Summary
Bacterial transformation and processing of diatom-derived organic matter (OM) is extremely important for the cycling of production and energy in marine ecosystems; this process contributes to the production of microbial food webs. In this study, a cultivable bacterium (Roseobacter sp. SD-R1) from the marine diatom Skeletonema dohrnii were isolated and identified. A combined Fourier transform ion cyclotron resonance mass spectrometry (FT-ICR MS)/untargeted metabolomics approach was used to synthesize the results of bacterial transformation with dissolved OM (DOM) and lysate OM (LOM) under warming and acidification through laboratory experiments. Roseobacter sp. SD-R1 had different preferences for the conversion of molecules in S. dohrnii-derived DOM and LOM treatments. The effects of warming and acidification contribute to the increased number and complexity of molecules of carbon, hydrogen, oxygen, nitrogen, and sulfur after the bacterial transformation of OM. The chemical complexity generated by bacterial metabolism provides new insights into the mechanisms that shape OM complexity.
Subject areas: Earth sciences, Global change, Marine geochemistry
Graphical abstract
Highlights
-
•
The key roles of algae-associated bacteria in the transformation of algae-derived OM
-
•
Bacteria have different preferences for the conversion of compounds in algae-derived OM
-
•
Warming and acidification affect microbial transformation of organic matter
Earth sciences; Global change; Marine geochemistry
Introduction
Phytoplanktons, especially marine diatoms, are major primary producers in the marine environment that excrete a wide range of metabolites and account for approximately 40% of marine primary productivity.1 In the microcosm, diatoms release various organic matter (OM) components into the diffusive boundary layer that surrounds individual cells.2 There is a large amount of matter/energy transformation in the diffusive boundary layer.3,4 Bacteria are essentially promoting the production of phytoplankton-derived dissolved organic matter (DOM).5 Bacteria can use and transform 10–50% of the photosynthetic products of phytoplankton.4 In comparison, lysate organic matter (LOM) from phytoplankton forms a pool of OM that is created extracellularly through metabolic excretion and adsorption.6 LOM offers significant protection to algal cells from adverse environmental conditions.7 Algae and bacteria have been widely recognized to play an important role in microbial carbon pumps (MCP).8 Numerous studies have shown that carbon fluxes between diatoms and bacterial taxa are important for substance cycling in the ocean and lead to multiple interactions between different aquatic biota.9,10,11,12 Intuitively, heterotrophic bacteria have been shown to rely on OM to support their growth.13,14 This mutually beneficial relationship between algae and bacteria facilitates a continuous utilization of OM. The bioreactivity and chemical composition of OM are changed after microbial transformation resulting in small molecules (low-molecular-weight, <10 kDa) that are very resistant to further microbial consumption.
Cell wall polymers of marine phytoplankton are considered one of the major components of recalcitrant DOM (RDOM).15 Incubation experiments have also shown that heterotrophic bacteria in the marine environment can convert sugars and protein-like substances into highly diverse components.5 Furthermore, organic compounds containing a series of richly carboxylated and thickened alicyclic rings are called carboxyl-rich alicyclic molecules (CRAM).16 Owing to their recalcitrant chemical structure and complexity, CRAM compounds are resistant to rapid degradation by microorganisms.16 The conversion of these compounds depends on the different bacterial groups and their specific oxidative capacity; thus, they contribute notably to the sequestration of carbon in the ocean.17
The coastal ocean is an important sink for anthropogenic carbon dioxide (CO2).18 In recent decades, the transitional exploitation of natural resources by humans has led to an increase in global temperatures and CO2 concentrations. With the persistent increase in the anthropogenic emission of CO2 and the consequent transmission to the ocean, seawater chemistry has tended toward acidification, resulting in ecological disasters.19 By the end of the century, atmospheric CO2 is expected to rise to 800–1000 ppm, whereas the pH of surface seawater will drop by 0.4–0.5 units.20 The impacts of ocean acidification will not be experienced in isolation; thus, synergistic interactions with other potential stressors, such as rising seawater temperatures (4–5.8°C), will likely exacerbate the microbial response to acidification. Previous studies have shown that warming and acidification lead to changes in the physiological, biochemical, and metabolic pathways in diatoms.21,22 In addition, warming and pressurization increase the vertical carbon flux in the ocean, subsequently affecting polysaccharide degradation rates, protein production, and enzyme activity.23 Owing to the significant difficulties and challenges of studying pure algal–bacterial interactions in the real-world environment, the vast majority of studies currently rely on laboratory-controlled co-culture systems of phytoplankton with individual bacteria. Although this approach enriches our knowledge of algal–bacterial interactions, an adequate model is urgently needed into the microbial complexity found in natural algal layers.
Members of the marine Roseobacter clade are major participants in the global carbon and sulfur cycles. Molecular-based approaches targeting 16S rRNA genes demonstrate that the Roseobacter clade is one of the major groups of marine Proteobacteria and is widely distributed in a variety of aquatic environments.24,25 Roseobacters account for 20% of bacterial cells in coastal waters, and dominate the microbial communities associated with a variety of marine algae.26 Because of their large genome sizes, versatile metabolic pathways, and regulatory circuits, Roseobacters have been considered classical patch-associated bacteria.27,28 Members of the Roseobacter clade share >89% identity of the 16S rRNA genes.29 Roseobacter sp. can transform in both the free and attached states, using both biologically high molecular weight substances and active low molecular weight carbon sources, which are present in all periods of algal growth.29 Here, Roseobacter sp. SD-R1 (average abundance, 4.62 × 102 cells mL−1) from the marine diatom, Skeletonema dohrnii, were isolated and identified. The isolated bacterial strains (Roseobacter sp. SD-R1) shared 91%–99% sequence similarity with the valid genus (Roseobacter sp.) from GenBank, according to the results (Figure S1). This work is based on laboratory-based studies over a 30-day time period, in which the preferences and transformation of Roseobacter sp. SD-R1 for S. dohrnii-derived OM under warming and acidification. We further characterized the variation in OM via the processing of Roseobacter sp. SD-R1 using FT-ICR MS and untargeted metabolomics. From chemical and metabolic point of view, the focus was on exploring the effect of utilization of algal-derived OM by epiphytic bacteria based on MCP theory. Further, we hypothesized that different OM characteristics are significantly associated with bacterial transformation and that they would exhibit different patterns in these linkages. It should be clarified that the results obtained from this experiment provide only theoretical and data support for the utilization and transformation of algal-derived OM by a single strain of bacteria. In brief, warming and acidification affect the bacterial response to algal-derived OM transformation.
Results
OM production and FT-ICR MS characterization
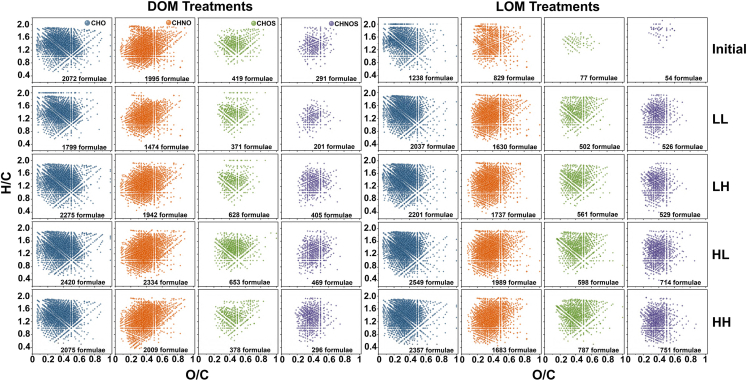
As shown in Figure 1, 4777 and 2198 molecules were detected in the DOM and LOM, respectively, at the initial stage of the experiment. Carbon–hydrogen–oxygen (CHO) compounds were the main OM molecules detected (DOM, 43.37%; LOM, 56.32%). This phenomenon applied to all treatment groups. For DOM samples, the total number of molecular formulae in the LH (5250 formulae) and HL (4876 formulae) treatment groups was higher than that in initial material (4777 formulae); LL (3845 formulae) and HH (4758 formulae) groups had lower number of molecules than the initial material. For LOM samples, the total number of molecular formulae in the four treatments (LL, 4695; LH, 5028; HL, 5950; and HH, 5578 formulae) was much greater after 30 days’ incubation than in the initial material. In both the DOM and LOM treatments, the addition of Roseobacter sp. SD-R1 increased the number of compounds for all the CHO, CHNO, CHOS, and CHNOS species. In addition, the numbers of different CHO, CHNO, CHOS, and CHNOS formulae were increased in LH, HL, and HH treatments, suggesting that the elevated temperature and acidification increased the chemical complexity of DOM and LOM. Notably, the number of CHOS and CHNOS formulae increased by about 10% in the LOM treatments, suggesting that bacteria promoted the transformation of the important fraction of organosulfur compounds under elevated temperature and acidification conditions (Figure S2).
Figure 1.
