Abstract
Radical S-adenosylmethionine (SAM) enzymes use a common catalytic core for diverse transformations. While all radical SAM enzymes bind a Fe4S4 cluster via a characteristic tri-cysteine motif, many bind additional metal cofactors. Recently reported structures of radical SAM enzymes that use methylcobalamin or additional iron-sulfur clusters as cosubstrates show that these auxiliary units are anchored by N- and C-terminal domains that vary significantly in size and topology. Despite this architectural diversity, all use a common surface for auxiliary cofactor docking. In the sulfur insertion and metallocofactor assembly systems evaluated here, interaction with iron-sulfur cluster assembly proteins or downstream scaffold proteins is an important component of catalysis. Structures of these complexes represent important new frontiers in structural analysis of radical SAM enzymes.
1. Introduction
Radical SAM enzymes are versatile catalysts found throughout all three kingdoms of life [1,2]. All deploy SAM in initiating remarkably diverse transformations that are generally radical-mediated [2]. These catalysts additionally bind a Fe4S42+ cluster, typically via a CX3CX2C motif found within an α6β6 core fold [2,3]. SAM coordinates the fourth iron. After one-electron reduction of the iron-sulfur cluster by a protein partner or chemical reductant, SAM is reductively cleaved to generate a 5′-deoxyadenosyl 5′-radical (5′-dA•). The 5′-dA• intermediate is a potent oxidant capable of initiating hydrogen atom transfer (HAT) reactions with substrates or co-substrates, generally from unactivated carbons. Recent efforts to trap and characterize the 5′-dA• [4-6] have revealed that this key intermediate may be initially caged as an organometallic complex, Ω, containing an Fe-C5′ covalent bond between the 5′-dA• and the unique iron of the Fe4S4 cofactor [4]. Studies of synthetic models of Ω suggest that it might function to confer selectivity in reaction outcome for radical SAM enzymes by sequestering the 5′-dA• until substrates can be properly positioned for catalysis [7].
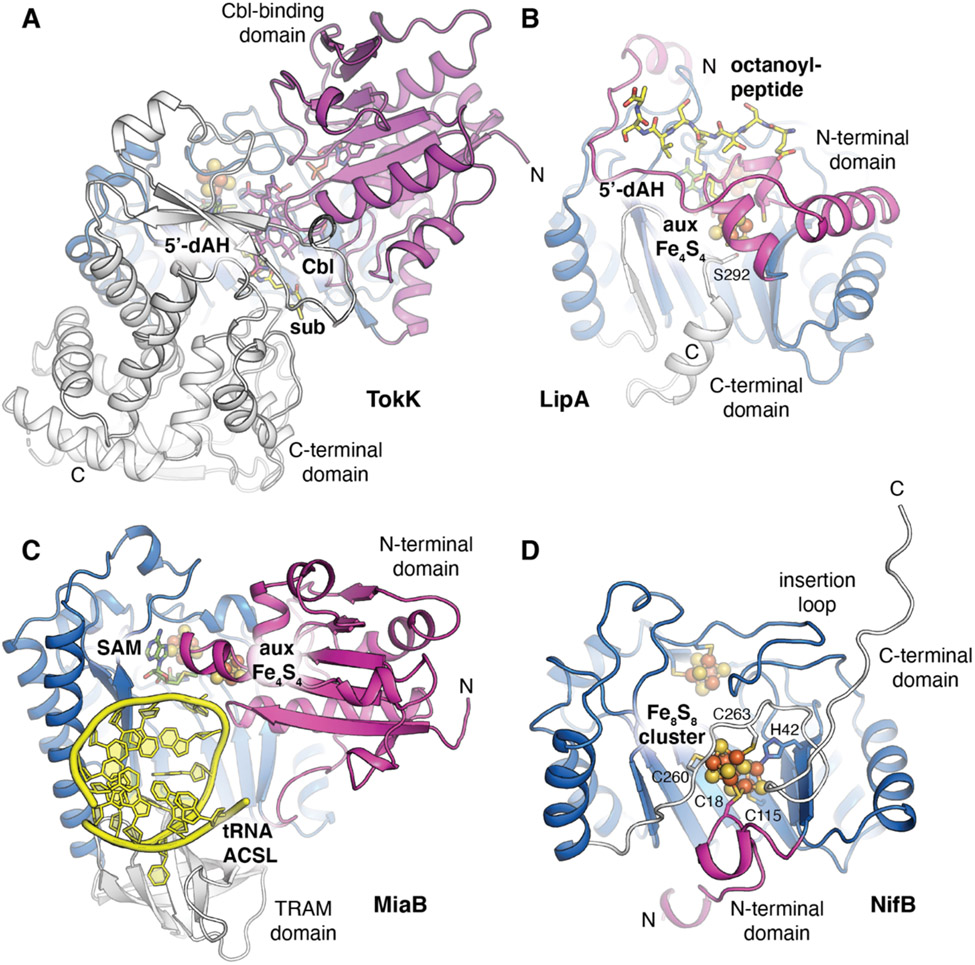
Many radical SAM enzymes bind additional transitional metals [8]. A number have been structurally characterized, including those with one or more additional iron-sulfur clusters [9-18], individual metal ions [19], or complex cofactors such as methylcobalamin [20]. Structural characterization of radical SAM enzymes containing auxiliary cofactors that are proposed to perform simple tasks, such as electron transfer [9,10,14,15] or substrate binding [16], reveals that dedicated domains are required to bind and utilize these entities [21]. Here we describe insights from recent structural characterization of four radical SAM enzymes that use additional metallocofactors for complex functionalization of substrates (Fig. 1). These include TokK, one of the first structurally characterized radical SAM enzyme that performs radical transformations involving a cobalamin (Cbl) cofactor [22]. Here the auxiliary cofactor serves as a functional group donor in a chemically challenging carbon methylation. We also review biochemical and structural analyses of two radical SAM sulfur insertion enzymes, lipoic acid synthase (LipA) [12] and MiaB [23]. Finally, we discuss structures of NifB, a radical SAM enzymes involved in complex cofactor biosynthesis. NifB is a methylase that fuses two auxiliary [4Fe-4S] clusters and facilitates insertion of an interstitial carbide [24,25] in construction of the nitrogenase active site cofactor [26]. Despite their structurally distinct auxiliary motifs and divergent reactivity, all of these enzymes use a common surface of the radical SAM core fold for docking additional cofactors. Several enzymes summarized here additionally employ unusual ligands to their auxiliary cofactors, and this strategy is likely key in enabling the chemically challenging reactivities of these systems.
