Abstract
Gliomas remain a clinical challenge, common and fatal. Treatment of glioblastoma remains elusive, and researchers have focused on discovering new mechanisms and drugs. It has been well established that the expression of voltage-gated sodium channels (VGSCs) is abnormally increased in numerous malignancies and, in general, is rarely expressed in the corresponding normal tissues. This suggests that ion channel activity appears to be associated with malignant progression of tumors. VGSCs remain largely unknown as to how their activity leads to an increase in cancer cell activity or invasiveness. Certain sodium ion channel subtypes (for instance, Nav1.5 and Nav1.7) are associated with metastasis and invasion in cancers including breast and colorectal cancers. A previous study by the authors explored the expression of certain ion channels in glioma, but there are few studies related to Nav1.6. The current study aimed to elucidate the expression and role of Nav1.6 in glioma and to screen potential drugs for the treatment of glioma by virtual screening and drug sensitivity analysis. Nav1.6 relative expression of mRNA and protein was determined by reverse transcription-quantitative PCR and western blot analysis. Cell proliferation was determined by Cell Counting Kit-8 assay. Cell migration was assessed by cellular wound healing assay. Cell invasion and apoptosis were detected by Transwell cell invasion assay and flow cytometry. Last but not least, FDA-approved drugs were screened using virtual screening, molecular docking and NCI-60 drug sensitivity analyses based on the expression and structure of Nav1.6. In glioma cells, Nav1.6 was significantly upregulated and expressed mostly in the cytoplasm and cell membrane; its expression was positively correlated with pathological grade. A172 and U251 cells exhibited reduced proliferation, migration and invasion when Nav1.6 expression was knocked down, and apoptosis was increased. TNF-α (100 pg/ml) acting on glioma cells was found to upregulate the expression level of Nav1.6, and TNF-α was involved in the process of Nav1.6 promoting malignant progression of glioma. Finally, certain FDA-approved drugs were identified by virtual screening and drug sensitivity analysis. In conclusion, the present study demonstrated the expression and role of Nav1.6 in glioma and identified several FDA-approved drugs that are highly correlated with Nav1.6 and could be candidate drugs for patients with glioma.
Keywords: glioma, Nav1.6, voltage-gated sodium channel, TNF-α, drug screening
Introduction
The treatment of gliomas remains a challenge for neurosurgeons (1). There remains a need to explore the molecular mechanisms of malignant gliomas and to find new therapeutic strategies and drug targets. Metastasis and invasion are important causes of cancer mortality. Numerous studies have shown that voltage-gated sodium channels (VGSCs) are aberrantly expressed in a variety of metastatic tumors (2-7). High expression of VGSCs has the potential to enhance cancer cell invasion and metastasis. It was found that several types of ion channels are distributed on the nervous system and glial cells, and certain sodium channels were found to play a role in the physiological progression of glial cells (8,9). Previous studies have shown that neonatal isoform Nav1.5 is highly expressed in high-grade gliomas but is also intimately linked to their biological behavior, including invasion, proliferation and migration (10-12). This suggests that VGSCs are associated with the development of gliomas.
VGSCs are directly or indirectly involved in various physiological processes by regulating the transport of sodium ions across membranes (13). VGSCs consist of a core α subunit and auxiliary β subunits (14). A total of 10 different isoforms have been identified depending on the α subunit. The distribution and function of the Navs varies among the different isoforms (15). The α-subunit is the main determinant of the physiological activity of VGSC and can function partially on its own, but is only fully functional when linked to the auxiliary subunit β-subunit by disulfide or non-covalent bonds. The sodium channel is widely expressed in the nervous system, but the encoded proteins, structures and electrophysiological properties vary between isoforms (16). The Nav1.6 gene encoded by sodium voltage-gated channel alpha subunit 8 (SCN8A) is widely expressed in the central and peripheral nervous systems. The expression and role of Nav1.6 have been verified in various mouse neurological diseases and animal models (17-20), and it has been reported that Nav1.6 is highly expressed in human colorectal and oral squamous carcinomas, and its knockdown significantly affects the invasive and metastatic ability of cancer cells (7). However, the specific mechanisms affecting tumor invasion and metastasis remain unclear (21,22).
Currently, the expression of Nav1.6 in glioma and its effects and mechanisms on glioma development have not been reported. In the present study, the expression of Nav1.6 in different grades of gliomas was firstly described and the role and mechanism of Nav1.6 isoforms in gliomas were explored. Finally, certain FDA-approved drugs were identified based on the expression and structure of Nav1.6.
Materials and methods
Cells and tissues
The glioma cell lines HEB, U87 MG (cat. no. HTB-14; glioblastoma of unknown origin; verified by STR analysis), A172 and U251 were obtained from the Shanghai Cell Bank and glioma pathological tissues were obtained from the Department of Neurosurgery, The First Affiliated Hospital of China Medical University (Shenyang, China). Ages ranged from 33 to 76, including six male and six female patients. Inclusion criteria were patients diagnosed with glioma and sufficient tumor tissue available for study. All tissues and specimens were obtained with the approval (approval no. 20190142) and endorsement of the Ethics Committee of China Medical University (Shenyang, China). The date range of tissue collection was between July 17, 2019 and August 12, 2021. Written informed consent was obtained from each tumor tissue donor for the use of the tumor tissue and clinical data for future research.
Cell culture and transfection
Glioma cells A172 and U251 were cultured in DMEM sugar-free medium (Gibco; Thermo Fisher Scientific, Inc.) containing 10% fetal bovine serum (Gibco; Thermo Fisher Scientific, Inc.) and 1% double anti-bodies (Gibco; Thermo Fisher Scientific, Inc.). All cells were cultured at 37°C in an atmosphere containing 5% CO2. Glioma A172 and U251 cell lines in logarithmic growth phase were received and transfection was performed at 37°C for 24 h using Lipofectamine® 3000 reagent according to the manufacturer's instructions (Invitrogen; Thermo Fisher Scientific, Inc.). The concentration of transfected small interfering (si)RNAs was 50 nM. The efficiency of transfection was verified by qPCR and western blotting (WB). The sequences of the siRNAs used are shown in Table SI.
RNA extraction and reverse transcription-quantitative PCR (RT-qPCR)
The corresponding tissues or cells (1×107) were received lysed by TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.); RNA was extracted by chloroform, precipitated by isopropanol and lysed in the presence of DEPC in water to inactivate RNAse enzymes. RNA purification was performed according to the instructions of the corresponding reverse transcription kit Shanghai Yeasen Biotechnology Co., Ltd. Hifair® III 1st Strand cDNA Synthesis SuperMix (cat. no. 11141ES60; Shanghai Yeasen Biotechnology Co., Ltd.) was used for qPCR. The thermocycling conditions for the RT-qPCR reaction were as follows: 25°C for 5 min, 55°C for 15 min, and 85°C for 5 min for the reverse transcription step, and 95°C for 5 min for the pre-denaturation step followed by one cycle; 95°C for 10 sec for the denaturation step followed by 40 cycles; and 60°C for 30 sec for the extension step followed by 40 cycles. The reference gene was β-actin, quantified by 2−ΔΔCq method (23). The sequences of the primers used are shown in Table SI.