Van Krevelen diagrams of composition of OM molecules under warming and/or acidification conditions
The assigned formulae in the composition of CHO (blue), CHNO (orange), CHOS (green), and CHNOS (purple) series in the DOM treatments (left) and the LOM treatments (right). Bubble size represents the log-normalized intensity of m/z peaks. Initial, initial value; LL, 26°C and 400 ppm; LH, 26°C and 1000 ppm; HL, 30°C and 400 ppm; HH, 30°C and 1000 ppm.
Highly unsaturated and phenolic compounds, as well as aliphatic compounds were present at a high percentage after all treatments (Figure S2 and S3). At the end of the 30-day incubation experiment, we observed a significant change in the proportion of different compounds. Specifically, in the DOM samples, the proportion of highly unsaturated and phenolic compounds decreased from 66.36% to 43.90%–60.70% and aliphatic compounds increased from 22.00% to 27.49–29.89%. For LOM samples, the opposite change in compound composition to that of DOM samples was observed, highly unsaturated and phenolic compounds increased from 43.90% to 52.74–53.45%, also there was a slight increase in condensed polycyclic compounds (i.e., from 2.73% to 4.56–5.07%). Conversely, the proportion of aliphatic compounds decreased from 46.04% to 32.64–33.50%. For the proportion of condensed polycyclic compounds and aromatic compounds, the change was not significant. These phenomena suggest that bacteria have different preferences for different compounds in OM.
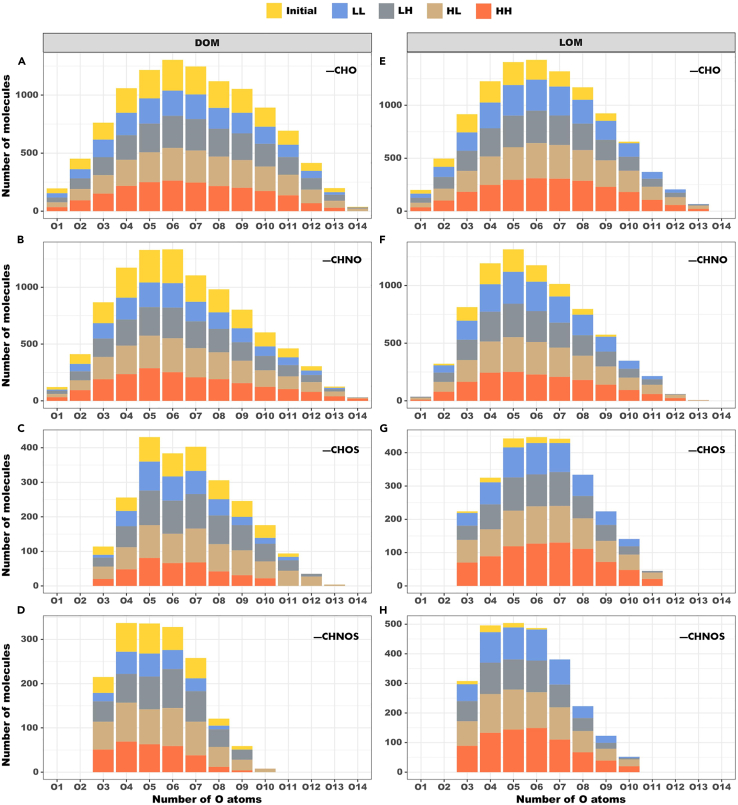
As shown in Figure 2, all molecules have O1 to O14 subgroups, which are classified according to the number of O atoms in molecules. Compared with CHOS and CHNOS compounds, the subgroups of O atoms are predominant and relatively abundant in the compounds of CHO and CHNO. As for these 14 subgroups, the number of compounds in DOM samples was generally greater than that in LOM samples. The O14 subgroup only appeared in the CHO and CHNO compounds of the DOM samples, but contained only an extremely small number of molecules. Subgroups with larger numbers of oxygen atoms (e.g., O12 to O14) are commonly found in DOM, suggesting that CHO compounds contain a lower proportion of oxygen in DOM samples, whereas most compounds in LOM samples contain a higher proportion of oxygen. In the LOM samples of CHOS and CHNOS, which initially contained only subgroups of O3–O7, four treatment groups presented subgroups of O8–O10 after 30 days’ incubation. This finding likely indicates that the production of more oxygen-containing compounds was because of the involvement of Roseobacter sp. SD-R1.
Figure 2.
Classification of CHO, CHNO, CHOS, and CHNOS compounds into subgroups according to the number of oxygen atoms in OM molecules
(A–D) DOM treatments; (E–H) LOM treatments. Initial, initial value; LL, 26°C and 400 ppm; LH, 26°C and 1000 ppm; HL, 30°C and 400 ppm; HH, 30°C and 1000 ppm.
Overview of metabolic profiles with untargeted metabolomics
To obtain information related to the effect of temperature increase and acidification on bacterial transformation of algal OM (including DOM and LOM) from a metabolomics perspective, 40 samples were analyzed by LC–MS-based untargeted metabolomics. All samples were preliminary analyzed by principal component analysis (PCA), and samples from the same OM sources were clustered together (Figure S4). The R2X value of PCA was 0.504, which showed a considerable separation between different treatments, and there was also bias within groups. Moreover, the PLS-DA and OPLS-DA models demonstrate the same results in the positive ion mode (Figure S4). The R2 and Q2 values were both close to 1, which revealed that the model was both stable and effective.
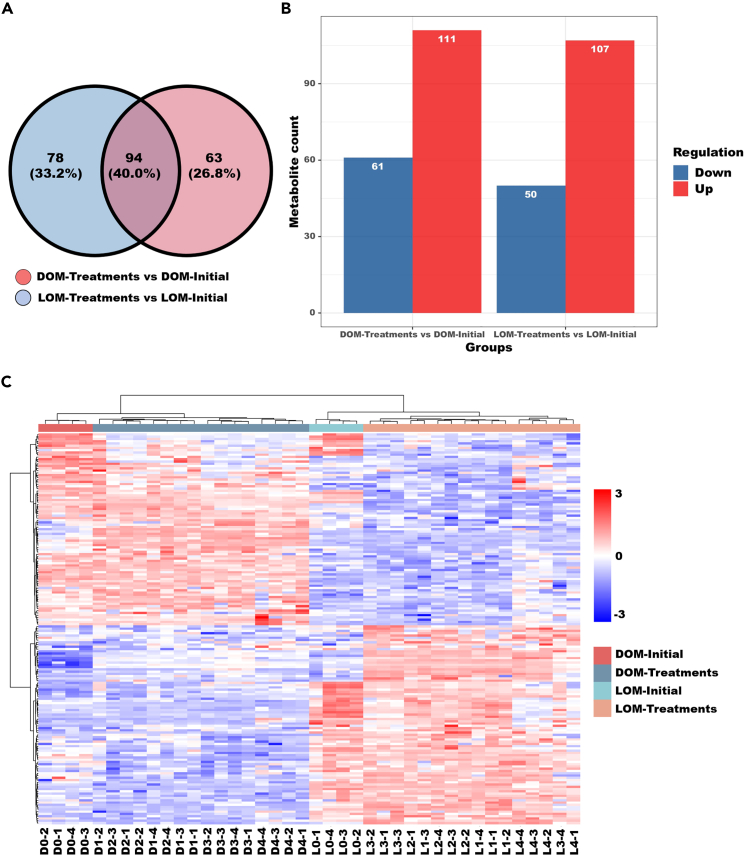
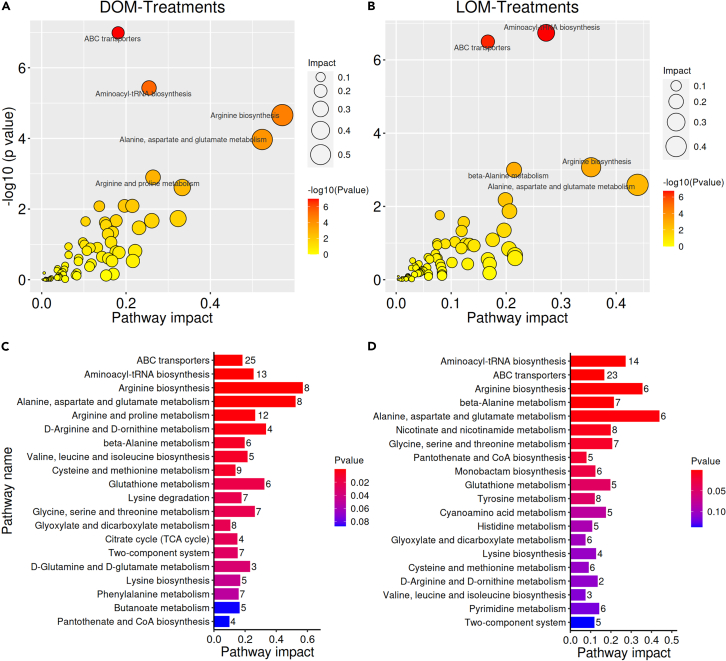
The relationships between differential metabolite groups were displayed as Venn diagrams; 78 (33.2%) metabolite counts were identified in DOM-Treatments versus DOM-Initial, and 63 (26.8%) metabolite counts were identified in LOM-Treatments versus LOM-Initial, with 94 (40.0%) overlapping counts (Figure 3A). Overall, 111 upregulated and 61 downregulated metabolites were identified between DOM-Treatments versus DOM-Initial, while there was 107 upregulated and 50 downregulated metabolites between LOM-Treatments versus LOM-Initial (Figure 3B). To compare the metabolomes among different treatments, a heatmap was generated to present the metabolic patterns under different experimental -conditions. Agglomerated hierarchical clustering analysis of differential metabolites among the different treatments showed that the expression patterns of the DOM were highly distinct from those of the LOM, but the expression patterns of elevated temperature and acidification-treated DOM and LOM samples were similar within groups (Figure 3C). Pathway enrichment analysis aims to elucidate the specific changes in the metabolic process of the bacterial transformation of OM. As shown in Figure 4, we screened out the top five major metabolic pathways for the DOM and LOM groups. The top five most impacted pathways in the DOM groups were (1) ABC transporters; (2) aminoacyl-tRNA biosynthesis; (3) arginine biosynthesis; (4) alanine, aspartate, and glutamate metabolism; and (5) arginine and proline metabolism. The top five most impacted pathways in the LOM groups were: (1) aminoacyl-tRNA biosynthesis; (2) ABC transporters; (3) arginine biosynthesis; (4) beta-alanine metabolism; and (5) alanine, aspartate, and glutamate metabolism. They affected similar metabolic pathways; the only differences were the arginine and proline metabolism (DOM groups) and beta-alanine metabolism (LOM groups). In addition, the contribution of metabolites detected under the arginine biosynthesis and alanine, aspartate, and glutamate metabolism pathways was the highest in DOM and LOM, respectively.
