Abstract
The physical association of bacteria during conjugation mediated by the IncPα plasmid RP4 was investigated. Escherichia coli mating aggregates prepared on semisolid medium were ultrarapidly frozen using copper block freezing, followed by freeze substitution, thin sectioning, and transmission electron microscopy. In matings where the donor bacteria contained conjugative plasmids, distinctive junctions were observed between the outer membranes of the aggregates of mating cells. An electron-dense layer linked the stiffly parallel outer membranes in the junction zone, but there were no cytoplasmic bridges nor apparent breaks in the cell walls or membranes. In control experiments where the donors lacked conjugative plasmids, junctions were not observed. Previous studies have shown that plasmid RP4 carries operons for both plasmid DNA processing (Tra1) and mating pair formation (Tra2). In matings where donor strains carried Tra2 only or Tra2 plus the pilin-processing protease TraF, junctions were found but they were shorter and more interrupted than the wild type. If the donor strain had the pilin gene knocked out (trbC), junctions were still found. Thus, it appears that the electron-dense layer between the outer membranes of the conjugating cells is not composed of pilin.
Conjugation mediated by the IncPα plasmid RP4 (also known as RK2 and RP1) has become an important model system for several reasons. The promiscuity of RP4 transfer between diverse taxa is analogous to the movement of plasmid-borne antibiotic resistance genes in clinical situations (2, 12, 20, 25). Horizontal gene transfers resulting from the movement of plasmids are important from evolutionary and ecological perspectives (24). As well, RP4 is theoretically interesting due to its similarities to other type IV secretion systems of bacteria, especially the Agrobacterium VirB operon (18, 27).
Most models of the physical association of bacteria during conjugation are based on F plasmid-mediated conjugation, as reviewed by Silverman (23). The thick, flexible F pili have been described as the “universal” mating type; this type is correlated with mating on either liquid or solid substrates (8). Earlier studies proposed pilus binding followed by the formation of wall-wall contacts in mating pairs or aggregates (1, 26). More recently, cryofixation and transmission electron microscopy (TEM) of thin sections of donor and recipient bacteria were used to observe tightly appressed regions of the outer membranes termed conjugational junctions (6).
In contrast to the F plasmid system, RP4 produces thin, rigid pili which detach easily from the bacteria (3). While RP4 transfer genes have been characterized (9, 11, 13, 16, 19), studies of the structural association between bacterial cells in RP4-mediated matings have been lacking. To use conventional microscopy techniques to study solid-substrate matings is difficult, as traditional chemical fixation methods have employed immersion in liquid, which could disrupt cell-cell associations.
Transfer of RP4 requires the coordinated activity of the Tra1 operon, which codes for the DNA processing machinery, and the Tra2 operon, which produces mating pair formation functions (reviewed in references 17 and 22). The criteria used to define a gene as involved in mating pair formation have been that it mobilizes RSF1010 (a non-self-transmissible plasmid), allows production of pili, and propagates donor-specific phage (13). The objective of this study was to define directly the structural nature of mating pair formation between Escherichia coli strains during RP4-mediated mating. Cryofixation was used to preserve the mating cells for electron microscopy; this technique allows preservation by solidification of existing cellular water rather than the immersion and gradual chemical cross-linking employed in conventional electron microscopic preparation. To test whether the DNA processing of RP4 triggers junction formation, donor strains lacking the Tra1 region were examined for intercellular junctions. Such strains had normal pili, as they contained all the Tra2 genes and the Tra1-encoded protease (TraF) necessary for normal pilus production (7, 14). As RP4-mediated conjugation is approximately 4 orders of magnitude greater on solid substrate than on liquid, the F plasmid-style mechanism of cell-cell contact by pilus retraction seems unlikely. To explore the role of the pilus in mating, we therefore looked at intercellular interactions in which the donor strain lacked pilin (trbC).
MATERIALS AND METHODS
All cells were grown in YT medium (19) at 37°C in rotating liquid cultures or on agar plates. To improve the proportion of well-frozen cells, 0.3 M glucose was included in the medium as a cryoprotectant (5). The cells were grown in the high-glucose medium overnight, and it was used in all subsequent phases of the experiments to avoid large changes in the osmotic environment. The efficiency of conjugation in the presence of 0.3 M glucose was not significantly different from that of matings in medium lacking glucose (data not shown). The donor strains were E. coli HB101 containing either no plasmid (nonmating control), RP4, or various derivatives of RP4 (Table 1); the recipient strains were E. coli SCS1 (Rifr). These strains are morphologically indistinguishable, a disadvantage, but these matings exactly reproduced the conditions used in earlier biochemical, molecular, and physiological studies (4, 7, 9, 11, 13, 14, 19).
TABLE 1.
