Abstract
Lysyl oxidase acts as both a matrix modifying enzyme and an oncogene suppressor. It is synthesized as a 50-kDa proenzyme, secreted, and processed into an ~30 kDa mature, active enzyme and an 18-kDa propeptide. The tumor suppressive effect of lysyl oxidase appears to be exerted within the cell, so the subcellular localization of protein forms was investigated. Propeptide-specific antibody detected 50-kDa proenzyme in cytoplasmic and nuclear extracts of non-transformed mouse fibroblasts, but free 18-kDa propeptide was not detected in any extract. Antibody to epitope near the N-terminus of mature lysyl oxidase detected the proenzyme product in non-transformed cells, and a 30-kDa cytoplasmic protein in both non-transformed and transformed cells. RNA interference reduced the expression of lysyl oxidase mRNA and 50-kDa proenzyme in non-transformed cells, but had no effect on 30-kDa protein, indicating that although this protein displays a lysyl oxidase epitope, it is not derived from lysyl oxidase message. The absence of both free 18-kDa propeptide and mature lysyl oxidase within non-transformed cells suggests that cellular reversion after restoration of lysyl oxidase gene expression is mediated by the 50-kDa proenzyme within cells.
Keywords: Tumor suppressor, Lysyl oxidase, Antibody, RNA interference, Interferon
1. Introduction
Lysyl oxidase (LOX) is a secreted enzyme that catalyzes the oxidative deamination of ε-amino lysines in collagen and elastin fibers. The subsequent condensation of the modified lysine residues creates the cross-linked fibers characteristic of the mature forms of these proteins in the extracellular matrix. Increased LOX activity is found in fibrotic disease of the lung, in atherosclerosis [1], and in in vitro models of rat hepatic fibrosis [2]. Type IX Ehlers–Danlos syndrome (occipital horn syndrome) is characterized by a decrease in LOX protein and mRNA, and LOX activity is decreased in Menkes syndrome [3,4]. Inhibition of LOX enzyme activity by the inhibitor β-aminoproprionitrile results in lathyrism [5].
As well as a protein-modifying enzyme, LOX was identified as a tumor suppressor [6,7]. LOX mRNA expression is almost completely lost in mouse NIH 3T3 cells after transformation by c-H-ras, and LOX mRNA is restored in interferon (IFN)β-reverted cells. LOX mRNA levels are reduced significantly in rodent cells transformed by a variety of other oncogenes, including v-raf, v-fes, [6] v-src, c-met, v-fms, v-fos, and c-trk (S. Contente, unpublished observation). Decreased LOX expression has been reported in ductal breast carcinoma cells [8], in prostate tumors [9], and in bronchogenic carcinoma [10]. LOX expression is lost or decreased in human breast and prostate tumor tissue sections studied by in situ hybridization (R.M. Friedman, D.L. Buchhagen, unpublished observation). LOX promoter methylation and loss of heterozygosity have been found in human gastric cancers [11]. LOX is expressed in human cells that do not normally synthesize collagen, such as breast ductal epithelium, colonic epithelium, prostatic epithelium, and neurons (S. Contente, unpublished observation), suggesting an additional function for this protein.
LOX is synthesized as a 50-kDa proenzyme (proLOX) that is secreted and then cleaved in the extracellular space by bone morphogenetic protein 1 to form an active enzyme of about 30 kDa [12–16]. However, antibody-reactive, enzymatically active 32-kDa LOX was reported in nuclei isolated from neonatal rat smooth muscle cells and NIH 3T3 cells [17], and a 30-kDa mature LOX was reported in the cytoplasm of human epithelial cells [18]. Purified bovine aorta 32-kDa protein, labeled with rhodamine, re-entered vascular smooth muscle cells and NIH 3T3 cells, and concentrated within nuclei [19]. LOX has been reported to use histone H1 as a substrate in vitro [20], and to interact specifically with histones H1 and H2 in vitro [21].
A possible nuclear role for LOX is supported by the report that over expression of a 32-kDa transgene in COS-7 cells activated transcription of a human collagen promoter in a luciferase reporter plasmid [22]. In addition, injected recombinant 32-kDa LOX also prevented the p21-H-ras-induced meiotic maturation of Xenopus laevis oocytes [23]. Recently, the18-kDa LOX propeptide, which is generated upon proteolysis of 50-kDa proLOX in the extracellular space, was expressed in bacteria and found to inhibit cell cycle progression and growth in soft agar of ras-transformed cells [24], and the invasive phenotype of a Her-2/neu cell line was inhibited after introduction of propeptide or proLOX constructs [25]. The18-kDa propeptide was reported to localize differently in MC3T3-E1 osteoblasts based on their stage of differentiation [26].
We prepared affinity purified, polyclonal antisera raised against peptides to determine the intracellular distribution of proenzyme, mature enzyme, and propeptide in non-transformed and ras-transformed mouse fibroblasts. Peptide sequences were located within the propeptide portion of proenzyme or at the amino terminus of mature processed enzyme. Only 50-kDa proenzyme, and not mature LOX or propeptide, was found within non-transformed cells. Although a 30-kDa cytoplasmic protein was recognized by the antibody to mature LOX in non-transformed cells, this protein was unaffected by siRNA that did affect proLOX expression. In addition, this 30-kDa cytoplasmic protein also appeared in transformed cells that do not express LOX. This protein, therefore, cannot be derived from the LOX gene. The distribution of proLOX, and the absence of propeptide and mature enzyme in any cell compartment suggest that the reversion observed after restoration of LOX gene expression in ras-transformed cells is associated with the appearance of 50-kDa proLOX in the nucleus, and that the action of the 18-kDa propeptide region is exerted as a part of the proenzyme.
2. Materials and methods
2.1. Antibodies
Rabbit polyclonal anti-LOpep3 [27] was affinity purified (Quality Controlled Biologicals, Hopkinton, MA) against 170KYSDDNPYYNYYDTYCR186, the immunogenic peptide. This sequence is located near the N-terminus of mature LOX, except that residue 185E was replaced by cysteine. Residues 170–186 (numbering refers to mouse protein) are identical in mouse and human LOX, and there is no identity with known human or mouse lysyl oxidase-like proteins [28–33]. Affinity-purified anti-LOpep3 was used at a 1:2000 dilution. Rabbit anti-LOpep4 serum was prepared and affinity purified against peptide C-141ASPQPPQLSNLRPPS155 (Quality Controlled Biologicals, Hopkinton, MA). This sequence is located within the 18-kDa propeptide region of proLOX. Affinity-purified antiserum was used at a 1:20,000 dilution. Polyclonal antibodies to other LOX epitopes were obtained from Novus Biologicals, Littleton, CO (NB100–2527, NB100–2528, NB 100–41090, NB110–59729, NB110–41568), and from Santa Cruz Biotechnology, Santa Cruz, CA (E-19 and V-20). Of these antibodies, only NB100–2527 and NB110–41568 bound specifically to in vitro translated LOX products. The other preparations were not used further. Secondary antibodies and loading control/compartment-specific antibodies were obtained from AbCam, Inc., Cambridge, MA, and Santa Cruz Biotechnology, Santa Cruz, CA. Antibody beads were created using 10 μl of affinity purified anti-LOpep3 and 100 μl of protein A Dynabeads (Invitrogen, Carlsbad, CA) according to the manufacturer’s instructions. Forty μg of protein extract was incubated with the antibody beads for 1 h at 4 °C, and the beads were captured by magnet. Supernatant was removed and concentrated using a Microcon YM-10 cartridge (Millipore, Bedford, MA) prior to gel analysis.
2.2. Construction of LOX expression clones and transcription–translation of products
The Gateway System (Invitrogen) was used to generate entry clones of LOX size species in pDONR221. PCR fragments representing propeptide (A22-G162), proenzyme (A22-Y411), and mature LOX (D163-Y411) were generated from plasmid clone Nco12, which contains a full-length mouse LOX cDNA. PCR primers contained attB1or attB2 sequences as specified in the Gateway manual, along with appropriate start or stop codons plus 18–25 bp of LOX-specific nucleotides to create the desired protein. Entry clones pENTRLO18, pENTRLO30, and pENTRLO50 were subsequently recombined with mammalian expression vector pcDNA3.2-V5Dest or bacterial expression vector pDEST14, and used for subsequent experiments. Constructs in mammalian expression vectors were expressed by in vitro transcription and translation using TNT Coupled Wheat Germ Extract System (Promega Corp., Madison, WI). pDEST14-LO18 was transfected into Escherichia coli BL21-A1 competent cells, and resultant ampicillin resistant colonies were cultured and induced with L-arabinose according to the Gateway protocol. Cells were collected by centrifugation after 4 h of induction, suspended in 1×SDS-PAGE sample buffer (Invitrogen), boiled for 5 min, and centrifuged briefly. 10 μl of supernatant was used for gel electrophoresis.
