Abstract
In Saccharomyces cerevisiae, the phospholipid biosynthetic genes are transcriptionally regulated in response to inositol and choline. This regulation requires the transcriptional activator proteins Ino4p and Ino2p, which form a heterodimer that binds to the UASINO element. We have previously shown that the promoters of the INO4 and INO2 genes are among the weakest promoters characterized in yeast. Because little is known about the promoters of weakly expressed yeast genes, we report here the analysis of the constitutive INO4 promoter. Promoter deletion constructs scanning 1,000 bp upstream of the INO4 gene identified a small region (−58 to −46) that is absolutely required for expression. S1 nuclease mapping shows that this region contains the transcription start sites for the INO4 gene. An additional element (−114 to −86) modestly enhances INO4 promoter activity (fivefold). Thus, the region required for INO4 transcription is limited to 68 bp. These studies also found that INO4 gene expression is not autoregulated by Ino2p and Ino4p, despite the presence of a putative UASINO element in the INO4 promoter. We further report that the INO4 steady-state transcript levels and Ino4p levels are regulated twofold in response to inositol and choline, suggesting a posttranscriptional mechanism of regulation.
Transcriptional regulation is a mechanism, in many organisms, by which to coordinate the expression of multiple genes in response to various cellular conditions. Specific cis and trans elements mediate the control of transcription. In Saccharomyces cerevisiae, an upstream activating sequence (UAS) located within 1,400 bp of the translational start site dictates the binding of activator proteins (11). Once bound to the UAS element, activator proteins interact with the general transcriptional machinery located at the TATA box (−60 to −120 bp). Therefore, it is the interactions between promoter elements and transcription factors that direct the timing and strength of gene expression. However, much of what is known about yeast promoters comes from studies of genes expressed in easily detectable quantities. With the complete genome sequence available and the advent of genome-wide approaches to the study of gene expression, it is becoming clear that there are a large number of weakly expressed genes that play an important role in the regulation of various processes in yeast. However, little is known about the cis and trans elements necessary for weakly expressed genes, although it is clear from studies on the GAL4 promoter that novel cis-acting elements are required for expression. For example, studies on the GAL4 promoter have identified an upstream essential sequence element that is required for basal transcription but is not interchangeable with a TATA element (10). Here we present an analysis of the INO4 promoter to further our understanding of the cis-acting elements necessary for weak promoter expression.
In S. cerevisiae, transcriptional regulation of the phospholipid biosynthetic genes is mediated by the INO4 and INO2 genes (9). Low levels of chloramphenicol acetyltransferase (CAT) activity from INO4-cat and INO2-cat promoter fusions places these genes into a small group of yeast promoters which drive weak gene expression (5). Sequence analysis of these two genes suggests that they belong to the basic helix-loop-helix (bHLH) family of transcriptional activators (15, 20). Consistent with this observation, the products of the INO4 and INO2 genes form a heterodimer that binds to a canonical bHLH binding site termed the UASINO element (C/AATGTGAAAT) (3, 27). The first six base pairs of this element contain the canonical bHLH binding site (5′ CANNTG 3′) (1). The UASINO element is found in the promoters of the coordinately regulated phospholipid biosynthetic genes INO1, CHO1, CHO2, and OPI3 (9). Transcription of these genes is derepressed when inositol and choline are absent from the growth medium and repressed in the presence of inositol and choline. The UASINO element is both necessary and sufficient to confer inositol-and-choline-mediated regulation (6, 17, 25).
Ino2p contains a transcriptional activation domain, which can activate a reporter gene when artificially tethered to DNA (27). On the other hand, it has been suggested that Ino4p cannot activate transcription on its own (27). This suggests that Ino4p is responsible for recruiting Ino2p to the UASINO element that enables the transcriptional activation domain of Ino2p to activate transcription of the phospholipid biosynthetic genes. Strains containing mutant alleles of INO2 or INO4 are unable to derepress INO1 expression, resulting in inositol auxotrophy (9, 13). Repression of the phospholipid genes in response to inositol-and-choline supplementation is mediated by the OPI1 gene, which encodes a leucine zipper protein. Strains containing a mutant allele of the OPI1 gene overexpress INO1, resulting in an overproduction-and-excretion-of-inositol phenotype (Opi+) (14).
A previous study showed that an INO2-cat reporter gene is regulated by inositol and choline (5). Addition of inositol and choline repressed INO2-cat gene expression 12-fold. This regulation required both the INO2 and INO4 genes, showing that INO2 is regulated in the same manner as the phospholipid biosynthetic genes. However, an INO4-cat gene was expressed constitutively with respect to inositol and choline and did not require the INO2 gene (5). This suggested that the mechanism controlling INO4 expression might be different from that of the phospholipid biosynthetic genes and the INO2 gene.
In the present study, we set out to define the cis elements necessary for INO4 gene expression. We identified two elements of the INO4 promoter that are necessary for full expression. One of these elements maps to the same region as the transcriptional start site. We also found that expression of the native INO4 gene, and its protein product, is modestly regulated by inositol and choline. Our INO4-cat reporter gene fusions suggest that this regulation does not occur at the level of transcription initiation.
MATERIALS AND METHODS
Strains and growth conditions.
The yeast strains used in this study were BRS1001 (MATa ade2-1 his3-11,15 leu2-3, 112 can1-100 ura3-1 trp1-1), BRS2001 (MATa ade2-1 his3-11,15 leu2-3,112 can1-100 ura3-1 trp1-1 ino2Δ::TRP1), BRS2004 (MATα ade2-1 his3-11,15 leu2-3,112 can1-100 ura3-1 trp1-1 ino4Δ::LEU2), BRS2005 (MATa ade2-1 his3-11,15 leu2-3,112 can1-100 ura3-1 trp1-1 opi1Δ::LEU2), Null20 (MATa his3-11,15 leu2-3,112 ura3-1 ino4Δ::LEU2), SH307 (MATα his3-11,15 leu2Δ1 ura3-52 ino4Δ::LEU2), and YB588 (MATa ade2 ade3 his3 leu2 ura3 trp1 nmt1-451D ino4Δ::HIS3). All cultures were grown at 30°C in synthetic medium either supplemented with 75 μM inositol and 1 mM choline or lacking inositol and choline (16).
