Abstract
The Corynebacterium glutamicum mutant KY9714, originally isolated as a lysozyme-sensitive mutant, does not grow at 37°C. Complementation tests and DNA sequencing analysis revealed that a mutation in a single gene of 1,920 bp, ltsA (lysozyme and temperature sensitive), was responsible for its lysozyme sensitivity and temperature sensitivity. The ltsA gene encodes a protein homologous to the glutamine-dependent asparagine synthetases of various organisms, but it could not rescue the asparagine auxotrophy of an Escherichia coli asnA asnB double mutant. Replacement of the N-terminal Cys residue (which is conserved in glutamine-dependent amidotransferases and is essential for enzyme activity) by an Ala residue resulted in the loss of complementation in C. glutamicum. The mutant ltsA gene has an amber mutation, and the disruption of the ltsA gene caused lysozyme and temperature sensitivity similar to that in the KY9714 mutant. l-Glutamate production was induced by elevating growth temperature in the disruptant. These results indicate that the ltsA gene encodes a novel glutamine-dependent amidotransferase that is involved in the mechanisms of formation of rigid cell wall structure and in the l-glutamate production of C. glutamicum.
Coryneform bacteria are rod-shaped, nonsporulating, gram-positive bacteria that are widely distributed in nature. The nonpathogenic species Corynebacterium glutamicum, Brevibacterium lactofermentum, and Brevibacterium flavum are used for fermentative production of amino acids such as l-glutamic acid (16), l-lysine (21), and l-threonine (26). Recently, B. lactofermentum and B. flavum were reclassified as C. glutamicum (18).
C. glutamicum was originally isolated as a glutamate-producing bacterium (16, 30). Induction of glutamate excretion by C. glutamicum requires biotin limitation (27) or treatment with penicillin (22) or a fatty acid ester surfactant (28). Since these treatments correlated with alterations in cell surface structure, it had been thought that glutamate leaks passively through a membrane. Recently, several findings that do not agree with the leakage model have been reported (9, 12, 15). However, the mechanism of the glutamate production and excretion of C. glutamicum is still unknown.
Coryneform bacteria have thick cell walls consisting of two layers. The inner layer, which consists mainly of peptidoglycan, invaginates to form the septum. The outer layer, which consists mainly of mycolic acid, breaks after cell division, resulting in postfission movement and the cell arrangements characteristic of these bacteria. C. glutamicum shows high tolerance to lysis by lytic enzymes, such as egg white lysozyme, that catalyze hydrolysis of the β-1,4 glycoside bond between the N-acetylglucosamine and N-acetylmuramic acid of peptidoglycan. This is probably because the outer layer, mainly consisting of mycolic acid, gives enough resistance against cell lysis to this bacterium and/or functions as a permeability barrier.
As the first step in analyzing the cell wall structure of coryneform bacteria and the relationship between glutamate production and the treatments that affect cell surface integrity, we analyzed lysozyme-sensitive mutants of C. glutamicum.
MATERIALS AND METHODS
Bacterial strains, growth condition, and plasmids.
C. glutamicum wild-type strain KY9611 and lysozyme-sensitive mutant KY9714 were obtained from Kyowa Hakko Kogyo Co., Ltd. (Tokyo, Japan). The method for isolation of lysozyme-sensitive mutants was described by Katsumata et al. (13). Escherichia coli K-12 strain JM109 [recA1 endA1 gyrA96 thi hsdR17 supE44 relA1 Δ(lac-proAB)/F′ (traD36 proAB+ lacIq lacZΔM15)] (33) was used for recombinant DNA procedures. For the preparation of plasmid DNA for the transformation of C. glutamicum, JM110 [dam dcm supE44 hsdR17 thi leu rpsL1 lacY galK galT ara tonA thr tsx Δ(lac-proAB)/F′ (traD36 proAB+ lacIq lacZΔM15)] (33) was used to escape from the restriction system of C. glutamicum. E. coli ER strain (asnA31 asnB32 thi) (3, 31) was used as an asparagine auxotroph mutant.
Cells were grown in Lennox (L) medium (1% Bacto Peptone, 0.5% yeast extract, 0.5% NaCl, 0.1% glucose [pH 7.0]) at 30 or 37°C. Growth of the liquid cultures was monitored by measuring the optical density at 660 nm (OD660). For an asparagine auxotrophy check, M9 minimal medium (per liter: 6 g of Na2HPO4, 3 g of KH2PO4, 0.5 g of NaCl, 1 g of NH4Cl, 0.25 g of MgSO4 · 7H2O, 2 g of glucose, 1 mg of thiamine · HCl, and 1.1 g of CaCl2 [pH 7.0]) with or without 200 mg of asparagine per liter was used. L or M9 plates were solidified by adding 1.5% agar. For l-glutamic acid production by C. glutamicum, cells were cultured in basal salt (BS) medium [per liter: 5 g of (NH4)2SO4, 5 g of urea, 2 g of KH2PO4, 2 g of K2HPO4, 0.25 g of MgSO4 · 7H2O, 0.01 g of FeSO4 · 7H2O, 0.01 g of MnSO4 · 4–5H2O, 0.01 g of CaCl2 · 2H2O, 0.03 mg of ZnSO4 · 7H2O, 0.1 mg of H3BO4, 0.07 mg of CoCl2 · 6H2O, 0.03 mg of CuCl2 · 2H2O, 0.01 mg of NiCl2, 0.1 mg of NaMo2O4 · 2H2O, 50 g of glucose, and 200 μg of biotin (pH 7.0)] (12). Antibiotics were used at the following concentrations for selection of cells carrying plasmids: 10 or 5 μg of kanamycin ml−1 for C. glutamicum and 20 μg of kanamycin ml−1, 25 μg of ampicillin ml−1, or 40 μg of chloramphenicol ml−1 for E. coli.