Figure 3.
Statistics of differential metabolites among the different treatments
(A) Venn diagram showing the overlap among metabolite counts of DOM-Initial versus DOM-Treatments and of LOM-Initial versus LOM-Treatments (A).
(B) Metabolite counts between the different treatments; red represents upregulated, and blue represents downregulated (B).
(C) Agglomerate hierarchical clustering analysis of the differential metabolites among the different treatments (C). Comparing the DOM-Treatments and LOM-Treatments, including D1 and L1 (LL), D2 and L2 (LH), D3 and L3 (HL), and D4 and L4 (HH) treatments. Four biological replicates per group.
Figure 4.
Analysis of differential metabolites among the different treatments
(A–D) Bubble diagram of metabolic pathways (A and B). Bar graph of metabolic pathway (C and D).
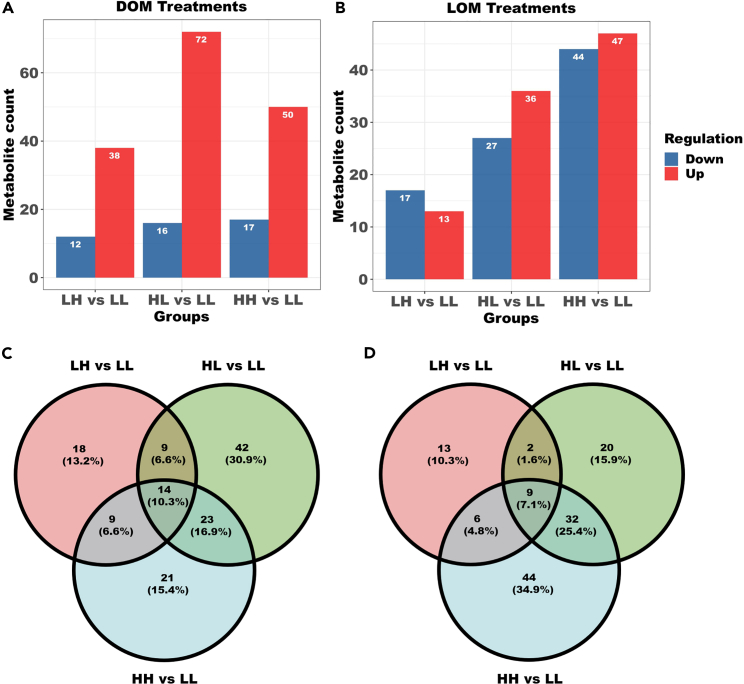
We further investigated the regulation of metabolite counts between groups of DOM and LOM treatments (Figure 5). We found that in DOM treatments, HL versus LL contained the most metabolite counts and had 72 upregulated metabolite counts (Figure 5A), suggesting that warming causes most metabolite counts to be increased. In contrast, LH versus LL had the fewest metabolite counts with only 38 upregulated and 12 downregulated, suggesting that acidification has less impact in the bacterial conversion of DOM. HH versus LL resulted in 50 upregulated and 17 downregulated metabolite counts (Figure 5B), indicating that the simultaneous stress of warming and acidification moderately increased the up- and down-regulated metabolite counts. In the LOM treatments, we found that HH versus LL contained 91 metabolite counts, including 47 upregulated and 44 downregulated metabolite counts. This indicates that the stress of both warming and acidification moderately increased the upregulated and downregulated metabolite counts, which can seriously affect the transformation of LOM by bacteria. The metabolite counts of LOM treatments were also affected when only warming or acidification alone was applied. Of interest, the number of downregulated metabolite counts was higher than the number of upregulated in the case of acidification. The Venn diagram presents further information on the proportion of the overlapping metabolic counts between different groups (Figures 5C and 5D).
Figure 5.
Comparison of differential metabolites among the different treatments
Metabolic counts (A and B) and Venn diagram (C and D) between different treatment groups. LL, 26°C and 400 ppm; LH, 26°C and 1000 ppm; HL, 30°C and 400 ppm; HH, 30°C and 1000 ppm.
Discussion
Diatoms are the algae that are most affected by changes in ocean climate.30 Under stressed conditions such as warming and acidification, phytoplankton releases large amounts of OM (e.g., DOM and LOM). Warming and acidification have been demonstrated to favor bacterial growth, which might be owing to two factors: (1) The favorable effect of temperature on bacteria, with more bacterial cells at higher temperatures,31 and (2) the availability of more enriched DOM for bacteria. Put simply, phytoplankton produces labile DOC, which heterotrophic bacteria can convert to recalcitrant DOC.32 Acidification affects the respiration of microorganisms and thus the metabolism and energy cost of bacteria.33 It has been shown that part of Roseobacter tend to exhibit a stronger competitive advantage for survival in carbon-limited marine environments and influence changes in the storage and ecodynamics of organic carbon sources in the marine environment.29,34 Most previous studies have investigated the dynamics of DOM production by phytoplankton and bacteria, as well as performing experiments in which many different carbon sources are added.5,35,36,37 The transformation of algal-derived OM by single heterotrophic strain has received little attention, especially, the bacteria were isolated from the phycosphere. As is well-known, the relationship between microalgae and bacteria are varied and complex. We speculate some specific bacterial species in the microbial community play a dominant role in the transformation of algal-derived OM and whether temperature and acidification have a positive or negative effect on the transformation of OM by heterotrophic bacteria. The present study uses diatom-derived DOM and LOM as the sole carbon sources to develop a chemical molecular and metabolic perspective to investigate the transformation of OM by culturable marine bacteria under warming and acidification conditions. It provides a new perspective for observing the transformation process of diatom-derived DOM and LOM by single heterotrophic strain.
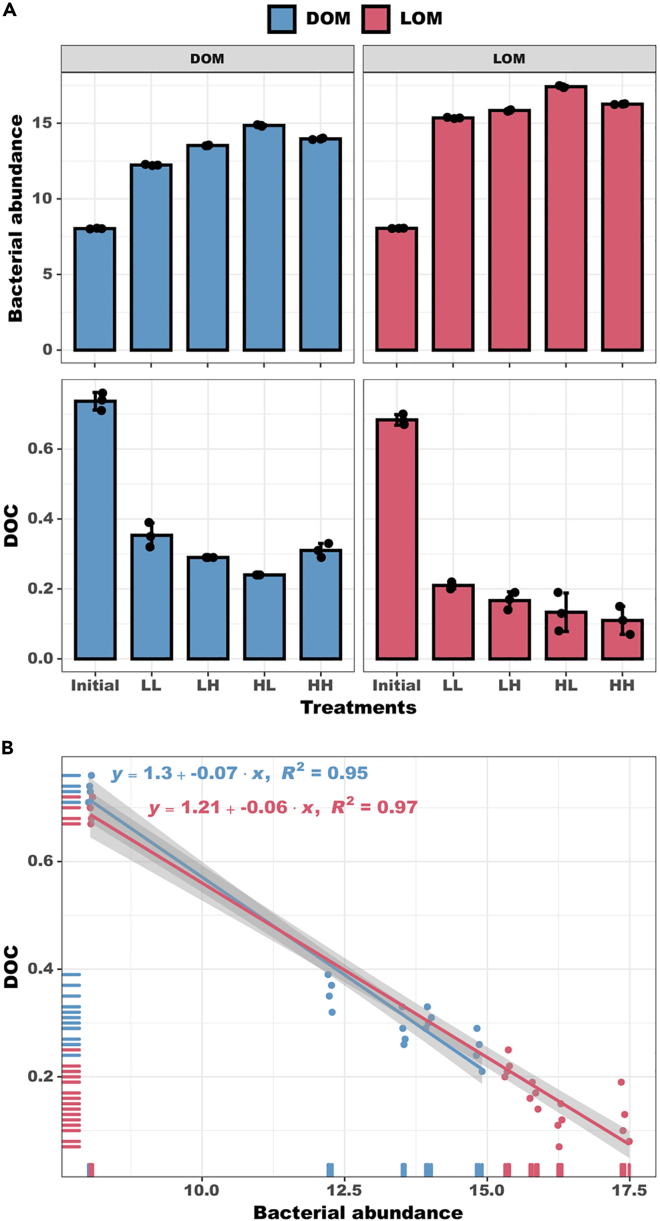
Owing to the addition of bacteria, a considerable portion of the bioavailable fraction of OM was reduced by bacterial respiration and absorption. In the present study, the significant negative correlation (p< 0.05) between bacterial abundance and different carbon pools (i.e., DOM and LOM) suggested a strong link between bacterial growth and activity (Figure 6), with bacteria actively transforming organic compounds. The DOC concentration showed a distinct decrease (net decreases of 0.28–0.65 mg L−1), suggesting the OM components were consumed (i.e., consumption was higher than production). The OM can be used as a substrate for the remineralization of carbon by heterotrophic microorganisms. Microorganisms use enzymes to catalyze OM into smaller compounds that can be transported to the environment across bacterial cell membranes.38 Organic carbon is then incorporated into the biomass or excreted as DOC in the form of metabolic products.38
Figure 6.
