Abstract
Physician progress notes are frequently organized into Subjective, Objective, Assessment, and Plan (SOAP) sections. The Assessment section synthesizes information recorded in the Subjective and Objective sections, and the Plan section documents tests and treatments to narrow the differential diagnosis and manage symptoms. Classifying the relationship between the Assessment and Plan sections has been suggested to provide valuable insight into clinical reasoning. In this work, we use a novel human-in-the-loop pipeline to classify the relationships between the Assessment and Plan sections of SOAP notes as a part of the n2c2 2022 Track 3 Challenge. In particular, we use a clinical information model constructed from both the entailment logic expected from the aforementioned Challenge and the problem-oriented medical record. This information model is used to label named entities as primary and secondary problems/symptoms, events and complications in all four SOAP sections. We iteratively train separate Named Entity Recognition models and use them to annotate entities in all notes/sections. We fine-tune a downstream RoBERTa-large model to classify the Assessment-Plan relationship. We evaluate multiple language model architectures, preprocessing parameters, and methods of knowledge integration, achieving a maximum macro-F1 score of 82.31%. Our initial model achieves top-2 performance during the challenge (macro-F1: 81.52%, competitors’ macro-F1 range: 74.54%−82.12%). We improved our model by incorporating post-challenge annotations (S&O sections), outperforming the top model from the Challenge. We also used Shapley additive explanations to investigate the extent of language model clinical logic, under the lens of our clinical information model. We find that the model often uses shallow heuristics and nonspecific attention when making predictions, suggesting language model knowledge integration requires further research.*
Keywords: natural language processing, language modeling, entailment, SOAP notes, Intensive care unit, electronic health record
Graphical Abstract

1. Introduction
Statement of Significance
| Problem or Issue | No systems exist that model the relationship between sections of SOAP notes. The extent to which biomedical language models (LMs) perform clinical reasoning with respect to a clinical information model is also poorly understood. |
| What is Already Known | LMs have achieved high performance on sentence-level clinical entailment benchmarks, but SOAP note section-level entailment has not been investigated. |
| What this Paper Adds | This study proposes an information model-based pipeline to model SOAP note section entailment and uses problem-based named entities to investigate LM clinical logic. We also demonstrate the use of an explainable AI method in investigating LM reasoning, identifying key differences between physician and LM decision making. |
A common approach to clinical documentation is the Subjective, Objective, Assessment, and Plan (SOAP) structure, which was first espoused by Weed [35] to aid in clinical reasoning and communication. The SOAP note is oriented around medical problems identified by the physician, qualified by the subjective information gained from the patient and objective information gained from clinical investigations such that they can be assessed and a plan for care put in place. The Assessment section of the note synthesizes and evaluates the information recorded in the Subjective and Objective (S&O) sections to arrive at a differential diagnosis or identify active problems. In the Plan section, tests and treatments needed to narrow the differential diagnosis or provide care are documented. Often, however, there is not an explicit link to Assessment content in the Plan section. Determining the relationship between the Assessment and Plan subsections of the SOAP note would help to model the entanglement of clinical reasoning and documentation, modeling the inferences made following information processing and synthesis necessary to perform the actions of clinical care. Recognizing the significance of these relationships, Track 3 of the 2022 n2c2 Challenge entitled ”Progress Note Understanding: Assessment and Plan Reasoning” tasked participants with developing a model to predict relationships between Assessment and Plan note subsections [8]. The Challenge organizers released a novel set of annotations of SOAP section headings and relations between sections defined by Gao et al. [8] for a patient cohort from MIMIC-III [15]. In this paper we use this newly released corpus and present a top-ranking transformer-based pipeline for the Track 3 Challenge, incorporating a clinical information model into the prediction task.
1.1. Related Work
Biomedical relation classification has seen significant progress in recent years, particularly with the advent of transformer-based neural language models (LMs) pretrained on biomedical text such as PubMed Central and MIMIC-III [24]. Most existing tasks focus on relation extraction (RE) using biomedical text, which requires document-level entity recognition prior to relation classification.[17, 4, 12] Few of these methods focus on relation extraction from clinical narratives [10, 34, 28]. In the clinical setting, in addition to classic BERT-based methods, improvements in clinical RE incorporate semantic type information [37] or external knowledge through the use of knowledge graphs [30]. Some methods also leverage LMs such as the Longformer that can incorporate longer-ranged context from clinical notes [19] or utilize multi-task learning paradigms to improve classification [23]. Despite these improvements in relation extraction, Gao et al. [8] present a novel dataset for document-level relation classification over clinical narratives, a setting that hasn’t been previously explored. Moreover, there does not exist a method that incorporates an explicit clinical information model in a neural relation classification system, as we propose in this work.
The theoretical basis for our clinical information model was both the problem-oriented nature of the SOAP note [36] and the classification criteria provided by the n2c2 Challenge [8]. This information model was qualified by the use of entailment graphs to identify the semantic structure of problem-oriented entities (primary and secondary signs, symptoms, complications) relevant to classifying a relationship between the Assessment and Plan subsections.
To distinguish between primary and secondary problems or note complications, events, or organ areas, we label all four note sections using a custom annotation scheme, and train RoBERTa-based Named Entity Recognition (NER) LMs using spacy (details in Section 2.3). Using the trained NER models, we label the text with entity-specific token tags, and further train another biomedical RoBERTa model on the n2c2 Track 3 Challenge data (predicting relationship classes between assessment and plan sections of a SOAP note). We then use an explainable AI method known as SHAP (SHapley Additive exPlanations) to investigate the extent of clinical logic in entailment and classification prediction, analyzing the nature of errors. We find that while primary and secondary problems are accurately recognized and tagged, the entailment LMs often ignores expected clinical logic and entities as described relative to the classification task. Our pipeline without S&O sections achieves top-2 performance (macro-F1: 81.52%), and our final pipeline (macro-F1: 82.31%) outperforms the best model from the Challenge [7] (challenge macro-F1 range: 74.54%−82.12%). We release all code on GitHub2 and all models on the Hugging Face Model Hub 3 for use by the community.
2. Methods
2.1. n2c2 Track 3 Challenge Dataset
To address the n2c2 Track 3 Challenge, we develop a novel human-in-the-loop pipeline to classify the relationships between the Assessment and Plan subsections of ICU SOAP notes from the MIMIC-III v1.3 database [15]. The data released as part of the Challenge was drawn from a uniform random sample across 84 different note types, including surgery, medicine, cardiovascular, neurology, and trauma daily progress notes. The Challenge organizers excluded admission notes, discharge summaries, and progress notes written by healthcare providers that do not use the SOAP format (e.g. social work notes). SOAP notes were further filtered to exclude those that only included system-wide assessments as these notes do not follow the problem-oriented format indicative of a SOAP note. The final dataset was annotated using 8 criteria, assigned to 4 different labels: DIRECT, INDIRECT, NEITHER, NOT RELEVANT. Relation labels indicate whether a Plan subsection addresses: the primary diagnoses or problems in the Assessment (DIRECT ), an adverse event or consequence from the primary diagnosis or secondary problem (e.g., comorbidity) in the Assessment (INDIRECT ), a problem or diagnosis not mentioned in the progress note (NEITHER); or does not address a problem or diagnosis listed (NOT RELEVANT ) (see Gao et al. [8] for additional note filtering details and a full table of criteria and labels for annotation).