2.3. Cell culture and transfection
NIH 3T3 is a fibroblast cell line derived from Swiss mouse. RS485 is NIH 3T3 transformed by c-H-ras [34]. Mouse cell lines and mouse primary cells were cultured in DMEM+ Glutamax-1 supplemented with 10% fetal bovine serum (GIBCO). All cells were cultured at 37 °C in a humidified 5% CO2 atmosphere. Mouse primary embryo fibroblast cells PMEF-NL were obtained from Chemicon International (Temicula, CA), and mouse primary fibroblast cells TGM-1 were cultured using shared tissue from 1-day old FVB/N mice. For transfections, 5 μg of endonuclease-free plasmid DNA was introduced into 3.6×105 RS485 cells using Lipofectamine (Invitrogen) for 5 h at 37 °C, 5% CO2, after which DNA complexes were removed and replaced with DMEM-10% FBS. Cells were split 1:20 after 48 h, and selection with G418 (500 μg/ml) was started 24 h after that. Individual G418 resistant clones selected 10 days later were expanded up into 75 cm2 flasks. For soft agar assays, 3×104 cells mixed into 3 ml of warm 0.35% agar were overlaid onto a base of 5 ml of 0.7% agar in a 60 mm gridded dish. After 10 days of incubation at 37 °C, dishes were stained with 1 ml 0.005% crystal violet for about 1 h. Colonies were counted in ten random 4 mm2 areas and averaged. For RNA interference, NIH 3T3 cells were stably transfected with each of four Sure Silencing shRNA plasmids for Mouse Lox (SuperArray Bioscience Corp, Frederick, MD). Cultures were selected for neomycin resistance and individual cells were cloned out. Cellular RNA was analyzed by northern blot with LOX DNA probes as previously described [27].
2.4. Protein preparation and immunoblot analyses
Whole cell lysates were prepared using M-PER, and cytoplasmic and nuclear extracts were prepared using NE-PER (Pierce Biotechnology, Rockford, IL). Protein concentration was determined using BCA Protein Assay kit (Pierce). Purified bovine heart lysyl oxidase was a gift from Dr. Herbert Kagan, Department of Biochemistry, Boston University School of Medicine. Total, cytoplasmic or nuclear cell extracts (20 μg) were denatured in NuPage sample buffer and electrophoresed on NuPage 4%–12% Bis–Tris gels in 3-(N-morpholino) propanesulfonic acid (MOPS)-SDS buffer (Invitrogen, Carlsbad, CA) according to the manufacturer’s instructions. MagicMark™ XP Western Protein Standard (Invitrogen) was used for direct visualization of protein standard bands by secondary antibodies. Electrophoresed proteins were transferred to Hybond-P membranes (GE Healthcare) at 35 V for 60 min at 4 °C in a Novex Xcell blot module, or at 23 V for 4 min in an iBlot Gel Transfer Device (Invitrogen). Membranes were rinsed in TBST (0.1% Tween-20 in 1.5 M NaCl, 100 mM Tris, pH 8.0) and blocked overnight with rocking at 4 °C in a blocking buffer of 5% nonfat dry milk in TBST. Membranes were incubated with primary antibody in blocking buffer with rocking for 2 h at room temperature or at 4 °C overnight, washed three times for 15 min each at room temperature with TBST, incubated with secondary antibody in blocking buffer for 45 min at room temperature with rocking, and washed as described. Membranes were developed with ECL Plus (GE Healthcare, Piscataway, NJ) for 5 min. Digital images were obtained using an LAS-1000 Plus Luminescent Image Analyzer (Fujifilm Co., Stamford, CT). Film images were obtained using Hyperfilm ECL (GE Biosciences), developed in a Kodak M35A X-OMAT processor. Membranes to be reprobed were stripped 15–30 min at 50 °C in Restore (Pierce Biotechnology, Rockford, IL).
3. Results
3.1. Validation of peptide antibody preparations
The specificity of peptide antisera was verified by immunoblot against in vitro transcription–translation products derived from subcloned cDNAs for LOX proenzyme (50-kDa), mature enzyme (30-kDa), or propeptide (18-kDa). Propeptide product isolated from a bacterial expression system was also used (Fig. 1A). Anti-LOpep3 (this laboratory), which recognizes an epitope at the N-terminus of mature LOX, and anti-LOX2527 (Novus), which recognizes an epitope in the central area of LOX, each detected in vitro products derived from proenzyme and mature enzyme cDNA clones. As expected, these antisera did not react with 18-kDa propeptide product (not shown). Anti-LOpep4 (this laboratory) and anti-LOX41568 (Novus), which were raised to different epitopes in the propeptide region, detected products from the 50-kDa proenzyme and 18-kDa propeptide clones. These antibodies did not recognize the 30-kDa product of the mature enzyme subclone, as expected. Anti-LOpep3 and anti-LOX2527 each recognized a 30-kDa protein in a preparation of active LOX purified from bovine tissue (Fig. 1B). Thus, LOX enzyme isolated from animal tissue does exhibit both these epitopes and mature LOX in cell extracts would be expected to react with both of these antibodies. Each of these four antibodies recognized the expected epitopes in engineered products and, in the case of mature LOX epitopes, the antibodies also recognized LOX isolated from bovine tissue. It was expected that these antibodies could be used to provide valid data on LOX expression in studies of cell extracts. However, as shown below, three of the four antibodies also recognize cellular proteins that, while close in size to that expected for mature LOX or propeptide, are in fact not derived from proLOX.
Fig. 1.
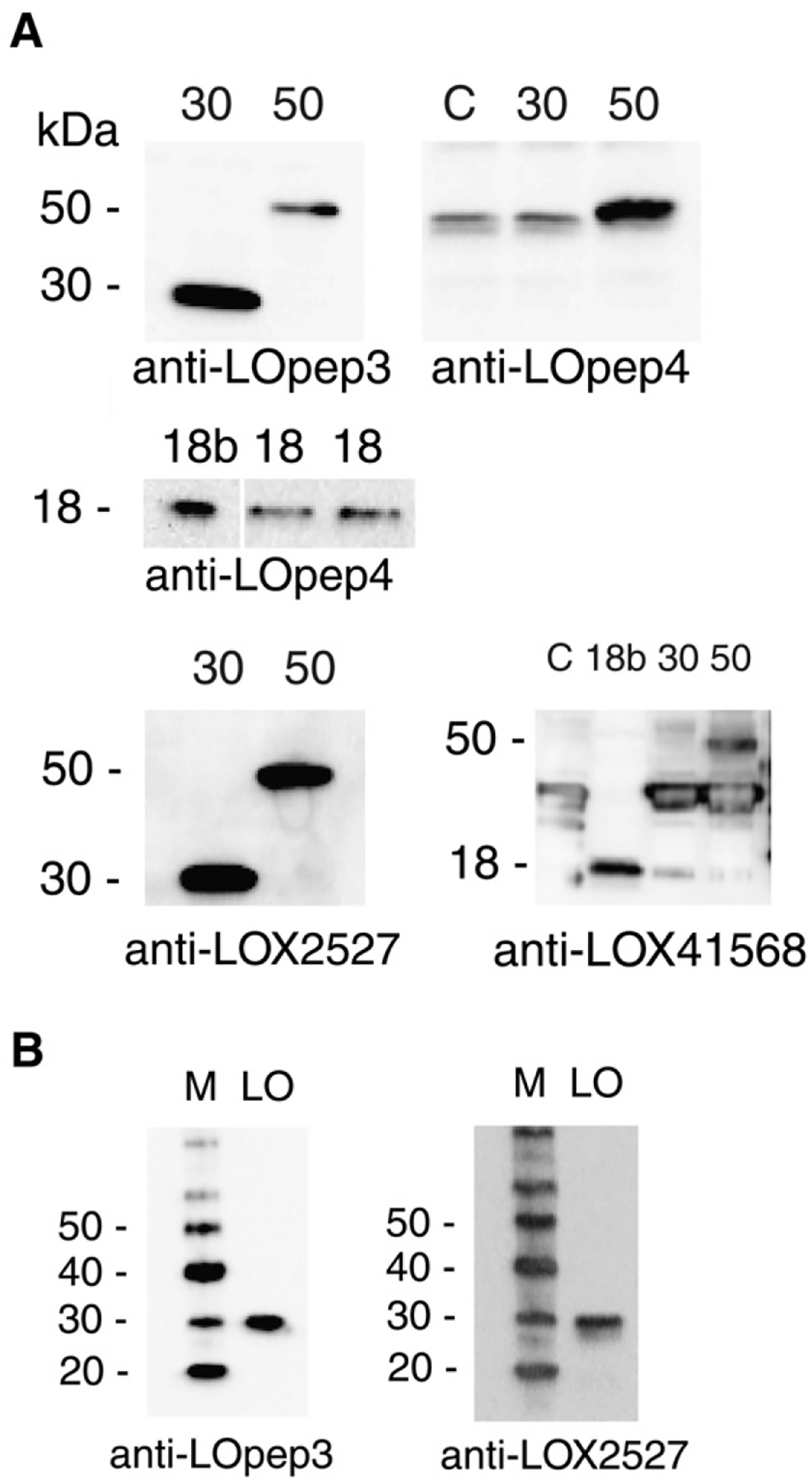
Specificity of anti-peptide antibodies. A. Protein blots of in vitro translated products of LOX expression clones and bacterial expression product probed with affinity purified antibodies. C, no DNA control translation; 30, product from mature LOX clone; 50, product from proenzyme clone; 18, in vitro translation of propeptide clone; 18b, propeptide clone bacterial product. Anti-LOpep4 binds non-specifically to a wheat germ protein slightly smaller than 50-kDa, and anti-LOX41568 binds non-specifically to a wheat germ product of about 40-kDa. B. Enzymatically active bovine heart LOX protein probed with antibodies to mature LOX epitopes. LO, 25 ng bovine LOX preparation; M, protein size markers.