Comparison of bacterial abundance and DOC concentration among the different treatments
Bacterial abundance (×105cells mL−1) and DOC concentration (mg L−1) (A) in the different treatment groups. Correlation analysis between bacterial abundance and DOC concentration (B). Error bars represent the standard error for four replicates. LL, 26°C and 400 ppm; LH, 26°C and 1000 ppm; HL, 30°C and 400 ppm; HH, 30°C and 1000 ppm.
Transformation of organic matter from an FT-ICR MS perspective
The detailed DOM and LOM characterization analyzed by FT-ICR MS was displayed in van Krevelen diagrams. Owing to the effects of warming and acidification, we found an increased the number of molecular formulae for all the CHO, CHNO, CHOS, and CHNOS formulae classes. This suggested that warming and acidification lead to an increase in the chemical complexity of the final bacterial transformation of DOM and LOM. After 30 days’ incubation, the number of N-rich and S-rich DOM-produced molecules (CHNO, CHOS, and CHNOS) were reduced in the LL and HH groups, suggesting that the simultaneous stress of warming and acidification affects the conversion of N and S elements by Roseobacter sp. SD-R1. For LOM, however, it is intuitive to note a significant increase in the number of molecules of CHOS and CHNOS, but it cannot be excluded that the number of molecules of both CHO and CHNO also increases. In particular, elevated temperature increases the number and complexity of molecules more than the effects of acidification. The overall molecular formula patterns in the DOM and LOM treatments were similar in both groups after 30 days, but the numbers of identified CHNO and CHOS formulae in the LOM treatments were still higher compared with the LL group. Hence, the transformation of LOM by Roseobacter sp. SD-R1 was more complete than that of DOM, and the diversity and complexity of OM chemical composition was increased.
The VK plot shows similarities in the chemical characteristics of DOM and LOM at the end of the incubation period. Despite DOM and LOM being different substrates, Roseobacter sp. are still able to convert them into similar OM molecules. It appears that bacteria are still capable of further transforming algal-derived OM under elevated temperature and acidification conditions. Long-term experiments conducted by adding simple substrates (e.g., glucose, glutamate, or a mixture of oligosaccharides and oligopeptides) have shown that OM after culture experiments has similar characteristics to OM in the real environment.39 This may mean that most OM in natural aquatic systems is relatively stable and contains components that are not readily available to bacteria. From another point of view, the increase in bacterial abundance indicates that the OM component is used by bacteria to transform substances required for their own growth and further transformed into more recalcitrant OM for release into the aquatic environment. However, such a process occurs over a relatively short timescale. The MCP concept emphasizes the importance of microbial activity in the conversion of unstable OM to recalcitrant OM.40 However, our study confirms that warming and acidification accelerate this process, but that more studies are needed to confirm this idea.
CHO compounds probably include carboxyl and/or hydroxyl functional groups in the natural environment.41,42 However, in this study, the number of CHO compounds in DOM samples (initial, 2,072) was obviously higher than that in LOM samples (initial, 1,238). CHNO compounds showed a similar shift after elevated temperature and acidification conditions. Compared with the LL group, the number of CHNO compounds in the HL and HH groups was increased from 1,995 to 2,234 and 2,009, respectively, after elevated temperature. However, the number of molecules in LH decreased at the end of the 30-day experiment compared to the initial period, with only 1,942 molecules. Furthermore, the numbers of CHO compounds in three treatment groups were increased.
The above phenomena suggest that warming and acidification have significant effects on CHNO compounds in both DOM and LOM groups, probably caused by dimerization and polymerization reactions. CHOS compounds accounted for a small proportion of OM in the initial phase, especially in LOM (only 3.5%). The number of CHOS molecular formulae increased dramatically from 77 to 561–787 after elevated temperature and acidification. In contrast, for DOM samples in which CHOS molecules increased only when warming (HL) or acidification (LH) was increased alone, CHOS decreased instead under the combined effect of warming and acidification. The reasons for these changes may be that the hydrolysis of sulfate groups may be an important degradation pathway in acidic medium for DOM samples. CHNOS, as a whole, changed in a similar way to CHOS compounds. The general increase in CHNOS was because compounds containing more heteroatoms and having greater potential biomolecular diversity are more stable in elevated temperature and acidification conditions.
Although FT-ICR MS differentiates the chemical compositional diversity of DOM, it cannot predict the chemical structure of each formula (Figure S5). The limitations of the current analysis of DOM include extraction biases and machine reproducibility.16,43 Although all samples were analyzed in broadband mode, most of the peaks were generally distributed over a narrow mass range between m/z 300 and 450 (approximately 43.48%–47.37%) in the spectra. These ranges displayed a similar pattern to those in previous OM studies.44,45 There were 3845–5250 and 2198–5950 molecular formulae identified in the DOM and LOM treatments, respectively. AImod index (modified aromaticity index) and DBE/C have been widely used to estimate the aromaticity of OM in aquatic environments.46 The AImod index in the LH, HL, and HH treatments was slightly higher than that for LL treatment and the initial material. This result suggests that degradation and transformation can occur in low aromatic labile OM by the addition of heterotrophic bacteria, thereby resulting in the increased aromaticity of DOM and LOM. Moreover, elevated temperature and acidification conditions will facilitate and accelerate this process.