The dataset includes a total of 5897 pairs of Assessment and Plan subsections, split into train, development, and test sets (4633 (78.57%), 597 (10.12%), 667 (11.31%) examples, respectively). The demographics of the patient cohort are shown in Table 1, along with its most prevalent diagnoses in Table 2. As mentioned by Gao et al. [8], the cohort includes an oversampling of medical (as opposed to surgical, etc.) notes, as this was the largest service area in the hospital. Following the Challenge, the organizers also released additional annotations which label the SOAP section spans from the entire note (not just assessment/plan sections), under the assumption that including S&O context would improve prediction. We incorporate these additional labels and test this hypothesis.
Table 1:
Demographic characteristics of n2c2 MIMIC-III cohort
| n | Overall 5897 | Direct 1403 | Indirect 1596 | Neither 1882 | Not Relevant 1016 | |
|---|---|---|---|---|---|---|
| ELECTIVE | 325 (5.5) | 72 (5.1) | 77 (4.8) | 110 (5.8) | 66 (6.5) | |
| ADMISSIONTYPE, n (%) | EMERGENCY | 5461 (92.6) | 1314 (93.7) | 1491 (93.4) | 1726 (91.7) | 930 (91.5) |
| URGENT | 111 (1.9) | 17 (1.2) | 28 (1.8) | 46 (2.4) | 20 (2.0) | |
| DEAD/EXPIRED | 696 (11.8) | 165 (11.8) | 195 (12.2) | 231 (12.3) | 105 (10.3) | |
| DISCH-TRAN TO PSYCH HOSP | 70 (1.2) | 17 (1.2) | 20 (1.3) | 18 (1.0) | 15 (1.5) | |
| HOME | 1349 (22.9) | 349 (24.9) | 330 (20.7) | 394 (20.9) | 276 (27.2) | |
| HOME HEALTH CARE | 1397 (23.7) | 310 (22.1) | 397 (24.9) | 448 (23.8) | 242 (23.8) | |
| HOSPICE-HOME | 57 (1.0) | 15 (1.1) | 13 (0.8) | 18 (1.0) | 11 (1.1) | |
| HOSPICE-MEDICAL FACILITY | 19 (0.3) | 5 (0.4) | 2 (0.1) | 8 (0.4) | 4 (0.4) | |
| DISCHARGEJLOCATION, n (%) | ICF | 11 (0.2) | 3 (0.2) | 3 (0.2) | 3 (0.2) | 2 (0.2) |
| LEFT AGAINST MEDICAL ADVI | 103 (1.7) | 18 (1.3) | 24 (1.5) | 43 (2.3) | 18 (1.8) | |
| LONG TERM CARE HOSPITAL | 704 (11.9) | 158 (11.3) | 211 (13.2) | 225 (12.0) | 110 (10.8) | |
| OTHER FACILITY | 23 (0.4) | 5 (0.4) | 8 (0.5) | 7 (0.4) | 3 (0.3) | |
| REHAB/DISTINCT PART HOSP | 325 (5.5) | 83 (5.9) | 85 (5.3) | 97 (5.2) | 60 (5.9) | |
| SHORT TERM HOSPITAL | 63 (1.1) | 13 (0.9) | 23 (1.4) | 19 (1.0) | 8 (0.8) | |
| SNF | 1080 (18.3) | 262 (18.7) | 285 (17.9) | 371 (19.7) | 162 (15.9) | |
| Government | 77 (1.3) | 22 (1.6) | 21 (1.3) | 15 (0.8) | 19 (1.9) | |
| Medicaid | 586 (9.9) | 151 (10.8) | 139 (8.7) | 184 (9.8) | 112 (11.0) | |
| INSURANCE, n (%) | Medicare | 3913 (66.4) | 891 (63.5) | 1097 (68.7) | 1304 (69.3) | 621 (61.1) |
| Private | 1294 (21.9) | 334 (23.8) | 326 (20.4) | 376 (20.0) | 258 (25.4) | |
| Self Pay | 27 (0.5) | 5 (0.4) | 13 (0.8) | 3 (0.2) | 6 (0.6) | |
| **TO | 9 (0.2) | 1 (0.1) | 5 (0.3) | 2 (0.1) | 1 (0.1) | |
| *BUL | 15 (0.3) | 5 (0.4) | 3 (0.2) | 5 (0.3) | 2 (0.2) | |
| AMER | 11 (0.2) | 4 (0.3) | 4 (0.3) | 2 (0.1) | 1 (0.1) | |
| African | 12 (0.2) | 5 (0.4) | 2 (0.1) | 2 (0.1) | 3 (0.3) | |
| CAPE | 45 (0.8) | 9 (0.6) | 17 (1.1) | 12 (0.6) | 7 (0.7) | |
| LANGUAGE, n (%) | ENGL | 5220 (88.9) | 1237 (88.5) | 1394 (87.7) | 1686 (90.0) | 903 (89.1) |
| East Asian | 67 (1.1) | 18 (1.3) | 20 (1.3) | 16 (0.9) | 13 (1.3) | |
| Middle Eastern | 30 (0.5) | 6 (0.4) | 10 (0.6) | 9 (0.5) | 5 (0.5) | |
| PTUN | 134 (2.3) | 29 (2.1) | 42 (2.6) | 37 (2.0) | 26 (2.6) | |
| RUSS | 149 (2.5) | 39 (2.8) | 47 (3.0) | 42 (2.2) | 21 (2.1) | |
| Romance | 183 (3.1) | 45 (3.2) | 46 (2.9) | 61 (3.3) | 31 (3.1) | |
| AMERICAN INDIAN | 24 (0.4) | 5 (0.4) | 3 (0.2) | 13 (0.7) | 3 (0.3) | |
| ASIAN | 162 (2.7) | 40 (2.9) | 43 (2.7) | 50 (2.7) | 29 (2.9) | |
| BLACK | 703 (11.9) | 167 (11.9) | 207 (13.0) | 226 (12.0) | 103 (10.1) | |
| HISPANIC/LATINO | 153 (2.6) | 36 (2.6) | 36 (2.3) | 53 (2.8) | 28 (2.8) | |
| ETHNICITY, n (%) | MIDDLE EASTERN | 10 (0.2) | 2 (0.1) | 4 (0.3) | 3 (0.2) | 1 (0.1) |
| OTHER | 108 (1.8) | 22 (1.6) | 32 (2.0) | 37 (2.0) | 17 (1.7) | |
| PATIENT DECLINED TO ANSWER | 23 (0.4) | 3 (0.2) | 8 (0.5) | 5 (0.3) | 7 (0.7) | |
| UNABLE TO OBTAIN | 68 (1.2) | 15 (1.1) | 22 (1.4) | 21 (1.1) | 10 (1.0) | |
| UNKNOWN/NOT SPECIFIED | 120 (2.0) | 29 (2.1) | 32 (2.0) | 39 (2.1) | 20 (2.0) | |
| WHITE | 4526 (76.8) | 1084 (77.3) | 1209 (75.8) | 1435 (76.2) | 798 (78.5) | |
| GENDER, n (%) | F | 2771 (47.0) | 676 (48.2) | 744 (46.6) | 897 (47.7) | 454 (44.7) |
| M | 3126 (53.0) | 727 (51.8) | 852 (53.4) | 985 (52.3) | 562 (55.3) |
Table 2:
Top 15 most common diagnoses in n2c2 MIMIC-III cohort
| ICD-9 Prefix | Code Description | Overall | Direct | Indirect | Neither | Not Relevant |
|---|---|---|---|---|---|---|
| 03 | Other Bacterial Diseases | 845 | 196 | 256 | 264 | 129 |