Figure 1.
Reactions catalyzed by radical SAM enzymes with auxiliary metallocofactors discussed in this review.
2.1. Cobalamin-dependent radical SAM methylase, TokK.
TokK methylates an unactivated carbon within the β-lactam core of asparenomycin A, a potent carbapenem antibiotic [27,28]. The products generated by TokK suggest that the enzyme uses a 5′-dA• to activate its substrate via HAT. The resulting substrate radical then attacks the methyl substituent of methyl-Cbl. TokK can perform serial methylations of its substrate [28], adding up to three methyl units to elaborate the C6 position of the β-lactam. This modification enhances the potency of carbapenems by preventing inactivation by β-lactamases [29].
TokK employs an N-terminal Rossmann-fold domain (Fig. 2) to dock the Cbl cofactor adjacent to the radical SAM domain [22], as observed previously in the structure of OxsB [20]. Cbl sits on top of the β2 secondary structure in the radical SAM core (Fig. 3), ~10 Å away from the SAM binding site. The enzyme also contains a third C-terminal domain that does not interact with Cbl but instead forms a substrate binding channel. TokK binds Cbl in the base-off conformation, an approach that allows for modulation of the coordination environment of the Co(III) ion by the protein environment. TokK lacks a lower axial ligand to the Co ion, using W76 to block solvent access underneath the cofactor. This arrangement likely renders the Co-C bond in MeCbl more reactive by preventing coordination with a sixth ligand.
Figure 2.
A comparison of the domain architectures of radical SAM enzymes with auxiliary metallocofactors. The enzymes are colored by domain with the radical SAM core fold shown in blue. Substrates are shown as yellow sticks and cosubstrate SAM or its cleavage product (5′-dAH) are shown as green sticks. (A) Cobalamin-(Cbl)-dependent radical SAM enzyme, TokK, involved in carbapenem (sub) methylation (PDB ID 7KDY). (B) Lipoyl synthase, LipA, involved in sulfur insertion into a protein-linked fatty acid substrate (PDB ID 5EXK). (C) RNA modification enzyme, MiaB, implicated in methylthiolation of an adenine base in the anticodon stem loop (ACSL) of tRNA (PDB ID 7MJV). (D) Nitrogenase cofactor biosynthesis enzyme, NifB, involved in carbide and sulfur insertion to generate the Fe8S9C NifB-co product (PDB ID 7BI7). Selected amino acids are shown in stick format.
Figure 3.
Comparison of the metallocofactor and/or cosubstrate binding sites within the α6β6 radical SAM core domain reveals a common auxiliary cofactor binding site near β2 adjacent to the radical SAM Fe4S4 cluster. (C) TokK binds Cbl near the substrate pocket opposite the SAM cosubstrate binding site (PDB ID 7KDY). (D) LipA binds an auxiliary Fe4S4 cluster adjacent to the octanoyl-peptide binding site (PDB ID 5EXK). (E) NifB binds a Fe8S8 cluster near the N-terminal side of the radical SAM core fold (PDB ID 7B17). (F) MiaB binds an auxiliary Fe4S4 cluster near the SAM binding site above the tRNA docking interface (PDB ID 7MJV).
