Abstract
Two energy-generating hydrogenases enable the aerobic hydrogen bacterium Ralstonia eutropha (formerly Alcaligenes eutrophus) to use molecular hydrogen as the sole energy source. The complex synthesis of the nickel-iron-containing enzymes has to be efficiently regulated in response to H2, which is available in low amounts in aerobic environments. H2 sensing in R. eutropha is achieved by a hydrogenase-like protein which controls the hydrogenase gene expression in concert with a two-component regulatory system. In this study we show that the H2 sensor of R. eutropha is a cytoplasmic protein. Although capable of H2 oxidation with redox dyes as electron acceptors, the protein did not support lithoautotrophic growth in the absence of the energy-generating hydrogenases. A specifically designed overexpression system for R. eutropha provided the basis for identifying the H2 sensor as a nickel-containing regulatory protein. The data support previous results which showed that the sensor has an active site similar to that of prototypic [NiFe] hydrogenases (A. J. Pierik, M. Schmelz, O. Lenz, B. Friedrich, and S. P. J. Albracht, FEBS Lett. 438:231–235, 1998). It is demonstrated that in addition to the enzymatic activity the regulatory function of the H2 sensor is nickel dependent. The results suggest that H2 sensing requires an active [NiFe] hydrogenase, leaving the question open whether only H2 binding or subsequent H2 oxidation and electron transfer processes are necessary for signaling. The regulatory role of the H2-sensing hydrogenase of R. eutropha, which has also been investigated in other hydrogen-oxidizing bacteria, is intimately correlated with a set of typical structural features. Thus, the family of H2 sensors represents a novel subclass of [NiFe] hydrogenases denoted as the “regulatory hydrogenases.”
Molecular hydrogen is frequently used as an energy source by diverse prokaryotic organisms. Many of these bacterial and archaeal species harbor multiple hydrogenases which mediate heterolytic cleavage of H2 into 2 H+ and 2 e−. [NiFe] hydrogenases are the most dominant enzymes, representing a fairly conserved family of proteins, composed of at least a large active site-containing subunit and a small electron-transferring subunit which bears one to three FeS clusters (1, 2, 18).
The facultative chemolithoautotrophic proteobacterium Ralstonia eutropha H16 (formerly Alcaligenes eutrophus [7]) harbors two energy-generating [NiFe] hydrogenases, a membrane-bound enzyme (MBH) and a cytoplasmic enzyme (SH). The MBH is primarily involved in electron transport-coupled phosphorylation, whereas the SH is able to reduce NAD and thus provides the cell with reducing equivalents (38, 40). The composition of the MBH resembles the prototype of [NiFe] hydrogenases whose atomic structure has been resolved by X-ray analysis (50). The two subunits of the R. eutropha MBH, encoded by hoxK and hoxG, are anchored to the outer face of the cytoplasmic membrane via a b-type cytochrome (4). The SH, encoded by hoxF, hoxU, hoxY, and hoxH, contains an FeS-flavoprotein in addition to the hydrogenase moiety (30). Mutants disrupted in either one of the two hydrogenases maintain their ability to grow on H2, which indicates that the two enzymes can replace each other physiologically (23).
The hydrogenase-related genes of R. eutropha are organized in the MBH and the SH operons, which are regulated coordinately (42). The MBH operon comprises 10 MBH-specific genes in addition to a set of accessory genes whose products are involved in the complex posttranslational maturation of the hydrogenases and the regulation of both the MBH and the SH operon (5, 11, 24, 41). The SH operon harbors the structural genes of the NAD-reducing hydrogenase together with a set of accessory genes which code for maturation proteins (45, 47, 52).
Hydrogenase gene expression is controlled by the major transcription factor HoxA, a member of the NtrC family of response regulators (12). HoxA binds specifically at the upstream regions of the MBH and SH operons and activates transcription in concert with the ς54-containing RNA polymerase (42, 54). Transcription activation by HoxA is stimulated by at least two environmental signals: the presence of molecular hydrogen and/or limitation of organic carbon and energy sources (25, 42). Recognition of molecular hydrogen by cells of R. eutropha is mediated by a complex signal transduction system consisting of the proteins HoxB and HoxC which share features of [NiFe] hydrogenases, and HoxJ, a histidine protein kinase which has autophosphorylation capacity with ATP as the phosphoryl donor (25, 26). Deletions in hoxB or hoxC of R. eutropha prevent hydrogenase from being synthesized, whereas a knockout of hoxJ leads to H2-independent high-level hydrogenase gene expression. The data suggest a model in which HoxBC functions as a hydrogen receptor which interacts either directly or indirectly with the sensor kinase HoxJ. Furthermore, unlike in most other two-component regulatory systems, the autophosphorylation-active kinase acts negatively on hydrogenase gene expression. This observation indicates that the HoxJ-mediated phosphorylation of the response regulator HoxA blocks hydrogenase gene transcription. The negative effect of HoxJ is released by HoxBC, provided H2 is present (25). Proteins similar to HoxBC, designated HupUV, have been identified in Rhodobacter capsulatus and Bradyrhizobium japonicum. Mutant analysis revealed that these proteins play a pivotal role in the H2-dependent regulation in these organisms. These results led to the conclusion that the HupUV proteins act as a hydrogen sensor (6, 15).
To study the mechanism of H2-signal transduction in more depth, the interacting partners of the system have to be isolated and characterized in vitro. Because previous attempts to overproduce active [NiFe] hydrogenases heterologously in Escherichia coli had not been successful, we constructed a novel expression vector for the native host R. eutropha. With the aid of this vector, we achieved an efficient expression of hoxBC and show that the resulting protein catalyzes H2 oxidation. The inspection of mutants revealed that the physiological role of HoxBC is H2 sensing and not the generation of energy for growth on H2. Thus, the third hydrogenase of R. eutropha is denoted as the “regulatory hydrogenase” (RH). Both the enzymatic activity and the regulatory function of the RH protein are strictly dependent on the availability of nickel in the medium, showing that nickel is essential for H2 sensing.
MATERIALS AND METHODS
Strains and plasmids.
The strains and plasmids used in this study are listed in Table 1. Strains with the initials HF were derived from R. eutropha H16 (wild type). R. eutropha HF433 harbors an active H2-sensing signal transduction chain, including the sensor kinase HoxJ. R. eutropha H16 is a natural variant in which the H2-dependent signal transduction is interrupted by a glycine-to-serine exchange at position 422 in HoxJ (HoxJG422S [25]). The newly isolated strains R. eutropha HF375, a derivative of strain H16, and HF459, a derivative of strain HF433, carry in-frame deletions in the nickel permease gene hoxN. R. eutropha HF500 bears in-frame deletions in hoxG, hoxH, and hoxC, resulting in an MBH− SH− RH− phenotype. Strain HF371 (31) harbors the inactive sensor kinase HoxJG422S in addition to in-frame deletions in hoxG and hoxH and was used as host for plasmid-based overexpression of hoxB and hoxC. E. coli JM109 was used as a host in standard cloning procedures (53). E. coli S17-1 (43) served as a donor in conjugative transfers.