| 42 | Other Forms Of Heart Disease (myocarditis, endocarditis) | 570 | 112 | 152 | 206 | 100 |
| 41 | Ischemic Heart Disease, Diseases Of Pulmonary Circulation | 561 | 131 | 135 | 172 | 123 |
| 57 | Other Diseases Of Digestive System (Chronic liver disease, Cholelithiasis) | 410 | 93 | 111 | 137 | 69 |
| 99 | Other And Unspecified Effects Of External Causes, Complications Of Surgical And Medical Care | 338 | 85 | 96 | 103 | 54 |
| 51 | Other Diseases Of Respiratory System (Pneumothorax, Pleurisy, pulmonary fibrosis) | 331 | 75 | 87 | 120 | 49 |
| 48 | Pneumonia And Influenza | 297 | 73 | 74 | 106 | 44 |
| 56 | Other Diseases Of Intestines And Peritoneum (Intestinal obstruction, diverticula) | 148 | 31 | 51 | 40 | 26 |
| 53 | Diseases Of Esophagus, Stomach, And Duodenum | 145 | 31 | 45 | 38 | 31 |
| 78 | Symptoms | 141 | 35 | 35 | 51 | 20 |
| 25 | Diseases Of Other Endocrine Glands | 122 | 35 | 37 | 34 | 16 |
| 58 | Diseases Of The Genitourinary System | 120 | 25 | 39 | 39 | 17 |
| 27 | Other Metabolic Disorders And Immunity Disorders | 119 | 27 | 24 | 51 | 17 |
| 50 | Pneumoconioses And Other Lung Diseases Due To External Agents | 115 | 29 | 34 | 33 | 19 |
| 59 | Other Diseases Of Urinary System (Calculus, cystitis) | 105 | 29 | 23 | 38 | 15 |
2.2. Theoretical Model of Clinical Information
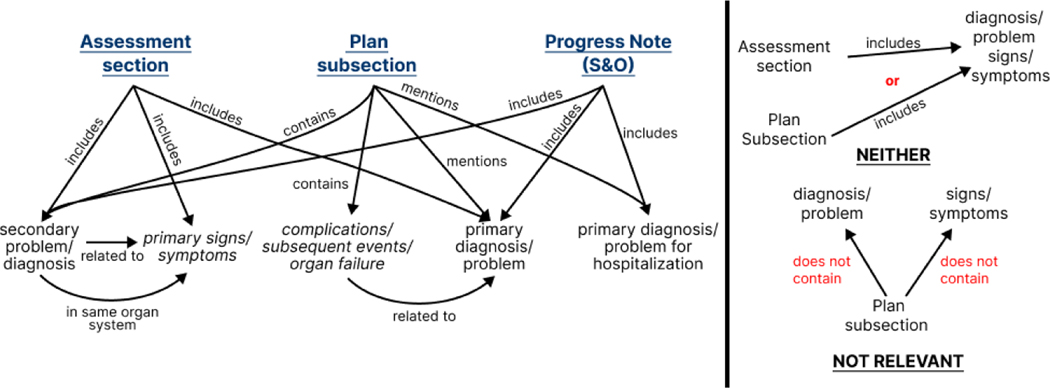
With the relationship categories as a target, a pipeline approach was developed building upon a clinical annotation scheme that we believed would improve the LM’s clinical entailment by explicitly tagging problem-oriented entities relevant to the patient’s admission. This annotation scheme is linked to a theoretical model of problem-oriented medical information which relates to the entailment of clinical problems within the medical record. The entailment of clinical problems can be described by graphs derived from the n2c2 Challenge annotation criteria (examples are shown in Supplemental Figure S1). Consolidating these individual graphs gives rise to a complete model of clinical information, within the context of SOAP note section entailment (Figure 1). The two negative classes, NEITHER and NOT RELEVANT, are not integrated into the information model for legibility, but it is important to note that all nodes overlap between these subgraphs and the larger information model graph.
Figure 1:
Clinical information model derived from n2c2 annotation guidelines. In blue, we highlight the four sections of the SOAP note. All other nodes in this graph act as named entities to be annotated.
In order to define our final annotation scheme, we perform several other preprocessing steps. Certain entailment relations are defined by negative relations within the graph, such as the ones found in the INDIRECT relation in Supplemental Figure S1b. We opt to convert these negative relations into positive ones by defining the concept of a secondary diagnosis/problem, which we define as any diagnosis that is not the predominant reason for the ICU visit. To capture negative labeling criteria, we add the secondary problem label to our clinical information model, as shown. We also separate out the two entities (complications/subsequent events/organ failure and primary signs/symptoms) such that each of these 5 concepts act as independent entities. These steps lead us to 8 total annotation labels: primary and secondary problems, signs, symptoms, and complications, events, and organ failure, all related to the primary problem. We discuss the specific label sets assigned to each SOAP note section in the following section. The results of the annotation using this information model and subsequent training are shown in Section 3.
2.3. NER Annotation and Model Training
Using the clinical information model described in Section 2.2, we identify several common key named entities which revolve around medical problems that we assign as NER labels to each SOAP note section; namely, we annotate the Assessment sections with the primary and secondary problems, as well as signs and symptoms. We annotate the Plan subsections with the primary problem, and the complications, events, and organ failures, each related to the primary problem. We also annotate the Subjective and Objective sections using the Assessment annotation scheme, as these sections capture primary information from the patient within the clinical encounter rather than inferred or evaluated secondary information (as we would expect with complications or events related to a primary problem). That said, progress notes which refer to previous problems replicated in the S&O sections may pose a challenge and this mismatch is described in Section 3 below.
