Abstract
We report the curing of the 1,360-kb megaplasmid pRme2011a from Sinorhizobium meliloti strain Rm2011. With a positive selection strategy that utilized Tn5B12-S containing the sacB gene, we were able to cure this replicon by successive rounds of selecting for deletion formation in vivo. Subsequent Southern blot, Eckhardt gel, and pulsed-field gel electrophoresis analyses were consistent with the hypothesis that the resultant strain was indeed missing pRme2011a. The cured derivative grew as well as the wild-type strain in both complex and defined media but was unable to use a number of substrates as a sole source of carbon on defined media.
Rhizobia are soil bacteria that are able to induce nitrogen-fixing nodules on the roots of leguminous plants. The formation and maintenance of a nitrogen-fixing nodule are determined by the expression and regulation of plant and bacterial genes (16, 28). Many of the bacterial genes necessary for an effective symbiotic association are plasmid borne (1, 2, 3, 6, 13, 19, 24, 26, 31, 39, 40).
Sinorhizobium meliloti typically contains two megaplasmids of approximately 1,400 and 1,600 kb (2, 3, 8, 9, 19, 24, 26, 37). In strain RCR2011 these plasmids have been termed pRme2011a and pRme2011b, respectively (also referred to as pRmeSU47a and pRmeSU47b or as pSymA and pSymB). Collectively, these plasmids comprise approximately 40% of the genome of this strain (23, 37). Although this represents a substantial proportion of this bacterial genome, relatively few traits have been ascribed to these replicons. The larger of these megaplasmids, pRme2011b, has been shown to carry determinants for exopolysaccharide synthesis, thiamine biosynthesis, high-affinity phosphate transport, and dicarboxylic acid transport (4, 12, 19, 22, 26, 39, 40). The construction of a genetic map facilitated the genetic characterization of this replicon, leading to the identification and localization of several catabolic loci (11, 12, 14).
In contrast, pRme2011a is not as well characterized. The phenotypes ascribed to this replicon are restricted to a region comprising less than one-third of the plasmid and are primarily involved in nodulation and nitrogen fixation (3, 5, 6, 7, 15, 30, 31, 32). The majority of pRme2011a is still considered cryptic. In an effort to characterize this replicon genetically, we have attempted to create large deletions by using a positive selection strategy which has been previously used successfully on the smaller plasmids of Rhizobium leguminosarum (24, 25). Here we present data which show that repetitive rounds of Tn5B12-S mutagenesis with selection for deletion can be used successfully to cure the nod-nif megaplasmid, pRme2011a, of strain Rm2011. Physical analysis using pulsed-field gel electrophoresis and Southern blot analysis of the putatively cured strain are consistent with the hypothesis that the nod-nif plasmid has been cured.
Deletion and curing of pRme2011a.
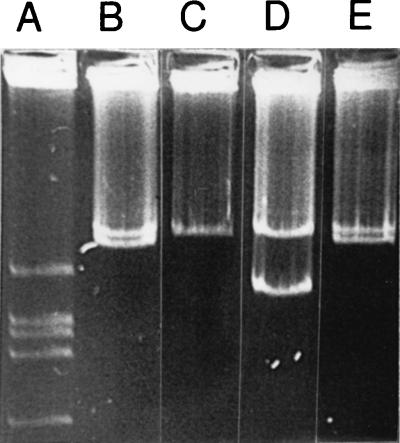
Creation of a derivative of Rm2011 which has been cured of pRme2011a was achieved in two steps. Rm2011-14 carrying Tn5B12-S insert 14, which was previously demonstrated to be in pRme2011a (25), was plated onto TY (tryptone-yeast extract) agar containing 5% sucrose. Resultant colonies were screened by Eckhardt gel electrophoresis (18, 24, 25) for putative deletions. The colony which carried the largest deletion was designated SmA146. From Eckhardt gels, using VF39SM as a size standard, it was estimated that this deletion reduced pRme2011a to approximately 600 kb (Fig. 1). This deletion was designated Δ14-6, and the replicon carrying this deletion was designated pRme2011aΔ14-6.
