Abstract
The severe acute respiratory syndrome coronavirus (SARS-CoV)-2 responsible for the global COVID-19 pandemic has caused almost 760 million confirmed cases and 7 million deaths worldwide, as of end-February 2023. Since the beginning of the first COVID-19 case, several virus variants have emerged: Alpha (B1.1.7), Beta (B135.1), Gamma (P.1), Delta (B.1.617.2) and then Omicron (B.1.1.529) and its sublineages. All variants have diversified in transmissibility, virulence, and pathogenicity. All the newly emerging SARS-CoV-2 variants appear to contain some similar mutations associated with greater "evasiveness" of the virus to immune defences. From early 2022 onward, several Omicron subvariants named BA.1, BA.2, BA.3, BA.4, and BA.5, with comparable mutation forms, have followed. After the wave of contagions caused by Omicron BA.5, a new Indian variant named Centaurus BA.2.75 and its new subvariant BA.2.75.2, a second-generation evolution of the Omicron variant BA.2, have recently been identified. From early evidence, it appears that this new variant has higher affinity for the cell entry receptor ACE-2, making it potentially able to spread very fast. According to the latest studies, the BA.2.75.2 variant may be able to evade more antibodies in the bloodstream generated by vaccination or previous infection, and it may be more resistant to antiviral and monoclonal antibody drug treatments. In this manuscript, the authors highlight and describe the latest evidences and critical issues have emerged on the new SARS-CoV-2 variants.
Keywords: SARS-CoV-2, Omicron, Variants, Vaccines, COVID-19, Antiviral drugs, Epidemiology, Monoclonal antibodies, Mutations, Pandemic, Spike protein, Public health, Coronavirus, B.1.1.529, VOC, VOI
Introduction
SARS-CoV-2 variants
SARS-CoV-2 is responsible for the current ongoing global pandemic of coronavirus disease 2019 (COVID-19) (https://www.who.int/emergencies/diseases/novel-coronavirus-2019). SARS-CoV-2 is a member of the Coronavirus (CoV) family. Coronaviruses (CoVs) are a large family of respiratory viruses with an envelope and a genome consisting of single-stranded positive-stranded RNA that can cause mild to moderate illness, from the common cold to severe respiratory syndromes (Peeri et al. 2020; Zhu et al. 2020). Coronaviruses are common in many animal species (such as camels and bats) but in some cases, albeit rarely, they can evolve and infect humans. SARS-CoV-2 penetrates the host cell through fusion of the viral envelope with the cell membrane or through membrane fusion within the endosome after endocytosis (Petrovszki et al. 2022). One of the major receptors for endocellular penetration utilized by SARS-CoV-2 is angiotensin-converting enzyme 2 (ACE2), which is widely expressed in pulmonary pneumocytes, intestinal cells, and myocardiocytes. COVID-19 disease can be asymptomatic, mild, severe to cause patient death (Balse and Hatem 2021). Typical symptoms of COVID-19 such as dyspnoea, fever, cough, and fatigue can be followed by numerous complications including pneumonia, myocarditis, and kidney injury, resulting in some cases of death (Rosa et al. 2021). To avoid a larger number of deaths caused by COVID-19, a tremendous effort has been made throughout the scientific world to make vaccines and antiviral treatments available as quickly as possible (Vitiello and Ferrara 2021; Vitiello et al. 2021). COVID-19 was declared a global pandemic by the World Health Organization (WHO) on March 11, 2020. As of 28 February 2023, data recorded by the WHO indicate 758. 390.564 confirmed cases, 6.859.093 associated deaths, and over 13.228 million vaccine doses administered (Table 1) (https://www.who.int/emergencies/diseases/novel-coronavirus-2019).
Table 1.
Confirmed SARS-CoV-2 cases, associated deaths, and COVID-19 vaccine doses administered in the WHO Regions, 28 February 2023
| WHO Region | Cumulative cases (%) | Cumulative deaths (%) | COVID-19 vaccine doses administered (%) |
|---|---|---|---|
| Europe | 273.261.618 (36) | 2.193.638 (32) | 1.709.075.705 (13) |
| Western Pacific | 201.263.891 (27) | 405.481 (6) | 4.658.304.367 (35) |
| Americas | 190.340.817 (25) | 2.931.281 (43) | 2.099.825.828 (16) |
| South-East Asia | 60.767.139 (8) | 803.851 (12) | 3.314.003.488 (25) |
| Eastern Mediterranean | 23.258.662 (3) | 349.534 (5) | 877.196.471 (7) |
| Africa | 9.497.673 (1) | 175.295 (3) | 570.207.767 (4) |
| Global | 758.390.564 (100) | 6.859.093 (100) | 13.228.613.626 (100) |
Over one third of the cases reported worldwide was registered in the WHO Europe Region (36%), followed by the Western Pacific (27%) and the Americas (25%) Regions. The Europe Region also contributed to over one third (32%) of the COVID-19 associated deaths, while the Americas registered almost half (43%) of the cumulative number of deaths occurred globally. The Western Pacific was the WHO Region with the highest number of COVID-19 vaccine doses administered, together with the South-East Asia Region (25%) (Table 1). Since the beginning of the first detected positive SARS-CoV-2 cases, numerous variants of the virus have occurred and alternated: Alpha (B1.1.7), Beta (B135.1), Gamma (P.1), Delta (B.1.617.2) and now Omicron (B.1.1.529) and subvariants. On May 31, 2021, the WHO announced easy-to-say labels for SARS-CoV-2 variants of concern (VOC) and variants of interest (VOI) that do not concern their origin. However, the established nomenclature systems for naming and tracking SARS-CoV-2 genetic lineages by GISAID, Nextstrain and Pango are still used by scientists. VOC are all those variants of SARS-CoV-2 with clear evidence suggesting a possible significant impact on the severity of COVID-19 and consequently on the epidemiological situation, while VOI are all the variants with still preliminary evidence and high uncertainty indicating that they could imply a significant impact on the transmissibility and severity of COVID-19. Most mutations in the various SARS-CoV-2 variants have had minimal impact on certain properties of the virus, such as ease of spread, transmissibility, associated