3.2. ProLOX, but not free propeptide, is found within non-transformed and primary mouse fibroblast cells
The antibody anti-LOpep4, which we prepared to the propeptide region residues 141–155, detected 50-kDa protein in cytoplasmic and nuclear extracts of NIH 3T3 and primary mouse fibroblasts (PMEF, TGM-1) (Fig. 2A). The protein in nuclear extracts migrated slightly slower, suggesting that nuclear proLOX may be modified. There was no 50-kDa protein present in ras-transformed RS485, which is consistent with the lack of LOX mRNA in this cell line [6,7]. However, no proteins corresponding to the 18-kDa propeptide fragment were detected by anti-LOpep4 in any cell lines. In cell line 62, a revertant of RS485 induced with IFNβ and retinoic acid [35], the expression of 50-kDa protein was restored in both cytoplasm and nucleus (not shown). As with the other cell lines, no 18-kDa propeptide was detected in the revertant. The lack of 18-kDa propeptide in both NIH 3T3 and RS485 was verified using immunoblots prepared with three times the usual amount of extract and probed with anti-LOpep4 (Fig. 2B). This also verified the lack of 50-kDa protein in RS485 extracts. Additionally, material was prepared as described [26] from the entire nuclei pellet obtained from cultured cells; immunoblot analysis of this material with anti-LOpep4 revealed no 18-kDa propeptide in either NIH 3T3 or RS485 (not shown).
Fig. 2.
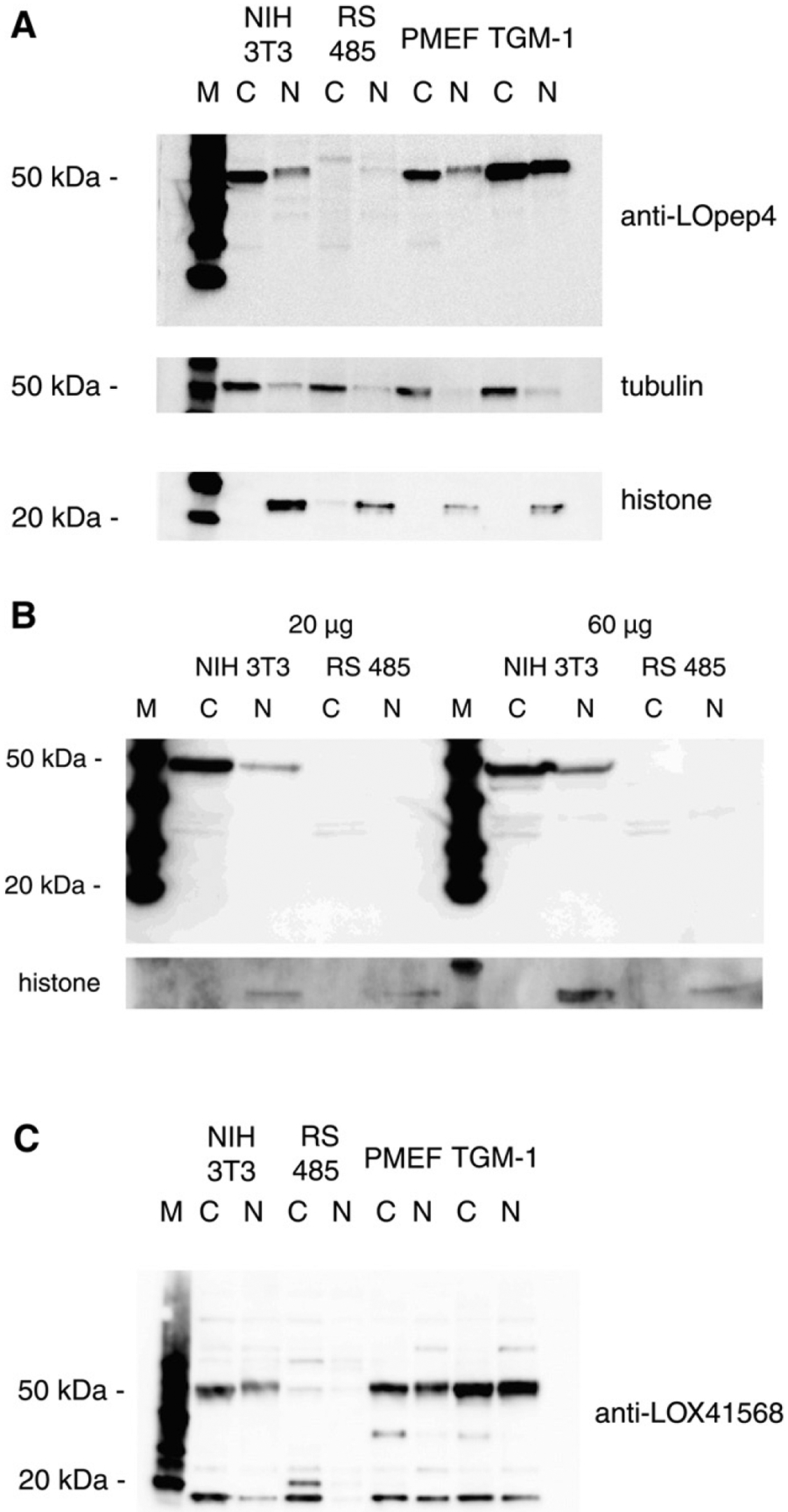
18-kDa propeptide is absent in cells. A. Cytoplasmic (C) and nuclear (N) extracts of non-transformed (NIH 3T3), ras-transformed (RS485) and primary mouse fibroblast cells PMEF and TGM-1 probed with propeptide antibody anti-LOpep4. Cytoplasm-specific (tubulin) and nuclear-specific (histone) antibodies were used for loading and compartment controls. M, protein size markers. B. Twenty and 60 μgram of cytoplasmic (C) and nuclear (N) extracts of NIH 3T3 and RS485 probed with propeptide antibody anti-LOpep4. M, protein size markers. C. Extracts as in A. probed with propeptide antibody anti-LOX 41568.
As other propeptide antibodies were reported to detect free 18-kDa in cells, we used anti-LOX41568 to verify the lack of 18-kDa propeptide in mouse cell extracts. This is a commercially available antibody, raised to propeptide region residues 78–115, which does recognize bacterial propeptide product (Fig. 1A). This antibody detected 50-kDa proenzyme in extracts of non-transformed cells, but not in RS485 extracts, as expected (Fig. 2C). However, this antibody did recognize an 18-kDa protein in both non-transformed and transformed cells. This protein, however, although of the expected size, cannot be the LOX propeptide because it is abundant in the transformed cells that lack LOX mRNA and 50-kDa proenzyme. In addition, as both anti-LOpep4 and anti-LOX41568 recognize 18-kDa propeptide synthesized in bacteria, both should recognize any propeptide present in cells, and this is not the case. Therefore, we conclude that there is no free18-kDa propeptide within cells.
3.3. Mature LOX is not found within cells
Anti-LOpep3 was prepared to the sequence KYSDDNPYYNYYDTYCR, located at the N-terminus of mature LOX. This antibody detected both a 50-kDa and a 30-kDa protein in cytoplasmic extracts of NIH 3T3 and primary mouse fibroblasts (TGM-1) (Fig. 3A). Based on the published literature that mature LOX is found within cells, this would appear to be an expected result. Several studies have been performed using antibody prepared to LOX peptide KYSDDNPYYNYYDTYERPRPGG [18,36–38], which is nearly identical to the pep3 sequence. This antibody also recognizes a 50-kDa and a 30-kDa protein [18,36], and it is reasonable to conclude that both antibodies recognize the same two proteins. However, cytoplasmic extracts of ras-transformed RS485 probed with anti-LOpep3 had no 50-kDa protein, but did have abundant 30-kDa protein (Fig. 3A). As LOX message is severely down regulated in RS485 and these cells have no LOX enzyme activity [6,7], little, if any, LOX protein was expected to appear in these transformed cells. Therefore, while the lack of 50-kDa protein was not surprising, the presence of an abundant 30-kDa protein, in the cytoplasm of transformed cells, was unexpected. Nuclear extracts of NIH 3T3 and TGM-1 did contain 50-kDa protein, although it was more abundant in TGM-1. No 50-kDa protein was observed in nuclear extracts of RS485, and 30-kDa protein did not appear in nuclear extracts of any cell lines. Therefore, anti-LOpep3 recognizes, in addition to proLOX in cytoplasmic and nuclear extracts, a cytoplasmic protein that could be mature LOX, but which appeared in cells where mature LOX should not be present.