FIG. 1.
Eckhardt gel showing plasmid profiles of S. meliloti strains with deletions in pRme2011a. Lanes: A, R. leguminosarum LRS39501 (24) (size standard); B, Rm2011; C, SmA818; D, SmA146; E, Rm2011. All lanes are from the same gel, but some intervening lanes showing derivatives with smaller deletions have been removed for clarity.
To reduce the size of the residual replicon in SmA146, the strain was subjected to a further round of transposon mutagenesis, utilizing Tn5B12-S. Single colonies were conjugally mated with Agrobacterium tumefaciens UBAPF2 to identify those colonies which carried a Tn5B12-S insertion on pRme2011aΔ14-6. Eckhardt gel electrophoresis analysis was carried out on putative transconjugants to ensure that the proper plasmid had been transferred. S. meliloti colonies which had a Tn5B12-S insertion on pRme2011aΔ14-6 were grown overnight in TY broth and plated onto TY agar containing 5% sucrose. Three colonies of 300 were found to be neomycin sensitive and no longer sensitive to sucrose. Of these, one appeared to be missing pRm2011aΔ14-6 (Fig. 1). This strain was designated SmA818.
SmA818 has deletions of all known genetic markers.
It has been shown previously that the deletion Δ14-6 did not carry the nodPQ region (35). To determine the extent of the deletion of Δ14-6 in SmA146 and to verify that SmA818 did not contain sequences associated with pRme2011a, Southern blot analysis was carried out on genomic DNA from these strains. Probes used corresponded to regions associated with pRme2011a (6, 15, 27, 33, 34).
The results of these analyses showed that strains SmA146 and SmA818 did not contain sequences homologous to nifHDK, fixLJ, fixG, or syrB (Table 1). SmA146 did, however, contain sequences that hybridized to the fixN probe (Table 1). In S. meliloti strain Rm2011, fixN is reiterated on pRme2011a (31). Further analysis revealed that the hybridizing fragment in SmA146 corresponded to the fixN′ region, which is located close to a locus necessary for trigonelline catabolism (7). Since SmA146 did not contain nifHDK sequences but did contain fixN′ sequences, this suggests that one end point of the pRme2011Δ14-6 deletion was between these two regions. As genes encoding trigonelline catabolism (trc) are found between these two markers, it was of interest to determine whether SmA146 could utilize trigonelline as a sole carbon source. The results showed that SmA146 could not use this compound as a sole carbon source (Table 2). Together these data suggest that one end point of the Δ14-6 deletion is between the trc and fixN′ loci and that it extends approximately 750 kb (Fig. 2).
TABLE 1.
Southern analysis of deletion and cured derivatives of Rm2011
| Strain | Relevant genotype | Presence of hybridizing bands in region probeda
|
|||||
|---|---|---|---|---|---|---|---|
| nifHDK | fixG | fixN | fixN′ | syrB | Ser/GABA | ||
| Rm2011 | Wild type | + | + | + | + | + | + |
| SmA146 | Δ14-6 | − | − | − | + | − | − |
| SmA818 | pRm2011a cured | − | − | − | − | − | − |
TABLE 2.
Phenotypes associated with pRm2011a deletions and cured derivativea
| Strain | Relevant genotype | Nod | Growth with carbon source
|
||||
|---|---|---|---|---|---|---|---|
| Glc | GABA | Gly/Ser | Ino | Trig | |||
| Rm2011 | Wild type | + | + | + | + | + | + |
| SmA146 | Δ14-6 | − | − | − | − | − | − |
| SmA818 | pRm2011a cured | − | − | − | − | − | − |
Designations are as follows: +, wild-type growth; −, no growth. Abbreviations: Nod, nodulation; Glc, gluconate (15 mM); Gly/Ser, glycine (10 mM) or serine (10 mM); Ino, inosine (10 mM); Trig, trigonelline (5 mM).