Plasmids used in this study and their relevant properties
| Plasmid(s) in donor strain HB101 | Properties | Antibiotic resistance carried | Transfera | Junctions detected |
|---|---|---|---|---|
| RP4 | Wild-type | Kanamycin | + | + |
| pML123, pVWDG23110Δ0.2 | Tra1 + Tra2 | Chloramphenicol, kanamycin | + | + |
| pML123, pWP471 | Tra2 + protease TraF (pili processed) | Chloramphenicol, ampicillin | − | ± |
| pML123 | Tra2 only (pili unprocessed) | Chloramphenicol | − | ± |
| pML123mtrbC45 | Pilin− mutant (MURFI linker in trbC) | Chloramphenicol | − | + |
| None | — | None | − | − |
To prepare samples for electron microscopy, the protocol used for quantitative filter matings was followed (19). Overnight cultures from frozen stocks were grown in YT medium containing the appropriate antibiotics for the plasmid(s) carried by each strain (Table 1). An aliquot of overnight culture was diluted in fresh medium and then allowed to grow to the exponential phase. The cell density was adjusted to an optical density at 600 nm (OD600) of 0.1, and then donor and recipients were mixed 1:1. The mixture was filtered onto a polycarbonate filter (Nucleopore; 25-mm diameter, 0.4-μm pore) and immediately transferred to a nutrient agar plate (no selection) for 20 min at 37°C.
The samples were then ready for cryofixation by copper block slamming. The agar plates containing the mating filters were maintained at 37°C using warm packs in a Styrofoam chest beside the freezing apparatus. Squares (3 by 2 mm) were dissected from the filter with a razor and placed on a Parafilm-coated foam pad attached to a metal specimen holder. The specimen was immediately covered with a small cap containing a moist filter paper in its top to maintain a humid environment and avoid air-drying. The sample was quickly moved to the plunging arm of the copper block freezer (Reichert MM80), which contained a gold-plated copper block with a mirrorlike finish, precooled to liquid nitrogen temperature (−196°C). Alternatively, squares of Nucleopore filter were loaded onto copper hats and frozen in a high-pressure freezer (Balzers HPM 010). Following freezing by either method, the samples were freeze-substituted in 2% OsO4 in anhydrous acetone containing 8% dimethoxypropane for 2 to 3 days at −80°C, followed by embedding in Epon. Following poststaining (2% aqueous uranyl acetate for 15 min, and Reynold's lead citrate for 5 min), the samples were examined and photographed with a Zeiss EM10C in TEM mode.
For immunolabeling, the samples were substituted in acetone alone and embedded at −20°C in LRGold resin. Sections of 70 nm were mounted on copper or nickel grids. Nickel grids were floated on blocking agent (5% nonfat dry milk in Tris-buffered saline [TBS] or 3% bovine serum albumin in TBS), followed by incubation in primary antibody (polyclonal antibodies to TrbC) diluted to 1:75. After a 30-s rinse in a stream of TBST (TBS with 0.1% Tween 20) buffer, the grids were floated on a drop of buffer for 10 min in preparation for secondary antibody-colloidal gold conjugate label (protein A–15-nm gold; Ted Pella). Unbound secondary antibody was removed with another 30-s rinse and 10-min incubation in TBST, and the samples were prepared for poststaining by washing off all traces of buffer with warm distilled water to avoid uranyl acetate precipitation. To test for nonspecific binding of the primary antibody, cells which did not contain the protein of interest (no-plasmid controls) were labeled, and only background levels of label were observed. Similarly, the fidelity of the secondary antibody-gold complex was tested by treating sections with buffer instead of primary antibody. Lacking its specific primary antibody, secondary antibody-gold binding was at sparse (background) levels.
In all matings, junctions tended to occur in patches, and the number of junctions found was highly variable. Therefore, it was important to sample as many filters as possible for each of the various strains mated. Careful sampling meant discarding all filters which were suspected of air drying or significant cooling, so generally five to seven samples could be frozen, per strain, in the 20 to 60 min after mating commenced, per experiment. The quality of freezing varied from experiment to experiment (between 10 and 80% of the filters were well frozen). For each strain tested, three to seven independent experiments were done, so the total number of well-frozen filters examined for each treatment was between 10 and 30.
