Abstract
The double-strand break (DSB) repair pathway called microhomology-mediated end-joining (MMEJ) is thought to be dependent on DNA polymerase theta (Polθ) and occur independently of nonhomologous end-joining (NHEJ) factors. An unresolved question is whether MMEJ is facilitated by a single Polθ-mediated end-joining pathway or consists of additional undiscovered pathways. We find that human X-family Polλ, which functions in NHEJ, additionally exhibits robust MMEJ activity like Polθ. Polλ promotes MMEJ in mammalian cells independently of essential NHEJ factors LIG4/XRCC4 and Polθ, which reveals a distinct Polλ-dependent MMEJ mechanism. X-ray crystallography employing in situ photo-induced DSB formation captured Polλ in the act of stabilizing a microhomology-mediated DNA synapse with incoming nucleotide at 2.0 Å resolution and reveals how Polλ performs replication across a DNA synapse joined by minimal base-pairing. Last, we find that Polλ is semisynthetic lethal with BRCA1 and BRCA2. Together, these studies indicate Polλ MMEJ as a distinct DSB repair mechanism.
DSBs are potentially lethal DNA lesions that are repaired primarily by homologous recombination (HR) and NHEJ1,2. HR occurs during S/G2 cell cycle phases and maintains genome integrity owing to its use of a homologous DNA donor template1. NHEJ, which can act throughout the cell cycle, is a low fidelity DSB repair pathway due to minimal DNA end processing and lack of utilization of a DNA donor template.2 More recently, the error-prone DSB repair pathway MMEJ, or alternative end-joining (alt-EJ), has been discovered3–5. Like HR, MMEJ acts on 3′ ssDNA overhangs generated by 5′−3′ resection of DSBs, but typically utilizes small tracts (2−8 bp) of microhomology for DNA synapsis and repair and results in chromosome translocations and large indels6–10. In contrast, NHEJ functions on DSBs containing blunt or very short 5′ or 3′ ssDNA overhangs and does not require microhomology2.
Although MMEJ (alt-EJ) was reported as a distinct end-joining mechanism in mammalian cells in 20083, thus far only a few proteins have been assigned to this pathway. The error-prone A-family Polθ was shown to promote MMEJ in mammalian cells and invertebrates6–9,11,12. Thus, MMEJ is also referred to as Polθ-mediated end-joining (TMEJ)12. Although Polθ plays an important role in MMEJ, residual MMEJ type DSB repair signatures have been observed in Polθ-deficient cells13–17. Thus, an important question that arises is whether MMEJ is facilitated by a single Polθ-mediated end-joining (TMEJ) process, or consists of additional undiscovered MMEJ pathways.
Results
Polλ exhibits similar MMEJ activity as Polθ
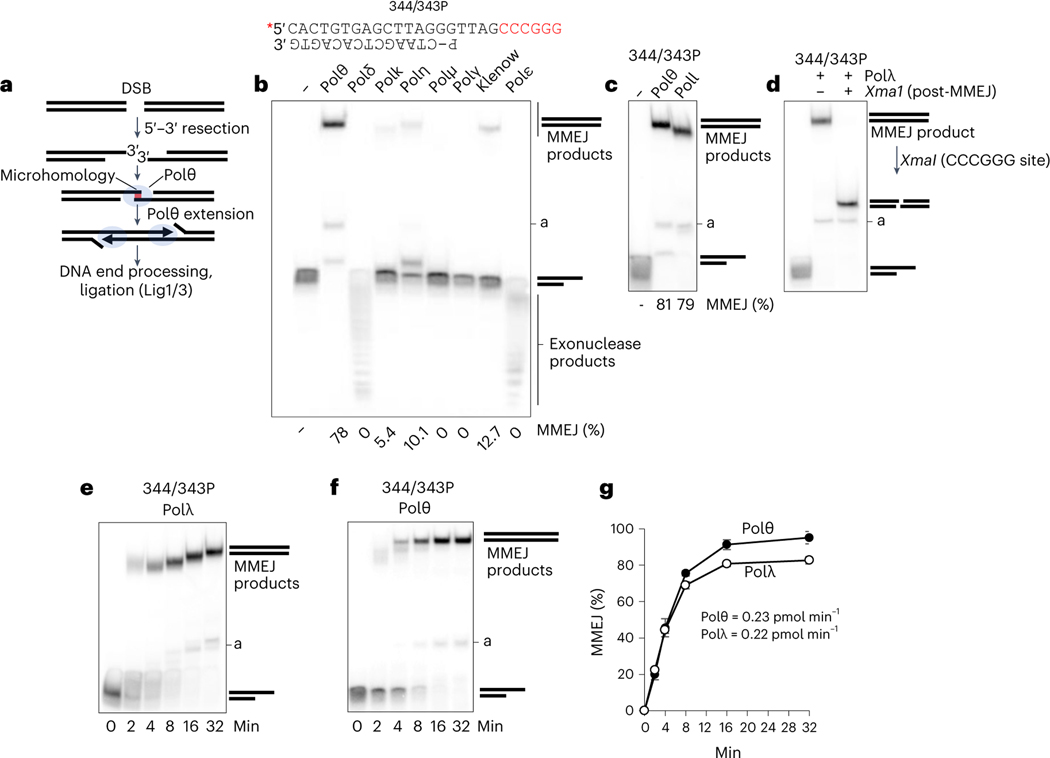
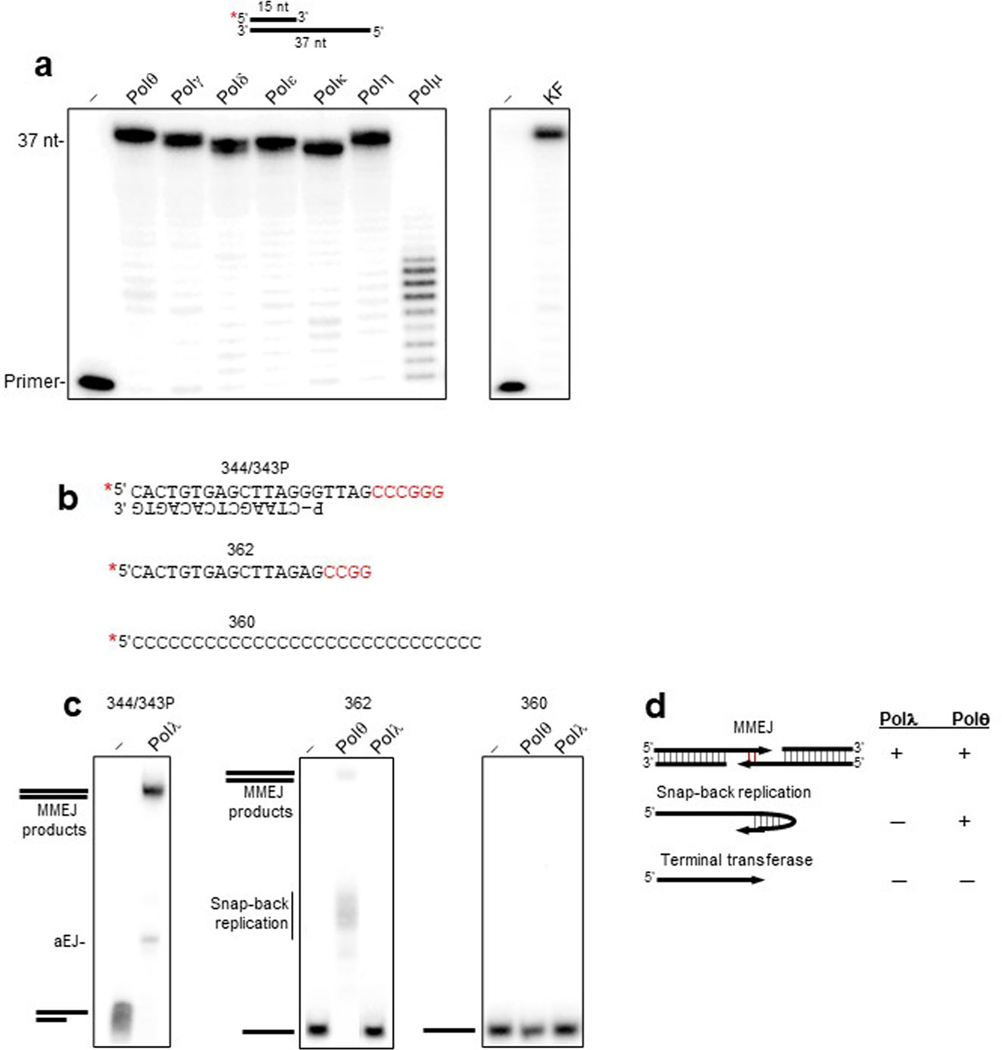
To initially probe whether Polθ independent MMEJ mechanisms exists, multiple human and yeast DNA polymerases (Pols) were screened for MMEJ activity in vitro. As a positive control for MMEJ activity, we utilized the polymerase domain expressed by POLQ (herein referred to as Polθ)11. The previously characterized MMEJ assay uses a preannealed partial ssDNA (pssDNA) radiolabeled DNA template containing a 12 nt 3′ ssDNA overhang with a 6 nt palindrome microhomology tract (Fig. 1b)11. Following 5′−3′ DNA resection of a DSB, Polθ promotes synapsis between 3′ ssDNA overhangs via microhomology-mediated base-pairing, then extends the partially paired strands by using the opposing overhang as a template in trans (Fig. 1a)11,12. In the absence of sufficient 3′ terminal microhomology, Polθ preferentially performs snapback replication via formation of a microhomology-mediated hairpin loop11,18. MMEJ was performed at 37 °C by incubating Polθ with the pssDNA (Fig. 1b, top), deoxyribonucleoside triphosphates (dNTPs), MgCl2, and standard buffer conditions for 45 min in the absence of a ligase. Reactions were terminated by EDTA and proteinase K, which degrades protein while leaving DNA intact. The DNA was resolved in a nondenaturing polyacrylamide gel run at room temperature then visualized by phosphorimager. These MMEJ assay and gel conditions were previously shown to detect and quantitate MMEJ products facilitated by the ability of Polθ to promote microhomology-mediated DNA synapse followed by extension of the minimally paired 3′ ssDNA overhangs11. Consistent with previous research, Polθ performed MMEJ efficiently as indicated by the top band (Fig. 1b, lane 2)11,18. We attribute the intermediate faint band to abortive end-joining. We next screened multiple eukaryotic Pols for MMEJ activity using identical conditions. Replicative B-family Pols δ and ε exhibited exonuclease activity on the pssDNA (Fig. 1b). A-family Polγ and X-family Polμ showed no MMEJ activity (Fig. 1b). A-family Klenow fragment—a homolog of Polθ—and Y-family Pols η and κ showed minimal MMEJ activity (Fig. 1b). All Pols exhibited activity on a traditional primer-template as expected (Extended Data Fig. 1a).