Plasmid constructions.
A nested set of INO4 promoter deletions to be fused to the cat reporter gene was created by PCR using appropriate primers (Table 1). The 5′-terminal deletion PCRs used the 3′ primer INO4-BamHI along with the 5′ primers INO4-1000 through INO4-E3 (Table 1). The 3′-terminal promoter deletions used the 5′ primer INO4-A along with the 3′ primers INO4-F through INO4-I. The individual PCR products were inserted into pGEM-T (Promega, Madison, Wis.). Each deletion fragment was excised from the pGEM-T derivative by digestion with BglII and BamHI and inserted into pBM2015 (10). The pBM2015 derivatives were transformed into yeast as previously described (5).
TABLE 1.
Oligonucleotides used in this study
| INO4-BamHI (0) | 5′-GGGGGGATCCTATTGCTTTTCTCTT-3′ |
|---|---|
| INO4-1000 | 5′-AGATCTGATGCATCGCCGACC-3′ |
| INO4-750 | 5′-AGATCTTGGATTATAGTTGTT-3′ |
| INO4-A (−496) | 5′-GGGGAGATCTAAGCTTTAGTGTCGA-3′ |
| INO4-B (−396) | 5′-AGATCTTGTAGTCCAAGCCAG-3′ |
| INO4-C (296) | 5′-AGATCTTTTAGTAGCATCAGG-3′ |
| INO4-D (−186) | 5′-AGATCTGATTAGTCGTCTTCT-3′ |
| INO4-UAS (−125) | 5′-AGATCTTATTCACATGTTTTTC-3′ |
| INO4-E (−114) | 5′-AGATCTTTTTTCTCACCTTAA-3′ |
| INO4-E1 (−86) | 5′-AGATCTTTGCCTTTGCGAAAT-3′ |
| INO4-E2 (−58) | 5′-AGATCTAGATAAGCTTGACGA-3′ |
| INO4-E3 (−30) | 5′-AGATCTGCGGCTTGAACTAAA-3′ |
| INO4-F (−46) | 5′-GGATCCGTCAAGCTTATCTTG-3′ |
| INO4-G (−186) | 5′-GGATCCTATTTATAACAAAGA-3′ |
| INO4-H (−201) | 5′-GGATCCTATTTGTTCACTATA-3′ |
| INO4-I (−346) | 5′-GGATCCGGAGATCCTTGTACA-3′ |
| INO4-3′ | 5′-CCCGGGTTAATTGTT-3′ |
| INO4-HA 5′ | 5′-ATGACGAAAGATCTTAAGGAGATACAA-3′ |
| INO4-HA 3′ | 5′-TATCTCCTTAAGATCTTTCGTCATTAT-3′ |
| INO4-PE | 5′-GCCAGTTCACCCTTTATCTCC-3′ |
Plasmids derived from YCp50 were used to test the various promoter deletions for the ability to direct INO4 expression. PCR was performed using the various 5′ deletion primers with the 3′ primer INO4-3′ to create the nested set of promoter deletions upstream of the entire INO4 open reading frame (ORF). These PCR products were cloned into the pGEM-T vector. SalI-SphI fragments were isolated from the pGEM-T derivatives and cloned into YCp50.
A YCp50-INO4-HA construct was created by mutational PCR. PCR was used to introduce a BglII site three codons into the INO4 ORF. To do this, a PCR using the primers INO4-3′ and INO4-HA 5′ created an 851-bp product containing the INO4 ORF and a 5′ BglII site. A second PCR with the primers INO4-A and INO4-HA 3′ created a 500-bp product containing the INO4 promoter and a 3′ BglII site. The PCR products were digested with BglII and ligated to create a product which contained the INO4 promoter and ORF with a BglII site immediately downstream of the start codon. The ligated fragment was used as a template for another round of PCR containing the primers INO4-A and INO4-3′, resulting in a 1,350-bp product. This PCR fragment was cloned into pGEM-T. The pGEM-T derivative was partially digested with BglII, and a 120-bp BglII fragment containing three tandem copies of the hemagglutinin (HA) epitope (provided by Susan Henry, Carnegie Mellon University) was inserted. A SalI-SphI fragment was isolated from the pGEM-T derivative and cloned into YCp50. All promoter constructs and the YCp50-INO4HA construct was confirmed by DNA sequencing. The plasmid pTA-INO4 contains the entire INO4 ORF, starting at the translational initiator codon and extending past the translational stop codon.
CAT assays.
Five-milliliter cultures were grown to 50 to 60 Klett units in appropriate media. Assays were conducted as previously described (5). Units of CAT activity were defined as counts per minute measured in the organic phase and expressed as a percentage of the total counts per minute (percent conversion) divided by the amount of protein assayed (in micrograms) and the time of incubation (in hours).
RNA analysis.
RNA was isolated from yeast by a glass bead disruption and hot phenol extraction procedure (8). RNA probes (cRNA) for Northern (RNA) hybridizations were synthesized with the Gemini II Core System (Promega, Madison, Wis.) from plasmids linearized with a restriction enzyme and transcribed with an RNA polymerase as follows (shown as plasmid, restriction enzyme, RNA polymerase) for the probe indicated in parentheses: pTA-INO4, SalI, T7 (INO4); pJH310, HinDIII, T7 (INO1); and pAB309Δ, EcoRI, SP6 (TCM1). Northern hybridizations were performed as previously described (14), results were visualized by autoradiography, and gene-specific counts per minute were quantitated using a Betascope 603 Blot Analyzer (Beta-gen, Waltham, Mass.).