RESULTS
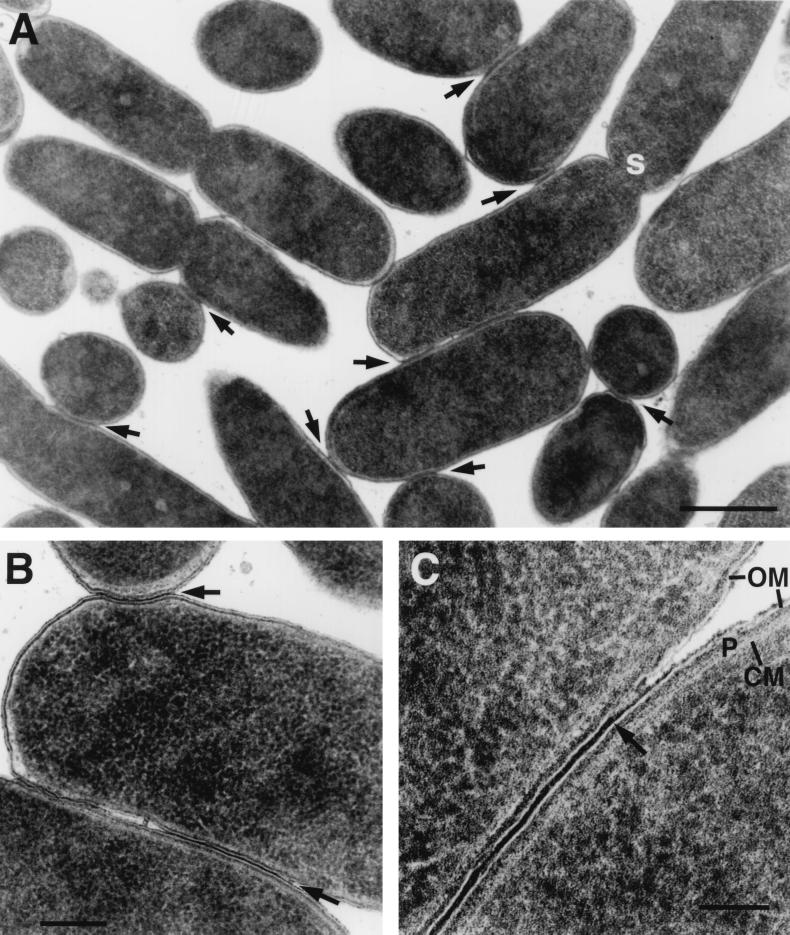
During RP4-mediated filter mating, cells formed close contact zones where their outer membranes were closely appressed (Fig. 1, 2B, and 3A and B) resembling the conjugational junctions described for F plasmid mating by Dürrenberger et al. (6). An electron-dense layer, the origin of which is unknown, linked the outer membranes. There were no apparent breaks in the membranes or cell walls and no cytoplasmic bridges in these regions. In addition to the electron-dense layer linking the cells in the junctions, the outer membranes were strictly parallel in the junctions. The outer membranes came into closer proximity in junctions (only 14 nm separated the centers of the bilayers of the outer membranes) compared with the closest approach of 35 to 40 nm in controls (donors lacking plasmids or just recipients) (Table 2). When junctions were at the poles of the cells, they formed spot attachments of about 150 to 200 nm, but when the junctions were along the lengths of the cell, they formed large oval patches of membranes that extended for up to 1.5 μm (1,500 nm). The variations in morphology, as well as sectioning effects, give rise to large variances in measurements of junction lengths (Table 3). Junctions in wild-type RP4 tended to be shorter than in the two-plasmid system, where Tra1 and Tra2 were carried on separate plasmids (pML123 and pVWDG23110Δ0.2). This may reflect the deletion of the RP4 control elements korA, korB, and trbA, resulting in overexpression of transfer machinery.
FIG. 1.
High-pressure frozen/freeze-substituted E. coli during RP4-mediated conjugation on solid substrate (filter mating). The donor contained RP4-Tra1 and RP4-Tra2 [E. coli HB101(pML123, pVWDG23110Δ0.2)], and the recipient was E. coli SCS1 (Rifr). (A) Lower magnification of mating cell aggregates. Arrows indicate junctions; s indicates the forming septum. Bar, 0.5 μm. (B) Junctions between pairs of bacteria (arrows). Bar, 0.25 μm. (C) High magnification of junction showing electron-dense area between outer membranes (arrow), lightly staining outer membrane (OM), dense periplasmic gel (P), and lightly staining inner cytoplasmic membrane (CM). Bar, 100 nm.
FIG. 2.