Recent studies have shown that DOM in different water masses has different properties and contains thousands of molecular formulas.47 In addition, Zheng et al.48 using two sorbents, solid-phase extracted (SPE) DOM from coastal, epipelagic, and deep-sea water and incubated it for six months. It is emphasized that intrinsically RDOM (especially CRAMs) likely has a relatively high contribution to SPE-DOM in the ocean. In the present experiment, CRAMs were predominant in OM; initially, 76.39% and 55.91% were detected in DOM and LOM treatments, respectively. Of interest, the relative number of CRAMs gradually decreased in DOM with the effects of warming and acidification, but it remained stable in LOM. This phenomenon suggests that the elevated temperature and acidification diminish the efficiency of bacterial conversion of CRAMs in DOM molecules, while not affecting the conversion of CRAMs in LOM molecules. In addition, higher DBE, DBE/C, and DBE/O suggest a higher degree of unsaturation (by considering the number of π bonds and rings), a higher density of C=C double bonds, and a lower number of C=O bonds, respectively.49 Overall, there was no significant change in values between each treatment group, and it is notable that in the LOM group, after 30 days of testing, the values were generally higher compared with the initial period, indicating an increase in the degree of unsaturation and the density of C=C double bonds and a decrease in the number of C=O bonds. The molecular lability boundary (MLB) showed completely opposite trends in the DOM and LOM groups, indicating that the proportion of unstable organic compounds increased and decreased in the DOM and LOM groups, respectively. Notably, the compounds in the DOM samples had higher O/C and lower H/C ratios than the LOM samples. Recent research has shown that substances with high O/C and low H/C ratios are likely to be highly oxygenated compounds.50 In general, more aromatic compounds with high O/C and low H/C are present in the aquatic environment in dissolved forms. The previous experiments showed that marine bacteria can convert glucose, glutamic acid, oligosaccharides, and algae extracts into highly diverse recalcitrant components.51 These exometabolites directly released by bacteria on the utilization of labile substrates ought to have intrinsic properties, rendering them resistant to decomposition and therefore allowing them to remain recalcitrant for a long time.52 The microbial carbon pump concept suggests that the successive processing of labile DOM leads to the formation of recalcitrant DOM.8,37,53
In the marine environment, the sunlit surface waters of the oceans are highly productive, and most of the carbon fixed in photosynthesis is remineralized by bacteria in the upper ocean.35 A tiny portion of the organic carbon used by bacteria escapes quick remineralization and manifests as a slowly cycling store of highly complex liquid molecules.35 The exometabolites that are directly produced by bacteria during the usage of labile substrates may have intrinsic features that make them resistant to degradation because the substrates in our investigation were solely derived from algal sources.39 On a far shorter period than ocean mixing, bacteria produce chemically complex and slowly cycling metabolites that accumulate in marine OM.54 Therefore, it is anticipated that changes in primary production will closely reflect changes in the production of these persistent molecules, and that the efficiency of carbon sequestration by marine bacteria in the microbial carbon pump may be on a par with the flux of particles into deep-sea sediments.8,37,53 Bacteria are the key drivers of these processes, and it appears that they also play a significant role in carbon sequestration by producing a variety of complex organic compounds that may endure for thousands of years.55
Microbial metabolic perturbations in different OM treatments
In the current experiments, bacterial growth kinetics differed between substrates (DOM and LOM) and differences in the structural and compositional characteristics of the final OM were observed. The reasons for these differences may reflect bacterial cell differentiation and metabolite exchange promoted by the different substrates. Although the overall metabolic pathways were broadly similar between treatment groups, changes in the metabolite expression of certain metabolic pathways were still affected. For DOM treatment groups (Figure S6), we found that acidification had a more significant effect on tryptophan metabolism, leading to the upregulation of 2-aminophenol, indole, picolinic acid, L-formylkynurenine (Figure S7, Table S1). The effect of warming on arginine biosynthesis was more pronounced, leading to the upregulation of L-glutamic acid and oxoglutaric acid and the downregulation of citrulline, L-arginine, and ornithine (Figure S8, Table S2). The simultaneous warming and acidification had more significant effects on D-arginine and D-ornithine metabolism, in which 1-pyrroline-2-carboxylic acid was upregulated and L-arginine and ornithine were downregulated (Figure S9, Table S3). Amino acids are the structural units of the proteins and polypeptides, and also serve as precursors for the synthesis of various metabolites with multiple functions in microbial processes.56 The types of these amino acids are up/downregulated, implying an acceleration of the amino acid synthesis and/or degradation of proteins.
For LOM treatments (Figure S10), the effect of acidification on purine metabolism was more pronounced, leading to an upregulation of cyclic GMP and a downregulation of guanine and 2′-deoxyguanosine (Figure S11, Table S4). Such nucleobase accumulation after acidification treatments could be related to an acceleration of their synthesis, their respective nucleoside/nucleotide degradation and salvage for reincorporation into nucleotides. Warming had a significant effect on beta-alanine metabolism, resulting in the upregulation of L-histidine, beta-alanyl-L-arginine, L-arginine, and beta-alanyl-L-lysine, and the downregulation of spermidine and N-acetyl-beta-alanine (Figure S12, Table S5). Pyrimidine and purine nucleotides are the structural units of the nucleic acids DNA and RNA. Therefore, the present results suggest an acceleration of DNA and RNA synthesis and turnover. This is in good agreement with the amino acid upregulation, as amino acids serve as precursors for a wide variety of metabolites including purine and pyrimidine nucleotides. The combined involvement of warming and acidification resulted in the upregulation of L-lysine, L-leucine, L-histidine, and deoxyuridine, and the downregulation of D-xylose, 2′-deoxyguanosine, D-fructose, sorbitol, and deoxycytidine (Figure S13, Table S6). We found D-xylose and D-fructose in ABC transports, belonging to sugar metabolism, indicating that warming and acidification reduced the bacterial transformation of sugar substances. In brief, the alteration of amino acid metabolism/biosynthesis, purine metabolism, and sugar metabolism suggests the activation of catabolic processes to accelerate/reduce the restoration of energy balance in microbial cells under warming and acidification.
Conclusions
This study reports a general exploration of the results of bacterial transformation of DOM and LOM from a chemical molecular and metabolic perspective under warming and acidification conditions. Bacterial growth kinetics were affected and differences in the structural and compositional characteristics of the OM compounds were observed. There was a significant negative correlation between bacterial abundance and the concentration of different carbon sources, while bacterial transformation of LOM was higher than that of DOM. Moreover, warming and acidification accelerated the efficiency of bacterial transformation of OM molecules. Bacterial metabolites expressed different patterns because of different substrates promoting cellular differentiation and metabolite exchange of microorganisms. In general, the important role of bacteria in the microbial carbon pump was further verified, as well as the preferences and transformation characteristics of the bacteria for various molecular compounds.
Limitations of the study
Because the culture experiments of single-strain are difficult to simulate in situ environments, we believe the reliability of the experimental results. Nevertheless, this study only examined two temperatures and pCO2 concentrations, which has certain restrictions, particularly the limited number of temperature and acidification treatments that cannot accurately reflect the effect of both conditions on the OM transformation by bacteria. Consequently, we have grounds to assume that different temperature/acidification spans could yield different results. This study also provides a new perspective for further study of bacterial transformation of OM. In the future, we will continue to explore how algae-associated microorganisms manipulate the global carbon cycle and respond to changes in the marine environment.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Biological samples | ||
| Skeletonema dohrnii | Natural population from the Xiamen Harbor in offshore China | None |
| Bacterial strains | ||
| Roseobacter sp. SD-R1 | NCBI | PRJNA784381 |
| Software and algorithms | ||
| Fourier-transform ion cyclotron resonance mass spectrometry | Bruker | 7.0 T Solarix 2XR |
| Thermo Vanquish | Thermo | Vanquish |
| Thermo Q Exactive HF-X | Thermo | Q Exactive HF-X |
| Microscope | Olympus | BX51 |
| Accuri C6 flow cytometer | Erembodegem | BD Biosciences |
| Total organic carbon analyzer | Shimadzu | TOC-L |
| Origin 2022b | OriginLab | www.originlab.com |
| R (v. 4.1.2) | R Foundation for Statistical Computing | |
Resource availability
Lead contact
Further information and requests for any resources should be directed to and will be evaluated by the lead contact, Jun Sun (phytoplankton@163.com) to determine whether they can be made available on a case-by-case basis without identifiers.
Materials availability
This study did not generate new unique reagents.
Experimental model and subject details
This work has not involved the use of human subjects or samples, nor has it use experimental models that require reporting of experimental model and subject details.
Method details
Diatom isolation, culture, and growth
Skeletonema dohrnii, a diatom species that is widely distributed in offshore China, was isolated from the Xiamen Harbour.57 All cultures were maintained in artificial seawater (ASW) medium in transparent conical flasks and incubated in growth chambers at 25°C, with 100 μmol photons m−2 s−1, and a 14:10-h light:dark cycle. All cultures were subcultured at least three times in ASW medium to ensure that S. dohrnii was adapted to the current environment. Algal growth was monitored using a hemocytometer, and enumerated under a microscope (BX51; Olympus, Japan).
Isolation and identification of bacterial strains
Bacterial strains were isolated from the S. dohrnii by the gradient dilution method and then cultured in transparent conical flasks (including 2216E medium, 200 mL) in a shaking incubator (26°C, 160 rpm). The TIANamp Bacteria DNA Kit (Tiangen-Biotech, Beijing, China) was used to extract the genomic DNA of epiphytic bacteria. Sanger sequencing of the 16S rRNA gene is used for the identification of bacteria. Using bacterial genomes and the closest related sequences from GenBank, a phylogenetic tree (Neighbor-Joining tree) was created, and genetic distances were determined (Figure S1).
Flow cytometric analysis of Roseobacter sp. SD-R1
Roseobacter sp. SD-R1 was cultivated in 250-mL transparent conical flasks that had been pre-combusted (450°C, 5 h). Bacterial abundance was measured using an Accuri C6 flow cytometer (BD Biosciences, Erembodegem, Belgium) as previously described.58 Samples were stained with 0.01% SYBR Green I for 30minat 37°C in the dark.59 Each sample was injected with 1 μm fluorescent beads (Polyscience, Warrington, PA, USA) as an internal standard, and measured at a flow rate of 0.25μL s−1 for 1 min.
Determination of dissolved organic carbon
Dissolved organic carbon (DOC) concentrations were determined using a TOC-L total organic carbon analyzer (Shimadzu, Kyoto, Japan) in accordance with standard practices. To prevent any carbon contamination, all glass products were acid-washed, rinsed with ultrapure water, and pre-combusted (450°C for 5 h). All DOC samples were gravimetrically filtered using pre-combusted (450°C for 5 h) GF/F glass fiber filters (0.7 μm pore size, 47 mm diameter, Whatman).