TABLE 1.
Strains, plasmids, and oligonucleotides used in this study
| Strain, plasmid, or oligonucleotide | Relevant characteristic(s) or sequence | Source or reference |
|---|---|---|
| Strains | ||
| A. eutrophus | ||
| H16 | MBH+ SH+ RH+, HoxJG422S | DSM428, ATCC 17699 |
| HF371 | hoxGΔ hoxHΔ; MBH− SH− RH+, HoxJG422S | 31 |
| HF375 | hoxNΔ; HoxN− HoxJG422S | This study |
| HF433 | MBH+ SH+ RH+ | 25 |
| HF459 | hoxNΔ; HoxN− | This study |
| HF500 | hoxGΔ hoxHΔ hoxCΔ; MBH− SH− RH−, HoxJG422S | This study |
| E. coli | ||
| JM109 | F′ traD36 lacIq, Δ(lacZ)M15 proA+B+/e14− (McrA−) Δ(lac-proAB) thi gyrA96 (Nalr) endA1 hsdR17(rK− mK+) relA1 supE44 recA1 | 53 |
| S17-1 | Tra+recA pro thi hsdR, chr:RP4-2 | 43 |
| Plasmids | ||
| LITMUS 29 | AprlacZ′, ColE1 ori | New England Biolabs |
| pLO2 | Kmr, sacB, RP4 oriT, ColE1 ori | 27 |
| pNEB193 | AprlacZ′, ColE1 ori | New England Biolabs |
| pQE-30 | Ampr, T5 promoter, ColE1 ori | Qiagen Inc. |
| pCH128 | Derivative of pSUP202 carrying hoxF | 13 |
| pCH231 | pBluescript KS(+) with a 2.2-kb HindIII-XhoI fragment containing hoxN | 14 |
| pCH394 | pACYC177 with a 2.38-kb HindIII-SmaI fragment containing hoxB | O. Lenz and B. Friedrich |
| pCH352 | pBluescript KS(+) with a 9.6-kb HindIII/BamHI fragment containing ′hoxA, hoxB, hoxC, hoxJ, and hoxN | K. Marin and B. Friedrich |
| pCH591 | LITMUS 29 with a 0.26-kb HindIII-NdeI fragment containing the SH promoter (PSH) | This study |
| pCH592 | pNEB193 with a 0.68-kb NdeI-EcoRI fragment containing hoxB′ | This study |
| pCH593 | pCH592 with a 3.3-kb PstI fragment containing ′hoxB, hoxC, hoxJ′ | This study |
| pCH594 | pCH591 with a 2.5-kb NdeI fragment from pCH593 | This study |
| pCH615 | 0.8-kb Eco47III fragment containing ′hoxJ into PmeI-digested pLO3 | 25 |
| pCH644 | pLO1 with a 2.45-kb PstI fragment containing hoxCΔ | 25 |
| pCH655 | 1.5-kb PspI (Klenow-treated) fragment containing hoxC into Ecl136II-digested pQE-30 | This study |
| pCH658 | 0.45-kb FspI-SmaI fragment from pCH231 into EcoRI-digested (Klenow-treated) pCH231 | This study |
| pCH659 | pLO2 with a 1.5-kb SalI fragment containing hoxNΔ | This study |
| pEDY305 | RK2 ori, Tcr, Mob+, promoterless lacZ gene | E. Schwartz and B. Friedrich |
| pEDY309 | Derivative of pEDY305 with a modified multiple cloning site | This study |
| pGE151 | Derivative of pRK404 | 24 |
| pGE301 | Φ(hoxK′-′lacZ), Tcr | 27 |
| pGE377 | pEDY309 with a 2.8-kb HindIII-XbaI fragment containing PSH-hoxB-hoxC | This study |
| pGE378 | 2.2-kb PvuII-Ecl136II containing Plac-hoxA into SwaI-digested pGE377 | This study |
| pGE400 | 1.95-kb SmaI-Ecl136II fragment containing hoxA into Ecl136II-digested pGE151; Plac-hoxA | This study |
| Oligonucleotides | ||
| 189 | GGTGGAAGGGGTGGCCG | This study |
| 340 | CGCCATGGCATATGGTCTCCTCCTTACTAATGTTCGCa | This study |
| 341 | CGCTGGCACAAGCTTGC | This study |
| 342 | TGGAGGACATATGAACGCGCCTGTATGTACCa | This study |
| 372 | CTCTAGAGGAGATCTCACAAGCTTCGAACTAGTTAGCTAGAT | This study |
| 373 | CGATCTAGCTAACTAGTTCGAAGCTTGTGAGATCTCCTCTAGAGGTAC | This study |
Mutagenic oligonucleotide (underlined residues represent altered nucleotides)
Plasmid pCH591 contains the modified SH promoter region and an NdeI site at the ATG start codon of the first gene hoxF of the SH operon (Table 1). pCH591 was constructed by amplification of the hoxF upstream region from pCH128 using the mutagenic primer 340 and the primer 341. The amplification product was cut with HindIII and NcoI, and the resulting 0.26-kb fragment was inserted into HindIII-NcoI-digested LITMUS 29, yielding pCH591. An NdeI site at the ATG start codon of hoxB was introduced as follows. pCH394 was used as the template for amplification of the 5′ region of hoxB with primer 189 and the mutagenic oligonucleotide 342. A 0.68-kb NdeI-EcoI fragment of the amplified product was inserted into pNEB193, resulting in pCH592. Subsequently, a 3.3-kb PstI fragment from pCH352 was inserted into PstI-digested pCH592, resulting in plasmid pCH593 containing the tandemly arranged genes hoxB and hoxC. A 2.5-kb NdeI fragment of pCH593 was cloned into pCH591 yielding pCH594 which harbors the hoxB and hoxC genes under control of the SH promoter (Fig. 1).
FIG. 1.
Construction of the hoxBC overexpression vector pGE378. Genes are marked by bold arrows. R. eutropha-derived genes are marked by hatched arrows. The orientations of PSH and Plac are given by arrows. Stop codons in all three reading frames downstream of the multiple cloning site (MCS) from pEDY309 are underlined. Ttrp, trp terminator; Tfd, phage fd terminator.
For overexpression of the hoxB and hoxC genes in the native host R. eutropha, we constructed the broad-host-range vector pEDY309 by modification of the multiple cloning region of pEDY305 (Table 1). pEDY305 was digested with KpnI-ClaI and used to introduce the polynucleotide kinase-treated hybridization product of oligonucleotides 372 and 373. Subsequently, the 2.8-kb HindIII-XbaI fragment of pCH594 was inserted into pEDY309, yielding plasmid pGE377. Finally, a hoxA-containing 2.2-kb PvuII-Ecl136II fragment, originating from pGE400, was ligated into SwaI-cut pGE377 to yield the hoxB-hoxC overexpression vector pGE378 (Fig. 1).