WB
Protein quantification was performed using a BCA kit (cat. no. WB6501; NCM Biotech; http://www.ncmbio.com/) according to the manufacturer's instructions. Phosphatase inhibitor (1%) and protease inhibitor were added during cell lysis (cat. no. P002; NCM Biotech). Proteins were separated using SDS-PAGE. A 7.5% gel (cat. no. P2011; NCM Biotech) was used, and 20 µg of protein was loaded per lane. This was performed in Tris-glycine buffer system. The separated proteins were subsequently transferred to 0.45-µm PVDF membrane (Merck KGaA) and Skim milk powder closed at room temperature for two h. Next, the membranes were rinsed three times with TBST prepared with 0.1% Tween 20 for 8 min each. The membranes were incubated overnight at 4°C with primary antibodies against SCN8A (1:800; cat. no. ab230654; Abcam) and β-actin (1:5,000; cat. no. bs-0061R; BIOSS). Following the primary incubation, membranes were incubated at room temperature with HRP-labeled goat anti-rabbit IgG secondary antibody (1:10,000; cat. no. bs-0295G; BIOSS). Membranes were washed with TBST for 10 min each time. Luminescence was performed, developed and images of the protein bands were captured for analysis using the ECL chemiluminescence kit (NCM). Densitometric analysis was performed using ImageJ software version 1.52a (National Institutes of Health).
Immunohistochemistry
Paraffin sections of 5-µm thickness were first dewaxed with xylene for 30 min after 1 h in an oven at 60°C. Subsequently, the sections were placed sequentially in a gradient alcohol and the tissue sections were washed in water to remove residual alcohol and dewaxing agent while the sections were hydrated. Sections were immersed in citrate buffer and placed in a microwave oven for antigen repair, then washed in PBS. Blocking treatment was performed using 5% bovine serum albumin for 1 h at room temperature. Sections were washed with PBS and incubated with 3% hydrogen peroxide. Deparaffinized sections were incubated overnight at 4°C with primary antibodies against SCN8A (1:100) and subsequently with HRP-conjugated goat anti-rabbit IgG secondary antibody (1:1,000) for 1 h at room temperature. The slides were washed and stained with DAB for color development. Color development was terminated by tap water. Hematoxylin counterstaining followed, using hydrochloric acid for 1% alcohol fractionation. Tap water was used to reverse the blue, followed by alcohol dehydration (gradient concentration) and dewaxing with 95% alcohol. Next, xylene dehydration, sealing and drying of the film was performed. Tissues were observed under a light microscope and images were captured.
Cellular experiments
Cell Counting Kit-8 (CCK-8) cell proliferation assay
A172 and U251 cells were inoculated into 96-well plates at a density of 1,500 and 2,500 per well, respectively. CCK-8 assay was performed following the manufacturer's protocol (BIOSS). Each well was spiked with 10 µl of CCK-8 reagent and incubated at 37°C for 2 h, followed by detection at 450 nm using a multifunctional enzyme marker (BioTek Instruments, Inc.).
Wound healing assay
A172 and U251 cells were spread into six-well plates at a density of 2×105 and 3×105 per well and treated with mitomycin (1 µg/ml) after spreading. Cells were scratched with a sterile gun tip. Cells were serum-starved (FBS <5%) during the assay. Images were captured at different time points (0, 12 and 24 h) using an inverted fluorescence microscope ECLIPSE Ts2 (Nikon Corporation). Data analysis was performed using GraphPad Prism 7.
Transwell cell invasion assay
Matrigel (cat. no. 356234; Corning, Inc.) was placed overnight at 4°C and then diluted 1:8 and wrapped on the surface of the upper chamber wrapped in Transwell bottom membrane. Cells (4×105) were seeded in the upper chamber containing 200 µl of serum-free medium and the lower chamber was filled with 600 µl of medium containing 10% FBS. Cells were fixed with 4% paraformaldehyde for 20 min, followed by staining with 0.1% crystal violet for 15 min at 25°C and inverted microscopy observation.
Apoptosis measurement by flow cytometry
Analysis of apoptosis was conducted using the PE Annexin V Apoptosis Detection kit (cat. no. 559763; BD Biosciences) according to the manufacturer's instructions. Apoptotic cells were subsequently detected on LSRFortessa (BD Biosciences) and analyzed using FACSDiva software (Version 6.2; BD Biosciences). Early apoptosis plus late apoptosis was used to assess the apoptosis of cells.
Virtual screening and drug sensitivity analysis
The FDA-approved and pharmacopeial drugs were downloaded from TargetMol (24), which contains 2858 drugs and their structures, with full drug names and drug IDs shown in Table SII. The protein structure for SCN8A was obtained from the AlphaFold Protein Structure Database (accession no. Q9UQD0) (25,26), and the binding sites were identified by the GHECOM algorithm (27); virtual screening and molecular docking were performed using UCSF DOCK 6.9 (28). Finally, docking conformations were demonstrated using PyMol (Version 2.3) and interaction forces were analysed and demonstrated by Ligplus (29). A total of 23808 genetic data (RNA: RNA-seq) and 24359 compounds' data (Compound activity: DTP NCI-60, which includes 218 drugs that have been approved by the FDA, and 574 drug molecules in clinical trials) have been identified in NCI-60 cell lines, and were obtained from the CellMiner database (30). The results of drugs that have undergone clinical trials and FDA approval were selected to ensure the reliability of the analysis. P<0.05 was used as a cut-off in screening the results based on Pearson's correlation coefficients between each gene expression and each drug.
Statistical analysis
All data are presented as the mean ± standard error of the mean (M ± SEM). Unpaired Student's t-test was used to determine the statistical significance between the two groups. The results were analyzed using GraphPad Prism (version 7.0; Dotmatics). Depending on the design of the experiment, the data were analyzed using one-way ANOVA followed by Tukey's post hoc test. P<0.05 was considered to indicate a statistically significant difference.
Results
Nav1.6 mRNA and protein expression are higher in human glioma cells and tissues than in normal glial cells
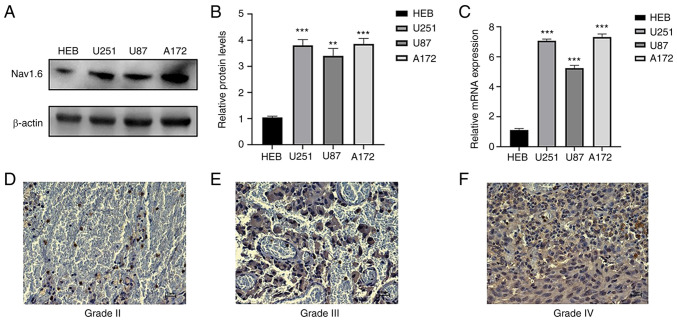
The expression of Nav1.6 mRNA and protein in glioma cell lines U251, U87 A172 and normal glial cells HEB was detected by RT-qPCR and WB. It was demonstrated that Nav1.6 was expressed in both normal and glioma cell lines, and the expression in glioma cell lines U251 and A172 was significantly higher than that in normal glial cells (Fig. 1A-C), therefore U251 and A172 cell lines were subsequently used for knockdown experiments. WB results also showed that the relative protein expression of Nav1.6 in glioma cell lines U251, U87, and A172 was several times higher than that in normal glial tissues. To investigate whether the expression level of Nav1.6 correlated with the malignant progression of glioma, immunohistochemical experiments were further performed using paraffin sections of clinically collected glioma tissues of different grades. The results revealed that Nav1.6 was mainly expressed on the cytoplasm and cell membrane, and the expression level of Nav1.6 was significantly higher in high-grade tissues compared with lower-grade glioma tissues (Fig. 1D-F). In summary, it was evident that the expression level of Nav1.6 is increased in glioma and positively correlates with the pathological grade of glioma.