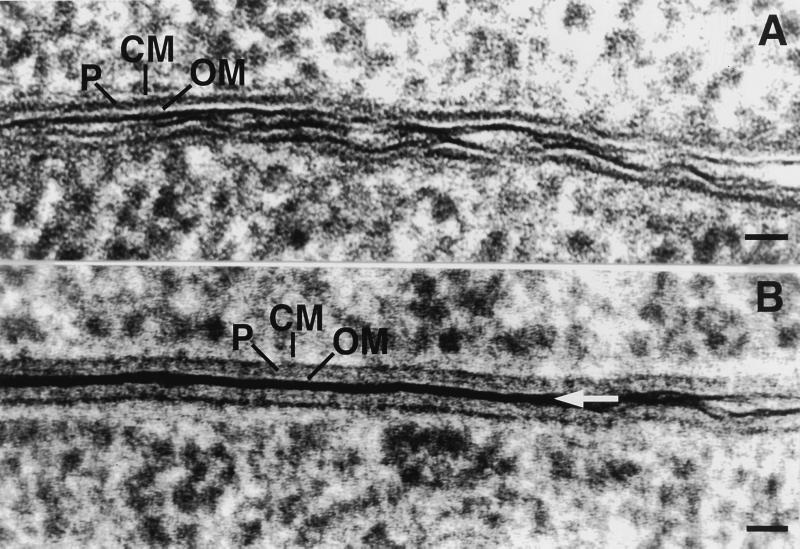
Copper block frozen/freeze-substituted E. coli during (A) no-plasmid control “mating” and (B) RP4-mediated conjugation on solid substrate (filter matings). (A) Bacteria lacking a conjugative plasmid were used as controls [E. coli HB101 (null) × E. coli SCS1 (Rifr)]. (B) Donor contained RP4-Tra1 and RP4-Tra2 [E. coli HB101(pML123, pVWDG23110Δ0.2)], and recipient was E. coli SCS1 (Rifr). Straight, closely appressed outer membranes are typical of junctions between bacteria in RP4-mediated mating with electron-dense material between outer membranes (arrow). Bar, 25 nm.
FIG. 3.
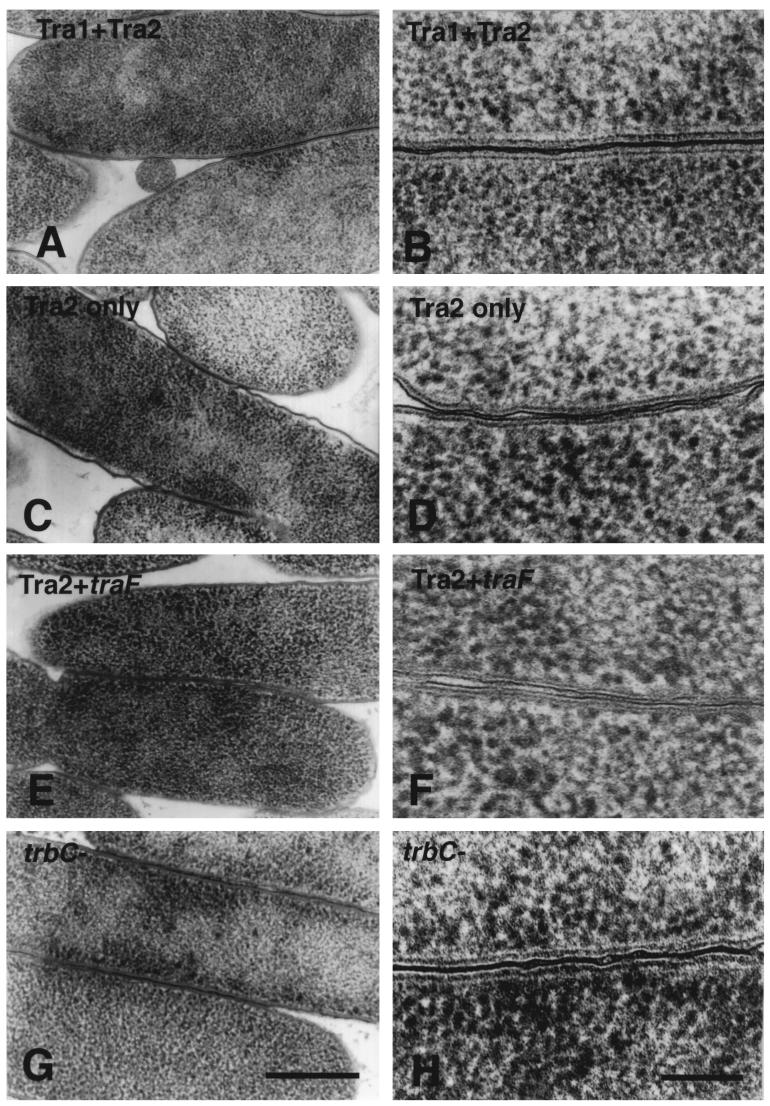
Copper block frozen/freeze-substituted E. coli. Cell-cell associations in intraspecific E. coli filter matings with various donor strains containing an RP4 equivalent or RP4 fragments. (A and B) Reconstituted RP4-mediated mating. Tra1 plus Tra2 conveys all necessary DNA-processing and transfer functions [E. coli HB101(pML123, pVWDG23110Δ0.2) × E. coli SCS1 (Rifr)] to form normal junctions and the areas of electron density between outer membranes seen in high magnification in B. (C and D) Donor carrying mating pair formation genes but not plasmid DNA-processing enzymes (Tra2 only, lacks normal pili). Cells form contact junctions, unlike null controls, but at high magnification as in D, these junctions had gaps where the electron-dense area linking them was interrupted. (E and F) Donor carrying mating pair formation genes for Tra2 plus TraF, necessary for pilin assembly, lacking the DNA-processing complex. Cells were in close contact, but as in C, gaps were observed at higher magnification. (G and H) Donors with a mutant trbC gene, lacking pili. Cells form junctions that are similar to the positive controls seen in A, but with some gaps. (A, C, E, G) Bar, 0.5 μm. (B, D, F, H) Bar, 100 nm.