The arrangement of reactive groups in the active site of TokK was revealed by characterization of the enzyme bound to its carbapenam substrate and coproducts of the SAM cleavage reaction, 5′-dA and methionine [22]. Whereas several other Cbl-dependent radical SAM enzymes had been structurally characterized previously [20,30], none contained the primary substrate bound in a productive conformation. The carbapenem threads through a long channel at the interface between the three domains of TokK [22]. The β-lactam binds adjacent to the cobalamin, with C6 position ~4 Å away from the top ligand of the Co ion and the 5′-C of the 5′-dA. The arrangement demands that methyl transfer occur at the bottom face of the β-lactam, consistent with the known stereochemistry of the C6 substituent. The structure additionally implies that the TokK active site architecture primarily serves to position substrates and cofactors for radical methyl addition with very little intervention from other amino acids.
In this structure (and nearly all others of Cbl-dependent radical SAM enzymes) [20,22,30], the top ligand of cobalamin is modeled as a solvent-derived water or hydroxide, rather than the native methyl component of cobalamin. This substitution could alter the internal water network near the top ligand, thereby modifying the electrostatic environment of the substrate binding pocket. Incorporation of the bona fide Cbl methyl ligand could enable solution of additional structures with substrates bound. Interestingly, concomitant with the recent report of the TokK enzyme substrate complex, Mmp10, a peptide-modifying Cbl-dependent radical SAM enzyme was characterized in complex with its substrate [31]. This structure revealed analogous distances between reactive groups to those found in TokK. Mmp10 has a distinctive domain architecture compared to other Cbl-dependent radical SAM enzymes, including a different fold for the Cbl-binding domain and a different docking site within the radical SAM core for the Cbl cofactor. Mmp10 was reportedly cocrystallized with MeCbl, which may have promoted substrate incorporation in this system. In the absence of substrate and SAM, Mmp10 appears to bind an unusual Tyr ligand to the unique iron of the radical SAM cluster. The authors suggest that this interaction could be important for re-methylation of Cbl, a phenomenon that is not well understood in any Cbl-dependent radical SAM enzyme. Additional structural and spectroscopic characterization of these unusual interactions will be critical for experimental validation of this proposal.
2.2. Sulfur insertion radical SAM enzymes with auxiliary iron-sulfur clusters.
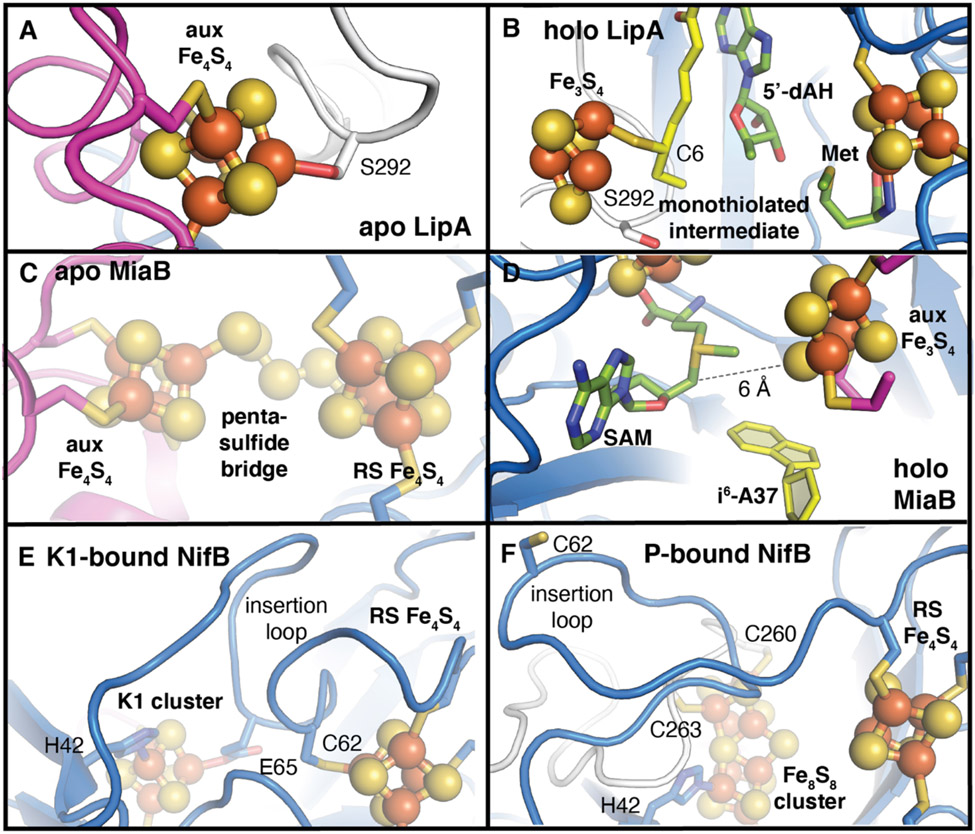
All radical SAM enzymes that insert sulfur into unactivated carbon centers contain additional iron-sulfur clusters that are proposed to serve as the sulfur source in these enzymes [8]. Several recent landmark studies by Booker and colleagues [12,32,33] have provided the comprehensive experimental evidence for such a role in the vitamin biosynthetic enzyme, LipA. LipA converts a carrier-protein-linked octanoyl side chain to lipoyl cofactor [34], which contains two thiol substituents at C6 and C8 (Fig. 1). The transformation proceeds as two distinct SAM-dependent transformations, with a 5′-dA• intermediate targeting C6 initially. Arrest of the reaction after the first step yields a cross-linked enzyme-substrate complex [32]. Analysis of this form of the enzyme by Mössbauer spectroscopy [32] and x-ray crystallography [12] revealed conversion of the auxiliary Fe4S4 cofactor to a Fe3S4 complex (Fig. 4). Comparison to structures solved in the absence of substrate shows that the lost iron in the auxiliary cluster was originally coordinated by a serine side chain located in a conserved C-terminal sequence motif. These interactions yield weaker bonds between the cofactor and the protein, likely enabling cofactor disassembly for sulfur donation.