Plasmids pHSG298 and pHSG398 (TaKaRa Shuzo Co., Ltd., Kyoto, Japan) and pMW119 (Nippon Gene Co., Ltd., Tokyo, Japan) were used as E. coli vectors, and pC2 (8) was used as an E. coli-C. glutamicum shuttle vector.
Lysozyme sensitivity and temperature sensitivity checks.
For the temperature sensitivity check, 10-fold serial dilutions of cultures were spotted onto L plates, and the plates were incubated at 30 or 37°C for 24 h. For the lysozyme sensitivity check, L plates containing 50 μg of lysozyme ml−1 were used and plates were incubated at 30°C.
Cloning of the ltsA gene.
Chromosomal DNA of the C. glutamicum wild-type strain KY9611 was isolated following the method described by Sambrook et al. (23), with the concentration of lysozyme modified to 2.5 μg ml−1 (32). Chromosomal DNA was digested with the restriction enzyme EcoRI and then ligated with EcoRI-digested shuttle vector pC2, which is composed of E. coli vector pHSG298 and B. lactofermentum cryptic plasmid pBL1 (8). The ligated mixture was directly introduced into KY9714 mutant cells by electroporation, and transformants were selected on L plates containing 5 μg of kanamycin ml−1 at the restrictive temperature 37°C.
DNA sequencing.
DNA sequencing was carried out by the methods of Sanger et al. (24) using the Thermo Sequenase fluorescent-labeled primer-cycle sequencing kit with 7-deaza-dGTP (Amersham Pharmacia Biotech UK Ltd.) with an automated DNA sequencer, DSQ-1000L (Shimadzu Co., Kyoto, Japan).
Construction of ltsA (C2A) mutant gene.
The mutant ltsA gene in which the N-terminal second cysteine residue was replaced by alanine residue, named ltsA (C2A), was amplified by PCR using primers 5′-TTTGAATTCAGGAGGATTTTTCATTCATGGCAGGC-3′ and 5′-CTCCTTCAACTAAAGAGGATCCGTT-3′. The PCR products were cloned into the EcoRI and BamHI sites of shuttle vector pC2. Since these PCR products do not include their own promoter sequence, the ltsA (C2A) mutant gene was expressed from the E. coli lac promoter on shuttle vector pC2. The resulting plasmid was named pLtsA-C2A. Plasmid pLtsA carrying the wild-type gene of the same construct was prepared similarly using primers 5′-TTTGAATTCAGGAGGATTTTTCATT-3′ and 5′-CTCCTTCAACTAAAGAGGATCCGTT-3′.
PCR was carried out using the LA PCR kit, version 2.1 (TaKaRa Shuzo Co., Ltd.) and a programmable thermal cycler, PTC-100 (MJ Research, Watertown, Mass.).
In vitro protein synthesis.
For in vitro protein synthesis, the isolated fragments were recloned on E. coli vector plasmid pMW119. In vitro protein synthesis using the system of De Vries and Zubay (5) was performed with the E. coli S30 extract system for circular DNA (Promega Co., Madison, Wis.) and l-[4,5-3H]leucine (2.22 to 4.44 TBq mmol−1; Moravek Biochemicals, Inc., Brea, Calif.).
Determination of a mutation point of the ltsA gene in KY9714.
A fragment of about 0.8 kb encompassing SacI and AgeI sites in the ltsA gene was amplified from the chromosomal DNA of KY9611 (wild-type) or KY9714 (mutant) by PCR using primers 5′-TGAACCCGACCGCATGCCAATGACT-3′ and 5′-TCCAAGGTCGACATGATCTTAGGAA-3′. PCR products were cloned into SphI-SalI sites of pHSG398. Three clones independently isolated were sequenced for both wild-type and mutant genes.
Construction of an ltsA disruptant.