Fig. 1 |. Polλ exhibits MMEJ activity similar to that of Polθ.
a, Schematic of MMEJ. Following 5′−3′ DNA resection, Polθ promotes microhomology-mediated synapsis of 3′ ssDNA overhangs then performs strand extension. b, Schematic of pssDNA 344/343P. Red text, microhomology; P, phosphate (top). Nondenaturing gel showing MMEJ activity by the indicated Pols. MMEJ (%) is shown at the bottom; a, abortive end-joining. c, Nondenaturing gel showing MMEJ activity by the indicated Pols. d, Nondenaturing gel showing XmaI digestion of the MMEJ product formed by Polλ. e,f, Nondenaturing gels showing a time course of MMEJ by Polλ (e) and Polθ (f). Experiments in b−f were all repeated with three independent samples and all yielded similar results. g, Scatter plot showing the relative rates of MMEJ by Polθ and Polλ; n = 3 independent samples, ± s.d. Relative steady-state rates of DNA joined by MMEJ are indicated.
We next examined X-family Polλ for MMEJ activity. Polλ promotes gap filling during NHEJ, is related to Polμ, which also functions in NHEJ2,19,20, and has been implicated in base excision repair21,22. Polλ promoted efficient MMEJ, similar to Polθ (Fig. 1c). Polθ MMEJ products were slightly larger than those of Polλ, which we attribute to the terminal transferase activity of Polθ23. As a control, the addition of XmaI resulted in endonucleolytic cleavage of the MMEJ product (Fig. 1d). This demonstrates the presence of the XmaI CCCGGG/GGG CCC double-strand recognition sequence formed by MMEJ, similar to previous studies11. Polθ and Polλ exhibited nearly identical relative rates of MMEJ, suggesting a similar mechanism of DNA end-joining (Fig. 1e–g).
Polλ specializes in MMEJ of DNA with short 3′ ssDNA overhangs
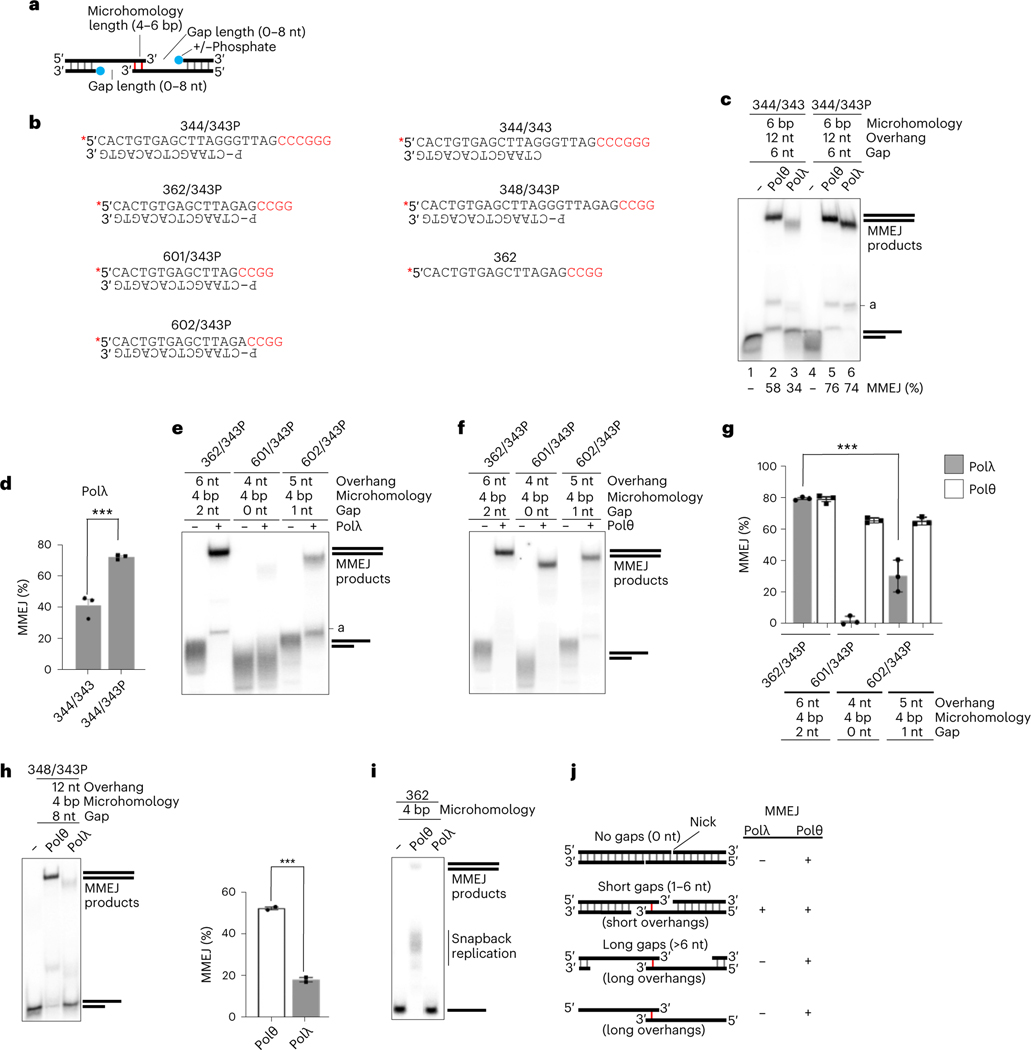
We next characterized the MMEJ template requirements of Polλ compared with those of Polθ to potentially distinguish their respective MMEJ mechanisms. Microhomology, gap length and 5′-phopshate presence were probed (Fig. 2a). Polθ MMEJ is known to be stimulated by the 5′ terminal phosphate on the resected strand11. Both Polθ and Polλ MMEJ were stimulated by the 5′-phosphate, indicating similar involvement of their respective lyase domains, which bind the 5′-phosphate adjacent to a ssDNA gap (Fig. 2c,d)19,24. Indeed, stimulation of Polλ gap filling by the presence of a 5′-phosphate was demonstrated in previous studies25. Polθ and Polλ exhibited identical MMEJ activity with 4 bp microhomology and a 2 nt gap (Fig. 2e–g). A 1 nt gap significantly reduced Polλ MMEJ, whereas Polθ MMEJ was reduced only slightly (Fig. 2e–g). Further reducing the gap length to 0 nt abolished Polλ MMEJ, but did not affect Polθ MMEJ (Fig. 2e–g). Polλ exhibited limited MMEJ (17%) on pssDNA, enabling a 8 nt gap, whereas Polθ showed significantly higher MMEJ (53%) on this template (Fig. 2h).
Fig. 2 |. Polλ specializes in MMEJ of DNA with short 3′ ssDNA overhangs.
a, Schematic showing pssDNA modifications. b, Schematic of pssDNA and ssDNA templates. Red text, microhomology; asterisk, radiolabel; P, 5′-phosphate. c, Nondenaturing gel showing MMEJ by Polθ and Polλ on the indicated templates with and without 5′-phosphate on the resected strand. MMEJ (%) indicated. Experiment was repeated with three independent samples and all yielded similar results. d, Bar plot showing MMEJ (%) by Polλ on the indicated templates. Data represent means; n = 3 independent samples, ± s.d. *P < 0.05, **P < 0.01, ***P < 0.001. Statistical significance from two-sample t-test between 344/343 and 344/343P, P = 0.0008. e,f, Nondenaturing gels showing MMEJ by Polλ (e) and Polθ (f) on the indicated templates. Experiment was repeated with three independent samples and all yielded similar results. g, Bar plot showing MMEJ (%) by Polλ and Polθ on the indicated templates. Data represent means; n = 3 independent samples, ± s.d. *P < 0.05, **P < 0.01, ***P < 0.001. Statistical significance from two-sample t-test between 362/343P and 602/343P for Polλ, P = 0.0005. h, Nondenaturing gel showing MMEJ activity by Polλ and Polθ on the indicated template. Bar plot showing MMEJ (%) by Polλ and Polθ on the indicated template (right). Data represent means; n = 3 independent samples, ± s.d. *P < 0.05, **P < 0.01, ***P < 0.001. Statistical significance from two-sample t-test between Polθ and Polλ, P = 0.0006. i, Nondenaturing gel showing MMEJ and snapback replication activities by Polλ and Polθ on the indicated template. j, Summary table comparing the respective MMEJ activities of Polλ and Polθ on different templates.
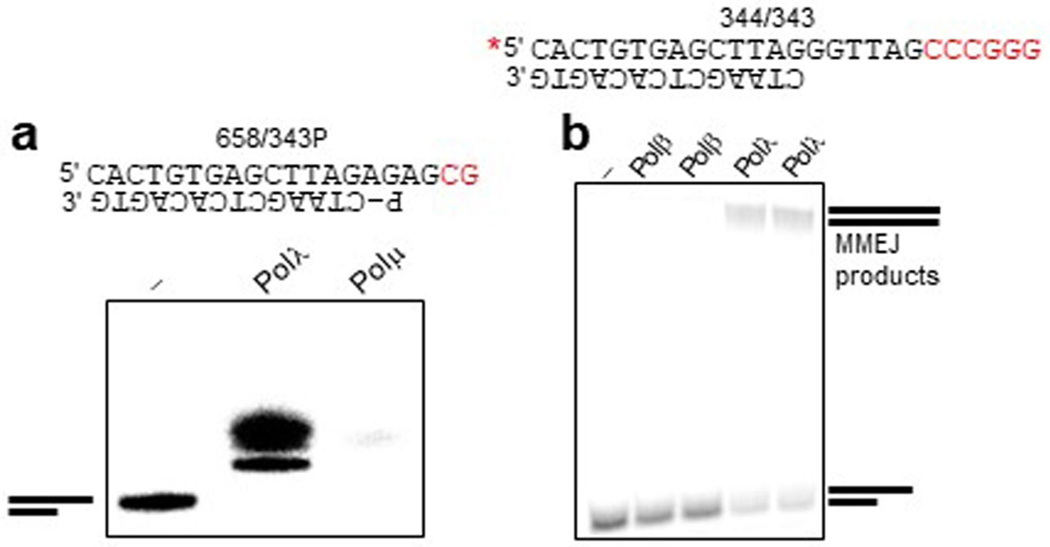
We compared Polλ and Polθ MMEJ of ssDNA, which models DSBs with long 3′ ssDNA overhangs. The polymerase domain of Polθ predominately performs snapback replication on ssDNA when intramolecular microhomology enables hairpin loop formation (Extended Data Fig. 1b–d)18. Full-length Polθ containing a N-terminal superfamily 2 helicase promotes proficient MMEJ on ssDNA substrates, which suggests Polθ acts on long 3′ ssDNA overhangs formed during S-phase18. As expected, Polθ polymerase performed primarily snapback replication on ssDNA. Polλ, however, showed no ssDNA extension activity (Fig. 2i and Extended Data Fig. 1b–d). Hence, ssDNA extension activity is not a requirement for Polλ MMEJ. Polθ performs minimal MMEJ on pssDNA with 2 bp microhomology11. Polλ is also capable of extending pssDNA with 2 bp microhomology, whereas Polμ is not (Extended Data Fig. 2a). Although a previous report implicated X-family Polβ in alt-EJ26, Polβ failed to perform MMEJ under identical conditions as used for Polλ (Extended Data Fig. 2b). These data demonstrate that Polλ exhibits robust MMEJ across DNA synapses with relatively short gaps and overhangs, whereas Polθ can tolerate both long and short gaps and overhangs (Fig. 2j).