disease severity, virulence, pathogenicity, associated vaccine antibody response, and efficacy of antiviral treatments (Vitiello et al. 2022). From early 2022 onward, several BA.1, BA.2, BA.3, BA.4, and BA.5 subvariants with very similar mutation forms have followed one another. After the wave of contagions caused by Omicron BA.5, a new Indian variant named Centaurus BA.2.75 and its new subvariant BA.2.75.2, a second-generation evolution of Omicron variant BA.2, have recently been identified (Shaheen et al. 2022). From early evidence, it appears that this new variant has higher affinity for the cell entry receptor ACE-2 making it potentially able to spread very fast. According to the latest studies, the BA.2.75.2 variant may be able to evade more antibodies in the bloodstream generated by vaccination or previous infection, and it may be more resistant to antiviral and monoclonal antibody drug treatments. Recently, updated mRNA vaccines were licensed against subvariants BA.4, and BA.5 (https://www.ema.europa.eu/en/human-regulatory/overview/public-health-threats/coronavirus-disease-covid-19/covid-19-latest-updates). The new updated mRNA vaccine helps to create a broader immune response and improve the strength and duration of protection against circulating variants. Epidemiologic studies are underway to test whether the new SARS-CoV-2 subvariants may have greater transmissibility and pathogenicity than their predecessor variants. In addition, epidemiological evidence is showing that SARS-CoV-2 infection is associated with a range of persistent symptoms impacting everyday functioning, known as post-COVID-19 condition or long COVID (Han et al. 2022). What might be the impact of infection with the new SARS-CoV-2 subvariants BA.4, and BA.5 in an individual with post-COVID-19 condition or long COVID symptoms? Are the licensed SARS-CoV-2 antivirals, such as remdesivir, PF-07321332/ritonavir, molnupiravir, equally effective against the new subvariants? In the epidemiological week 6 to 12 February 2023, pooled recombinant variant sequences accounted for 41.5% of all the collected sequences, XBB.1.5 accounting for 32.6%, BA.5 and its descendent lineages 31.8%, BA.2 and its descendent lineages 13.7%, XBF 1.2%. In the same week, Omicron BA.1, BA.3 and BA.4 variants and their descendent lineages all accounted for < 1% prevalence; unassigned sequences (all presumably Omicron awaiting descendent lineage assignment) accounted for 12.9% of the shared sequences (https://www.ecdc.europa.eu/en/covid-19/country-overviews).
Omicron subvariants
In recent months, multiple lineages of the omicron SARS-CoV-2 variant have been identified globally, i.e. the BA.1 and BA.2 subvariants, the BA.2.12.1 subvariant which is the dominant strain in the United States currently, BA.4 and BA.5 are dominant in South Africa, and lastly the Centaurus BA2.75 and BA2.75.2 subvariants. These variants can partially evade the antibody response induced by licensed vaccines (Cele et al. 2022; Liu et al. 2022). A country overview of the prevalence of several Omicron variants assigned as VOC or VOI in EU/EEA countries, and other variants, registered in epidemiological week 8, 2023, is shown in Table 2 (European Centre and for Disease Prevention and Control 2023a).
Table 2.
Prevalence (%) of the Omicron variants and others detected in the EU/EEA countries, epidemiological week 8, 2023 (European Centre and for Disease Prevention and Control 2023a)
| Country | SARS-CoV-2 circulating variants in EU/EEA (%) | |||||||
|---|---|---|---|---|---|---|---|---|
| BQ.1 | XBB.1.5 | BA.2.75 | BA.5 | BA.2 | XBB | BA.4 | Other | |
| Austria | 23.6 | 45.5 | 22.2 | 4.6 | 1.2 | 2.9 | 0.1 | |
| Belgium | 44.1 | 25.1 | 22.7 | 3.4 | 4.7 | |||
| Bulgaria | ||||||||
| Croatia | 48.9 | 6 | 14.9 | 27.3 | 0.9 | 1.4 | 0.6 | |
| Cyprus | ||||||||
| Czechia | 15 | 70 | 10 | 5 | ||||
| Denmark | 22.9 | 33.6 | 35.5 | 6.6 | 0.6 | 0.8 | ||
| Estonia | 15.4 | 8.5 | 29.9 | 19.7 | 26.5 | |||
| Finland | 53.6 | 16.3 | 6 | 10.2 | 6.6 | 7.2 | ||
| France | 48.4 | 33.9 | 10.4 | 4.9 | 0.3 | 2 | ||
| Germany | 23.2 | 52.1 | 24.6 | 0.1 | ||||
| Greece | 43.2 | 7.9 | 24.5 | 8.6 | 15.1 | 0.7 | ||
| Hungary | 22.5 | 32.5 | 45 | |||||
| Iceland | 4.3 | 52.2 | 43.5 | |||||
| Ireland | 12.9 | 46.9 | 27.1 | 4.4 | 0.2 | 8.5 | ||
| Italy | 54.9 | 20.3 | 11.2 | 10.4 | 1.2 | 1.7 | 0.1 | 0.1 |
| Latvia | 33.8 | 64.4 | 1.6 | 0.2 | ||||
| Liechtenstein | ||||||||
| Lithuania | ||||||||
| Luxembourg | 30 | 35 | 11.2 | 7.5 | 14.2 | 2.1 | ||
| Malta | 100 | |||||||
| The Netherlands | 30.3 | 30.5 | 20.3 | 5.3 | 0.6 | 9.6 | 0.4 | 3 |
| Norway | 18.8 | 12.5 | 31.2 | 6.2 | 25 | 6.2 | ||
| Poland | 48.5 | 14.9 | 17.9 | 15.7 | 0.7 | 2.2 | ||
| Portugal | 48.3 | 24.1 | 27.6 | |||||
| Romania | 34.3 | 2 | 12.6 | 31.8 | 16.2 | 2 | 1 | |
| Slovakia | ||||||||
| Slovenia | 38.2 | 11.8 | 35.3 | 14.7 | ||||
| Spain | 38.5 | 49.3 | 8.8 | 2 | 0.7 | 0.7 | ||
| Sweden | 22.7 | 15.8 | 10 | 30.4 | 12.1 | 8.9 | ||
The VOC (Table 3) and VOI (Table 4) circulating at the global level are listed in the tables below, together with the mutation sites identified in the spike protein of each variant, the date of first detection, and the estimated impact on transmissibility, immunity and severity of disease.
Table 3.
SARS-CoV-2 Variants of Concern (VOC) and spike mutations of interest identified globally, as of 23 February 2023 (https://www.ecdc.europa.eu/en/covid-19/country-overviews)
| WHO label | Lineage | Spike mutations of interest | Date of first detection | Impact on | ||
|---|---|---|---|---|---|---|
| Transmissibility | Immunity | Severity | ||||
| Omicron | BA.2 | G142D, N211I, Δ212, V213G, G339D, S371F, S373P, S375F, T376A, D405N, R408S, K417N, N440K, S477N, T478K, E484A, Q493R, Q498R, N501Y, Y505H, D614G, H655Y, N679K, P681H, N764K, D796Y, Q954H, N969K | November 2021 | Increased | Increased | Reduced |
| Omicron | BA.4 | L452R, F486V, R493Q | January 2022 | No evidence | Increased | No evidence |
| Omicron | BA.5 | L452R, F486V, R493Q | February 2022 | No evidence | Increased | Unclear |
Table 4.
