Abstract
The sequences of the 16S rRNA and haloalkane dehalogenase (dhaA) genes of five gram-positive haloalkane-utilizing bacteria isolated from contaminated sites in Europe, Japan, and the United States and of the archetypal haloalkane-degrading bacterium Rhodococcus sp. strain NCIMB13064 were compared. The 16S rRNA gene sequences showed less than 1% sequence divergence, and all haloalkane degraders clearly belonged to the genus Rhodococcus. All strains shared a completely conserved dhaA gene, suggesting that the dhaA genes were recently derived from a common ancestor. The genetic organization of the dhaA gene region in each of the haloalkane degraders was examined by hybridization analysis and DNA sequencing. Three different groups could be defined on the basis of the extent of the conserved dhaA segment. The minimal structure present in all strains consisted of a conserved region of 12.5 kb, which included the haloalkane-degradative gene cluster that was previously found in strain NCIMB13064. Plasmids of different sizes were found in all strains. Southern hybridization analysis with a dhaA gene probe suggested that all haloalkane degraders carry the dhaA gene region both on the chromosome and on a plasmid (70 to 100 kb). This suggests that an ancestral plasmid was transferred between these Rhodococcus strains and subsequently has undergone insertions or deletions. In addition, transposition events and/or plasmid integration may be responsible for positioning the dhaA gene region on the chromosome. The data suggest that the haloalkane dehalogenase gene regions of these gram-positive haloalkane-utilizing bacteria are composed of a single catabolic gene cluster that was recently distributed worldwide.
Hydrolytic dehalogenation by haloalkane dehalogenases is the most important step in the biodegradation of 1-halo-n-alkanes and α,ω-dihalo-n-alkanes. Haloalkane dehalogenases active against long-chain haloaliphatic compounds are present in the gram-positive haloalkane-utilizing bacterial strains Y2, isolated in the United Kingdom (16), m15-3, isolated in Japan (25), HA1, isolated in Switzerland (20), GJ70, isolated in The Netherlands (7), and NCIMB13064, isolated in the United Kingdom (2). These enzymes are different from the extensively studied haloalkane dehalogenase (DhlA) found in several gram-negative 1,2-dichloroethane-utilizing bacteria of the genera Xanthobacter and Ancylobacter (8, 23). DhlA is able to dehalogenate 1,2-dichloroethane, which is a poor substrate for other haloalkane dehalogenases, whereas the enzymes isolated from the gram-positive strains exhibit higher activity toward long-chain mono- and dihalogenated substrates.
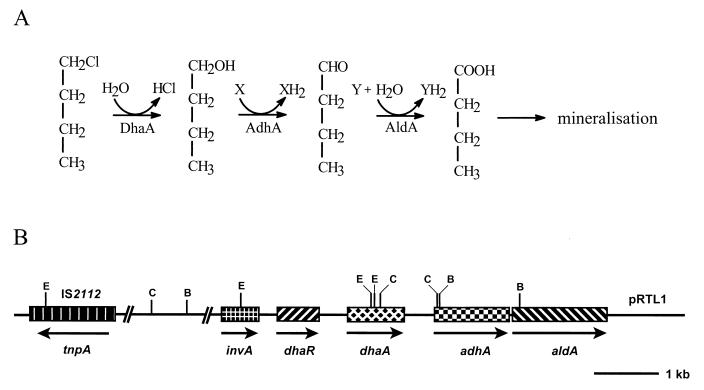
Rhodococcus sp. strain NCIMB13064 has been used to study the genetics of haloalkane metabolism in gram-positive bacteria. It is host to the self-transmissible 100-kb plasmid pRTL1 (11), which carries the genes (dhaA, adhA, and aldA) for the initial steps in the aerobic degradation of 1-chlorobutane and several other 1-halo-n-alkanes (Fig. 1). The dhaA gene codes for a haloalkane dehalogenase that hydrolytically converts 1-chlorobutane to n-butanol (12). Based on amino acid sequence similarities, the adhA and aldA genes, which are located downstream of dhaA (Fig. 1B), were proposed to encode the alcohol and aldehyde dehydrogenases that catalyze the oxidative conversion of n-butanol to n-butyric acid (15a). The latter compound is further metabolized by β-oxidation to acetate, which can serve as a growth substrate for many bacteria (2). The 13-kb DNA fragment of pRTL1 that has been sequenced harbors two additional genes, dhaR and invA, and the insertion sequence IS2112 (Fig. 1B). The dhaR gene product putatively acts as a repressor-type regulator for dhaA expression (15a), and IS2112 was shown to be involved in several genome rearrangements that resulted in the loss of haloalkane dehalogenase activity in strain NCIMB13064 (10). The invA gene encodes a putative protein that shares extensive similarity with the DNA invertase Pin of Escherichia coli (50%) and Hin of Salmonella enterica serovar Typhimurium (48%) (15a). The Pin and Hin invertases are responsible for the inversion of a specific DNA fragment that can serve as a genetic switch between the expression of alternative sets of genes (4). However, it is not known whether the putative invertase gene invA plays an analogous role in regulating haloalkane metabolism in Rhodococcus sp. strain NCIMB13064.
FIG. 1.
(A) Initial enzymatic steps in the degradation of 1-chlorobutane by Rhodococcus sp. strain NCIMB13064. Below each conversion step, the responsible enzyme is indicated (DhaA, haloalkane dehalogenase; AdhA, alcohol dehydrogenase; AldA, aldehyde dehydrogenase). (B) Genetic organization of the haloalkane-degradative genes dhaA, adhA, and aldA, the regulatory gene dhaR, the putative invertase gene invA, and insertion element IS2112 on plasmid pRTL1 in Rhodococcus sp. strain NCIMB13064 (as determined by DNA sequencing [15a]; accession no. L49435). Arrows indicate the direction of transcription. Solid line, noncoding pRTL1 DNA. The positions of BamHI, ClaI, and EcoRI restriction sites are important for explaining the hybridization results obtained in this study and are indicated by B, C, and E, respectively.