Solid-phase extraction and FT-ICR MS analysis
Pre-treatment of OM samples was performed, as described by Dittmar et al.,43using SPE with PPL cartridges filled with a styrene-divinylbenzene polymer (Bond Elut PPL, 200 mg; Agilent, USA). FT-ICR MS was used to determine the molecular composition of DOM and LOM components in the exometabolome. The analysis of all SPE samples were performed using a 7.0 T Solarix 2XR FT-ICR MS (Bruker, USA) equipped with an electrospray ionization source; the instrument was located in the School of Earth System Science, Tianjin University, Tianjin, China. We referenced a standard method for the specific parameter settings and operational procedure of the machine.60 In this study, the formula assignments were based on the following numbers of atoms: C5–37, H3–55, O1–14, N0–2, and S0-1 (the mass accuracy was <0.2 ppm and the signal-to-noise ratio was ≥6). In addition, the formulas present in the combustion samples were repeated for further analysis. The molecular composition of the DOM was visualized by van Krevelen (VK) diagrams. Various molecular formulas were plotted based on their H/C and O/C atomic ratios to allow the unique H/C and O/C values for each DOM formula to be shown as previously described.43,60 The OM molecular function groups can be classified by modified aromaticity index (AImod), calculated by 1 + C−0.5O−S−0.5N−0.5P−0.5H)/(C−0.5O−S−N−P).61 To calculate the double bond equivalent (DBE) associated with elemental assignment, it should be executed for each component according to the formula (2C−H+N+P+2)/2.16 Here, various molecular groups were operationally assigned as16: polycyclic aromatics compounds (AImod> 0.67), which include condensed combustion-derived dissolved black carbon if C > 15, (2) aromatic compounds (0.5< AImod ≤ 0.67), (3) highly unsaturated and phenolic compounds (AImod ≤ 0.5 and H/C < 1.5), (4) aliphatic compounds (AImod ≤ 0.5 and H/C ≥ 1.5), and (5) carboxylic-rich alicyclic compounds (CRAM) (formulas with DBE/C = 0.3–0.68, DBE/H = 0.2–0.95, and DBE/O = 0.77–1.75).35 Here, CRAMs represent a class of potentially recalcitrant components identified in OM.62
Untargeted LC–MS-based metabolomics analysis
Ultra-high-performance liquid chromatography in combination with mass spectrometry (UHPLC-MS) was performed on a Vanquish UHPLC System (Thermo Fisher Scientific, USA) connected to a Q Exactive HF-X MS (Thermo Fisher Scientific, USA). The liquid chromatography analysis was performed on a Vanquish UHPLC System (Thermo Fisher Scientific, USA). Chromatography was performed on an ACQUITY UPLC HSS T3 (150 × 2.1 mm, 1.8 μm) (Waters, Milford, MA, USA). The column was maintained at 40°C. The flow rate and injection volume were set at 0.25 mL min−1 and 2 μL, respectively. For LC-ESI (+)-MS analysis, the mobile phases consisted of 0.1% formic acid in acetonitrile (v/v) and 0.1% formic acid in water (v/v). Separation was conducted under the following gradient: 0–1 min, 2% C; 1–9 min, 2%–50% C; 9–12 min, 50%–98% C; 12–13.5 min, 98% C; 13.5–14 min, 98%–2% C; 14–20 min, 2% C. For LC-ESI (−)-MS analysis, the analytes were eluted with acetonitrile and ammonium formate (5 mM). Separation was conducted under the following gradient: 0–1 min, 2% A; 1–9 min, 2%–50% A; 9–12 min, 50%–98% A; 12–13.5 min, 98% A; 13.5–14 min, 98%–2% A; 14–17 min, 2% A.63 Mass spectrometric detection of metabolites was performed on Q Exactive HF-X (Thermo Fisher Scientific, USA) with an ESI ion source. Simultaneous MS1 and MS/MS (Full MS-ddMS2 mode, data-dependent MS/MS) acquisition was used. The parameters were as follows: sheath gas pressure, 30 arb; aux gas flow, 10 arb; spray voltage, 3.50 kV and −2.50 kV for ESI(+) and ESI(−), respectively; capillary temperature, 325°C; MS1 range, m/z 81–1000; MS1 resolving power, 60000 FWHM; number of data-dependent scans per cycle, 8; MS/MS resolving power, 15000 FWHM; normalized collision energy, 30%; dynamic exclusion time, automatic.64
Experimental setup
In this study, all incubation experiments were performed in transparent conical flasks that were precleaned with an acid washed and with Milli-Q water. S. dohrnii cells were cultivated in artificial seawater (ASW) medium, and after reaching the degradation growth phase (algal concentration approximately (7.16 ± 0.12) × 106 cells mL−1), the microalgae liquid was filtered through a 0.2-μm polycarbonate membrane (Millipore, USA) to remove particles. Then, the filtrate, which was regarded as the DOM fraction, was placed in a pre-combusted (450°C, 5 h) 1-L conical flask. To obtain the LOM fraction, the microalgae liquid was centrifuged at 5000 rpm for 5 min. The supernatant was removed, resuspended with 0.05% NaCl solution, and heated at 60°C for 30 min. The extracted solutions were then centrifuged at 7000 rpm for 20 min and the supernatant was considered to be the LOM fraction. The LOM samples were filtered through a 0.2-μm polycarbonate membrane (Millipore, United States) to prevent the introduction of particulate matter.
To assess the effects of warming and acidification on the transformation of diatom-derived OM by Roseobacter sp. SD-R1, two different temperatures (26°C and 30°C, i.e., low and high temperature) and pCO2 conditions (400 ppm and 1000 ppm, i.e., low and high pCO2) were used in this work. The following four treatments were tested: (i) 26°C + 400 ppm (LL), (ii) 26°C + 1000 ppm (LH), (iii) 30°C + 400 ppm (HL), and (iv) 30°C + 1000 ppm (HH). Under the above conditions, Roseobacter sp. SD-R1 Was added into 1-L conical flasks with different OM fractions (including the DOM and LOM fractions), which were gently bubbled with CO2, and the gas flow rate (0.5 L min−1) was controlled using a Bronkhorst mass flow controller. The initial abundance of Roseobacter sp. in each treatment group was approximately (8.03 ± 0.07) × 105 cells mL−1. The whole experiment lasted 30 days, with samples collected on the initial (day 0) and last (day 30) day. Four independent replicates were analyzed per experimental group (i.e., 32 conical flasks for 16 DOM treatment groups and 16 LOM treatment groups).
Quantification and statistical analysis
All experiments were performed under dark conditions and the mean ± standard deviation was determined. All statistical analyses were performed in RStudio for R version 4.2.1.
Acknowledgments
This research was financially supported by the National Key Research and Development Project of China (2019YFC1407800), the National Natural Science Foundation of China (41876134), the Changjiang Scholar Program of Chinese Ministry of Education (T2014253), and supported by State Key Laboratory of Biogeology and Environmental Geology, China University of Geosciences (No. GKZ21Y645 and GKZ22Y656) to Jun Sun.
Author contributions
Y.L.: Conceptualization, Data curation, Writing – original draft, Writing – review and editing. C.M.: Sample Testing. J.S.: Conceptualization, funding acquisition, Writing – review and editing.
Declaration of interests
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Inclusion and diversity
We support inclusive, diverse, and equitable conduct of research.
Published: May 4, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2023.106812.
Supplemental information
Data and code availability
All data will be shared upon request to the lead contact. No standardized datatype data were generated in this study. This study did not generate new code. Any additional analysis information for this work is available by request to the lead contact.
