Abstract—
Vaccination is the most efficient way to prevent infectious diseases. mRNA-based vaccines is a new approach to vaccine development, which have several very useful advantages over other types of vaccines. Since mRNA encodes only the target antigen there is no potential risk of infection as in the case with attenuated or inactivated pathogens. The mode of action of mRNA-vaccines implies that their genetic information is expressed only in the cytosol, leaving very little possibility of mRNA integration into the host’s genome. mRNA-vaccines can induce specific cellular and humoral immune responses, but do not induce the antivector immune response. The mRNA-vaccine platform allows for easy target gene replacement without the need to change the production technology, which is important to address the time lag between the epidemic onset and vaccine release. The present review discusses the history of mRNA vaccines, mRNA vaccine production technology, ways to increase mRNA stability, modifications of the cap, poly(A)-tail, coding and noncoding parts of mRNA, target mRNA vaccine purification from byproducts, and delivery methods.
Keywords: RNA, mRNA vaccines, chemically modified nucleotides, untranslated 5'- and 3'-regions, mRNA delivery methods
CONTENTS
INTRODUCTION
HISTORY OF mRNA VACCINES
TYPES OF RNA-BASED VACCINES
WAYS TO INCREASE mRNA STABILITY
Cap Structure
Poly(A) Sequence at the 3'-end of mRNA- the Poly(A)-Tail
Non-Coding Regions in mRNA
mRNA Coding Sequence Optimization
Modified Nucleosides in mRNA
IVT-mRNA PURIFICATION FROM THE dsRNA
IN VIVO mRNA DELIVERY
Lipid Nanoparticles
Polycationic Polymers
Physical Delivery Methods
CONCLUSIONS
REFERENCES
INTRODUCTION
Vaccination is the most efficient way to prevent infectious diseases. One new approach to vaccine development is mRNA-based vaccines. mRNA vaccines have several very useful advantages over other types of vaccines. First, mRNA vaccines are safe. mRNA encodes only the target antigen, therefore there is no potential risk of infection, as it is the case with attenuated and inactivated vaccines, and the immune system load is lower as well. Second, the mode of action of mRNA-vaccines implies that their genetic information is expressed only in the cytosol with no need to enter the nucleus. Hence, the probability of mRNA integration into the host organism genome is extremely low. In addition, mRNA vaccines, like DNA vaccines, are capable of inducing cellular and humoral immune responses. Finally, mRNA is a minimal genetic vector, therefore immunisation does not induce an antivector immune response, and mRNA vaccines can be used repeatedly. Moreover, mRNA is easily eliminated from the body, while mRNA half-life in vivo can be regulated by modifying its structural elements and using different cell delivery methods [1].
Given well-developed production infrastructure, mRNA-based vaccine manufacturing is fast, inexpensive, scalable, and rather routine. The mRNA-vaccine platform allows for easy target gene replacement without changing the production technology. The abovementioned advantages make it possible to address a very important issue in the prevention of a number of viral diseases, namely the issue of the time lag between the epidemic onset and vaccine release.
The review focuses on the technology of mRNA vaccine production, including the history of mRNA vaccines, ways to improve mRNA stability, modifications of the cap, poly(A)-tail, coding and noncoding parts of mRNA, target mRNA vaccine purification from by-products, and delivery methods.
HISTORY OF mRNA VACCINES
The first papers reporting biologically active mRNA production were published in 1984 by Krieg, Melton, Maniatis, and Green. They used T7 RNA polymerase to obtain the biologically active mRNA, the method which is still used today. Among these papers, there is a report by Krieg et al. (1984), who injected the obtained mRNA into the frog egg cells and showed that it worked in exactly the same way as the naturally obtained mRNA [2]. In 1987, Melton demonstrated that mRNA can be used to both activate and inhibit protein synthesis. Both Melton and Krieg considered synthetic mRNAs as first of all a research tool, which could be used to study gene function and activity, i.e., they did not see mRNA as a platform to construct vaccines.
In 1990, Wolff et al. showed that it is possible to express synthetic mRNA in animals [3]. In their work, the authors injected mRNAs encoding chloramphenicol acetyltransferase, luciferase and β-galactosidase reporter genes into mice and demonstrated the presence of the corresponding protein products in the animals. A subsequent study conducted in 1992 demonstrated that the introduction of the vasopressin-coding mRNA into the hypothalamus induced physiological effects in rats [4]. Once it had been demonstrated that synthetic mRNAs are able to provide for protein synthesis in the body, the reports on studying them as preventive and therapeutic agents started to appear. However, the researchers encountered a number of problems including mRNA physical instability, its immunostimulatory properties and difficulties in penetrating cell membranes [5]. For this reason, many scientists focused on DNA because of its high natural stability and the simplicity of in vitro production. At the same time, DNA vaccines have a number of drawbacks such as low immunogenicity and the risk of integration into the host cell genome. mRNA vaccines do not have these drawbacks, that is why mRNA studies also continued [6, 7].
In 2000, Hoerr et al. reported that a direct mRNA injection could induce an immune response in mice [8]. The same year, Hoerr founded the CureVac company, where the first human mRNA trials took place. The company’s lead scientist Steve Pascolo was the first research subject: he administered the mRNA to himself [9].
The invaluable contribution to the development of mRNA vaccines as a platform was made by Katalin Kariko. In 1999, she coauthored a paper with Drew Weissman, which described the work on the mRNA-based HIV-1 vaccine development. Unfortunately, the synthesised mRNAs caused massive inflammatory reactions when injected into mice [10]. The authors soon found out the underlying cause of this phenomenon. It turned out that the synthetic mRNA activated a number of cell receptors known as Toll-like receptors, which are the first to respond to the entry of a pathogen’s RNA [11]. In 2005, Kariko et al. reported that the inclusion of the uridine analog pseudouridine in the mRNA prevented the experimental mRNA from being identified as a foreign RNA by the immune system [12].
Few scientists understood the therapeutic value of these modified nucleotides at that time, however, it was not long before the scientific world recognized their potential [13]. In 2008, the major pharmaceutical companies Novartis and Shire established mRNA research divisions with the former company focusing on vaccines, and the latter, on therapeutics. At this time, the BioNTech and Moderna companies also joined the work on the development of mRNA-based technologies. By 2019, Moderna developed nine candidate mRNA vaccines against the infectious diseases caused by the coronavirus, respiratory syncytial virus, human metapneumovirus, cytomegalovirus, influenza virus, Epstein–Barr, virus human immunodeficiency virus, Zika virus and Nipah virus. Approximately the same range of vaccines has been developed by the German company BioNTech. None of the vaccines under development have been licensed [14].
The practical use of mRNA-based vaccines was given a strong impetus in the early 2020 after the outbreak of the COVID-19 pandemics. For example, the mRNA-1273 prototype vaccine against Severe Acute Respiratory Syndrome Coronavirus-2 (SARS-CoV-2) was created by the American mRNA vaccine developers-Moderna Inc. in collaboration with the National Institute of Allergy and Infectious Diseases (NIAID)- in an unprecedentedly short time. It took only 63 days from the selection of a viral nucleotide sequence for vaccine production to the first phase of clinical trials. BioNTech in collaboration with the Pfizer pharmaceutical company, also created an mRNA vaccine against COVID-19 within approximately the same time frame [15].
As a result of research performed over the past decades, the stability of mRNA and the efficiency of its delivery have been greatly improved. By combining different modifications of mRNA structural elements and its cell delivery methods, the immunogenicity of mRNA molecules can be significantly improved, making this approach promising for vaccine development.
TYPES OF RNA-BASED VACCINES
Currently, there are two types of RNA vaccines: the ones based on nonreplicating mRNA and the ones based on self-amplifying RNA.
Nonreplicating mRNA vaccine is an mRNA molecule which is produced by in vitro transcription using the plasmid DNA encoding the target immunogene as a template. In addition to the coding sequence, the obtained mRNA contains the cap at the 5'-end, the untranslated regions (UTRs) and the poly-A sequence (poly(A)-tail) at the 3'-end, which are necessary for effective translation, mRNA protection against exonucleases and proper splicing of the transcript. Note that the first vaccines against SARS-CoV-2 approved for human use were created on the basis of nonreplicating mRNAs [16, 17]. However, the issues of mRNA instability and inefficient delivery have not yet been completely resolved.