Polλ promotes MMEJ independently of Polθ and NHEJ factors
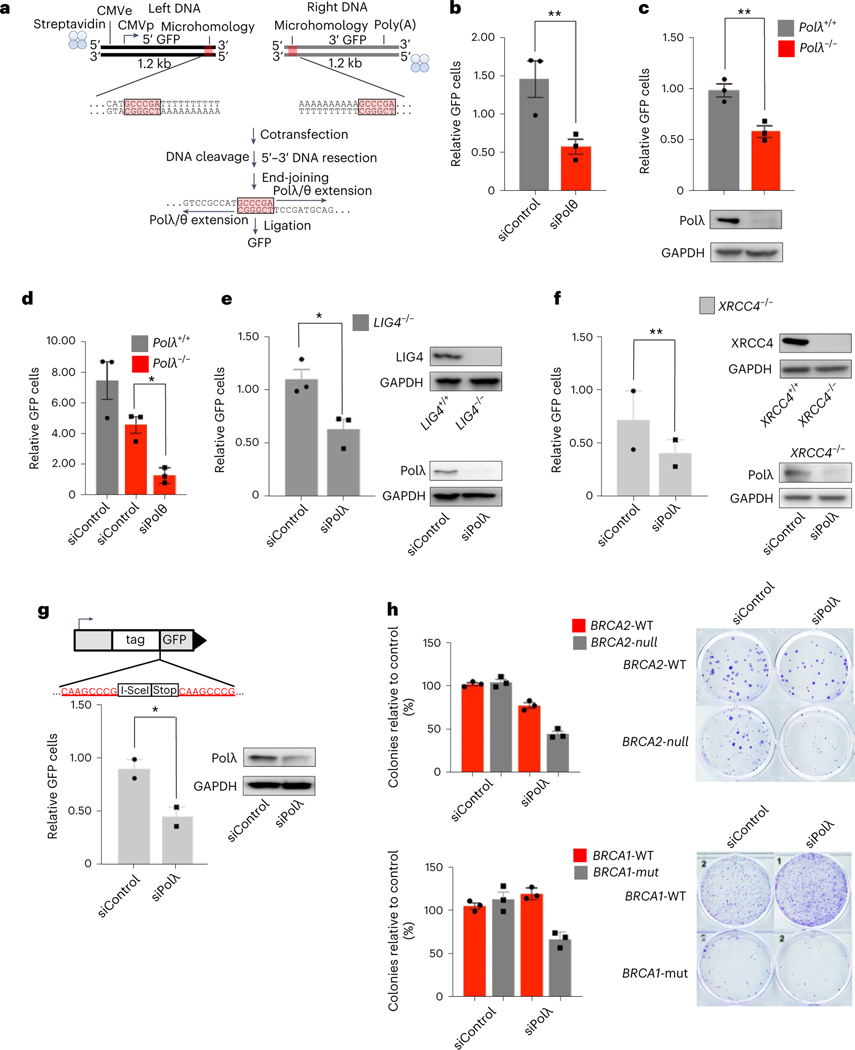
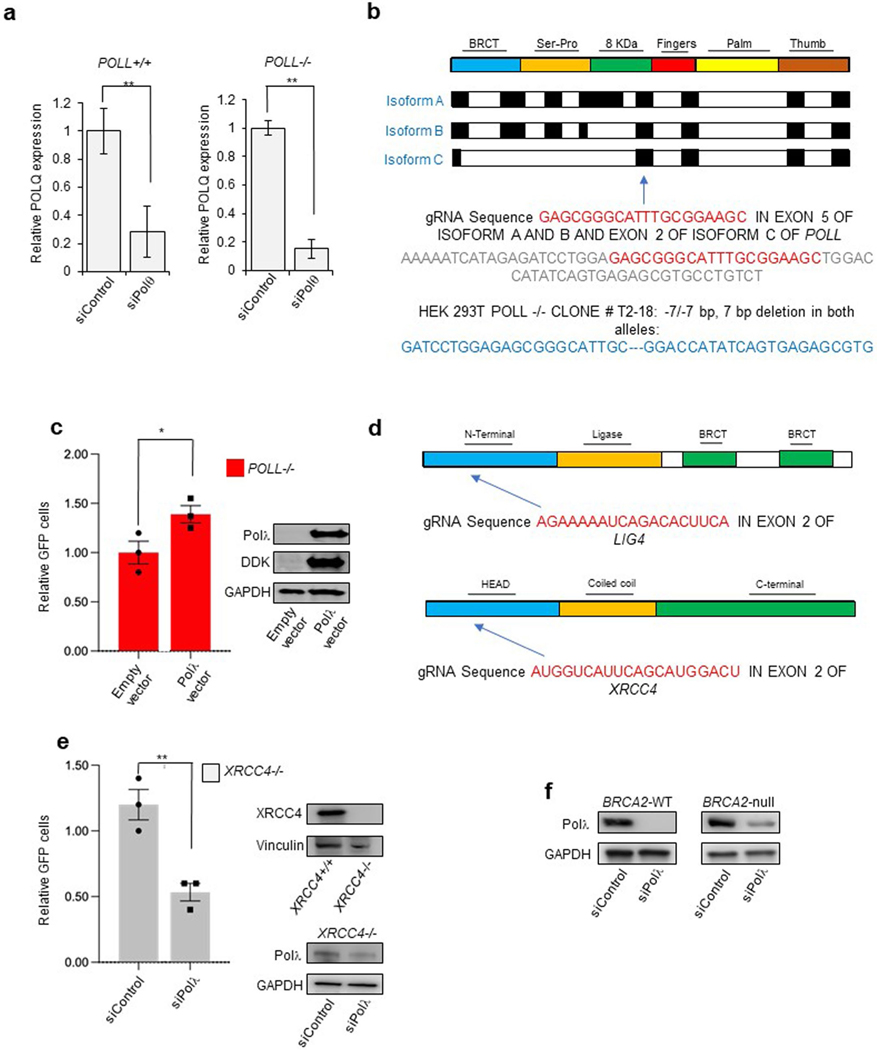
Next, the ability of Polλ to promote MMEJ in a biological setting was tested using MMEJ GFP reporter assays. We recently developed a MMEJ GFP reporter assay containing 5′ DNA-protein conjugates that demonstrated a significant reduction in MMEJ in Polθ-deficient cells13. We slightly modified the reporter assay to examine Polλ MMEJ in mammalian cells. The bipartite DNA reporter includes separate upstream (left DNA) and downstream (right DNA) portions of a linear GFP expression construct that include 6 bp of overlapping sequence, which serves as microhomology in close proximity to the internal break (Fig. 3a). For this reporter, 5′−3′ DNA resection in cells is required to generate the 3′ ssDNA overhangs that expose the complementary (microhomologous) bases. A deletion of the terminal AT-rich sequence tracts is also required for MMEJ repair and activation of the green fluorescent (GFP) expression construct in cells (Fig. 3a). The upstream and downstream terminal ends of the left and right constructs, respectively, contained streptavidin-biotin linkages to suppress nuclease degradation of DNA nonproximal to the microhomology as in previous studies13. The left and right DNA were transfected into 293T cells, then 72 h posttransfection cells were harvested and GFP(%) determined via fluorescent-activated cell sorting (FACS) of live cells. A mCherry expression vector was used as a control for normalizing transfection efficiency. As a positive control, we demonstrated that knockdown of Polθ via siRNA significantly reduced GFP cells, which confirms a main function for Polθ in MMEJ (Fig. 3b and Extended Data Fig. 3a)13. We next generated POLλ−/− 293T cells via CRISPR−Cas9 (Fig. 3c and Extended Data Fig. 3b), and determined whether these cells exhibit reduced MMEJ compared with POLλ+/+ 293T cells using the reporter assay. The results showed a significant reduction of GFP POLλ−/− cells, which demonstrates that Polλ promotes cellular MMEJ, similar to Polθ (Fig. 3c). As a control, we showed that Polλ expression rescued MMEJ in POLλ−/− cells, indicated by a significant increase in GFP (Extended Data Fig. 3c). siRNA knockdown of Polθ in POLλ−/− cells significantly reduced GFP, indicating that Polλ and Polθ have a nonepistatic relationship in MMEJ (Fig. 3d and Extended Data Fig. 3a).
Fig. 3 |. Polλ promotes MMEJ independently of Polθ and NHEJ factors.
a, Top, schematic of MMEJ reporter containing 5′-streptavidin-biotin linkages. Middle, internal termini of left and right MMEJ reporter DNA constructs. Bottom, schematic of MMEJ reporter assay. b−f, Bar plots showing relative GFP frequencies following cotransfection of left and right MMEJ reporter DNA constructs, with immunoblots showing abundance of protein shown in c and e−g. b, GFP+ frequencies are shown relative to nontargeting siRNA (siControl = 1) in wildtype HEK293T cells; n = 3; P = 0.01. c, GFP+ frequencies relative to POLλ+/+ 293T cells (POLλ+/+ = 1). n = 3, P = 0.01. d, Same as in b in POLλ−/− 293T cells. n = 3, P = 0.03. e, GFP+ frequencies relative to nontargeting siRNA (siControl = 1). n = 3, P = 0.04. f, GFP+ frequencies relative to nontargeting siRNA (siControl = 1) in XRCC4−/− 293T cells. Data represent means. n = 2, P = 0.04. g, MMEJ GFP reporter assay. Schematic of GFP reporter assay (top). Bar plot of percentage of GFP cells following transient expression of I-SceI and cotransfection of either Polλ siRNA or Control siRNA. n = 2, P = 0.04. h, Bar plots showing percentage of colonies relative to control after siRNA transfection in DLD1 BRCA2−/− or DLD1 Parental cells (top), in MDA-MB-436 BRCA1 mut or MDA-MB-231 cells (bottom). Percentage of colonies are normalized to nontargeting siRNA (siControl = 100). n = 1. Colony images are on the right. In b and c−g, GFP+ frequencies are normalized to transfection efficiency. Data represent means. ‘n’ denotes number of independent experiments with triplicates for each condition, ± s.e.m. *P < 0.05, **P < 0.01, ***P < 0.001. Statistical significance was measured from two-sample t-test and P values are indicated.