Figure 1.
Detection of Nav1.6 expression levels in glioma cells and tissues. (A and B) Detection of protein expression levels of Nav1.6 in different glioma cell lines using western blotting. (C) Detection of Nav1.6 mRNA expression levels in different glioma cell lines by reverse transcription-quantitative PCR. (D-F) Expression of Nav1.6 in different grades of glioma tissues using immunohistochemical analysis (scale bar, 20 µm). **P<0.01 and ***P<0.001.
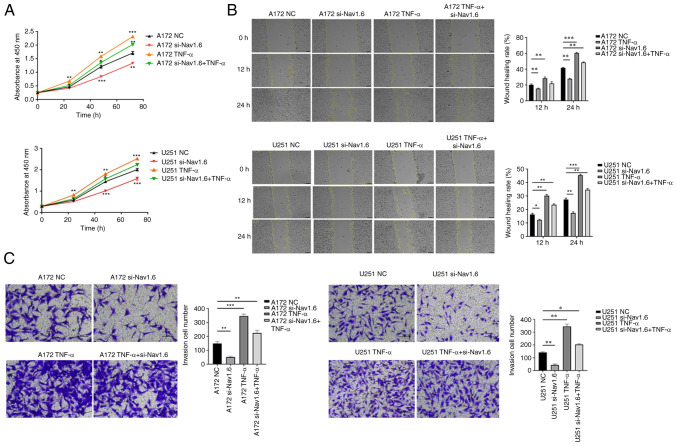
Knockdown of Nav1.6 inhibits the proliferation, migration and invasive ability of glioma cells and promotes apoptosis of glioma cells
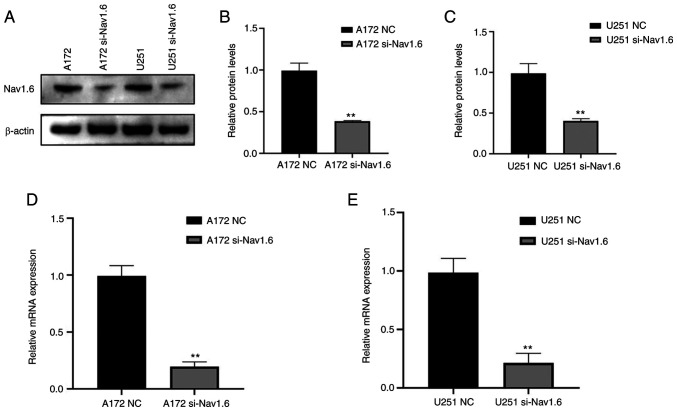
The glioma U251 and A172 cell lines were transfected with si-Nav1.6 and si-NC, and the efficiency of transfection was verified by WB (Fig. 2A-C) and RT-qPCR (Fig. 2D and E). It was identified that the mRNA and protein levels of Nav1.6 in both U251 and A172 cells were significantly decreased after transfection with si-Nav1.6 compared with the NC group, and the knockdown effect was significant.
Figure 2.
Nav1.6 protein and mRNA expression levles in glioma cells after transfection with si-Nav1.6. (A-C) Nav1.6 protein expression in glioma cells after si-Nav1.6 transfection using western blotting. (D and E) Nav1.6 mRNA expression in glioma cells after si-Nav1.6 transfection. **P<0.01. si-, small interfering; NC, negative control.
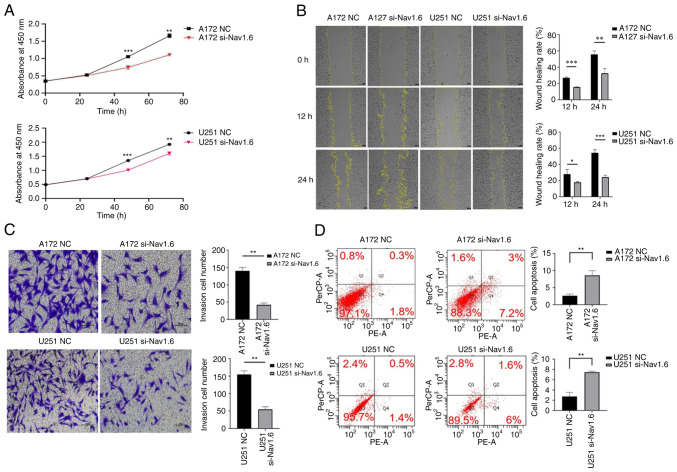
Several assays were conducted to detect the effect of si-Nav1.6 on glioma cells, including proliferation, wound healing, cell invasion and flow cytometric analysis apoptosis assay. The experimental data demonstrated that there was no significant difference in cell proliferation between A172 si-Nav1.6 and U251 si-Nav1.6 within 24 h compared with the control group, and transfection of si-Nav1.6 significantly inhibited the proliferation of glioma A172 and U251 cells after 24 h (P<0.01, Fig. 3A). In addition, it was revealed that si-Nav1.6 wound healing and invasive abilities of A172 and U251 cells (Fig. 3B and C, respectively). Flow cytometric analysis of apoptosis assay showed that the transfection of si-Nav1.6 promoted the apoptotic rate of glioma A172 and U251 cells (Fig. 3D).
Figure 3.
Effect of transfection with si-Nav1.6 on the biological activity of glioma cells. (A) Cell Counting Kit-8 cell proliferation assay showed that transfection with si-Nav1.6 reduced the proliferation of glioma A172 and U251 cells. (B) Wound healing assays revealed that transfection with si-Nav1.6 decreased glioma A172 and U251 cell migration (scale bar, 100 µm). (C) Transwell cell invasion assay demonstrated that transfection with si-Nav1.6 reduced the invasion of glioma A172 and U251 cells (scale bar, 50 µm). (D) Flow cytometric analysis of apoptosis assay showed that transfection of si-Nav1.6 promoted the apoptotic rate of glioma A172 and U251 cells. *P<0.05, **P<0.01 and ***P<0.001. si-, small interfering; NC, negative control.
TNF-α induces the expression of Nav1.6 and induces the proliferation, migration and invasion of glioma cells
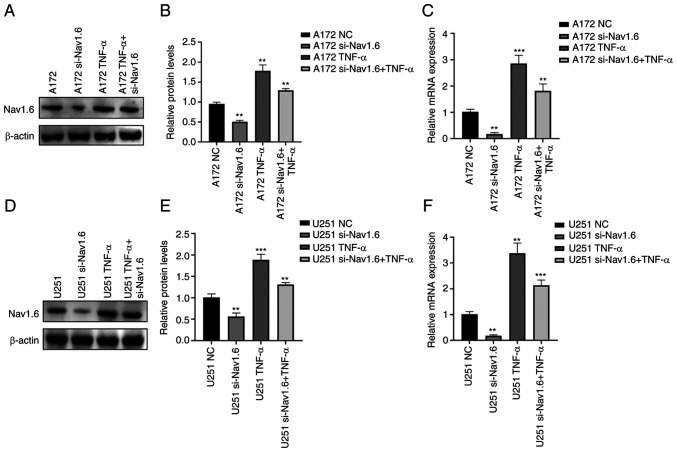
Through literature review and analysis of relevant data, it was found that TNF-α can affect Nav1.6 expression and has been verified at the electrophysiological level (31), but it has still not been studied in tumor cells. Therefore, the effect of TNF-α on Nav1.6 expression was studied at the cellular level. First, the optimal concentration and optimal time of TNF-α action were mapped by pre-experiments. The effect of TNF-α on Nav1.6 was detected at concentrations of 1, 10, 100 and 1,000 pg/ml, and the final treatment was selected at an action concentration of 100 pg/ml. The relative expression levels of mRNA and protein of Nav1.6 were significantly increased after the action of TNF-α (100 pg/ml) on A172 and U251 cells. TNF-α action in cell lines transfected with si-Nav1.6 significantly increased the protein and mRNA expression of Nav1.6, but was slightly lower than the direct action of TNF-α (Fig. 4A-F). This suggested that TNF-α induced the protein and mRNA expression of Nav1.6 in glioma cells and that si-Nav1.6 partially blocked the induction of TNF-α.