Although the haloalkane dehalogenases of strains Y2, m15-3, HA1, and GJ70 have been characterized (7, 16, 20, 25), nothing is known about the genetics of haloalkane degradation in these strains. The biochemical characteristics of the haloalkane dehalogenases isolated from these strains, however, closely resemble those of the haloalkane dehalogenase (DhaA) from Rhodococcus sp. strain NCIMB13064. Furthermore, the amino-terminal sequences of the dehalogenases from strains Y2 and HA1 are identical to that of DhaA. This apparent homology raises interesting questions about the evolution and distribution of these enzymes since the organisms were isolated from geographically separate locations. To determine the degree of homology between the haloalkane dehalogenases and to obtain insight in their evolution and distribution, we analyzed the genetic organization and genomic location of the haloalkane dehalogenase gene region in each of the strains Y2, m15-3, HA1, and GJ70 and in another gram-positive haloalkane-degrading bacterium, strain TB2, newly isolated from an industrial site in the United States. In addition, we determined the phylogenetic affiliation of the different haloalkane degraders by 16S rRNA gene sequence analysis. The results indicate that the analyzed gram-positive haloalkane degraders are members of the genus Rhodococcus, and despite the fact that they were isolated independently from different continents, they all possess a haloalkane catabolic gene cluster highly similar to the one found on plasmid pRTL1 in Rhodococcus sp. strain NCIMB13064.
MATERIALS AND METHODS
Haloalkane-utilizing bacteria used in this study.
The bacterial strains screened for the presence of the dhaA gene are listed in Table 1. Strains Y2, m15-3, HA1, and GJ70 were isolated from widely separated sites by different research groups (6, 16, 19, 24). Strains NCIMB13064 and 170 (formerly known as Pseudomonas cichorii 170) have been investigated previously and were shown to possess identical dhaA genes (12, 14). Strain TB2 was isolated in our laboratory from a soil sample obtained from a trichloropropane-polluted site in the United States and is capable of growth with 1-chlorobutane as the sole carbon and energy source.
TABLE 1.
List of haloalkane-utilizing strains carrying the dhaA gene
| Straina | Originb | Reference(s) or source |
|---|---|---|
| Y2 | UK; industrial site exposed to haloalkanes | 16 |
| NCIMB13064 | UK; industrial site exposed to chlorinated alkanes | 2, 12 |
| m15-3 | Japan | 24, 25 |
| HA1 | Switzerland | 19, 20 |
| GJ70 | The Netherlands; sludge contaminated with pesticides | 6, 7 |
| TB2 | USA; industrial site exposed to 1,2,3-trichloropropane | This study |
| P. pavonaceae 170 | The Netherlands; soil exposed to 1,3-dichloropropene | 14 |
On the basis of the 16S rRNA gene sequence analysis performed in this study, strains Y2, NCIMB13064, m15-3, HA1, GJ70, and TB2 should be classified as strains of Rhodococcus erythropolis.
UK, United Kingdom; USA, United States.
PCR amplification, cloning, and sequencing of putative dhaA genes.
Total genomic DNA was isolated from nutrient broth-grown cells by a phenol extraction procedure described elsewhere (14) and was directly used as the template for PCR amplification. Putative dhaA genes were amplified with the same primers that were successfully used to amplify the dhaA gene from Pseudomonas pavonaceae 170 (14): 5′-AAAATCGCCATGGCAGAAATCGGTA-3′ and 5′-TGGACATCGGACCATGGCGTGAACC-3′ (NcoI sites are underlined). The amplification reaction mixture (100 μl) contained standard Taq amplification buffer, 250 μM (each) deoxyribonucleotide triphosphate, 100 ng of each primer, 100 ng of genomic DNA, and 2 U of Taq DNA polymerase (Boehringer GmbH, Mannheim, Germany). The cycling parameters were 94°C for 5 min followed by 30 cycles of 94°C for 60 s, 58°C for 60 s, and 72°C for 90 s, with a final elongation step of 72°C for 10 min. Samples (10 μl) of the reaction mixtures were subjected to electrophoresis in 0.8% agarose gels, and PCR products were stained with ethidium bromide.
The PCR-amplified dhaA genes of strains GJ70 and Y2 were directly cloned in the TA cloning vector pCR2.1 according to the recommendations of the supplier (Invitrogen, Leek, The Netherlands). PCR products obtained for strains TB2, m15-3, and HA1 were cloned in the NcoI site behind the T7 promoter in the expression vector pGEF+ as described before (14). Both DNA strands of the cloned dehalogenase genes were sequenced by the dideoxy chain termination method (18), and nucleotide sequences were aligned by using LALIGN (Institut de Génétique Humaine, Montpellier, France).
16S rRNA gene sequence analysis.
The 16S rRNA genes were amplified with the oligonucleotide primers that were described by Marchesi et al. (13): 63f (5′-CAGGCCTAACACATGCAAGTC-3′) and 1387r (5′-GGGCGG(A/T)GTGTACAAGGC-3′) (numbering based on the E. coli 16S rRNA gene [1]). The amplification reaction mixture contained 50 mM KCl, 10 mM Tris-HCl (pH 8.3), 2.5 mM MgCl2, 0.01% bovine serum albumin, 0.01% gelatin, 0.05% Triton X-100, 200 μM (each) deoxyribonucleotide triphosphate, 0.5 μM (each) primer, 100 ng of genomic DNA, and 1 to 2.5 U of Taq DNA polymerase. The cycling parameters were 94°C for 2 min followed by 30 cycles of 92°C for 30 s, 55°C for 30 s, and 75°C for 45 s, with a final elongation step of 75°C for 5 min.