Fig. 3.
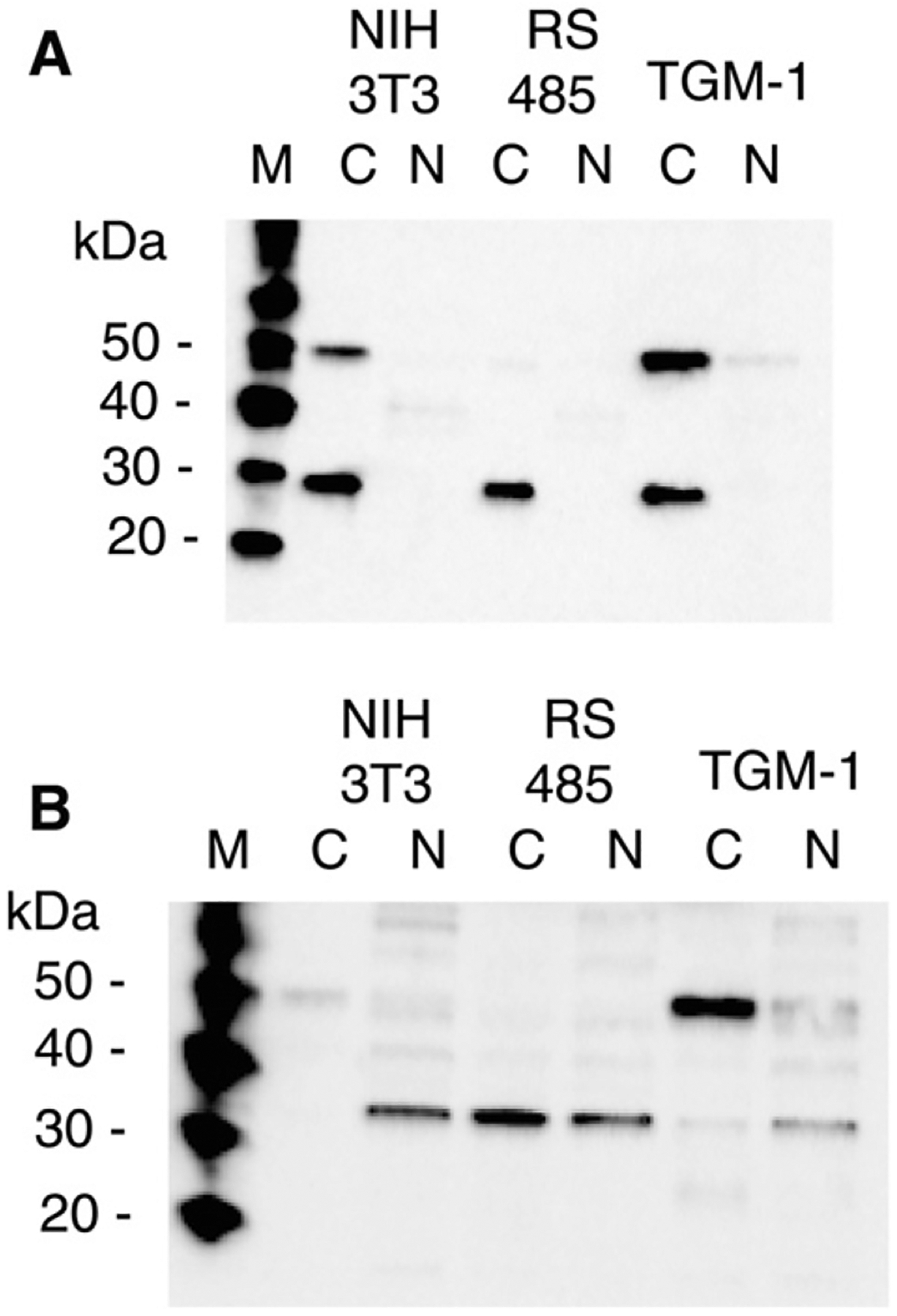
Antibodies to epitopes within mature LOX detect different cellular proteins. Cytoplasmic (C) and nuclear (N) extracts of non-transformed (NIH 3T3), ras-transformed (RS485) and primary mouse fibroblast (TGM-1) cells were probed with A. anti-LOpep3 (N-terminal epitope) and B. anti-LOX 2527 (central epitope). M, protein size markers.
A second antibody to mature LOX, anti-LOX2527, detected 50-kDa proLOX in NIH 3T3 and TGM-1 cell extracts, but not in RS485 cell extracts (Fig. 3B). This was consistent with the expression pattern observed for proLOX with anti-LOpep3, and was as expected: both antibodies recognized the in vitro translated LOX proenzyme. However, anti-LOX2527 also detected a 32-kDa protein in transformed and non-transformed cell extracts. This protein had a different compartmental distribution. It was predominantly nuclear in non-transformed cells, and abundant in both compartments of transformed cells. The 30-kDa protein recognized by anti-LOpep3 was found only in cytoplasmic extracts. The 32-kDa protein was not recognized by anti-LOpep3, and the 30-kDa cytoplasmic protein was not recognized by anti-LOX2527. This indicates that the 30-kDa protein displays the N-terminal LOX epitope, but not the central epitope, and that the 32-kDa protein displays the central epitope but not the N-terminal epitope. It was previously shown that both antibodies did recognize purified bovine LOX (Fig. 1C), thus neither cellular protein can be mature LOX, even though each displays at least one LOX epitope. The presence of both of these small cellular proteins in transformed cells, which do not express proLOX, additionally supports the conclusion that neither of these small cellular proteins is true mature LOX. Analysis of secreted LOX using both antibodies (Section 3.5) also supports this conclusion.
3.4. RNA interference for LOX does not affect 18-kDa or 30-kDa expression
Fig. 4A shows RNA analysis of four clones obtained after stable transfection of NIH 3T3 with mouse LOX shRNA plasmid #2 or #3. Two size species of LOX mRNA always appear in normal mouse cells, as there are two, well-separated polyadenylation signals that are utilized in this message [39]. There are reduced amounts of LOX mRNA in the shRNA clones (lanes 3–6) compared to an NIH 3T3 control (lane 1), and there is almost no LOX mRNA in RS485 (lane 2). The LOX mRNA signals were quantitated by densitomtery, and normalized to the GAPDH signals. The LOX mRNA remaining in the shRNA clones was 19%, 15%, 23% and 22% of that expressed in NIH 3T3. RS485 expresses 2% of LOX mRNA compared to NIH 3T3. Fig. 4B presents western analysis of protein extracted from the shRNA clones. Probing with anti-LOpep4 revealed a reduction of 50-kDa proenzyme product in these clones to 6%, 6%, 15% and 19% of that observed in NIH 3T3, but probing with anti-LOpep3 showed that the amount of 30-kDa protein in the shRNA clones was not reduced, and that this protein was actually more abundant in shRNA clones than in NIH 3T3. Thus the 30-kDa cytoplasmic protein cannot be derived from the LOX gene and it is not mature LOX. Proenzyme product in shRNA extracts as detected by anti-LOX41568 was reduced to 40–62% of the amount present in NIH 3T3, and no proenzyme was detected in RS485 (not shown). The amounts of the 18-kDa protein detected in shRNA extracts and RS485 were 70–95% of the amount of this protein found in NIH 3T3. These findings confirm that this 18-kDa protein, although recognized by antibody anti-LOX41568, cannot be LOX propeptide.
Fig. 4.