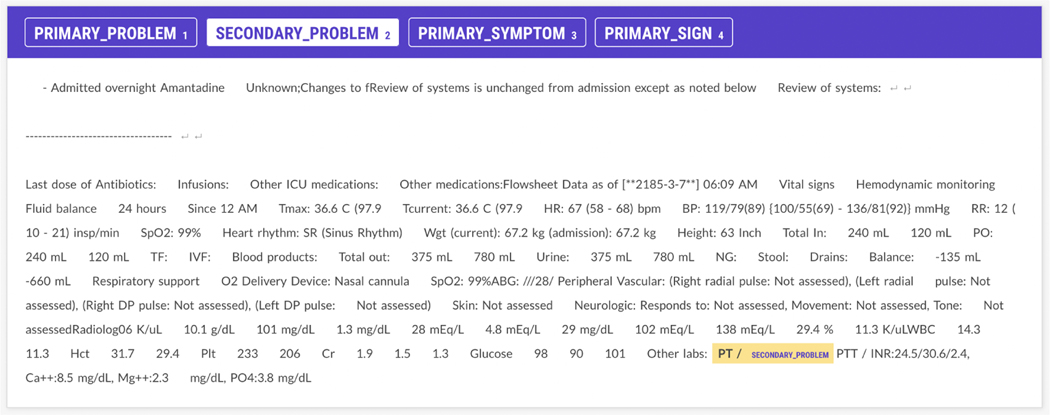
Accurate labeling of relationships therefore depends upon differentiation of primary versus secondary problems and labeling adverse events or consequences of primary problems. Moreover, linking these problems across the Assessment and Plan subsections must account for concept abbreviation (”congestive heart failure” vs. ”CHF”) and semantically similar clinical descriptions (”congestive heart failure” vs ”diastolic failure”). We hypothesize that we can use the aforementioned annotation scheme to annotate a subset of the n2c2 Challenge dataset to improve our downstream Challenge task. The annotation guidelines and rules were initially developed and trialed by a board certified generalist (emergency medicine) physician (RAT). Two medical students (AG, DC) were trained on the annotation scheme. For the annotation task, 240 Assessment and 250 Plan progress notes from the original n2c2 Track 3 Challenge data were randomly selected and uploaded into a Prodigy server environment (see Figure 2). Each medical student annotator labeled all selected notes across the two sections. Discrepancies between the annotators were adjudicated by the board certified physician. To evaluate inter-annotator agreement, we use the F-measure (before adjudication; F=0.6200), as Cohen’s kappa has been shown to be poorly defined for NER tasks [13].
Figure 2:
Screenshots of Prodigy Assessment and Plan subsection annotation interfaces with a DIRECT entailment relation
In addition to annotating and developing NER models for the Assessment and Plan subsections, we also annotate 135 Subjective and 135 Objective note sections using the method described above. We hypothesize that including additional context from the SOAP note will improve Challenge task prediction. We also recognize that certain annotation criteria expect information from the entire progress note (see Supplemental Figure S1b).
To support human annotation, we first annotate 100 Assessment and Plan subsections manually using Prodigy, and then use spacy-transformers 4 to fine-tune a general domain RoBERTa-base model [21] pretrained on OntoNotes 5 [26] for both the Assessment and Plan section NER tagging. We employ spacy’s [3] human-in-the-loop training paradigm to perform iterative annotation and model training. The initial model suggests NER labels for annotators to correct. Once the full set of annotations are corrected, we fine-tune a new general domain RoBERTa-base model from scratch using all annotations, split into train and test sets (192 (80%) and 48 (20%)), respectively. For the S&O model, we start with the pretrained Assessment model and further fine-tune it on annotations from concatenated Subjective and Objective note sections using a 90/10 train and test set split (122 (90%), 13 (10%)). The use of an iterative process is intended to reduce annotator cognitive load and speed up the annotation process.
Figure 2 illustrates some of the difficulty in employing our information model in SOAP note annotation. In Figure 2a, we see that the Assessment and Plan subsection have a DIRECT relationship linking the primary problem of 3-vessel disease (3VD) to the primary symptom of recurrent chest pain within the Plan subsection (Figure 2b). However, 3VD in the Plan subsection could also be considered the problem with recurrent chest pain being a symptom, though it was not annotated this way, as shown. The ambiguity associated with selecting a single primary problem among related conditions/symptoms arises due to our Plan subsection annotation scheme, which doesn’t make the distinction between primary and secondary symptoms, like the Assessment scheme does. We also see that to properly identify this entailment, acronyms such as EF (ejection fraction) and CABG (coronary artery bypass graft) must be disambiguated, necessitating the use of biomedical LMs as shown in Table 4. Finally, in tagging Coronary Artery Bypass Graft Surgery (CABG) as an Event Related to Primary Problem, the model fails to perform implicit temporal reasoning, assuming CABG is a physiological consequence following 3VD, instead of a intervention planned for the future. Despite some misalignment between our model of clinical information and the entailment task, we are able to effectively label several named entity types (see Section 3).
Table 4:
Macro-F1 scores for all pipeline choices. Underlined values for each subtable indicate the best performing model for a particular set of experiments. The F1 score in bold is the best performing model across all experiments.
| Experiment | Model | Evaluation Set Macro-FI | Test Set Macro-F1 |
|---|---|---|---|
| Model Architecture | GPT Neo-1.7B | 24.70 | 13.11 |
| SapBERT | 77.46 | 78.4 | |
| Biomed-RoBERTa-Base | 80.34 | 79.36 | |
| Biomed-RoBERTa-Large | 81.71 | 80.77 | |
|
| |||
| Knowledge Integration | Pipeline | 81.52 | 81.59 |
| Pipeline (NER at end) | 81.26 | 82.18 | |
|
| |||
| Text Preprocessing | Pipeline + Abbreviations | 80.41 | 80.92 |
| Pipeline + DelD + Abbvs | 80.30 | 80.68 | |
| Pipeline + Weighted Loss | 81.34 | 81.59 | |
| Pipeline + DelD | 81.17 | 81.55 | |
|
| |||
| S&O Integration | Pipeline + S&O | 82.31 | 81.73 |
| Pipeline + S&O tagged | 79.70 | 84.05 | |
2.4. Entailment Model Pipeline
Following the annotation of entity spans within the assessment and plan note subsections, we then perform a set of experiments to determine the best downstream model choices for note subsection relation prediction. We first test three different model architectures. As a baseline, we fine-tune the Biomedical RoBERTa-Base model (Biomed-RoBERTa-Base) from Lewis et al. [18], pre-trained on PubMed, PMC, and MIMIC-III with a byte-pair encoding (BPE) vocabulary learned from PubMed. We also train the larger, 355M parameter version of this model (Biomed-RoBERTa-Large). We only focus on biomedical RoBERTa due to the evidence of significant improvement from in-domain pretraining and vocabulary [33]. Due to the knowledge-intensive nature of this task, we also test SapBERT [20], a PubMedBERT model whose embedding space has been realigned to equate embeddings of UMLS synonyms, encouraging better named entity disambiguation. Finally, we test a generative LM, GPT-NEO 127M [2]. While we were not able to fine-tune this model due to computational constraints, we test its in-context performance against the fine-tuned BERT-based models.
