.T. (2018) Inflammation as a central mechanism in Alzheimer’s disease. Alzheimers Dement (N Y), 4, 575–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ebanks, B., Ingram, T.L. and Chakrabarti, L. (2020) ATP synthase and Alzheimer’s disease: putting a spin on the mitochondrial hypothesis. Aging (Albany NY), 12, 16647–16662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Cho, S.-J., Park, M.H., Han, C., Yoon, K. and Koh, Y.H. (2017) VEGFR2 alteration in Alzheimer’s disease. Sci. Rep., 7, 17713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Carpanini, S.M., Torvell, M. and Morgan, B.P. (2019) Therapeutic inhibition of the complement system in diseases of the central nervous system. Front. Immunol., 10, 362. 10.3389/fimmu.2019.00362/full. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Quintela-López, T., Ortiz-Sanz, C., Serrano-Regal, M.P., Gaminde-Blasco, A., Valero, J., Baleriola, J., Sánchez-Gómez, M.V., Matute, C. and Alberdi, E. (2019) Aβ oligomers promote oligodendrocyte differentiation and maturation via integrin β1 and Fyn kinase signaling. Cell Death Dis., 10, 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kircheis, R., Haasbach, E., Lueftenegger, D., Heyken, W.T., Ocker, M. and Planz, O. (2020) NF-κB pathway as a potential target for treatment of critical stage COVID-19 patients. Front. Immunol., 11. 10.3389/fimmu.2020.598444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kwee, T.C. and Kwee, R.M. (2020) Chest CT in COVID-19: what the radiologist needs to know. Radiographics, 40, 1848–1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Qiao, Y., Wang, X.-M., Mannan, R., Pitchiaya, S., Zhang, Y., Wotring, J.W., Xiao, L., Robinson, D.R., Wu, Y.-M., Tien, J.C.-Y.et al. (2021) Targeting transcriptional regulation of SARS-CoV-2 entry factors ACE2 and TMPRSS2. Proc. Natl. Acad. Sci. U S A, 118, e20211450118. https://www.pnas.org/content/118/1/e2021450118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Tremblay, M.-E., Madore, C., Bordeleau, M., Tian, L. and Verkhratsky, A. (2020) Neuropathobiology of COVID-19: the role for glia. Front. Cell. Neurosci., 14, 363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Liu, M., Guo, S., Hibbert, J.M., Jain, V., Singh, N., Wilson, N.O. and Stiles, J.K. (2011) CXCL10/IP-10 in infectious diseases pathogenesis and potential therapeutic implications. Cytokine Growth Factor Rev., 22, 121–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Reusch, N., De Domenico, E., Bonaguro, L., Schulte-Schrepping, J., Baßler, K., Schultze, J.L. and Aschenbrenner, A.C. (2021) Neutrophils in COVID-19. Front. Immunol., 12, 652470. 10.3389/fimmu.2021.652470/full. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Hampel, H., Caraci, F., Cuello, A.C., Caruso, G., Nisticò, R., Corbo, M., Baldacci, F., Toschi, N., Garaci, F., Chiesa, P.A.et al. (2020) A path toward precision medicine for neuroinflammatory mechanisms in Alzheimer’s disease. Front. Immunol., 11, 456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Landhuis, E. (2021) Could the immune system be key to Alzheimer’s disease? Know. Mag. Ann. Rev. https://knowablemagazine.org/article/health-disease/2021/could-immune-system-be-key-alzheimers-disease. [Google Scholar]
- 48. Parham, P. (2014) The Immune System. Garland Science, New York. [Google Scholar]