References
- 1.Tréguer P., Bowler C., Moriceau B., Dutkiewicz S., Gehlen M., Aumont O., Bittner L., Dugdale R., Finkel Z., Iudicone D., et al. Influence of diatom diversity on the ocean biological carbon pump. Nat. Geosci. 2018;11:27–37. doi: 10.1038/s41561-017-0028-x. [DOI] [Google Scholar]
- 2.Bell W., Mitchell R. Chemotactic and growth responses of marine bacteria to algal extracellular products. Biol. Bull. 1972;143:265–277. doi: 10.2307/1540052. [DOI] [Google Scholar]
- 3.Seymour J.R., Amin S.A., Raina J.B., Stocker R. Zooming in on the phycosphere: the ecological interface for phytoplankton–bacteria relationships. Nat. Microbiol. 2017;2:17065–17112. doi: 10.1038/nmicrobiol.2017.65. [DOI] [PubMed] [Google Scholar]
- 4.Amin S.A., Parker M.S., Armbrust E.V. Interactions between diatoms and bacteria. Microbiol. Mol. Biol. Rev. 2012;76:667–684. doi: 10.1128/mmbr.00007-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu Y., Kan J., He C., Shi Q., Liu Y.X., Fan Z.C., Sun J. Epiphytic bacteria are essential for the production and transformation of algae-derived carboxyl-rich alicyclic Molecule (CRAM)-like DOM. Microbiol. Spectr. 2021;9 doi: 10.1128/spectrum.01531-21. e0153121–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McIntyre A.M., Guéguen C. Binding interactions of algal-derived dissolved organic matter with metal ions. Chemosphere. 2013;90:620–626. doi: 10.1016/j.chemosphere.2012.08.057. [DOI] [PubMed] [Google Scholar]
- 7.Pereira S., Zille A., Micheletti E., Moradas-Ferreira P., De Philippis R., Tamagnini P. Complexity of cyanobacterial exopolysaccharides: composition, structures, inducing factors and putative genes involved in their biosynthesis and assembly. FEMS Microbiol. Rev. 2009;33:917–941. doi: 10.1111/j.1574-6976.2009.00183.x. [DOI] [PubMed] [Google Scholar]
- 8.Jiao N., Herndl G.J., Hansell D.A., Benner R., Kattner G., Wilhelm S.W., Kirchman D.L., Weinbauer M.G., Luo T., Chen F., et al. Microbial production of recalcitrant dissolved organic matter: long-term carbon storage in the global ocean. Nat. Rev. Microbiol. 2010;8:593–599. doi: 10.1038/nrmicro2386. [DOI] [PubMed] [Google Scholar]
- 9.Azam F., Malfatti F. Microbial structuring of marine ecosystems. Nat. Rev. Microbiol. 2007;5:782–791. doi: 10.1021/ac200464q. [DOI] [PubMed] [Google Scholar]
- 10.Diner R.E., Schwenck S.M., McCrow J.P., Zheng H., Allen A.E. Genetic manipulation of competition for nitrate between heterotrophic bacteria and diatoms. Front. Microbiol. 2016;7:880. doi: 10.3389/fmicb.2016.00880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Petrou K., Baker K.G., Nielsen D.A., Hancock A.M., Schulz K.G., Davidson A.T. Acidification diminishes diatom silica production in the Southern Ocean. Nat. Clim. Chang. 2019;9:781–786. doi: 10.1038/s41558-019-0557-y. [DOI] [Google Scholar]
- 12.Blain S., Rembauville M., Crispi O., Obernosterer I. Synchronized autonomous sampling reveals coupled pulses of biomass and export of morphologically different diatoms in the Southern Ocean. Limnol. Oceanogr. 2021;66:753–764. doi: 10.1002/lno.11638. [DOI] [Google Scholar]
- 13.Falkowski P.G., Fenchel T., Delong E.F. The microbial engines that drive Earth's biogeochemical cycles. Science. 2008;320:1034–1039. doi: 10.1126/science.1153213. [DOI] [PubMed] [Google Scholar]
- 14.Smriga S., Fernandez V.I., Mitchell J.G., Stocker R. Chemotaxis toward phytoplankton drives organic matter partitioning among marine bacteria. Proc. Natl. Acad. Sci. USA. 2016;113:1576–1581. doi: 10.1073/pnas.1512307113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Benner R., Kaiser K. Abundance of amino sugars and peptidoglycan in marine particulate and dissolved organic matter. Limnol. Oceanogr. 2003;48:118–128. doi: 10.4319/lo.2003.48.1.0118. [DOI] [Google Scholar]
- 16.Hertkorn N., Benner R., Frommberger M., Schmitt-Kopplin P., Witt M., Kaiser K., Kettrup A., Hedges J.I. Characterization of a major refractory component of marine dissolved organic matter. Geochim. Cosmochim. Acta. 2006;70:2990–3010. doi: 10.1016/j.gca.2006.03.021. [DOI] [Google Scholar]
- 17.Legendre L., Rivkin R.B., Weinbauer M.G., Guidi L., Uitz J. The microbial carbon pump concept: potential biogeochemical significance in the globally changing ocean. Prog. Oceanogr. 2015;134:432–450. doi: 10.1016/j.pocean.2015.01.008. [DOI] [Google Scholar]
- 18.Cao Z., Yang W., Zhao Y., Guo X., Yin Z., Du C., Zhao H., Dai M. Diagnosis of CO2 dynamics and fluxes in global coastal oceans. Natl. Sci. Rev. 2020;7:786–797. doi: 10.1093/nsr/nwz105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.DeVries T., Holzer M., Primeau F. Recent increase in oceanic carbon uptake driven by weaker upper-ocean overturning. Nature. 2017;542:215–218. doi: 10.1038/nature21068. [DOI] [PubMed] [Google Scholar]
- 20.Riebesell U., Gattuso J.P. Lessons learned from ocean acidification research. Nat. Clim. Chang. 2015;5:12–14. doi: 10.1038/nclimate2456. [DOI] [Google Scholar]
- 21.O'Brien P.A., Morrow K.M., Willis B.L., Bourne D.G. Implications of ocean acidification for marine microorganisms from the free-living to the host-associated. Front. Mar. Sci. 2016;3:47. doi: 10.3389/fmars.2016.00047. [DOI] [Google Scholar]
- 22.Thangaraj S., Sun J. Transcriptomic reprogramming of the oceanic diatom Skeletonema dohrnii under warming ocean and acidification. Environ. Microbiol. 2021;23:980–995. doi: 10.1111/1462-2920.15248. [DOI] [PubMed] [Google Scholar]
- 23.Grossart H.P., Allgaier M., Passow U., Riebesell U. Testing the effect of CO2 concentration on the dynamics of marine heterotrophic bacterioplankton. Limnol. Oceanogr. 2006;51:1–11. doi: 10.4319/lo.2006.51.1.0001. [DOI] [Google Scholar]
- 24.Eilers H., Pernthaler J., Peplies J., Glöckner F.O., Gerdts G., Amann R. Isolation of novel pelagic bacteria from the German Bight and their seasonal contributions to surface picoplankton. Appl. Environ. Microbiol. 2001;67:5134–5142. doi: 10.1128/aem.67.11.5134-5142.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Selje N., Simon M., Brinkhoff T. A newly discovered Roseobacter cluster in temperate and polar oceans. Nature. 2004;427:445–448. doi: 10.1038/nature02272. [DOI] [PubMed] [Google Scholar]
- 26.Moran M.A., Belas R., Schell M.A., González J.M., Sun F., Sun S., Binder B.J., Edmonds J., Ye W., Orcutt B., et al. Ecological genomics of marine Roseobacters. Appl. Environ. Microbiol. 2007;73:4559–4569. doi: 10.1128/AEM.02580-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Newton R.J., Griffin L.E., Bowles K.M., Meile C., Gifford S., Givens C.E., Howard E.C., King E., Oakley C.A., Reisch C.R., et al. Genome characteristics of a generalist marine bacterial lineage. ISME J. 2010;4:784–798. doi: 10.1038/ismej.2009.150. [DOI] [PubMed] [Google Scholar]
- 28.Luo H., Moran M.A. How do divergent ecological strategies emerge among marine bacterioplankton lineages? Trends Microbiol. 2015;23:577–584. doi: 10.1016/j.tim.2015.05.004. [DOI] [PubMed] [Google Scholar]
- 29.Buchan A., González J.M., Moran M.A. Overview of the marine Roseobacter lineage. Appl. Environ. Microbiol. 2005;71:5665–5677. doi: 10.1128/aem.71.10.5665-5677.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hinder S.L., Hays G.C., Edwards M., Roberts E.C., Walne A.W., Gravenor M.B. Changes in marine dinoflagellate and diatom abundance under climate change. Nat. Clim. Chang. 2012;2:271–275. doi: 10.1038/nclimate1388. [DOI] [Google Scholar]
- 31.Apple J.K., Del Giorgio P.A., Kemp W.M. Temperature regulation of bacterial production, respiration, and growth efficiency in a temperate salt-marsh estuary. Aquat. Microb. Ecol. 2006;43:243–254. doi: 10.3354/ame043243. [DOI] [Google Scholar]
- 32.Stoderegger K., Herndl G.J. Production and release of bacterial capsular material and its subsequent utilization by marine bacterioplankton. Limnol. Oceanogr. 1998;43:877–884. doi: 10.4319/lo.1998.43.5.0877. [DOI] [Google Scholar]
- 33.Siu N., Apple J.K., Moyer C.L. The effects of ocean acidity and elevated temperature on bacterioplankton community structure and metabolism. Open J. Ecol. 2014;04:434–455. doi: 10.4236/oje.2014.48038. [DOI] [Google Scholar]