The hoxN in-frame deletion allele was constructed as follows. A pCH231-derived 0.45-kb FspI-SmaI fragment containing the 3′ region of hoxN was inserted into EcoRI-digested, end-polished pCH231. The resulting plasmid, designated pCH658, contains a hoxN allele in which 768 of 903 bp (85%) were deleted. For recombination into R. eutropha the hoxNΔ allele was subcloned as a 1.5-kb SalI-fragment into pLO2, yielding pCH659. The fusion sites in the hoxNΔ allele and in the PCR amplification products were verified by sequencing.
Plasmid pCH655, which was used for overexpression of hoxC in E. coli, was constructed by insertion of a 1.5-kb PspI (Klenow-treated) fragment harboring the hoxC sequence without the first two codons into the Ecl136II site of pQE-30.
Media and growth conditions.
Strains of R. eutropha were grown in nutrient broth (NB), in a modified Luria broth (LB) with 0.25% sodium chloride (LSLB), or in mineral salts medium as described previously (12). Synthetic media for heterotrophic growth contained 0.4% (wt/vol) fructose (FN) or 0.2% (wt/vol) fructose and 0.2% (vol/vol) glycerol (FGN). Cultivation under lithoautotrophic conditions was done in mineral salts medium under an atmosphere of hydrogen, carbon dioxide, and oxygen (8:1:1, vol/vol/vol). Sucrose-resistant segregants of sacB-harboring strains were selected on LSLB plates containing 15% (wt/vol) sucrose (27).
E. coli strains were grown in LB medium. Solid medium contained 1.2% (wt/vol) agar. Antibiotics were supplemented with the following: kanamycin (400 μg/ml) and tetracycline (15 μg/ml) for R. eutropha and kanamycin (25 μg/ml), tetracycline, (15 μg/ml), and ampicillin (100 μg/ml) for E. coli.
Gene replacement.
The hoxNΔ allele was reintroduced into R. eutropha H16 via conjugation using the suicide vector pCH659. The allelic exchange procedure was based on the conditionally lethal sacB gene (27). The resultant sucrose-resistant isolates were screened for the presence of the desired mutation by amplification of the respective target sites as previously described (5). Deletion-carrying isolates were identified on the basis of the altered electrophoretic mobility of the amplification products. The resulting hoxNΔ strain HF375 served as the recipient for the hoxJ-containing suicide vector pCH615 to generate the isogenic hoxNΔ hoxJ strain HF459. Strain HF500, a derivative of HF371 (31), was isolated by the same recombination technique using pCH644 which contains the hoxCΔ allele (25).
Cell fractionation.
Cells were disrupted in a French pressure cell, and the resulting crude extract was separated into soluble and membrane fractions as described earlier (19). Cytoplasmic and periplasmic fractions were separated by a modified version (4) of the procedure of Probst and Schlegel (36).
Immunogenic techniques.
E. coli JM109 cells harboring the hoxC expression plasmid pCH655 were grown in LB medium at 30°C to an optical density at 600 nm (OD600) of 0.8. Expression of His6-HoxC was induced by adding IPTG (isopropyl-β-d-thiogalactopyranoside) to a final concentration of 1 mM. Cells were harvested after 3 h of induction and disrupted by two passages through a French pressure cell. Inclusion bodies which contained most of the His6-HoxC protein were treated with 6 M guanidinium-HCl. Subsequently, the His6-HoxC protein was purified using the Ni-nitrilotriacetic acid Spin Kit (Qiagen, Inc.) according to the manufacturer's instructions. Purified His6-HoxC was used as antigen for immunization of rabbits (BioGenes GmbH, Berlin, Germany). Proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), transferred to Protran BA85 nitrocellulose membranes (Schleicher and Scheull), and identified immunologically according to a standard protocol (46). HoxC was detected with anti-HoxC serum, diluted 1:1,000, and an alkaline phosphatase-labeled goat anti-rabbit immunoglobulin G (Dianova, Hamburg, Germany).
Labeling with 63NiCl2.
Labeling of hydrogenases with 63NiCl2 was essentially performed as previously described (5). Cells were grown in FGN medium in the presence of 120 nM of 63NiCl2 (6.38 mCi/ml; Amersham-Buchler). Soluble extracts were prepared and subjected to native PAGE. Gels were run in a continuous buffer system consisting of 90 mM Tris, 80 mM borate, and 2.5 mM EDTA (pH 8.3) at 200 V and 4°C for 2,500 V-h. After electrophoresis the gels were dried under vacuum and subjected to autoradiography using a SI 550 storage PhosphorImager (Molecular Dynamics).
Assays.
Hydrogenase assays were performed with cells grown heterotrophically in FGN medium. SH (hydrogen-NAD+ oxidoreductase; EC 1.12.1.2) activity was determined by photometric recording of the H2-dependent NAD reduction in the soluble fraction (39). MBH (hydrogen-acceptor oxidoreductase EC 1.18.99.1) and RH activities were photometrically measured in the membrane fraction using methylene blue as an electron acceptor (38). Amperometric H2 uptake measurements using an H2 electrode and methylene blue as an electron acceptor were done as previously described (35). For in-gel chromogenic detection of hydrogenase activity (5), soluble extracts were resolved on native PAGE gels as described above. The gels were subsequently incubated in H2-saturated 50 mM potassium phosphate buffer (pH 5.5) containing 0.09 mM phenazine methosulfate and 0.06 mM nitroblue tetrazolium under an atmosphere of H2 at 30°C. O2 uptake assays were performed with whole cells using a Clark electrode (Rank Brothers Model 10). O2 consumption was recorded amperometrically in 2.7 ml of H2-saturated potassium phosphate buffer (50 mM, pH 7.0) at 30°C. Then, 200 μl of O2-saturated water was added, and the reaction was started by the addition of 100 μl of cell suspension which was previously adjusted to an OD436 of 11. H2-independent O2 consumption was monitored in N2-saturated potassium phosphate buffer. β-Galactosidase assays were performed as described previously (54), and the activities (in units) were calculated according to the Miller method (33) except that cell density was measured at 436 nm. The level of protein in extracts was determined by the method of Lowry et al. (28).
RESULTS AND DISCUSSION
The RH and homologous H2-sensing proteins form a subclass of [NiFe] hydrogenases.