Self-amplifying RNAs (saRNAs) are replicons which include the gene encoding the target immunogene as well as the viral genome elements which enable target RNA replication. As a rule, DNA or RNA viruses can replicate their DNA in the cytosol and penetrate into the nucleus after entering the cell. It has been also demonstrated that some viruses are characterised by cytosolic replication and expression [18]. The resulting replicons are not capable of forming infectious viral particles after infecting the host cell, however, the RNA encoding the target immunogene is capable of self-amplification [19]. Thus, the production of the encoded immunogene is increased as compared to nonreplicating mRNAs. Due to the sustained production of the encoded immunogene, saRNAs in lower doses demonstrate the same level of immunogenicity as nonreplicating mRNAs. Most saRNAs are derived from the single-stranded positive-sense alphaviruses such as Venezuelan equine encephalitis virus, Sindbis virus and Semliki forest virus [20, 21]. A number of problems associated with the use of viral vectors to produce vaccines should be mentioned. These include the immunogenicity of the vector itself, which can induce an unwanted immune response and interfere with subsequent vaccine booster injections using the same viral replicon. As is the case with live attenuated vaccines, replication-capable alphaviral vectors can bear a risk of viral reactivation [22–24].
In this review, we will focus on the production and delivery of nonreplicating mRNA vaccines.
WAYS TO INCREASE mRNA STABILITY
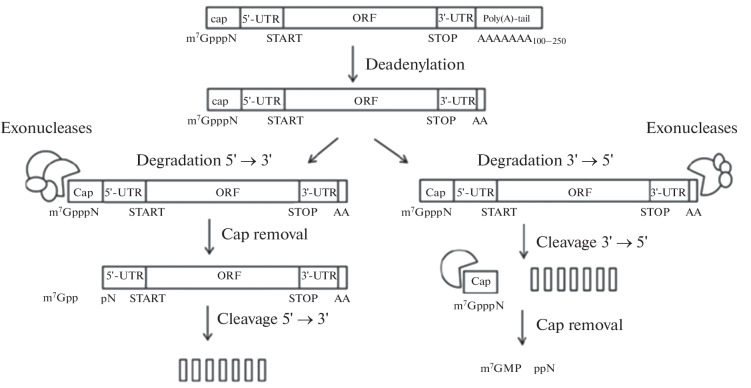
It is known that the mature eukaryotic mRNA consists of five important parts, which include the cap at the 5'-end (m7GpppN (N is any nucleotide)), the 5'-untranslated region (5'-UTR), the open reading frame (ORF), the 3'-untranslated region (3'-UTR) and the 3'-tail consisting of 100–250 adenyl residues (poly(A)-tail), the length of which varies in different cell types [25]. The main cause of mRNA instability as compared to DNA is the presence of the hydroxyl group at the 2'-carbon atom of ribose, which facilitates hydrolytic degradation of RNA molecules. The innate immune system is able to recognise foreign RNA, thereby triggering its degradation. The main pathway of mRNA degradation in eukaryotic cells is shown in Fig. 1.
Fig. 1.
Schematic representation of mRNA degradation. mRNA degradation occurs in the cytoplasm within the ribonucleic complexes called P-bodies, which contain 5'-3'-exonucleases, decapping and deadenylating enzymes. Once the poly(A)-tail is shortened to 12 residues or less, mRNA degradation occurs through cap cleavage and 5'→3' or 3'→5' cleavage [21]. Endonucleases (not shown in the figure) may also be involved in mRNA degradation.
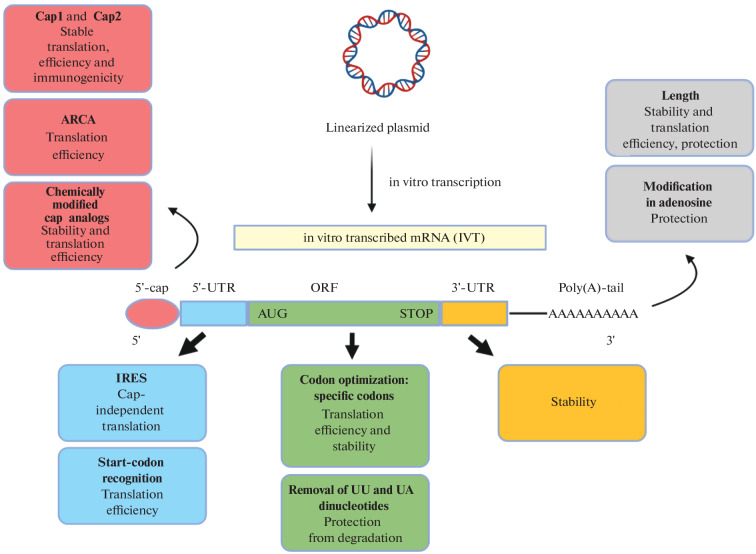
Thus, in its canonical version exogenous mRNA is unstable and low-immunogenic. For this reason, different modifications of each of the mRNA elements are used when creating mRNA vaccines. Figure 2 shows schematically the key structural elements of the in vitro transcribed (IVT) mRNA which can be subject to modifications. It is known that certain chemical modifications can increase the efficiency of mRNA translation in the cell. For example, in vitro mRNA production often uses modified 5'-cap analogs, such as Anti-Reverse Cap Analog (ARCA) and CleanCap®, which ensure proper cap attachment. Modifications of each of these elements are discussed in more detail below [5].
Fig. 2.
Schematic outline of in vitro transcribed (IVT) modified mRNA production: 5'-UTR, 5'-untranslated region, 3'-UTR, 3'‑untranslated region, ORF, open reading frame, ARCA, Anti-Reverse Cap Analog, IRES, Internal Ribosome Entry Site, AUG, start-codon and STOP, stop codon [5].
Cap Structure
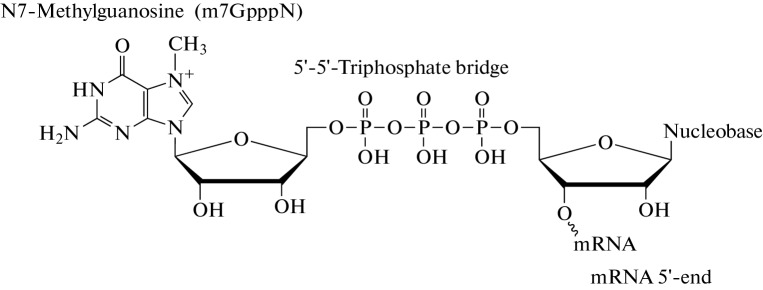
Native eukaryotic mRNA has a 5'-cap structure (Fig. 3). Cap is the m7GpppN structure which contains a modified nucleoside N7-methylguanosine (m7G) and is linked to the first transcribed nucleotide via a 5'-5'-triphosphate bridge. Cap plays an important role in normal mRNA functioning, including in mRNA stabilisation during translation, splicing, polyadenylation and nuclear export and in mRNA protection from exonucleases. Capping takes place in the nucleus, when the first 20–30 nucleotides of mRNA are transcribed, as a result of three sequential enzymatic reactions: phosphate group cleavage from the 5'-terminal nucleotide of the transcript by the RNA triphosphatase, then guanosine monophosphate (GMP) residue transfer onto the phosphate group of the 5'-terminal nucleotide by the guanyltransferase and methylation of the guanine residue in the guanosine triphosphate (GTP) with the formation of N7-methylguanosine by the guanyl-N7-methyltransferase.
Fig. 3.
Cap structure (cap0). Cap is a N7-methylguanosine ribonucleotide linked by a 5'-5'-triphosphate bridge to the first nucleotide residue in the transcript.
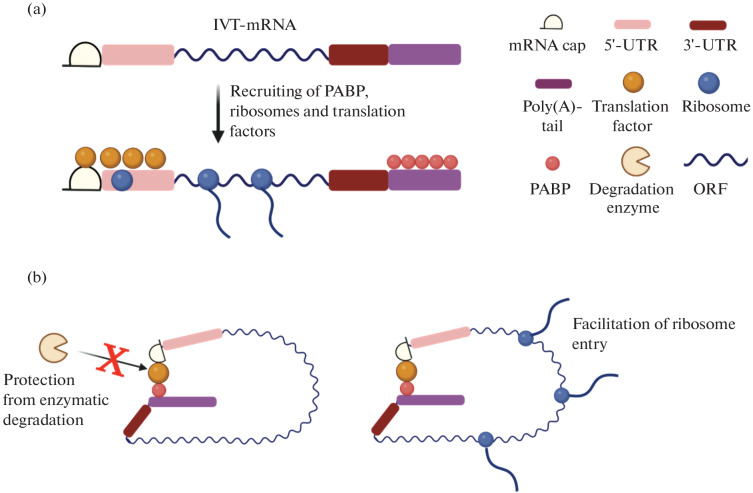
The cap structure participates in mRNA translation by recruiting translation initiation factors (e.g., 4E (eIF4E), which is critical for the initiation of translation) and by forming the mRNA closed loop model (Fig. 4). In addition, cap interacts with the cap-binding proteins (CBPs) required for the nuclear export of mRNA [26].
Fig. 4.