After determining the best model architecture to solve the relation classification task, we also sought to determine the impact of two different knowledge integration methods: either by surrounding text spans with entity tags (as seen in the Assessment section of Figure 5) or by concatenating a list of entities to the end of the raw SOAP note text. Next we perform an ablation study over various text preprocessing steps and training parameters. We tested several approaches such as dictionary-based abbreviation expansion (Pipeline + Abbreviations), MIMIC-III de-identification tag replacement (Pipeline + DeID), and weighted loss (Pipeline + Weighted Loss). For abbreviation disambiguation, we use the Meta-Inventory from Grossman Liu et al. [9], which synthesizes a number of sources including UMLS and Wikipedia. We test de-identification tag replacement as we noticed in preliminary analysis that Biomed-RoBERTa poorly tokenizes MIMIC de-ID tags, so we experiment replacing them with placeholders. Following the preprocessing experiments, we investigate the impact of introducing additional context through the integration of the Subjective and Objective sections.
Figure 5:
In this example, the model correct identified a DIRECT relation based on a directly referred sign/symptom. However, when we look at the attention of the tokens over the Assessment text, we see a very nonspecific relationship between all problems, signs, and symptoms mentioned and Hypotension in the Plan subsection. While the model identifies certain relevant clinical features (including low BP in the S&O sections) much of its attention is fairly nonspecific, implying a lack of clinical reasoning in decision making.
All models were trained with their default parameters from Huggingface transformers v4.25.1 [38], except for Biomed-RoBERTa, which was trained for 15 epochs and a larger learning rate of 1e-5. We train models on the original Challenge dataset, as well as integrate additional S&O context following the Assessment and Plan subsections. To do so, the S&O sections were directly concatenated to the end of the Plan Subsection text. All models used the default maximum sequence length of 512 tokens. For those note pairs that extended beyond this length, the Plan Subsection was truncated. Visual inspection demonstrated less relevant context later in the section than the text that would be truncated in the Assessment section (e.g. dot phrases, physician signatures). These experiments result in our final pipeline (as shown in Figure 3). We present the results for all experiments in Section 3.
Figure 3:
Task pipeline including NER pipeline and classification model
3. Results
We report the results of training RoBERTa-base NER models on notes labeled with our information model annotation schemes across all four sections of the SOAP note in Table 3. Using the Assessment annotation scheme, we see that the trained model identified and tagged primary and secondary problems better than signs and symptoms. However, this isn’t the case when the Assessment scheme is applied to the S&O sections. NER in the S&O sections performed the worst across all entities from any section of the note, though symptom recognition was relatively more effective when compared with the Assessment sections. Finally, the model trained using the Plan section annotation scheme performed best in identifying the primary problem in that section, but performs significantly worse in identifying inferred or evaluated secondary information (i.e. complications/organ failure).
Table 3:
Assessment, Plan section, and Subjective & Objective NER Model Training Results After Adjudication. We use the same annotation scheme for both the Assessment and the Subjective & Objective (S&O) sections.
| Entity | Precision | Recall | Macro-Fl | |
|---|---|---|---|---|
| Assessment | Primary Problem | 63.83 | 46.88 | 54.05 |
| Primary Symptom | 48.48 | 66.67 | 56.14 | |
| Primary Sign | 21.74 | 26.32 | 23.81 | |
| Secondary Problem | 78.20 | 75.36 | 76.75 | |
|
| ||||
| Plan Subsection | Problem | 90.24 | 88.1 | 89.16 |
| Complication RTP | 40.00 | 10.00 | 16.00 | |
| Event RTP | 25.00 | 12.50 | 16.67 | |
| Organ Failure RTP | 100.00 | 33.33 | 50.00 | |
|
| ||||
| S&O | Primary Problem | 53.85 | 38.89 | 45.16 |
| Primary Symptom | 42.86 | 12.50 | 19.35 | |
| Primary Sign | 12.50 | 12.50 | 12.50 | |
| Secondary Problem | 20.00 | 12.50 | 15.38 | |
In Table 4 we show performance metrics for all experiments regarding pipeline choices. All Pipeline experiments used Biomed-RoBERTa as that performed the best among all model architectures. Similarly, when test- ing S&O integration, we used Biomed-RoBERTa with the Assessment and Plan subsections tagged inline, as this performed better than other text preprocessing choices. We find that fine-tuning the Biomed-RoBERTa model on the direct output of the spacy NER models and including the raw S&O section text performs the best, achieving a macro F1 of 82.31% on the validation set and 81.73% on the test set, as shown at the end of Table 4. We also notice that both preprocessing, training choices (weighted loss), and knowledge integration methods have minimal impacts on performance. We only see marginal improvement when the text is tagged with the NER models, yet combined with additional SOAP note context, we outperform the best model of the Challenge [7].
3.1. Qualitative Assessment of Annotations
RAT also performed a qualitative assessment of complications during annotation for all four SOAP sections. Annotators found annotating primary and secondary problems in the Assessment section to be straightforward, although in some cases, additional context from the S&O sections was required to distinguish the primary cause for hospitalization. Further, while annotating the S&O sections, annotators identified malformed text, auto-populated templates, and text-based tables which could not classified into an annotation category. Many S&O sections, particularly unconventional ones such as radiology, did not follow the problem-based SOAP note paradigm, making annotation using our primary/secondary problem annotation scheme difficult, as shown in Figure 4.
Figure 4:
An image of the Subjective & Objective in Prodigy with limited context that makes problem, sign, and symptom identification difficult.
In this example, the Subjective section (above the dashed line) simply refers to a previous note. Inclusion of a previous review of systems would allow for some annotation using our information model. Similarly, the list of medication, laboratory results, and vitals in the Objective section are difficult to label using the Assessment annotation schema, especially without the full clinical narrative found from other sections of the health record.
4. Discussion
In this work, we describe the results of our participation in the n2c2 2022 Track 3 Challenge involving classification of progress note SOAP section relations in the MIMIC-III corpus. The data provided by the Challenge annotators used 8 criteria to classify Assessment and Plan subsections pairs into 4 relations: DIRECT, INDIRECT, NOT RELEVANT, NEITHER. Following the Challenge, organizers also provided span annotations splitting sections into Subjective/Objective/Assessment/Plan subsections. We develop an approach that incorporates a clinical information model into LM-based relation classification by using a custom problem-based NER annotation scheme derived from clinical entailment graphs. We demonstrate that training a Biomedical RoBERTa-large model and including NER annotations of the Assessment and Plan subsections and the raw Subjective & Objective sections performs better than all other pipeline setups. Our results show that this model performs better than the best performing model of the Challenge [7]. As shown in Supplementary Figure S2, AUROC and AUPRC curves demonstrate slight variations in performance between all four classes, though all perform fairly well. Moreover, we develop models that begin to differentiate between primary and secondary problems in clinical notes and explore potential methods of integrating entity-based knowledge into LMs.
Considerable work has been done in designing NLP tasks to integrate aspects of clinical reasoning into LMs [28, 14, 6, 32]. However, despite substantial progress on certain benchmarks [25, 27], LMs have been shown to poorly model clinical reasoning, relying on shallow heuristics and demonstrating an inability to generalize to out-of-sample clinical concepts [11]. Therefore, continued examination of various facets of the problem, including Track 3 of the n2c2 2022 Challenge presented by Gao et al. [8] and discussed in this work, help advance the science. Additionally, a core component of improving LM performance in knowledge-intensive tasks is the integration of external knowledge alongside clinical logic [39, 29, 31]. By using an entailment model which represents clinical information and logic simultaneously, we demonstrate in this work that integrating named-entity based knowledge is non-trivial. We also present a new scheme for annotating clinical problems with respect to SOAP section relationships to facilitate explainable analysis: an information model structured around clinical entailment. Despite our results outperforming the top models on the Challenge task, we identify a number of gaps during error analysis.