Amplification products were cloned into plasmid pTAg as specified by the manufacturer (R&D Systems, Abingdon, United Kingdom). Cloned 16S rRNA genes were cycle sequenced using the Amersham Thermo Sequenase cycle sequencing kit with 7-deaza-dGTP and IRD-41-labeled primers that annealed to the M13 universal and reverse sites of the pTAg vector. Sequencing reaction mixtures were run on an automatic sequencing machine (Li-Cor 4000L; MWG-Biotech). Both DNA strands were sequenced to ensure accuracy. Phylogenetic analysis of the newly determined 16S rRNA gene sequences was done essentially as described before (15).
Plasmid visualization.
Rhodococcus strains were grown in 10 ml of yeast extract-peptone medium (11) at 28°C to an optical density at 600 nm of 0.9 to 1.0. Cultures were diluted (1:1) with 0.9% NaCl, after which cells from 2 ml of culture were collected in 1.5-ml tubes. Cell pellets were resuspended in 100 μl of 20% Ficoll (approximate molecular weight, 400,000) in 1× Tris-acetate (TAE) buffer (17). Resuspended cells were mixed with 5 μl of fresh lysozyme solution (18% Ficoll, 0.18% bromophenol blue, lysozyme [2.4 mg/ml], and RNase [0.24 mg/ml] in TAE) and incubated for 10 min at room temperature. Then, 15 μl of 2% sodium dodecyl sulfate (SDS)–10% Ficoll in TAE was added, and preparations were incubated at 60°C for 20 to 30 min. The mixtures were subsequently transferred into the wells of a 0.9% vertical agarose slab, from which the TAE buffer was carefully removed. Mixtures were overlaid with 50 μl of 2% SDS–5% Ficoll in TAE, after which the wells were sealed with melted agarose. Electrophoresis was performed in TAE buffer for 40 min at 1 V/cm and then at 5 to 6 V/cm until completion. Plasmid DNA was stained with ethidium bromide.
Southern hybridizations.
DNA fragments or intact plasmids were separated on an agarose gel and subsequently transferred to a Hybond-N+ membrane (Amersham Pharmacia Biotech) or to a positively charged nylon membrane (Boehringer GmbH) by diffusion blotting (17). The membranes were treated according to the instructions of the manufacturer. Parts of the dhaA gene or of IS2112 were amplified by PCR and were purified using the Qiaquick PCR purification kit (Qiagen) or by using the GFX PCR DNA and gel band purification kit (Amersham). These PCR fragments were labeled with digoxigenin-11-dUTP using the nonradioactive DNA labeling and detection kit from Boehringer GmbH or with fluorescein-11-dUTP by using the gene images random prime labeling module of Amersham and were used as hybridization probes. Hybridization was carried out at 62 to 68°C, and detection of the hybridization signals was performed according to the manufacturer's protocol.
The complete dhaA gene was amplified by using the primers and conditions described above. An internal part of the dhaA gene (550 bp) was obtained by using the following oligonucleotide primers: f (5′-GACGACCACGTCCGCTACC-3′) and r (5′-CCGATGTCCACTGTCTTGC-3′). The IS2112 hybridization probe (nucleotide positions 17 to 1108 of the tnpA open reading frame [ORF]) was generated with primers f (5′-CCCACGCATGGGTCG-3′) and r (5′-CGACGCGCCAAGGCG-3′). Conditions used for the amplification of IS2112 have been described previously (10).
Cloning and sequencing of the dhaA gene regions of strains GJ70 and HA1.
To obtain the dhaA gene regions of strains GJ70 and HA1, genomic libraries of these strains were constructed using the cosmid vector pLAFR3 (21). Total DNA was partially digested with Sau3A, and fragments of 15 to 25 kb in size were isolated and ligated into the BamHI site of pLAFR3. Ligation mixtures were packaged in vitro by using the DNA packaging kit of Boehringer GmbH. E. coli HB101 was transduced with these packaging mixtures (17), and colonies were selected on LB agar plates without NaCl (LBZ) (15) containing tetracycline (12.5 μg/ml). Tetracycline-resistant colonies were screened for dehalogenase activity by monitoring halide production upon incubation with 1,2-dibromoethane as described before (15).
Recombinant cosmids encoding haloalkane dehalogenase activity were isolated and directly used as the template for sequencing DNA regions upstream of the invA gene. Oligonucleotide primers were designed on the basis of the known sequence of the dhaA gene region of Rhodococcus sp. strain NCIMB13064 (accession no. L49435). DNA sequencing was performed as described elsewhere (15a), and newly determined sequences for strains GJ70 and HA1 were aligned with the known sequence of the dhaA gene region of strain NCIMB13064 by using the program LALIGN.
Nucleotide sequence accession numbers.
The 16S rRNA gene sequences of strains TB2, m15-3, HA1, Y2, NCIMB13064, and GJ70 have been submitted to the DDBJ/EMBL/GenBank databases under accession no. AJ250924, AJ250925, AJ250926, AJ250927, AJ250928, and AJ250929, respectively. The nucleotide sequence data of the regions upstream of the invA gene in strains GJ70 and HA1 have been submitted under accession no. AJ250982 and AJ250983, respectively.
RESULTS AND DISCUSSION
16S rRNA gene sequence analysis.
The haloalkane-utilizing bacterial strains Y2, NCIMB13064, m15-3, HA1, GJ70, and TB2 were isolated by different research groups from geographically distinct locations (Table 1). The first four strains were previously identified as Rhodococcus erythropolis Y2 (16), Rhodococcus rhodochrous NCIMB13064 (2), Corynebacterium sp. strain m15-3 (25), and Arthrobacter sp. strain HA1 (19). Strain GJ70 was previously identified as a gram-positive actinomycete-like organism (7). In order to establish precisely the phylogenetic affiliation of these haloalkane-utilizing bacteria, we determined their 16S rRNA gene sequences.