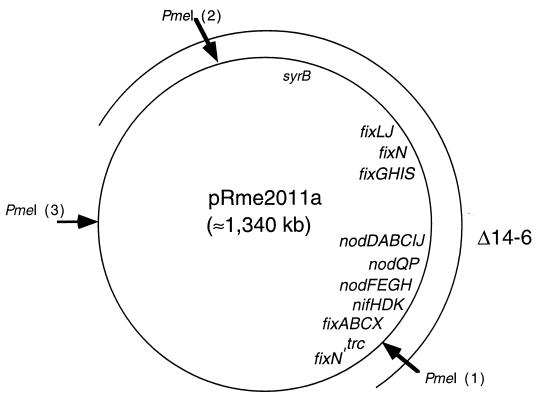
FIG. 2.
Schematic representation of pRme2011a showing relative positions of known genetic markers. The approximate position of syrB::Tn5 in strain MB101 (5) was determined by conjugal-transfer experiments similar to those which were described for pRmeSU47b (11). The oriT used for these experiments was that of Ω30::Tn5-11 from Rm5420 (which is linked to nifH) (18). The direction of transfer was determined to be clockwise, using fixJ2.3::Tn5 from strain GMI 5704 as a marker for conjugal transfer (15). The arc shown represents the approximate position of Δ14-6. The end points of this deletion are undefined. The large arrows indicate the positions of PmeI restriction sites (23). PmeI fragment 5 extends between sites 1 and 2, fragment 6 extends between sites 1 and 3, and fragment 7 extends between sites 2 and 3. All three fragments are missing in strain SmA818.
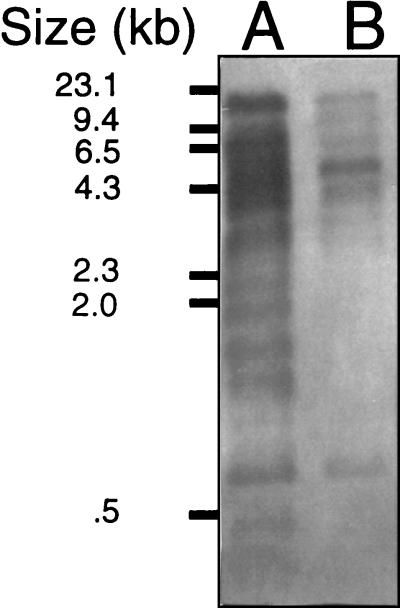
In an effort to confirm that pRme2011Δ14-6 had indeed been cured from SmA818, plasmid DNA from SmA146 carrying pRme2011Δ14-6 was isolated from a preparative Eckhardt gel, labeled, and used to probe Southern blots containing DNA from Rm2011, SmA146, and SmA818. Consistent with the hypothesis that pRme2011a was cured in SmA818, the results showed a large reduction in the number, and in most cases the intensity, of hybridizing fragments when Rm2011 and SmA818 were compared (Fig. 3). We note that it has previously been shown that a great deal of reiteration exists in the S. meliloti and other rhizobial genomes (20). Moreover, it has also been shown that Rm2011 contains at least five different insertion elements, four of which are highly reiterated (as many as 11 or 12 copies) within the genome (36). It has also been shown that 18% of the sequence of the nodulation plasmid of Sinorhizobium strain NGR234 shows homology to insertion elements (21). The high proportion of reiterated sequences in the rhizobia may explain the presence of the few remaining hybridizing bands seen in SmA818 (Fig. 3). It is also theoretically possible that small segments of plasmid pRme2011a do remain in strain SmA818, integrated into either the chromosome or plasmid pRme2011b.
FIG. 3.
Southern blot analysis of SmA818. Equal amounts of EcoRI-restricted DNA from Rm2011 and SmA818 were electrophoresed, blotted to a nylon membrane, and probed with pRme2011Δ14-6 DNA which was isolated from a preparative Eckhardt gel. Lanes: A, Rm2011; B, SmA818.
Genomic analysis of SmA146 and SmA818 using pulsed-field gel electrophoresis.