The regulatory region of the megaplasmid-borne hydrogenase gene complex in R. eutropha has previously been extended by three additional open reading frames (ORFs), designated hoxB, hoxC, and hoxJ (25, 26). The ORFs fill a gap between hoxA, the response regulator gene, and hoxN, the nickel permease gene. Database searches revealed similarity of the hoxJ product to histidine protein kinases (26) and of HoxB and HoxC to [NiFe] hydrogenases, in particular to a small group of proteins which are present in aerobic H2-oxidizing bacteria (Fig. 2). The closest relatives are the HoxB and HoxC proteins of Alcaligenes hydrogenophilus (26) and the HupU and HupV proteins of R. capsulatus (15) and B. japonicum (6), with sequence identities ranging from 53 to 79%.
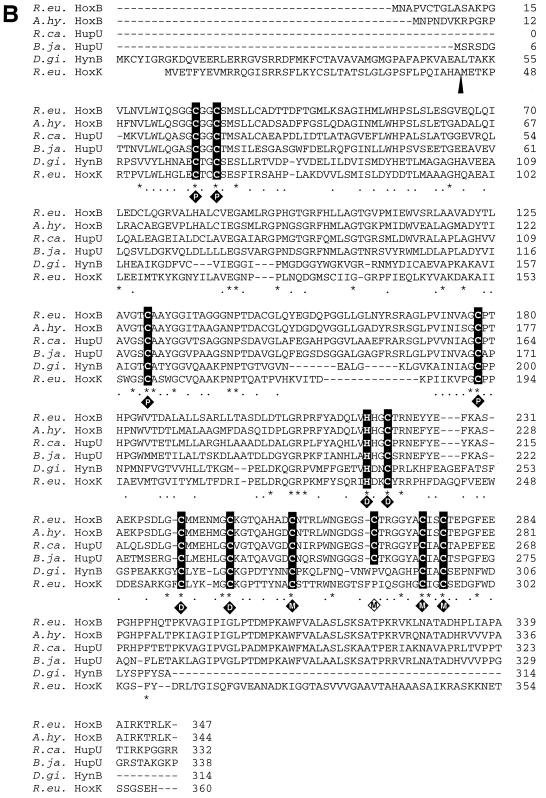
FIG. 2.
Alignment of H2-sensing and standard [NiFe] hydrogenase subunits. Identical and similar amino acids are marked by asterisks and dots, respectively. (A) The five stretches of conservative amino acids identified in [NiFe] hydrogenase large subunits (2) are shown in boldface letters above the sequences (elements 1 to 5). The two cysteine pairs which coordinate nickel and iron at the catalytic center of the D. gigas hydrogenase are highlighted. The cleavage site of C-terminal proteolysis of HynA is marked by an arrowhead. (B) The cysteine and histidine residues which coordinate the proximal (P), medial (M), and distal (D) iron sulfur clusters in the small subunit HynB of D. gigas are highlighted in addition to Cys267 in HoxB of R. eutropha. The cleavage site of the N-terminal leader peptide in HynB is marked by an arrowhead. Abbreviations: R.eu, R. eutropha; A.hy., A. hydrogenophilus; B.ja., B. japonicum; R.ca., R. capsulatus; D.gi., D.gigas.
HoxB and HoxC and their close relatives show typical signatures of standard [NiFe] hydrogenases (Fig. 2), represented by the prototypic periplasmic [NiFe] hydrogenase from Desulfovibrio gigas (51). The product of hoxC displays the conserved amino acid motifs which are considered as essential elements for the coordination of the NiFe cofactor (Fig. 2A) (2). The N-terminal RGxE motif (element 1) shows the regular spacing of 16 residues to the metal binding motif RxCGxCxxxH (element 2). It is worth noting, however, that the highly conserved histidine residue of element 2 is replaced by a glutamine residue in all HoxC-like proteins known so far (Fig. 2A). The conserved signature of the more variable element 3 is restricted to only two of five histidine residues. In element 4, GxxxPRGxxxxH, which is oriented to the C-terminal end of the polypeptide, an alanine substitutes for the highly conserved proline residue, whereas the C-terminal NiFe coordination site DPCxxCxxH (element 5) shows the perfect consensus of standard hydrogenases. Moreover, HoxC-like proteins terminate at a histidine residue and are devoid of a C-terminal extension (Fig. 2A). This observation suggests that, unlike the situation in most [NiFe] hydrogenases (29), the regulatory proteins do not undergo a proteolytic cleavage prior to metallocenter insertion and subunit oligomerization. The lack of a C-terminal tail has also been reported for the CO-induced hydrogenase from Rhodospirillum rubrum and for the Ech hydrogenase from Methanosarcina barkeri. However, these enzymes are related to hydrogenase 3 of E. coli and are considered to be involved in H2 evolution rather than in H2 sensing (17, 32).
HoxB and its homologues have potential coordination sites for three FeS clusters similar to the small HynB subunit of D. gigas (Fig. 2B). Four cysteines (Cys27, -30, -130, and -178) correlating with the ligands of the proximal [4Fe-4S] cluster (P), are present in the N-terminal region of HoxB. Three cysteine residues (Cys220, -240, and -247) and one histidine (His217) coincide with the ligands of the distal [4Fe-4S] cluster (D) in HynB. Interestingly, HoxB and its close relatives reveal four instead of three conserved cysteines (Cys256, -267, -274, and -277) for binding the putative medial FeS cluster (M), suggesting that the common [3Fe-4S] center might be replaced by a [4Fe-4S] cluster in this group of proteins. Most notably, HoxB and its homologues lack the N-terminal leader sequence which directs the export of periplasmic and membrane-bound hydrogenases (3, 34). This observation points to a cytoplasmic location of these proteins. Another interesting structural feature of HoxB-like proteins is a C-terminal peptide of 54 to 55 amino acids which is not present in the periplasmic HynB protein. Although the small subunits of membrane-bound hydrogenases also carry an extension of about 50 amino acids, including a stretch of 22 hydrophobic amino acids and a highly conserved histidine residue, the primary structure is clearly distinct from the tail of the HoxB-type proteins (Fig. 2B). It has been shown that the C-terminal domain plays a pivotal role in anchoring the membrane-bound hydrogenases to the membrane and in coupling the proteins to the primary electron acceptors, the b-type cytochromes (4, 21). In analogy, it seems likely that the C-terminal domain of HoxB-like proteins links the regulatory hydrogenases to their specific partners, namely, the histidine protein kinases HoxJ and HupT, respectively (16, 25, 48). We now have evidence that signal transduction is mediated by the RH protein in direct contact with the sensor kinase HoxJ (T. Buhrke and B. Friedrich, unpublished results).
It is interesting to note that HoxJ and the related kinases from A. hydrogenophilus (26), B. japonicum (48), and R. capsulatus (16) belong to a superfamily of proteins that contain PAS domains. PAS modules are found in Bacteria, Archaea, and Eukarya, predominantly in proteins that are involved in signal transduction (44). Since PAS domains monitor changes in light, redox potential, oxygen, and the general energy status of the cell, the HoxJ sensor kinase is the favorite candidate to receive and convert the signal from the RH.