- 49. Java, A., Apicelli, A.J., Liszewski, M.K., Coler-Reilly, A., Atkinson, J.P., Kim, A.H.J. and Kulkarni, H.S.. The complement system in COVID-19: friend and foe. JCI Insight, 5(15), e140711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hammad, A., Westacott, L. and Zaben, M. (2018) The role of the complement system in traumatic brain injury: a review. J. Neuroinflammation, 15, 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Marshe, V.S., Maciukiewicz, M., Hauschild, A.-C., Islam, F., Qin, L., Tiwari, A.K., Sibille, E., Blumberger, D.M., Karp, J.F., Flint, A.J.et al. (2021) Genome-wide analysis suggests the importance of vascular processes and neuroinflammation in late-life antidepressant response. Transl. Psychiatry, 11, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Heffron, A.S., McIlwain, S.J., Amjadi, M.F., Baker, D.A., Khullar, S., Armbrust, T., Halfmann, P.J., Kawaoka, Y., Sethi, A.K., Palmenberg, A.C., et al. (2021) The landscape of antibody binding in SARS-CoV-2 infection. PLOS Biology, 19, e3001265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Hu, J., Li, C., Wang, S., Li, T. and Zhang, H. (2021) Genetic variants are identified to increase risk of COVID-19 related mortality from UK biobank data. Hum. Genom., 15, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Pairo-Castineira, E., Clohisey, S., Klaric, L., Bretherick, A.D., Rawlik, K., Pasko, D., Walker, S., Parkinson, N., Fourman, M.H., Russell, C.D.et al. Genetic mechanisms of critical illness in COVID-19. Nature. 591, 92–98. [DOI] [PubMed] [Google Scholar]
- 55. Hou, Y., Zhao, J., Martin, W., Kallianpur, A., Chung, M.K., Jehi, L., Sharifi, N., Erzurum, S., Eng, C. and Cheng, F.. New insights into genetic susceptibility of COVID-19: an ACE2 and TMPRSS2 polymorphism analysis. BMC Med. 18, 216. 10.1186/s12916-020-01673-z#citeas. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Kong, Y., Han, J., Wu, X., Zeng, H., Liu, J. and Zhang, H. (2020) VEGF-D: a novel biomarker for detection of COVID-19 progression. Crit. Care, 24, 373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. AD Knowledge Portal -. https://adknowledgeportal.synapse.org/.
- 58. Agora. https://agora.adknowledgeportal.org/genes/comparison.
- 59. Rustenhoven, J., Smith, A.M., Smyth, L.C., Jansson, D., Scotter, E.L., Swanson, M.E.V., Aalderink, M., Coppieters, N., Narayan, P., Handley, R.et al. PU.1 regulates Alzheimer’s disease-associated genes in primary human microglia. Mol. Neurodegener. 13, 44. 10.1186/s13024-018-0277-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Muraoka, S., Jedrychowski, M.P., Iwahara, N., Abdullah, M., Onos, K.D., Keezer, K.J., Hu, J., Ikezu, S., Howell, G.R., Gygi, S.P.et al. Enrichment of neurodegenerative microglia signature in brain-derived extracellular vesicles isolated from Alzheimer’s disease mouse models. J. Proteome Res., 20, 1733–1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Mathys, H., Davila-Velderrain, J., Peng, Z., Gao, F., Mohammadi, S., Young, J.Z., Menon, M., He, L., Abdurrob, F., Jiang, X.et al. Single-cell transcriptomic analysis of Alzheimer’s disease. Nature. 570, 332–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Guo, J., Cai, Y., Ye, X., Ma, N., Wang, Y., Yu, B. and Wan, J. (2019) MiR-409-5p as a regulator of neurite growth is down regulated in APP/PS1 murine model of Alzheimer’s disease. Front. Neurosci., 13. 10.3389/fnins.2019.01264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Gabbouj, S., Ryhänen, S., Marttinen, M., Wittrahm, R., Takalo, M., Kemppainen, S., Martiskainen, H., Tanila, H., Haapasalo, A., Hiltunen, M.et al. (2019) Altered insulin Signaling in Alzheimer’s disease brain – special emphasis on PI3K-Akt pathway. Front. Neurosci., 13, 629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Zhou, Y., Zhou, B., Pache, L., Chang, M., Khodabakhshi, A.H., Tanaseichuk, O., Benner, C. and Chanda, S.K. (2019) Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun, 10, 1523. https://pubmed.ncbi.nlm.nih.gov/30944313/. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Kumar, S., Ambrosini, G. and Bucher, P.. SNP2TFBS - a database of regulatory SNPs affecting predicted transcription factor binding site affinity. Nucleic Acids Res., 45(D1). https://pubmed.ncbi.nlm.nih.gov/27899579/. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Radcliffe, S. (2022) COVID-19 may cause brain changes, even with mild infection. Healthline. https://www.healthline.com/health-news/covid-19-may-cause-brain-changes-even-with-mild-infection. [Google Scholar]