SARS-CoV-2 Variants of Interest (VOI) and spike mutations identified globally, as of 23 February 2023 (https://www.ecdc.europa.eu/en/covid-19/country-overviews)
| WHO label | Lineage + additional mutations | Spike mutations of interest | Year and month first detected | Impact on | ||
|---|---|---|---|---|---|---|
| Transmissibility | Immunity | Severity | ||||
| Omicron | BA.2.75 | May 2022 | Unclear | Similar to baseline | No evidence | |
| Omicron | BQ.1 | K444T, N460K | Not available | Baseline | Baseline | Baseline |
| Omicron | XBB | N460K, F490S | Not available | Similar to baseline | Increased | No evidence |
| Omicron | XBB.1.5 | N460K, S486P, F490S | Not available | Increased | Increased | Similar to baseline |
BA.2
The Delta variant of SARS-CoV-2 was rapidly replaced by the Omicron variant and subvariants due to natural selection. Several lineages, BA.1, BA.2, have resulted from the Omicron variate. Omicron subvariant BA.2 shares 32 mutations with BA.1 and 28 different ones and is about 1.5 and 4.2 times more infectious than BA.1 and Delta, respectively. Epidemiological data immediately indicated a greater transmissibility and spread of BA.2 than BA.1 (European Centre and for Disease Prevention and Control 2023a). Molecular studies showed 31 amino acid changes in the spike region compared to the reference strain Wuhan/Hu-1/2019, specifically 7 changes in the N-terminal domain (NTD), including substitutions and deletions (T19L, Δ24-27S, G142D, V213G); 16 substitutions in the receptor-binding domain (RBD) (G339D, S371F, S373P, S375F, T376A, D405N, R408S, K417N, N440K, S477N, T478K, E484A, Q493R, Q498R, N501Y and Y505H), 3 substitutions near the furin cleavage site (H655Y, N679K and P681H), and 4 substitutions in the S2 region (N764K, D796Y, Q954H and N969K) (Lyngse et al. 2022). Studies have shown that in the face of greater immune evasiveness, the BA.2 variant is comparable to BA.1 in terms of pathogenicity (Kawaoka et al. 2022; Halfmann et al. 2022; Shuai et al. 2022). However, an interesting study showed that in a paediatric population sample, Omicron SARS-CoV2 BA.2 can cause severe illness in unvaccinated and hospitalised children who have had low exposure to previous SARS-CoV-2 variants in the past 2 years (Suzuki et al. 2022). However, evidence has shown no difference in severity for individuals infected with subvariant BA.2 and BA.1 Omicron, considering the risk of severe clinical outcomes and length of hospital stay, indicating that the reduced COVID-19 severity associated with the Omicron variant compared to the Delta variant persisted after the emergence and establishment of the BA.2 subvariant (Tso et al. 2022).
BA.4/BA.5
The two lineages BA.4 and BA.5 of the Omicron variant were first identified in South Africa (Lewnard et al. 2022) and then rapidly spread to different countries around the world. At the end of 2022, according to data from the WHO and the European Centre for disease prevention and control (ECDC), and as also shown in Table 1, subvariants BA.4 and BA.5 are the most widespread in EU/EEA countries. Variants BA.4 and BA.5 caused a surge in COVID-19 positive cases in South Africa, rapidly replacing the previous variants BA.1 and BA.2, despite the large number of people vaccinated and already recovered from a previous COVID-19 infection. However, despite the surge in cases, no increase in the number of deaths has been reported (Callaway 2022a). From a molecular point of view, the BA.4 and BA.5 variants have spike region mutations at positions L452R, F486V, R493Q, as shown in Table 2. These mutations are responsible for the increased spread and transmissibility of the virus. In particular, the spike region mutation L452R is responsible for the increased transmissibility because it promotes both the viral attachment to the human cells and mechanisms of viral escape (European Centre and for Disease Prevention and Control 2023b). In addition, the spike region mutation F486V could help the variants to escape the human immune system. The symptoms and severity of COVID-19 caused by subvariants BA.4 and BA.5 are not yet fully known. However, it is important to emphasise that the evidence does not show an increase in the number of deaths caused by these subvariants. Underlying this may probably be several factors such as mass vaccination and natural immunity resulting from a large Omicron wave (Islam et al. 2022). In addition to the assumption of increased immune evasion by BA.4 and BA.5, an interesting study showed that these subvariants substantially evade neutralising antibodies induced by both vaccination and infection, and that furthermore, neutralising antibody titres against subvariant BA.4 or BA.5 are lower than the titres against subvariants BA.1 and BA.2, suggesting that the omicron variant of SARS-CoV-2 has continued to evolve with increasing escape ability from neutralisation. All of these described findings may explain the spikes caused by subvariants BA.4 and BA.5 in populations with high frequencies of vaccination and previous BA.1 or BA.2 infection (Cao et al. 2022). However, in the face of increased immune evasion, the evidence does not demonstrate increased severity or mortality caused by BA.4 BA.5. infected individuals had a similar odd of hospitalisation and severe outcome compared to BA.1-infected individuals (Callaway 2022b). Probably the extremely high population immunity, and specifically the elicited T-cell responses, could explain the low severity observed despite the emergence of additional Omicron lineages with increased transmissibility and immune escape. An important aspect to consider is that the spike region mutations L452Q, L452R and F486V are located in the receptor binding domain (RBD) of the spike protein, the main target of monoclonal antibody therapies. To date, the efficacy of monoclonal antibodies against BA.4 and BA.5 subvariants is unknown. An interesting study that used live-virus focus reduction neutralisation testing (FRNT) showed that monoclonal antibody casirivimab lost neutralising activity against BA.4, and BA.5, instead imdevimab retained neutralising activity against these VOC (Hachmann et al. 2022; Takashita et al. 2022).