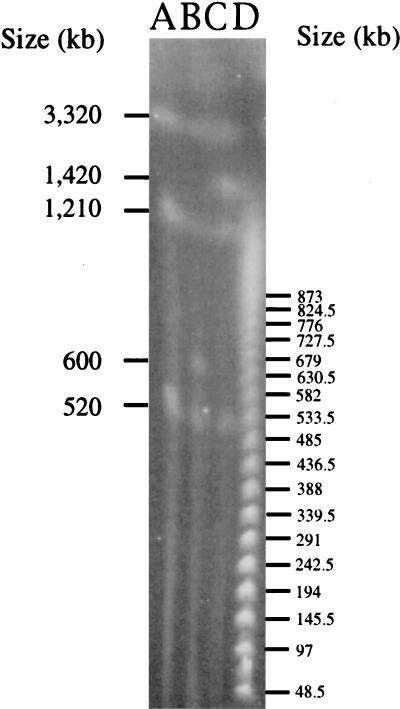
To verify that genomic rearrangements resulting in insertion of portions of pRme2011a into another replicon had not occurred during the deletion process that generated strains SmA146 and SmA818, the genomes of these strains were analyzed by pulsed-field gel electrophoresis using restriction enzymes that cut the S. meliloti genome infrequently (23, 37). Using the enzymes I-CeuI, PacI, PmeI, and SwaI, we ascertained that the chromosomal fragments in SmA818 and SmA146 had mobilities indistinguishable from those of the wild type, Rm2011. Furthermore, there were no detectable changes in the sizes and restriction patterns of bands known to correspond to pRme2011b. Pulsed-field analysis using PacI and SwaI, both of which cut pRme2011a once, revealed the absence of a 1,400-kb band in SmA818 (Fig. 4). Moreover, analysis using PmeI, which yields seven PmeI fragments in the wild-type strain, resulted in only four discernible fragments in SmA818; bands 5, 6, and 7, which comprise pRme2011a (23), were absent (data not shown). In strain SmA146, only a single 600-kb PmeI fragment remained, suggesting that a single PmeI site [labeled PmeI (3) in Fig. 2] remained in the form of the pRme2011a replicon with the deletion. The presence of a single PmeI site in pRme2011aΔ14-6 is consistent with the mapping of the deletion end point between fixN′ and trc, both of which are found on PmeI fragment 6 to the left of PmeI fragment 5 (Fig. 2), and with the Southern analysis, which shows that fixN′ remains in SmA146. Together these data strongly suggest that plasmid pRme2011a is missing completely in strain SmA818.
FIG. 4.
Pulsed-field gel profiles of PacI-digested S. meliloti strains. Lanes: A, SmA818; B, SmA146; C, Rm2011; D, concatemers, molecular size markers.
Phenotypic characterization of SmA818.
Growth experiments were performed to characterize SmA818. These experiments demonstrated that SmA818 and Rm2011 have identical doubling times when grown either in Vincent's minimal medium (38) containing succinate (15 mM) or glucose (15 mM) as a sole carbon source or in complex media (TY).
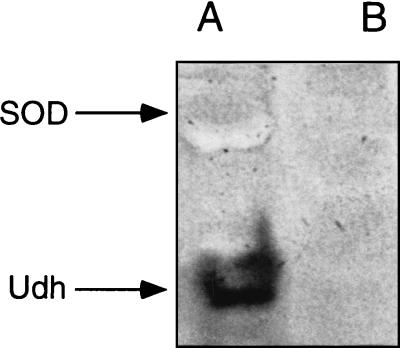
It has been shown previously that unidentified dehydrogenase activity was encoded by pRme2011a (14). To provide corroborating evidence that pRme2011a was indeed cured, cell extracts of Rm2011, SmA818, and A. tumefaciens UBAPF2(pRme2011a) were prepared, separated on native polyacrylamide gel electrophoresis gels, and stained for unidentified dehydrogenase (14). The data show that SmA818 did not have this activity, whereas Rm2011 and UBAPF2(pRme2011a) both had an unidentified dehydrogenase band of activity (Fig. 5).
FIG. 5.