TABLE 2.
Distances between outer membranesa
| Donor × recipient | Mean distance between outer membranes at zone of closest approach (nm) ± SE | No. of junctions measured |
|---|---|---|
| E. coli HB101 (Tra1 + Tra2) × E. coli SCS1 (Rifr) | 14 ± 1 | 32 |
| E. coli HB101 (null) × E. coli SCS1 (Rifr) | 35 ± 4 | 14 |
| E. coli SCS1 (Rifr) × E. coli SCS1 (Rifr) | 39 ± 4 | 15 |
The distances between outer membranes in junction zones (or the region of closest proximity for bacteria in control samples) in RP4-mediated matings (Tra1 plus Tra2) versus plasmidless controls (null) or recipient × recipient “pseudo-matings.” Actively conjugating bacteria were tightly appressed, as evidenced by the small mean distance between outer membranes.
TABLE 3.
Features of conjugative junctionsa
| Donor strain plasmid(s) | Mean junction length (nm) ± SE (no. of junctions measured) | Median junction length (nm) | Mean no. of interruptions along junction length ± SE (no. of junctions measured) |
|---|---|---|---|
| RP4 | 193 ± 41 (22) | 117 | 1.1 ± 0.4 (22) |
| Tra1 + Tra2 | 281 ± 41 (50) | 188 | 0.9 ± 0.3 (10) |
| Tra2 alone (pili unprocessed) | 86 ± 22 (21) | 42 | 2.3 ± 1.0 (6) |
| Tra2 + TraF (pili processed) | 111 ± 17 (17) | 104 | 2.3 ± 0.4 (7) |
| Pilin-minus (trbC) | 199 ± 29 (71) | 99 | 1.7 ± 0.4 (20) |
| Null | 25 ± 5 (18) | 18 | NA |
The features of conjugative junctions in RP4-mediated matings (and in matings where the donor carried different RP4 transfer components) were determined. Conjugational junction lengths are defined as the areas of close approach between outer membranes with an electron-dense area filling the intercellular space (including interruptions). The number of interruptions per junction (defined as areas where outer membranes deviated from a straight, tightly appressed course and lacked electron density between outer membranes) was determined along the length of a junction. NA, not applicable.
The junctions were not found in control mixtures of plasmid-free cells (nonmating controls) (Fig. 2A and Table 2) or in recipient × recipient pseudomating experiments (Table 2). Small areas of electron density did occur between cells in these samples, but they were very small compared to the extensive junction lengths in mating cells (Table 3). Samples were frozen using either high-pressure freezing (Fig. 1) or copper block freezing (Fig. 2 and 3). Regardless of the freezing technique, junctions were never observed in control, plasmidless donor mating experiments, and conversely, junctions were observed in every case that conjugating cells were examined. The results from replicate experiments were completely consistent, but the variation of the number of junctions found in each combination of cells was high.
To test whether the DNA-processing functions of the relaxosome are necessary for junction formation, donor strains lacking the Tra1 operon but with intact mating pair formation functions and pili (Tra2 plus TraF) were examined. These strains contained the assembly machinery necessary for normal pilus production but have been shown to be transfer deficient (13). When cryofixed samples were assayed by TEM, mating cells were more closely associated than control samples (Fig. 3E and F). At high magnification, the junctions had more gaps along their length (Fig. 3F versus 3B) and the junctions were shorter than in the RP4 wild type or Tra1-Tra2 two-plasmid system (Table 3).
In strains that lack Tra1 functions (Tra2 alone), pili could not be assembled normally due to the loss of the TrbC maturation protease TraF (13). When the Tra2 donors were used in matings, some junctions were found. These junctions often contained interruptions or small gaps along the length of the long nonpolar junction (Fig. 3C and D). These junctions also tended to be shortest and have the most interruptions (Tables 3 and 4).
To look at donors that lacked pili but did not contain residual unprocessed pilin, pilin-negative mutants were used in matings. The pilin gene (trbC) was disrupted by the addition of a MURFI linker (amber codons in all three reading frames and a central XbaI site, minimizing polar effects on downstream genes) (13). Donors lacking trbC (Fig. 3G and H) formed junctions which appeared similar to wild-type junctions and the positive-control, two-plasmid (Tra1 plus Tra2)-containing donor matings. The junctions were long, as were those mediated by Tra1 plus Tra2, but they were more often interrupted (Table 3). This suggests that incomplete processing of pilin can affect junction formation.