A PstI-SalI fragment of about 1.2 kb, in which both the N-terminal and C-terminal portions of the ltsA gene were truncated, was cloned into the PstI-SalI sites of the E. coli vector plasmid pHSG298. The constructed plasmid, pSLP2, was introduced into the C. glutamicum wild-type strain KY9611 by electroporation, and kanamycin-resistant colonies were selected at 30°C on L plates containing 10 μg of kanamycin ml−1. In kanamycin-resistant colonies, pSLP2 should be integrated into the host chromosome by homologous recombination at the ltsA locus because pSLP2 cannot replicate in C. glutamicum. The chromosomal disruption of the ltsA gene was checked by PCR using primer 5′-TGAACCCGACCGCATGCCAATGACT-3′, which anneals to the ltsA gene sequence, and primer 5′-CAGGAAACAGCTATGAC-3′, which anneals to the pHSG298 sequence (data not shown).
Measurement of l-glutamate production and glucose consumption.
Overnight cultures of KY9761, KY9714, or the ltsA disruptant were inoculated in BS medium and cells were grown at 30°C to an OD660 of about 0.4. Then cultures were shifted to 30, 35, or 37°C and incubated for 24 h. After cells were removed by centrifugation, concentrations of l-glutamate and glucose in the medium were measured with a Biotech-Analyzer AS-210 (Sakura Seiki, Tokyo, Japan) using glutamate oxidase sensor or glucose oxidase sensor. In the case of penicillin induction, 1 μM penicillin G was added to the culture of KY9611 at an OD660 of about 0.4 and cells were cultured at 30°C for 24 h.
Nucleotide sequence accession number.
The nucleotide sequence data reported in this paper will appear in the DDBJ/EMBL/GenBank nucleotide sequence databases with accession number AB029550.
RESULTS
Physiological characterization of the C. glutamicum lysozyme-sensitive mutant KY9714.
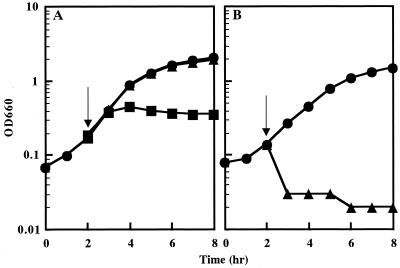
We determined growth patterns of the C. glutamicum KY9714 mutant strain, which was originally isolated as a lysozyme-sensitive mutant following N-methyl-N′-nitro-N-nitrosoguanidine mutagenesis (13). It is known that coryneform bacteria, such as C. glutamicum, are highly resistant to lysozyme. Growth of a wild-type strain of C. glutamicum, KY9611, was not affected by treatment with 12.5 μg of lysozyme ml−1. Growth was inhibited by treatment with 400 μg of lysozyme ml−1 but the turbidity of the culture did not decrease, indicating that cells so treated did not lyse (Fig. 1A). This was confirmed by microscopic observation. On the other hand, growth of the mutant strain KY9714 was inhibited by treatment with 12.5 μg of lysozyme ml−1. The turbidity of the culture decreased soon after the addition of lysozyme, indicating that cells were lysing (Fig. 1B).
FIG. 1.
Effect of lysozyme on growth of C. glutamicum wild-type strain KY9611 and the lysozyme-sensitive mutant strain KY9714. KY9611 (A) and KY9714 (B) were incubated in L broth at 30°C, and lysozyme was added to the cultures at the time indicated by arrows. Growth was monitored by measuring OD660. Circles, no addition; triangles, 12.5 μg of lysozyme ml−1; and squares, 400 μg of lysozyme ml−1.
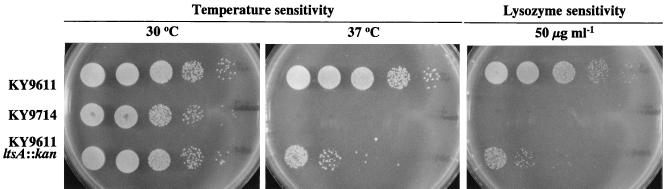
Strain KY9714 did not grow at 37°C on L plates (Fig. 2). Temperature sensitivity of the mutant was rescued when 20% sucrose was added, although colonies were smaller than those of the wild-type strain. Moreover, lysozyme sensitivity of the mutant was also recovered by 20% sucrose (data not shown). These results suggest that defects in the mutant cells caused by treatment with lysozyme and by culturing at the restricted temperature were similar, that is, they resulted in a defect of cell wall structure. A similar phenomenon is also observed with E. coli temperature-sensitive mutants deficient in cell wall synthesis (20).
FIG. 2.
Temperature sensitivity and lysozyme sensitivity of C. glutamicum KY9714 and the ltsA disruptant. Serial dilutions (1/10 each) of cultures of C. glutamicum wild-type strain KY9611, the lysozyme-sensitive mutant strain KY9714, and the ltsA disruptant were spotted onto L plates or L plates containing 50 μg of lysozyme ml−1 and then incubated at 30°C (L plates alone and with lysozyme) or 37°C (L plates alone).