Unidentified dehydrogenase (Udh) and SOD activities in S. meliloti. Cell extracts of Rm2011 and SmA818 were run on nondenaturing polyacrylamide gels. The gel was stained as previously described (8). Lanes: A, Rm2011; B, SmA818.
When native gels containing Rm2011 and SmA818 extracts were stained for prolonged periods, an achromatic band(s) was often observed. Native gels stained for superoxide dismutase (SOD) or tetrazolium oxidase activities yielded a negative band on a dark background (17). Interestingly, a negative staining band also appeared to be missing in SmA818 (Fig. 5). Under our growth conditions, we were able to resolve three bands of SOD activity in the wild-type strain (data not shown). SmA818 was found to be missing one of these bands. Transfer of pRme2011a to A. tumefaciens resulted in the appearance of a band of SOD activity with an Rf similar to that of the band missing in SmA818 (data not shown). This suggests that pRme2011a carries a determinant(s) that influences the expression of SOD activity. We note that plasmid-borne SOD activity has previously been reported for R. leguminosarum (1).
To reveal possible catabolic defects present in a strain lacking pRme2011a, SmA818 was screened for potential phenotypes using Biolog plates, which enabled screening for utilization of 96 carbon sources simultaneously. Putative phenotypes were confirmed by streaking SmA818, SmA146, and Rm2011 on defined media containing the carbon source being tested (Table 2). This analysis shows that SmA818 was unable to catabolize inosine, γ-aminobutyric acid (GABA), gluconate, glycine, and serine as sole carbon sources (Table 2). Further analysis of the glycine and serine phenotypes suggested that both of these phenotypes map to one locus and that one of the genes involved may be a transport protein responsible for the uptake of these amino acids. Moreover, cosmids containing the Gly-Ser utilization region also complemented GABA utilization. These phenotypes, however, were shown to be genetically distinct (I. J. Oresnik and M. F. Hynes, unpublished data).
The megaplasmids of S. meliloti are essentially stable, and conventional methods for plasmid curing have proven to be unsuccessful for these replicons (2, 3, 24). Conventional mutagenesis and screening for phenotypes are often unsuccessful due to reiterated DNA sequences which are found in Rhizobium (20, 31, 34, 35). Methods which have utilized curing or the construction of defined deletions have been useful in studying cryptic replicons in Rhizobium (1, 2, 3, 6, 12, 24, 25, 31). In this work we provide evidence for the curing of pRme2011a. This is, to the best of our knowledge, the largest replicon cured to date in any bacterium. By curing this plasmid, we have demonstrated that pRme2011a does not carry single-copy genes which are essential for cell viability or for the maintenance of this strain on normal lab media. Thus, by definition, pRme2011a is a plasmid and not a minichromosome as others have suggested (29). Extensive probing using markers localized to the characterized regions of pRme2011a has shown that the corresponding DNA is absent in strain SmA818. Pulsed-field analyses of SmA818 have been consistent with the hypothesis that, in this strain, pRme2011a has been cured. Judging by the size of the pulsed-field electrophoresis fragments in SmA818, it appears unlikely that genomic rearrangements larger than 40 kb occurred in either of the remaining replicons as a consequence of either the deletion or curing events used to generate this strain.
Surprisingly, although almost one-quarter of the genome of Rm2011 has been removed, the strain still exhibits growth rates that are identical to that of the wild type on defined and complex media. This in itself is remarkable, considering that this replicon is inherently stable and that large deletions in this plasmid are rarely isolated. This suggests that there must be reasons other than viability for the maintenance of this plasmid. Studies addressing these issues as well as more detailed phenotype determination will presumably be more fruitful following the complete determination of the genome sequence of Rm1021, which is currently under way (10).
Acknowledgments
We thank J. Batut, S. R. Long, M. Barnett, T. Finan, and T. Charles for strains and probes that were used in this study. We are also very grateful to S. R. Long and M. Barnett for comments on the manuscript. The advice, support, and encouragement of P. Boistard, in whose laboratory a small part of the initial work was done, are gratefully acknowledged.