Figure 4.
Zoomed-in views of the auxiliary iron-sulfur clusters in the sulfur-/carbide-insertion enzymes show the unusual metal binding motifs employed in the apo forms of each enzyme. (A, B) LipA uses a Ser ligand to the auxiliary Fe4S4 cluster (PDB ID 5EXJ) to facilitate formation of a monothiolated intermediate in which C6 of the octanoyl substrate side chain has added to a sulfide of the auxiliary cluster (PDB ID 5EXK). (C, D) MiaB contains a pentasulfide bridge between the unique irons of the auxiliary and radical SAM Fe4S4 clusters (PDB ID 7MJZ). Binding of SAM and the tRNA substrate results in a complex poised for methylation of sulfide in the auxiliary cluster and subsequent transfer to C2 of i6-A37 (PDB ID 7MJV). (E, F) In K1-bound NifB, an insertion loop in the radical SAM core contributes Glu and Cys ligands to the K1 and radical SAM Fe4S4 clusters, respectively (PDB ID 6Y1X). These unusual coordination interactions are disrupted upon occupancy of the enzyme with a Fe8S8 cluster (PDB ID 7B17).
A second comprehensive structural study of a radical SAM sulfur insertion enzyme was published earlier this year [23]. Methylthiotransferase, MiaB, responsible for synthesis of a methylthio substituent at C2 of a hypermodified adenine base in transfer RNA [35], was characterized bound to an RNA substrate in several different stages of its reaction cycle [23]. MiaB contains an auxiliary Fe4S4 cluster bound within a large N-terminal domain (Fig. 2). As in TokK and LipA, this cofactor docks against the N-terminal side of the radical SAM core (Fig. 3). Surprisingly, the structure and size of this domain differs significantly between LipA and MiaB. In LipA, the auxiliary cluster is coordinated by much smaller domains at the N- and C-termini (Fig. 2). However, MiaB shares with LipA an unusual cluster binding motif in its apo form. The auxiliary Fe4S4 cluster has a unique iron site that is linked to the radical SAM cluster via a polysulfide bridge in the apo state but becomes open upon SAM and RNA binding (Fig. 4).
Structures of MiaB with a small RNA fragment bound reveal a substrate binding site located directly underneath the two Fe4S4 cluster cofactors, approximately equidistant from the 5′-C of the 5′-dAH and one of the sulfide ions of the auxiliary cluster. The structures are consistent with a two-step mechanism for methylthiolation. In a first step, SAM methylates the auxiliary cluster at the outermost sulfide ion, yielding a Fe3S4 intermediate [36]. Attack of the methylated thiol upon the RNA substrate yields a transient enzyme-substrate intermediate that is proposed to be resolved by reductive cleavage of a second equivalent of SAM.
Observation of auxiliary cluster degradation during catalysis would, in theory, limit LipA and MiaB to single turnover reactions. Recently, two iron-sulfur cluster carrier proteins, NfuA and IscU, were shown to reconstitute the degraded auxiliary cluster in the LipA reaction [33,37], enabling multiple turnovers. Structural characterization of these carrier proteins in complex with LipA (and other sulfur insertion enzymes) will be an important next step in understanding the mechanism of iron-sulfur cluster reconstitution. Many other open questions remain about the structure and metal binding mode of the carrier proteins themselves. NfuA has been characterized by NMR spectroscopy, providing information about its overall fold [38], but little insight into how it binds its iron-sulfur cluster substrate and transfers it to target enzymes. Several crystal structures of IscU bound to a Fe2S2 cluster have been reported [39,40]. While the cluster binding motif remains ambiguous, the Fe2S2 unit is most likely coordinated by three cysteines and either an aspartic acid or a histidine at the fourth site. Observation of site differentiation in both the auxiliary cofactor of the target enzyme (LipA) and one of its partner iron-sulfur cluster carrier proteins suggests that this feature could play a role in cluster assembly and degradation.
