Abstract
Significant interindividual and intraindividual variations on cytochrome P450 (CYP)-mediated drug metabolism exist in the general population globally. Genetic polymorphisms are one of the major contribution factors for interindividual variations, but epigenetic mechanisms mainly contribute to intraindividual variations, including DNA methylation, histone modifications, microRNAs, and long non-coding RNAs. The current review provides analysis of advanced knowledge in the last decade on contributions of epigenetic mechanisms to intraindividual variations on CYP-mediated drug metabolism in several situations, including (1) ontogeny, the developmental changes of CYP expression in individuals from neonates to adults; (2) increased activities of CYP enzymes induced by drug treatment; (3) increased activities of CYP enzymes in adult ages induced by drug treatment at neonate ages; and (4) decreased activities of CYP enzymes in individuals with drug-induced liver injury (DILI). Furthermore, current challenges, knowledge gaps, and future perspective of the epigenetic mechanisms in development of CYP pharmacoepigenetics are discussed. In conclusion, epigenetic mechanisms have been proven to contribute to intraindividual variations of drug metabolism mediated by CYP enzymes in age development, drug induction, and DILI conditions. The knowledge has helped understanding how intraindividual variation are generated. Future studies are needed to develop CYP-based pharmacoepigenetics to guide clinical applications for precision medicine with improved therapeutic efficacy and reduced risk of adverse drug reactions and toxicity.
SIGNIFICANCE STATEMENT
Understanding epigenetic mechanisms in contribution to intraindividual variations of CYP-mediated drug metabolism may help to develop CYP-based pharmacoepigenetics for precision medicine to improve therapeutic efficacy and reduce adverse drug reactions and toxicity for drugs metabolized by CYP enzymes.
Introduction
Drug metabolism mediated by cytochrome P450 (P450 or CYP) enzymes has a direct impact on therapeutic efficacy as well as adverse drug reactions (ADRs) and toxicity for many small chemical drugs currently used. Among the 57 identified P450 members in the human P450 superfamily, approximately 20 members in subfamily CYP1, 2, 3, and 4 are involved in metabolism of xenobiotics, including clinical used small chemical drugs (Esteves et al., 2021). These CYP enzymes contribute approximately 80% phase I drug metabolizing reactions (Evans and Relling, 1999) and are primarily response to metabolize 50% to 60% clinical used drugs (Zanger and Schwab, 2013; Song et al., 2021). Quantity and quality of these CYP enzymes in the body can directly impact clinical outcomes of the 50% to 60% clinical used drugs metabolized by them.
Significant interindividual variations in CYP-mediated drug metabolism exist in the general population globally. Differences in some CYP enzyme activities can reach up to nearly 100-fold between rapid and poor metabolizer groups (Tracy et al., 2016). Genetic polymorphisms in some CYP genes have been implicated as one of the major contributors to the interindividual variations, resulting in the development of pharmacogenetics and pharmacogenomics to predict drug metabolism phenotypes based on genotypes of the CYP genes (Ingelman-Sundberg et al., 2007; Ingelman-Sundberg and Sim, 2010). Vast data have been documented in various CYP genes, such as CYP1A2 (Aklillu et al., 2003; Thorn et al., 2012; Koonrungsesomboon et al., 2018), 2C9 (Rosemary and Adithan, 2007; Daly et al., 2017), 2C19 (Bohanec et al., 2009; Boulenc et al., 2012; S.J. Lee, 2013; Gong et al., 2018; Pratt et al., 2018), 2D6 (Bertilsson et al., 2002; Lee et al., 2009; Taylor et al., 2020), and 3A5 (Uno et al., 2010; Lamba et al., 2012), as well as P450 oxidoreductase (Hart et al., 2008; Pandey and Sproll, 2014). Genetic tests on some CYP polymorphisms have been suggested or even mandated in the Food and Drug Administration’s drug labeling for some drugs with a potential risk to develop severe ADRs and toxicity in a special population associated with some CYP polymorphisms (Kitzmiller et al., 2011).
In addition to the interindividual variations, intraindividual variability on CYP-mediated drug metabolism also exists in the general population globally. The intraindividual variability refers to variability of CYP-mediated drug metabolism within a same individual with same genetic information (Shibasaki et al., 2013). The variability can happen for an individual across different ages from neonate, infant, child, adolescence, to adult, which is normally referred as ontogeny of CYP enzymes (Hines, 2007; Thakur et al., 2021). The variability can also happen for a same individual at different conditions, such as increased activities of CYP enzymes induced by drug treatment (Hakkola et al., 2020) or decreased activities of CYP enzymes in the liver at a drug-induced liver injury (DILI) condition (Bao et al., 2020). The intraindividual variations on CYP-mediated drug metabolism have a direct impact on clinical outcomes for the treatment of some drugs for an individual at different conditions. The supporting evidence can come from alterations of drug metabolism in individuals at different ages between pediatric and adult (Fernandez et al., 2011), in individuals between health and disease conditions, such liver failure (Diep et al., 2017), and in individuals taking medications, which can result in alterations of drug metabolism, for example, induction and inhibition of CYP expression (Hakkola et al., 2020).
Because intraindividual variability on CYP-mediated drug metabolism refers to variations in the same individuals with same DNA sequences, genetic polymorphisms in the CYP genes cannot contribute to the variations. The contribution factors must be beyond DNA sequences, normally referring to epigenetics (Bonasio et al., 2010). Epigenetics is the molecular events in response to the establishment and maintenance of physiologic states that are essential for the cells to remember past events, such as developmental alterations, the external stimulation for example drug induction, or challenges, for instance, DILI. The most-studied epigenetic mechanisms include DNA methylation, histone modifications, and various RNA-mediated processes by noncoding RNAs (ncRNAs), such as small ncRNAs (sncRNAs) and long ncRNAs (lncRNAs) (Gibney and Nolan, 2010). Accumulated data in the past decade have shown that epigenetic mechanisms contribute to the intraindividual variations of CYP-mediated drug metabolism in various manners. This article aims to provide the key recent advanced knowledge, challenges and knowledge gaps, and future perspectives in the following situations: (1) ontogeny, the developmental changes of CYP expression in individuals from neonates to adults; (2) increased activities of CYP enzymes induced by drug treatment; (3) increased activities of CYP enzymes in adult ages induced by drug treatment at neonate ages; and (4) decreased activities of CYP enzymes in individuals with DILI.
Key Recent Advanced Knowledge
Epigenetic Mechanisms Contribute to Intraindividual Variations of CYP Enzymes through Age
Ontogeny of CYP Enzymes in Humans
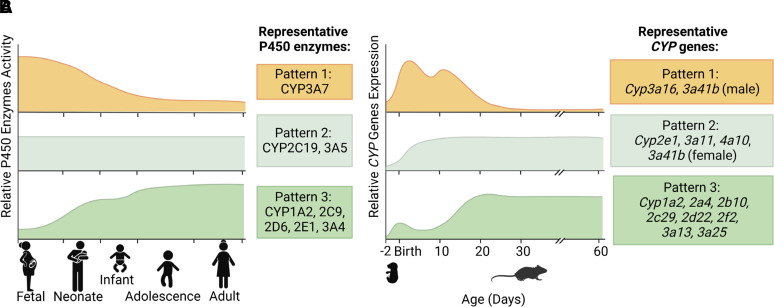
Gene expression and protein activities of the drug-metabolizing CYP enzymes are not consistently without changes after birth. The developmental changes of hepatic expression of CYP enzymes have been studied in the liver samples from various populations through ages from fetus, neonate, infant, child, adolescence, to adult (de Wildt et al., 1999; Hines and McCarver, 2002; Koukouritaki et al., 2004; Blake et al., 2005; Hines, 2007; Stevens et al., 2008; Sadler et al., 2016; Meier et al., 2018). Three major patterns of developmental changes have been identified, which are summarized in Fig. 1A. The CYP enzymes in Pattern 1 show a high expression level during the first three trimesters of fetal life, remain a constant level or gradually decrease around birth, and stay at a low expression level or even silent around 2 years after birth. A representative CYP enzyme in this pattern is CYP3A7 (de Wildt et al., 1999). The CYP enzymes in Pattern 2 do not show significant changes throughout ages with the representative enzymes of CYP2C19 and CYP3A5 (Koukouritaki et al., 2004). The CYP enzymes in Pattern 3 usually express at a very low level in the gestation period, rapidly increase within the 2 years after birth, especially in the first 6 months, and reach a high level in adults. The CYP enzymes in this pattern include CYP2C9, 2D6, 2E1, and 3A4 (Stevens et al., 2008; Lang et al., 2021). Ontogeny of CYP enzymes also exists in small intestine (Kiss et al., 2021) and kidney (Bueters et al., 2020) with similar patterns identified in liver.
Fig. 1.
Ontogenic patterns of CYP expression in human liver (A) and mouse liver (B).
The intraindividual changes of CYP activities through developmental ages provide an explanation on variations in response to drugs between children and adults and are important for accurate prediction of drug pharmacokinetics and toxicity in children (Thakur et al., 2021). For example, clearance of cisapride and sildenafil, CYP3A4 substrates, in neonates and infants showed a trend of increase with age (Kearns et al., 2003; Mukherjee et al., 2009). Newborns have also higher tolerance to acetaminophen (APAP)-induced liver injury (AILI) than older children and adults, because of the immaturity of the CYP1A2, 2E1, and 3A4 enzymes, which produce fewer toxic metabolites of APAP (Penna and Buchanan, 1991; de Wildt et al., 2014). A developmental switch from CYP3A7 in fetuses and neonates to CYP3A4 in adults occurs in the human liver (Lacroix et al., 1997; He et al., 2016), which can lead to differences in drug metabolism mediated by the CYP3A enzymes between pediatric and adult patients.
Ontogeny of CYP Enzymes in Experimental Animal Species
Understanding expression patterns of the CYP genes and figuring out what mechanisms can influence their expression levels and enzyme activities are the fundamental questions in the field of drug metabolism. However, due to ethical, moral, and technical constraints that may arise from human fetuses, studies of the mechanisms in control of expression of CYP enzymes are restricted in human liver at the individual levels during development and maturation. Use of experimental animal models is critical to address these fundamental questions.
Mice, rats, and pigs are commonly used as experimental animal model species for CYP enzymes. Ontogeny of the CYP enzymes during liver development and maturation was studied in mice (Hart et al., 2009; Cui, et al., 2012a), rats (Alcorn et al., 2007; Cherala et al., 2007; de Zwart et al., 2008), and pigs (Millecam et al., 2018). Similarly, expression of mouse CYP enzymes during liver maturation was also classified into three patterns (Hart et al., 2009; Cui, et al., 2012a; Peng et al., 2012), which is illustrated in Fig. 1B. The Cyp genes in Pattern 1 are expressed during the perinatal period but almost disappear by 30 days of age. Its representative genes are Cyp3a16 and male 3a41b. The Cyp genes in Pattern 2 increase rapidly after birth until peaking at day 5, including Cyp2e1, 3a11, 4a10, and female 3a41b. The Cyp genes that belong to Pattern 3, including Cyp1a2, 2a4, 2b10, 2c29, 2d22, 2f2, 3a13, and 3a25, remain at low expression levels between day 10 to day 15, but they increase significantly to a stable high level on day 20 (Hart et al., 2009). RNA sequencing (RNA-seq) was applied to reveal the dynamic changes of abundance of the CYP messenger RNAs (mRNAs) during liver development, which further confirmed the developmental patterns in mice (Peng et al., 2012). At the same time, RNA-seq results helped identifying different alternative transcripts expressed at the same time points (Peng et al., 2012). A developmental switch of expression of CYP mRNAs was also observed in mouse liver with CYP3A16 as the fetal and neonatal CYP3A and CYP3A11 as the adult CYP3A (Hart et al., 2009). Besides CYPs, other non-CYP phase I enzymes (Peng et al., 2013), phase II enzymes (Lu et al., 2013), and transporters (Cui, et al., 2012b) were also reported to have similar mRNA expression patterns as the CYP mRNAs in mice. The phenotypes of ontogenic expression patterns of the CYP enzymes in mice provide a tool for determining underlying mechanisms in control of CYP expression during liver development and maturation.