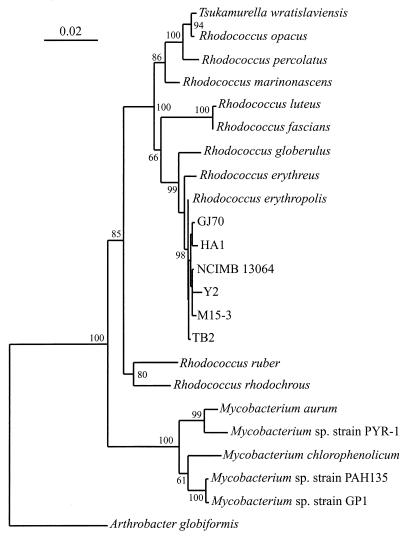
A comparison of the newly determined 16S rRNA gene sequences revealed less than 1% sequence divergence and clearly places the six haloalkane degraders within the same genus. Phylogenetic analysis was conducted by comparing the 16S rRNA gene sequences for these strains with other 16S rRNA gene sequences available in the GenBank database and in the Ribosomal Database Project. This analysis revealed that the haloalkane degraders are members of the genus Rhodococcus and are all most closely related to R. erythropolis (Fig. 2); the pairwise sequence identities were >99%. This high 16S sequence identity indicates that the six haloalkane degraders should be classified as new strains of R. erythropolis or a closely related species. The results thus show that strains m15-3 and HA1 were previously identified incorrectly (19, 25) and that, like strains GJ70 and TB2, they should be (re)assigned to the genus Rhodococcus (26).
FIG. 2.
Phylogenetic tree based on 16S rRNA gene sequence analysis, illustrating the relationships of the six haloalkane-degrading strains GJ70, HA1, NCIMB13064, Y2, m15-3, and TB2 to the most closely related bacteria. Base positions 54 to 1368 (numbering based on the E. coli 16S rRNA gene) were included in the analysis. Scale bar, 0.02 fixed mutation per site. Bootstrap values were derived from 500 analyses. A sequence from Arthrobacter globiformis was used as the outgroup.
Sequence identity among dhaA genes of haloalkane-utilizing bacteria.
Haloalkane dehalogenases active against long-chain haloaliphatic compounds were previously isolated from strains Y2 (16), m15-3 (25), HA1 (20), and GJ70 (7). On the basis of their substrate specificities and their identical N-terminal amino acid sequences, these four dehalogenases have been classified as one group of related enzymes (3). The biochemical characteristics and amino-terminal sequences of these dehalogenases suggest that they closely resemble the haloalkane dehalogenase (DhaA) that had been found in strain NCIMB13064 (12) and recently also in P. pavonaceae 170 (14). To determine the extent of sequence similarity between the haloalkane dehalogenase genes of strains Y2, m15-3, HA1, and GJ70 and the dhaA genes that are present in strains NCIMB13064 and 170, the putative dhaA genes of the first four strains were amplified with primers designed for the 5′ and 3′ ends of the dhaA gene (12). In addition, we also screened the newly isolated strain TB2 for the presence of the dhaA gene.
For the five strains analyzed here, PCR amplification with dhaA-specific primers consistently produced a 0.9-kb DNA fragment corresponding to the size of the dhaA gene (data not shown). DNA sequencing revealed that the PCR-amplified haloalkane dehalogenase genes of strains Y2, m15-3, HA1, GJ70, and TB2 were completely identical to the dhaA genes of strains NCIMB13064 and 170. Since the lineage of the dhaA gene is less diverged (100% sequence identity) than that of the hosts, as determined by 16S rRNA gene sequence analysis (see above), the dhaA gene has probably spread among these bacterial strains by lateral transfer. Thus the haloalkane dehalogenases isolated and characterized from these chloroalkane-utilizing bacteria (2, 7, 14, 16, 25) are all identical to the dehalogenase that was first described in strain m15-3 by Yokota et al. (25).
Genetic organization of the dhaA gene regions in haloalkane-utilizing Rhodococcus strains.
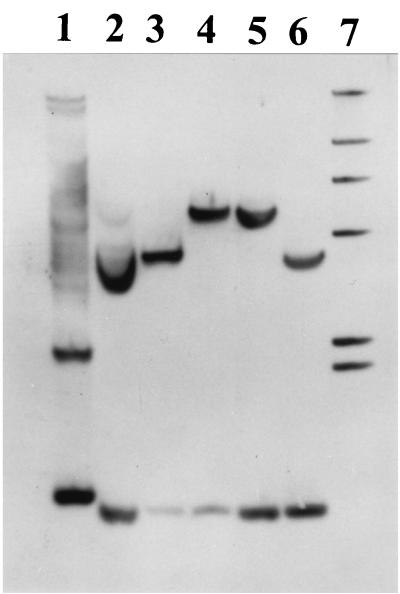
Southern blot hybridization analysis under high-stringency conditions with dhaA- and IS2112-specific probes was used to determine the extent of the conserved dhaA segments in strains m15-3, HA1, Y2, TB2, and GJ70, compared to the sequenced dhaA gene region of strain NCIMB13064 (Fig. 1B). When the complete dhaA gene was used as the probe, BamHI-digested DNA of strains HA1, m15-3, NCIMB13064, Y2, and TB2 revealed single hybridization signals at about 5.6 kb (data not shown). For BamHI-digested DNA of strain GJ70, a single hybridization signal at about 9 kb was found. We examined the regions flanking the dhaA gene by digesting the genomic DNAs with EcoRI or ClaI. Since the dhaA gene has two restriction sites for EcoRI close to each other (Fig. 1B), only two hybridizing bands were expected, and these were found. EcoRI-digested DNA gave hybridization signals at about 9 and 2 kb for all strains (data not shown). The 2-kb DNA band was identical in size to the EcoRI restriction fragment that includes the dhaR gene in strain NCIMB13064 (Fig. 1B). These results suggest that the six haloalkane degraders possess a conserved DNA segment of at least 11 kb, including a 1.5-kb region upstream, and an 8.5-kb region downstream, of the dhaA gene. Since the dhaA gene has one cutting site for ClaI (Fig. 1B), two DNA bands hybridized with the dhaA gene probe (Fig. 3). One fragment (0.9 kb) was found in DNA from all six strains, which is in agreement with a conserved region downstream of the dhaA gene (Fig. 1B). The size of the second DNA fragment, however, was different for some strains: 5.4 kb for strains m15-3, NCIMB13064, and TB2; 4 kb for strains HA1 and Y2; and 3.5 kb for strain GJ70. These results imply that the DNA regions upstream of the conserved 11-kb dhaA gene segment (upstream of the EcoRI site in the invA gene) in strains HA1, Y2, and GJ70 are different from those in strains NCIMB13064, m15-3, and TB2.