- 34.Oz A., Sabehi G., Koblízek M., Massana R., Béjà O. Roseobacter-like bacteria in Red and Mediterranean Sea aerobic anoxygenic photosynthetic populations. Appl. Environ. Microbiol. 2005;71:344–353. doi: 10.1128/aem.71.1.344-353.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lechtenfeld O.J., Hertkorn N., Shen Y., Witt M., Benner R. Marine sequestration of carbon in bacterial metabolites. Nat. Commun. 2015;6:6711–6718. doi: 10.1038/ncomms7711. [DOI] [PubMed] [Google Scholar]
- 36.Lian J., Zheng X., Zhuo X., Chen Y.L., He C., Zheng Q., Lin T.H., Sun J., Guo W., Shi Q., et al. Microbial transformation of distinct exogenous substrates into analogous composition of recalcitrant dissolved organic matter. Environ. Microbiol. 2021;23:2389–2403. doi: 10.1111/1462-2920.15426. [DOI] [PubMed] [Google Scholar]
- 37.Cai R., Jiao N. Recalcitrant dissolved organic matter and its major production and removal processes in the ocean. Deep-Sea Res., Part A I. 2023;191:103922. doi: 10.1016/j.dsr.2022.103922. [DOI] [Google Scholar]
- 38.Arnosti C. Microbial extracellular enzymes and the marine carbon cycle. Ann. Rev. Mar. Sci. 2011;3:401–425. doi: 10.1146/annurev-marine-120709-142731. [DOI] [PubMed] [Google Scholar]
- 39.Koch B.P., Kattner G., Witt M., Passow U. Molecular insights into the microbial formation of marine dissolved organic matter: recalcitrant or labile? Biogeosciences. 2014;11:4173–4190. doi: 10.5194/bg-11-4173-2014. [DOI] [Google Scholar]
- 40.Jiao N.j., Azam F. Microbial carbon pump and its significance for carbon sequestration in the ocean. Microbial Carbon Pump in the Ocean. 2011;31:43–45. [Google Scholar]
- 41.Cech N.B., Enke C.G. Practical implications of some recent studies in electrospray ionization fundamentals. Mass Spectrom. Rev. 2001;20:362–387. doi: 10.1002/mas.10008. [DOI] [PubMed] [Google Scholar]
- 42.Lin P., Rincon A.G., Kalberer M., Yu J.Z. Elemental composition of HULIS in the Pearl River Delta Region, China: results inferred from positive and negative electrospray high resolution mass spectrometric data. Environ. Sci. Technol. 2012;46:7454–7462. doi: 10.1021/es300285d. [DOI] [PubMed] [Google Scholar]
- 43.Dittmar T., Koch B., Hertkorn N., Kattner G. A simple and efficient method for the solid-phase extraction of dissolved organic matter (SPE-DOM) from seawater. Limnol Oceanogr. Methods. 2008;6:230–235. doi: 10.4319/lom.2008.6.230. [DOI] [Google Scholar]
- 44.Koch B.P., Ludwichowski K.U., Kattner G., Dittmar T., Witt M. Advanced characterization of marine dissolved organic matter by combining reversed-phase liquid chromatography and FT-ICR-MS. Mar. Chem. 2008;111:233–241. doi: 10.1016/j.marchem.2008.05.008. [DOI] [Google Scholar]
- 45.Bae E., Yeo I.J., Jeong B., Shin Y., Shin K.H., Kim S. Study of double bond equivalents and the numbers of carbon and oxygen atom distribution of dissolved organic matter with negative-mode FT-ICR MS. Anal. Chem. 2011;83:4193–4199. doi: 10.1021/ac200464q. [DOI] [PubMed] [Google Scholar]
- 46.Song J., Li M., Jiang B., Wei S., Fan X., Peng P. Molecular characterization of water-soluble humic like substances in smoke particles emitted from combustion of biomass materials and coal using ultrahigh-resolution electrospray ionization Fourier transform ion cyclotron resonance mass spectrometry. Environ. Sci. Technol. 2018;52:2575–2585. doi: 10.1021/acs.est.7b06126. [DOI] [PubMed] [Google Scholar]
- 47.Seidel M., Vemulapalli S.P.B., Mathieu D., Dittmar T. Marine dissolved organic matter shares thousands of molecular formulae yet differs structurally across major water masses. Environ. Sci. Technol. 2022;56:3758–3769. doi: 10.1021/acs.est.1c04566. [DOI] [PubMed] [Google Scholar]
- 48.Zheng X., Cai R., Yao H., Zhuo X., He C., Zheng Q., Shi Q., Jiao N. Experimental insight into the enigmatic persistence of marine refractory dissolved organic matter. Environ. Sci. Technol. 2022;56:17420–17429. doi: 10.1021/acs.est.2c04136. [DOI] [PubMed] [Google Scholar]
- 49.Roth V.N., Dittmar T., Gaupp R., Gleixner G. Latitude and pH driven trends in the molecular composition of DOM across a north south transect along the Yenisei River. Geochim. Cosmochim. Acta. 2013;123:93–105. doi: 10.1016/j.gca.2013.09.002. [DOI] [Google Scholar]
- 50.Han H., Feng Y., Chen J., Xie Q., Chen S., Sheng M., Zhong S., Wei W., Su S., Fu P. Acidification impacts on the molecular composition of dissolved organic matter revealed by FT-ICR MS. Sci. Total Environ. 2022;805:150284. doi: 10.1016/j.scitotenv.2021.150284. [DOI] [PubMed] [Google Scholar]
- 51.Ogawa H., Amagai Y., Koike I., Kaiser K., Benner R. Production of refractory dissolved organic matter by bacteria. Science. 2001;292:917–920. doi: 10.1126/science.1057627. [DOI] [PubMed] [Google Scholar]
- 52.Castillo C.R., Sarmento H., Álvarez-Salgado X.A., Gasol J.M., Marraséa C. Production of chromophoric dissolved organic matter by marine phytoplankton. Limnol. Oceanogr. 2010;55:446–454. doi: 10.4319/lo.2010.55.1.0446. [DOI] [Google Scholar]
- 53.Jiao N., Cai R., Zheng Q., Tang K., Liu J., Jiao F., Wallace D., Chen F., Li C., Amann R., et al. Unveiling the enigma of refractory carbon in the ocean. Natl. Sci. Rev. 2018;5:459–463. doi: 10.1093/nsr/nwy020. [DOI] [Google Scholar]
- 54.Stuiver M., Quay P.D., Ostlund H.G. Abyssal water carbon-14 distribution and the age of the world oceans. Science. 1983;219:849–851. doi: 10.1126/science.219.4586.849. [DOI] [PubMed] [Google Scholar]
- 55.Buesseler K.O., Lamborg C.H., Boyd P.W., Lam P.J., Trull T.W., Bidigare R.R., Bishop J.K.B., Casciotti K.L., Dehairs F., Elskens M., et al. Revisiting carbon flux through the ocean's twilight zone. Science. 2007;316:567–570. doi: 10.1126/science.1137959. [DOI] [PubMed] [Google Scholar]
- 56.Bromke M.A. Amino acid biosynthesis pathways in diatoms. Metabolites. 2013;3:294–311. doi: 10.3390/metabo3020294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gu H., Zhang X., Sun J., Luo Z. Diversity and seasonal occurrence of Skeletonema (Bacillariophyta) species in Xiamen Harbour and surrounding seas, China. Cryptogam. Algol. 2012;33:245–263. doi: 10.7872/crya.v33.iss3.2012.245. [DOI] [Google Scholar]
- 58.Moens F., Weckx S., De Vuyst L. Bifidobacterial inulin-type fructan degradation capacity determines cross-feeding interactions between bifidobacteria and Faecalibacterium prausnitzii. Int. J. Food Microbiol. 2016;231:76–85. doi: 10.1016/j.ijfoodmicro.2016.05.015. [DOI] [PubMed] [Google Scholar]
- 59.Marie D., Partensky F., Jacquet S., Vaulot D. Enumeration and cell cycle analysis of natural populations of marine picoplankton by flow cytometry using the nucleic acid stain SYBR Green I. Appl. Environ. Microbiol. 1997;63:186–193. doi: 10.1128/aem.63.1.186-193.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.He C., Zhang Y., Li Y., Zhuo X., Li Y., Zhang C., Shi Q. In-house standard method for molecular characterization of dissolved organic matter by FT-ICR mass spectrometry. ACS Omega. 2020;5:11730–11736. doi: 10.1021/acsomega.0c01055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Herzsprung P., Hertkorn N., von Tümpling W., Harir M., Friese K., Schmitt-Kopplin P. Understanding molecular formula assignment of Fourier transform ion cyclotron resonance mass spectrometry data of natural organic matter from a chemical point of view. Anal. Bioanal. Chem. 2014;406:7977–7987. doi: 10.1007/s00216-014-8249-y. [DOI] [PubMed] [Google Scholar]
- 62.D'Andrilli J., Cooper W.T., Foreman C.M., Marshall A.G. An ultrahigh-resolution mass spectrometry index to estimate natural organic matter lability. Rapid Commun. Mass Spectrom. 2015;29:2385–2401. doi: 10.1002/rcm.7400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zelena E., Dunn W.B., Broadhurst D., Francis-McIntyre S., Carroll K.M., Begley P., O’Hagan S., Knowles J.D., Halsall A., HUSERMET Consortium. Wilson I.D., Kell D.B. Development of a robust and repeatable UPLC−MS method for the long-term metabolomic study of human serum. Anal. Chem. 2009;81:1357–1364. doi: 10.1021/ac8019366. [DOI] [PubMed] [Google Scholar]
- 64.Want E.J., Masson P., Michopoulos F., Wilson I.D., Theodoridis G., Plumb R.S., Shockcor J., Loftus N., Holmes E., Nicholson J.K. Global metabolic profiling of animal and human tissues via UPLC-MS. Nat. Protoc. 2013;8:17–32. doi: 10.1038/nprot.2012.135. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data will be shared upon request to the lead contact. No standardized datatype data were generated in this study. This study did not generate new code. Any additional analysis information for this work is available by request to the lead contact.