Figure 4.
Expression of Nav1.6 after TNF-α action on glioma A172 and U251 cells. (A and B) Nav1.6 protein expression after TNF-α action on glioma A172 cells. (C) Nav1.6 mRNA expression after TNF-α action in glioma A172 cells. (D and E) Nav1.6 protein expression after TNF-α action in glioma U251 cells. (F) Nav1.6 mRNA expression after TNF-α action in glioma U251 cells. **P<0.01 and ***P<0.001. si-, small interfering; NC, negative control.
To determine the effects of TNF-α on glioma cells, CCK-8, wound healing and Transwell invasion assays were performed. Proliferation, migration, and invasion of glioma cells were significantly enhanced, and TNF-α also resisted the effect of si-Nav1.6 and increased the level of Nav1.6 expression after transfection with siRNA, but si-Nav1.6 also partially blocked the induction of TNF-α (Fig. 5A-C). The association of Nav1.6 with the development and malignant progression of gliomas was also further confirmed by the action of TNF-α.
Figure 5.
Effect of TNF-α on glioma biological activity after action on glioma A172 and U251 cells. (A) TNF-α induced the proliferation of A172 and U251 cells. (B) TNF-α induced the migration of A172 and U251 cells (scale bar, 100 µm). (C) TNF-α induced A172 and U251 cell invasion (scale bar, 50 µm). *P<0.05, **P<0.01 and ***P<0.001. si-, small interfering; NC, negative control.
A virtual screening and sensitivity analysis of drugs
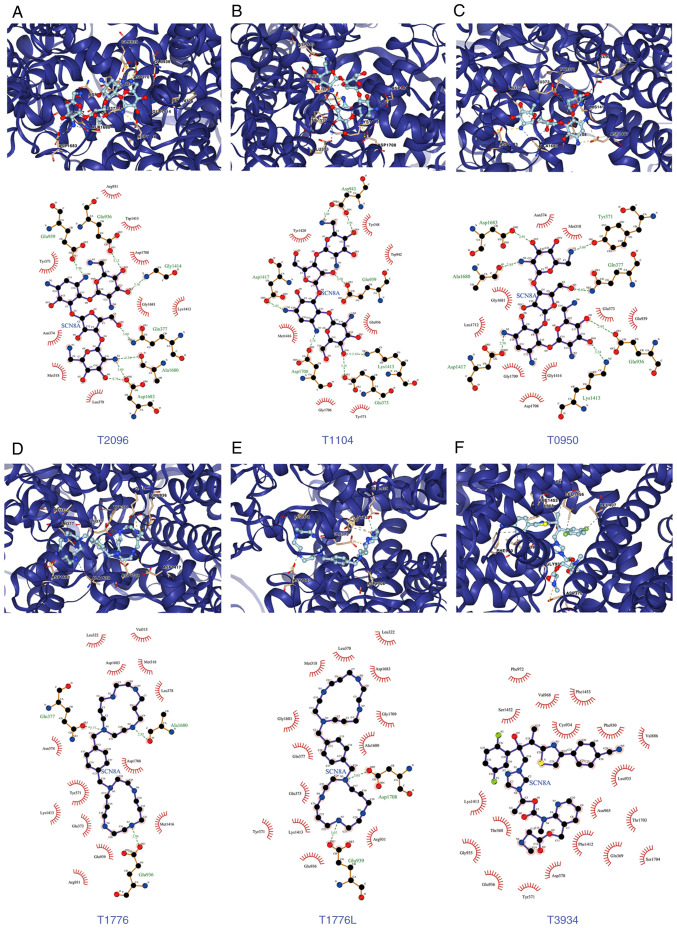
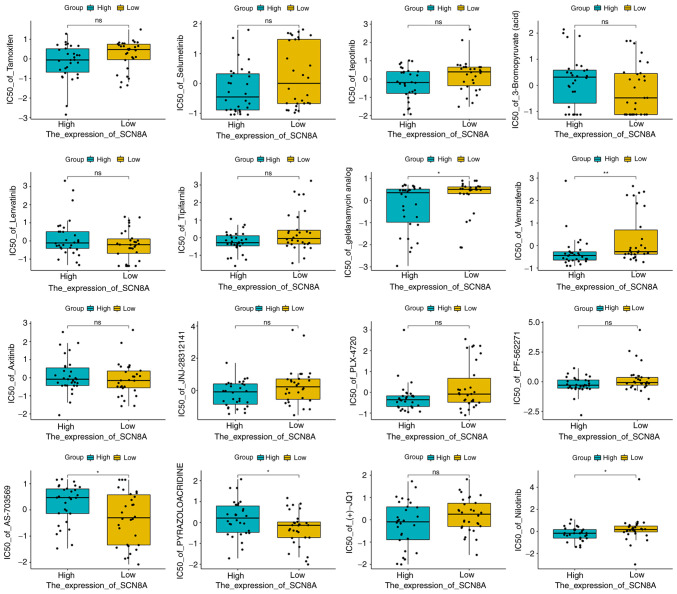
Based on the aforementioned analysis, in order to explore the drugs targeting SCN8A, molecular docking and virtual screening against FDA-approved drugs was conducted based on the 3D structure of the protein. A list of the top 10 scoring drug molecules is shown in Table I and a summary of the scores is presented in Table SII. The conformation and interaction force analysis of the top six scoring drugs can be observed in Fig. 6, and the results indicated that Neomycin Sulphate B (T2096), Paromomycin Sulfate (T1104), Neomycin sulfate (T0950), Plerixafor 8HCl (T1776 and T1776L) and Isavuconazonium sulfate (T3934) showed stronger binding to SCN8A. The results of NCI-60 drug sensitivity analysis similarly revealed that the sensitivity of 23 drugs correlated with the expression of SCN8A. A total of 3 of these drugs [Tamoxifen (TAM), Selumetinib and tepotinib] demonstrated a significant correlation (Table II, Fig. 7). In addition, different expression levels of SCN8A similarly affected the half maximal inhibitory concentration values of the drugs (Fig. 8). Finally, it is worth mentioning that numerous of the screened drugs already have therapeutic value in non-tumor and other oncologic diseases, and based on the analysis of the aforementioned results, they could also be potential drugs for the treatment of glioma.
Table I.