This work was supported by an NSERC grant to M.F.H.
ADDENDUM IN PROOF
A high-resolution physical map of pRme2011a (pSymA) has very recently been published (F. Barloy-Hubler et al. J. Bacteriol. 182:1185–1189, 2000). This paper suggested that genes for the production of the siderophore rhizobactin mapped to the middle of the undeleted region in plasmid pRme2011aΔ14-6, and we have confirmed that strain SmA818 does not produce rhizobactin. It is also of note that at least four copies of ISRm2011-2 map to the undeleted region of pRme2011a in strain SmA416.
REFERENCES
- 1.Baldani J I, Weaver R W, Hynes M F, Eardly B D. Utilization of carbon substrates, electrophoretic enzyme patterns, and symbiotic performance of plasmid-cured clover rhizobia. Appl Environ Microbiol. 1992;58:2308–2314. doi: 10.1128/aem.58.7.2308-2314.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Banfalvi Z, Kondorosi E, Kondorosi A. Rhizobium meliloti carries two megaplasmids. Plasmid. 1985;13:129–138. doi: 10.1016/0147-619x(85)90065-4. [DOI] [PubMed] [Google Scholar]
- 3.Banfalvi Z, Sakanyan V, Koncz C, Kiss A, Dusha I, Kondorosi A. Location of nitrogen fixation genes on a high molecular weight plasmid of R. meliloti. Mol Gen Genet. 1981;184:318–325. doi: 10.1007/BF00272925. [DOI] [PubMed] [Google Scholar]
- 4.Bardin S, Dan S, Osteras M, Finan T M. A phosphate transport system is required for symbiotic nitrogen fixation by Rhizobium meliloti. J Bacteriol. 1996;178:4540–4547. doi: 10.1128/jb.178.15.4540-4547.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barnett M J, Long S R. Identification and characterization of a gene on Rhizobium meliloti pSyma, syrB, that negatively affects syrM expression. Mol Plant-Microbe Interact. 1997;10:550–559. doi: 10.1094/MPMI.1997.10.5.550. [DOI] [PubMed] [Google Scholar]
- 6.Batut J, Terzaghi B, Ghérardi M, Huguet M, Terzaghi E, Garnerone A-M, Boistard P, Huguet T. Localization of a symbiotic fix region on Rhizobium meliloti pSym megaplasmid more than 200 kilobases from the nod-nif region. Mol Gen Genet. 1985;199:232–239. [Google Scholar]
- 7.Boivin C, Camut S, Malpica C A, Truchet G, Rosenberg C. Rhizobium meliloti genes encoding catabolism of trigonelline are induced under symbiotic conditions. Plant Cell. 1990;2:1157–1170. doi: 10.1105/tpc.2.12.1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burkardt B, Burkardt H J. Visualization and exact molecular weight determination of Rhizobium meliloti megaplasmid. J Mol Biol. 1984;175:213–218. doi: 10.1016/0022-2836(84)90475-3. [DOI] [PubMed] [Google Scholar]
- 9.Burkardt B, Schillik D, Pühler A. Physical characterization of Rhizobium meliloti megaplasmids. Plasmid. 1987;17:13–25. doi: 10.1016/0147-619x(87)90004-7. [DOI] [PubMed] [Google Scholar]
- 10.Capela D, Barlowy-Hubler F, Gatuis M, Gouzy J, Galibert F. A high-density physical map of Sinorhizobium meliloti 1021 chromosome derived from bacterial artificial chromosome library. Proc Natl Acad Sci USA. 1999;96:9357–9362. doi: 10.1073/pnas.96.16.9357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Charles T C, Finan T M. Genetic map of Rhizobium meliloti megaplasmid pRmeSU47b. J Bacteriol. 1990;172:2469–2476. doi: 10.1128/jb.172.5.2469-2476.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Charles T C, Finan T M. Analysis of a 1600-kilobase Rhizobium meliloti megaplasmid using defined deletions generated in vivo. Genetics. 1991;127:5–20. doi: 10.1093/genetics/127.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Charles T C, Newcomb W, Finan T M. ndvF, a novel locus located on megaplasmid pRmeSU47b (pEXO) of Rhizobium meliloti, is required for normal nodule development. J Bacteriol. 1991;173:3981–3992. doi: 10.1128/jb.173.13.3981-3992.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Charles T C, Singh R S, Finan T M. Lactose utilization and enzymes encoded by megaplasmids in Rhizobium meliloti SU47: implications for population studies. J Gen Microbiol. 1990;136:2497–2502. [Google Scholar]