For the S1 nuclease digestion assay, BRS1001 transformed with pJA201 (contains the entire INO4 gene) and BRS2004 were grown in the appropriate media. RNA was isolated as described above. A single-stranded probe was created using asymmetric PCR (12). A standard 100-μl PCR mixture contained 1 μg of the INO4-PE oligonucleotide and 0.3 ng of the INO4-A oligonucleotide. The single-stranded PCR product was purified by gel electrophoresis and labeled at the 5′ terminus with [γ-32P]ATP using T4 polynucleotide kinase (Gibco BRL, Grand Island, N.Y.) (22). Approximately 10 ng of labeled probe was coprecipitated with 20 μg of total yeast RNA. The pellet was resuspended in 10 μl of S1 hybridization buffer [80% formamide, 0.4 M NaCl, 0.04 M piperazine-N,N′-bis(2-ethanesulfonic acid) (PIPES; pH 6.6), 0.1 mM EDTA]. The hybridization reaction was overlaid with mineral oil, denatured at 95°C for 3 min, and hybridized overnight at 46°C. Next, 300 μl of S1 digestion buffer (0.28 M NaCl, 0.03 M sodium acetate [pH 4.5], 4.5 mM ZnCl2, salmon sperm DNA at 20 μg/ml, S1 nuclease at 1,200 U/ml (Boehringer Mannheim, Indianapolis, Ind.) was added and the reaction mixture was incubated at 15°C for 1 h. The digestion reaction was terminated by addition of 50 μl of 2.5 M ammonium acetate–50 mM EDTA. The reaction mixture was then extracted with an equal volume of phenol-chloroform-isoamyl alcohol (25:24:1), and the digestion products were precipitated. The pellet was resuspended in 10 μl of gel loading dye (10 mM NaOH, 95% formamide, 0.05% bromophenol blue, 0.05% xylene cyanol), denatured at 95°C for 5 min, and fractionated through a 4% denaturing polyacrylamide gel. A sequencing ladder generated using the INO4-PE primer was used as the standard.
Western blots.
Preparation of whole-cell extracts of S. cerevisiae was performed as previously described (18), and 50 μg of yeast extract was separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Proteins were transferred to nitrocellulose by electroblotting at 150 mA for 2 h. The blot was stained with Ponceau to confirm equal loading and transfer of proteins. The blot was incubated in BLOTTO (100 mM Tris, 1.5 M NaCl, 0.5% Tween 40, 5% powder milk) overnight at 4°C with constant shaking. Anti-HA antibody (Boehringer Mannheim) was added to a final dilution of 1/250 and incubated for 1 h at room temperature with shaking. The blot was then washed in BLOTTO five times for 5 min each time. An alkaline phosphatase-conjugated anti-immunoglobulin G antibody (Zymed Laboratory Inc., San Francisco, Calif.) was added to BLOTTO at a final dilution of 1/1,000 and incubated for 1 h at room temperature. The blot was washed in BLOTTO five times for 5 min each time. The blot was suspended in alkaline phosphate buffer (100 mM Tris, 100 mM NaCl, 5 mM MgCl2). Nitroblue tetrazolium–5-bromo-4-chloro-3-indolylphosphate (BCIP) (Promega) was added in a 2:1 ratio to the buffer and allowed to react for 1 h. The reaction was stopped by the addition of stop buffer (200 mM Tris [pH 8.0], 5 mM EDTA).
RESULTS
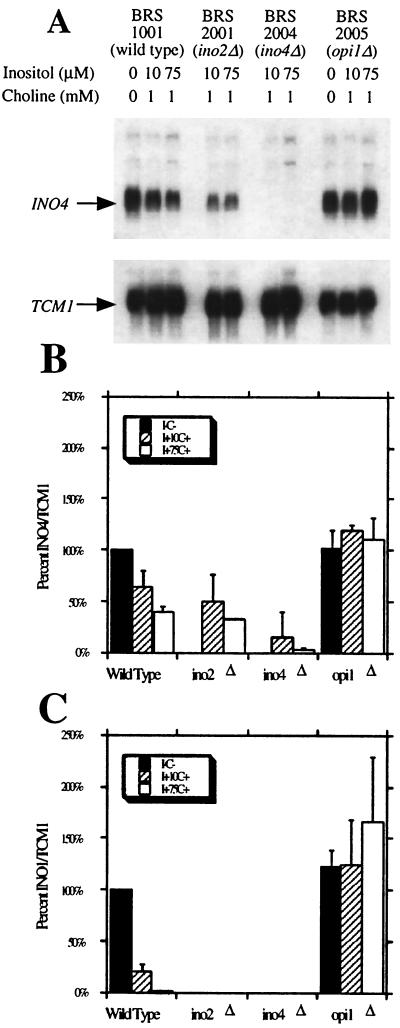
Previous results obtained by using an INO4 promoter fusion to the cat reporter gene suggested that INO4 expression is unresponsive to inositol and choline (5). Until this report, we had been unable to detect the INO4 transcript using Northern blot hybridization. In part, this is due to the low abundance of the INO4 transcript, which was overcome by generating a high-specific-activity cRNA probe. In addition, the INO4 sequences included in the probe itself are also important since the ORF and 3′ untranslated region are required. Probes containing the INO4 ORF or parts of the ORF are not sufficient to detect the INO4 transcript (J. M. Lopes, unpublished data). To determine if the INO4-cat reporter accurately reflected expression of the native INO4 gene, steady-state transcript levels were quantitated by Northern blot hybridization. RNA was isolated from strains grown in medium that normally represses (with 75 μM inositol and 1 mM choline), partially derepresses (with 10 μM inositol and 1 mM choline), or completely derepresses (lacking inositol and choline) most of the phospholipid biosynthetic structural genes (9, 13). An INO4-specific cRNA probe recognized a single major RNA of approximately 600 nucleotides (Fig. 1A). Quantitation of this transcript revealed that steady-state INO4 levels are reduced by 60% in a wild-type strain in response to inositol-and-choline supplementation (Fig. 1B). Repression of the phospholipid biosynthetic genes in response to inositol and choline is known to require the negative regulatory protein encoded by OPI1. Like the phospholipid biosynthetic genes, the 60% reduction of INO4 expression by inositol and choline is dependent on a wild-type allele of OPI1. However, unlike the phospholipid biosynthetic genes, regulation of INO4 by inositol and choline does not require a functional copy of INO2 (9) (Fig. 1A and B). To ensure that the ino2Δ strain lacked the INO2 gene function, we quantified expression of the INO1 transcript. As expected, INO1 expression was eliminated in the ino2Δ mutant strain (Fig. 1C).
