Abstract
The gene (ybeN) coding for nicotinate mononucleotide adenylyltransferase, an NAD(P) biosynthetic enzyme, has been identified and overexpressed in Escherichia coli. This enzyme catalyzes the reversible adenylation of nicotinate mononucleotide and shows product inhibition. The rate of adenylation of nicotinate mononucleotide is at least 20 times faster than the rate of adenylation of nicotinamide mononucleotide.
The cofactors flavin adenine dinucleotide, coenzyme A, and NAD(P) are all adenylated. However, the genes coding for the corresponding adenylyltransferases have been difficult to identify because the adenylated reaction products are not taken up by bacteria; therefore, auxanographic screening of mutants is impossible. The flavin adenine dinucleotide synthetase gene (ribF) and the phosphopantetheine adenylyltransferase gene (kdtB or coaD) were identified by using sequence information derived from the purified proteins (5; K. Kitatsuji, S. Ishino, S. Teshiba, and M. Arimoto, 1993, European patent application). While the Escherichia coli nicotinic acid mononucleotide (NAMN) adenylyltransferase has been partially purified (3), the corresponding gene (nadD) has not yet been identified in any microorganism (Fig. 1A). The gene for the closely related enzyme, nicotinamide mononucleotide (NMN) adenylyltransferase, involved in the salvage of NMN, has been cloned from several sources (4, 7–11).
FIG. 1.
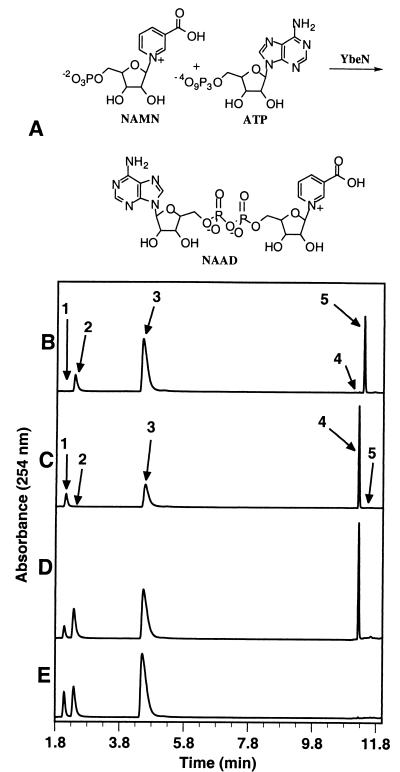
(A) The YbeN-catalyzed adenylation reaction. (B and C) HPLC analyses of the YbeN-catalyzed adenylation reactions: (B) 1.5 mM NAMN, reaction time is 15 min; (C) 1.5 mM NMN, reaction time is 40 min. (D and E) HPLC analysis of the competitive adenylation of NAMN and NMN: (D) the reaction mixture at t = 5 min; (E) the reaction mixture at t = 0 min. Labeled peaks: 1, NAMN; 2, NMN; 3, ATP; 4, NAAD; 5, NAD.
Mutations in nadD have been previously identified in Salmonella typhimurium as 6-amino-nicotinamide-resistant mutants which had elevated levels of intracellular NAMN and reduced levels of NAMN adenylyltransferase activity (6). Most of these mutants were temperature-sensitive lethal mutants, and all mapped between lip and leuS. There are four unidentified open reading frames in this region of the E. coli genome, one of which, ybeN, had sequence homology with Rv2421c from Mycobacterium tuberculosis. Rv2421c had low sequence homology to the recently identified NMN adenylyltransferase (YLR328W) from Saccharomyces cerevisiae (4). All three genes contained a good match for the nucleotidyl transferase consensus sequence (GXFXXXHXGH) (2), which suggested that ybeN was a reasonable candidate for the NAMN adenylyltransferase. Surprisingly, a BLAST search indicated that YbeN did not show strong sequence similarity to other bacterial gene products. In this communication, we report the overexpression of ybeN and the demonstration that the gene product catalyzes the adenylation of NAMN (Fig. 1A).
Materials.
A dehydrated form of Luria-Bertani broth was purchased from Gibco BRL (Gaithersburg, Md.). Ampicillin and isopropyl-β-d-thiogalactopyranoside (IPTG) were obtained from Jersey Lab and Glove Supply (Livingston, N.J.). Tris, agarose, EDTA, agar, dithiothreitol, 5-bromo-4-chloro-3-indolyl-β-d-galacto-pyranoside (X-Gal), and sodium dodecyl sulfate-polyacrylamide gel electrophoresis molecular weight markers were from Sigma (St. Louis, Mo.). Sodium chloride was from Fisher (Pittsburgh, Pa.). NAMN, NMN, and NAD were from Aldrich-Sigma Chemical Co. (St. Louis, Mo.). Acrylamide and N,N-methylenebisacrylamide (37.5:1) were purchased from Bio-Rad Laboratories (Hercules, Calif.).