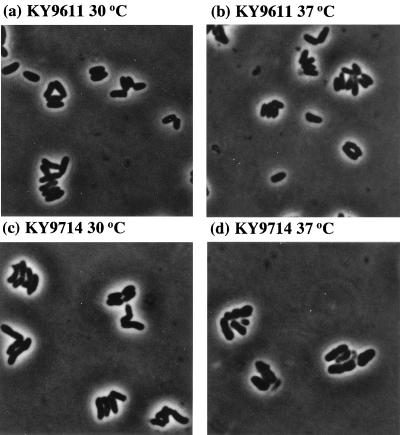
Upon phase-contrast microscopy, the wild-type strain KY9611 showed a normal rod shape at both 30 and 37°C. On the other hand, the KY9714 mutant strain became little fat rods or club-shaped rods even at the permissive temperature (30°C), and they became swollen at the restrictive temperature (37°C) (Fig. 3). These morphologies are typical of temperature-sensitive mutants of C. glutamicum (14).
FIG. 3.
Morphology of C. glutamicum wild-type strain KY9611 (a and b) and the lysozyme-sensitive mutant strain KY9714 (c and d). Cells grown in L broth at 30°C (a and c) and 37°C (b and d) were observed. Phase-contrast microphotographs are shown.
Considering these results, we speculate that the KY9714 mutant has a defect(s) in cell wall structure.
Cloning of a gene involved in the lysozyme and temperature sensitivity of the C. glutamicum KY9714 mutant.
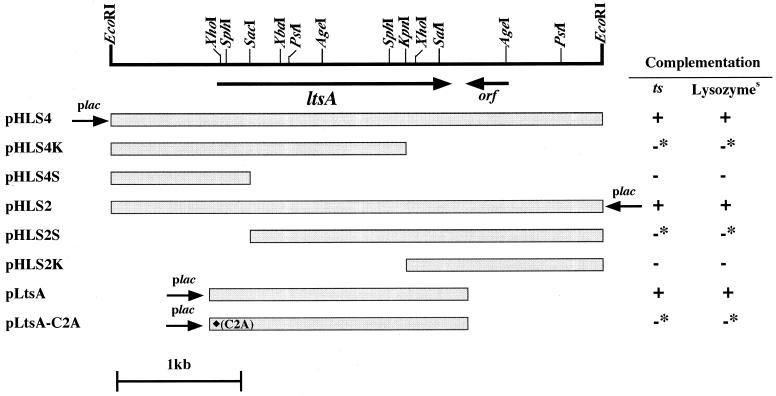
We shotgun cloned a gene involved in the lysozyme and temperature sensitivity of a KY9714 mutant. An EcoRI library of wild-type C. glutamicum chromosome DNA in the E. coli-C. glutamicum shuttle vector pC2 (8) was introduced directly into KY9714, and temperature-resistant clones were selected at 37°C as described in Materials and Methods. Two clones, pHLS2 and pHLS4, which carried the same EcoRI fragment of about 4 kb in different orientations, were isolated. These plasmids could complement not only the temperature sensitivity but also the lysozyme sensitivity of the KY9714 mutant (Fig. 4).
FIG. 4.
Summary of complementation of the lysozyme-sensitive mutant strain KY9714. Lysozyme sensitivity (Lysozymes) and temperature sensitivity (ts) were checked as described in Materials and Methods. +, complemented; −, not complemented; ∗, low-frequency complementation, probably by homologous recombination between plasmids and chromosome; ⧫, replacement of the second cysteine residue by alanine.
Several subclones of pHLS2 and pHLS4 did not complement the phenotypes of the KY9714 mutant (Fig. 4). However, two constructs, pHLS4K, carrying an approximately 2.5-kb EcoRI-KpnI fragment, and pHLS2S, carrying an approximately 3-kb SacI-EcoRI fragment, generated transformants that showed the wild-type phenotype, although at a low frequency (Fig. 4). This was probably because homologous recombination between the fragments on the plasmids and the chromosomal DNA of the mutant took place. These results indicate that the gene which complemented both phenotypes of the KY9714 mutant encompasses both the SacI and KpnI sites and that the mutation point of KY9714 is located within an approximately 1.3-kb SacI-KpnI fragment. Since the complementation patterns of lysozyme sensitivity and temperature sensitivity were identical, it seemed that a single mutation is responsible for both phenotypes of the KY9714 mutant.
Nucleotide sequencing analysis and homology search.
The nucleotide sequence of the cloned EcoRI fragment was determined by the dideoxy method (24). The EcoRI fragment contained two open reading frames (ORFs). The longer one (1,920 bp), which contained both SacI and KpnI sites, encodes a protein of 640 amino acid residues with a predicted molecular mass of 72,443 Da, and the shorter one (342 bp) encodes a protein of 114 amino acids with a molecular mass of 12,094 Da and is downstream of the longer ORF in the opposite direction. Judging from the results of complementation tests, we concluded that the longer ORF is the gene that complemented the phenotypes of the KY9714 mutant, and we designated it ltsA. This was confirmed by a complementation test with pLtsA carrying only the ltsA gene and by determination of the mutation point as mentioned below.