Association of DNA Methylation with Ontogeny of CYP Enzymes
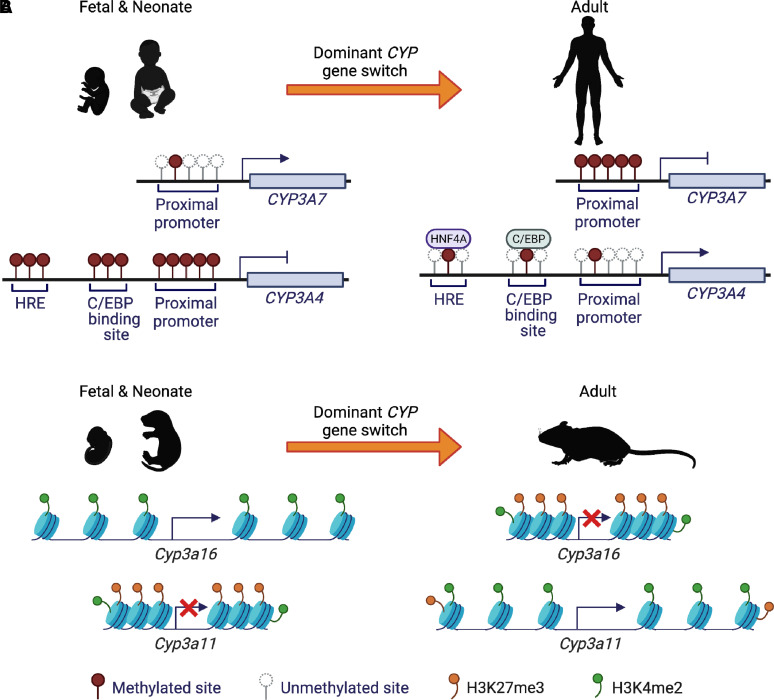
The epigenetic mechanism of DNA methylation has been investigated for its contribution to the ontogenic expression patterns of the CYP enzymes in liver during development and maturation. DNA methylation transfers a methyl group onto the C5 position of the cytosine in a CpG dinucleotide to form 5-methylcytosine (5mC). The 5mC can be further converted to 5-hydroxymethylcytosine. DNA methylation regulates gene expression by inhibiting the binding of transcription factor(s) to their DNA response elements in the genome or by recruiting proteins involved in gene repression (Moore et al., 2013). Analysis of DNA methylation landscape has revealed the roles of DNA methylation in the regulation of drug-metabolizing enzymes, including P450 enzymes (Habano et al., 2015). Developmental changes in DNA methylation in the proximal promoter of the CYP3A4 and 3A7 genes in pediatric livers were observed, which were correlated with ontogenic changes in expression of mRNAs for these two enzymes (Vyhlidal et al., 2016). The dynamics changes of DNA methylation in the hepatic CYP3A4 promoter between fetal and adult human livers and their effect on DNA binding by hepatocyte nuclear factor 4 alpha (HNF4A) and CCAAT-enhancer-binding proteins could partially explain the observed intraindividual variation of CYP3A4 expression in the hepatic developmental shift (Kacevska et al., 2012). Figure 2A illustrates a possible mechanism of DNA methylation in the CYP3A locus associated with the ontogenic expression of the CYP3A7 and 3A4 enzymes in liver during development and maturation.
Fig. 2.
Epigenetic mechanisms contribute to ontogenic switch of CYP3As from fetal and neonate to adult. (A) During human liver development, DNA methylation levels in the proximal promoter region of CYP3A7 change from low levels in newborns to high levels in adults, leading to inhibition of transcriptional expression of CYP3A7 in adults. Conversely, DNA methylation levels in the proximal promoter region of CYP3A4 gene change from high levels in newborns to low levels in adults. At the same time, decreased methylation levels in the HNF4A response element and CCAAT-enhancer-binding protein (C/EBP) binding site contribute to HNF4A and C/EBP bind to these sites, respectively. All these events result in the promotion expression of CYP3A4 in adults. As a result, the dominant CYP gene switches from CYP3A7 in newborns to CYP3A4 in adults in human. (B) During mouse liver development, H3K4me2 levels at the Cyp3a16 locus alter from high levels in neonates to low levels in adults, while H3K27me3 show the opposite changing trend, which leads to the chromatin condensation and finally represses CYP3A16 expression. In contrast, H3K4me2 levels at the Cyp3a11 locus alter from low levels in neonates to high levels in adults, while H3K27me3 decreases from neonates to adults, which leads to the chromatin decondensation and finally promotes CYP3A16 expression. As a result, the dominant Cyp gene switches from Cyp3a16 in newborns to Cyp3a11 in adults in mice.
The mouse livers display significant global decreases in 5mC and hydroxymethylcytosine with age. Furthermore, the level of 5mC increased at two regulatory regions of the mouse Cyp2e1 gene with age, which was associated with decreased expression of its mRNA and protein (Kronfol et al., 2020). DNA methyltransferase 1 (DNMT1), a DNA methylation writer enzyme, plays a critical role in maintenance of postnatal liver histogenesis under homeostasis and stress conditions (Kaji et al., 2016). A conditional knockout of Dnmt1 gene in postnatal mouse liver caused global DNA hypomethylation, enhanced DNA damage response, and initiated a progressive inability to maintain tissue homeostasis. In conclusion, status of DNA methylation in the genomic regions of the CYP/Cyp genes (human/mouse) has been associated with the ontogeny of CYP/Cyp gene expression in human/mouse liver.
Association of Histone Modifications with Ontogeny of CYP Enzymes
Histone modification is another epigenetic mechanism associated with the ontogenic expression of the CYP enzymes in liver during development and maturation. Histones are frequently altered with various modifications (Karlić et al., 2010; Bannister and Kouzarides, 2011). These histone modifications have impact in various chromatin-dependent biologic processes, including regulation of gene transcription (Wang, et al., 2009a; Karlić et al., 2010). Histone modification landscapes have been defined with certain modifications associated with distinct functions in activation or inactivation of gene expression (Wang et al., 2008, 2009b).
Histone modifications have been associated with ontogeny of CYP enzymes. With a focus on the developmental switch of Cyp3a16 and 3a11 genes in mouse liver, dynamic changes of an active modification, histone 3 lysine 4 dimethylation (H3K4me2), and an inactive modification, histone 3 lysine 27 trimethylation (H3K27me3), were associated with the developmental changes of expression of these mouse CYP mRNAs (Li et al., 2009). The high expression levels of CYP3A16 mRNA in neonatal mouse livers and CYP3A11 mRNA in adult livers were associated with high levels of H3K4me2, while the low levels of CYP3A16 mRNA in adult liver coincided with the low level of H3K4me2 and high level of H3K27me3 around the Cyp3a16 locus. Figure 2B demonstrates a possible mechanism of histone modifications in the mouse Cyp3a locus associated with the ontogenic expression of the CYP3A16 and 3A11 mRNAs in liver during development and maturation. The dynamic changes of H3K4me2 and H3K27me3 were also found in human liver samples in a Chinese Han population, which are associated with the developmental switch of expression of CYP3A7 and CYP3A4 enzymes (He et al., 2016). Another example of association of histone modifications to the ontogenic expression of CYP enzymes is CYP2E1. The status of histone 3 lysine 9 acetylation in the 5′-untranslated region of the mouse Cyp2e1 gene showed a dynamic change through age, which was correlated with the changes of intrinsic clearance of the CYP2E1 specific probe drug chlorzoxazone (Kronfol et al., 2020). In addition to the CYP genes, histone modifications are also involved in the regulation of developmental expression of a phase II enzyme UDP-glucuronosyltransferase 1A1 (UGT1A1) in human liver (Nie et al., 2017). The discussed examples provided here are solid evidence to support the association between histone modifications and expression of CYP enzymes in a dynamic way during liver development.
Ontogeny of Epigenetic Modifiers
Status of epigenetic modifications in certain regions of chromatin is established by the epigenetic modifier enzymes, including writers, erasers, and readers. The dynamic patterns of DNA methylation are maintained by DNA methyltransferases (writers), DNA demethylases (erasers), and methyl-CpG binding proteins (readers) (Moore et al., 2013). The histone modifications of acetylation and methylation are determined by histone acetyltransferases and methyltransferases (writers), deacetylases and demethylases (erasers), and histone modification binding proteins (readers). Dynamic changes of mRNAs of epigenetic modifier genes in mouse liver during postnatal maturation were established by RNA-seq (Lu et al., 2012). The presented data in the study suggest that ontogenic changes of epigenetic modifiers may play important roles in determining the addition or removal of corresponding epigenetic signatures during liver development. How dynamic changes of histone modification writers, erasers, and readers are controlled during development is a question that needs to be addressed in future studies.
Association of ncRNAs with Ontogeny of CYP Enzymes
RNA-mediated processes are other epigenetic mechanisms, including ncRNAs. Based on size, ncRNAs can be classified as sncRNAs (<200 bp) and lncRNAs (>200 bp) (Hombach and Kretz, 2016). The main classes of sncRNAs include microRNA (miRNAs), small interfering RNAs, and piwi-interacting RNAs (Zhang et al., 2019). Vast literatures support the involvement of miRNAs in the regulation of drug metabolism and disposition (Yu, 2009; Yu and Pan, 2012; Tezcan et al., 2022). Age-related changes of miRNA expression have been studied in human liver samples between the fetal and pediatric developmental periods (Burgess et al., 2015). The data support the role of the age-dependent miRNAs in regulating drug-metabolizing enzyme genes, including several CYP genes. Age-related changes in miRNA expression also influence ontogeny of a phase II enzyme of glutathione transferase zeta 1 in human liver (Jahn et al., 2020). miRNAs as a major epigenetic mechanism in posttranscriptional regulation of drug-metabolizing enzymes has been referred as miRNA pharmacoepigenetics, which may serve as a new strategy for development of more effective therapy (Yu et al., 2016).
lncRNAs have not shown to be involved in the determination of ontogenic expression of CYP enzymes, which may be due to the lack of enough knowledge in this area. However, an attempt has tried to establish developmental programming of lncRNAs during postnatal liver maturation in mice (Peng et al., 2014). Like coding RNAs, lncRNAs also displayed three major ontogenic patterns: enriched at neonatal, adolescent, or adult ages. Neighboring coding and lncRNAs showed the trend to exhibit highly correlated ontogenic expression patterns, implying the potential importance of lncRNAs in liver maturation. Further studies are needed to determine whether lncRNAs are involved in epigenetic regulation of ontogeny of CYP enzymes. H19 is one of the most abundantly expressed lncRNAs in mouse fetal livers but differently expressed during liver maturation to an undetectable level in adult liver (Pope et al., 2017a); however, it did not affect the developmental expression of the Cyp genes in the H19 knockout mice (Pope et al., 2017b). Even though there is no direct in vivo evidence to address the problem of whether lncRNAs regulate developmental expression of CYP enzymes, some in vitro studies still give promising indications. The lncRNA CUDR (cancer upregulated drug resistant) has been reported to enhance the differentiation of human embryonic stem cells into hepatocytes-like cells by the decrease of H3K27me3 (Gui et al., 2015). A novel lncRNA regulator of hepatic lineages (lnc-RHL) is essential for the differentiation from human bipotent progenitor cells to hepatocytes (Prabhakar et al., 2021). It is believed that CYP expressions are time-dependent during liver development and liver cell lines share similar characteristics with liver organs. As a result, it can be reasonably inferred that exploring lncRNAs to regulate developmental expression of CYP enzymes is still advisable.
In summary, drug metabolism mediated by the CYP enzymes mainly occur in the liver, which is the organ immature at neonatal ages (Grijalva and Vakili, 2013). Postnatal maturation is critical for the liver to transit its functions from hematopoietic in prenatal and neonatal to metabolism in adults (Suzuki et al., 2019). During the liver maturation process, expression of the CYP enzymes shows ontogenic changes with three distinct patterns. Epigenetic mechanisms, including DNA methylation, histone modifications, and miRNAs have been associated with the developmental regulation of the CYP expression, which could explain the distinct patterns of some CYP enzymes, particularly for the developmental switch of the CYP3A members in both human and mice. Epigenetic writers, erasers, and readers are the key element proteins to build epigenetic signatures in the CYP genes to fulfill the developmental regulation of gene expression. In relation to the demonstrated role of epigenetics in developmental biology (Skinner, 2011), next questions are which factors control the writers, erasers, and readers to establish the developmental program and whether there are critical windows of development that are more responsive to environmental factors to modify the developmental patterns.