Closed mRNA loop model. (a) In vitro transcribed mRNA (IVT-mRNA) can recruit translation initiation factors which bind to the 5'-cap and 5'-untranslated region (5'-UTR), promoting mRNA entry into the ribosome and mRNA translation and also recruit the poly(A)-binding protein (PABP) to the poly(A)-tail; (b) in the closed loop model, strong interaction between the translation factors on both sides of the mRNA induces the formation of a stable loop, which protects the transcripts from RNA degrading enzymes and facilitates mRNA reentry into the ribosome thereby enhancing translation [22].
In addition to cap0, two new 5'-cap structures, namely, cap1 and cap2, and the enzymes involved in their synthesis have been identified in recent years. In these cases, m7G-specific 2'-O-methyltransferase (2'-O-MTase) methylates the second or third ribonucleotide at the 2'-O-ribose position, generating cap1 or cap2 structures, respectively. It has been demonstrated that cap1 masks mRNA from the cytosolic sensors RIG-I and MDA5, which trigger the activation of the interferon type I signalling pathway resulting in mRNA degradation. Hence, cap1 mRNA is less affected by the innate immune response. Consequently, cap1 mRNA translation and target protein production in the body are more efficient than in the case of cap0 mRNA [27].
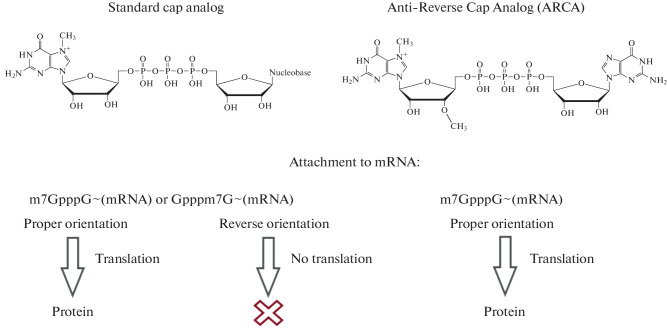
It is important to note that the body’s immune system recognises uncapped RNA as a foreign RNA, since the presence of the cap distinguishes cellular mRNA from viral mRNA [5]. Incorrectly capped or uncapped (5'-ppp or 5'-pp) mRNAs are recognised by the Pattern Recognition Receptors (PRRs) such as RIG-I and IFIT which trigger the synthesis of type I interferons ultimately leading to RNA degradation. Therefore, in order to match the chemical structure of eukaryotic mRNAs, synthetic RNA transcripts should be capped. In the case of a vaccine, it is important to achieve the maximum efficiency of mRNA capping (including by purifying the final product) to avoid hyperactivation of innate immunity by remaining uncapped or incorrectly capped products [28]. Capping can be performed either after transcription is completed, i.e., posttranscriptionally (for instance, using a recombinant enzyme from the smallpox virus), or during transcription, i.e., cotranscriptionally (by introducing cap analogs such as ARCA and CleanCap® into the reaction mixture).
In practical terms, cotranscriptional capping reduces the number of steps involved in mRNA production and decreases the number of enzymes used in the work. These factors are crucial for reducing the cost of mRNA production [29]. At the same time, cotranscriptional capping also has a number of limitations. On the one hand, not all synthesized mRNA molecules are capped due to competition between the cap analog and guanosine triphosphate (GTP), which acts as the initiator nucleotide. As a consequence, uncapped RNAs can induce the unwanted immune response. The strategy to reduce the immune response in this case is to remove triphosphates from the 5'-end of uncapped IVT-RNA using phosphatase. On the other hand, there is a risk of cap analog incorporation in the reverse orientation, which prevents mRNA binding with the cap-binding proteins (CBPs) and subsequent translation [30].
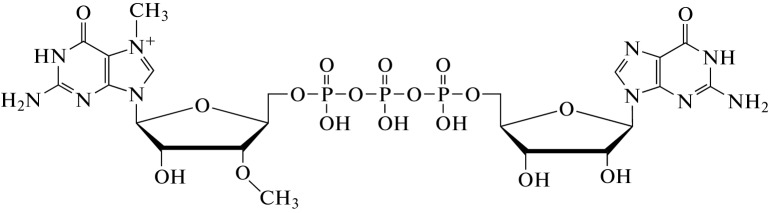
To address these issues and to increase the efficiency of the in vitro transcribed mRNA translation, chemical modifications (e.g., attachment of methyl groups) can be introduced into the 3'- or 2'-position of the cap analogs. These modifications prevent reverse cap orientation and improve the quality of the synthesized mRNA. For example, there is an Anti-Reverse Cap Analog (ARCA), which contains a modified cap structure representing the m7,3'-OGpppG dinucleotide (Fig. 5) [31]. ARCA contains a methyl group attached to the 3'-OH of the m7G nucleotide, which ensures proper cap attachment during RNA synthesis [5].
Fig. 5.
The Structure of Anti-Reverse Cap Analog (ARCA).
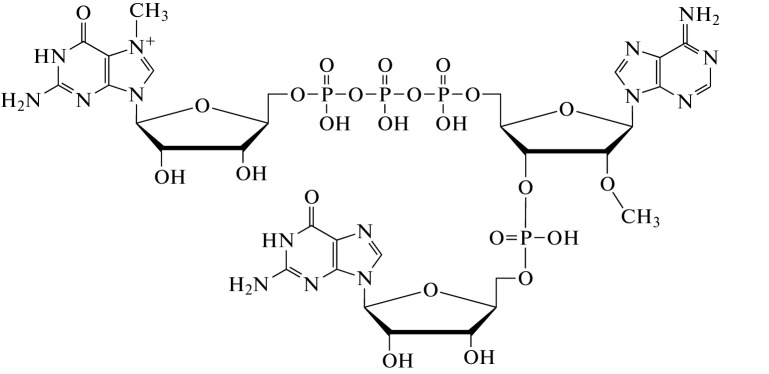
In addition, the introduction of additional cap modifications and elongated 5'-5'-phosphate bridges in ARCA have also been reported to increase the translation efficiency and mRNA stability. The principle behind ARCA is presented in Fig. 6 [32].
Fig. 6.
Principle of Anti-Reverse Cap Analog (ARCA) action.
The ARCA structure has currently been improved, and CleanCap®, a second-generation antireverse cap analog, which can be used to incorporate the cap1 structure into the mRNA, has been developed. Its use allows us to achieve higher yields of capped mRNA (up to 95%) compared to the use of the first-generation ARCA. The structure of the second-generation CleanCap cap analog® is shown in Fig. 7 [29]. CleanCap® is used by BioNTech/Pfizer in the production of the BNT162b1 and BNT162b2 mRNA vaccines [33, 34].
Fig. 7.
The structure of the second-generation CleanCap analog®.
Poly(A) Sequence at the 3'-end of mRNA- the Poly(A)-tail
The poly(A)-tail is one of the key elements of efficient translation and increased mRNA stability [35]. In mammalian cells, the most actively translated mRNAs contain 100–250 adenosine residues [36]. The minimal poly(A)-tail length at which exogenous mRNA can be translated is 20 adenosine residues [37]. The role of the poly(A)-tail in translation consists in that it binds with numerous polyadenosyl-binding proteins (PABP), which in their turn bind with the eukaryotic translation initiation factor 4G (eIF4G). As mentioned above, the ring structure with the cap-eIF4E-eIF4G-PABP-poly(A) closed loop is formed, which facilitates ribosome binding and protects mRNA from nuclease degradation [38].
In the in vitro transcribed mRNA, the poly(A)-tail can be obtained in two ways. The first way is to add adenosine residues to the 3'-end by the enzymatic synthesis using poly(A)-polymerase, which allows up to 200 adenosine residues to be added to the mRNA molecule. However, in this case, the resulting product is heterogeneous. The second way is to attach the poly(A)-tail by means of matrix synthesis. In this case, a fragment with a certain number of thymidine residues is incorporated into the DNA matrix. Usually, due to the instability of the poly(A)-sequence in the matrix, no more than 120 adenosine residues can be added using matrix synthesis [39].
Noncoding Regions in mRNA
The coding region is flanked by the noncoding regions (UTRs) at both the 5'- and 3'-ends. These regions do not encode any protein, although their sequence, length and secondary structures are important for the regulation of mRNA translation. It is well-known that 5'-UTR is involved in the translation initiation, whereas 3'-UTR affects mRNA stability and elongation efficiency [5].
Most eukaryotic mRNAs contain decay signals in the 3'-untranslated region (3'-UTR), which affect their stability. AU-rich sequences in the 3'-UTR have been reported to be involved in poly(A)-tail removal. mRNA half-life increases when AU-rich sequences are replaced by the 3'-UTR sequences from stable mRNAs [39]. For example, the presence of the specific human α-globin or β-globin mRNA sequences (or two copies of the 3'-UTR from the human β-globin gene [40]) in this region increases IVT-mRNA stability and consequently the duration of protein production period.