The 10 drugs with the highest docking scores to Nav1.6.
| ID | Grid Score | Grid_vdw | Grid_es | Int_energy |
|---|---|---|---|---|
| T2096 | −122.5961 | −67.5617 | −55.0345 | 14.1862 |
| T1104 | −122.1724 | −69.007 | −53.1654 | 19.9233 |
| T0950 | −122.1169 | −70.7967 | −51.3203 | 15.3193 |
| T1776 | −121.5208 | −71.2886 | −50.2322 | 4.5184 |
| T1776L | −120.1207 | −69.3475 | −50.7733 | 4.7753 |
| T3934 | −112.7299 | −102.7981 | −9.9318 | 31.7494 |
| T1013 | −111.8229 | −68.5774 | −43.2455 | 12.3296 |
| T4509 | −111.4996 | −110.4062 | −1.0934 | 24.5032 |
| T2119 | −111.4223 | −106.4619 | −4.9605 | 22.766 |
| T1670 | −110.7266 | −107.2315 | −3.4951 | 54.6025 |
Figure 6.
Docking conformations and interaction forces of Nav1.6 with improved scoring drugs. (A) T2096 and Nav1.6, (B) T1104 and Nav1.6, (C) T0950 and Nav1.6, (D) T1776 and Nav1.6, (E) T1776L and Nav1.6 and (F) T3934 and Nav1.6. Upper panels: PyMol showed docking conformation, yellow dashed lines for ionic bonds, blue dashed lines for hydrogen bonds, residues are identified by labels; The bottom half of the figure demonstrated the interaction forces between the drug molecule (middle) and the amino acid residues associated with it, hydrogen bonds are shown in green dashed lines, and amino acids forming hydrogen bonds are indicated in green names.
Table II.
An analysis of correlations between SCN8A expression and the sensitivity of FDA-approved drugs.
| Gene | Drug | Correlation | P-value |
|---|---|---|---|
| SCN8A | Tamoxifen | −0.3877366 | 0.00220652 |
| SCN8A | Selumetinib | −0.3460751 | 0.0067579 |
| SCN8A | tepotinib | −0.3302933 | 0.00995474 |
| SCN8A | 3-Bromopyruvate (acid) | 0.32957993 | 0.01012599 |
| SCN8A | Lenvatinib | 0.32851997 | 0.01038515 |
| SCN8A | Tipifarnib | −0.3153499 | 0.0141166 |
| SCN8A | geldanamycin analog | −0.3099119 | 0.01596459 |
| SCN8A | Vemurafenib | −0.3057577 | 0.01751239 |
| SCN8A | Axitinib | 0.30044955 | 0.01967502 |
| SCN8A | JNJ-28312141 | −0.2939791 | 0.02261459 |
| SCN8A | PLX-4720 | −0.2898328 | 0.02468725 |
| SCN8A | PF-562271 | −0.2885017 | 0.02538575 |
| SCN8A | AS-703569 | 0.27787558 | 0.03158141 |
| SCN8A | PYRAZOLOACRIDINE | 0.27702531 | 0.03212751 |
| SCN8A | (+)-JQ1 | −0.2765262 | 0.03245173 |
| SCN8A | Nilotinib | −0.2744798 | 0.0338097 |
| SCN8A | VINORELBINE | −0.273364 | 0.0345699 |
| SCN8A | ARRY-162 | −0.2731846 | 0.03469338 |
| SCN8A | RO-5126766 | −0.2696654 | 0.03719198 |
| SCN8A | EMD-534085 | −0.2692616 | 0.037488 |
| SCN8A | Amonafide | 0.26430171 | 0.04128567 |
| SCN8A | HYPOTHEMYCIN | −0.2622113 | 0.04297867 |
| SCN8A | Pimasertib | −0.2599519 | 0.04487232 |
SCN8A, sodium voltage-gated channel alpha subunit 8.
Figure 7.
Scatter plot of SCN8A expression in relation to drug sensitivity. SCN8A, sodium voltage-gated channel alpha subunit 8.
Figure 8.
Box plot of drug sensitivity in SCN8A-high and SCN8A-low expression groups. *P<0.05. SCN8A, sodium voltage-gated channel alpha subunit 8; IC50, half maximal inhibitory concentration.
Discussion
Metastatic and aggressive progression of tumors is the main reason for the poor prognosis and low survival rate of most patients with malignant tumors. Therefore, it is particularly important to explore the specific mechanisms of metastasis and invasion of malignant tumors. In 2017, the electron microscopic structure of eukaryotic VGSCs was first reported by Pan et al (32) using cryo-electron microscopy. This also provided a basis for further understanding of the VGSCs. VGSCs only started to be studied in gliomas in 2002, and researchers initially detected and elaborated the expression of different ion isoforms in gliomas, but no systematic study of tumor cells has been performed yet (33). The aberrant expression of ion channels in gliomas also suggests that VGSCs have the potential to become glioma-related biomarkers.
VGSCs are composed of core α and β subunits (34). The α-subunit is a key determinant of the activity and function of VGSCs, whereas the β-subunit can regulate channel expression, homeostasis and kinetic properties by binding to the α-subunit. α-subunit can function intact by binding to the β-subunit. α-subunit and β-subunit interactions can enhance the internal repair capacity of the channel, improve channel stability and kinetic properties, and increase the expression of the channel. Even the α subunits of two VGSCs can interact to regulate the activity of ion channels, with the direct mutual region located at the N terminus (35). Sodium channels are key initiators of electrical signals in all animals, and electrical signals play a crucial importance in a range of basic physiological activities including neural activity and muscle contraction. In humans, there are nine known subtypes of VGSCs that function in different organs and physiological processes. In the nervous system, sodium channels are widely distributed. Abnormalities in sodium channels contribute to the progression of neurological disorders, muscular disorders and cardiac ailments. This also appears to be consistent with the currently known distribution of ion channel subtypes, further validating the association of VGSCs with tumor progression.
In the present experiment, it was verified that Nav1.6 is highly expressed in gliomas. This is also consistent with previous studies and the previous experimental results (Wang et al, unpublished data) of the authors' group. Previous studies also showed that Nav1.6 was associated with malignant progression of glioma, and inhibition of its expression reduced the metastatic and invasive potential of glioma, which was also consistent with the hypothesis of the current study. There are few studies on Nav1.6 in tumors, and certain of the rare studies are in colorectal and cervical cancers (7,36). The majority of the studies involve disease models, mouse-rat models and electrophysiology. Extending the studies in colorectal and cervical cancer to other cellular and cancer models will also help advance the understanding and mechanistic studies of sodium channels. In addition, Nav1.6 isoforms have also been proposed as a therapeutic target for the treatment of cancer, as they have been shown to play a role in promoting tumor growth and migration. Targeting Nav1.6 isoforms with specific inhibitors, such as siRNAs or drugs, has been shown to reduce the growth and spread of cancer cells in vitro and in animal models (37). However, while these findings are promising, further research is needed to determine the usefulness of Nav1.6 isoforms as diagnostic markers and therapeutic targets in the treatment of clinical metastatic tumors. This may involve further studies in larger animal models and clinical trials in humans to determine the safety and efficacy of targeting Nav1.6 isoforms for the treatment of cancer. Overall, Nav1.6 isoforms appear to be promising as diagnostic markers and therapeutic targets in the treatment of clinical metastatic tumors, but further research is required to determine their usefulness in clinical practice.