The blunt cloning vector pSTBlue-1, the Perfectly Blunt Cloning kit, the overexpression strain Tuner(DE3), and the T7 promoter based overexpression plasmid pET-16b(+) were purchased from Novagen (Madison, Wis.). All restriction endonucleases and T4 DNA ligase were purchased from New England Biolabs (Beverly, Mass.). Platinum Pfx DNA polymerase was purchased from Gibco BRL. Automated DNA Fluorescence Sequencing was performed at the Cornell BioResource Center.
Molecular cloning.
Standard methods were used for DNA restriction endonuclease digestion, ligation, and transformation (1, 12). Plasmid DNA was purified with the Wizard Plus SV DNA miniprep kit (Promega, Madison, Wis.). DNA fragments were separated by agarose gel electrophoresis, were excised, and were purified with the QIAEX II Gel Extraction Kit (QIAGEN, Valencia, Calif.). E. coli DH5α was used as a recipient for transformations during plasmid construction and for plasmid propagation and storage. A Perkin-Elmer GeneAmp PCR System 2400 was used for PCR.
The PCR to amplify the open reading frame corresponding to ybeN used E. coli B genomic DNA as the template and 5′-TTA TCG ACG GTT CAT ATG AAA TCT TTA CAG GCT CTG TTT GGC-3′ (inserts an NdeI site at the start of the open reading frame) and 5′-CAG TCG CAT CAT GCC CTC GAG AAC GAC AGG TAT CAG CG-3′ (inserts an XhoI site downstream of the open reading frame) as the primer pair. The PCR product was purified by the QIAquick PCR purification kit (QIAGEN). The Novagen Perfectly Blunt Cloning kit was used to blunt the fragment ends and ligate the PCR fragment into pSTBlue-1. Plasmid DNA from eight white colonies was purified and screened for the presence of the insert. A representative insert-containing plasmid was designated pCLK1039. The open reading frame was then removed from pCLK1039 by digestion with NdeI and XhoI and was ligated into similarly digested pET-16b. Colonies were screened for the presence of the insert and a representative insert-containing plasmid was designated pCLK1040. The PCR-derived DNA was sequenced and found to contain three silent mutations when compared to the database sequence for E. coli K-12.
Overexpression and purification of YbeN.
The overexpression strain pCLK1040/Tuner(DE3) was grown in Luria-Bertani broth supplemented with 200 μg of ampicillin per ml at 37°C in a shaker at 200 rpm until the absorbance at 600 nm was approximately 0.4. The temperature was then adjusted to 15°C. Once the absorbance at 600 nm was 0.6, overexpression was induced by adding IPTG to 1.0 mM, and growth was continued for 9 h. Cells were harvested by centrifugation (6,100 × g, 30 min). Cells from 0.5 liters of media were resuspended in 30 ml of binding buffer (5 mM imidazole, 500 mM NaCl, 20 mM Tris, pH 7.9) and were lysed by a combination of lysozyme treatment and sonication (Heat Systems Ultrasonics model W-385 sonicator equipped with a 0.5-in. tip on a 5-s cycle, 50% duty for 3 min), and the lysate was cleared by centrifugation (27,000 × g, 30 min). Nickel column affinity chromatography was performed as described in the pET System Manual (Novagen). The yield of purified protein was 10 mg.
Enzymatic assays.
The NAMN and NMN adenylyltransferase assays were carried out in 100 mM Tris-HCl, pH 8.0, with 2.0 mM MgCl2. The substrates (0.25 or 1.5 mM) and enzyme (1 μg/100 μl) were incubated with 2 mM ATP at 37°C for the appropriate time interval, and 20-μl aliquots were analyzed by high-pressure liquid chromatography (HPLC). The substrate concentration for the reverse reaction was 10 mM sodium pyrophosphate and 0.1 mM NAAD. NAD was identified by comigration with a reference sample, and NAAD was purified by HPLC and identified by nuclear magnetic resonance spectroscopy.
HPLC assay.
HPLC was performed on a Hewlett-Packard 1100 by using a Supelcosil LC-18-T 15-cm by 4.6-mm column (Supelco, Bellefonte, Pa.). Buffer A contained 100 mM KH2PO4 (aqueous), pH 7.5. Buffer B contained 80% 100 mM KH2PO4 (aqueous), pH 7.5, and 20% methanol. Elution conditions were 100% buffer A for 0 to 7 min, 100% buffer A to 100% buffer B in 7 to 8 min, and 100% buffer B for 8 to 13 min. Flow rate was 1 ml/min. Under these conditions, the following compounds were readily separated (retention times in parentheses): NAMN (2.08 min), NMN (2.38 min), ATP (4.54 min), NAAD (11.24 min), and NAD (11.45 min).
Enzymatic assay of YbeN.