BA.2.75 and BA.2.75.2
SARS-CoV-2 Omicron variant BA.2.75 was first detected in India in May 2022, then it spread rapidly to other countries such as Japan, and United States (Wolter et al. 2022). Subsequently, on July 19, 2022, the WHO classified this variant as a VOC under close monitoring. BA.2.75 shows higher immunogenicity profile and virulence than other Omicron variants, and different susceptibility to anti-SARS-CoV-2 monoclonal antibodies. Compared to BA.2, BA.2.75 consists of 14 amino acid substitutions, including 9 substitutions in the protein S. In an interesting study, it was shown that, BA.2.75 exhibits an approximately 57-fold higher binding affinity to the ACE-2 membrane receptor compared to the BA.5 variant and an approximately 3000-fold higher receptor binding affinity compared to the Alpha variant (B.1.1.7) (Hirotsu and Omata 2023). Sheward and colleagues (Zappa et al. 2022) also showed that in the spike protein of Omicron subvariant BA.2.75 there are 9 additional mutations compared to variant BA.2 namely, G339H, G446S, G257S, I210V, F157L, R493Q, N460K, W152R, and K147E (Fig. 1).
Fig. 1.
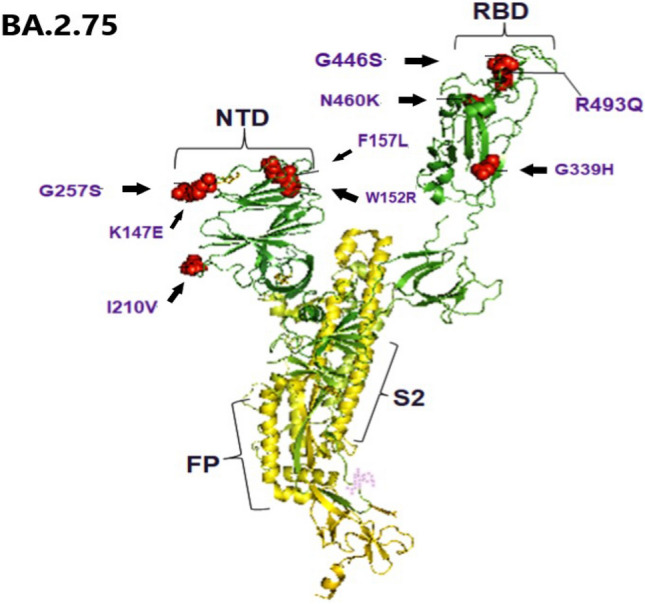
Illustration of the three-dimensional structure of the spike protein from SARS-CoV-2 Omicron subvariant BA.2.75. Nine additional mutations are highlighted in the spike protein subvariant BA.2.75 compared with the BA.2 variant, namely G339H, G446S, G257S, I210V, F157L, R493Q, N460K, W152R and K147E. RBD: receptor-binding domain; FP: N-terminal hydrophobic fusion peptide; S2 subunit of spike protein
In view of the mutations present at the spike protein, the target site of anti-SARS-CoV-2 monoclonal antibodies, and the antibody response induced by stimulation following administration of anti-COVID-19 mRNA vaccines, several studies have investigated the possible immune escape of the BA.2.75 with respect to the vaccine-induced antibody response, and with respect to anti-SARS-CoV-2 antibodies. Important evidence has shown that the use of the monoclonal antibody cilgavimab has an approximately 11-fold reduced efficacy and potency compared with the B.1 variant, while the monoclonal antibody tixagevimab showed overlapping potency. In addition, for the monoclonal antibody bebtelovimab, an approximately sevenfold reduction against BA.2.75 was demonstrated, while the first anti-SARS-CoV-2 monoclonal antibodies that came on the market, such as casivirimab and imdevimab, showed no neutralizing activity against BA.2.75 (Sheward et al. 2022a). Recently, it has been noted that the Omicron SARS-CoV-2 subvariant BA.2.75 is further mutating into a new subvariant called BA.2.75.2. The first cases of infection caused by this new subvariant BA.2.75.2 have been reported in India (Sheward et al. 2022b). Times and colleagues reported that this subvariant is more transmissible than the others and it is going to be established as the dominant Omicron subvariant. However, no scientific reports to this effect have been published to date. Currently, the BA.2.75.2 subvariant is present in eight countries, including Germany, India, Chile, England, Singapore, and Spain. Global monitoring of BA.2.75.2 is immediately needed, because as in the past few months Omicron subvariant BA.2.75 had replaced previous subvariants BA.4 and BA.5, now BA.2.75.2 appears to be relatively more transmissible and thus may have high chances of rapid spread in various countries (Times 2022). An important study was conducted to investigate resistance to serum antibodies and monoclonal antibody agents against the BA.2.75.2 subvariant. The study demonstrated significantly greater resistance of BA.2.75.2 than BA.2.75 against serum antibodies suggesting that it effectively evades current humoral immunity in the test population (Times 2022).
BQ.1
BQ.1 is a sublineage of BA.5, which carries spike mutations in some key antigenic sites, including K444T and N460K. Since 20 October 2022, ECDC have been designated BQ.1 and its sub-lineages as VOI. BQ.1 was monitored as it showed a significant spread advantage over other circulating Omicron sublineages in Europe and the US probably due to immune escape advantage over other circulating Omicron sublineages associated to those mutations. Therefore, the main warrant is a possible higher reinfection risk, while there is no epidemiologic data suggesting an increase in disease severity. Protection by vaccines (both the index and the recently introduced bivalent vaccines) against infection may be reduced, but no major impact on protection against the severe disease is evident so far.
XBB
XBB is a recombinant of BA.2.10.1 and BA.2.75 sublineages. Along with BQ.1 variants, XBB variants are the most antibody-resistant variants known. There has been a broad increase in prevalence of XBB in the global genomic surveillance, not associated consistently with an increase in new infections. The current data do not suggest there are substantial differences in disease severity for XBB infections. The prevalence of XBB, a subvariant of BA.2, is rapidly increasing in several countries, including the US and India However, early evidence suggests a higher reinfection risk, as compared to other circulating Omicron sublineages. Depending on the regional immune landscape as affected by the size and timing of previous Omicron waves, as well as the COVID-19 vaccination coverage, XBB variant could drive new infection waves. Moreover, the XBB variant has additional substitutions in the spike (S) protein receptor binding domain, which is the main target of vaccines and therapeutic monoclonal antibodies for coronavirus disease 2019 (COVID-19). These subvariants may therefore be more immune-evasive than BA.5 and BA.2. Recent evidence suggests that the Omicron XBB subvariant has superior immune evasion capabilities compared to earlier Omicron variants, including BA.5 and BA.2. This evidence reinforces the need to identify new therapeutic monoclonal antibodies for COVID-19.