The deduced amino acid sequence of the ltsA gene product shows significant similarity to the glutamine-dependent asparagine synthetases of various organisms, such as E. coli (accession number J05554), humans (M27396), the yeast Saccharomyces cerevisiae (Z48675), and the pea Pisum sativum (X52179). However, the ltsA gene was unable to complement asparagine auxotrophy of the E. coli asnA asnB double mutant strain ER (data not shown). Glutamine-dependent asparagine synthetase belongs to purF-type glutamine-dependent amidotransferase (25, 29). Its N-terminal second cysteine residue is absolutely required for enzyme activity in the amidotransfer reaction (1, 11, 34).
To confirm that the ltsA gene product is a member of the purF-type amidotransferase family, we replaced the N-terminal second cysteine residue of the LtsA protein by alanine residue, as described in Materials and Methods. The mutant ltsA gene, named ltsA (C2A), could not complement the temperature-sensitive growth and the lysozyme sensitivity of the KY9714 mutant, although recombinants with wild-type phenotypes were generated at low frequency, as in the case of pHLS4K and pHLS2S (Fig. 4). These results strongly suggest that the ltsA gene does not encode an asparagine synthetase but encodes another glutamine-dependent amidotransferase activity which is probably responsible for the synthesis of cell wall components of C. glutamicum.
The gene product of the shorter ORF shows homology with the hypothetical proteins Rv2204c of Mycobacterium tuberculosis (4), YadR of E. coli (7), and HI1723 of Haemophilus influenzae (6), etc., but the functions of these gene products have not been determined.
We examined the gene products of the cloned EcoRI fragment and its deletion derivatives by in vitro protein synthesis using an E. coli S30 extract system (5). The results indicate that the 61-kDa protein corresponds to the product of ltsA gene, although the measured molecular mass of this protein is a little lower than that calculated from the nucleotide sequence (72,443 Da). The 15-kDa product corresponds to that of the shorter ORF, but the molecular mass was a little higher than that calculated from the nucleotide sequence (12,094 Da) (data not shown).
Identification of mutation of ltsA gene in KY9714 mutant and ltsA gene disruption.
Judging from the results of recombinational complementation of the KY9714 mutant by using several deletion derivatives of pHLS2 and pHLS4, strain KY9714 has a mutation(s) in the 0.8-kb SacI-AgeI fragment encoding the N-terminal portion of the LtsA protein (data not shown). We amplified by PCR the corresponding DNA region and sequenced it as described in Materials and Methods. Surprisingly, the codon TGG of Trp132 was changed to a stop codon (TAG), that is, an amber mutation. This means that the KY9714 mutant produces only the N-terminal 1/5 portion of the LtsA protein. This G:C→A:T transition is as expected from the mutation spectrum caused by N-methyl-N′-nitro-N-nitrosoguanidine.
To confirm whether disruption of the ltsA gene actually causes lysozyme and temperature sensitivity, we constructed a disruptant of the ltsA gene by homologous recombination between the chromosomal ltsA gene and the truncated ltsA gene on a plasmid. Plasmid pSLP2, which is composed of the internal fragment of the ltsA gene and E. coli vector pHSG298, was introduced directly into C. glutamicum wild-type strain KY9611 and kanamycin-resistant colonies selected at 30°C. Because pSLP2 cannot replicate in C. glutamicum, in kanamycin-resistant colonies pSLP2 should be integrated into the host chromosome by homologous recombination at the ltsA locus, generating an N-terminally truncated ltsA gene and a C-terminally truncated ltsA gene on the chromosome. The chromosomal disruption of the ltsA gene was checked by PCR (data not shown). The ltsA disruptant could grow at 30°C and showed a lysozyme- and temperature-sensitive phenotype similar to that of the KY9714 mutant, although the phenotype was somewhat leakier than that of KY9714 (Fig. 2). This is probably because the disruptant produced a truncated protein, which probably has a residual activity, much longer (about 9/10 of LtsA), than that produced by KY9714 (about 1/5 of LtsA).
Glutamic acid production by ltsA mutant and disruptant.