FIG. 1.
Quantitative analysis of INO4 and INO1 mRNAs. (A) Representative Northern blot hybridization showing INO4 transcript levels. TCM1, a constitutively expressed ribosomal protein gene, was used to normalize for loading variations. The relevant genotypes of the strains used are shown in parentheses. (B) Bar values represent ratios of INO4 to TCM1 counts per minute quantified using a Betascope 603 Blot Analyzer (Beta-gen). (C) Bar values represent ratios of INO1 to TCM1 counts per minute. Cells were grown in complete synthetic medium supplemented with 75 μM inositol and 1 mM choline (I+75C+) or 10 μM inositol and 1 mM choline (I+10C+) or lacking inositol and choline (I−C−). The values shown are averages of at least three separate trials and are normalized to that of the wild type without inositol and choline (100%).
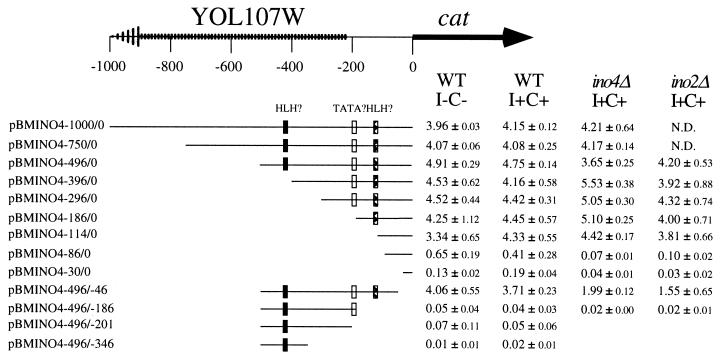
It is interesting that the modest regulation of steady-state INO4 levels in response to inositol and choline was not observed with the INO4-cat reporter gene (5). One possible explanation for this difference is that promoter sequences absent from the INO4-cat reporter gene are required for the response to inositol and choline. The original INO4-cat gene contained 500 bp of the INO4 promoter (5). To ensure that all of the potential promoter elements were included, two additional INO4-cat constructs were created that contained 750 and 1,000 bp upstream of the INO4 translational start site. However, these constructs also failed to exhibit a response to inositol-and-choline supplementation (Fig. 2). It seems unlikely that the INO4 promoter would encompass sequences farther upstream than 1,000 bp, since this would include most of a divergent ORF, YOL107W. The lack of regulation of the INO4-cat gene suggests that INO4 expression may be regulated at the level of mRNA stability by inositol and choline. This possibility was explored in greater detail as described below.
FIG. 2.
Expression of INO4-cat fusions in the wild type (WT) (BRS1001), ino4Δ (BRS2004), and ino2Δ (BRS2001) strains. Transformants containing the various promoter deletions were grown in uracil-lacking synthetic medium either containing 75 μM inositol and 1 mM choline (I+C+) or lacking inositol and choline (I−C−). Extracts from the yeast transformants were prepared and assayed for CAT activity. The values shown are averages of at least three separate trials. YOL107W is a divergent ORF. The locations of two putative bHLH binding sites and a potential TATA box are shown. N.D., not determined.
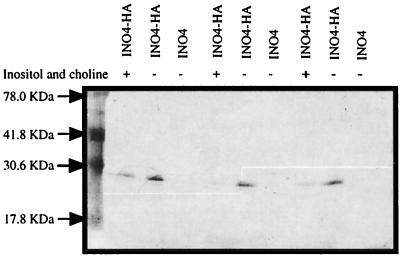
The possibility that inositol and choline may also exert regulation at the level of mRNA stability prompted us to examine Ino4p levels under repressing and derepressing conditions. Toward this end, we tagged Ino4p at the N terminus with three tandem copies of the HA epitope. This fusion was inserted into the YCp50 (YCp50-INO4HA) vector and subsequently transformed into an ino4Δ mutant strain (BRS2004). Yeast extracts were prepared from strains transformed with either YCp50-INO4 or YCp50-INO4HA grown in medium that normally represses and derepresses the phospholipid biosynthetic genes. Western blotting using an anti-HA antibody revealed that Ino4p protein levels are decreased in the presence of inositol and choline (Fig. 3). This result is consistent with the steady-state INO4 mRNA levels.
FIG. 3.
Western blot analysis of HA-tagged Ino4p. This representative Western blot shows Ino4p levels in cells grown in complete synthetic medium either lacking (−) or supplemented with (+) 75 μM inositol and 1 mM choline.