Epigenetic Mechanisms Are Involved in Intraindividual Variations of CYP Enzymes Induced by Drug Treatment
Induction of CYP Expression by Drug Treatment
Expression of some CYP genes can be induced at transcription levels by drug treatment, including CYP1A2, 2B6, 2C9, 2C19, 2D6, 2E1, 3A4, and 3A7 genes, resulting in clinically significant drug-drug interactions (DDIs) that can cause unanticipated therapeutic failures, ADRs, and toxicity (Lee et al., 2006; Amacher, 2010; Hakkola et al., 2020). The effect of induction on CYP transcription can simply increase the amount of CYP enzymes present and speed up the metabolism of a coadministrated drug. Several dozens of drugs have been indicated to be able to induce CYP expression, including some commonly used drugs, such as rifampicin, phenobarbital, carbamazepine, phenytoin, efavirenz, ritonavir, lorlatinib, mitotane, and ivosidenib (Hachad et al., 2010; Hakkola et al., 2020). Drug induction on CYP enzyme activities is one of the main causes of DDIs and result in intraindividual variations on CYP-mediated drug metabolism in comparison with no induction. Numerous DDI databases have been developed with searchable information on lists of drugs as clinical inducers for CYP-mediated metabolites (Hachad et al., 2010; Andersson et al., 2013; Xiong et al., 2022). Understanding molecular mechanisms underlying the induction of CYP gene expression at transcriptional and translational levels is critical for explanation of intraindividual variations caused by drug induction.
Induction of CYP Expression by Drugs through Activation of Xenobiotic Sensors
The “xenobiotic sensors” refer to nuclear receptors of pregnane X receptor (PXR) and constitutive androstane receptor (CAR), which were discovered or characterized in the 1990s. A recent review has summarized a journey of rewards of xenobiotic receptors for their discovery stories (Xie, 2023). PXR and CAR are transcription factors and have since been defined as master regulators of xenobiotic responses through their transcriptional regulation of drug-metabolizing enzymes and transporters, including CYPs (Poudel et al., 2022). Activation of PXR and CAR is also associated with the development of numerous metabolic diseases (Zhang et al., 2023). At a molecular level, when a drug that can induce CYP expression is taken up into a metabolic cell, it can serve as a ligand to bind to either PXR or CAR in its ligand binding domain. Binding of PXR or CAR by the drug can result in a configuration change and activation of PXR or CAR in cytoplasm and subcellular translocation into the nucleus (Men and Wang, 2023). The DNA binding domain of PXR or CAR can recognize its DNA response elements in the promoter regions of some CYP genes to facilitate transcriptional regulation (Jana and Paliwal, 2007). The following questions, which need to be addressed, are (1) how PXR and CAR facilitate transcriptional regulation of CYP expression after binding to the CYP promoter regions and (2) whether epigenetic mechanisms of DNA methylation, histone modifications, and lncRNAs are involved in the PXR- or CAR-mediated transcription regulation of CYP genes. These questions have been investigated.
Roles of DNA Methylation and Histone Modifications in Induction of CYP Expression by Drug Treatment
DNA methylation and histone modifications have been shown to play an important role in the induction of CYP expression by drug treatment through the activation of PXR or CAR. The Cyp2b10, a CAR target gene, was concomitantly DNA hypomethylated and transcriptionally activated in a liver tissue-specific manner following treatment with phenobarbital, which can activate CAR (Lempiäinen et al., 2011).
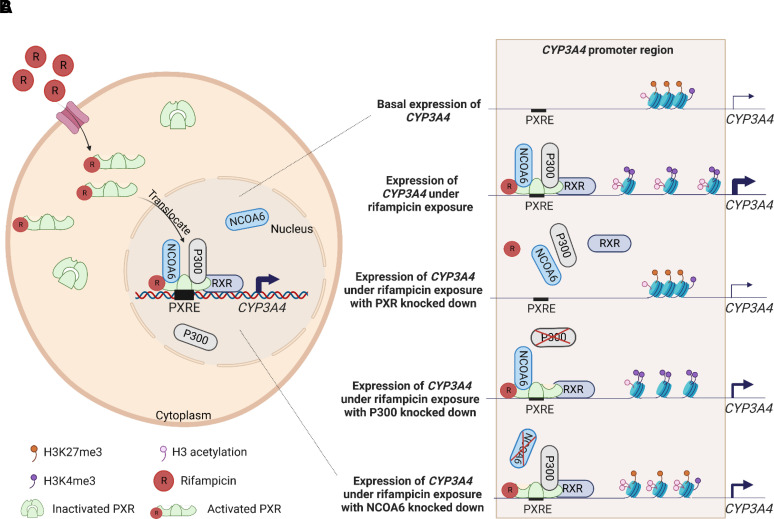
Treatment with phenobarbital resulted in a switch of histone modifications from a repressive form of H3K27me3 to active forms of H3K4me2 and H3 acetylation at the promoter of the Cyp2b10 gene in the liver of B6C3F1 mice (Lempiäinen et al., 2011). The role of histone modifications in the CYP3A4 promoter regions in the induced expression of CYP3A4 enzyme by rifampicin was studied in LS174T cells (Yan et al., 2017). In the treatment with rifampicin, not only did the CYP3A4 mRNA level increase but also H3K4me3 and H3 acetylation increased in the CYP3A4 promoter, while H3K27me3 decreased. The changes of histone modifications could be explained by enhanced recruitment of histone modification writers and erasers of nuclear receptor coactivator 6, containing H3K4 methyltransferase and H3K27 demethylase, as well as p300, a histone acetyltransferase to the CYP3A4 promoter by PXR after rifampicin treatment. This resulted in elevated levels of H3K4me3 and H3 acetylation as well as reduced levels of H3K27me3 in the CYP3A4 promoter to decondense the chromatin, further contributing to the induction of CYP3A4 mRNA expression (Yan et al., 2017). A possible molecular mechanism is illustrated to explain the transcriptional regulation of CYP3A4 expression by activation of PXR with rifampicin in a cell (Fig. 3A) and p300 (methyltransferase) as well as nuclear receptor coactivator 6 (H3K4 methyltransferase and H3K27 demethylase) in knockdown experiments (Fig. 3B). Furthermore, in the HepaRG cell line, PXR and CYP gene expression was reduced after knocking down histone 3 lysine 9 methyl transferase enzyme (G9a), another histone modification writer (Pande et al., 2020). In an in vivo mouse model, increasing H3K4me3 enrichment and at the same time decreasing the H3K27me3 enrichment in the PXR response elements (PXREs) of the Cyp3a11 gene based on the treatment of pregnenolone 16α-carbonitrile (PCN), an activated ligand of PXR in rodents, could upregulate CYP3A11 mRNA expression (Wang et al., 2019a).
Fig. 3.
Histone modifications in rifampicin-induced expression of CYP3A4 in human liver cells. (A) Rifampicin can get into cytoplasm through an uptake transporter and bind to an inactivated PXR to stimulate it to become an activated form. Activated PXR with rifampicin translocates into nucleus to form a heterodimer with retinoid X receptor (RXR) and to bind to the PXRE in the CYP3A4 promoter, recruiting nuclear receptor coactivator 6 (NCOA6) and p300 to promote CYP3A4 transcription. (B) Compared with basal level, rifampicin can enhance expression of CYP3A4 through rifampicin/PXR/RXR/NCOA6/p300 complex binding, which causes increased H3K4me3 and H3 acetylation as well as decreased H3K27me3 at the promoter of CYP3A4 to decondense the chromatin. When under the situation of rifampicin exposure with PXR knocked down, no activated PXR can translocate into nucleus to form a transcription factor complex and bind to the PXRE so the expression of CYP3A4 is the same as basal level. When under the situation of rifampicin exposure with P300 knocked down, only NCOA6 and activated PXR can interact with the PXRE, leading to increased H3K4me3 and decreased H3K27me3 but no change of H3 acetylation at the promoter of CYP3A4. In this condition, chromatin of CYP3A4 is limited decondensed to small boost its expression. When under the situation of rifampicin exposure with NCOA6 knocked down, only P300 and activated PXR can interact with the PXRE, leading to increased H3 acetylation but no change of H3K4me3 and H3K27me3 at the promoter of CYP3A4. In this condition, chromatin of CYP3A4 is limited decondensed to small boost its expression.
Collectively, the presented data support that DNA methylation and histone modifications as two epigenetic mechanisms can be dynamically altered in the genomic regions of some CYP genes to facilitate induction of expression of CYP enzymes in response to drug treatment.
Roles of lncRNAs in Induction of CYP Expression by Drug Treatment
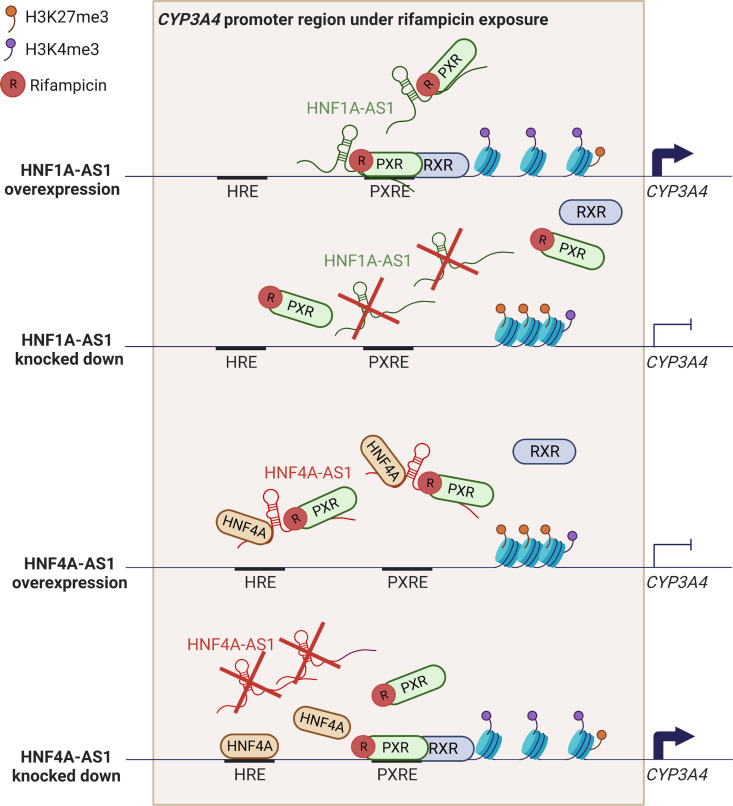
Implication on drug metabolism and disposition by lncRNAs is a recent hot topic (Wang et al., 2020). Several lncRNAs have been demonstrated to play a critical role in the induced expression of CYP enzymes by drug treatment. A transcriptional regulatory network was identified to control both basal and induced expression of numerous CYP enzymes by rifampicin and phenobarbital in HepaRG cells (Chen et al., 2018). The key members in the transcriptional regulatory network contain transcription factors of hepatic nuclear factor 1 alpha (HNF1A), HNF4A, PXR, and CAR as well as lncRNAs HNF1A antisense 1 (HNF1A-AS1) and HNF4A antisense 1 (HNF4A-AS1). HNF1A-AS1 and HNF4A-AS1 are two antisense lncRNA genes physically located at the immediate neighbor of the HNF1A and HNF4A genes, respectively. The antisense lncRNAs and transcription factors show concordant expression patterns in different tissues (Chen, et al., 2020a). Further studies in Huh7 cells and human liver samples confirmed that HNF1A-AS1 can positively regulate the CYPs in the liver at both basal and drug-induced levels (Wang et al., 2019b), whereas HNF4A-AS1 is able to negatively regulate the CYP expression via altering histone modifications of H3K4me3 and H3K27me3 (Wang et al., 2021). Figure 4 illustrates a possible mechanism of lncRNAs HNF1A-AS1 and HNF4A-AS1 in the regulation of CYP expression under a drug-induced condition.
Fig. 4.
Possible mechanism of HNF1A-AS1 and HNF4A-AS1 in the regulation of rifampicin-induced expression of CYP3A4. HNF1A-AS1 can act as a scaffold to recruit activated PXR and guide it to the PXRE, where PXR forms a heterodimer with RXR to boost CYP3A4 expression. When overexpressing HNF1A-AS1, more activated PXR can be led to the PXRE so that H3K4me3 level increases and H3K27me3 decreases. CYP3A4 expresses more because of chromatin decondensation in its gene promoter region. On the contrary, knocking down HNF1A-AS1 can destroy the guidance for PXR to the PXRE, which leads to decreased H3K4me3 and increased H3K27me3 to compress the chromatin in the promoter of CYP3A4 and repress its expression. HNF4A-AS1 exhibits a different regulation mechanism of CYP3A4. It can function as a central binding platform for silencing HNF4A and PXR to inhibit CYP3A4 expression. When overexpressing HNF4A-AS1, more HNF4A and PXR are captured by HNF4A-AS1 and silenced, so they cannot bind to the HNF4A response element and the PXRE, which decreases H3K4me3 and increases H3K27me3 to inhibit CYP3A4. On the opposite, knocking down HNF4A-AS1 can destroy the central platform of HNF4A and PXR, which leads to more HNF4A and PXR binding to their binding sites and increase H3K4me3 and decrease H3K27me3 to decondense the chromatin in the promoter of CYP3A4 and promote its expression.