Preliminary immunogold labeling studies were done using antibodies against various mating pair formation proteins. None of the Tra2 proteins tested were localized exclusively in junctions (data not shown). For example, anti-TrbB labeled the entire envelope, anti-TrbC labeled the envelope and extracellular filaments, and anti-TrbE, in contrast, gave a cytoplasmic label pattern.
DISCUSSION
During conjugation, plasmid DNA must be transferred from donor to recipient bacteria, crossing the membranes as well as cell walls of the participants. The lack of apparent membrane fusions or cytoplasmic bridges in the conjugational junctions observed here and by Dürrenberger et al. (6) suggests that the structural link between mating bacteria is a subtle anatomical feature. If a multiprotein complex of Tra2 products is spanning the envelopes in these regions, the complex may be contained within the thickness of the TEM section (70 nm). One informative approach could be to open the membranes through the junction zones by freeze-fracture treatment and see if the distribution of intramembrane particles is correlated with the presence of active transfer, i.e., whether “rafts” of transfer protein complexes analogous to gap junctions of mammalian cells could be observed. Fractionation by differential gradient centrifugation of the membranes of donor cells has revealed a fraction of intermediate density between those of the cytoplasmic and outer membranes, containing all the Tra2 components except TrbB (11). Perhaps this fraction is the biochemical footprint of the precursor for the junction structures observed in this study.
Conjugation on solid substrate is more difficult to observe under a light microscope, as opposed to a liquid system such as F plasmid-mediated mating, where the series of conjugation events in a mating cycle such as cell aggregation, aggregate stabilization, and cell separation have been described (1, 26). In the F plasmid system, traN-deficient cells were blocked in aggregate stabilization, leading to the suggestion that this protein is the “glue” in the electron-dense region between the outer membranes (6). However, there is no homolog to TraN in the RP4 system, so the identity of the electron-dense region between mating cells remains elusive. Silverman (23) considered F-pilin to be the only protein abundant enough in the envelope to account for the conjugational junctions observed by Dürrenberger et al. (6). Perhaps the electron-dense regions are generated by quantitatively few, unidentified transfer proteins which act as rivets to bring the lipopolysaccharides into close proximity. From our results, we can draw no conclusions about the nature of the electron-dense material characterizing the junctions except to eliminate the pilin as a likely candidate in the RP4 system.
The presence of junctions in matings mediated by donors lacking functional pili (Tra1 alone, with unprocessed pili; trbC mutants) suggests that pili are not essential for close contact to occur in RP4-mediated matings. The higher efficiency of RP4 transfer on solid substrates than in liquid may reflect a diminished role for the pili in bringing the cells together (as in F-pilus-mediated mating). The role of the pili may be in stabilizing the junctions, as the junctions were smaller and more interrupted in donors lacking pili. As well, it is possible that the pilin is involved in correctly targeting a transfer protein needed for junction stabilization and DNA transfer. While, in this study, we show that these trbC donor-mediated junctions look morphologically like the wild type, it has been shown that actual DNA transfer does not occur under these conditions (13). If there is a small-diameter transport bridge, rich in pilin, we did not detect it in our preliminary immunogold experiments. We never saw clusters of antipilin (or any other anti-Tra protein tested) antibody along the lengths of the junctions, but these sites, depending on their dimensions and sensitivity to the experimental processing, may not bind more than one antibody molecule. Perhaps if one could biochemically isolate junctions and then attempt labeling experiments with Fab fragments of antipilin, the conditions would allow such labeling.
The nonmating controls were never observed to contain the morphologically distinct long junctions joining the cells. This strongly suggests that the junctions found were the result of plasmid-mediated changes to the envelope. In this study, the appearance of donor cells alone was not tested, so we cannot comment on whether surface exclusion processes would prevent or alter the conjugative junction formation.
It is not known why the RP4-mediated matings (and its equivalent two-plasmid system Tra1 plus Tra2) had highly variable amounts of the characteristic junctions present from sample to sample. The number of junctions found in a sample could have varied depending on the time of freezing (between 20 and 60 min after mating began), the area of filter (randomly) chosen for sectioning, or the proportion of donors and recipients in a given area. Whatever the basis of this variability, it meant that a statistically significant sample size for meaningful measurement of the number of junctions in each mating combination would have been prohibitively large.