Through an extensive literature review, it was found that Cheng et al (31) indicated that TNF-α upregulated Nav1.6 currents in retinal cells; Ding et al (38) also found that the TNF-α/STAT3 signalling pathway upregulated Nav1.6 expression in the dorsal root ganglion. However, the regulation within the tumor remains unclear, thus the role of TNF-α in this regard was investigated in the present study. As a classical inflammatory cytokine, TNF-α binds to multiple receptors within the nervous system and exerts multiple functions. TNF-α does not have a single effect on tumor cells, but can both promote and inhibit cell proliferation and induce apoptosis (39). In the present study, it was demonstrated that TNF-α promotes the production of Nav1.6 protein and mRNA, and si-Nav1.6 could partially block the induction of TNF-α in glioma, suggesting that TNF-α is associated with malignant progression of tumors in glioma.
Subsequently, a number of drugs that have potential therapeutic value for gliomas were screened. After the initial screening was completed, there were certain interesting and surprising findings during the further analysis of drug feasibility and possible relevant mechanisms. The available literature on several drugs is consistent with the current experimental results, and this has greatly increased the authors' confidence in these drugs. Among the numerous studies on the mechanisms of VGSCs regulating tumor metastasis, the most likely mechanisms are the following: The inward flow of sodium ions affects the PH and calcium ion concentrations, which carry hydrogen ions out of the cell into the extracellular matrix by the action of the sodium-hydrogen exchanger, and the activation of cysteine proteases during acidification accelerates the degradation of the extracellular matrix and thus promotes the PH-dependent tumor cell metastasis (40). There is also another sodium-calcium exchanger (NCX) where ion exchange leads to changes in intracellular calcium ion concentration and PH as well as changes in the cellular microenvironment. The altered calcium ion concentration promotes the formation of pedicles/endopods, which indirectly affects the cell migration and metastasis process (41-43). Among the results of virtual screening based on the structure of SCN8A, T0950 (trade name Neomycin sulfate) is an FDA-approved aminoglycoside antibiotic and phospholipase C inhibitor known to be commercially available. It can inhibit bacteria, cell proliferation and angiogenesis, and more importantly, it is also a commercially available calcium channel blocker (44). As a drug targeting Nav1.6, it also plays a role in the blockade of calcium ions. This appears to be consistent with the mechanistic hypothesis that currently exists. Blocking the inward flow of sodium ions affects the action of NCX, somehow blocking calcium ions as well. And as early as 1999, Mikkelsen et al (45) found that it also has an effect on pH. Perhaps there is also a relationship with the inhibition of the action of the sodium-hydrogen exchanger. These previous studies suggested that the drug has far-reaching research prospects and the reliability of drug screening is further enhanced. The specific mechanism is to be verified and explored in depth in the authors' future study.
Meanwhile, it was found that T1716 (Trade name Echinacoside), possesses a variety of biological activities. In vitro experiments have revealed that Echinacoside behaves as an antioxidant and an effective free radical scavenger in vitro. TNF-α-induced apoptosis in SH-SY5Y neuronal cells is reversed by this agent. This also supports the relationship between Nav1.6 and TNF-α in the current study. Echinacoside can block Nav1.6 expression and act on TNF-α-induced apoptosis. In addition, it was recently reported that Echinacoside can also effectively inhibit the Wnt/β-catenin signaling pathway (46). Echinacoside plays a neuroprotective role by activating Trk receptor and its downstream signaling pathway (47). This provides a pathway for further research on the pathway and mechanism. It also raises the authors' expectations that Echinacoside can play a key role in neurological tumors as well.
In the subsequent NCI-60 drug sensitivity analysis, the sensitivity of 23 drugs was correlated with the expression of SCN8A. A total of 3 of these drugs (TAM, Selumetinib and tepotinib) exhibited significant correlations (P<0.01). Of particular interest was the strongest correlation, TAM, which is a selective estrogen receptor modulator, widely used as a treatment and prevention of breast cancer. Different concentrations of TAM exert different effects, with low concentrations of (0.1-1 µM) inducing cell cycle arrest and drug concentrations (>5 µM) inducing apoptosis in breast cancer cells. A total of 5 µM TAM rapidly induces sustained ERK1/2 activation in ER+ breast cancer cells (MCF-7 and T47D) (48). It has also been identified that TAM can also induce knockout in CreER (T2) transgenic mice (49). By contrast, it is carcinogenic in the human uterus and rat liver (50). These experiments have demonstrated that TAM exerts different effects in different tumors and diseases and has a large application potential. This has been confirmed by extensive clinical trial studies. Certain studies that completed clinical phase IV have fully demonstrated the clinical promise of TAM in the treatment of breast cancer (NCT00537771), infertility (NCT02690870), vaginal bleeding (NCT04933240) and endometrium (NCT03060304); several studies in clinical phase II [NCT00108069 (51), NCT00492687, NCT04765098, NCT00024336, NCT00541138 and NCT00006388] suggested that TAM has great potential in the treatment of gliomas. TAM has been shown to cross the blood-brain barrier, which renders it a promising drug for the treatment of brain tumors (52). The exact mechanism by which TAM crosses the blood-brain barrier is not well understood. It is considered that TAM may cross the blood-brain barrier through passive diffusion, transcytosis, or facilitated transport (53). The exact absorption percentage of TAM in the brain is not well characterized, but it is considered to be low due to the presence of the blood-brain barrier. To further understand the potential of TAM in the treatment of gliomas, animal experiments need be performed. These experiments can enable to determine the efficacy and safety of TAM in treating brain tumors, as well as to further understand the mechanism by which TAM crosses the blood-brain barrier. Animal experiments may involve the use of animal models of gliomas, including mice or rats, and can help to evaluate the efficacy and safety of TAM in treating brain tumors. These experiments can also aid to determine the optimal dosing regimen for TAM, as well as to identify any potential side effects of the drug. Overall, animal experiments can provide valuable information about the potential of TAM in the treatment of gliomas and can help to expand the understanding of the mechanism by which TAM crosses the blood-brain barrier.
These existing studies demonstrated the therapeutic value and potential of these screened drugs as treatment for glioma in non-oncologic diseases and other oncologic diseases. It is considered that in the next studies the authors' group will be able to fully refine and fill certain of the gaps of currently unstudied drugs and provide a new possibility for the treatment of glioma.
In conclusion, the present study first demonstrated the expression and role of Nav1.6 in glioma, laying the foundation for further investigation of specific mechanisms, and several FDA-approved drugs that may be glioma treatment candidate drugs were screened.
Supplementary Data
Acknowledgments
The authors would like to thank Mr Zhihong Zong from the Department of Biochemistry and Molecular Biology, School of Basic Medical Sciences, China Medical University for providing the experimental space and technical support.
Funding Statement
The present study was supported by the National Natural Science Foundation of China (grant no. 31100770) and the Shenyang Natural Science Foundation (grant no. 18-013-0-60).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
AY, WJ, LJY, OSW and ZXD designed the study. AY, HXD, GJT and ZXD performed the experimental work. AY, ZXD, WJ, OSW and LJY performed the data analysis. AY, ZXD, HXD, GJT, LJY, OSW and WJ produced the main drafts of the text and figures. AY, ZXD and WJ confirm the authenticity of all raw data. All authors have reviewed, corrected, read and approved the final manuscript.