Importantly, MMEJ (alt-EJ) is defined as occurring independently of essential NHEJ factors. Because Polλ is thought to function primarily in NHEJ, we sought to determine whether Polλ MMEJ activity occurs independently of essential NHEJ factors Ligase 4 (LIG4) and XRCC4. We generated LIG4−/− 293T cells via CRISPR−Cas9 (Fig. 3e and Extended Data Fig. 3d), and tested whether Polλ promotes MMEJ in these NHEJ-deficient cells. Knockdown of Polλ via siRNA significantly reduced GFP positive LIG4−/− cells (Fig. 3e), demonstrating that Polλ promotes MMEJ independently of LIG4. Knockdown of Polλ also significantly reduced GFP-positive XRCC4-deficient 293T cells (Fig. 3f and Extended Data Fig. 3d) and XRCC4-deficient HCT116 cells (Extended Data Fig. 3e).
To further investigate Polλ MMEJ activity, we probed its ability to promote the repair of another MMEJ reporter that was originally used to identify MMEJ/alt-EJ in mammalian cells3. Here, 8 bp microhomology tracts within a GFP expression construct chromosomally integrated in mouse embryonic stem cells (mESCs) are separated by a stop codon and rare I-SceI endonuclease recognition site (Fig. 3g). Transfection of an I-SceI expression vector induces a site-specific DSB and subsequent GFP activation via MMEJ. Importantly, previous studies demonstrated that MMEJ activation of this GFP reporter occurs independently of NHEJ3. Consistent with the results above for the bipartite MMEJ reporter, siRNA knockdown of Polλ significantly reduced MMEJ of the chromosomally integrated reporter, as indicated by a significant reduction of GFP. Hence, these data further confirm the ability of Polλ to facilitate MMEJ independently of NHEJ factors.
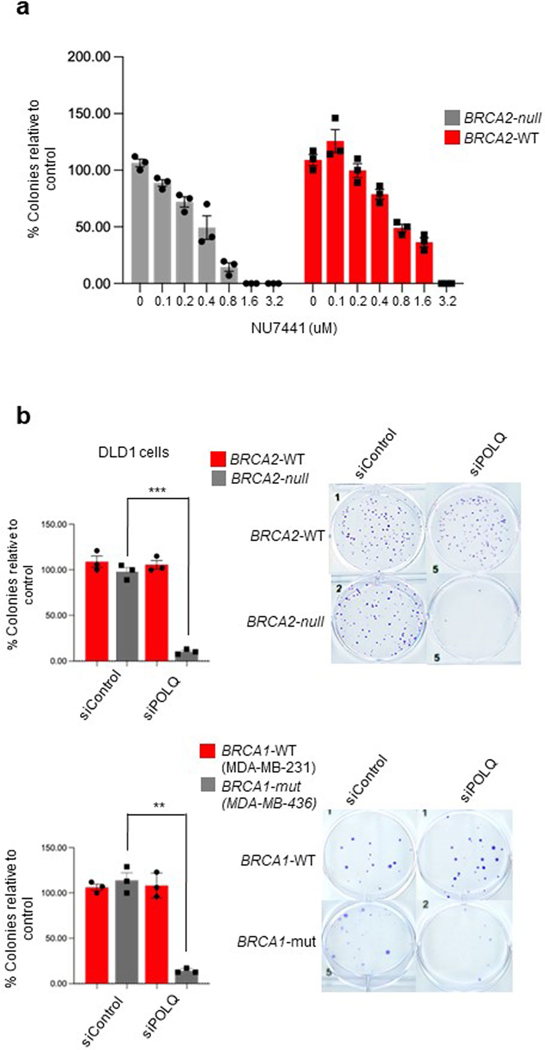
Since Polλ promotes MMEJ in addition to NHEJ, and has been implicated in base excision repair, we investigated whether it contributes to the proliferation of BRCA1/2-mutant cells, like Polθ6,12,27,28. Although suppression of NHEJ via inhibition of DNA-dependent protein kinase (DNA-PK) failed to induce synthetic lethality in BRCA1/2-mutant cells in recent studies29, and herein (Extended Data Fig. 4a), we envisaged that Polλ may promote the survival of HR-deficient cells due to its NHEJ-independent DNA repair activities. Suppression of Polλ expression via siRNA caused a substantial reduction in clonogenic survival of BRCA2-null cells; a modest reduction in survival was observed for BRCA2-wildtype (WT) cells (Fig. 3h). Knockdown of Polλ also substantially reduced the survival of BRCA1-mutant cells (Fig. 3h). By comparison, knockdown of Polθ induced near complete synthetic lethality in BRCA1/2-deficient cells (Extended Data Fig. 4b). Although Polλ is not absolutely essential for the survival of BRCA1/2-deficient cells, a semisynthetic lethal interaction is observed between Polλ and BRCA1/2, which suggests that Polλ contributes to compensatory DSB repair in HR-deficient cells, albeit to a lesser degree than Polθ.
Structural basis for Polλ MMEJ activity
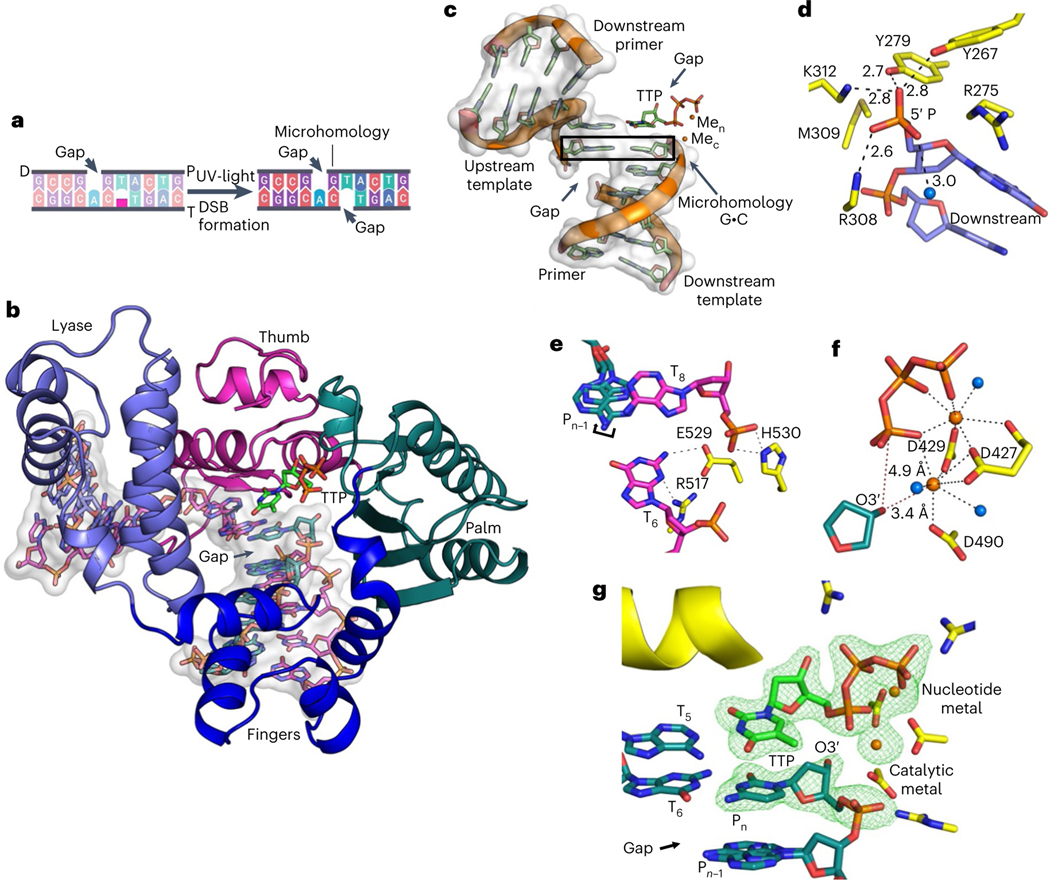
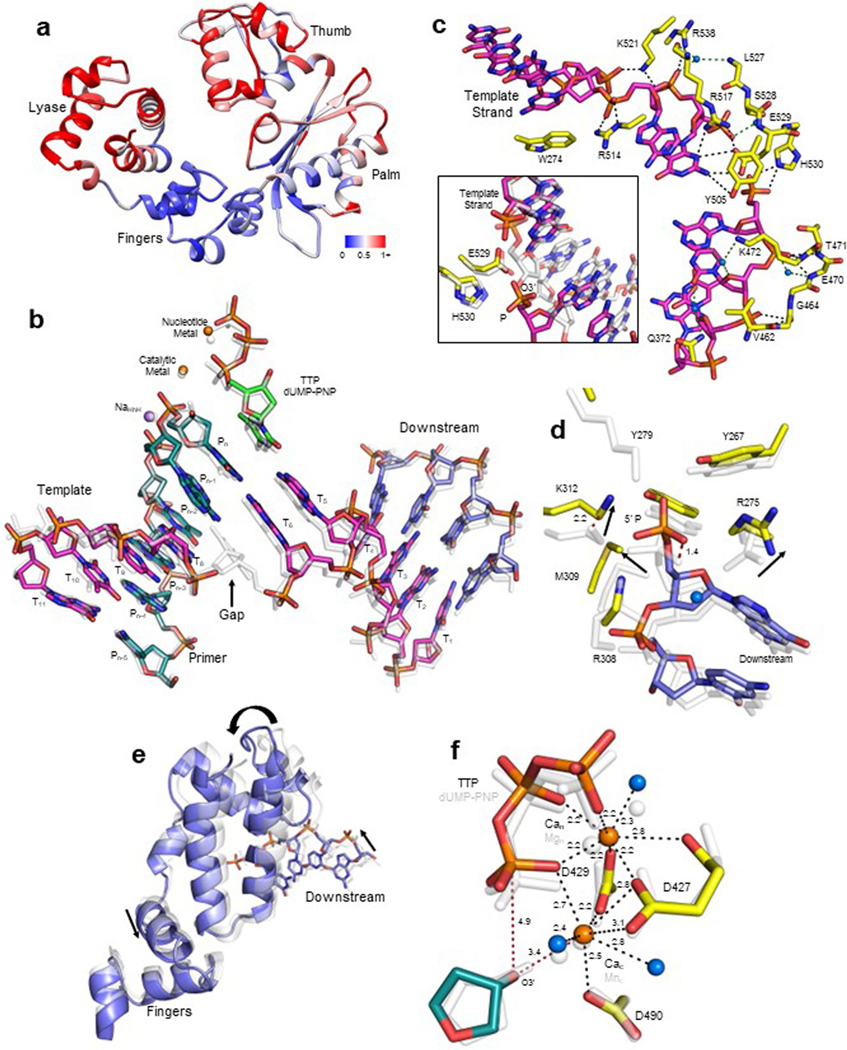
Finally, we sought to elucidate the molecular basis of Polλ MMEJ activity by employing X-ray crystallography. Attempts at growing crystals of Polλ bound to a MMEJ substrate with a short 3′-ssDNA overhang containing small tracts of microhomology were unsuccessful likely due to transient binding by the polymerase. An in situ photocleavage method was developed that employed UV-irradiation of a photocleavable nucleo tide embedded in the template strand to induce a site-specific DSB break within the crystal (Fig. 4a). This approach allowed us to solve a 2.0 Å structure of Polλ caught in the act of stabilizing a MMEJ DNA synapse mediated by a single base pair of microhomology while binding the incoming (TTP) (Fig. 4b). The structure reveals how Polλ mediates productive stabilization of a MMEJ synapse. The Polλ stabilized MMEJ junction contains four strands of polymerase-bound substrate DNA comprised of 1-nt gaps in both primer and template strands bridged by a single G-C base pair between the gaps, and bound TTP (Fig. 4c). The structure is similar to a previously determined precatalytic structure of Polλ bound to DNA containing a single nucleotide gap and a TTP analog (Extended Data Fig. 5a)30. Our structure is also similar to recently solved Polλ structures on NHEJ DNA repair intermediates paired by 1−2 base pairs31. However, an important distinction between our structure and recently solved structures of Polλ bound to NHEJ DNA intermediates is the presence of gaps on both sides of the microhomology-mediated DNA synapse, which is more representative of a MMEJ intermediate (Fig. 4a,c). The MMEJ DNA intermediate adopted a characteristic conformation displaying a bend of around 90° and bridged by a single G-C base pair (Fig. 4c and Extended Data Fig. 5b).