In summary, drug-induced expression of CYP enzymes results in intraindividual variability of CYP-mediated drug metabolism and DDIs. The induction is mediated by activation of transcription factors such as PXR and CAR. Status of DNA methylation and histone modifications is normally altered in the CYP genes due to the recruitment of related epigenetic writers and erasers by PXR or CAR. lncRNAs may play a key role in the connection of PXR or CAR with the epigenetic writers and erasers to the regions of CYP genes.
In addition to the induction of CYP expression, transcriptional suppression of CYP expression by endogenous and exogenous chemical compounds is also widely observed (Riddick et al., 2004; Hakkola et al., 2020). Lipopolysaccharide (LPS) could downregulate CYP3A expression in mouse liver by transcriptional suppression (Taneja et al., 2019). Furthermore, LPS also was shown to be able to repress expression of several histone-modifying enzymes of HDAC1, HDAC3, and EZH2 (Taneja et al., 2019), implying that histone modifications may be involved in the suppression of CYP3A expression by LPS. However, molecular mechanisms have not been fully illustrated yet in as much detail as the studies in the induction of CYP expression by epigenetic mechanisms.
Epigenetic Mechanisms Are Involved in Intraindividual Variations of CYP-mediated Drug Metabolism in Adults Received Drug Exposure at Neonatal Ages
Impact of Drug Treatment at Neonatal Ages on Intraindividual Variability of Drug Metabolism and Drug-Drug Interactions in Adults
The impact of drug treatment at neonatal ages on variability of drug metabolism and DDIs in adults has been recently discussed (Piekos et al., 2017). Many neonates and infants are administrated drugs, which may result in DDIs. Hospitalized pediatric patients are often administrated numerous medications simultaneously, and 41% have potential DDIs (Feinstein et al., 2015). Neonatal exposure to antibiotic drugs may impair child growth (Uzan-Yulzari et al., 2021), and some adverse consequences can be both short and long term (Cotten, 2016).
The effects of early life exposure to specific drugs, nutrients, and environmental toxicants on metabolic and detoxification processes in the adult life have been documented in animal models starting from early 1980s. Exposure of newborn rats to pharmacologically active compounds, such as phenobarbital, may permanently alter the metabolism of carcinogens and increase the risk of developing cancer (Faris and Campbell, 1981). Neonatal exposure to rats with methadone, a synthetic opioid agonist for chronic pain release, alters sex differentiation of hepatic monooxygenase of testosterone in adult rats (Lui et al., 1981). Neonatal exposure to phenobarbital results in increased P450-dependent activities in adult male and female rats (Bagley and Hayes, 1983, 1985). The induction of CYP expression in adult rats by phenobarbital exposure at neonatal age has been observed in different CYP subfamilies, including CYP2B1 and 2B2 (Agrawal and Shapiro, 1996), 2C7 (Agrawal and Shapiro, 2000), 2C11, 3A1, and 3A2 (Agrawal and Shapiro, 2003). This phenomenon has been referred as neonatal imprinting of phenobarbital on overinduction of constituent CYP expression in adult rats (Haake and Safe, 1988; Agrawal and Shapiro, 2003).
A similar phenomenon on long-term impact of CYP expression by neonatal exposure to drugs has also been observed in mice in later studies (Tien et al., 2015, 2017; Piekos et al., 2018). The dose of phenobarbital and age of treatment at early life have been identified as two key factors for the persistent induction of CYP enzymes in adult mouse liver (Tien et al., 2015). Only a dose over a certain threshold of phenobarbital could result in persistent induction of CYPs in adults. A sensitive age window in early life was identified for persistent induction. When mice were older than the age window, no matter how high doses were used, exposure to phenobarbital did not cause long-term induction of CYP expression. Phenobarbital treatment at a neonatal age can also result in decreased efficacy in acid suppression in adult mouse stomach by omeprazole, a proton pump inhibitor drug, due to increased metabolism of omeprazole by induced CYP enzymes (Tien et al., 2017). Omeprazole is known to be metabolized by CYP enzymes, mainly by CYP2C19 and 3A4 in human liver (Ishizaki and Horai, 1999). Phenytoin, a voltage-gated sodium channel blocker, is also known to be a CYP inducer by activation of CAR in both humans and mice (Wang et al., 2004; Jackson et al., 2006). Neonatal exposure to phenytoin in mice could also result in either short- or long-term consequences of alterations of CYP-mediated drug metabolism (Piekos et al., 2018). These data indicate there are underlying molecular mechanisms involved in the induced expression of CYP enzymes in adult life, which can remember a drug exposure in early neonatal life.
Roles of Histone Modifications in Induction of CYP Expression in Adults by Drug Exposure in Neonatal Age
A concept of epigenetic programming in fetal and neonatal development has been established to indicate that environment-induced epigenetic modifications and their associated gene expression activity are relatively stable to be transmitted after birth until adults, which determines the physiologic phenotypes in adults from fetal and neonatal development (Zhu et al., 2019). Changes of the established epigenetic programming in chromatin structure, DNA methylation, and histone modifications in different tissues following neonatal exposure to specific environmental xenobiotics may result in permanent alterations of the existing epigenetic programming for a long-term impact on physiologic phenotypes in adult life.
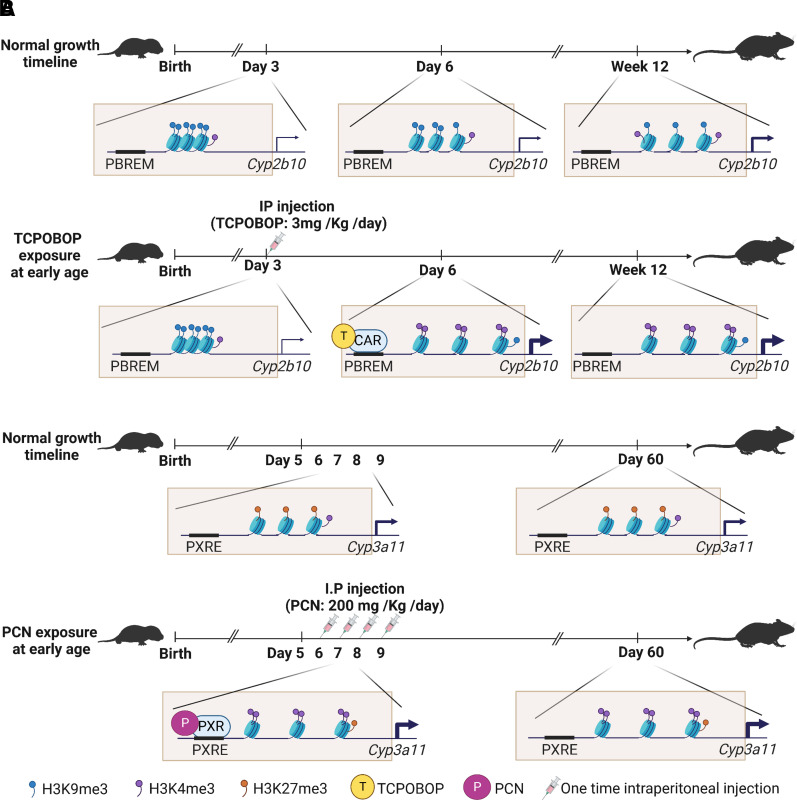
A connection of the neonatal exposure of chemical compounds and permanent induction of CYP enzymes in adults to activation of nuclear receptors and alterations of epigenetic programming has been connected. A synthetic compound, 1,4-bis-[2-(3,5,-dichlorophyridyloxy)] benzene (TCPOBOP) is an agonist with a high potency to active murine CAR and has been widely used to study molecular mechanisms of induction of murine CYP enzymes in vivo (Baskin-Bey et al., 2007). Administration of TCPOBOP to neonatal mice at day 5 after birth was found to selectively activate CAR, leading to the permanent induction of CYP2B10 and 2C37 mRNA expression in adult mice at week 12 (Chen et al., 2012). By focusing the histone modifications in the promoter region of Cyp2b10 gene, the study detected an increased level of H3K4me3 and a decreased level of H3K9me3, which correlated to the increased level of CYP2B10 mRNA expression in adult mouse liver. Such changes of histone modifications and correlation to CYP2B10 expression could not be observed in CAR knockout mice, implying that the persistent epigenetic changes in adult by neonatal exposure were dependent on CAR activation. These observations indicate that activation of CAR by TCPOBOP at a neonatal age may produce a modified epigenetic memory and reprogramming that persists into adult life and favors the induction of CYP expression. Figure 5A illustrates a possible epigenetic mechanism of histone modifications in induction of Cyp2b10 expression in both neonate and adult ages by phenobarbital exposure in a neonatal age of day 3 in mouse liver.
Fig. 5.
Possible mechanism of histone modifications in induction of CYP expression in adults by drug exposure in neonatal age. (A) During the normal growth timeline, a high level of H3K9me3 and a low level of H3K4me3 in the promoter of Cyp2b10 cause chromatin condensed and repress its expression at day 3 and day 6. While at week 12, H3K9me3 drops a little, meanwhile H3K4me3 level increases a little, thus, the chromatin is decondensed to a certain level to increase CYP2B10 expression. This developmental pattern matches pattern 3 in Fig. 1B. After TCPOBOP injected at day 3, TCPOBOP activates CAR and bind to phenobarbital-responsive enhancer module to decrease H3K9me3 level and increase H3K4me3 level to decondense the chromatin in the promoter, resulting in increased CYP2B10 expression at day 6. Even at week 12 without TCPOBOP treatment, Cyp2b10 still keeps the same histone modification level and mRNA expression level as day 6. These observations indicate that activation of CAR by TCPOBOP at a neonatal age may produce a modified epigenetic memory and programming that persists into adult life, which favors the induction of CYP expression. (B) During the normal growth timeline, the constant level of H3K27me3 and H3K4me3 in the promoter of Cyp3a11 makes a constant expression throughout the age. This developmental pattern matches pattern 2 in Fig. 1B. When under the situation of PCN consecutively injected from day 6 to 9, PCN activates PXR and binds to the PXRE to decrease H3K27me3 level and increase H3K4me3 level to decondense the chromatin in the promoter, resulting in increased CYP3A11 expression. In day 60, Cyp3a11 still keeps the same histone modification level and mRNA expression level as day 6. Neonatal exposure to PCN in mice resulted in persistently decreased H3K27me3 level and increased H3K4me3 level to induce CYP3A11 expression in adult liver.
A change of epigenetic memory is also involved in the persistent alterations of CYP enzymes in adult mice by neonatal exposure to PCN, a murine PXR ligand (Wang et al., 2019a). PCN is a species-specific agonist to rodent PXR, not human PXR, and can induce the expression of the CYP3A enzymes in rodent liver, intestine, and kidney as well as primary cultures of rodent hepatocytes (Kliewer et al., 1998). Neonatal exposure to PCN in mice resulted in persistently induced expression levels of CYP1A2, 2B10, 3A11, and UGT1A1 mRNAs in adult liver. The inducibility of CYP3A11 by PCN was also seen in the primary hepatocytes derived from adult mice who received PCN treatment at a neonatal age. Mechanistically, an enhanced level of H3K4me3 and a reduced level of H3K27me3 in the PXRE of the Cyp3a11 promoter were correlated to the persistent induction of CYP3A11 expression by neonatal PCN treatment, demonstrating that epigenetic memory is involved in the process (Wang et al., 2019a). Figure 5B illustrates a possible epigenetic mechanism of histone modifications in induction of Cyp3a11 expression in an adult age at day 60 by PCN exposure at neonatal ages between day 6 and 9 in mouse liver.
In summary, the activation of PXR and CAR by exposure to their potent ligands at neonatal ages can result in alterations of histone modifications as a change of epigenetic memory during the postnatal liver maturation, leading to a persistent induction of CYP expression in adult liver and causing changes of CYP-mediated drug metabolism with impact on the efficacy of drugs metabolized by CYP enzymes, such as omeprazole.
The phenomenon and concept of long-term impact of drug metabolism in adult life by neonatal exposure to drugs, which have capabilities to induce CYP expression by altering epigenetic memory, are established mainly in studies using animal models of mice and rats. Whether this phenomenon is also true for humans has not been supported by clinical studies, which require a long time (at least more than 20 years) to follow patients’ clinical outcomes who received drug treatment at neonatal ages. Current studies in animal models may stimulate such study design for long-term health of drug uses in pediatric populations.