4.1. Explainability Analysis
While we tried to integrate clinical knowledge into pre-trained biomedical LMs, we notice little improvement when including our entity tagged data. In particular, we used SHAP values [22] to visualize the impact of particular text spans on entailment. Through our error analysis, we identify three limitations in our pipeline: a reliance on shallow relation heuristics, nonspecific attention over S&O sections, and an overall lack of clinical reasoning, all potentially leading to the limited improvements seen in Section 3 from our NER pipeline approach.
As seen from Table 4 and the ROC curves in Supplemental Figure S2, while our model performs well on average, our NER annotation scheme and use of subsequent annotations does not significantly improve results. One reason for this is that often our model relies on syntactic mention-level heuristics for relation prediction. For instance, many DIRECT relations have a primary problem surface form in both the Assessment and Plan subsection. The surface form of a concept is the form in which the concept appears in the text. Similarly, INDIRECT relations often also feature matching surface forms, but are often preceded by text like PMH or history of. However, we notice that many incorrect classifications are INDIRECT relations that have no matching surface forms across the two sections, as seen in Figure 6.
Figure 6:
Model incorrectly labels a note as a NEITHER relation without the Subjective & Objective context, but then adjusts its predict to the correct one with additional context.
While our NER models did not significantly improve prediction, we can still use the annotations to investigate evidence of clinical reasoning performed by the model. For instance, in Figure 5, we see that the model correctly identifies a DIRECT relation which was probably labeled using the criteria, ”Plan subsection contains a problem/diagnosis related to the primary signs/symptoms in the Assessment section.” The model focuses on the occurrence of a matching surface form in both the Assessment and Plan subsection. It further attends to the vitals section of the Objective section, though it doesn’t solely focus on blood pressure (BP), but also includes temperature, heart rate (HR), and respiratory rate (RR), implying that the model’s attention is fairly nonspecific. In Figure 6, we further see evidence of the lack of intrinsic clinical reasoning in entailment prediction. Here we show the same document, with and without the Subjective & Objective sections included. Before the S&O sections are used, the model incorrectly predicts a NEITHER relation, due to the lack of the surface form heuristic. However, once additional context in added, the model adjusts its predict to the correct INDIRECT relation. However, it focuses on general vitals, instead of identifying particular indications of hypothyroidism (such as the Levothyroxine prescription), implying that the model does not employ conventional clinical reasoning to makes its prediction and the inclusion of the S&O components simply provide additional differentiating text to attend over. Reviewing these two examples, we see that our NER models work well for certain concepts (e.g. primary problems, signs, symptoms) and additional context improves prediction. However, our entity tagging did not positively impact entailment due to the model’s seemingly more semantic, rather than clinical, reasoning used in prediction.
A limitation of explainability analysis using this dataset is the lack of the specific annotation criteria used by the Challenge organizers to label note relations. In the example in Figure 6, there is no clear indication in the Assessment section of the nature of the patient’s hypothyroidism. Reviewing the guidelines from Gao et al. [8], we see that an INDIRECT relation can be caused by a Plan subsection containing (1) complications/subsequent events or organ failure related to the primary diagnosis, (2) other listed diagnoses/problems from the overall Progress Note or in the Assessment section that are not part of the primary diagnosis, or (3) a diagnosis/problem that is not previously mentioned but closely related (i.e., same organ system) to the primary diagnosis. Hypothyroidism in a hypotensive patient with a past medical history of cancer presenting with retroperitoneal (RP) bleeding does not seem to fall into either category (1) or (3), as the hypothyroidism is neither a consequence nor in the same organ system as the RP bleed. Hypothyroidism has been shown to be associated with myelodysplastic syndrome (MDS) which could explain an INDIRECT relation [16]. However, this is only true for autoimmune causes of hypothyroidism, such as Hashimoto’s thyroiditis, and the etiology of this patients hypothyroidism is unknown so it is impossible justify the relation with the information given. It is likely the annotators used the mention of Levothyroxine to tag this relation as a category (2) INDIRECT relation, but without these finer grained labels, further interpretation must be speculative.
4.2. Implications and Future Work
We design a novel NER annotation scheme that identifies primary and secondary signs and symptoms across SOAP notes from the MIMIC-III corpus, enabling contextual tagging of clinical problems. While our NER models perform adequately, at least with regards to primary and secondary problems in the Assessment and the primary problem in the Plan section, this tagging and subsequent integration into a downstream pipeline does not significantly improve results. Based on our error analysis, we hypothesize that our methods of knowledge integration do not improve token-level embedding representations with respect to clinical reasoning. However, even if our models were able to more effectively integrate knowledge, we see that often SOAP note structures are poorly organized and often do not contain the full information discovered during the clinical encounter in each note, making information extraction (the NER task) and entailment (the relation classification task) difficult. Leveraging additional annotations, particularly from a single institution, to integrate clinical information may also limit generalizability to other institutions. This limitation applies to the Challenge, given that annotations were only provided for critical care notes from a single institution in Boston [15]. Selecting a more nationally representative sample of text for training and testing of both the NER and relation classification models would allow for better generalizability. Additionally, a more robust annotation framework, along with multi-institution annotators, would be necessary to ensure the generalizability of the human-in-the-loop component of the pipeline.
Given the nature of progress notes and evolving care over the course of the encounter, a complete recapitulation of knowledge about the patient in every SOAP progress note seems redundant and a cause of note bloat, a driver of physician burnout [5]. However, interpreting partially reported information from evolving progress notes necessitates better language modeling of the medical record beyond the note, keeping in mind the multi-modal and temporal nature of the documentation of a clinical encounter. Such clinically informed natural language understanding would improve clinical reasoning and entailment problems, such as the one we tackle in this work.
To further refine our pipeline, we plan to continue annotations for our upstream NER models to ensure that we minimize noise and error propagation to downstream steps. This will require upsampling documents that contain less frequent labels (e.g. Event Related to Problem). It may also include defining additional entities in our information model, particularly for the S&O sections, that better align with the conventional recording of observations in SOAP notes. We also plan to test other entity-based knowledge integration methods such as TekGen [1], which uses a generative LM to convert KG triples to natural sentences to further fine-tune an LM.
5. Conclusion
This work presents a series of experiments addressing the challenge of the classification of relationships between SOAP Assessment and Plan sections. We investigate model architectures, methods of knowledge integration, and text preprocessing choices. We find that an NER pipeline method is the optimal solution for submission to the n2c2 2022 Track 3 Challenge. We further refine this work by integrating the full progress note (S&O sections) upon reflection during the 2022 AMIA Annual Symposium. We demonstrate that a human-in-the-loop approach, combining medical knowledge and diagnostic entity tagging with pre-trained linguistic information contained in LMs, is the most effective method for progress note understanding. We also begin to address the challenge of separating primary and secondary problems, as well as recognizing clinical consequences to these diagnoses in SOAP notes. Finally, we use an explainable AI method to identify and interpret some reasoning gaps by biomedical LMs in note entailment. We release both our entailment model and NER tagging models for future use by the community.