Fig. 4 |. Structural basis for Polλ MMEJ activity.
a, A photocleavable nucleotide (pink) was converted into a nucleotide gap within the Polλ-DNA crystal via UV light, resulting in a MMEJ synapse bridged by a single G-C base pair. T, template strand; P, primer strand; D, downstream strand. b, Structure of Polλ:MMEJ synapse with incoming nucleotide. Polymerase subdomains, lyase (magenta), fingers (blue), palm (green) and thumb (purple), are shown as cartoon. DNA (purple, template strand; cyan, primer strand; magenta, downstream strand) and TTP are shown in stick representation. Atomic volume for the DNA is shown as a transparent white surface. c, Overall DSB substrate conformation. Microhomology is outlined with a black rectangle. TTP and DNA bases are shown in stick representation with the phosphate backbone as cartoon. Orange spheres, catalytic (Mec) and nucleotide (Nac) metals. Atomic volume is shown as white transparent surface. d, Downstream 5′-phosphate coordination of the polymerase-bound microhomology. Sidechains (yellow) and downstream DNA (magenta) are shown in stick representation. Lyase domain sidechain 5′ phosphate coordination is shown with black dashes; distances (Å) are indicated. Sidechains are shown in yellow stick representation. Blue sphere, water molecule. e, Template strand gap stabilization. Stabilizing sidechain interactions with the template marginal phosphates are shown with black dashes. A curved arrow indicates alternate conformations of the base of the primer strand nucleotide opposite the template strand gap. f, Key active site distances and metal coordination. Coordination (black) and key distances (red) are indicated with dashes. g, Active site conformation is consistent with catalysis. Simulated annealing (Fo−Fc) density is shown for TTP, primer terminal (Pn) nucleotide and active site metal atoms. Helix N that stacks with the incoming nucleotide is shown as a yellow cartoon.
The lyase (8 kDa) subdomain binds the 5′-phosphate and downstream primer and stabilizes the MMEJ synapse (Fig. 4d and Extended Data Fig. 5d,e), which is consistent with 5′-phosphate stimulation of Polλ MMEJ (Fig. 2c). The fingers and palm subdomains primarily stabilize the primer strand, and the palm and thumb subdomains stabilize the template strand (Fig. 4b). Template strand gap stabilization is achieved by interactions with the phosphates on the margins of the microhomology gap (Fig. 4e and Extended Data Fig. 5c). Interestingly, His530 stabilizes the backbone phosphate of the downstream template strand gap marginal nucleotide (T8) in our structure (Fig. 4e). Yet, in a Polλ NHEJ intermediate structure containing a nick at this particular position, His530 interacts with the 3′-hydroxyl of a deoxyribose (Extended Data Fig. 5c)31. Additionally, Arg517 and Glu529 stabilize the nucleotide base of the upstream (T6) template strand (Fig. 4e). Arg517 is an active site residue that forms part of α-helix N, which influences active site conformation and nucleotide insertion fidelity.
Interestingly, the base of the primer strand nucleotide (Pn−1) opposite the template strand gap could be modeled in both the anti- and syn-conformations, and thus likely influences primer strand conformation and metal coordination in the active site (Fig. 4f and Extended Data Fig. 5f). The catalytic and nucleotide metal sites display coordination of a divalent metal ion consistent with active site metal coordination by most other polymerases. A divalent metal ion in this site has not been previously observed in the Polλ ground state ternary complex. This is significant since the catalytic metal allows activation of primer terminal O3′ for reaction with Pα to enable nucleotide insertion. Overall, the active site is in a conformation consistent with catalysis on this MMEJ synapse (Fig. 4g). Hence, these structural observations strongly support a role for Polλ in microhomology-mediated DNA synapse stabilization and productive nucleotide insertion across a MMEJ junction, consistent with our biochemical and cell biology findings.
Discussion
Our report characterizes a previously undiscovered MMEJ mechanism that is promoted by Polλ, but occurs independently of NHEJ factors LIG4/XRCC4 and Polθ. Future studies are necessary to elucidate how this mechanism of Polλ-dependent MMEJ is regulated and to identify associated end-joining factors. For example, although MMEJ was reported to occur independently of Ku70, and Ku70/Ku80 shows very weak binding to DSBs with 3′ ssDNA overhangs5,10, the possibility exists that these NHEJ factors may influence Polλ MMEJ choice following initial DSB formation3. It will also be interesting to determine whether PARP1, MRN-CtIP and LIG1/3 play direct roles in Polλ dependent MMEJ. Important considerations are whether the rate of Polλ MMEJ is significantly slowed in the absence of LIG4 (ref. 32), and whether other ligases (LIG1/3) can substitute for one another to facilitate this form of MMEJ. Mechanistically, our structural studies reveal how Polλ stabilizes a minimally paired DNA synapse to enable productive nucleotide insertion across a MMEJ junction. The enhancement of template strand gap stabilization by specific residue interactions suggests that Polλ evolved to efficiently perform replication across a minimally paired DNA synapse. Although the 1 bp MMEJ junction formed in the crystal structure is substantially shorter than the microhomology lengths used in cellular assays, we note that Polλ interacts with the MMEJ junction and incoming nucleotide in a manner that is poised for phosphodiester bond formation. Thus, the structure of the Polλ MMEJ intermediate complex supports productive DNA synthesis across the minimally paired ssDNA overhangs and is consistent with the efficient MMEJ activity of Polλ in vitro.
Consistent with our findings, recent studies further support the ability of Polλ to perform DNA synapsis and deoxyribonucleotide insertion across short 3′ ssDNA overhangs joined by 1−2 bp of microhomology31. In contrast to previous studies, our report characterizes the activity of Polλ on DNA end-joining substrates containing significantly longer 3′ ssDNA overhangs, gaps and microhomology tracts that were designed to model MMEJ intermediates. For example, we find that Polλ performs MMEJ of DNA substrates containing 5−12 nt ssDNA overhangs, 1−8 nt gaps and 4−6 bp of microhomology in a similar fashion to Polθ, the prototypical MMEJ polymerase. Interestingly, both Polλ and Polθ possess efficient extension of mismatched base pairs, suggesting their shared promiscuous activities contribute to MMEJ33,34. Importantly, our cellular studies support the ability of Polλ to promote DSB repair by utilizing 6−8 bp of microhomology along 3′ ssDNA overhangs generated by 5′−3′ resection. Hence, our report demonstrates that the end-joining activity of Polλ is not limited to DNA breaks containing short (<4 nt) ssDNA overhangs and the minimal (1−2 bp) microhomology that is more typical of NHEJ. Thus, our findings reveal that the biochemical activity of Polλ was selected to accommodate a wider variety of end-joining substrates than previously recognized. Finally, the observation that Polλ contributes to the survival of BRCA1/2-deficient cells demonstrates the potential of this multifunctional polymerase as a DNA damage response drug target.
Online content
Any methods, additional references, Nature Portfolio reporting summaries, source data, extended data, supplementary information, acknowledgements, peer review information; details of author contributions and competing interests; and statements of data and code availability are available at https://doi.org/10.1038/s41594-022-00895-4.
Methods
In vitro MMEJ assay
The indicated 5′ 32P-γ-ATP radiolabeled pssDNA or ssDNA template (50 nM) was mixed with 20 μM dNTPs and 2 mM ATP, in a 40 μl volume of reaction buffer solution (25 mM Tris-HCl pH 8.8, 1 mM DTT, 0.01% NP-40, 10% glycerol, 10 mM MgCl2). MMEJ reactions were initiated by adding 20 nM of the indicated polymerase and incubating at 37 °C. Reactions were terminated after 40 min, unless indicated otherwise, by the addition of 10 μl 5× stop buffer (0.1 M Tris-HCl pH 7.5, 80 mM EDTA, 10 mg ml−1 proteinase K, 0.5% SDS) and incubating at 37 °C for 15−30 min. DNA was resolved in nondenaturing 12% polyacrylamide gels run at 20 W to maintain low temperature. MMEJ (%) was determined by dividing the intensity of the upper band (MMEJ products) by the sum of the intensities of all apparent bands in the respective lane. Bar plot data is represented as mean of MMEJ (%), n = 3, ± s.d.