XBB.1.5
The Omicron XBB.1.5 variant is a sublineage of XBB, which is a recombinant form of two BA.2 sublineages. Most of the recent sequences have been reported from the United States of America, but this variant is also described in the United Kingdom, Canada, Germany, Austria and Denmark. Based on the rapid increase in proportion in the north-east part of the United States of America, on 24 February 2023 the WHO’s Technical Advisory Group on Virus Evolution (TAG-VE) assessed that XBB.1.5 may contribute to increase case incidence, at least in the United States of America (Sheward et al. 2022c). Indeed, there is high-strength of evidence for increased risk of transmission XBB.1.5. Besides, it is shown to be equally immune evasive as XBB.1, and one of the Omicron subvariant with the highest immune escape reported, as of February 2023. So far there is limited data available globally, with a low number of cases associated with XBB.1.5 is still low in many countries. Severity assessments are ongoing, with no early signals of increased severity observed or any mutation known to be associated with potential change in severity. Therefore, the available information does not suggest that XBB.1.5 has additional public health risks compared to the Omicron variants circulating currently. Besides the VOC and the VOI, some more variants named Variants Under Monitoring (VUM) could have properties similar to a VOC and in the process to be assessed with evidence-based data. As of 23 February 2023, VUM include five Omicron variants: BA.2.3.20, BF.7, XBC, BN.1, CH.1.1. Moreover, XAY lineage has not been labelled by the WHO yet. The country where these variants were first detected is unknow, as well as data are missing regarding their impact on transmission, immunity and disease severity (https://www.ecdc.europa.eu/en/covid-19/country-overviews).
Discussion
SARS-CoV-2 continues to spread at the regional and global levels. As of 28 February 2023, the global cumulative incidence reached 758.390.564 confirmed cases and 6.859.093 associated deaths, despite 13.228.613.626 vaccine doses administered globally (WHO 2023a; Real-time dashboard 2019). The highest number of new 28-day deaths was reported from the United States of America (12 111 new deaths; − 17%), followed by China (5915 new deaths; − 91%), Japan (4818 new deaths; − 52%), Brazil (2186 new deaths; − 24%), and the UK (2027 new deaths; − 48%) (WHO 2023a; Real-time dashboard 2019). Despite this decreasing trend for both new cases and deaths, local and national prevalence surveys indicated that the number of reported COVID-19 cases have been underestimated in respect of the true burden of global infections and reinfections (WHO 2023b; Cohen et al. 2022; Coronavirus (COVID-19) Infection Survey, UK 2022). The general reduction in testing and delays in reporting in many countries contribute to this underestimation. Besides, different countries have distinct immune profiles because their histories of COVID-19 waves and vaccination rates differed. Moreover, most of the western countries are doing little to control SARS-CoV-2 spread currently, therefore the spread of subvariants is driven almost entirely by the level of population immunity. As of 26 February 2023, 60.559 SARS-CoV-2 sequences were shared through GISAID globally, in the last 28 days. Among these, 60.521 sequences (99.9%) were VOC; among them, over one third was represented by XBB.1.5 and another one third by BA.5 and its descendent lineages. As of 3 March 2023, Omicron BA.2, BA.4 and BA.5 variants are not circulating in the European Region. Therefore, ECDC and WHO stated that these variants are no longer considered VOC, reflecting the current improved epidemiological situation (WHO 2023d). Indeed, during the week ending on 26 February 2023, the hospital admission rate for the EU/EEA was 2.4 per 100.000 population (data reported by 15 countries, country range 0.0–10.0), compared to 2.9 (country range 0.0–8.9) in the previous week; the ICU admission rate in the EU/EEA was 0.2 per 100.000 population (data reported by 13 countries, country range 0.0–0.5), compared to 0.2 (country range 0.1–0.4) in the previous week; the 14-day COVID-19 death rate was 4.5 per million population (data reported by 25 EU/EEA countries, country range 0.0–16.9), compared to 5.5 (country range 0.0–19.6) in the previous week. The complexity of the molecular, biological, pathophysiological mechanisms and subsequent clinical manifestations of SARS-CoV-2 infection is still being studied and not fully understood. In addition, as already extensively described above, the continuous emergence of SARS-CoV-2 variant forms that differ from their predecessors in terms of virulence, transmissibility and pathogenicity makes the scenario even more complex. In parallel with all these considerations, there has been a continuous evolution of the most appropriate drug therapy to be used in the various stages of the disease, whether at home or in hospital. However, it should be emphasised that for some therapeutic interventions included in certain treatment schemes, clinical evidence is still being gathered to determine the most correct timing of administration. Numerous pharmacological therapeutic opportunities are currently available using monoclonal and antiviral antibodies directed against SARS-CoV-2. For both types of treatment, the greatest efficacy is observed with early administration with respect to the onset of symptoms, possibly within 72 h (Parikh et al. 2020). Currently, the indication for therapy with both monoclonal antibodies and antivirals is to individuals with mild-to-moderate disease who are not hospitalised and not on oxygen therapy and who have risk factors for the development of severe COVID-19. Authorised antivirals for the treatment of COVID-19 include remdesivir, paxlovid (nirmatrelvir + ritonavir); monoclonal antibodies include bamlanivimab, etesevimab, casirivimab, imdevimab, sotrovimab, tixagevimab and cilgavimab (Ferrara et al. 2021). Two immunomodulators baricitinib and tocilizumab are also approved for the treatment of COVID-19 infection. However, research efforts to find new active and effective compounds or to extend the use of compounds already on the market for other indications are continuing (Conti et al. 2021; Jorgensen et al. 2020). To date, numerous clinical trials are underway to identify new compounds with antiviral activity against SARS-CoV-2 or capable of managing the hyperinflammatory state of the infection. Among the numerous molecules in trials for the treatment of COVID-19, masitinib is an oral tyrosine kinase inhibitor. Recent studies have shown the anti-severe SARS-CoV-2 activity of masitinib through inhibition of the major protease enzyme (Mpro), an important pharmacological target for blocking coronavirus replication. Masitinib analogues are also being tested to identify more tolerable compounds (Mouffak et al. 2021). Other innovative compounds being tested are inhibitors of transmembrane serine protease 2 (TMPRSS2). This protein is an important mediator in SARS-CoV-2 viral infection. Other innovative compounds under investigation are inhibitors of transmembrane serine protease 2 (TMPRSS2) (Gurung et al. 2023; Mantzourani et al. 2022). This protein is an important mediator of SARS-CoV-2 viral infection. SARS-CoV-2 uses ACE-2 and TMPRSS2 for endocellular penetration. Currently, the main drugs targeting TMPRSS2 tested against COVID-19 are camostat mesylate and nafamostat mesylate. Numerous in vitro experiments confirm their efficacy against SARS-CoV-2 infection. In addition, several inhibitors of the viral RNA-dependent RNA polymerase (RdRp) of SARS-CoV-2 are under investigation. RdRp plays a central role in viral replication and is therefore an important target for the development of antiviral approaches (Hoffmann et al. 2020). Moreover, it must be remembered that the lack of a solid rationale and the absence of evidence of efficacy in the treatment of patients with SARS-CoV-2 viral infection alone do not allow the use of antibiotics to be recommended, either alone or in combination with other drugs. Unjustified use of antibiotics may, moreover, lead to the emergence and spread of antimicrobial resistance that could compromise the response to the future therapies.