The outstanding biological role of H2-sensing hydrogenases (6, 15, 25), obviously based on a series of individual structural features, places this group of regulatory proteins into a distinct subclass of [NiFe] hydrogenases. Public databases provided evidence for the occurrence of this group of hydrogenases in Rhodobacter sphaeroides (20) and Oligotropha carboxydovorans (37). Moreover, one of the three putative hydrogenases of Aquifex aeolicus shows characteristics of HoxBC-like proteins, pointing to a possible H2-sensing process in this phylogenetically ancient bacterium (10). Thus, regulatory hydrogenases may be more common than originally envisaged.
The RH of R. eutropha is a soluble H2-oxidizing protein located in the cytoplasm.
The structural similarity to hydrogenases and its role in H2-sensing implied the fundamental questions whether the RH catalyzes H2 oxidation and if so, whether the hydrogenase activity is necessary for its regulatory function. It was not possible to discriminate the RH activity in wild-type cells clearly from the MBH- and SH-derived hydrogenase activities. To exclude assay interferences the MBH− SH− RH+ strain HF371, which carries in-frame deletions in the large subunit genes of the MBH (hoxGΔ) and the SH (hoxHΔ), was grown under heterotrophic, hydrogenase-derepressing conditions and then tested for H2-oxidizing activity. Soluble extracts of this mutant showed a low level of H2 uptake activity (1.5 nmol H2/min/mg of protein) measured amperometrically with methylene blue as the electron acceptor. However, the activity was clearly above the background level (<0.5 nmol of H2/min/mg of protein) of extracts derived from strain HF500, which is disrupted in all three hydrogenase proteins. Notably, the MBH− SH− RH+ strain HF371 was not able to grow autotrophically with H2 as the energy source. These results indicate that the RH is either formed at an extremely low level and/or that the protein exhibits only poor hydrogenase activity, a finding which is in agreement with its regulatory role.
To determine the cellular localization of the RH protein, cells of the MBH− SH− RH+ strain HF371 were separated into the cytoplasmic, periplasmic, and membrane fractions. In order to detect the RH immunologically a hexahistidine-tagged variant of the HoxC protein was purified from E. coli to raise a polyclonal antiserum. Immunoblots developed with this antiserum gave a faint band corresponding to a 52-kDa protein which was exclusively present in the cytoplasmic fraction (data not shown). The size of this protein was in good agreement with the molecular mass of 52.4 kDa predicted for HoxC. A HoxC signal was absent in extracts of the control strain HF500, which lacks all three hydrogenases, showing that the antiserum is specific for HoxC (data not shown). These immunological data are completely in line with the prediction for a cytoplasmic location of the RH deduced from the primary sequence. Since dihydrogen is a freely diffusible molecule, there is no need for the cell to anchor the H2-sensing protein to the membrane.
Homologous overproduction of the RH protein.
To get further insight into the biochemical properties of the H2 sensor, the intracellular level of the protein had to be increased. This was achieved by overexpressing the native hoxB and hoxC genes in R. eutropha under the control of the SH promoter (PSH), which directs transcription of the SH operon (42, 54). The HoxA-controlled, homologous system has the advantage that the Hyp proteins, which are required for metallocenter assembly (11), are potentially available for RH maturation. Moreover, PSH is a well-characterized, relatively strong promoter, and the putative ribosome-binding site of the first gene hoxF of the SH operon is in perfect agreement with the consensus in E. coli (42, 54).
The construction of the expression vector is based on three steps, which are described in detail in Materials and Methods. (i) The native hoxB and hoxC genes were tandemly fused to a modified SH promoter region yielding plasmid pCH594 (Fig. 1). (ii) A fragment containing the PSh-hoxBC fusion was transferred to the broad-host-range vector pEDY309 that replicates stably in R. eutropha. (iii) To enhance transcription from PSH, a copy of the hoxA activator gene, governed by the lac promoter, was inserted into pGE377, resulting in the expression vector pGE378 (Fig. 1). Ongoing research in our laboratory showed that the vector system is also suitable for a general application (3, 8, 10). A moderate expression of the cloned genes in slowly growing cells obviously prevents the occurrence of toxic effects and the formation of inclusion bodies.
To estimate the effectiveness of the overexpression system, pGE378 was introduced into strain HF371. The resulting transconjugant was grown under hydrogenase-derepressing conditions, and the cells were fractionated into membrane, cytoplasmic, and periplasmic extracts. Immunological analysis showed that the level of HoxC was enhanced significantly and that the protein was located in the cytoplasm (data not shown). A 40-fold increase in hydrogenase activity (58.6 nmol H2/min/mg of protein) was obtained with HF371(pGE378) in comparison with the control strain HF371(pEDY309) (1.5 nmol of H2/min/mg of protein). Nevertheless, even the enhanced RH activity did not support autotrophic growth of the strain with H2 as the sole energy source, again indicating that the RH is not coupled to an energy-generating electron transport process. This conclusion is consistent with the observation that the O2 uptake rates of the strains HF500 (MBH− SH− RH−, HF371 (MBH− SH− RH+), and HF371(pGE378) (MBH− SH− RH++) remained constant at a basal level (30 nmol of O2/min/mg of protein) upon addition of H2, whereas the O2 uptake rate of the MBH- and SH-harboring wild-type cells increased significantly under these conditions from 30 to 120 nmol of O2/min/mg of protein. The results do not unambiguously show that the RH has no potential for providing energy for growth, since the experiment did not exclude the possibility that the RH is linked to an unknown electron transport component that was not overexpressed by the vector system used.
The function of the RH is nickel dependent.
The structural similarity with [NiFe] hydrogenases and the ability to react with H2 raised the question as to whether the enzymatic and the regulatory functions of the RH depend on nickel. The RH-overproducing strain HF371(pGE378) was grown under hydrogenase-derepressing conditions in the presence of various concentrations of NiCl2. Soluble extracts were prepared, and the proteins were separated by native PAGE and subjected to a hydrogenase-specific activity staining assay with phenazine methosulfate as the electron acceptor. The RH activity strictly correlated with the addition of nickel to the growth medium (Fig. 3A). In the presence of 1 μM NiCl2 the dye reaction was very intense and did not occur with cells cultivated under nickel starvation. The quantitative RH data obtained from extracts with methylene blue as the electron acceptor (Fig. 3A) supported the conclusions drawn from the staining assay. The occurrence of double bands in native PAGE gels indicates that the RH displays different conformations. We observed that the ratio of the two bands varied with respect to the preparation. From gel filtration experiments we obtained evidence that the slowly migrating band correlates with a tetramer consisting of two RH moieties and that the rapidly migrating band correlates with the sole RH dimer (M. Bernhard and B. Friedrich, unpublished results).