Supplementary Material
Highlights.
The extent to which biomedical language models perform clinical reasoning with respect to a clinical information model is poorly understood.
Neural language models have achieved high performance on sentence-level clinical entailment benchmarks, but entailment on section-level clinical narratives, such as SOAP notes, has not been investigated.
This study proposes an information model-based pipeline to model SOAP note section entailment and uses problem-based named entities to investigate LM clinical logic.
The paper demonstrates the use of an explainable AI method in investigating LM reasoning, identifying key differences between physician and LM decision making.
6. Acknowledgements
We thank Meliha Yetisgen, Ozlem Uzuner, Diwakar Mahajan, and Yanjun Gao for organizing the n2c2 2022 Track 3 Challenge and providing us with the data, as well as helpful feedback during model development and paper submission. We also thank the other members of the DECILE Lab at Yale School of Medicine for their feedback.
Funding:
This work was supported in part by the National Institutes of Health [grant number T15LM007056] and the National Institute of Diabetes And Digestive And Kidney Diseases of the NIH [grant number T35DK104689]. This publication was also made possible by the Yale School of Medicine Fellowship for Medical Student Research.
*.
- SOAP
Subjective, Objective, Assessment, Plan
- LM
Language model
- S&O
Subjective and Objective
- NER
Named Entity Recognition
- SHAP
SHapley Additive exPlanations
Footnotes
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Vimig Socrates reports financial support was provided by National Institutes of Health. Aidan Gilson reports financial support was provided by National Institute of Diabetes and Digestive and Kidney Diseases. Aidan Gilson reports financial support was provided by Yale School of Medicine.
Vimig Socrates, David Chartash, Kevin Lopez: Conceptualization, Methodology Andrew Taylor, David Chartash: Supervision. Vimig Socrates: Software, Validation. Aidan Gilson, Ling Chi, Andrew Taylor, David Chartash: Data Annotation. All Authors: Writing- Original draft preparation, Writing- Reviewing and Editing.
Conflict of Interest Statement
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Agarwal Oshin, Ge Heming, Shakeri Siamak, and Rami Al-Rfou. Knowledge graph based synthetic corpus generation for knowledge-enhanced language model pre-training. arXiv preprint arXiv:2010.12688, 2020. [Google Scholar]
- [2].Black Sid, Gao Leo, Wang Phil, Leahy Connor, and Biderman Stella. GPT-Neo: Large Scale Autoregressive Language Modeling with Mesh-Tensorflow, March 2021. URL 10.5281/zenodo.5297715. [DOI]
- [3].Adriane Boyd. explosion/spaCy: v2.3.9: Compatibility with NumPy v1.24+, December 2022. URL 10.5281/zenodo.7445599. [DOI]
- [4].Bravo Àlex, Piñero Janet, Queralt-Rosinach Núria, Rautschka Michael, and Furlong Laura I. Extraction of relations between genes and diseases from text and large-scale data analysis: implications for translational research. BMC bioinformatics, 16:1–17, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Downing N Lance, Bates David W, and Longhurst Christopher A. Physician burnout in the electronic health record era: are we ignoring the real cause? Annals of Internal Medicine, 169(1):50–51, 2018. [DOI] [PubMed] [Google Scholar]
- [6].Gao Yanjun, Dligach Dmitriy, Miller Timothy, Caskey John, Sharma Brihat, Churpek Matthew M, and Afshar Majid. Dr. bench: Diagnostic reasoning benchmark for clinical natural language processing. arXiv preprint arXiv:2209.14901, 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Gao Yanjun, Dligach Dmitriy, Miller Timothy, Churpek Matthew M., and Uzuner Majid Afshar Ozlemand. Progress note understanding:assessment and plan reasoning n2c2 track 3 overview. American Medical Informatics Association Annual Symposium, 2022. URL https://n2c2.dbmi.hms.harvard.edu/2022-amia-workshop. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Gao Yanjun, Dligach Dmitriy, Miller Timothy, Tesch Samuel, Laffin Ryan, Churpek Matthew M., and Afshar Majid. Hierarchical Annotation for Building A Suite of Clinical Natural Language Processing Tasks: Progress Note Understanding, 2022. URL http://arxiv.org/abs/2204.03035. [PMC free article] [PubMed]
- [9].Liu Lisa Grossman, Grossman Raymond H, Mitchell Elliot G, Weng Chunhua, Natarajan Karthik, Hripcsak George, and Vawdrey David K. A deep database of medical abbreviations and acronyms for natural language processing. Scientific Data, 8(1):1–9, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Henry Sam, Buchan Kevin, Filannino Michele, Stubbs Amber, and Uzuner Ozlem. 2018 n2c2 shared task on adverse drug events and medication extraction in electronic health records. Journal of the American Medical Informatics Association, 27(1):3–12, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Herlihy Christine and Rudinger Rachel. Mednli is not immune: Natural language inference artifacts in the clinical domain. arXiv preprint arXiv:2106.01491, 2021. [Google Scholar]
- [12].Herrero-Zazo Maŕıa, Segura-Bedmar Isabel, Mart´ınez Paloma, and Declerck Thierry. The ddi corpus: An annotated corpus with pharmacological substances and drug–drug interactions. Journal of biomedical informatics, 46(5):914–920, 2013. [DOI] [PubMed] [Google Scholar]
- [13].Hripcsak George and Rothschild Adam S. Agreement, the f-measure, and reliability in information retrieval. Journal of the American medical informatics association, 12(3):296–298, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Jin Di, Pan Eileen, Oufattole Nassim, Weng Wei-Hung, Fang Hanyi, and Szolovits Peter. What disease does this patient have? a large-scale open domain question answering dataset from medical exams. arXiv preprint arXiv:2009.13081, 2020. [Google Scholar]
- [15].Johnson Alistair EW, Pollard Tom J, Shen Lu, Lehman Li-wei H, Feng Mengling, Ghassemi Mohammad, Moody Benjamin, Szolovits Peter, Celi Leo Anthony, and Mark Roger G. Mimic-iii, a freely accessible critical care database. Scientific data, 3(1):1–9, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Komrokji Rami S, Kulasekararaj Austin, Al Ali Najla H, Shahram Kordasti, Bart-Smith Emily, Craig Benjamin M, Padron Eric, Zhang Ling, Lancet Jeffrey E, Pinilla-Ibarz Javier, et al. Autoimmune diseases and myelodysplastic syndromes. American journal of hematology, 91(5):E280–E283, 2016. [DOI] [PubMed] [Google Scholar]