The consensus Kozak sequence located in the 5'-UTR also plays an important role in the translation initiation. The Kozak sequence, defined as RCCAUGG, where R is a purine (A or G), is considered to be the preferred sequence for translation initiation. In this sequence, some nucleotides are more important that the other, in particular, the nucleotides in positions –3 and +4 relative to the adenosine of the AUG start codon. To increase the efficiency of the AUG start codon recognition, the nucleotide in the +4 position should be G, and the nucleotide in the –3 position should be A or G [5].
mRNA Coding Sequence Optimization
The codon composition of the region encoding the protein sequence may also have an impact on the translation efficiency and mRNA stability. The open reading frame (ORF) can be modified at the codon level by regulating the translation elongation rate or by finding the optimal mRNA secondary structure [28]. There are different strategies for codon optimisation, for example, replacing several rare codons with the more frequently occurring ones for the same amino acid. Another strategy is the optimised use of dicodons, or, in other words, the use of pairs of codons which together ensure optimal translation. It has been demonstrated that reducing the number of UU and UA dinucleotides in the ORF protects the IVT-mRNA from the decapping enzymes [41, 42]. The third strategy is to use ORF sequence with the same codon ratios as found in the naturally occurring highly expressed genes [5].
Optimal codons located next to the starting codon increase the elongation rate, which leads to higher levels of mRNA translation. In contrast, rare codons are the cause of a low elongation rate, which leads to ribosomes stacking on the mRNA molecule. This disturbance of elongation allows the DEAD-Box RNA helicase to bind to the transcript and thus accelerates mRNA decay after 5'-decapping. At the same time, a high elongation rate is not always desirable. Sometimes it can interfere with the proper folding of the protein encoded by the mRNA, as it was shown for the codon-optimised firefly luciferase mRNA, which lost 50% of its activity [43]. In such cases, rare codons can provide for the lower translation rate and, therefore, proper protein folding, which is critical in getting the right antigen conformation. Therefore, depending on the target antigen, different strategies should be used to optimise the codon composition of the target mRNA. Optimisation of all codons is an appropriate approach in the case of the linear epitope-based mRNA vaccines. In contrast, complex antigens may require slow translation rates ensuring normal folding of the domains within the protein and proper epitope conformation. In any case, it is recommended to avoid using rare codons in both strategies in order to optimise protein biosynthesis [28].
Optimisation is also often performed by recoding the terminal codons from U to C. All U nucleotides in the mRNA vaccines are usually replaced with N1-methylpseudouridine or pseudouridine, which is described in more detail in the next section. These modified nucleotides may be complementary to all other nucleotides, which in some cases may lead to noncanonical base pairing and disruption of the primary structure of the encoded protein. For example, GAΨ encoding Asp can be recognized by the tRNAGlu anticodon, which leads to a nonsynonymous substitution. There is no such problem for the codons with the terminal cytosine [44].
Modified Nucleosides in mRNA
It should be taken into account that when mRNA is introduced into the body it may be recognized as foreign and induce the unwanted innate immune response, leading to mRNA degradation and inflammatory reactions. Both DNA and RNA stimulate the innate immune response in mammals by interacting with the Pattern Recognition Receptors (PRRs), including Toll-like receptors (TLRs) and cytoplasmic RNA sensors such as the Retinoic Acid Induced Protein I (RIG-I). Uridine residues are known to activate TLR7, while GU- and AU-rich RNA chains activate TLR7 and TLR8 [5]. Thirteen TLRs have been identified, four of which (TLR3 for the double-stranded RNA (dsRNA), TLR7 and TLR8 for the U-rich single-stranded RNA and TLR9 for the DNA CpG motif) are involved in nucleic acid recognition. It has been reported that IVT-mRNA preparations induce a strong TNF-α response in dendritic cells (DCs) [45].
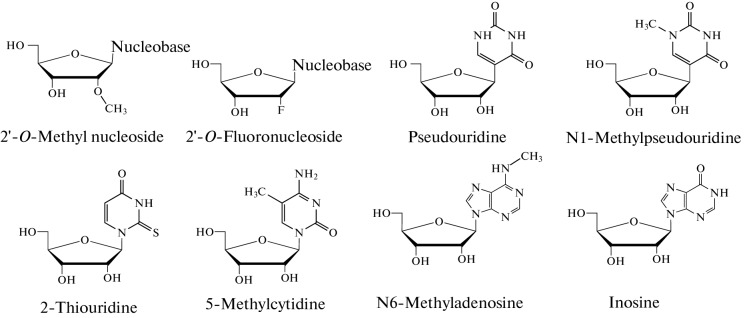
The use of modified nucleosides (Fig. 8) in the in vitro transcription is thought to significantly suppress the TLR-mediated dendritic cell (DC) activation, however, the effects of the modified nucleosides on the TLR-independent immune response have not yet been studied. Modified nucleosides can enhance mRNA vaccine efficiency in two different ways. First, they prevent dsRNA formation during in vitro transcription, and second, they prevent Pattern Recognition Receptor (PRR) activation when the mRNA is introduced into the body [28]. Since many modified nucleosides including pseudouridine, N1-methylpseudouridine, 2-thiouridine, 5-methylcytidine, 6-methyladenosine, inosine, and 2'-O-methylated nucleosides as the 5'-end cap are present in the mammalian RNA these nucleosides may be used to reduce the unwanted immune response to the injected mRNA [46]. The approved vaccines mRNA-1273 (Moderna) and BNT162b2 (BioNTech/Pfizer) take advantage of N1-methylpseudouridine [16, 17].
Fig. 8.
The structures of modified nucleosides.
At the same time, there is another approach to creating mRNA vaccines, which does not use modified nucleosides. CureVac has applied their mRNA synthesis technology to the development of a SARS-CoV-2 vaccine specially designed to achieve maximum protein production and balanced immune activation. The CureVac technology includes the optimisation of the coding part for codon composition and the untranslated regions in the mRNA, and poly(A)-tail modification. In particular, the GC content in the coding part was increased, untranslated regions from the highly expressed and stable mRNAs of the known genes were included into the mRNA, and the “histone stem–loop” sequence was used instead of the classical poly(A)-tail. In such a way, the CVnCoV and CV2CoV mRNA vaccines against SARS-CoV-2 were developed. They consist of the mRNA encapsulated in the lipid nanoparticles encoding the full-length S-protein with two proline (S-2P) mutations [47, 48]. However, the trials have revealed that the efficiency of these vaccines was much lower than that of the mRNA vaccines containing modified nucleosides [49].
IVT-mRNA PURIFICATION FROM THE dsRNA
Contamination with the dsRNAs formed during transcription may be one of the causes of innate immunity activation in response to the injection of in vitro synthesized mRNAs. T7 polymerase has been shown to frequently produce byproducts, dsRNAs, which can activate the RIG-I and MDA5 cytosolic sensors. dsRNAs are formed as a result of the hybridisation of the sense transcript and its fully complementary antisense transcript. Antisense RNA is produced by the promoter-independent initiation of transcription from the 3'-end (–) of the DNA matrix [42].
It has been demonstrated that mRNA purification from dsRNAs can lead to more than a 100-fold increase in protein production in human dendritic cells [50].
Purification using reversed-phase high performance liquid chromatography (RP HPLC) can remove the dsRNAs produced during the in vitro transcription from the final product. This prevents innate immunity activation and mRNA degradation in the body, which, in the long run, increases target protein production. However, RP HPLC purification is very expensive, needs the use of sophisticated equipment and consumables, is difficult to scale, and requires the disposal of hazardous wastes [51].
A purification method is known which allows to avoid the problems associated with RP HPLC and is able to eliminate up to 90% of dsRNA. It is assumed that dsRNA selectively binds with cellulose in the ethanol-containing buffer. This method is cheap, fast and scalable. It is suitable for purifying large quantities of mRNA using fast protein liquid chromatography (FPLC) without producing toxic or hazardous wastes. Importantly, comparable translation levels of the mRNAs purified using RP HPLC and cellulose were observed after their intravenous injection into mice [50].
IN VIVO mRNA DELIVERY
To be fully functional, mRNA must avoid extracellular degradation by nucleases, remain intact, and enter the cell. Since cellular uptake of individual nucleic acids is inefficient, various options for their delivery using both viral and nonviral delivery systems have been proposed.
Nonviral delivery of mRNA include the approaches that can be divided into two groups: (1) mRNA delivery encapsulated in liposomes or in various polycationic polymers; (2) mechanical mRNA delivery across the cell membrane using electroporation, gene guns, ultrasound, or high-pressure injection [52]. These methods can be used both in vivo and in vitro.