Epigenetic Mechanisms are Associated with Susceptibility of DILI Mediated by CYP Enzymes
Role of CYP Activities in DILI
DILI is very common, can be caused by nearly all classes of medications (David and Hamilton, 2010), and contributes to nearly 60% of acute liver failure annually in the United States with APAP as the leading causing drug (W.M. Lee, 2013). APAP-induced liver injury (AILI) remains a global issue. In the United States, AILI contributes to more than 50% of acute liver failure and nearly 20% of the liver transplant cases (Yoon et al., 2016). The onset and extent of DILI can vary significantly in different patients or the same patients in different conditions using identical drugs. CYP-mediated drug metabolism plays an important role in the susceptibility of DILI by impacting either drug or metabolite concentrations in hepatocyte cells (Corsini and Bortolini, 2013). Either poorly metabolized drugs or rapidly metabolized metabolites can result in overaccumulation of toxic compounds in hepatocyte cells to cause DILI. In addition to genetic polymorphisms in the CYP genes, the poor or rapid drug metabolism can contribute to the alterations of epigenetic mechanisms.
Role of Epigenetic Mechanisms in the CYP Genes in Susceptibility of DILI
The status of DNA methylation in the CYP gene regions has been associated with onset and extent of DILI. After the treatment of HepaRG cells with rifampicin, the DNA methylation levels in the CYP2D6 and 2E1 gene regions were proportionally associated with the onset and extent of rifampicin-induced DILI as measured by cell viability, glutathione content, and caspase activities (Wei et al., 2020). The levels of DNA methylation in the CpG islands in the Cyp1a1 and 1b1 promoter regions were also associated with severe levels of isoniazid-induced liver injury in rats (Li et al., 2018).
ncRNAs also play an important role in DILI, particularly AILI. A comprehensive review has summarized the roles of ncRNAs in AILI, including numerous miRNAs, such as miR-122, miR-125b, and miR-223 (Chowdhary et al., 2021). APAP is metabolized by CYP1A2, 2E1, and 3A4 to form a metabolite, N-acetyl-p-benzo-quinone imine, which is the primary toxic compound leading to AILI (Manyike et al., 2000). Activities of CYP1A2, 2E1, and 3A4 have direct impact on the susceptibility of AILI. lncRNAs HNF1A-AS1 and HNF4A-AS1 have been demonstrated to be involved in the regulation of both basal and induced expression of CYP1A2, 2E1, and 3A4 by drugs in either a positive or negative manner, respectively. By knocking down HNF1A-AS1 and HNF4A-AS1 in HepaRG cells, susceptibility of AILI was altered, but in different directions with decreased AILI toxicity in HNF1A-AS1 knockdown and increased AILI in HNF4A-AS1 knockdown (Chen, et al., 2020b). lncRNAs HNF1A-AS1 and HNF4A-AS1 may work together to control the balance of CYP enzyme activities in liver. Imbalanced HNF1A-AS1 and HNF4A-AS1 may drive the liver to be either more sensitive or more resistant to AILI by affecting APAP biotransformation to N-acetyl-p-benzo-quinone imine mediated by APAP metabolism. More lncRNAs were explored under the APAP-treated condition, and the RNA-seq results of APAP-treated HepaRG cells showed the correlation between lncRNA expression and APAP toxicity. Among them, LINC00844 was significantly downregulated in a concentration-dependent manner in response to APAP exposure, and it showed the positive relation with the mRNA levels of CYP3A4, 2E1, SULT2A1, PXR, and HNF4A (Li et al., 2020). Another lncRNA, LINC00574, has been proven to coordinately regulate expression of a phase II drug-metabolizing enzyme UGT2B15 by interacting with miRNA miR-129 in HepaRG cells treated with APAP (Yu et al., 2020).
In addition to APAP, knockdown and overexpression of lncRNAs HNF1A-AS1 and HNF4A-AS1 have also been shown to impact susceptibility of ritonavir-induced cytotoxicity in different directions in Huh7 and HepG2 cells via alterations of CYP3A4 expression and enrichment of PXR binding as well as status of H3K4me3 and H3K27me3 in the CYP3A4 promoter (Wang et al., 2022). DDI of ritonavir with rifampicin can also be impacted by lncRNAs HNF1A-AS1 and HNF4A-AS1 (Wang et al., 2022).
On the one hand, CYP activities had a direct impact on susceptibility of DILI, particularly AILI, whereas, on the other hand, the levels of AILI could also alter CYP activities in an age-dependent manner, which was demonstrated in mouse livers (Bao et al., 2020). Alterations of CYP enzyme activities also happen throughout whole liver repair and regeneration processes after AILI and result in a critical time window with a high risk of ADRs and toxicity for drug therapy mediated by CYP metabolism (Bao et al., 2022). Dose justification may be needed to reduce the risk of ADRs and toxicity, which has been demonstrated by the dose justification of midazolam’s anesthesia property (Bao et al., 2022).
In conclusion, the growing evidence in the past decade has proven that epigenetic mechanisms of DNA methylation, histone modifications, and ncRNAs play critical roles in the regulation of CYP expression in various conditions related to age development, drug induction, and DILI to contribute intraindividual variations of CYP-mediated drug metabolism with direct impact on efficacy and ADRs/toxicity of drug therapies. However, this area also faces strong challenges and knowledge gaps for clinical applications with the existing known knowledge.
Current Challenges, Knowledge Gaps, and Future Perspective
Some fundamental knowledge gaps still exist that prevent a better understanding of the underlying mechanisms in control of epigenetic alterations in different conditions. The status of epigenetic signatures of DNA methylation and histone modifications in the genomic regions of CYP genes is determined by epigenetic writers, erasers, and readers; however, it is still unknown which factors are upstream determining factors to control the writers, erasers, and readers to add or remove certain epigenetic signatures under special conditions. A more detailed understanding of epigenetic modifications is necessary for elucidating complex biologic processes in control of CYP expression and ultimately for improvement of CYP-mediated drug metabolism for treatment of diseases (Hyun et al., 2017). Such knowledge gaps should be filled in future studies.
lncRNAs may serve as mediator factors to bring transcription factors and histone modification writers and erasers to some specific genomic regions to facilitate transcription regulation of CYP expression. It is still unclear how lncRNAs are involved in the processes. The functions of lncRNAs often rely on specific, typically short, conserved elements as functional motifs to facilitate their major biologic functions (Ross and Ulitsky, 2022). A current challenge and future direction are to identify functional motifs in the lncRNAs involved in the regulation of CYP expression, such as HNF1A-AS1 and HNF4A-AS1, and to define their biologic functions.
lncRNAs should not exist as linear structures in cells. How do lncRNAs form secondary and tertiary structures? Are these structures of lncRNAs critical for their physiologic functions? Crystal structures of proteins have provided powerful tools to understand biologic functions of proteins (McPherson and Gavira, 2014). Crystal structures have been used to reveal CYP protein functions in interaction with their substrates in CYP2B6 (Gay et al., 2010; Roberts et al., 2023) as well as CYP3A4 and 3A5 (Hsu et al., 2018). lncRNAs tend to acquire complex secondary and tertiary structures, and in many cases their functions are dependent on structural conservation rather than primary nucleotide sequences (Graf and Kretz, 2020). The 3D shape and topology of full-length native lncRNAs have been visualized recently, which requires synergistic integration of computational, biochemical, and biophysical approaches (Chillón and Marcia, 2020). Determination of secondary and tertiary structures of lncRNAs involved in the regulation of CYP expression, such as HNF1A-AS1 and HNF4A-AS1, is a critical step to fill in the knowledge gaps in understanding lncRNA functions to interact with other partners in control of CYP expression in different conditions.
Alternative splicing of lncRNAs has been implicated in increasing lncRNA gene utilization to increase the variety of RNA transcripts. Alternative transcripts of lncRNAs plays a significant regulatory role in human diseases (Chen et al., 2021). Multiple alternative transcripts have been annotated in both human and mouse genomes for some CYP-associated lncRNAs, such as HNF1A-AS1 and HNF4A-AS1. However, these annotated alternative transcripts have not been experimentally proven yet. Some alternative transcripts from the same genes have completely different RNA sequences, implicating that they may have different biologic functions. Future validation and characterization of the alternative transcripts of the CYP-associated lncRNAs, such as HNF1A-AS1 and HNF4A-AS1, should be conducted to fill the knowledge gap: which particular alternative transcript regulates CYP expression under different conditions?
A new research area in pharmacoepigenetics and pharmacoepigenomics is under development to address how to apply the current knowledge of epigenetics for clinical applications (Majchrzak-Celińska and Baer-Dubowska, 2017; Hack et al., 2019). However, pharmacoepigenetics has much stronger challenges than pharmacogenetics for clinical applications. In pharmacogenetics, changes of CYP-mediated drug metabolism caused by genetic polymorphisms can be predicted based on DNA sequence information detected from DNA samples isolated from specimens, such as blood and oral mucosal. In contrast to pharmacoepigenetics, not like genetic information, epigenetic information is tissue specific and not easily observed or diagnosed based on measurement of biomarkers in patient specimen samples, such as blood, urine, and oral mucosal. Development of epigenetic biomarkers to produce relevant information for diagnosis, prognosis, and therapy optimization in routine clinical treatment is challenging (García-Giménez et al., 2017). A future direction is to fill the knowledge gap by developing clinical approaches to assess epigenetic alterations in biologic specimens to predict intraindividual variations of CYP-mediated drug metabolism for improving therapeutic efficacy and avoiding ADRs and toxicity.
Conclusions
Epigenetic mechanisms of DNA methylation, histone modifications, and ncRNAs have been proven to contribute to intraindividual variations of drug metabolism mediated by CYP enzymes in age development, drug induction, and DILI conditions. The knowledge has helped in the understanding of how intraindividual variations are generated. Future studies are needed to develop CYP-based pharmacoepigenetics to guide clinical applications for precision medicine with improved therapeutic efficacy and reduced risk of ADRs and toxicity.
Abbreviations
- 5mC
5-methylcytosine
- ADR
adverse drug reaction
- AILI
APAP-induced liver injury
- APAP
acetaminophen
- CAR
constitutive androstane receptor
- CYP or P450
cytochrome P450
- DDI
drug-drug interaction
- DILI
drug-induced liver injury
- HNF1A
hepatic nuclear factor 1 alpha
- HNF1A-AS1
HNF1A antisense 1
- HNF4A
hepatocyte nuclear factor 4 alpha
- HNF4A-AS1
lncRNA HNF4A antisense 1
- H3K4me2
histone 3 lysine 4 dimethylation
- H3K9me3
histone 3 lysine 9 trimethylation
- H3K27me3
histone 3 lysine 27 trimethylation
- lncRNA
long noncoding RNA
- LPS
lipopolysaccharide
- mRNA
messenger RNA
- miRNA
microRNA
- ncRNA
noncoding RNA
- PCN
pregnenolone 16α-carbonitrile
- PXR
pregnane X receptor
- PXRE
PXR response elements
- sncRNA
small ncRNA
- RNA-seq
RNA sequencing
- TCPOBOP
1, 4-bis-[2-(3, 5, -dichlorophyridyloxy)] benzene
Authorship Contributions
Participated in research design: Jin, Zhong.
Performed data analysis: Jin, Zhong.
Wrote or contributed to the writing of the manuscript: Jin, Zhong.
Footnotes
This study was supported by the National Institutes of Health National Institute of General Medical Sciences [Grant R35-GM140862] (to X-b.Z.).
No author has an actual or perceived conflict of interest.