Transcriptional regulation of the phospholipid biosynthetic genes and the INO2 regulatory gene is dependent on both the INO2 and INO4 genes. However, INO4 transcription does not require INO2 (Fig. 1A and B). This is consistent with a previous report showing that expression of an INO4-cat promoter fusion did not require the INO2 gene (5). Genes that are responsive to inositol and choline are typically dependent on both the INO2 and INO4 genes for derepression. However, some genes have been found which require INO4 but not INO2 for their expression (9). In fact, this laboratory and others have reported that INO4-cat and INO4-lacZ reporter gene expression does not require INO2 but does require INO4 (5, 26). Surprisingly, in the present study, we observed that the level of CAT activity in an ino4Δ strain (BRS2004) was equivalent to that in the wild-type strain, suggesting that INO4 is not required for INO4-cat expression (Fig. 2).
An explanation for this difference presented itself when we noticed that in the earlier study we had employed a different ino4Δ strain, Null20 (5). Curiously, Null20 and BRS2004 both originated as ino4Δ mutant spores isolated from the same tetrad and are predicted to be isogenic (J. Ambroziak, personal communication). To determine if the two ino4Δ strains behaved differently, we retransformed the INO4-cat construct into the Null20 strain and assayed for CAT activity. As seen in the earlier report, no CAT activity was observed in the Null20 transformants. Therefore, the discrepancy lay within the ino4Δ strains. One of the strains may have obtained a second mutation that alters expression of the INO4-cat reporter. To determine the correct INO4-cat phenotype, we assayed for CAT activity in two additional, independently isolated ino4Δ mutant strains with entirely different genetic histories: SH307 (kindly provided by Miriam Greenberg, Wayne State University) and YB588 (kindly provided by Steven Cok, Washington University) The CAT expression levels in these two strains were comparable to the levels in strain BRS2004. We measured INO4-cat expression in multiple ino4Δ strains and found the following mean levels of CAT activity (± the standard deviations): BRS2004, 4.75 ± 0.14 U; SH307, 2.97 ± 0.53 U; YB588, 3.15 ± 0.84 U; Null20, 0.02 ± 0.01 U. These results support the conclusion that INO4 is not autoregulated.
Transcriptional regulation of the phospholipid biosynthetic genes, and the INO2 regulatory gene, in response to inositol and choline is mediated by a UASINO element. The INO4 promoter contains two putative bHLH binding sites, with the proximal bHLH binding site resembling a UASINO element. This prompted us to delineate the region(s) of the INO4 promoter that is required for gene expression. To do this, we created a nested set of INO4 promoter deletions fused to the cat reporter gene. The constructs were integrated at the GAL4 locus in single copy, and all integrations were confirmed by Southern blotting. Cultures were grown in media with or without both inositol and choline and assayed for CAT activity. This assay demonstrated that the −86 to −46 region of the INO4 promoter is required to drive expression of the cat gene in an inositol-and-choline-independent manner (Fig. 2). As shown above, expression from the complete INO4 promoter does not require INO2 or INO4 (Fig. 2). However, the possibility remained that elements within the promoter require INO2 and INO4 to maintain appropriate INO4 expression levels. Therefore, the promoter deletions were assayed in ino2Δ and ino4Δ mutant strains under completely repressing conditions. The results confirmed that the −86 to −46 region of the INO4 promoter is necessary and sufficient for INO4 expression (Fig. 2). This also demonstrated that the two putative bHLH binding sites within the INO4 promoter are not functional and not dependent on the INO2 and INO4 genes.
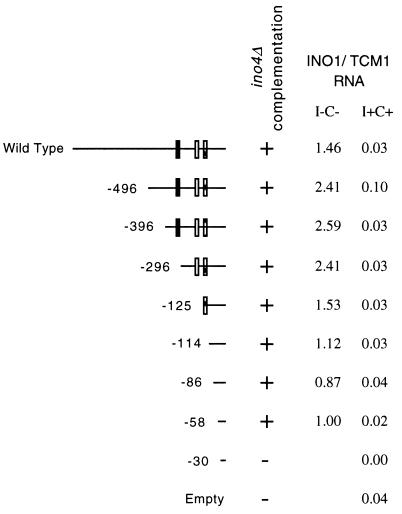
Because it is possible that integration of the INO4-cat constructs at the GAL4 locus could artificially alter expression of the INO4-cat gene, we assayed for the ability of the INO4 promoter deletions to drive expression of the INO4 gene. PCR products that contained the various promoter deletions upstream of the entire INO4 ORF were created. The PCR products were ligated into the YCp50 vector and transformed into ino4Δ mutant strain BRS2004. Expression of the INO4 gene from the various promoter deletions resulted in complementation of the inositol auxotrophy of BRS2004. This assay provided further proof that the −58 to −46 region of the INO4 promoter is absolutely required to express INO4 at levels sufficient to complement the inositol auxotrophy of an ino4Δ mutant strain (Fig. 4).
FIG. 4.
Complementation of ino4Δ (BRS2004) inositol auxotrophy by various YCp50-INO4 promoter deletions. Respective transformants were plated on uracil-lacking synthetic medium containing inositol and choline and replica plated to uracil-lacking synthetic medium also lacking inositol and choline. Growth is indicated by a plus sign, and absence of growth is indicated by a minus sign. Quantitation of steady-state INO1 mRNA transcript levels by Northern blot hybridization in the wild-type strain (BRS1001) and an ino4Δ mutant strain (BRS2004) transformed with the YCp50-INO4 promoter deletion constructs is shown. Strains were grown in uracil-lacking synthetic medium either containing 75 μM inositol and 1 mM choline (I+C+) or lacking inositol and choline (I−C−). TCM1 was used to normalize for loading variations. The values shown represent ratios of INO1 to TCM1 expression levels. The locations of two putative bHLH binding sites and a potential TATA box are shown as in Fig. 2.
Placement of the deletion constructs on a centromeric plasmid may cause overexpression of the INO4 gene. This may alter the regulation of the INO1 gene, potentially leading to the abnormal expression of INO1. To address this issue, we measured the steady-state INO1 transcript levels in these strains by Northern blot hybridization. The regulation of INO1 transcript levels in the transformants was comparable to the regulation observed in a wild-type strain (compare Fig. 4 to Fig. 1C).