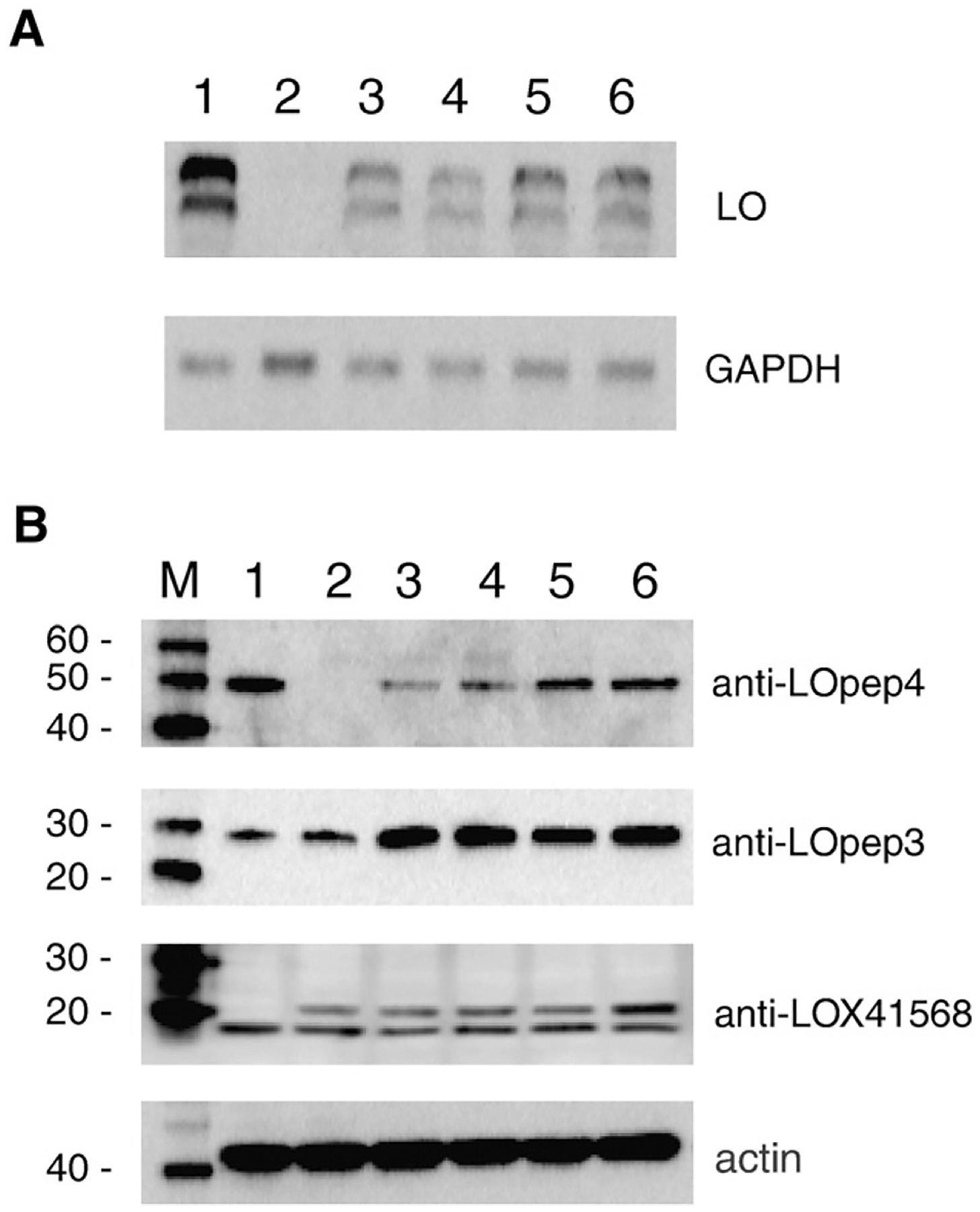
RNA interference reduces LOX message and proenzyme protein. A. RNA blot probed with LOX DNA probe. GAPDH used as loading control. 1. NIH 3T3; 2. RS485; 3–6. shRNA clones 2–2A-2, 3–1A-4, 3–1B-14, and 3–1B-17. B. Total protein extracts probed with the indicated antibody. Actin used as loading control. M, protein size markers. 1. NIH 3T3; 2. RS485; 3–6. shRNA clones 2–2A-2, 3–1A-4, 3–1B-14, and 3–1B-17.
3.5. Protein secreted by NIH 3T3 does bind both anti-LOpep3 and anti-LOX2527
Conditioned media from NIH 3T3, which contains enzymatically active, mature LOX enzyme, was assayed by immunoblot with each antibody (Fig. 5). Each antibody recognized a secreted protein slightly larger than 30 kDa in conditioned media. In NIH 3T3 cellular extract run in parallel, it is clear that the small cellular proteins detected are either slightly larger or slightly smaller than the secreted protein. The secreted protein was not detected, by either antibody, in conditioned media from transformed cells (not shown). This was consistent with the lack of LOX in transformed cells. This >30 kDa extracellular protein must therefore be mature LOX.
Fig. 5.
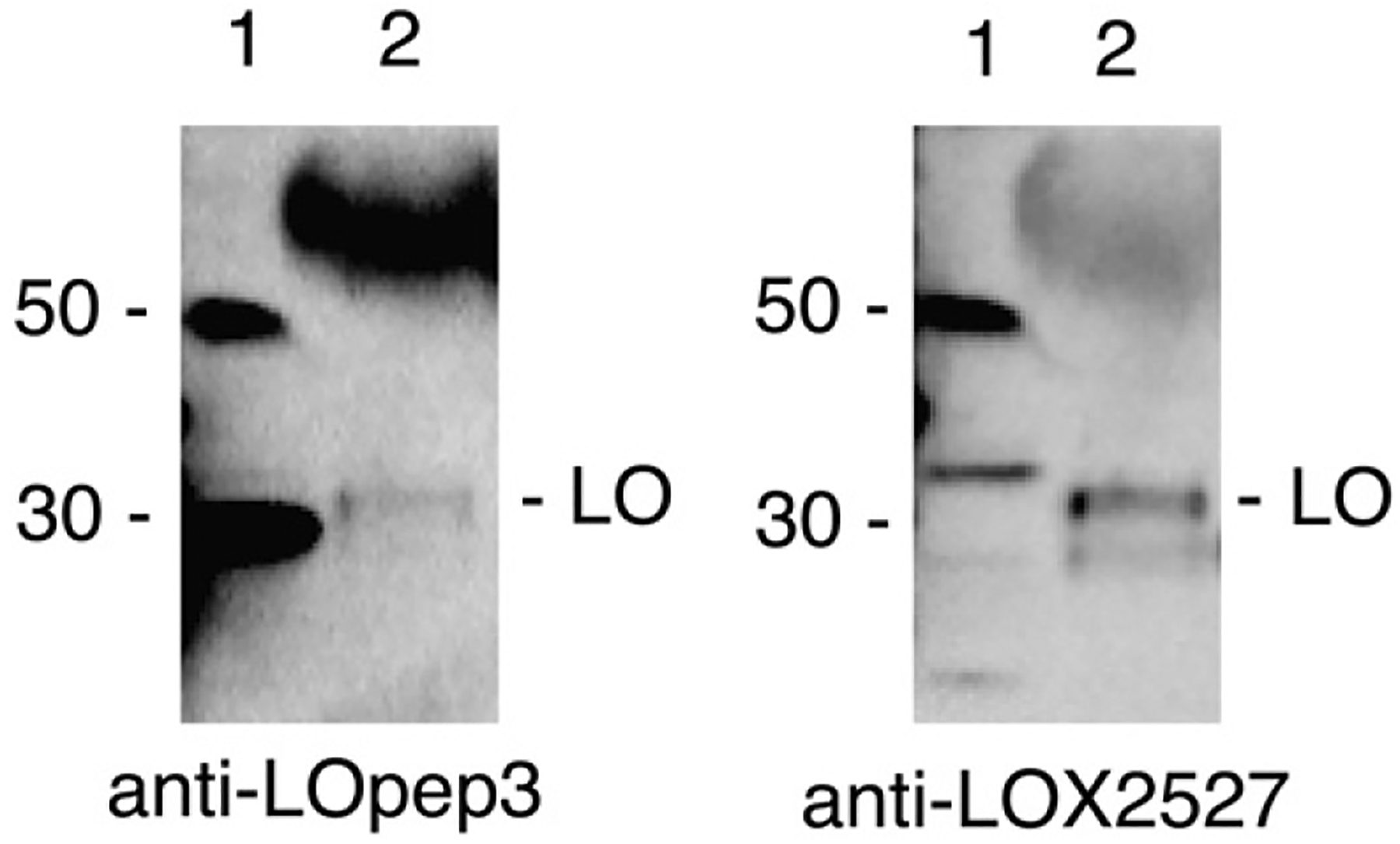
Mature LOX appears only in conditioned media. Conditioned media from NIH 3T3 cells was blotted and probed with anti-LOX2527 and anti-LOpep3. Lane 1, NIH 3T3 cell extract; lane 2, 30 μl of NIH 3T3 conditioned media, concentrated from 0.1 ml on Centricon YM-10 column. The strongly hybridizing band at ~55 kDa in lane 2 is immunoglobulin derived from the fetal bovine serum added to cell culture media.