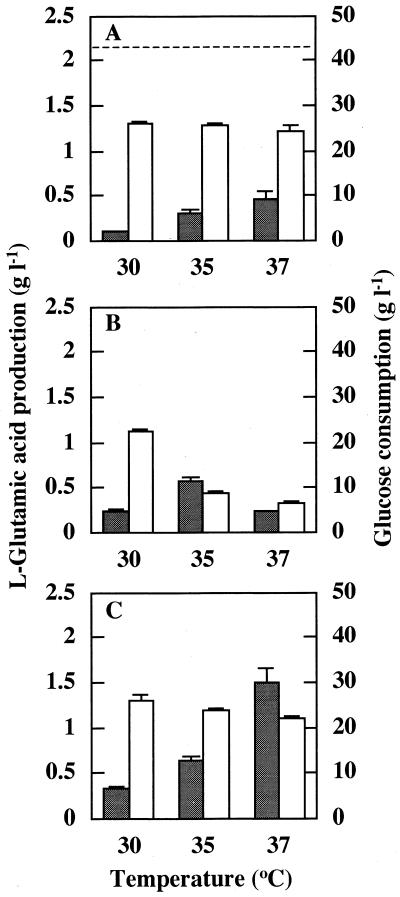
It is well known that l-glutamate production by C. glutamicum is induced by treatment with penicillin, an inhibitor of cell wall peptidoglycan synthesis. However, its mechanism has not been clarified. To examine whether the ltsA gene is involved in l-glutamate production by this organism, we studied l-glutamate production by the KY9714 mutant and the ltsA disruptant at various temperatures (Fig. 5). Both the KY9714 mutant and the disruptant began to produce l-glutamate when the growth temperature was raised. The ltsA disruptant produced an especially large amount of l-glutamate at 37°C (Fig. 5C), almost comparable to that produced by the wild-type strain treated with penicillin (Fig. 5A). In the case of the KY9714 mutant, l-glutamate production was higher at 35 than at 37°C (Fig. 5B). The differences in the effect of temperature on l-glutamate production between the KY9714 mutant and the disruptant are probably due to the extent of leakiness of the phenotypes. Since growth yields as well as glucose consumptions of KY9714 at higher temperatures were much lower than those of the wild-type and the disruptant (Fig. 5), a severe defect of the ltsA gene function may inhibit not only cell growth but also intracellular metabolism due to a loss of cell integrity.
FIG. 5.
l-Glutamate production by C. glutamicum ltsA mutants. Cells of C. glutamicum wild-type strain KY9611 (A), KY9714 (B), and the ltsA disruptant (C) were grown in BS medium at 30, 35, and 37°C for 24 h. After the cells were removed, concentrations of l-glutamate and glucose in the medium were measured by using glutamate oxidase or glucose oxidase sensor. Doubling times were 123 min (30°C), 132 min (35°C), and 156 min (37°C) in KY9611 and 126 min (30°C), 168 min (35°C), and 279 min (37°C) in the ltsA disruptant. The growth rate of KY9714 was 129 min at 30°C. After the temperature shift-up, the growth rate gradually decreased, and the growth stopped after several generations. Final growth yields (OD660) were 3.81 (30°C), 3.70 (35°C), and 3.67 (37°C) in KY9614; 3.49 (30°C), 1.66 (35°C), and 0.83 (37°C) in KY9714; and 3.68 (30°C), 3.35 (35°C), and 3.15 (37°C) in the ltsA disruptant. Closed bars indicate the production of l-glutamate, and open bars indicate the glucose consumption. Means plus standard deviations are indicated (n = 3). The horizontal dashed line in graph A indicates the level of l-glutamate produced by the wild-type strain KY9611 treated with 1 μM penicillin G for 24 h.
DISCUSSION
C. glutamicum was originally isolated as an l-glutamic acid producer (16, 30). However, C. glutamicum does not excrete l-glutamic acid under normal growth conditions. Treatment with penicillin or surfactants, which decreases cell surface integrity and often causes cell lysis in bacteria, induces production of l-glutamic acid in this microorganism (22, 28). It is also known that C. glutamicum is highly resistant to treatment with lysozyme. We assume that this nonlytic phenotype is closely related to the mechanism of l-glutamic acid production by C. glutamicum. To study the relationship between cell wall structure and l-glutamic acid production, we analyzed C. glutamicum mutant strain KY9714, which was originally isolated as a lysozyme-sensitive mutant. We isolated a novel gene, ltsA, which encodes a purF-type glutamine-dependent amidotransferase and complements the lysozyme sensitivity and temperature sensitivity of KY9714.
The ltsA gene product shows significant sequence similarity to glutamine-dependent asparagine synthetases of various organisms that catalyze the transfer of the γ amino residue of glutamine to the carboxyl residue of aspartate (19). Glutamine-dependent asparagine synthetase belongs to the purF-type amidotransferase (25, 29). The purF-type amidotransferases have 12 conserved amino acid residues in the glutamine amide transfer domain of about 200 amino acid residues (2). These 12 amino acid residues are also found in LtsA protein. The N-terminal cysteine residue is absolutely required for enzyme activity of the amidotransfer reaction (1, 11, 34). Replacement of this cysteine residue by alanine results in loss of complementation ability, indicating that the LtsA is indeed a purF-type amidotransferase. However, the C. glutamicum ltsA gene does not complement asparagine auxotrophy of the E. coli mutant strain ER (asnA asnB double mutant). Since it was reported that even the human gene encoding glutamine-dependent asparagine synthetase could complement asparagine auxotrophy of E. coli mutant JE6279, a derivative of the ER strain (10), it is very likely that the LtsA protein is not an asparagine synthetase. Genome projects revealed that M. tuberculosis and Bacillus subtilis have the putative asnB genes (4, 17). However, these putative gene products show greater similarity to the LtsA than to asparagine synthetases such as E. coli AsnB (data not shown). Therefore, it is likely that these putative genes encode proteins with functions similar to those of the C. glutamicum ltsA gene product and not to asparagine synthetase activity.