Cell lines
Mouse embryonic stem cells with MMEJ reporter was a kind gift from J. Stark (City of Hope) and were generated in previous studies as described3. The cells were cultured in ES medium on either MEF feeders or gelatinized plates. ES medium contains DMEM (Gibco) supplemented with 15% FBS (Cytivia), 2 mM l-glutamine (Sigma), 100 U ml−1 Penicillin (Sigma), 0.1 μg ml−1 streptomycin (Sigma), 0.1 mM nonessential amino acids (Invitrogen), leukemia inhibitory factor (LIF), 2-β-mercaptoethanol (Gibco) and penicillin/streptomycin (Sigma). POLλ−/− HEK293T cells were generated by CRISPR−Cas9 engineering and were purchased from Genscript. Lig4 null HEK293T cells were generated by CRISPR−Cas9 engineering. DNA plasmid (5 μg) (pX458 containing LIG4 short guide RNA sequence) was transfected into HEK293T cells using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. At 48 h after transfection, cells were harvested and genomic DNA was isolated using a DNA extraction kit (Qiagen) according to the manufacturer’s instructions. Genomic DNA fragments of LIG4 around the short guide RNA site were amplified by PCR. PCR products were used for Surveyor nuclease mutation detection assays using the SURVEYOR mutation detection kit (Transgenomic) according to manufacturer’s protocol. Single cells were sorted by flow cytometry (BD FACSAria). Single cell clones were expanded and screened for and knockout by western blotting. A XRCC4 null HEK293T pool was generated by CRISPR−Cas9 engineering and were purchased from Synthego. HEK293T cells were cultured in DMEM supplemented with 10% FBS, 2 mM l-glutamine, nonessential amino acids and penicillin/streptomycin. XRCC4 null HCT116 was a kind gift from E. Hendrickson (University of Minnesota) and were generated in previous studies35. DLD1 BRCA2−/− and DLD1 Parental were obtained from Horizon discovery. MDA 436 BRCA1 mut and MDA 231 (used as WT control for MDA 436) cells were obtained from the American Type Culture Collection. DLD1 BRCA2−/−, DLD1 Parental, MDA-MB-436 BRCA1 mut and MDA-MB-231 were cultured in RPMI supplemented with 10% FBS, 2 mM l-glutamine, nonessential amino acids and penicillin/streptomycin.
Bipartite MMEJ GFP reporter assay
The methods described below were derived from a previously published article13. HEK293T and HCT116 cells (1 × 104) were plated on a 24 -well plate and transfected 24 h later with 0.25 μg of each of the indicated left- and right-flanking DNA GFP constructs using Lipofectamine 2000 (Invitrogen). To measure transfection efficiency, 100 ng WT linear mcherry expression construct (pCAGGS-mcherry) was transfected simultaneously along with left- and right-flanking DNA GFP constructs. For overexpression of Polλ WT, 1 × 104 HEK293T cells were plated and, after 24 h, 200 ng either Polλ WT-Myc-DDK-tagged (Origene catalog no. RC230157) or Empty vector control plasmid was transfected using lipofectamine 2000. At 24 h after transfection of the Polλ or empty vector control plasmid, 0.25 μg each of left and the right flank of GFP, along with 100 ng of pCAGGS-mcherry was transfected using Lipofectamine 2000. For siRNA experiments, cells were transfected with 20 pmol siRNA along with 0.25 μg each of left and right flank of GFP, 100 ng mCherry. GFP-positive cell frequencies were measured 3 days after transfection by flow cytometry using Becton Dickinson 5 Laser LSRII in independent replicates and corrected for transfection efficiency and background events. BD FACS DIVA was used for analysis. Data are represented as the mean and s.e.m. of three independent experiments, with at least duplicates per experiment. Statistical analysis was carried out by paired t-test.
Synthesis of DNA substrates for bipartite MMEJ GFP reporter assay
PCR was performed according to recommended conditions for the Phusion High-Fidelity DNA Polymerase (New England BioLabs) with 10 ng of the pCMV-GFP plasmid in 1× Phusion HF Buffer. PCR for the left-flank DNA with 6 bp of internal microhomology (PCR1.6I.SA1) was performed with forward primer RP500B and RP506. PCR for the right-flank DNA with 6 bp of internal microhomology (PCR2.6I.SA1) was performed with primers RP507 and RP503B. Following PCR, left- or right-flank DNA constructs were pooled together and digested with DpnI (New England Biolabs) in 1× CutSmart buffer and then purified via QIAGEN QIAquick PCR Purification Kit. Purified PCR was then conjugated to Strepatividin (Sigma) at 110 ng ml−1 of PCR and 0.8 mg ml−1 Streptavidin in 10 mM Tris-HCl pH 7.5, 100 mM NaCl at 37 °C for 1 h. Conjugation was confirmed by resolution in a 0.8% agarose gel stained with ethidium bromide.
Chromosomal MMEJ GFP assay
mESCs (2 × 105) harboring the chromosomal MMEJ reporter were transfected in suspension in 24-well plates with 0.5 μg pCMV-3×-NLS-I-SceI or 0.5 μg control vector pCMV-3×-NLS using Lipofectamine 2000 (Invitrogen). GFP+ frequencies were measured 3 days posttransfection by FACS using Becton Dickinson 5 Laser LSRII in triplicate and corrected for transfection efficiency and background events. (Transfection efficiency was measured simultaneously by parallel transfection with 0.05 μg WT GFP expression vector.) For siRNA experiments, cells were transfected with 1 μl 20 μM (that is, 20 pmol) siRNA plus 0.3 μg pcDNA3β−myc NLS-I-SceI (or control vector) per well.
Clonogenic assay
DLD1 BRCA2−/−, DLD1 Parental, MDA 436 BRCA1 mut and MDA 231 were plated on 24-well plates and transfected with 20 pmol or either siControl or siPolλ or siPolθ 24 h after plating using Lipofectamine RNAimax (Invitrogen). Cells were subjected to a second round of transfection on day 3, and on day 5 they were plated on six-well plates at 1,000 cells per well for clonogenic assay. Medium was replaced every 2 days until the colonies were ready for staining in 10−12 days. Medium was removed from plates and cells were rinsed with PBS. Fixation was carried out with Acetic acid:methanol (1:7) for 30 min followed by staining of colonies with 0.5% crystal violet for 2 h at room temperature. The plates were rinsed with water and left to dry overnight at room temperature.
Purification of Polλ for X-ray crystallography
Truncated human Polλ (residues 242−575 (ref. 14) was overexpressed in BL21(DE3)CodonPlus-RIL cells (Invitrogen) and purified as described previously36. The construct employed included a modified loop1 and C543A mutations to improve crystallizability. Briefly, cells were resuspended in lysis buffer (25 mM Tris-HCl pH 7.5, 0.35 M NaCl, 10% glycerol, 1 mM DTT, 1 mM EDTA) and lysed by sonication on ice. Genomic DNA was precipitated by addition of 0.1% polyethylene imine. The supernatant was purified in tandem by Q Sepharose-heparin chromatography in lysis buffer and eluted with a linear gradient to 1 M NaCl. Pooled fractions were then purified on a MonoS HR 10/10 column (linear gradient, 0.1−1 M NaCl) after overnight dialysis in purification buffer (25 mM Tris-HCl pH 7.5, 0.1 M NaCl, 10% glycerol, 1 mM DTT, 1 mM EDTA). Fractions resulting from size exclusion chromatography on a Superdex 200 26/600 column were pooled and dialyzed overnight in purification buffer lacking glycerol and EDTA. Pol λ was concentrated to 16 mg ml−1, flash frozen in liquid nitrogen and stored at −80 °C.
X-ray crystallography
X-ray crystallography was performed as described previously36. An 11-mer template oligonucleotide (5′-CGGCAG(PC)ACTG-3′; (PC) is a photocleavable 1-(2-nitrophenyl)propane-1,3-diol spacer) was annealed with a 6-mer upstream (5′-CAGTAC-3′) oligonucleotide and a 5′-phosphorylated downstream 4-mer (5′-pGCCG-3′) oligonucleotide in a 1:1:1 ratio to create a duplex DNA with a single nucleotide gap. DNA synthesis was performed by Biosynthesis Inc.
Oligonucleotides were dissolved in 100 mM Tris-HCl pH 7.5, heated to 95 °C for 5 mins and then cooled down to 4 °C at a rate of 1 °C min−1. Crystals of Polλ were grown as follows: Polλ (16 mg ml−1) was mixed with annealed DNA in a 1:2 ratio and incubated at 4 °C for 2 h, then the mixture was incubated for an additional 2 h in the presence of 2 mM dTTP and 10 mM CaCl2. Crystallization plates were set up by mixing Polλ-DNA-dNTP ternary complex with well solution (20 mM bicine pH 7.5, 14−20% polypure PEG, 300 mM Na-K tartrate) in sitting drop format. Crystals were irradiated at 365 nm in cryosolution consisting of artificial mother liquor for 2 h.
Data collection was performed at the Advanced Photon Source (Argonne National Laboratory) on the ID22 beamline (SER-CAT (Southeast Regional Collaborative Access Team)) using the Eiger 16 M detector at 1.00 Å. Data were processed using DIALS37 and StarAniso (http://staraniso.globalphasing.org/cgi-bin/staraniso.cgi). The structure of Polλ bound to a DSB was determined using molecular replacement employing a previous structure of Polλ (PDB id 3UPQ)38. Rfree flags were taken from the starting model. Refinement was carried out using the PHENIX software package39 and iterative model building was done with Coot40. Simulated annealing omit density maps are shown as a green mesh in the figures and were generated by deleting the region of interest and performing a refinement with simulated annealing. The figures were prepared with PyMol. Crystallographic statistics are shown in Table 1.
Table 1 |.
Crystallographic statistics
| PDB ID | |
|---|---|
| Data collection | |
| Space group | P212121 |
| Cell dimensions | |
| a, b, c (Å) | 56.135 |
| 62.774 | |
| 140.193 | |
| α, β, γ (°) | 90, 90, 90 |
| Resolution (Å)a | 50−2.04 |
| (2.11 − 2.04) | |
| I /σIa | 10.6 (1.6) |
| Completeness (spherical)a | 94.0 (68.5) |
| Completeness (ellipsoid)a | 97.3 (98.6) |
| Redundancya | 13.3 (13.8) |
| CC(1/2)a | 0.93 (0.59) |
| Unique reflections (no.)a | 32,337 |
| Refinement | |
| Resolution (Å) | 50−2.04 |
| Reflections (no.) | 25,392 |
| Rwork / Rfree | 0.22 / 0.25 |
| Atoms (no.) | |
| Protein / DNA | 2,347 / 432 |
| Water / ligands | 150 / 55 |
| B-factors | |
| Protein / DNA | 46.3 / 34.3 |
| Water / ligands3 | 38.3 / 35.5 |
| Wilson B | 43.2 |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.002 |
| Bond angles (°) | 0.523 |
Data in the highest resolution shell are shown in parentheses.