3.6. Antibodies do bind to the same 50-kDa protein
To verify that the antibodies used were recognizing the same 50-kDa protein, NIH 3T3 cytoplasmic extract was absorbed with magnetic bead-linked anti-LOpep3, and the proteins remaining in the supernatant were analyzed by immunoblot with anti-LOpep3, anti-LOpep4, and anti-LOX-2527 (Fig. 6). The 50-kDa band was not detected by anti-LOpep3 in the absorbed material, indicating that the 50-kDa protein was successfully removed from the extract by the antibody-linked beads. The 30-kDa protein, although recognized on immunoblots as a denatured protein, did not bind to anti-LOpep3 in its native form and was not adsorbed onto the beads. Thus, this protein appeared in both lanes when anti-LOpep3 was used to probe the blot. Anti-LOpep4 and anti-LOX 2527 did not detect any 50-kDa protein in the absorbed material, indicating that all three antibodies recognize one 50-kDa protein, which does represent LOX proenzyme. The slight shadow above the 50-kDa position in the absorbed material lane represents IgG released from the beads during incubation with extract.
Fig. 6.
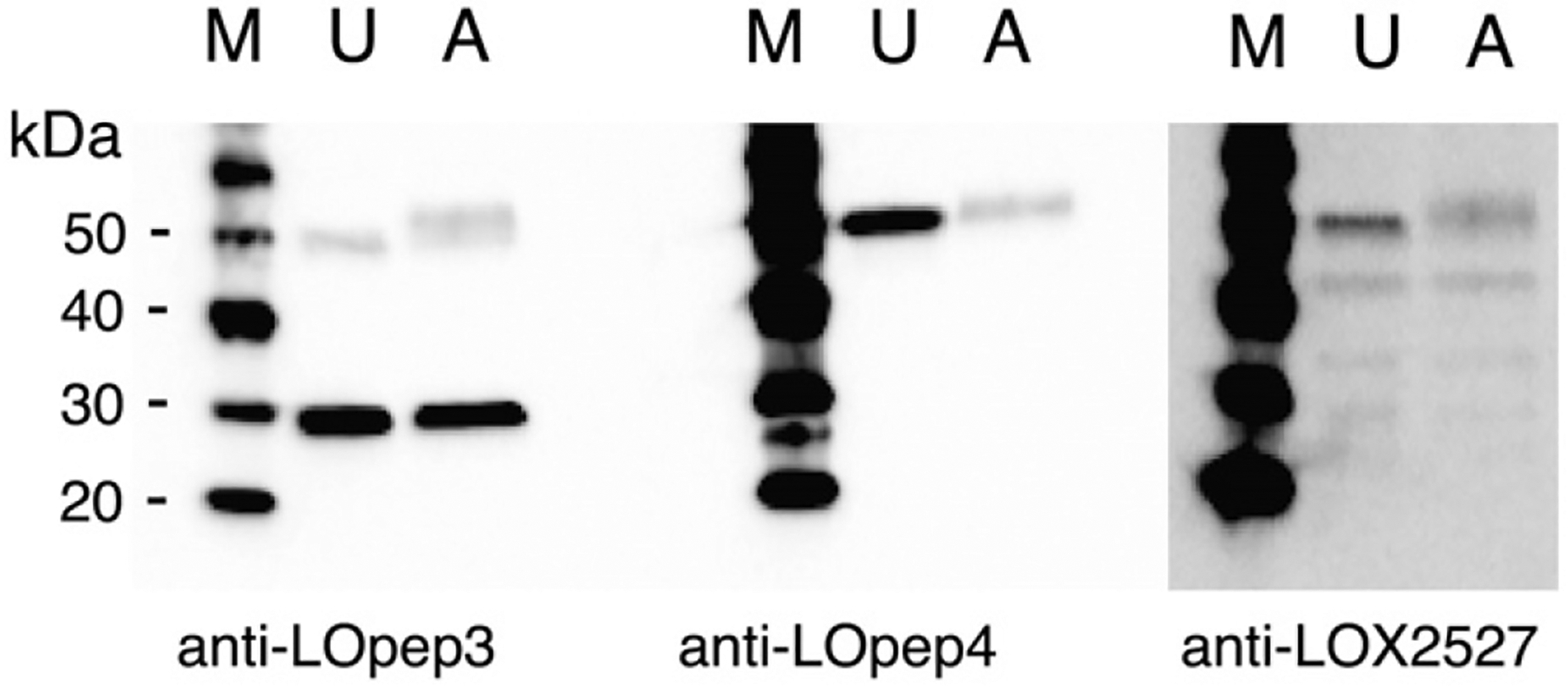
50-kDa protein displays each LOX epitope. NIH 3T3 cytoplasmic extract was absorbed against magnetic bead-linked anti-LOpep3, and untreated extract (U) and absorbed extract (A) were probed with the indicated LOX antibody. M, protein size markers.
3.7. Proenzyme expression can inhibit growth of ras-transformed cells
Expression constructs of 50-kDa proenzyme, 18-kDa propeptide, or control plasmid were transfected into RS485 cells and geneticin-resistant clones were selected. Two of the 18 resistant clones obtained with the proenzyme construct expressed a 50-kDa protein (Fig. 7), and each of these had a 99% reduction in colony formation in soft agar as compared to RS485. The colonies that did form were significantly smaller than control RS485 colonies, and were similar in size to the few colonies that are observed with NIH 3T3 (95% reduction in colony formation compared to RS485). Revertant LO50–3 exhibits two extra bands, of about 35- and 40-kDa in cytoplasmic extracts. In ras-revertants generated by both IFN-RA treatment and by stable transfection with proLOX expression clones, we consistently see these extra bands in the cytoplasm along with the restoration of 50-kDa proLOX. Thus, we do not believe these bands are an artifact of either treatment. The extra bands disappear, along with 50-kDa proLOX, when revertants are retransformed by treatment with 5-azacytidine (T.J. Yeh, unpublished results).
Fig. 7.
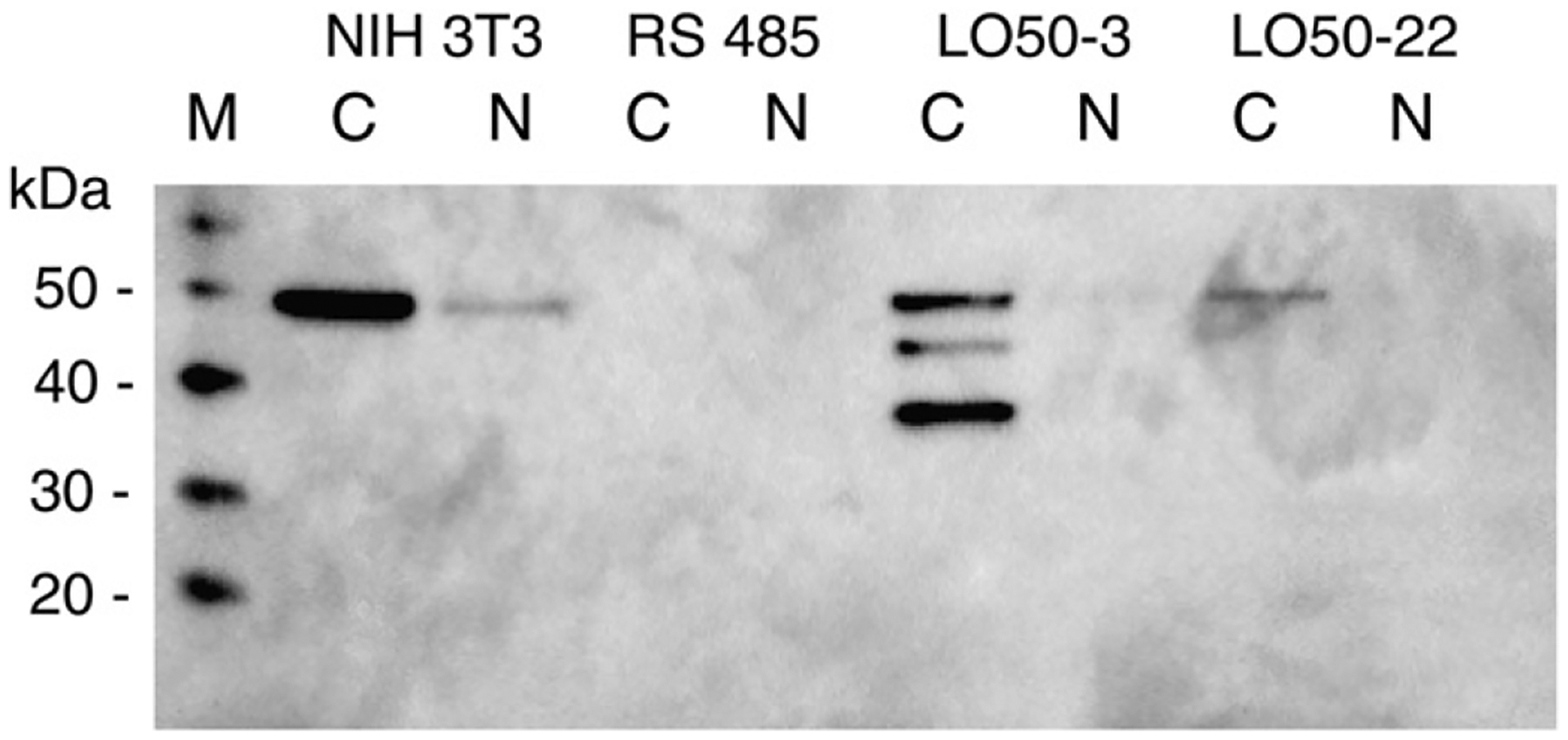
Expression of proenzyme in cells reverted by transfection. Cytoplasmic (C) and nuclear (N) extracts of non-transformed, ras-transformed, and reverted lines LO50–3 and LO50–22 (reverted by transfection with proenzyme expression construct), were probed with anti-LOpep4. M, protein size markers.