FIG. 3.
(A) In-gel RH activity staining. Soluble extracts prepared from cells grown in FGN medium in the presence of various NiCl2 concentrations were separated on 4 to 15% native PAGE gels (40 μg of protein in each lane). The gel was soaked for 3 h in H2-saturated 50 mM potassium phosphate buffer (pH 7.0) containing 0.09 mM phenazine methosulfate and 0.06 mM nitroblue tetrazolium under a hydrogen atmosphere. Methylene blue reducing activity (lower part of the figure) was measured photometrically in soluble extracts at pH 7.0 using methylene blue as an electron acceptor. Lane 1, HF371(pEDY309); lanes 2 to 4, HF371(pGE378). (B) 63Ni incorporation into the RH in vivo. Cells were grown in FGN medium in the presence of 120 nM 63NiCl2. Soluble extracts were separated in a 4 to 15% native PAGE gel (200 μg of protein on each lane). Lane 1, H16(pEDY309); lane 2, H16(pGE378); lane 3, HF371(pEDY309); lane 4, HF371(pGE378). RH++, RH overproduction.
Preliminary Fourier transform infrared (FTIR) spectroscopy analyses have indicated that the active site of the RH is similar to that of standard [NiFe] hydrogenases (35). To determine whether the RH is a nickel-containing protein, we tested the accumulation of 63Ni by autoradiography (5, 11). In fact, the soluble extract of the overproducing strain HF371(pGE378) developed a strong 63Ni signal, and a faint signal occurred below the strong one (Fig. 3B, lane 4). The migration behavior and, to a lesser extent, the relative intensity of the two signals correlated with the two bands obtained in the activity staining assay (Fig. 3A, lane 3). Only traces of a 63Ni signal were recognized with cells which produced the RH at a normal level (Fig. 3B, lane 3). Interestingly, the RH-specific signal decreased in extracts of cells containing intact MBH and SH proteins, indicating competition of the three hydrogenases for 63Ni or for the pleiotropically acting Hyp proteins which are required for metallocenter assembly (11, 29).
Experiments to test whether the H2-sensing function is also nickel dependent are not trivial since only a low intracellular level of the RH is probably instrumental in the regulatory process. Therefore, traces of nickel might be sufficient for the regulatory function of the RH. In order to demonstrate a correlation between nickel supply and the level of hydrogenase induction, we had to expose the cells to conditions of severe nickel limitation. This was achieved by using R. eutropha HF459, a derivative of strain HF433, which carries a deletion in the high-affinity nickel permease gene hoxN, and by addition of the chelating agent nitrilotriacetic acid. Since the MBH and SH promoters are regulated coordinately (42), β-galactosidase activity was monitored representatively with a plasmid-based Φ(hoxK′-′lacZ) fusion. The result is illustrated in Fig. 4 and shows a clear correlation between β-galactosidase activity and supplementation of NiCl2 to the medium. As expected, the expression level was low when H2 was omitted, even with nickel excess. The conclusion that nickel is essential for H2 recognition by the RH protein is confirmed by the behavior of strain HF375, in which the H2-dependent signal transduction is interrupted due to a lesion in the sensor kinase (Table 1). This strain displayed high-level hydrogenase gene expression independently of H2 and hence independently of nickel (Fig. 4).
FIG. 4.
Hydrogenase gene expression in the presence of various NiCl2 concentrations. The R. eutropha strains HF459 (HoxN−; black bars) and HF375 (HoxN− HoxJ−; white bars), each containing the plasmid-based Φ(hoxK′-′lacZ) fusion pGE301, were grown in FGN medium containing 10 μM nitrilotriacetic acid at the nonpermissive temperature of 37°C until they reached an OD436 of 7. At time zero the cells were shifted to 30°C and 10% (vol/vol) H2 (+H2) or 10% (vol/vol) N2 (−H2) was added. After further incubation for 5 h the cells were collected, and the β-galactosidase activity was determined.
It is obvious from these results that the regulatory function of the RH relies on the availability of nickel in the medium. For HupUV of B. japonicum it was proposed that the protein acts as a sensor of nickel and/or of oxygen and hydrogen (6). The content of hydrogenase-specific mRNA in B. japonicum correlated with the addition of nickel to the medium showing that nickel acts as an effector of transcriptional regulation (22). Our experiments revealed that only trace amounts of the metal were required for the H2-sensing function of the RH in R. eutropha, suggesting that nickel is a tightly bound component of the active site rather than an effector of regulation.
The results of this study, together with previous findings regarding the electron paramagnetic resonance and FTIR properties of the RH (35) and the H2-binding capacity of the HupUV protein from R. capsulatus (49), indicate that H2 recognition is based on a nickel-iron-containing active site similar to that of standard [NiFe] hydrogenases, including two CN− groups and one CO molecule at the iron site (35). The capability of the sensors to oxidize H2 does not yet allow the conclusion that H2 binding is intimately connected with a redox reaction. Thus, elucidation of the underlying mechanism of H2 signal transduction is a fascinating subject of current research in our laboratory.
ACKNOWLEDGMENTS
O. Lenz and L. Kleihues contributed equally to this work.
We are grateful to Edward Schwartz for providing plasmid pEDY305 and for critical reading of the manuscript.
This work was supported by the Deutsche Forschungsgemeinschaft and by the Fonds der Chemischen Industrie.