The ybeN gene product was overexpressed as a soluble 24.5-kDa protein in good yield (20 mg/liter of culture). YbeN catalyzed the complete conversion of 0.25 mM solutions of NAMN and NMN to NAAD and NAD, respectively. When the reaction was carried out using 1.5 mM NAMN, the reaction stopped when the product concentration reached 1.0 mM (Fig. 1B and C). The addition of fresh enzyme did not result in additional conversion to product; however, the reaction could be reinitiated by fourfold dilution. This suggests that NAMN adenylyltransferase is susceptible to product inhibition. NadD was able to catalyze the complete conversion of 0.1 mM NAAD to ATP and NAMN when 10 mM pyrophosphate was provided.
In a competition reaction containing 1.5 mM NAMN and 1.5 mM NMN, only NAAD is formed. This demonstrates that NAMN is adenylated more than 20 times faster than NMN (Fig. 1D and E). This result stands in contrast to the adenylation catalyzed by E. coli NadR, a bifunctional protein that regulates NAD biosynthesis and catalyzes the adenylation of NMN 170 times faster than the adenylation of NAMN (7).
Conclusions. Our experiments demonstrate that YbeN catalyzes the reversible adenylation of NAMN to give NAAD (Fig. 1A). We therefore propose that ybeN be renamed nadD.
Acknowledgments
We gratefully acknowledge the support of this research by Hoffmann-La Roche and by a grant from the National Institutes of Health (DK44083).
ADDENDUM IN PROOF
While this paper was being reviewed, we learned that Andrei Osterman at Integrated Genomics had also identified the function of YbeN, using the WIT program (A. Osterman, personal communication).
REFERENCES
- 1.Ausubel F M, Brent R, Hingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley and Sons; 1987. [Google Scholar]
- 2.Bork P, Holm L, Eugene K V, Sander C. The cytidylyltransferase superfamily: identification of the nucleotide-binding site and fold prediction. Proteins Struct Funct Genet. 1995;22:259–266. doi: 10.1002/prot.340220306. [DOI] [PubMed] [Google Scholar]
- 3.Dahmen W J, Webb B, Preiss J. The deamidodiphosphopyridine nucleotide and diphosphopyridine nucleotide pyrophosphorylases of Escherichia coli and yeast. Arch Biochem Biophys. 1967;120:440–450. doi: 10.1016/0003-9861(67)90262-7. [DOI] [PubMed] [Google Scholar]
- 4.Emanuelli M, Carnevali F, Lorenzi M, Raffaelli N, Amici A, Ruggieri S, Magni G. Identification and characterization of YLR328W, the Saccharomyces cerevisiae structural gene encoding NMN adenylyltransferase. Expression and characterization of the recombinant enzyme. FEBS Lett. 1999;455:13–17. doi: 10.1016/s0014-5793(99)00852-2. [DOI] [PubMed] [Google Scholar]
- 5.Geerlof A, Lewendon A, Shaw W V. Purification and characterization of phosphopantetheine adenylyltransferase from Escherichia coli. J Biol Chem. 1999;274:27105–27111. doi: 10.1074/jbc.274.38.27105. [DOI] [PubMed] [Google Scholar]
- 6.Hughes K T, Ladika D, Roth J R, Olivera B M. An indispensable gene for NAD biosynthesis in Salmonella typhimurium. J Bacteriol. 1983;155:213–221. doi: 10.1128/jb.155.1.213-221.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Raffaelli N, Lorenzi T, Mariani P L, Emanuelli M, Amici A, Ruggieri S, Magni G. The Escherichia coli NadR regulator is endowed with nicotinamide mononucleotide adenylyltransferase activity. J Bacteriol. 1999;181:5509–5511. doi: 10.1128/jb.181.17.5509-5511.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Raffaelli N, Emanuelli M, Pisani F M, Amici A, Lorenzi T, Ruggieri S, Magni G. Identification of the archaeal NMN adenylyltransferase gene. Mol Cell Biochem. 1999;193:99–102. [PubMed] [Google Scholar]
- 9.Raffaelli N, Lorenzi T, Amici A, Emanuelli M, Ruggieri S, Magni G. Synechocystis sp. slr0787 protein is a novel bifunctional enzyme endowed with both nicotinamide mononucleotide adenylyltransferase and ‘Nudix’ hydrolase activities. FEBS Lett. 1999;444:222–226. doi: 10.1016/s0014-5793(99)00068-x. [DOI] [PubMed] [Google Scholar]
- 10.Raffaelli N, Pisani F M, Lorenzi T, Emanuelli M, Amici A, Ruggieri S, Magni G. Characterization of nicotinamide mononucleotide adenylyltransferase from thermophilic archaea. J Bacteriol. 1997;179:7718–7723. doi: 10.1128/jb.179.24.7718-7723.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Raffaelli N, Amici A, Emanuelli M, Ruggieri S, Magni G. Pyridine dinucleotide biosynthesis in archaebacteria: presence of NMN adenylyltransferase in Sulfolobus solfataricus. FEBS Lett. 1994;355:233–236. doi: 10.1016/0014-5793(94)01195-8. [DOI] [PubMed] [Google Scholar]
- 12.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]