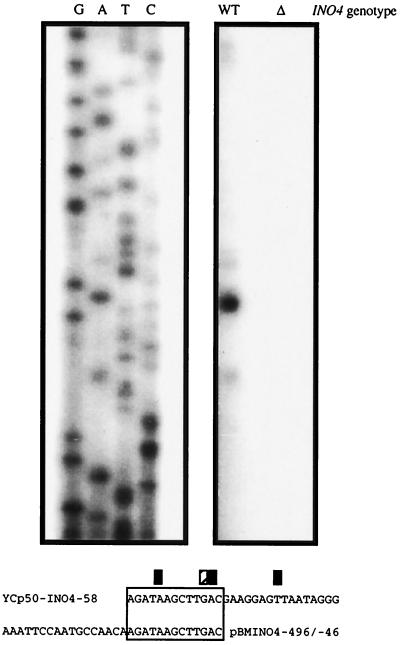
To aid in the interpretation of the promoter deletion studies, we mapped the INO4 transcriptional start site using an S1 nuclease assay. However, native INO4 expression levels were too low to be detected by this assay. Therefore, wild-type strain BRS1001 was transformed with pJA201, a multicopy plasmid containing the entire INO4 gene driven by its own promoter (2). Overexpression of the INO4 gene in this transformed strain allowed detection of the INO4 initiation sites. The assay revealed one major start site at −48 and two additional minor transcript start sites at −54 and −47 (Fig. 5). Therefore, the transcriptional start sites of INO4 fall within the minimal region of the promoter required for expression as defined by the INO4-cat deletion studies and the YCp50-INO4 derivatives.
FIG. 5.
S1 nuclease digestion assay of INO4 mRNA. A single-stranded PCR probe was hybridized to total cellular mRNA from the wild type (WT) strain (BRS1001) transformed with pJA201, and an ino4Δ mutant strain (BRS2004). BRS1001 was grown in leucine-lacking synthetic medium containing 75 μM inositol and 1 mM choline, while BRS2004 was grown in complete synthetic medium containing 75 μM inositol and 1 mM choline. S1 digestion fragments were run on a denaturing polyacrylamide gel next to the INO4 sequence that was generated with the same primer used to synthesize the probe. Different exposures of the sequencing ladder (1 day) and S1 digestion products (4 days) are shown. Nucleotide assignment of INO4 mRNA start sites. The stippled box represents the major start site. The solid boxes represent minor start sites. Sequences refer to the exact nucleotides present in the respective promoter deletions.
The INO4-cat promoter deletion studies also identified a potential regulatory element. Deletion of the region from −114 to −86 resulted in a 5- to 10-fold decrease in cat expression from the INO4 promoter (Fig. 2). Yet, the YCp50 derivatives containing INO4 promoter sequences to −86 and −58 were still able to complement the inositol auxotrophy of BRS2004 (Fig. 4). This suggested that in spite of the 5- to 10-fold decrease in INO4, as seen in the CAT assay, there is sufficient Ino4p present to regulate its target genes.
DISCUSSION
Because little is known about elements necessary to direct expression from a weak promoter in yeast, we determined the element(s) necessary for INO4 expression. We created various promoter deletions within the INO4 promoter and fused them upstream of the cat reporter gene. These constructs did reveal that the region from −86 to −46 bp upstream of the INO4 translational start site is absolutely necessary for INO4 expression (Fig. 2). To eliminate the possibility of artifacts from the INO4-cat constructs, we inserted the INO4 promoter deletions fused upstream of the entire INO4 ORF into YCp50. We assayed the deletions for the ability to complement the inositol auxotrophy of an ino4Δ strain. These experiments supported the INO4-cat construct data demonstrating that the −58 to −46 region of the INO4 promoter is required for expression. An S1 nuclease digestion assay demonstrated that the major INO4 transcriptional start site, along with two minor start sites, lies within the −58 to −46 region (Fig. 5). Therefore, various INO4 promoter deletions and the S1 nuclease assay have shown that INO4 contains an initiator element and no TATA box. To our knowledge, INO4 is the first yeast gene that requires an initiator element but contains no TATA box. However, this phenomenon is very prevalent in the housekeeping genes of higher eukaryotes. Housekeeping genes are constitutively expressed genes with promoters that usually do not contain TATA boxes or other regulatory elements (7).
The INO4 promoter study also identified a region that is required for full INO4-cat expression. Deletion of nucleotides −114 to −86 resulted in an 80 to 90% decrease in cat expression (Fig. 2). However, YCp50-INO4 constructs lacking these sequences still complemented the inositol auxotrophy and maintained proper INO1 regulation (Fig. 4). This suggested that INO4 expression is in excess of what is needed to regulate the phospholipid biosynthetic genes. This idea is also supported by the fact that overexpression of Ino4p, but not Ino2p, does not result in elevated levels of the Ino2p/Ino4p/UASINO complex in mobility shift assays (19).
Sequence analysis illuminated two potential bHLH binding sites and a TATA-like element. However, the promoter deletions demonstrated that none of these elements were necessary for INO4-cat expression. Studies with in vitro-transcribed and -translated Ino2p and Ino4p have shown that they can bind to the proximal putative INO4 bHLH binding site which most closely resembles the UASINO element (28). Also, this element, when fused to a CYC1-lacZ reporter, was sufficient to confer regulation by inositol and choline. However, it has been observed that certain UASINO elements are functional in the CYC1-lacZ reporter system (6, 25) but not functional in a native context (4). We have demonstrated that neither of the INO4 bHLH promoter elements is required for expression or regulation (Fig. 2 and 4).