WHO and the international competent authorities are still monitoring variants to detect signals of potential VOC or VOI and to assess the risk posed to global public health. National authorities can designate other variants of local concern or interest. Public health measures that can reduce transmission through scientific and proven disease control strategies, including infection prevention and control (IPC) measures, as well as avoid introduction into animal populations, are crucial to reduce the occurrence of mutations that can impact the health of the general public and all the human activities. The WHO continues to recommend to strengthen epidemiologic and microbiologic surveillance and sequencing capacities.
Conclusions
Despite over 13 billion of COVID-19 vaccine doses administered, SARS-CoV-2 infections and deaths continue to impact human health and activities globally. The main health indicators improved in the last 28 days in all the six WHO regions, except for the Eastern Mediterranean Region where − 22% of cases, but + 18% of deaths, have been registered. However, the continued emergence of SARS-CoV-2 variants requires constant close monitoring and assessment of their virulence, transmissibility and pathogenicity. Recently, WHO updated the COVID-19 guidelines on masks, treatments and clinical management (WHO 2023c). On the other hand, a number of pharmacological therapeutic opportunities are currently available, e.g. monoclonal antibodies and inhibitors, antivirals, immunomodulators, with the greatest efficacy observed with early administration soon after the onset of symptoms. Besides, the research for new effective compounds continues worldwide. Global coordination and fast sharing of accurate data remains crucial for an up-to-date view of the SARS-CoV-2 pandemic trend, building emergency preparedness and response capacity.
Author contributions
Conceptualization, MS, AV; methodology, MS, AV, GR; investigation, AV, MS, SC, AZ, FF; writing—original draft preparation, MS, AV; writing—review and editing, SC, AZ, FF, MB, RL, PS, GR; visualization, SC, MB, CC, RL, AP, PS, GR. All the authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Availability of data and materials
Full availability of data and materials.
Declarations
Conflict of interest
All the authors declare no conflict of interest.
Other
All the authors declare that the opinions expressed are of a personal nature and do not in any way commit the responsibility of the Administrations to which they belong.
Ethical approval
Not applicable.
Consent to participate
Not applicable.
Consent to publish
The authors consent to the publication of the manuscript.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Michela Sabbatucci, Email: m.sabbatucci@sanita.it.
Antonio Vitiello, Email: avitiello@hotmail.it.
Salvatore Clemente, Email: s.clemente@sanita.it.
Andrea Zovi, Email: zovi.andrea@gmail.com.
Mariarosaria Boccellino, Email: mariarosaria.boccellino@unicampania.it.
Francesco Ferrara, Email: frafer85@gmail.com.
Carla Cimmino, Email: carlacimmino81@gmail.com.
Roberto Langella, Email: roberto.langella87@gmail.com.
Annarita Ponzo, Email: annari.ponzo@gmail.com.
Paola Stefanelli, Email: paola.stefanelli@iss.it.
Giovanni Rezza, Email: g.rezza@sanita.it.
References
- Balse E, Hatem SN. Do cellular entry mechanisms of SARS-Cov-2 affect myocardial cells and contribute to cardiac injury in COVID-19 patients? Front Physiol. 2021;12:630778. doi: 10.3389/fphys.2021.630778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callaway E. New Omicron relatives BA.4 and BA.5 offer hints about the future of SARS-CoV-2. Nature. 2022;605:204–206. doi: 10.1038/d41586-022-01240-x. [DOI] [PubMed] [Google Scholar]
- Callaway E. What Omicron's BA.4 and BA.5 variants mean for the pandemic. Nature. 2022;606(7916):848–849. doi: 10.1038/d41586-022-01730-y. [DOI] [PubMed] [Google Scholar]
- Cao Y, Yisimayi A, Jian F, Song W, Xiao T, Wang L, Du S, Wang J, Li Q, Chen X, Yu Y, Wang P, Zhang Z, Liu P, An R, Hao X, Wang Y, Wang J, Feng R, Sun H, Zhao L, Zhang W, Zhao D, Zheng J, Yu L, Li C, Zhang N, Wang R, Niu X, Yang S, Song X, Chai Y, Hu Y, Shi Y, Zheng L, Li Z, Gu Q, Shao F, Huang W, Jin R, Shen Z, Wang Y, Wang X, Xiao J, Xie XS. BA.2.12.1, BA.4 and BA.5 escape antibodies elicited by Omicron infection. Nature. 2022;608(7923):593–602. doi: 10.1038/s41586-022-04980-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cele S, Jackson L, Khoury DS, et al. Omicron extensively but incompletely escapes Pfizer BNT162b2 neutralization. Nature. 2022;602:654–656. doi: 10.1038/s41586-021-04387-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen C, Kleynhans J, von Gottberg A, et al. SARS-CoV-2 incidence, transmission, and reinfection in a rural and an urban setting: results of the PHIRST-C cohort study, South Africa, 2020–21. Lancet Infect Dis. 2022;22(6):821–834. doi: 10.1016/S1473-3099(22)00069-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conti P, Pregliasco FE, Calvisi V, Al C, Gallenga CE, Kritas SK, Ronconi G. Monoclonal antibody therapy in COVID-19. J Biol Regul Homeost Agents. 2021;35(2):423–427. doi: 10.23812/Conti_Edit_35_2_1. [DOI] [PubMed] [Google Scholar]
- Coronavirus (COVID-19) Infection Survey, UK: 4 November 2022—Office for National Statistics. https://www.ons.gov.uk/releases/coronaviruscovid19infectionsurveyuk4november2022. Accessed 21 Nov 2022