- 67. Corces, M.R., Shcherbina, A., Kundu, S., Gloudemans, M.J., Frésard, L., Granja, J.M., Louie, B.H., Eulalio, T., Shams, S., Bagdatli, S.T.et al. (2020) Single-cell epigenomic analyses implicate candidate causal variants at inherited risk loci for Alzheimer’s and Parkinson’s diseases. Nat. Genet., 52, 1158–1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Gupta, C., Xu, J., Jin, T., Khullar, S., Liu, X., Alatkar, S., Cheng, F. and Wang, D. (2022) Single-cell network biology characterizes cell type gene regulation for drug repurposing and phenotype prediction in Alzheimer’s disease. PLoS Comput. Biol., 18, e1010287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Li, Y., Xiao, X., Chen, H., Chen, Z., Hu, K. and Yin, D. (2020) Transcription factor NFYA promotes G1/S cell cycle transition and cell proliferation by transactivating cyclin D1 and CDK4 in clear cell renal cell carcinoma. Am. J. Cancer Res., 10, 2446. [PMC free article] [PubMed] [Google Scholar]
- 70. Lu, T., Aron, L., Zullo, J., Pan, Y., Kim, H., Chen, Y., Yang, T.-H., Kim, H.-M., Drake, D., Liu, X.S.et al. (2014) REST and stress resistance in ageing and Alzheimer’s disease. Nature, 507, 448–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Maezawa, I., Jenkins, D.P., Jin, B.E. and Wulff, H. (2012) Microglial KCa3.1 channels as a potential therapeutic target for Alzheimer’s disease. Int. J. Alzheimers Dis., 2012, 868972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Magno, L., Lessard, C.B., Martins, M., Lang, V., Cruz, P., Asi, Y., Katan, M., Bilsland, J., Lashley, T., Chakrabarty, P.et al. (2019) Alzheimer’s disease phospholipase C-gamma-2 (PLCG2) protective variant is a functional hypermorph. Alzheimers Res. Ther., 11, 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Jiang, J., Wang, C., Qi, R., Fu, H. and Ma, Q. (2020) scREAD: a single-cell RNA-Seq database for Alzheimer’s disease. iScience, 23, 101769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Guo, Z., Peng, X., Li, H.-Y., Wang, Y., Qian, Y., Wang, Z., Ye, D., Ji, X., Wang, Z., Wang, Y.et al. (2019) Evaluation of peripheral immune dysregulation in Alzheimer’s disease and vascular dementia. J. Alzheimers Dis., 71, 1175–1186. [DOI] [PubMed] [Google Scholar]
- 75. Resource.PsychEncode. (2018). http://resource.psychencode.org/.
- 76. The Cancer Genome Atlas Program - National Cancer Institute. https://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga (2018).
- 77. Coyle, P.K. (2011) Dissecting the immune component of neurologic disorders: a grand challenge for the 21st century. Front. Neurol., 2, 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Polan, S. (2021) Schizophrenia second only to age as greatest risk factor for COVID-19 death. NYU Langone News. https://nyulangone.org/news/schizophrenia-second-only-age-greatest-risk-factor-covid-19-death. [Google Scholar]
- 79. Langfelder, P. and Horvath, S. (2008) WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics, 9, 559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Blalock, E.M., Geddes, J.W., Chen, K.C., Porter, N.M., Markesbery, W.R. and Landfield, P.W. (2004) Incipient Alzheimer’s disease: microarray correlation analyses reveal major transcriptional and tumor suppressor responses. Proc. Natl. Acad. Sci. U S A, 101, 2173–2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Davis, S. and Meltzer, P.S. (2007) GEOquery: a bridge between the Gene Expression Omnibus (GEO) and BioConductor. Bioinformatics, 23, 1846–1847. [DOI] [PubMed] [Google Scholar]
- 82. Carlson, M. (2016) hgu133a.db: Affymetrix Human Genome U133 Set annotation data (chip hgu133a). R package version 3.2.3. Bioconductor. http://bioconductor.org/packages/hgu133a.db/.