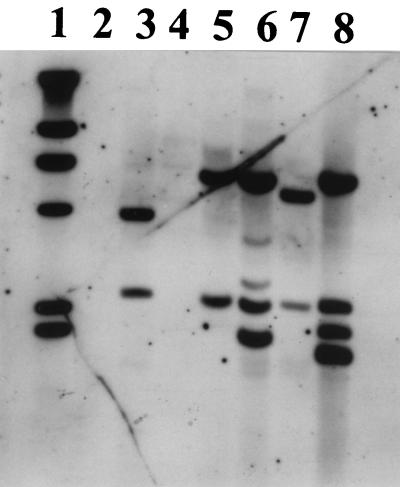
FIG. 3.
Results of a Southern hybridization experiment with a dhaA gene probe and ClaI-digested total DNA of strains GJ70 (lane 2), HA1 (lane 3), m15-3 (lane 4), NCIMB13064 (lane 5), and Y2 (lane 6). ClaI-digested total DNA of the dhaA-carrying gram-negative bacterium P. pavonaceae 170 (14) was used as a positive control (lane 1). Lane 7, digoxigenin-labeled DNA markers of 23.1, 9.4, 6.7, 4.4, 2.3, and 2.0 kb. Hybridization signals identical in size to the hybridization signals obtained for strains NCIMB13064 and m15-3 were also observed for strain TB2 (result not shown).
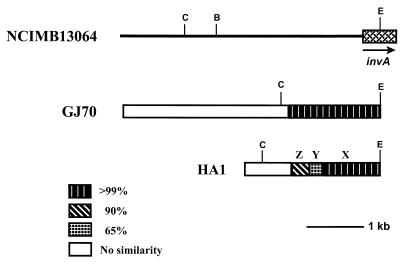
To compare the DNA regions upstream of the EcoRI site in the invA gene in strains HA1 and GJ70 in more detail with that in strain NCIMB13064 and to determine whether the conserved DNA fragments in these strains include the entire invA gene, we cloned and sequenced the corresponding regions from strains HA1 and GJ70. The NCIMB13064 and GJ70 sequences were still very similar (>99% sequence identity) for approximately 1.7 kb upstream of the EcoRI site in the invA gene and then abruptly became completely unrelated (Fig. 4). When the corresponding region of strain HA1 was compared to that of strain NCIMB13064, three conserved DNA segments having up to 35% differences were found. The first approximately 1,000 nucleotides upstream of the EcoRI site in the invA gene in strain HA1 were still very similar (>99% sequence identity) to the corresponding region in strain NCIMB13064. This segment (Fig. 4) is followed by a segment of approximately 200 nucleotides that shows 35% sequence divergence and a segment of approximately 300 nucleotides that differs from the corresponding segment in strain NCIMB13064 in about 10% of the positions. No further similarity between the two sequences was found. These results show that, compared to the sequenced dhaA gene region of strain NCIMB13064, the conserved DNA fragments in strains GJ70 and HA1 extend to approximately 1.7 and 1.5 kb, respectively, beyond the EcoRI site in the invA gene and thus include the complete invA gene. Since the sequence similarity between strains HA1 and NCIMB13064 vanishes before the BamHI site that is located upstream of the invA gene (Fig. 4), the localization of the dhaA genes in both strains on BamHI restriction fragments of approximately the same size (about 5.6 kb; see above) must be coincidental.
FIG. 4.
Schematic overview of the sequenced DNA regions upstream of the EcoRI site in the invA gene of strains NCIMB13064, GJ70, and HA1. DNA segments in strain HA1 that differ from the corresponding region in strain NCIMB13064 in about 0 to 1 (X), 10 (Z), or 35% (Y) of their positions are indicated. Open boxes, regions in strains HA1 or GJ70 that show no sequence similarity to the corresponding region in strain NCIMB13064; B, C, and E, BamHI, ClaI, and EcoRI restriction sites, respectively.
The genetic organization of the dhaA gene regions was further examined by performing hybridization experiments with BamHI- or EcoRI-digested DNA and an IS2112-specific probe. Since IS2112 has no restriction site for BamHI (Fig. 1B), a single hybridizing band corresponds to each copy of IS2112. Two copies of IS2112 were present in strains NCIMB13064 and TB2, whereas one copy was present in strains GJ70, m15-3, and Y2 (results not shown). No sequences homologous to IS2112 were found in strain HA1. We examined the regions flanking IS2112 by digesting the genomic DNAs with EcoRI. Since IS2112 has one cutting site for this enzyme (Fig. 1B), two hybridizing DNA bands were found for each copy of IS2112 (Fig. 5). Hybridizing bands at about 6 and 2.3 kb were found in DNA from strains m15-3, NCIMB13064, and TB2. The 6-kb DNA band was identical in size to the EcoRI restriction fragment that includes the DNA region upstream of the tnpA ORF of IS2112 in strain NCIMB13064 (Fig. 1B). Since in strain m15-3 only one copy of IS2112 is present, the hybridizing band at about 2.3 kb corresponds to the DNA region downstream of this copy of IS2112. These results indicate that the regions flanking the IS2112 sequence that is present approximately 6 kb upstream of the dhaA gene in strain NCIMB13064 (Fig. 1B) are conserved in strains m15-3 and TB2. The second copy of IS2112 in strain NCIMB13064 does not contain an EcoRI restriction site, indicating that this sequence is not perfectly conserved. This is in agreement with the results of Kulakov and coworkers (10), who demonstrated that the second copy of IS2112 in strain NCIMB13064 is an iso-IS2112 sequence containing 23 nucleotide substitutions. In total DNA of strain Y2 the 2.3-kb hybridizing DNA band was present, whereas the hybridization pattern for strain GJ70 was completely different (Fig. 5).