References
- Agrawal AK, Shapiro BH (2000) Latent overexpression of hepatic CYP2C7 in adult male and female rats neonatally exposed to phenobarbital: a developmental profile of gender-dependent P450s. J Pharmacol Exp Ther 293:1027–1033. [PubMed] [Google Scholar]
- Agrawal AK, Shapiro BH (1996) Phenobarbital induction of hepatic CYP2B1 and CYP2B2: pretranscriptional and post-transcriptional effects of gender, adult age, and phenobarbital dose. Mol Pharmacol 49:523–531. [PubMed] [Google Scholar]
- Agrawal AK, Shapiro BH (2003) Phenobarbital-imprinted overinduction of adult constituent CYP isoforms. Pharmacology 68:204–215. [DOI] [PubMed] [Google Scholar]
- Aklillu E, Carrillo JA, Makonnen E, Hellman K, Pitarque M, Bertilsson L, Ingelman-Sundberg M (2003) Genetic polymorphism of CYP1A2 in Ethiopians affecting induction and expression: characterization of novel haplotypes with single-nucleotide polymorphisms in intron 1. Mol Pharmacol 64:659–669. [DOI] [PubMed] [Google Scholar]
- Alcorn J, Elbarbry FA, Allouh MZ, McNamara PJ (2007) Evaluation of the assumptions of an ontogeny model of rat hepatic cytochrome P450 activity. Drug Metab Dispos 35:2225–2231. [DOI] [PubMed] [Google Scholar]
- Amacher DE (2010) The effects of cytochrome P450 induction by xenobiotics on endobiotic metabolism in pre-clinical safety studies. Toxicol Mech Methods 20:159–166. [DOI] [PubMed] [Google Scholar]
- Andersson ML, Böttiger Y, Lindh JD, Wettermark B, Eiermann B (2013) Impact of the drug-drug interaction database SFINX on prevalence of potentially serious drug-drug interactions in primary health care. Eur J Clin Pharmacol 69:565–571. [DOI] [PubMed] [Google Scholar]
- Bagley DM, Hayes JR (1983) Neonatal phenobarbital administration results in increased cytochrome P450-dependent monooxygenase activity in adult male and female rats. Biochem Biophys Res Commun 114:1132–1137. [DOI] [PubMed] [Google Scholar]
- Bagley DM, Hayes JR (1985) Neonatal phenobarbital imprinting of hepatic microsomal enzymes in adult rats: modulation by neonatal testosterone presence. Toxicol Appl Pharmacol 79:227–235. [DOI] [PubMed] [Google Scholar]
- Bannister AJ, Kouzarides T (2011) Regulation of chromatin by histone modifications. Cell Res 21:381–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao Y, Phan M, Zhu J, Ma X, Manautou JE, Zhong XB (2022) Alterations of cytochrome P450-mediated drug metabolism during liver repair and regeneration after acetaminophen-induced liver injury in mice. Drug Metab Dispos 50:694–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao Y, Wang P, Shao X, Zhu J, Xiao J, Shi J, Zhang L, Zhu H-J, Ma X, Manautou JE, et al. (2020) Acetaminophen-induced liver injury alters expression and activities of cytochrome P450 enzymes in an age-dependent manner in mouse liver. Drug Metab Dispos 48:326–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baskin-Bey ES, Anan A, Isomoto H, Bronk SF, Gores GJ (2007) Constitutive androstane receptor agonist, TCPOBOP, attenuates steatohepatitis in the methionine choline-deficient diet-fed mouse. World J Gastroenterol 13:5635–5641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertilsson L, Dahl M-L, Dalén P, Al-Shurbaji A (2002) Molecular genetics of CYP2D6: clinical relevance with focus on psychotropic drugs. Br J Clin Pharmacol 53:111–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blake MJ, Castro L, Leeder JS, Kearns GL (2005) Ontogeny of drug metabolizing enzymes in the neonate. Semin Fetal Neonatal Med 10:123–138. [DOI] [PubMed] [Google Scholar]
- Bohanec Grabar P, Grabnar I, Rozman B, Logar D, Tomšič M, Šuput D, Trdan T, Peterlin Mašič L, Mrhar A, Dolžan V (2009a) Investigation of the influence of CYP1A2 and CYP2C19 genetic polymorphism on 2-Cyano-3-hydroxy-N-[4-(trifluoromethyl)phenyl]-2-butenamide (A77 1726) pharmacokinetics in leflunomide-treated patients with rheumatoid arthritis. Drug Metab Dispos 37:2061–2068. [DOI] [PubMed] [Google Scholar]
- Bonasio R, Tu S, Reinberg D (2010) Molecular signals of epigenetic states. Science (1979) 330:612–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulenc X, Djebli N, Shi J, Perrin L, Brian W, Van Horn R, Hurbin F (2012) Effects of omeprazole and genetic polymorphism of CYP2C19 on the clopidogrel active metabolite. Drug Metab Dispos 40:187–197. [DOI] [PubMed] [Google Scholar]
- Bueters R, Bael A, Gasthuys E, Chen C, Schreuder MF, Frazier KS (2020) Ontogeny and cross-species comparison of pathways involved in drug absorption, distribution, metabolism, and excretion in neonates (review): kidney. Drug Metab Dispos 48:353–367. [DOI] [PubMed] [Google Scholar]
- Burgess KS, Philips S, Benson EA, Desta Z, Gaedigk A, Gaedigk R, Segar MW, Liu Y, Skaar TC (2015) Age-related changes in microRNA expression and pharmacogenes in human liver. Clin Pharmacol Ther 98:205–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Liu Y, Min J, Wang H, Li F, Xu C, Gong A, Xu M (2021) Alternative splicing of lncRNAs in human diseases. Am J Cancer Res 11:624–639. [PMC free article] [PubMed] [Google Scholar]
- Chen L, Bao Y, Jiang S, Zhong X-B (2020a) The roles of long noncoding RNAs HNF1α-AS1 and HNF4α-AS1 in drug metabolism and human diseases. Noncoding RNA 6:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Bao Y, Piekos SC, Zhu K, Zhang L, Zhong X-B (2018) A transcriptional regulatory network containing nuclear receptors and long noncoding RNAs controls basal and drug-induced expression of cytochrome P450s in HepaRG cells. Mol Pharmacol 94:749–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Wang P, Manautou JE, Zhong XB (2020b) Knockdown of long noncoding rnas hepatocyte nuclear factor 1α antisense RNA 1 and hepatocyte nuclear factor 4α antisense RNA 1 alters susceptibility of acetaminophen-induced cytotoxicity in HepaRG cells. Mol Pharmacol 97:278–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W-D, Fu X, Dong B, Wang Y-D, Shiah S, Moore DD, Huang W (2012) Neonatal activation of the nuclear receptor CAR results in epigenetic memory and permanent change of drug metabolism in mouse liver. Hepatology 56:1499–1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherala G, Shapiro BH, D’mello AP (2007) Effect of perinatal low protein diets on the ontogeny of select hepatic cytochrome p450 enzymes and cytochrome p450 reductase in the rat. Drug Metab Dispos 35:1057–1063. [DOI] [PubMed] [Google Scholar]
- Chillón I, Marcia M (2020) The molecular structure of long non-coding RNAs: emerging patterns and functional implications. Crit Rev Biochem Mol Biol 55:662–690. [DOI] [PubMed] [Google Scholar]
- Chowdhary V, Biswas P, Ghoshal K (2021) Role of noncoding RNAs in acetaminophen-induced liver injury. Gene Expr 20:179–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corsini A, Bortolini M (2013) Drug-induced liver injury: the role of drug metabolism and transport. J Clin Pharmacol 53:463–474. [DOI] [PubMed] [Google Scholar]
- Cotten CM (2016) Adverse consequences of neonatal antibiotic exposure. Curr Opin Pediatr 28:141–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui JY, Gunewardena SS, Yoo B, Liu J, Renaud HJ, Lu H, Zhong XB, Klaassen CD (2012a) RNA-Seq reveals different mRNA abundance of transporters and their alternative transcript isoforms during liver development. Toxicol Sci 127:592–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui JY, Renaud HJ, Klaassen CD (2012b) Ontogeny of novel cytochrome P450 gene isoforms during postnatal liver maturation in mice. Drug Metab Dispos 40:1226–1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daly AK, Rettie AE, Fowler DM, Miners JO (2017) Pharmacogenomics of CYP2C9: functional and clinical considerations. J Pers Med 8:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David S, Hamilton JP (2010) Drug-induced liver injury. US Gastroenterol Hepatol Rev 6:73–80. [PMC free article] [PubMed] [Google Scholar]
- de Wildt SN, Kearns GL, Leeder JS, van den Anker JN (1999) Cytochrome P450 3A: ontogeny and drug disposition. Clin Pharmacokinet 37:485–505. [DOI] [PubMed] [Google Scholar]
- de Wildt SN, Tibboel D, Leeder JS (2014) Drug metabolism for the paediatrician. Arch Dis Child 99:1137–1142 [DOI] [PubMed] [Google Scholar]
- de Zwart L, Scholten M, Monbaliu JG, Annaert PP, Van Houdt JM, Van den Wyngaert I, De Schaepdrijver LM, Bailey GP, Coogan TP, Coussement WC, et al. (2008) The ontogeny of drug metabolizing enzymes and transporters in the rat. Reprod Toxicol 26:220–230. [DOI] [PubMed] [Google Scholar]
- Diep U, Chudow M, Sunjic KM (2017) Pharmacokinetic changes in liver failure and impact on drug therapy. AACN Adv Crit Care 28:93–101. [DOI] [PubMed] [Google Scholar]
- Esteves F, Rueff J, Kranendonk M (2021) The central role of cytochrome P450 in xenobiotic metabolism—a brief review on a fascinating enzyme family. J Xenobiot 11:94–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans WE, Relling MV (1999) Pharmacogenomics: translating functional genomics into rational therapeutics. Science 286:487–491. [DOI] [PubMed] [Google Scholar]
- Faris RA, Campbell TC (1981) Exposure of newborn rats to pharmacologically active compounds may permanently alter carcinogen metabolism. Science (1979) 211:719–721. [DOI] [PubMed] [Google Scholar]
- Feinstein J, Dai D, Zhong W, Freedman J, Feudtner C (2015) Potential drug-drug interactions in infant, child, and adolescent patients in children’s hospitals. Pediatrics 135:e99–e108. [DOI] [PubMed] [Google Scholar]
- Fernandez E, Perez R, Hernandez A, Tejada P, Arteta M, Ramos JT (2011) Factors and mechanisms for pharmacokinetic differences between pediatric population and adults. Pharmaceutics 3:53–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Giménez JL, Seco-Cervera M, Tollefsbol TO, Romá-Mateo C, Peiró-Chova L, Lapunzina P, Pallardó FV (2017) Epigenetic biomarkers: current strategies and future challenges for their use in the clinical laboratory. Crit Rev Clin Lab Sci 54:529–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gay SC, Shah MB, Talakad JC, Maekawa K, Roberts AG, Wilderman PR, Sun L, Yang JY, Huelga SC, Hong W-X, et al. (2010) Crystal structure of a cytochrome P450 2B6 genetic variant in complex with the inhibitor 4-(4-chlorophenyl)imidazole at 2.0-A resolution. Mol Pharmacol 77:529–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibney ER, Nolan CM (2010) Epigenetics and gene expression. Heredity 105:4–13. [DOI] [PubMed] [Google Scholar]
- Gong J, Hansen L, Iacono L (2018) Clinical pharmacokinetics and the impact of genetic polymorphism on a CYP2C19 substrate, BMS-823778, in healthy subjects. Drug Metab Dispos 46:316–325. [DOI] [PubMed] [Google Scholar]
- Graf J, Kretz M (2020) From structure to function: route to understanding lncRNA mechanism. BioEssays 42:e2000027. [DOI] [PubMed] [Google Scholar]
- Grijalva J, Vakili K (2013) Neonatal liver physiology. Semin Pediatr Surg 22:185–189. [DOI] [PubMed] [Google Scholar]
- Gui X, Li H, Li T, Pu H, Lu D (2015) Long noncoding RNA CUDR regulates HULC and β-catenin to govern human liver stem cell malignant differentiation. Mol Ther 23:1843–1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haake JM, Safe SH (1988) Neonatal phenobarbital imprinting of rat hepatic microsomal testosterone hydroxylations. J Biochem Toxicol 3:309–319. [DOI] [PubMed] [Google Scholar]
- Habano W, Kawamura K, Iizuka N, Terashima J, Sugai T, Ozawa S (2015) Analysis of DNA methylation landscape reveals the roles of DNA methylation in the regulation of drug metabolizing enzymes. Clin Epigenetics 7:105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hachad H, Ragueneau-Majlessi I, Levy RH (2010) A useful tool for drug interaction evaluation: the University of Washington Metabolism and Transport Drug Interaction Database. Hum Genomics 5:61–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hack LM, Fries GR, Eyre HA, Bousman CA, Singh AB, Quevedo J, John VP, Baune BT, Dunlop BW (2019) Moving pharmacoepigenetics tools for depression toward clinical use. J Affect Disord 249:336–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakkola J, Hukkanen J, Turpeinen M, Pelkonen O (2020) Inhibition and induction of CYP enzymes in humans: an update. Arch Toxicol 94:3671–3722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart SN, Cui Y, Klaassen CD, Zhong XB (2009) Three patterns of cytochrome P450 gene expression during liver maturation in mice. Drug Metab Dispos 37:116–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart SN, Wang S, Nakamoto K, Wesselman C, Li Y, Zhong XB (2008) Genetic polymorphisms in cytochrome P450 oxidoreductase influence microsomal P450-catalyzed drug metabolism. Pharmacogenet Genomics 18:11–24. [DOI] [PubMed] [Google Scholar]