- da Rosa Mesquita R, Francelino Silva Junior LC, Santos Santana FM, Farias de Oliveira T, Campos Alcântara R, Monteiro Arnozo G, Rodrigues da Silva Filho E, Galdino Dos Santos AG, Oliveira da Cunha EJ, Salgueiro de Aquino SH, Freire de Souza CD. Clinical manifestations of COVID-19 in the general population: systematic review. Wien Klin Wochenschr. 2021;133(7–8):377–382. doi: 10.1007/s00508-020-01760-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- European Centre for Disease Prevention and Control (2023a) Country overview report: week 8 2023a. Produced on 1 March 2023a at 18.00. https://covid19-country-overviews.ecdc.europa.eu/variants_of_concern.html
- European Centre for Disease Prevention and Control (2023b) Epidemiological update: SARS‐CoV‐2 Omicron sub‐lineages BA.4 and BA.5.2022
- Ferrara F, Porta R, Daiuto V, Vitiello A. Remdesivir and COVID-19. Ir J Med Sci. 2021;190(3):1237–1238. doi: 10.1007/s11845-020-02401-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurung AB, Ali MA, Aljowaie RM, Almutairi SM, Sami H, Lee J. Masitinib analogues with the N-methylpiperazine group replaced—a new hope for the development of anti-COVID-19 drugs. J King Saud Univ Sci. 2023;35(1):102397. doi: 10.1016/j.jksus.2022.102397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hachmann NP, Miller J, Collier AY, Ventura JD, Yu J, Rowe M, Bondzie EA, Powers O, Surve N, Hall K, Barouch DH. Neutralization escape by SARS-CoV-2 Omicron subvariants BA.2.12.1, BA.4, and BA.5. N Engl J Med. 2022;387(1):86–88. doi: 10.1056/NEJMc2206576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halfmann PJ et al (2022) SARS-CoV-2 Omicron virus causes attenuated disease in mice and hamsters. Nature. 10.1038/s41586-022-04441-6. [PMC free article] [PubMed] [CrossRef] [Google Scholar] [DOI] [PMC free article] [PubMed]
- Han Q, Zheng B, Daines L, Sheikh A. Long-term sequelae of COVID-19: a systematic review and meta-analysis of one-year follow-up studies on post-COVID symptoms. Pathogens. 2022;11(2):269. doi: 10.3390/pathogens11020269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirotsu Y, Omata M. Detection of the omicron BA.2.75 subvariant in Japan. J Infect. 2023;86(1):e5–e7. doi: 10.1016/j.jinf.2022.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M, Kleine-Weber H, Schroeder S, et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181(2):271–280. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- https://www.ecdc.europa.eu/en/covid-19/country-overviews
- https://www.ema.europa.eu/en/human-regulatory/overview/public-health-threats/coronavirus-disease-covid-19/covid-19-latest-updates
- https://www.who.int/emergencies/diseases/novel-coronavirus-2019
- Islam MR, Shahriar M, Bhuiyan MA. The latest Omicron BA.4 and BA.5 lineages are frowning toward COVID-19 preventive measures: a threat to global public health. Health Sci Rep. 2022;5(6):e884. doi: 10.1002/hsr2.884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen SCJ, Tse CLY, Burry L, Dresser LD. Baricitinib: a review of pharmacology, safety, and emerging clinical experience in COVID-19. Pharmacotherapy. 2020;40(8):843–856. doi: 10.1002/phar.2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaoka Y, Uraki R, Kiso M, Iida S, Imai M, Takashita E, Kuroda M, Halfmann P, Loeber S, Maemura T, Yamayoshi S, Fujisaki S, Wang Z, Ito M, Ujie M, Iwatsuki-Horimoto K, Furusawa Y, Wright R, Chong Z, Ozono S, Yasuhara A, Ueki H, Sakai Y, Li R, Liu Y, Larson D, Koga M, Tsutsumi T, Adachi E, Saito M, Yamamoto S, Matsubara S, Hagihara M, Mitamura K, Sato T, Hojo M, Hattori SI, Maeda K, Okuda M, Murakami J, Duong C, Godbole S, Douek D, Watanabe S, Ohmagari N, Yotsuyanagi H, Diamond M, Hasegawa H, Mitsuya H, Suzuki T (2022) Characterization and antiviral susceptibility of SARS-CoV-2 Omicron/BA.2. Res Sq [Preprint]. 10.21203/rs.3.rs-1375091/v1. Update in: Nature. 2022 May 16
- Lewnard JA, Hong VX, Patel MM, Kahn R, Lipsitch M, Tartof SY. Clinical outcomes associated with SARS-CoV-2 Omicron (B.1.1.529) variant and BA.1/BA.1.1 or BA.2 subvariant infection in Southern California. Nat Med. 2022;28(9):1933–1943. doi: 10.1038/s41591-022-01887-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Iketani S, Guo Y, et al. Striking antibody evasion manifested by the omicron variant of SARS-CoV-2. Nature. 2022;602:676–681. doi: 10.1038/s41586-021-04388-0. [DOI] [PubMed] [Google Scholar]
- Lyngse F et al (2022) Transmission of SARS-CoV-2 Omicron VOC subvariants BA.1 and BA.2: evidence from Danish households. medRxiv. 10.1101/2022.01.28.22270044. [CrossRef] [Google Scholar]
- Mantzourani C, Vasilakaki S, Gerogianni VE, Kokotos G. The discovery and development of transmembrane serine protease 2 (TMPRSS2) inhibitors as candidate drugs for the treatment of COVID-19. Expert Opin Drug Discov. 2022;17(3):231–246. doi: 10.1080/17460441.2022.2029843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouffak S, Shubbar Q, Saleh E, El-Awady R. Recent advances in management of COVID-19: a review. Biomed Pharmacother. 2021;143:112107. doi: 10.1016/j.biopha.2021.112107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parikh S, O’Laughlin K, Ehrlich HY, et al. Point prevalence testing of residents for SARS-CoV-2 in a subset of Connecticut nursing homes. JAMA. 2020;324(11):1101–1103. doi: 10.1001/jama.2020.14984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peeri NC, Shrestha N, Rahman MS, Zaki R, Tan Z, Bibi S, Baghbanzadeh M, Aghamohammadi N, Zhang W, Haque U. The SARS, MERS and novel coronavirus (COVID-19) epidemics, the newest and biggest global health threats: what lessons have we learned? Int J Epidemiol. 2020;49(3):717–726. doi: 10.1093/ije/dyaa033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrovszki D, Walter FR, Vigh JP, Kocsis A, Valkai S, Deli MA, Dér A. Penetration of the SARS-CoV-2 spike protein across the blood–brain barrier, as revealed by a combination of a human cell culture model system and optical biosensing. Biomedicines. 2022;10(1):188. doi: 10.3390/biomedicines10010188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Real-time dashboard. Coronavirus disease 2019. https://covid19.sph.hku.hk/dashboard. Accessed 5 Mar 2023