Five other proenzyme clones had a faint signal for 50-kDa protein, but colony formation in soft agar for these clones was unaffected compared to RS485. There was no free 18-kDa propeptide detected in any of the clones created with the proenzyme construct. Of 27 resistant lines obtained with propeptide construct, none expressed an 18-kDa product. Four randomly chosen clones from this group showed no reduction of colony formation in soft agar. All 13 control plasmid clones were negative for 50-kDa or 18-kDa products, and none of the four randomly chosen control clones had reduced colony formation in soft agar.
4. Discussion
LOX functions both as an enzyme that cross links extracellular matrix proteins and as a tumor suppressor. Recent work suggests that the tumor suppressor function of LOX is exerted inside the cell, most likely in the nucleus. Our studies show that only the 50-kDa proenzyme is present in cytoplasmic and nuclear extracts of primary mouse fibroblasts and cell line NIH 3T3, and that, in contrast to previously published results [17,18,26], mature LOX and free 18-kDa propeptide are not found within cells.
The lack of proLOX in the ras-transformed cell line, the reappearance of this protein in mouse revertant lines induced by interferon-retinoic acid combination treatment, and the lack of mature LOX or LOX propeptide within non-transformed cells strongly suggest that it is the proenzyme form of the protein that is responsible for reversion. The 18-kDa propeptide region of LOX, whether supplied as a bacterially produced protein, or introduced into cells as an expression construct, was shown to inhibit transformed cell growth and tumor formation [24,25]. While we were able to generate two lines of transformed RS485 cells expressing 50-kDa proenzyme, with resultant inhibition of cell growth in soft agar, none of the cell lines transfected with the 18-kDa propeptide construct expressed the propeptide protein. Therefore, we were unable to verify that the propeptide is sufficient for inhibiting cell growth in ras-transformants.
We did not detect the 18-kDa propeptide as a discrete entity in either cytoplasmic or nuclear extracts of primary cells or non-transformed cell lines using our propeptide antibody anti-LOpep4. This antibody was successful in detecting 50-kDa proLOX in cell extracts, as well as in vitro translated proLOX or 18-kDa propeptide, and an 18-kDa propeptide clone expressed in bacteria. Propeptide antibody anti-LOX41568, raised to a different epitope, also detected the in vitro translated LOX products as expected. However, while this antibody detects 50-kDa proLOX in non-transformed cells, it also detects an 18-kDa protein in non-transformed and transformed cells. RNA interference for LOX reduced the amount of proLOX, but not the amount of 18-kDa protein detected with anti-LOX41568, indicating that the 18-kDa protein recognized by anti-LOX41568 is not LOX propeptide. Thus, we believe that the failure of our propeptide region antibody, anti-LOpep4, to detect 18-kDa propeptide in these cell extracts reflects the absence of 18-kDa propeptide in these extracts. Accordingly, while the tumor suppressor activity in proLOX can be mapped to the propeptide region, it is the entire 50-kDa proLOX, not a separate 18-kDa propeptide fragment, which functions in the nucleus as a growth regulator when ras-transformed mouse fibroblasts are reverted by restoration of lysyl oxidase gene expression. The 18-kDa protein did appear as a doublet in RS485 and LOX shRNA clones, so it is possible that expression of this protein is related to the reduced expression of LOX.
A 30-kDa cytoplasmic protein that was recognized by antibody to mature LOX N-terminus, and a 32-kDa protein recognized by antibody to a central LOX epitope were both abundant in ras-transformed cells as well as in non-transformed cells. This suggested that neither of these two small proteins was mature LOX, as ras-transformed cells are transcriptionally down regulated for LOX expression, have no LOX enzymatic activity, and do not express proenzyme. RNA interference studies confirmed that 30-kDa cytoplasmic protein is not derived from the LOX gene. The 30-kDa protein was upregulated in the LOX shRNA clones. The 30-kDa, 32-kDa and 18-kDa proteins are abundantly expressed in non-transformed cells as well, and many proteins are upregulated after ras transformation. These proteins could be LOX related; each certainly contains an epitope that reacts with one of the LOX antibodies. But, at present, it is clear only that these proteins are not derived from LOX mRNA.
We have observed consistently that the mature region antibodies do not bind proLOX as well as do the propeptide region antibodies. We have determined that both mature region antibodies and our propeptide antibody (LOpep4) do bind to the same 50-kDa proLOX protein (Fig. 6). As each mature region antibody also cross-reacts with a different cellular protein, we do not plan to use these preparations for any future in situ studies of LOX. We do hope to develop a completely specific antibody for mature LOX.
The finding that mature LOX is not an intracellular protein underscores the importance of using independent antisera to at least two epitopes for unambiguous identification of proteins. Previous conclusions about the presence and role of mature LOX and free propeptide within cells should be evaluated in light of the findings presented here. The three small, non-LOX proteins are abundant in transformed cell lines, and each binds with one of three anti-LOX preparations. In situ studies using such preparations could lead to incorrect conclusions about the expression of LOX or propeptide in tumor tissues.
Acknowledgements
This work was supported by USUHS grant C074OL (SC).
References
- [1].Kagan HM, Characterization and regulation of lysyl oxidase, in: Mecham RP (Ed.), Biology of extracellular matrix: a series, Regulation of matrix accumulation, vol. 1, Academic Press, Orlando, 1986, pp. 321–398. [Google Scholar]
- [2].Wakasaki H, Ooshima A, Synthesis of lysyl oxidase in experimental hepatic fibrosis, Biochem Biophys Res Commun 166 (1990) 1201–1204. [DOI] [PubMed] [Google Scholar]
- [3].Kuivaniemi H, Peltonen L, Kivirikko KI, Type IX Ehlers-Danlos syndrome and Menkes syndrome: the decrease in lysyl oxidase activity is associated with a corresponding deficiency in the enzyme protein, Am J Hum Genet 37 (1985) 798–808. [PMC free article] [PubMed] [Google Scholar]
- [4].Kemppainen R, Hamalainen ER, Kuivaniemi H, Tromp G, Pihlajaniemi T, Kivirikko KI, Expression of mRNAs for lysyl oxidase and type III procollagen in cultured fibroblasts from patients with the Menkes and occipital horn syndromes as determined by quantitative polymerase chain reaction, Arch Biochem Biophys 328 (1996) 101–106. [DOI] [PubMed] [Google Scholar]
- [5].Barrow MV, Simpson CF, Miller EJ, Lathyrism: a review, Q Rev Biol 49 (1974) 101–128. [DOI] [PubMed] [Google Scholar]
- [6].Contente S, Kenyon K, Rimoldi D, Friedman RM, Expression of gene rrg is associated with reversion of NIH 3T3 transformed by LTR-c-H-ras, Science 249 (1990) 796–798. [DOI] [PubMed] [Google Scholar]
- [7].Kenyon K, Contente S, Trackman PC, Tang J, Kagan HM, Friedman RM, Lysyl oxidase and rrg messenger RNA, Science 253 (1991) 802. [DOI] [PubMed] [Google Scholar]
- [8].Peyrol S, Raccurt M, Gerard F, Gleyzal C, Grimaud JA, Sommer P, Lysyl oxidase gene expression in the stromal reaction to in situ and invasive ductal breast carcinoma, Am J Pathol 150 (1997) 497–507. [PMC free article] [PubMed] [Google Scholar]