Western blot analyses
Cells were harvested using 0.25% Trypsin-EDTA (Gibco, catalog no. 25200056) and washed in PBS, then lysed in 1× RIPA Lysis and Extraction Buffer (Thermo Scientific, catalog no. 89901) containing Halt Phosphatase Inhibitor Cocktail (Thermo Scientific, catalog no. 78420). Protein concentration was measured using a Pierce BCA Protein Assay Kit (Thermo Scientific, catalog no. 23225). The proteins were separated by electrophoresis using 7.5% Mini-PROTEAN TGX Precast Protein Gels (Bio-Rad, catalog no. 4561023) and transferred to Immuno Blot PVDF Membrane (Bio-Rad, catalog no. 1620177). Membranes were blocked using 5%-milk-TBST for 1 h at room temperature and washed with TBST before antibody incubation. Primary antibodies were incubated overnight at 4 °C in 5%-milk-TBST. DDK was detected using Clone OTI4C5, Anti-DDK (FLAG) monoclonal antibody (OriGene, catalog no. TA50011-30) diluted 1:2,000. Polλ was detected using DNA Polλ rabbit polyclonal antibody (Novus Bio, catalog no. NB100-81665) diluted 1:1,000. Lig4 was detected using DNA Ligase IV (D5N5N) rabbit monoclonal antibody (Cell Signaling Technology, catalog no. 14649) diluted 1:2,000. Gapdh was detected using Gapdh (14C10) rabbit monoclonal antibody (Cell Signaling Technology, catalog no. 2118) diluted 1:4,000. Xrcc4 was detected using XRCC4 rabbit polyclonal antibody (Invitrogen, catalog no. PA576068) diluted 1:5,000. Vinculin was detected using Vinculin mouse monoclonal antibody (Invitrogen, catalog no. PA576068) diluted 1:5,000. Membranes were washed with TBST before secondary antibody incubation for 1 h at room temperature using respective Goat anti-rabbit IgG (H+L) HRP (Invitrogen, catalog no. 32466) or Goat anti-Mouse IgG (H+L) HRP (Invitrogen, catalog no. 31430)-tagged secondary antibody diluted 1:5,000 in 5%-milk-TBST. Membranes were washed using TBST and then treated with Amersham ECL Prime Western Blotting Detection Reagent (Cytiva, catalog no. RPN2232) for 5 min and images were obtained using the Bio-Rad ChemiDoc Imaging System.
Proteins
Polθ, Polλ, Polδ, Polμ and Polβ were purified as described11,41–43. Klenow fragment 3′−5′ exo minus was purchased from New England Biosciences (NEB). Polγ exo minus was a gift from B. Copeland (NIEHS). Polκ was purchased from Enzymax. Polε was a gift from M. O’Donnell (Rockefeller University).
Recombinant human Polη was expressed and purified as follows. Polη expression plasmid was purchased from GenScript, and contains the cDNA sequence corresponding to amino acids 1−711 inserted into a pET32a backbone. BL21-CodonPlus(DE3)-RIL competent cells (Agilent Technologies) were used as expression strain. Polη was expressed and purified as described44,45, with a minor change: instead of MonoS column and buffer C, dialysis was performed before concentration with 2 l of the following buffer (25 mM sodium phosphate (pH 7.4), 10% glycerol, 200 mM NaCl, 5 mM β-mercaptoethanol). Identified protein fractions were analyzed by SDS−PAGE gels. Selected fractions containing highly purified polymerase were aliquoted and frozen and stored at −80 °C until further use.
DNA (5′- 3′)
RP500B - /5Biosg/GCTAGCCAGTCAGTGGGCCCGC
RP506 - AAAAAAAAAATCG GGC ATG GCG GAC TTG AAG AAG TCG
RP507- AAAAAAAAAAGCCCGAAGGCTACGTCCAGGAGCG
RP503B - /5Biosg/TGATTACGCCAAGTTAATTAAGGACGTCCT CCTGCTGG
CRISPR gRNA sequence (5′- 3′)
Human Xrcc4 − AUGGUCAUUCAGCAUGGACU
Human Polλ − GAGCGGGCATTTGCGGAAGC
siRNA (5′- 3′)
Human Polθ − SMARTpool: CAACAACCCUUAUCGUAAA, CGACUAAGAUAGAUCAUUU, ACACAGUAGGCGAGAGUAU, CCUUAAGACUGUAGGUACU
Human Polλ − SMARTpool: CCAUCGGCCUGAAGCAUUA, GAAGCUGGACCAUAUCAGU, GAACGUAUGCCCAGGGAGG, GAGAAUGGUCAGCAACAGA
Mouse Polλ − SMARTpool: GCAUGAGCCUGUCGGAGCA, CGGGAAUUGGCAAGCGGAU, GAGCAGACGGUCCGGAUAU, AGAGUGGGCAUCUGCGGAA
RT-qPCR
A portion of cells from MMEJ reporter assays or clonogenic assays performed with siRNA was used for RNA extraction. RNA was extracted using High Capacity cDNA Reverse Transcription Kit (Thermo Fisher Scientific, catalog no. 4368814). Analysis of first strand cDNA was by Power SYBR Green PCR master mix (Applied Biosystems, catalog no. 4367659). Quant Studio 12 K flex system (Thermo Fisher) was used for RT-qPCR. Primers used for RT-qPCR:
Polθ − sense: 5′-GCCAGGGTTCTCTATGCTTC;
Polθ − antisense: 5′-TCTTCAACTGCTTCCTCTTCC
Actin − sense: 5′-TGACCCAGATCATGTTTGAGACCTTCA;
Actin − antisense: 5′-GGAGTCCATCACAATGCCTGTGG
Statistics and reproducibility
Two-sample t-test was used in Figs. 2 and 3 and Extended Data Figs. 3 and 4. Data are presented as mean ± s.e.m. of results from at least two independent experiments with triplicates for each condition. Comparisons between two groups (POLλ+/+ versus POLλ−/−, siControl versus siPolλ, siControl versus siPolθ) were analyzed by two-tailed Student’s t-test. Significance was assumed at P < 0.05. Asterisks in the figures indicate significance, *P < 0.05, **P < 0.01, ***P < 0.001. Statistically significant P values and number of replicates are indicated in the figure legends.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Extended Data
Extended Data Fig. 1 |. Controls for Pol activity on primer-templates and substrates containing 3’ ssDNA.
a, Denaturing gels showing extension of the indicated primer-template by the indicated Pols using identical conditions. Polμ is known to require a downstream ssDNA strand for optimal activity primer extension activity along a short gap. 20 nM Pol concentrations were used. b, Schematic of DNA templates. Microhomology indicated as red text. c, Non-denaturing gel showing Polλ MMEJ as a positive control for its activity (left panel). Denaturing gels showing extension of the indicated templates in the presence of all 4 dNTPs by the indicated Pols (middle and right panel). 20 nM Pol concentrations were used. d, Schematic showing the respective activities of Polθ and Polλ on the indicated templates. although both enzymes perform MMEJ (top), only Polθ exhibits ssDNA extension due to its snap-back replication activity.
Extended Data Fig. 2 |. Controls for Pol activity on MMEJ substrates.
a, Denaturing gel showing extension of a pssDNA substrate containing 2 bp of microhomology in the presence of dCTP. Polλ but not Polμ performs addition of dCMP on the indicated substrate. Microhomology indicated as red text. 20 nM Pol concentrations were used. b, Non-denaturing gel showing MMEJ activity by the indicated Pols on the indicated pssDNA containing 6 bp of microhomology (red text). Polλ performs MMEJ whereas Polβ does not. Reactions were performed in duplicate. 20 nM Pol concentrations were used.
Extended Data Fig. 3 |. Supplemental data for protein expression, genetic engineering, and MMEJ activity.
a. RT qPCR analysis of Polθ expression. mRNA levels were corrected with internal control for Actin in siRNA-treated cells used in Fig. 3b, d as well as normalized to non-targeting siRNA (siControl = 1). Data represent mean. n = 1 experiment with triplicate for each condition ±SEM. b. gRNA sequence used to generate POLL−/− HEK293T cells via CRISPR-Cas9 engineering. Schematic representation of three isoforms of human Polλ with protein domains as well as location of gRNA sequence (red) is indicated. The genome sequence flanking the gRNA sequence (red) is shown in gray. POLL −/− clone # T2 was generated by CRISPR-Cas9 engineering and carries 7 bp deletion in both alleles. Sequence of the region harboring the 7 bp deletion is indicated in blue. c. Bar plot showing relative GFP following overexpression of indicated plasmids and co- transfection of left and right MMEJ reporter DNA constructs in HEK293T cells. GFP+ frequencies are normalized to transfection efficiency. Data represent mean. n = 1 experiment with triplicates for each condition, +/− s.e.m. Bottom panel: Immunoblot showing abundance of protein. d. gRNA sequence used to generate LIG4 −/− HEK293T cells (top) and XRCC4 −/− HEK293T cells (bottom) via CRISPR-Cas9 engineering. Schematic representation of human Lig4 (top) and Xrcc4 (bottom) with protein domains as well as location of gRNA sequence is indicated (red). e. Same as in Fig. 3f in XRCC4−/− HCT116 cells. Data represent mean. n = 1 experiment with triplicate for each condition, +/− s.e.m. Bottom panel: Immunoblot showing abundance of protein. f. Western blot of Polλ (top) and Gapdh (bottom) following transfection of either Polλ siRNA or siControl in DLD1 BRCA2+/+ (left) and DLD1 BRCA2 −/− cells (right).
Extended Data Fig. 4 |. Analysis of synthetic lethality interactions in BRCA1/2-deficient cells.
a. Plot showing percentage of colonies relative to control after treatment with indicated concentrations of DNA-PK inhibitor (NU-7441) in DLD1 BRCA2 −/− or DLD1 Parental cells. Data represent mean. n = 1 experiment with triplicate for each condition, ± s.e.m. b. Bar plots showing percentage of colonies relative to control after siRNA transfection with either siControl or siPolθ in DLD1 BRCA2 −/− or DLD1 Parental cells (top), in MDA 436 BRCA1 mut or MDA 231 cells (bottom). Percentage of colonies are normalized to non-targeting siRNA (siControl = 100). Data represent mean. n = 1 experiment with triplicate for each condition, ± s.e.m. Colony images are on the right.