- Shaheen N, Mohamed A, Soliman Y, Abdelwahab OA, Diab RA, Desouki MT, Rababah AA, Khaity A, Hefnawy MT, Swed S, Shaheen A, Elfakharany B, Shoib S. Could the new BA.2.75 sub-variant lead to another COVID-19 wave in the world?—Correspondence. Int J Surg. 2022;105:106861. doi: 10.1016/j.ijsu.2022.106861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheward DJ, Kim C, Fischbach J, Muschiol S, Ehling RA, Björkström NK, et al. Evasion of neutralising antibodies by omicron sublineage BA.2.75. Lancet Infect Dis. 2022;22(10):1421–1422. doi: 10.1016/S1473-3099(22)00524-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheward DJ, Kim C, Fischbach J, Muschiol S, Ehling RA, Björkström NK, KarlssonHedestam GB, Reddy ST, Albert J, Peacock TP, Murrell B. Evasion of neutralising antibodies by omicron sublineage BA.2.75. Lancet Infect Dis. 2022;22(10):1421–1422. doi: 10.1016/S1473-3099(22)00524-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheward DJ, Kim C, Fischbach J, Sato K, Muschiol S, Ehling RA, Björkström NK, KarlssonHedestam GB, Reddy ST, Albert J, Peacock TP, Murrell B. Omicron sublineage BA.2.75.2 exhibits extensive escape from neutralising antibodies. Lancet Infect Dis. 2022;22(11):1538–1540. doi: 10.1016/S1473-3099(22)00663-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuai H et al (2022) Attenuated replication and pathogenicity of SARS-CoV-2 B.1.1.529 Omicron. Nature. 10.1038/s41586-022-04442-5. [PubMed] [CrossRef] [Google Scholar] [DOI] [PubMed]
- Suzuki R et al (2022) Attenuated fusogenicity and pathogenicity of SARS-CoV-2 Omicron variant. Nature. 10.1038/s41586-022-04462-1. [PMC free article] [PubMed] [CrossRef] [Google Scholar] [DOI] [PMC free article] [PubMed]
- Takashita E, Yamayoshi S, Simon V, van Bakel H, Sordillo EM, Pekosz A, Fukushi S, Suzuki T, Maeda K, Halfmann P, Sakai-Tagawa Y, Ito M, Watanabe S, Imai M, Hasegawa H, Kawaoka Y. Efficacy of antibodies and antiviral drugs against Omicron BA.2.12.1, BA.4, and BA.5 subvariants. N Engl J Med. 2022;387(5):468–470. doi: 10.1056/NEJMc2207519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Times TE (2022) Omicron's new 'BA.2.75.2' variant growing quickly in India, becoming more transmissible, immune evasive: Scientists. https://economictimes.indiatimes.com/news/pune-news/omicrons-new-ba-2-75-2-variant-growing-quickly-in-india-becoming-more-transmissible-immune-evasive-scientists/articleshow/94128543.cms
- Tso WWY, Kwan MYW, Wang YL, Leung LK, Leung D, Chua GT, Ip P, Fong DYT, Wong WHS, Chan SHS, Chan JFW, Peiris M, Lau YL, Rosa Duque JS. Severity of SARS-CoV-2 Omicron BA.2 infection in unvaccinated hospitalized children: comparison to influenza and parainfluenza infections. Emerg Microbes Infect. 2022;11(1):1742–1750. doi: 10.1080/22221751.2022.2093135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitiello A, Ferrara F. Brief review of the mRNA vaccines COVID-19. Inflammopharmacology. 2021;29(3):645–649. doi: 10.1007/s10787-021-00811-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitiello A, Ferrara F, Troiano V, La Porta R. COVID-19 vaccines and decreased transmission of SARS-CoV-2. Inflammopharmacology. 2021;29(5):1357–1360. doi: 10.1007/s10787-021-00847-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitiello A, Ferrara F, Auti AM, Di Domenico M, Boccellino M. Advances in the Omicron variant development. J Intern Med. 2022;292(1):81–90. doi: 10.1111/joim.13478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO (2023a) XBB.1.5 updated risk assessment, 24 February 2023. https://www.who.int/docs/default-source/coronaviruse/22022024xbb.1.5ra.pdf
- WHO (2023b) Coronavirus disease (COVID-19) Weekly epidemiological updates and monthly operational updates. https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports. Accessed 5 Mar 2023
- WHO (2023c) WHO updates COVID-19 guidelines on masks, treatments and patient care. https://www.who.int/news/item/13-01-2023-who-updates-covid-19-guidelines-on-masks--treatments-and-patient-care
- Statement on the update of WHO’s working definitions and tracking system for SARS-CoV-2 variants of concern and variants of interest. Accessed 16 March 2023. https://www.who.int/news/item/16-03-2023d-statement-on-the-update-of-who-s-working-definitions-and-tracking-system-for-sars-cov-2-variants-of-concern-and-variants-of-interest [PMC free article] [PubMed]
- Wolter N, Jassat W, Walaza S, Welch R, Moultrie H, Groome MJ, Amoako DG, Everatt J, Bhiman JN, Scheepers C, Tebeila N, Chiwandire N, du Plessis M, Govender N, Ismail A, Glass A, Mlisana K, Stevens W, Treurnicht FK, Subramoney K, Makatini Z, Hsiao NY, Parboosing R, Wadula J, Hussey H, Davies MA, Boulle A, von Gottberg A, Cohen C. Clinical severity of SARS-CoV-2 Omicron BA.4 and BA.5 lineages compared to BA.1 and Delta in South Africa. Nat Commun. 2022;13(1):5860. doi: 10.1038/s41467-022-33614-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zappa M, Verdecchia P, Angeli F. Knowing the new Omicron BA.2.75 variant (‘Centaurus’): a simulation study. Eur J Intern Med. 2022;105:107–108. doi: 10.1016/j.ejim.2022.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z, Lian X, Su X, Wu W, Marraro GA, Zeng Y. From SARS and MERS to COVID-19: a brief summary and comparison of severe acute respiratory infections caused by three highly pathogenic human coronaviruses. Respir Res. 2020;21(1):224. doi: 10.1186/s12931-020-01479-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Full availability of data and materials.