Ethics approval and consent to participate
All tissues and specimens were obtained with the approval and endorsement of the Ethics Committee of China Medical University (Shenyang, China). Written informed consent was provided by all patients for the use of their glioma tissue in the present study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Lapointe S, Perry A, Butowski NA. Primary brain tumours in adults. Lancet. 2018;392:432–446. doi: 10.1016/S0140-6736(18)30990-5. [DOI] [PubMed] [Google Scholar]
- 2.Luo Q, Wu T, Wu W, Chen G, Luo X, Jiang L, Tao H, Rong M, Kang S, Deng M. The functional role of voltage-gated sodium channel Nav1.5 in metastatic breast cancer. Front Pharmacol. 2020;11:1111. doi: 10.3389/fphar.2020.01111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Altamura C, Greco MR, Carratu MR, Cardone RA Desaphy JF. Emerging roles for ion channels in ovarian cancer: Pathomechanisms and pharmacological treatment. Cancers (Basel) 2021;13:668. doi: 10.3390/cancers13040668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brummelhuis IS, Fiascone SJ, Hasselblatt KT, Frendl G Elias KM. Voltage-gated sodium channels as potential biomarkers and therapeutic targets for epithelial ovarian cancer. Cancers (Basel) 2021;13:5437. doi: 10.3390/cancers13215437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brackenbury WJ. Voltage-gated sodium channels and metastatic disease. Channels (Austin) 2012;6:352–361. doi: 10.4161/chan.21910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu J, Tan H, Yang W, Yao S, Hong L. The voltage-gated sodium channel Nav1.7 associated with endometrial cancer. J Cancer. 2019;10:4954–4960. doi: 10.7150/jca.31544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lin S, Lv Y, Xu J, Mao X, Chen Z, Lu W. Over-expression of Nav1.6 channels is associated with lymph node metastases in colorectal cancer. World J Surg Oncol. 2019;17:175. doi: 10.1186/s12957-019-1715-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Black JA, Liu S, Waxman SG. Sodium channel activity modulates multiple functions in microglia. Glia. 2009;57:1072–1081. doi: 10.1002/glia.20830. [DOI] [PubMed] [Google Scholar]
- 9.Takayasu T, Kurisu K, Esquenazi Y, Ballester LY. Ion channels and their role in the pathophysiology of gliomas. Mol Cancer Ther. 2020;19:1959–1969. doi: 10.1158/1535-7163.MCT-19-0929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang J, Ou SW, Wang YJ, Kameyama M, Kameyama A, Zong ZH. Analysis of four novel variants of Nav1.5/SCN5A cloned from the brain. Neurosci Res. 2009;64:339–347. doi: 10.1016/j.neures.2009.04.003. [DOI] [PubMed] [Google Scholar]
- 11.Guan G, Zhao M, Xu X, Boczek T, Mao X, Li Z, Gao Q, Li J, Zhao D, Niu W, et al. Abnormal changes in voltage-gated sodium channels subtypes NaV1.1, NaV1.2, NaV1.3, NaV1.6 and CaM/CaMKII pathway in low-grade astrocytoma. Neurosci Lett. 2018;674:148–155. doi: 10.1016/j.neulet.2018.03.047. [DOI] [PubMed] [Google Scholar]
- 12.Xing D, Wang J, Ou S, Wang Y, Qiu B, Ding D, Guo F, Gao Q. Expression of neonatal Nav1.5 in human brain astrocytoma and its effect on proliferation, invasion and apoptosis of astrocytoma cells. Oncol Rep. 2014;31:2692–2700. doi: 10.3892/or.2014.3143. [DOI] [PubMed] [Google Scholar]
- 13.Mao W, Zhang J, Korner H, Jiang Y, Ying S. The emerging role of voltage-gated sodium channels in tumor biology. Front Oncol. 2019;9:124. doi: 10.3389/fonc.2019.00124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu L, Ding X, Wang T, Mou S, Sun H, Hou T. Voltage-gated sodium channels: Structures, functions, and molecular modeling. Drug Discov Today. 2019;24:1389–1397. doi: 10.1016/j.drudis.2019.05.014. [DOI] [PubMed] [Google Scholar]
- 15.Wang J, Ou SW, Wang YJ. Distribution and function of voltage-gated sodium channels in the nervous system. Channels (Austin) 2017;11:534–554. doi: 10.1080/19336950.2017.1380758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Diaz-Garcia A, Varela D. Voltage-Gated K(+)/Na(+) channels and scorpion venom toxins in cancer. Front Pharmacol. 2020;11:913. doi: 10.3389/fphar.2020.00913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schreiber JM, Tochen L, Brown M, Evans S, Ball LJ, Bumbut A, Thewamit R, Whitehead MT, Black C, Boutzoukas E, et al. A multi-disciplinary clinic for SCN8A-related epilepsy. Epilepsy Res. 2020;159:106261. doi: 10.1016/j.eplepsyres.2019.106261. [DOI] [PubMed] [Google Scholar]
- 18.Bennett DL, Clark AJ, Huang J, Waxman SG, Dib-Hajj SD. The role of Voltage-gated sodium channels in pain signaling. Physiol Rev. 2019;99:1079–1151. doi: 10.1152/physrev.00052.2017. [DOI] [PubMed] [Google Scholar]
- 19.Alrashdi B, Dawod B, Schampel A, Tacke S, Kuerten S, Marshall JS, Côté PD. Nav1.6 promotes inflammation and neuronal degeneration in a mouse model of multiple sclerosis. J Neuroinflammation. 2019;16:215. doi: 10.1186/s12974-019-1622-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Menezes LFS, Sabia Junior EF, Tibery DV, Carneiro LDA, Schwartz EF. Epilepsy-Related Voltage-gated sodium channelopathies: A review. Front Pharmacol. 2020;11:1276. doi: 10.3389/fphar.2020.01276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Angus M, Ruben P. Voltage gated sodium channels in cancer and their potential mechanisms of action. Channels (Austin) 2019;13:400–409. doi: 10.1080/19336950.2019.1666455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Besson P, Driffort V, Bon E, Gradek F, Chevalier S, Roger S. How do voltage-gated sodium channels enhance migration and invasiveness in cancer cells? Biochim Biophys Acta. 2015;1848:2493–2501. doi: 10.1016/j.bbamem.2015.04.013. [DOI] [PubMed] [Google Scholar]
- 23.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 24.FDA-Approved & Pharmacopeia Drug Library. https://www.targetmol.com/compound-library/fda_approved_&_pharmacopeia_drug_library Accessed March 8, 2022.