The facts that the C. glutamicum KY9714 mutant has an amber mutation in the ltsA gene and that an ltsA disruptant could be constructed indicated that the ltsA gene is not essential for growth at 30°C. It seems that the ltsA gene product is required for formation or maintenance of the rigid cell wall structure of C. glutamicum. Swollen cell morphology of the mutant at the restrictive temperature and the suppression of temperature sensitivity and lysozyme sensitivity by 20% sucrose also support this notion. The ltsA gene product may catalyze the amido-transfer reaction from glutamine onto some unknown cell surface component(s), which is now under investigation.
The ltsA disruptant produced a significant amount of l-glutamate at high temperature, nearly comparable to that obtained by penicillin treatment. Cell morphology and suppression of the mutant phenotypes by 20% sucrose strongly suggest that the ltsA gene is involved in cell wall formation in C. glutamicum. A defect of the ltsA gene function resulted in l-glutamate production, leading us to expect that further investigation of this gene may give us a clue to the relationship between cell wall synthesis and l-glutamate production. Strains used for industrial production of glutamic acid have been constructed by classical genetic methods, that is, repeated mutagenesis and selection of improved strains. Mutations of these strains that contribute to glutamate production have not been elucidated to date. Analysis of the ltsA locus in these strains would be of interest. It is also possible that introduction of ltsA mutations into the industrial strains would improve their glutamate production.
ACKNOWLEDGMENTS
We thank M. Schaechter for critical reading of the manuscript and Kyowa Hakko Kogyo Co., Ltd. for providing C. glutamicum strains.
This work was partly supported by a Grant-in-Aid for Scientific Research on Priority Areas (08277104) and a Grant-in-Aid for Scientific Research (A) (11356003) from the Ministry of Education, Science, Sports, and Culture of Japan and a research grant from the Japan Bioindustry Association.
REFERENCES
- 1.Boehlein S K, Richards N G J, Schuster S M. Glutamine-dependent nitrogen transfer in Escherichia coli asparagine synthetase B. Searching for the catalytic triad. J Biol Chem. 1994;269:7450–7457. [PubMed] [Google Scholar]
- 2.Boehlein S K, Richards N G J, Walworth E S, Schuster S M. Arginine 30 and asparagine 74 have functional roles in the glutamine dependent activities of Escherichia coli asparagine synthetase B. J Biol Chem. 1994;269:26789–26795. [PubMed] [Google Scholar]
- 3.Cedar H, Schwartz J H. The asparagine synthetase of Escherichia coli. I. Biosynthetic role of the enzyme, purification, and characterization of the reaction products. J Biol Chem. 1969;244:4112–4121. [PubMed] [Google Scholar]
- 4.Cole S T, Brosch R, Parkhill J, Garnier T, Churcher C, et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 5.De Vries J K, Zubay G. DNA-directed peptide synthesis. II. The synthesis of the α-fragment of the enzyme β-galactosidase. Proc Natl Acad Sci USA. 1967;57:1010–1012. doi: 10.1073/pnas.57.4.1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fleischmann R D, Adams M D, White O, Clayton R A, Kirkness E F, et al. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 7.Fujita N, Mori H, Yura T, Ishihama A. Systematic sequencing of the Escherichia coli genome: analysis of the 2.4–4.1 min (110,917–193,643 bp) region. Nucleic Acids Res. 1994;22:1637–1639. doi: 10.1093/nar/22.9.1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goyal D, Wachi M, Kijima N, Kobayashi M, Yukawa H, Nagai K. A cryptic plasmid pBL1 from Brevibacterium lactofermentum causes growth inhibition and filamentation in Escherichia coli. Plasmid. 1996;36:62–66. doi: 10.1006/plas.1996.0033. [DOI] [PubMed] [Google Scholar]
- 9.Gutmann M, Hoischen C, Krämer R. Carrier-mediated glutamate secretion by Corynebacterium glutamicum under biotin limitation. Biochim Biophys Acta. 1992;1112:115–123. doi: 10.1016/0005-2736(92)90261-j. [DOI] [PubMed] [Google Scholar]
- 10.Heeke G V, Schuster S M. Expression of human asparagine synthetase in Escherichia coli. J Biol Chem. 1989;264:5503–5509. [PubMed] [Google Scholar]
- 11.Heeke G V, Schuster S M. The N-terminal cysteine of human asparagine synthetase is essential for glutamine-dependent activity. J Biol Chem. 1989;264:19475–19477. [PubMed] [Google Scholar]
- 12.Hoischen C, Krämer R. Membrane alteration is necessary but not sufficient for effective glutamate secretion in Corynebacterium glutamicum. J Bacteriol. 1990;172:3409–3416. doi: 10.1128/jb.172.6.3409-3416.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Katsumata R, Oka T, Fruya A. Japan patent H01-3475. 1991. [Google Scholar]