2.3. Radical SAM enzymes in nitrogenase cofactor biosynthesis.
Radical SAM enzymes with auxiliary cofactors are additionally involved in complex cofactor assembly [2]. For example, a precursor to FeMo-co of nitrogenase is synthesized upon the radical SAM enzyme, NifB [41]. The core of mature FeMo-co contains two Fe4S3 subunits connected via three μ2 sulfide ions and one μ6 carbide ion [26]. The early stages of FeMo-co assembly require two proteins, NifB and NifEN [42,43]. NifB fuses two simple Fe4S4 clusters (K1 and K2) and inserts an additional sulfide and carbide to form a Fe8S9C product (Fig. 1), also known as NifB-co. NifB-co is subsequently transferred to scaffold protein NifEN for further maturation to give the active FeMo-co.
Two recently reported x-ray crystal structures of NifB provide initial insight into how two simple Fe4S4 clusters (K1 and K2) might be converted into NifB-co [24,25]. Fajardo et al. solved a crystal structure of NifB (PDB ID: 6Y1X) that shows the relative locations of the radical SAM iron-sulfur cluster and one of the two auxiliary [4Fe-4S] clusters, K1 [24]. The K2 cluster is missing in this dataset, along with >20 amino acids at the C-terminus of NifB. The K1 cluster is coordinated by two Cys side chains, a His, and a Glu. These ligands are found on the N-terminal side of the radical SAM core or in NifB-specific structural features (Fig. 2) that include a small N-terminal domain and insertion loop between β1 and β2 of the radical SAM fold. K1 binds near β2 in the core, similar to the auxiliary cluster binding sites in TsrM, MiaB, and LipA (Fig. 3).
The radical SAM cluster of NifB also exhibits unusual four-Cys coordination. Three ligands are provided by the predicted CX3CX2C cluster signature motif, but a fourth Cys (C62) is distinct to NifB, and binds to the iron that is typically “open” for SAM to bind (Fig. 4). C62 is in the insertion loop that also contains the Glu ligand to K1. The radical SAM binding motif is reminiscent of that of TsrM, which uses a Glu to bind the fourth iron. However, unlike TsrM, NifB is a dual-purpose enzyme. In NifB, SAM is used both as a methyl donor and for hydrogen atom abstraction via reductive cleavage [43]. Methyl donation by SAM provides the interstitial carbide in NifB-co. But NifB produces both S-adenosylhomocysteine (SAH) and 5′-dAH as byproducts, indicating that a second SAM molecule is subsequently used to produce the 5′-dA•. The exact mechanism of carbide insertion by NifB remains unknown. The structure of NifB shows that the distance between the radical SAM cluster and K1 [24] is 12 Å, likely too far for direct methyl transfer to K1. The authors propose that the K2 cluster must therefore be the initial target for methylation and it probably resides in between the two iron-sulfur clusters observed in the structure.
A second structure of NifB was reported by Kang et al. [25], purported to contain the radical SAM cluster and both K1 and K2 clusters (PDB ID 7JMB). This structure would seem to show the K2 binding site and coordinating side chains. However, due to modest resolution (3.0 Å) and poor fit of the auxiliary cluster models to both omit and anomalous difference electron density maps, information about these features of the active site is limited. Refinement of this NifB structure by another research group [44] indicated that the electron density assigned to the auxiliary cofactors instead likely corresponds to a P-cluster-like fused Fe8S8 cluster with a μ2-bridging sulfide ion (PDB ID: 7BI7). The significance of this cluster form as a possible intermediate in the NifB reaction remains unclear, although the bridging sulfide resides proximal to the radical SAM iron-sulfur cluster (Fig. 4), consistent with its initial target for methylation. Occupancy of the K2 binding site is accompanied by ordering of the C-terminus, which could provide additional ligands for binding a second auxiliary cluster. The insertion loop also changes conformation to move away from the cavity between the radical SAM cluster and K1. Important open questions remain about the oxidation states of the K1 and K2 clusters, both in the x-ray crystal structures and during catalysis. Finally, the SAM binding site remains to be elucidated in any structure of NifB. Given the dual role for SAM in NifB, structures in complex with this cosubstrate will be particularly important for understanding the mechanism of carbide insertion and cluster fusion.
3. Conclusions and outlook
Our analysis of recently reported x-ray crystal structures of radical SAM methylases and sulfur insertion enzymes highlights a common binding site for their auxiliary iron-sulfur cluster and cobalamin cofactors (or cosubstrates). All use the N-terminal side of the α6β6 radical SAM core fold to anchor the auxiliary moiety opposite the SAM binding site. Interestingly, this general binding location is shared by several other radical SAM enzymes that use auxiliary iron-sulfur clusters for electron transfer or substrate binding such as MoaA [16] and SuiB [14] (Fig. 2). Many of the enzymes discussed here also require interface with iron-sulfur-cluster assembly proteins. Use of a common auxiliary cluster binding site could provide a common docking surface for partner proteins. Structural characterization of these protein-protein complexes remains an important goal for future work. Additionally, we show that auxiliary domains at the N- and C-termini contribute important ligands to the auxiliary cofactors in these systems. The auxiliary metal binding motifs often include labile ligands that are likely responsible for the unique reactivities of the sulfur and carbon modification reactions catalyzed by this subset of radical SAM enzymes.