The E. coli fixed by copper block freezing had the ultrastructure typical of bacteria that had been well preserved by cryofixation, as previously described (10, 15). Well-frozen samples were characterized by envelopes with smooth, intact, evenly spaced membranes and even cytoplasmic density. The density of staining of the cell membrane was lower in copper block frozen samples (Fig. 2), while in high-pressure frozen samples, the membranes were more strongly stained (Fig. 1). There is no theoretical reason for the different techniques for solidifying water during cryofixation to result in differential staining, but the more complicated sample loading with the high-pressure freezer may result in an increased salt concentration resulting from slight air drying during loading. In many cases, samples were discarded if the filter containing the bacteria had obviously air dried before freezing. This problem was easily distinguished because it resulted in a solid mass of cells with no intercellular spaces. We are confident that we can differentiate between the mating junctions and cells that are squeezed together in a nonspecific manner. In well-frozen, non-air-dried samples, the cells were all consistently separated by a minimum of 35 nm, as in control cells, except for areas of mating aggregates where junctions occurred. This was clearly different from air-dried masses, in which every cell was compacted against its neighbor. It is theoretically possible that the junctions detected in this study are the result of partial air drying, but if so, one would expect to find them in all combinations of bacteria. The fact that the junctions are not found in control matings where the donor strain lacks conjugative plasmid contraindicates this possibility. The low staining properties of the cell (inner) membranes in this study are similar to those previously described (10); the authors commented that adjacent membrane leaflets were so tightly appressed to the periplasm “as to render them indistinguishable, even by optical densitometry.” There was also an obvious asymmetry of the outer membrane, with the lipopolysaccharide layer retaining the osmium and uranyl ions. As noted above, the electron-dense deposit found between the cells in the junction zones could be the result of overlap of the lipopolysaccharide coats.
RP4 is a model conjugative plasmid, and analyses of its function have increased our understanding of antibiotic resistance gene spread (reviewed in references 22 and 25). The notion of limited host range has been challenged with the demonstration of intergeneric and interkingdom conjugative plasmid transfer in any system where appropriate replicons permit plasmid maintenance in the recipient (2, 18, 20). While transfer is possible between diverse taxa, the frequency of transfer is often so low that Waters (25) has made the distinction between “essential components” of conjugative systems that make any transfer feasible and the “auxillary components” which in some way enhance the frequency of effective transfer. In RP4-bearing E. coli, the changes in membrane physiology allowing the formation of conjugational junctions may be one such essential component. The altered and characteristic electrophysiological behavior of such membranes has been demonstrated (4). The promiscuity of RP4 transfer could reflect a change in membrane physiology which allows binding to a variety of cell walls or membranes from diverse taxa.
ACKNOWLEDGMENTS
The technical assistance of Michelle Tao, Yeen Ting Hwang, and Susan Shinn is gratefully acknowledged. We thank Fernando de la Cruz for his expert critiques of this work and Tom Giddings for high-pressure freezing assistance.
E.L.'s work was supported by the Deutsche Forschungsgemeinschaft; J.E.D.'s work supported by B. C. Health Research Foundation and the Canadian Natural Sciences and Engineering Research Council.
REFERENCES
- 1.Achtman M. Mating aggregates in Escherichia coli conjugation. J Bacteriol. 1975;123:505–515. doi: 10.1128/jb.123.2.505-515.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bates S, Cashmore A M, Wilkins B M. IncP plasmids are unusually effective in mediating conjugation of Escherichia coli and Saccharomyces cerevisiae. J Bacteriol. 1998;180:6538–6543. doi: 10.1128/jb.180.24.6538-6543.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bradley D E. Morphological and serological relationships of conjugative pili. Plasmid. 1980;2:632–636. doi: 10.1016/0147-619x(80)90005-0. [DOI] [PubMed] [Google Scholar]
- 4.Daugelavicius R, Bamford J K H, Grahn A M, Lanka E, Bamford D. The IncP plasmid-encoded cell envelope-associated DNA transfer complex increases cell permeability. J Bacteriol. 1997;179:5195–5202. doi: 10.1128/jb.179.16.5195-5202.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dubochet J, McDowall A W, Mege B, Schmid E N, Lickfeld K G. Electron microscopy of frozen-hydrated bacteria. J Bacteriol. 1983;155:381–390. doi: 10.1128/jb.155.1.381-390.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dürrenberger M B, Villiger W, Bächi T. Conjugational junctions: morphology of specific contacts in conjugating Escherichia coli bacteria. J Struct Biol. 1991;107:146–156. doi: 10.1016/1047-8477(91)90018-r. [DOI] [PubMed] [Google Scholar]