The bHLH region of Ino4p shares homology with the bHLH region of the mammalian protein Max (20). Max forms heterodimers with Myc, Mad, and Mxi-1 to regulate genes required for cell proliferation and differentiation. Myc levels increase in cells undergoing proliferation, while Mad levels increase in cells as they differentiate (1). Max is constitutively expressed (1). Therefore, it is Myc and Mad concentrations that are rate limiting for heterodimer formation and ultimately responsible for determining which cell program is expressed. The similarity between the sequence and expression of Ino4p and Max suggests that Ino4p, like Max, may bind other bHLH proteins in yeast. Results of a two-hybrid study suggest that this is, in fact, true (K. A. Robinson and J. M. Lopes, unpublished data). By binding to bHLH proteins other than Ino2p, it is possible that Ino4p is responsible for regulating genes other than the phospholipid biosynthetic genes. Results obtained by using a whole-genome array suggest that there are genes independent of the phospholipid biosynthetic genes, which are each regulated positively and negatively by INO4 (Robinson and Lopes, unpublished data). Therefore, the INO4 pattern of expression suggests that it is a housekeeping gene in yeast.
A previous report from this laboratory (5) and data presented here (Fig. 2) showed that the INO4-cat construct is expressed constitutively. Another report using INO4-lacZ suggested that INO4 is autoregulated in response to inositol and choline (26). Because of this conflict, we decided to look at the in vivo steady-state levels of the INO4 mRNA transcript. Northern analysis demonstrated that the INO4 transcript is regulated two- to threefold in response to inositol and choline (Fig. 1A and B). Consistent with the steady-state INO4 mRNA levels, we found that the Ino4p levels are also regulated in response to inositol and choline. To eliminate the possibility that the constitutive expression of the INO4-cat construct was the result of omitted upstream regulatory sequences, we created additional INO4-cat constructs. Constructs containing sequences 750 and 1,000 bp upstream of the INO4 translational start site still elicited constitutive CAT activity (Fig. 2). One possible cause of the discrepancy is that INO4 is regulated at the level of mRNA stability. INO4 would not be the first gene in the phospholipid biosynthetic pathway to be regulated at the level of mRNA stability in response to inositol and choline. INO1 and CHO2 transcript stability is regulated 50 to 60% by inositol and choline (J. Yates and J. M. Lopes, unpublished data). In fact, the threefold regulation seen with the INO4-lacZ gene may result from mRNA stability, since the INO4-lacZ construct is a translational fusion (26).
There are currently four methods for measuring mRNA half-life (21). One method, labeling of cells to steady state (or by pulse-chase) in vivo, has not been successfully used with low-abundance transcripts. We attempted to determine the half-life of the INO4 transcript using thiolutin and by the use of a yeast temperature-sensitive RNA polymerase (rpb1-1) mutant. Incubation with thiolutin resulted in rapid degradation of the INO4 transcript, which made it impossible to detect the transcript by Northern blot hybridization. Experiments using the rpb1-1 mutant strain yielded the same result as the thiolutin assay. Moreover, the rpb1-1 mutant is an inositol auxotroph (23, 24), which would make it difficult to exclude the possibility that artifacts resulted from the auxotrophy. A final method for determining mRNA half-life requires placing the gene under the control of the tightly regulated strong GAL1 promoter (21). Therefore, the increase in transcript numbers may facilitate detection by Northern blot hybridization. However, in the case of INO4, this would create gross overexpression of the INO4 transcript, well beyond physiological levels.
Transcriptional regulation of the phospholipid biosynthetic genes in response to inositol and choline is mediated by INO2 and INO4 (9). However, INO4 expression does not require INO2 or INO4 (Fig. 1 and 2). Nevertheless, as is the case with the coordinately regulated phospholipid biosynthetic genes, INO4 is constitutively overexpressed in an opi1Δ strain (Fig. 1A and B). This suggests that OPI1 may regulate both transcription initiation and mRNA stability in response to inositol and choline.
ACKNOWLEDGMENTS
We thank Miriam Greenberg and Steven Cok for providing strains. We also thank Kyle Gardenour and Mohan Kaadige for helpful discussions.
This work was supported by an American Cancer Society grant (RPG-97-002-01-CNE) to J.M.L.
REFERENCES
- 1.Amati B, Land K. Myc-Max-Mad: a transcription factor network controlling cell-cycle progression. Curr Opin Genes Dev. 1994;4:102–108. doi: 10.1016/0959-437x(94)90098-1. [DOI] [PubMed] [Google Scholar]
- 2.Ambroziak J. Ph.D. thesis. Pittsburgh, Pa: Carnegie Mellon University; 1994. [Google Scholar]
- 3.Ambroziak J, Henry S A. INO2 and INO4 gene products, positive regulators of phospholipid biosynthesis in Saccharomyces cerevisiae, form a complex that binds to the INO1 promoter. J Biol Chem. 1994;269:15344–15349. [PubMed] [Google Scholar]
- 4.Anderson M S, Lopes J M. Carbon source regulation of PIS1 gene expression in Saccharomyces cerevisiae involves the MCM1 gene and the two-component regulatory gene, SLN1. J Biol Chem. 1996;271:26596–26601. doi: 10.1074/jbc.271.43.26596. [DOI] [PubMed] [Google Scholar]