Acknowledgements.
Work by the authors on radical SAM enzymes has been supported by the National Institutes of Health (GM119707), the National Science Foundation (MCB-1158486), and the Howard Hughes Medical Institute.
Footnotes
Statement of competing interests. The authors declare no competing interests.
References
• of special interest
•• of outstanding interest
- 1.Holliday GL, Akiva E, Meng EC, Brown SD, Calhoun S, Pieper U, Sali A, Booker SJ, Babbitt PC: Atlas of the radical SAM superfamily: divergent evolution of function using a “plug and play" domain. Methods Enzymol 2018, 606:1–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Broderick JB, Duffus BR, Duschene KS, Shepard EM: Radical S-adenosylmethionine enzymes. Chem Rev 2014, 114:4229–4317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vey JL, Drennan CL: Structural insights into radical generation by the radical SAM superfamily. Chem Rev 2011, 111:2487–2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •4. Horitani M, Shisler K, Broderick WE, Hutcheson RU, Duschene KS, Marts AR, Hoffman BM, Broderick JB: Radical SAM catalysis via an organometallic intermediate with an Fe-[5'-C]-deoxyadenosyl bond. Science 2016, 352:822–825. The first spectroscopic report of an organometallic transient state (Ω) in which the 5' carbon of a deoxyadenosyl moiety forms a bond with the unique iron site of the [4Fe-4S] cluster.
- 5.Sayler RI, Stich TA, Joshi S, Cooper N, Shaw JT, Begley TP, Tantillo DJ, Britt RD: Trapping and electron paramagnetic resonance characterization of the 5’-dAdo radical in a radical S-adenosylmethionine enzyme reaction with a non-native substrate. ACS Cent Sci 2019, 5:1777–1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang H, McDaniel EC, Impano S, Byer AS, Jodts RJ, Yokoyama K, Broderick WE, Broderick JB, Hoffman BM: The elusive 5’-deoxyadenosyl radical: captured and characterized by electron paramagnetic resonance and electron nuclear double resonance spectroscopies. J Am Chem Soc 2019, 141:12139–12146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brown AC, Suess DLM: Reversible formation of alkyl radicals at [Fe4S4] clusters and its implications for selectivity in radical SAM enzymes. J Am Chem Soc 2020, 142:14240–14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lanz ND, Booker SJ: Auxiliary iron-sulfur cofactors in radical SAM enzymes. Biochim Biophys Acta 2015, 1853:1316–1334. [DOI] [PubMed] [Google Scholar]
- 9.Goldman PJ, Grove TL, Booker SJ, Drennan CL: X-ray analysis of butirosin biosynthetic enzyme BtrN redefines structural motifs for AdoMet radical chemistry. Proc Natl Acad Sci U S A 2013, 110:15949–15954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goldman PJ, Grove TL, Sites LA, McLaughlin MI, Booker SJ, Drennan CL: X-ray structure of an AdoMet radical activase reveals an anaerobic solution for formylglycine posttranslational modification. Proc Natl Acad Sci U S A 2013, 110:8519–8524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harmer JE, Hiscox MJ, Dinis PC, Fox SJ, Iliopoulos A, Hussey JE, Sandy J, Van Beek FT, Essex JW, Roach PL: Structures of lipoyl synthase reveal a compact active site for controlling sequential sulfur insertion reactions. Biochem J 2014, 464:123–133. [DOI] [PubMed] [Google Scholar]
- ••12. McLaughlin MI, Lanz ND, Goldman PJ, Lee KH, Booker SJ, Drennan CL: Crystallographic snapshots of sulfur insertion by lipoyl synthase. Proc Natl Acad Sci U S A 2016, 113:9446–9450. The first crystallographic capture of an intermediate in the lipoyl synthase reaction. This structure shows that the enzyme obtains its sulfur atoms by cannibalizing its auxiliary Fe4S4 cluster.