Lipid Nanoparticles
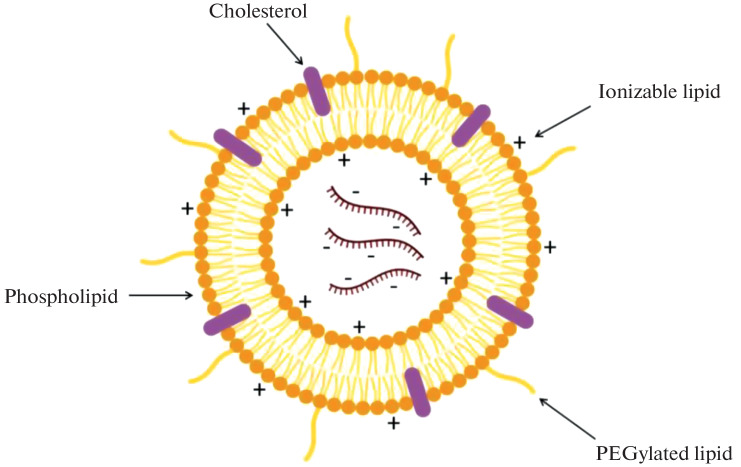
Lipid nanoparticles (LNPs) are one of the most commonly used mRNA delivery systems. LNPs often consist of four components (Fig. 9): (1) the ionizable cationic lipid that provides for the self-assembly of particles and facilitates endosomal release of the mRNA into the cytoplasm; (2) lipid-conjugated polyethylene glycol (PEG); (3) cholesterol, the stabilizing agent, and (4) phospholipids, which maintain the two-layer lipid structure [53, 54]. The level and duration of mRNA-LNP vaccine translation in vivo can be partially controlled by changing the route of its administration. It has been shown that intradermal, intramuscular, and subcutaneous injection of mRNA-LNP complexes leads to prolonged protein production at the injection site [55].
Fig. 9.
Lipid nanoparticle structure.
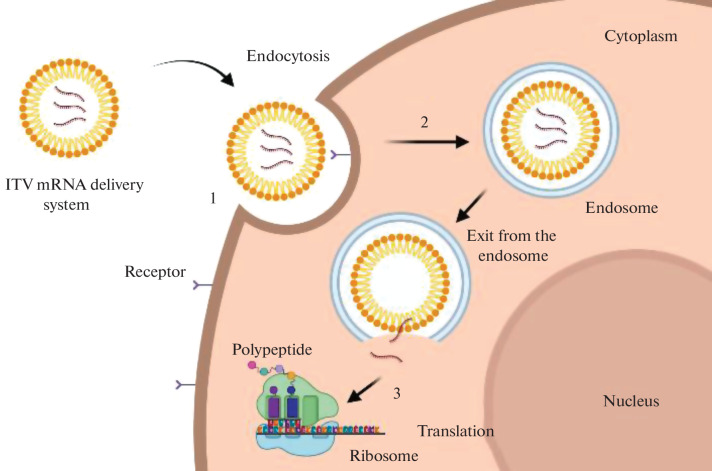
As it is shown in Fig. 10, the mRNA delivery system should interact with the target cell and penetrate into the cytoplasm through the cytoplasmic membrane, and then provide for the mRNA release into the cytoplasm so that it could reach the ribosomes.
Fig. 10.
Intracellular barriers for in vitro transcribed (IVT) mRNA delivery: (1) interaction between the delivery system and the cell membrane, (2) endocytosis, (3) exit from the endosome and mRNA release to initiate translation. Endocytosis is a mechanism by which extracellular components and fragments of the plasma membrane internalize with the formation of the endocytic vesicle. This process involves vesicles with the internal pH of ~5 known as endosomes which develop from the early endosomes to the late endosomes before fusing with the intracellular organelles called lysosomes. In such a way, particles entering the cell via endocytosis are captured by the endosomes and eventually appear in the lysosomes, where active enzymatic degradation takes place [5, 46].
The delivery system components may bind to the cell surface via the electrostatic interactions between the components and the membrane surface. Binding can be made more efficient by incorporating ligands capable of interacting with the specific receptors on the cell surface into the delivery systems [56].
The key mechanism by which mRNA delivery systems enter the cell is endocytosis. It involves many complex processes which determine the intracellular localisation of mRNA. mRNAs get inside the endosomes as a result of cell membrane invagination. Endosomes mature and fuse with lysosomes, where the acidic environment and the presence of hydrolytic enzymes may destroy the delivery system and the nucleic acid. Therefore, the delivery system components should provide for the optimal time interval between the mRNA exit from the endosomes and the nucleic acid degradation, as it is crucial for the successful mRNA work [5].
The mechanisms underlying mRNA release from the artificial lipid nanoparticles into the cytoplasm are not yet completely understood. It has been shown that LNPs are internalised via a process involving both clathrin-dependent endocytosis and macropinocytosis [57].
Currently, there are approved mRNA vaccines against COVID-19 that deliver the mRNA encoding the SARS-CoV-2 S-protein using LNP. These include the mRNA-1273 (Moderna) and BNT162b2 (BioNTech/Pfizer) vaccines [58]. The LNP composition of these vaccines is shown in Table 1. The structural formulas of the components included in the LNP are shown in Fig. 11.
Table 1.
Lipid nanoparticle composition in the mRNA-1273 (Moderna) and BNT162b2 (BioNTech/Pfizer) vaccines [58]
| Vaccine | mRNA-1273 | BNT162b2 |
|---|---|---|
| Manufacturer | Moderna | BioNTech/Pfizer |
| mRNA dose, µg | 100 | 30 |
| Components | SM-102 | ALC-0315 |
| Distearoylphosphatidylcholine | Distearoylphosphatidylcholine | |
| Cholesterol | Cholesterol | |
| DMG-PEG2000 | ALC-0159 | |
| Ionizable cationic lipids : neutral lipids : cholesterol : PEGylated lipids (molar ratio, %) | 50 : 10 : 38.5 : 1.5 | 46.3 : 9.4 : 42.7 : 1.6 |
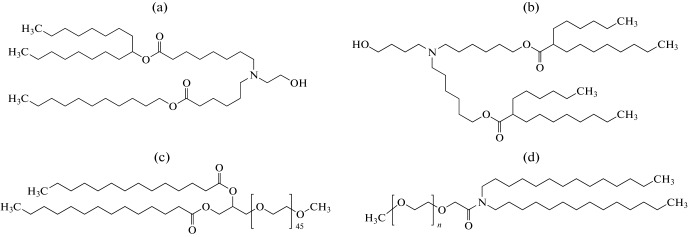
Fig. 11.
The structures of the LNP components: (a) SM-102—(heptadecane-9-yl-8-((2-hydroxyethyl)(6-oxo-6-(undecyloxy)hexyl)amino)octanoate)); (b) ALC-0315—((4-hydroxybutyl)azanediyl)bis(hexane-6,1-diyl)bis(2-hexyl decanoate); (c) DMG-PEG2000—1-monomethoxypolyethylene glycol-2,3-dimyristylglycerol with polyethylene glycol with the average molecular weight of 2000; (d) ALC-0159—2-[(polyethylene glycol)-2000]-N,N-ditetradecylacetamide.
The candidate mRNA vaccines CVnCoV (CureVac) and ARCoV (Walvax), which are in the clinical trials, use LNPs of different composition for the delivery (Table 2) [59, 60].
Table 2.
| Vaccine | CVnCoV | ARCoV |
|---|---|---|
| Manufacturer | CureVac | Walvax |
| mRNA dose, µg | 12 | 5, 10, 15, 20 and 25 |
| Components |
Cationic Lipid (Acuitas Therapeutics) Phospholipid Cholesterol PEG-lipid conjugate |
Cationic lipid (no data) Dystearoylphosphatidylcholine Cholesterol PEG-lipid conjugate |
| Ionizable cationic lipids : neutral lipids : cholesterol : PEGylated lipids (molar ratio, %) | 50 : 10 : 38.5 : 1.5 | 50 : 10 : 38.5 : 1.5 |
The main issue in the mRNA delivery using lipid nanoparticles lies in the nature of the lipids [61]. In particular, positively charged lipid particles can bind to the negatively charged proteins and nucleic acids and adhere to the cell surface, which destabilises the plasma membrane and causes side effects in the vaccinated patients [62, 63]. Lipid components can induce immune responses after systemic or local administration. For example, PEGylated lipids can stimulate the complement system. In addition, antibodies against PEG can lead to the rapid elimination of PEGylated nanoparticles from the body. This can alter the bioavailability and biodistribution of the preparation encapsulated into the PEGylated nanoparticles and thus cause side effects. Additionally, cationic and ionizable lipids have been reported to stimulate the secretion of proinflammatory cytokines and reactive oxygen species. Although the immunogenicity of these lipids is not yet fully understood, the complement system and Toll-like receptors may be involved in the activation of the innate immunity. The cytotoxicity of lipid materials also poses a problem. The in vivo use of lipid nanoparticles has been reported to cause liver and lung damage in rodents, which may be related to their cytotoxicity and the induction of proinflammatory factors [64].