REFERENCES
- 1.Adams M W. Biochemical diversity among sulfur-dependent, hyperthermophilic microorganisms. FEMS Microbiol Rev. 1994;15:261–277. doi: 10.1111/j.1574-6976.1994.tb00139.x. [DOI] [PubMed] [Google Scholar]
- 2.Albracht S P. Nickel hydrogenases: in search of the active site. Biochim Biophys Acta. 1994;1188:167–204. doi: 10.1016/0005-2728(94)90036-1. [DOI] [PubMed] [Google Scholar]
- 3.Bernhard M, Friedrich B, Siddiqui R A. Ralstonia eutropha TF93 is blocked in tat-mediated protein export. J Bacteriol. 2000;182:581–588. doi: 10.1128/jb.182.3.581-588.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bernhard M, Benelli B, Hochkoeppler A, Zannoni D, Friedrich B. Functional and structural role of the cytochrome b subunit of the membrane-bound hydrogenase complex of Alcaligenes eutrophus H16. Eur J Biochem. 1997;248:179–186. doi: 10.1111/j.1432-1033.1997.00179.x. [DOI] [PubMed] [Google Scholar]
- 5.Bernhard M, Schwartz E, Rietdorf J, Friedrich B. The Alcaligenes eutrophus membrane-bound hydrogenase gene locus encodes functions involved in maturation and electron transport coupling. J Bacteriol. 1996;178:4522–4529. doi: 10.1128/jb.178.15.4522-4529.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Black L K, Fu C, Maier R J. Sequence and characterization of hupU and hupV genes of Bradyrhizobium japonicum encoding a possible nickel-sensing complex involved in hydrogenase expression. J Bacteriol. 1994;176:7102–7106. doi: 10.1128/jb.176.22.7102-7106.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brim H, Heyndrickx M, de Vos P, Wilmotte A, Springael D, Schlegel H G, Mergeay M. Amplified rDNA restriction analysis and further genotypic characterisation of metal-resistant soil bacteria and related facultative hydrogenotrophs. Syst Appl Microbiol. 1999;22:258–268. doi: 10.1016/S0723-2020(99)80073-3. [DOI] [PubMed] [Google Scholar]
- 8.Cramm R, Pohlmann A, Friedrich B. Purification and characterization of the single-component nitric oxide reductase from Ralstonia eutropha H16. FEBS Lett. 1999;460:6–10. doi: 10.1016/s0014-5793(99)01315-0. [DOI] [PubMed] [Google Scholar]
- 9.Deckert G, Warren P V, Gaasterland T, Young W G, Lenox A L, Graham D E, Overbeek R, Snead M A, Keller M, Aujay M, Huber R, Feldman R A, Short J M, Olsen G J, Swanson R V. The complete genome of the hyperthermophilic bacterium Aquifex aeolicus. Nature. 1998;392:353–358. doi: 10.1038/32831. [DOI] [PubMed] [Google Scholar]
- 10.Degen O, Kobayashi M, Shimizu S, Eitinger T. Selective transport of divalent cations by transition metal permeases: the Alcaligenes eutrophus HoxN and the Rhodococcus rhodochrous NhlF. Arch Microbiol. 1999;171:139–145. doi: 10.1007/s002030050691. [DOI] [PubMed] [Google Scholar]
- 11.Dernedde J, Eitinger T, Patenge N, Friedrich B. hyp gene products in Alcaligenes eutrophus are part of a hydrogenase maturation system. Eur J Biochem. 1996;235:351–358. doi: 10.1111/j.1432-1033.1996.00351.x. [DOI] [PubMed] [Google Scholar]
- 12.Eberz G, Friedrich B. Three trans-acting regulatory functions control hydrogenase synthesis in Alcaligenes eutrophus. J Bacteriol. 1991;173:1845–1854. doi: 10.1128/jb.173.6.1845-1854.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eberz G, Hogrefe C, Kortlüke C, Kamienski A, Friedrich B. Molecular cloning of structural and regulatory hydrogenase genes (hox) of Alcaligenes eutrophus H16. J Bacteriol. 1986;168:636–641. doi: 10.1128/jb.168.2.636-641.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eitinger T, Friedrich B. Cloning, nucleotide sequence, and heterologous expression of a high-affinity nickel transport gene from Alcaligenes eutrophus. J Biol Chem. 1991;266:3222–3227. [PubMed] [Google Scholar]
- 15.Elsen S, Colbeau A, Chabert J, Vignais P M. The hupTUV operon is involved in negative control of hydrogenase synthesis in Rhodobacter capsulatus. J Bacteriol. 1996;178:5174–5181. doi: 10.1128/jb.178.17.5174-5181.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Elsen S, Richaud P, Colbeau A, Vignais P M. Sequence analysis and interposon mutagenesis of the hupT gene, which encodes a sensor protein involved in repression of hydrogenase synthesis in Rhodobacter capsulatus. J Bacteriol. 1993;175:7404–7412. doi: 10.1128/jb.175.22.7404-7412.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fox J D, He Y, Shelver D, Roberts G P, Ludden P W. Characterization of the region encoding the CO-induced hydrogenase of Rhodospirillum rubrum. J Bacteriol. 1996;178:6200–6208. doi: 10.1128/jb.178.21.6200-6208.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Friedrich B, Schwartz E. Molecular biology of hydrogen utilization in aerobic chemolithotrophs. Annu Rev Microbiol. 1993;47:351–383. doi: 10.1146/annurev.mi.47.100193.002031. [DOI] [PubMed] [Google Scholar]
- 19.Friedrich C G, Bowien B, Friedrich B. Formate and oxalate metabolism in Alcaligenes eutrophus. J Gen Microbiol. 1979;115:185–192. [Google Scholar]
- 20.Gomelsky M, Kaplan S. Isolation of regulatory mutants in photosynthesis gene expression in Rhodobacter sphaeroides 2.4.1 and partial complementation of a PrrB mutant by the HupT histidine-kinase. Microbiology. 1995;141:1805–1814. doi: 10.1099/13500872-141-8-1805. [DOI] [PubMed] [Google Scholar]
- 21.Gross R, Simon J, Theis F, Kröger A. Two membrane anchors of Wolinella succinogenes hydrogenase and their function in fumarate and polysulfide respiration. Arch Microbiol. 1998;170:50–58. doi: 10.1007/s002030050614. [DOI] [PubMed] [Google Scholar]
- 22.Kim H, Maier R J. Transcriptional regulation of hydrogenase synthesis by nickel in Bradyrhizobium japonicum. J Biol Chem. 1990;265:18729–18732. [PubMed] [Google Scholar]
- 23.Kömen R, Schmidt K, Friedrich B. Hydrogenase mutants of Alcaligenes eutrophus H16 show alterations in the electron transport system. FEMS Microbiol Lett. 1992;75:173–178. doi: 10.1016/0378-1097(92)90399-9. [DOI] [PubMed] [Google Scholar]
- 24.Kortlüke C, Horstmann K, Schwartz E, Rohde M, Binsack R, Friedrich B. A gene complex coding for the membrane-bound hydrogenase of Alcaligenes eutrophus H16. J Bacteriol. 1992;174:6277–6289. doi: 10.1128/jb.174.19.6277-6289.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lenz O, Friedrich B. A novel multicomponent regulatory system mediates H2 sensing in Alcaligenes eutrophus. Proc Natl Acad Sci USA. 1998;95:12474–12479. doi: 10.1073/pnas.95.21.12474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lenz O, Strack A, Tran-Betcke A, Friedrich B. A hydrogen-sensing system in transcriptional regulation of hydrogenase gene expression in Alcaligenes species. J Bacteriol. 1997;179:1655–1663. doi: 10.1128/jb.179.5.1655-1663.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lenz O, Schwartz E, Dernedde J, Eitinger M, Friedrich B. The Alcaligenes eutrophus H16 hoxX gene participates in hydrogenase regulation. J Bacteriol. 1994;176:4385–4393. doi: 10.1128/jb.176.14.4385-4393.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lowry O H, Rosebrough N J, Farr A L, Randall R J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 29.Maier T, Böck A. Nickel incorporation into hydrogenases. In: Hausinger R P, Eichhorn G L, Marzilli L G, editors. Advances in inorganic biochemistry: mechanisms of metallocenter assembly. New York, N.Y: VHC Publishers, Inc.; 1996. pp. 173–192. [Google Scholar]