Extended Data Fig. 5 |. Supporting data for the structural basis of Polλ MMEJ activity.
a. Structural comparison of Polλ bound to a microhomology-mediated DNA synapse and a single nucleotide gap (PDB id 2PFO). Differences (0−4.2 Å) in backbone Cα positioning are displayed as a heatmap colored from blue (0 Å) to white (0.5 Å) to red (1+Å) mapped onto the structure of the double strand break bound pol λ in cartoon representation. b. Overlay of the microhomology substrate containing gaps in template and primer strands with a single nucleotide gap substrate (PDB id 2PFO, transparent gray). Incoming TTP (green) or dUMPNPP (transparent gray) and downstream (magenta), primer (cyan), template (purple) strands are shown in stick representation. The orange spheres are active site metal ions and the purple sphere is a sodium atom. c. Template strand interactions in DSB bound Polλ. Template strand (magenta) and sidechains (yellow) are shown in stick representation. Key interactions are shown with black (side chains) or green (water) dashes. Waters are shown as blue spheres. Inset. Comparison of template strand positioning and gap marginal nucleotide interactions in structures of Polλ with a nick in the template strand (white transparent sticks, PDB id 7M0D31) and a gap in the same position (yellow sidechains, purple DNA). d. Differences in downstream 5′ phosphate coordination overlaid with the structure of a single nucleotide gap (PDB id 2PFO, transparent gray). e. Lyase domain and downstream primer shift compared to the structure with a single nucleotide gap (PDB id 2PFO). Protein backbone and DNA are shown in magenta cartoon and stick representation, respectively. f. Metal coordination in the active site. Shown is an overlay with a structure of Polλ bound to a single nucleotide gap and a non-hydrolyzable nucleotide (PDB id 2PFO, transparent gray).
Acknowledgements
This report is dedicated to Samuel H. Wilson who passed away during the course of these studies. We thank B. Copeland (National Institute of Environmental Health Sciences) for providing Polκ and M. O’Donnell (Rockefeller University) for providing recombinant yeast Polε. We are grateful to Dr. Wilson (NIEH) for providing recombinant human Polλ, Polμ, and Polβ. This research was supported by National Institute of Health grants 1R01GM130889 and 1R01GM137124 to R.T.P., and DOD W81XWH-18-1-0148 and Career Enhancement Program grant from the Yale Head and Neck Cancer SPORE to S.A.
Footnotes
Additional information
Extended data is available for this paper at https://doi.org/10.1038/s41594-022-00895-4.
Supplementary information The online version contains supplementary material available at https://doi.org/10.1038/s41594022-00895-4.
Peer review information Nature Structural & Molecular Biology thanks the anonymous reviewers for their contribution to the peer review of this work. Peer reviewer reports are available. Primary Handling Editors: Beth Moorefield, Florian Ullrich and Carolina Perdigoto, in collaboration with the Nature Structural & Molecular Biology team.
Competing interests
R.T.P. is a cofounder and chief scientific officer of Recombination Therapeutics, LLC. The other authors declare no competing interests.
Data availability
X-ray crystallography structure of Polλ with microhomology-mediated DNA synapse was deposited with protein data bank under accession code PDB 2PFO. Source data are provided with this paper.
References
- 1.Moynahan ME & Jasin M. Mitotic homologous recombination maintains genomic stability and suppresses tumorigenesis. Nat. Rev. Mol. Cell Biol 11, 196–207 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhao B, Rothenberg E, Ramsden DA & Lieber MR The molecular basis and disease relevance of non-homologous DNA end joining. Nat. Rev. Mol. Cell Biol 21, 765–781 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bennardo N, Cheng A, Huang N. & Stark JM Alternative-NHEJ is a mechanistically distinct pathway of mammalian chromosome break repair. PLoS Genet. 4, e1000110 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yan CT et al. IgH class switching and translocations use a robust non-classical end-joining pathway. Nature 449, 478–482 (2007). [DOI] [PubMed] [Google Scholar]
- 5.Boboila C. et al. Alternative end-joining catalyzes robust IgH locus deletions and translocations in the combined absence of ligase 4 and Ku70. Proc. Natl Acad. Sci. USA 107, 3034–3039 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mateos-Gomez PA et al. Mammalian polymerase θ promotes alternative NHEJ and suppresses recombination. Nature 518, 254–257 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yousefzadeh MJ et al. Mechanism of suppression of chromosomal instability by DNA polymerase POLQ. PLoS Genet. 10, e1004654 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Koole W. et al. A polymerase θ-dependent repair pathway suppresses extensive genomic instability at endogenous G4 DNA sites. Nat. Commun. 5, 3216 (2014). [DOI] [PubMed] [Google Scholar]
- 9.Chan SH, Yu AM & McVey M. Dual roles for DNA polymerase θ in alternative end-joining repair of double-strand breaks in Drosophila. PLoS Genet. 6, e1001005 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wyatt DW et al. Essential roles for polymerase θ-mediated end joining in the repair of chromosome breaks. Mol. Cell 63, 662–673 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kent T, Chandramouly G, McDevitt SM, Ozdemir AY & Pomerantz RT Mechanism of microhomology-mediated end-joining promoted by human DNA polymerase θ. Nat. Struct. Mol. Biol. 22, 230–237 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ramsden DA, Carvajal-Garcia J. & Gupta GP Mechanism, cellular functions and cancer roles of polymerase-θ-mediated DNA end joining. Nat. Rev. Mol. Cell Biol. 23, 125–140 (2022). [DOI] [PubMed] [Google Scholar]
- 13.Chandramouly G. et al. Polθ promotes the repair of 5′-DNA-protein crosslinks by microhomology-mediated end-joining. Cell Rep. 34, 108820 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hussmann JA et al. Mapping the genetic landscape of DNA double-strand break repair. Cell 184, 5653–5669 e5625 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carvajal-Garcia J. et al. Mechanistic basis for microhomology identification and genome scarring by polymerase θ. Proc. Natl Acad. Sci. USA 117, 8476–8485 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kamp JA, van Schendel R, Dilweg IW & Tijsterman M. BRCA1-associated structural variations are a consequence of polymerase θ-mediated end-joining. Nat. Commun. 11, 3615 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Feldman T. et al. Recurrent deletions in clonal hematopoiesis are driven by microhomology-mediated end joining. Nat. Commun. 12, 2455 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Black SJ et al. Molecular basis of microhomology-mediated end-joining by purified full-length Polθ. Nat. Commun. 10, 4423 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bebenek K, Pedersen LC & Kunkel TA Structure-function studies of DNA polymerase λ. Biochemistry 53, 2781–2792 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lieber MR The mechanism of human nonhomologous DNA end joining. J. Biol. Chem. 283, 1–5 (2008). [DOI] [PubMed] [Google Scholar]
- 21.Braithwaite EK et al. DNA polymerases β and λ mediate overlapping and independent roles in base excision repair in mouse embryonic fibroblasts. PLoS ONE 5, e12229 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Braithwaite EK et al. DNA polymerase λ mediates a back-up base excision repair activity in extracts of mouse embryonic fibroblasts. J. Biol. Chem. 280, 18469–18475 (2005). [DOI] [PubMed] [Google Scholar]
- 23.Kent T, Mateos-Gomez PA & Pomerantz RT Polymerase θ is a robust terminal transferase that oscillates between three different mechanisms during end-joining. eLife 5, e13740 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Prasad R. et al. Human DNA polymerase θ possesses 5’-dRP lyase activity and functions in single-nucleotide base excision repair in vitro. Nucleic Acids Res. 37, 1868–1877 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Crespan E, Czabany T, Maga G. & Hubscher U. Microhomology- mediated DNA strand annealing and elongation by human DNA polymerases λ and β on normal and repetitive DNA sequences. Nucleic Acids Res. 40, 5577–5590 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ray S. et al. DNA polymerase β participates in DNA end-joining. Nucleic Acids Res. 46, 242–255 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zatreanu D. et al. Polθ inhibitors elicit BRCA-gene synthetic lethality and target PARP inhibitor resistance. Nat. Commun. 12, 3636 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ceccaldi R. et al. Homologous-recombination-deficient tumours are dependent on Polθ-mediated repair. Nature 518, 258–262 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fok JHL et al. AZD7648 is a potent and selective DNA-PK inhibitor that enhances radiation, chemotherapy and olaparib activity. Nat. Commun. 10, 5065 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Garcia-Diaz M, Bebenek K, Krahn JM, Pedersen LC & Kunkel TA Role of the catalytic metal during polymerization by DNA polymerase λ. DNA Repair (Amst.) 6, 1333–1340 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kaminski AM et al. Analysis of diverse double-strand break synapsis with Polλ reveals basis for unique substrate specificity in nonhomologous end-joining. Nat. Commun. 13, 3806 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Han L. & Yu K. Altered kinetics of nonhomologous end joining and class switch recombination in ligase IV-deficient B cells. J. Exp. Med. 205, 2745–2753 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Picher AJ et al. Promiscuous mismatch extension by human DNA polymerase λ. Nucleic Acids Res. 34, 3259–3266 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Seki M. & Wood RD DNA polymerase θ (POLQ) can extend from mismatches and from bases opposite a (6–4) photoproduct. DNA Repair (Amst.) 7, 119–127 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Oh S, Wang Y, Zimbric J. & Hendrickson EA Human LIGIV is synthetically lethal with the loss of Rad54B-dependent recombination and is required for certain chromosome fusion events induced by telomere dysfunction. Nucleic Acids Res. 41, 1734–1749 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jamsen JA, Shock DD & Wilson SH Watching right and wrong nucleotide insertion captures hidden polymerase fidelity checkpoints. Nat. Commun. 13, 3193 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Beilsten-Edmands J. et al. Scaling diffraction data in the DIALS software package: algorithms and new approaches for multi-crystal scaling. Acta Crystallogr. D Struct. Biol. 76, 385–399 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gosavi RA, Moon AF, Kunkel TA, Pedersen LC & Bebenek K. The catalytic cycle for ribonucleotide incorporation by human DNA Pol λ. Nucleic Acids Res. 40, 7518–7527 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Adams PD et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Emsley P, Lohkamp B, Scott WG & Cowtan K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Howard MJ, Horton JK, Zhao ML & Wilson SH Lysines in the lyase active site of DNA polymerase β destabilize nonspecific DNA binding, facilitating searching and DNA gap recognition. J. Biol. Chem. 295, 12181–12187 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jamsen JA et al. Structural basis for proficient oxidized ribonucleotide insertion in double strand break repair. Nat. Commun. 12, 5055 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Oertell K. et al. A transition-state perspective on Y-family DNA polymerase η fidelity in comparison with X-family DNA polymerases λ and β. Biochemistry 58, 1764–1773 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Beardslee RA, Suarez SC, Toffton SM & McCulloch SD Mutation of the little finger domain in human DNA polymerase η alters fidelity when copying undamaged DNA. Environ. Mol. Mutagen. 54, 638–651 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Suarez SC, Beardslee RA, Toffton SM & McCulloch SD Biochemical analysis of active site mutations of human polymerase η. Mutat. Res. 745–746, 46−54 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
X-ray crystallography structure of Polλ with microhomology-mediated DNA synapse was deposited with protein data bank under accession code PDB 2PFO. Source data are provided with this paper.