These issues are addressed by using various modifications of the lipid nanoparticle components [65], but this increases the cost of LNPs production and makes their composition more complex. Another drawback of lipid nanoparticles is that they are sensitive to freezing and thawing, therefore, they must be stored and shipped at –80°C, which makes their use for mass vaccination challenging [66, 67].
Polycationic Polymers
Polymeric materials are not as widely used for nucleic acid delivery as lipids are. Compared to lipids, polymeric materials pose a number of additional problems, such as difficult biodegradation of polymers with large molecular weight. Nevertheless, the research is carried out aimed at resolving these problems. For example, low-molecular-weight poly(ethylenimine) (PEI) modified with fatty acid chains is used for mRNA delivery to reduce the toxicity of high-molecular-weight PEI. Poly(glycoamidoamine) polymers modified with fatty acid chains, such as TarN3C10, which contains the tartrate backbone (consisting of esters and tartaric acid salts), have demonstrated their effectiveness in erythropoietin mRNA delivery in mice [68].
Polymethacrylates with amine-bearing side chains, polyaspartamides with oligoaminoethylene side chains, and polyacrylic acids amidized with tetramine with alternating ethylpropylethyl spacers can also deliver mRNAs into the cells. Self-degradable esters have been reported, which the authors have named Charge-Altering Releasable Transporters (CARTs). These polymers are capable of releasing mRNA after rearrangement followed by degradation at pH 7.4. Biodegradable aminopolyesters (APEs) are known to be able to selectively deliver mRNA to tissues [69].
Chitosan, a biodegradable biocompatible polymer which represents a chitin derivative obtained by removing the acetate part of chitin, is also an interesting subject of research. Chitosan contains chemical functional groups which can be modified to achieve specific goals, making it a polymer with a huge range of potential applications. Nanoparticles based on chitosan and its derivatives usually possess a positive surface charge and mucoadhesive properties which makes them able to attach to mucous membranes and release a drug [70].
Dendrimers such as poly(amidoamine) (PAMAM) or polypropyleneimine derivatives have been studied for nucleic acid delivery. PAMAM dendrimers modified with fatty acid chains have been synthesised to deliver small interfering RNAs, which were subsequently used to develop the intramuscularly administered self-replicating mRNA vaccine platform for the expression of the Ebola virus, H1N1 influenza, Toxoplasma gondii and Zika virus antigens [71]. PAMAM dendrimers can improve water solubility, stability, targeting and pharmacokinetics of the drugs. Owing to their versatility dendrimers can be used as an alternative platform for next-generation drug delivery [72]. However, a number of problems arise when using this delivery platform. As long as repetitive dendrimer units form tree-like branching, their enzymatic biodegradation can be hindered due to steric factors leading to the toxicity associated with the tissue accumulation of these materials [73].
Along with the known agents, other materials for mRNA delivery are being developed that can ensure the safety and efficacy of the vaccine as well as facilitate its storage and transportation. In particular, a polyglucin-spermidine (PGS) conjugate has been obtained at the State Research Center of Virology and Biotechnology “Vector” to serve as the carrier for the mRNA-RBD vaccine encoding the receptor-binding domain (RBD) of the SARS-CoV-2 S-protein [74].
PGS contains only two components, polyglucin and spermidine, and allows the nucleic acid to be lyophilised and stored for long periods at positive temperatures. It has been demonstrated that PGS-encapsulated DNA vaccine can be stored for at least two years at 4°C not losing its specific activity. PGS conjugate is the component of a HIV-1 DNA vaccine, and its safety has been confirmed by preclinical studies and phase I clinical trials [75, 76].
The important advantage of PGS components is their biodegradability and safety for humans. Polyglucin (a glucose polymer with the molecular weight of 40 000) is not toxic for humans and acts as a certified plasma substitute with the hemodynamic activity, which is used to restore the circulating blood volume. The polyglucin envelope has been demonstrated to protect the yeast dsRNA from degradation by serum nucleases [77]. Spermidine is a naturally occurring polyamine found in all living organisms; it plays an important role in maintaining cellular homeostasis and is involved in many biological processes, including cell growth and proliferation, DNA and RNA stabilisation and translation regulation [78, 79]. In addition, the low cost of the components and possibility to lyophilise and store at 4°C give the PGS conjugate additional technological advantages in vaccine production and transportation.
It is thought that the mRNA-PGS complex enters the antigen-presenting cells by endocytosis owing to its size, which is comparable with the average size of viral particles (100–200 nm). In addition, packaging in PGS protects the mRNA from degradation by nucleases, which ultimately results in increased immune response. The studies have demonstrated that the proposed polyglucin-spermidine polycationic conjugate may be regarded as a promising and safe means of mRNA vaccine delivery, in particular, for mRNA vaccines against SARS-CoV-2 [80].
Physical Delivery Methods
Various physical manipulations are used to increase the efficiency of direct transfection [52, 81]. For the direct delivery of nucleic acids into the cells, both in vivo and in vitro, electroporation, gene guns, ultrasound, or high-pressure injection can be used. Electroporation is one of the most effective mRNA delivery methods. Since mRNA does not require nuclear localisation, gentle electrical pulses can be used to reduce cellular toxicity. Another advantage of electroporation is the direct delivery of mRNA into the cytosol, which can prevent the unwanted immune response [82].
Other delivery methods different from those discussed above are currently being investigated. Although much success has been achieved in this area, there is an idea that the combination of different mRNA delivery systems will be the most effective.
CONCLUSIONS
mRNA vaccines have become a promising platform for creating the means of infectious disease prevention since they offer significant advantages over other types of vaccines. First of all, it is safety: mRNAs, unlike classical viral vaccines, are noninfectious and have low reactogenicity. mRNA vaccines can efficiently activate the specific cellular and humoral immune responses, but do not induce the antivector immune response. A significant advantage of mRNA-vaccines is their rapid, inexpensive, scalable and routine production, ensuring high yields of the desired product under in vitro conditions. The mRNA vaccine platform allows for easy target gene replacement without changing the production technology, which is important for addressing the time lag between the epidemic outbreak and vaccine release.
The present review discussed the main mRNA vaccines modifications aimed at enhancing their efficiency and delivery ways. Modifications of the cap, poly(A)-tail, and coding and noncoding parts of the mRNA are used to increase stability. In addition, an important step is the purification of the target mRNA vaccine from the by-products (dsRNA). Both viral and nonviral delivery systems are used to deliver the mRNA into the cells. Nonviral delivery systems include mRNA encapsulation in liposomes (LNP) and various polycationic polymers, as well as mRNA delivery across the cell membrane using physical methods, including electroporation, gene guns, ultrasound, and high-pressure injection.
Owing to the advances in enhancing the stability and efficiency of mRNA vaccine translation and in increasing the efficiency of mRNA delivery into the cells, mRNA vaccines have become a promising tool which could be widely used to prevent viral diseases caused by such agents as coronavirus, influenza viruses, human immunodeficiency virus, etc.
FUNDING
The work was supported by the State Task to the State Research Center of Virology and Biotechnology “Vector”, Federal Service for Surveillance on Consumer Rights Protection and Human Welfare.
COMPLIANCE WITH THE ETHICAL STANDARDS
The authors declare that there is no conflicts of interest.
The paper does not report any studies involving animals or human performed by the authors.
Footnotes
Abbreviations: 3'-UTR, 3'-untranslated region; 5'-UTR, 5'-untranslated region; ARCA, Anti-Reverse Cap Analog; CBP, Cap-Binding Proteins; COVID-19, Coronavirus Infection Disease 2019; CPP, Cell-Penetrating Peptides; IVT, in vitro Transcribed; LNP, lipid nanoparticles; ORF, open reading frame; PAMAM, polyamidoamine; PEI, polyethyleneimine; PGS, polyglucine-spermidine conjugate (spermidine – N1-(3-aminopropyl)butane-1,4-diamine); PRR, pattern-recognition receptors; PABP, poly(A)-binding proteins; RIG-I, retinoic acid-induced protein I; SARS-CoV-2—Severe Acute Respiratory Syndrome Coronavirus-2; TLR, Toll-like receptors; dsRNA, double-stranded RNA; RP HPL, Reversed-Phase High-Performance Liquid Chromatography; PEG, polyethylene glycol (poly(oxyethylene)); saRNA, self-amplifying RNA.
Corresponding author; phone: +7 (383) 363-47-10, 22-29.