FIG. 5.
Results of a Southern hybridization experiment with an IS2112-specific probe and EcoRI-digested total DNA of strains GJ70 (lane 3), HA1 (lane 4), m15-3 (lane 5), NCIMB13064 (lane 6), Y2 (lane 7), and TB2 (lane 8). Lane 1 contained the same markers as lane 7 in Fig. 3. EcoRI-digested total DNA of P. pavonaceae 170 was used as a negative control (lane 2).
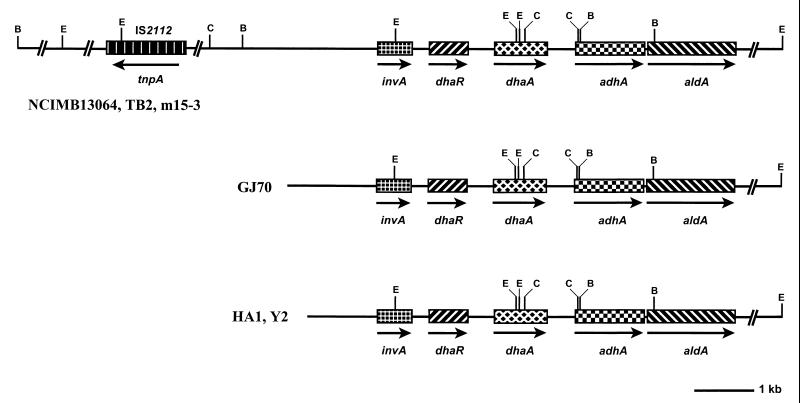
The hybridization and sequence data were used to construct physical maps of the dhaA gene regions in the six chloroalkane degraders. In summary, three classes that differ from each other with respect to the extent of the conserved dhaA gene segment could be defined (Fig. 6). The minimal structure present in all strains consists of a conserved region of 12.5 kb, including the regulatory gene dhaR and the haloalkane-degradative genes dhaA, adhA, and aldA, as well as the putative invertase gene invA. The presence of this highly conserved haloalkane catabolic gene cluster in all chloroalkane-degrading strains strongly suggests that these strains may have obtained this gene cluster as a preassembled unit from a common ancestral bacterial strain. It is noteworthy that the sequence similarity between strains NCIMB13064, m15-3, and TB2, isolated from contaminated sites in the United Kingdom, Japan, and the United States, respectively, extends to at least 22 kb (Fig. 6).
FIG. 6.
Physical maps of the dehalogenase gene regions of the six Rhodococcus strains isolated from widely separated geographical locations. Arrows indicate the direction of transcription. B, C, and E, BamHI, ClaI, and EcoRI restriction sites, respectively.
Genomic localization of the dhaA gene region.
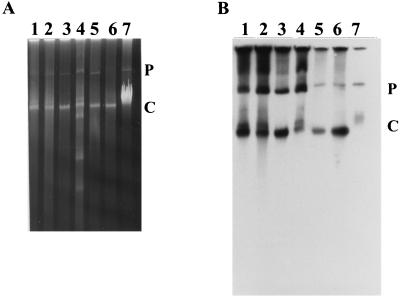
Since the haloalkane catabolic gene cluster of strain NCIMB13064 is located on a self-transmissible plasmid (pRTL1) (11, 15a), it is reasonable to propose that plasmid transfer has played a role in the dissemination of this gene cluster among the Rhodococcus strains. Using a direct lysis method, which prevents shearing of plasmid DNA, plasmids were detected in all analyzed strains (Fig. 7A). Plasmids of the same size, slightly smaller than 80-kb plasmid pRTL2 (11) of strains NCIMB13064 and S92 (a derivative of strain NCIMB13064) (11), were detected in strains Y2, TB2, m15-3, GJ70, and HA1. In strains TB2 and HA1, plasmids comparable in size to the 100-kb plasmid pRTL1 of strains NCIMB13064 and S92 were also present. In strain GJ70, additional smaller plasmids were detected (Fig. 7A).
FIG. 7.
Visualization of plasmids of haloalkane-degrading Rhodococcus strains. Agarose gel (A) and autoradiogram (B) of the same gel after Southern hybridization with the dhaA gene as the probe. Lane 1, strain Y2; lane 2, strain TB2; lane 3, strain m15-3; lane 4, strain GJ70; lane 5, strain HA1; lane 6, strain NCIMB13064; lane 7, strain S92 (derivative of strain NCIMB13064) (11). C and P, chromosomal and plasmid DNA, respectively.
In order to determine the location of the haloalkane catabolic gene cluster in the six Rhodococcus strains, chromosomal and plasmid DNA was hybridized with a dhaA-specific probe. In all strains (except S92), hybridization to both plasmid and chromosomal DNA was observed (Fig. 7B). The dhaA probe hybridized with plasmids of the same size (approximately 70 kb) in strains Y2, TB2, m15-3, and GJ70. For strain HA1, hybridization was detected with a plasmid whose size was the same as that of pRTL1 (compare lanes 5 to 7 in Fig. 7B). The hybridization signals observed above the plasmid bands in strains Y2, TB2, m15-3, and GJ70 are probably due to hybridization with the different forms of the same plasmid.
The occurrence of the highly conserved haloalkane catabolic gene cluster on plasmids ranging in size from 70 to 100 kb might suggest that a single mobile plasmid may have been transferred between different hosts and subsequently has undergone insertions or deletions (or vice versa). A similar phenomenon was also observed by Herrick and coworkers (5), who found that a naphthalene dioxygenase gene (nahAc) was present on different-size plasmids of naphthalene-degrading bacteria isolated from a coal tar-contaminated field site. Restriction fragment length polymorphism patterns and hybridization analysis indicated that all these plasmids were closely related to each other and to the naphthalene catabolic plasmid (pDTG1) of Pseudomonas putida NCIB 9816-4, indicating that a pDTG1-like plasmid is the mobile genetic element responsible for transferring naphthalene catabolic genes among bacteria in situ (22). Further investigations are necessary to determine whether the haloalkane catabolic gene cluster is located on pRTL1-like plasmids in all Rhodococcus strains analyzed here or on other plasmid backbones.