- 83. Project TB. (2015) hgu133acdf: hgu133acdf. R package version 2.18.0. Bioconductor. http://bioconductor.org/packages/hgu133acdf/.
- 84. Gautier, L., Cope, L., Bolstad, B.M., Irizarry, R.A. (2004) affy—analysis of Affymetrix GeneChip data at the probe level. Bioinformatics, 20(3), 307–315. ISSN 1367-4803. [DOI] [PubMed] [Google Scholar]
- 85. Fan, F.Y. (2013) RMA Normalization for Microarray Data. https://felixfan.github.io/RMA-Normalization-Microarray/.
- 86. Ihaka, R. and Gentleman, R. (1993) R: The R Project for Statistical Computing. https://www.r-project.org/.
- 87. Bennett, D.A., Buchman, A.S., Boyle, P.A., Barnes, L.L., Wilson, R.S. and Schneider, J.A. (2018) Religious Orders Study and Rush Memory and Aging Project. Journal of Alzheimer’s Disease, 64, S161–S189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Fullard, J.F., Hauberg, M.E., Bendl, J., Egervari, G., Cirnaru, M.-D., Reach, S.M., Motl, J., Ehrlich, M.E., Hurd, Y.L. and Roussos, P. (2018) An atlas of chromatin accessibility in the adult human brain. Genome Res., 28, 1243–1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Jung, I., Schmitt, A., Diao, Y., Lee, A.J., Liu, T., Yang, D., Tan, C., Eom, J., Chan, M., Chee, S.et al. (2019) A compendium of promoter-centered long-range chromatin interactions in the human genome. Nat. Genet., 51, 1442–1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Carlson, M., Maintainer, B.P. (2015) TxDb.Hsapiens.UCSC.hg19.knownGene: Annotation package for TxDb object(s). R package version 3.2.2. Bioconductor. http://bioconductor.org/packages/TxDb.Hsapiens.UCSC.hg19.knownGene/.
- 91. Rödelsperger, C., Köhler, S., Schulz, M.H., Manke, T., Bauer, S. and Robinson, P.N. (2009) Short ultraconserved promoter regions delineate a class of preferentially expressed alternatively spliced transcripts. Genomics, 94, 308–316. [DOI] [PubMed] [Google Scholar]
- 92. García-Ruiz, S., Gil-Martínez, A.L., Cisterna, A., Jurado-Ruiz, F., Reynolds, R.H., NABEC (North America Brain Expression Consortium), Cookson, M.R., Hardy, J., Ryten, M. and Botía, J.A. (2021) CoExp: A Web Tool for the Exploitation of Co-expression Networks. Frontiers in Genetics, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Botía, J.A., Vandrovcova, J., Forabosco, P., Guelfi, S., D’Sa, K., Hardy, J., Lewis, C.M., Ryten, M., Weale, M.E. and The United Kingdom Brain Expression Consortium (2017) An additional k-means clustering step improves the biological features of WGCNA gene co-expression networks. BMC Syst. Biol., 11, 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Raudvere, U., Kolberg, L., Kuzmin, I., Arak, T., Adler, P., Peterson, H. and Vilo, J. (2019) g:Profiler: a web server for functional enrichment analysis and conversions of gene lists (2019 update). Nucleic Acids Research, 47, W191–W198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Winter, D., Chamberlain, S. and Guangchun, H. (2020) rentrez: ‘Entrez’ in R.
- 96. GeneSets - Bader Lab @ The University of Toronto. https://baderlab.org/GeneSets.
- 97. Rouillard, A.D., Gundersen, G.W., Fernandez, N.F., Wang, Z., Monteiro, C.D., McDermott, M.G., Ma’ayan, A. (2016) The harmonizome: a collection of processed datasets gathered to serve and mine knowledge about genes and proteins. Database (Oxford). https://maayanlab.cloud/Harmonizome/. [DOI] [PMC free article] [PubMed]
- 98. Grote, S., Prüfer, K. and Kelso, J. (2016) ABAEnrichment: an R package to test for gene set expression enrichment in the adult and developing human brain. Bioinformatics. 32(20), 3201–3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Gutierrez-Sacristan, A., Hernandez-Ferrer, C., Gonzalez, J., Furlong, L. (2022) psygenet2r: psygenet2r - An R package for querying PsyGeNET and to perform comorbidity studies in psychiatric disorders. R package version 1.30.0.