Translated by E. Martynova
REFERENCES
- 1.Pardi N., Hogan M.J., Porter F.W., Weissman D. Nat. Rev. Drug Discov. 2018;17:261–279. doi: 10.1038/nrd.2017.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Melton D.A., Krieg P.A., Rebagliati M.R., Maniatis T., Zinn K., Green M.R. Nucleic Acids Res. 1984;12:7035–7056. doi: 10.1093/nar/12.18.7035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wolff J.A., Malone R.W., Williams P., Chong W., Acsadi G., Jani A., Felgner P.L. Science. 1990;247:1465–1468. doi: 10.1126/science.1690918. [DOI] [PubMed] [Google Scholar]
- 4.Jirikowski G.F., Sanna P.P., Maciejewski-Lenoir D., Bloom F.E. Science. 1992;255:996–998. doi: 10.1126/science.1546298. [DOI] [PubMed] [Google Scholar]
- 5.Gómez-Aguado I., Rodríguez-Castejón J., Vicente-Pascual M., Rodríguez-Gascón A., Solinís M.Á. Nanomaterials. 2020;10:364. doi: 10.3390/nano10020364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kwon H., Kim M., Seo Y., Moon Y.S., Lee H.J., Lee K., Lee H. Biomaterials. 2018;156:172–193. doi: 10.1016/j.biomaterials.2017.11.034. [DOI] [PubMed] [Google Scholar]
- 7.Goryaev, A.A., Savkina, M.V., Obukhov, Yu.I., Merkulov, V.A., and Olefir, Yu.V., Bioprep.: Profil., Diagn., Lech., 2019, vol. 19, no. 72–80. 10.30895/2221-996X-2019-19-2-72-80
- 8.Hoerr I., Obst R., Rammensee H.-G., Jung G. Eur. J. Immunol. 2000;30:1–7. doi: 10.1002/1521-4141(200001)30:1<1::AID-IMMU1>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 9.Probst J., Weide B., Scheel B., Pichler B.J., Hoerr I., Rammensee H.-G., Pascolo S. Gene Ther. 2007;14:1175–1180. doi: 10.1038/sj.gt.3302964. [DOI] [PubMed] [Google Scholar]
- 10.Kariko K., Kuo A., Barnathan E.S. Gene Ther. 1999;6:1092–1100. doi: 10.1038/sj.gt.3300930. [DOI] [PubMed] [Google Scholar]
- 11.Karikó K., Ni H., Capodici J., Lamphier M., Weissman D. J. Biol. Chem. 2004;279:12542–12550. doi: 10.1074/jbc.M310175200. [DOI] [PubMed] [Google Scholar]
- 12.Karikó K., Buckstein M., Ni H., Weissman D. Immunity. 2005;23:165–175. doi: 10.1016/j.immuni.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 13.Warren L., Manos P.D., Ahfeldt T., Loh Y.H., Li H., Ebina W., Mandal P.K., Smith Z.D., Meissner A., Daley G.Q., Brack A.S., Collins J.J., Cowan C., Schlaeger T.M., Rossi D.J. Cell Stem Cell. 2010;7:618–630. doi: 10.1016/j.stem.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moderna. Product Pipeline. www.modernatx.com/pipeline.
- 15.Dolgin E. Nature. 2021;597:318–324. doi: 10.1038/d41586-021-02483-w. [DOI] [PubMed] [Google Scholar]
- 16.Corbett K.S., Edwards D.K., Leist S.R., Abiona O.M. Nature. 2020;586:567–571. doi: 10.1038/s41586-020-2622-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vogel A.B., Kanevsky I., Che Y., Swanson K.A., Muik A. Nature. 2021;592:283–289. doi: 10.1038/s41586-021-03275-y. [DOI] [PubMed] [Google Scholar]
- 18.Bitzer M., Armeanu S., Lauer U.M., Neubert W.J. J. Gene Med. 2003;5:543–553. doi: 10.1002/jgm.426. [DOI] [PubMed] [Google Scholar]
- 19.Bloom K., Berg F., Arbuthnot P. Gene Ther. 2021;28:117–129. doi: 10.1038/s41434-020-00204-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mu Z., Haynes B.F., Cain D.W. Vaccines. 2021;9:134. doi: 10.3390/vaccines9020134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Melo M., Porter E., Zhang Y., Silva M., Li N., Dobosh B., Liguori A., Skog P., Landais E., Menis S., Sok D., Nemazee D., Schief W.R., Weiss R., Irvine D.J. Mol. Ther. 2019;27:2080–2090. doi: 10.1016/j.ymthe.2019.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lundstrom K. Viruses. 2021;13:317. doi: 10.3390/v13020317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bulcha J.T., Wang Y., Ma H., Tai P.W.L., Gao G. Sign. Transduct. Target Ther. 2021;6:1–24. doi: 10.1038/s41392-021-00487-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ghosh S., Brown A.M., Jenkins C., Campbell K. Appl. Biosaf. 2020;25:7–18. doi: 10.1177/1535676019899502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Youn H., Chung J.K. Expert Opin. Biol. Ther. 2015;15:1337–1348. doi: 10.1517/14712598.2015.1057563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ogino T., Green T.J. Viruses. 2019;11:504. doi: 10.3390/v11060504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ramanathan A., Robb G.B., Chan S.H. Nucleic Acids Res. 2016;44:7511–7526. doi: 10.1093/nar/gkw551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Linares-Fernandez S., Lacroix C., Exposito J.Y., Verrier B. Trends Mol. Med. 2020;26:311–323. doi: 10.1016/j.molmed.2019.10.002. [DOI] [PubMed] [Google Scholar]
- 29.Henderson J.M., Ujita A., Hill E., Yousif-Rosales S., Smith C., Ko N., McReynolds T., Cabral C.R., Escamilla-Powers J.R., Houston M.E. Curr. Protoc. 2021;1:e39. doi: 10.1002/cpz1.39. [DOI] [PubMed] [Google Scholar]
- 30.Pasquinelli A.E., Dahlberg J.E., Lund E. RNA. 1995;1:957–967. [PMC free article] [PubMed] [Google Scholar]
- 31.Stepinski J., Wandell C., Stolarski R., Darzynkiewicz E., Rhoads R.E. RNA. 2001;7:1486–1495. [PMC free article] [PubMed] [Google Scholar]
- 32.Strenkowska M., Kowalska J., Lukaszewicz M., Zuberek J., Su W., Rhoads R.E., Darzynkiewicz E., Jemielity J. New J. Chem. 2010;34:993–1007. doi: 10.1039/b9nj00644c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sahin U., Muik A., Derhovanessian E., Vogler I., Kranz L.M. Nature. 2020;586:594–599. doi: 10.1038/s41586-020-2814-7. [DOI] [PubMed] [Google Scholar]
- 34.Pascolo S. Viruses. 2021;13:270. doi: 10.3390/v13020270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chang H., Lim J., Ha M., Kim V.N. Mol. Cell. 2014;53:1044–1052. doi: 10.1016/j.molcel.2014.02.007. [DOI] [PubMed] [Google Scholar]
- 36.Li B., Zhang X., Dong Y. WIREs Nanomed. . Nanobiotechnol. 2019;11:e1530. doi: 10.1002/wnan.1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jalkanen A.L., Coleman S.J., Wilusz J. Semin. Cell Dev. Biol. 2014;34:24–32. doi: 10.1016/j.semcdb.2014.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Newbury S.F. Biochem. Soc. Trans. 2006;34:30–34. doi: 10.1042/bst20060030. [DOI] [PubMed] [Google Scholar]
- 39.Klausner R.D., Rouault T.A., Harford J.B. Cell. 1993;72:19–25. doi: 10.1016/0092-8674(93)90046-S. [DOI] [PubMed] [Google Scholar]
- 40.Linares-Fernández S., Moreno J., Lambert E. Mol. Ther. Nucleic Acids. 2021;26:945–956. doi: 10.1016/j.omtn.2021.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Al-Saif M., Khabar K.S.A. Mol. Ther. 2012;20:954–959. doi: 10.1038/mt.2012.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mauro V.P., Chappell S.A. Trends Mol. Med. 2014;20:604–613. doi: 10.1016/j.molmed.2014.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Spencer P.S., Siller E., Anderson J.F., Barral J.M. J. Mol. Biol. 2012;422:328–335. doi: 10.1016/j.jmb.2012.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xia X. Vaccines. 2021;9:734. doi: 10.3390/vaccines9070734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yamamoto A., Kormann M., Rosenecker J., Rudolph C. Eur. J. Pharm. Biopharm. 2009;71:484–489. doi: 10.1016/j.ejpb.2008.09.016. [DOI] [PubMed] [Google Scholar]
- 46.Mu X., Greenwald E., Ahmad S., Hur S. Nucleic Acids Res. 2018;46:5239–5249. doi: 10.1093/nar/gky177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rauch S., Roth N., Schwendt K., Fotin-Mleczek M., Mueller S.O., Petsch B. Vaccines. 2021;6:1–9. doi: 10.1038/s41541-021-00311-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gebre M.S., Rauch S., Roth N., Yu J., Chandrashekar A., Mercado N.B., He X., Liu J., McMahan K., Martinot A., Martinez D.R., Giffin V., Hope D., Patel S., Sellers D., Sanborn O., Barrett J., Liu X., Cole A.C., Pessaint L., Valentin D., Flinchbaugh Z., Yalley-Ogunro J., Muench J., Brown R., Cook A., Teow E., Andersen H., Lewis M.G., Boon A.C.M., Baric R.S., Mueller S.O., Petsch B., Barouch D.H. Nature. 2022;601:410–414. doi: 10.1038/s41586-021-04231-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.CureVac. RNA—Revolution für das Leben. www.curevac.com/en/2021/06/16/curevac-provides-update-on-phase-2b-3-trial-of-first-generation-covid-19-vaccine-candidate-cvncov/.