- 5.Ashburner B P, Lopes J M. Autoregulated expression of the yeast INO2 and INO4 helix-loop-helix activator genes effects cooperative regulation on their target genes. Mol Cell Biol. 1995;15:1709–1715. doi: 10.1128/mcb.15.3.1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bacchawat N, Ouyang Q, Henry S A. Functional characterization of the inositol-sensitive upstream activation sequence of yeast. J Biol Chem. 1995;270:25087–25095. doi: 10.1074/jbc.270.42.25087. [DOI] [PubMed] [Google Scholar]
- 7.Dynan W S. Modularity in promoters and enhancers. Trends Genet. 1986;2:196–197. [Google Scholar]
- 8.Elion E A, Warner J R. The major promoter element of rRNA transcription. Cell. 1984;39:663–673. doi: 10.1016/0092-8674(84)90473-2. [DOI] [PubMed] [Google Scholar]
- 9.Greenberg M L, Lopes J M. Genetic regulation of phospholipid biosynthesis in Saccharomyces cerevisiae. Microbiol Rev. 1996;60:1–20. doi: 10.1128/mr.60.1.1-20.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Griggs D W, Johnston M. Promoter elements determining weak expression of the GAL4 regulatory gene of Saccharomyces cerevisiae. Mol Cell Biol. 1993;13:4999–5009. doi: 10.1128/mcb.13.8.4999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guarente L. Regulatory proteins in yeast. Annu Rev Genet. 1987;21:425–452. doi: 10.1146/annurev.ge.21.120187.002233. [DOI] [PubMed] [Google Scholar]
- 12.Gyllensten U B, Erlich H A. Generation of single-stranded DNA by the polymerase chain reaction and its application to direct sequencing of the HLA-DQA locus. Proc Natl Acad Sci USA. 1988;85:7652–7656. doi: 10.1073/pnas.85.20.7652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Henry S A, Patton-Vogt J L. Genetic regulation of phospholipid metabolism: yeast as a model eukaryote. Prog Nucleic Acid Res Mol Biol. 1998;61:133–179. doi: 10.1016/s0079-6603(08)60826-0. [DOI] [PubMed] [Google Scholar]
- 14.Hirsch J P, Henry S A. Expression of the Saccharomyces cerevisiae inositol-1-phosphate synthase (INO1) gene is regulated by factors that affect phospholipid synthesis. Mol Cell Biol. 1986;6:3320–3328. doi: 10.1128/mcb.6.10.3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hoshizaki D K, Hill J E, Henry S A. The Saccharomyces cerevisiae INO4 gene encodes a small, highly basic protein required for derepression of phospholipid biosynthetic enzymes. J Biol Chem. 1990;265:4736–4745. [PubMed] [Google Scholar]
- 16.Kelly B L, Greenberg M L. Characterization and regulation of phosphatidylglycerolphosphate phosphatase in Saccharomyces cerevisiae. Biochim Biophys Acta. 1990;1046:144–150. doi: 10.1016/0005-2760(90)90181-v. [DOI] [PubMed] [Google Scholar]
- 17.Koipally J, Ashburner B P, Bachhawat N, Gill T, Hung G, Henry S A, Lopes J M. Functional characterization of the repeated UASINO element in the promoters of the INO1 and CHO2 genes of yeast. Yeast. 1996;12:653–665. doi: 10.1002/(SICI)1097-0061(19960615)12:7%3C653::AID-YEA953%3E3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 18.Lopes J M, Henry S A. Interaction of trans and cis regulatory elements in the INO1 promoter of Saccharomyces cerevisiae. Nucleic Acids Res. 1991;19:3987–3994. doi: 10.1093/nar/19.14.3987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nikoloff D M, Henry S A. Functional characterization of the INO2 gene of Saccharomyces cerevisiae. J Biol Chem. 1994;269:7402–7411. [PubMed] [Google Scholar]
- 20.Nikoloff D M, McGraw P, Henry S A. The INO2 gene of Saccharomyces cerevisiae encodes a helix-loop-helix protein that is required for activation of phospholipid biosynthesis. Nucleic Acids Res. 1992;20:3253. doi: 10.1093/nar/20.12.3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Parker R, Herrick D, Peltz S W, Jacobson A. Measurement of mRNA decay rates in Saccharomyces cerevisiae. Methods Enzymol. 1990;194:415–523. doi: 10.1016/0076-6879(91)94032-8. [DOI] [PubMed] [Google Scholar]
- 22.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. pp. 10.59–10.67. [Google Scholar]
- 23.Scafe C, Martin C, Nonet M, Podos S, Okamura S, Young R A. Conditional mutants occur predominantly in highly conserved residues of RNA polymerase II subunits. Mol Cell Biol. 1990;10:1270–1275. doi: 10.1128/mcb.10.3.1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Scafe C, Nonet M, Young R A. RNA polymerase II mutants defective in transcription of a subset of genes. Mol Cell Biol. 1990;10:1010–1016. doi: 10.1128/mcb.10.3.1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schüller H J, Richter K, Hoffmann B, Ebbert R, Schweizer E. DNA binding site of the yeast heteromeric Ino2p/Ino4p basic helix-loop-helix transcription factor: structural requirements as defined by saturation mutagenesis. FEBS Lett. 1995;370:149–152. doi: 10.1016/0014-5793(95)00818-t. [DOI] [PubMed] [Google Scholar]
- 26.Schüller H J, Schorr R, Hoffmann B, Schweizer E. Regulatory gene INO4 of yeast phospholipid biosynthesis is positively autoregulated and functions as a trans-activator of fatty acid synthase genes FAS1 and FAS2 from Saccharomyces cerevisiae. Nucleic Acids Res. 1992;20:5955–5961. doi: 10.1093/nar/20.22.5955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schwank S, Ebbert R, Rautenstraß K, Schweizer E, Schüller H J. Yeast transcriptional activator INO2 interacts as an Ino2p/Ino4p basic helix-loop-helix heteromeric complex with the inositol/choline responsive element necessary for expression of the phospholipid biosynthetic genes in Saccharomyces cerevisiae. Nucleic Acids Res. 1995;23:230–237. doi: 10.1093/nar/23.2.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schwank S, Hoffmann B, Schüller H J. Influence of gene dosage and autoregulatory genes INO2 and INO4 on inositol/choline-repressible gene transcription in yeast Saccharomyces cerevisiae. Curr Genet. 1997;31:462–468. doi: 10.1007/s002940050231. [DOI] [PubMed] [Google Scholar]