- 100. Jain, A. and Tuteja, G. (2018) TissueEnrich: tissue-specific gene enrichment analysis. Bioinformatics. 35(11), 1966–1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Wu, T., Hu, E., Xu, S., Chen, M., Guo, P., Dai, Z., Feng, T., Zhou, L., Tang, W., Zhan, L.et al. clusterProfiler 4.0: A Universal Enrichment Tool for Interpreting Omics Data: The Innovation. https://www.cell.com/the-innovation/fulltext/S2666-6758(21)00066-7?_returnURL=https%3A%2F%2Flinkinghub.elsevier.com%2Fretrieve%2Fpii%2FS2666675821000667%3Fshowall%3Dtrue. [DOI] [PMC free article] [PubMed]
- 102. Yu, G., Wang, L., Han, Y. and He, Q. (2012) clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS: A Journal of Integrative Biology, 16, 284–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Zhang, X., Lan, Y., Xu, J., Quan, F., Zhao, E., Deng, C., Luo, T., Xu, L., Liao, G. and Yan, M. (2019) et al, CellMarker: a manually curated resource of cell markers in human and mouse. Nucleic Acids Res., 47, D721–D728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Safieh, M., Korczyn, A.D. and Michaelson, D.M. (2019) ApoE4: an emerging therapeutic target for Alzheimer’s disease. BMC Med., 17, 64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Wang, D., Liu, S., Warrell, J., Won, H., Shi, X., Navarro, F.C.P., Clarke, D., Gu, M., Emani, P., Yang, Y.T.et al. (2018) Comprehensive functional genomic resource and integrative model for the human brain. Science. 362(6420). https://science.sciencemag.org/content/362/6420/eaat8464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Jin, T., Rehani, P., Ying, M., Huang, J., Liu, S., Roussos, P. and Wang, D. (2021) scGRNom: a computational pipeline of integrative multi-omics analyses for predicting cell-type disease genes and regulatory networks. Genome Medicine, 13, 95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Tan, G. (2021) TFBSTools: Software Package for Transcription Factor Binding Site (TFBS) Analysis. Bioconductor version: Release (3.12).
- 108. Schep, A. and University, S. (2021) motifmatchr: Fast Motif Matching in R. motifmatchr: Fast Motif Matching in R. Bioconductor version: Release (3.12).
- 109. Groeneveld, C., Robertson, G., Wang, X., Fletcher, M., Markowetz, F., Meyer, K. and Castro, M.. RTN: RTN: Reconstruction of Transcriptional regulatory Networks and analysis of regulons, Bioconductor version: Release (3.12), 2021.
- 110. Ament, S., Shannon, P. and Richards, M. (2021) trena: Fit Transcriptional Regulatory Networks Using Gene Expression, Priors, Machine Learning, Bioconductor version: Release (3.12).
- 111. Huynh-Thu, V.A., Aibar, S. and Geurts, P.. GENIE3: GEne Network Inference with Ensemble of trees, Bioconductor version: Release (3.12), 2021.
- 112.KEGG PATHWAY Database. https://www.genome.jp/kegg/pathway.html.
- 113. Luo, W. (2021) pathview: a tool set for pathway based data integration and visualization. Bioconductor version: Release (3.12).
- 114. Overmyer, K.A., Shishkova, E., Miller, I.J., Balnis, J., Bernstein, M.N., Peters-Clarke, T.M., Meyer, J.G., Quan, Q., Muehlbauer, L.K., Trujillo, E.A., et al. (2021) Large-Scale Multi-omic Analysis of COVID-19 Severity. Cell Syst, 12, 23–40.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Love, M., Ahlmann-Eltze, C., Forbes, K., Anders, S. and Huber, W. (2021) DESeq2: Differential gene expression analysis based on the negative binomial distribution. Bioconductor version: Release (3.12).