- 13.Berkovitch F, Nicolet Y, Wan JT, Jarrett JT, Drennan CL: Crystal structure of biotin synthase, an S-adenosylmethionine-dependent radical enzyme. Science 2004, 303:76–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Davis KM, Schramma KR, Hansen WA, Bacik JP, Khare SD, Seyedsayamdost MR, Ando N: Structures of the peptide-modifying radical SAM enzyme SuiB elucidate the basis of substrate recognition. Proc Natl Acad Sci U S A 2017, 114:10420–10425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grove TL, Himes PM, Hwang S, Yumerefendi H, Bonanno JB, Kuhlman B, Almo SC, Bowers AA: Structural insights into thioether bond formation in the biosynthesis of sactipeptides. J Am Chem Soc 2017, 139:11734–11744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hanzelmann P, Schindelin H: Crystal structure of the S-adenosylmethionine-dependent enzyme MoaA and its implications for molybdenum cofactor deficiency in humans. Proc Natl Acad Sci U S A 2004, 101:12870–12875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dinis P, Suess DL, Fox SJ, Harmer JE, Driesener RC, De La Paz L, Swartz JR, Essex JW, Britt RD, Roach PL: X-ray crystallographic and EPR spectroscopic analysis of HydG, a maturase in [FeFe]-hydrogenase H-cluster assembly. Proc Natl Acad Sci U S A 2015, 112:1362–1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rohac R, Martin L, Liu L, Basu D, Tao L, Britt RD, Rauchfuss TB, Nicolet Y: Crystal structure of the [FeFe]-hydrogenase maturase HydE bound to complex-B. J Am Chem Soc 2021, 143:8499–8508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dowling DP, Bruender NA, Young AP, McCarty RM, Bandarian V, Drennan CL: Radical SAM enzyme QueE defines a new minimal core fold and metal-dependent mechanism. Nat Chem Biol 2014, 10:106–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bridwell-Rabb J, Zhong A, Sun HG, Drennan CL, Liu HW: A B12-dependent radical SAM enzyme involved in oxetanocin A biosynthesis. Nature 2017, 544:322–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grell TA, Goldman PJ, Drennan CL: SPASM and twitch domains in S-adenosylmethionine (SAM) radical enzymes. J Biol Chem 2015, 290:3964–3971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••22. Knox HL, Sinner EK, Townsend CA, Boal AK, Booker SJ: Structure of a B12-dependent radical SAM enzyme in carbapenem biosynthesis. Nature 2022, 602:343–348. This paper describes one of the first X-ray crystal structures of a Cbl-dependent (Class B) radical SAM enzyme in complex with its substrate. This work reveals that the active site primarily functions to position the Cbl, substrate, and SAM cosubstrate for radical methylation chemistry.
- ••23. Esakova OA, Grove TL, Yennawar NH, Arcinas AJ, Wang B, Krebs C, Almo SC, Booker SJ: Structural basis for tRNA methylthiolation by the radical SAM enzyme MiaB. Nature 2021, 597:566–570. This paper reports the first x-ray crystal structures of MiaB bound to its tRNA substrate. Multiple structures of reactant and intermediate states of catalysis give further support to use of the auxiliary cluster as the sulfur source and are consistent with a two-step reaction mechanism in which SAM methylates the auxiliary cluster then transfers a methyl-thio group to the RNA substrate.
- ••24. Fajardo AS, Legrand P, Paya-Tormo LA, Martin L, Pellicer Marti Nez MT, Echavarri-Erasun C, Vernede X, Rubio LM, Nicolet Y: Structural insights into the mechanism of the radical SAM carbide synthase NifB, a key nitrogenase cofactor maturating enzyme. J Am Chem Soc 2020, 142:11006–11012. The first reported X-ray structure of NifB with the K1 cluster bound. The structure showed that the unique iron in the radical SAM Fe4S4 cluster is coordinated by a cysteine in an insertion loop, a motif that potentially controls the dual reactivity of SAM in NifB. While structures lack both SAM and the K2 cluster, they suggest that K2 is the initial target for reaction with SAM due to the long distance between K1 and the radical SAM Fe4S4 cluster.
- •25. Kang W, Rettberg LA, Stiebritz MT, Jasniewski AJ, Tanifuji K, Lee CC, Ribbe MW, Hu Y: X-Ray crystallographic analysis of NifB with a full complement of clusters: structural insights into the radical SAM-dependent carbide insertion during nitrogenase cofactor assembly. Angew Chem Int Ed Engl 2021, 60:2364–2370. This paper describes x-ray crystal structures NifB with three Fe4S4 clusters modeled. The radical SAM cluster was coordinated by three Cys, with the unique iron open. Adjacent electron density is modeled as the K1- and K2-cluster, each coordinated by one His and two Cys. However, a subsequent study showed that this feature models best as a Fe8S8 species of unknown significance in the NifB reaction
- 26.Einsle O, Rees DC: Structural enzymology of nitrogenase enzymes. Chem Rev 2020, 120:4969–5004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li R, Lloyd EP, Moshos KA, Townsend CA: Identification and characterization of the carbapenem MM 4550 and its gene cluster in Streptomyces argenteolus ATCC 11009. Chembiochem 2014, 15:320–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •28. Sinner EK, Lichstrahl MS, Li R, Marous DR, Townsend CA: Methylations in complex carbapenem biosynthesis are catalyzed by a single cobalamin-dependent radical S-adenosylmethionine enzyme. Chem Commun (Camb) 2019, 55:14934–14937. This manuscript established the activity of Cbl-dependent radical SAM methylase TokK, showing that the enzyme is capable of performing mutlple serial methylations upon a carbapenam substrate.