- 15.David M, Daveran M L, Batut J, Dedieu A, Domergue O, Ghai J, Hertig C, Boistard P, Kahn D. Cascade regulation of nif gene expression in Rhizobium meliloti. Cell. 1988;54:671–683. doi: 10.1016/s0092-8674(88)80012-6. [DOI] [PubMed] [Google Scholar]
- 16.Dénarié J, Débellé F, Rosenberg C. Signaling and host range variation in nodulation. Annu Rev Microbiol. 1992;46:497–531. doi: 10.1146/annurev.mi.46.100192.002433. [DOI] [PubMed] [Google Scholar]
- 17.Eardly B D, Materon L A, Smith N H, Johnson D A, Rumbaugh M D, Selander R K. Genetic structure of natural populations of the nitrogen-fixing bacterium Rhizobium meliloti. Appl Environ Microbiol. 1990;56:187–194. doi: 10.1128/aem.56.1.187-194.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eckhardt T. A rapid method for the identification of plasmid deoxyribonucleic acid in bacteria. Plasmid. 1978;1:584–588. doi: 10.1016/0147-619x(78)90016-1. [DOI] [PubMed] [Google Scholar]
- 19.Finan T M, Kunkel B, De Vos G F, Signer E R. Second symbiotic megaplasmid in Rhizobium meliloti carrying exopolysaccharide and thiamine synthesis genes. J Bacteriol. 1986;167:66–72. doi: 10.1128/jb.167.1.66-72.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Flores M, González V, Brom S, Martínez E, Piñero D, Romero D, Dávila G, Palacios R. Reiterated DNA sequences in Rhizobium and Agrobacterium spp. J Bacteriol. 1987;169:5782–5788. doi: 10.1128/jb.169.12.5782-5788.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Freiberg C, Fellay R, Bairoch R A, Broughton W J, Rosenthal A, Perret X. Molecular basis of symbiosis between Rhizobium and legumes. Nature. 1997;387:394–401. doi: 10.1038/387394a0. [DOI] [PubMed] [Google Scholar]
- 22.Glazebrook J, Walker G C. A novel exopolysaccharide can function in place of the calcofluor-binding exopolysaccharide in nodulation of alfalfa by Rhizobium meliloti. Cell. 1989;56:661–672. doi: 10.1016/0092-8674(89)90588-6. [DOI] [PubMed] [Google Scholar]
- 23.Honeycutt R J, McClelland M, Sobral B W S. Physical map of the genome of Rhizobium meliloti 1021. J Bacteriol. 1993;175:6945–6952. doi: 10.1128/jb.175.21.6945-6952.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hynes M F, McGregor N F. Two plasmids other than the nodulation plasmid are necessary for formation of nitrogen-fixing nodules by Rhizobium leguminosarum. Mol Microbiol. 1990;4:567–571. doi: 10.1111/j.1365-2958.1990.tb00625.x. [DOI] [PubMed] [Google Scholar]
- 25.Hynes M F, Quandt J, O'Connell M P, Pühler A. Direct selection for curing and deletion of Rhizobium plasmids using transposons carrying the Bacillus subtilis sacB gene. Gene. 1989;78:111–120. doi: 10.1016/0378-1119(89)90319-3. [DOI] [PubMed] [Google Scholar]
- 26.Hynes M F, Simon R, Müller P, Niehaus K, Labes M, Pühler A. The two megaplasmids of Rhizobium meliloti are involved in the effective nodulation of alfalfa. Mol Gen Genet. 1986;202:356–362. [Google Scholar]