- 25.Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, Tunyasuvunakool K, Bates R, Žídek A, Potapenko A, et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596:583–589. doi: 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Varadi M, Anyango S, Deshpande M, Nair S, Natassia C, Yordanova G, Yuan D, Stroe O, Wood G, Laydon A, et al. AlphaFold protein structure database: Massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 2022;50:D439–D444. doi: 10.1093/nar/gkab1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kawabata T. Detection of multiscale pockets on protein surfaces using mathematical morphology. Proteins. 2010;78:1195–1211. doi: 10.1002/prot.22639. [DOI] [PubMed] [Google Scholar]
- 28.Allen WJ, Fochtman BC, Balius TE, Rizzo RC. Customizable de novo design strategies for DOCK: Application to HIVgp41 and other therapeutic targets. J Comput Chem. 2017;38:2641–2663. doi: 10.1002/jcc.25052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wallace AC, Laskowski RA, Thornton JM. LIGPLOT: A program to generate schematic diagrams of protein-ligand interactions. Protein Eng. 1995;8:127–134. doi: 10.1093/protein/8.2.127. [DOI] [PubMed] [Google Scholar]
- 30.Reinhold WC, Sunshine M, Liu H, Varma S, Kohn KW, Morris J, Doroshow J, Pommier Y. CellMiner: A web-based suite of genomic and pharmacologic tools to explore transcript and drug patterns in the NCI-60 cell line set. Cancer Res. 2012;72:3499–3511. doi: 10.1158/0008-5472.CAN-12-1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cheng S, Wang HN, Xu LJ, Li F, Miao Y, Lei B, Sun X, Wang Z. Soluble tumor necrosis factor-alpha-induced hyperexcitability contributes to retinal ganglion cell apoptosis by enhancing Nav1.6 in experimental glaucoma. J Neuroinflammation. 2021;18:182. doi: 10.1186/s12974-021-02236-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pan X, Li Z, Zhou Q, Shen H, Wu K, Huang X, Chen J, Zhang J, Zhu X, Lei J, et al. Structure of the human voltage-gated sodium channel Nav1.4 in complex with β1. Science. 2018;362:eaau2486. doi: 10.1126/science.aau2486. [DOI] [PubMed] [Google Scholar]
- 33.Schrey M, Codina C, Kraft R, Beetz C, Kalff R, Wölfl S, Patt S. Molecular characterization of voltage-gated sodium channels in human gliomas. Neuroreport. 2002;13:2493–2498. doi: 10.1097/00001756-200212200-00023. [DOI] [PubMed] [Google Scholar]
- 34.Catterall WA, Lenaeus MJ, Gamal El-Din TM. Structure and pharmacology of voltage-gated sodium and calcium channels. Annu Rev Pharmacol Toxicol. 2020;60:133–154. doi: 10.1146/annurev-pharmtox-010818-021757. [DOI] [PubMed] [Google Scholar]
- 35.Clatot J, Ziyadeh-Isleem A, Maugenre S, Denjoy I, Liu H, Dilanian G, Hatem SN, Deschênes I, Coulombe A, Guicheney P, Neyroud N. Dominant-negative effect of SCN5A N-terminal mutations through the interaction of Na(v)1.5 α-subunits. Cardiovasc Res. 2012;96:53–63. doi: 10.1093/cvr/cvs211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hernandez-Plata E, Ortiz CS, Marquina-Castillo B, Medina-Martinez I, Alfaro A, Berumen J, Rivera M, Gomora JC. Overexpression of NaV 1.6 channels is associated with the invasion capacity of human cervical cancer. Int J Cancer. 2012;130:2013–2023. doi: 10.1002/ijc.26210. [DOI] [PubMed] [Google Scholar]
- 37.Yang Y, Luo Z, Hao Y, Ba W, Wang R, Wang W, Ding X, Li C. mTOR-mediated Na+/Ca2+ exchange affects cell proliferation and metastasis of melanoma cells. Biomed Pharmacother. 2017;92:744–749. doi: 10.1016/j.biopha.2017.05.104. [DOI] [PubMed] [Google Scholar]
- 38.Ding HH, Zhang SB, Lv YY, Ma C, Liu M, Zhang KB, Ruan XC, Wei JY, Xin WJ, Wu SL. TNF-α/STAT3 pathway epigenetically upregulates Nav1.6 expression in DRG and contributes to neuropathic pain induced by L5-VRT. J Neuroinflammation. 2019;16:29. doi: 10.1186/s12974-019-1421-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lei Q, Gu H, Li L, Wu T, Xie W, Li M, Zhao N. TNIP1-mediated TNF-α/NF-κB signalling cascade sustains glioma cell proliferation. J Cell Mol Med. 2020;24:530–538. doi: 10.1111/jcmm.14760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Onganer PU, Djamgoz MB. Small-cell lung cancer (human): Potentiation of endocytic membrane activity by voltage-gated Na(+) channel expression in vitro. J Membr Biol. 2005;204:67–75. doi: 10.1007/s00232-005-0747-6. [DOI] [PubMed] [Google Scholar]
- 41.Reddy Chichili VP, Xiao Y, Seetharaman J, Cummins TR, Sivaraman J. Structural basis for the modulation of the neuronal voltage-gated sodium channel NaV1.6 by calmodulin. Sci Rep. 2013;3:2435. doi: 10.1038/srep02435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bulling A, Brucker C, Berg U, Gratzl M, Mayerhofer A. Identification of voltage-activated Na+ and K+ channels in human steroid-secreting ovarian cells. Ann N Y Acad Sci. 1999;868:77–79. doi: 10.1111/j.1749-6632.1999.tb11275.x. [DOI] [PubMed] [Google Scholar]
- 43.Anderson KJ, Cormier RT, Scott PM. Role of ion channels in gastrointestinal cancer. World J Gastroenterol. 2019;25:5732–5772. doi: 10.3748/wjg.v25.i38.5732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huang K, Wang X, Liu Y, Zhao Y. CRAC channel is inhibited by neomycin in a Ptdlns(4,5)P2-independent manner. Cell Biochem Funct. 2015;33:97–100. doi: 10.1002/cbf.3088. [DOI] [PubMed] [Google Scholar]
- 45.Mikkelsen NE, Brännvall M, Virtanen A, Kirsebom LA. Inhibition of RNase P RNA cleavage by aminoglycosides. Proc Natl Acad Sci USA. 1999;96:6155–6160. doi: 10.1073/pnas.96.11.6155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tang C, Gong L, Lvzi X, Qiu K, Zhang Z, Wan L. Echinacoside inhibits breast cancer cells by suppressing the Wnt/β-catenin signaling pathway. Biochem Biophys Res Commun. 2020;526:170–175. doi: 10.1016/j.bbrc.2020.03.050. [DOI] [PubMed] [Google Scholar]
- 47.Zhu M, Lu C, Li W. Transient exposure to echinacoside is sufficient to activate Trk signaling and protect neuronal cells from rotenone. J Neurochem. 2013;124:571–580. doi: 10.1111/jnc.12103. [DOI] [PubMed] [Google Scholar]
- 48.Zheng A, Kallio A, Härkönen P. Tamoxifen-induced rapid death of MCF-7 breast cancer cells is mediated via extracellularly signal-regulated kinase signaling and can be abrogated by estrogen. Endocrinology. 2007;148:2764–2777. doi: 10.1210/en.2006-1269. [DOI] [PubMed] [Google Scholar]
- 49.Feil S, Valtcheva N, Feil R. Inducible Cre mice. Methods Mol Biol. 2009;530:343–363. doi: 10.1007/978-1-59745-471-1_18. [DOI] [PubMed] [Google Scholar]
- 50.Rondón-Lagos M, Rangel N, Di Cantogno LV, Annaratone L, Castellano I, Russo R, Manetta T, Marchiò C, Sapino A. Effect of low doses of estradiol and tamoxifen on breast cancer cell karyotypes. Endocr Relat Cancer. 2016;23:635–650. doi: 10.1530/ERC-16-0078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Odia Y, Kreisl TN, Aregawi D, Innis EK, Fine HA. A phase II trial of tamoxifen and bortezomib in patients with recurrent malignant gliomas. J Neurooncol. 2015;125:191–195. doi: 10.1007/s11060-015-1894-y. [DOI] [PubMed] [Google Scholar]
- 52.Lien EA, Solheim E, Ueland PM. Distribution of tamoxifen and its metabolites in rat and human tissues during steady-state treatment. Cancer Res. 1991;51:4837–4844. [PubMed] [Google Scholar]
- 53.Novick AM, Scott AT, Neill Epperson C, Schneck CD. Neuropsychiatric effects of tamoxifen: Challenges and opportunities. Front Neuroendocrinol. 2020;59:100869. doi: 10.1016/j.yfrne.2020.100869. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.