- 29.Fisher JF, Mobashery S: Three decades of the class A beta-lactamase acyl-enzyme. Curr Protein Pept Sci 2009, 10:401–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Knox HL, Chen PY, Blaszczyk AJ, Mukherjee A, Grove TL, Schwalm EL, Wang B, Drennan CL, Booker SJ: Structural basis for non-radical catalysis by TsrM, a radical SAM methylase. Nat Chem Biol 2021, 17:485–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••31. Fyfe CD, Bernardo-Garcia N, Fradale L, Grimaldi S, Guillot A, Brewee C, Chavas LMG, Legrand P, Benjdia A, Berteau O: Crystallographic snapshots of a B12-dependent radical SAM methyltransferase. Nature 2022, 602:336–342. This manuscript reports one of the first structures of a Cbl-dependent radical SAM enzyme in complex with its primary substrate. It shows a different domain architecture and Cbl binding site when compared with TokK.
- 32.Lanz ND, Rectenwald JM, Wang B, Kakar ES, Laremore TN, Booker SJ, Silakov A: Characterization of a radical intermediate in lipoyl cofactor biosynthesis. J Am Chem Soc 2015, 137:13216–13219. [DOI] [PubMed] [Google Scholar]
- ••33. McCarthy EL, Booker SJ: Destruction and reformation of an iron-sulfur cluster during catalysis by lipoyl synthase. Science 2017, 358:373–377. The first report of efficient reconstitution of a radical SAM sulfur insertion enzyme by iron-sulfur cluster assembly proteins, NfuA and IscU. This work demonstrates how enzymes that degrade their auxiliary Fe4S4 clusters as a sulfur source can act catalytically with iron-sulfur cluster carrier proteins.
- 34.Cronan JE: Assembly of lipoic acid on its cognate enzymes: an extraordinary and essential biosynthetic pathway. Microbiol Mol Biol Rev 2016, 80:429–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Esberg B, Leung HC, Tsui HC, Bjork GR, Winkler ME: Identification of the miaB gene, involved in methylthiolation of isopentenylated A37 derivatives in the tRNA of Salmonella typhimurium and Escherichia coli. J Bacteriol 1999, 181:7256–7265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •36. Zhang B, Arcinas AJ, Radle MI, Silakov A, Booker SJ, Krebs C: First step in catalysis of the radical S-adenosylmethionine methylthiotransferase MiaB yields an intermediate with a [3Fe-4S](0)-like auxiliary cluster. J Am Chem Soc 2020, 142:1911–1924. The first report of spectroscopic and biochemical evidence for conversion of an auxiliary [4Fe-4S]2+ cluster in MiaB to a Fe3S4-like cluster during the methylation step of catalysis. This study shows that the auxiliary Fe4S4 cluster acts as the direct sulfur source during catalysis.
- 37.McCarthy EL, Rankin AN, Dill ZR, Booker SJ: The A-type domain in Escherichia coli NfuA is required for regenerating the auxiliary [4Fe-4S] cluster in Escherichia coli lipoyl synthase. J Biol Chem 2019, 294:1609–1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cai K, Liu G, Frederick RO, Xiao R, Montelione GT, Markley JL: Structural/functional properties of human NFU1, an intermediate [4Fe-4S] carrier in human mitochondrial iron-sulfur cluster biogenesis. Structure 2016, 24:2080–2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •39. Kunichika K, Nakamura R, Fujishiro T, Takahashi Y: The structure of the dimeric state of IscU harboring two adjacent [2Fe-2S] clusters provides mechanistic insights into cluster conversion to [4Fe-4S]. Biochemistry 2021, 60:1569–1572. This paper describes a recently reported X-ray crystal structure of dimeric IscU.
- 40.Shimomura Y, Wada K, Fukuyama K, Takahashi Y: The asymmetric trimeric architecture of [2Fe-2S] IscU: implications for its scaffolding during iron-sulfur cluster biosynthesis. J Mol Biol 2008, 383:133–143. [DOI] [PubMed] [Google Scholar]
- 41.Ribbe MW, Hu Y, Hodgson KO, Hedman B: Biosynthesis of nitrogenase metalloclusters. Chem Rev 2014, 114:4063–4080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fay AW, Blank MA, Rebelein JG, Lee CC, Ribbe MW, Hedman B, Hodgson KO, Hu Y: Assembly scaffold NifEN: A structural and functional homolog of the nitrogenase catalytic component. Proc Natl Acad Sci U S A 2016, 113:9504–9508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •43. Wiig JA, Hu Y, Chung Lee C, Ribbe MW: Radical SAM-dependent carbon insertion into the nitrogenase M-cluster. Science 2012, 337:1672–1675. A radiolabeling study showing that the interstitial carbide in the nitrogenase FeMo-co originates from the methyl group of SAM and it is inserted into a precursor of the active cofactor by the assembly enzyme, NifB.
- ••44. Jenner LP, Cherrier MV, Amara P, Rubio LM, Nicolet Y: An unexpected P-cluster like intermediate en route to the nitrogenase FeMo-co. Chem Sci 2021, 12:5269–5274. In this paper, previously reported NifB electron density in Ref. 26 was reprocessed to show that a [Fe8S8] cluster is the best fit to density previously modeled as K1 and K2.