- 30.Massanz C, Schmidt S, Friedrich B. Subforms and in vitro reconstitution of the NAD-reducing hydrogenase of Alcaligenes eutrophus. J Bacteriol. 1998;180:1023–1029. doi: 10.1128/jb.180.5.1023-1029.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Massanz C, Fernandez V M, Friedrich B. C-terminal extension of the H2-activating subunit, HoxH, directs maturation of the NAD-reducing hydrogenase in Alcaligenes eutrophus. Eur J Biochem. 1997;245:441–448. doi: 10.1111/j.1432-1033.1997.t01-3-00441.x. [DOI] [PubMed] [Google Scholar]
- 32.Meuer J, Bartoschek S, Koch J, Künkel A, Hedderich R. Purification and catalytic properties of Ech hydrogenase from Methanosarcina barkeri. J Biochem. 1999;265:325–335. doi: 10.1046/j.1432-1327.1999.00738.x. [DOI] [PubMed] [Google Scholar]
- 33.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1972. [Google Scholar]
- 34.Nivière V, Wong S-L, Voordouw G. Site-directed mutagenesis of the hydrogenase signal peptide consensus box prevents export of a β-lactamase fusion protein. J Gen Microbiol. 1992;138:2173–2183. doi: 10.1099/00221287-138-10-2173. [DOI] [PubMed] [Google Scholar]
- 35.Pierik A J, Schmelz M, Lenz O, Friedrich B, Albracht S P J. Characterization of the active site of a hydrogen sensor from Alcaligenes eutrophus. FEBS Lett. 1998;438:231–235. doi: 10.1016/s0014-5793(98)01306-4. [DOI] [PubMed] [Google Scholar]
- 36.Probst I, Schlegel H G. Respiratory components and oxidase activities in Alcaligenes eutrophus. Biochim Biophys Acta. 1976;440:412–428. doi: 10.1016/0005-2728(76)90075-x. [DOI] [PubMed] [Google Scholar]
- 37.Santiago B, Meyer O. Purification and molecular characterization of the H2 uptake membrane-bound NiFe-hydrogenase from the carboxidotrophic bacterium Oligotropha carboxidovorans. J Bacteriol. 1997;179:6053–6060. doi: 10.1128/jb.179.19.6053-6060.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schink B, Schlegel H G. The membrane-bound hydrogenase of Alcaligenes eutrophus. I. Solubilization, purification and biochemical properties. Biochem Biophys Acta. 1979;567:315–324. doi: 10.1016/0005-2744(79)90117-7. [DOI] [PubMed] [Google Scholar]
- 39.Schneider K, Schlegel H G, Cammack R, Hall D O. The iron sulfur centers of soluble hydrogenase from Alcaligenes eutrophus. Biochim Biophys Acta. 1979;578:445–461. doi: 10.1016/0005-2795(79)90175-2. [DOI] [PubMed] [Google Scholar]
- 40.Schneider K, Schlegel H G. Purification and properties of the soluble hydrogenase from Alcaligenes eutrophus H16. Biochim Biophys Acta. 1976;452:66–80. doi: 10.1016/0005-2744(76)90058-9. [DOI] [PubMed] [Google Scholar]
- 41.Schwartz E, Buhrke T, Gerischer U, Friedrich B. Positive transcriptional feedback controls hydrogenase expression in Alcaligenes eutrophus H16. J Bacteriol. 1999;181:5684–5692. doi: 10.1128/jb.181.18.5684-5692.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schwartz E, Gerischer U, Friedrich B. Transcriptional regulation of Alcaligenes eutrophus hydrogenase genes. J Bacteriol. 1998;180:3197–3204. doi: 10.1128/jb.180.12.3197-3204.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Simon R, Priefer U, Pühler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram-negative bacteria. Bio/Technology. 1983;1:717–743. [Google Scholar]
- 44.Taylor B L, Zhulin I B. PAS domains: internal sensors of oxygen, redox potential, and light. Microbiol Mol Biol Rev. 1999;63:479–506. doi: 10.1128/mmbr.63.2.479-506.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thiemermann S, Dernedde J, Bernhard M, Schröder W, Massanz C, Friedrich B. Carboxy-terminal processing of the soluble, NAD-reducing hydrogenase of Alcaligenes eutrophus requires the hoxW gene product. J Bacteriol. 1996;178:2368–2374. doi: 10.1128/jb.178.8.2368-2374.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of protein from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4357. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tran-Betcke A, Warnecke U, Böcker C, Zaborosch C, Friedrich B. Cloning and nucleotide sequences of the genes for the subunits of NAD-reducing hydrogenase of Alcaligenes eutrophus H16. J Bacteriol. 1990;172:2920–2929. doi: 10.1128/jb.172.6.2920-2929.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Van Soom C, Lerouge I, Vanderleyden J, Ruiz-Argueso T, Palacios J M. Identification and characterization of hupT, a gene involved in negative regulation of hydrogen oxidation in Bradyrhizobium japonicum. J Bacteriol. 1999;181:5085–5089. doi: 10.1128/jb.181.16.5085-5089.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vignais P M, Dimon B, Zorin N A, Colbeau A, Elsen S. HupUV proteins of Rhodobacter capsulatus can bind H2: evidence from the H-D reaction. J Bacteriol. 1997;179:290–292. doi: 10.1128/jb.179.1.290-292.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Volbeda A, Charon M-H, Piras C, Hatchikan E C, Frey M, Fontecilla-Camps J C. Crystal structure of the nickel-iron hydrogenase from Desulfovibrio gigas. Nature. 1995;373:580–587. doi: 10.1038/373580a0. [DOI] [PubMed] [Google Scholar]
- 51.Voordouw G, Menon N K, Le Gall J, Choi E S, Peck H D, Przybyla A E. Analysis and comparison of nucleotide sequences encoding the genes for [NiFe] and [NiFeSe] hydrogenases from Desulfovibrio gigas and Desulfovibrio baculatus. J Bacteriol. 1989;171:2894–2899. doi: 10.1128/jb.171.5.2894-2899.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wolf I, Buhrke T, Dernedde J, Pohlmann A, Friedrich B. Duplication of hyp genes involved in maturation of [NiFe] hydrogenases in Alcaligenes eutrophus H16. Arch Microbiol. 1998;170:451–459. doi: 10.1007/s002030050666. [DOI] [PubMed] [Google Scholar]
- 53.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 54.Zimmer D, Schwartz E, Tran-Betcke A, Gewinner P, Friedrich B. Temperature tolerance of hydrogenase expression in Alcaligenes eutrophus is conferred by a single amino acid exchange in the transcriptional activator HoxA. J Bacteriol. 1995;177:2373–2380. doi: 10.1128/jb.177.9.2373-2380.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]