- [17].Krallinger Martin, Rabal Obdulia, Akhondi Saber A, Pérez Martın Pérez, Santamaría Jesús, Rodríguez Gael Pérez, Tsatsaronis Georgios, Intxaurrondo Ander, López José Antonio, Nandal Umesh, et al. Overview of the biocreative vi chemical-protein interaction track. In Proceedings of the sixth BioCreative challenge evaluation workshop, volume 1, pages 141–146, 2017. [Google Scholar]
- [18].Lewis Patrick, Ott Myle, Du Jingfei, and Stoyanov Veselin. Pretrained language models for biomedical and clinical tasks: Understanding and extending the state-of-the-art. In Proceedings of the 3rd Clinical Natural Language Processing Workshop, pages 146–157, 2020. [Google Scholar]
- [19].Li Yikuan, Wehbe Ramsey M, Ahmad Faraz S, Wang Hanyin, and Luo Yuan. Clinical-longformer and clinical-bigbird: Transformers for long clinical sequences. arXiv preprint arXiv:2201.11838, 2022. [Google Scholar]
- [20].Liu Fangyu, Shareghi Ehsan, Meng Zaiqiao, Basaldella Marco, and Collier Nigel. Self-alignment pretraining for biomedical entity representations. arXiv preprint arXiv:2010.11784, 2020. [Google Scholar]
- [21].Liu Yinhan, Ott Myle, Goyal Naman, Du Jingfei, Joshi Mandar, Chen Danqi, Levy Omer, Lewis Mike, Zettlemoyer Luke, and Stoyanov Veselin. Roberta: A robustly optimized bert pretraining approach. arXiv preprint arXiv:1907.11692, 2019. [Google Scholar]
- [22].Lundberg Scott M and Lee Su-In. A unified approach to interpreting model predictions. In Guyon I, Luxburg UV, Bengio S, Wallach H, Fergus R, Vishwanathan S, and Garnett R, editors, Advances in Neural Information Processing Systems 30, pages 4765–4774. Curran Associates, Inc., 2017. URL http://papers.nips.cc/paper/7062-a-unified-approach-to-interpreting-model-predictions.pdf. [Google Scholar]
- [23].Mulyar Andriy, Uzuner Ozlem, and McInnes Bridget. Mt-clinical bert: scaling clinical information extraction with multitask learning. Journal of the American Medical Informatics Association, 28(10):2108–2115, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Peng Yifan, Yan Shankai, and Lu Zhiyong. Transfer learning in biomedical natural language processing: an evaluation of bert and elmo on ten benchmarking datasets. arXiv preprint arXiv:1906.05474, 2019. [Google Scholar]
- [25].Phan Long N, James T Anibal Hieu Tran, Chanana Shaurya, Bahadroglu Erol, Peltekian Alec, and Altan-Bonnet Grégoire Scifive: a text-to-text transformer model for biomedical literature. arXiv preprint arXiv:2106.03598, 2021. [Google Scholar]
- [26].Pradhan Sameer, Moschitti Alessandro, Xue Nianwen, Hwee Tou Ng Anders Björkelund, Uryupina Olga, Zhang Yuchen, and Zhong Zhi. Towards robust linguistic analysis using OntoNotes. In Proceedings of the Seventeenth Conference on Computational Natural Language Learning, pages 143–152, Sofia, Bulgaria, August 2013. Association for Computational Linguistics. URL https://aclanthology.org/W13-3516. [Google Scholar]
- [27].Kanakarajan Kamal raj, Kundumani Bhuvana, and Sankarasubbu Malaikannan. Bioelectra: pretrained biomedical text encoder using discriminators. In Proceedings of the 20th Workshop on Biomedical Language Processing, pages 143–154, 2021. [Google Scholar]
- [28].Romanov Alexey and Shivade Chaitanya. Lessons from natural language inference in the clinical domain. arXiv preprint arXiv:1808.06752, 2018. [Google Scholar]
- [29].Rosset Corby, Xiong Chenyan, Phan Minh, Song Xia, Bennett Paul, and Tiwary Saurabh. Knowledge-aware language model pretraining. arXiv preprint arXiv:2007.00655, 2020. [Google Scholar]
- [30].Roy Arpita and Pan Shimei. Incorporating medical knowledge in bert for clinical relation extraction. In Proceedings of the 2021 conference on empirical methods in natural language processing, pages 5357–5366, 2021. [Google Scholar]
- [31].Safavi Tara and Koutra Danai. Relational world knowledge representation in contextual language models: A review. arXiv preprint arXiv:2104.05837, 2021. [Google Scholar]
- [32].Singhal Karan, Azizi Shekoofeh, Tu Tao, S Sara Mahdavi, Wei Jason, Chung Hyung Won, Scales Nathan, Tanwani Ajay, Cole-Lewis Heather, Pfohl Stephen, et al. Large language models encode clinical knowledge. arXiv preprint arXiv:2212.13138, 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Tinn Robert, Cheng Hao, Gu Yu, Usuyama Naoto, Liu Xiaodong, Naumann Tristan, Gao Jianfeng, and Poon Hoifung. Fine-tuning large neural language models for biomedical natural language processing. arXiv preprint arXiv:2112.07869, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Uzuner Özlem, South Brett R, Shen Shuying, and DuVall Scott L. 2010 i2b2/va challenge on concepts, assertions, and relations in clinical text. Journal of the American Medical Informatics Association, 18(5):552–556, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Weed Lawrence L. Medical records that guide and teach. New England Journal of Medicine, 278(12):593–600, 1968. [DOI] [PubMed] [Google Scholar]
- [36].Weed Lawrence L. Medical records, medical education, and patient care: the problem-oriented record as a basic tool. Press of Case Western Reserve University, 1969. [PubMed] [Google Scholar]
- [37].Wei Qiang, Ji Zongcheng, Si Yuqi, Du Jingcheng, Wang Jingqi, Tiryaki Firat, Wu Stephen, Tao Cui, Roberts Kirk, and Xu Hua. Relation extraction from clinical narratives using pre-trained language models. In AMIA annual symposium proceedings, volume 2019, page 1236. American Medical Informatics Association, 2019. [PMC free article] [PubMed] [Google Scholar]
- [38].Wolf Thomas, Debut Lysandre, Sanh Victor, Chaumond Julien, Delangue Clement, Moi Anthony, Cistac Pierric, Rault Tim, Louf Remi, Funtowicz Morgan, Davison Joe, Shleifer Sam, Patrick von Platen Clara Ma, Jernite Yacine, Plu Julien, Xu Canwen, Le Scao Teven, Gugger Sylvain, Drame Mariama, Lhoest Quentin, and Rush Alexander. Transformers: State-of-the-art natural language processing. In Proceedings of the 2020 Conference on Empirical Methods in Natural Language Processing: System Demonstrations, pages 38–45, Online, October 2020. Association for Computational Linguistics. doi: 10.18653/v1/2020.emnlp-demos.6. URL https://aclanthology.org/2020.emnlp-demos.6. [DOI] [Google Scholar]
- [39].Yamada Ikuya, Asai Akari, Shindo Hiroyuki, Takeda Hideaki, and Matsumoto Yuji. Luke: deep contextualized entity representations with entity-aware self-attention. arXiv preprint arXiv:2010.01057, 2020. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.