- 7.Eisenbrandt R, Kalkum M, Lai E-M, Lurz R, Kado C I, Lanka E. Conjugative pili of IncP plasmids, and the Ti plasmid T pilus are composed of cyclic subunits. J Biol Chem. 1999;274:22548–22555. doi: 10.1074/jbc.274.32.22548. [DOI] [PubMed] [Google Scholar]
- 8.Frost L S. Conjugative pili and pilus-specific phages. In: Clewell D B, editor. Bacterial conjugation. New York, N.Y: Plenum Publishing Corp.; 1993. pp. 189–221. [Google Scholar]
- 9.Giebelhaus L A, Frost L S, Lanka E, Gormley I P, Davies J E, Leskiw B. The Tra2 core of the IncP plasmid RP4 is required for intergeneric mating between Escherichia coli and Streptomyces lividans. J Bacteriol. 1996;178:6378–6381. doi: 10.1128/jb.178.21.6378-6381.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Graham L L, Harris R, Villiger W, Beveridge T J. Freeze-substitution of gram-negative eubacteria: general cell morphology and envelope profiles. J Bacteriol. 1991;173:1623–1633. doi: 10.1128/jb.173.5.1623-1633.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grahn A M, Haase J, Bamford D H, Lanka E. Components of the RP4 conjugative transfer apparatus form an envelope structure bridging inner and outer membranes of donor cells: implications for related macromolecule transport systems. J Bacteriol. 2000;182:1564–1574. doi: 10.1128/jb.182.6.1564-1574.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guiney D G. Broad host range conjugation and mobilizable plasmids in gram-negative bacteria. In: Clewell D B, editor. Bacterial conjugation. New York, N.Y: Plenum Publishing Corp.; 1993. pp. 75–97. [Google Scholar]
- 13.Haase J, Lurz R, Grahn A M, Bamford D H, Lanka E. Bacterial conjugation mediated by plasmid RP4: RSF1010 mobilization, donor-specific phage propagation, and pilus production require the same Tra2 core components of a proposed DNA transport complex. J Bacteriol. 1995;177:4779–4791. doi: 10.1128/jb.177.16.4779-4791.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haase J, Lanka E. A specific protease encoded by the conjugative DNA transfer systems of IncP and Ti plasmids is essential for pilus synthesis. J Bacteriol. 1997;179:5728–5735. doi: 10.1128/jb.179.18.5728-5735.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hobot J A, Villiger W, Escaig J, Maeder M, Ryter A, Kellenberger E. Shape and fine structure of nucleoids observed on sections of ultrarapidly frozen and cryosubstituted bacteria. J Bacteriol. 1985;162:960–971. doi: 10.1128/jb.162.3.960-971.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krause S, Bárcena M, Pansegrau W, Lurz R, Carazo J M, Lanka E. Sequence related protein export NTPases encoded by the conjugative transfer regions of RP4 and by the cag pathogenicity island of Helicobacter pylori share similar hexameric ring structures. 2000. Proc. Natl. Acad. Sci. USA, in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lanka E, Wilkins B M. DNA processing reactions in bacterial conjugation. Annu Rev Biochem. 1995;64:141–169. doi: 10.1146/annurev.bi.64.070195.001041. [DOI] [PubMed] [Google Scholar]
- 18.Lessl M, Lanka E. Common mechanisms in bacterial conjugation and Ti-mediated T-DNA transfer to plant cells. Cell. 1994;77:321–324. doi: 10.1016/0092-8674(94)90146-5. [DOI] [PubMed] [Google Scholar]
- 19.Lessl M, Balzer D, Weyrauch K, Lanka E. The mating pair formation system of plasmid RP4 defined by RSF1010 mobilization and donor-specific phage propagation. J Bacteriol. 1993;175:6415–6425. doi: 10.1128/jb.175.20.6415-6425.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mazodier P, Davies J E. Gene transfer between distantly related bacteria. Annu Rev Genet. 1991;25:147–171. doi: 10.1146/annurev.ge.25.120191.001051. [DOI] [PubMed] [Google Scholar]
- 21.Miller J H. Short course in bacterial genetics. Plainview, N.Y: Cold Spring Harbor Press; 1992. [Google Scholar]
- 22.Pansegrau W, Lanka E. Enzymology of DNA transfer by conjugative mechanisms. Prog Nucleic Acid Res Mol Biol. 1996;54:197–251. doi: 10.1016/s0079-6603(08)60364-5. [DOI] [PubMed] [Google Scholar]
- 23.Silverman P M. Towards a structural biology of bacterial conjugation. Mol Microbiol. 1997;23:423–429. doi: 10.1046/j.1365-2958.1997.2411604.x. [DOI] [PubMed] [Google Scholar]
- 24.Syvanen M. In search of horizontal gene transfer. Nat Biotech. 1999;17:833. doi: 10.1038/12781. [DOI] [PubMed] [Google Scholar]
- 25.Waters V L. Conjugative transfer in the dissemination of β-lactam and aminoglycoside resistance. Frontiers Biosci. 1999;4:433–456. doi: 10.2741/waters. [DOI] [PubMed] [Google Scholar]
- 26.Willetts N, Skurray R. Structure and function of the F factor and mechanism of conjugation. In: Neidhardt F C, Ingraham J L, Low K B, Magasanik B, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella typhimurium: cellular and molecular biology. Washington, D.C.: American Society for Microbiology; 1987. pp. 1110–1133. [Google Scholar]
- 27.Winans S C, Burns D L, Christie P J. Adaptation of a conjugal transfer system for the export of pathogenic macromolecules. Trends Microbiol. 1996;4:64–68. doi: 10.1016/0966-842X(96)81513-7. [DOI] [PMC free article] [PubMed] [Google Scholar]