Kulakova et al. (11) demonstrated two important characteristics of plasmid pRTL1 that account for the mobility of the dhaA catabolic unit within the genome of strain NCIMB13064: first, the ability of the entire pRTL1 plasmid to integrate into the bacterial chromosome, and second, the spontaneous deletion of a 20-kb fragment of pRTL1 and the subsequent integration of this fragment into the chromosome. Both events lead to the loss of haloalkane dehalogenase activity by the host cells, and in both cases the corresponding derivative strains can revert to the original pheno- and genotypes. Insertion sequence IS2112 was shown to be involved in these genome rearrangements, although no direct link between the rearrangements and IS2112 transposition could be demonstrated (10). Based on these observations, it is likely that plasmid integration and/or transposition events may also be responsible for positioning the haloalkane catabolic gene cluster on the chromosomes of the other Rhodococcus strains analyzed in this study. Integration and excision of an entire catabolic plasmid or catabolic gene unit could play an important role in gene persistence and (enhanced) expression of catabolic genes in environments where substrate resources fluctuate (9, 11).
Since all gram-positive haloalkane degraders analyzed in this study were identified as rhodococci, it seems that there is a reservoir for haloalkane catabolic genes in members of this genus. However, it should be noted that these haloalkane-degrading strains were isolated by enrichment from (undiluted) environmental samples using high haloalkane concentrations. If Rhodococcus strains are preferentially isolated under these conditions because of their rapid growth, this could explain why most of the haloalkane-degrading bacteria isolated from natural soil samples exhibit surprising uniformity in their phenotypes and cell structures (G. J. Poelarends, J. E. T. van Hylckama Vlieg, T. Bosma, and D. B. Janssen, unpublished data; this study). We thus may have analyzed only a small branch of cultivable gram-positive bacteria capable of degrading (synthetic) haloalkanes. In recent work, however, we reduced the possible bias during batch enrichment by directly plating bacteria from diluted environmental samples on selective plates (Poelarends et al., unpublished data). Haloalkane-degrading bacteria could be easily isolated, and most of them had the typical Rhodococcus morphology and contained the dhaA gene. Since Rhodococcus strains were easily isolated from the natural environment, they are clearly of ecological importance.
In summary, we have shown that the haloalkane dehalogenase gene regions of the analyzed gram-positive haloalkane-utilizing bacteria are composed of a single catabolic gene cluster distributed worldwide. The high degree of sequence similarity, the conservation of gene order, and the absence of inserted or deleted ORFs within this gene cluster in all strains suggest that the distribution process occurred recently, possibly as the result of the widespread use of synthetic haloalkanes in industry and agriculture. The data also suggest that transfer of the haloalkane catabolic gene cluster has occurred over large geographical distances, which is difficult to interpret without invoking long-distance distribution mechanisms for microorganisms. This group of bacteria should prove to be an interesting subject for studying microbial evolution and gene transfer.
ACKNOWLEDGMENTS
This study was supported by the Life Sciences Foundation, which is subsidized by the Netherlands Organization for Scientific Research, and by the EC Environment and Climate Research Program contract ENV4-CT95-0086.
We thank P. Terpstra (Biomedical Technology Centre, University of Groningen, Groningen, The Netherlands) for his assistance in DNA sequencing. We thank the Czech Collection of Microorganisms (Masaryk University, Brno, Czech Republic), T. Leisinger (Swiss Federal Institute of Technology, ETH, Zürich, Switzerland), and T. Omori (University of Tokyo, Tokyo, Japan) for providing different chloroalkane-degrading bacteria.
REFERENCES
- 1.Brosius J, Palmer J L, Kennedy H P, Noller H F. Complete nucleotide sequence of a 16S ribosomal RNA gene from Escherichia coli. Proc Natl Acad Sci USA. 1978;75:4801–4805. doi: 10.1073/pnas.75.10.4801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Curragh H, Flynn O, Larkin M J, Stafford T M, Hamilton J T G, Harper D B. Haloalkane degradation and assimilation by Rhodococcus rhodochrous NCIMB 13064. Microbiology. 1994;140:1433–1442. doi: 10.1099/00221287-140-6-1433. [DOI] [PubMed] [Google Scholar]
- 3.Damborsky J, Nyandoroh M G, Nemec M, Holoubek I, Bull A T, Hardman D J. Some biochemical properties and the classification of a range of bacterial haloalkane dehalogenases. Biotechnol Appl Biochem. 1997;26:19–25. [PubMed] [Google Scholar]
- 4.Glasgow A C, Hughes K T, Simon M I. Bacterial DNA inversion systems. In: Berg D E, Howe M M, editors. Mobile DNA. Washington, D.C.: American Society for Microbiology; 1989. pp. 637–659. [Google Scholar]