- 27.Kahn D, David M, Domergue O, Daveran M-L, Ghai J, Hirsch P R, Batut J. Rhizobium meliloti fixGHI sequence predicts involvement of a specific cation pump in symbiotic nitrogen fixation. J Bacteriol. 1989;171:929–939. doi: 10.1128/jb.171.2.929-939.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Long S R. Rhizobium symbiosis: Nod factors in perspective. Plant Cell. 1996;8:1885–1898. doi: 10.1105/tpc.8.10.1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mercado-Blanco J, Toro N. Plasmids in rhizobia: the role of nonsymbiotic plasmids. Mol Plant-Microbe Interact. 1996;9:535–545. [Google Scholar]
- 30.Ogawa J, Long S R. The Rhizobium meliloti groELc locus is required for regulation of early nod genes by the transcription activator NodD. Genes Dev. 1995;9:714–729. doi: 10.1101/gad.9.6.714. [DOI] [PubMed] [Google Scholar]
- 31.Renalier M-H, Batut J, Ghai J, Terzaghi B, Gherardi M, David M, Garnerone A-M, Vasse J, Truchet G, Huguet T, Boistard P. A new symbiotic cluster on the pSym megaplasmid of Rhizobium meliloti 2011 carries a functional fix gene repeat and a nod locus. J Bacteriol. 1987;169:2231–2238. doi: 10.1128/jb.169.5.2231-2238.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rosenberg C, Boistard P, Dénarié J, Casse-Delbart F. Genes controlling early and late functions in symbiosis are located on a megaplasmid in R. meliloti. Mol Gen Genet. 1981;184:326–333. doi: 10.1007/BF00272926. [DOI] [PubMed] [Google Scholar]
- 33.Ruvkun G B, Sundaresan V, Ausubel F M. Directed transposon Tn5 mutagenesis and complementation analysis of Rhizobium meliloti symbiotic nitrogen fixation genes. Cell. 1982;29:551–559. doi: 10.1016/0092-8674(82)90171-4. [DOI] [PubMed] [Google Scholar]
- 34.Schlüter A, Patschkowski T, Quandt J, Selinger L B, Weidner S, Krämer M, Zhou L, Hynes M F, Priefer U B. Functional and regulatory analysis of the two copies of the fixNOQP operon of Rhizobium leguminosarum strain VF39. Mol Plant-Microbe Interact. 1997;10:605–616. doi: 10.1094/MPMI.1997.10.5.605. [DOI] [PubMed] [Google Scholar]
- 35.Schwedock J S, Long S R. Rhizobium meliloti genes involved in sulfate activation: the two copies of nodPQ and a new locus, saa. Genetics. 1992;132:899–909. doi: 10.1093/genetics/132.4.899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Simon R, Hötte B, Klauke B, Kosier B. Isolation and characterization of insertion sequence elements from gram-negative bacteria by using new broad-host-range, positive selection vectors. J Bacteriol. 1991;173:1502–1508. doi: 10.1128/jb.173.4.1502-1508.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sobral B W S, Honeycutt R J, Atherly A G, McClelland M. Electrophoretic separation of the three Rhizobium meliloti replicons. J Bacteriol. 1991;173:5173–5180. doi: 10.1128/jb.173.16.5173-5180.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vincent J M. A manual for the practical study of root-nodule bacteria (IBP handbook 15). Oxford, England: Blackwell Scientific Publications; 1970. [Google Scholar]
- 39.Watson R J, Chan Y-K, Wheatcroft R, Yang A-F, Han S. Rhizobium meliloti genes required for C4-dicarboxylate transport and symbiotic nitrogen fixation are located on a megaplasmid. J Bacteriol. 1988;170:927–934. doi: 10.1128/jb.170.2.927-934.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yarosh O K, Charles T C, Finan T M. Analysis of C4 dicarboxylate transport genes in Rhizobium meliloti. Mol Microbiol. 1989;3:813–823. doi: 10.1111/j.1365-2958.1989.tb00230.x. [DOI] [PubMed] [Google Scholar]