- 50.Kariko K., Muramatsu H., Ludwig J., Weissman D. Nucleic Acids Res. 2011;39:e142. doi: 10.1093/nar/gkr695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Baiersdörfer M., Boros G., Muramatsu H., Mahiny A., Vlatkovic I., Sahin U., Karikó K. Mol. Ther. Nucleic Acids. 2019;15:26–35. doi: 10.1016/j.omtn.2019.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li S., Ma Z. Curr. Gene Ther. 2001;1:201–226. doi: 10.2174/1566523013348814. [DOI] [PubMed] [Google Scholar]
- 53.Guan S., Rosenecker J. Gene Ther. 2017;24:133–143. doi: 10.1038/gt.2017.5. [DOI] [PubMed] [Google Scholar]
- 54.Reichmuth A.M., Oberli M.A., Jaklenec A., Langer R., Blankschtein D. Ther. Delivery. 2016;7:319–334. doi: 10.4155/tde-2016-0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bahl K., Senn J.J., Yuzhakov O., Bulychev A., Brito L.A., Hassett K.J., Laska M.E., Smith M., Almarsson O., Thompson J., Ribeiro A., Watson M., Zaks T., Ciaramella G. Mol. Ther. 2017;25:1316–1327. doi: 10.1016/j.ymthe.2017.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Varkouhi A.K., Scholte M., Storm G., Haisma H.J. J. Controlled Release. 2011;151:220–228. doi: 10.1016/j.jconrel.2010.11.004. [DOI] [PubMed] [Google Scholar]
- 57.Gilleron J., Querbes W., Zeigerer A., Borodovsky A., Marsico G., Schubert U., Manygoats K., Seifert S., Andree C., Stoter M., Epstein-Barash H., Zhang L., Koteliansky V., Fitzgerald K., Fava E., Bickle M., Kalaidzidis Y., Akinc A., Maier M., Zerial M. Nat. Biotechnol. 2013;31:638–646. doi: 10.1038/nbt.2612. [DOI] [PubMed] [Google Scholar]
- 58.Li M., Li Y., Li S., Jia L., Wang H., Li M., Deng J., Zhub A., Mab L., Lib W., Yua P., Zhu T. Eur. J. Med. Chem. 2022;227:113910. doi: 10.1016/j.ejmech.2021.113910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schoenmaker L., Witzigmann D., Kulkarni J.A., Verbeked R., Kerstenae G., Jiskootae W., Crommelin D.J.A. Int. J. Pharm. 2021;601:120586. doi: 10.1016/j.ijpharm.2021.120586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen G.L., Li X.F., Dai X.H., Li N., Cheng M.L., Huang Z. Lancet Microbe. 2022;3:E193–E202. doi: 10.1016/S2666-5247(21)00280-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pardi N., Tuyishime S., Muramatsu H., Kariko K., Mui B.L., Tam Y.K., Madden T.D., Hope M.J., Weissman D. J. Controlled Release. 2015;217:345–351. doi: 10.1016/j.jconrel.2015.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yi XueH. Guo, P., Wen, W.-C., and Lun, WongH. Curr. Pharm. Des. 2015;21:3140–3147. doi: 10.2174/1381612821666150531164540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sedic M., Senn J.J., Lynn A., Laska M., Smith M., Platz S.J., Bolen J., Hoge S., Bulychev A., Jacquinet E., Bartlett V., Smith P.F. Vet. Pathol. 2018;55:341–354. doi: 10.1177/0300985817738095. [DOI] [PubMed] [Google Scholar]
- 64.Hou X., Zaks T., Langer R., Dong Y. Nat. Rev. Mater. 2021;6:1078–1094. doi: 10.1038/s41578-021-00358-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhuang X., Qi Y., Wang M., Yu N., Nan F., Zhang H., Tian M., Li C., Lu H., Jin N. Vaccines. 2020;8:123. doi: 10.3390/vaccines8010123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chang H.I., Yeh M.K. Int. J. Nanomed. 2012;7:49. doi: 10.2147/IJN.S26766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ball R.L., Bajaj P., Whitehead K.A. Int. J. Nanomed. 2017;12:305. doi: 10.2147/IJN.S123062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dong Y., Dorkin J.R., Wang W., Chang P.H., Webber M.J., Tang B.C., Yang J., Abutbul-Ionita I., Danino D., DeRosa F., Heartlein M., Langer R., Anderson D.G. Nano Lett. 2016;16:842–848. doi: 10.1021/acs.nanolett.5b02428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kowalski P.S., Palmiero U.C., Huang Y., Rudra A., Langer R., Anderson D.G. Adv. Mater. 2018;30:1801151. doi: 10.1002/adma.201801151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mohammed M.A., Syeda J.T.M., Wasan K.M., Wasan E.K. Pharmaceutics. 2017;9:53. doi: 10.3390/pharmaceutics9040053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Moura L.I.F., Malfanti A., Peres C., Matos A.I., Guegain E., Sainz V., Zloh M., Vicent M.J., Florindo H.F. Mater. Horiz. 2019;6:1956–1973. doi: 10.1039/c9mh00628a. [DOI] [Google Scholar]
- 72.Chauhan A.S. Molecules. 2018;23:938. doi: 10.3390/molecules23040938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Islam M.A., Xu Y., Tao W., Ubellacker J.M., Lim M. Nat. Biomed. Eng. 2018;2:850–864. doi: 10.1038/s41551-018-0284-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Borgoyakova M.B., Karpenko L.I., Rudometov A.P., Volosnikova E.A., Merkuleva I.A., Starostina E.V., Zadorozhny A.M., Isaeva A.A., Nesmeyanova V.S., Shanshin D.V., Baranov K.O., Volkova N.V., Zaitsev B.N., Orlova L.A., Zaykovskaya A.V., Pyankov O.V., Danilenko E.D., Bazhan S.I., Shcherbakov D.N., Taranin A.V., Ilyichev A.A. Int. J. Mol. Sci. 2022;23:2188. doi: 10.3390/ijms23042188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lebedev L.R., Karpenko L.I., Poryvaeva V.A., Azaev M.S., Ryabchikova E.I., Gileva I.P., Ilyichev A.A. Mol. Biol. 2000;34:413–417. doi: 10.1007/BF02759674. [DOI] [PubMed] [Google Scholar]
- 76.Karpenko L.I., Bazhan S.I., Bogryantseva M.P., Ryndyuk N.N., Ginko Z.I., Kuzubov V.I., Lebedev L.R., Kaplina O.N., Reguzova A.Yu., Ryzhikov A.B., Usova S.V., Oreshkova S.F., Nechaeva E.A., Danilenko E.D., Ilyichev A.A. Russ. J. Bioorg. Chem. 2016;42:170–182. doi: 10.1134/S1068162016020060. [DOI] [Google Scholar]
- 77.Singh D.V., Singh R., Sodhi S.P.S. Vet. Res. Commun. 2005;29:421–430. doi: 10.1007/s11259-005-1434-x. [DOI] [PubMed] [Google Scholar]
- 78.Perepelytsya S., Ulicny J., Laaksonen A., Mocci F. Nucleic Acids Res. 2019;47:6084–6097. doi: 10.1093/nar/gkz434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lightfoot H.L., Hall J. Nucleic Acids Res. 2014;42:11275–11290. doi: 10.1093/nar/gku837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Karpenko L.I., Rudometov A.P., Sharabrin S.V., Shcherbakov D.N., Borgoyakova M.B., Bazhan S.I., Volosnikova E.A., Rudometova N.B., Orlova L.A., Pyshnaya I.A., Zaitsev B.N., Volkova N.V., Azaev M.Sh., Zaykovskaya A.V., Pyankov O.V., Ilyichev A.A. Vaccines. 2021;9:76. doi: 10.3390/vaccines9020076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ponsaerts P., Der Sar S.V., Van Tendeloo V.F.I., Jorens P.G, Berneman Z.N., Singh P.B. Cloning Stem Cells. 2004;6:211–216. doi: 10.1089/clo.2004.6.211. [DOI] [PubMed] [Google Scholar]
- 82.Campillo-Davo D., De Laere M., Roex G., Versteven M., Flumens D., Berneman Z.N., Van Tendeloo V.F.I., Anguille S., Lion E. Pharmaceutics. 2021;13:396. doi: 10.3390/pharmaceutics13030396. [DOI] [PMC free article] [PubMed] [Google Scholar]