- 5.Herrick J B, Stuart-Keil K G, Ghiorse W C, Madsen E L. Natural horizontal transfer of a naphthalene dioxygenase gene between bacteria native to a coal tar-contaminated field site. Appl Environ Microbiol. 1997;63:2330–2337. doi: 10.1128/aem.63.6.2330-2337.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Janssen D B, Jager D, Witholt B. Degradation of n-haloalkanes and α,ω-dihaloalkanes by wild-type and mutants of Acinetobacter sp. strain GJ70. Appl Environ Microbiol. 1987;53:561–566. doi: 10.1128/aem.53.3.561-566.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Janssen D B, Gerritse J, Brackman J, Kalk C, Jager D, Witholt B. Purification and characterization of a bacterial dehalogenase with activity toward halogenated alkanes, alcohols and ethers. Eur J Biochem. 1988;171:67–72. doi: 10.1111/j.1432-1033.1988.tb13759.x. [DOI] [PubMed] [Google Scholar]
- 8.Janssen D B, Pries F, van der Ploeg J, Kazemier B, Terpstra P, Witholt B. Cloning of 1,2-dichloroethane degradation genes of Xanthobacter autotrophicus GJ10 and expression and sequencing of the dhlA gene. J Bacteriol. 1989;171:6791–6799. doi: 10.1128/jb.171.12.6791-6799.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ka J O, Tiedje J M. Integration and excision of a 2,4-dichlorophenoxyacetic acid-degradative plasmid in Alcaligenes paradoxus and evidence of its natural intergeneric transfer. J Bacteriol. 1994;176:5284–5289. doi: 10.1128/jb.176.17.5284-5289.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kulakov L A, Poelarends G J, Janssen D B, Larkin M J. Characterization of IS2112, a new insertion sequence from Rhodococcus, and its relationship with mobile elements belonging to the IS110 family. Microbiology. 1999;145:561–568. doi: 10.1099/13500872-145-3-561. [DOI] [PubMed] [Google Scholar]
- 11.Kulakova A N, Stafford T M, Larkin M J, Kulakov L A. Plasmid pRTL1 controlling 1-chloroalkane degradation by Rhodococcus rhodochrous NCIMB13064. Plasmid. 1995;33:208–217. doi: 10.1006/plas.1995.1022. [DOI] [PubMed] [Google Scholar]
- 12.Kulakova A N, Larkin M J, Kulakov L A. The plasmid-located haloalkane dehalogenase gene from Rhodococcus rhodochrous NCIMB13064. Microbiology. 1997;143:109–115. doi: 10.1099/00221287-143-1-109. [DOI] [PubMed] [Google Scholar]
- 13.Marchesi J R, Sato T, Weightman A J, Martin T A, Fry J C, Hiom S J, Dymock D, Wade W G. Design and evaluation of useful bacterium-specific PCR primers that amplify genes coding for 16S rRNA. Appl Environ Microbiol. 1998;64:795–799. doi: 10.1128/aem.64.2.795-799.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Poelarends G J, Wilkens M, Larkin M J, van Elsas J D, Janssen D B. Degradation of 1,3-dichloropropene by Pseudomonas cichorii 170. Appl Environ Microbiol. 1998;64:2931–2936. doi: 10.1128/aem.64.8.2931-2936.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Poelarends G J, van Hylckama Vlieg J E T, Marchesi J R, Freitas dos Santos L M, Janssen D B. Degradation of 1,2-dibromoethane by Mycobacterium sp. strain GP1. J Bacteriol. 1999;181:2050–2058. doi: 10.1128/jb.181.7.2050-2058.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15a.Poelarends G J, Kulakov L A, Larkin M J, van Hylckama Vlieg J E T, Janssen D B. Roles of horizontal gene transfer and gene integration in evolution of 1,3-dichloropropene- and 1,2-dibromoethane-degradative pathways. J Bacteriol. 2000;182:2191–2199. doi: 10.1128/jb.182.8.2191-2199.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sallis P J, Armfield S J, Bull A T, Hardman D J. Isolation and characterization of a haloalkane halidohydrolase from Rhodococcus erythropolis Y2. J Gen Microbiol. 1990;136:115–120. doi: 10.1099/00221287-136-1-115. [DOI] [PubMed] [Google Scholar]
- 17.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 18.Sanger F, Nicklen S, Coulsen A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Scholtz R, Schmuckle A, Cook A M, Leisinger T. Degradation of eighteen 1-monohaloalkanes by Arthrobacter sp. strain HA1. J Gen Microbiol. 1987;133:267–274. [Google Scholar]
- 20.Scholtz R, Leisinger T, Suter F, Cook A M. Characterization of 1-chlorohexane halidohydrolase, a dehalogenase of wide substrate range from an Arthrobacter sp. J Bacteriol. 1987;169:5016–5021. doi: 10.1128/jb.169.11.5016-5021.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Staskawicz B, Dahlbeck D, Keen N, Napoli C. Molecular characterization of cloned avirulence genes from race 0 and race 1 of Pseudomonas syringae pv. glycinea. J Bacteriol. 1987;169:5789–5794. doi: 10.1128/jb.169.12.5789-5794.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stuart-Keil K G, Hohnstock A M, Drees K P, Herrick J B, Madsen E L. Plasmids responsible for horizontal transfer of naphthalene catabolism genes between bacteria at a coal tar-contaminated site are homologous to pDTG1 from Pseudomonas putida NCIB 9816-4. Appl Environ Microbiol. 1998;64:3633–3640. doi: 10.1128/aem.64.10.3633-3640.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van den Wijngaard A J, van der Kamp K W H J, van der Ploeg J, Pries F, Kazemier B, Janssen D B. Degradation of 1,2-dichloroethane by Ancylobacter aquaticus and other facultative methylotrophs. Appl Environ Microbiol. 1992;58:976–983. doi: 10.1128/aem.58.3.976-983.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yokota T, Fuse H, Omori T, Minoda Y. Microbial dehalogenation of haloalkanes mediated by oxygenase or halidohydrolase. Agric Biol Chem. 1986;50:453–460. [Google Scholar]
- 25.Yokota T, Omori T, Kodama T. Purification and properties of haloalkane dehalogenase from Corynebacterium sp. strain m15-3. J Bacteriol. 1987;169:4049–4054. doi: 10.1128/jb.169.9.4049-4054.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yoon J H, Lee J S, Shin Y K, Park Y H, Lee S T. Reclassification of Nocaridioides simplex ATCC 13260, ATCC 19565, and ATCC 19566 as Rhodococcus erythropolis. Int J Syst Bacteriol. 1997;47:904–907. doi: 10.1099/00207713-47-3-904. [DOI] [PubMed] [Google Scholar]