- 116. scikit-learn: Machine Learning in Python — scikit-learn 0.24.1 documentation. https://scikit-learn.org/stable/.
- 117. Srinivasan, K., Friedman, B.A., Etxeberria, A., Huntley, M.A., van derBrug, M.P., Foreman, O., Paw, J.S., Modrusan, Z., Beach, T.G., Serrano, G.E.et al. (2020) Alzheimer’s patient microglia exhibit enhanced aging and unique transcriptional activation. Cell Rep., 31,(13), 107843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Jansen, I.E., Savage, J.E., Watanabe, K., Bryois, J., Williams, D.M., Steinberg, S., Sealock, J., Karlsson, I.K., Hägg, S., Athanasiu, L.et al. (2019) Genome-wide meta-analysis identifies new loci and functional pathways influencing Alzheimer’s disease risk. Nat. Genet., 51, 404–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Kunkle, B., Grenier-Boley, B., Sims, R., Bis, J., Damotte, V., Naj, A., Boland, A., Vronskaya, M., van derLee, S., Amlie-Wolf, A.et al. (2019) Genetic meta-analysis of diagnosed Alzheimer's disease identifies new risk loci and implicates Aβ, tau, immunity and lipid processing. Nat. Genet., 51, 414–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Wightman, D.P., Jansen, I.E., Savage, J.E., Shadrin, A.A., Bahrami, S., Holland, D., Rongve, A., Børte, S., Winsvold, B.S., Drange, O.K.et al. (2021) A genome-wide association study with 1,126,563 individuals identifies new risk loci for Alzheimer’s disease. Nat. Genet., 53, 1276–1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Pan UKBB | Pan UKBB . https://pan.ukbb.broadinstitute.org/.
- 122. Bellenguez, C., Fahri Küçükali, F., Jansen, I.E., Kleineidam, L., Moreno-Grau, S., Amin, N., Naj, A.C., Campos-Martin, R., Grenier-Boley, B., Andrade, V.et al. (2022) New insights into the genetic etiology of Alzheimer’s disease and related dementias. Nat. Genet., 54, 412–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. COVID-19 Host Genetics Initiative. https://www.covid19hg.org/results/r7/ (accessed Nov 10, 2022).
- 124. Coetzee, S.G. and Hazelett, D.J. (2021) motifbreakR: A Package for Predicting the Disruptiveness of Single Nucleotide Polymorphisms on Transcription Factor Binding Sites, Bioconductor version: Release (3.12).
- 125. The GTEx Consortium, Aguet, F., Anand, S., Ardlie, K.G., Gabriel, S., Getz, G.A., Graubert, A., Hadley, K., Handsaker, R.E., Huang, K.H.et al. (2020) The GTEx consortium atlas of genetic regulatory effects across human tissues. Science, 369, 1318–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Patel, D., Zhang, X., Farrell, J.J., Chung, J., Stein, T.D., Lunetta, K.L. and Farrer, L.A. (2021) Cell-type-specific expression quantitative trait loci associated with Alzheimer disease in blood and brain tissue. Transl. Psychiatry, 11, 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Bryois, J., Calini, D., Macnair, W., Foo, L., Urich, E., Ortmann, W., Iglesias, V.A., Selvaraj, S., Nutma, E., Marzin, M.et al. (2022) Cell-type-specific cis-eQTLs in eight human brain cell types identify novel risk genes for psychiatric and neurological disorders. Nat. Neurosci., 25, 1104–1112. [DOI] [PubMed] [Google Scholar]
- 128. Zeng, B., Bendl, J., Kosoy, R., Fullard, J.F., Hoffman, G.E. and Roussos, P. (2022) Multi-ancestry eQTL meta-analysis of human brain identifies candidate causal variants for brain-related traits. Nat. Genet., 54, 161–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Machiela, M.J. and Chanock, S.J. (2015) LDlink: a web-based application for exploring population-specific haplotype structure and linking correlated alleles of possible functional variants. Bioinformatics, 31, 3555–3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Our analysis (codes and data including diagnosis information) is open-source available at https://github.com/daifengwanglab/ADSNPheno and our functional genomics resource for AD is available at https://adsnpheno.shinyapps.io/AlzheimersDisease_SNPheno.