- 14.Kijima N, Goyal D, Takada A, Wachi M, Nagai K. Induction of only limited elongation instead of filamentation by inhibition of cell division in Corynebacterium glutamicum. Appl Microbiol Biotechnol. 1998;50:227–232. [Google Scholar]
- 15.Kimura E, Abe C, Kawahara Y, Nakamatsu T. Molecular cloning of a novel gene, dtsR, which rescues the detergent sensitivity of a mutant derived from Brevibacterium lactofermentum. Biosci Biotechnol Biochem. 1996;60:1565–1570. doi: 10.1271/bbb.60.1565. [DOI] [PubMed] [Google Scholar]
- 16.Kinoshita S, Udaka S, Shimono M. Studies on the amino acid fermentation. Part I. Production of L-glutamic acid by various microorganisms. J Gen Appl Microbiol. 1957;3:193–205. [PubMed] [Google Scholar]
- 17.Kunst F, Ogasawara N, Moszer I, Albertini A M, Alloni G, et al. The complete genome sequence of the Gram-positive bacterium Bacillus subtilis. Nature. 1997;390:249–256. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- 18.Liebl W, Ehrmann M, Ludwig W, Schleifer K H. Transfer of Brevibacterium divaricatum DSM 20297T, “Brevibacterium flavum” DSM 20411, “Brevibacterium lactofermentum” DSM 20412 and DSM 1412, and Corynebacterium lilium DSM 20137T to Corynebacterium glutamicum and their distinction by rRNA gene restriction patterns. Int J Syst Bacteriol. 1991;41:255–260. doi: 10.1099/00207713-41-2-255. [DOI] [PubMed] [Google Scholar]
- 19.Milman H A, Cooney D A. Partial purification and properties of L-asparagine synthetase from mouse pancreas. Biochem J. 1979;181:51–59. doi: 10.1042/bj1810051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miyakawa T, Matsuzawa H, Matsuhashi M, Sugino Y. Cell wall peptidoglycan mutants of Escherichia coli K-12: existence of two clusters of genes, mra and mrb, for cell wall peptidoglycan biosynthesis. J Bacteriol. 1972;112:950–958. doi: 10.1128/jb.112.2.950-958.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nakayama K, Kitada S, Kinoshita S. Studies on lysine fermentation. I. The control mechanism on lysine accumulation by homoserine and threonine. J Gen Appl Microbiol. 1961;7:145–154. [Google Scholar]
- 22.Nunheimer T D, Birnbaum J, Ihnen E D, Demain A L. Product inhibition of the fermentative formation of glutamic acid. Appl Microbiol. 1970;20:215–217. doi: 10.1128/am.20.2.215-217.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 24.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Scofield M A, Lewis W S, Schuster S M. Nucleotide sequence of Escherichia coli asnB and deduced amino acid sequence of asparagine synthetase B. J Biol Chem. 1990;265:12895–12902. [PubMed] [Google Scholar]
- 26.Shiio I, Nakamori S. Microbial production of L-threonine. Part II. Production by α-amino-β-hydroxyvaleric acid resistant mutants of glutamate producing bacteria. Agric Biol Chem. 1970;34:448–456. [Google Scholar]
- 27.Shiio I, Otsuka S, Takahashi M. Effect of biotin on the bacterial formation of glutamic acid. I. Glutamate formation and cellular permeability of amino acids. J Biochem. 1962;51:56–62. doi: 10.1093/oxfordjournals.jbchem.a127500. [DOI] [PubMed] [Google Scholar]
- 28.Takinami K, Yoshii H, Tsuri H, Okada H. Biochemical effects of fatty acid and its derivatives on L-glutamic acid fermentation. Part III. Biotin-Tween 60 relationship in the accumulation of L-glutamic acid and the growth of Brevibacterium lactofermentum. Agric Biol Chem. 1965;29:351–359. [Google Scholar]
- 29.Tso J Y, Hermodson M A, Zalkin H. Glutamine phosphoribosylpyrophosphate amidotransferase from cloned Escherichia coli purF. NH2-terminal amino acid sequence, identification of the glutamine site, and trace metal analysis. J Biol Chem. 1982;257:3532–3536. [PubMed] [Google Scholar]
- 30.Udaka S. Screening method for microorganisms accumulating metabolites and its use in the isolation of Micrococcus glutamicus. J Bacteriol. 1960;79:754–755. doi: 10.1128/jb.79.5.754-755.1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.von Meyenburg K, Hansen F G, Nielsen L D, Riise E. Origin of replication, oriC, of the Escherichia coli chromosome on specialized transducing phages λasn. Mol Gen Genet. 1978;160:287–295. doi: 10.1007/BF00332972. [DOI] [PubMed] [Google Scholar]
- 32.Wachi M, Wijayarathna C D, Teraoka H, Nagai K. A murC gene from coryneform bacteria. Appl Microbiol Biotechnol. 1999;51:223–228. doi: 10.1007/s002530051385. [DOI] [PubMed] [Google Scholar]
- 33.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 34.Zalkin H. The amidotransferases. Adv Enzymol Relat Areas Mol Biol. 1993;66:203–309. doi: 10.1002/9780470123126.ch5. [DOI] [PubMed] [Google Scholar]