- [9].Ren C, Yang G, Timme TL, Wheeler TM, Thompson TC, Reduced lysyl oxidase messenger RNA levels in experimental and human prostate cancer, Cancer Res 58 (1998) 1285–1290. [PubMed] [Google Scholar]
- [10].Woznick AR, Braddock AL, Dulai M, Seymour ML, Callahan RE, Welsh RJ, Chmielewski GW, Zelenock GB, Shanley CJ, Lysyl oxidase expression in bronchogenic carcinoma, Am J Surg 189 (2005) 297–301. [DOI] [PubMed] [Google Scholar]
- [11].Kaneda A, Wakazono K, Tsukamoto T, Watanabe N, Yagi Y, Tatematsu M, Kaminishi M, Sugimura T, Ushijima T, Lysyl oxidase is a tumor suppressor gene inactivated by methylation and loss of heterozygosity in human gastric cancers, Cancer Res 64 (2004) 6410–6415. [DOI] [PubMed] [Google Scholar]
- [12].Trackman PC, Bedell-Hogan D, Tang J, Kagan HM, Post-translational glycosylation and proteolytic processing of a lysyl oxidase precursor, J Biol Chem 267 (1992) 8666–8671. [PubMed] [Google Scholar]
- [13].Cronshaw AD, Fothergill-Gilmore LA, Hulmes DJ, The proteolytic processing site of the precursor of lysyl oxidase, Biochem J 306 (1995) 279–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Panchenko MV, Stetler-Stevenson WG, Trubetskoy OV, Gacheru SN, Kagan HM, Metalloproteinase activity secreted by fibrogenic cells in the processing of prolysyl oxidase. Potential role of procollagen C-proteinase, J Biol Chem 271 (1996) 7113–7119. [DOI] [PubMed] [Google Scholar]
- [15].Uzel MI, Scott IC, Babakhanlou-Chase H, Palamakumbura AH, Pappano WN, Hong HH, Greenspan DS, Trackman PC, Multiple bone morphogenetic protein 1-related mammalian metalloproteinases process pro-lysyl oxidase at the correct physiological site and control lysyl oxidase activation in mouse embryo fibroblast cultures, J Biol Chem 276 (2001) 22537–22543. [DOI] [PubMed] [Google Scholar]
- [16].Giampuzzi M, Botti G, Cilli M, Gusmano R, Borel A, Sommer P, Di Donato A, Down-regulation of lysyl oxidase-induced tumorigenic transformation in NRK-49F cells characterized by constitutive activation of ras proto-oncogene, J Biol Chem 276 (2001) 29226–29232. [DOI] [PubMed] [Google Scholar]
- [17].Li W, Nellaiappan K, Strassmaier T, Graham L, Thomas KM, Kagan HM, Localization and activity of lysyl oxidase within nuclei of fibrogenic cells, Proc Natl Acad Sci USA 94 (1997) 12817–12822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Jansen MK, Csiszar K, Intracellular localization of the matrix enzyme lysyl oxidase in polarized epithelial cells, Matrix Biol 26 (2007) 136–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Nellaiappan K, Risitano A, Liu G, Nicklas G, Kagan HM, Fully processed lysyl oxidase catalyst translocates from the extracellular space into nuclei of aortic smooth-muscle cells, J Cell Biochem 79 (2000) 576–582. [DOI] [PubMed] [Google Scholar]
- [20].Kagan HM, Williams MA, Calaman SD, Berkowitz EM, Histone H1 is a substrate for lysyl oxidase and contains endogenous sodium borotritide-reducible residues, Biochem Biophys Res Commun 115 (1983) 186–192. [DOI] [PubMed] [Google Scholar]
- [21].Giampuzzi M, Oleggini R, Di Donato A, Demonstration of in vitro interaction between tumor suppressor lysyl oxidase and histones H1 and H2: definition of the regions involved, Biochim Biophys Acta 1647 (2003) 245–251. [DOI] [PubMed] [Google Scholar]
- [22].Giampuzzi M, Botti G, Di Duca M, Arata L, Ghiggeri G, Gusmano R, Ravazzolo R, Di Donato A, Lysyl oxidase activates the transcription activity of human collagene III promoter. Possible involvement of Ku antigen, J Biol Chem 275 (2000) 36341–36349. [DOI] [PubMed] [Google Scholar]
- [23].Di Donato A, Lacal JC, Di Duca M, Giampuzzi M, Ghiggeri G, Gusmano R, Micro-injection of recombinant lysyl oxidase blocks oncogenic p21-Ha-Ras and progesterone effects on Xenopus laevis oocyte maturation, FEBS Lett 419 (1997) 63–68. [DOI] [PubMed] [Google Scholar]
- [24].Palamakumbura AH, Jeay S, Guo Y, Pischon N, Sommer P, Sonenshein GE, Trackman PC, The propeptide domain of lysyl oxidase induces phenotypic reversion of ras-transformed cells, J Biol Chem 279 (2004) 40593–40600. [DOI] [PubMed] [Google Scholar]
- [25].Min C, Kirsch KH, Zhao Y, Jeay S, Palamakumbura AH, Trackman PC, Sonenshein GE, The tumor suppressor activity of the lysyl oxidase propeptide reverses the invasive phenotype of Her-2/neu-driven breast cancer, Cancer Res 67 (2007) 1105–1112. [DOI] [PubMed] [Google Scholar]
- [26].Guo Y, Pischon N, Palamakumbura AH, Trackman PC, Intracellular distribution of the lysyl oxidase propeptide in osteoblastic cells, Am J Physiol Cell Physiol 292 (2007) C2095–2102. [DOI] [PubMed] [Google Scholar]
- [27].Yeh TJ, Contente S, Friedman RM, Transformation of revertant murine cells by 5-azacytidine results in rapid inhibition of lysyl oxidase expression, Acta Microbiol Immunol Hung 52 (2005) 433–442. [DOI] [PubMed] [Google Scholar]
- [28].Kenyon K, Modi WS, Contente S, Friedman RM, A novel human cDNA with a predicted protein similar to lysyl oxidase maps to chromosome 15q24-q25, J Biol Chem 268 (1993) 18435–18437. [PubMed] [Google Scholar]
- [29].Saito H, Papaconstantinou J, Sato H, Goldstein S, Regulation of a novel gene encoding a lysyl oxidase-related protein in cellular adhesion and senescence, J Biol Chem 272 (1997) 8157–8160. [DOI] [PubMed] [Google Scholar]
- [30].Jang W, Hua A, Spilson SV, Miller W, Roe BA, Meisler MH, Comparative sequence of human and mouse BAC clones from the mnd2 region of chromosome 2p13, Genome Res 9 (1999) 53–61. [PMC free article] [PubMed] [Google Scholar]
- [31].Maki JM, Kivirikko KI, Cloning and characterization of a fourth human lysyl oxidase isoenzyme, Biochem J 355 (2001) 381–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Maki JM, Tikkanen H, Kivirikko KI, Cloning and characterization of a fifth human lysyl oxidase isoenzyme: the third member of the lysyl oxidase-related subfamily with four scavenger receptor cysteine-rich domains, Matrix Biol 20 (2001) 493–496. [DOI] [PubMed] [Google Scholar]
- [33].Ito H, Akiyama H, Iguchi H, Iyama K, Miyamoto M, Ohsawa K, Nakamura T, Molecular cloning and biological activity of a novel lysyl oxidase-related gene expressed in cartilage, J Biol Chem 276 (2001) 24023–24029. [DOI] [PubMed] [Google Scholar]
- [34].Chang EH, Furth ME, Scolnick EM, Lowy DR, Tumorigenic transformation of mammalian cells induced by a normal human gene homologous to the oncogene of Harvey murine sarcoma virus, Nature 297 (1982) 479–483. [DOI] [PubMed] [Google Scholar]
- [35].Friedman RM, Yeh A, Gutman P, Contente S, Kenyon K, Reversion by deletion of transforming oncogene following interferon-beta and retinoic acid treatment, J Interferon Cytokine Res 17 (1997) 647–651. [DOI] [PubMed] [Google Scholar]
- [36].Li PA, He Q, Cao T, Yong G, Szauter KM, Fong KS, Karlsson J, Keep MF, Csiszar K, Up-regulation and altered distribution of lysyl oxidase in the central nervous system of mutant SOD1 transgenic mouse model of amyotrophic lateral sclerosis, Brain Res Mol Brain Res 120 (2004) 115–122. [DOI] [PubMed] [Google Scholar]
- [37].Hayashi K, Fong KS, Mercier F, Boyd CD, Csiszar K, Hayashi M, Comparative immunocytochemical localization of lysyl oxidase (LOX) and the lysyl oxidase-like (LOXL) proteins: changes in the expression of LOXL during development and growth of mouse tissues, J Mol Histol 35 (2004) 845–855. [DOI] [PubMed] [Google Scholar]
- [38].Payne SL, Fogelgren B, Hess AR, Seftor EA, Wiley EL, Fong SF, Csiszar K, Hendrix MJ, Kirschmann DA, Lysyl oxidase regulates breast cancer cell migration and adhesion through a hydrogen peroxide-mediated mechanism, Cancer Res 65 (2005) 11429–11436. [DOI] [PubMed] [Google Scholar]
- [39].Contente S, Csiszar K, Kenyon K, Friedman RM, Structure of the mouse lysyl oxidase gene, Genomics 16 (1993) 395–400. [DOI] [PubMed] [Google Scholar]