- He H, Nie Y-L, Li J-F, Meng X-G, Yang W-H, Chen Y-L, Wang S-J, Ma X, Kan Q-C, Zhang L-R (2016) Developmental regulation of CYP3A4 and CYP3A7 in Chinese Han population. Drug Metab Pharmacokinet 31:433–444. [DOI] [PubMed] [Google Scholar]
- Hines RN (2007) Ontogeny of human hepatic cytochromes P450. J Biochem Mol Toxicol 21:169–175. [DOI] [PubMed] [Google Scholar]
- Hines RN, McCarver DG (2002) The ontogeny of human drug-metabolizing enzymes: phase I oxidative enzymes. J Pharmacol Exp Ther 300:355–360. [DOI] [PubMed] [Google Scholar]
- Hombach S, Kretz M (2016) Non-coding RNAs: classification, biology and functioning, in Non-coding RNAs in Colorectal Cancer. Advances in Experimental Medicine and Biology (Slaby O, Calin G, eds), pp 3–17, Springer, Cham. [DOI] [PubMed] [Google Scholar]
- Hsu M-H, Savas U, Johnson EF (2018) The X-ray crystal structure of the human mono-oxygenase cytochrome P450 3A5-ritonavir complex reveals active site differences between P450s 3A4 and 3A5. Mol Pharmacol 93:14–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyun K, Jeon J, Park K, Kim J (2017) Writing, erasing and reading histone lysine methylations. Exp Mol Med 49:e324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingelman-Sundberg M, Sim SC (2010) Pharmacogenetic biomarkers as tools for improved drug therapy; emphasis on the cytochrome P450 system. Biochem Biophys Res Commun 396:90–94. [DOI] [PubMed] [Google Scholar]
- Ingelman-Sundberg M, Sim SC, Gomez A, Rodriguez-Antona C (2007) Influence of cytochrome P450 polymorphisms on drug therapies: pharmacogenetic, pharmacoepigenetic and clinical aspects. Pharmacol Ther 116:496–526. [DOI] [PubMed] [Google Scholar]
- Ishizaki T, Horai Y (1999) Review article: cytochrome P450 and the metabolism of proton pump inhibitors—emphasis on rabeprazole. Aliment Pharmacol Ther 13:27–36. [DOI] [PubMed] [Google Scholar]
- Jackson JP, Ferguson SS, Negishi M, Goldstein JA (2006) Phenytoin induction of the cyp2c37 gene is mediated by the constitutive androstane receptor. Drug Metab Dispos 34:2003–2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahn SC, Gay LA, Weaver CJ, Renne R, Langaee TY, Stacpoole PW, James MO (2020) Age-related changes in miRNA expression influence GSTZ1 and other drug metabolizing enzymes. Drug Metab Dispos 48:563–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jana S, Paliwal J (2007) Molecular mechanisms of cytochrome p450 induction: potential for drug-drug interactions. Curr Protein Pept Sci 8:619–628. [DOI] [PubMed] [Google Scholar]
- Kacevska M, Ivanov M, Wyss A, Kasela S, Milani L, Rane A, Ingelman-Sundberg M (2012) DNA methylation dynamics in the hepatic CYP3A4 gene promoter. Biochimie 94:2338–2344. [DOI] [PubMed] [Google Scholar]
- Kaji K, Factor VM, Andersen JB, Durkin ME, Tomokuni A, Marquardt JU, Matter MS, Hoang T, Conner EA, Thorgeirsson SS (2016) DNMT1 is a required genomic regulator for murine liver histogenesis and regeneration. Hepatology 64:582–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlić R, Chung HR, Lasserre J, Vlahoviček K, Vingron M (2010) Histone modification levels are predictive for gene expression. Proc Natl Acad Sci USA 107:2926–2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearns GL, Robinson PK, Wilson JT, Wilson-Costello D, Knight GR, Ward RM, van den Anker JN; Pediatric Pharmacology Research Unit Network (2003) Cisapride disposition in neonates and infants: in vivo reflection of cytochrome P450 3A4 ontogeny. Clin Pharmacol Ther 74:312–325. [DOI] [PubMed] [Google Scholar]
- Kiss M, Mbasu R, Nicolaï J, Barnouin K, Kotian A, Mooij MG, Kist N, Wijnen RMH, Ungell A-L, Cutler P, et al. (2021) Ontogeny of small intestinal drug transporters and metabolizing enzymes based on targeted quantitative proteomics. Drug Metab Dispos 49:1038–1046. [DOI] [PubMed] [Google Scholar]
- Kitzmiller JP, Groen DK, Phelps MA, Sadee W (2011) Pharmacogenomic testing: relevance in medical practice: why drugs work in some patients but not in others. Cleve Clin J Med 78:243–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliewer SA, Moore JT, Wade L, Staudinger JL, Watson MA, Jones SA, McKee DD, Oliver BB, Willson TM, Zetterström RH, et al. (1998) An orphan nuclear receptor activated by pregnanes defines a novel steroid signaling pathway. Cell 92:73–82. [DOI] [PubMed] [Google Scholar]
- Koonrungsesomboon N, Khatsri R, Wongchompoo P, Teekachunhatean S (2018) The impact of genetic polymorphisms on CYP1A2 activity in humans: a systematic review and meta-analysis. Pharmacogenomics J 18:760–768. [DOI] [PubMed] [Google Scholar]
- Koukouritaki SB, Manro JR, Marsh SA, Stevens JC, Rettie AE, McCarver DG, Hines RN (2004) Developmental expression of human hepatic CYP2C9 and CYP2C19. J Pharmacol Exp Ther 308:965–974. [DOI] [PubMed] [Google Scholar]
- Kronfol MM, Jahr FM, Dozmorov MG, Phansalkar PS, Xie LY, Aberg KA, McRae M, Price ET, Slattum PW, Gerk PM, et al. (2020) DNA methylation and histone acetylation changes to cytochrome P450 2E1 regulation in normal aging and impact on rates of drug metabolism in the liver. Geroscience 42:819–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacroix D, Sonnier M, Moncion A, Cheron G, Cresteil T (1997) Expression of CYP3A in the human liver—evidence that the shift between CYP3A7 and CYP3A4 occurs immediately after birth. Eur J Biochem 247:625–634. [DOI] [PubMed] [Google Scholar]
- Lamba J, Hebert JM, Schuetz EG, Klein TE, Altman RB (2012) PharmGKB summary: very important pharmacogene information for CYP3A5. Pharmacogenet Genomics 22:555–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang J, Vincent L, Chenel M, Ogungbenro K, Galetin A (2021) Impact of hepatic CYP3A4 ontogeny functions on drug-drug interaction risk in pediatric physiologically-based pharmacokinetic/pharmacodynamic modeling: critical literature review and ivabradine case study. Clin Pharmacol Ther 109:1618–1630. [DOI] [PubMed] [Google Scholar]
- Lee MD, Ayanoglu E, Gong L (2006) Drug-induced changes in P450 enzyme expression at the gene expression level: a new dimension to the analysis of drug-drug interactions. Xenobiotica 36:1013–1080. [DOI] [PubMed] [Google Scholar]
- Lee S-J (2013) Clinical application of CYP2C19 pharmacogenetics toward more personalized medicine. Front Genet 3:318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S-J, Lee SS, Jung H-J, Kim H-S, Park S-J, Yeo C-W, Shin J-G (2009) Discovery of novel functional variants and extensive evaluation of CYP2D6 genetic polymorphisms in Koreans. Drug Metab Dispos 37:1464–1470. [DOI] [PubMed] [Google Scholar]
- Lee WM (2013) Drug-induced acute liver failure. Clin Liver Dis 17:575–586, viii. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lempiäinen H, Müller A, Brasa S, Teo S-S, Roloff T-C, Morawiec L, Zamurovic N, Vicart A, Funhoff E, Couttet P, et al. (2011) Phenobarbital mediates an epigenetic switch at the constitutive androstane receptor (CAR) target gene Cyp2b10 in the liver of B6C3F1 mice. PLoS One 6:e18216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Wu L, Knox B, Chen S, Tolleson WH, Liu F, Yu D, Guo L, Tong W, Ning B (2020) Long noncoding RNA LINC00844-mediated molecular network regulates expression of drug metabolizing enzymes and nuclear receptors in human liver cells. Arch Toxicol 94:1637–1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Cui Y, Hart SN, Klaassen CD, Zhong XB (2009) Dynamic patterns of histone methylation are associated with ontogenic expression of the Cyp3a genes during mouse liver maturation. Mol Pharmacol 75:1171–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Li Y, Zheng G, Zhu L, Wang J, Mu S, Ren Q, Feng F (2018) Cytochrome P450 1A1 and 1B1 promoter CpG island methylation regulates rat liver injury induced by isoniazid. Mol Med Rep 17:753–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H, Cui JY, Gunewardena S, Yoo B, Zhong XB, Klaassen CD (2012) Hepatic ontogeny and tissue distribution of mRNAs of epigenetic modifiers in mice using RNA-sequencing. Epigenetics 7:914–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H, Gunewardena S, Cui JY, Yoo B, Zhong XB, Klaassen CD (2013) RNA-sequencing quantification of hepatic ontogeny and tissue distribution of mRNAs of phase II enzymes in mice. Drug Metab Dispos 41:844–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lui EM, Gregson J, Lucier GW (1981) Altered sex differentiation of hepatic ethylmorphine N-demethylation in the male rat following neonatal methadone exposure. Pediatr Pharmacol (New York) 1:187–195. [PubMed] [Google Scholar]
- Majchrzak-Celińska A, Baer-Dubowska W (2017) Pharmacoepigenetics: an element of personalized therapy? Expert Opin Drug Metab Toxicol 13:387–398. [DOI] [PubMed] [Google Scholar]
- Manyike PT, Kharasch ED, Kalhorn TF, Slattery JT (2000) Contribution of CYP2E1 and CYP3A to acetaminophen reactive metabolite formation. Clin Pharmacol Ther 67:275–282. [DOI] [PubMed] [Google Scholar]
- McPherson A, Gavira JA (2014) Introduction to protein crystallization. Acta Crystallogr F Struct Biol Commun 70:2–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier R, Bi C, Gaedigk R, Heruth DP, Ye SQ, Leeder JS, Fridley BL (2018) Ontogeny-related pharmacogene changes in the pediatric liver transcriptome. Pharmacogenet Genomics 28:86–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Men S, Wang H (2023) Phenobarbital in nuclear receptor activation: an update. Drug Metab Dispos 51:210–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millecam J, De Clerck L, Govaert E, Devreese M, Gasthuys E, Schelstraete W, Deforce D, De Bock L, Van Bocxlaer J, Sys S, et al. (2018) The ontogeny of cytochrome P450 enzyme activity and protein abundance in conventional pigs in support of preclinical pediatric drug research. Front Pharmacol 9:470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore LD, Le T, Fan G (2013) DNA methylation and its basic function. Neuropsychopharmacology 38:23–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee A, Dombi T, Wittke B, Lalonde R (2009) Population pharmacokinetics of sildenafil in term neonates: evidence of rapid maturation of metabolic clearance in the early postnatal period. Clin Pharmacol Ther 85:56–63. [DOI] [PubMed] [Google Scholar]
- Nie Y-L, Meng X-G, Liu J-Y, Yan L, Wang P, Bi H-Z, Kan Q-C, Zhang L-R (2017) Histone modifications regulate the developmental expression of human hepatic UDP-glucuronosyltransferase 1A1. Drug Metab Dispos 45:1372–1378. [DOI] [PubMed] [Google Scholar]
- Pande P, Zhong XB, Ku WW (2020) Histone methyltransferase G9a regulates expression of nuclear receptors and cytochrome P450 enzymes in HepaRG cells at basal level and in fatty acid induced steatosis. Drug Metab Dispos 48:1321–1329. [DOI] [PubMed] [Google Scholar]
- Pandey AV, Sproll P (2014) Pharmacogenomics of human P450 oxidoreductase. Front Pharmacol 5:103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng L, Cui JY, Yoo B, Gunewardena SS, Lu H, Klaassen CD, Zhong X-B (2013) RNA-sequencing quantification of hepatic ontogeny of phase-I enzymes in mice. Drug Metab Dispos 41:2175–2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng L, Paulson A, Li H, Piekos S, He X, Li L, Zhong XB (2014) Developmental programming of long non-coding RNAs during postnatal liver maturation in mice. PLoS One 9:e114917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng L, Yoo B, Gunewardena SS, Lu H, Klaassen CD, Zhong X-B (2012) RNA sequencing reveals dynamic changes of mRNA abundance of cytochromes P450 and their alternative transcripts during mouse liver development. Drug Metab Dispos 40:1198–1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penna A, Buchanan N (1991) Paracetamol poisoning in children and hepatotoxicity. Br J Clin Pharmacol 32:143–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piekos S, Pope C, Ferrara A, Zhong XB (2017) Impact of drug treatment at neonatal ages on variability of drug metabolism and drug-drug interactions in adult life. Curr Pharmacol Rep 3:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piekos SC, Chen L, Wang P, Shi J, Yaqoob S, Zhu H-J, Ma X, Zhong XB (2018) Consequences of phenytoin exposure on hepatic cytochrome P450 expression during postnatal liver maturation in mice. Drug Metab Dispos 46:1241–1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pope C, Mishra S, Russell J, Zhou Q, Zhong X-B (2017a) Targeting H19, an imprinted long non-coding RNA, in hepatic functions and liver diseases. Diseases 5:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pope C, Piekos SC, Chen L, Mishra S, Zhong X-B (2017b) The role of H19, a long non-coding RNA, in mouse liver postnatal maturation. PLoS One 12:e0187557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poudel S, Huber AD, Chen T (2022) Regulation of nuclear receptors PXR and CAR by small molecules and signal crosstalk: roles in drug metabolism and beyond. Drug Metab Dispos DOI: 10.1124/dmd.122.000858 [published ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prabhakar B, Lee S, Bochanis A, He W, Manautou JE, Rasmussen TP (2021) lnc-RHL, a novel long non-coding RNA required for the differentiation of hepatocytes from human bipotent progenitor cells. Cell Prolif 54:e12978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratt VM, Del Tredici AL, Hachad H, Ji Y, Kalman LV, Scott SA, Weck KE (2018) Recommendations for clinical CYP2C19 genotyping allele selection: a report of the Association for Molecular Pathology. J Mol Diagn 20:269–276. [DOI] [PubMed] [Google Scholar]
- Riddick DS, Lee C, Bhathena A, Timsit YE, Cheng P-Y, Morgan ET, Prough RA, Ripp SL, Miller KKM, Jahan A, et al. (2004) Transcriptional suppression of cytochrome P450 genes by endogenous and exogenous chemicals. Drug Metab Dispos 32:367–375. [DOI] [PubMed] [Google Scholar]
- Roberts AG, Stevens JC, Szklarz GD, Scott EE, Kumar S, Shah MB, Halpert JR (2023) Four decades of cytochrome P450 2B research: from protein adducts to protein structures and beyond. Drug Metab Dispos 51:111–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosemary J, Adithan C (2007) The pharmacogenetics of CYP2C9 and CYP2C19: ethnic variation and clinical significance. Curr Clin Pharmacol 2:93–109. [DOI] [PubMed] [Google Scholar]
- Ross CJ, Ulitsky I (2022) Discovering functional motifs in long noncoding RNAs. Wiley Interdiscip Rev RNA 13:e1708. [DOI] [PubMed] [Google Scholar]
- Sadler NC, Nandhikonda P, Webb-Robertson B-J, Ansong C, Anderson LN, Smith JN, Corley RA, Wright AT (2016) Hepatic cytochrome P450 activity, abundance, and expression throughout human development. Drug Metab Dispos 44:984–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibasaki H, Hosoda K, Goto M, Suzuki A, Yokokawa A, Ishii K, Furuta T (2013) Intraindividual and interindividual variabilities in endogenous cortisol 6β-hydroxylation clearance as an index for in vivo CYP3A phenotyping in humans. Drug Metab Dispos 41:475–479. [DOI] [PubMed] [Google Scholar]
- Skinner MK (2011) Role of epigenetics in developmental biology and transgenerational inheritance. Birth Defects Res C Embryo Today 93:51–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y, Li C, Liu G, Liu R, Chen Y, Li W, Cao Z, Zhao B, Lu C, Liu Y (2021) Drug-metabolizing cytochrome P450 enzymes have multifarious influences on treatment outcomes. Clin Pharmacokinet 60:585–601. [DOI] [PubMed] [Google Scholar]
- Stevens JC, Marsh SA, Zaya MJ, Regina KJ, Divakaran K, Le M, Hines RN (2008) Developmental changes in human liver CYP2D6 expression. Drug Metab Dispos 36:1587–1593. [DOI] [PubMed] [Google Scholar]
- Suzuki T, Kikuguchi C, Nishijima S, Nagashima T, Takahashi A, Okada M, Yamamoto T (2019) Postnatal liver functional maturation requires Cnot complex-mediated decay of mRNAs encoding cell cycle and immature liver genes. Development 146:dev168146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taneja G, Maity S, Jiang W, Moorthy B, Coarfa C, Ghose R (2019) Transcriptomic profiling identifies novel mechanisms of transcriptional regulation of the cytochrome P450 (Cyp)3a11 gene. Sci Rep 9:6663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor C, Crosby I, Yip V, Maguire P, Pirmohamed M, Turner RM (2020) A review of the important role of CYP2D6 in pharmacogenomics. Genes (Basel) 11:1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tezcan T, Kotiloğlu SÖ, Akyüzlü DK (2022) The involvement of miRNAs in CYP450 gene expression: a brief review of the literature. J Res Pharm 26:243–254. [Google Scholar]
- Thakur A, Parvez MM, Leeder JS, Prasad B (2021) Ontogeny of drug-metabolizing enzymes. Methods Mol Biol 2342:551–593. [DOI] [PubMed] [Google Scholar]
- Thorn CF, Aklillu E, Klein TE, Altman RB (2012) PharmGKB summary: very important pharmacogene information for CYP1A2. Pharmacogenet Genomics 22:73–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tien Y-C, Liu K, Pope C, Wang P, Ma X, Zhong XB (2015) Dose of phenobarbital and age of treatment at early life are two key factors for the persistent induction of cytochrome P450 enzymes in adult mouse liver. Drug Metab Dispos 43:1938–1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tien Y-C, Piekos SC, Pope C, Zhong X-B (2017) Phenobarbital treatment at a neonatal age results in decreased efficacy of omeprazole in adult mice. Drug Metab Dispos 45:330–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tracy TS, Chaudhry AS, Prasad B, Thummel KE, Schuetz EG, Zhong XB, Tien Y-C, Jeong H, Pan X, Shireman LM, et al. (2016) Interindividual variability in cytochrome P450-mediated drug metabolism. Drug Metab Dispos 44:343–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uno Y, Matsushita A, Osada N, Uehara S, Kohara S, Nagata R, Fukuzaki K, Utoh M, Murayama N, Yamazaki H (2010) Genetic variants of CYP3A4 and CYP3A5 in cynomolgus and rhesus macaques. Drug Metab Dispos 38:209–214. [DOI] [PubMed] [Google Scholar]
- Uzan-Yulzari A, Turta O, Belogolovski A, Ziv O, Kunz C, Perschbacher S, Neuman H, Pasolli E, Oz A, Ben-Amram H, et al. (2021) Neonatal antibiotic exposure impairs child growth during the first six years of life by perturbing intestinal microbial colonization. Nat Commun 12:443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vyhlidal CA, Bi C, Ye SQ, Leeder JS (2016) Dynamics of cytosine methylation in the proximal promoters of CYP3A4 and CYP3A7 in pediatric and prenatal livers. Drug Metab Dispos 44:1020–1026. [DOI] [PubMed] [Google Scholar]
- Wang H, Faucette S, Moore R, Sueyoshi T, Negishi M, LeCluyse E (2004) Human constitutive androstane receptor mediates induction of CYP2B6 gene expression by phenytoin. J Biol Chem 279:29295–29301. [DOI] [PubMed] [Google Scholar]
- Wang P, Chen S, Wang Y, Wang X, Yan L, Yang K, Zhong X-B, Han S, Zhang L (2021) The long noncoding RNA hepatocyte nuclear factor 4α antisense RNA 1 negatively regulates cytochrome P450 enzymes in Huh7 cells via histone modifications. Drug Metab Dispos 49:361–368. [DOI] [PubMed] [Google Scholar]
- Wang P, Liu G, Nie Y, Han S, Li J, Zhong X-B, Zhang L (2019a) Epigenetic memory is involved in the persistent alterations of drug-processing genes in adult mice due to PCN-activated PXR during early life. Toxicol Sci kfz177:10.1093/toxsci/kfz177. [DOI] [PubMed] [Google Scholar]
- Wang X, Yu Y, Wang P, Yang K, Wang Y, Yan L, Zhong XB, Zhang L (2022) Long noncoding RNAs hepatocyte nuclear factor 4A antisense RNA 1 and hepatocyte nuclear factor 1A antisense RNA 1 are involved in ritonavir-induced cytotoxicity in hepatoma cells. Drug Metab Dispos 50:704–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Fang Z, Hong M, Yang D, Xie W (2020) Long-noncoding RNAs (lncRNAs) in drug metabolism and disposition, implications in cancer chemo-resistance. Acta Pharm Sin B 10:105–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Yan L, Liu J, Chen S, Liu G, Nie Y, Wang P, Yang W, Chen L, Zhong X, et al. (2019b) The HNF1α-regulated LncRNA HNF1α-AS1 is involved in the regulation of cytochrome P450 expression in human liver tissues and Huh7 cells. J Pharmacol Exp Ther 368:353–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Schones DE, Zhao K (2009a) Characterization of human epigenomes. Curr Opin Genet Dev 19:127–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Zang C, Cui K, Schones DE, Barski A, Peng W, Zhao K (2009b) Genome-wide mapping of HATs and HDACs reveals distinct functions in active and inactive genes. Cell 138:1019–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Zang C, Rosenfeld JA, Schones DE, Barski A, Cuddapah S, Cui K, Roh T-Y, Peng W, Zhang MQ, et al. (2008) Combinatorial patterns of histone acetylations and methylations in the human genome. Nat Genet 40:897–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y, Huai C, Zhou C, Gao Y, Chen L, Zhou W, Wei M, Qin S (2020) A methylation functional detection hepatic cell system validates correlation between DNA methylation and drug-induced liver injury. Pharmacogenomics J 20:717–723. [DOI] [PubMed] [Google Scholar]
- Xie W (2023) Xenobiotic receptors: a journey of rewards. Drug Metab Dispos 51:207–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong G, Yang Z, Yi J, Wang N, Wang L, Zhu H, Wu C, Lu A, Chen X, Liu S, et al. (2022) DDInter: an online drug-drug interaction database towards improving clinical decision-making and patient safety. Nucleic Acids Res 50(D1):D1200–D1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L, Wang Y, Liu J, Nie Y, Zhong X-B, Kan Q, Zhang L (2017) Alterations of histone modifications contribute to pregnane X receptor-mediated induction of CYP3A4 by rifampicin. Mol Pharmacol 92:113–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon E, Babar A, Choudhary M, Kutner M, Pyrsopoulos N (2016) Acetaminophen-induced hepatotoxicity: a comprehensive update. J Clin Transl Hepatol 4:131–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu A-M (2009) Role of microRNAs in the regulation of drug metabolism and disposition. Expert Opin Drug Metab Toxicol 5:1513–1528. [DOI] [PubMed] [Google Scholar]
- Yu A-M, Pan Y-Z (2012) Noncoding microRNAs: small RNAs play a big role in regulation of ADME? Acta Pharm Sin B 2:93–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu A-M, Tian Y, Tu M-J, Ho PY, Jilek JL (2016) MicroRNA pharmacoepigenetics: posttranscriptional regulation mechanisms behind variable drug disposition and strategy to develop more effective therapy. Drug Metab Dispos 44:308–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu D, Chen J, Chen S, Xu L, Wu L, Li D, Luo J, Jin Y, Zhao Y, Knox B, et al. (2020) Coordinated regulation of UGT2B15 expression by long noncoding RNA LINC00574 and hsa-miR-129-5p in HepaRG cells. Drug Metab Dispos 48:297–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanger UM, Schwab M (2013) Cytochrome P450 enzymes in drug metabolism: regulation of gene expression, enzyme activities, and impact of genetic variation. Pharmacol Ther 138:103–141. [DOI] [PubMed] [Google Scholar]
- Zhang J, Jia Q, Li Y, He J (2023) The function of xenobiotics receptors in metabolic diseases. Drug Metab Dispos 51:237–248. [DOI] [PubMed] [Google Scholar]
- Zhang P, Wu W, Chen Q, Chen M (2019) Non-coding RNAs and their integrated networks. J Integr Bioinform 16: 20190027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z, Cao F, Li X (2019) Epigenetic programming and fetal metabolic programming. Front Endocrinol (Lausanne) 10:764. [DOI] [PMC free article] [PubMed] [Google Scholar]